Abstract
The complex reservoir of metabolite-producing bacteria in the gastrointestinal tract contributes tremendously to human health and disease. Bacterial composition, and by extension gut metabolomic composition, is undoubtably influenced by the use of modern antibiotics. Herein, we demonstrate that polymyxin B, a last resort antibiotic, influences the production of the genotoxic metabolite colibactin from adherent-invasive Escherichia coli NC101. Colibactin can promote colorectal cancer through DNA double stranded breaks and interstrand crosslinks. While the structure and biosynthesis of colibactin have been elucidated, chemical-induced regulation of its biosynthetic gene cluster and subsequent production of the genotoxin by E. coli are largely unexplored. Using a multi-omic approach, we identified that polymyxin B stress enhances the abundance of colibactin biosynthesis proteins (Clb’s) in multiple pks+ E. coli strains, including pro-carcinogenic AIEC: NC101, the probiotic strain: Nissle 1917, and the antibiotic testing strain: ATCC 25922. Expression analysis via qPCR revealed that increased transcription of clb genes likely contributes to elevated Clb protein levels in NC101. Enhanced production of Clb’s by NC101 under polymyxin stress matched an increased production of the colibactin prodrug motif, a proxy for the mature genotoxic metabolite. Furthermore, E. coli with heightened tolerance for polymyxin induced greater mammalian DNA damage, assessed by quantification of γH2AX staining in cultured intestinal epithelial cells. This study establishes a key link between the polymyxin B stress response and colibactin production in pks+ E. coli. Ultimately, our findings will inform future studies investigating colibactin regulation and the ability of seemingly innocuous commensal microbes to induce host disease.
Graphical Abstract
INTRODUCTION
Microbiota community structure and repertoire of microbial-derived metabolites in the gastrointestinal (GI) tract carries tremendous implications in human health and disease.1–4 Antibiotics can dramatically shift microbial community dynamics, killing susceptible bacteria and leaving resistant ones to flourish.5,6 Enterobacteriaceae, including Escherichia coli, are often overrepresented in the gut following antibiotic treatment.6,7 E. coli are key influencers of health and disease, with strains ranging from commensal to pathogenic depending on the presence of virulence genes encoded within each strain. The striking influence E. coli have on GI disease has led to a rapidly expanding field focused on characterizing the dynamic relationships between intestinal E. coli metabolites and their impact on host phenotype. Proteomic changes, including the production of virulence factors, can enhance the ability of E. coli to adapt to new niches in the gut, disrupt microbiota homeostasis, and trigger or exacerbate intestinal diseases.8
The bacterial microbiota and associated metabolites of patients with inflammatory bowel diseases (IBD), Crohn’s disease and ulcerative colitis, differ significantly compared to non-IBD counterparts.9–13 Crohn’s disease patients in particular often harbor a high abundance of adherent-invasive E. coli (AIEC) in their microbiota and at the mucosally-adherent niche.14,15 AIEC strains lack typical virulence genes but are defined by their enhanced ability to adhere to and invade intestinal epithelial cells and survive and replicate in macrophages.16 Importantly, AIEC have been linked to the pathogenesis of IBD and inflammation-associated colorectal cancer (CRC).17 CRC patients harbor high levels of mucosally-adherent E. coli18 and a significantly high proportion of CRC patients harbor E. coli strains capable of producing the genotoxin colibactin.4,19
Colibactin is one of the most intriguing microbial-produced metabolites stemming from resident intestinal E. coli strains harboring a 54-kb polyketide synthase gene cluster known as the pks island.20,21 Colibactin alkylates host DNA through two distinct cyclopropane warheads, causing double strand breaks and interstrand crosslinks.22–28 The pks cluster has been shown to induce cell cycle arrest and activate DNA repair machinery in mammalian cells.20,29 In epithelial cells in contact with pks+ E. coli, DNA damage is commonly characterized by γH2AX foci, megalocytosis, and activation of DNA damage signaling cascades.30 pks+ E. coli promote CRC in multiple models,4,31–33 accelerate cancer progression,19 and are enriched in human CRC tissues.4,31 In addition, colibactin-producing E. coli imprint a unique carcinogenic mutational signature in primary tumors and metastases of many human CRC patients.34–36 Taken together, these data provide a direct link between the presence of pks+ E. coli and DNA damage patterns that drive CRC development.
The pks island is a hybrid nonribosomal peptide synthetase – polyketide synthase (NRPS-PKS) gene cluster consisting of nineteen genes (clbA – clbS), eighteen of which are required to elicit genotoxicity.20 Direct measurement of the mature metabolite is extremely difficult due to its instability, however, the expression and abundance of colibactin’s biosynthetic machinery likely contribute greatly to the cellular levels of colibactin and extent of DNA damage in the host. For example, in vivo upregulation of clb genes has been found in urinary tract infections37 and in states of intestinal inflammation and developing cancer.31 Therefore, understanding the regulation of the pks island is key to understanding propensity for genotoxicity.
Recent studies suggest growth conditions are vital to the expression of clb genes. Concentration of iron,38 aeration,21 and media composition39,40 have been shown to impact clb gene regulation in vitro. Microbial factors that mediate these changes in expression include the ferric uptake regulator (Fur) and the small regulatory non-coding RNA RyhB,38,41 polyphosphate kinase (PPK),42 stress-response protein Hsp90,43 and the spermidine biosynthetic cluster.40 Additionally, ClbR was identified as a key transcriptional regulator of the pks island.39 Although several conditions influencing clb gene transcription and colibactin production have been identified, chemical-induced regulation of the biosynthetic machinery is poorly characterized.
The effects of antibiotics on microbial communities undisputedly have significant consequences on the metabolic activity of its inhabitants.44,45 Polymyxin antibiotics are non-ribosomally synthesized cyclic peptides containing a fatty acid tail that facilitates interaction with the outer membrane of Gram-negative pathogens, resulting in increased membrane permeabilization and cell death.46 Polymyxin B (PmB) is one of two polymyxins used clinically, both which are classified as “reserve” antibiotics by the World Health Organization.47 According to the 2017 Medicare Part D Prescriber data, polymyxins are still prescribed for many bacterial infections, notably in internal medicine and patients with chronic lung infections.48
Herein, E. coli strains evolved in the presence of sustained sub-inhibitory pressure with PmB demonstrate increased antibiotic resistance and elevated genotoxicity attributed to colibactin production. Label-free quantitative proteomics was used to characterize the altered proteomic landscape in the evolved E. coli strains and shows increased abundances of colibactin biosynthetic machinery in three separate evolutions of E. coli NC101, a pro-carcinogenic AIEC strain.4,49 Further, increased transcription of the pks island and production of a colibactin prodrug motif are captured in the evolved strains even after antibiotic pressure is released, culminating in heightened genotoxicity confirmed through in vitro γH2AX assays quantifying DNA damage response. Two additional strains of pks+ E. coli were evolved in PmB, and also displayed increased levels of colibactin’s biosynthetic machinery. These findings demonstrate the indirect biomolecular consequences of antibiotic exposure in the GI tract and subsequent deleterious effects on the host.
RESULTS AND DISCUSSION
In vitro evolution of E. coli NC101 with polymyxin B leads to increased resistance
Adherent-invasive Escherichia coli (AIEC) are gut-resident pathobionts that lack traditional virulence factors and are linked to IBD and CRC. E. coli NC101 (NC101) is a pro-carcinogenic AIEC strain harboring the pks island and has become a model organism for studying intestinal inflammation and colibactin-induced genotoxicity.4,31,50 Three separate lineages of NC101 were passaged into sub-inhibitory concentrations of PmB (0.5 μg mL−1). Once significant growth was observed, lineages were inoculated into fresh media containing a 100% increase in PmB concentration. This was performed iteratively until all three lineages grew in 8 μg mL−1 PmB (Figure 1A). Minimum inhibitory concentrations were assessed by broth microdilution assays, and strains exhibited roughly a 4x increase in PmB resistance when compared to the PmB sensitive parent strain (Figure 1B). These evolved lineages are denoted as NC101 PmB008R-[1–3] (Table 1). Growth curves revealed somewhat different growth rates between the evolved strains and WT NC101 (Figure S1).
Figure 1.
