Abstract
The cell cycle arrest and proapoptotic functions of p53 are under tight control by Mdm2. After stress activation of p53 by nontranscriptional mechanisms, transcription of the mdm2 gene results in increased synthesis of Mdm2 and down-regulation of p53. Disruption of this autoregulatory loop has profound effects on cell survival and tumorigenesis. We show that a defective p53-Mdm2 autoregulatory loop results from inactivation of a basal transcription factor, TAFII250, in tsBN462 cells. We found that Mdm2 expression rescues the temperature-sensitive phenotype of tsBN462 cells, as shown by activation of cell cycle-regulated gene promoters (B-myb, cyclin A, and cdc25C), increased cell growth and DNA synthesis, and inhibition of apoptosis. These effects of Mdm2 are mediated by p53. Exogenous Mdm2 expression apparently complements endogenous Mdm2 synthesis in tsBN462 cells, which is reduced compared to that in the equivalent parental cells with wild-type TAFII250, BHK21. Expression of wild-type TAFII250 in tsBN462 stimulates and prolongs the synthesis of Mdm2 and rescues the temperature-sensitive phenotype. The TAFII250 rescue is blocked by inhibition of Mdm2-p53 interactions. We also show that Mdm2 promoter activation, after transfer to the nonpermissive temperature, is attenuated in cells with mutant TAFII250. The temperature-sensitive phenotype apparently results from inefficient inhibition of heat-induced p53 by reduced Mdm2 synthesis due to low mdm2 promoter activity. These results raise the possibility that the p53-Mdm2 autoregulatory loop could guard against transcriptional defects in cells.
The tumor suppressor p53 responds to cellular stresses and protects against tumor formation and faulty development (for reviews, see references 1a, 2, 24–26, 44, and 60). There are many inducers of p53, including heat shock, genotoxins, hypoxia, cytokines, growth factors, metabolic changes, loss of anchorage, cell-cell interactions, and activated oncogenes. p53 protein levels and activity are increased by a variety of mechanisms. p53 regulates the transcription of downstream effectors that are involved in cell cycle inhibition (e.g., the cyclin-dependent kinase inhibitor p21WAF1/CIP1), apoptosis (e.g., Bax), and its own inhibition (Mdm2) (for reviews, see references 14, 16, 39, and 46). Increased mdm2 gene transcription produces Mdm2 protein, which complexes with p53, inhibits its ability to activate transcription, exports it from the nucleus, and stimulates its degradation by the ubiquitin pathway. Mdm2-mediated inhibition of p53 is critical. In mice, inactivation of the mdm2 gene results in a lethality that is rescued by inactivation of the p53 gene (27, 40). Furthermore, a tumor suppressor, ARF-INK4a, regulates the p53-Mdm2 loop by inhibiting Mdm2-mediated down-regulation of p53. Mdm2 has p53-independent functions, in that it interacts physically and/or functionally with several regulators of the cell cycle (pRb, E2F1-DP1, and p107) and has various effects in mice when it is overexpressed in the absence of p53. Nevertheless, the p53-Mdm2 regulatory circuit is fundamentally important in regulating the apoptotic properties of p53.
We have found that inactivation of the general transcription factor TAFII250 affects the p53-Mdm2 circuit. TAFII250 is a component of TFIID, a complex that binds to the core promoter and mediates transcriptional activation from regulatory proteins to the basal machinery (3). tsBN462 hamster cells (41) express a temperature-sensitive TAFII250 (CCG1) mutant (G690E), whose inactivation at the nonpermissive temperature leads to cell cycle arrest (52) and apoptosis (49). We found that a defect generated by TAFII250 inactivation at the nonpermissive temperature is a faulty p53-Mdm2 autoregulatory circuit resulting from reduced mdm2 promoter activity, mRNA synthesis, and protein production. Exogenous Mdm2 expression rescues the phenotype by a p53-dependent mechanism. Rescue by wild-type TAFII250 expression is inhibited by disrupting the p53-Mdm2 regulatory circuit. These results show that the Mdm2-p53 regulatory circuit is sensitive to a defect in a transcription factor and raise the possibility that having a transcriptional step in an apoptotic autoregulatory circuit is a manner of eliminating cells with transcriptional defects.
MATERIALS AND METHODS
Cell culture and transfections.
tsBN462 cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal calf serum and penicillin-streptomycin at 32°C in 5% CO2. They were transfected by the calcium phosphate method (5) with 20 μg of plasmid DNA in 1 ml of precipitate per 9-cm-diameter dish containing different amounts of expression vector (0.1 to 1 μg/ml), 1 μg of reporters per ml, 1 μg of pSG5-LacZ (internal control for transfection efficiency) per ml, and the appropriate quantities of empty expression vectors and pEMBL18. Transfections were performed in duplicate for each condition within any single experiment and were repeated several times using different plasmid preparations for each construct. The cells were freshly plated at 50% confluence 2 h before transfection, incubated with precipitated DNA for 16 h, washed, and then incubated at 39°C for 24 h or the indicated time (33).
Recombinants.
The following recombinants were used: p.fos.lcf (human c-fos −711/+45) (38); cyclin D1 (D1Δ-973pXP2) luc (20); E2 luc, three copies of the E2F motif (31) in pGL3 (Promega) (1); pCMV-Mdm2 (10); pEIA-SV (63); pSG5-LacZ (63); B-myb wt (Mus −303/+100) luc (36); B-myb mE2F (TTGGCGGG to TTGtatGG) (36); B-myb mCHR (ATAGGAAGTG to ATAGGcctGTG) luc (36); cyclin A (−214/+100) luc (67); cyclin A mCDE (G to T at −33) luc (67); cyclin A mCHR (−26/−23 mutated to GGTC) (67); C290 (cdc25C −290/+121) luc (67); C290R1.T7 (mCDE, TGGCGGA to TGGCtGA) luc (67); C290m−6/−3 (−6/−3 mutated to GGTC) luc (67); pCDNAHA-E2F1(1-368)RB(379-792) (53); Zn (10); pCMV Hdm2, pCMV Hdm2 G58A, and pCMV Hdm2 V75A (13); pBC-IP3.2 (63); pcDNA3-myc-ARF (66); p53 R175H (pC53-Cx22AN3) (21); p53 R273H (pC53-4.2N3) (21); pXJ41- TAFII250 (33); pEGFP-C1 (Clontech); pGKneo (63); pHOOK-1 (Invitrogen); WAF1-luc, a gift from B. Vogelstein (12); pCON and pGUP, a gift from J. Shay (15); pG13 Py luc (wild type) and mG15 Py luc, a gift from B. Vogelstein (29); Mdm2-luc, containing the mouse mdm2 gene p53-responsive promoter in pGL2 basic (28), a gift from M. Oren; hamster mdm2 cDNA (4); mouse WAF1 cDNA, a gift from B. Vogelstein (12); pSFFV-mBcl2, a gift from S. Korsmeyer; and pSG5-hBcl2, a gift from H. Gronemeyer.
Luciferase assays.
The cells were harvested in phosphate-buffered saline (PBS)–3 mM EDTA, and extracts were prepared by three sequential freeze-thaw cycles in 10 mM potassium phosphate–1 mM dithiothreitol, with vortexing between cycles. The extracts were centrifuged at 10,000 × g for 10 min at 4°C, and the supernatants were used for the assays. Luciferase was measured in duplicate as described previously (33) in 100 mM potassium phosphate (pH 7.8)–1 mM dithiothreitol–15 mM MgSO4–5 mM ATP–0.2 mM luciferin, using a Monolight 2010 luminometer.
Flow cytometry.
Cells were analyzed by flow cytometry as described previously (10).
Cell clones.
The cells were transfected in 9-cm plates with 20 μg of total DNA (100 ng of pCMVneo-based recombinants that express Hdm2, Hdm2 D68A, Hdm2 G58A, Hdm2 V75A, and p53 R175H; or pXJ41-TAFII250 plus 0.5 μg of pGKneo). At 48 h after transfection, the cells were seeded at a 1:4 dilution and cultured for 2 to 3 weeks in medium containing 1 mg of G418 per ml, and individual clones were isolated and expanded.
Purification of transfected cells.
The cells were split and, after 2 h, transfected with 1 μg of pHOOK-1, 1 μg of pEGFP-C1, and 10 μg of Hdm2 or TAFII250 expression vectors. At 18 h after transfection, the cells were harvested in PBS–3 mM EDTA, resuspended in 1 ml of complete medium, mixed with 10 μl of washed Capture-Tec beads (1.5 × 106 beads), and incubated for 60 min at 30°C on a slow rotator. The tubes were placed next to a strong magnet, and the medium was removed. The beads were resuspended in 1 ml of complete medium, vortexed gently, and reattracted to the magnet; these steps were carried out twice. Green fluorescence protein (GFP)-expressing cells in the supernatants were detected by fluorescence microscopy. Selected cells were resuspended in complete medium, incubated for 2 h at 32°C in 3.5-cm plates, washed, and then incubated in low-serum medium for 14 h. The cells were transferred to 39°C and harvested at different times (0, 2, 4, 6, 8, and 12 h), and cell extracts were analyzed by Western blotting as described previously (33).
S-phase detection by BrdU labeling.
The cell clones were plated at 40% confluence in six-well dishes. After 6 h, they were washed, incubated in low-serum (0.05% fetal calf serum) medium for 24 h at 32°C, and induced at 39°C for 12 h. Bromodeoxyuridine (BrdU) (5 μM) in fresh culture medium was added, and incubation was continued for 1 h at 39°C. The cells were washed twice in cold PBS, fixed in 3.7% paraformaldehyde in PBS for 20 min at 4°C, washed three times with PBS, permeabilized with methanol for 10 min at −20°C, washed three times with PBS, incubated in 0.25% Triton X-100 for 5 min, saturated with 0.5% bovine serum albumin (BSA) in PBS for 10 min, washed three times with PBS, incubated in 1.5 N HCl for 10 min at room temperature, washed twice for 3 min each with PBS and once with 0.5% BSA in PBS, and incubated with mouse anti-BrdU for 30 min and, after three PBS washes, with goat anti-mouse immunoglobulin G Texas red for 30 min (Becton Dickinson). The cells were stained with 10 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml for 10 min, washed twice with PBS and once with 0.5% BSA in PBS, rinsed rapidly with water, mounted with Mowiol, and observed by immunofluorescence microscopy.
Western blotting.
Protein extraction, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, and antibodies were as described previously (33).
Northern blotting.
RNA was extracted with Trizol reagent (Gibco-BRL). Total RNA (40 μg) was electrophoresed on 6% formaldehyde–1% agarose gels, transferred to Hybond-N+ membranes (Amersham-Pharmacia Biotech), hybridized with random-prime-labeled probes (hamster mdm2, mouse p21WAF1/CIP1, and human 36B4) at 60°C, and washed (6). The membranes were analyzed with a PhosphorImager instrument (Molecular Dynamics).
RESULTS
Mdm2 stimulates the activity of cell cycle-regulated promoters.
