Abstract
We developed and evaluated a prototype automated specimen preparation instrument for the specific capture of hepatitis C virus (HCV) RNA with probes and magnetic bead-fluid separation. HCV RNA was isolated from serum by lysis of virus particles with a chaotropic agent, followed by hybridization of the RNA with biotinylated probes and capture of the hybridized RNA with streptavidin-coated paramagnetic particles. After washing of the hybrid-particle complexes to remove nonspecifically bound materials, the particles were resuspended in a specimen diluent and were then ready for amplification and detection with a fully automated PCR system (COBAS AMPLICOR; Roche Diagnostic Systems). The analytical sensitivity in the dilution series was 33 copies per ml or greater. Comparison of the test results with those obtained by a manual method based on organic extraction and precipitation of RNA (SepaGene RV-R; Sanko Junyaku Co., Ltd.) showed 93% (49 of 53 samples) sensitivity and 100% (12 of 12 samples) specificity. There was 94% overall agreement between results. When RNA was extracted by the manual method from serum containing 103 or 105 copies of HCV per ml in the presence of heparin, there was an inhibitory effect on detection of both HCV RNA and the internal control. In contrast, when RNA was extracted from the serum by the automated method, there was no inhibitory effect. This inhibitory effect of heparin on the manual method was also observed for a series of serum specimens from a hemodialysis patient, but the inhibitory effect was eliminated by the automated specimen preparation method. In summary, a fully automated RNA extraction system for PCR detection of HCV RNA by use of specific capture with probes and magnetic bead-fluid separation was shown to have performance similar to that of the conventional manual method. In addition, it successfully eliminated the inhibitory effect of the heparin in the serum and permitted the detection of HCV RNA in serum samples from a hemodialysis patient. The prototype automated RNA extraction system is suitable as a totally automated system, starting with RNA extraction to detection of HCV, if it was combined with the fully automated COBAS AMPLICOR PCR system.
Advances in molecular biology and biotechnology have facilitated analyses for detection of DNA or RNA sequences (16). New technological advances have led to the automation of major portions of the assay process (2, 8). Automated systems have been developed for amplification and detection of the nucleic acid sequences of infectious agents by PCR. An example of this is the COBAS AMPLICOR system (Roche Diagnostic Systems, Branchburg, N.J.), which amplifies target nucleic acid sequences, captures the biotinylated and amplified products on oligonucleotide-coated paramagnetic microparticles, and detects the products with an avidin-horseradish peroxidase conjugate system (3, 7). The remaining portion of the process to be automated is extraction of nucleic acid from clinical specimens.
Extraction of RNA from serum specimens for laboratory use has been widely performed with commercially available kits. However, they are not very user-friendly when one is handling numerous samples at once, and the extraction efficiency varies among samples. Moreover, those kits cannot eliminate some inhibitors of enzymatic amplification that may be present in clinical specimens and that may cause false-negative PCR results (1, 14). Heparin is a potent inhibitor that may be present in blood specimens. It has been used as an anticoagulant for blood collection and hemodialysis and for the treatment of disseminated intravascular coagulation. Because the presence of heparin in a sample makes the interpretation of results difficult, sera from patients undergoing hemodialysis may not be appropriate for testing by PCR (15). Since a high prevalence of hepatitis C virus (HCV) infection has been reported in patients undergoing hemodialysis (6, 13), accurate assays for the detection of HCV RNA in those patients need to be developed. In the study described here, we developed and evaluated a prototype automated specimen preparation instrument for the specific capture of HCV RNA with probes and magnetic bead-fluid (B-F) separation. In particular, we addressed the questions of whether the extraction system could eliminate the inhibitory effects of heparin and whether it was suitable for automated RNA extraction followed by detection of HCV with the fully automated COBAS AMPLICOR PCR system.
MATERIALS AND METHODS
Clinical specimens.
The serum specimens used in this study were obtained from 65 patients referred to Tokai University Hospital for chronic liver diseases. To determine the limit of detection of the assay, serial dilutions of serum from a patient containing 530 copies of HCV per ml were prepared in HCV-negative serum. To assess the effect of heparin, a series of serum samples was obtained from a hemodialysis patient who was anticoagulated with 3,400 U of heparin. When needed, HCV RNA was quantitatively measured by the AMPLICOR HCV MONITOR test (Roche Diagnostic Systems) (5). All samples were separated from clots within 4 h of collection, divided into aliquots, and stored at −80°C until the RNA was extracted.