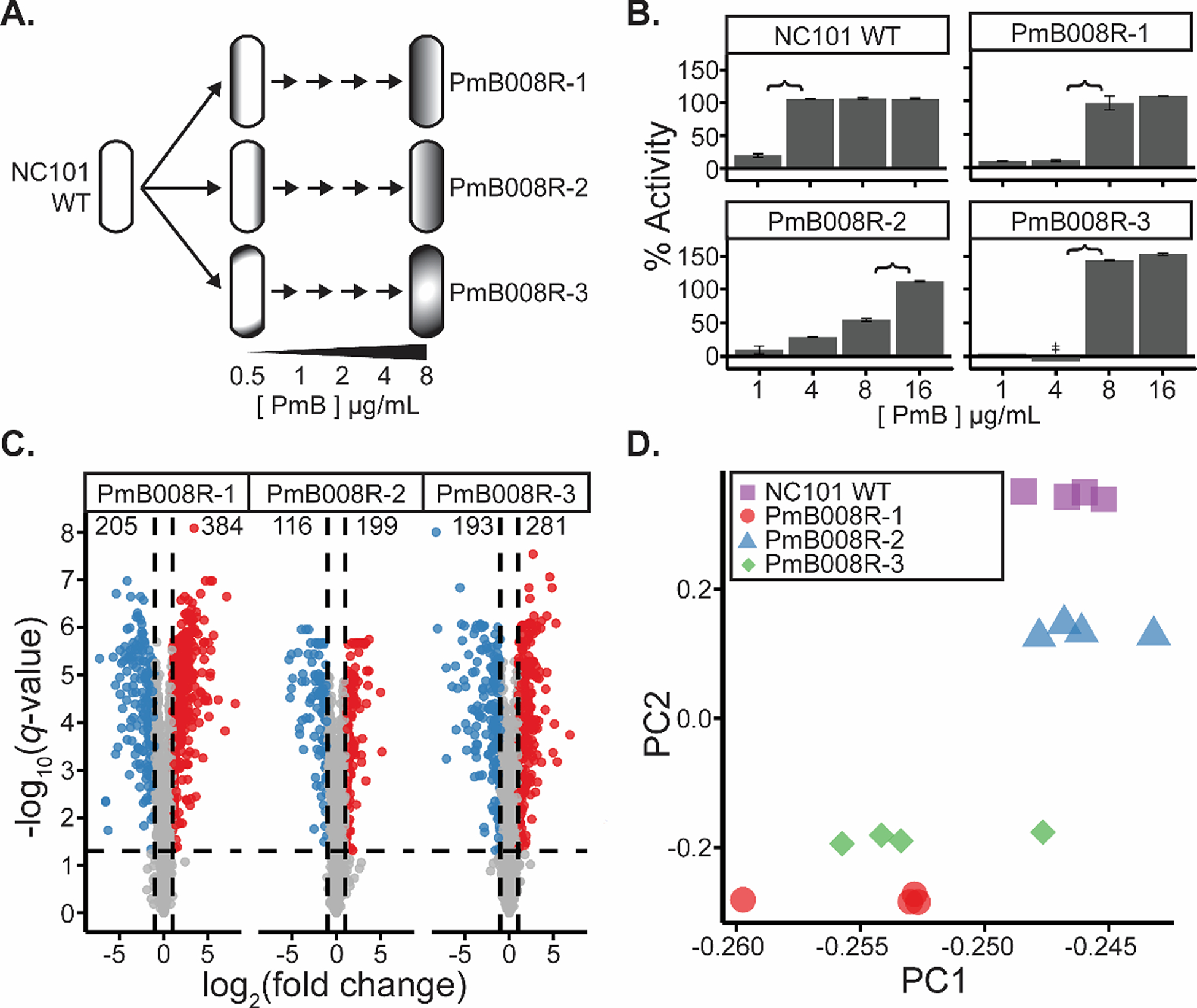
Differentiation of WT and polymyxin B evolved E. coli cultures. a) Schematic of in vitro E. coli evolution. From a parent culture, three lineages of E. coli were evolved with increasing pressure of polymyxin B (PmB). Lineages were first passaged into 0.5 μg mL−1 PmB, and once significant growth was observed, cultures were passaged into fresh media containing a 100% increase in PmB concentration. This was repeated until the three lineages were able to grow in 8 μg mL−1 PmB. These strains were denoted as NC101 PmB008R-[1–3] b) Antibiotic activity assays of NC101 WT and NC101 PmB008R-[1–3] cultures (n = 3) after growth in varying concentrations of PmB (in μg mL−1). % activity represents the efficacy of PmB in each concentration compared to ampicillin and water controls, and is calculated based on formula (1). Error bars represent +/− 1 standard deviation away from the mean value. ‡ represents activity of −66 ± 43%. Brackets represent estimated the expected range of the minimum inhibitory concentration for each of the isolates. c) Volcano plots of proteomic changes in NC101 PmB008R-[1–3] compared to NC101 WT (n = 4). Vertical lines represent the cutoff used to determine significant proteomic changes (|log2(FC)| > 1). Horizontal lines represent the threshold used to determine significant false discovery rates determined by an FDR-corrected two-sided t-test (−log10(q-value) > 1.3). d) Principal component analysis of proteomic replicates of each strain analyzed.
Table 1.
Escherichia coli strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| NC101 Wild-type (WT) | Classical murine-derived model AIEC | Kim, S.C. et al. 2005 |
| NC101 PmB008R-1 | First PmB evolved isolate of NC101 WT | This study |
| NC101 PmB008R-2 | Second PmB evolved isolate of NC101 WT | This study |
| NC101 PmB008R-3 | Third PmB evolved isolate of NC101 WT | This study |
| NC101 Δpks | NC101 strain lacking the entire pks island necessary for colibactin production | Arthur, J.C. et al. 2012 |
| Nissle 1917 | Commonly used probiotic strain | - |
| ATCC 25922™ | Commonly used antibiotic susceptibility testing strain | https://www.atcc.org/products/all/25922.aspx |
To assess dependence of the pks island on the ability of NC101 to evolve to PmB, five independent lineages of NC101 and NC101 Δpks, an isogenic strain lacking the pks island, were evolved in parallel. Each lineage was passaged into a 100% increased concentration of PmB every 24 hours until they could no longer survive. After 12 days of passaging, two of the five NC101 strains were able to grow in 16 μg mL−1 PmB, and three of the five NC101 Δpks strains were able to grow in 16 μg mL−1 PmB (Figure S2). These results suggest that increased PmB resistance is independent of pks expression.
Genomic and proteomic characterization of polymyxin resistant pks+ E. coli
Because lineages evolved and grew at different rates (Figure S1), it was postulated that different mechanisms for resistance were employed and/or genetic variation emerged across strains during serial passaging. Whole genome sequencing revealed distinct genetic mutations in each of the strains (Table 2), suggesting that the strains took separate evolutionary paths when evolving PmB resistance. As such, subsequent analysis of each lineage was treated separately.
Table 2.
Mutations occurring in each of the evolved Escherichia coli lineages.
| Lineage | # mutation sites | Locations |
|---|---|---|
| NC101 PmB008R-1 | 2 |
|
| NC101 PmB008R-2 | 5 |
|
| NC101 PmB008R-3 | 2 |
|
Label-free quantitative (LFQ) proteomics51–57 revealed protein abundance changes in NC101 PmB008R-[1–3] resistant strains compared to the NC101 WT control. Overall, 2,003 proteins were quantified, representing ~41% of the total E. coli proteome (Table S1). When compared to WT, 384, 199, and 281 proteins had significantly higher abundances in NC101 PmB008R-[1–3], while 205, 116, and 193 proteins were at significantly lower abundance in the evolved strains, respectively (Figure 1C). While the differently evolved strains share many common proteomic shifts when compared to NC101 WT, reduction to principle components further emphasizes that distinct differences in protein expression and resistance mechanisms are employed due to genetic variation and/or divergent evolutionary paths in NC101 PmB008R-[1–3] (Figure 1D). Proteins were sorted by function and many known polymyxin resistance mechanisms employed by E. coli were revealed in the data (Figure 2, Table S2).
Figure 2.
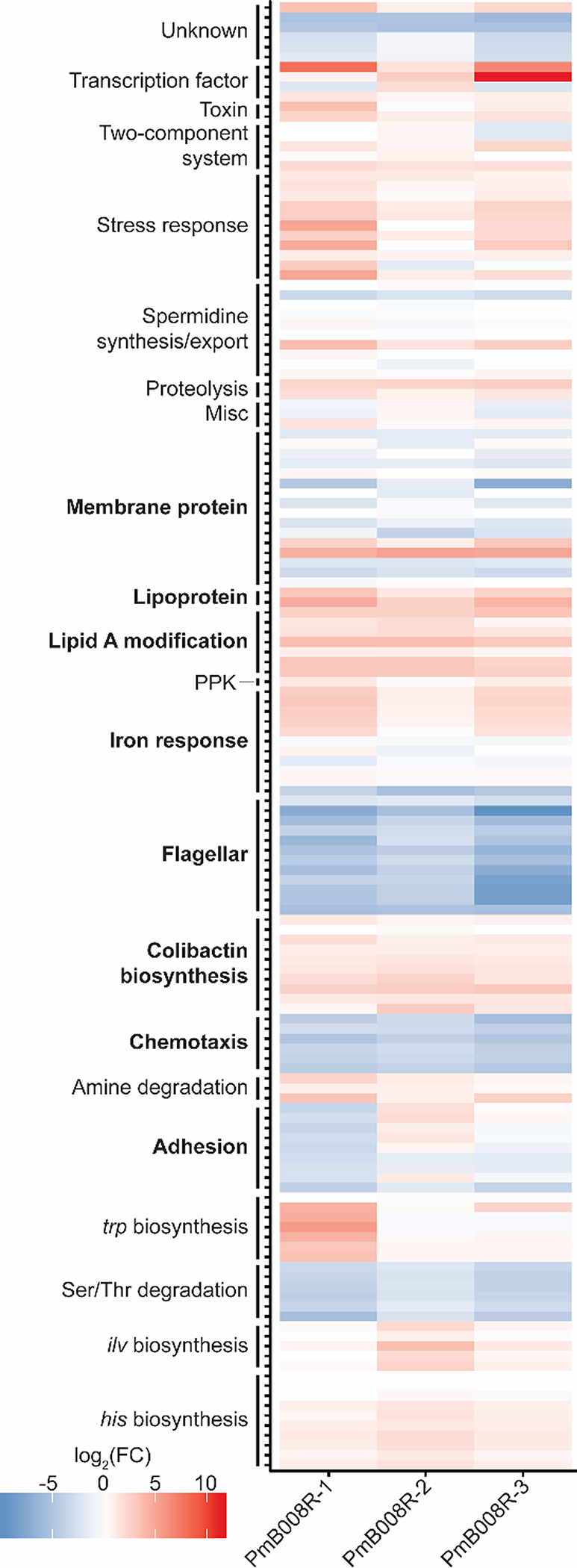
Heatmap showing cellular pathways with significantly changing proteins across evolved strains. Colors depict log2(FC) relative to NC101 WT, as represented in the figure key. Bolded pathways depict proteins important protein FC values determined through manual annotation. A complete list of protein accessions in this table is supplied in Table S2.
Concurrent with commonly identified PmB response mechanisms, proteins involved in Lipid A modification, chemotaxis/flagellar synthesis, cellular adhesion, membrane proteins/lipoproteins, and general stress response proteins were also found significantly changed in this study (Figure 2). Modification of Lipid A, a major component of the bacterial lipopolysaccharide (LPS), is one of the best studied resistance mechanisms of E. coli against PmB. To support previous findings, proteins involved in the attachment of phosphoethanolamine (PEtN) and/or 4-amino-4-deoxy-beta-L-arabinose (L-Ara4N) moieties to the 1’ and 4’ headgroups on lipid A were found in higher abundance for PmB resistant strains in this study (Figure S3). Cellular motility is also commonly affected by antimicrobial resistance. As such, we found a significant decrease in flagellar/chemotaxis-related proteins and decreased motility in the PmB evolved strains. These data link the proteomics results to known physiological effects induced by PmB (Figure S4).
