The spread of antimicrobial resistance (AR) is a public health threat in both high-income countries (HICs) and low- and middle-income countries (LMICs). Multidrug-resistant organisms (MDROs), particularly gram-negative bacteria, are of critical concern with several having been identified by the United States Centers for Disease Control and Prevention and the World Health Organization (WHO) as priority pathogens for control and research [1, 2]. Essential to the task of mitigating the spread of AR is the development and use of robust surveillance systems to measure and track the incidence, prevalence, and spread of AR as policies and interventions for its prevention and control are introduced and evaluated.
However, current MDRO surveillance in LMICs is limited by its reliance on data from incident clinical infections. While this methodology has been a cornerstone of AR surveillance in both HICs and LMICs, including the WHO’s Global Antimicrobial Resistance Surveillance System (GLASS), it is highly dependent on healthcare-seeking behavior and utilization. In LMICs, access to healthcare services may be limited; clinical laboratories are often underdeveloped or poorly resourced; and the cost of culture and antibiotic susceptibility testing (AST) is high and frequently borne by patients. In contrast, proceeding directly to empiric antibiotic treatment without diagnostic testing is less expensive. These factors lead to poor diagnostic stewardship, such that cultures are often not obtained, obtained only after antibiotic therapy, or obtained only after initial empiric therapy fails [3, 4]. For example, unpublished program evaluation data from 2 hospitals participating in a bloodstream infection surveillance system in India (2018) showed that only 24%–39% of intensive care unit patients with febrile episodes received blood cultures (personal communication, Paul Malpiedi).
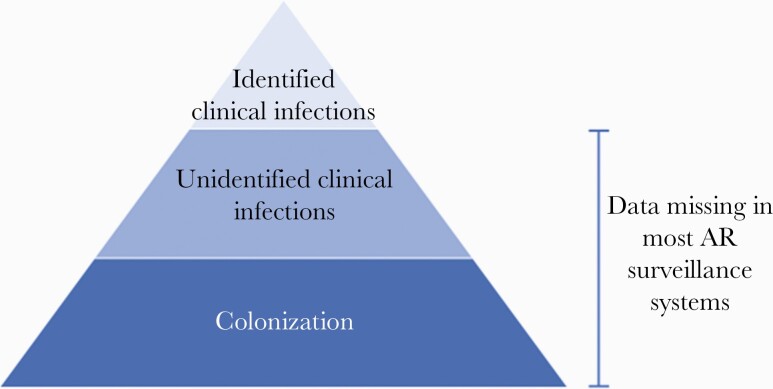
These factors lead to ascertainment bias within surveillance systems that may overrepresent the prevalence of resistance and undercount incidence of infection, including some types of resistant infections (eg, those that are covered by empiric treatment) [5]. This approach also misses persons who are colonized, but not infected, with MDROs and contribute to the overall reservoir of resistance. The extent to which AR surveillance systems miss potentially actionable MDRO data can be visualized as a surveillance pyramid (Figure 1). While attempts to improve underlying issues of diagnostic stewardship and laboratory capacity are worthy long-term endeavors, achieving more robust AR surveillance cannot wait for such large-scale change. We submit that incorporating measures of MDRO colonization into existing global AR surveillance efforts, in effect getting to the bottom of the AR surveillance pyramid, is an important complementary approach to existing AR surveillance efforts, such as WHO GLASS, and can inform AR control and containment activities, particularly in LMICs.
Figure 1.
Visualizing the “missingness” of antimicrobial resistance (AR) data within the antibiotic resistance surveillance pyramid.
Colonization increases the risk of infection with many clinically significant MDROs, such as carbapenem-resistant Enterobacterales (CRE), extended-spectrum β-lactamase–producing Enterobacterales, and methicillin-resistant Staphylococcus aureus (MRSA), among others [6, 7]. Thus, persons colonized with MDROs represent the pool of individuals at high risk for developing MDRO infections over time. Colonized persons are also an important source of transmission of MDROs to other people, and the prevalence of colonization is an important public health metric even if most colonized individuals never develop clinical infection [8, 9]. From the perspective of augmenting AR surveillance and public health response, the importance of MDRO colonization data can be thought of in 3 main areas: (1) addressing bias in existing AR surveillance systems; (2) tracking emergence and spread of MDROs, and (3) building toward a better understanding of AR within the One Health schema.
