Graphical abstract
Abbreviations: ARDS, Acute respiratory distress syndrome; COVID-19, Coronavirus disease of 2019; CLR, C-type lectin like receptor; DAMP, Damage-associated molecular pattern; DG, Degranulation; EC, Endothelial cell; ITIM, Immunoreceptor tyrosine-based inhibitory motifs; MAPK, Mitogen-activated protein kinase; NET, Neutrophil extracellular trap; NLR, NOD-like receptor; PAMP, Pathogen-associated molecular pattern; PRR, Pattern recognition receptor; RAGE, Receptor for advanced glycation end products; RB, Respiratory burst; RLR, RIG-I-like receptor; ROS, Reactive oxygen species; TAK, Transforming growth factor-β activated kinases; TLR, Toll-like receptor
Keywords: DAMP, TLR/PRR, Siglecs, CD24, CD24Fc, ARDS, HMGB1, Neutrophils, Endothelial Dysfunction, Hypercoagulation
Abstract
The host response to SARS-CoV-2, the virus that causes COVID-19, is highly heterogeneous, ranging from mild/asymptomatic to severe. The moderate to severe forms of COVID-19 often require hospitalization, are associated with a high rate of mortality, and appear to be caused by an inappropriately exaggerated inflammatory response to the virus. Emerging data confirm the involvement of both innate and adaptive immune pathways both in protection from SARS-CoV-2, and in driving the pathology of severe COVID-19. In particular, innate immune cells including neutrophils appear to be key players in the inflammation that causes the vicious cycle of damage and inflammation that underlies the symptomatology of severe COVID-19. Several recent studies support a link between damage and inflammation, with damage-associated molecular patterns (DAMPs) playing a key role in the pathology of severe COVID-19. In this review, we put into perspective the role of DAMPs and of components of the DAMP-signaling cascade, including Siglecs and their cognate ligands CD24 and CD52, in COVID-19. Further, we review clinical data on proposed therapeutics targeting DAMP pathways to treat SARS-CoV-2 infection and the regulation of these signaling cascades in COVID-19. We also discuss the potential impact of DAMP-mediated inflammation in other indications related to COVID-19, such as ARDS, endothelial dysfunction, hypercoagulation, and sepsis.
1. Introduction
The ongoing COVID-19 pandemic has had immeasurable impact on human lives globally since the disease was first described in December 2019, in Wuhan, China. As of September 2021, over 200 million documented cases of COVID-19 and over 4 million fatalities have been reported around the world [1]. COVID-19 is caused by a novel coronavirus, SARS-CoV-2, belonging to the genus Betacoronavirus, which also includes the common cold viruses HKU1, NL63, OC43, as well as the more pathogenic viruses SARS-CoV and MERS-CoV. The disease has highly heterogeneous effects on patients, ranging from asymptomatic infection to severe multi-organ damage leading to death. Patients at risk of developing severe disease include the elderly, and patients with chronic health conditions like obesity and cardiovascular disease [2]. Beyond the acute effects of SARS-CoV-2 infection, long-lasting symptoms are common in convalescent patients that survive the initial infection, a syndrome known as “long COVID” or post-acute sequelae of COVID-19 (PASC) [3], [4], [5].
The disruptive impact of the COVID-19 pandemic has triggered an unprecedented global wave of scientific innovation that has resulted in the rapid discovery and development of multiple vaccines and therapeutics to prevent and treat COVID-19. The first vaccine for the prevention of COVID-19, Gam-COVID-Vac, was approved for emergency use by the Russian Ministry of Health in August 2020, just 5 months after the disease was declared a pandemic. The mRNA-based vaccine from Pfizer, tozinameran, earned the first emergency use approval for a COVID-19 vaccine in the west, specifically in the UK, in December 2020. Since then, over a dozen different vaccines have been approved for clinical use in almost 200 countries [6], [7], [8]. In addition to vaccines, a handful of therapeutics have been approved, fully or for emergency use, for treatment of COVID-19 in different jurisdictions: the broad anti-inflammatory dexamethasone; the antivirals remdesivir and favipiravir; the anti-SARS-CoV-2 antibodies bamlanivimab/etesevimab, casirivimab/imdevimab, sotrovimab, and regdanvimab; the anti-IL-6 antibody tocilizumab; and convalescent plasma [9], [10]. Dozens of additional vaccines and therapies are in late-stage clinical development, promising to add to the armamentarium of available remedies for this devastating disease.
In addition to driving the development of new vaccines and therapeutics, the ongoing scientific surge has enabled a deep, if still incomplete, understanding of SARS-CoV-2 infection and COVID-19 pathology. Within weeks of the discovery of SARS-CoV-2, it was demonstrated that the key host receptor for the virus is ACE2 and that the virus binds to this protein on the surface of human cells via its spike glycoprotein [11]. Viral attachment to host cells leads to internalization of the virus, viral replication, cell death, and release of nascent virions that can infect neighboring cells, perpetuating the replication cycle. While other potential receptors have been described for SARS-CoV-2, ACE2 appears to be the primary receptor, and cells that do not express ACE2 are generally resistant to infection [12]. This is consistent with the observation that the virus primarily infects epithelial cells of the lung and gut which express ACE2, although it has also been suggested that SARS-CoV-2 can infect other cell types within the body, including cardiomyocytes, neurons, endothelial cells, and some leukocytes [13], [14], [15], [16], [17], [18], [19].
In most patients, infection with SARS-CoV-2 is associated with a rapid, often pre-symptomatic, increase in viral load, triggering an immune response that effectively clears the virus within days and is associated with relatively mild symptoms that resemble the flu – mild respiratory distress, fever, and body aches. In other patients, in particular those with underlying risk factors like age and comorbidities, the disease can progress to more severe symptoms including respiratory failure, and in some cases death [20], [21]. Severe COVID-19 is thought to result from a harmful self-perpetuating cycle of hyperinflammation and tissue/cellular damage due to an inappropriately extreme immune response, sometimes associated with cytokine storm and/or ARDS. Much of the work seeking to characterize COVID-19 pathology has focused on the adaptive immune response which is undoubtedly important in pathogen clearance and in establishing immune memory, as demonstrated by the high level of efficacy achieved with COVID-19 vaccines. However, the innate immune response also plays an important role in perpetuating the hyper-inflamed state that contributes to severe forms of COVID-19. Recent data have implicated monocytes, neutrophils, and other myeloid cells, in COVID-19 pathology. In this review, we discuss this expanding appreciation for the role played by the innate immune system in moderate to severe forms of COVID-19, and highlight evidence to support that damage-associated molecular patterns (DAMPs) may be key players in the cycle of damage and inflammation that underlies severe forms of the disease. Furthermore, we suggest that our evolving understanding of COVID-19 pathology will inform future treatment options not only for COVID-19 but also for other diseases, infectious or not, which exhibit similar mechanisms of disease.
2. Immune mechanisms in COVID-19
SARS-CoV-2 is a respiratory virus whose human-to-human transmission is predominantly mediated through droplets and aerosols that are formed during speaking, coughing, and sneezing. Thus, mucosal tissues of the mouth and respiratory tract are among the first to come into contact with the virus. These tissues are also the most vulnerable for viral infection owing to their high expression of ACE2. Infection of the host cells leads to viral replication and release of viral particles associated with cell and tissue damage and release of DAMPs. In severe disease, it is thought that these molecules trigger an inflammatory immune cascade characterized by a vicious cycle of immune cell activation and further loss of tissue integrity.
Patients with severe COVID-19 present with inflammatory foci in the lungs that are detectable macroscopically by X-ray. In these patients, SARS-CoV-2 has typically infected the ACE2-expressing epithelial cells in the lower airway, triggering an immune response that ultimately leads to hospitalization, and possibly to the need for intensive care and intubation [22]. In addition to respiratory symptoms, COVID-19 patients can also develop thrombotic complications, pulmonary embolism and problems associated with increased coagulation, reminiscent of disseminated intravascular coagulation observed in sepsis patients. Patients exhibit elevated D-dimer levels and widespread alveolar capillary microthrombi [23], [24], [25], altered platelet-immune cell interactions [26], [27] and the presence of megakaryocytes in affected lungs. The clinical course of COVID-19 is driven by the host immune response, which can range from appropriate and protective, to uncontrolled and highly dysfunctional, and everywhere in between.
The normal and protective host response to SARS-CoV-2 begins with recognition of pathogen-associated molecular patterns (PAMPs), such as viral single-stranded RNA, by innate immune cells through pattern recognition receptors (PRRs) including toll-like receptors (TLRs), C-type lectin receptors (CLRs), NOD-like receptors (NLRs), RIG-I-like receptors (RLRs) and melanoma differentiation-associated protein 5 (MDA-5). This leads to activation of immune cells, secretion of pro-inflammatory cytokines and establishment of a chemokine gradient to recruit additional innate and adaptive immune cells to the site of inflammation [28]. Similarly, activation of resident myeloid cells leads to their mobilization and migration to the draining lymph node where they present viral antigens to T and B cells, orchestrating the protective adaptive immune response, including the establishment of immune memory. If the immune response is successful, patients typically recover within 7 to 14 days post-infection. The protective nature of an appropriate immune response to SARS-CoV-2 is exemplified in vaccinated individuals who have circulating neutralizing antibodies and memory lymphocytes specific for the viral capsid protein which can fend off infection, or at least lessen the severity and duration of disease.