Colibactin regulation in PmB resistant pks+ E. coli
The colibactin pks island consists of 19 genes (Figure 3A), 18 of which are critical for genotoxicity.20 Across the three evolved strains, 10 of the 19 Clb enzymes were quantified via LFQ-proteomics, 9 of which were significantly increased in at least one of the evolved strains (Figure 3B). Significant proteomic upregulation of Clb proteins indicates that PmB resistance activates the pks island. Additionally, we released antibiotic pressure by culturing one evolved strain, PmB008R–2, for three days in media without PmB and observed a persistent increase in Clb enzyme abundance compared to similarly-treated WT NC101 controls (Table S3).
Figure 3.
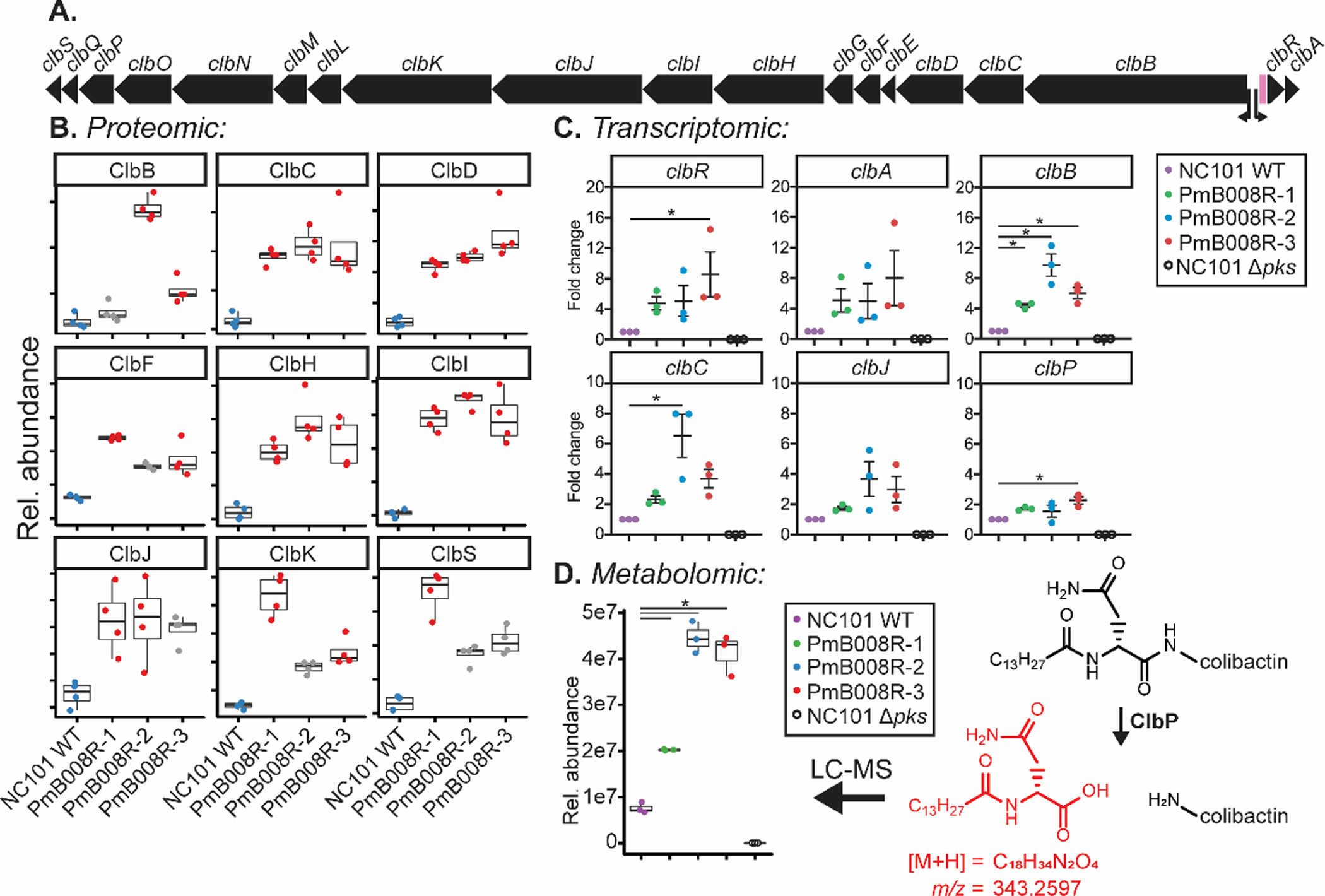
Regulation of the pks island in evolved E. coli lineages. a) Layout of the pks island in the E. coli NC101 genome. Transcription start sites for clbB and clbA are shown with arrows. The variable numbers of tandem repeats (VNTR) region is depicted in pink. b) Relative abundance of pks island gene products that were significantly changing in at least one evolved strain, each dot represents a single replicate (n = 4). Data points in red represent conditions with a significant difference compared to the WT condition (blue). Data points in gray represent conditions that were not significantly changing based on two criteria: log2(FC) > 1 and −log10(q-value) > 1.3 based on an FDR-corrected two-sided t-test. Center lines in the box plots represent mean values for the respective conditions, surrounded by the first and third quartiles. Whiskers extend from the hinge to the highest and lowers values that is within 1.5 * the inner quartiles. c) qPCR data representing transcriptional regulation of clb genes of the WT and evolved strains in the absence of polymyxin B. Each dot represents a single biological replicate (n = 3). Lines are at mean +/− SEM. Significance was noted at p < 0.05 by one-way ANOVA d) Changes in the colibactin prodrug cleavage motif (red) upon colibactin activation by ClbP in the WT and evolved strains in the absence of polymyxin B. Each dot represents a single biological replicate (n = 3). Significance was noted at p < 0.05 by a two-sided t-test. Center lines in the box plots represent mean values for the respective conditions, surrounded by the first and third quartiles. Whiskers extend from the hinge to the highest and lowers values that is within 1.5 * the inner quartiles.
Consistent with upregulation of Clb proteins, a plethora of proteins from cellular pathways previously implicated in colibactin biosynthesis were found differentially regulated in this study, such as iron response proteins,38,41 polyphosphate kinase (PPK),42 and spermidine synthesis/export40 (Figure 2, Table S2). Intriguingly, the Ser/Thr degradation pathway (tdc pathway) was markedly decreased across all conditions. Serine is a substrate incorporated into colibactin and precolibactins.27 Thus, the downregulation of the tdc operon could facilitate an increase in available Ser for ClbDEFGH.
We next determined if differences in Clb protein expression were linked to transcriptional changes and downstream metabolite production. Transcription of the Clb cluster was assessed using qPCR to measure clb gene expression affected by known transcription start sites.58 Primers used in this study are listed in Table S4 and include clbA, clbB, clbC, clbJ, clbP, and clbR. Each strain was removed from PmB pressure, preserved as a glycerol stock, passaged onto agar plates, expanded in liquid culture overnight, and then grown for 4 h in LB before extracting RNA and evaluating gene expression. Evolved strains showed significant increases in clbR, clbB, clbC, and clbP expression versus WT (Figure 3C). NC101 Δpks served as a control for primer specificity. As iron availability has previously been shown to contribute to pks island production38, we wanted to examine the expression levels of iron response genes in evolved strains compared to WT NC101. In WT NC101, iron chelation increases expression of irp1 and decreases expression of bcsA.59,60 Therefore, monitoring of irp1 and bcsA transcript levels served as a readout for iron responsive genes (Figure S5). As expected, iron depletion of WT NC101 induced clear upregulation of irp1 and downregulation of bcsA. However, this expression pattern was not observed in the evolved strains, suggesting that evolved strains were not regulating clb gene expression simply through decreased iron availability. In summary, these data show that transcriptional control of Clb enzyme abundance is involved in the upregulation of Clb proteins upon polymyxin-resistance.
Elevated production of the colibactin pro-drug motif and genotoxicity
Direct detection of mature colibactin is elusive. Hence, the colibactin prodrug motif, cleaved by the ClbP peptidase in the periplasm during the last step in colibactin maturation has served as an indirect measurement of colibactin production.38,41,61 Bacterial supernatants from NC101 WT, NC101 PmB008R-[1–3], and NC101 Δpks were analyzed for the production of this prodrug motif. Concurrent with proteomic and transcriptomic findings, an increase in the relative abundance of the metabolite was present in PmB-evolved strains (Figure 3D).
Increased transcripts, protein, and metabolite levels related to the pks island provide strong evidence for enhanced functional effects of the colibactin biosynthetic gene cluster in the PmB-resistant bacterial strains. Assessment of DNA damage is crucial to determine if these increases could result in increased DNA damage and eukaryotic host phenotype. To confirm increased production and delivery of the genotoxic metabolite, we measured the levels of phosphorylated histone H2AX (γH2AX) as a proxy for colibactin-mediated DNA damage in the host. The γH2AX assay is commonly used to characterize colibactin-associated DNA damage.20,29 As we have previously described4, cells of the intestinal epithelial cell line IEC-6 were exposed to the parental and evolved NC101 strains. Staurosporine exposure served as a control for apoptosis, which induces pan-nuclear staining that is distinguished from the punctate staining of DNA damage-induced γH2AX foci Although γH2AX quantitation is traditionally based on counting γH2AX+ cells (> 3 foci), this experiment showed little quantitative difference when applying this method (Figure S6), despite a discernible qualitative difference in γH2AX signal intensity (Figure 4A). The intensity of γH2AX staining is linked to the extent of DNA damage.62 As such, the extent of DNA damage in each nucleus was quantified from the total fluorescence of γH2AX proteins (Figure 4B). A significant increase in intensity of γH2AX staining in IEC-6 cells treated with evolved E. coli strains was seen when compared to the NC101 WT strain. This result demonstrates the ability of PmB-resistant E. coli strains to elicit an increased genotoxic effect in eukaryotic cells that underlies colibactin’s pro-carcinogenic activity.
Figure 4.