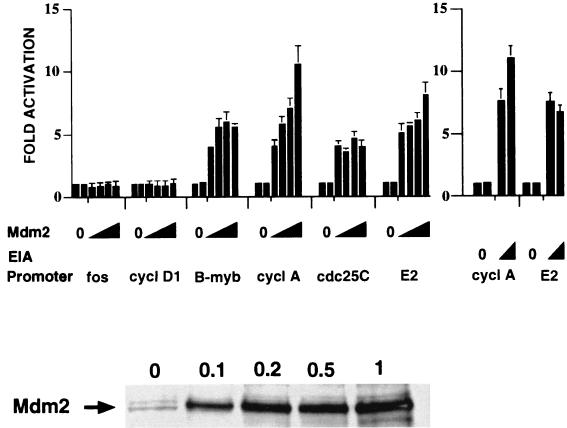
We have shown previously that Mdm2 expression stimulates the activity of the cyclin A promoter in tsBN462 cells at the nonpermissive temperature (33). tsBN462 cells express a mutated temperature-sensitive TAFII250, whose inactivation at the nonpermissive temperature specifically reduces cyclin A promoter activity (61). We investigated whether Mdm2 activation is specific for the cyclin A promoter or whether other cell cycle-regulated promoters may also be stimulated (Fig. 1). tsBN462 cells were transfected as described previously (33) with luciferase reporters containing promoter sequences from the c-fos (38), cyclin D1 (20), B-myb, cyclin A, cdc25C (35), and adenovirus E2 (31) genes. The amounts of Mdm2 expression vector were titrated to ensure that optimum conditions for each reporter were being compared. Basal promoter activity was measured in duplicate in each titration, since this reference activity is used to calculate activation by different amounts of Mdm2 (see two bars for zero values in the figures). Mdm2 was shown to be expressed by Western blotting of transfected-cell extracts with antibodies raised against mouse Mdm2 (antibody 365 [Fig. 1, lower panel]; 0.1, 0.2, 0.5, and 1.0 show the amounts, in micrograms, of Mdm2 expression vector were used in the transfection). Mdm2 expression had no detectable effect on the activities of the c-fos and cyclin D1 promoters, whereas it stimulated the B-myb, cyclin A, and cdc25C promoters between 5- and 10-fold under optimum conditions. We compared the effects of Mdm2 with those of the adenovirus E1A oncoprotein, which efficiently activates the cyclin A promoter (65). E1A expression stimulated the cyclin A promoter about 11-fold, which is similar to the maximum effect of Mdm2, indicating that Mdm2 is an efficient activator of the cell cycle regulated promoters. The three Mdm2-activated promoters are regulated by E2F, suggesting that Mdm2 ultimately activates E2F under these conditions. Indeed, Mdm2 expression stimulated by sevenfold the activity of a reporter containing three E2F motifs from the adenovirus E2 gene promoter. Mdm2 activation was relatively efficient, since it was comparable to the effects of E1A. The effects were not general for any cell cycle regulator, since we found that expression of pRB or p107 did not affect cyclin A or E2F promoter activity under these conditions (data not shown). Furthermore, Mdm2 expression did not affect cyclin A and E2F promoter activity at the permissive temperature (32°C) (data not shown), showing that the effects of Mdm2 were observed only when TAFII250 was inactivated at the nonpermissive temperature.
FIG. 1.
Activation of cell cycle-responsive promoters by Mdm2 expression in tsBN462 cells. The cells were transfected in six-well dishes with luciferase reporters containing promoters from the fos, cyclin D1 (cycl D1), B-myb, cyclin A (cycl A), cdc25C, and adenovirus E2 genes (1 μg) and increasing amounts of expression vectors for Mdm2 (pCMV-Mdm2; 0.1, 0.2, 0.5, and 1 μg) or E1A (0.5 and 1 μg). After transfection the cells were transferred to 39°C, and 24 h later the luciferase activities were measured. Luciferase activities were corrected for variations in transfection efficiency by using the internal control (pSG5-LacZ; 1 μg/ml). They are presented relative to the control (set to 1) that was included in duplicate in each experiment. The fold activations are the averages from three independent experiments, and the error bars indicate the standard deviations. The Western blot in the lower panel was probed with rabbit anti-Mdm2 antibody no. 365 raised against a mouse Mdm2-derived peptide (33). The arrow indicates the Mdm2-specific band migrating at a molecular weight of around 90,000. The lane markers 0, 0.1, 0.2, 0.5, and 1 correspond to the amounts (in micrograms) of transfected pCMV-Mdm2.
Mdm2 activation of the B-myb, cyclin A, and cdc25C promoters is mediated by their cell cycle-regulated elements.
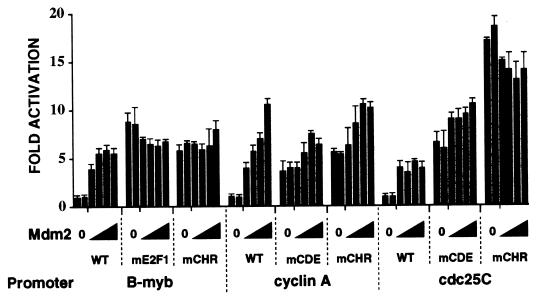
We localized the sequences required for Mdm2 activation of the B-myb, cyclin A, and cdc25C promoters. Initial deletion analysis of the cyclin A promoter indicated that the cell cycle-dependent elements are involved (data not shown). The cell cycle-dependent elements control the variation of promoter activity during the cell cycle. When the cell cycle is inhibited, the elements repress promoter activity, such that mutation of the sites activates the promoters (68). The cell cycle-dependent elements are bipartite and include E2F and CHR motifs in the B-myb promoter (36) and CDE and CHR motifs in both the cyclin A and cdc25C promoters (67). We found that in the absence of exogenous Mdm2, point mutation of each of the motifs in the three promoters stimulated promoter activity 5- to 15-fold (Fig. 2; compare 0 values for the wild-type [WT] and mutated [mE2F1, mCHR, and mCDE] promoters). These results show that the B-myb, cyclin A, and cdc25C promoters are repressed at the nonpermissive temperature in tsBN462 cells. We then studied whether the cell cycle-dependent elements mediated stimulation by Mdm2. We found that the mutated promoters were insensitive to Mdm2 expression (Fig. 2). There were small residual effects of Mdm2, suggesting that the point mutations do not completely inactivate the elements. The activities of the wild-type B-myb and cyclin A promoters in the presence of Mdm2 were quite similar to the activities of the mutated promoters, in agreement with the conclusion that Mdm2 acts through the cell cycle-dependent motifs. The mechanisms of regulation of the cell cycle-dependent elements are complex, involving common as well as promoter-specific factors (68). It is therefore not surprising that there are quantitative differences in the response to Mdm2. In particular, Mdm2 is less efficient than CHR mutation in activating the cdc25C promoter. However, the results show that Mdm2 activation of the B-myb, cyclin A, and cdc25C promoters is mediated by the cell cycle-dependent motifs.
FIG. 2.
Identification of the Mdm2-inducible elements in the cell cycle-responsive promoters. tsBN462 cells were transfected in six-well dishes with luciferase reporters containing either wild-type or point-mutated promoters from the B-myb, cyclin A, and cdc25C genes (1 μg) and increasing amounts of the Mdm2 expression vector (pCMV-Mdm2; 0.1, 0.2, 0.5, and 1 μg). After transfection the cells were transferred to 39°C, and 24 h later the luciferase activities were measured. Luciferase activities were corrected for variations in transfection efficiency by using the internal control (pSG5-LacZ; 1 μg/ml). They are presented relative to the control (set to 1) that was transfected in duplicate in each experiment. The fold activations are the averages from three independent experiments, and the error bars indicate the standard deviations.
Mdm2 activation of the cyclin A promoter is mediated by E2F.
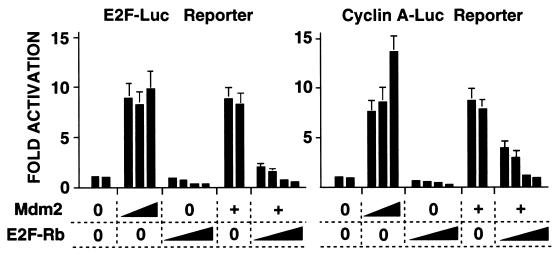
The above results show that both simple (E2) and more complex (B-myb, cyclin A, and cdc25C) E2F motifs mediate Mdm2 activation of transcription. To provide evidence that E2F is involved in activation of the cyclin A promoter, we used a trans-dominant inhibitor of E2F, an E2F1-Rb chimera which has the Rb pocket in place of the activation domain of E2F1 (53). Expression of the E2F-Rb chimera in tsBN462 cells at the nonpermissive temperature inhibited both the basal and Mdm2-induced activities of the E2F reporter (Fig. 3, left panel). The chimera also inhibited the basal and Mdm2-induced activity of the cyclin A promoter (right panel). These results are consistent with those above (Fig. 1 and 2), and provide further evidence that Mdm2 activation of the cyclin A promoter is mediated by E2F factors.
FIG. 3.
Inhibition of E2F blocks Mdm2 activation of the cyclin A promoter reporter. tsBN462 cells were transfected in six-well dishes with either the E2F or the cyclin A Luc reporters (1 μg) and increasing amounts of expression vectors for Mdm2 (0.05, 0.2, and 2 μg, or 0.2 μg where indicated by +) and/or E2F-RB (0.1, 0.3, 1, and 3 μg). After transfection the cells were transferred to 39°C, and 24 h later the luciferase activities were measured. Luciferase activities were corrected for variations in transfection efficiency by using the internal control (pSG5-LacZ; 1 μg/ml). They are presented relative to the control (set to 1) that was included in duplicate in each experiment. The fold activations are the averages from three independent experiments, and the error bars indicate the standard deviation.
Mdm2-p53 interactions are required for Mdm2 activation of the cyclin A promoter.
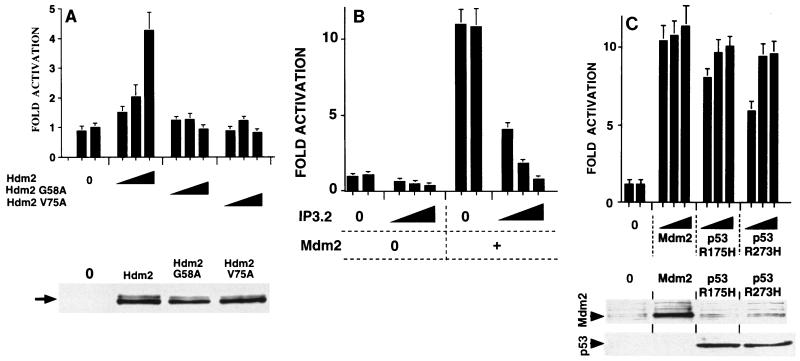
The major function of Mdm2 is to down-regulate p53. The N-terminal region of Mdm2 is required for complex formation and inhibition of p53. We studied whether p53 is involved in Mdm2 activation of the cyclin A promoter by using a variety of approaches. Initially, we used various N-terminal mutants of Mdm2 and Hdm2, which are inhibited in their ability to form complexes and inactivate p53. Zn, an N-terminal deletion mutant of Mdm2 (10), had no detectable effect on cyclin A promoter activity (data not shown). Two-point mutants of Hdm2, G58A and V75A (13), were also inactive, in contrast to Hdm2 (Fig. 4A; the mutants were efficiently expressed [lower panel]). We also investigated whether inhibitors of p53 interactions with Mdm2 would block activation of the cyclin A promoter. IP3.2 is a peptide homologue of p53, which has a much higher affinity than p53 for Mdm2 and efficiently and specifically dissociates the p53-Mdm2 in vivo (63). Expression of IP3.2 inhibited Mdm2 induced activation of the cyclin A promoter (Fig. 4B). It also inhibited the basal activity of the cyclin A promoter, but quantitatively to a much smaller extent. The effect was specific, since a mutated IP3.2 peptide, with greatly reduced affinity for Mdm2 (63), had no effect (data not shown). Expression of ARF, which prevents inhibition of p53 by Mdm2, blocked activation of the cyclin A promoter by exogenous Mdm2 (data not shown). Taken together, these results show that Mdm2 activation of cyclin A promoter activity is mediated by its inhibitory effects on p53. They predict that other inhibitors of p53 activity would stimulate cyclin A promoter activity. Mutant p53 proteins, such as p53 R175H and R273H, are trans-dominant inhibitors of wild-type p53, probably due to interactions with other proteins. Expression of either p53 R175H or R273H activated cyclin A promoter activity with efficiencies comparable to that of Mdm2 (Fig. 4C). Western blots with DO1, a human p53-specific antibody that recognizes only exogenous p53, showed that the p53 mutants were efficiently expressed (Fig. 4C, lower panel). These results all led to the conclusion that inhibition of p53 is the route by which Mdm2 expression leads to cyclin A promoter activation in tsBN462 cells at the nonpermissive temperature.