In a subset of patients, however, the adaptive immune response fails to clear the virus and eventually peters out due to T cells exhaustion and an inadequate B cells/antibody (humoral) response [29]. Persistence of the viral infection leads to continued activation of the innate immune system and production of pro-inflammatory cytokines [30], which can lead to systemic vascular inflammation and aberrant coagulation, among other non-pulmonary symptoms. A reduced type I interferon (IFN-I) response may contribute to the inability of the host to clear the virus, leading to the uncontrolled inflammatory response that drives severe COVID-19 [31], [32], consistent with the higher levels of autoimmune antibodies specific for type I IFNs observed in these patients [33], [34]. However, the role of type I interferon (IFN-I) response in severe COVID-19 patients still remains unclear since some reports show a robust IFN-I response in severe COVID-19 [35], [36], [37].
3. Innate immunity in COVID-19
Much of the available data on the role of the adaptive and innate immune responses in COVID-19 has been gleaned from patient cohort studies utilizing state-of-the-art multi-omics approaches [30], [38], [39], [40], [41], [42], [43]. Initial immunophenotyping studies indicated lymphopenia and increased abundance of neutrophils in severe COVID-19 [44], [45], [46]. Deeper bulk and single-cell transcriptomics approaches identified increased expansion of plasmablasts, megakaryocytes and increased erythropoiesis [39]; basophil depletion, alterations in non-neutrophil myeloid cells including monocytes, macrophages, dendritic cells (DCs), and B/T cell phenotypes [38], [41]; and increase in inflammatory macrophages and altered epithelial-immune cell interactions [43] in severe patients. Overall, these data paint a picture wherein the adaptive immune response driven by CD4+/CD8+ T cells and antibody production is crucial in controlling SARS-CoV-2 infection, whereas severe disease seems to result in part from an altered/dysfunctional and insufficient adaptive response. In such cases, the innate immune response, normally the first line of defense in infection, plays an outsized role, perhaps as a compensatory mechanism for an inadequate adaptive response.
In line with this, early studies into the pathology of COVID-19 showed a systemic increase of a variety of pro-inflammatory cytokines and chemokines (including IL-6, IL-7, IL-10, TNF-α, and IP10) [47]. Furthermore, mononuclear phagocytes are elevated in bronchoalveolar fluid (BALF) of severe COVID-19 patients, pointing to activation and dysregulation of monocytes and macrophages in COVID-19-associated hyperinflammation [[48], [49]]. Peripheral blood is enriched in pro-inflammatory CD14highCD16high monocytes, and has abnormally low levels of non-classical CD14lowCD16high monocytes, in COVID-19 patients confined to intensive care [[30], [50], [51], [52]]. In addition to this inflammatory dysfunction, altered host metabolic processes are observed in severe COVID-19. Specifically, a recent study has shown that increasing disease severity in COVID-19 correlates with differential abundance and metabolic programming of hyperactive T cell subpopulations and two metabolically distinct monocyte subsets. In addition, results from this study have identified acetoacetate and α-ketobutyrate as markers for predicting disease outcome in individuals diagnosed with COVID-19 [53]. Further, the products of purine metabolism, nicotinate and nicotinamide metabolism, tryptophan metabolism, TCA cycle, and arginine metabolism are all also associated with higher disease severity [[54], [55], [56], [57]].
Beyond the studies outlined above, most early assessments of the immune response to SARS-CoV-2 largely overlooked innate immune cells - in particular neutrophils - possibly due to technical challenges (for instance, the study of neutrophils requires freshly drawn blood) as well as to the justified race to study the adaptive immune response with the goal of developing vaccines and identifying neutralizing antibodies as potential treatments for patients. However, evidence from recent studies has begun to implicate neutrophils and neutrophil extracellular trap (NET) formation (NETosis) in the pathophysiology of inflammation, coagulopathy, organ damage, and immunothrombosis associated with severe COVID-19 [[58], [59], [60], [61], [62]]. Indeed, activated neutrophils - rather than platelets - appear to be the drivers of coagulation, through a mechanism mediated by NETs and reminiscent of sepsis [[63], [64]]. As the primary innate immune effectors, it is not surprising that neutrophils also play a critical role in ARDS, characteristic of COVID-19. Neutrophils are armed with an arsenal of microbicidal effectors, including cell damaging reactive oxygen species (ROS) generated by the Rac2/NADPH oxidase complex [[65], [66]]. Additionally, they possess a wealth of anti-microbial enzymes (e.g. neutrophil elastase, cathepsin-G, myeloid peroxidase, matrix metallopeptidases, and peptides (e.g. LL-37, bactericidal permeability increasing protein (BPI)) stored in cytoplasmic granules [[65], [66]]. While these are normally deployed through local degranulation (DG) and respiratory burst (RB) within phagosomes to destroy an internalized pathogen, the excessive DG/RB that occurs the context of inflammatory diseases leads to release of these mediators into the extracellular space, where they can cause collateral damage to host tissues, including the lungs, blood vessels, and others [[67], [68], [69], [70], [71], [72], [73], [74], [75], [76]].
In addition to their more classical functionalities, neutrophils can also cause damage through NETosis, a recently described alternative effector modality in these cells. NETosis involves the progressive re-organization and extrusion of nuclear material, leading to the formation of NETs made of DNA fibers that ensnare pathogens [[77], [78], [79], [80], [81]]. While NETosis likely evolved as a protective mechanism, NETs have also been associated with several pathological conditions ranging from infectious diseases to inflammatory disorders including systemic lupus erythematosus, rheumatoid arthritis, small vessel vasculitis, gout, and cardiovascular disease. For instance, a randomized clinical trial identified an association between markers of NETs and poor outcome in community-acquired pneumonia [82]. Overall, it is thought that NETs are involved in mediating the crosstalk between cells of the innate and adaptive immune systems and can induce localized tissue damage independent of the infecting organism, thus perpetuating a vicious circle of damage and inflammation. Furthermore, NETs are a driver of coagulation in bacterial sepsis, endocarditis, and pneumonia [83], owing to their ability to facilitate thrombus formation by activated platelets.
Recent data show that NETs may play a role in the pathology of COVID-19. Indeed, treatment of healthy neutrophils with serum from COVID-19 patients triggered the release of NETs, and SARS-CoV-2 has been shown to stimulate neutrophils to release NETs via interactions with ACE2 [[58], [59]]. Different constituents of NETs along with other factors such as oxidative stress, excessive immune signaling, and increased alveolar epithelial cell necrosis contribute to release of endogenous DAMPs, severe hypoxia, and eventually ARDS, in patients with severe COVID-19 [[84], [85], [86], [87], [88], [89]]. While DAMPs normally act as key bridging molecules between immune and non-immune cells during the cycle of tissue injury and immune resolution, under pathological conditions, including in COVID-19, they can amplify the innate and adaptive immune responses by directly activating various cell subsets, leading to further inflammation and tissue/cell damage.
4. DAMP signaling in COVID-19
Similar to PAMPs, DAMPs act through various cell-surface and intracellular PRRs such as membrane-bound TLRs and CLRs; cytoplasmic NLRs, RLRs, MDA5, cyclic GMP–AMP synthase (cGAS), absent in melanoma 2 (AIM2), or through non-classical transmembrane proteins such as receptor for advanced glycation end products (RAGE), triggering receptors expressed on myeloid cells (TREMs), G-protein-coupled receptors (GPCRs), transient receptor potential (TRP) and P2X7 receptor (P2X7R) channels [28]. DAMP-PRR signaling triggers the activation of canonical myeloid differentiation primary response gene 88 (MyD88) cascade proteins including IL-1 receptor-associated kinases (IRAKs), transforming growth factor-β activated kinases (TAK), TAK binding proteins (TABs), mitogen-activated protein kinases (MAPKs), and IκB kinase (IKK) isoforms. This results in translocation of NF-κB to the nucleus, transcription of various pro-inflammatory mediators, and regulation of several cellular processes including apoptosis, proliferation, adhesion, and angiogenesis [[90], [91]]. PRR-mediated activation of NF-κB signaling also leads to transcriptional upregulation of intracellular inflammasome genes; DAMPs can bind to and activate NLRP3, leading to caspase-1 autoproteolysis and activation, and cleavage of pro-IL-1β and pro-IL-18 to their active forms, IL-1β and IL-18. Pro-inflammatory signaling by NLRP3 inflammasome results either in cellular death by pyroptosis or activation of downstream processes such as recruitment of immune cell populations, immune surveillance, and cell proliferation [92]. In this way, DAMP-mediated localized inflammatory cell death and signaling can further extend to the vasculature, leading to barrier disintegration and leakage of inflammatory mediators, thus triggering a cycle of cell injury, amplified inflammation, and dysregulation of cellular processes.
Recent plasma proteomic studies have revealed increases in DAMPs including circulating mitochondrial DNA, HMGB1 and S100 proteins in moderate to severe COVID-19 patients [[40], [42], [47], [90], [93], [94], [95], [96]]. Based on their known pro-inflammatory effects, summarized above, high levels of circulating DAMPs are likely to play an exacerbative role in COVID-19. Indeed, it can be surmised that DAMPs play a critical role in driving the uncontrolled immune response associated with COVID-19, as demonstrated in part by the protective effects of Paquinimod, a specific inhibitor of S100A8/A9 which can reduce pathological inflammatory signaling by neutrophils and re-establish an optimal anti-viral response against COVID-19 [97]. In addition, increased HMGB1 in severe COVID-19 patients has been shown to promote ACE2 expression via the RAGE receptor in alveolar epithelial cells, thereby facilitating viral entry into cells [93]. In some instances, DAMPs may play a protective role in disease, such as in the case of elevated levels of alarmins such as S100A8 and S100A9 that are correlated with an anti-viral immune responses [[97], [98]]. Nevertheless, the totality of the data point toward a harmful role of DAMPs in COVID-19. This is further supported by studies implicating other components of DAMP-mediated signaling, including DAMP receptors, and associated signaling components, as discussed below.
