Abstract
PCR and culture were comparatively evaluated for their abilities to demonstrate Francisella tularensis in wound specimens from tularemia patients during an outbreak in Sweden in 1998. For transport of the specimens used for PCR, a buffer solution containing a nuclease inhibitor was used, and for transport of the specimens used for culture, a commercial transport system was selected after experimental comparison of various systems. Of 40 patients with culture- and/or serology-verified ulceroglandular tularemia, PCR detected F. tularensis DNA in 30 (75%) patients, whereas culture detected bacterial growth in 25 (62%) patients. Compared to data from a previous study, the present inclusion of a nuclease inhibitor in the transport medium did not improve the sensitivity of the PCR, whereas the sensitivity of the culture procedure was significantly increased by selection of the system used for transport. Among eight patients with clinically suspected tularemia but with negative serology and culture, specimens from four patients showed detectable DNA. In three of these patients the diagnosis was verified by the demonstration of an F. tularensis-specific T-cell response in vitro. In conclusion, PCR was more sensitive than culture for demonstration of F. tularensis in wound specimens. Besides, we showed that tularemia may proceed without development of serum antibodies, and in these patients, PCR may be of special importance for verification of the diagnosis.
Francisella tularensis is endemic throughout the Northern Hemisphere and causes outbreaks of tularemia in various mammals including rodents, lagomorphs, and humans. In humans, the clinical presentation depends on the route of entrance of the bacteria. The ulceroglandular form of the disease is acquired either by direct contact with an infected animal or by vector transmission. Patients typically present with fever, enlarged and tender lymph nodes, and an ulcer at the place of entry (4, 5). The skin lesion is usually slight, and the appearance of an infected insect bite need not actually differ from that of a noninfected bite.
F. tularensis is highly virulent, and in diagnostic work involving culture procedures, nonvaccinated staff are at high risk of acquiring clinical disease (3). In most clinical laboratories, serology is the only diagnostic test used. Exceptions are patients with septicemia in whom tularemia may be diagnosed more or less accidentally by growth of F. tularensis in blood cultures. Consequently, some work is performed to optimize the blood culture procedure for F. tularensis (11, 12). There is, however, little experience with the use of culture of wound specimens in clinical diagnostic work. The optimal way of sampling and the optimal handling of wound specimens during transport are unknown, and therefore, the potential efficacy of the procedure is also unknown.
Rapid methods for the identification of F. tularensis such as the immunofluorescence assay and the enzyme-linked immunosorbent assay for the detection of antigen and the RNA hybridization assay have been tried but have so far not been included in routine diagnostics (9; M. Forsman, K. Kuoppa, A. Sjöstedt, and A. Tärnvik, Letter, Eur. J. Clin. Microbiol. Infect. Dis. 9:784–785, 1990; A. Tärnvik, S. Löfgren, L. Öhlund, and G. Sandström, Letter, Eur. J. Clin. Microbiol. 6:318–319, 1987). We recently introduced PCR for the demonstration of F. tularensis in wound specimens (16). The method showed a high degree of specificity, and by use of spiked samples, a sensitivity of 102 bacteria was demonstrated. In an outbreak of ulceroglandular tularemia in Sweden in 1995, F. tularensis DNA was successfully amplified from wound specimens from 29 of 40 patients. In that study, specimens were sent in saline. When various methods for treatment of the specimens prior to the PCR analysis were compared, the best success was achieved by use of a protocol that included the nuclease inhibitor guanidine thiocyanate as the lysis agent.
The use of PCR for the direct diagnosis of ulceroglandular tularemia is thus highly promising, and more work on the conditions that might influence the assay seems to be warranted. By inclusion of a nuclease inhibitor in the transport medium, we addressed in the present study the question of whether degradation during transport might adversely affect the outcome of PCR. As regards culture, we compared various transport systems by experimental inoculation and storage. When in 1998 a new outbreak of ulceroglandular tularemia occurred in the same geographic region as the 1995 outbreak (16), we compared the sensitivity of PCR with that of culture.
MATERIALS AND METHODS
Bacteria.
F. tularensis live vaccine strain (LVS) (ATCC 29684) was supplied by the U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, Frederick, Md. A virulent strain of F. tularensis (strain FSC200) was isolated from a patient during the 1998 outbreak of ulceroglandular tularemia in central Sweden. F. tularensis strains were handled under biosafety level 3 laboratory conditions. Pseudomonas sp. strain CF600 was kindly provided by Victoria Shingler, Umeå University, Umeå, Sweden.