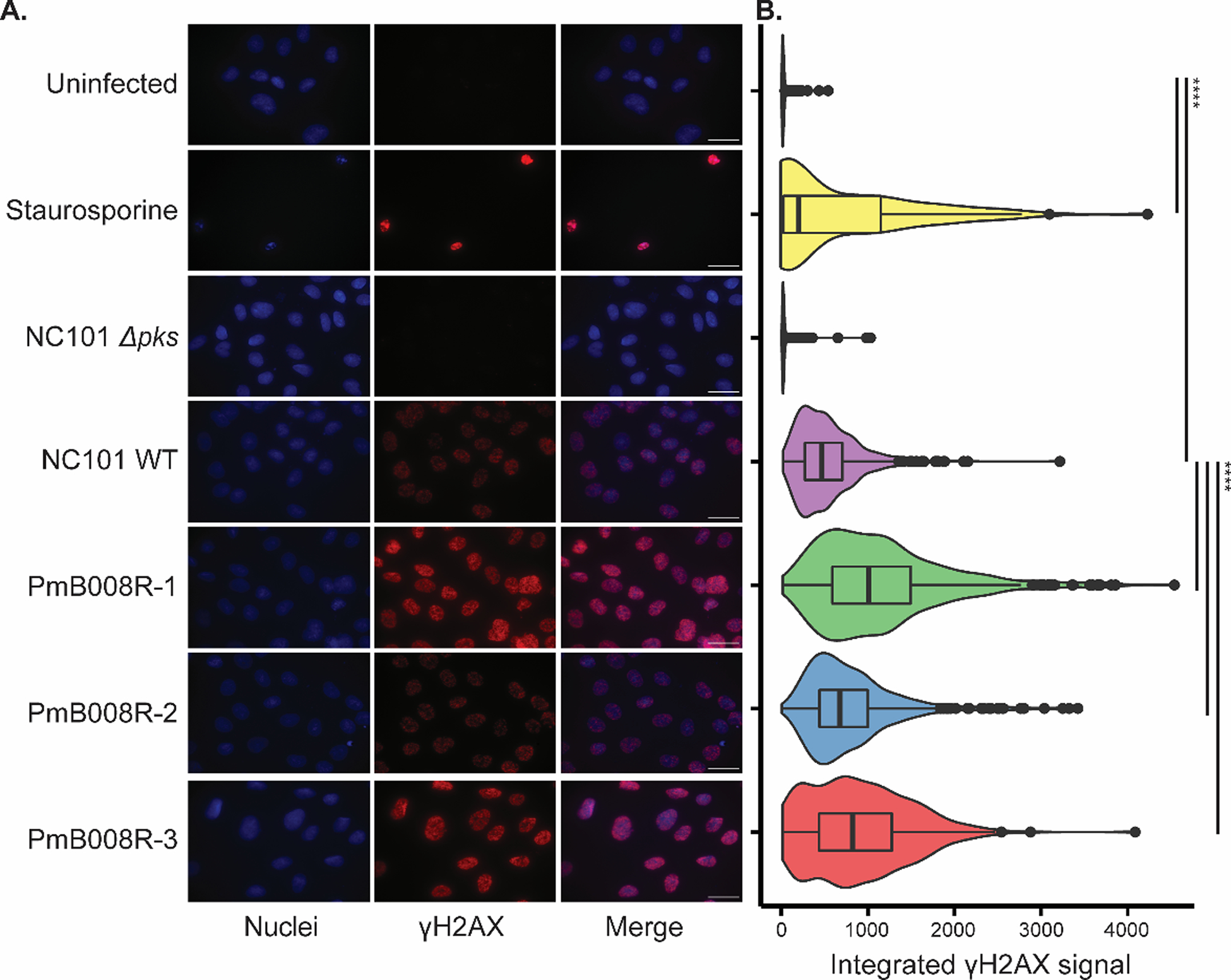
Genotoxicity of E. coli NC101 and evolved lineages. a) Representative microscopy images from IEC-6 rat epithelial cells infected with the following strains of NC101 at a multiplicity of infection (MOI) = 100: NC101 WT, NC101 Δpks (colibactin-deficient), PmB evolved isolates PmB008R-[1–3]. Uninfected samples show baseline staining, while Staurosprine treated samples serve as a positive control for pan-nuclear γH2AX staining indicating apoptosis, rather than punctate staining from DNA damage. Nuclei (Blue, DAPI channel) and γH2AX (Red, TxRed channel) are shown for each group. Scale bars = 25 μM. b) Density plots of quantified γH2AX signal intensity. Significance (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001), denoted by lines to the right of density plots, was determined via t-test with Bonferonni correction. Center lines in the box plots represent mean values for the respective conditions, surrounded by the first and third quartiles. Whiskers extend from the hinge to the highest and lowers values that is within 1.5 * the inner quartiles. Outlier data beyond the end of the whiskers are plotted as points.
Upregulation of Clb enzymes in multiple pks+ E. coli strains
Various pks+ strains have been used to study the effects and regulation of colibactin. Other than NC101, E. coli Nissle 1917 has been of great interest due to its common use as a probiotic.63–65 E. coli ATCC 25922 (ATCC 25922) is another pks+ strain most notably used for antibiotic susceptibility testing in labs, but has also recently been associated with pks genotoxicity.66 Each of these strains were passaged into 4 μg mL−1 PmB, and their proteomes were analyzed for changes in Clb abundance relative to untreated controls of each respective parent culture (Figure 5). Intriguingly, increased Clb enzyme abundance was observed in all strains, suggesting a conserved mechanism for pks regulation under PmB stress and resistance.
Figure 5.
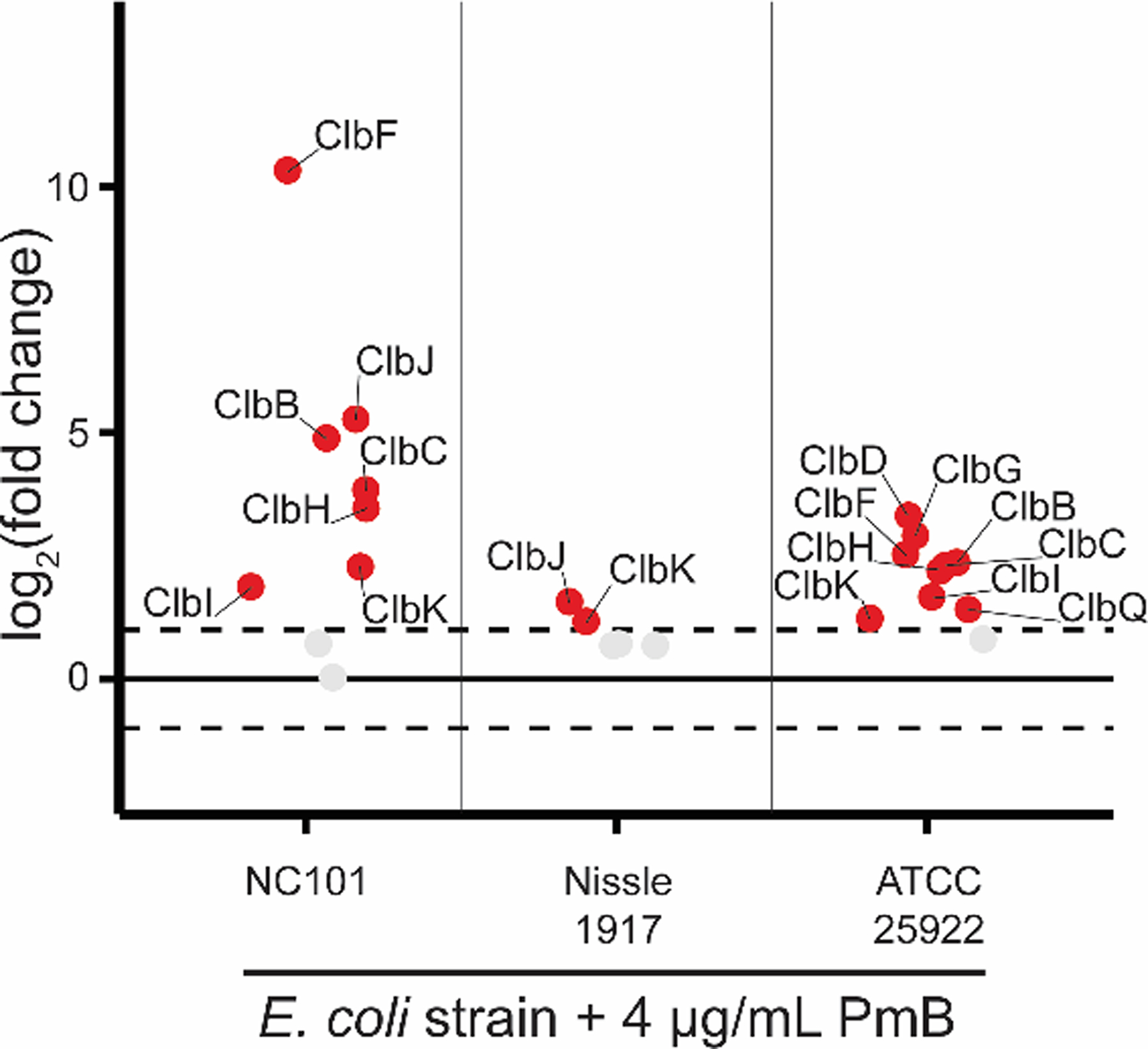
Quantitative proteomics of Clb enzymes in PmB treated pks+ E. coli strains. Data depict log2(FC) of Clb enzymes from NC101 (n = 4), Nissle 1917 (n = 4), and ATCC 25922 (n = 3) relative to untreated cultures of each strain, respectively. Proteins highlighted in red and labelled were in significantly higher abundance in PmB treated cultures compared to WT cultures meeting the significance criteria of log2(FC) > 1 and −log10(q-value) > 1.3 based on an FDR-corrected two-sided t-test.
CONCLUSIONS
A number of resistance mechanisms in response to polymyxins have been observed in Gram-negative bacteria despite their efficacy as bactericidal agents.67 In addition to known PmB resistance mechanisms (lipid A modification, chemotaxis/flagellar synthesis, cellular adhesion, membrane proteins/lipoproteins, etc.), we observed an increased abundance of the enzymes responsible for producing colibactin in E. coli evolved to resist PmB. Transcript, metabolite, and phenotypic assessments confirm production and colibactin-induced DNA damage in three evolved lineages of E. coli NC101. Whole genome sequencing revealed multiple mutations in each of the three lineages. While this study doesn’t directly assess the influence of each mutation on the regulation of the gene cluster, it is possible that the functionality of the mutated proteins, and the pathways they participate in, impact the regulation of the pks island. Overall, the proteomics data represent a rich and complex list of proteins differentially regulated in this state of antibiotic stress. Colibactin regulation is likely influenced by a multitude of the proteins detected in this study. Importantly, our lab and others can leverage this novel dataset to inform future studies that mechanistically dissect the regulation of colibactin production.
