Abstract
T cell therapies require the removal and culture of T cells ex vivo to expand several thousand-fold. However, these cells often lose the phenotype and cytotoxic functionality for mediating effective therapeutic responses. The extracellular matrix (ECM) has been used to preserve and augment cell phenotype; however, it has not been applied to cellular immunotherapies. Here, a hyaluronic acid (HA)-based hydrogel is engineered to present the two stimulatory signals required for T-cell activation—termed an artificial T-cell stimulating matrix (aTM). It is found that biophysical properties of the aTM—stimulatory ligand density, stiffness, and ECM proteins—potentiate T cell signaling and skew phenotype of both murine and human T cells. Importantly, the combination of the ECM environment and mechanically sensitive TCR signaling from the aTM results in a rapid and robust expansion of rare, antigen-specific CD8+ T cells. Adoptive transfer of these tumor-specific cells significantly suppresses tumor growth and improves animal survival compared with T cells stimulated by traditional methods. Beyond immediate immunotherapeutic applications, demonstrating the environment influences the cellular therapeutic product delineates the importance of the ECM and provides a case study of how to engineer ECM-mimetic materials for therapeutic immune stimulation in the future.
Keywords: adoptive T cell therapy, artificial matrix, extracellular matrix, hydrogel, immunotherapy, mechanotransduction, T cell stimulation
T lymphocytes are increasingly targeted and utilized in immunotherapies with the success of checkpoint blockade, adoptive cell transfer (ACT), and chimeric antigen receptor (CAR) T-cell therapy.[1] For both ACT and CAR T cell therapies, T cells must be removed from patients, cultured and stimulated ex vivo, and then reinjected into patients for cancer immunotherapy.[2] This presents two major challenges. First, the number of T cells needed is very large, so they are cultured for 6–8 weeks at a time, by which time the cells’ functionality and phenotype to mediate effective killing and long-term memory may have been lost or altered.[3-6] By improving the quality or phenotype and functionality ex vivo, therapeutic outcomes can also be improved significantly.[7] Second, antigen-specific stimulations utilize antigen-presenting cells that may be immunosuppressed and are often dysfunctional, or nonspecific stimulation from synthetic surfaces through CD3 can result in expansion of irrelevant and potentially harmful clones.[8-10]
There are several approaches to address these challenges including altering composition of cytokine cocktails, signaling pathway inhibitors, and feeder cells.[11] Additionally, the two signals necessary to stimulate the T cell receptor and costimulatory molecules have been conjugated to synthetic materials: inorganic or polymeric particles[12-15] and surfaces.[16-18 While current synthetic T cell stimulation platforms are helpful in efficiently enriching and activating antigen-specific T cells,[19-21] providing cell membrane-mimetic materials,[16] and acting as in vitro or in vivo stimulators,[15] however, none provide environmental cues similar to what T cells encounter in the lymphoid organs, such as the spleen or lymph node.
The extracellular matrix (ECM) is an important regulator of cellular function, including gene expression, differentiation, migration, proliferation, and morphology.[22-25] T lymphocytes primarily reside in the lymphoid organs. These unique microenvironments enable rapid communication, cell differentiation, and allow antigen-specific cells to expand thousands-fold in response to infection.[26-29] Even though it is well demonstrated that cells are influenced by ECM properties such as composition, stiffness, and bioactive cues that create unique microenvironments suited to the function of each cell and tissue,[30] the role of ECM on T cell activation has not been investigated. Furthermore, bioengineering approaches have developed ex vivo culture environments with control over matrix properties for cell and tissue engineering applications.[31-38] We hypothesize that a biomimetic, engineered artificial T cell stimulating matrix (aTM) can improve the functionality and phenotype of ex vivo stimulation of T cells for therapeutic applications.
Here we generated aTM hydrogels from ECM-based materials with tunable stiffness and two types of key signaling molecules for T-cell stimulation of murine or human cells. This approach also provides ligands from the hydrogel matrix to ECM receptors on the T cell—contributing a potential additional signaling component[39]—in contrast to other T-cell stimulating materials. Additionally, the stiffness of the hydrogel matrix can be tuned allowing effective mechanotransduction required for effective T cell receptor (TCR) signaling. We examine these unique biophysical properties and study them mechanistically for producing more functional antigen-specific T cells and assess their efficacy in preclinical models of tumor immunotherapy.
We formed ECM hydrogels by cross-linking thiolated hyaluronic acid (HA) with polyethylene glycol diacrylate (PEGDA) (Figure S1, Supporting Information). HA is a linear polysaccharide and is the only nonsulfated glycosaminoglycan found distributed throughout the ECM, including lymphoid tissues[40] that impacts cell motility and adhesion, differentiation, gene expression, and proliferation.[41-43] Furthermore, we chose HA as an ECM mimic because it can be easily modified through tunable chemistry—which enables the addition of adhesive ligands,[44,45] conjugation of drugs or growth factors,[46] and control of the elastic modulus and porosity of the hydrogel.[47,48]
We engineered this material into an antigen-presenting material by conjugating the signals (Signal 1 and Signal 2) needed for T cell activation directly to the scaffold (Figure 1A). We first used anti-CD3 and anti-CD28 antibodies for polyclonal wild-type B6 murine CD8+ T cell expansion. This presents a unique approach to use the biophysical properties of hydrogels to influence the potency of a stimulatory environment, which we term an aTM (artificial T-cell stimulating matrix).
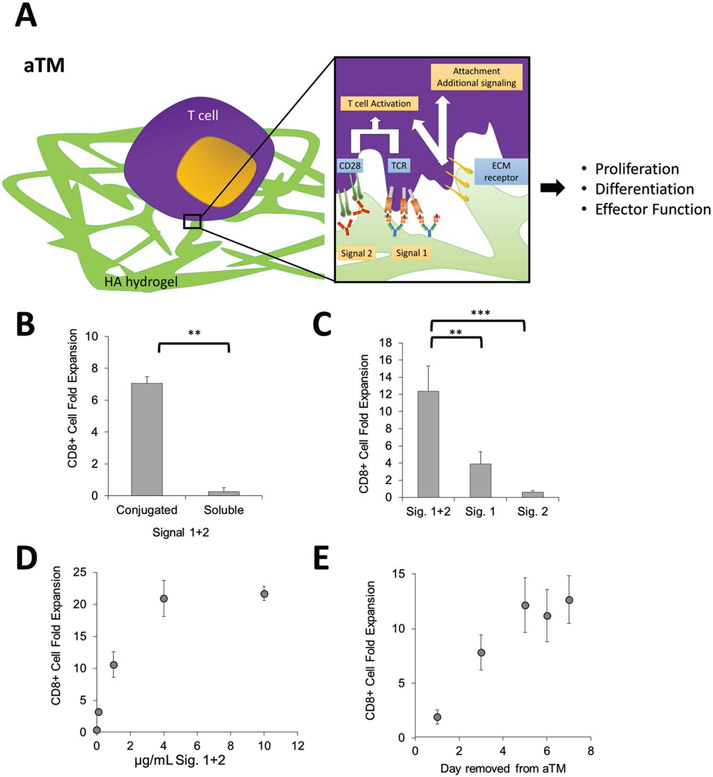
Figure 1.
An artificial T cell stimulating matrix (aTM) is engineered by conjugating T cell stimulating signals to a hydrogel. A) Schematic of aTM made from conjugating Signals 1 and 2 to a hyaluronic acid hydrogel. Attachments of Signal 1 and 2 enable effective T cell stimulation that leads to T cell proliferation, differentiation, and effector function. Receptors bind to ECM hydrogel and also contribute to attachment and T cell signaling. B–D) B6 CD8+ T cell fold expansion measured after 7 d of stimulation of the antigen-specific T cells on the hydrogels with Signals 1 + 2 conjugated or soluble (error bars show s.e.m.; **p < 0.005, n = 4, Student’s t-test, two-tailed) (B), conjugated together or alone (error bars show standard error of the mean (s.e.m.); **p < 0.005, ***p < 0.0005, n = 5–7, one-way ANOVA with Tukey’s post-test) (C), and at varying amounts of Signals 1 + 2, n = 5 (D). E) Day 7 CD8+ T cell fold expansion measured after 7 d of stimulation of the antigen-specific T cells on the aTM. T cells were removed from aTM on the day noted and cultured on TCP until day 7.
Postconjugation, nearly all (at least 85%) of the stimulatory signals conjugated remained attached to the scaffold postgelation (Figure S2A, Supporting Information). Direct conjugation of Signals 1 and 2 at 1 μg mL−1 (i.e., aTM) mediated about sevenfold polyclonal T cell proliferation, whereas the same hydrogel substrate with soluble Signals 1 and 2 showed little proliferation (Figure 1B). Substrates with only Signal 1 or 2 conjugated resulted in much lower T cell activation and proliferation (Figure 1C). Investigating different ratios of Signal 1 to Signal 2 at 1 μg mL−1 revealed that Signal 1 concentration was most critical, and that optimal T cell expansion occurred at a 1:1 ratio, which is used in all subsequent studies (Figure S2B,C, Supporting Information). There are a number of different co-stimulatory molecules, which may also provide T cell co-stimulation. Incorporation of a different co-stimulatory molecule anti-41BB at various ratios to anti-CD28 did not further enhance CD8+ T cell activation on the aTM, and also demonstrated the necessity of inclusion of anti-CD28 as a co-stimulatory signal for early activation of CD8+ T cells (Figure 2A, Supporting Information).
The density of T cell stimulating signals is an important parameter to control and optimize. We and others have shown previously on particle and planar surfaces that effective T cell stimulation is observed when the interligand spacing is maintained below 75–150 nm.[12,49,50] As we increased the concentration of stimulatory ligands on the aTM, the amount of CD8+ T cell proliferation increased, though it plateaued at around 20-fold expansion when 4 μg mL−1 of Signals 1 and 2 was used (Figure 1D). We estimated the surface density of the signals attached to the surface of the aTM for each concentration (Figure S2E, Supporting Information). Our findings estimate that the spacing for ligands need to only be at least 500 nm apart (corresponding to 1 μg mL−1), which is larger than previously reported values, potentially due to the fact that signals may not be evenly distributed across hydrogel surface and could be clustered on ECM polymers, or that compliant surfaces require less dense arrays of signal. Additionally, the viability of T cells decreased beyond 4 μg mL−1 of Signals 1 and 2 (Figure S3A, Supporting Information), and thus less than 4 μg mL−1 or less were used for subsequent studies with a cell concentration of 0.1 × 106 cells mL−1 (Figure S3B, Supporting Information). aTM also effectively stimulated CD8+ T cells from splenocyte starting populations at similar densities of Signals 1 and 2 (Figure S3C, Supporting Information). Finally, CD8+ T cells required at least five days to be fully stimulated on the aTM surface, where suboptimal activation was observed when cells were removed from the hydrogels on days 1 and 3 (Figure 1E), indicating a need for dynamic engagement of conjugated stimulatory molecules.
