Abstract
CD8+ T cells are critical mediators of adaptive immunity but may also exhibit innate-like properties such as surface expression of NKG2C, an activating receptor typically associated with natural killer (NK) cells. We demonstrate that, similar to NK cells, NKG2C+TCRαβ+CD8+ T cells are associated with prior human cytomegalovirus (HCMV) exposure. In addition to expressing several NK cell markers such as CD56 and KIR, NKG2C+CD8+ T cells are oligoclonal and do not upregulate PD-1 even in response to persistent activation. Furthermore, we found that NKG2C+CD8+ T cells from some individuals exhibited strong effector function against leukemia cells and HCMV-infected fibroblasts, which was dictated by both NKG2C and TCR specificity. Transcriptomic analysis revealed that the transcription factor BCL11B, a regulator of T cell developmental fate, is significantly downregulated in NKG2C+CD8+ T cells when compared to conventional NKG2C−CD8+ T cells. BCL11B deletion in conventional CD8+ T cells resulted in the emergence of a similar innate-like CD56+CD94+DAP12+NKG2C+CD45RA+CCR7−PD-1−/low T cell population with activity against HLA-E+ targets. Based on their intrinsic capacity to recognize diseased cells coupled with lack of PD-1 induction, NKG2C+CD8+ T cells represent a lymphocyte population that resides at the boundary between innate and adaptive immunity, presenting an attractive alternative for cellular therapy, including CAR T-based therapies.
One sentence summary:
NKG2C+CD8+ T cells, found in 40% of HCMV-seropositive individuals, have lost BCL11B expression and adopt an innate-like identity.
Introduction
Few pathogens leave as large an imprint on the immune system as human cytomegalovirus (HCMV)(1), which infects all populations with a 50-100% penetrance(2). In healthy individuals, natural killer (NK) and T cells play complementary roles in maintaining HCMV in a latent state. HCMV downregulates classical HLA class I expression on the infected cell, impairing T cell recognition, but potentially increasing its susceptibility to NK cell surveillance(3). T cell responses are critical for resolving and preventing HCMV disease(1). Conventional T cell immunity against HCMV includes cytotoxic targeting of infected cells followed by clonal expansion, differentiation, establishment of memory, and recall of virus-specific subsets(4). Because of the latent nature of HCMV infection, it is likely that certain HCMV antigens are periodically shed, resulting in lifelong accumulation of virus-specific T cells, with one or a few clones capable of populating up to 30% of the entire T cell memory pool(5).
In addition to T cell responses, cytomegalovirus-specific NK cell responses have been identified in both mice and humans, and in the former are characterized by increased cytotoxicity and early production of IFN-γ, followed by clonal expansion and recall of virus-specific subsets(6). In humans, the non-classical HLA-E molecule is the ligand for both the activating CD94/NKG2C and the inhibitory CD94/NKG2A heterodimers, which are nearly mutually exclusive in their cell surface expression. HLA-E expression is increased by HCMV infection(7) through stabilizing presentation of HCMV-derived UL40 peptides(8), potentially explaining the expansion of NK cells expressing NKG2C(9, 10). NKG2C+ NK cells are educated by self-HLA-specific inhibitory killer Ig-like receptors (KIR) and expand during primary and secondary infection, but only in some individuals(11). NKG2C expression marks a subset of mature (CD57+) NK cells labeled “adaptive NK”, which are deficient in the FcεRIγ chain, the transcription factor PLZF, and the signaling molecules Syk and Eat-2(11). Adaptive NK cells display a heightened response to HCMV, triggered by antibody-dependent cellular cytotoxicity, but can still be inhibited through KIR engagement of HLA class I ligands and are hyporesponsive to a broad set of targets(12).
Over the past two decades, the distinction between innate and adaptive immunity has become blurred. In mice, bacterial and viral pathogens can stimulate innate-like T cells(13). In humans, some individuals harbor TCRαβ+CD8+ T cells expressing molecules typically associated with NK phenotypes: CD56, ILT2, KIR(14, 15), and NKG2A or NKG2C receptors(10, 16). NK-like TCRαβ+CD8+ T cells seem to be HLA-E-restricted with a strong bias in TCR repertoire(17). Stimulation with HLA-E-expressing targets leads to proliferation and cytotoxicity of NKG2C+ T cells, illuminating alternative pathways for T cell activation(16). A potential clinical benefit to the NKG2C+ T cell population has been observed in humans infected with Mycobacterium leprae, where NKG2C marks a subset of CD8+ T cells with superior antimicrobial capacity compared to other CD8+ T cell subsets(18).
Here, we show that in HCMV-seropositive individuals, an expanded population of CD8+ T cells expresses NKG2C and acquires an NK-like genetic signature and phenotype. These NKG2C+CD8+ cells downregulate genes associated with a T cell identity, the most prominent of which encodes the transcription factor BCL11B. NKG2C+CD8+ T cells have a restricted TCR repertoire with preferential use of the Vβ chain TRBV-14. NKG2C+CD8+ T cells can be activated by HLA-E engagement and, in those individuals with TRBV-14 TCR restriction, demonstrate high efficiency killing of a set of tumor cells via both the TCR and NKG2C engagement. Despite expansion in response to chronic stimulation, this population does not upregulate PD-1. Together, our data suggest that in humans, HCMV exposure gives rise to the expansion of a BCL11BlowNKG2C+CD8+ T cell population with an NK phenotype and broad immune surveillance capacity. The innate ability of these cells to respond to infected target cells and tumor cell lines reveals an opportunity for adoptive cellular therapy.