Bacterial transport systems.
Four different transport systems were compared. First, a modified variant of the nonnutrient Stuart medium intended for transport of gonococci (13) has been used for several decades for transport of bacterial specimens and was used during the 1995 outbreak of ulceroglandular tularemia (16). Second, a modified Thayer-Martin medium (14) routinely used in the laboratory for culture of F. tularensis was evaluated. It was supplemented with 7.5 mg of colistin (Sigma, St. Louis, Mo.) per liter, 2.5 mg of amphotericin B (Bristol-Myers Squibb, New Brunswick, N.J.) per liter, 0.5 mg of lincomycin (Upjohn, Kalamazoo, Mich.) per liter, 4 mg of trimethoprim (Duchefa, Haarlem, The Netherlands) per liter, and 10 mg of ampicillin (Duchefa) per liter as described previously (2). Sterile saline (3-ml volumes) was also evaluated as a transport medium. Sterile cotton-tipped wood applicators (Selefatrade AB, Spånga, Sweden) were used for inoculation of F. tularensis in all these media. Finally, we assessed a commercial transport system designed for transport of common anaerobic and aerobic pathogens (7, 10); J. L. Perry, D. R. Ballou, and J. L. Salyer, Letter, J. Clin. Microbiol. 35:3367–3368, 1997). The system included a rayon-tipped plastic applicator and a tube containing Amies agar with charcoal (Copan Italia, Brescia, Italy).
Patient samples.
In August and September 1998, tularemia occurred along the Ljusnan River in central Sweden. Through general practitioners, wound and blood samples were obtained from patients with suspected ulceroglandular tularemia. Forty-eight patients (ages 1 to 83 years; mean age, 38.8 years; 22 females and 26 males) were included, but only 1 patient had received antibiotic treatment before the day of the visit. Material was collected from the ulcer by use of two applicators. A cotton-tipped applicator, intended for use in PCR analysis, was transported in a sterile tube containing 1.0 ml of a guanidine isothiocyanate-containing buffer (8), a solution previously shown under experimental conditions to preserve F. tularensis DNA for at least 1 month without a loss of amplifiable material (16). A rayon-tipped applicator was sent in Amies agar with charcoal for use in the culture diagnostic method. The samples arrived at the laboratory after 1 to 3 days of transport. Blood samples were drawn from each patient while the patient was in the acute phase of infection and 3 to 6 weeks after the onset of disease for assay of F. tularensis-specific antibodies. Before a patient was considered seronegative, at least four serum samples were analyzed over a period of ≥2 months.
Culture diagnosis of tularemia.
After inoculation on a modified Thayer-Martin agar plate (14) at 37°C in 5% CO2 for 6 days, growth of F. tularensis was confirmed by slide agglutination with a commercial antiserum (Difco Laboratories, Augsburg, Germany) and DNA amplification by PCR.
Enzyme immunoassay.
For enzyme immunoassay, microplates were coated with an F. tularensis carbohydrate-protein complex (14), and serum samples, diluted 1/500, were assayed as described previously (18). Samples from a large number of subjects who denied that they had previously had tularemia or tularemia vaccination showed mean ± standard deviation enzyme-linked immunosorbent assay values of 0.320 ± 0.084 for immunoglobulin G (IgG) antibodies and 0.099 ± 0.011 for IgM antibodies, and on that basis, an IgG value of ≥0.76 and an IgM value of ≥0.40 were considered to confirm the diagnosis.
Preparation of DNA and PCR analysis.