In stem cells, matrix stiffness modulates cell function through mechanotransduction signaling mechanisms.[31,51] Secondary lymphoid tissue is a soft tissue and the stiffness has been reported to be between 0.1 to 2 kPa.[52,53] To control the mechanical stiffness of our hydrogel within this range, we altered the amount of cross-linker, varying the elastic modulus from 0.2 to 3 kPa (Figure 2A,B). A softer aTM (0.5 kPa) stimulated CD8+ T cell proliferation more effectively than the stiffer aTM (3 kPa) as determined by the dilution of the proliferation dye, carboxyfluorescein succinimidyl ester (CFSE) (Figure 2C). Greater than 80% of the CD8+ T cells divided past the first and second generation when stimulated on the 0.5 kPa aTM, while the majority of T cells on 3 kPa aTM did not divide at all (Figure S4, Supporting Information). We further probed the spectrum of substrate stiffness to determine the optimal range for T cell stimulation. aTMs with a stiffness below 1 kPa were more effective at stimulating CD8+ T cell expansion, where we observed a dramatic decrease in T cell expansion occurs with aTM greater than 1 kPa (Figure 2D). We further evaluated the properties of these gels through scanning electron microscopy (SEM) imaging, rheometry following cell-incubation, and fluorescent staining to determine conjugation efficiency of stimulatory antibodies. As expected, the gross morphological comparison between the two gels showed that the 0.5 kPa gels were more porous than the 3 kPa gels (Figure S5, Supporting Information). The integrity of gel stiffness after 7 d of incubation was maintained with minor changes in stiffness of both the 3 kPa and 0.5 kPa gels (Figure S6, Supporting Information). Finally, we found that similar densities of stimulatory signal were conjugated to the surface of the different stiffness aTM through fluorescent secondary antibody staining (Figure S7, Supporting Information).
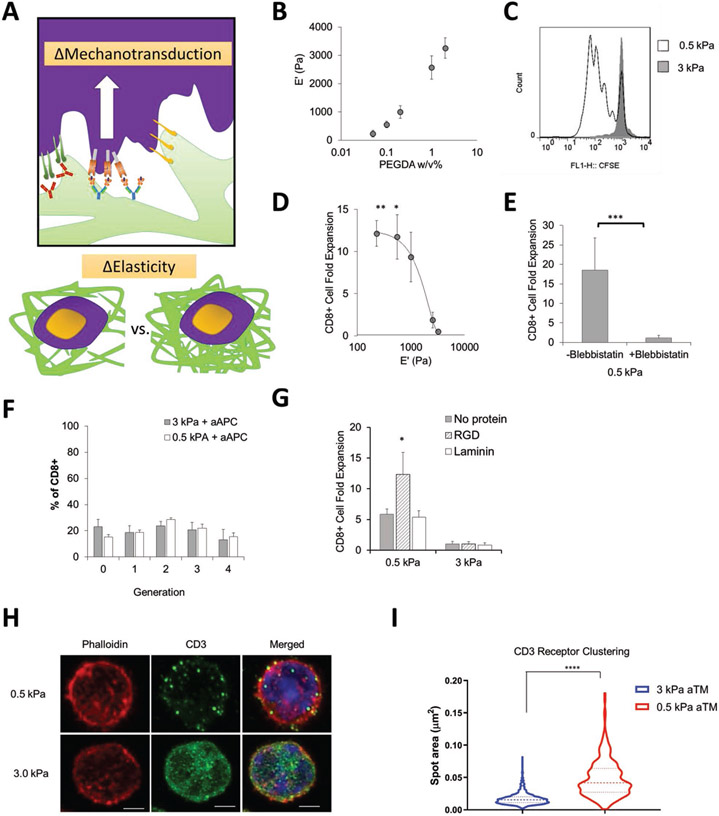
Figure 2.
Tuning the stiffness of the aTM impacts T cell stimulation. A) Schematic illustrating hypothesis that tuning stiffness of aTM may change the ability for cell mechanotransduction. B) Elastic modulus measured by rheometry with varying PEGDA cross-linker weight percent (error bars show s.e.m., n = 3). C) CFSE proliferation dye dilution measured after 3 d of stimulation of T cells comparing a stiff (3 kPa) and soft 0.5 kPa) aTM. D) CD8+ T cell fold expansion measured after 7 d of stimulation of the T cells on aTMs with varying stiffness (error bars show s.e.m.; *p < 0.05, **p < 0.005, n = 4–12, one-way ANOVA with Tukey’s post-test). E) CD8+ T cell fold expansion measured for T cells stimulated on soft aTMs (0.5 kPa) with or without blebbistatin (error bars show s.e.m.; ***p < 0.0005, n = 4, Student’s t-test, two-tailed). F) Quantitation of percentage of T cells in each divisional generation based on CFSE proliferation dye dilution with T cells stimulated on HA hydrogels of different stiffness with aAPC (error bars show s.e.m, n = 4–8). G) CD8+ T cell fold expansion measured after 7 d of stimulation of T cells on the aTMs with either laminin and RGD attached (error bars show s.e.m.; *p < 0.05, n = 3–6, one-way ANOVA with Tukey’s post-test). H) Airyscan super-resolution imaging of phalloidin and CD3 of CD8+ T cells cultured on either soft or stiff aTM (scale bar = 2 μm.), I) where a total of 515 spots were analyzed from 16 cells in the 0.5 kPa condition and 1580 spots were analyzed from 13 cells in the 3 kPa condition. (Error bars show s.e.m.; ****p < 0.0001, Student’s t-test, two-tailed).
It was surprising to observe such a dramatic increase in T cell expansion at stiffness below 1 kPa. We hypothesized that this expansion was a result of stiffness-dependent TCR signaling through the conjugated stimulatory signals more so than T cell-ECM interactions, as mechanotransduction may play a role in TCR signaling due to the motile nature of interacting cognate T cells and antigen presenting cells.[54-58]
To evaluate our hypothesis of the role of mechanotransduction of TCR stimulating signals, we performed additional experiments where we utilized myosin inhibitors, decoupled signaling components from the hydrogel, macro and microscale visualized cellular attachment, and added further cell-adhesive molecules. First, we added a blebbistatin, a myosin II inhibitor important in the role of T cell mechanotransduction,[18,59] to the culture of a 0.5 kPa aTM. In the presence of the inhibitor, CD8+ T cell expansion is abolished even with the same amount of stimulatory ligand present (Figure 2E).
Next, we decoupled the stimulatory agent from the matrix and stimulated with cognate artificial antigen presenting cells (aAPC) on different stiffness of hydrogels. The nanoparticle aAPC contain both Signals necessary for CD8+ T cell activation—Signal 1: peptide loaded major histocompatibility complex (pMHC) and Signal 2: anti-CD28 costimulatory antibody (Figure S8, Supporting Information). There were no differences in the CFSE proliferation assay or in resultant cell phenotype between soft (0.5 kPa) and stiff (3 kPa) HA hydrogels (Figure 2F, Figure S9, Supporting Information). This demonstrates that the mechanotransduction is independent of ECM-cell adhesion receptor interactions traditionally investigated, but instead dependent upon TCR signaling when stimulatory ligands are attached to HA hydrogels.
Furthermore, we visualized the interaction of the T cells and the aTM hydrogels or HA hydrogels (without Signals 1 and 2) with light video microscopy (Figure S10, Videos S1-S4, Supporting Information). After 24 h, only CD8+ T cells remained attached to the soft (0.5 kPa) aTM hydrogel with both stimulatory signals conjugated. Whereas CD8+ T cells did not attach to stiff aTM (3 kPa) or soft (0.5 kPa) hydrogels without Signals 1 and 2 attached.
Adding cell-adhesive ligands has been shown to increase cell attachment to surfaces.[60] We included additional ECM-binding proteins to the aTM scaffold such as laminin and cyclic RGD, a sequence derived from ECM-binding proteins to determine whether this might improve engagement and stimulation on stiff hydrogels. Even providing cell-adhesive ligands did not help stiff aTMs (3 kPa) stimulate antigen-specific PMEL CD8+ T cells, whereas RGD further increased T cell proliferation on soft (0.5 kPa) aTM surfaces resulting in effective expansion (Figure 2G, Figure S11, Supporting Information).
Finally, the ability for TCR clustering is a critical aspect to effective T cell activation,[12,61,62] and we hypothesized that the ability of surface clustering of stimulatory signals may be influenced by the stiffness of the hydrogel aTM. To test this hypothesis we performed super resolution microscopy and stained for actin and CD3, a component of the TCR. Interestingly, we observed significant differences in the CD3 clustering on T cells cultured on the 0.5 kPa aTM versus the 3 kPa aTM with little difference in actin co-localization (Figure 2H). Indeed by analyzing the CD3 cluster size, T cells cultured on the softer aTM had significantly greater CD3 cluster area of 0.05 μm2 compared to 0.02 μm2 for that of the T cells stimulated on the stiffer aTM (Figure 2I).
Taken together, these data indicate that the role of mechanical stimulation is mediated through the TCR and the stimulatory ligands conjugated to the matrix, yet cannot be overcome with adding additional cell-attachment sequences. This enhanced mechanotransduction at lower stiffness could be due to a number of reasons. First, softer hydrogels which are more compliant and may enable enhanced clustering of neighboring attached Signal 1 molecules on the hydrogel—shown to promote superior TCR signaling, which we observe in Figure 2H,I.[12,63] Second, traditionally T cells are stimulated by surfaces which have stimulatory signals nonspecifically adsorbed to surfaces, whereas our signals are chemically attached to the matrix, which may contribute to more effective mechanotransduction. Third, the stiffness range of the aTM more closely matches the stiffness of the secondary lymphoid tissue. To our knowledge this represents the lowest stiffness of a surface, orders of magnitude less than T cells have been stimulated with, where traditionally the surface is a stiff (≈MPa) plastic.[17,18] At softer hydrogel surfaces, we are approaching the appropriate level of resistance that a T cell may observe at a cellular level, where researchers have shown that the minimum adhesion strength to antigen presenting cells to be around 90 Pa.[54,64] In conclusion the stiffness of the aTM is critical to enhance mechanotransduction TCR signaling for effective T cell expansion.
