SUMMARY
RBFOX2, which has a well-established role in alternative splicing, is linked to heart diseases. However, it is unclear whether RBFOX2 has other roles in RNA processing that can influence gene expression in muscle cells, contributing to heart disease. Here, we employ both 3′-end and nanopore cDNA sequencing to reveal a previously unrecognized role for RBFOX2 in maintaining alternative polyadenylation (APA) signatures in myoblasts. RBFOX2-mediated APA modulates mRNA levels and/or isoform expression of a collection of genes, including contractile and mitochondrial genes. Depletion of RBFOX2 adversely affects mitochondrial health in myoblasts, correlating with disrupted APA of mitochondrial gene Slc25a4. Mechanistically, RBFOX2 regulation of Slc25a4 APA is mediated through consensus RBFOX2 binding motifs near the distal polyadenylation site, enforcing the use of the proximal polyadenylation site. In sum, our results unveil a role for RBFOX2 in fine-tuning expression of mitochondrial and contractile genes via APA in myoblasts relevant to heart diseases.
Graphical Abstract
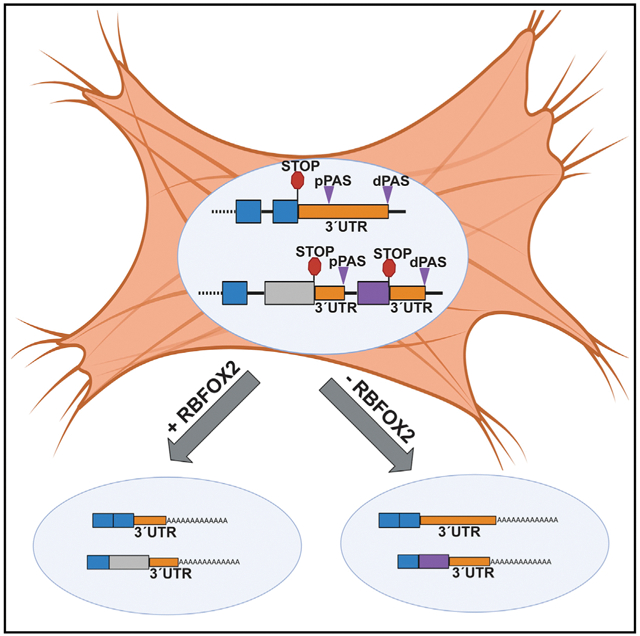
In brief
Cao et al. show that depletion of RBFOX2 in rat myoblasts modulates alternative polyadenylation patterns of mitochondrial and contractile genes. RBFOX2 loss adversely affects mitochondrial health.
INTRODUCTION
RBFOX2 belongs to a family of RNA binding proteins including RBFOX1 and RBFOX3(neuron specific). RBFOX2 has a well-characterized role in alternative splicing (AS) regulation of pre-mRNAs that can affect gene expression and function. RBFOX2 controls AS by binding to a highly conserved motif, (U)GCAUG, in intronic and/or exonic regions of pre-mRNAs (Huang et al., 2012; Lovci et al., 2013; Sun et al., 2012; Yeo et al., 2009) and regulates AS in a large complex with other splicing regulators (Damianov et al., 2016). RBFOX2 has been linked to congenital heart defects (Homsy et al., 2015; Verma et al., 2016), heart failure (Wei et al., 2015), cardiac complications of diabetes (Nutter et al., 2016, 2017), and cardiac arrhythmias in myotonic dystrophy (Misra et al., 2020). RBFOX2 has been shown to be important for skeletal muscle development and function (Berberoglu et al., 2017; Gallagher et al., 2011; Runfola et al., 2015; Singh et al., 2014, 2018). Because of RBFOX2’s important role in AS, the majority of studies have focused on identification of RBFOX2-regulated AS events in heart and skeletal muscle (Runfola et al., 2015; Singh et al., 2018; Verma et al., 2016). However, it remains unclear how a loss of RBFOX2 contributes to heart and skeletal muscle defects. Recent studies have suggested that RBFOX2 binds within the 3′UTR of transcripts (Lovci et al., 2013; Wang et al., 2008; Weyn-Vanhentenryck et al., 2014) in proximity to poly(A) sites (PASs) (Wang et al., 2008). The consequences of this binding are unknown, and a role for RBFOX2 in cleavage and polyadenylation [poly(A)] of pre-mRNAs has not yet been demonstrated.
Mammalian 3′-end processing involves the cleavage of the pre-mRNA near the PASs followed by poly(A) of the cleaved RNA. This process is complex and requires both cis-acting elements in the RNA and trans-acting proteins that bind to these elements and regulate cleavage and poly(A). More than 50% of genes in the human genome present more than one PAS and, thus, can generate multiple mRNA isoforms defined by different 3′ termini (Tian et al., 2005). Alternative poly(A) (APA) is defined by the differential usage of these PASs in a given mRNA (Shi, 2012). APA can regulate the generation of multiple gene isoforms and impact mRNA expression and homeostasis (Derti et al., 2012). There are two major types of APA: tandem APA and splicing APA. Tandem APA is mediated by the preferential usage of tandem PASs within a 3′UTR. This type of APA can affect 3′UTR length, modulating the presence of cis-acting regulatory elements and/or microRNA binding sites that can control mRNA stability, localization, or translation (Di Giammartino et al., 2011). Splicing APA can lead to the generation of multiple protein isoforms with distinct functions and properties via AS of the terminal exons (TEs) that have PASs (Cooke et al., 1999). Tight regulation of APA is essential for normal growth and development, and, as a result, aberrant APA has been linked to many disease states including heart failure, cancer, and muscle disease (Batra et al., 2014; Creemers et al., 2016; Masamha et al., 2014).
In this study, our goal was to investigate whether RBFOX2 influences APA decisions in H9c2 myoblasts. We used long-read nanopore cDNA sequencing and poly(A)-ClickSeq (PAC-seq) (Routh et al., 2017), which allow precise genome-wide determination of APA and can quantify gene expression and isoform generation. We identified 233 genes with APA changes upon RBFOX2 depletion in H9c2 myoblasts. In total, 43% of these genes also exhibited mRNA level changes. Further analysis showed that 136 genes that displayed 3′UTR length changes in RBFOX2-depleted myoblasts are known targets of micro-RNAs, suggesting that modulation of 3′UTR length can impact protein production. We identified RBFOX2 as a regulator of both tandem and splicing APA, impacting gene expression and isoform generation. Importantly, our results identified mitochondrial gene Slc25a4 and contractile gene Tropomyosin 1 as APA targets of RBFOX2. Investigating how RBFOX2 impacts APA-mediated rat Slc25a4 expression revealed that RBFOX2 binding motifs located <79 nt from the distal PAS in Slc25a4 are inhibitory for PAS usage, negatively modulating Slc25a4 protein levels. Moreover, the loss of Slc25a4 APA regulation in RBFOX2-depleted cells correlated with a striking increase in mitochondrial membrane potential and swelling. In summary, we report a role for RBFOX2 in tuning APA patterns and mitochondrial gene expression.
RESULTS
RBFOX2 depletion alters global alternative poly(A) patterns of pre-mRNAs in H9c2 myoblasts
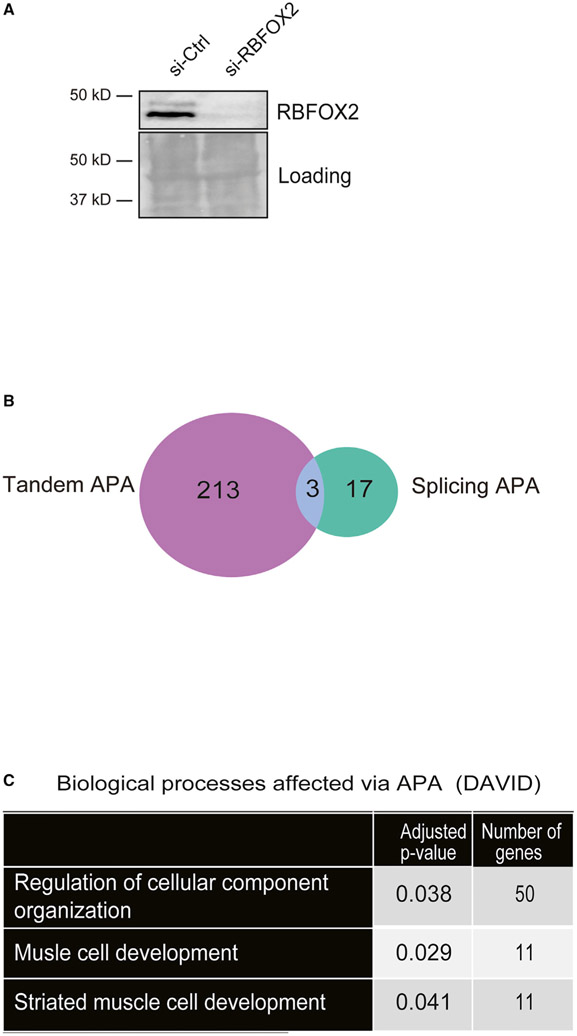
RBFOX2 binds to the 3′UTR of transcripts (Lovci et al., 2013; Wang et al., 2008; Weyn-Vanhentenryck et al., 2014) near the PASs (Wang et al., 2008), suggesting a potential role in APA. H9c2 cells obtained from embryonic rat hearts have been well characterized and used for heart and skeletal muscle studies (Hescheler et al., 1991; Kimes and Brandt, 1976; Mejía-Alvarez et al., 1994; Ménard et al., 1999; van der Putten et al., 2002; Wang et al., 1999). Moreover, we have previously shown that H9c2 myoblasts successfully recapitulated RBFOX2-mediated AS regulation observed in mouse hearts (Nutter et al., 2016), making it a good cell culture model to study RBFOX2 function in vitro. Thus, we tested whether RBFOX2 influences APA patterns in rat H9c2 myoblasts. We treated H9c2 cells with scrambled or Rbfox2-specific small interfering RNAs (siRNAs) and validated knockdown (KD) of RBFOX2 by western blot (WB) (Figure 1A). We then isolated RNA from these cells and performed PAC-seq (Routh et al., 2017) and analyzed the data using the differential-poly(A)-clustering (DPAC) pipeline (Routh, 2019). We detected 101,338 unique PASs with at least 5 unique reads mapping from at least one sample. Multiple PASs occurring within 10 nt were clustered together to yield 81,035 unique PASs. We further filtered out all PASs that did not map to 3′UTRs already annotated in the UCSC RefSeq database. After instituting these filters, the remaining 22,768 PASs mapped to 10,964 unique mRNA transcripts, of which 5,317 contained multiple PASs and 556 contained multiple TEs. DPAC reported 233 APA changes in RBFOX2-depleted myoblasts based on the criteria that differential PAS usage was reported if an individual PAS has a fold change of >1.5 and results with an independent hypothesis weighted (Ignatiadis et al., 2016) multiple testing p-adjusted value of < 0.1 (Figure 1B; Data S1).