RNA extraction.
HCV RNA was isolated from serum with a prototype instrument for an automated system consisting of the reagents developed by Roche Molecular Systems (Pleasanton, Calif.) and a robotic processor developed by Precision System Science Co. Ltd. (Tokyo, Japan) (Fig. 1). Briefly, HCV RNA was isolated from 300 μl of serum by lysis of virus particles with 500 μl of guanidinium thiocyanate solution at 60°C for 20 min. The RNA was hybridized with biotinylated probes (KY78) that were specific to the 5′ untranslated region of the HCV genome (11) and that were identical to the downstream primer for amplification. The hybridized RNA was then captured with streptavidin-coated paramagnetic particles. The internal control (40 copies) was introduced into the specimen during the lysis reaction. After washing of the hybrid-particle complexes to remove nonspecifically bound materials, the particles were resuspended in 50 μl of a specimen diluent and were then ready for amplification and detection by the COBAS AMPLICOR HCV test.
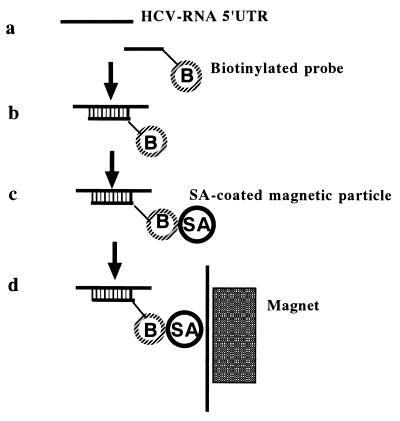
FIG. 1.
Assay format. (a) Virus is lysed to release nucleic acid. (b) Nucleic acid is bound to biotinylated (B) probe. (c) Biotinylated (B) probe is captured onto streptavidin (SA)-coated paramagnetic particles. (d) Paramagnetic particles are separated and washed on a magnet. 5′ UTR, 5′ untranslated region.
RNA was also isolated from 300 μl of serum by a manual method based on guanidinium thiocyanate lysis and isopropanol precipitation (SepaGene RV-R; Sanko Junyaku Co., Ltd., Tokyo, Japan). Briefly, 300 μl of guanidinium thiocyanate solution was added to the specimen, and the phases were then separated by adding 300 μl of sodium acetate and 600 μl of chloroform-agglutination solution, followed by centrifugation at 12,000 × g for 15 min. The RNA was precipitated from the upper phase with isopropanol and was resuspended in 50 μl of a specimen diluent. The internal control (20 copies) was added to the specimen diluent prior to amplification.
COBAS AMPLICOR HCV test.
The COBAS AMPLICOR HCV test has been described previously (3, 7). The optical density of each reaction mixture at 660 nm was measured. Optical density readings of >0.200 for HCV RNA were considered positive, and those of 0.100 to 0.200 were considered equivocal. Optical density readings of >0.200 for the internal control were considered positive.
All samples were extracted once, and a single amplification and detection were performed with each extract unless stated otherwise. One positive control and two negative controls provided with the kit were run with each batch of patient specimens.
Interference by heparin.
To assess the effect of heparin, 3 to 13 U of sodium heparin per ml was added to a serum sample containing 103 or 105 copies of HCV per ml. The RNA was extracted from the serum by the SepaGene RV-R method and the specific capture with probes and magnetic B-F separation. To assess the effect of heparin treatment in vivo, a series of serum specimens containing 5.1 × 105 copies of HCV per ml, as quantified by the AMPLICOR HCV MONITOR test, was serially obtained from a patient before and at 1, 3, and 5 h after hemodialysis in which 3,400 U of heparin was used as an anticoagulant. RNA was extracted from the serum, and then HCV RNA was amplified by the COBAS AMPLICOR HCV test. To demonstrate the presence of inhibitors, the series of serum samples obtained after hemodialysis from the hemodialysis patient was diluted 1:10 in HCV-negative serum, and RNA was then extracted from the serum by the SepaGene RV-R method.