Beyond biophysical cues such as stiffness, the ECM can provide molecular signaling cues via cellular receptor activation. We were particularly curious to how the base HA hydrogel affects both T cell functionality and phenotype, as T cells express CD44—a ligand specific for HA.[42,65] For T cells, CD44 has primarily been utilized as a marker for cellular phenotype and not examined as a co-stimulatory molecule.[66-69] To investigate how the HA hydrogel contributes to T cell activation and signaling, we decoupled the other unique biophysical variable—having the T cell stimulatory signals attached to the surface—by utilizing aAPC for T cell stimulation. In this manner we could mechanistically study differences in T cell signaling directly due to the HA hydrogel.
To examine the influence of stimulatory environment, transgenic PMEL CD8+ T cells and cognate aAPC were co-incubated and then either plated onto ECM-mimic hydrogels (HA) or the traditional tissue culture plate (TCP) wells (Figure 3A). Interestingly, CD8+ T cells that were cultured on HA hydrogel surfaces demonstrate much higher antigen-specific T cell proliferation as indicated by CFSE dilution after three days of culture (Figure 3B). In fact, there are significantly more T cells that have reached the second, third, and fourth generations when compared to the T cells cultured on traditional tissue culture plates, where the majority of the cells have not yet divided (Figure S12, Supporting Information). There was no inherent signaling or activation without stimulatory aAPC with no effects on cell viability on the HA surface (Figure S13, Supporting Information). Moreover, including soluble HA also increased the percent of CD8+ T cells to divide (≈35%) as compared to the tissue culture plate without hydrogel (≈15%), but not as much when it is cross-linked into a hydrogel (≈65%) (Figure 3C). Therefore, the benefit of the HA to early CD8+ T cell proliferation is partially mediated through direct interaction with cross-linked HA in combination with TCR signaling.
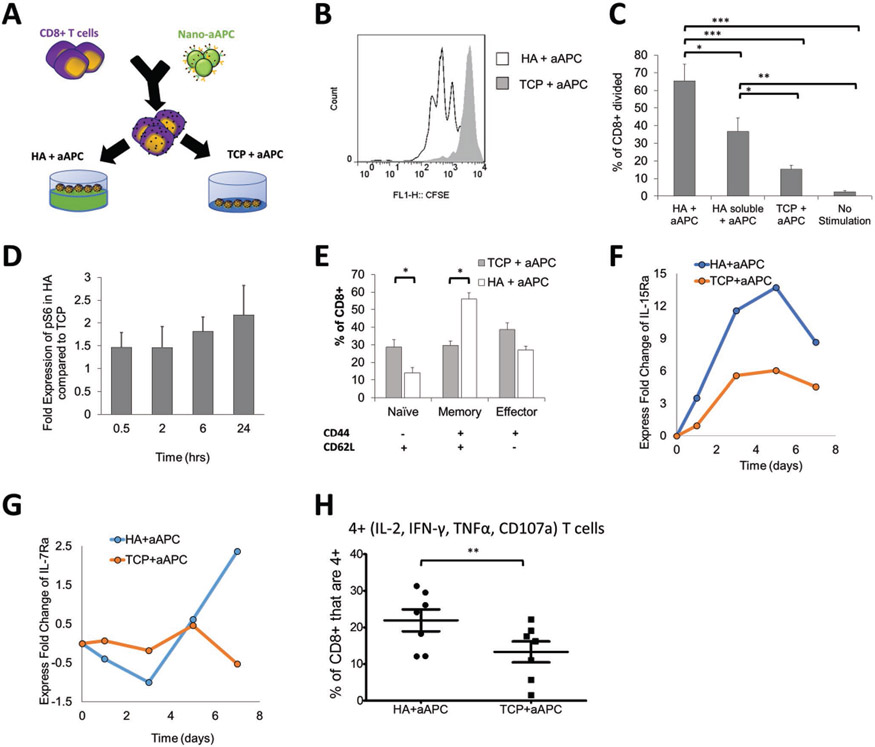
Figure 3.
Stimulated T cells are influenced by additional signaling from the HA hydrogel. A) Schematic showing experimental setup testing the difference between activating antigen-specific CD8+ T cells with nanoparticle artificial antigens presenting cells (aAPC) on HA hydrogel versus a tissue culture plate (TCP). B) CFSE proliferation dye dilution measured after 3 d of stimulation of antigen-specific T cells stimulated by the same dose of aAPC on either TCP or on HA hydrogel surface. C) Percent of CD8+ T cells that have divided by day 3 as measured by CFSE proliferation dye dilution (error bars show s.e.m., *p < 0.05, **p < 0.005, ***p < 0.0005 n = 7, one-way ANOVA with Tukey’s post-test). D) Time course experiment using p-S6 (S240/S244) as the read out for mTORC1 activation. This relative fold-change pattern represents three independent experiments using phospho-flow cytometry. E) Phenotypic markers (CD62L, CD44) measured by flow cytometry after 7 d of stimulation with aAPC on different surfaces (error bars show s.e.m.; *p < 0.05, n = 7, Student’s t-test, two-tailed). F,G) Time course experiment detecting fold change of F) IL15Ra (CD215) and G) IL7Ra (CD127). Geometric means of each data point are compared first with their isotype controls followed by the baseline control. Data represents two independent experiments. H) T cells positive for all four cytokine and functional molecules (IL-2, IFN-γ, TNFα, CD107a) were measured by flow cytometry after 7 d of stimulation (error bars show s.e.m.; *p < 0.05, n = 7, Paired t-test, two-tailed).
We investigated the role of HA in signaling and inducing greater early expansion of CD8+ T cells in the hydrogel condition. Exploring key signaling pathways related to T cell activation and proliferation, we identified a significant increased expression of p-S6K1 and p-S6 (Figure 3D, Figure S14, Supporting Information), consistent with upregulation of mTORC1 (mammalian target of rapamycin complex 1), and a downregulation of p-AKT (indicative of mTORC2) under the HA culture condition compared to the TCP condition (Figure 14, Supporting Information). Furthermore, CD44-signaling has been shown to trigger Ras-Erk signaling in other cell types,[43] and Ras-Erk and PI3K-mTOR pathways have been shown to crosstalk and compensate each other in T cells.[70] Indeed we observe a significant amount of p-S6 signal may come from CD44-induced Ras-Erk signaling (Figure S15A,B, Supporting Information). It has been shown previously that CD44 directly interacts with Src family proteins like Lck.[71] Lck is involved in early phosphorylation the ITAM domains of TCRζ chain and CD3 complex for T cell activation upstream of both Ras-Erk and PI3k-mTOR signaling.[72] We observed a higher level of phosphorylation of p-Src family protein at Y416 (which includes Y934 in p-Lck as an activation signal), with equal amounts of p-Lck Y505 (inhibiting signal) and total Lck, (Figure S16, Supporting Information). Taken together, the interaction of T cells with HA clearly influences early T cell priming and activation with noted contributions of enhanced Lck phosphorylation and potential crosstalk between Ras-Erk and PI3K-mTOR pathways, where mTOR is an important integrator of immune cues for robust T cell activation and phenotype skewing.[73]
To investigate the long-term role of ECM-mimic hydrogels in enhancing CD8+ T cell expansion and hemostasis, we looked at the expression of CD44 after stimulation on the two different surfaces. CD44 is expressed by CD8+ T cells in concordance with changes in phenotype, where CD44 remains upregulated in activated and memory cells.[66-69] We also stained for CD62L to separate CD8+ T cell phenotypes influenced by the HA hydrogel—naïve, memory, and effector.
Stimulation on the HA hydrogel resulted in significantly higher percentage of CD44+ T cells (Figure 3E). This increase was associated with nearly double the percentage of memory T cells (CD62L+, CD44+) and a decrease in effector T cells though not statistically significant. Consistent with an increase in memory-like cells, we observed a global upregulation of IL-15Ra (Figure 3F) and a conditional upregulation of IL-7Ra (Figure 3G) at late contraction phases under HA condition, confirmed at the mRNA level (Figure S17, Supporting Information). Both IL-7Ra and IL-15Ra are receptors for cytokines that trigger memory T cell homeostasis,[74] and thus an upregulation of both receptors may induce memory T cell formation. In conclusion, these both inform that HA can contribute to increased proliferation, and also demonstrate that the ECM hydrogel surface influences the final phenotype of the cells.
Memory T cells have an increased persistence and potential for proliferation, with stem-cell like qualities,[75] and have been shown effective in anticancer responses for ACT.[76] To test the function and quality of the resultant CD8+ T cells in vitro, we examined how well they co-produced multiple cytokines and cytolytic molecules,[77] which is associated with successful immune responses in infection and cancer.[3,78-80] In chronic infections and after extended stimulations such as in ACT and in the cancer microenvironment, CD8+ T cells will continue to differentiate and become “exhausted” or less potent.[3,5,81,82] We observed a higher percentage of CD8+ T cells that are copositive for all functionality markers—IFN-γ, TNF-α, IL-2, and CD107a—on the HA hydrogel, when compared to the TCP condition (Figure 3H). Thus, the HA hydrogel ECM environment enhances early cell proliferation, memory cell induction, and functional T cell generation.
To show that aTM is also capable of stimulating human CD8+ T cells, we attached antihuman CD3 (Signal 1) and antihuman CD28 (Signal 2) to the HA hydrogels. We observed a similar increase in the fold expansion when increasing the density of the Signal 1 and 2 to 4 μg mL−1 (≈25-fold CD8+ T cell expansion in 1 week) on 0.5 kPa aTM, but beyond this value, the fold proliferation of the cells dramatically decreased where little to no expansion was detected in the 25 μg mL−1 condition (Figure 4A), with minimal CFSE dilution (Figure S18, Supporting Information). Nevertheless, phenotypic studies revealed that the cells are still proliferating at this dose, albeit at lower frequency (Figure 4B). Interestingly, this indicates control over phenotype independent of cell proliferation. Additionally, we verified the interaction of the T cells with the aTM across stimulatory signal densities through light video microscopy. We observed a higher fraction of cells bound to the aTM matrix over the first hour of attachment (Figure S19, Supporting Information), similar to what we have observed with murine T cells (Figure S10, Supporting Information). After 3 d of stimulation, we also noted macroscopically more punctate, smaller cell clusters in the 25 μg mL−1 aTMs than in the 4 μg mL−1 condition, potentially indicating the antibody density may prevent beneficial multi-cellular interactions from forming due to an inhibition of migration necessary for expansion (Figure S20, Supporting Information).