Figure 1. RBFOX2 depletion in H9c2 myoblasts leads to altered APA patterns determined by PAC-seq.
(A) A representative WB showing efficient knockdown (KD) of RBFOX2 in H9c2 myoblasts. Ponceaustained membrane was used to monitor protein loading in each lane.
(B) The number of genes undergoing tandem APA or splicing APA in RBFOX2 KD H9c2 myoblasts.
(C) Gene Ontology (GO) analysis of genes that undergo APA changes in RBFOX2-depleted myoblasts.
We investigated what types of APA were affected upon RBFOX2 depletion in H9c2 cells. Out of 233 APA events, 213 were tandem APA, 17 were splicing APA, and 3 displayed both splicing and tandem APA (Figure 1B; Data S1). Using the Gene Ontology (GO) biological function analysis of these APA events, we found an enrichment of genes involved in striated muscle development and regulation of cell organization (Figure 1C; Data S1). This is consistent with RBFOX2’s involvement in congenital heart defects (Homsy et al., 2015; Verma et al., 2016) and muscle development (Singh et al., 2014, 2018).
RBFOX2-mediated splicing APA is important for expression of rat Tropomyosin 1 muscle-specific isoforms
Based on the GO analysis of RBFOX2-regulated APA events, we first investigated genes that have roles in striated muscle development that are impacted by RBFOX2-mediated splicing APA (Data S1). Of the 17 splicing APA changes identified in RBFOX2-depleted myoblasts, Tropomyosin 1 gene (Tpm1) has an essential role in striated muscle development and function and undergoes a dramatic splicing APA change in RBFOX2-depleted cells (Data S1). Tpm1 is an essential gene required for myofibril organization (Thomas et al., 2010), muscle contraction (Wolska and Wieczorek, 2003), and cardiac development (England et al., 2017). Tpm1 has very complex AS and APA patterns and many isoforms that are tissue specific and developmentally regulated (Gooding and Smith, 2008). Thus, we further studied splicing APA regulation of rat Tpm1 by RBFOX2.
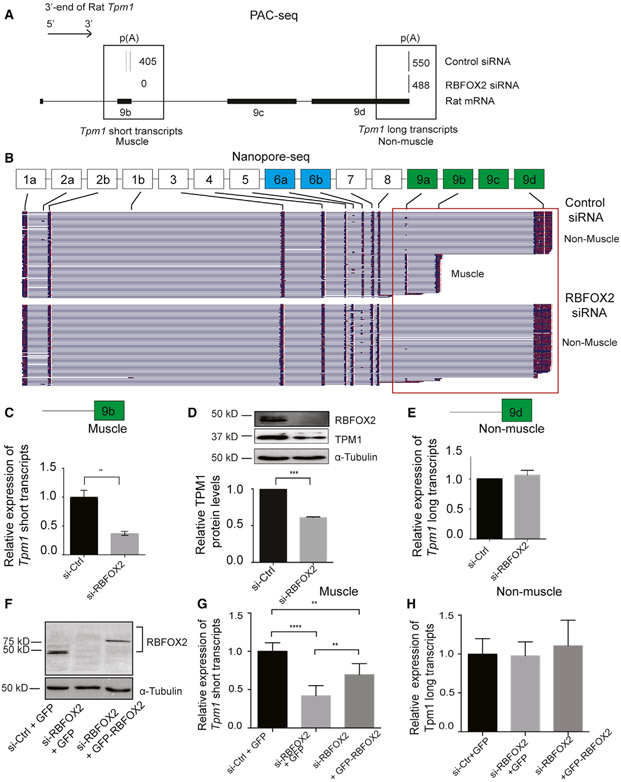
PAC-seq identified two PASs located in two different last exons (exon 9b and exon 9d) that are utilized in H9c2 myoblasts (Figure 2A). Tpm1 gene isoforms ending with exon 9b are important for muscle contraction in muscle cells (Lin et al., 2008) (short or muscle-specific transcripts), whereas gene isoforms ending with exon 9d are ubiquitously expressed to support cytoskeleton function (long or non-muscle transcripts). The usage of PASs in muscle-specific exon 9b diminished in RBFOX2 KD myoblasts (Figure 2A).
Figure 2. RBFOX2 regulates expression levels of muscle-specific isoforms of rat Tpm1 via splicing APA in H9c2 myoblasts.
(A) The poly(A) usage of Tpm1 transcripts in control and RBFOX2-depleted H9c2 cells determined by PAC-seq (n = 3). The number of reads mapped to Tpm1 transcripts was labeled within black boxes. PASs located in either TE 9b or TE 9d were utilized in H9c2 myoblasts.
(B) Full-length Tpm1 short (muscle) or long (non-muscle) isoforms identified by nanopore sequencing in control versus RBFOX2-depleted H9c2 myoblasts (n = 3). See also Figures S2 and S3
(C) Relative mRNA levels of Tpm1 muscle-specific (short) transcripts that end with exon 9b in control and RBFOX2-depleted H9c2 cells were determined by qRT-PCR. mRNA levels in control cells were normalized to 1. Data represent means ± SD. Statistical significance was calculated using t test to compare two different groups in three independent experiments (n = 3). **p = 0.0079.
(D) Relative protein levels of TPM1 in control and RBFOX2-depleted H9c2 cells determined by WB. α-tubulin was used as a loading control. Protein levels in control cells were normalized to 1. Data represent means ± SD. Statistical significance was calculated using t test to compare two different groups in three independent experiments (n = 3). ***p = 0.0001.
(E) Relative mRNA levels of Tpm1 non-muscle transcripts (long) in control and RBFOX2-depleted H9c2 cells. mRNA levels in control cells were normalized to 1. Data represent means ± SD. No statistical significance was found using t test in three independent experiments (n = 3).
(F) RBFOX2 protein expression levels in (1) scrambled-siRNA-treated, (2) RBFOX2-siRNA-treated, and (3) RBFOX2-siRNA-treated H9c2 cells ectopically expressing GFP or GFP-RBFOX2.
(G) Ectopic expression of GFP-RBFOX2 partially rescued splicing APA change in Tpm1 caused by RBFOX2 depletion. Relative expression levels of muscle-specific Tpm1 (short) transcripts in scrambled-siRNA-treated, RBFOX2-siRNA-treated, or RBFOX2-siRNA-treated H9c2 myoblasts expressing GFP or GFP-RBFOX2. mRNA levels in control cells (1) were normalized to 1. Data represent means ± SD. Statistical significance was calculated using one-way ANOVA to compare three different groups in three independent experiments (n = 3). p value for si-Ctrl-+GFP versus si-RBFOX2+GFP is ****p < 0.0001; for si-RBFOX2+ GFP versus si-RBFOX2+ GFP-RBFOX2 is **p = 0.0056; for si-Ctrl+GFP versus si-RBFOX2+ GFP-RBFOX2 is **p = 0.0027.
(H) Relative expression levels of non-muscle Tpm1 (long) transcripts in control, RBFOX2-depleted, or RBFOX2-depleted cells expressing GFP or GFP-RBFOX2. mRNA levels in (1) cells were normalized to 1. Data represent means ± SD. Statistical significance was calculated using one-way ANOVA to compare three different groups in three independent experiments (n = 3).
To confirm the results obtained by PAC-seq, we used nanopore cDNA sequencing as an orthogonal approach to investigate full-length Tpm1 isoforms and their expression in response to changes in RBFOX2 levels. We performed nanopore cDNA sequencing in control versus RBFOX2 KD cells and obtained high-quality and full-length nanopore reads that were mapped to the rn6 genome using Minimap2 (Li, 2018), yielding an average of 550,000 mapped reads spanning 3,000–4,300 unique mRNAs (Figure S1; Table S1). Reads were mapped to specific rat Tpm1 exons (Figure 2B, black rectangles), identifying 22 different full-length Tpm1 transcripts in control H9c2 myoblasts (Figure S2). These Tpm1 transcripts ended with different TEs (exon 9a, exon 9b, or exon 9d) in control cells (representative images shown in Figure 2B). Upon depletion of RBFOX2, we observed a strong reduction in the diversity of Tpm1 isoforms, with only nine different full-length transcripts that ended almost exclusively with exon 9d (Figures 2B and S3), consistent with reduced PAS usage in exon 9b in RBFOX2 KD cells (Figure 2A). These indicate that RBFOX2 KD reduces expression of muscle-specific isoforms that end with exon 9b.
To further validate APA changes in muscle-specific (short) isoforms of Tpm1, we designed primers spanning Tpm1 exon7 and exon 9b or exon 7 and exon 9d (Table S2). The levels of Tpm1 muscle-specific transcripts were dramatically decreased in RBFOX2-depleted cells (Figure 2C). Consistent with reduced levels of muscle-specific Tpm1 isoforms, TPM1 protein levels were reduced by 45% in RBFOX2 KD cells (Figure 2D). Importantly, mRNA levels of Tpm1 non-muscle (long) transcripts did not change upon RBFOX2 KD (Figure 2E).