COLONIZATION DATA CAN ADDRESS BIAS IN EXISTING AR SURVEILLANCE
Accurate measurement of MDRO colonization is a powerful way to understand the landscape of MDROs in a community or healthcare facility, particularly when ascertainment bias is a factor. Actively assessing and measuring colonization (eg, through point [single time point] or periodic [over a time interval] prevalence surveys) is independent of diagnostic testing and, if done among community-dwellers, independent of healthcare-seeking behavior. A similar approach utilizing chest radiographs followed by targeted sputum culture has been used for more than a decade for estimating burden of Mycobacterium tuberculosis (TB) in countries where routine surveillance estimates are unreliable [10]. This WHO-endorsed approach has been successful in providing robust TB estimates in LMICs and is used to guide elimination and control efforts.
COLONIZATION DATA CAN HELP TO TRACK THE EMERGENCE AND SPREAD OF AR
Just as capturing asymptomatic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections is critical to assess and track spread of SARS-CoV-2 variants, tracking colonizing MDROs is critical to gain a full picture of circulating strains and elucidate their transmission dynamics. Evaluating colonization prevalence, along with relevant epidemiologic data, can improve our understanding of how humans acquire MDROs, and how resistance genes and plasmids are shared among bacteria. As the number of persons colonized with MDROs at any given point in time far outnumber the individuals who are infected, colonizing MDRO isolates may also be a more sensitive indicator for early identification of novel or emerging resistance mechanisms. Such early identification is critical for public health entities charged with implementing regional or national control strategies, including the development of diagnostic assays or even informing therapeutics to target emerging mechanisms before they become widespread.
COLONIZATION DATA CAN HELP BUILD A BETTER UNDERSTANDING OF AR WITHIN THE ONE HEALTH SCHEMA
In the One Health schema, MDROs causing human colonization and infection can either originate from, or are amplified in, animals or other environmental sources. While the final transmission event resulting in human colonization may be human-to-human, environment-to-human, or animal-to-human, thorough characterization of the colonization prevalence and investigation of risk factors associated with colonization is critical to inform how AR develops, is selected for, and spreads to people from other sources. This may include, in addition to better prevalence estimates of colonizing MDROs among humans, sequencing and comparing colonizing isolates from humans and environmental or animal sources.
EXPERIENCE WITH USING MDRO COLONIZATION DATA FOR AR SURVEILLANCE
Over the past 5 years, the coauthors of this editorial have implemented Antibiotic Resistance in Communities and Hospitals (ARCH) studies in 6 countries: Bangladesh, Botswana, Chile, Guatemala, India, and Kenya. The ARCH studies aim to evaluate the population-based prevalence of clinically important MDROs. They were conceptualized to address the challenges in obtaining valid estimates of MDRO prevalence outlined here, using methodology that has been successful for TB prevalence internationally and MRSA colonization within the United States [11, 12]. In brief, these studies leveraged community cohorts and hospitals in the same geographic area to collect and test stool and nasal swabs for MDRO colonization [13]. Demographic and epidemiologic risk factor data were collected via standardized questionnaires. Chromogenic agar was used as selective media to culture specimens; isolates underwent identification and AST confirmation. MDROs of interest will undergo whole genome sequencing to identify the underlying genetic mechanisms of resistance.
Findings from ARCH sites have already demonstrated the value of systematically collected MDRO colonization data. High prevalence of MDRO colonization among hospitalized patients and geographically co-located communities has been shown [14]. In 2018, ARCH data from Chile demonstrated the presence of New Delhi metallo-β-lactamase (NDM)–producing CRE in colonization samples before it was commonly identified in clinical samples at a study hospital. Since then, the recovery of NDM among CRE isolates at the hospital has increased 6-fold. NDM has also been identified in additional Enterobacterales species (personal communication, Rafael Araos). While other experiences are needed to understand the value of colonization data, it seems evident that identifying circulating strains prior to infection can be invaluable to help laboratories and hospitals prepare to detect and prevent emerging resistant strains.
However, further work is needed to move from ARCH as a research initiative to incorporating MDRO colonization into existing AR surveillance efforts. Using colonization data to inform real-time public health efforts to contain CRE deserves further exploration, as such data could be enormously helpful to notify public health officials of novel or emerging carbapenemase-producing genes that might require additional diagnostic testing or capacity. Repeat periodic prevalence surveys of MDRO colonization could track the impact of prevention and control measures, such as antibiotic stewardship campaigns, as well as look at the impact of naturally occurring events, such as the coronavirus disease 2019 pandemic. Finally, additional experience can inform how to compare data across settings and regions, as well characterize the relationship between prevalence of colonization and risk for MDRO infections.