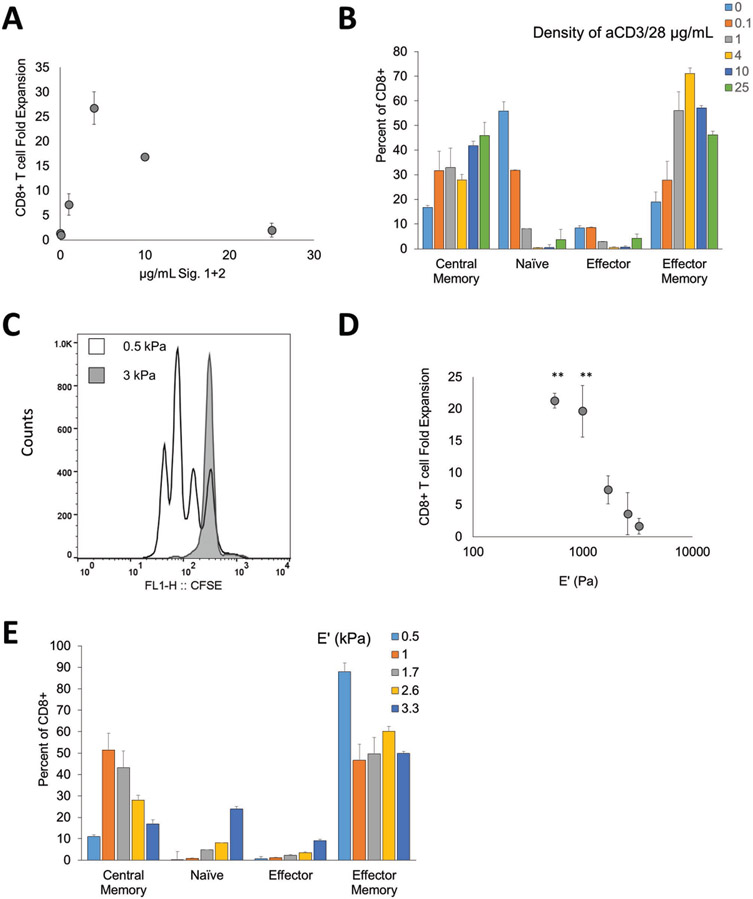
Figure 4.
Artificial T cell stimulating matrix hydrogels provide effective stimulation to human CD8+ T cells. A) CD8+ T cell fold expansion measured after 7 d of stimulation by aTM with Signals 1 + 2 (anti-CD3 and anti-CD28) conjugated at varying amounts, n = 3 independent donors. B) Phenotype of CD8+ T cells after culture on aTM surfaces of varying Signals 1 + 2 amounts defined by CD45RA and CD62L (error bars show s.e.m). C) CFSE proliferation dye dilution measured after 3 d of stimulation of CD8+ T cells comparing a stiff (3 kPa) and soft (0.5 kPa) aTM, n = 3 independent donors. D) CD8+ T cell fold expansion measured after 7 d of stimulation on aTMs with varying stiffness (error bars show s.e.m.; **p < 0.01, n = 3 independent donors, one-way ANOVA with Dunnett’s post-test comparing to 3 kPa condition). E) Phenotype of CD8+ T cells after culture on aTM surfaces of varying stiffness defined by CD45RA and CD62L (error bars show s.e.m; n = 3 independent donors).
Similar matrix stiffness-dependent effects were observed where more effective stimulation (>20 fold expansion in 1 week) is observed on aTM hydrogels with an elastic modulus less than 1 kPa (Figure 4C,D; Figures S21 and 22, Supporting Information). By changing the stiffness of the aTM, we observed differences in phenotype even within conditions that have similar fold expansions (Figure 4E). For example, the 0.5 and 1 kPa aTMs both provided nearly 20-fold expansion, but the 1 kPa aTM generated a more balanced ratio of central memory to effector memory CD8+ T cells than the 0.5 kPa aTM. In summary, this demonstrates that we can create an aTM that stimulates and polarizes human CD8+ T cells for potential ACT therapy.
A main goal of ACT is to be able expand rare (frequency of 1 in 105 to 106 CD8+ T cells), antigen-specific CD8+ T cells to high numbers that are functional. Because of the difficulty in obtaining and activating these cells, most studies investigate the antigen-specific activation of T cells from transgenic mice or the nonspecific activation of endogenous T cells. This limits clinical relevance because of lack of translatability and the monoclonality of these T cells. In contrast, here we investigated an optimized version of the aTM in the setting of activating rare antigen-specific CD8+ T cells. Instead of using nonspecific Signal 1 (anti-CD3), we conjugate antigen-specific Signal 1 (pMHC: Kb-SIY) to aTM with costimulatory anti-CD28, and compared stimulating conditions similar to those used in Figure 3 with T cells mixed with aAPC cultured on HA hydrogel or TCP surface.
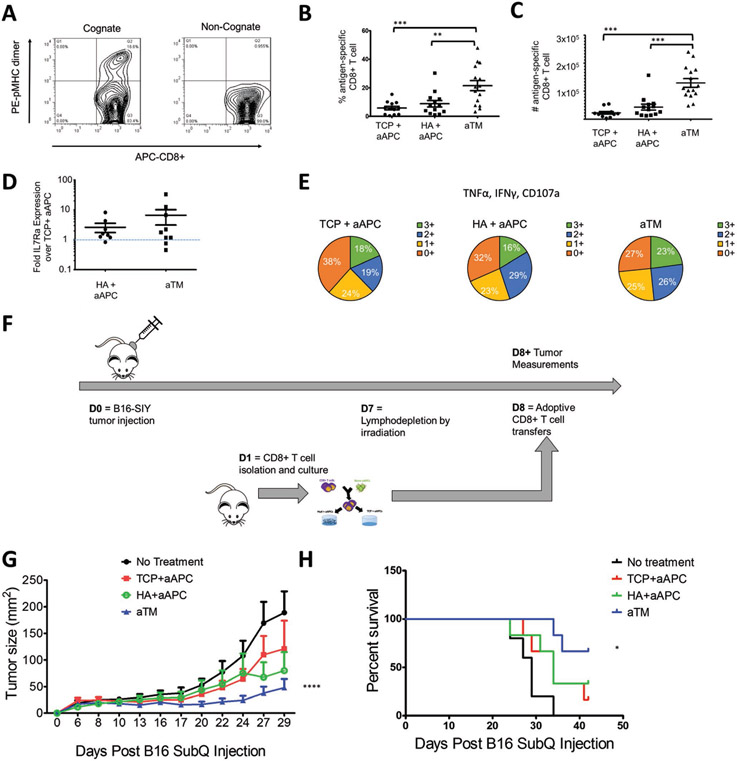
After 7 d of stimulation, we determined the antigen-specificity of the cultures and found that an average of 22% of the CD8+ T cells were antigen-specific from aTM cultures (Figure 5A). Indeed, we observed more than double the percentage of antigen-specific cells and more than quadruple the total number of antigen-specific cells expanded on aTM (Figure 5B,C). This highlights the importance of studying endogenous T cell activation where now differences are much larger between groups than when studying with transgenic cells or in nonspecific activation. Such a drastic increase in cell number, even between the aTM and HA + aAPC groups where the only difference was the location of the stimulatory signals, is quite surprising. Therefore, the combination of both engaging the TCRs and CD28 from stimulatory signals conjugated to the soft hydrogel and direct interaction with the hyaluronic acid ECM hydrogel surface represents important progress in activating antigen-specific T cells effectively.
Figure 5.
aTM stimulates a greater number and percent of functional antigen-specific CD8+ T cells that provide more effective tumor treatment. A,B) Percentage of antigen-specific T cells after 7 d of stimulation is determined by staining with cognate and noncognate antigen-loaded peptide major histocompatibility complex (pMHC) and anti-CD8a. B,C) Percentages (B) and numbers (C) of antigen-specific CD8+ T cells stimulated by aTM, or by aAPC on either TCP or HA hydrogel surface (error bars show s.e.m.; **p < 0.01, ***p < 0.001, n = 12–15, one-way ANOVA with Tukey’s post-test). D) Fold IL7Ra expression on antigen-specific CD8+ T cells from HA+ aAPC and aTM compared to IL7Ra expression on antigen-specific CD8+ T cells from TCP + aAPC (error bars show s.e.m., n = 8–9). E) T cell functionality was measured by the number of functional molecules co-expressed by each antigen-specific cell (IFN-γ, TNFα, CD107a) after 7 d of stimulation (n = 5–7). F) Murine melanoma therapeutic in vivo model for adoptively transferred cells. G) Tumor size measurements indicate that adoptive T cells from aTM stimulation significantly delayed tumor growth. Significance measured by two-way ANOVA with Bonferroni post-test (p < 0.0001) and H) significantly extended survival. Significance measured by log-rank test (p = 0.05, n = 5–6).
Since we had observed differences in the IL7Ra expression and functionality of the CD8+ T cells cultured on the HA hydrogel, we also probed antigen-specific cells after 7 d of stimulation for these markers. We again observed an increase in both the IL7Ra (Figure 5D) and the functionality associated with an increase in the percent of SIY+ T cells that were positive for multiple cytokines and degranulation markers (Figure 5E; Figure S23, Supporting Information), and was confirmed by an in vitro killing assay (Figure 24, Supporting Information). This is significant because it shows that aTM is capable of generating higher numbers of functional antigen-specific CD8+ T cells. We also confirmed that these findings were consistent with CD8+ T cells isolated from mice with established tumors (Figure S25, Supporting Information), and that we could expand human antigen-specific T cells with aTM specific for CMV+ CD8+ human T cells (Figure 26, Supporting Information).
Finally, we tested the in vivo activity of aTM-stimulated and expanded T cells in an ACT model where T cells were transferred into mice with established B16-SIY melanoma tumors (Figure 5F). T cells stimulated by aTM significantly reduced tumor growth as compared to T cells on other surfaces and no treatment controls. Even on day 29, tumors in the group receiving aTM-stimulated T cells were stable below 50 mm2 (Figure 5G; Figure S27, Supporting Information). This treatment also resulted in improved survival rate. By day 40, none among the no treatment group, 16% of TCP + aAPC treated mice, 33% of HA+ aAPC treated group survived, in contrast, 66% of mice survived after receiving aTM-stimulated T cells at the same dose (Figure 5H). Furthermore, there was no significant difference in the percentage of transferred cells 21 d after transfer (Figure S28, Supporting Information). Thus, T cells stimulated on the aTM had significantly increased efficacy compared to those stimulated with traditional methods. In conclusion, the aTM offers a unique combination of an ECM cue and stiffness-mediated mechanical signaling through the TCR. It was only when these two parameters were combined and optimized which resulted in effective antigen-specific expansion, phenotype skewing, and effective control of an established, aggressive, immunosuppressive murine melanoma.