We performed rescue experiments by ectopically expressing GFP-tagged RBFOX2 in RBFOX2 KD H9c2 cells. Rbfox2 siRNA treatment caused effective KD of RBFOX2 in H9c2 cells, and GFP-RBFOX2 protein was expressed at lower levels than the endogenous RBFOX2 due to targeting by RBFOX2-specific siRNAs (Figure 2F, lanes 3 versus 1). Even low levels of GFP-RBFOX2 partially rescued the expression of muscle-specific Tpm1 transcripts (Figure 2G). GFP-RBFOX2 expression did not affect expression of non-muscle Tpm1 isoforms (Figure 2H). Importantly, Tpm1 regulation by RBFOX2 was specific because we did not observe dramatic changes in APA patterns of rat Tpm2, Tpm3, and Tpm4 in RBFOX2 KD myoblasts (Figure S4).
These data show that RBFOX2 controls expression of muscle-specific isoforms of essential contractile gene Tpm1 via splicing APA. Moreover, our results also demonstrate that nanopore sequencing is an effective method in revealing complex and coordinated APA and AS patterns and validating AS and APA variants.
RBFOX2-mediated tandem APA alters 3′UTR length and mRNA levels
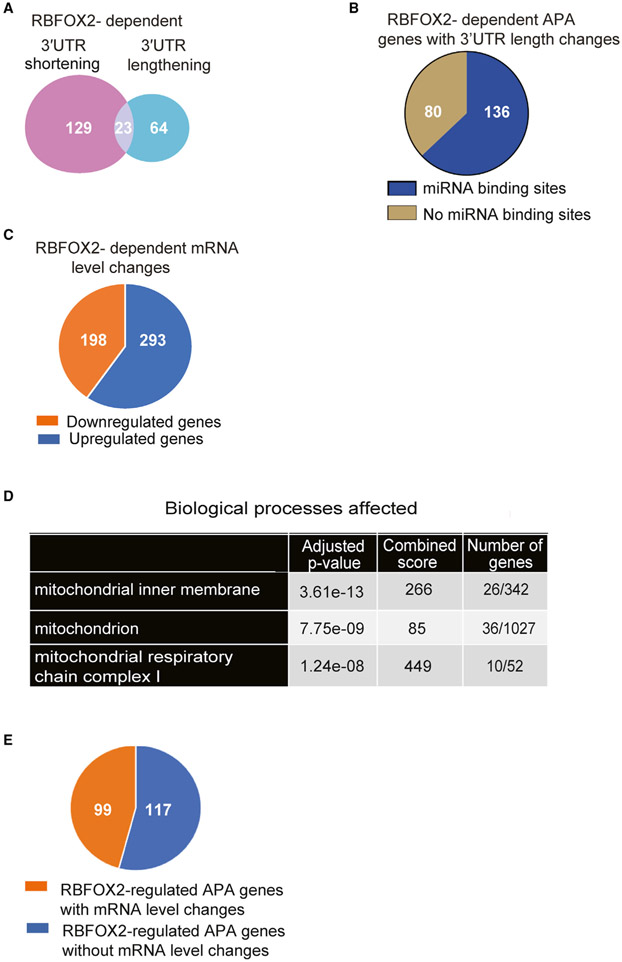
Having established splicing-APA-mediated regulation of Tpm1 pre-mRNA by RBFOX2, we sought to identify tandem APA events in genes that may be critical for muscle development and function. We found that the majority of the APA changes in RBFOX2-depleted cells were tandem APA (Figure 1B), which can modulate the 3′UTR length and, in turn, gene expression levels. Analysis of PAC-seq data revealed that 129 out of 216 (59%) APA changes (Data S1) led to 3′UTR shortening (Data S2), and 64 out of 216 (29.6%) led to 3′UTR lengthening (Data S2) in RBFOX2-depleted cells (Figure 3A). There was a smaller subset that included 23 genes that contained 3 or more PASs, giving rise to complex changes and resulting in both lengthening and shortening of 3′UTRs in RBFOX2 KD cells due to the modulation of levels of both the most distal and most proximal PASs (dPASs and pPASs, respectively) relative to an intermediate PAS (Figure 3A; Data S2). It has been well established that 3′UTRs harbor binding sites for microRNAs that can influence mRNA translation and/or stability. We used TargetScan to determine whether genes with 3′UTR length changes harbor known targets of microRNAs. We found that out of 216 tandem APA events that display 3′UTR length changes in RBFOX2 KD myoblasts, the majority (136 genes) displayed microRNA binding sites in their 3′UTRs (Figure 3B; Data S3). These results suggest that changes in 3′UTR length mediated via RBFOX2 depletion could affect microRNA-mediated regulation of these genes.
Figure 3. RBFOX2-regulated tandem APA modulates 3′UTR length and affects mRNA levels.
(A) The number of genes that undergo tandem APA changes generating shorter or longer 3′UTRs in RBFOX2 KD H9c2 myoblasts.
(B) Enrichr TargetScan analysis of genes that display 3′UTR length changes upon RBFOX2 depletion revealed known microRNA binding sites within the majority of these genes.
(C) Gene expression changes in RBFOX2 KD H9c2 myoblasts.
(D) GO analysis of genes that are downregulated in RBFOX2-depleted H9c2 myoblasts.
(E) The number of genes that exhibit both APA and gene expression changes in RBFOX2 KD H9c2 myoblasts.
Given that 3′UTR length can alter mRNA stability, we wondered if genes that exhibit APA changes upon RBFOX2 KD also display expression level changes. There were 491 mRNA level changes (fold change > 1.5, p-adjust < 0.1) in RBFOX2 KD myoblasts (Figure 3C; Data S4). GO analysis (Kuleshov et al., 2016) of genes that are downregulated in RBFOX2 KD myoblasts revealed mitochondria, mitochondrial membrane, and mitochondrial respiratory chains as affected processes (Figure 3D; Data S4). There were 26 mitochondrial genes that were downregulated in RBFOX2-depleted myoblasts (Data S4). Importantly, our analysis of differential gene expression also revealed that 99 of the 216 (46%) genes with tandem APA changes in RBFOX2-depleted myoblasts exhibited changes in mRNA levels (Figure 3E). Collectively, these data indicate that RBFOX2-mediated regulation of tandem APA events impacts both 3′UTR length and gene expression but also uncovered an enrichment of target genes involved in mitochondrial function.
RBFOX2-mediated tandem APA regulates 3′UTR length and protein levels of mitochondrial gene Slc25a4 that encodes for ANT1
To better understand how RBFOX2 impacts mitochondrial gene expression, we analyzed mitochondrial genes that displayed APA changes and are identified in our GO analysis to be important for muscle development (Data S1). Our DPAC analysis revealed that Slc25a4 undergoes a significant APA change in RBFOX2-depleted cells (Data S1). Slc25a4 gene is the ATP/ADP translocator 1 (ANT1) critical for energy production in the mitochondria. It has been shown that ablation of Slc25a4 (ANT1) in mice causes cardiomyopathy associated with defects in mitochondrial respiration (Graham et al., 1997). Loss-of-function mutations in Slc25a4 are linked to hypertrophic cardiomyopathy and skeletal muscle myopathy in humans (King et al., 2018; Körver-Keularts et al., 2015).
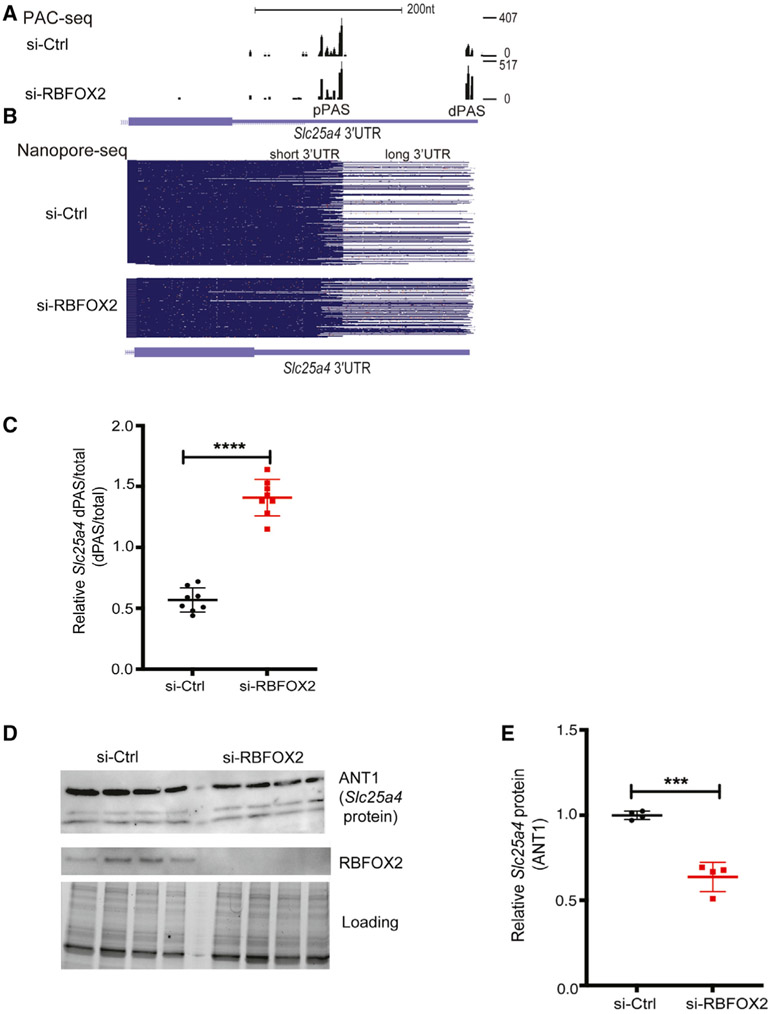
PAC-seq identified a tandem APA change in Slc25a4 in RBFOX2 KD cells (Data S1). Slc25a4 has two major PASs (dPAS and pPAS) in its 3′UTR (Figure 5A). While a dPAS was minimally used in control cells, its usage was favored in RBFOX2 KD cells (Figure 4A). This result from PAC-seq was consistent with the nanopore sequencing data that identified mostly Slc25a4 transcripts with long 3′UTR generated via dPAS usage in RBFOX2 KD H9c2 cells (Figure 4B, bottom panel). These results were reproducible in three different control and RBFOX2 KD myoblasts (Figure S5).