Antibiotic regulation of clb genes in this study provide an orthogonal view to the current field, as most studies probing pks regulation have assessed iron availability and media composition. Instead, we approached the question of colibactin regulation by asking what is the microbiological reason by which E. coli would harbor such an energy intensive biosynthetic gene cluster? Surely the pks+ island and colibactin production is not maintained in E. coli as a beneficial effort to induce cancer in their host fifty+ years after colonization. We hypothesized that colibactin serves as a response mechanism against environmental stressors. Antibiotics are not only a common stressor of intestinal-resident bacteria, but are often naturally produced by neighboring bacteria of a particular shared niche. Our work suggests that colibactin is induced by PmB in pro-carcinogenic intestinal E. coli to 1) defend against the antibiotic or 2) colibactin itself serves an antibiotic function against neighboring bacteria in a shared niche. The potential antibiotic activity of colibactin is supported in a recent paper where pks+ E. coli show antibiotic activity against strains of Staphylococcus.68 Further, increased levels of Clb enzymes were found in two other pks+ E. coli strains under polymyxin stress; Nissle 1917 and ATCC 25922. This provides strong evidence that Clb expression under PmB stress is not limited to AIEC or intestinal-resident strains, but is applicable to other B2 group E. coli harboring the pks island.
Previous studies have characterized shifts in the microbiota composition following antibiotic treatment,5,6 however this study highlights that altered behavior of individual strains can cause detrimental side effects to the host. These findings may be especially relevant for patients exposed to antibiotics for chronic conditions (e.g., intestinal diseases and chronic lung infection9,10,48,69,70). Polymyxin antibiotics are commonly used for lung infections (Table S5) and intravenous delivery of the antibiotic can traverse a number of areas in the body, including the GI-tract.70 Repeated and/or continuous exposure to polymyxins in this context could alter the functional capabilities of the gut microbiota. Cystic fibrosis (CF) patients are commonly prescribed PmB antibiotics for treatment of lung infections, and it is well documented that the overall risk for CF patients to develop CRC is 5–10x higher.71, 72 Consistent exposure to polymyxins could provoke pks+ E. coli production of colibactin, inducing colibactin signature mutagenesis, and later enhance CRC development, even after antibiotic treatment is finished.
In the last 15 years, colibactin has been at the forefront of microbial metabolite studies, and much is still left to uncover about this molecule and its regulation. While measurement of the colibactin prodrug motif cleavage by ClbP serves as a sufficient proxy for colibactin, direct quantitation of the mature metabolite may be important in determining the precise delivery mechanism in host epithelial cells. Another major challenge is determining precise regulatory mechanisms of the colibactin pks island. This study contributes to the rapidly expanding knowledge of colibactin regulation and may be used to inform future studies probing these central questions.
METHODS
Bacterial strains:
All bacterial strains used in this study are listed in Table (1).
In vitro evolution of pks+ E. coli NC101 with polymyxin B
E. coli was streaked onto Mueller-Hinton agar plates and incubated for 16 h at 37°C. Isolates were inoculated into 5 mL of Mueller-Hinton broth and incubated for 16 h at 37°C, 250 rpm. A single parent culture was back-diluted into three different culture tubes containing bacteria to create three lineages. Resistant E. coli lineages were generated by serial passaging with increasing concentrations of (PmB) beginning with 0.5 μg mL−1 PmB grown in 7 mL MHB. Cultures were grown at 37°C, 250 rpm, to an OD600 ~ 1.0 (log-phase) and inoculated into cultures containing a 100% increased concentration of PmB and a final OD600 = 0.01. For the PmB008R-[1–3] strains, this was repeated until all cultures reached PmB concentrations of 8 μg mL−1. Aliquots of log-phase cells were harvested at each antibiotic increment before transferring to the next generation, pelleted by centrifugation, flash frozen and stored at −80°C for future proteomic experiments. For the cultures used in the comparison across strains (NC101, Nissle 1917, and ATCC 25922), analysis stopped at 4 μg mL−1 PmB and bacterial aliquots were prepped the same as described above.
Long-term evolution of NC101 and NC101 Δpks against polymyxin B
E. coli NC101 and NC101 Δpks were streaked onto Mueller-Hinton agar plates and incubated for 16 h at 37°C. Isolates were inoculated into 5 mL of Mueller-Hinton broth and incubated for 16 h at 37°C, 250 rpm. Overnight cultures were back-diluted to an OD600 = 0.02 and 200 μL of culture was added into two different wells of a 96-well plate. To one of the wells, 0.5 μg mL−1 PmB was added, and the other was left untreated. Plates were incubated at 37°C for 24 h. Following incubation, OD600 measurements were taken for each culture. If the culture of the treated well was OD600 > 0.4, then 4 μL of culture was transferred into two new wells, one containing the same concentration of antibiotic and one containing a 100% increase in antibiotic. If the culture of the treated well was OD600 < 0.4, then 4 μL of non-treated culture was transferred into two wells containing the same treatments. This process was repeated for 12 days with increasing concentrations of PmB as needed.
Colibactin regulation in evolved cultures with released antibiotic pressure
From glycerol stocks of NC101 WT and NC101 PmB008R–2, cultures were streaked out onto Mueller-Hinton agar plates and incubated overnight at 37°C. Colonies from each plate were inoculated into 5 mL of MHB media and incubated overnight. Overnight cultures were back-diluted to an OD600 = 0.02 in MHB void of PmB. When cultures reached mid-log phase, aliquots were taken for proteomics, and bacteria were passaged to fresh media. This was continued for a total of three passages. Aliquots were flash frozen and stored at −80°C for proteomic analysis.
Whole genome sequencing of evolved strains
Glycerol stocks of evolved strains, NC101 PmB008R-[1–3], were streaked on Mueller-Hinton agar plates and sent to the Microbial Genome Sequencing Center for genomic DNA extraction and Illumina 2×150 paired end whole genome sequencing on the NextSeq 550. Sequencing files were imported as paired reads into CLC Genomics Workbench and mapped to the reference E. coli NC101 genome (SAMN16810910). A consensus sequence was extracted and imported into Geneious Prime for alignment. Samples had average coverage of 78x for PmB00R–1, 80x for PmB00R–2, and 78x for PmB00R–3. Repository information for these assembled sequences can be found in Table S6.
Growth curve
E. coli strains were streaked onto LB agar plates and single colonies were inoculated into 5 mL of Mueller-Hinton broth. Cultures were grown at 37°C and 250 rpm for 18 h. The density of each strain was determined by OD600 measurement, and all strains were normalized to OD600 = 0.25 at t = 0. Four replicates of each strain were grown at 37°C and 250 rpm for 9 h in a 24-well plate. OD600 measurements were taken throughout the 9 h.
Antibacterial assay
E. coli strains were streaked onto LB agar plates and single colonies were inoculated into 5 mL of Mueller-Hinton broth. Cultures were grown at 37°C and 250 rpm for 18 h and normalized to OD600 = 0.25 and grown for 1 h. Cultures were then back-diluted to OD600 = 0.125, and plated in triplicate on a 96-well plate with increasing concentrations of polymyxin B. Water and ampicillin were used as negative and positive controls, respectively. Plates were incubated at 37°C and 250 rpm for 5 h and OD600 measurements were taken. Activity in each well was calculated relative to the positive and negative controls according to Equation 1.
| (1) |
Proteomics
Lysis and protein extraction:
Cell pellets were resuspended in 2 mL of lysis buffer (100 mM Tris-HCl, 0.1% (v/v) TritonX-100, pH 8.0). Cells were lysed via sonication using a E220 Focused-ultrasonicator (Covaris) for 2 m at 200 cycles/burst, 150 W power, and 13% duty cycle or by shaking in a thermocycler for 30 m at 25 °C (850 RPM). Following lysis, protein was precipitated in 5:1 cold 100 mM ammonium acetate in methanol:cell lysate. Samples were incubated at −20°C for 30 min and proteins were pelleted by centrifugation for 10 min at 3,220 RCF. Precipitated protein was resuspended in 4 M urea, 100 mM Tris-HCl, pH 8 and quantified with a CB-X assay (Biosciences) against BSA standards. Proteins (50–100 μg) were reduced (10 mM dithiothreitol, 30 min, 25°C) and alkylated (30 mM iodoacetamide, 45 min, 25°C, dark). Reduced and alkylated proteins were precipitated with 10:1 cold acetone:sample and collected via centrifugation (20,000 RPM, 10 m). Precipitated protein was resuspended in 2 M urea, 100 mM Tris-HCl, pH 8. Reduced and alkylated proteins were enzymatically digested with Trypsin gold (1:50 enzyme:protein) (Promega) for 16 h, shaking at 850 rpm at 25 °C. Samples were acidified using trifluoroacetic acid (TFA) to a pH < 3, desalted using 50 mg C18 Sep Paks (Waters), and dried in a CentriVap (Labconco).
LC-MS/MS analysis:
Samples were resuspended to 200–250 ng/μL in 0.1% (v/v) TFA. Peptides were analyzed using nanoLC-ESI- MS/MS platform consisting of an Acquity M-class UPLC (Waters) coupled to a Q Exactive HF-X (Thermo-Fisher) mass spectrometer. Mobile phase A consisted of water with 0.1% (v/v) formic acid (FA) and mobile phase B consisted of acetonitrile with 0.1% (v/v) FA. Approximately 800 ng of sample was injected onto a C18 trap column (100 Å, 5 μm, 180 μm × 20 mm; Waters) with a flow rate of 5 μL/min for 3 min using 99% (v/v) A and 1% (v/v) B. Peptides were separated on a HSS T3 C18 column (100 Å, 1.8 μm, 75 μm × 250 mm; Waters) using an increasing linear gradient of 5–40% (v/v) B over 90 min, then 85% (v/v) B for 5 min before returning to 5% (v/v) B in 2 min and re-equilibrating for 13 min. The mass spectrometer was operated in positive polarity mode. MS survey scans were collected from 350–2000 m/z at 120,000 resolution. MS/MS scans for the top 20 features were collected using NCE = 28 at 30,000 resolution. Dynamic exclusion for precursor m/z was set to a 10 s window.