5. Siglec signaling in COVID-19
The innate immune machinery has evolved to modulate PAMP/DAMP-PRR signaling pathways at transcriptional, post-transcriptional and post-translational levels [[99], [100]]. For instance, phosphatases SHP-1 and SHP-2 mediate inhibition of PRR signaling by selectively dephosphorylating different components of NF-κB and MAPK pathways [[101], [102]]. The sialic acid–binding immunoglobulin-like lectins (Siglecs) are one group of proteins that utilize SHP-1 to act as a checkpoint on the innate immune system. Indeed, the majority of Siglecs are primarily expressed on innate immune cells, and contain immunoreceptor tyrosine-based inhibitory motifs (ITIMs) or ITIM-like regions in their intracellular domains, through which they suppress DAMP-mediated NF-κB signaling [103] .
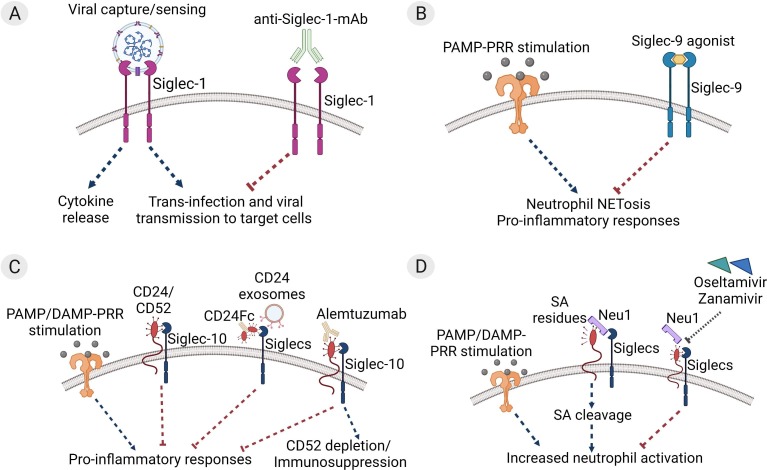
Results from recent studies highlight the potential significance of Siglecs in COVID-19 (Fig. 1 ). For instance, sialylated secreted glycoproteins from SARS-CoV-2 can bind to and activate host Siglecs thus downregulating the antiviral response [104]. Furthermore, SARS-CoV-2 viral spike proteins contain α2,6 and α2,3 linked sialic acids that enable their interaction with Siglec-1, Siglec-3, Siglec-9, and Siglec-10, facilitating their entry into host immune cells [[19], [105]], a phenomenon which can be blocked with an anti-Siglec-1 monoclonal antibody [19] (Fig. 1 A). On the other hand, Siglecs appear to play a protective role in the context of severe COVID-19 by tamping down the uncontrolled inflammation that drives pathology. For example, Siglec-9 has been shown to suppress neutrophil innate immune responses, and transcriptomic studies have identified upregulation of Siglec-9 in neutrophils of severe COVID-19 patients [[30], [106], [107]]. Additionally, a Siglec-9 agonist was found to be effective in inhibiting cellular activation and excessive NETosis in neutrophils from patients with COVID-19 [108] (Fig. 1 B). This suggests an interplay between biological regulation of Siglecs and the cycle of injury perpetuated by aberrant NETosis and increased DAMPs such as HMGB1 and S100 proteins in severe COVID-19.
Fig. 1.
Therapeutic targeting of Siglecs in COVID-19. (A) Viral capture and uptake by Siglec-1 on myeloid antigen presenting cells (APCs) leads to cytokine storm and viral propagation. Anti-Siglec-1 monoclonal antibodies (mAbs) can block uptake of SARS-CoV-2 and inhibit trans-infection of target cells expressing ACE2/TMPRSS2 [19]. (B) SARS-CoV-2 pathogen/damage-associated molecular pattern (PAMP and DAMP)-mediated pattern recognition receptor (PRR) stimulation leads to production of pro-inflammatory neutrophil extracellular traps (NETosis), that propagate the hyperinflammatory cascade in COVID-19. Synthetic Siglec-9 agonists can trigger clustering of Siglec-9 receptors on neutrophils and suppress NETosis and associated inflammation in COVID-19 [108]. (C) Association of CD24 or CD52 with Siglec-10 inhibits PAMP/DAMP-PRR-mediated inflammatory responses. CD24Fc and CD24 exosomes may act through Siglec stimulation to protect against severe COVID-19 [[118], [119]]. Treatment with anti-human CD52 antibody is associated with mild COVID-19 symptoms and may also act through Siglec stimulation [116]. (D) Increased levels of neuraminidase 1 (Neu1) enzyme in severe COVID-19 may prevent protective Siglec activation through cleavage of sialic acid (SA) residues on CD24/CD52. Treatment with Neu inhibitors Oseltamivir or Zanamivir reduces SA shedding and neutrophil overactivation in COVID-19 patients [128].
6. Regulation of DAMP-PRR-Siglec signaling cascade in COVID-19
Under homeostatic conditions, endogenous DAMPs signal via PRRs and non-PRRs to trigger downstream innate immune responses and production of pro-inflammatory cytokines through activation of either the NF-κB or NLRP3 inflammasome cascades [28]. Therefore, modulating the activity of DAMP ligands, receptors, and/or NF-κB/NLRP3 inflammasome signaling components, may be beneficial in various disease states, including severe COVID-19. One possible approach involves activation of the Siglecs, either directly, or through their cognate ligands CD24 and CD52. CD24 and CD52 are related proteins with similar genomic organization, structures, and functionalities. For example, CD24 has been shown to associate with Siglec-10 and downregulate HMGB1-mediated NF-κB inhibition, while soluble CD52 binding to Siglec-10 inhibits T cell receptor-associated kinase phosphorylation and T cell activation [[109], [110]]. Interestingly, recent clinical data support a potential role for CD24 and CD52 in COVID-19 (Fig. 1 C).
Alemtuzumab, an anti-human CD52 antibody, was designed to deplete CD52+ lymphocytes in lymphocyte-mediated diseases such as multiple sclerosis (MS) and graft-versus-host disease [[111], [112]]. While patients treated with alemtuzumab have varying levels of immunodeficiency and an increased risk of infection, case reports indicate that alemtuzumab-treated multiple sclerosis (MS) patients developed mild COVID-19 disease [[113], [114], [115], [116], [117]]. It has not been determined whether milder symptoms are due to CD52 depletion-induced immunosuppression or to Siglec-dependent down-regulation of inflammation. Given its role in inhibition of DAMP signaling and the increased levels of DAMPs in COVID-19, a targeted increase in CD52 signaling, perhaps through dosing of CD52 itself in some form, could be beneficial in the context of severe COVID-19. Indeed, such an approach was used recently with the related protein CD24. Results from an interim analysis of a Phase III clinical trial with CD24Fc, a protein consisting of two molecules of CD24 attached to a single human IgG1 Fc, showed a decreased risk of respiratory failure and death compared with the placebo group, in moderate to severe COVID-19 patients [118]. Another clinical study from the Tel-Aviv Sourasky Medical Center evaluated the efficacy of inhaled CD24-containing exosomes in patients with moderate/severe COVID-19 disease. The results showed that 29 out of 30 patients treated with this therapy fully recovered from disease within three to five days, although no placebo control arm was included in this study [119]. While both CD24 and CD52 drive inhibition of DAMP-mediated inflammation, existing literature suggests that these two proteins could be acting on different immune subsets - CD24 on myeloid populations [[120], [121], [122]] and CD52 on lymphocyte populations [[110], [123], [124], [125]]. Overall, available clinical data are consistent with a protective effect of CD24 and CD52 in COVID-19, though additional studies are required to confirm these beneficial effects, and to better understand the mechanism of protection. Given the multiple known biological effects of CD24 and CD52 – including, but not limited to, activation of Siglecs - any beneficial effects of increasing CD24/CD52-mediated signaling in severe COVID-19 must be weighed against the potential harmful effects of targeting this complex biology.
The role of sialic acid residues in Siglec binding and activation could also be harnessed as a potential therapeutic approach in COVID-19 [[126], [127]]. Neuraminidase (Neu) enzymes, which are expressed at higher levels in the respiratory tract of severe COVID-19 patients [43], cleave sialic acid residues which enhances ROS production and NETosis by inflammatory neutrophils in COVID-19, and these effects can be blocked by the Neu inhibitors oseltamivir or zanamivir (Fig. 1 D) [128]. Similarly, the Neu inhibitor peramivir, in combination with an anti-HMGB1 antibody, attenuated immune signaling and improved survival in an influenza-induced pneumonia mouse model [129].
In addition to targeting Siglec biology, blocking the TLRs, which act as receptors for both PAMPs and DAMPs, is being studied as a way of tamping down inflammation in several diseases including COVID-19 [[130], [131], [132]]. Additionally, cytokine blockers - for example the anti-IL-6R antibodies tocilizumab (Actemra) and sarilumab (Kevzara) and the anti-IL-6 antibody siltuximab (Sylvant) - are also being tested for efficacy COVID-19 patients. Results from these studies are mixed and indicate that while anti-IL6 drugs/TLR therapeutics may have a marginal effect on mortality in severe cases of COVID-19, the timing of treatment relative to infection onset appears to be critical for efficacy [[133], [134], [135]]. Furthermore, IL-6 is only one of several proinflammatory mediators released in response to DAMP-mediated inflammation, highlighting the need to identify targets that are higher up in the inflammation cascade, including, perhaps, the DAMPs themselves.
In summary, available data suggest that therapeutic targeting of the pro-inflammatory DAMP-PRR and anti-inflammatory Siglec pathways is promising for the treatment of severe COVID-19, although additional studies are required to validate this approach, to identify therapeutic targets within these pathways that will appropriately balance benefit and risk, and to inform about patient stratification and timing of therapy.