Herein we have engineered an aTM. By considering the native T cell biology we designed the aTM through mimicking critical features of both the natural ECM and the antigen presenting cells. We conjugated T-cell stimulating molecules to develop the first ECM-based T cell activation biomaterial. The density of the signal attached and stiffness were key biophysical parameters engineered that influenced the ability for both murine and human CD8+ T cells to interact and be stimulated by the aTM. Additionally, we utilized an HA as an ECM molecule and found that it provides an additional signaling component influencing both the activation and polarization of T cells. By fine-tuning these biophysical properties, the aTM produced four times as many functional, therapeutic antigen-specific CD8+ T cells than current stimulation materials—resulting in more effective tumor inhibition. Additional work will need to be done to continue to study the underlying signaling implications for why biophysical properties of the aTM confer improved T cell activation. This has implications for adoptive T cell and CAR T cell therapies, where large numbers of high quality antigen-specific T cells are needed.[83-85] Engineering the environment with ECM modulation represents a new approach to control T cell activation, where previously researchers have focused on cytokine cocktails, and generating artificial cells using particles or scaffolds instead of environmental cues.[12,14,16] Finally, besides creating an ex vivo environment for T cell activation, the aTM has the potential to be applied for direct T cell activation in vivo, thus eliminating the need for ex vivo T cell manipulation.[86]
Experimental Section
Mice:
B6, 2C, and PMEL transgenic mice were maintained per guidelines approved by the Johns Hopkins University’s Institutional Review Board.
Human Subjects:
For human studies, the ethical committee of the Johns Hopkins University approved this study, and all healthy volunteers gave written informed consent (Human IRB protocol number: NA_00027947).
Reagents:
Soluble MHC-Ig dimers loaded with peptides (pMHC-Ig) and artificial antigen presenting cells (aAPC) were produced in-house as described.[12,19]
Hydrogel Preparation:
Thiol-modified HA (ESI BIO, Alameda, CA, USA) was resuspended with 1 mL sterile dH2O and incubated at 37 °C for 30 min until completely dissolved to form 1% HA solution in 1 × phosphate-buffered saline (PBS). To form hydrogels, HA was plated immediately after getting mixed with PEGDA with a molecular weight of 3400 (Laysan Bio, Arab, AL, USA) cross-linker at a 4:1 volume ratio to fully cover the well. Plated hydrogels were incubated for a minimum of 1 h prior to cell culture.
Preparation of aTM:
HA solution was prepared as previously described. Anti-CD3 and anti-CD28 antibodies were purchased respectively from BioXCell (145-2C11; West Lebanon, NH, USA) and BioLegend (37.51; San Diego, CA, USA). Antibody and MHC-Ig dimers were partially reduced with 100 × 10−3 m dithiothreitol (DTT) for 30 min at room temperature to expose free thiol groups and thoroughly washed through a centrifugation filtration with a 50 kDa MWCO filter. PEGDA cross-linker was added to reduced MHC-Ig dimers, anti-CD3 and anti-CD28 antibody solutions to a final concentration of 0.5% PEGDA in preparation for cross-linking of HA and conjugation of the signaling cues. This results in at least a 200-fold excess of PEGDA to antibody ratio, which prevents further thiol oxidation and ensures long-term storage of antibody-PEGDA conjugates and that antibody can be attached effectively to the thiolated HA hydrogel. Prior to hydrogel formation, MHC-Ig dimers or anti-CD3 antibodies and anti-CD28 co-stimulatory signals with 0.5% PEGDA were added to the HA solution to directly attach signals on HA through thiol-diene chemistry. The HA-antibody solution was then mixed with PEGDA cross-linker at a 4:1 ratio to be plated. The aTM was allowed to form within flat-bottomed tissue culture plates to form a complete layer for at least 1 h prior to washing three times with 1 × PBS to remove any unbound stimulatory signal and cells were subsequently plated. To investigate the effects of ECM protein attachment, cyclic RGD (sequence: CCRRGDWLC), which was synthesized by solid phase methods as described previously,[87] or laminin (ThermoFisher) was added to make an HA solution with protein concentration of respectively 100 × 10−6 m and 20 μg mL−1, prior to the hydrogel formation. To investigate the stiffness effects of aTMs, the PEGDA cross-linker concentration was changed from a final concentration of 0.05 wt/vol% to 2 wt/vol%.
Characterization of aTM:
To evaluate the mechanical stiffness of aTMs, elastic moduli of hydrogels were measured using Ares G2 oscillatory shear rheometer. First, HA solution was mixed with varying PEGDA cross-linker concentrations to a final volume of 200 μL and placed immediately on the stationary lower plate of the rheometer. The shear storage modulus, G′, and the sheer loss modulus, G″, were recorded during in situ hydrogel formation over 1 h at 37 °C. The elastic modulus, E′, was calculated by E′ = 2G′(1+γ) where γ is the Poisson’s ratio. For HA hydrogels, γ was assumed to be 0.5 because the Poisson’s ratio of incompressible materials was ≈0.5 and the hydrogels were used under low strain conditions.[47]
For SEM, hydrogels were cross-linked overnight at 37 °C and subsequently flash frozen in liquid nitrogen for 5 min. Samples were placed in the FreeZone 4.5 Benchtop (LABCONCO) freeze dry system for 72 h for complete dehydration of samples. Samples were coated using a Desk III (Denton Vacuum) Au/Pd sputter coater for 2 min at 25 mA before imaging in LEO 435 VP SEM.
To evaluate the conjugation to the surface of the hydrogel, Both HA hydrogels with no antibodies attached and HA hydrogels with 10 μg mL−1 anti-CD3, anti-CD28 were stained in black 96-well half area wells. Clone G192-1 was stained with FITC-anti-Armenian and Syrian Hamster IgG and clone G94-56 (BD Pharminogen) was stained with FITC-anti-Armenian and Syrian Hamster IgG1 for 5 min at 4 °C. Then the surfaces of the hydrogels were washed time times with PBS and then the fluorescence per well was read on a fluorescent plate reader and compared to a standard curve of the fluorescent antibodies titrated down the plate. To estimate the density of the ligand on the surface of the aTM, the thickness observed by cells was assumed to be 1 μm, and then the density was calculated based off the total mass of Signal 1 and 2 within this slab and then dividing by the surface area.
CD8+ T Lymphocyte Isolation:
Murine cells were obtained from adult mouse lymph nodes and spleens. Obtained cells were treated with ACK lysing buffer to lyse red blood cells and filtered through cell strainers to isolate splenocytes. PBMCs from healthy donors were isolated by Ficoll-Paque PLUS gradient centrifugation (GE Healthcare). CD8+ T lymphocytes were then isolated from splenocytes or PBMCs by negative selection using CD8+ isolation kits and magnetic columns from Miltenyi Biotech (Auburn, CA, USA) according to the manufacturer’s protocol. PBMCs were obtained from blood drawn from healthy males and females per JHU IRB approved protocols.
Ex Vivo T Cell Culture and Activation:
For ex vivo T cell expansion, isolated CD8+ T cells were cultured in the T cell culture media (RPMI supplemented with L-glutamine, nonessential amino acids, vitamin solution, sodium pyruvate, β-mercaptoethanol, 10% fetal bovine serum, ciproflaxin, and a cocktail of T cell growth factors as described previously[88]). In the case of human T cell expansion, 10% AB serum was used instead of 10% fetal bovine serum. On day 3 or 4 of culture, cells were fed with half the volume of the initial T cell culture media with twice the concentration of T cell growth factor cocktail.
For activation with aAPC, T cells were cocultured with a concentration of 75 × 10−12 m bound pMHC-Ig on the aAPC and then plated on respective surfaces. For stimulation on the aTM, cells were plated on the surfaces aTM with concentrations of the stimulatory antibody (either anti-CD3 and anti-CD28 or pMHC-Ig and anti-CD28) conjugated to the HA hydrogel.
T Cell Proliferation Assay:
CD8+ T cells were isolated as previous described and resuspended in 1 mL T cell culture media. Cells were mixed with 1 μL CellTrace carboxyfluorescein succinimidyl ester (CFSE) dye (ThermoFisher) in 1 mL T cell culture media per 3 million cells and incubated at 37 °C for 20 min. CFSE stained cells were washed with 50 mL T cell culture media to remove unstained dye and plated. On day 3 of culture, cells were harvested and stained with a 1:100 PBS solution of APC-conjugated rat antimouse CD8a, clone 53–6.7 (BD Pharmingen) for 15 min at 4 °C. The CFSE fluorescence intensity was measured using BD FACSCalibur flow cytometer. Cell proliferation was analyzed using FlowJo with diluted CFSE fluorescence peaks signifying population after each round of cell division. A subset of the cells were allowed to expand for 7 d and viable cells were counted with a hemocytometer to determine fold expansion. Images of cell cultures were taken with an Olympus IX71 inverted light microscope at a 4 × magnification on day 3 of cultures.
Video Cell Microscopy:
Imaging began immediately after seeding cell onto hydrogel surface in a 24-well plate. Epiflourecent images taken every 2 min for 1 h at four locations in each condition. (Zeiss Axio Observer Z1 with LD Plan-Neofluar 203/0.4 Korr Ph2 and AxioCamHR3 camera). Incubation was maintained at 37 °C and 5% CO2 throughout the experiment. Cells were tracked using the “Spots” function in Imaris 9 (Bitplane). The faction of cells bound to the hydrogel surface over the 1 h period was computed at each location. Cells moving less than 1.4 μm min−1 were considered to be bound.
Time Course Experiments:
Two million purified CD8+ T cells from either PMEL or 2C mice were cultured on HA or TCP conditions. The cells were collected at designated time points. These cells were frozen down in liquid nitrogen for western blots, stored in TRIzol for mRNA detection, or PFA fixed for phospho-flow. For drugs, the final concentration of rapamycin (mTORC1 inhibitor), U-0126 (Erk1/2 inhibitor), blebbistatin, and anti-CD44 (KM201) were 0.1 × 10−6, 10 × 10−6, 100 × 10−6 m, and 5 μg mL−1, respectively. Half-volumes of T cell culture media is added every other day to keep cells in good condition. For western blot and rt-PCR, live cells that have been cultured for more than 24 h were first purified using Ficoll-Paque followed by the procedure mentioned above.