Bioinformatics analysis:
Raw files were automatically aligned in Progenesis QI for Proteomics (Nonlinear Dynamics, version 2.0) and peptides were identified in Mascot (Matrix Science, version 2.5.1). Strain specific databases were imported and used for identification of each strain, respectively. Sequences for common laboratory contaminants (www.thegpm.org/cRAP; 116 entries) were appended to each of the databases prior to searching. MGF files were exported from Progenesis QI and searched using precursor/fragment tolerance of 15 ppm/0.02 Da, trypsin specificity, two possible missed cleavages, fixed modification cysteine carbamidomethylation, and variable modifications of methionine oxidation and protein N-term acetylation. Significant peptide identifications above the homology threshold were adjusted using the embedded Mascot percolator algorithm and uploaded to Progenesis for peak matching before exporting protein measurements. Label-free quantitative proteomic quantitation was used to compare normalized abundances of Clb enzymes in treated vs. non-treated conditions. A students t-test with a Benjamini-Hochberg FDR correction was used to determine proteins that were significantly changing between WT and evolved conditions (q-value < 0.05). A list of proteins involved in processes of interest and significantly changing in > 1 evolved strain were manually curated and compiled and shown in Figure 2.
Motility
Bacterial isolates, selected from Mueller-Hinton agar plates, were grown overnight in Mueller-Hinton broth (MHB) at 37°C with shaking at 220 rpm. Strains were diluted to OD600 = 0.5 in MHB and 1 μL was used to inoculate the center of Mueller-Hinton soft agar plates (0.25% (w/v) agar). Plates were incubated at 37° for 10 h. The diameters of motility swarm spots were measured and representative images recorded.
Quantitative real-time PCR (qPCR)
RNA extraction:
Bacteria were sub-cultured for 4hr in LB broth. 1mL of culture was recovered, spun down, and resuspended in lysozyme stock. RNA was extracted using the Qiagen RNeasy Mini Kit (#74104). After eluting in 30 μL molecular water, RNA concentration was analyzed using a NanoDrop. Then, 30 μL RNA (below 100 ng μL−1) was transferred to a new tube and subject to a second DNase treatment with Ambion DNase mix (#1906) at 37°C for 25 m. DNase was inactivated using the kit-provided DNase inactivation reagent. 350 μL RLT+BME (from Qiagen RNeasy kit) was added to RNA solution, mixed with 250 μL 100% ethanol and loaded onto a RNeasy mini column. All remaining steps of the Qiagen extraction protocol were performed before eluting in 30 μL molecular water. RNA quality was assessed by gel electrophoresis and samples were stored at −80°C.
cDNA synthesis:
cDNA was synthesized from 1 μg total RNA, using the SuperScript II RNase H-Reverse Transcriptase (Invitrogen, #18064-014) and manufacturer protocol. Samples were diluted 1:10 in molecular water. To confirm the absence of genomic DNA contamination, minus-reverse transcriptase control samples were generated in-tandem.
qPCR:
Single reactions included: 6.5 SyGreen Blue Lo-ROX (Genesee Sci. #17-505B), 0.375 μL each of 10 μM forward and reverse primers, 2.5 μL molecular water, and 3 μL diluted cDNA. Samples were run with the following conditions: 95°C for 10 m, followed by 40 cycles at 95°C for 15 s, 48°C for 15 s, and 72°C for 15 s. Data was collected using QuantStudio 6 Flex software. Fold change was calculated using the Delta-Delta Ct method. All data were normalized to housekeeping gene, 16S, and compared to WT NC101 in untreated media. Melting curves and gels of cDNA products were assessed to ensure specificity of the amplified products. Primers used in this study are listed in Table S4.
Metabolite detection
Bacterial growth and metabolite extraction:
1 mL replicate cultures (n = 3) from the table below were grown to OD600 = ~0.5–0.7 and harvested into microcentrifuge tubes. Bacterial supernatant was harvested following centrifugation, and 600 μL of supernatant was dried down via vacuum centrifugation.
LC-MS analysis and quantitation:
Dried supernatants were resuspended in 50 μL of H2O + 0.1% (v/v) trifluoroacetic acid. 10 μL of resuspended samples were injected onto an H-class UPLC (Waters) coupled to a Q Exactive HF-X. Mobile phase A consisted of water with 0.1% (v/v) FA and mobile phase B consisted of acetonitrile with 0.1% (v/v) FA. Samples were analyzed on an C18 column (130 Å, 1.7 μm, 2.1 mm × 50 mm; Waters) over a 9 m gradient from 0–100% (v/v) B with a flow rate of 0.5 mL min−1. The mass spectrometer was operating in positive mode, set to scan with a 2 m/z isolation window from 342.2597 – 344.2597 at 60,000 resolution. Runs were processed in Quan browser and area under the curve was used to quantify relative abundances of the prodrug motif.
γH2AX immunofluorescence assay
Genotoxicity was evaluated using the established in vitro γH2AX assay.4
Media composition:
IEC-6 media – DMEM high glucose (Gibco #11995–06) with 5% (w/v) heat-inactivated FBS (Corning #35–015-CV), 1X L-glutamine (Life Tech. In. #25030081),1 U mL−1 insulin (Life Tech. In. #12595014), and 1X Penicillin-Streptomycin (Life Tech. In. #15140122). Infection media – DMEM with 25 mM Hepes and 5% (w/v) FBS.
Bacterial infection:
IEC-6 rat epithelial cells were maintained according to ATCC standards in IEC-6 media. Cells were dissociated using trypsin/EDTA (0.25% (w/v), Life Tech. In. #25200056) at 37°C for 5 m. Cells were enumerated, plated on 8-well chamber slides (Lab-Tek #177445) at 1×105 cells per well in 400 μl total volume IEC-6 media, and allowed to adhere overnight.
The next day, select E. coli strains (WT NC101, NC101 Δpks, and NC101 PmB008R-[1–3]) were diluted 1:10 in LB broth and sub-cultured for 2 h at 37°C with shaking at 200 rpm to reach log phase. IEC-6 cells were washed twice with 1X phosphate buffered saline (PBS) and infected with log phase E. coli at a multiplicity of infection (MOI) = 100 in 400 μL infection media. MOI was determined based on culture optical density at OD600. To validate infection dose, prepared bacterial infection media was serial diluted and plated on LB agar plates. Uninfected IEC-6 cells were used to evaluate baseline γH2AX production. Staurosporine treated (1 μM; Sigma, #56942) cells were used as a positive control for γH2AX production.4 All treated and untreated IEC-6 cells were maintained at 37°C and 5% CO2 for 4 h.
Immunofluorescence staining:
Cells were fixed immediately in 250 μL, 4% (v/v) methanol-free formaldehyde in PBS (Thermo, #28908) for 10 m at room temperature (RT). Fixed cells were rinsed with 1X PBS, then blocked with blocking buffer (2.5 mL 10X PBS and 2.5 mL Normal Goat Serum brought to 25 mL in ddiH2O) for 60 m at RT. Primary γH2AX rabbit polyclonal antibody (clone 20E3, Cell Signaling, #9718) was diluted 1:200 in dilution buffer (0.4g Bovine Serum Albumin in 40mL 1X PBS + 120 μL Triton X-100) and added to cells at 4°C overnight. The next day, cells were washed with 1X PBS and secondary antibody (anti-Rabbit IgG (H+L) AlexaFluor 594; FisherSci, #A-11037) was added at 1:500 in dilution buffer for 1hr at RT. Cells were washed in 1X PBS, and Hoescht 33342 staining dye (abcam #ab228551) was added at 1:10,000 in PBS for 8 m at RT. Cells were washed with 1XPBS, and slides were mounted in Fluoromount-G (Southern Biotech) with a #1.5 coverslip. Stained slides were set to dry for a minimum of 2 h in the dark.
Image acquisition and analysis:
Fluorescence microscopy was performed using an Olympus BX61 upright wide field microscope with Hg lamp. Images were captured using a Hamamatsu ORCA-R2 (C10600–10B) camera and Improvision’s Volocity Software. Separate grayscale images were recorded for Hoescht (Nuclei, blue) and AlexaFluor 594 (γH2AX, red) - excitation(ex) and emission(em) filters were as follows: 1) DAPI (blue) ex: 377/50 em: 447/60 and 2)TxRed (red) ex: 562/40; em: 624/40. Images were acquired with 20X/0.50 UPlanFLN and 60X/1.42 Oil PLanApo N objective lenses. Imaging parameters were set to avoid saturation of pixels and exposure was optimized to prevent photobleaching. All images in a single biological replicate, containing essential controls, were collected with the same acquisition settings. Six randomized images were taken per each technical replicate. In total, we collected images from 3–4 biological replicates with 1–2 technical replicates per sample group (n = 3–4). %γH2AX+ cells were quantified based on equation (2).
| (2) |
Quantification of γH2AX signal intensity:
Nuclear regions were segmented based on DAPI staining using Cell Profiler73 image analysis software (v. 3.1.9). Briefly, images were smoothed using a Gaussian Filter, background corrected (Convex Hull background model), thresholded using adaptive Otsu algorithm and separated using an intensity–based algorithm (de–clumping). Intensity of fluorescence signal in γH2AX channel was measured within nuclear regions and rings surrounding nuclei. Median value of signal within rings was subtracted from the nuclear intensity for local background correction.
Statistical Analysis
All data in this study was performed in sufficient biological replicates (n = 3 – 4). Proteomic experiments leveraged a student’s t-test with a Benjamini-Hochberg FDR correction to determine proteins that were significantly changing between control and experimental conditions (|log2(fold-change)| > 1 and q-value < 0.05). Protein measurements for each of the samples were transformed for PCA analysis. qPCR analysis was analyzed for significance (p<0.05) using a one-way ANOVA; Δpks was excluded from statistical analysis because it served primarily as a control for primer specificity. Metabolite quantitation was examined using a two-sided t-test (p<0.05). The γH2AX assays were performed in biological triplicate and technical duplicates. Significance (p<0.05) of %γH2AX+ cells was assessed by one-way ANOVA. Significance of γH2AX signal intensity was evaluated with a t-test with Bonferroni correction (p<0.05).