RESULTS
Assay performance.
The analytical sensitivity based on testing of the serial dilutions was 33 copies of HCV per ml or greater (Table 1). Comparison of test results with those obtained by the manual method showed 93% (49 of 53 samples) sensitivity and 100% (12 of 12 samples) specificity. There was 94% overall agreement between the results obtained by the automated method and those obtained by the manual method (Table 2).
TABLE 1.
Limit of detection of automated specimen preparation method
| No. of viral copies/ml | No. of samples with positive results/total no. tested | % Agreement |
|---|---|---|
| 265 | 2/2 | 100 |
| 132 | 6/6 | 100 |
| 66 | 6/6 | 100 |
| 33 | 6/6 | 100 |
| 16.6 | 4/6 | 67 |
| 0 | 0/6 | 100 |
TABLE 2.
Comparison of extraction methods for detection of HCV RNA
| SepaGene RV-R method result | No. of samples with the following automated extraction resulta:
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 49 | 4 | 53 |
| Negative | 0 | 12 | 12 |
| Total | 49 | 16 | 65 |
Sensitivity, 93% (49 of 53 samples); specificity, 100% (12 of 12 samples); agreement, 94% (61 of 65 samples).
Elimination of inhibitory effect of heparin.
Heparin was added at up to 13 U/ml to serum containing 103 or 105 copies of HCV per ml, and then RNA was extracted. When RNA was extracted from both of these serum samples by the manual method, the PCR results were negative (Table 3). Analysis of the results for the internal control indicated that there was PCR inhibition for the PCR-negative samples. In contrast, when RNA was extracted by the automated method, there was a lack of inhibition.
TABLE 3.
Inhibitory effects of heparin on PCRa
| No. of viral copies/ml | Extraction method | RNA | Test results (A660) for serum samples with the following heparin concn (U/ml):
|
||
|---|---|---|---|---|---|
| 3 | 6.5 | 13 | |||
| 103 | Manual | HCV | − (0.028) | − (0.022) | − (0.018) |
| IC | − (0.014) | − (0.009) | − (0.012) | ||
| Automated | HCV | + (3.992) | + (3.992) | + (3.992) | |
| IC | + (3.993) | + (3.993) | + (3.993) | ||
| 105 | Manual | HCV | − (0.042) | − (0.018) | − (0.019) |
| IC | − (0.012) | − (0.010) | − (0.007) | ||
| Automated | HCV | + (3.995) | + (3.995) | + (3.995) | |
| IC | + (3.993) | + (3.993) | + (3.843) | ||
Heparin was added to serum containing 103 or 105 copies of HCV per ml. RNA was extracted by the conventional manual method (the SepaGene RV-R method) and the automated method based on probe capture and magnetic B-F separation, and the HCV RNA was amplified with the COBAS AMPLICOR system. Data are means of duplicate assays. IC, internal control.
Elimination of inhibitory effects on PCR with serum from a hemodialysis patient.
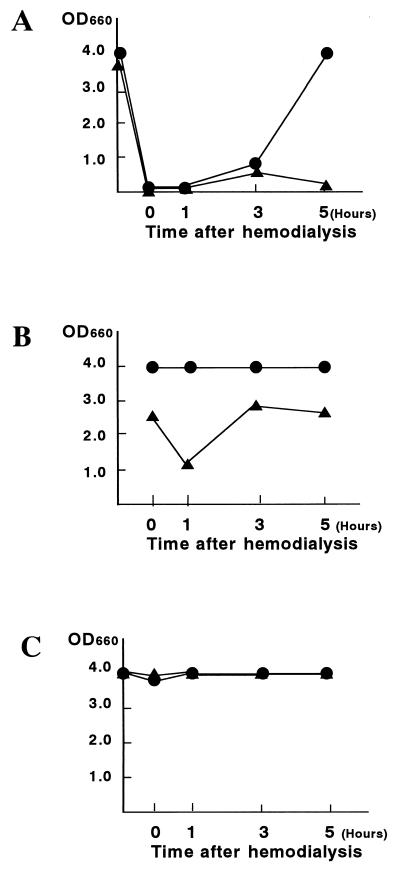
When RNA was extracted by the manual method from serum obtained at various times after hemodialysis, the HCV PCR assay was negative at least until 3 h after hemodialysis (Fig. 2A). Analysis of the results for the internal control indicated that there was PCR inhibition for these PCR-negative samples. Dilution of the sample 1:10 and a repeat assay produced positive PCR results for both HCV RNA and the internal control (Fig. 2B). In contrast, with extraction by the automated method, the inhibitory effect on detection of both HCV RNA and the internal control was abolished for the serum sample obtained immediately after hemodialysis (Fig. 2C).