A 450-μl volume was collected from each patient sample that had been suspended in a guanidine isothiocyanate-containing buffer (8), and the volume was further diluted with the buffer to a final volume of 900 μl. DNA was prepared as described previously (8, 16) and was dissolved in 12 μl of H2O for amplification in a multiplex PCR with primer pairs specific to the gene encoding a 17-kDa lipoprotein of F. tularensis LVS and to the human β-actin gene. The 17-kDa gene primers TUL4-435 and TUL4-863 (16) yield an F. tularensis-specific, 0.4-kb fragment (17), whereas the human β-actin gene primers yield a 0.2-kb fragment. In each PCR analysis, heat-killed F. tularensis and H2O were included as controls. The reaction mixture contained (at a final concentration) a mixture of deoxynucleoside triphosphates (Pharmacia Biotech, Uppsala, Sweden) at a concentration of 200 μM, each primer (Pharmacia Biotech) at a concentration of 0.8 μM, 3 mM MgCl2, and 1 U of thermostable Taq polymerase in 25 μl of Taq reaction buffer (Advanced Biotechnologies, London, United Kingdom). To each reaction mixture, 3 μl of sample was added for denaturation at 94°C for 3 min in a DNA Progene thermal cycler (Techne, Cambridge, United Kingdom), followed by amplification for 30 cycles. Each cycle consisted of denaturation at 94°C for 30 s, primer annealing at 65°C for 30 s, and primer extension at 72°C for 1 min. After a final extension at 72°C for 5 min, the tubes were stored at 4°C until a 5-μl portion of each reaction mixture was subjected to electrophoresis in a 2% agarose gel. After ethidium bromide staining, the amplified gene products were visualized with UV light.
Competitive PCR.
A 1,141-bp fragment of the Francisella 16S rRNA gene (rDNA) was amplified from patient samples by use of a genus-specific primer pair, primers F5 and F11 (6). For use as an internal standard, a 673-bp DNA fragment was generated from the 2,3-dioxygenase-encoding gene (dmpB gene) of Pseudomonas sp. strain CF600 (1) with the composite primers F11dmpF and F5dmpR. The primer F11dmpF (5′-TACCAGTTGGAAACGACTGTATCGACGAGGACTGCCTGAA) consisted of primer F11 (boldface letters) appended to the 5′ end of bases 259 to 278 of the dmpB gene, and F5dmpF (CCTTTTTGAGTTTCGCTCCCCAGCCAGGTCACGGGCTT) consisted of primer F5 (boldface letters) appended to the 5′ end of bases 892 to 874 of the dmpB gene. These primers thus generated a fragment flanked by the binding sequences for primers F5 and F11. To ascertain that Francisella 16S rDNA and the internal standard construct were amplified with similar efficiencies, an equal molar amount of the respective target was amplified separately. Aliquots were removed after cycles 10, 15, 20, 25, and 30, and the quantity of each amplicon was plotted in relation to the number of cycles. In three experiments, no significant differences in the slope coefficients for the linear portions of two curves were found. The amount of 16S rDNA on ethidium bromide-stained agarose gels was estimated by identifying the dilution of the competitor fragment that after amplification showed the same intensity as the amplicon of the sample DNA after correction for the different lengths of the two fragments.
Assay of proliferative T-cell response.
Peripheral blood mononuclear cells were prepared from heparinized blood by centrifugation on a Ficoll-Metrizoate gradient (Lymphoprep; NYCOMED AS, Oslo, Norway), and cultures were established. Each culture (200 μl) contained 3 × 105 mononuclear cells. The culture medium consisted of RPMI-HEPES (GIBCO Laboratories, Grand Island, N.Y.) supplemented with 15% pooled human serum, 100 μg of gentamicin per ml, and 2 mM l-glutamine. Concanavalin A (10 μg/ml) or heat-killed F. tularensis (105 cells/ml) was used as a stimulating agent. In preliminary experiments, these concentrations were found to be optimal. To estimate the proliferative response, five replicate cultures were incubated at 37°C for 4 days, pulsed for 6 h with 1 μCi of [3H]thymidine, and harvested. The proliferative indices were calculated by dividing the mean for cultures with and without stimulating agent. Concanavalin A-stimulated cell cultures all showed proliferative indices of >3.
Recovery of F. tularensis in various transport systems.
To compare the various transport media, a virulent strain (strain FSC200) of F. tularensis, which was isolated from a Swedish patient in 1998 and passaged only once, and live vaccine strain F. tularensis LVS (ATCC 29684) were used. Bacteria were grown on modified Thayer-Martin medium and were suspended in saline at a density of 107 cells/ml. Portions of 0.1 ml were collected from the suspension by inserting a tipped applicator for 5 s. The applicator was stored in a transport medium at room temperature for a given period of time, removed, and rotated vigorously for 60 s in 200 μl of saline for determination of viable counts. For PCR analysis, a 50-μl portion was transferred to a tube containing 1.0 ml of the guanidine isothiocyanate-containing buffer. The DNA of the lysed bacteria was prepared as described previously (8) and was subjected to PCR with TUL4-435 and TUL4-863 as primers. Applicators inoculated in saline were used as negative controls. After amplification, a 5-μl portion of each reaction mixture was subjected to electrophoresis in a 2% agarose gel. The amplified gene products were visualized with UV light after ethidium bromide staining.