Safety statement
No unexpected or unusually high safety hazards were encountered in experiments described.
Supplementary Material
Acknowledgments
This work was supported by NIH-NIGMS under award R01 GM125814 to L.M.H., NIH-NIDDK under award R01 DK124617 to J.C.A, and an Innovation Award from UNC Lineberger Comprehensive Cancer Center (J.C.A and L.M.H). P.W.S. acknowledges support from the NSF Graduate Research Fellowship program. S.J.B acknowledges NIH Biophysics training grant support (T32 GM008570). The Q Exactive HF-X mass spectrometer was funded via an NSF Major Research Instrumentation award (CHE-1726291). The Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
The authors would also like to thank Cassandra J. Barlogio for microbiological support, Dr. Christopher A. Broberg for his help conceptualizing early stages of experiments, Dr. Pablo Ariel for his imaging expertise, E. Diane Weatherspoon for assistance with metabolite detection, Torhera A. Durand and Hannah R. Hall for their contributions to transcriptional analysis, Kenji S.Truman for retrieval and analysis of the Medicare Provider Utilization and Payment Data, and Dr. Azcarate-Peril’s lab for kindly providing E. coli Nissle 1917.
Footnotes
Supporting Information
Supporting Information Available: This material is available free of charge via the internet.
The authors declare no competing financial interests.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortiom via PRIDE74 partner repository under the dataset identifier PXD025088 (doi: 10.6019/PXD025088). Metabolite data is submitted to the mass spectrometry interactive virtual environment (MassIVE) under the dataset identifier MSV000087101. Whole genome sequencing for E. coli NC101 WT strains is summarized in Table S6, and was deposited to GeneBank under accession SAMN16810910 and is publicly accessible. Evolved lineages (NC101 PmB008R-[1–3]) have been submitted as a BioProject to NCBI under the accession PRJNA717334.
References
- 1.Sharon G, Cruz NJ, Kang DW, Gandal MJ, Wang B, Kim YM, Zink EM, Casey CP, Taylor BC, Lane CJ, Bramer LM, Isern NG, Hoyt DW, Noecker C, Sweredoski MJ, Moradian A, Borenstein E, Jansson JK, Knight R, Metz TO, Lois C, Geschwind DH, Krajmalnik-Brown R, and Mazmanian SK (2019) Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice, Cell. 177, 1600–1618 e1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, Anders RA, Giardiello FM, Wick EC, Wang H, Wu S, Pardoll DM, Housseau F, and Sears CL (2018) Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria, Science. 359, 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL, Crabtree-Hartman E, Sand IK, Gacias M, Zhu Y, Casaccia P, Cree BAC, Knight R, Mazmanian SK, and Baranzini SE (2017) Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models, Proc. Natl. Acad. Sci. U.S.A 114, 10713–10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, and Jobin C (2012) Intestinal inflammation targets cancer-inducing activity of the microbiota, Science. 338, 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dethlefsen L, Huse S, Sogin ML, and Relman DA (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing, PLoS Biol. 6, e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isaac S, Scher JU, Djukovic A, Jimenez N, Littman DR, Abramson SB, Pamer EG, and Ubeda C (2017) Short- and long-term effects of oral vancomycin on the human intestinal microbiota, J. Antimicrob. Chemother 72, 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, Dore J, and Other Members of the, I. T. (2010) Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics, J. Pediatr. Gastroenterol. Nutr 51, 77–84. [DOI] [PubMed] [Google Scholar]
- 8.Bien J, Sokolova O, and Bozko P (2012) Role of Uropathogenic Escherichia coli Virulence Factors in Development of Urinary Tract Infection and Kidney Damage, Int. J. Nephrol 2012, 681473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, and Pace NR (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases, Proc. Natl. Acad. Sci. U.S.A 104, 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavelle A, and Sokol H (2020) Gut microbiota-derived metabolites as key actors in inflammatory bowel disease, Nat. Rev. Gastroenterol. Hepatol 17, 223–237. [DOI] [PubMed] [Google Scholar]
- 11.Burisch J, Jess T, Martinato M, Lakatos PL, and EpiCom E (2013) The burden of inflammatory bowel disease in Europe, J. Crohns. Colitis 7, 322–337. [DOI] [PubMed] [Google Scholar]
- 12.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, Sauk JS, Wilson RG, Stevens BW, Scott JM, Pierce K, Deik AA, Bullock K, Imhann F, Porter JA, Zhernakova A, Fu J, Weersma RK, Wijmenga C, Clish CB, Vlamakis H, Huttenhower C, and Xavier RJ (2019) Gut microbiome structure and metabolic activity in inflammatory bowel disease, Nat. Microbiol 4, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, Martinez X, Varela E, Sarrabayrouse G, Machiels K, Vermeire S, Sokol H, Guarner F, and Manichanh C (2017) A microbial signature for Crohn’s disease, Gut. 66, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, and Colombel JF (1998) Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease, Gastroenterology. 115, 1405–1413. [DOI] [PubMed] [Google Scholar]
- 15.Darfeuille–Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, and Colombel JF (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease, Gastroenterology. 127, 412–421. [DOI] [PubMed] [Google Scholar]
- 16.Darfeuille-Michaud A (2002) Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn’s disease, Int. J. Med. Microbiol 292, 185–193. [DOI] [PubMed] [Google Scholar]
- 17.Camprubi-Font C, and Martinez-Medina M (2020) Why the discovery of adherent-invasive Escherichia coli molecular markers is so challenging?, World J. Biol. Chem 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, Weber J, and Lochs H (1998) Association between intraepithelial Escherichia coli and colorectal cancer, Gastroenterology. 115, 281–286. [DOI] [PubMed] [Google Scholar]
- 19.Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, and Bonnet R (2013) High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer, PLoS One. 8, e56964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, and Oswald E (2006) Escherichia coli induces DNA double-strand breaks in eukaryotic cells, Science. 313, 848–851. [DOI] [PubMed] [Google Scholar]
- 21.Homburg S, Oswald E, Hacker J, and Dobrindt U (2007) Expression analysis of the colibactin gene cluster coding for a novel polyketide in Escherichia coli, FEMS Microbiol. Lett 275, 255–262. [DOI] [PubMed] [Google Scholar]
- 22.Bleich RM, and Arthur JC (2019) Revealing a microbial carcinogen, Science. 363, 689–690. [DOI] [PubMed] [Google Scholar]
- 23.Bossuet-Greif N, Vignard J, Taieb F, Mirey G, Dubois D, Petit C, Oswald E, and Nougayrede JP (2018) The Colibactin Genotoxin Generates DNA Interstrand Cross-Links in Infected Cells, mBio. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Stornetta A, Villalta PW, Wilson MR, Boudreau PD, Zha L, Balbo S, and Balskus EP (2019) Reactivity of an Unusual Amidase May Explain Colibactin’s DNA Cross-Linking Activity, J. Am. Chem. Soc 141, 11489–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vizcaino MI, and Crawford JM (2015) The colibactin warhead crosslinks DNA, Nat. Chem 7, 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carra A, Brennan CA, Chun E, Ngo L, Samson LD, Engelward BP, Garrett WS, Balbo S, and Balskus EP (2019) The human gut bacterial genotoxin colibactin alkylates DNA, Science. 363, 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue M, Kim CS, Healy AR, Wernke KM, Wang Z, Frischling MC, Shine EE, Wang W, Herzon SB, and Crawford JM (2019) Structure elucidation of colibactin and its DNA cross-links, Science. 365. 1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue M, Shine E, Wang W, Crawford JM, and Herzon SB (2018) Characterization of Natural Colibactin-Nucleobase Adducts by Tandem Mass Spectrometry and Isotopic Labeling. Support for DNA Alkylation by Cyclopropane Ring Opening, Biochemistry. 57, 6391–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, and Nougayrede JP (2010) Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells, Proc. Natl. Acad. Sci. U.S.A 107, 11537–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez LR, Bleich RM, and Arthur JC (2021) Microbiota Effects on Carcinogenesis: Initiation, Promotion, and Progression, Annu Rev Med 72, 243–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arthur JC, Gharaibeh RZ, Muhlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, Fodor AA, and Jobin C (2014) Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer, Nat. Commun 5, 4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, Sauvanet P, Darcha C, Dechelotte P, Bonnet M, Pezet D, Wodrich H, Darfeuille-Michaud A, and Bonnet R (2014) Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype, Gut. 63, 1932–1942. [DOI] [PubMed] [Google Scholar]
- 33.Tomkovich S, Yang Y, Winglee K, Gauthier J, Muhlbauer M, Sun X, Mohamadzadeh M, Liu X, Martin P, Wang GP, Oswald E, Fodor AA, and Jobin C (2017) Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis, Cancer. Res 77, 2620–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arthur JC (2020) Microbiota and colorectal cancer: colibactin makes its mark, Nat. Rev. Gastroenterol. Hepatol 17, 317–318. [DOI] [PubMed] [Google Scholar]
- 35.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, Paganelli FL, Geurts MH, Beumer J, Mizutani T, Miao Y, van der Linden R, van der Elst S, Genomics England Research C, Garcia KC, Top J, Willems RJL, Giannakis M, Bonnet R, Quirke P, Meyerson M, Cuppen E, van Boxtel R, and Clevers H (2020) Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli, Nature. 580, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dziubanska-Kusibab PJ, Berger H, Battistini F, Bouwman BAM, Iftekhar A, Katainen R, Cajuso T, Crosetto N, Orozco M, Aaltonen LA, and Meyer TF (2020) Colibactin DNA-damage signature indicates mutational impact in colorectal cancer, Nat. Med 26, 1063–1069. [DOI] [PubMed] [Google Scholar]