Results
NKG2C+CD8+ T cells are associated with HCMV seropositivity in healthy donors
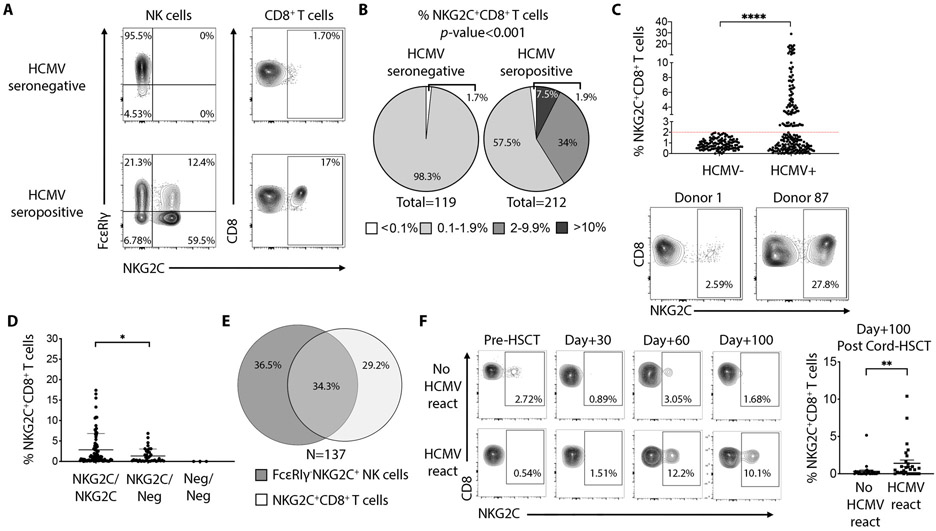
To quantify the NKG2C+CD8+ T cell population in humans, we analyzed peripheral blood from 331 healthy subjects. Of these individuals, 212 (64%) were HCMV-seropositive. In line with previous findings(9, 16), we observed in some individuals that both T cells (TCRαβ) and NK cells express NKG2C. The presence of an “adaptive” FcεRIγ−NKG2C+ NK cell population, was associated with HCMV-seropositivity of the donor (Fig. 1A). Likewise, among T cells, NKG2C+CD8+ T cells were also associated with HCMV-seropositivity. 41.5% of HCMV-seropositive donors exhibited an expanded population of NKG2C+CD8+ T cells (Figure 1B), as defined by a frequency threshold greater than 2% of total CD8+ T cells. All donors (except for three individuals lacking KLRC2, the gene encoding NKG2C) had a detectable NKG2C+CD8+ T cell population, however this frequency never surpassed 2% of the total CD8+ T cell population in HCMV-seronegative individuals. In contrast, 41.5% of HCMV-seropositive individuals exhibited NKG2C+CD8+ T cells with frequencies ranging from 2.6% to 27.8% of the total CD8+ T cell population (Fig. 1C). The highest population frequency was observed in individuals homozygous for KLRC2 (Fig. 1D). Among all HCMV-seropositive donors tested, 137 (65%) displayed expansion of a NKG2C+ population within NK or T cells. Of these individuals, 36.5% had expansion in NK cells only, 29.2% in CD8+ T cells only and 34.3% in both lymphocyte populations (Fig. 1E). The age of the donor did not influence the magnitude of the population (Fig. S1A). Notably, in individuals with an expanded NKG2C+CD8+ T cell population, NKG2C expression was absent on CD8+ T cells specific for the HCMV-derived antigen pp65 (Fig. S1B). When analyzed for specific reactivity against the five most recognized HCMV open reading frames (ORFs) by CD8+ T cells(5), NKG2C+CD8+ T cells showed no specific activation (Fig. S1C). Therefore, the presence of a CD8+ T cell population expressing NKG2C was independent of the presence of an NKG2C+ NK population. Furthermore, the expansion of the population is variable and age-independent between individuals, and antigen specificity among NKG2C+CD8+ T cells is not related to the most common HCMV peptides that dominate canonical CTL response(5, 19, 20).
Fig. 1. Prevalence of NKG2C+CD8+ T cells in HCMV seropositive individuals.
(A) FACS plots showing the frequency of FcεRIγ−NKG2C+ NK cells and NKG2C+CD8+ T cells in one representative HCMV-seronegative and HCMV-seropositive donor. (B) Pie charts show the prevalence of NKG2C+CD8+ T cells among HCMV-seronegative and HCMV-seropositive individuals according to frequency of the corresponding lymphocyte subset. P-value was calculated using a χ2 test. (C) Graph showing the frequency of NKG2C+CD8+ T cells among HCMV-seronegative (N=119) and HCMV-seropositive individuals (n=212). Statistical analysis was performed by Mann-Whitney test. Dot plots show the minimum and maximum frequency of NKG2C+ cells among total CD8+ T cells found in donors who display a detectable population (above 2% of total CD8+ T cells). (D) Graph showing the frequency of NKG2C+CD8+ T cells among individuals homozygous (NKG2C/NKG2C), heterozygous (NKG2C/Neg) or negative (Neg/Neg) for the KLRC2 gene. (E) Venn diagram showing the frequencies of healthy HCMV-seropositive donors (N=137) having FcεRIγ−NKG2C+ NK cells, NKG2C+CD8+ T cells or both lymphocyte subsets. (F) FACS plots showing the frequency of NKG2C+CD8+ T cells present after cord-blood HSCT in two representative patients, experiencing HCMV reactivation or not. Graph on the right show cumulative analysis of 59 patients, 29 with HCMV reactivation between Day+30 and Day+60 post HSCT. Statistical analysis was performed by Mann-Whitney test. ** P-value < 0.01, **** P-value < 0.0001.
To provide further evidence of the association between NKG2C+CD8+ T cells and HCMV infection, we retrospectively examined patients who received allogeneic hematopoietic cell transplantation with umbilical cord blood, a clinical setting with a common incidence of HCMV reactivation (Table S1). Frequent and systematic surveillance post-transplantation for viral reactivation coincidentally allowed us to detect the emergence and expansion of NKG2C+CD8+ T cells, and to define a temporal relationship with HCMV reactivation. We identified clear expansion of NKG2C+CD8+ T cells in patients following documented HCMV reactivation, but no expansion was observed in patients who did not experience HCMV reactivation (Fig. 1F). From the serology studies of healthy individuals and from the immune reconstitution data from transplant recipients, we conclude that the expansion of an NKG2C+CD8+ T cell population is associated with HCMV infection.
NKG2C+CD8+ T cells are PD-1-negative and resemble NK cells
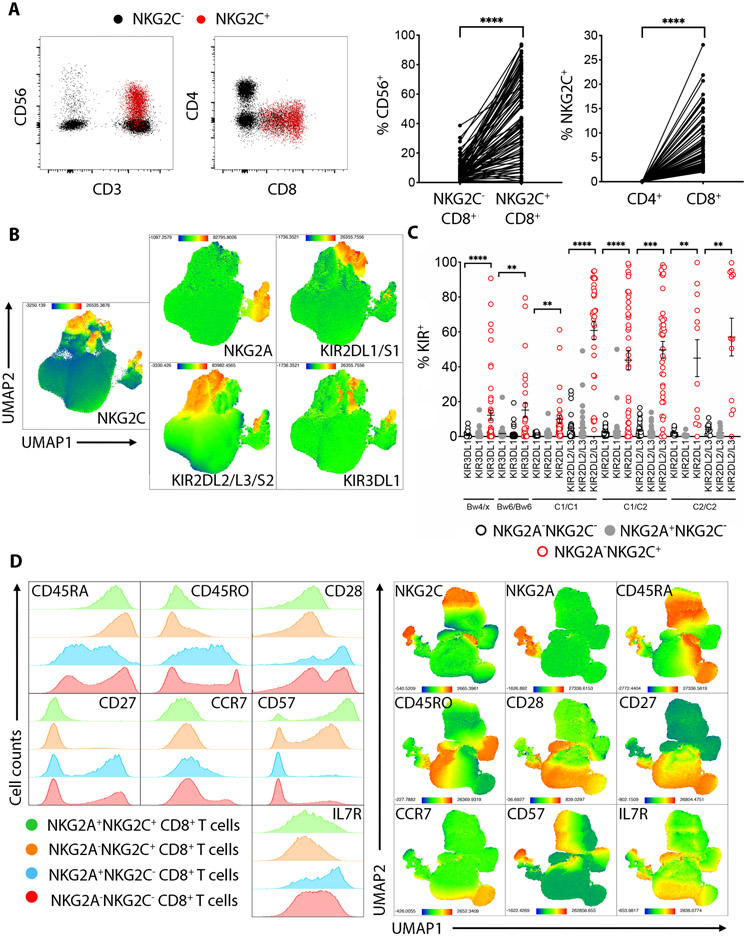
Further phenotyping of NKG2C+CD8+ T cells revealed that in the majority of donors tested, NKG2C+CD8+ T cells co-expressed CD56. We found no NKG2C-expressing cells among CD4+ T cells (Fig. 2A). When analyzed for inhibitory receptor surface expression, NKG2C+CD8+ T cells contained a higher frequency of KIR2DL1/S1-, KIR2DL2/L3/S2- and KIR3DL1-expressing cells compared to conventional NKG2C−CD8+ T cells and to NKG2A+CD8+ T cells (Fig. 2B-C). There was no evidence for skewing of KIR expression based on self-HLA class I ligands, except a slight bias for higher KIR2DL1 expression in individuals homozygous for HLA-C allotypes expressing the C2 epitope (C2/C2) compared to individuals homozygous for the C1 epitope (C1/C1).