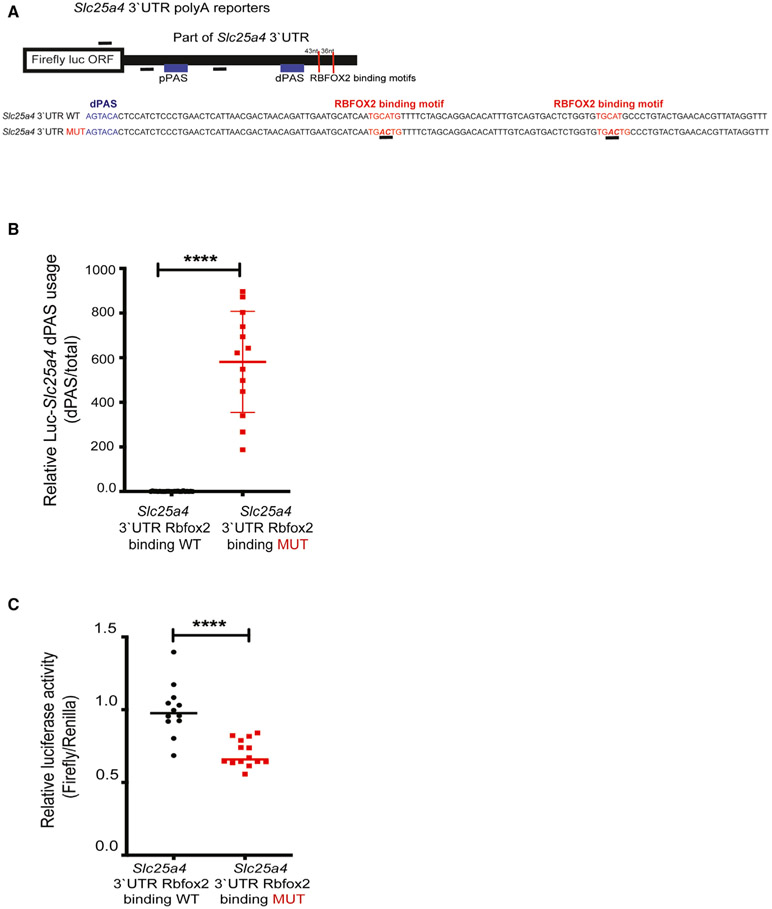
Figure 5. RBFOX2 binding sites near dPASs are critical for APA regulation of the Slc25a4 gene.
(A) Cartoon representation of luciferase-Slc25a4 poly(A) reporter constructs that harbor both pPASs and dPASs of Slc25a4 in the 3′UTR with or without mutated RBFOX2 binding motifs (underlined) located downstream of the dPAS.
(B) qRT-PCR analysis of dPAS usage when normalized to total mRNA levels in HEK293 cells expressing WT or RBFOX2 binding site mutant poly(A) luciferase-Slc25a4 3′UTR constructs. n = 24 for WT, n = 13 for mutant samples from three independent experiments, ****p < 0.0001.
(C) Relative firefly luciferase levels in HEK293 cells expressing WT or RBFOX2 binding site mutant poly(A) luciferase-Slc25a4 3′UTR constructs. n = 12 for WT samples, n = 14 for mutant samples from three independent experiments, ****p < 0.0001.
Figure 4. RBFOX2 regulates tandem APA and expression of essential mitochondrial gene Slc25a4.
(A) Slc25a4 3′UTR lengthening mediated via tandem APA in RBFOX2-depleted H9c2 myoblasts identified by PAC-seq. The number of reads mapped to Slc25a4 transcripts is shown on the right side. See also Figure S5.
(B) Nanopore sequencing analysis of full-length Slc25a4 transcripts with different 3′UTR lengths in control versus RBFOX2 KD myoblasts. See also Figure S5.
(C) RT-PCR analysis of endogenous Slc25a4 dPAS/total mRNA ratio in RBFOX2 KD myoblasts ectopically expressing GFP or GFP-RBFOX2. n = 8, ****p < 0.0001 (three independent experiments). See Table S2 for primer information.
(D) WB analysis of ANT1 protein (Slc25a4) in control versus RBFOX2 KD myoblasts determined by Bio-Rad ChemiDoc Imager. Even protein loading was monitored by imaging stain-free gels. ANT1 protein levels were normalized to loading control, and fold change in ANT1 protein levels was quantified using Bio-Rad ChemiDoc software. n = 4, ***p = 0.0002 (three independent experiments).
To validate the APA changes in Slc25a4 transcripts, we designed primers to detect long Slc25a4 transcripts generated via dPAS usage and total Slc25a4 transcripts (long and short) (Table S2). The relative Slc25a4 dPAS/total mRNA ratio was increased by 2-fold in RBFOX2 KD cells (Figure 4C), consistent with the PAC-seq and nanopore sequencing data indicating increased dPAS usage (Figures 4A and 4B). Because increased dPAS usage generates Slc25a4 transcripts with long 3′UTRs, which can negatively impact mRNA stability and/or translation, we checked Slc25a4 protein (ANT1) levels. We found that ANT1 protein levels were downregulated by almost 2-fold in RBFOX2 KD myoblasts (Figures 4D and 4E), correlating well with the presence of Slc24a5 transcripts with long 3′ UTR (Figures 4B and 4C). These results indicate that RBFOX2 is critical for Slc25a4 expression.
RBFOX2 binding motifs near PASs are critical for APA-mediated regulation of Slc25a4 expression
To better understand how RBFOX2 regulates Slc25a4 expression via APA, we examined 3′UTR of rat Slc25a4 and found two RBFOX2 binding consensus sites: one 43 nt away and another 79 nt away from the dPAS of Slc25a4 (Figure 5A). To test whether RBFOX2 binding motifs near the dPAS are functionally important for dPAS usage and Slc25a4 expression, we generated luciferase-Slc25a4 3′UTR poly(A) heterologous reporters. While generating these constructs, we inserted part of the Slc25a4 3′UTR that contains pPAS, dPAS, and RBFOX2 binding sites, downstream of the firefly luciferase open reading frame. We also removed the SV40 PAS within the plasmid (Figure 5A). In this way, luciferase mRNA will be cleaved and polyadenylated using the PASs within the Slc25a4 3′UTR.
Using this luciferase reporter, we mutated the two RBFOX2 consensus “TGCATG” motifs (Figure 5A, in red and underlined) near the dPAS that have been shown to abrogate RBFOX2 binding (Lovci et al., 2013). We tested the relative dPAS usage as well as luciferase activity of the wild-type (WT) construct and compared it to the mutant counterpart that disrupted the RBFOX2 binding site (Figure 5A, mutant sequence underlined). Mutating RBFOX2 binding motifs in luciferase-Scl25a4 3′UTR poly(A) reporter resulted in a significant increase in dPAS usage (Figure 5B, WT versus MUT) and a decrease in firefly luciferase activity (Figure 5C, WT versus MUT). Firefly luciferase activity was downregulated in RBFOX2 binding site mutants, correlating with increased usage of dPAS generating a longer 3′UTR (Figure 5B), which can reduce mRNA stability and/or translation. These results (Figures 5B and 5C) are in agreement with the results from PAC-seq, nanopore reads, and RT-PCR validations and by WB in RBFOX2-depleted myoblasts (Figure 4). Altogether, these results suggest that RBFOX2 binding sites close to dPAS repress dPAS usage and modulate Slc25a4 expression levels. To investigate how RBFOX2 regulates tandem APA globally, we determined where the RBFOX2 binding sites are located with respect to the annotated PASs identified in H9c2 myoblasts. We performed metagene analysis using the available ENCODE RBFOX2-cross-linking immunoprecipitation (CLIP) sequencing data (ENCODE Project Consortium, 2012; Davis et al., 2018) and determined the distribution of RBFOX2 CLIP peaks 1,000 nt upstream and downstream of annotated PASs that are identified in H9c2 cells. RBFOX2 binding sites were found both upstream and downstream of all PASs detected in H9c2 cells close to the PASs (labeled as position 0) (Figure S6). Notably, RBFOX2 binding site distribution was predominantly upstream of the PASs undergoing APA changes (upregulated and downregulated) in RBFOX2 KD cells (Figure S6), suggesting that RBFOX2 binding near PASs may influence PAS usage.
RBFOX2 depletion adversely impacts mitochondrial health
Since loss of Slc25a4 (ANT1) is linked to cardiomyopathy and skeletal muscle myopathy, and mitochondrial defects and Slc25a4 (ANT1) protein levels are downregulated in RBFOX2 KD myoblasts, we investigated the status of mitochondria in RBFOX2-depleted cells. Mitochondrial function is essential for heart and muscle function, and it declines in human heart failure and diabetic hearts (Rosca and Hoppel, 2013), in which RBFOX2 is implicated. In addition, mitochondrial defects have been identified in hypoplastic left heart syndrome (Karamanlidis et al., 2011; Liu et al., 2017) in which RBFOX2 mutations have been identified (Homsy et al., 2015; Verma et al., 2016). The role of RBFOX2 in mitochondrial health is unclear.