FIG. 4.
Involvement of p53 in the effects of Mdm2 on cyclin A promoter activity. tsBN462 cells were transfected in six-well dishes with the cyclin A Luc reporter (1 μg) and increasing amounts of expression vectors for Hdm2 and mutants with single-amino-acid substitutions in the p53 binding domain (0.1, 0.5, and 2 μg) (A), IP3.2 (pBC-IP3.2, 0, 0.05, 0.1 and 0.5 μg; made up to 0.5 μg with pBC) and Mdm2 (0 and 0.5 μg) (B), and Mdm2, p53 R175H, and R273H (0, 0.1, 0.5, and 2 μg) (C). After transfection the cells were transferred to 39°C, and 24 h later the luciferase activities were measured. Luciferase activities were corrected for variations in transfection efficiency by using the internal control (pSG5-LacZ; 1 μg/ml). They are presented relative to the control (set to 1) that was included in duplicate in each experiment. The fold activations are the averages from three independent experiments, and the error bars indicate the standard deviations. The Western blots in the lower panels were probed with the IF2 mouse monoclonal antibody against Mdm2 (Ab-1; Calbiochem OP46 (A) and rabbit anti-Mdm2 antibody no. 365 raised against a mouse Mdm2-derived peptide (33) and the DO1 mouse monoclonal against human p53 (C). Arrows indicate the corresponding specific bands of the expected mobility. Extracts from transfections with the highest levels of the corresponding expression vectors are shown.
Mdm2 expression affects the cell cycle.
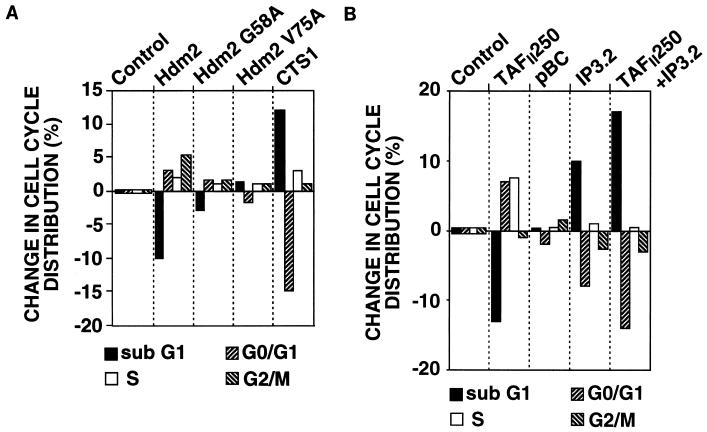
In the promoter studies above, we found that the effects of Mdm2 are mediated by cell cycle-dependent elements and involve p53, a regulator of the cell cycle and of apoptosis. The possibility that Mdm2 expression affects the cell cycle at the nonpermissive temperature was investigated by flow cytometry. tsBN462 cells were transfected with vectors that express Hdm2 and the cell surface marker CD20, and after 24 h at 39°C, the cells were harvested and analyzed by flow cytometry as described previously (10). The percentage of transfected cells (expressing CD20) in each phase of the cell cycle was calculated and then subtracted from the value for the control cells transfected with the corresponding empty vectors (Fig. 5; the values for the controls, i.e., zero, are shown simply for illustration, to indicate that they are the reference values). In the control, the cell cycle distribution was 38% sub-G0/G1, 44% G0/G1, 9% S, and 9% G2/M. Hdm2 expression decreased the number of cells in sub-G0/G1 and increased the number of cells in the G1, S, and G2/M phases (Fig. 5A). Similar results were obtained with Mdm2 (not shown). Hdm2 with point mutations that inhibit p53 binding (G58A and V75A) had much reduced effects on the cell cycle. CTS1, a chimeric tumor suppressor derived from p53 that is resistant to inhibition by Mdm2 and mutant p53 (7), increased the sub-G0/G1 population and decreased the G0/G1 population, as expected from its ability to efficiently induce apoptosis. At earlier and later time points, the cell cycle distributions observed with the different expressed proteins resulted in changes consistent with the conclusions drawn from the 24-h time point (data not shown). These results indicate that a major consequence of Mdm2 expression in tsBN462 cells is to overcome the effects of p53. They suggest that p53 plays an important role in the response of tsBN462 cells to inactivation of TAFII250 and that Mdm2 is limiting in the response.
FIG. 5.
Effect of Mdm2, TAFII250, and inhibiting Mdm2-p53 interactions on the cell cycle. tsBN462 cells were transfected in 9-cm dishes with vectors that express CD20 (5 μg, all samples) and, in addition, Hdm2 (10 μg), Hdm2 G58A (10 μg), Hdm2 V75A (10 μg), and CTS1 (10 μg) (A) and TAFII250 (10 μg), pBC (10 μg, control for IP3.2), IP3.2 (pBC-IP3.2 10 μg), and TAFII250 plus IP3.2 (10 μg of each) (B); after 24 h at 39°C the cells were harvested and analyzed by flow cytometry. The results presented are the average of two values, each of which was derived from two independently transfected plates. One representative experiment of three is shown. The percentage of transfected cells in each phase of the cell cycle was calculated and subtracted from the control transfected with empty vector. The control (zero change) is presented as small bars for illustration only.
To investigate these possibilities, we studied the effect of TAFII250 expression (Fig. 5B). After 24 h at the nonpermissive temperature, TAFII250 decreased the number of cells in sub-G0/G1, increased the number in G0/G1 and S, and had little effect on the number in G2/M. Globally, these effects are similar to Hdm2 expression. We then studied the consequences of inhibition of p53-Mdm2 interactions by IP3.2. Expression of IP3.2 mainly increased the sub-G0/G1 content and decreased the G0/G1 content, similar to the effects of CTS1. The control vector for IP3.2, pBC, had little effect. These results show that the p53-Mdm2 regulatory circuit is partially functional in the response to heat inactivation of TAFII250. We then tested whether the effects of TAFII250 expression could be overcome by inhibition of p53-Mdm2 interactions. IP3.2 expression overcame the effects of TAFII250, leading to an even greater increase in the sub-G0/G1 content and decrease in the G0/G1 content than that due to IP3.2 alone. These results suggest that heat inactivation of TAFII250 leads to an incomplete response at the level of Mdm2 to heat-induced p53. Exogenous Mdm2 expression complements endogenous Mdm2.
Rescue of tsBN462 cells by expression of Mdm2.
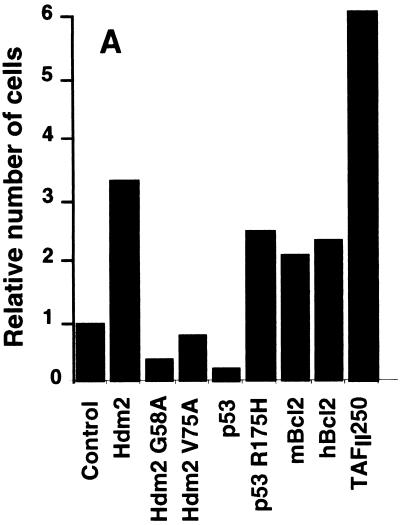
We investigated Mdm2 rescue of TAFII250-inactivated cells by using two selection techniques. In the first approach, cells were selected by incubation at the nonpermissive temperature following transfection. In the second method, stable clones were first isolated using G418 selection at the permissive temperature and then the clones expressing exogenous protein were analyzed at the higher temperature.
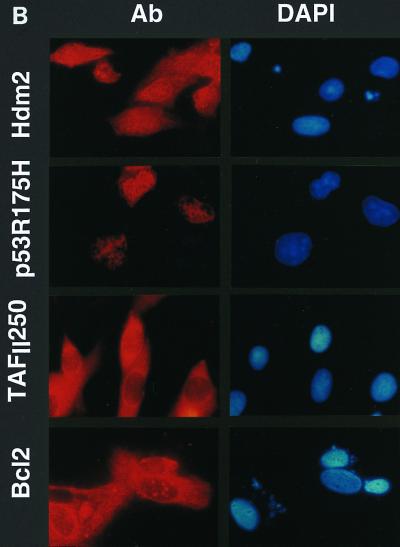
The temperature selection assays were performed essentially as described by O'Brien and Tjian (42). tsBN462 cells were transfected with expression vectors and an internal control that produces β-galactosidase (β-Gal). In parallel, cells were transfected uniquely with an expression plasmid for GFP, to estimate the proportion of cells that had been transfected. At 24 h after transfection, aliquots of the cells from each transfection were lysed and analyzed for β-Gal activity. The separate plates expressing GFP were observed by fluorescence microscopy. About 10% of the cells were found to be GFP positive (data not shown). The remaining cells, which were not processed for β-Gal measurements, were transferred to the nonpermissive temperature for 7 days. The number of viable cells was then counted, adjusted for transfection efficiency, and plotted relative to the number of surviving cells transfected with the empty vector (Fig. 6A). Protein expression in the surviving cells was verified by immunocytochemistry with antibodies specific for the exogenous proteins and a fluorescently labeled secondary antibody (Fig. 6B [representative fields are shown]). The corresponding proteins were readily detectable, whereas control cells gave only low levels of background staining (data not shown). These results show that the surviving cells express the exogenous proteins. We reproducibly found, in several independent experiments, that Hdm2 increased the number of surviving cells about threefold, which was about half the efficiency of TAFII250 (Fig. 6A). Similar results were obtained with mouse Mdm2 (data not shown). Hdm2 mutants that cannot interact with p53 did not lead to rescue (Hdm2 G58A and V75A). In fact, Hdm2 G58A reproducibly decreased cell survival, suggesting that it acts as a trans-dominant inhibitor of endogenous Mdm2, as has been observed in other systems (30). Wild-type p53 decreased cell survival, whereas mutant p53 R175H increased survival, consistent with its ability to inhibit endogenous p53. The apoptosis inhibitor Bcl2, from both mouse and human origins, rescued to a similar extent to Hdm2 and less efficiently than TAFII250. The similar efficiencies of rescue by Hdm2 and Bcl2 suggest that they act by related mechanisms, i.e., inhibition of apoptosis. The lower efficiency of Bcl2 and Hdm2 than TAFII250 is consistent with inhibition of apoptosis being insufficient to rescue cells from the loss of a transcription factor that is involved in other functions.