Modifications to improve cost and acceptability outside of the research setting are also critical, such as identifying inexpensive but reliable selective culture methods, considering alternative methods for community sampling, and addressing the need for informed consent for colonization screening in countries where it is required. However, even if costs are reduced, financing the surveys will remain a need. Similar large-scale studies undertaken to measure TB prevalence have been funded by donors and global technical partners. If consensus is achieved around the utility of colonization surveys for global AR surveillance, financing by a similar consortium of donors, or even via national governments, could be a feasible avenue to support this critical work.
CONCLUSIONS
Building robust AR surveillance systems continues to be an important global public health priority to combat the spread of AR worldwide. AR surveillance for clinical infections will continue to be critical to inform AR prevention and control efforts. However, to clearly elucidate and effectively respond to AR challenges, additional surveillance efforts are needed. These can complement existing approaches by getting to the bottom of the surveillance pyramid through incorporation of systematically collected MDRO colonization data as part of the surveillance system. In places with capacity, incorporating identification of genetic mechanisms of resistance as preferred surveillance targets could also allow for a more complete view of growing resistance threats. MDRO colonization data can be a vital adjunct to clinical-isolate AR surveillance systems and is critical in forming a complete picture of AR locally and across regions.
Notes
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: CDC; 2019. [Google Scholar]
- 2. World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva, Switzerland: WHO; 2017. [Google Scholar]
- 3. Hemlock C, Luby SP, Saha S, et al. Utilization of blood culture in South Asia for the diagnosis and treatment of febrile illness. Clin Infect Dis 2020; 71(Suppl 3):S266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gebretekle GB, Haile Mariam D, Abebe W, et al. Opportunities and barriers to implementing antibiotic stewardship in low and middle-income countries: lessons from a mixed-methods study in a tertiary care hospital in Ethiopia. PLoS One 2018; 13:e0208447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rempel OR, Laupland KB.. Surveillance for antimicrobial resistant organisms: potential sources and magnitude of bias. Epidemiol Infect 2009; 137:1665–73. [DOI] [PubMed] [Google Scholar]
- 6. Nurjadi D, Eichel VM, Tabatabai P, et al. Surveillance for colonization, transmission, and infection with methicillin-susceptible Staphylococcus aureus in a neonatal intensive care unit. JAMA Netw Open 2021; 4:e2124938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin Q, Wang Y, Yu J, et al. Bacterial characteristics of carbapenem-resistant Enterobacteriaceae (CRE) colonized strains and their correlation with subsequent infection. BMC Infect Dis 2021; 21:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hagel S, Makarewicz O, Hartung A, et al. ESBL colonization and acquisition in a hospital population: the molecular epidemiology and transmission of resistance genes. PLoS One 2019; 14:e0208505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tschudin-Sutter S, Frei R, Dangel M, et al. Rate of transmission of extended-spectrum beta-lactamase-producing Enterobacteriaceae without contact isolation. Clin Infect Dis 2012; 55:1505–11. [DOI] [PubMed] [Google Scholar]
- 10. Glaziou P, Falzon D, Floyd K, Raviglione M.. Global epidemiology of tuberculosis. Semin Respir Crit Care Med 2013; 34:3–16. [DOI] [PubMed] [Google Scholar]
- 11. Gorwitz RJ, Kruszon-Moran D, McAllister SK, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis 2008; 197:1226–34. [DOI] [PubMed] [Google Scholar]
- 12. World Health Organization. Tuberculosis Prevalence Surveys: A Handbook. Geneva, Switzerland: WHO Press; 2011. [Google Scholar]
- 13. Sharma A, Luvsansharav U-O, Paul P, et al. Multi-country cross-sectional study of colonization with multidrug-resistant organisms in communities and hospitals: protocol and methods. BMC Public Health 2021; 21:1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Araos Bralic R, Peters A, Sanches F, et al. Colonization with antibiotic-resistant gram-negative bacteria in population-based hospital and community settings in Chile. Infect Control Hosp Epidemiol 2020; 41(Suppl 1):S175–76. [Google Scholar]