Fig. 2. Phenotype of NKG2C+CD8+ T cells.
(A) Dot plots showing CD56, CD4 and CD8 expression on NKG2C+CD8+ T cells (red) or NKG2C−CD8+ T cells (black). Graphs on the right show cumulative analysis from 88 donors. Statistical significance was calculated using paired student-t test. (B) UMAP plots showing the cumulative expression of NKG2C, NKG2A and KIR receptors in total CD8+ T cells from 88 donors. (C) Graph showing the frequency of NKG2C+CD8+ T cells expressing the indicated KIR receptors compared to NKG2C−CD8+ T cells or NKG2A+CD8+ T cells, segregated by donor-expressed KIR ligands HLA-C1, -C2, -Bw4 and Bw6. Statistical analysis was performed by student-t test comparing the NKG2C−CD8+ or the NKG2A+CD8+ vs NKG2C+CD8+ for each KIR within the indicated HLA haplotype. (D) Left: FACS histograms show staining of the indicated markers in one representative individual. Right: UMAP plots show the expression of the indicated markers in total CD8+ T cells, cumulative of 6 different individuals. ** P-value < 0.01, *** P-value < 0.001, **** P-value < 0.0001.
Assessment of T cell differentiation markers revealed that the NKG2C-expressing population is CD45RA+CD45RO−CD28−CD27−CCR7−CD57+IL7Rlow/−, potentially resembling terminally differentiated TEMRA cells. The CD8+ population co-expressing NKG2A and NKG2C exhibited a phenotype similar to the NKG2C+ cells (Fig. 2D). In contrast, NKG2A+CD8+ T cells resembled central memory T cells (CD45RA−CD45RO+CD28+CD27+CCR7+CD57−IL7R+). Interestingly, the small fraction (<2%) of NKG2C+ cells from HCMV-seronegative individuals retained CD27 surface expression, suggesting that this population had not been exposed to antigen-induced activation and expansion (Fig. S2A-B). Of note, the total NK population (predominantly composed by CD56dim cells) exhibited the same expression pattern as the NKG2C+CD8+ T cells for the T cell differentiation markers described above, suggesting that the NKG2C+CD8+ T population is either a true TEMRA population or that it is NK-like (Fig. S2C). The frequency of NKG2C+CD8+ T cells expressing KIR2DL1/S1, KIR2DL2/L3/S2 or KIR3DL1 was comparable to frequencies found in NK cells and much higher than in the naïve (CD45RA+CCR7+) or TEMRA (CD45RA+CCR7−) populations (Fig. S2D-E).
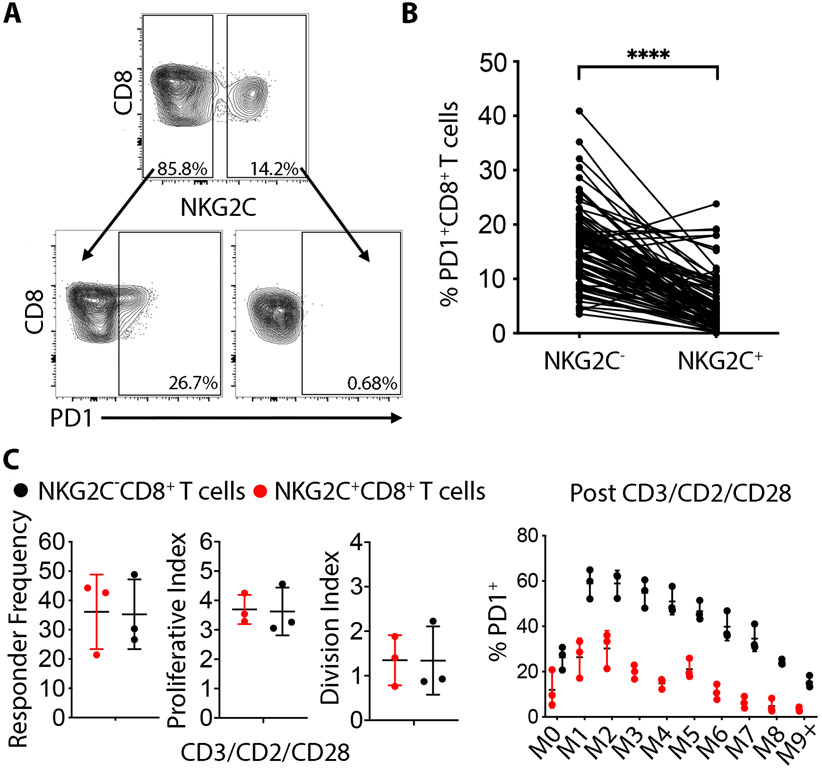
Because classic TEMRA cells are a terminally differentiated population that is more likely than other T cell populations to express exhaustion markers and to have impaired proliferative capacity(21), we investigated if NKG2C+CD8+ T cells exhibited the same features. We found that the overwhelming majority of NKG2C+CD8+ T cells were negative for the immune checkpoint molecule PD-1 at rest, a notable distinction when compared to their NKG2C−CD8+ T cell counterparts (Fig. 3A-B). Despite differences in surface expression of PD-1, NKG2C+CD8+ cells display similar proliferation kinetics to NKG2C−CD8+ T cells after one week of CD3/CD2/CD28 stimulation in vitro. Nevertheless, the majority of NKG2C+CD8+ T cells remained PD-1low in every mitotic generation, in contrast to conventional CD8+ T cells which upregulated PD-1 expression upon activation (Fig. 3C). Lack of PD-1 expression despite repetitive activation suggests the NKG2C+CD8+ T cell population may therefore phenotypically resemble NK cells more than TEMRA.
Fig. 3. NKG2C+CD8+ T cells are PD-1 negative at rest and do not upregulate PD-1 even after strong stimulation.
(A) NKG2C− and NKG2C+CD8+ T cells from the same representative HCMV-seropositive donor were stained for PD-1 at rest. (B) Cumulative analysis for PD-1 expression on NKG2C−CD8+ and NKG2C+CD8+ T cells in 88 HCMV-seropositive donors. Statistical analysis was performed by paired student-t test comparing the NKG2C− vs NKG2C+CD8+ within the same individuals. (C) Left: Total CD8+ T cells were CTV labelled and stimulated for 7 days with beads against CD3/CD2/CD28 and proliferation indexes were analyzed separately for NKG2C− and NKG2C+CD8+ T cells monitoring CTV dilutions. Right: PD-1 expression was analyzed on every mitotic generation following 7 days stimulation with beads against CD3/CD2/CD28 in 3 independent donors. **** P-value < 0.0001.