Western Blot:
Frozen cells were lysed in a radioimmunoprecipitation assay (RIPA) buffer-based mixture containing proteinase inhibitor, PMSF (phenylmethane sulfonyl fluoride), sodium pyrophosphate, sodium fluoride, sodium orthovanadate, and β-glycerophosphate to inhibit phosphatases. Then, protein samples underwent standard western blot procedure with 1–2 h of incubation in 5% milk, overnight incubation in primary antibodies (in 4% BSA), and 1 h incubation in secondary antibodies. Films were imaged in a UVP BioSpectrum Imaging System, analyzed in UVP VisionWorks and quantified in ImageJ. Antibodies used include:
| Target | Cat# | Vendor |
|---|---|---|
| p-S6 (S240/244) | 2215 | Cell Signaling |
| p-Erk (Y202/204) | 4695 | Cell Signaling |
| Beta-actin | 4970 | Cell Signaling |
| p-S6K1 (T389) | 9234 | Cell Signaling |
| p-AKT (S473) | 3787 | Cell Signaling |
| Total Lck | 2752 | Cell Signaling |
| p-Lck (Y505) | 2751 | Cell Signaling |
| p-Src (Y416) | 2101 | Cell Signaling |
RT-PCR:
Cells were kept in TRIzol in −80 °C for storage. mRNA was purified using Zymo Quick-RNA MiniPrep Kit. Then, reaction mix was prepared based on standard RT-PCR protocol. Probes were from TaqMan FAM/MGB probes with VIC/TAMRA Eukaryotic 18S rRNA as an endogenous control (ThermoFisher). Samples were run in quintuplicate in Applied Biosystems StepOnePlus Real-Time PCR system and analyzed using Excel.
Phosphorylation Flow Cytometry:
Cells were first stained with Live/Dead stain and then were fixed using BD Phosflow Fix Buffer I at room temperature for 10 min. After washing, cells were permeabilized using ice cold BD Phosflow Perm Buffer II for 30 min on ice. Samples were then stained with a solution of FACS wash buffer with 1:50 PE conjugated rat antimouse CD8a, clone 53 6.7 (BD Pharmingen), and a 1:100 Rabbit anti-Phospho S6 Ribosomal Protein (Ser235/236), clone D57.2.2E, or Rabbit IgG Isotype Control, clone DA1E (Cell Signaling Technology, Danvers, Massachusetts) for 45 min at room temperature. Samples were then washed with FACS wash buffer and then stained with a solution of FACS wash buffer with 1:250 of Alexa Fluor 647-conjugated Goat S22 anti-Rabbit IgG, polyclonal (ThermoFisher) for 45 min at room temperature. Samples were washed and resuspended with FACS wash buffer and read on a BD FACSCalibur.
T Cell Phenotype Assay:
On day 7 of culture, the numbers of cells were counted using hemocytometer. After counting, less than 500 000 cells were collected and stained with a 1:100 PBS solution of APC-conjugated rat antimouse CD8a, clone 53–6.7 (BD Pharmingen), PE-conjugated rat antimouse CD62L, clone MEL-14 (BD Pharmingen), PerCP-conjugated rat antimouse CD44, clone IM7 (Biolegend), and 1:1000 of LIVE/DEAD Fixable Green Dead Cell Stain (ThermoFisher) for 15 min at 4 °C. Cells were then washed with FACS wash buffer to be read on BD FACSCalibur flow cytometer and analyzed using FlowJo to measure the population of naïve T cells (CD62L+CD44−), effector T cells (CD62L−CD44+), and memory T cells (CD62L+CD44+). For human phenotype experiments, the same protocol was used except, the cells were instead stained with a 1:100 PBS solution of APC-conjugated antihuman CD45RA, Clone HI100 (Biolegend), PE-conjugated antihuman CD62L, clone DREG-56 (Biolegend), PerCP-conjugated antihuman CD8a, clone SK-1 (Biolegend), and 1:1000 of LIVE/DEAD Fixable Green Dead Cell Stain for 15 min at 4 °C. Cells were then washed with FACS wash buffer to be read on BD FACSCalibur flow cytometer and analyzed using FlowJo to measure the population of naïve T cells (CD62L−CD45RA−), effector T cells (CD62L−CD45RA+), central memory T cells (CD62L+CD45RA−), and effector memory T cells (CD62L−CD45RA−).
For analysis of IL-7Ra and IL-15Ra expression on cells, on day 7 of culture, the numbers of cells were counted using hemocytometer. After counting, samples were divided into four tubes (less than 500 000 cells per tube) and stained with a 1:100 PBS solution of APC-conjugated rat antimouse CD8a, clone 53–6.7 (BD Pharmingen), 1:1000 of LIVE/DEAD Fixable Green Dead Cell Stain (ThermoFisher), and either PE-conjugated rat antimouse IL7Ra, clone A7R34 (Biolegend), or isotype control PE-conjugated Rat IgG2a, κ Isotype Ctrl, clone RTK2758 (Biolegened), or PE-conjugated rat antimouse IL15Ra, clone DNT15Ra (eBioscience), or isotype control PE-conjugated Rat IgG1, κ Isotype Ctrl, clone eBRG1 (eBioscience), for 15 min at 4 °C. Cells were then washed with FACS wash buffer to be read on BD FACSCalibur flow cytometer and analyzed using FlowJo.
T Cell Cytokine Functionality Assay:
On day 7 of culture, ≈500 000 CD8+ T cells were isolated from each condition and separated into restimulation and no-stimulation groups in 100 μL T cell culture media. To inhibit protein transport, 10 μL solution of 1:50 FITC anti-CD107a, 1:350 BD GolgiStop Protein Transport Inhibitor (BD Biosciences), and 1:350 BD GolgiPlug Protein Transport Inhibitor (BD Biosciences) in PBS was added to the samples. For the restimulation group, microparticle Dyanl-based aAPC were added at a 1:1 ratio. Both groups were incubated at 37 °C for 6 h. After incubation, cells were washed and stained with 1:100 PBS solution of PerCP-conjugated antimouse CD8a, clone 53–6.7 (Biolegend) and 1:1000 of LIVE/DEAD AmCyan Fixable Aqua Dead Cell Stain (ThermoFisher) at 4 °C for 30 min. Cells were then fixed and permeabilized with 100 μL BD Cytofix/Cytoperm Fixation and Permeabilization Solution (BD Biosciences) overnight. To analyze intracellular cytokines, cells were washed with 1× BD PERM/Wash buffer with 2% BSA the following day and stained with 1:100 solution of PE-conjugated rat antimouse IFN-γ, clone XMG1.2 (BD Pharmingen), APC-conjugated rat antimouse IL2, clone JES6-5H4 (BD Pharmingen), and PE-Cy7-conjugated rat antimouse TNFα, clone MP6-XT22 (Biolegend) in PERM/Wash buffer with 2% BSA at 4 °C for 1 h. Stained cells were read on BD LSR II flow cytometer and analyzed by subtracting cytokine positive cells in the nostimulation group from the restimulation group using FlowJo.
For antigen-specific cells a similar assay was used with the following modifications. Instead of a restimulation, cells were simply stained with 1 μg of either cognate or noncognate biotinylated pMHC-Ig dimer for 1 h at 4 °C. After washing, samples were stained with a 1:350 ratio of PE-labeled streptavidin (BD Pharmingen). Then 10 μL solution of 1:50 FITC anti-CD107a, 1:350 BD GolgiStop Protein Transport Inhibitor (BD Biosciences), and 1:350 BD GolgiPlug Protein Transport Inhibitor (BD Biosciences) in PBS was added to the samples and incubated for 37 °C for 6 h. Cells were then washed and stained with 1:100 PBS solution of PerCP-conjugated antimouse CD8a, clone 53–6.7 (Biolegend) and 1:1000 of LIVE/DEAD AmCyan Fixable Aqua Dead Cell Stain (ThermoFisher) at 4 °C for 30 min. Cells were then fixed and permeabilized with 100 μL BD Cytofix/Cytoperm Fixation and Permeabilization Solution (BD Biosciences) overnight. Cells were then washed with 1× BD PERM/Wash buffer with 2% BSA and stained with 1:100 solution of APC-conjugated rat antimouse IFN-γ, clone XMG1.2 (BD Pharmingen) and PE-Cy7-conjugated rat antimouse TNFα, clone MP6-XT22 (Biolegend) in PERM/Wash buffer with 2% BSA at 4 °C for 1 h. Stained cells were read on BD LSR II flow cytometer.
In Situ Staining and Super-Resolution Microscopy:
CD8+ T cells were added to the surface of the aTM hydrogels and allowed to culture at 37 °C for 1 h. Gels were fixed with 4% PFA for 20 min at room temperature, followed by permeabilization, and staining with Alexafluor phalloidin-564 and CD3 (Novus Biologicals). The secondary antibody used was Alexafluor-488. Gels were mounted on coverslips and imaged using the Zeiss 800 confocal microscope equipped with an AiryScan detector. Airyscan super-resolution images were processed using Zen software. Quantification of CD3 spot area was performed in FIJI/ImageJ using the analyze particles function.
Expansion of Rare Antigen-Specific T cells:
B6 CD8+ T cells were stimulated on aTM surfaces as described previously for 7 d. To detect antigen-specific CD8+ T cells, cells were stained with 1 μg of either cognate or noncognate biotinylated pMHC-Ig dimer, with a 1:100 ratio of APC-conjugated rat antimouse CD8a, clone 53–6.7 (BD Pharmingen) in FACS wash buffer for 1 h at 4 °C. Samples were washed and then stained with a 1:350 ratio of PE-labeled streptavidin (BD Pharmingen) and a 1:1000 ratio of LIVE/DEAD Fixable Green Dead Cell Stain (ThermoFisher) in PBS for 15 min at 4 °C. Cells were then washed and read on a BD FACSCalibur. Percent antigen-specific cells were calculated by subtracting the percent gated in cognate stained CD8+ T cells from noncognate stained CD8+ T cells. Number of antigen-specific cells was determined from multiplying the percent of antigen-specific cells by the number counted following cell harvest. Detection of antigen-specific human cells was done similarly, except instead of staining with biotynlated dimer, the antigen-specific cells were stained with purchased PE-labeled tetramer (MBL International, Woburn, MA) for 30 min at room temperature, then washed and stained with APC-conjugated antihuman CD8a, clone SK-1 (Biolegend), and 1:1000 of LIVE/DEAD Fixable Green Dead Cell Stain for 15 min at 4 °C.
For expansion of rare T cells from tumor-experienced mice, mice were injected with 2 × 106 B16-SIY melanoma tumor cells expressing the SIY antigen and tumors were allowed to grow until on average were around 100 mm2. CD8+ T cells were then harvested from the lymph nodes and spleens as previously described and expansion and detection were performed as previously described.