FIG. 2.
Elimination of inhibitory effects on PCR of serum from a hemodialysis patient by the automated specific capture with probes and magnetic B-F separation. HCV RNA-positive serum was obtained from a hemodialysis patient, who was anticoagulated with 3,400 U of heparin, at 0, 1, 3, and 5 h following hemodialysis. Serum samples were diluted 1:10 in HCV RNA-negative serum. RNA was extracted from undiluted (A) and diluted (B) aliquots by the manual method (the SepaGene RV-R method) and then amplified for HCV RNA and the internal control with the COBAS AMPLICOR system. HCV RNA was extracted from undiluted aliquots by the automated extraction method based on probe capture and magnetic B-F separation, and then amplified for HCV RNA with the COBAS AMPLICOR system (C). Data are means of duplicate assays. ●, HCV RNA; ▴, internal control.
DISCUSSION
In the study described in this report, we developed and evaluated a prototype automated specimen preparation instrument for the specific capture of HCV RNA with probes and magnetic B-F separation. Extracted RNA was successfully used in an automated PCR assay for the detection of HCV RNA with the COBAS AMPLICOR system. The analytical sensitivity of the automated extraction procedure was similar to that of the SepaGene RV-R method. There was 94% overall agreement between the results obtained by the automated method and those obtained by the manual method. The automated RNA extraction system would be suitable as a totally automated system starting with RNA extraction to detection of HCV if it was combined with a fully automated PCR system.
An automated system for PCR assays provides improvements not only in labor efficiency but also in the accuracy of results. Major problems with PCR assays are false-positive results because of carryover contamination of previously amplified products and false-negative results because of the amplification inhibitors present in clinical specimens (4, 12). In the COBAS AMPLICOR system, the use of dUTP and uracil-N-glycosylase reduces the risk of carryover contamination (9). With regard to total quality control, false-negative PCR results become more problematic. In particular, the inhibitory effect of heparin on PCR has been problematic with samples from hemodialysis patients (1). When RNA was extracted from sera by the SepaGene RV-R method, there was an inhibitory effect on the detection of both HCV RNA and the internal control at least until 3 h after hemodialysis. The inhibitory effect of heparin was successfully eliminated when RNA was extracted by the automated probe capture and magnetic B-F separation method. This extraction system would be applicable to PCR assays of serum specimens from hemodialysis patients.
One of the major advantages of nucleic acid amplification over conventional methods for the diagnosis of an infectious disease is sensitive detection of agents directly from clinical specimens. However, clinical specimens may contain a variety of amplification inhibitors such as heparin, hemoglobin, heme, urea, and so on (4, 12). The nature of many inhibitors is still unknown. Therefore, it is desirable to use an extraction method that can eliminate inhibitors as much as possible and monitor the efficacy of extraction. The extraction method used in the present study is theoretically suitable for eliminating any inhibitors in the serum, because it is based on specific capture with probes and magnetic B-F separation. On the other hand, conventional manual methods such as the SepaGene RV-R method are not able to eliminate heparin, which is thought to bind to nucleic acids (1). In the present study, the internal control was demonstrated to be useful for monitoring for the presence of inhibitors in a patient under hemodialysis as well as in an in vitro interference study. It could be used to determine which samples yield invalid results and therefore need to be retested (10). Since many kinds of clinical specimens, from body fluids to tissues, are subjected to tests for direct detection of infectious agents, development of automated systems is desirable for nucleic acid extraction procedures that can be applied to a variety of clinical specimens, in addition to serum specimens.