RESULTS
Survival of F. tularensis in various transport media.
The survival of F. tularensis after storage in modified Stuart medium, saline, or Amies agar with charcoal was tested for two different strains of F. tularensis, a recent patient isolate, strain FSC200, and the live vaccine strain, strain LVS. Irrespective of the strain used, storage in saline or Stuart medium resulted in a significant reduction in bacterial numbers within 4 h, and 2 days after inoculation of 1.5 × 106 or 5.5 × 106 organisms, no viable bacteria were demonstrated (Table 1). In contrast, Amies agar with charcoal (Copan) preserved bacterial viability for 1 week, and after 7 days bacterial numbers had decreased ≤1 log10. Irrespective of the medium, PCR analysis performed after 7 days of storage showed the presence of F. tularensis DNA. Control samples containing saline showed no visible amplicons. Thayer-Martin medium modified by the addition of antibiotics was repeatedly found to preserve bacterial viability as effectively as Amies agar with charcoal (data not shown).
TABLE 1.
Survival of F. tularensis in transport media
| Strain | Transport medium | Survival (no. of bacteria) at the following times postinoculationa:
|
||||
|---|---|---|---|---|---|---|
| 1 h | 4 h | 48 h | 96 h | 168 h | ||
| FSC200b | Saline | 9.4 × 104 | 4.9 × 102 | 0 | 0 | 0 |
| Modified Stuart | 4.1 × 105 | 9.2 × 104 | 0 | 0 | 0 | |
| Copan | 6.2 × 105 | 4.5 × 105 | 4.4 × 105 | 3.9 × 105 | 1.8 × 105 | |
| ATCC 29684c | Saline | 8.6 × 102 | 9 × 100 | 0 | 0 | 0 |
| Modified Stuart | 4.5 × 104 | 1.7 × 103 | 0 | 0 | 0 | |
| Copan | 3.8 × 105 | 2.2 × 105 | 3.3 × 105 | 4.1 × 105 | 2.7 × 105 | |
The bacteria were inoculated in transport medium and were stored at room temperature for the indicated period of time. Each value represents the mean number of bacteria in duplicate samples.
The inoculum was ∼5.5 × 106. The strain was isolated from a wound sample of a patient during the 1998 outbreak along the Ljusnan River in Sweden.
The inoculum was ∼1.5 × 106. The strain was the live vaccine strain F. tularensis LVS.
Culture and PCR analysis of wound specimens.
Wound specimens from 48 patients with clinically suspected tularemia were sent in Amies agar with charcoal for culture and in guanidine isothiocyanate-containing buffer for PCR. For 40 patients the diagnosis was confirmed by serology and/or culture. F. tularensis was isolated from 25 (62%) of these 40 patients, and F. tularensis DNA was successfully amplified from 30 (75%) of these 40 patients (Table 2). β-Actin DNA was amplifiable from all 30 F. tularensis DNA-containing specimens. For 4 of 10 F. tularensis DNA-negative specimens, however, β-actin DNA was not detected, indicating that these specimens may not have contained significant amounts of biological material. Thus, F. tularensis DNA was detected in 30 (83%) of 36 patients from whose samples β-actin DNA was amplified.
TABLE 2.
Sensitivities of various methods for diagnosis of tularemia during an outbreak in Sweden in 1998
| Clinical condition | No. of samples positive/total no. of samples investigated (%)
|
|||
|---|---|---|---|---|
| PCR amplification of gene for:
|
Culture | Serology | ||
| β-Actin | 17-kDa lipoprotein | |||
| Tularemia confirmed by culture or seroconversion | 36/40 (90) | 30/40 (75) | 25/40 (62) | 30/31 (97) |
| Tularemia suspected but serology and culture negativea | 8/8 (100) | 4/8 (50) | 0/8 (0) | 0/8 (0) |
All patients lacked serum antibodies to F. tularensis when the serum was tested while the patient was in the acute phase of infection and 3 to 5 weeks after the onset of disease.
T-cell response to F. tularensis of patients with detectable F. tularensis DNA but negative serology and culture.