For analysis of IL7Ra of antigen-specific T cells, a similar process was used. Cells were stained with 1 μg of either cognate or noncognate biotinylated pMHC-Ig dimer, with a 1:100 ratio of PerCP-conjugated rat antimouse CD8a, clone 53–6.7 (BD Pharmingen) in FACS wash buffer for 1 h at 4 °C. Samples were washed and then stained with a 1:350 ratio of PE-labeled streptavidin (BD Pharmingen), either APC-conjugated rat antimouse IL7Ra, clone A7R34 (Biolegend) or isotype control APC-conjugated Rat IgG2a, κ Isotype Ctrl, clone RTK2758 (Biolegend), and a 1:1000 ratio of LIVE/DEAD Fixable Green Dead Cell Stain (ThermoFisher) in PBS for 15 min at 4 °C.
In Vitro Killing Assay:
Target cells were harvested from splenocytes of B6 mice. 20 × 106 splenocytes were labeled with a high concentration of CFSE (5 × 10−6 m) and another was labeled with a low concentration of CFSE (0.05 × 10−6 m) in 1 mL of PBS at 37 °C for 10 min (Invitrogen, Eugene, OR). Media with fetal bovine serum (FBS) was added to quench the reaction and allowed to incubate at 37 °C for another 5 min and then washed with media. The CFSE-high cells were then incubated at 37 °C for 1 h with 1 × 10−6 m SIY peptide in media without serum. The cells were then washed and mixed at a 1:1 ratio with control nontarget, CFSE-low splenocytes. This mixed population was added to CD8+ T cells which had been stimulated for 7 d at a 1:1 ratio and allowed to incubate for 18 h at 37 °C in a cell incubator. Then cells were washed, stained with a 1:1000 of LIVE/DEAD AmCyan Fixable Aqua Dead Cell Stain (ThermoFisher) at 4 °C for 30 min. The cells were washed and read on a BD LSRII flow cytometer. The percent killing was calculated as follows: % of in vivo killing = 100 – ([(% specific peptide pulsed cells in treatment/% unspecific B6 cells in treatment)/(% specific peptide pulsed in no treatment controls/% unspecific B6 cells in no treatment controls)] × 100).
Therapeutic Adoptive Transfer of T Cells:
On day 0, B6 mice were injected with 2 × 106 B16-SIY melanoma tumor cells expressing the SIY antigen. On day 1, CD8+ T cells were isolated from wild-type B6 mice and cultured for 7 d to produce stimulated T cells for adoptive transfer. On day 7, mice were given a central dose of 500 cGy, which induces transient lymphopenia similar standard approaches within adoptive immunotherapy.[89] On day 8, T cells cultured ex vivo were harvested and adoptively transferred intravenously in volumes of 100 μL. For every 3 mice receiving treatment, 1 B6 spleen was used for CD8+ T cell isolation and stimulation. This resulted in each mouse receiving 500 000 stimulated CD8+ T cells. Tumor sizes were measured using calipers and multiplying the longest measured length by the perpendicular direction of the tumor. Mice were sacrificed once tumors grew larger than 200 mm2. For studies involving persistence of cells, Thy1.1+ B6 donor mice were used. On day 21 blood, spleen, and lymph nodes were harvested from recipient mice and stained with a 1:100 ratio of APC-conjugated rat antimouse CD8a, clone 53–6.7 (BD Pharmingen) and 1:100 ratio of Alexa Fluor 488-conjugated mouse antimouse CD90.1 (Thy1.1), clone OX-7 (Biolegend) in FACS wash buffer for 15 min at 4 °C.
Supplementary Material
Acknowledgements
J.W.H. thanks the NIH Cancer Nanotechnology Training Center at the Johns Hopkins Institute for NanoBioTechnology, the National Science Foundation Graduate Research Fellowship (DGE-1232825), and the ARCS foundation for fellowship support. This work was partially supported by a Johns Hopkins University Discovery Award from the Office of the Vice Provost for Research and funded by support from the National Institutes of Health (5R21CA185819-02). H.C.P. and S.G. acknowledge the IIC core: NIH SIG award #1s10OD020152-01A1 and the T32: 5T32CA153952-08. A.K.F. acknowledges NIGMS 3T32GM007309 and A.J.E. acknowledges NCI U54 CA2101732. J.W.H., Y.D., J.W.C., H.Q.M., and J.P.S. designed the studies and wrote the manuscript. J.W.H., Y.D., J.W.C, and S.S. performed the experiments. X.L. and C.C. assisted with HA hydrogel preparation and physical property characterizations. H.C.P., J.W.H., and S.G. performed and analyzed super-resolution microscopy experiments. A.K.F., J.W.H., and A.J.E. performed and analyzed light video microscopy experiments. C.B. helped with in vivo experiments. J.W.H., Y.D., J.W.C, S.S., H.Q.M., and J.P.S. analyzed the data.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
John W. Hickey, Department of Biomedical Engineering, School of Medicine, Baltimore, MD 21218, USA, Institute for Cell Engineering, School of Medicine, Baltimore, MD 21205, USA, Department of Pathology, School of Medicine, Baltimore, MD 21287, USA, Translational Tissue Engineering Center, Baltimore, MD 21287, USA, Institute for NanoBioTechnology, Baltimore, MD 21218, USA
Yi Dong, Graduate Program in Immunology, School of Medicine, Baltimore, MD 21205, USA.
Jae Wook Chung, Department of Chemical and Biomolecular Engineering, Whiting School of Engineering, Baltimore, MD 21218, USA.
Sebastian F. Salathe, Department of Biology, Krieger School of Arts and Sciences, Baltimore, MD 21218, USA
Hawley C. Pruitt, Institute for NanoBioTechnology, Baltimore, MD 21218, USA, Department of Chemical and Biomolecular Engineering, Whiting School of Engineering, Baltimore, MD 21218, USA
Xiaowei Li, Translational Tissue Engineering Center, Baltimore, MD 21287, USA, Department of Materials Science and Engineering, Whiting School of Engineering, Baltimore, MD 21218, USA.
Calvin Chang, Department of Biomedical Engineering, School of Medicine, Baltimore, MD 21218, USA, Translational Tissue Engineering Center, Baltimore, MD 21287, USA.
Andrew K. Fraser, Department of Biomedical Engineering, School of Medicine, Baltimore, MD 21218, USA, Department of Cell Biology and Center for Cell Dynamics, School of Medicine, Baltimore, MD 21205, USA
Catherine A. Bessell, Graduate Program in Immunology, School of Medicine, Baltimore, MD 21205, USA
Andrew J. Ewald, Department of Biomedical Engineering, School of Medicine, Baltimore, MD 21218, USA, Department of Cell Biology and Center for Cell Dynamics, School of Medicine, Baltimore, MD 21205, USA, Department of Oncology, School of Medicine, Baltimore, MD 21205, USA
Sharon Gerecht, Department of Biomedical Engineering, School of Medicine, Baltimore, MD 21218, USA, Institute for NanoBioTechnology, Baltimore, MD 21218, USA, Department of Chemical and Biomolecular Engineering, Whiting School of Engineering, Baltimore, MD 21218, USA, Physical Sciences-Oncology Center, Baltimore, MD 21218, USA.
Hai-Quan Mao, Translational Tissue Engineering Center, Baltimore, MD 21287, USA, Institute for NanoBioTechnology, Baltimore, MD 21218, USA.
Jonathan P. Schneck, Institute for Cell Engineering, School of Medicine, Baltimore, MD 21205, USA, Department of Pathology, School of Medicine, Baltimore, MD 21287, USA, Department of Medicine, School of Medicine, Johns Hopkins University, Baltimore, MD 21205, USA
References
- [1].Hickey JW, Kosmides AK, Schneck JP, Int. Rev. Cell Mol. Biol 2018, 341, 277. [DOI] [PubMed] [Google Scholar]
- [2].Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME, Nat. Rev Cancer 2008, 8, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, Rosenberg SA, Blood 2009, 114, 1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pollack SM, Jones RL, Farrar EA, Lai IP, Lee SM, Cao J,Pillarisetty VG, Hoch BL, Gullett A, Bleakley M, J. Immunother. Cancer 2014, 2, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wherry EJ, Nat. Immunol 2011, 12, 492. [DOI] [PubMed] [Google Scholar]
- [6].Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, Von Andrian UH, Ahmed R, Nat. Immunol 2003, 4, 225. [DOI] [PubMed] [Google Scholar]
- [7].Hinrichs CS, Borman Z. a, Gattinoni L, Yu Z, Burns WR, Klebanoff C. a, Johnson L. a, Kerkar SP, Yang S, Muranski P,Palmer DC, Scott CD, Morgan RA, Robbins PF, Rosenberg SA, Restifo NP, Huang J, Blood 2011, 117, 808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Della Bella S, Gennaro M, Vaccari M, Ferraris C, Nicola S, Riva A, Clerici M, Greco M, Villa ML, Br. J. Cancer 2003. , 89, 1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Satthaporn S, Robins A, Vassanasiri W, El-Sheemy M, Jibril JA, Clark D, Valerio D, Eremin O, Cancer Immunol. Immunother 2004, 53, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ye F, Yu Y, Hu Y, Lu W, Xie X, J. Exp. Clin. Cancer Res 2010, 29, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rosenberg SA, Restifo NP, Science 2015, 348, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hickey JW, Vicente FP, Howard GP, Mao H-Q, Schneck JP, Nano Lett. 2017, 17, 7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Oelke M, Maus MV, Didiano D, June CH, Mackensen A, Schneck JP, Nat. Med 2003, 9, 619. [DOI] [PubMed] [Google Scholar]
- [14].Fadel TR, Sharp FA, Vudattu N, Ragheb R, Garyu J, Kim D,Hong E, Li N, Haller GL, Pfefferle LD, Justesen S, Harold KC, Fahmy TM, Nat. Nanotechnol, 2014, 9, 639. [DOI] [PubMed] [Google Scholar]