7. Impact of DAMP signaling on other dysregulated processes in COVID-19 and in related disorders
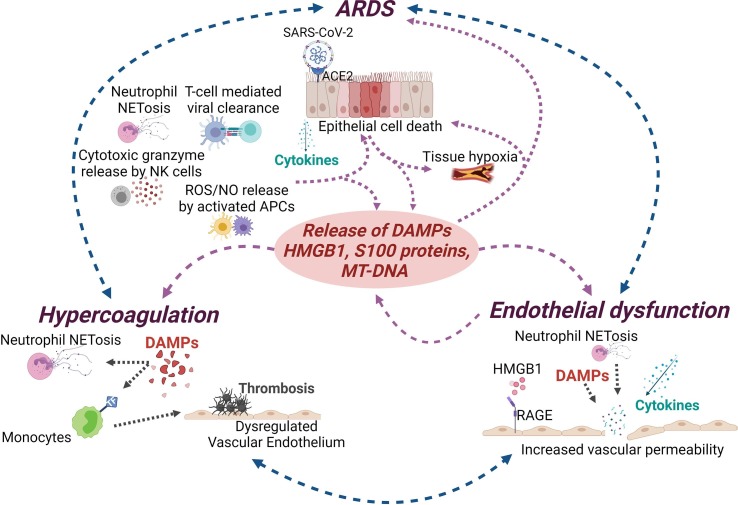
Severe COVID-19 is associated not only with pulmonary symptoms/ARDS, but also with systemic complications including endothelial dysfunction and hypercoagulability [[136], [137], [138]]. Vascular endothelial cells (ECs) express ACE2, making them direct targets for SARS-CoV-2 infection [[139], [140], [141]]. In addition, excessive inflammation associated with COVID-19 ARDS leads to increased pro-inflammatory cytokine signaling and NETosis which results in activation of ECs, and ultimately, endothelial dysfunction. Activated ECs, in turn, increase NET formation, leading to a positive feedback loop that further propagates EC dysfunction [[142], [143]]. This damage to the vascular endothelium causes platelet aggregation, resulting in a prothrombotic phenotype and increased coagulation. In addition, breakdown of the endothelium due to NETosis in the intravascular and perivascular space destabilizes the EC barrier leading to vascular leakage [[80], [81], [143], [144], [145], [146], [147], [148], [149], [150]]. The mechanism of endothelial damage and vascular leakage in COVID-19 can be in part surmised from studies in sepsis, which show many of the same hallmarks as severe COVID-19. For instance, in sepsis, circulating neutrophils undergo ‘intravascular priming’ coupled to microvascular sequestration, and this increased prevalence of primed neutrophils, as well as neutrophil clustering, correlates with leak and severity of disease [[67], [151], [152], [153], [154], [155], [156], [157], [158]]. Vascular damage-induced tissue hypoxia and thrombosis-induced ischemic injury/ROS production also leads to the release of DAMPs, thus fueling the cycle of inflammatory, coagulative, and dysregulated cellular responses. This constant source of DAMPs can impact immunothrombosis and thrombus formation in multiple ways; while DAMPs act on neutrophils to induce formation of NETs, they also act on monocytes to induce expression of tissue factor (TF) [159]. Moreover, different DAMPs can have different effects on immune and non-immune cell subsets, including endothelial cells. For instance, HMGB1 has been shown to induce RAGE-dependent cytokine production in endothelial cells as well as platelets, leading to barrier dysfunction and increased coagulation [[160], [161], [162]], while another DAMP, S100A9, drives thrombus formation and vascular injury, in mouse models [163].
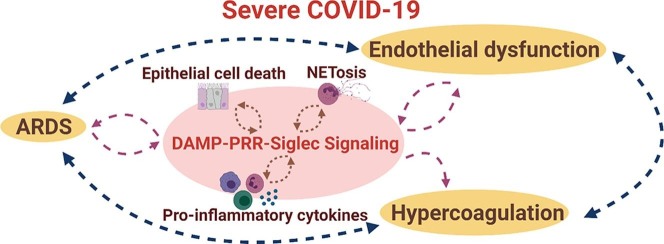
Taken together, existing data highlight the impact of DAMPs on multiple pathophysiological aspects of SARS-CoV-2 infection and emphasize the potential benefits of inhibiting DAMP signaling in severe COVID-19 and related disorders (Fig. 2 ).
Fig. 2.
Impact of DAMPs on COVID-19 and associated disorders. Damage-associated molecular patterns (DAMPs) act as a central driver of the feedback loop between cell/tissue damage and hyperactivation of the innate immune response, thus playing a central role in COVID-19 associated complications. SARS-CoV- 2 infection and the resulting inflammation can cause epithelial cell death and further release of cytokines and DAMPs that can lead to acute respiratory distress syndrome (ARDS). DAMP-mediated inflammation can also cause damage to the vascular endothelium, platelet activation, thrombosis, and prolonged inflammation resulting in vascular leakage, and hypercoagulation.
8. Future perspectives
Impaired immune cell function leading to prolonged uncontrolled inflammation is the hallmark of severe COVID-19 pathology. For lack of better options, one common strategy used to control ongoing inflammation is administration of broad-spectrum corticosteroids. Although these drugs ameliorate clinical symptoms in critically ill patients if administered in a timely manner, the resulting immunosuppression can lead to an increased risk of infections. These complications necessitate a careful weighing of the risk–benefit-ratio and optimization of dose, timing, and duration when it comes to administration of steroids. Therefore, further research including additional clinical trials will be crucial to evaluate the safety and efficacy of broad interventional therapies targeting inflammation in severe COVID-19.
Like other pathogens, SARS-CoV-2 drives innate immune responses not only through generation of PAMPs that activate PRRs, but also through the generation of endogenous DAMPs. While DAMPs can have a beneficial effect by contributing to anti-viral inflammatory responses, their continuous release perpetuates overaction of innate/adaptive immune cells and cytokine storm, leading to several adverse complications such as ARDS, endothelial barrier dysfunction, and increased coagulation. Thus, targeted modulation of DAMP signaling, and associated pathway proteins may be an effective tool in modulating the complex immunological networks and inflammation associated with severe COVID-19. A handful of therapeutics targeting DAMP-mediated signaling have been clinically evaluated in COVID-19 patients, with promising preliminary outcomes. While their development will require additional investigation, targeting DAMPs in COVID-19 and related disorders could address the issues associated with broader anti-inflammatory approaches.
CRediT authorship contribution statement
Upasana Parthasarathy: Conceptualization, Writing – original draft, Writing – review & editing. Roberta Martinelli: Conceptualization, Writing – original draft, Writing – review & editing. Elisabeth H. Vollmann: Writing – original draft. Katharine Best: Writing – original draft. Alex G. Therien: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
All authors are employees of Merck & Co., Inc.
Acknowledgements
Figure illustrations were created using BioRender.com.
References
- 1.World Health Organization (WHO) | Coronavirus (COVID-19) Dashboard. https://covid19whoint/. 2021.
- 2.Stefan N., Birkenfeld A.L., Schulze M.B. Global pandemics interconnected - obesity, impaired metabolic health and COVID-19. Nature reviews. Endocrinology. 2021;17(3):135–149. doi: 10.1038/s41574-020-00462-1. [DOI] [PubMed] [Google Scholar]
- 3.Sudre C.H., Murray B., Varsavsky T., Graham M.S., Penfold R.S., Bowyer R.C., et al. Attributes and predictors of long COVID. Nature medicine. 2021;27(4) doi: 10.1038/s41591-021-01292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Lancet Facing up to long COVID. Lancet (London, England) 2020;396(10266):1861. doi: 10.1016/S0140-6736(20)32662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proal A.D., VanElzakker M.B. Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Frontiers in microbiology. 2021;12 doi: 10.3389/fmicb.2021.698169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J.H., Marks F., Clemens J.D. Looking beyond COVID-19 vaccine phase 3 trials. Nature medicine. 2021;27(2):205–211. doi: 10.1038/s41591-021-01230-y. [DOI] [PubMed] [Google Scholar]
- 7.U.S. Food & Drug Administration (FDA) | COVID-19 Vaccines. https://wwwfdagov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines. 2021. [PubMed]
- 8.World Health Organization (WHO) | COVID-19 vaccine tracker and landscape. https://wwwwhoint/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. 2021.
- 9.National Institute of Health (NIH) | Anti-SARS-CoV-2 Monoclonal Antibodies; COVID-19 Treatment Guidelines. https://wwwcovid19treatmentguidelinesnihgov/therapies/anti-sars-cov-2-antibody-products/anti-sars-cov-2-monoclonal-antibodies/. 2021.
- 10.COVID-19 Treatment Guidelines. 2021 https://wwwcovid19treatmentguidelinesnihgov/management/clinical-management/hospitalized-adults–therapeutic-management/ [Google Scholar]
- 11.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X.i., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schäfer R., Spohn G., Bechtel M., Bojkova D., Baer P.C., Kuçi S., Seifried E., Ciesek S., Cinatl J. Human Mesenchymal Stromal Cells Are Resistant to SARS-CoV-2 Infection under Steady-State, Inflammatory Conditions and in the Presence of SARS-CoV-2-Infected Cells. Stem cell reports. 2021;16(3):419–427. doi: 10.1016/j.stemcr.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science (New York, NY). 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sungnak W., Huang N.i., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature medicine. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Geng X., Tan Y., Li Q., Xu C., Xu J., Hao L., Zeng Z., Luo X., Liu F., Wang H. New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;127:110195. doi: 10.1016/j.biopha.2020.110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullen C.K., Hogberg H.T., Bahadirli-Talbott A., Bishai W.R., Hartung T., Keuthan C., et al. Infectability of human BrainSphere neurons suggests neurotropism of SARS-CoV-2. ALTEX. 2020;37(4) doi: 10.14573/altex.2006111. [DOI] [PubMed] [Google Scholar]
- 17.Bojkova D, Wagner JUG, Shumliakivska M, Aslan GS, Saleem U, Hansen A, et al. SARS-CoV-2 infects and induces cytotoxic effects in human cardiomyocytes. Cardiovascular research. 2020;116(14). [DOI] [PMC free article] [PubMed]
- 18.Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet (London, England). 2020;395(10234). [DOI] [PMC free article] [PubMed]
- 19.Perez-Zsolt D., Muñoz-Basagoiti J., Rodon J., Elousa M., Raïch-Regué D., Risco C., et al. Siglec-1 on dendritic cells mediates SARS-CoV-2 trans-infection of target cells while on macrophages triggers proinflammatory responses. BioRxiv 443572 [Preprint]. Available from. 2021 doi: 10.1101/2021.05.11.443572. [DOI] [Google Scholar]
- 20.Guan W.-J., Ni Z.-y., Hu Y.u., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-l., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L.i., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S. Clinical Characteristics of Coronavirus Disease 2019 in China. The New England journal of medicine. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention (CDC) | COVID-19 Mortality Overview. NVSS - Provisional Death Counts for COVID-19 - Executive Summary. https://wwwcdcgov/nchs/covid19/mortality-overviewhtm. 2021.