NKG2C+CD8+ T cells acquire NK cell transcriptional features and downregulate a BCL11B-dependent transcriptional program.
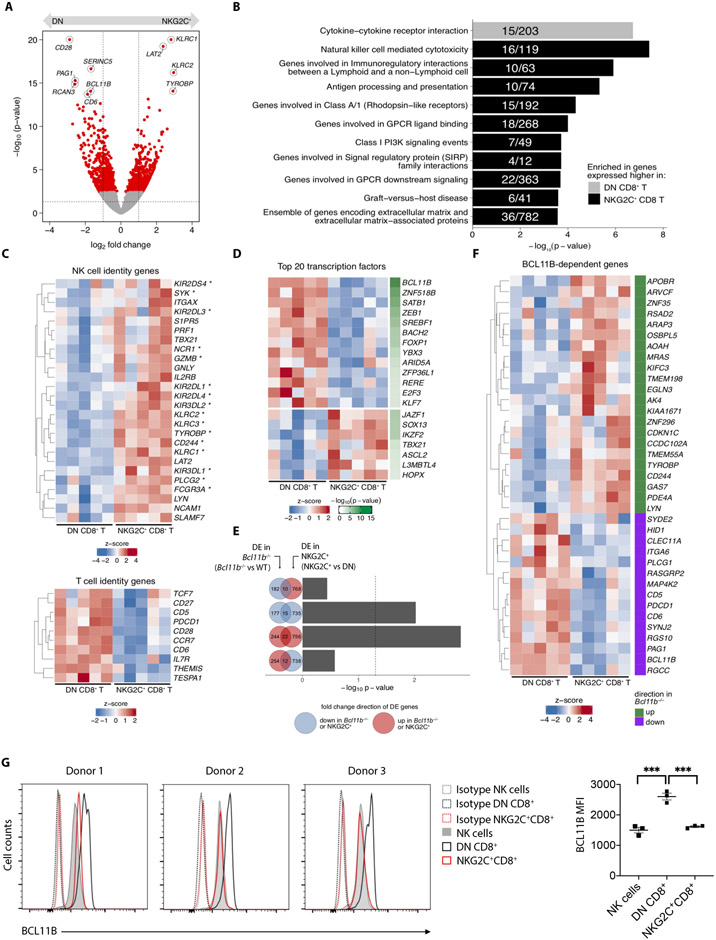
The intriguing mixture of NK-like features within the NKG2C+CD8+ T cell subset implies these cells may possess a transcriptional signature distinct from conventional CD8+ T cells. We performed whole transcriptome analysis on paired samples of NKG2A−NKG2C+CD8+ T cells (NKG2C+CD8+) and NKG2A−NKG2C−CD8+ T cells (DN CD8+ T) obtained from the peripheral blood of five healthy HCMV-seropositive donors. NKG2C+ and DN CD8+ T cells defined transcriptionally distinct populations (Fig. 4A). Gene set enrichment analysis revealed that NKG2C+CD8+ T cells are enriched for transcription of genes associated with the "NK cell-mediated cytotoxicity" pathway, indicating that NKG2C+CD8+ T cells have a strong NK cell-like signature (Fig. 4B). When the analysis was focused specifically on genes commonly associated with NK or T cell identity, NKG2C+CD8+ T cells demonstrated higher expression of genes classically associated with NK cells and lower expression of genes associated with T cells when compared to DN CD8+ T cells (Fig. 4C). The finding of a prominent NK signature enrichment validates the phenotypic similarities between NK cells and NKG2C+CD8+ T cells and suggests shared biological functions.
Fig. 4. NKG2C+CD8+ T cells acquire NK cell transcriptional features and downregulate a BCL11B-dependent transcriptional program.
NKG2A−NKG2C+ (NKG2C+CD8+) or NKG2A−NKG2C− (DN) CD8+ T cells were sorted from PBMC of 5 different healthy donors and processed for RNA sequencing. (A) Volcano plot depicts log2 fold change on x-axis and -log10(p-value) (calculated by Wald tests) on y-axis. Red dots highlight DE genes when comparing NKG2C+ versus DN CD8+ T cells. Highlighted are top 10 DE genes ranked by FDR-corrected p-value. Dotted vertical and horizontal lines mark +/− 1 and p = 0.05, respectively. (B) Bar plots display -log10 (p-value) of gene set enrichment in genes higher in DN (gray) or higher in NKG2C+ (black) CD8+ T cells. Fractions within bar plots indicate number of DE genes found within the total gene set. (C) Heatmaps show expression of curated NK-cell identity genes (top) and T-cell identity genes (bottom). Asterisks indicate genes found within the “Natural killer cell mediated cytotoxicity” gene set displayed in (B). (D) Heatmap shows expression of top 20 DE genes that encode transcription factors. (E) Bar plots display -log10 (p-value) calculated by hypergeometric tests evaluating overrepresentation of BCL11B-dependent genes (e.g. DE genes comparing Bcl11b−/− to WT) among NKG2C+ versus DN DE genes. Tests were performed using any combination of gene sets that were either upregulated (red circle) or downregulated (blue circle) in either comparison, as represented by Venn diagrams. For example, the top bar plot depicts the -log10 (p-value) calculated from testing whether the number of genes that are significantly downregulated in Bcl11b−/− cells (blue circle) are overrepresented among the genes that are significantly upregulated in NKG2C+ cells (red circle). Absolute numbers of their differences and intersection are indicated. (F) Heatmap shows expression of BCL11B-dependent genes that are also DE in NKG2C+ versus DN comparison. Green marks genes that are upregulated upon BCL11B-deficiency, while purple marks genes that are downregulated. (G) Histograms show intracellular staining for BCL11B in three different donors, gated on total NK cells, DN or NKG2C+CD8+ T cells. Dotted histograms represent isotype controls. Graph on the right shows cumulative analysis from 3 independent donors. Statistical significance was calculated by one-way ANOVA with Turkey’s multiple comparison test. DE = differentially expressed; FDR = false discovery rate. *** P-value < 0.001
Among the top 10 differentially expressed (DE) genes between the two populations, was the gene encoding the transcription factor (TF) BCL11B (Fig. 4A). Among TFs, BCL11B was found to be the highest DE gene (Fig. 4D). Bcl11b, expressed in all T cell subsets and progenitors in mice, initiates progenitor cell commitment to the T cell lineage prior to TCR expression and regulates key processes of T cell function and survival(22). As a master regulator of T cell fate, Bcl11b controls a transcriptional program that actively prevents development of lymphocytes towards an innate NK-like phenotype(23). The finding that NKG2C+CD8+ T cells in humans exhibit significantly decreased BCL11B transcripts suggests that this cell population has lost T cell identity during development.