In eight patients with clinically suspected tularemia, neither serology of samples obtained over ≥12 weeks nor wound culture confirmed the diagnosis (Table 3). F. tularensis DNA was successfully amplified from wound samples from four of these patients. To more fully investigate the latter patients with regard to laboratory-based evidence of tularemia, the in vitro T-cell stimulation test was performed. In three of the patients, a recall T-cell response to F. tularensis was demonstrated ∼5 months after onset of disease (Table 4). Stimulatory indices were >3.0 times higher than the background value, whereas it was <2.3 times higher for seven reference subjects who lacked a history of tularemia or tularemia vaccination. The fourth patient showed a stimulatory index of 1.0. The latter patient showed no typical signs of tularemia, and specific antibiotic treatment was not instituted.
TABLE 3.
Summary of laboratory results from the 1998 tularemia outbreak
| Serology result | PCR result | Culture result | No. of patients |
|---|---|---|---|
| + | − | − | 9 |
| + | + | − | 6 |
| + | + | + | 14 |
| + | − | + | 1 |
| − | + | + | 1 |
| NDa | + | + | 9 |
| − | + | − | 4 |
| − | − | − | 4 |
ND, not determined.
TABLE 4.
T-cell response to F. tularensis of patients showing detectable F. tularensis DNA in wound specimens but no evidence of tularemia by culture or serology
| Age (yr) | Sex | Serum antibodies to F. tularensisa | F. tularensis DNAb | Time of institution of antibiotics (days after onset) | T-cell response to F. tularensisc |
|---|---|---|---|---|---|
| 45 | Male | − | + | 2 | 8.8 |
| 67 | Female | − | + | 2 | 9.8 |
| 42 | Female | − | + | 1 | 3.1 |
| 59 | Female | − | + | Not instituted | 1.0 |
Not detected in any of at least four serum samples collected over a period of ≥3 months.
Detectable DNA in wound specimen.
After 4 days of incubation with heat-killed F. tularensis, T cells were pulsed with [3H]thymidine. An index representing the ratio of the mean incorporation of five cultures with and without antigen is shown. Reference subjects without any previous history of exposure to F. tularensis (n = 7) all showed indices of <2.3.
Quantitation of F. tularensis DNA in wound samples.
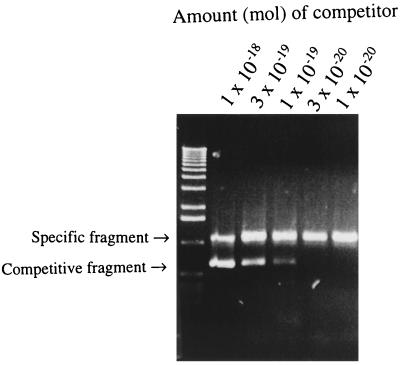
A competitive PCR was used to assess the amount of DNA present in six specimens (three PCR- and culture-positive specimens and three PCR-positive and culture-negative specimens) obtained during the 1995 tularemia outbreak. For all investigated samples, the estimated amount of DNA varied from 106 to 107 genomic equivalents per specimen. Results of a representative analysis are shown in Fig. 1.
FIG. 1.
Competitive PCR analysis for quantitative estimation of F. tularensis 16S rDNA in wound specimens from tularemia patients. Results of a representative analysis are shown. The amount of 16S rDNA was estimated by identifying the dilution of competitor fragment showing the same intensity as the amplicon of the sample DNA after amplification.
DISCUSSION
Although the nutritional requirements of F. tularensis have been known for decades, there has been little experience with the use of culture for the routine diagnosis of tularemia. Due to the high degree of virulence of the organism, culture of wound specimens is normally avoided (5). A more general use of culture for the diagnosis of tularemia has been reported only from Scandinavia, where the less virulent subspecies F. tularensis subsp. holarctica is endemic (4, 16). In one of the latter studies (16), the sensitivity of the culture procedure was found to be as low as 25%. In that study a modified Stuart medium was used for transport, but it was found to be suboptimal in the present study. According to the present experiments with spiked samples, a commercial transport medium and a medium recommended for culture were more preservative. When the commercial medium was used as the transport medium for specimens from the present outbreak, a sensitivity of 62% was recorded. The transport medium thus seemed to be important for the success of culture of F. tularensis from wound specimens.