FIG. 6.
Survival of tsBN462 cells selected at the nonpermissive temperature. (A) The survival assay, using selection at the nonpermissive temperature, was performed essentially as described by O'Brien and Tjian (42). tsBN462 cells were transfected with 5 μg (9-cm plates) of vectors expressing nothing (control), Hdm2, Hdm2 G58A, Hdm2 V75A, p53, p53 R175H, mBcl2, hBcl2, and TAFII250, as indicated, and 1 μg of pSG5-lacZ (all plates). Parallel plates were transfected with 0.5 μg of pEGFP. After 16 h in the presence of the precipitate, the cells were washed, incubated for 24 h, trypsinized, and replated at half the density. An aliquot was removed to measure β-Gal activity (to correct for variations in transfection efficiency). The plates transfected with pEGFP were observed under a fluorescence microscope. The cells were incubated overnight at 32°C and then for 7 days at the nonpermissive temperature (39°C). The medium was changed every 2 days. To measure the number of surviving cells, they were trypsinized and the viable cells that excluded trypan blue were counted with a Bürker slide. The numbers of cells were adjusted for variations in transfection efficiency and expressed relative to the control empty vector. There were approximately 106 cells per 9-cm plate for TAFII250 at the end of the experiment. Similar results were obtained in two independent experiments (±10%). (B) Following selection the cells were fixed, stained with specific antibodies (Hdm2, IF2; p53, DO1; TAFII250, anti-HA, 12CA5; Bcl2, sc-509 [Santa Cruz]) followed by Cy3-labeled secondary antibodies and DAPI (nuclei). The cells were photographed under a fluorescence microscope. Control plates incubated with the specific antibodies and labeled secondary antibodies gave low levels of background fluorescence (not shown). The antibodies are specific for exogenous proteins.
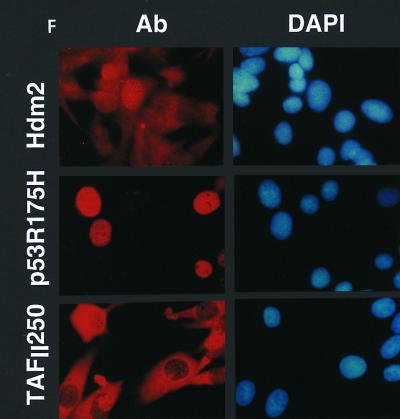
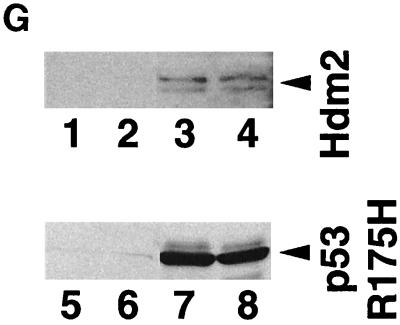
Stable clones of tsBN462 cells were isolated at the permissive temperature following transfection with vectors that confer G418 resistance and express TAFII250, Hdm2, Hdm2 G58A, Hdm2 V75A, or nothing from the control empty vector. Following selection with G418, at least six independent clones were picked, expanded, and analyzed for cell growth (Fig. 7A and B), cell cycle distribution (Fig. 7C and D), and BrdU incorporation (Fig. 7E). The results for several representative clones are shown in the figures. The clones were shown to express the corresponding exogenous proteins by immunocytochemistry (Fig. 7F) or Western blotting (Fig. 7G) with antibodies specific for the exogenous proteins (Hdm2, IF2; p53 R175H, DO1; TAFII250, anti-HA).
FIG. 7.
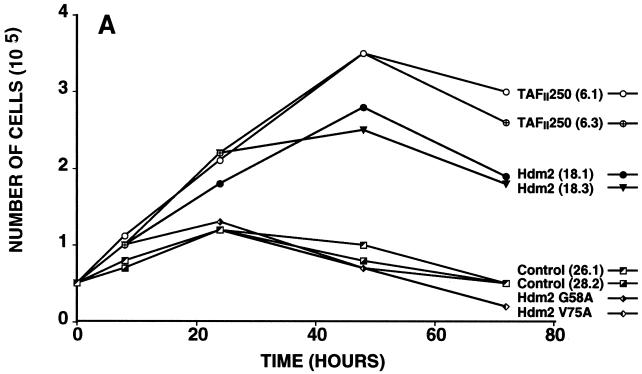
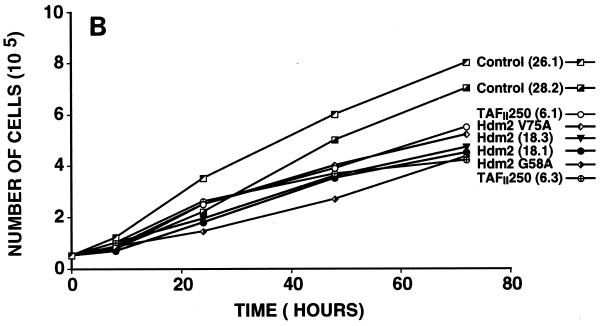
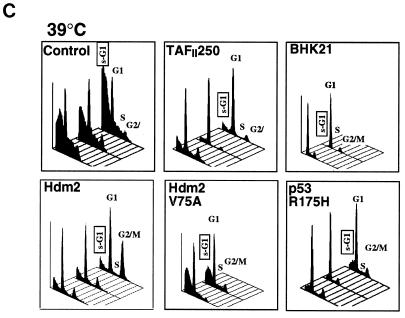
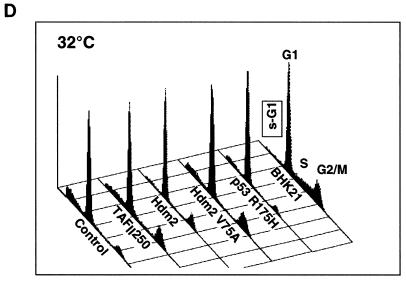
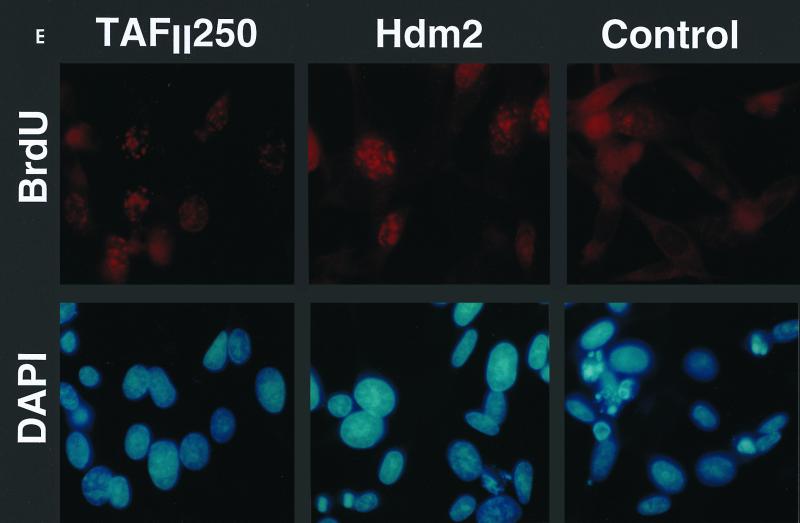
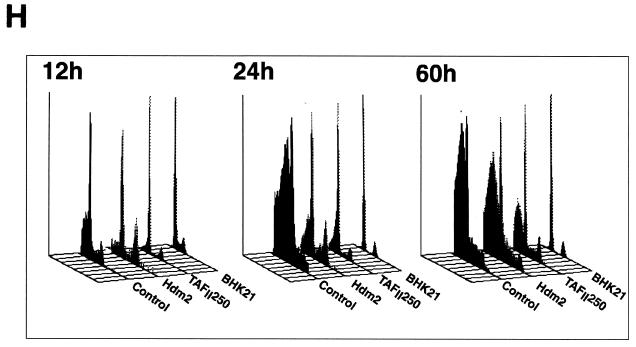
Effect of TAFII250, Hdm2, and p53 R175H expression on cell growth, cell cycle distribution, DNA synthesis, and long-term survival. Stable G418-resistant clones were established that express TAFII250, Hdm2, Hdm2 G58A, Hdm2 V75A, or p53 R175H or contain the empty pCMV expression vector. (A and B) The cells were plated in six-well dishes (5 × 104 per well) and incubated at 32°C for 2 h before being incubated at either 39°C (A) or 32°C (B). At the indicated times, the cells were collected by trypsinization and viable cells (that exclude trypan blue) were counted in a Bürker cell. The clone numbers are indicated in parentheses. (C and D) Exponentially growing cells were incubated at 39°C (C) or 32°C (D) for 24 h and analyzed by flow cytometry. The scans for independent clones are shown in panel C (Control, clones 26.1, 28.2, and 28.3; TAFII250, clones 6.1, 6.2, and 6.3; BHK21, two plates analyzed separately; Hdm2, clones 18.1, 18.2, and 18.3; Hdm2 V75A, clones 24.1 and 23.1; p53 R175H, clones 12.1, 12.2, and 11.1). One representative clone for each vector is shown in panel D. s-G1, sub-G0/G1 (boxed and rotated); G1, G0/G1. (E) The clones were incubated for 12 h at 39°C, labeled with BrdU for 1 h, fixed, and incubated with anti-BrdU (Becton-Dickinson) and Texas red anti-mouse (Jackson) antibodies, and DAPI. Fluorescent cells in 10 different fields containing 10 to 50 cells were counted. The proportions of BrdU-positive cells were as follows: TAFII250 (clones 6.1 and 6.3), 33% ± 5%; Hdm2 (clones 17.2 and 18.2), 35% ± 4%; Control (clones 27.2 and 28.3), 4% ± 2%. Representative photographs are shown. (F) Cells were fixed, stained with specific antibodies (Hdm2, IF2; p53R175H, DO1; TAFII250, anti-HA, 12CA5) followed by Cy3-labeled secondary antibodies and DAPI (nuclei). The cells were photographed under a fluorescence microscope. Control clones incubated with the specific antibodies and labeled secondary antibodies gave low levels of background fluorescence (not shown), as expected for antibodies that are specific for human or viral proteins. One representative clone for each is shown. (G) Western blots of clones established with the selectable marker alone (lanes 1, 2, 5, and 6) or with expression vectors for Hdm2 (lanes 3 and 4) or p53R175H (lanes 7 and 8) and probed with antibodies against Hdm2 (Ab-1, Calbiochem OP46 [lanes 1 to 4]) or p53 (DO1 [lanes 5 to 8]) followed by enhanced chemiluminescence (ECL) (Amersham). (H) Exponentially growing cells were incubated at 39°C for 12, 24, and 60 h and analyzed by flow cytometry. One representative clone for each vector is shown.