To determine if BCL11B loss in human NKG2C+CD8+ T cells results in a transcriptional program that diverts T cells to an NK fate, we converted the murine genes known to be differentially expressed between Bcl11b-knockout and wild-type CD8+ T cells(22) to their human orthologs and compared them to the DE genes between NKG2C+CD8+ and conventional (DN CD8+) T cells (Fig. 4E). Using this approach, we find that the pattern of Bcl11b-dependent genes, both upregulated and downregulated, is recapitulated in humans when NKG2C+CD8+ T cells are compared to conventional CD8+ T cells (Fig. 4F). Thus, the induced adoption of an NK phenotype in T lymphocytes following deletion of Bcl11b in mice occurs naturally in the human NKG2C+CD8+ T cell population. We then confirmed by intracellular staining that NKG2C+CD8+ T cells have significantly less BCL11B protein compared to conventional CD8+ T cells. Moreover, the low levels of BCL11B in NKG2C+CD8+ T cells are comparable to those observed in NK cells from the same donors (Fig. 4G).
To assess how much of the unique gene signature observed in NKG2C+CD8+ T cells can be attributed to a putative TEMRA state or if loss of BCL11B has triggered “NK reprogramming”, we first derived an “NK signature,” comparing gene transcripts from NK cells (sorted on CD56+CD3−NKG2A−NKG2C−) with those of autologous DN CD8+ T cells. Compared to the DN CD8+ T cells, NKG2C+CD8+ T cells showed significantly higher expression of genes associated with an “NK signature” (Fig. S3A-B). We then derived a “TEMRA signature,” taking advantage of previously published transcriptome data from TEMRA cells(24) and comparing it to our newly generated RNA-seq datasets from DN CD8+ T cells and NKG2C+CD8+ T cells. We did not observe a clear pattern of expression for DE genes in the NKG2C+CD8+ vs. DN T cell population that are part of a TEMRA signature (Fig. S3B-C). Furthermore, NKG2C+CD8+ T cells exhibit significantly less BCL11B protein than classic NKG2C−CD45RA+CCR7−TEMRA cells, again closer to the low BCL11B expression seen in NK cells (Fig. S3D). Taken together, our data show that NKG2C+CD8+ T cells and NK cells appear phenotypically and transcriptionally closely related and that the dramatic downregulation of BLC11B in NKG2C+CD8+ T cells and loss of its downstream transcriptional effects likely account for its adoption of an NK-like identity.
BCL11B deletion promotes the in vitro generation of NKG2C+CD8+ T cells.
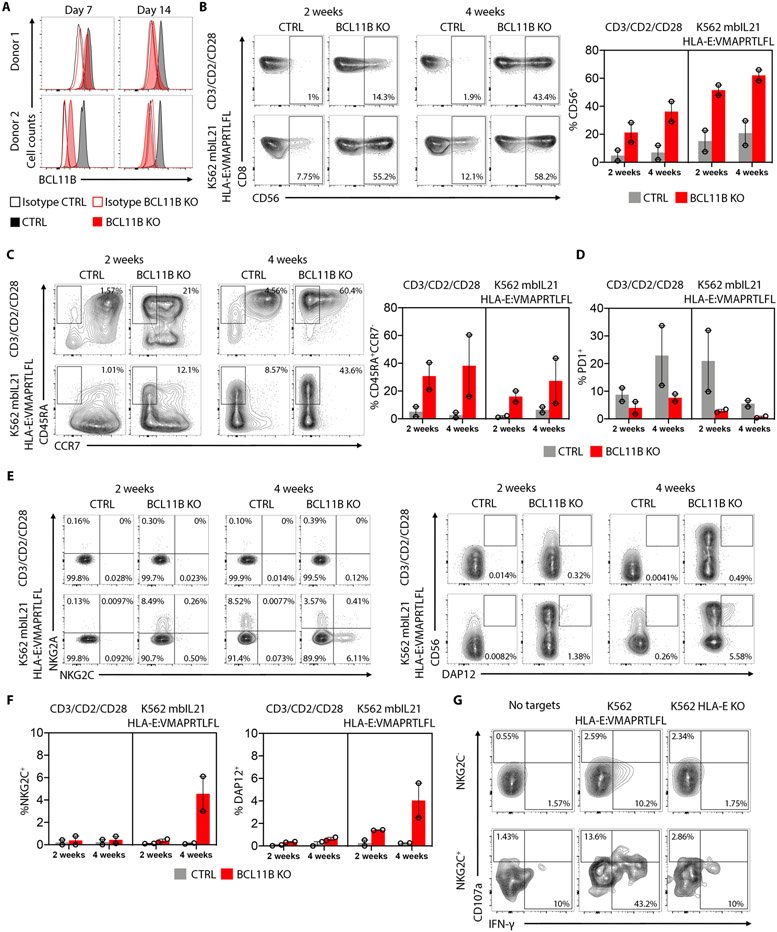
NKG2C+CD8+ T cells display a pronounced downregulation of BCL11B both at the transcription and protein level. To explore the potential role of BCL11B in the generation of the NKG2C+CD8+ T cells we isolated CD56−NKG2A−NKG2C−KIR−CD3+CD8+ T cells from two different HCMV-seronegative individuals and nucleofected these cells with CRISPR/Cas-9 ribonucleoproteins (RNPs) containing either sgRNAs targeting BCL11B for knockout (KO) or an irrelevant control sgRNA. Maximum protein loss, as measured by flow cytometry, was achieved after 14 days in culture for both donors (Fig. 5A). Nucleofected control and BCL11B KO cells were expanded using either CD3/CD2/CD28 beads or the K562 cell line expressing HLA-E with the HLA-G*01 leader sequence peptide (VMAPRTLFL) which is homologous to a high-affinity HCMV UL-40 peptide(8). The cell line also expresses membrane bound IL-21 (mbIL21) which enhances resting T cell proliferation in vitro and promotes antigen-specific CD8+ T cell expansion in vivo(25). Following deletion of BCL11B in conventional CD8+ T cells, we observed a striking increase in the frequency of cells expressing CD56, a surface marker classically associated with NK cells, when compared to the control cells under both stimulation conditions (Fig. 5B). Further analysis stratifying the cells for CD45RA and CCR7 expression showed a preferential expansion of CD45RA+CCR7− cells upon BCL11B deletion, not observed in the control cells (Fig. 5C). BCL11B KO cells also failed to upregulate PD-1 to the same extent as control cells following stimulation (Fig. 5D). Strikingly, after 4 weeks of stimulation with K562 mbIL21 HLA-E:VMAPRTLFL, NKG2C+ cells emerged in the CD8+ T cell population deleted for BCL11B. DAP12 is the adaptor molecule that associates with NKG2C to initiate the activation signal; not surprisingly, DAP12 was also induced on CD8+ T cells after 4 weeks of stimulation with K562 mbIL21 HLA-E:VMAPRTLFL. In contrast, NKG2C+ T cell expansion was not observed in control cells or in the BCL11B KO cells stimulated with CD3/CD2/CD28 beads (Fig. 5E-F). The BCL11B−NKG2C+ cells degranulated and produced IFN-γ against K562 HLA-E:VMAPRTLFL, in contrast to the BCL11B−NKG2C− cells, which demonstrated no evidence of activation by degranulation or by cytokine production (Fig. 5G). Our results provide evidence for a direct role of BCL11B downregulation in promoting an NK-like pathway of differentiation in human T cells, specifically inducing an NKG2C+CD8+ T cell phenotype that expands upon HLA-E ligand stimulation.