Due to hazards associated with culture, ulceroglandular tularemia seems to be a perfect target for gene-based pathogen identification. PCR for identification of the gene encoding a 17-kDa outer membrane lipoprotein allows the sensitive identification of F. tularensis. The gene is conserved among various strains of F. tularensis and shows no significant similarity to other prokaryotic or eukaryotic gene sequences in current gene banks. F. tularensis is not closely related to organisms known to be associated with human infection or colonization (6), thus further minimizing problems with interpretation of the findings.
According to the present results and those of a previous study (16), the sensitivity of PCR applied to wound specimens from patients with ulceroglandular tularemia is ∼75%. In principle, a failure to detect DNA may be due either to inefficient sampling or to degradation of DNA during transport. The present and previous results favor the first explanation. If degradation was the main problem, the present inclusion of a nuclease inhibitor in the transport medium would have been expected to result in an increased sensitivity. The sensitivity was, however, not higher than that found when samples were sent in saline (16). Moreover, some samples would have been expected to contain relatively small amounts of amplifiable DNA. When six samples were subjected to quantitative PCR, all were found to have large amounts of DNA (106 to 107 genomic equivalents). In line with this, we previously showed that the success of PCR amplification was not decisively affected by the time of transport (16).
If the transport is not the weak link of the present PCR application, difficulties associated with sampling may be the more important. Some of those specimens that lacked amplifiable F. tularensis DNA also lacked amounts of β-actin DNA sufficient for PCR detection, indicating that in those samples, no or very little biological material was present. By contrast, β-actin DNA was amplified from all 30 samples in which F. tularensis-specific DNA was detectable. Moreover, only 1 of the 10 PCR-negative samples was culture positive. It should be recalled that in some patients with tularemia, the ulcer is very slight, and it may even be difficult to distinguish between an infected and a noninfected mosquito bite (4). Thus, the sensitivity of the PCR may well be restricted by difficulty with obtaining representative material. To improve sensitivity, intense rubbing of the wound surface may be tried, and sampling from more than one lesion may also be attempted. When lesions are minute or dry, a skin biopsy might be useful.
In the present study we identified four patients with clinically suspected tularemia but with repeated negative serology results when their sera were tested up to 12 weeks after the onset of symptoms and with negative culture results. When subjected to the T-cell stimulation test, three of them showed a strong cell-mediated immune response to F. tularensis. In previous vaccination trials, such a lack of correlation between serological and cell-mediated responses to the organism has been demonstrated, and similar to the patients in the present study, some of the vaccinees showed a strong T-cell response but no antibodies (18, 19). In clinical studies of tularemia, there are virtually no reports of the absence of antibodies in patients who were monitored by serologic testing for several weeks. A problem, however, is that the presence of serum antibodies is often used as an inclusion criterion for a study, and seronegative patients with tularemia may thereby be excluded a priori. In the present study those three patients who failed to respond with detectable antibodies all received treatment within 2 days of the onset of disease. An inhibitory effect of the early institution of antibiotics on the serological response in tularemia has been suggested previously (15). In essence, PCR may verify some cases of tularemia that would otherwise escape detection.
Thus, under transport conditions believed to preserve the viability of F. tularensis, in the present study PCR showed a sensitivity of ∼75% and culture showed a sensitivity of 62%. Moreover, PCR allowed detection of some cases of tularemia that failed to be detected by culture or serology. The sensitivity of PCR may possibly be restricted by difficulty with obtaining representative material from all patients.
ACKNOWLEDGMENTS
We thank Stig Granström for expert advice on transport systems for culture specimens and Michal Kroca and Thorsten Johansson for help with lymphocyte stimulation assays.
Financial support was obtained from the Swedish Medical Research Council (grant 9485), Västerbottens Landsting, and the Medical Faculty, Umeå University, Umeå, Sweden.