- 22.Rendeiro A.F., Ravichandran H., Bram Y., Chandar V., Kim J., Meydan C., Park J., Foox J., Hether T., Warren S., Kim Y., Reeves J., Salvatore S., Mason C.E., Swanson E.C., Borczuk A.C., Elemento O., Schwartz R.E. The spatial landscape of lung pathology during COVID-19 progression. Nature. 2021;593(7860):564–569. doi: 10.1038/s41586-021-03475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rapkiewicz A.V., Mai X., Carsons S.E., Pittaluga S., Kleiner D.E., Berger J.S., Thomas S., Adler N.M., Charytan D.M., Gasmi B., Hochman J.S., Reynolds H.R. Megakaryocytes and platelet-fibrin thrombi characterize multi-organ thrombosis at autopsy in COVID-19: A case series. EClinicalMedicine. 2020;24:100434. doi: 10.1016/j.eclinm.2020.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lax S.F., Skok K., Zechner P., Kessler H.H., Kaufmann N., Koelblinger C., Vander K., Bargfrieder U., Trauner M. Pulmonary Arterial Thrombosis in COVID-19 With Fatal Outcome : Results From a Prospective, Single-Center, Clinicopathologic Case Series. Annals of internal medicine. 2020;173(5):350–361. doi: 10.7326/M20-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voeght A.D., Calmes D., Beck F., Sylvestre J.-B., Delvenne P., Peters P., et al. Thrombotic microvascular injury is not mediated by thrombotic microangiopathy despite systemic complement activation in Covid-19 patients. MedRxiv 20115873 [Preprint]. Available from. 2020 doi: 10.1101/2020.06.18.20115873. [DOI] [Google Scholar]
- 26.Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, Stubben C, et al. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136(11). [DOI] [PMC free article] [PubMed]
- 27.Leppkes M., Knopf J., Naschberger E., Lindemann A., Singh J., Herrmann I., Stürzl M., Staats L., Mahajan A., Schauer C., Kremer A.N., Völkl S., Amann K., Evert K., Falkeis C., Wehrfritz A., Rieker R.J., Hartmann A., Kremer A.E., Neurath M.F., Muñoz L.E., Schett G., Herrmann M. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58:102925. doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong T., Liu L., Jiang W., Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nature reviews Immunology. 2020;20(2):95–112. doi: 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- 29.Diao B.o., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L.i., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y.i., Wu Y., Chen Y. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Frontiers in immunology. 2019;11 doi: 10.3389/fimmu.2020.0082710.3389/fimmu.2020.00827.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., et al. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell. 2020;182(6) doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science (New York, NY). 2020;369(6504). [DOI] [PMC free article] [PubMed]
- 32.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181(5):1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez J., Mommert M., Mouton W., Pizzorno A., Brengel-Pesce K., Mezidi M., et al. Early nasal type I IFN immunity against SARS-CoV-2 is compromised in patients with autoantibodies against type I IFNs. The Journal of experimental medicine. 2021;218(10) doi: 10.1084/jem.20211211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., et al. Vol. 370. Science (New York; NY): 2020. (Autoantibodies against type I IFNs in patients with life-threatening COVID-19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J.S., Shin E.-C. The type I interferon response in COVID-19: implications for treatment. Nature reviews Immunology. 2020;20(10):585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.-S., Jeong S.J., Lee H.K., Park S.H., Park S.-H., Choi J.Y., Kim S.-H., Jung I., Shin E.-C. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Science immunology. 2020;5(49) doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z., Ren L., Zhang L.i., Zhong J., Xiao Y., Jia Z., Guo L.i., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y.u., Chen M., Gao Z., Yang J., Dong J., Liu B.o., Zhang X., Wang W., He K., Jin Q.i., Li M., Wang J. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell host & microbe. 2020;27(6):883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su Y., Chen D., Yuan D., Lausted C., Choi J., Dai C.L., et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell. 2020;183(6) doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernardes J.P., Mishra N., Tran F., Bahmer T., Best L., Blase J.I., et al. Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity. 2020;53(6) doi: 10.1016/j.immuni.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y.-M., Zheng Y., Yu Y., Wang Y., Huang Q., Qian F., Sun L., Song Z.-G., Chen Z., Feng J., An Y., Yang J., Su Z., Sun S., Dai F., Chen Q., Lu Q., Li P., Ling Y., Yang Z., Tang H., Shi L., Jin L.i., Holmes E.C., Ding C., Zhu T.-Y., Zhang Y.-Z. Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage. The EMBO journal. 2020;39(24) doi: 10.15252/embj.2020105896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laing A.G., Lorenc A., Barrio I.D.M.D., Das A., Fish M., Monin L., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nature medicine. 2020;26(10) doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 42.Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., Rameau P., Catelain C., Alfaro A., Dussiau C., Friedrich C., Sourdeau E., Marin N., Szwebel T.-A., Cantin D., Mouthon L., Borderie D., Deloger M., Bredel D., Mouraud S., Drubay D., Andrieu M., Lhonneur A.-S., Saada V., Stoclin A., Willekens C., Pommeret F., Griscelli F., Ng L.G., Zhang Z., Bost P., Amit I., Barlesi F., Marabelle A., Pène F., Gachot B., André F., Zitvogel L., Ginhoux F., Fontenay M., Solary E. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell. 2020;182(6):1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Müller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.-E., Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nature biotechnology. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y., Du X., Chen J., Jin Y., Peng L.i., Wang H.H.X., Luo M., Chen L., Zhao Y. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. The Journal of infection. 2020;81(1):e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y., Nie H.-X., Hu K.e., Wu X.-J., Zhang Y.-T., Wang M.-M., Wang T., Zheng Z.-S., Li X.-C., Zeng S.-L. Abnormal immunity of non-survivors with COVID-19: predictors for mortality. Infectious diseases of poverty. 2020;9(1) doi: 10.1186/s40249-020-00723-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao K., Li R., Wu X., Zhao Y., Wang T., Zheng Z., Zeng S., Ding X., Nie H. Clinical features in 52 patients with COVID-19 who have increased leukocyte count: a retrospective analysis. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical. Microbiology. 2020;39(12):2279–2287. doi: 10.1007/s10096-020-03976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L., Long X., Xu Q., Tan J., Wang G., Cao Y., Wei J., Luo H., Zhu H., Huang L., Meng F., Huang L., Wang N.a., Zhou X., Zhao L., Chen X., Mao Z., Chen C., Li Z., Sun Z., Zhao J., Wang D., Huang G., Wang W., Zhou J. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cellular & Molecular Immunology. 2020;17(9):992–994. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nature medicine. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 49.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nature reviews Immunology. 2020;20(6):355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang D, Guo R, Lei L, Liu H, Wang Y, Wang Y, et al. Frontline Science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. Journal of leukocyte biology. 2021;109(1). [DOI] [PMC free article] [PubMed]
- 51.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., et al. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. National Science. Review. 2020 doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell discovery. 2020;6(1) doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JW, Su Y, Baloni P, Chen D, Pavlovitch-Bedzyk AJ, Yuan D, et al. Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nature biotechnology. 2021. [DOI] [PMC free article] [PubMed]
- 54.Ansone L., Ustinova M., Terentjeva A., Perkons I., Birzniece L., Rovite V., et al. Tryptophan and arginine metabolism is significantly altered at the time of admission in hospital for severe COVID-19 patients: findings from longitudinal targeted metabolomics analysis. MedRxiv 21254699 [Preprint]. Available from. 2021 doi: 10.1101/2021.03.31.21254699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blasco H., Bessy C., Plantier L., Lefevre A., Piver E., Bernard L., Marlet J., Stefic K., Benz-de Bretagne I., Cannet P., Lumbu H., Morel T., Boulard P., Andres C.R., Vourc’h P., Hérault O., Guillon A., Emond P. The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Scientific reports. 2020;10(1) doi: 10.1038/s41598-020-73966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu J., Zhou M., Luo P., Yin Z., Wang S., Liao T., et al. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2021. Plasma metabolomic profiling of patients recovered from COVID-19 with pulmonary sequelae 3 months after discharge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B-W, Fan X, Cao W-J, Tian H, Wang S-Y, Zhang J-Y, et al. Systematic Discovery and Pathway Analyses of Metabolic Disturbance in COVID-19. Infectious Diseases & Immunity. 2021. [DOI] [PMC free article] [PubMed]