Fig. 5. NKG2C+CD8+ T cells can be generated in vitro by BCL11B deletion.
(A) FACS histograms showing BCL11B intracellular expression on CD8+ T cells electroporated with Cas/9 and control mRNA guides or BCL11B specific mRNA guides at the indicated time points. (B) FACS plots showing the frequency of CD8+CD56+ cells on the control and BCL11B KO cells after 2 or 4 weeks of expansion with CD3/CD2/CD28 beads or K562 mbIL21 HLA-E:VMAPRTLFL. Graph on the right shows cumulative analysis from 2 independent donors. (C) FACS plots showing the frequency of CD45RA+CCR7−CD8+ cells on the control and BCL11B KO cells after 2 or 4 weeks of expansion with CD3/CD2/CD28 beads or K562 mbIL21 HLA-E:VMAPRTLFL. Graph on the right shows cumulative analysis from 2 independent donors. (D) Cumulative analysis from 2 independent donors on PD-1+CD8+ T cells in control or BCL11B KO cells after 2 or 4 weeks of expansion with CD3/CD2/CD28 beads or K562 mbIL21 HLA-E:VMAPRTLFL. (E) FACS plots showing the frequency of NKG2A+ vs NKG2C+ and CD56+DAP12+CD8+ T cells on the control and BCL11B KO cells after 4 weeks of expansion with K562 mbIL21 HLA-E:VMAPRTLFL. (F) Cumulative analysis of 2 independent donors from the data shown in (E). (G) Representative flow cytometry plots showing degranulation (CD107a) and intracellular IFN-γ expression by K562 mbIL21 HLA-E:VMAPRTLFL-expanded pre-gated NKG2C+ or NKG2C−CD8+ T cells after 6 hours stimulation with the indicated target cells.
NKG2C+CD8+ T cells can be activated via NKG2C and TCR
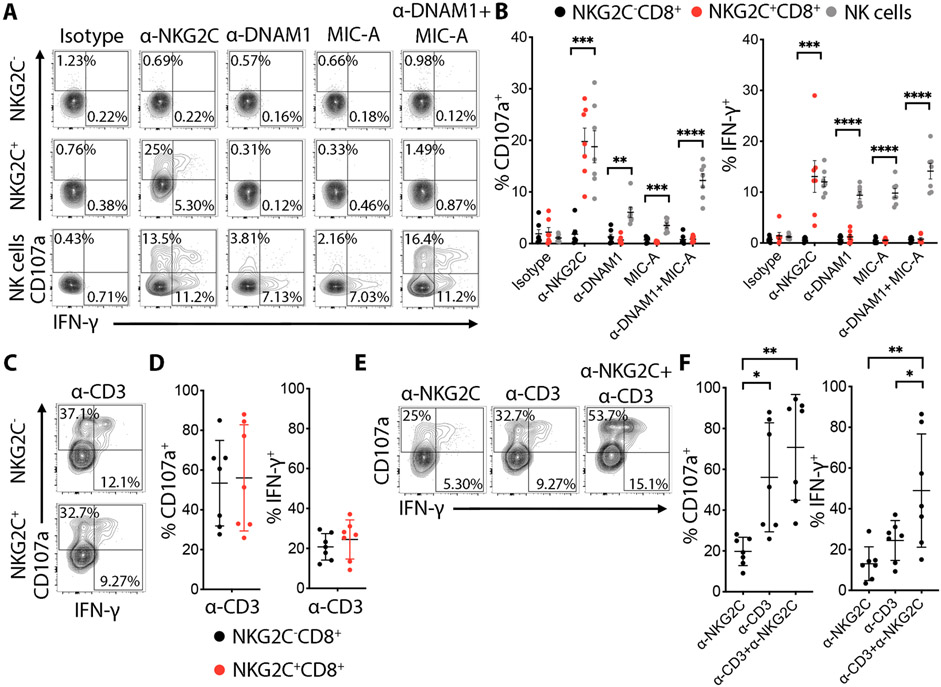
To assess whether NKG2C triggers effector function in CD8+ T cells, as it does in NK cells(26), we stimulated NKG2C by plate-bound antibody. We detected increased degranulation and IFN-γ production in NKG2C+CD8+ T cells, in contrast to NKG2C−CD8+ T cells where no significant increase was observed (Fig. 6A-B). Antibody triggering of the activating receptors DNAM-1 and NKG2D, highly expressed on NKG2C+CD8+ T cells (Fig.S2F), or a combination of both did not induce a response (Fig. 6A-B). CD3 stimulation induced high amounts of degranulation and IFN-γ production in both NKG2C− and NKG2C+CD8+ T cells, indicating that TCR signaling was equally functional in both populations (Fig. 6C-D). Importantly, even when CD3 was stimulated, NKG2C engagement could still increase the IFN-γ response among NKG2C+CD8+ T cells (Fig. 6E-F). These results demonstrate that NKG2C stimulation can induce cytotoxic and cytokine effector functions, highlighting an alternative non-antigen-restricted pathway for activation of this T cell population, in addition to their intact TCR signaling. Furthermore, co-triggering of NKG2C and CD3 enhances CD8+ T cell response beyond each individually, thereby diversifying the responsiveness and broadening the activation potential of the cell.
Fig. 6. NKG2C+CD8+ T cells express functional NKG2C that can cooperate with TCR activation in degranulation and IFN-γ production.
(A) Total PBMC cells were stimulated with the indicated plate-bound mAbs for 6 h. Representative flow cytometry plots showing degranulation (CD107a) and intracellular IFN-γ expression by pre-gated NKG2C+ or NKG2C−CD8+ T cells after triggering with plate-bound mAbs (10 μg/mL). (B) Graphs showing the percentage of CD107a+ and IFN-γ+ cells from 7 independent donors. Data are shown as mean ± SD. Statistical analysis was performed by t-test comparing the NKG2C−CD8+ vs NKG2C+CD8+ for each antibody with Holm-Sidak post-test correction. (C) FACS plots representing CD107a and intracellular IFN-γ expression by NKG2C− or NKG2C+CD8+ T cells after anti-CD3 (1 μg/mL) plate-bound stimulation. (D) Corresponding graphs showing the percentage of CD107a+ and IFN-γ+ cells from 7 independent donors. Data are shown as mean ± SD. (E) Representative FACS plots showing the percentage of CD107a+ cells after triggering of NKG2C+CD8+ T cells with plate-bound anti-CD3 or anti-NKG2C mAb alone or in combination. (F) Data are shown as the mean ± SD for 7 independent donors. Statistical significance was calculated by one-way ANOVA with Turkey’s multiple comparison test. * P-value ≤ 0.05, ** P-value < 0.01,***P-value ≤ 0.001, **** P-value < 0.0001.