For the growth experiments, equal numbers of cells were plated and incubated at 39°C (Fig. 7A) or 32°C (Fig. 7B), and at different times the viable cells were counted. At the nonpermissive temperature, there was a clear difference in the growth of the TAFII250 and Hdm2 cells compared to controls (Fig. 7A). The number of TAFII250 and Hdm2 cells continued to increase up to 48 h, whereas the number of control cells (empty vector, Hdm2 G58A, and Hdm2 V75A) increased up to 24 h and then decreased. These differences were not observed at 32°C, where all the cells grew at similar rates (Fig. 7B), showing that the effects were observed only at the nonpermissive temperature. Hdm2 and Mdm2 clones behaved similarly (data not shown).
For flow cytometry, the clones were analyzed after 24 h at the nonpermissive (Fig. 7C) or permissive (Fig. 7D) temperature. For each group, the cell cycle profiles for the different clones were very similar but not identical, as expected from their independent origin (Fig. 7C). The control clones at the nonpermissive temperature had a large proportion of cells in the sub-G0/G1 range (average, 55% ± 10%), indicating extensive apoptosis. The cell cycle profiles of the TAFII250 and Hdm2 clones showed a clear difference from those of the controls, with significantly fewer cells in the sub-G0/G1 population (17% ± 2% for TAFII250 and 24% ± 2% for Hdm2). BHK21, the parental cells for tsBN462, also had a smaller sub-G0/G1 population (4% ± 2%), as expected for cells expressing natural levels of wild-type TAFII250. Clones expressing Hdm2 with a mutation in the p53 binding site (V75A) had more sub-G0/G1 cells than did the Hdm2 clones (sub-G0/G1 population, 40% ± 5% compared to 24% ± 2%). Clones expressing mutant p53 (R175H) had fewer cells in the sub-G0/G1 population (19% ± 5%) than did the control clones. These results show that cells stably expressing Hdm2, TAFII250, and trans-dominant mutant p53 R175H are less sensitive to apoptosis (as measured by sub-G0/G1 cells) induced by incubation at the nonpermissive temperature, apparently through a p53-dependent mechanism. The effects were shown to be specific for the nonpermissive temperature. The different clones gave similar profiles, with low levels of sub-G0/G1 cells at the permissive temperature (Fig. 7D) (only one representative clone of at least three tested is shown; the proportion of cells in the sub-G0/G1 population were as follows: control, 14% ± 6%; TAFII250, 7% ± 3%; Hdm2, 9% ± 2%, Hdm2 V75A, 13% ± 3%; p53 R175H, 8% ± 3%, BHK21, 4% ± 2%).
DNA synthesis was measured by BrdU incorporation for 1 h, 12 h after the shift to the nonpermissive temperature. Cell morphology was examined by DAPI staining and fluorescence microscopy. The TAFII250 and Hdm2 clones had BrdU-positive nuclei with a normal appearance (representative fields are shown in Fig. 7E). In contrast, there were very few BrdU-positive cells in the control clones, with many condensed and fragmented nuclei detected by DAPI staining. The proportions of BrdU-positive cells were, on average, 33% ± 5% for TAFII250, 35% ± 4% for Hdm2, and 4% ± 2% for the controls. These results show that Mdm2 and p53 R175H expression can significantly reverse the temperature-sensitive phenotype of tsBN462 cells due to mutated TAFII250.
TAFII250 is expected to be more efficient than Hdm2 at rescuing the temperature-sensitive phenotype after longer incubation, since exogenous TAFII250 compensates for the defective endogenous protein whereas Hdm2 corrects for the loss of one particularly sensitive function, production of Hdm2. We used flow cytometry to monitor the effects of incubation for different times at the nonpermissive temperature. The sub-G0/G1 population was clearly detectable for the control clone after 12 h at 39°C and continued to increase after 24 and 60 h. It was detectable after 24 h for the Hdm2 clone and increased afterward, whereas it increased more slowly for the TAFII250 clone. Similar results were obtained with other clones and in growth experiments (data not shown). Clones expressing exogenous TAFII250 were not as resistant as the parental cell line, BHK21, to incubation at the nonpermissive temperature. Exogenous TAFII250 may not completely compensate for the defect in TAFII250 for a number of reasons (expression level, inefficient incorporation into a higher-order complex [TFIID], presence of defective TAFII250, etc.). These results show that Hdm2 is less efficient than TAFII250 at rescuing the temperature-sensitive phenotype, as expected from its ability to compensate for only one (but important) defect resulting from the inactivation of TAFII250.
Delayed kinetics of Mdm2 synthesis due to the sensitivity of the Mdm2 promoter to TAFII250 inactivation.
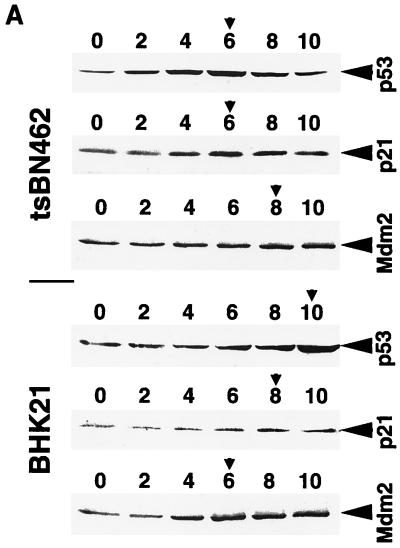
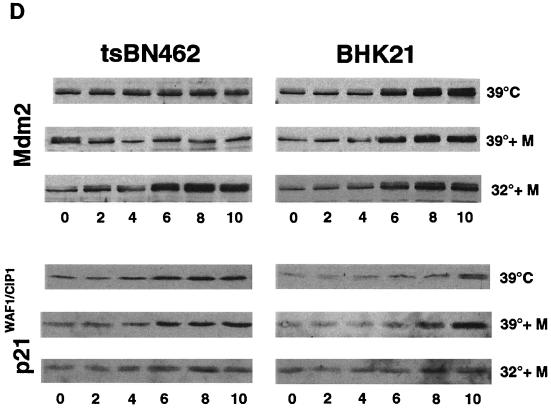
Our results suggest that endogenous Mdm2 expression is impaired in tsBN462 cells, such that p53 induction by heat at the nonpermissive temperature is not followed by adequate Mdm2 synthesis, resulting in excess p53 activity, cell cycle arrest, and apoptosis. To test this hypothesis, we transferred tsBN462 and BHK21 parental cells to the nonpermissive temperature and, at 2-h intervals, analyzed cell extracts by Western blotting for the expression of p53, Mdm2, and the product of another p53-responsive gene, p21WAF1/CIP1. Equal loading was verified by Ponceau S staining of membranes and blotting of RNA polymerase II large subunit (data not shown). We found that p53 levels rose rapidly in tsBN462 cells, reaching a maximum by 6 h (Fig. 8A; compare the zero time point with the peak of protein expression indicated by the arrowhead). p21WAF1/CIP1 levels followed p53 levels closely, also peaking at 6 h. In contrast, there was no significant increase in Mdm2 levels (similar results were obtained with two different antibodies against Mdm2, 2A10 and SMP14 [data not shown]). The kinetics were different in BHK21 cells, where p53 levels rose more slowly. The p53 level increased only slightly after 6 h, compared to the starting level, and continued to increase after 8 and 10 h. p21WAF1/CIP1 increase was also delayed, rising after 8 and 10 h. Strikingly, Mdm2 levels rose after 4 h and reached a maximum after 6 h, significantly faster than in tsBN462 cells. The faster rise in Mdm2 would account for the slower increase in p53 and p21WAF1/CIP1 in BHK21.
FIG. 8.
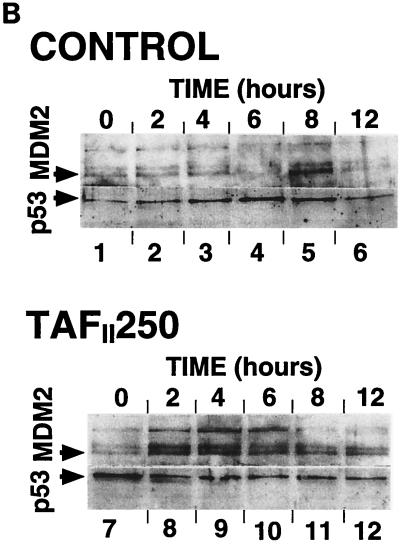
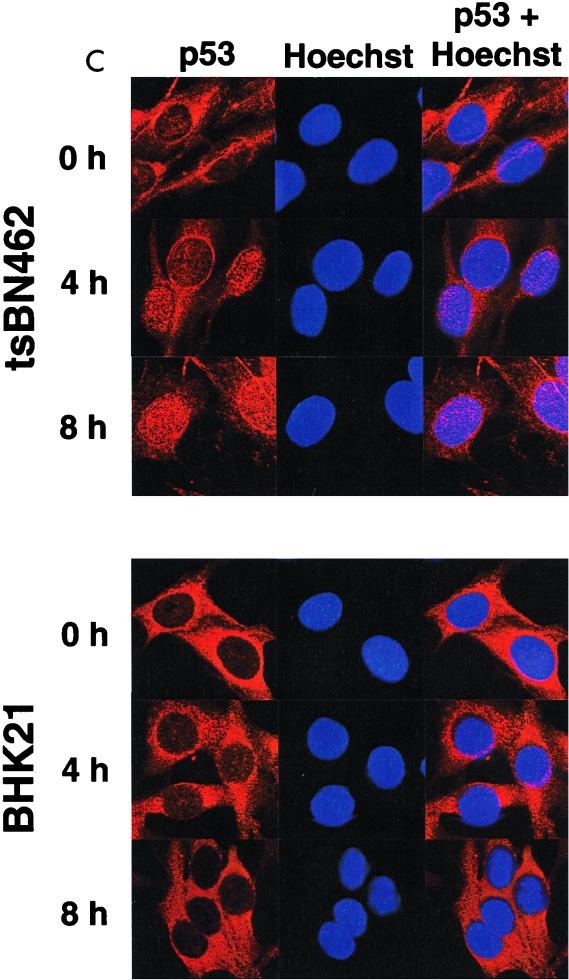
p53, p21WAF1/CIP1, and Mdm2 protein expression in tsBN462 and BHK21. (A) Exponentially growing tsBN462 and BHK21 cells were transferred to 39°C. At the indicated times (0, 2, 4, 6, 8, and 10 h), cell extracts were prepared by lysis in loading buffer and analyzed by SDS-PAGE (8% polyacrylamide), Western blotting, and ECL (Amersham), as described previously (33). The antibodies used were as follows: p53, PAb 240; p21WAF1/CIP1, C-19-G (sc-397-G [Santa Cruz]); Mdm2, 2A10 (similar results were obtained with SMP14 [data not shown]). Arrows pointing to the left indicate specific bands with the expected mobility. Arrowheads pointing down indicate the time point with the maximum level of expression. Ponceau S staining of the membranes and blotting for RNA polymerase II large subunit (a gift from M. Vigneron) were used to verify equal loading (not shown). (B) tsBN462 cells were transfected in 9-cm dishes with vectors that express HOOK (1 μg pHOOKTM-1) and either TAFII250 (10 μg) or nothing (Control) (10 μg). Transfected cells were isolated with the Capture-Tec kit (Invitrogen), replated, cultivated for 18 h, and then induced by heat shock at 39°C. Samples were collected at different times (0, 2, 4, 6, 8, and 12 h) and analyzed by SDS-PAGE (8% polyacrylamide), Western blotting, and ECL to detect Mdm2 (SMP14) and p53 (DO1). Ponceau S staining of the membranes was used to verify equal loading (not shown). (C) tsBN462 and BHK21 cells were transferred to 39°C, and at different times (0, 4, and 8 h) the cells were fixed, stained, and analyzed by confocal microscopy, as described previously (63). p53 was revealed with a rabbit antibody (no. 588) followed by Cy3-labeled goat anti-rabbit antibody (Jackson). Similar localizations were observed with PAb 240. Hoechst stains nuclei. Typical fields are shown. (D) Exponentially growing tsBN462 and BHK21 cells were incubated at 39°C or 32°C in the absence or presence (+M) of mitomycin C (5 μg/ml). At the indicated times (0, 2, 4, 6, 8, and 10 h), cell extracts were analyzed by SDS-PAGE (8% polyacrylamide) and Western blotting (antibodies: p21WAF1/CIP1, C-19-G [sc-397-G]; Mdm2, 2A10) and ECL. Protein quantification (Bio-Rad protein assay), Ponceau S staining of the membranes, and blotting for RNA polymerase II large subunit (a gift from M. Vigneron) were used to verify equal loading (not shown).