- 37.Hancock V, Seshasayee AS, Ussery DW, Luscombe NM, and Klemm P (2008) Transcriptomics and adaptive genomics of the asymptomatic bacteriuria Escherichia coli strain 83972, Mol. Genet. Genom 279, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tronnet S, Garcie C, Brachmann AO, Piel J, Oswald E, and Martin P (2017) High iron supply inhibits the synthesis of the genotoxin colibactin by pathogenic Escherichia coli through a non-canonical Fur/RyhB-mediated pathway, Pathog. Dis 75. [DOI] [PubMed] [Google Scholar]
- 39.Wallenstein A, Rehm N, Brinkmann M, Selle M, Bossuet-Greif N, Sauer D, Bunk B, Sproer C, Wami HT, Homburg S, von Bunau R, Konig S, Nougayrede JP, Overmann J, Oswald E, Muller R, and Dobrindt U (2020) ClbR Is the Key Transcriptional Activator of Colibactin Gene Expression in Escherichia coli, mSphere. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chagneau CV, Garcie C, Bossuet-Greif N, Tronnet S, Brachmann AO, Piel J, Nougayrede JP, Martin P, and Oswald E (2019) The Polyamine Spermidine Modulates the Production of the Bacterial Genotoxin Colibactin, mSphere. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tronnet S, Garcie C, Rehm N, Dobrindt U, Oswald E, and Martin P (2016) Iron Homeostasis Regulates the Genotoxicity of Escherichia coli That Produces Colibactin, Infect. Immun 84, 3358–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang-Fichaux M, Chagneau CV, Bossuet-Greif N, Nougayrede JP, Oswald E, and Branchu P (2020) The Polyphosphate Kinase of Escherichia coli Is Required for Full Production of the Genotoxin Colibactin, mSphere 5. e00414–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcie C, Tronnet S, Garenaux A, McCarthy AJ, Brachmann AO, Penary M, Houle S, Nougayrede JP, Piel J, Taylor PW, Dozois CM, Genevaux P, Oswald E, and Martin P (2016) The Bacterial Stress-Responsive Hsp90 Chaperone (HtpG) Is Required for the Production of the Genotoxin Colibactin and the Siderophore Yersiniabactin in Escherichia coli, J. Infect. Dis 214, 916–924. [DOI] [PubMed] [Google Scholar]
- 44.Davies J, Spiegelman GB, and Yim G (2006) The world of subinhibitory antibiotic concentrations, Curr. Opin. Microbiol 9, 445–453. [DOI] [PubMed] [Google Scholar]
- 45.Goh EB, Yim G, Tsui W, McClure J, Surette MG, and Davies J (2002) Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics, Proc. Natl. Acad. Sci. U.S.A 99, 17025–17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore RA, Bates NC, and Hancock RE (1986) Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin, Antimicrob. Agents. Chemother 29, 496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization Model List of Essential Medicines, 21st List, Geneva: World Health Organization; 2019. Licence: CC BY-NC-SA 3.0 IGO. https://apps.who.int/iris/bitstream/handle/10665/325771/WHO-MVP-EMP-IAU-2019.06-eng.pdf (accessed 2020-12-05). [Google Scholar]
- 48.Medicare Provider Utilization and Payment Data: Part D Prescriber. https://www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports/Medicare-Provider-Charge-Data/Part-D-Prescriber (accessed 2020-11-18)
- 49.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, and Sartor RB (2005) Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria, Gastroenterology. 128, 891–906. [DOI] [PubMed] [Google Scholar]
- 50.Mousa JJ, Yang Y, Tomkovich S, Shima A, Newsome RC, Tripathi P, Oswald E, Bruner SD, and Jobin C (2016) MATE transport of the E. coli-derived genotoxin colibactin, Nat. Microbiol 1, 15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hicks LM, Cahoon RE, Bonner ER, Rivard RS, Sheffield J, and Jez JM (2007) Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana, Plant Cell. 19, 2653–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alvarez S, Berla BM, Sheffield J, Cahoon RE, Jez JM, and Hicks LM (2009) Comprehensive analysis of the Brassica juncea root proteome in response to cadmium exposure by complementary proteomic approaches, Proteomics. 9, 2419–2431. [DOI] [PubMed] [Google Scholar]
- 53.Alvarez S, Galant A, Jez JM, and Hicks LM (2011) Redox-regulatory mechanisms induced by oxidative stress in Brassica juncea roots monitored by 2-DE proteomics, Proteomics. 11, 1346–1350. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Alvarez S, and Hicks LM (2012) Comprehensive comparison of iTRAQ and label-free LC-based quantitative proteomics approaches using two Chlamydomonas reinhardtii strains of interest for biofuels engineering, J. Proteome. Res 11, 487–501. [DOI] [PubMed] [Google Scholar]
- 55.Wang HX, Gau B, Slade WO, Juergens M, Li P, and Hicks LM (2014) The Global Phosphoproteome of Chlamydomonas reinhardtii Reveals Complex Organellar Phosphorylation in the Flagella and Thylakoid Membrane, Mol. Cell. Proteomics 13, 2337–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park JJ, Wang H, Gargouri M, Deshpande RR, Skepper JN, Holguin FO, Juergens MT, Shachar-Hill Y, Hicks LM, and Gang DR (2015) The response of Chlamydomonas reinhardtii to nitrogen deprivation: a systems biology analysis, Plant. J 81, 611–624. [DOI] [PubMed] [Google Scholar]
- 57.Rodrigues SP, Alvarez S, Werth EG, Slade WO, Gau B, Cahoon EB, and Hicks LM (2015) Multiplexing strategy for simultaneous detection of redox-, phospho- and total proteome – understanding TOR regulating pathways in Chlamydomonas reinhardtii, Anal. Methods 7, 7336–7344. [Google Scholar]
- 58.Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W, Oelschlaeger TA, Oswald E, Forestier C, Hacker J, and Dobrindt U (2009) Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae, Infect. Immun 77, 4696–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellermann M, Huh EY, Liu B, Carroll IM, Tamayo R, and Sartor RB (2015) Adherent-Invasive Escherichia coli Production of Cellulose Influences Iron-Induced Bacterial Aggregation, Phagocytosis, and Induction of Colitis, Infect. Immun 83, 4068–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellermann M, Gharaibeh RZ, Fulbright L, Dogan B, Moore LN, Broberg CA, Lopez LR, Rothemich AM, Herzog JW, Rogala A, Gordon IO, Rieder F, Brouwer CR, Simpson KW, Jobin C, Sartor RB, and Arthur JC (2019) Yersiniabactin-Producing Adherent/Invasive Escherichia coli Promotes Inflammation-Associated Fibrosis in Gnotobiotic Il10(−/−) Mice, Infect. Immun 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brotherton CA, and Balskus EP (2013) A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity, J. Am. Chem. Soc 135, 3359–3362. [DOI] [PubMed] [Google Scholar]
- 62.Lee Y, Wang Q, Shuryak I, Brenner DJ, and Turner HC (2019) Development of a high–throughput gamma-H2AX assay based on imaging flow cytometry, Radiat. Oncol 14, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kruis W, Fric P, Pokrotnieks J, Lukas M, Fixa B, Kascak M, Kamm MA, Weismueller J, Beglinger C, Stolte M, Wolff C, and Schulze J (2004) Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine, Gut. 53, 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malchow HA (1997) Crohn’s disease and Escherichia coli. A new approach in therapy to maintain remission of colonic Crohn’s disease?, J. Clin. Gastroenterol 25, 653–658. [DOI] [PubMed] [Google Scholar]
- 65.Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, and Axon AT (1999) Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial, Lancet. 354, 635–639. [DOI] [PubMed] [Google Scholar]
- 66.Kaewkod T, Tobe R, Tragoolpua Y, and Mihara H (2020) Medicinal plant extracts protect epithelial cells from infection and DNA damage caused by colibactin-producing Escherichia coli, and inhibit the growth of bacteria, J. Appl. Microbiol 130, 769–785. [DOI] [PubMed] [Google Scholar]
- 67.Olaitan AO, Morand S, and Rolain JM (2014) Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria, Front. Microbiol 5, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fais T, Cougnoux A, Dalmasso G, Laurent F, Delmas J, and Bonnet R (2016) Antibiotic Activity of Escherichia coli against Multiresistant Staphylococcus aureus, Antimicrob. Agents. Chemother 60, 6986–6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dayama G, Priya S, Niccum DE, Khoruts A, and Blekhman R (2020) Interactions between the gut microbiome and host gene regulation in cystic fibrosis, Genome. Med 12, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin YW, Zhou Q, Onufrak NJ, Wirth V, Chen K, Wang J, Forrest A, Chan HK, and Li J (2017) Aerosolized Polymyxin B for Treatment of Respiratory Tract Infections: Determination of Pharmacokinetic-Pharmacodynamic Indices for Aerosolized Polymyxin B against Pseudomonas aeruginosa in a Mouse Lung Infection Model, Antimicrob. Agents. Chemother 61. e00211–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamada A, Komaki Y, Komaki F, Micic D, Zullow S, and Sakuraba A (2018) Risk of gastrointestinal cancers in patients with cystic fibrosis: a systematic review and meta-analysis, Lancet. Oncol 19, 758–767. [DOI] [PubMed] [Google Scholar]
- 72.Maisonneuve P, Marshall BC, Knapp EA, and Lowenfels AB (2013) Cancer risk in cystic fibrosis: a 20-year nationwide study from the United States, J. Natl. Cancer. Inst 105, 122–129. [DOI] [PubMed] [Google Scholar]
- 73.McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, Wiegraebe W, Singh S, Becker T, Caicedo JC, and Carpenter AE (2018) CellProfiler 3.0: Next–generation image processing for biology, PLoS Biol. 16, e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, and Vizcaino JA (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data, Nucleic Acids Res. 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortiom via PRIDE74 partner repository under the dataset identifier PXD025088 (doi: 10.6019/PXD025088). Metabolite data is submitted to the mass spectrometry interactive virtual environment (MassIVE) under the dataset identifier MSV000087101. Whole genome sequencing for E. coli NC101 WT strains is summarized in Table S6, and was deposited to GeneBank under accession SAMN16810910 and is publicly accessible. Evolved lineages (NC101 PmB008R-[1–3]) have been submitted as a BioProject to NCBI under the accession PRJNA717334.