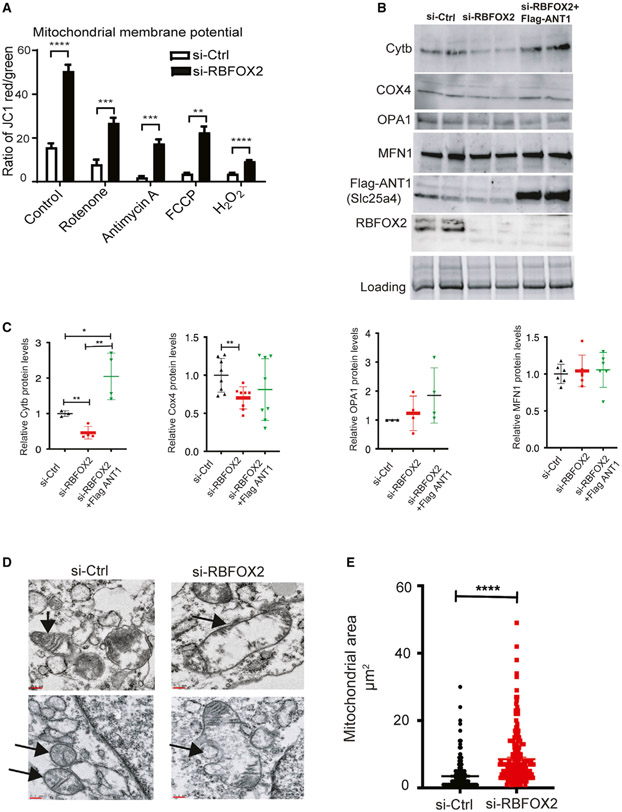
Mitochondrial membrane potential is important for respiration and mitochondrion viability (Zorova et al., 2018). This process is affected in RBFOX2-depleted myoblasts (Data S4). Therefore, we tested if RBFOX2 KD alters the mitochondrial membrane potential (Ψm), evaluated by the JC-1 assay (Perelman et al., 2012). Mitochondrial membrane potential was increased by 2.5-fold in RBFOX2 KD (versus control) cells (Figure 6A). We also used inhibitors of different complexes in the mitochondria to assess mitochondrial membrane changes. In response to rotenone (inhibits complex I), antimycin A (inhibits complex III), FCCP (mitochondrial oxidative phosphorylation uncoupler, depolarizes mitochondrial membrane potential), and H2O2 treatment, mitochondrial membrane potential was downregulated in control cells as expected. This was also the case in RBFOX2 KD cells; however, RBFOX2 KD cells maintained a higher baseline level of Ψm than was noted in control cells (Figure 6A). These results may suggest that increased mitochondrial membrane potential upon RBFOX2 deficiency might be to avoid oxidative injury.
Figure 6. RBFOX2 depletion in H9c2 myoblasts affects mitochondrial gene expression and health.
(A) Mitochondrial membrane potential (Ψm) was determined by JC-1 staining of the H9c2 cells transfected with scrambled or Rbfox2 siRNA. Shown are the ratios of fluorescence intensity of J-aggregates (red fluorescence) to J-monomers (green fluorescence). Statistical significance was calculated using unpaired t test to compare two different groups in five replicates of two independent experiments. Data represent means ± SD. For mitochondrial membrane potential in untreated control versus RBFOX2-KD H9c2 cells, ****p < 0.0001; in control versus RBFOX2-KD H9c2 cells treated with rotenone, ***p = 0.0004; in control versus RBFOX2-KD H9c2 cells treated with antimycin A, ***p = 0.0009; in control versus RBFOX2-KD H9c2 cells treated with FCCP, **p = 0.0017; in control versus RBFOX2-KD H9c2 cells treated with H2O2, ****p < 0.0001.
(B) Representative WB images of mitochondrial proteins cytochrome b (Cytb), cytochrome c oxidase 4 (COX4) (n = 3 for control and n = 4 for RBFOX2 KD samples from three independent experiments), OPA1 (n = 3 for control and n = 4 for RBFOX2 KD samples from three independent experiments), and MFN1 (n = 6 for each group from three independent experiments) in control siRNA or RBFOX2-siRNA-treated cells expressing empty vector or FLAG-ANT1 (n ≥ 3, three independent experiments).
(C) Quantification of mitochondrial protein levels in control or RBFOX2 KD H9c2 cells expressing empty vector or FLAG-ANT1 plasmid with respect to their respective loading controls. COX4 si-Ctrl versus si-Rbfox2 **p = 0.0068, Cytb si-Ctrl versus si-Rbfox2 **p = 0.0046, si-Ctrl versus si-Rbfox2+ FLAG-ANT1 *p = 0.033, si-RBFOX2 versus si-Rbfox2 +FLAG-ANT1 **p = 0.0020.
(D) Representative transmission electron microscopy images of mitochondria in control versus RBFOX2-KD H9c2 cells. Black arrows mark the mitochondria. n = 3 (three independent experiments). Scale bar: 0.1 micron.
(E) Surface area of mitochondria in control and RBFOX2-depleted myoblasts was calculated using ImageJ. n = 234 mitochondria for control, n = 216 mitochondria for RBFOX2 KD myoblasts from three independent KD experiments, ****p < 0.0001.
Mitochondrial fusion and fission play a key role in maintaining mitochondrial health. Mitofusin 1 (MFN1) is an outer membrane GTPase that mediates mitochondrial clustering and fusion. Dynamin-like OPA1 protein regulates mitochondrial fusion and cristae structure in the inner mitochondrial membrane (Song et al., 2009). These two proteins were used as markers to assess mitochondrial health and fusion. Our WB analysis revealed that RBFOX2 depletion did not overtly impact MFN1 or OPA1 expression (Figures 6B and 6C). However, when we examined mitochondrial proteins cytochrome c oxidase 4 (COX4, nuclear DNA coded) and cytochrome b (Cytb, mitochondrial DNA coded), which are part of the electron transport chain complexes, we found that both of these proteins were downregulated (Figures 6B and 6C). Importantly, exctopic expression of FLAG-ANT1 (Slc25a4 protein) in RBFOX2-depleted cells improved Cytb protein levels but did not significantly improve COX4 protein levels (Figures 6B and 6C).
To further determine the effect of RBFOX2 on mitochondrial health, we performed transmission electron microscopy to assess mitochondrial ultrastructure and morphology. We found that in RBFOX2 KD H9c2 cells, mitochondria were swollen, as seen in representative micrographs (Figure 6D, black arrows indicate mitochondria) and as also evident from increased mitochondrial area in RBFOX2-depleted cells (Figure 6E). These findings, overall, show that RBFOX2 is important for mitochondrial gene expression and health, which are relevant to its role in heart diseases.
DISCUSSION
RBFOX2 is implicated in cardiovascular diseases, congenital heart defects, and skeletal muscle development (Gallagher et al., 2011; Homsy et al., 2015; Nutter et al., 2016; Singh et al., 2014, 2018; Verma et al., 2016; Wei et al., 2015). However, it is not clear whether RBFOX2 has additional roles other than AS regulation that influences gene expression and function in muscle cells. In this study, we addressed this fundamentally important question utilizing state-of-the-art techniques and performing functional assays. We uncovered a critical role for RBFOX2 in maintaining APA patterns in myoblasts and provided insights into the functional consequences of APA changes mediated by RBFOX2 in myoblasts. In addition, our results revealed a critical role for RBFOX2 in regulating mitochondrial gene expression relevant to human heart diseases.
In this study, combined usage of PAC-seq and nanopore sequencing allowed us to precisely determine RBFOX2-regulated APA patterns and evaluate the consequences of RBFOX2-mediated APA changes. PAC-seq allowed us to sensitively detect PASs and simultaneously measure transcript abundance, while nanopore sequencing with MinION allowed us to simultaneously validate PAS usage, characterize exon inclusion across the entire mRNA transcript, and determine 3′UTR length. By combining PAC-seq and nanopore sequencing, we discovered a role for RBFOX2 in APA regulation. We identified 233 APA changes in RBFOX2 KD myoblasts and found that 46% of these APA changes impacted mRNA levels. APA regulation is linked to physiological processes including heart development (Nimura et al., 2016), cell proliferation in immune response (Sandberg et al., 2008), and embryonic and postnatal development (Mangone et al., 2010). Aberrant regulation of APA is linked to pathological conditions including heart failure, cancer, and muscular dystrophy (Batra et al., 2014; Creemers et al., 2016; Masamha et al., 2014). Here, we showed that RBFOX2-dependent APA events impacted essential contractile gene Tropomyosin 1. Our recent work using nanopore sequencing also revealed that RBFOX2 regulates AS of internal exons of Tropomyosin 1 regulated during rat heart development (Cao et al., 2021). By identifying developmentally regulated APA networks that impact muscle-specific gene expression and gene isoforms, we provide insights into the post-transcriptional regulation of these genes in the developing muscle.
AS can directly affect PAS usage and APA by regulating the splicing of exons that harbor PASs. RBFOX2 is a well-known regulator of AS. Despite its prominent role in AS, the majority of RBFOX2-induced APA changes were not mediated via splicing APA. Instead, they were mediated via tandem APA, which affected the length of 3′UTRs. We validated RBFOX2-mediated splicing APA and tandem APA using several different methods including PAC-seq, nanopore sequencing, and RT-PCR. We identified rat Slc25a4 gene as a target of RBFOX2 and found that RBFOX2 binding sites near PASs are important determinants of Slc25a4 dPAS usage and its expression levels. Mutating RBFOX2 binding motifs close to the dPAS increased dPAS usage, suggesting that RBFOX2 may act as a repressor of PAS usage when bound nearby.