NKG2C+CD8+ T cells display potent anti-tumor and anti-HCMV effector functions mediated by NKG2C and TCR specificity
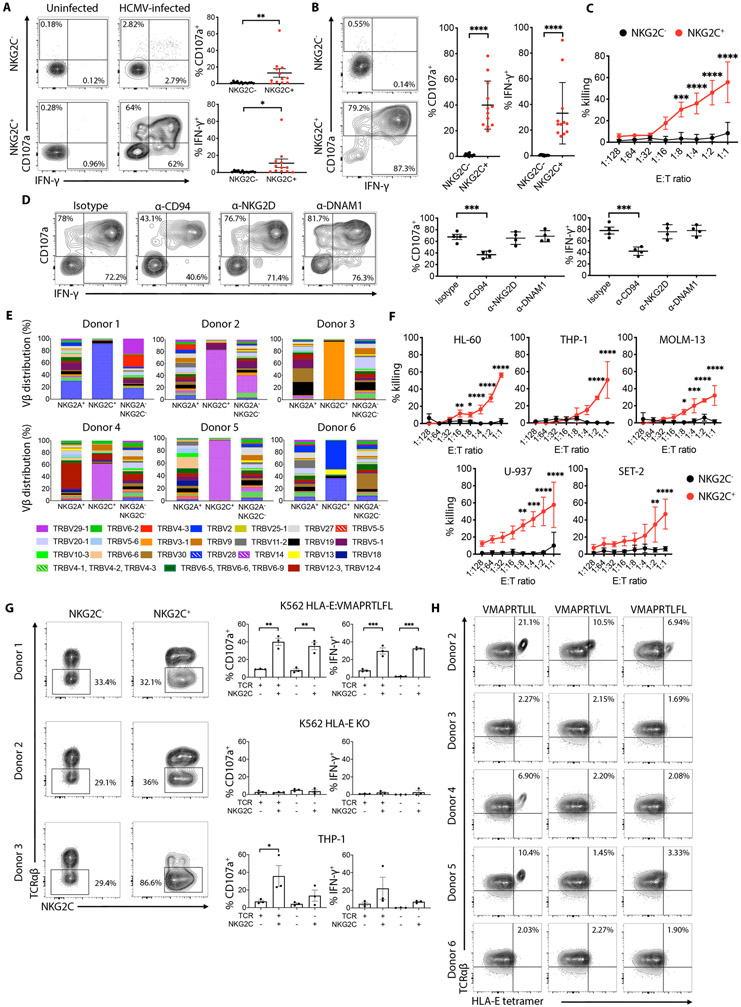
Because NKG2C+CD8+ T cells are expanded in HCMV-seropositive individuals, we assessed their activation against human fibroblasts (HFs) infected with the TB-40E HCMV strain(27). NKG2C+CD8+ T cells displayed strong reactivity to infected cells, compared to NKG2C−CD8+ T cells from the same individuals (Fig. 7A). NKG2C+CD8+ T cells were not reactive to uninfected cells. To determine if NKG2C+CD8+ T cells are capable of recognizing and killing target cells via NKG2C, we co-incubated them with K562 HLA-E:VMAPRTLFL. NKG2C+CD8+ T cells robustly responded, as measured by CD107a mobilization and IFN-γ production, whereas the NKG2C−CD8+ T cells did not (Fig. 7B). NKG2C+CD8+ T cells effectively killed K562 HLA-E:VMAPRTLFL compared to NKG2C−CD8+ T cells (Fig. 7C). Blocking NKG2C with an antibody against CD94, we observed a significant reduction in CD107a mobilization and IFN-γ production in the NKG2C+CD8+ T cell population against the target cell line, indicating contribution from the receptor (Fig. 7D). In contrast, blocking NKG2D or DNAM-1 receptors, did not have any effect (Fig. 7D). Complete blockade of NKG2C+CD8+ T cell functionality could not be achieved, however, suggesting that a separate receptor, possibly the TCR, also contributed to target cell recognition.
Fig. 7. NKG2C+CD8+ T cells anti-tumor and anti-HCMV effector functions are mediated by their NKG2C and TCR specificity.
(A) Representative flow cytometry plots showing degranulation (CD107a) and intracellular IFN-γ expression by pre-gated NKG2C+ or NKG2C−CD8+ T cells after 6 hours stimulation with uninfected or HCMV-infected human fibroblasts. Graphs on the right show cumulative analysis of CD107a+ and IFN-γ+ NKG2C+ or NKG2C−CD8+ T cells from 12 independent donors against uninfected or HCMV-infected human fibroblasts. Statistical significance was calculated using Wilcoxon matched-pairs signed rank test. (B) Representative flow cytometry plots showing degranulation (CD107a) and intracellular IFN-γ expression by pre-gated NKG2C+ or NKG2C−CD8+ T cells after 6 hours stimulation with K562 HLA-E:VMAPRTLFL target cells. Graph on the right show cumulative analysis of CD107a+ and IFN-γ+ NKG2C+ or NKG2C−CD8+ T cells from 12 independent donors against K562 HLA-E:VMAPRTLFL target cells. Statistical significance was calculated using Wilcoxon matched-pairs signed rank test. (C) In vitro cytotoxicity of FACS-sorted NKG2C+ or NKG2C−CD8+ T cells against K562 HLA-E:VMAPRTLFL target cells was assessed using a 6 hours bioluminescence assay. Results from 3 independent donors are shown. Statistic was calculated using a 2-way ANOVA comparing the mean of each effector to target (E:T) ratio between NKG2C− and NKG2C+CD8+ T cells. (D) Representative flow cytometry plots showing degranulation (CD107a) and intracellular IFN-γ expression by pre-gated NKG2C+CD8+ T cells against K562 HLA-E:VMAPRTLFL in the presence of the indicated blocking mAbs (10 μg/mL). Graphs on the right show cumulative analysis of 4 independent donors. Statistical significance was calculated using one-way ANOVA with Turkey’s multiple comparison test. (E) CD8+ T cell from 6 healthy donor PBMC were stained with a combination of different monoclonal antibodies against individual Vβ TCRs and analyzed by flow cytometry. Cells were pre-gated on NKG2A+NKG2C− (NKG2A+), NKG2A−NKG2C+ (NKG2C+) and NKG2A−NKG2C− CD8+ T cells. (F) In vitro cytotoxicity of FACS-sorted NKG2C+ or NKG2C−CD8+ T cells against the indicated AML target cell lines was assessed using a 6 hour bioluminescence assay. Results from 3 independent donors are shown. Statistic was calculated using a 2-way ANOVA comparing the mean of each E:T ratio between NKG2C− and NKG2C+CD8+ T cells (G) FACS plots showing TCRαβ expression on NKG2C+CD8+ or NKG2C−CD8+ T cells electroporated with Cas/9 and TRAC specific mRNA guides. Graphs on the right show cumulative analysis of CD107a+ and IFN-γ+ cells against the indicated targets. Statistical significance was calculated by one-way ANOVA with Turkey’s multiple comparison test. (H) NKG2C+CD8+ T cells from 5 donors were stained with the indicated HLA-E tetramers in the presence of a CD94 blocking antibody. ****P-value ≤ 0.0001, ***P-value ≤ 0.001, **P-value ≤ 0.01, *P-value ≤ 0.05.