We also tested whether expression of exogenous wild-type TAFII250 in tsBN462 would rescue Mdm2 expression. tsBN462 cells were transfected with pHOOK-1 (Invitrogen) and either the control or TAFII250 expression vectors. Transfected cells were selected by the Capture-Tec kit (Invitrogen) and plated, the temperature was raised to 39°C, and samples were taken at different times and analyzed for the expression of Mdm2 and p53 by Western blotting (Fig. 8B). In control cells, transfected with the empty vector, the kinetics were similar to those described above (Fig. 8A), with Mdm2 levels increasing after 8 h (lane 5) and p53 levels peaking at 6 h (lane 4). In TAFII250-transfected cells there was a rapid increase in Mdm2 levels, detectable by 2 h and reaching a maximum after 4 h (lane 9). These results show that restoring wild-type TAFII250 activity in tsBN462 cells stimulates endogenous Mdm2 expression. The kinetics of Mdm2 expression in the TAFII250-transfected cells was faster than in BHK21 cells, showing that the conditions in transfected cells did not entirely reproduce the conditions in BHK21 cells. However, the results with the transfected cells help confirm that the difference between tsBN462 and BHK21 cells is due to TAFII250 and Mdm2.
p53 is negatively regulated by Mdm2 by a mechanism involving nuclear export of p53 and degradation in the cytoplasm. The delay in Mdm2 synthesis in tsBN462 cells is expected to result in a rise in nuclear p53 levels in tsBN462 compared to BHK21 cells. To test this hypothesis, tsBN462 and BHK21 cells were incubated at the nonpermissive temperature and, at various times, the cells were fixed, stained with rabbit anti-p53 antibodies (no. 588) followed by Cy3-conjugated goat anti-rabbit (Jackson) and Hoechst to reveal nuclei, and analyzed by confocal microscopy (Fig. 8C). p53 was predominantly cytoplasmic before incubation at the nonpermissive temperature (0 h) in both cell types. p53 accumulated in the nuclei of tsBN462 cells, such that after 4 h, most of the cells had detectable levels of nuclear p53 (65% ± 5%, estimated by counting at least 200 cells in different fields) that persisted after 8 h (75% ± 5%). In contrast, in BHK21 cells there was no detectable increase in the level of nuclear p53, even after 8 h at the nonpermissive temperature. Similar results were obtained with a different anti-p53 antibody (PAb 240 [data not shown]). These results show that there is an increase in the level of nuclear p53 in tsBN462 cells that is not observed in BHK21 cells, as would be expected from reduced Mdm2 synthesis in the TAFII250 mutant cells.
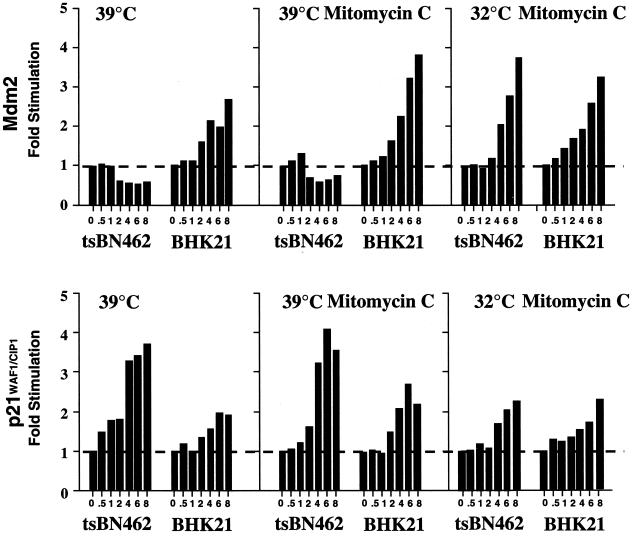
Heat shock may affect p53 properties in a different manner from that of other inducers. We studied the effects of a more established p53 activator, mitomycin C, on both protein (Fig. 8D) and RNA (see below) expression. We transferred tsBN462 and BHK21 cells to the nonpermissive temperature, treated them with mitomycin C, and analyzed cell extracts by Western blotting for Mdm2 and p21WAF1/CIP1. Mdm2 levels remained constant in tsBN462 cells, similar to those in cells that were not exposed to the DNA-damaging agent (Fig. 8D, compare 39°C and 39°C + M). Mdm2 levels increased in BHK21 cells with similar kinetics whether or not mitomycin C was present. At 32°C, mitomycin C induced Mdm2 in tsBN462 as well as BHK21 cells, showing that Mdm2 can be induced in the temperature-sensitive cell line. Mitomycin C did not increase p21WAF1/CIP1 induction in tsBN462 at 39°C. In BHK21 cells, p21WAF1/CIP1 was induced to slightly higher levels but the kinetics were still significantly slower than in tsBN462 cells under the same conditions. Comparing 32 with 39°C, in tsBN462 cells p21WAF1/CIP1 was less inducible by mitomycin C, as expected from the greater induction of Mdm2 at 32°C and consequent inhibition of p53. In BHK21 cells, p21WAF1/CIP1 was also less inducible by mitomycin C at 32°C despite the induction of Mdm2 under all conditions, suggesting that it resulted from a lack of the additional effect of heat shock. These results show that a classical activator of p53 is not capable of inducing Mdm2 in tsBN462 at the nonpermissive temperature and indicate that TAFII250 inactivation rather than the mechanism of p53 induction influences Mdm2 activation.
TAFII250 is a transcription factor, raising the possibility that the response of the mdm2 gene promoter to p53 is particularly sensitive to inactivation of TAFII250 compared to other p53-responsive promoters. To test this possibility, we analyzed RNA transcribed from the mdm2 and p21WAF1/CIP1 genes by Northern blotting (Fig. 9) and studied promoter activity by transfection of reporters (Fig. 10). tsBN462 and BHK21 cells were incubated at the nonpermissive temperature (39°C), and RNA was analyzed by Northern blotting (Fig. 9). The level of mdm2 RNA slightly but reproducibly decreased in tsBN462 cells whereas it increased in BHK21 cells over the 8-h period of analysis. In contrast, the level of p21WAF1/CIP1 RNA increased to a greater extent in tsBN462 than in BHK21 cells. Under more traditional conditions of p53 induction with mitomycin C, comparable patterns of mdm2 and p21WAF1/CIP1 induction were observed. mdm2 levels did not increase over the time course of the experiment in tsBN462 cells, even though there were slightly higher inductions of mdm2 in BHK21 cells and p21WAF1/CIP1 in both cell lines. At the permissive temperature (32°C), mdm2 was induced by mitomycin C in tsBN462 cells to a similar extent to that in BHK21 cells. Furthermore, p21WAF1/CIP1 induction was similar in both cell lines.
FIG. 9.
Mdm2 and p21WAF1/CIP1 RNA expression in tsBN462 and BHK21 cells. tsBN462 and BHK cells were split and incubated 24 h at 32°C. They were then (zero time point) transferred to 39°C and incubated in the absence or presence of mitomycin C (5 μg/ml) or kept at 32°C and treated with mitomycin C. At the indicated time points (0, 0.5, 1, 2, 4, 8, and 10 h), samples were processed for Northern blotting, hybridized with 32P-labelled Mdm2, p21WAF1/CIP1, and 36B4 (loading control) probes, and quantified by PhosphorImager analysis. The results are presented as fold stimulation relative to the zero time point. One representative experiment out of six is shown.
FIG. 10.
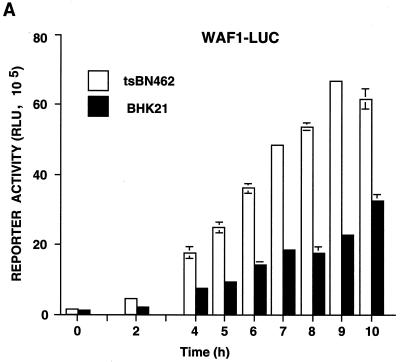
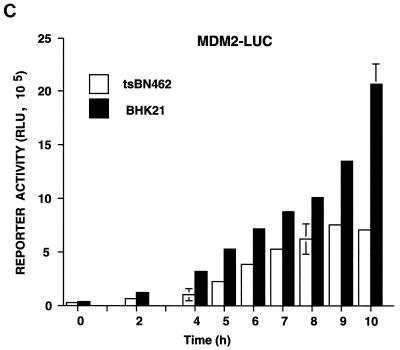
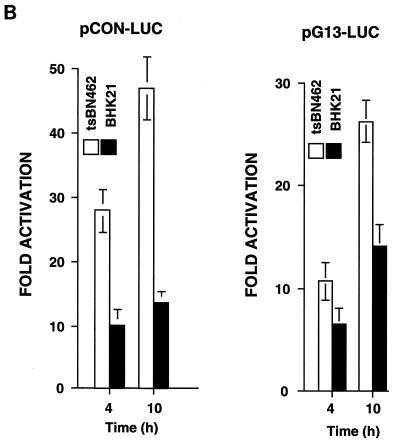
Kinetics of p21WAF1/CIP1, Mdm2, and p53 reporter activity in tsBN462 and BHK21 cells at the nonpermissive temperature. tsBN462 and BHK21 cells were transfected in six-well dishes with either the WAF1-luc (A), pCON-luc, pGUP-luc, pG13-luc, and mG15-luc (B), or Mdm2-luc (C) reporters (1 μg) and the internal control (pSG5-LacZ; 1 μg/ml). After transfection, the cells were transferred to 39°C, and luciferase activities were measured at various times. Luciferase activities at any particular time point were corrected for variations in transfection efficiency using the internal control. Fold activation (B) is the ratio of the activities of the reporters containing (pCON-luc and pG13-luc) and lacking (pGUP-luc and mG15-luc) functional p53 response elements. Similar results were obtained in three separate experiments.