In this study, we also focused on the functional consequences of RBFOX2-mediated APA and gene expression changes. We found that RBFOX2 is critical in maintaining mitochondrial gene expression (i.e., OPA1 and Slc25a4) and mitochondrial health. Importantly, we identified the Slc25a4 gene, which encodes for ANT1 protein as an APA target of RBFOX2. ANT1 is necessary for ATP/ADP translocation across the mitochondrial membrane during oxidative phosphorylation (King et al., 2018; Körver-Keularts et al., 2015). Mutations in this gene are linked to cardiomyopathy in human patients (King et al., 2018; Körver-Keularts et al., 2015). We found that RBFOX2 depletion affected Slc25a4 APA and its expression levels. Consistently, both Rbfox2 deletion in cardiomyocytes and loss of Slc25a4 function resulted in dilated cardiomyopathy (Wei et al., 2015; King et al., 2018; Körver-Keularts et al., 2015). Mitochondrial membrane potential is important for ATP production, which is required for heart and muscle function, and reduced mitochondrial function is a hallmark of failing hearts (Rosca and Hoppel, 2013). Importantly, we found profound changes in mitochondrial gene expression upon RBFOX2 depletion. Our findings demonstrate that RBFOX2 controls mitochondrial gene expression via APA, providing insights into how RBFOX2 may alter mitochondrial health in diseased hearts.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact and will be fulfilled by the lead contact, Dr. Muge N. Kuyumcu-Martinez (nmmartin@utmb.edu).
Materials availability
Luciferase-Slc25a4 3′UTR plasmid constructs generated for this study is available upon request from the lead contact.
Data and code availability
All raw sequencing data generated by Nanopore sequencing and PAC-Seq in this manuscript are deposited into the NCBI SRA database with project number PRJNA517125. Original western blot images have been deposited to Mendeley and will be publicly available. Electron microscopy data will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
H9c2 myoblasts (Rattus Norvegicus, obtained from embryonic rat heart tissue) and HEK293 (Homo sapiens, female embryonic kidney cells) were used for this study. H9c2 myoblast cells (ATCC CRL-1446) were cultured and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (ATCC 30-2002) supplemented with 10% fetal bovine serum (FBS, ATCC 30-2020) and 100 units/ml penicillin and streptomycin (Thermofisher Scientific 15140122).
HEK293 cells were maintained in DMEM (Corning 10-013) supplemented with 10% FBS (Corning 35-075), and 100 units/ml penicillin and streptomycin (Thermofisher Scientific 15140122). Both cell lines are grown at 37°C with 5% CO2.
METHOD DETAILS
siRNA treatment and rescue experiments
For siRNA-KD experiments, H9c2 cells were seeded at 106 cells per 100mm dish and transfected with scrambled siRNA (Thermofisher Scientific AM4611) or Rbfox2 siRNA (Thermofisher Scientific siRNA ID# s96620) at 20nM using Lipofectamine RNAiMAX (Thermofisher Scientific 13778150). Cells were harvested 72 hours post-transfection for RNA or protein extraction. For rescue experiments, 3X106 H9c2 cells were transfected with eGFP (Sigma-Aldrich), human GFP-RBFOX2 (Addgene, plasmid #63086), Flag-ANT1 (SinoBiological HG17969-NF), or empty vector (pcDNA 5) together with scrambled or Rbfox2 specific siRNAs by Neon Nucleofection System (Thermofisher Scientific) and harvested 48 hours post-transfection as we have described (Verma et al., 2013). pEGFP-RBFOX2 (transcript variant 3) was a gift from Nicolas Charlet-Berguerand (Addgene plasmid # 63086; http://addgene.org/63086; RRID:Addgene_63086).
RNA
RNA was extracted from cells using TRIzol (Thermofisher Scientific 15596-018) by following the manufacturer’s protocol with the exception that RNA was precipitated overnight at −70°C. For PAC-seq, the amount and quality of RNA were analyzed by using an Agilent bio-analyzer at the University of Texas Medical Branch Next-Generation Sequencing Core Facility. For nanopore sequencing, poly(A)+ RNA was enriched by using magnetic mRNA isolation kit according to the manufacturer’s protocol (New England biolabs S1550S).
Poly(A)-ClickSeq (PAC-seq)
The PAC-seq protocol is previously described by Routh et al. (2017). Three samples per treatment/biological condition were used for the PAC-seq analysis. Briefly, 2 μg of total RNA was used to synthesize cDNA through standard reverse transcription initiated by Illumina 4N_21T primer p7 and terminated by incorporation of azido-nucleotides (AzVTPs). Subsequently, the cDNA was cleaned by addition of RNase H (NEB) and purified through DNA Clean and Concentrator Kit (Zymo) before click-reaction, in which the 5′ hexynyl- “click-adapter” p5 was click-ligated with azido-terminated cDNA in the presence of copper-TBTA (Lumiprobe) and Vitamin C. After DNA purification, the clicked cDNA was mixed with indexing primer, universal primer complementary to p7 and One Taq Standard Buffer Master Mix for PCR amplification. PAC-seq libraries were submitted for 1x150 single end sequencing yielding between 14 and 25 million reads per sample. The output data was processed and mapped to the rattus norvegicus genome (rn6) and poly(A)-clusters (PACs) were generated using the DPAC pipeline (Routh, 2019), designed for automated analysis and annotation of poly(A)-targeted RNaseq libraries such as PAC-seq.
DPAC (Routh, 2019) uses DESeq2 (Love et al., 2014) to measure changes in PAC usage between control and RBFOX2 KD cells. Data processing, read mapping and poly(A)-site detection were performed using the default parameters of the Differential-Poly(A)-Clustering (DPAC) pipeline (Routh, 2019). PASs are defined as the exact nucleotide of the 3′UTR to poly(A) tract junction found within individual sequencing reads, while poly(A)-clusters (PACs) are defined as clusters of PASs when they are found within < 10nts of one another (default parameter in DPAC pipeline). The differential PAC usage was reported if an individual PAC comprises at least 5% of a gene’s PACs, has a fold change of > 1.5, and results with an Independent Hypothesis Weighted (Ignatiadis et al., 2016) multiple testing p-adjusted value of < 0.1.
Nanopore sequencing with MinION
For nanopore sequencing, two sets of control and RBFOX2 KD H9c2 cells were used. Total cellular RNA was first poly(A) enriched and then amplified using oligo-dT primers and template switching oligos using Oxford Nanopore Technologies (ONT) cDNA-PCR sequencing kit (PCS108) as prescribed by the manufacturer. Samples were multiplexed using ONT barcodes. Pooled samples were sequenced on R9.4 flow cells. Reads were demultiplexed and base-called using Albacore followed by mapping to the rat genome (rn6) using the splice function of minimap2 (Li, 2016). Transcript isoforms were identified using FLAIR (Tang et al., 2020).
JC-1 Mitochondrial membrane potential assay
H9c2 cells were seeded on to 96-well black flat bottom plates (Costar). After 24 hours, cells were incubated in DMEM media without FBS and phenol red at 37 °C/5% CO2 with 5 μM rotenone (inhibits complex I) or 30 μM antimycin A (inhibits complex III) for 24 hours. In some experiments, cells were also incubated with 250 μM trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP, uncoupler of membrane permeability and oxidative phosphorylation) or 400 μM H2O2 that was added during last 4 hours of incubation. Plates were washed with PBS and then 100 μl of 10 μg/ml JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide, Invitrogen, T3168) added. Cells were incubated at 37 °C for 15 minutes in dark, washed twice with PBS, and 100 μl PBS was added to each well. Plates were read in SpectraMax M2 (Molecular Devices) to measure red J-aggregates fluorescence (a sensitive marker of Ψm) at an excitation of 535 nm and emission of 595 nm and green J-monomers fluorescence (indicator of disruption of Ψm) at an excitation of 485 nm and emission of 535 nm. Ratio of red/green fluorescence was calculated and plotted on the graph.
RT-qPCR
Briefly, a master mix was set up by mixing 5 μL of cDNA, 3 μL of H2O, 2 μL of PCR gene specific primer (10X conc) and 10 μL of master mix (Roche 04707516001) in 20 μL reaction. The qRT-PCR was conducted using LightCycler 480 Instrument (Roche) using the following conditions: 95°C 10 s; 62°C 15 s; 72°C 10 s for 40 cycles. Melting curve was obtained to ensure single product. ΔCt method was adopted for quantification. qPCR quantifications were as described previously (Belanger et al., 2018; Nutter et al., 2016; Verma et al., 2016). For luciferase-Slc25a4 reporter qPCRs, forward primer was designed on the firefly luciferase ORF, and reverse primers were designed to bind Slc25a4 3′UTR in different locations (Table S2). Firefly-Slc25a4 heterologous mRNA generated via dPAS usage in comparison to total Firefly-Slc25a4 heterologous mRNA was normalized to renilla luciferase mRNA levels. Semiquantitative RT-PCR was used for determining Tpm1 (short and long) transcripts due to the generation of two distinct DNA bands representing short and long isoforms from the same PCR reaction. 2 μg of total RNA was used for cDNA synthesis using AMV reverse transcriptase (15 units/μg, Life Biosciences). PCR was performed using 5 μL of cDNA, 25 μM dNTPs, 100ng of each gene specific forward and reverse primer and 0.2 μL of Biolase Taq polymerase (Bioline) in a 20 μL reaction. All primer sequences are provided in Table S2.
Western blot
Cells were lysed in lysis buffer (10mM HEPES-KOH, pH7.5, 0.32 M Sucrose, 5 μM MG132, 5mM EDTA, 1.0% SDS, and proteinase inhibitor cocktail from Roche) and sonicated. Protein concentration was determined by using the Bicinchoninic acid assay (BCA, Sigma BCA1-1KT). 30 μg of protein was separated on 10% SDS-PAGE and transferred to a PVDF membrane (Immobilon-P, Millipore). The membrane was blocked with 5% dry fat-free milk solution in PBS containing 0.1% Tween (PBST) at RT for 1 hour. The membrane was cut into 2-3 pieces based on the molecular weight of the proteins of interest and each piece of membrane was incubated with indicated primary antibodies overnight at 4°C. The membranes were washed with PBST for three times and incubated with HRP-conjugated secondary antibody for 2 hours at RT followed by three washes using PBST for 15 minutes each. Immobilon Western chemiluminescent (Millipore WBKLS0500) kit was used to detect HRP activity of the secondary antibody. The membrane was then imaged using ChemiDoc Touch imaging system (Bio-Rad). ImageJ software was used for band intensity quantification. Primary antibodies used for this study are as follows: RBFOX2 (1:1000, Abcam, ab57154), TPM1 (1:1000, Cell Signaling, D12H4), α-tubulin (1:20000, Sigma-Aldrich, T6074), mitofusin (1:1000, Abcam, 57602), OPA1 (1:1000, Abcam, 157457), COX4 (1:1000, Abcam, ab96056), CYTB (1/100, Santa Cruz, sc-11436) and ANT1:Slc25a4 (1/750, Abcam, ab102032).