- 58.Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, et al. Neutrophil extracellular traps in COVID-19. JCI insight. 2020;5(11). [DOI] [PMC free article] [PubMed]
- 59.Veras FP, Pontelli MC, Silva CM, Toller-Kawahisa JE, Lima Md, Nascimento DC, et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. The Journal of experimental medicine. 2020;217(12). [DOI] [PMC free article] [PubMed]
- 60.Middleton EA, He X-Y, Denorme F, Campbell RA, Ng D, Salvatore SP, et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136(10). [DOI] [PMC free article] [PubMed]
- 61.Borges L, Pithon-Curi TC, Curi R, Hatanaka E. Excessive Neutrophils and Neutrophil Extracellular Traps in COVID-19. Frontiers in immunology. 2020;11. [DOI] [PMC free article] [PubMed]
- 62.Ng H., Havervall S., Rosell A., Aguilera K., Parv K., von Meijenfeldt F.A., Lisman T., Mackman N., Thålin C., Phillipson M. Circulating Markers of Neutrophil Extracellular Traps Are of Prognostic Value in Patients With COVID-19. Arteriosclerosis, thrombosis, and vascular biology. 2021;41(2):988–994. doi: 10.1161/ATVBAHA.120.315267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petito E, Falcinelli E, Paliani U, Cesari E, Vaudo G, Sebastiano M, et al. Association of Neutrophil Activation, More Than Platelet Activation, With Thrombotic Complications in Coronavirus Disease 2019. The Journal of infectious diseases. 2021;223(6). [DOI] [PMC free article] [PubMed]
- 64.Busch M.H., Timmermans S.A.M.E.G., Nagy M., Visser M., Huckriede J., Aendekerk J.P., de Vries F., Potjewijd J., Jallah B., Ysermans R., Oude Lashof A.M.L., Breedveld P.H., van de Poll M.C.G., van de Horst I.C.C., van Bussel B.C.T., Theunissen R.O.M.F.I.H., Spronk H.M.H., Damoiseaux J.G.M.C., ten Cate H., Nicolaes G.A.F., Reutelingsperger C.P., van Paassen P. Neutrophils and Contact Activation of Coagulation as Potential Drivers of COVID-19. Circulation. 2020;142(18):1787–1790. doi: 10.1161/CIRCULATIONAHA.120.050656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lacy P. Mechanisms of degranulation in neutrophils. Allergy, asthma, and clinical immunology : official journal of the Canadian Society of Allergy and Clinical Immunology. 2006;2(3):98–108. doi: 10.1186/1710-1492-2-3-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Almyroudis N.G., Grimm M.J., Davidson B.A., Rohm M., Urban C.F., Segal B.H. NETosis and NADPH oxidase: at the intersection of host defense, inflammation, and injury. Front Immunol. 2013;4:45. doi: 10.3389/fimmu.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown K.A., Brain S.D., Pearson J.D., Edgeworth J.D., Lewis S.M., Treacher D.F. Neutrophils in development of multiple organ failure in sepsis. Lancet. 2006;368(9530):157–169. doi: 10.1016/S0140-6736(06)69005-3. [DOI] [PubMed] [Google Scholar]
- 68.Levi M., van der Poll T. Endothelial injury in sepsis. Intensive care medicine. 2013;39(10):1839–1842. doi: 10.1007/s00134-013-3054-1. [DOI] [PubMed] [Google Scholar]
- 69.Berton G., Yan S.R., Fumagalli L., Lowell C.A. Neutrophil activation by adhesion: mechanisms and pathophysiological implications. International journal of clinical & laboratory research. 1996;26(3):160–177. doi: 10.1007/BF02592978. [DOI] [PubMed] [Google Scholar]
- 70.Albelda S.M., Smith C.W., Ward P.A. Adhesion molecules and inflammatory injury. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1994;8(8):504–512. [PubMed] [Google Scholar]
- 71.Lowell C.A., Berton G. Integrin signal transduction in myeloid leukocytes. Journal of leukocyte biology. 1999;65(3):313–320. doi: 10.1002/jlb.65.3.313. [DOI] [PubMed] [Google Scholar]
- 72.DiStasi M.R., Ley K. Opening the flood-gates: how neutrophil-endothelial interactions regulate permeability. Trends in immunology. 2009;30(11):547–556. doi: 10.1016/j.it.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nathan C., Sanchez E. Tumor necrosis factor and CD11/CD18 (beta 2) integrins act synergistically to lower cAMP in human neutrophils. J Cell Biol. 1990;111(5 Pt 1):2171–2181. doi: 10.1083/jcb.111.5.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suchard S.J., Boxer L.A. Exocytosis of a subpopulation of specific granules coincides with H2O2 production in adherent human neutrophils. J Immunol. 1994;152(1):290–300. [PubMed] [Google Scholar]
- 75.Mocsai A., Ligeti E., Lowell C.A., Berton G. Adhesion-dependent degranulation of neutrophils requires the Src family kinases Fgr and Hck. J Immunol. 1999;162(2):1120–1126. [PubMed] [Google Scholar]
- 76.Forsyth KevinD, Fitzpatrick MargaretM, Simpson AnnaC, Barratt T.M., Levinsky RolandJ. Neutrophil-mediated endothelial injury in haemolytic uraemic syndrome. Lancet. 1989;334(8660):411–414. doi: 10.1016/s0140-6736(89)90591-6. [DOI] [PubMed] [Google Scholar]
- 77.Fuchs T.A., Brill A., Wagner D.D. Neutrophil extracellular trap (NET) impact on deep vein thrombosis. Arterioscler Thromb Vasc Biol. 2012;32(8):1777–1783. doi: 10.1161/ATVBAHA.111.242859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Demers M., Wagner D.D. Neutrophil extracellular traps: A new link to cancer-associated thrombosis and potential implications for tumor progression. Oncoimmunology. 2013;2(2):e22946. doi: 10.4161/onci.22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martinod K, Wagner DD. Thrombosis: tangled up in NETs. Blood. 2013. [DOI] [PMC free article] [PubMed]
- 80.Brinkmann V., Zychlinsky A. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol. 2012;198(5):773–783. doi: 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yipp B.G., Kubes P. NETosis: how vital is it? Blood. 2013;122(16):2784–2794. doi: 10.1182/blood-2013-04-457671. [DOI] [PubMed] [Google Scholar]
- 82.Ebrahimi F., Giaglis S., Hahn S., Blum C.A., Baumgartner C., Kutz A., van Breda S.V., Mueller B., Schuetz P., Christ-Crain M., Hasler P. Markers of neutrophil extracellular traps predict adverse outcome in community-acquired pneumonia: secondary analysis of a randomised controlled trial. The European respiratory journal. 2018;51(4):1701389. doi: 10.1183/13993003.01389-201710.1183/13993003.01389-2017.Supp1. [DOI] [PubMed] [Google Scholar]
- 83.Zucoloto AZ, Jenne CN. Platelet-Neutrophil Interplay: Insights Into Neutrophil Extracellular Trap (NET)-Driven Coagulation in Infection. Frontiers in cardiovascular medicine. 2019;6. [DOI] [PMC free article] [PubMed]
- 84.Grasselli G, Tonetti T, Filippini C, Slutsky AS, Pesenti A, Ranieri VM. Pathophysiology of COVID-19-associated acute respiratory distress syndrome. The Lancet Respiratory Medicine. 2021;9(1). [DOI] [PMC free article] [PubMed]
- 85.Nailwal H., Chan F.-M. Necroptosis in anti-viral inflammation. Cell death and differentiation. 2019;26(1):4–13. doi: 10.1038/s41418-018-0172-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pouwels S.D., Heijink I.H., ten Hacken N.HT., Vandenabeele P., Krysko D.V., Nawijn M.C., van Oosterhout A.JM. DAMPs activating innate and adaptive immune responses in COPD. Mucosal immunology. 2014;7(2):215–226. doi: 10.1038/mi.2013.77. [DOI] [PubMed] [Google Scholar]
- 87.Land W.G. Role of DAMPs in respiratory virus-induced acute respiratory distress syndrome-with a preliminary reference to SARS-CoV-2 pneumonia. Genes and immunity. 2021;22(3):141–160. doi: 10.1038/s41435-021-00140-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huckriede J, Anderberg SB, Morales A, Vries Fd, Hultström M, Bergqvist A, et al. Evolution of NETosis markers and DAMPs have prognostic value in critically ill COVID-19 patients. Scientific reports. 2021;11(1). [DOI] [PMC free article] [PubMed]
- 89.Mikacenic C., Moore R., Dmyterko V., West T.E., Altemeier W.A., Liles W.C., Lood C. Neutrophil extracellular traps (NETs) are increased in the alveolar spaces of patients with ventilator-associated pneumonia. Critical care (London. England) 2018;22(1) doi: 10.1186/s13054-018-2290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fujishima S. Pathophysiology and biomarkers of acute respiratory distress syndrome. Journal of intensive care. 2014;2(1) doi: 10.1186/2052-0492-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aboudounya M.M., Heads R.J., Dozio E. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediators of inflammation. 2021;2021:1–18. doi: 10.1155/2021/8874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Freeman T.L., Swartz T.H. Targeting the NLRP3 Inflammasome in Severe COVID-19. Frontiers in immunology. 2020;11 doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen R., Huang Y., Quan J., Liu J., Wang H., Billiar T.R., Lotze M.T., Zeh H.J., Kang R., Tang D. HMGB1 as a potential biomarker and therapeutic target for severe COVID-19. Heliyon. 2020;6(12):e05672. doi: 10.1016/j.heliyon.2020.e05672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aceti A., Margarucci L.M., Scaramucci E., Orsini M., Salerno G., Di Sante G., Gianfranceschi G., Di Liddo R., Valeriani F., Ria F., Simmaco M., Parnigotto P.P., Vitali M., Romano Spica V., Michetti F. Serum S100B protein as a marker of severity in Covid-19 patients. Scientific reports. 2020;10(1) doi: 10.1038/s41598-020-75618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fan X, Song J-W, Wang S-Y, Cao W-J, Wang X-W, Zhou M-J, et al. Changes of Damage Associated Molecular Patterns in COVID-19 Patients. Infectious Diseases & Immunity. 2021. [DOI] [PMC free article] [PubMed]