For the promoter analysis, tsBN462 and BHK21 cells were transfected with reporters containing natural promoters from the mdm2 (Mdm2-luc) and p21WAF1/CIP1 (WAF1-luc) genes, as well as artificial promoters with p53-responsive promoters and their controls lacking functional elements (Fig. 10). The transfected cells were shifted to the nonpermissive temperature, and cell extracts were analyzed for luciferase activity at different times. The cotransfected internal control, pSG5-LacZ, was used to correct for variations in transfection efficiency at any particular time point. We found that WAF1-luc reporter activity increased with time in tsBN462 cells, reaching a maximum at around 9 h. Reporter activity was more than half maximum after 6 h, when p53 protein levels peaked (Fig. 10A and 8A). In BHK21 cells, WAF1-luc reporter activity was lower, as expected from the slower kinetics of p53 and p21WAF1/CIP1 induction. The differences were not due to transfection efficiency, since β-Gal activity was similar in both cell types. The p21WAF1/CIP1 promoter is complex, containing elements regulated by a variety of transcription factors besides p53. To measure p53 activity more directly, we used two reporters containing consensus p53 response elements (pCON-luc and pG13-luc) and measured the fold activation relative to that for control reporters lacking the p53 response elements (pGUP-luc and mG15-luc). We found that both reporter pairs gave a higher fold activation in tsBN462 than in BHK21 cells, both 4 and 10 h after transfer to the nonpermissive temperature (Fig. 10B). These results agree with the faster induction of p53 protein (Fig. 8A), nuclear accumulation of p53 (Fig. 8C), and higher WAF1-luc promoter activity (Fig. 10A) in tsBN462 cells. We next investigated Mdm2-luc reporter activity. Strikingly, Mdm2-luc promoter activity was higher in BHK21 than in tsBN462 cells, in apparent contrast with the lower p53 activity. The lower promoter activity is entirely consistent with the relatively lower Mdm2 mRNA and protein levels and faster rise in p53 activity in tsBN462 cells (due to the effect of the loop) and with the ability to rescue the temperature-sensitive phenotype by Mdm2 expression. They show that the mdm2 gene promoter is more sensitive to TAFII250 mutation than is another p53-responsive promoter, from the p21WAF1/CIP1 gene.
DISCUSSION
We have shown that Mdm2 can prevent apoptosis and cell cycle arrest induced in tsBN462 cells by the inactivation of TAFII250 at the nonpermissive temperature. Mdm2 overexpression stimulates cell cycle-dependent promoters by an E2F-dependent mechanism. This activation is most probably a result of inactivation of p53, since it is abrogated by mutation of the p53 binding site of Mdm2, by ARF expression, and by a peptide inhibitor of Mdm2-p53 interactions. Furthermore, p53 inhibition with mutant p53 proteins has the same effect as Mdm2 expression. Mdm2 rescues tsBN462 cells from apoptosis in a p53-dependent manner, as shown by flow cytometry, nuclear condensation, and cell morphology. We found that endogenous Mdm2 synthesis is both delayed and attenuated after TAFII250 inactivation in tsBN462 cells, compared to the same cells expressing exogenous wild-type TAFII250 or the parental BHK21 cells with wild-type TAFII250. In contrast, p53 synthesis is enhanced, as expected from the delayed synthesis of Mdm2. Finally, the Mdm2 promoter is less responsive than the p21WAF1/CIP1 promoter to p53 induction by heat in cells containing heat-sensitive TAFII250, whereas it is more responsive in the equivalent cells with wild-type TAFII250. Our results suggest the following sequence of events. Heat treatment of tsBN462 cells leads to the activation of p53 and inhibition of mutant TAFII250. The rising p53 levels do not efficiently induce Mdm2 synthesis, due to the decreased activity of the mdm2 gene promoter compared to that in cells expressing wild-type TAFII250. The levels of p53 continue to increase, resulting in the inhibition of cell cycle-dependent promoters through their cell cycle-dependent elements, cell cycle arrest and apoptosis. These results indicate that the p53-Mdm2 autoregulatory loop is sensitive to a transcription defect in the cell.
We found that Mdm2 expression stimulated three of the five promoters that are activated at different stages of the cell cycle. The c-fos promoter belongs to the immediate-early class that is rapidly activated by addition of serum to quiescent cells (58). The cyclin D1 promoter is most notably regulated by serum and growth factors (20). Interestingly, the cyclin D1 promoter is down-regulated by heat inactivation of TAFII250 (47, 50, 51, 56), indicating that Mdm2 may not directly overcome some consequences of TAFII250 inhibition. However, restoration of cyclin D levels appears to be less critical than inhibition of p53 for cell survival. The three promoters that are activated by Mdm2 are all controlled by E2F or related factors (11, 18, 68). Mdm2 appears to affect promoter activity through an E2F-dependent mechanism since the E2F-regulated elements of each promoter are required for the activation, a reporter containing only E2F elements is regulated in the same manner, and a trans-dominant inhibitor of E2F blocks the effects of Mdm2. The precise molecular mechanisms by which E2F is regulated by Mdm2 remain to be defined.
The DNA binding component of E2F is composed of heterodimers between the products of six E2F genes and two DP genes. These heterodimers form higher-order complexes, most notably with the three members of the retinoblastoma family, pRb, p107, and p130. A number of these factors have been implicated in the regulation of the B-myb, cyclin A, and cdc25C promoters (8, 19, 45, 48, 65). These promoters are also regulated by less well defined factors, notably CDF-1 (35). It was possible that Mdm2 targets some of these factors directly. Mdm2 interacts physically and/or functionally with pRb (23, 64), p107 (10), and E2F1-DP1 (37). The cell cycle-regulated elements of the B-myb, cyclin A, and cdc25C promoters are bipartite and more complex than minimal E2F DNA binding sites. We found that mutating the cdc25C CHR had a greater effect than Mdm2 expression on cdc25C promoter activity whereas mutation and Mdm2 expression had quantitatively similar effects on the other elements studied, raising the possibility that Mdm2 does not simply target a common component for all the promoters. Although we cannot exclude that some of the effects of Mdm2 are due to a direct effect on pRb and its interactions with E2F (64), our data indicate that the predominant effects of Mdm2 in this system are indirect and a consequence of p53 inactivation.
Mdm2 activation of the cell cycle-dependent promoters, as well as its other effects, appear to essentially result from inactivation of wild-type p53 expressed by tsBN462 cells (49). Deletion and point mutations in the p53 binding site of Mdm2 abolish its activity. These mutations leave intact the interaction sites for both pRB (23) and TAFII250 (33). Deletion of the C-terminal domain of Mdm2, which interacts with TAFII250 (33) and is involved in ubiquitination of p53 (22), is not sufficient to block the effects of Mdm2 (data not shown). Down-regulation of Mdm2 inhibition of p53, with either a peptide inhibitor of p53-Mdm2 interactions (63) or the tumor suppressor ARF, inhibit the effects of Mdm2. Finally, trans-dominant mutant p53s, which are thought to inhibit by contacting DNA through another protein, have effects similar to Mdm2. It is reasonable that E2F acts as a downstream effector of p53. We found that E2F reporter activity is down-regulated by exogenous p53 expression in tsBN462 cells (data not shown). These results are in agreement with previous studies showing that E2F is inhibited by p53 by both direct physical interactions (32, 43, 55) and indirectly through the cyclin-dependent protein kinase inhibitor p21WAF1/CIP1 (9).
The effects of TAFII250 inactivation in tsBN462 cells, as well as the similar tsBN13 cell line reported in other studies, are compatible with our findings. Heat treatment of these cells induces apoptosis (49), decreases cyclin D and cyclin A levels, and induces p21WAF1/CIP1 (47, 51, 56, 62). Inhibition of the cyclin D1 and cyclin A promoters was found to depend on both the core and upstream activator sequences. It is reasonable that p53 intervenes in at least some of these effects. Heat shock is a known inducer of p53. We found that a classical activator of p53, mitomycin C, did not alter the response to elevated temperature, as expected if heat alone was sufficient to efficiently activate p53. p53 activation is known to lead to induction of p21WAF1/CIP1, cell cycle arrest, and apoptosis. Interestingly, tsBN462 cells have recently been reported to be partially rescued by overexpression of D-type cyclins, E2F1, simian virus 40 large T antigen, and HPV16 E7, but the cells still undergo apoptosis (50). We found that Mdm2 blocks apoptosis whereas release of p53 by an inhibitor of interactions with Mdm2 stimulated apoptosis. These results indicate that apoptosis in tsBN462 cells is due to p53. The partial rescue by cyclin D, E2F1, simian virus 40 large T antigen, and HPV16 E7 could compensate for defects due to TAFII250 inactivation or could feed into the pathways affected by p53, or both. D-type cyclins in complex with cyclin-dependent kinases are known to phosphorylate and inhibit pRb, resulting in activation of E2F. E2F1 overexpression activates the cyclin A promoter, indicating one manner by which pRb pathway components could compensate for activation of p53.
TAFII250 has been considered to be a general transcription factor, yet its inactivation in tsBN462 cells has rather specific effects that are limited to a few genes (17, 34, 61). This may reflect the nature of the mutation, G690D, which may have a partial effect on this large multifunctional protein. The yeast homologue, TAFII145, plays a specialized role in transcription of G1/S genes and does not function as a general coactivator (54, 59). TAFII250 may also have predominant effects on the cell cycle.
The Mdm2-p53 autoregulatory loop plays a critical role in restraining the activity of p53. The importance of Mdm2 for inhibition of p53 activity is demonstrated by the rescue of the lethal phenotype of Mdm2−/− mice by inactivation of p53 (27, 40). A variety of cellular stresses increase p53 protein levels and its transcriptional activity. p53 then mediates both growth arrest and apoptosis. Little is known about the mechanisms leading to the choice between whether the cell undergoes apoptosis or stops growing. Our results indicate that a critical determinant could be a functional transcriptional response mediated by the p53-Mdm2 loop, which is sensitive to a transcriptional defect. Interestingly, the transcriptional step in the p53-Mdm2 loop is also targeted by the E1A oncoprotein. E1A activates p53, which selectively induces target genes such as p21WAF1/CIP1 but not Mdm2, leading to cell death. Mdm2 is not induced, since E1A titrates out p300, which is required for p53 regulation of Mdm2 expression (57). Transcription is a vital function, whose defects can lead to a variety of pathological consequences. Our results raise the possibility that the p53-Mdm2 loop may even be a general sensor of a variety of transcriptional defects in the cell.
ACKNOWLEDGMENTS
We thank M. Argentini, J. Bos, K. W. Chang, H. Gronemeyer, S. Korsmeyer, N. La Thangue, T. Lèveillard, A. Levine, Y. Lutz, R. Muller, M. Oren, J. Shay, L. Tora, M. Vigneron, B. Vogelstein, and W. Yarbrough for the gift of recombinants and antibodies; J.-L. Vonesch for expertise and help with confocal microscopy; and the IGBMC facilities staff for invaluable help.
We thank BioAvenir (Rhone-Poulenc), the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, the Ligue Nationale Française contre le Cancer, the Ligue Régionale (Haut-Rhin) contre le Cancer, and the Ligue Régionale (Bas-Rhin) contre le Cancer for financial assistance.