REFERENCES
- 1.Bartilson M, Shingler V. Nucleotide sequence and expression of the catechol 2,3-dioxygenase-encoding gene of phenol-catabolizing Pseudomonas CF600. Gene. 1989;85:233–238. doi: 10.1016/0378-1119(89)90487-3. [DOI] [PubMed] [Google Scholar]
- 2.Berdal B P, Söderlund E. Cultivation and isolation of Francisella tularensis on selective chocolate agar, as used routinely for the isolation of gonococci. Acta Pathol Microbiol Scand Sect B. 1977;85:108–109. doi: 10.1111/j.1699-0463.1977.tb01681.x. [DOI] [PubMed] [Google Scholar]
- 3.Burke D S. Immunization against tularemia: analysis of the effectiveness of live Francisella tularensis vaccine in prevention of laboratory-acquired tularemia. J Infect Dis. 1977;135:55–60. doi: 10.1093/infdis/135.1.55. [DOI] [PubMed] [Google Scholar]
- 4.Christenson B. An outbreak of tularemia in the northern part of central Sweden. Scand J Infect Dis. 1984;16:285–290. doi: 10.3109/00365548409070402. [DOI] [PubMed] [Google Scholar]
- 5.Evans M E, Gregory D W, Schaffner W, McGee Z A. Tularemia: a 30-year experience with 88 cases. Medicine (Baltimore) 1985;64:251–269. [PubMed] [Google Scholar]
- 6.Forsman M, Sandström G, Sjöstedt A. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int J Syst Bacteriol. 1994;44:38–46. doi: 10.1099/00207713-44-1-38. [DOI] [PubMed] [Google Scholar]
- 7.Hudspeth M K, Citron D M, Goldstein E J. Evaluation of a novel specimen transport system (Venturi Transystem) for anaerobic bacteria. Clin Infect Dis. 1997;25(Suppl. 2):S132–S133. doi: 10.1086/516198. [DOI] [PubMed] [Google Scholar]
- 8.Ibrahim A, Norlander L, Macellaro A, Sjöstedt A. Specific detection of Coxiella burnetii through partial amplification of 23S rDNA. Eur J Epidemiol. 1997;13:329–334. doi: 10.1023/a:1007385104687. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson K A, Söderlind O. Studies of the diagnosis of tularemia. In: Winblad S, editor. Contributions to microbiology and immunology. Vol. 2. 1973. pp. 224–230. Yersinia, Pasteurella and Francisella. A symposium. S. Karger, Basel, Switzerland. [Google Scholar]
- 10.Perry J L. Assessment of swab transport systems for aerobic and anaerobic organism recovery. J Clin Microbiol. 1997;35:1269–1271. doi: 10.1128/jcm.35.5.1269-1271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provenza J M, Klotz S A, Penn R L. Isolation of Francisella tularensis from blood. J Clin Microbiol. 1986;24:453–455. doi: 10.1128/jcm.24.3.453-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reary B W, Klotz S A. Enhancing recovery of Francisella tularensis from blood. Diagn Microbiol Infect Dis. 1988;11:117–119. doi: 10.1016/0732-8893(88)90080-6. [DOI] [PubMed] [Google Scholar]
- 13.Ringertz O. A modified Stuart medium for the transport of gonococcal specimens. Acta Pathol Microbiol Scand. 1960;74:371–380. [Google Scholar]
- 14.Sandström G, Tärnvik A, Wolf-Watz H, Löfgren S. Antigen from Francisella tularensis: nonidentity between determinants participating in cell-mediated and humoral reactions. Infect Immun. 1984;45:101–106. doi: 10.1128/iai.45.1.101-106.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saslaw S, Carhart S. Studies with tularemia vaccines in volunteers. III. Serological aspects following intracutaneous or respiratory challenge in both vaccinated and nonvaccinated volunteers. Am J Med Sci. 1961;241:689–699. [PubMed] [Google Scholar]
- 16.Sjöstedt A, Eriksson U, Berglund L, Tärnvik A. Detection of Francisella tularensis in ulcers of patients with tularemia by PCR. J Clin Microbiol. 1997;35:1045–1048. doi: 10.1128/jcm.35.5.1045-1048.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjöstedt A, Kuoppa K, Johansson T, Sandström G. The 17 kDa lipoprotein and encoding gene of Francisella tularensis LVS are conserved in strains of Francisella tularensis. Microb Pathog. 1992;13:243–249. doi: 10.1016/0882-4010(92)90025-j. [DOI] [PubMed] [Google Scholar]
- 18.Tärnvik A, Löfgren M L, Löfgren S, Sandström G, Wolf-Watz H. Long-lasting cell-mediated immunity induced by a live Francisella tularensis vaccine. J Clin Microbiol. 1985;22:527–530. doi: 10.1128/jcm.22.4.527-530.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tärnvik A, Löfgren S. Stimulation of human lymphocytes by a vaccine strain of Francisella tularensis. Infect Immun. 1975;12:951–957. doi: 10.1128/iai.12.5.951-957.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]