In summary, a fully automated RNA extraction system was developed. The system was based on the specific capture with probes and magnetic B-F separation, with the performance of the assay being comparable to that of the conventional manual method. In addition to that, it successfully eliminated inhibitors, such as heparin, from serum and can be used for the assay of serum from patients undergoing hemodialysis. The automated RNA extraction system is suitable for use as a totally automated system, starting with RNA extraction to detection of HCV, when combined with a fully automated PCR system.
ACKNOWLEDGMENT
We thank Takatoshi Kakuta of the Department of Internal Medicine, Tokai University School of Medicine, for providing patient materials.
REFERENCES
- 1.Beutler E, Gelbart T, Kuhl W. Interference of heparin with the polymerase chain reaction. BioTechniques. 1990;9:166. [PubMed] [Google Scholar]
- 2.Diamandis E P. Automation of molecular diagnostics. Clin Chem. 1996;42:7–8. [PubMed] [Google Scholar]
- 3.DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z G, Rosenstraus M. COBAS AMPLICOR™: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chem. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 4.Greenfield L, White T J. Sample preparation methods. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 122–137. [Google Scholar]
- 5.Ichijo T, Matsumoto A, Kobayashi M, Furihata K, Tanaka E. Quantitative measurement of HCV RNA in the serum: a comparison of three assays based on different principles. J Gastroenterol Hepatol. 1997;12:500–506. doi: 10.1111/j.1440-1746.1997.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 6.Jeffers L J, Perez G O, De Medina M D, Ortiz-Interian C J, Schiff E R, Reddy K R, Jimenez M, Bourgoignie J J, Vaamonde C A, Duncan R, Houghton M, Choo G-L, Kuo G. Hepatitis C infection in two urban hemodialysis units. Kidney Int. 1990;38:320–322. doi: 10.1038/ki.1990.203. [DOI] [PubMed] [Google Scholar]
- 7.Jungkind D, DiRenzo S, Beavis K G, Silverman N S. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J Clin Microbiol. 1996;34:2778–2783. doi: 10.1128/jcm.34.11.2778-2783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landegren U, Kaizer R, Caskey C T, Hood L. DNA diagnostics. Molecular techniques and automation. Science. 1988;242:229–237. doi: 10.1126/science.3051381. [DOI] [PubMed] [Google Scholar]
- 9.Longo M C, Berninger M S, Hartley J L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 10.Miyachi H, Masukawa A, Ohshima T, Fusegawa H, Hirose T, Impraim C, Ando Y. Monitoring of inhibitors of enzymatic amplification in polymerase chain reaction and evaluation of efficacy of RNA extraction for the detection of hepatitis C virus using the internal control. Clin Chem Lab Med. 1998;36:571–575. doi: 10.1515/CCLM.1998.098. [DOI] [PubMed] [Google Scholar]
- 11.Nolte F S, Thurmond C, Fried M W. Preclinical evaluation of AMPLICOR hepatitis C virus test for detection of hepatitis C virus RNA. J Clin Microbiol. 1995;33:1775–1778. doi: 10.1128/jcm.33.7.1775-1778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolfs A, Schuller I, Finckh U, Weber-Rolfs I. PCR: clinical diagnostics and research. Berlin, Germany: Springer-Verlag; 1992. pp. 51–67. [Google Scholar]
- 13.Silini E, Bono F, Cerino A, Piazza V, Solcia E, Mondelli M U. Virological features of hepatitis C virus infection in hemodialysis patients. J Clin Microbiol. 1993;31:2913–2917. doi: 10.1128/jcm.31.11.2913-2917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasa G, Uuskula M, Hirvonen A, Mikelsaar A-V. Optimization of PCR to yield successful amplification from heparin-contaminated DNA. Methods Mol Cell Biol. 1995;5:122–124. [Google Scholar]
- 15.Willems M, Moshage H, Nevens F, Fevery J, Yap S H. Plasma collected from heparinized blood is not suitable for HCV-RNA detection by conventional RT-PCR assay. J Virol Methods. 1993;42:127–130. doi: 10.1016/0166-0934(93)90184-s. [DOI] [PubMed] [Google Scholar]
- 16.Young F E. DNA probes: fruits of the new biotechnology. JAMA. 1987;229:2404–2406. doi: 10.1001/jama.258.17.2404. [DOI] [PubMed] [Google Scholar]