Transmission electron microscopy
To obtain ultrastructural analysis of cells in ultrathin sections, cells were fixed for at least 1 hour in a mixture of 2.5% formaldehyde prepared from paraformaldehyde powder, and 0.1% glutaraldehyde in 0.05M cacodylate buffer pH 7.3 to which 0.01% picric acid and 0.03% CaCl2 were added. Cell monolayers were washed in 0.1 M cacodylate buffer, were scraped off and processed further as a pellet. The pellets were post-fixed in 1% OsO4 in 0.1M cacodylate buffer pH 7.3 for 1 hour, washed with distilled water and en bloc stained with 2% aqueous uranyl acetate for 20 min at 60°C. The cell pellets were dehydrated in ethanol, processed through propylene oxide and embedded in Poly/Bed 812 (Polysciences, Warrington, PA). Ultrathin sections were cut on Leica EM UC7 ultramicrotome (Leica Microsystems, Buffalo Grove, IL), stained with lead citrate and examined in a JEM-1400 (JEOL USA, Peabody, MA) transmission electron microscope at 80 kV. Digital images were acquired with a bottom-mounted CCD camera Orius SC200 1 (Gatan, Pleasanton, CA). Mitochondrial size was quantified using ImageJ software.
Luciferase assays
The 3′end of rat Slc25a4 3′UTR sequence, which contains multiple poly(A) sites (pPAS and pPAS) and RBFOX2 binding sites downstream of dPAS were cloned into pmirGLO Dual-luciferase vector (Promega, USA) downstream of firefly luciferase open reading frame (ORF). SV40 polyA site was removed in this plasmid to allow only the usage of polyA sites provided within the Slc25a4 3′UTR. Two RBFOX2 binding motifs near the distal poly(A) sites in the 3′UTR of Slc25a4 gene were abolished by mutating “CA” to “AC” (Slc25a4 3′UTR RBFOX2 binding WT or Slc25a4 3′UTR RBFOX2 binding MUT). 1X106 HEK293 cells seeded in 6 well plates 18h before transfection and transfected with 750 ng DNA (Luciferase Slc25a4 3′UTR polyA reporter constructs) using X-tremeGene 9 (Sigma Aldrich, USA). Cells were lysed 24 hours after transfection and determined by the Dual luciferase assay kit (Promega, USA) and recorded using BioTek Cytation 5 plate reader. Firefly luciferase was normalized to renilla luciferase activity and compared between WT and mutant luciferase Slc25a4 3′UTR polyA reporter constructs.
Metagene analysis
For metagene analysis, annotations for PASs identified in the rn6 genome were lifted over to the hg19 genome using the ‘Lift Genome Annotations’ tools on the UCSC Genome Browser. Coverage of eCLIP sites, assigning each BED entry with a score coverage of one, was generated both up and downstream by 1000nt relative to each PAS or for select PASs as indicated in the figures. Significant difference in BED coverage in different regions of the metagene were calculated using Mann-Whitey U test.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical analysis for each experiment is detailed within each section of the Star Methods or in Figure legends. Prism 6 was used to plot the graphs and to perform all statistical analyses (GraphPad Software). Data represent means ± standard deviation. N = number of samples from control or treated cells. Statistical significance was calculated using t test to compare two different groups in three independent experiments. For two group comparisons, Student’s t test was used. Multiple groups were compared using one-way analyses of variance (ANOVAs) with Bonferroni’s post hoc test. P values < 0.05 are designated as significant. Significance is defined as *p < 0.05; **p < 0.01; ***p < 0.001. For PAC-seq, Nanopore seq, and Tpm1 RT-qPCR n = 3 samples from 3 independent experiments. For endogenous Slc25a4 RT-PCR validations, n = 8 samples from three independent experiments. For ANT1 protein (Slc25a4) quantifications n = 4 from three independent experiments. For RT-qPCR analysis of dPAS usage using pA Luciferase-Slc25a4 3′UTR reporter constructs, n = 6 from three independent experiments. For relative firefly luciferase quantifications n-5, from three independent experiments. For mitochondrial membrane potential quantifications, n = 10 (five replicates from two independent experiments). For mitochondrial protein level quantifications n = > 3 samples per treatment group, from three independent experiments. Transmission electron microscopy images of mitochondria n = 3 samples from three independent experiments. For quantification of surface area of mitochondria (n = 234 mitochondria for control, n = 216 mitochondria for RBFOX2 KD myoblasts, from 3 independent KD experiments. Exact p values and n numbers for each figure can be found in the Figure Legends section for each figure.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| RBFOX2 | Abcam | Cat# ab57154; RRID:AB_2285090 |
| TPM1 | Cell Signaling | Cat# 3910; RRID:AB_2205654 |
| α-tubulin | Sigma-Aldrich | Cat# T6074; RRID:AB_477582 |
| mitofusin | Abcam | Cat# 57602; RRID:AB_2142624 |
| OPA1 | Abcam | Cat# 157457; RRID:AB_2864313 |
| COX4 | Abcam | Cat# 110272; RRID:AB_10862891 |
| CYTB | Santa Cruz | Cat# sc-11436; RRID:AB_2088887 |
| ANT1:Slc25a4 | Abcam | Cat# ab102032; RRID:AB_10710263 |
| Critical commercial assays | ||
| Dual Luciferase Reporter Assay System | Promega USA | Cat# E1980 |
| Deposited data | ||
| Raw and deposited data (PAC seq Nanopore seq) | NCBI SRA | PRJNA517125 |
| Experimental models: Cell lines | ||
| H9c2 myoblasts (Rattus Norvegicus, obtained from embryonic rat heart tissue) | ATCC | Cat# CRL-1446 |
| HEK293 (Homo sapiens, female embryonic kidney cells) | ATCC | Cat# ACS-4500 |
| Oligonucleotides | ||
| Primers (See Table S2) | This paper | N/A |
| scrambled siRNA | Thermofisher Scientific | Cat# AM4611 |
| Rbfox2 siRNA | Thermofisher Scientific | Cat# s96620 |
| Recombinant DNA | ||
| human GFP-RBFOX2 (transcript variant 3) | pEGFP rbFOX2 was a gift from Nicolas Charlet-Berguerand | Addgene plasmid # 63086 ; http://addgene.org/63086 |
| Flag-ANT1 | SinoBiological | Cat# HG17969-NF |
| pcDNA 5 | Invitrogen | Cat# V6520-20 |
| Luciferease-Slc25a4 polyA reporter (see Star Methods) | This paper | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/index.html |
| Poly(A)CLICK-seq pipeline | Routh, 2019 | https://sourceforge.net/projects/DPAC-Seq |
Highlights.
Loss of RBFOX2 is linked to human heart diseases
RBFOX2 depletion in rat myoblasts alters alternative polyadenylation (APA) patterns
RBFOX2-mediated APA modulates contractile and mitochondrial gene expression
RBFOX2 loss impacts mitochondrial health
ACKNOWLEDGMENTS
This work was supported, in part, by UTMB Department of Biochemistry and Molecular Biology Bridging funds and grants from the National Institutes of Health/National Heart Lung Blood Institute (1R01HL135031), the UTMB John Sealy Memorial Endowment Pilot Award, and the American Heart Association (20TPA35490206) to M.N.K-M. The contents of the manuscript are solely the responsibility of the authors and do not necessarily represent the official views of NHLBI of NIH. J.C. is funded by a post-doctoral fellowship from the American Heart Association (18POST 3399018). N.J.G. is funded by grants from the National Institute of Allergy and Infectious Diseases (R01AI136031) of the National Institutes of Health. A.L.R. is supported by start-up funds from UTMB. E.J.W. acknowledges support of UTMB startup funds, and P.J. is supported by funds from the National Institutes of Health (R03CA223893). R.J.H. is supported by a fellowship from the American Heart Association Research Supplement to Promote Diversity in Science (2021AHA000DIVSUP0211476). The authors acknowledge the University of Texas Medical Branch Next Generation Sequencing Core Facility for providing RNA sequencing services. We thank Dr. Pei-Yong Shi for allowing us to use their plate readers.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109910.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY STATEMENT
One or more of the authors of this paper received support from a program designed to increase minority representation in science.