- 96.Scozzi D, Cano M, Ma L, Zhou D, Zhu JH, O'Halloran JA, et al. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI insight. 2021;6(4). [DOI] [PMC free article] [PubMed]
- 97.Guo Q., Zhao Y., Li J., Liu J., Yang X., Guo X., Kuang M., Xia H., Zhang Z., Cao L., Luo Y., Bao L., Wang X., Wei X., Deng W., Wang N., Chen L., Chen J., Zhu H., Gao R., Qin C., Wang X., You F. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell host & microbe. 2021;29(2):222–235.e4. doi: 10.1016/j.chom.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi H., Zuo Y., Yalavarthi S., Gockman K., Zuo M., Madison J.A., et al. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. Journal of Leukocyte Biology. 2020 doi: 10.1002/JLB.3COVCRA0720-359R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carpenter S., Ricci E.P., Mercier B.C., Moore M.J., Fitzgerald K.A. Post-transcriptional regulation of gene expression in innate immunity. Nature reviews Immunology. 2014;14(6):361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 100.Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nature reviews Immunology. 2016;16(1):35–50. doi: 10.1038/nri.2015.8. [DOI] [PubMed] [Google Scholar]
- 101.Hardin A.O., Meals E.A., Yi T., Knapp K.M., English B.K. SHP-1 inhibits LPS-mediated TNF and iNOS production in murine macrophages. Biochemical and biophysical research communications. 2006;342(2):547–555. doi: 10.1016/j.bbrc.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 102.An H., Zhao W., Hou J., Zhang Y., Xie Y., Zheng Y., Xu H., Qian C., Zhou J., Yu Y., Liu S., Feng G., Cao X. SHP-2 phosphatase negatively regulates the TRIF adaptor protein-dependent type I interferon and proinflammatory cytokine production. Immunity. 2006;25(6):919–928. doi: 10.1016/j.immuni.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 103.Crocker P.R., Paulson J.C., Varki A. Siglecs and their roles in the immune system. Nature reviews Immunology. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 104.Murch S.H. Common determinants of severe Covid-19 infection are explicable by SARS-CoV-2 secreted glycoprotein interaction with the CD33-related Siglecs, Siglec-3 and Siglec-5/14. Medical hypotheses. 2020;144:110168. doi: 10.1016/j.mehy.2020.110168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chiodo F., Bruijns S.C.M., Rodriguez E., Li R.J.E., Molinaro A., Silipo A., et al. Novel ACE2-Independent Carbohydrate-Binding of SARS-CoV-2 Spike Protein to Host Lectins and Lung Microbiota. BioRxiv 092478 [Preprint]. Available from. 2020 doi: 10.1101/2020.05.13.092478. [DOI] [Google Scholar]
- 106.Carlin AF, Uchiyama S, Chang Y-C, Lewis AL, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113(14). [DOI] [PMC free article] [PubMed]
- 107.Aschenbrenner AC, Mouktaroudi M, Krämer B, Oestreich M, Antonakos N, Nuesch-Germano M, et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome medicine. 2021;13(1). [DOI] [PMC free article] [PubMed]
- 108.Delaveris C.S., Wilk A.J., Riley N.M., Stark J.C., Yang S.S., Rogers A.J., Ranganath T., Nadeau K.C., Blish C.A., Bertozzi C.R. Synthetic Siglec-9 Agonists Inhibit Neutrophil Activation Associated with COVID-19. ACS central science. 2021;7(4):650–657. doi: 10.1021/acscentsci.0c01669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen G-Y, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science (New York, NY). 2009;323(5922). [DOI] [PMC free article] [PubMed]
- 110.Bandala-Sanchez E., Zhang Y., Reinwald S., Dromey J.A., Lee B.-H., Qian J., Böhmer R.M., Harrison L.C. T cell regulation mediated by interaction of soluble CD52 with the inhibitory receptor Siglec-10. Nature immunology. 2013;14(7):741–748. doi: 10.1038/ni.2610. [DOI] [PubMed] [Google Scholar]
- 111.Riechmann L., Clark M., Waldmann H., Winter G. Reshaping human antibodies for therapy. Nature. 1988;332(6162):323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- 112.Lowenstein H., Shah A., Chant A., Khan A. Different mechanisms of Campath-1H-mediated depletion for CD4 and CD8 T cells in peripheral blood. Transplant international : official journal of the European Society for Organ. Transplantation. 2006;19(11):927–936. doi: 10.1111/j.1432-2277.2006.00382.x. [DOI] [PubMed] [Google Scholar]
- 113.Fiorella C, Lorna G. COVID-19 in a multiple sclerosis (MS) patient treated with alemtuzumab: Insight to the immune response after COVID. Multiple sclerosis and related disorders. 2020;46. [DOI] [PMC free article] [PubMed]
- 114.Guevara C, Villa E, Cifuentes M, Naves R, Graziaa Jd. Mild COVID-19 infection in a patient with multiple sclerosis and severe depletion of T-lymphocyte subsets due to alemtuzumab. Multiple sclerosis and related disorders. 2020;44. [DOI] [PMC free article] [PubMed]
- 115.Fernández-Díaz E, Gracia-Gil J, García-García JG, Palao M, Romero-Sánchez CM, Segura T. COVID-19 and multiple sclerosis: A description of two cases on alemtuzumab. Multiple sclerosis and related disorders. 2020;45. [DOI] [PMC free article] [PubMed]
- 116.Matías-Guiu J, Montero-Escribano P, Pytel V, Porta-Etessam J, Matias-Guiu JA. Potential COVID-19 infection in patients with severe multiple sclerosis treated with alemtuzumab. Multiple sclerosis and related disorders. 2020;44. [DOI] [PMC free article] [PubMed]
- 117.Carandini T, Pietroboni AM, Sacchi L, Riz MAD, Pozzato M, Arighi A, et al. Alemtuzumab in multiple sclerosis during the COVID-19 pandemic: A mild uncomplicated infection despite intense immunosuppression. Multiple sclerosis (Houndmills, Basingstoke, England). 2020;26(10). [DOI] [PubMed]
- 118.ClinicalTrials.gov. CD24Fc as a Non-antiviral Immunomodulator in COVID-19 Treatment - Full Text View - ClinicalTrials.gov. https://clinicaltrialsgov/ct2/show/NCT04317040. 2021.
- 119.ClinicalTrials.gov. Evaluation of the Safety of CD24-Exosomes in Patients With COVID-19 Infection - Full Text View - ClinicalTrials.gov. https://clinicaltrialsgov/ct2/show/NCT04747574. 2021.
- 120.Barkal A.A., Brewer R.E., Markovic M., Kowarsky M., Barkal S.A., Zaro B.W., Krishnan V., Hatakeyama J., Dorigo O., Barkal L.J., Weissman I.L. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. 2019;572(7769):392–396. doi: 10.1038/s41586-019-1456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Toubai T, Hou G, Mathewson N, Liu C, Wang Y, Oravecz-Wilson K, et al. Siglec-G-CD24 axis controls the severity of graft-versus-host disease in mice. Blood. 2014;123(22). [DOI] [PMC free article] [PubMed]
- 122.Chen G.-Y., Chen X.i., King S., Cavassani K.A., Cheng J., Zheng X., Cao H., Yu H., Qu J., Fang D., Wu W., Bai X.-F., Liu J.-Q., Woodiga S.A., Chen C., Sun L., Hogaboam C.M., Kunkel S.L., Zheng P., Liu Y. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nature biotechnology. 2011;29(5):428–435. doi: 10.1038/nbt.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Samten B. CD52 as both a marker and an effector molecule of T cells with regulatory action: Identification of novel regulatory T cells. Cellular & molecular immunology. 2013;10(6):456–458. doi: 10.1038/cmi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bandala-Sanchez E, Bediaga NG, Goddard-Borger ED, Ngui K, Naselli G, Stone NL, et al. CD52 glycan binds the proinflammatory B box of HMGB1 to engage the Siglec-10 receptor and suppress human T cell function. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(30). [DOI] [PMC free article] [PubMed]
- 125.Bhamidipati K., Silberstein J.L., Chaichian Y., Baker M.C., Lanz T.V., Zia A., Rasheed Y.S., Cochran J.R., Robinson W.H. CD52 Is Elevated on B cells of SLE Patients and Regulates B Cell Function. Frontiers in immunology. 2021;11 doi: 10.3389/fimmu.2020.62682010.3389/fimmu.2020.626820.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pillai S., Netravali I.A., Cariappa A., Mattoo H. Siglecs and immune regulation. Annual review of immunology. 2012;30(1):357–392. doi: 10.1146/annurev-immunol-020711-075018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Duan S., Paulson J.C. Siglecs as Immune Cell Checkpoints in Disease. Annual review of immunology. 2020;38(1):365–395. doi: 10.1146/annurev-immunol-102419-035900. [DOI] [PubMed] [Google Scholar]
- 128.Formiga R.O., Amaral F.C., Souza C.F., Mendes D.A.G.B., Wanderley C.W.S., Lorenzini C.B., et al. Neuraminidase inhibitors rewire neutrophil function in vivo in murine sepsis and ex vivo in COVID-19. BioRxiv 379115 [Preprint] Available from. 2020 doi: 10.1101/2020.11.12.379115. [DOI] [Google Scholar]