- [15].Kosmides AKK, Meyer RAA, Hickey JWW, Aje K, Cheung KNN, Green JJJ, Schneck JPP, Biomaterials 2017, 118, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cheung AS, Zhang DKY, Koshy ST, Mooney DJ, Nat. Biotechnol 2018, 36, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC, Biophys. J 2012, 102, L5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].O’Connor RS, Hao X, Shen K, Bashour K, Akimova T, Hancock WW, Kam LC, Milone MC, J. Immunol 2012, 189, 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kosmides AK, Necochea K, Hickey JW, Schneck JP, Nano Lett. 2018, 18, 1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Perica K, Bieler JG, Schütz C, Varela JC, Douglass J, Skora A, Chiu YL, Oelke M, Kinzler K, Zhou S, ACS Nano 2015, 9, 6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hickey JW, Isser AY, Vicente FP, Warner SB, Mao H-Q, Schneck JP, Biomaterials 2018, 187, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Adams JC, Watt FM, Nature 1989, 340, 307. [DOI] [PubMed] [Google Scholar]
- [23].Watt FM, Curr. Opin. Cell Biol 1989, 1, 1107. [DOI] [PubMed] [Google Scholar]
- [24].Lynch MP, Stein JL, Stein GS, Lian JB, Exp. Cell Res 1995, 216, 35. [DOI] [PubMed] [Google Scholar]
- [25].Bissell MJ, Barcellos-hoff MH, J. Cell Sci 1987, 1987, 327. [DOI] [PubMed] [Google Scholar]
- [26].Gretz JE, Anderson AO, Shaw S, Immunol. Rev 1997, 156, 11. [DOI] [PubMed] [Google Scholar]
- [27].Ayroldi E, Cannarile L, Migliorati G, Bartoli A, Nicoletti I, Riccardi C, Blood 1995, 86, 2672. [PubMed] [Google Scholar]
- [28].Pozzi A, Wary KK, Giancotti FG, Gardner HA, J. Cell Biol 1998, 142, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Petrie HT, Nat. Rev. Immunol 2003, 3, 859. [DOI] [PubMed] [Google Scholar]
- [30].Engler AJ, Sen S, Sweeney HL, Discher DE, Cell 2006, 126, 677. [DOI] [PubMed] [Google Scholar]
- [31].Jaalouk DE, Lammerding J, Nat. Rev. Mol. Cell Biol 2009, 10, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Stuart MAC, Boehm H, Li B, Vogel V, Nat. Mater 2012, 11, 642. [DOI] [PubMed] [Google Scholar]
- [33].Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA, Cell Motil. Cytoskeleton 2005, 60, 24. [DOI] [PubMed] [Google Scholar]
- [34].Fu J, Wang Y-K, Yang MT, Desai RA, Yu X, Liu Z, Chen CS, Nat. Methods 2010, 7, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].DeLong SA, Moon JJ, West JL, Biomaterials 2005, 26, 3227. [DOI] [PubMed] [Google Scholar]
- [36].Lee S-H, Moon JJ, West JL, Biomaterials 2008, 29, 2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT,Weber FE, Fields GB, Hubbell JA, Proc. Natl. Acad. Sci. USA 2003, 100, 5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kloxin AM, Kasko AM, Salinas CN, Anseth KS, Science 2009, 324, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Föger N, Marhaba R, Zöller M, Eur. J. Immunol 2000, 30, 2888. [DOI] [PubMed] [Google Scholar]
- [40].Jackson DG, Immunol. Rev 2009, 230, 216. [DOI] [PubMed] [Google Scholar]
- [41].Toole BP, Nat. Rev. Cancer 2004, 4, 528. [DOI] [PubMed] [Google Scholar]
- [42].Entwistle J, Hall CL, Turley EA, J. Cell. Biochem 1996, 61, 569. [DOI] [PubMed] [Google Scholar]
- [43].Ponta H, Sherman L, Herrlich PA, Nat. Rev. Mol. Cell Biol 2003, 4, 33. [DOI] [PubMed] [Google Scholar]
- [44].Lei Y, Gojgini S, Lam J, Segura T, Biomaterials 2011, 32, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shu XZ, Ahmad S, Liu Y, Prestwich GD,J. Biomed. Mater. Res., Part A 2006, 79A, 902. [DOI] [PubMed] [Google Scholar]
- [46].Peattie RA, Nayate AP, Firpo MA, Shelby J, Fisher RJ,Prestwich GD, Biomaterials 2004, 25, 2789. [DOI] [PubMed] [Google Scholar]
- [47].Li X, Liu X, Cui L, Brunson C, Zhao W, Bhat NR, Zhang N, Wen X, FASEB J. 2013, 27, 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tan H, Ramirez CM, Miljkovic N, Li H, Rubin JP, Marra KG, Biomaterials 2009, 30, 6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Deeg J, Axmann M, Matic J, Liapis A, Depoil D, Afrose J,Curado S, Dustin ML, Spatz JP, Nano Lett. 2013, 13, 5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Matic J, Deeg J, Scheffold A, Goldstein I, Spatz JP, Nano Lett. 2013, 13, 5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Humphrey JD, Dufresne ER, Schwartz MA, Nat. Rev. Mol. Cell Biol 2014, 15, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hirsch S, Guo J, Reiter R, Papazoglou S, Kroencke T, Braun J, Sack I, Magn. Reson. Med 2014, 71, 267. [DOI] [PubMed] [Google Scholar]
- [53].Miyaji K, Furuse A, Nakajima J, Kohno T, Ohtsuka T, Yagyu K, Oka T, Omata S, Cancer 1997, 80, 1920. [DOI] [PubMed] [Google Scholar]
- [54].Kim ST, Takeuchi K, Sun Z-YJ, Touma M, Castro CE, Fahmy A, Lang MJ, Wagner G, Reinherz EL, J. Biol. Chem 2009, 284, 31028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li Y-C, Chen B-M, Wu P-C, Cheng T-L, Kao L-S, Tao M-H, Lieber A, Roffler SR, J. Immunol 2010, 184, 5959. [DOI] [PubMed] [Google Scholar]
- [56].Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang J-H, Wagner G, Reinherz EL, Proc. Natl. Acad. Sci. USA 2015, 112, 1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Huse M, Nat. Rev. Immunol 2017, 17, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liu B, Chen W, Evavold BD, Zhu C, Cell 2014, 157, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML, Nat. Immunol 2009, 10, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Massia SP, Hubbell JA, J. Cell Biol 1991, 114, 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Taylor MJ, Husain K, Gartner ZJ, Mayor S, Vale RD, Cell 2017, 169, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lee K-H, Holdorf AD, Dustin ML, Chan AC, Allen PM, Shaw AS, Science 2002, 295, 1539. [DOI] [PubMed] [Google Scholar]
- [63].Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ, Nat. Commun 2015, 6, 6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wülfing C, Sjaastad MD, Davis MM, Proc. Natl. Acad. Sci. USA 1998, 95, 6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Day AJ, Prestwich GD, J. Biol. Chem 2002, 277, 4585. [DOI] [PubMed] [Google Scholar]
- [66].Sanders ME, Makgoba MW, Sharrow SO, Stephany D, Springer TA, Young HA, Shaw S, J. Immunol 1988, 140, 1401. [PubMed] [Google Scholar]
- [67].Graham VA, Marzo AL, Tough DF, Eur. J. Immunol 2007, 37, 925. [DOI] [PubMed] [Google Scholar]
- [68].Budd RC, Cerottini J-C, MacDonald HR, J. Immunol 1987, 138, 1009. [PubMed] [Google Scholar]
- [69].Budd RC, Cerottini JC, Horvath C, Bron C, Pedrazzini T, Howe RC, MacDonald HR, J. Immunol 1987, 138, 3120. [PubMed] [Google Scholar]
- [70].Mendoza MC, Er EE, Blenis J, Trends Biochem. Sci 2011, 36, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lefebvre DC, Lai JCY, Maeshima N, Ford JL, Wong ASL, Cross JL, Johnson P, Mol. Immunol 2010, 47, 1882. [DOI] [PubMed] [Google Scholar]
- [72].Rossy J, Williamson DJ, Gaus K, Front. Immunol 2012, 3, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pollizzi KN, Patel CH, Sun I-H, Oh M-H, Waickman AT, Wen J, Delgoffe GM, Powell JD,J. Clin. Invest 2015, 125, 2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tan JT, Ernst B, Kieper WC, LeRoy E, Sprent J, Surh CD, J. Exp. Med 2002, 195, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Berger C, Jensen MC, Lansdorp PM, Gough M, Elliott C, Riddell SR, J. Clin. Invest 2008, 118, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Klebanof CA, Gattinoni L, Torabi-Parizi P, Kerstann K, Cardones AR, Finkelstein SE, Palmer DC, Antony PA, Hwang ST, Rosenberg SA, Proc. Natl. Acad. Sci. USA 2005, 102, 9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chiu Y-LL, Shan L, Huang H, Haupt C, Bessell C,Canaday DH, Zhang H, Ho YC, Powell JD, Oelke M, Margolick JB, Blankson JN, Griffin DE, Schneck JP, J. Clin. Invest 2014, 124, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Rodrigue-Gervais IG, Rigsby H, Jouan L, Sauvé D, Sékaly R-P,Willems B, Lamarre D, J. Immunol 2010, 184, 3134. [DOI] [PubMed] [Google Scholar]
- [79].Ciuffreda D, Comte D, Cavassini M, Giostra E, Bühler L, Perruchoud M, Heim MH, Battegay M, Genné D, Mulhaupt B, Eur. J. Immunol 2008, 38, 2665. [DOI] [PubMed] [Google Scholar]
- [80].Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D,Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin A-G, Douek D, Autran B, Appay V, J. Exp. Med, 2007, 204, 2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Blood 2006, 107, 4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Frebel H, Richter K, Oxenius A, Eur. J. Immunol 2010, 40, 654. [DOI] [PubMed] [Google Scholar]
- [83].Maus MV, Thomas AK, Leonard DGB, Allman D, Addya K, Schlienger K, Riley JL, June CH, Nat. Biotechnol 2002, 20, 143. [DOI] [PubMed] [Google Scholar]
- [84].Rosenberg SA, Spiess P, Lafreniere R, Science 1986, 233, 1318. [DOI] [PubMed] [Google Scholar]
- [85].Yee C, Thompson JA, Byrd D, Riddell SR, Roche P, Celis E, Greenberg PD, Proc. Natl. Acad. Sci. USA 2002, 99, 16168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Stephan SB, Taber AM, Jileaeva I, Pegues EP, Sentman CL, Stephan MT, Nat. Biotechnol 2015, 33, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Li X, Liu X, Josey B, Chou CJ, Tan Y, Zhang N, Wen X, Stem Cells Transl. Med 2014, 3, 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Oelke M, Moehrle U, Chen J-L, Behringer D, Cerundolo V, Lindemann A, Mackensen A, Clin. Cancer Res 2000, 6, 1997. [PubMed] [Google Scholar]
- [89].Wrzesinski C, Paulos CM, Kaiser A, Muranski P, Palmer DC,Gattinoni L, Yu Z, Rosenberg SA, Restifo NP, J. Immunother, 2010, 33, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.