REFERENCES
- Batra R, Charizanis K, Manchanda M, Mohan A, Li M, Finn DJ, Goodwin M, Zhang C, Sobczak K, Thornton CA, and Swanson MS (2014). Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell 56, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger K, Nutter CA, Li J, Tasnim S, Liu P, Yu P, and Kuyumcu-Martinez MN (2018). CELF1 contributes to aberrant alternative splicing patterns in the type 1 diabetic heart. Biochem. Biophys. Res. Commun 503, 3205–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberoglu MA, Gallagher TL, Morrow ZT, Talbot JC, Hromowyk KJ, Tenente IM, Langenau DM, and Amacher SL (2017). Satellite-like cells contribute to pax7-dependent skeletal muscle repair in adult zebrafish. Dev. Biol 424, 162–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Routh AL, and Kuyumcu-Martinez MN (2021). Nanopore sequencing reveals full-length Tropomyosin 1 isoforms and their regulation by RNA-binding proteins during rat heart development. J. Cell. Mol. Med 25, 8352–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C, Hans H, and Alwine JC (1999). Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol 19, 4971–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers EE, Bawazeer A, Ugalde AP, van Deutekom HW, van der Made I, de Groot NE, Adriaens ME, Cook SA, Bezzina CR, Hubner N, et al. (2016). Genome-Wide Polyadenylation Maps Reveal Dynamic mRNA 3′-End Formation in the Failing Human Heart. Circ. Res 118, 433–438. [DOI] [PubMed] [Google Scholar]
- Damianov A, Ying Y, Lin CH, Lee JA, Tran D, Vashisht AA, Bahrami-Samani E, Xing Y, Martin KC, Wohlschlegel JA, and Black DL (2016). Rbfox Proteins Regulate Splicing as Part of a Large Multiprotein Complex LASR. Cell 165, 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. (2018). The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, and Babak T (2012).A quantitative atlas of polyadenylation in five mammals. Genome Res. 22, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, and Manley JL (2011). Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England J, Granados-Riveron J, Polo-Parada L, Kuriakose D, Moore C, Brook JD, Rutland CS, Setchfield K, Gell C, Ghosh TK, et al. (2017). Tropomyosin 1: Multiple roles in the developing heart and in the formation of congenital heart defects. J. Mol. Cell. Cardiol 106, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher TL, Arribere JA, Geurts PA, Exner CRT, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, and Conboy JG (2011). Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol 359, 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding C, and Smith CWJ (2008). Tropomyosin exons as models for alternative splicing. Adv. Exp. Med. Biol 644, 27–42. [DOI] [PubMed] [Google Scholar]
- Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, and Wallace DC (1997). A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet 16, 226–234. [DOI] [PubMed] [Google Scholar]
- Hescheler J, Meyer R, Plant S, Krautwurst D, Rosenthal W, and Schultz G (1991). Morphological, biochemical, and electrophysiological characterization of a clonal cell (H9c2) line from rat heart. Circ. Res 69, 1476–1486. [DOI] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. (2015). De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Ou AC, Park J, Yu F, Yu B, Lee A, Yang G, Zhou A, and Benz EJ Jr. (2012). RBFOX2 promotes protein 4.1R exon 16 selection via U1 snRNP recruitment. Mol. Cell. Biol 32, 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis N, Klaus B, Zaugg JB, and Huber W (2016). Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13, 577–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanlidis G, Bautista-Hernandez V, Fynn-Thompson F, Del Nido P, and Tian R (2011). Impaired mitochondrial biogenesis precedes heart failure in right ventricular hypertrophy in congenital heart disease. Circ. Heart Fail 4, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimes BW, and Brandt BL (1976). Properties of a clonal muscle cell line from rat heart. Exp. Cell Res 98, 367–381. [DOI] [PubMed] [Google Scholar]
- King MS, Thompson K, Hopton S, He L, Kunji ERS, Taylor RW, and Ortiz-Gonzalez XR (2018). Expanding the phenotype of de novo SLC25A4-linked mitochondrial disease to include mild myopathy. Neurol. Genet 4, e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körver-Keularts IM, de Visser M, Bakker HD, Wanders RJ, Vansenne F, Scholte HR, Dorland L, Nicolaes GA, Spaapen LM, Smeets HJ, et al. (2015). Two Novel Mutations in the SLC25A4 Gene in a Patient with Mitochondrial Myopathy. JIMD Rep. 22, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2016). Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJC, Eppinga RD, Warren KS, and McCrae KR (2008). Human tropomyosin isoforms in the regulation of cytoskeleton functions. Adv. Exp. Med. Biol 644, 201–222. [DOI] [PubMed] [Google Scholar]
- Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, Peterson KA, Li Y, Schwartz MC, Reynolds WT, et al. (2017). The complex genetics of hypoplastic left heart syndrome. Nat. Genet 49, 1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al. (2013). Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol 20, 1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. (2010). The landscape of C. elegans 3’UTRs. Science 329, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, and Wagner EJ (2014). CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510, 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejía-Alvarez R, Tomaselli GF, and Marban E (1994). Simultaneous expression of cardiac and skeletal muscle isoforms of the L-type Ca2+ channel in a rat heart muscle cell line. J. Physiol 478, 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménaxrd C, Pupier S, Mornet D, Kitzmann M, Nargeot J, and Lory P (1999). Modulation of L-type calcium channel expression during retinoic acid-induced differentiation of H9C2 cardiac cells. J. Biol. Chem 274, 29063–29070. [DOI] [PubMed] [Google Scholar]
- Misra C, Bangru S, Lin F, Lam K, Koenig SN, Lubbers ER, Hedhli J, Murphy NP, Parker DJ, Dobrucki LW, et al. (2020). Aberrant Expression of a Non-muscle RBFOX2 Isoform Triggers Cardiac Conduction Defects in Myotonic Dystrophy. Dev. Cell 52, 748–763.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Yamamoto M, Takeichi M, Saga K, Takaoka K, Kawamura N, Nitta H, Nagano H, Ishino S, Tanaka T, et al. (2016). Regulation of alternative polyadenylation by Nkx2-5 and Xrn2 during mouse heart development. eLife 5, e16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutter CA, Jaworski EA, Verma SK, Deshmukh V, Wang Q, Botvinnik OB, Lozano MJ, Abass IJ, Ijaz T, Brasier AR, et al. (2016). Dysregulation of RBFOX2 Is an Early Event in Cardiac Pathogenesis of Diabetes. Cell Rep. 15, 2200–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutter CA, Jaworski E, Verma SK, Perez-Carrasco Y, and Kuyumcu-Martinez MN (2017). Developmentally regulated alternative splicing is perturbed in type 1 diabetic skeletal muscle. Muscle Nerve 56, 744–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman A, Wachtel C, Cohen M, Haupt S, Shapiro H, and Tzur A (2012). JC-1: alternative excitation wavelengths facilitate mitochondrial membrane potential cytometry. Cell Death Dis. 3, e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosca MG, and Hoppel CL (2013). Mitochondrial dysfunction in heart failure. Heart Fail. Rev 18, 607–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A (2019). DPAC: A Tool for Differential Poly(A)-Cluster Usage from Poly(A)-Targeted RNAseq Data. G3 (Bethesda) 9, 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A, Ji P, Jaworski E, Xia Z, Li W, and Wagner EJ (2017). Poly(A)-ClickSeq: click-chemistry for next-generation 3′-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res. 45, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfola V, Sebastian S, Dilworth FJ, and Gabellini D (2015). Rbfox proteins regulate tissue-specific alternative splicing of Mef2D required for muscle differentiation. J. Cell Sci 128, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, and Burge CB (2008). Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y (2012). Alternative polyadenylation: new insights from global analyses. RNA 18, 2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Xia Z, Bland CS, Kalsotra A, Scavuzzo MA, Curk T, Ule J, Li W, and Cooper TA (2014). Rbfox2-coordinated alternative splicing of Mef2d and Rock2 controls myoblast fusion during myogenesis. Mol. Cell 55, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Kolonin AM, Fiorotto ML, and Cooper TA (2018). Rbfox-Splicing Factors Maintain Skeletal Muscle Mass by Regulating Calpain3 and Proteostasis. Cell Rep. 24, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Ghochani M, McCaffery JM, Frey TG, and Chan DC (2009). Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell 20, 3525–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Fregoso O, and Krainer AR (2012). Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA 18, 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AD, Soulette CM, Baren M.J.v., Hart K, Hrabeta-Robinson E, Wu CJ, and Brooks AN (2020). Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun 11, 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Rajan S, Thurston HL, Masineni SN, Dube P, Bose A, Muthu V, Dube S, Wieczorek DF, Poiesz BJ, and Dube DK (2010). Expression of a novel tropomyosin isoform in axolotl heart and skeletal muscle. J. Cell. Biochem 110, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, and Lutz CS (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Putten HH, Joosten BJ, Klaren PH, and Everts ME (2002). Uptake of tri-iodothyronine and thyroxine in myoblasts and myotubes of the embryonic heart cell line H9c2(2-1). J. Endocrinol 175, 587–596. [DOI] [PubMed] [Google Scholar]
- Verma SK, Deshmukh V, Liu P, Nutter CA, Espejo R, Hung ML, Wang GS, Yeo GW, and Kuyumcu-Martinez MN (2013). Reactivation of fetal splicing programs in diabetic hearts is mediated by protein kinase C signaling. J. Biol. Chem 288, 35372–35386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SK, Deshmukh V, Nutter CA, Jaworski E, Jin W, Wadhwa L, Abata J, Ricci M, Lincoln J, Martin JF, et al. (2016). Rbfox2 function in RNA metabolism is impaired in hypoplastic left heart syndrome patient hearts. Sci. Rep 6, 30896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Watanabe M, Nakamura T, Kudo Y, and Ochi R (1999). Properties and expression of Ca2+-activated K+ channels in H9c2 cells derived from rat ventricle. Am. J. Physiol 276, H1559–H1566. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, and Burge CB (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Qiu J, Zhou Y, Xue Y, Hu J, Ouyang K, Banerjee I, Zhang C, Chen B, Li H, et al. (2015). Repression of the Central Splicing Regulator RBFox2 Is Functionally Linked to Pressure Overload-Induced Heart Failure. Cell Rep. 10, 1521–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. (2014). HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 6, 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolska BM, and Wieczorek DM (2003). The role of tropomyosin in the regulation of myocardial contraction and relaxation. Pflugers Arch. 446, 1–8. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, and Gage FH (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol 16, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorova LD, Popkov VA, Plotnikov EY, Silachev DN, Pevzner IB, Jankauskas SS, Babenko VA, Zorov SD, Balakireva AV, Juhaszova M, et al. (2018). Mitochondrial membrane potential. Anal. Biochem 552, 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data generated by Nanopore sequencing and PAC-Seq in this manuscript are deposited into the NCBI SRA database with project number PRJNA517125. Original western blot images have been deposited to Mendeley and will be publicly available. Electron microscopy data will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.