REFERENCES
- 1.Argentin, M., N. Barboule, and B. Wasylyk. The contribution of the RING finger domain of Mdm2 to cell cycle progression. Oncogene, in press. [DOI] [PubMed]
- 1a.Attardi L D, Jacks T. The role of p53 in tumour suppression: lessons from mouse models. Cell Mol Life Sci. 1999;55:48–63. doi: 10.1007/s000180050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates S, Vousden K H. Mechanisms of p53-mediated apoptosis. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 4.Chang K W, Laconi S, Mangold K A, Hubchak S, Scarpelli D G. Multiple genetic alterations in hamster pancreatic ductal adenocarcinomas. Cancer Res. 1995;55:2560–2568. [PubMed] [Google Scholar]
- 5.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conseiller E, Debussche L, Landais D, Venot C, Maratrat M, Sierra V, Tocque B, Bracco L. CTS1: a p53-derived chimeric tumor suppressor gene with enhanced in vitro apoptotic properties. J Clin Investig. 1998;101:120–127. doi: 10.1172/JCI1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. . (Erratum, 15:5846–5847.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimri G P, Nakanishi M, Desprez P Y, Smith J R, Campisi J. Inhibition of E2F activity by the cyclin-dependent protein kinase inhibitor p21 in cells expressing or lacking a functional retinoblastoma protein. Mol Cell Biol. 1996;16:2987–2997. doi: 10.1128/mcb.16.6.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubs-Poterszman M C, Tocque B, Wasylyk B. MDM2 transformation in the absence of p53 and abrogation of the p107 G1 cell-cycle arrest. Oncogene. 1995;11:2445–2449. [PubMed] [Google Scholar]
- 11.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 12.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 13.Freedman D A, Epstein C B, Roth J C, Levine A J. A genetic approach to mapping the p53 binding site in the MDM2 protein. Mol Med. 1997;3:248–259. [PMC free article] [PubMed] [Google Scholar]
- 14.Freedman D A, Wu L, Levine A J. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haines D S. The mdm2 proto-oncogene. Leuk Lymphoma. 1997;26:227–238. doi: 10.3109/10428199709051772. [DOI] [PubMed] [Google Scholar]
- 17.Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T. The CCG1/TAFII250 gene is mutated in thermosensitive G1 mutants of the BHK21 cell line derived from golden hamster. Gene. 1994;141:267–270. doi: 10.1016/0378-1119(94)90583-5. [DOI] [PubMed] [Google Scholar]
- 18.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 19.Henglein B, Chenivesse X, Wang J, Eick D, Brechot C. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc Natl Acad Sci USA. 1994;91:5490–5494. doi: 10.1073/pnas.91.12.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herber B, Truss M, Beato M, Muller R. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene. 1994;9:1295–1304. . (Erratum, 9:2105–2107.) [PubMed] [Google Scholar]
- 21.Hinds P W, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 22.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh J K, Chan F S, O'Connor D J, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 24.Hupp T R. Regulation of p53 protein function through alterations in protein-folding pathways. Cell Mol Life Sci. 1999;55:88–95. doi: 10.1007/s000180050272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janus F, Albrechtsen N, Dornreiter I, Wiesmuller L, Grosse F, Deppert W. The dual role model for p53 in maintaining genomic integrity. Cell Mol Life Sci. 1999;55:12–27. doi: 10.1007/s000180050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayaraman L, Prives C. Covalent and noncovalent modifiers of the p53 protein. Cell Mol Life Sci. 1999;55:76–87. doi: 10.1007/s000180050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones S N, Roe A E, Donehower L A, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 28.Juven T, Barak Y, Zauberman A, George D L, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- 29.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 30.Kubbutat M H, Ludwig R L, Levine A J, Vousden K H. Analysis of the degradation function of Mdm2. Cell Growth Differ. 1999;10:87–92. [PubMed] [Google Scholar]
- 31.La Thangue N B, Thimmappaya B, Rigby P W. The embryonal carcinoma stem cell Ela-like activity involves a differentiation-regulated transcription factor. Nucleic Acids Res. 1990;18:2929–2938. doi: 10.1093/nar/18.10.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee C W, Sorensen T S, Shikama N, La Thangue N B. Functional interplay between p53 and E2F through co-activator p300. Oncogene. 1998;16:2695–2710. doi: 10.1038/sj.onc.1201818. [DOI] [PubMed] [Google Scholar]
- 33.Leveillard T, Wasylyk B. The MDM2 C-terminal region binds to TAFII250 and is required for MDM2 regulation of the cyclin A promoter. J Biol Chem. 1997;272:30651–30661. doi: 10.1074/jbc.272.49.30651. [DOI] [PubMed] [Google Scholar]
- 34.Liu H T, Gibson C W, Hirschhorn R R, Rittling S, Baserga R, Mercer W E. Expression of thymidine kinase and dihydrofolate reductase genes in mammalian ts mutants of the cell cycle. J Biol Chem. 1985;260:3269–3274. [PubMed] [Google Scholar]
- 35.Liu N, Lucibello F C, Korner K, Wolfraim L A, Zwicker J, Muller R. CDF-1, a novel E2F-unrelated factor, interacts with cell cycle-regulated repressor elements in multiple promoters. Nucleic Acids Res. 1997;25:4915–4920. doi: 10.1093/nar/25.24.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N, Lucibello F C, Zwicker J, Engeland K, Muller R. Cell cycle-regulated repression of B-myb transcription: cooperation of an E2F site with a contiguous corepressor element. Nucleic Acids Res. 1996;24:2905–2910. doi: 10.1093/nar/24.15.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin K, Trouche D, Hagemeier C, Sorensen T S, La Thangue N B, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 38.Medema R H, Wubbolts R, Bos J L. Two dominant inhibitory mutants of p21ras interfere with insulin-induced gene expression. Mol Cell Biol. 1991;11:5963–5967. doi: 10.1128/mcb.11.12.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Momand J, Zambetti G P. Mdm-2: “big brother” of p53. J Cell Biochem. 1997;64:343–352. [PubMed] [Google Scholar]
- 40.Montes de Oca Luna R, Wagner D S, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 41.Nishimoto T, Sekiguchi T, Kai R, Yamashita K, Takahashi T, Sekiguchi M. Large-scale selection and analysis of temperature-sensitive mutants for cell reproduction from BHK cells. Somatic Cell Genet. 1982;8:811–824. doi: 10.1007/BF01543021. [DOI] [PubMed] [Google Scholar]
- 42.O'Brien T, Tjian R. Functional analysis of the human TAFII250 N-terminal kinase domain. Mol Cell. 1998;1:905–911. doi: 10.1016/s1097-2765(00)80089-1. [DOI] [PubMed] [Google Scholar]
- 43.O'Connor D J, Lam E W, Griffin S, Zhong S, Leighton L C, Burbidge S A, Lu X. Physical and functional interactions between p53 and cell cycle co-operating transcription factors, E2F1 and DP1. EMBO J. 1995;14:6184–6192. doi: 10.1002/j.1460-2075.1995.tb00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oren M, Rotter V. Introduction: p53--the first twenty years. Cell Mol Life Sci. 1999;55:9–11. doi: 10.1007/s000180050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Philips A, Huet X, Plet A, Le Cam L, Vie A, Blanchard J M. The retinoblastoma protein is essential for cyclin A repression in quiescent cells. Oncogene. 1998;16:1373–1381. doi: 10.1038/sj.onc.1201655. [DOI] [PubMed] [Google Scholar]
- 46.Piette J, Neel H, Marechal V. Mdm2: keeping p53 under control. Oncogene. 1997;15:1001–1010. doi: 10.1038/sj.onc.1201432. [DOI] [PubMed] [Google Scholar]
- 47.Rushton J J, Steinman R A, Robbins P D. Differential regulation of transcription of p21 and cyclin D1 conferred by TAF(II)250. Cell Growth Differ. 1997;8:1099–1104. [PubMed] [Google Scholar]
- 48.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sekiguchi T, Nakashima T, Hayashida T, Kuraoka A, Hashimoto S, Tsuchida N, Shibata Y, Hunter T, Nishimoto T. Apoptosis is induced in BHK cells by the tsBN462/13 mutation in the CCG1/TAFII250 subunit of the TFIID basal transcription factor. Exp Cell Res. 1995;218:490–498. doi: 10.1006/excr.1995.1183. [DOI] [PubMed] [Google Scholar]
- 50.Sekiguchi T, Nishimoto T, Hunter T. Overexpression of D-type cyclins, E2F-1, SV40 large T antigen and HPV16 E7 rescue cell cycle arrest of tsBN462 cells caused by the CCG1/TAF(II)250 mutation. Oncogene. 1999;18:1797–1806. doi: 10.1038/sj.onc.1202508. [DOI] [PubMed] [Google Scholar]
- 51.Sekiguchi T, Noguchi E, Hayashida T, Nakashima T, Toyoshima H, Nishimoto T, Hunter T. D-type cyclin expression is decreased and p21 and p27 CDK inhibitor expression is increased when tsBN462 CCG1/TAFII250 mutant cells arrest in G1 at the restrictive temperature. Genes Cells. 1996;1:687–705. doi: 10.1046/j.1365-2443.1996.00259.x. [DOI] [PubMed] [Google Scholar]
- 52.Sekiguchi T, Yoshida M C, Sekiguchi M, Nishimoto T. Isolation of a human X chromosome-linked gene essential for progression from G1 to S phase of the cell cycle. Exp Cell Res. 1987;169:395–407. doi: 10.1016/0014-4827(87)90200-x. [DOI] [PubMed] [Google Scholar]
- 53.Sellers W R, Rodgers J W, Kaelin W G., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen W C, Green M R. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 55.Sorensen T S, Girling R, Lee C W, Gannon J, Bandara L R, La Thangue N B. Functional interaction between DP-1 and p53. Mol Cell Biol. 1996;16:5888–5895. doi: 10.1128/mcb.16.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas A, White E. Suppression of the p300-dependent mdm2 negative-feedback loop induces the p53 apoptotic function. Genes Dev. 1998;12:1975–1985. doi: 10.1101/gad.12.13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker S S, Shen W C, Reese J C, Apone L M, Green M R. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 60.Wallace-Brodeur R R, Lowe S W. Clinical implications of p53 mutations. Cell Mol Life Sci. 1999;55:64–75. doi: 10.1007/s000180050270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang E H, Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 62.Wang E H, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wasylyk C, Salvi R, Argentini M, Dureuil C, Delumeau I, Abecassis J, Debussche L, Wasylyk B. p53 mediated death of cells overexpressing MDM2 by an inhibitor of MDM2 interaction with p53. Oncogene. 1999;18:1921–1934. doi: 10.1038/sj.onc.1202528. [DOI] [PubMed] [Google Scholar]
- 64.Xiao Z X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 65.Zerfass K, Spitkovsky D, Schulze A, Joswig S, Henglein B, Jansen-Durr P. Adenovirus E1A activates cyclin A gene transcription in the absence of growth factors through interaction with p107. J Virol. 1996;70:2637–2642. doi: 10.1128/jvi.70.4.2637-2642.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 67.Zwicker J, Lucibello F C, Wolfraim L A, Gross C, Truss M, Engeland K, Muller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwicker J, Muller R. Cell-cycle regulation of gene expression by transcriptional repression. Trends Genet. 1997;13:3–6. doi: 10.1016/s0168-9525(96)30112-1. [DOI] [PubMed] [Google Scholar]