- 129.Hatayama K., Nosaka N., Yamada M., Yashiro M., Fujii Y., Tsukahara H., Liu K., Nishibori M., Matsukawa A., Morishima T. Combined effect of anti-high-mobility group box-1 monoclonal antibody and peramivir against influenza A virus-induced pneumonia in mice. Journal of medical virology. 2019;91(3):361–369. doi: 10.1002/jmv.25330. [DOI] [PubMed] [Google Scholar]
- 130.Gambuzza M., Licata N., Palella E., Celi D., Foti Cuzzola V., Italiano D., Marino S., Bramanti P. Targeting Toll-like receptors: emerging therapeutics for multiple sclerosis management. Journal of neuroimmunology. 2011;239(1-2):1–12. doi: 10.1016/j.jneuroim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 131.Lloyd C. Chemokines in allergic lung inflammation. Immunology. 2002;105(2). [DOI] [PMC free article] [PubMed]
- 132.Piccinini A.M., Midwood K.S. DAMPening inflammation by modulating TLR signalling. Mediators of inflammation. 2010;2010:1–21. doi: 10.1155/2010/672395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52(6) doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Castelnovo L, Tamburello A, Lurati A, Zaccara E, Marrazza MG, Olivetti M, et al. Anti-IL6 treatment of serious COVID-19 disease: A monocentric retrospective experience. Medicine. 2021;100(1). [DOI] [PMC free article] [PubMed]
- 135.Patra R., Chandra Das N., Mukherjee S. Targeting human TLRs to combat COVID-19: A solution? Journal of medical virology. 2021;93(2):615–617. doi: 10.1002/jmv.26387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Evans PC, Rainger GE, Mason JC, Guzik TJ, Osto E, Stamataki Z, et al. Endothelial dysfunction in COVID-19: a position paper of the ESC Working Group for Atherosclerosis and Vascular Biology, and the ESC Council of Basic Cardiovascular Science. Cardiovascular research. 2020;116(14). [DOI] [PMC free article] [PubMed]
- 137.Klok F.A., Kruip M.J.H.A., van der Meer N.J.M., Arbous M.S., Gommers D.A.M.P.J., Kant K.M., Kaptein F.H.J., van Paassen J., Stals M.A.M., Huisman M.V., Endeman H. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thrombosis research. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mangalmurti N.S., Reilly J.P., Cines D.B., Meyer N.J., Hunter C.A., Vaughan A.E. COVID-19-associated Acute Respiratory Distress Syndrome Clarified: A Vascular Endotype? American journal of respiratory and critical care medicine. 2020;202(5):750–753. doi: 10.1164/rccm.202006-2598LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., Li W.W., Li V.W., Mentzer S.J., Jonigk D. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. The New England journal of medicine. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pons S., Fodil S., Azoulay E., Zafrani L. The vascular endothelium: the cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Critical care (London, England) 2020;24(1) doi: 10.1186/s13054-020-03062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends in immunology. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gupta AK, Joshi MB, Philippova M, Erne P, Hasler P, Hahn S, et al. Activated endothelial cells induce neutrophil extracellular traps and are susceptible to NETosis-mediated cell death. FEBS letters. 2010;584(14). [DOI] [PubMed]
- 143.Villanueva E., Yalavarthi S., Berthier C.C., Hodgin J.B., Khandpur R., Lin A.M., Rubin C.J., Zhao W., Olsen S.H., Klinker M., Shealy D., Denny M.F., Plumas J., Chaperot L., Kretzler M., Bruce A.T., Kaplan M.J. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. Journal of immunology (Baltimore, Md. 2011;187(1):538–552. doi: 10.4049/jimmunol.1100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wolfson R.K., Chiang E.T., Garcia J.G.N. HMGB1 induces human lung endothelial cell cytoskeletal rearrangement and barrier disruption. Microvascular research. 2011;81(2):189–197. doi: 10.1016/j.mvr.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu J., Zhang X., Pelayo R., Monestier M., Ammollo C.T., Semeraro F., Taylor F.B., Esmon N.L., Lupu F., Esmon C.T. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15(11):1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fuchs T.A., Brill A., Duerschmied D., Schatzberg D., Monestier M., Myers D.D., Wrobleski S.K., Wakefield T.W., Hartwig J.H., Wagner D.D. Extracellular DNA traps promote thrombosis. Proc Natl Acad Sci U S A. 2010;107(36):15880–15885. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McDonald B., Urrutia R., Yipp B., Jenne C., Kubes P. Intravascular neutrophil extracellular traps capture bacteria from the bloodstream during sepsis. Cell host & microbe. 2012;12(3):324–333. doi: 10.1016/j.chom.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 148.Massberg S., Grahl L., von Bruehl M.-L., Manukyan D., Pfeiler S., Goosmann C., Brinkmann V., Lorenz M., Bidzhekov K., Khandagale A.B., Konrad I., Kennerknecht E., Reges K., Holdenrieder S., Braun S., Reinhardt C., Spannagl M., Preissner K.T., Engelmann B. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat Med. 2010;16(8):887–896. doi: 10.1038/nm.2184. [DOI] [PubMed] [Google Scholar]
- 149.Kessenbrock K., Krumbholz M., Schönermarck U., Back W., Gross W.L., Werb Z., Gröne H.-J., Brinkmann V., Jenne D.E. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med. 2009;15(6):623–625. doi: 10.1038/nm.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Clark S.R., Ma A.C., Tavener S.A., McDonald B., Goodarzi Z., Kelly M.M., Patel K.D., Chakrabarti S., McAvoy E., Sinclair G.D., Keys E.M., Allen-Vercoe E., DeVinney R., Doig C.J., Green F.H.Y., Kubes P. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13(4):463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 151.Wagner J.G., Roth R.A. Neutrophil migration during endotoxemia. Journal of leukocyte biology. 1999;66(1):10–24. doi: 10.1002/jlb.66.1.10. [DOI] [PubMed] [Google Scholar]
- 152.Lewis S.M., Khan N., Beale R., Treacher D.F., Brown K.A. Depletion of blood neutrophils from patients with sepsis: treatment for the future? International immunopharmacology. 2013;17(4):1226–1232. doi: 10.1016/j.intimp.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 153.Brealey D., Singer M. Multi-organ dysfunction in the critically ill: effects on different organs. Journal of the Royal College of Physicians of London. 2000;34(5):428–431. [PMC free article] [PubMed] [Google Scholar]
- 154.Quaid G.A., Cave C., Robinson C., Williams M.A., Solomkin J.S. Preferential loss of CXCR-2 receptor expression and function in patients who have undergone trauma. Arch Surg. 1999;134(12):1367–1371. doi: 10.1001/archsurg.134.12.1367. discussion 71–2. [DOI] [PubMed] [Google Scholar]
- 155.Tellado J.M., Christou N.V. Critically ill anergic patients demonstrate polymorphonuclear neutrophil activation in the intravascular compartment with decreased cell delivery to inflammatory focci. Journal of leukocyte biology. 1991;50(6):547–553. doi: 10.1002/jlb.50.6.547. [DOI] [PubMed] [Google Scholar]
- 156.Nuytinck H.K., Offermans X.J., Kubat K., Goris J.A. Whole-body inflammation in trauma patients. An autopsy study. Arch Surg. 1988;123(12):1519–1524. doi: 10.1001/archsurg.1988.01400360089016. [DOI] [PubMed] [Google Scholar]
- 157.Thijs A., Thijs L.G. Pathogenesis of renal failure in sepsis. Kidney international Supplement. 1998;66:S34–S37. [PubMed] [Google Scholar]
- 158.Wyman T.H., Bjornsen A.J., Elzi D.J., Smith C.W., England K.M., Kelher M., Silliman C.C. A two-insult in vitro model of PMN-mediated pulmonary endothelial damage: requirements for adherence and chemokine release. American journal of physiology Cell physiology. 2002;283(6):C1592–C1603. doi: 10.1152/ajpcell.00540.2001. [DOI] [PubMed] [Google Scholar]
- 159.Ito T. PAMPs and DAMPs as triggers for DIC. Journal of intensive care. 2014;2(1) doi: 10.1186/s40560-014-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fiuza C, Bustin M, Talwar S, Tropea M, Gerstenberger E, Shelhamer JH, et al. Inflammation-promoting activity of HMGB1 on human microvascular endothelial cells. Blood. 2003;101(7). [DOI] [PubMed]
- 161.Treutiger C.J., Mullins G.E., Johansson A.-S.- M., Rouhiainen A., Rauvala H.M.E., Erlandsson-Harris H., Andersson U., Yang H., Tracey K.J., Andersson J., Palmblad J.E.W. High mobility group 1 B-box mediates activation of human endothelium. Journal of internal medicine. 2003;254(4):375–385. doi: 10.1046/j.1365-2796.2003.01204.x. [DOI] [PubMed] [Google Scholar]
- 162.Ahrens I., Chen Y.-C., Topcic D., Bode M., Haenel D., Hagemeyer C.E., Seeba H., Duerschmied D., Bassler N., Jandeleit-Dahm K.A., Sweet M.J., Agrotis A., Bobik A., Peter K. HMGB1 binds to activated platelets via the receptor for advanced glycation end products and is present in platelet rich human coronary artery thrombi. Thrombosis and haemostasis. 2015;114(11):994–1003. doi: 10.1160/TH14-12-1073. [DOI] [PubMed] [Google Scholar]
- 163.Kawano T., Shimamura M., Nakagami H., Iso T., Koriyama H., Takeda S., Baba K., Sasaki T., Sakaguchi M., Morishita R., Mochizuki H. Therapeutic Vaccine Against S100A9 (S100 Calcium-Binding Protein A9) Inhibits Thrombosis Without Increasing the Risk of Bleeding in Ischemic Stroke in Mice. Hypertension (Dallas. Tex. 2018;72(6):1355–1364. doi: 10.1161/HYPERTENSIONAHA.118.11316. [DOI] [PubMed] [Google Scholar]