Based on these results, we analyzed TCR Vβ usage by NKG2C+CD8+ T cells in six individuals using a panel of 24 TCR Vβ antibodies to stain the NKG2A+NKG2C−, NKG2A−NKG2C+ and NKG2A−NKG2C− CD8+ T cell subpopulations. In all individuals, the majority of the NKG2C+CD8+ T cells expressed one prominent TCR Vβ chain, with the population being strikingly dominant in four of the six individuals. In contrast, all other CD8+ T cell subpopulations, including those expressing NKG2A, demonstrated a diversity of TCR Vβ usage (Fig. 7E). Interestingly, in 50% of individuals, we observed a skewed distribution towards a single dominant Vβ chain: TRBV-14. To determine if other CD45RA+CCR7− populations, including canonical TEMRA cells, demonstrate a similarly narrow TCR Vβ chain repertoire, we analyzed TCR Vβ chain usage in two different individuals. We stratified the CD8+ T cell population into canonical TEMRA (CD45RA+CCR7−) and the NKG2C+CD8+ population, and compared both to naïve T cells (CD45RA+CCR7+). As expected, the NKG2C+CD8+ population exhibited the narrowest TCR Vβ chain repertoire (Fig. S4A). Interestingly, NKG2C+CD8+ T cells from HCMV-seronegative donors, in which they are found at a low frequency (<2%), exhibited a polyclonal repertoire comparable to that observed in NKG2C−CD8+ cells from the same individuals (Fig. S4B).
The sequence of CDR3 and the identity of the flanking V and J gene segments are widely used to classify TCR variants(28). To identify unique CDR3 regions, we took advantage of iPair next-generation sequencing technology to specifically amplify the productive α and β TCR transcripts across the CDR3 in the NKG2C+CD8+ T cells of three donors previously analyzed by flow cytometry for TCR Vβ usage. Sequencing of NKG2C+CD8+ T cell clones confirmed the predominance of single Vβ family usage within each donor (Table S2). Of the 21 productive CDR3 sequences identified in Donor 1, 19 (86%) showed TRBV-28 usage. For donors 4 and 5, we also observed predominance of one Vβ use, with 26 of 29 clones (90%) in donor 4 and 21 of 22 clones (95%) in donor 5 exhibiting TRBV-14 usage.
Given the difference in TCR usage across donors, we investigated whether this variability influenced recognition of a series of allogeneic acute myeloid leukemia (AML) cell lines. While NKG2C+CD8+ T cells from all donors tested were able to recognize K562 HLA-E:VMAPRTLFL (Fig. 7B-C), only those exhibiting TRBV-14 usage demonstrated significantly higher killing capacity against all six leukemia cell lines compared to their NKG2C−CD8+ T cell counterparts (Fig. 7F). CD94 blockade reduced CD107a mobilization and IFN-γ production by NKG2C+CD8+ T cells against the AML targets (Fig. S5A), all of which displayed HLA-E surface expression (Fig. S5B). The lack of complete abrogation of NKG2C+CD8+ T cell functionality with CD94 blockade suggests non-overlapping activation via a separate mechanism, such as the TCR. Recognition of the AML cells was specific to the NKG2C+CD8+ population, as NKG2C−CD8+ cells, both naïve (CD45RA+CCR7+) and canonical TEMRA (CD45RA+CCR7−), displayed no response to the tumor targets (Fig. S5C).
To determine if the TCR was involved in the recognition of AML target cells, we performed a TCR knockout using CRISPR/Cas-9 RNPs targeting the TRAC locus from the three broadly reactive donors exhibiting an expanded NKG2C+CD8+ T cell population. After 4 days in culture, a proportion of both NKG2C− and NKG2C+ cells had lost surface TCRαβ expression, as assessed by flow cytometry (Fig. 7G). Loss of TCRαβ did not lead to significant reduction in CD107a mobilization and IFN-γ production by NKG2C+TCRαβ−CD8+ T cells against the K562 HLA-E:VMAPRTLFL target cell line. This indicates that the intact NKG2C receptor on these cells was potently activated by the peptide VMAPRTLFL, as previously found for NKG2C+ NK cells(8), and that the TCR is not specific for this UL-40 derived HCMV sequence (Fig.7G). Loss of HLA-E expression on the target cells completely abrogated the functional response of the NKG2C+CD8+ T cells, confirming activation occurs through NKG2C:HLA-E engagement (Fig.7G). In contrast, loss of TCRαβ in NKG2C+CD8+ T cells led to partial abrogation of the reactivity against the AML cell line THP-1, indicating contribution of the TCR receptor to target cell recognition (Fig.7G).
To determine whether the TCR of NKG2C+CD8+ T cells has specificity for HLA-E-bound peptides, HLA-E*01:03 tetramers folded with VMAPRTLIL, VMAPRTLVL or VMAPRTLFL were used to stain CD8+ T cells from 5 different donors, in the presence of anti-CD94 mAb to prevent binding of the NKG2A/NKG2C receptors to the HLA-E tetramer. The VMAPRTLIL, VMAPRTLVL, and VMAPRTLFL peptides are present in the leader sequence of the UL40 protein derived from different HCMV strains, but they are also part of the leader sequence of various HLA class I alleles(29). The majority of TCR+ cells from donor 2, 4 and 5 showed binding to the HLA-E tetramer folded with the VMAPRTLIL sequence, while equivalent cells from donors 3 and 5 did not show significant tetramer binding (Fig. 7H). Of note, donors 2, 4 and 5 were the same donors that displayed a dominant TRBV-14 Vβ chain usage and broad recognition of several AML targets. Analysis of the HLA genotype of donors and target cells (Table S3) revealed that VMAPRTLIL is a non-self peptide for donors 2, 4 and 5, due to its absence in their HLA-C alleles. In contrast, donors with HLA-C alleles exhibiting the VMAPRTLIL peptide (donors 3 and 6) failed to generate CD8+ T cells specific for the self-peptide (Fig. 7H) and did not functionally react against the allogeneic AML targets. Therefore, in some individuals with expansion of NKG2C+CD8+ T cells, the TCR is HLA-E-restricted and recognizes HCMV-derived peptides but only if they are homologous to non-self HLA peptides.
Like NK cells, the NKG2C+CD8+ T cell population expresses KIR receptors (Fig. 2B-C). To investigate if functional inhibition is mediated by KIR, we co-incubated KIR2DL2/L3+NKG2C+CD8+ T cells with the HLA A-B-C-deficient cell line 721.221, and a 721.221 transfectant expressing the HLA-C1 ligand Cw*03. NKG2C+CD8+ T cells responded to wild-type 721.221 cells (Fig. S5D) via HLA-E recognition (Fig. S5E-F). Presence of the KIR2DL2/L3 cognate ligand HLA-Cw*03 on the target cell, however, abolished CD107a mobilization and IFN-γ production by NKG2C+CD8+ T cells, similar to KIR2DL+ NK cell response from the same individuals (Fig. S5D), indicating that KIR receptors are fully functional in the T cell and can mount an inhibitory signal upon binding to its HLA ligand.
Overall, TCR studies of the NKG2C+CD8+ T cell population establish that in some HCMV-seropositive individuals, the NK-like T cells undergo substantial clonal expansion in vivo, frequently bearing a TRBV-14-derived TCR that targets a non-self HLA-E-presented peptide. Combined with the broader recognition of HLA-E through co-expressed NKG2C, NKG2C+CD8+ T cells from these individuals display greater cytotoxic potential compared to conventional CD8+ T cells.
NKG2C+CD8+ T cells proliferate in response to HLA-E and represent a platform for CAR-T cell manipulation
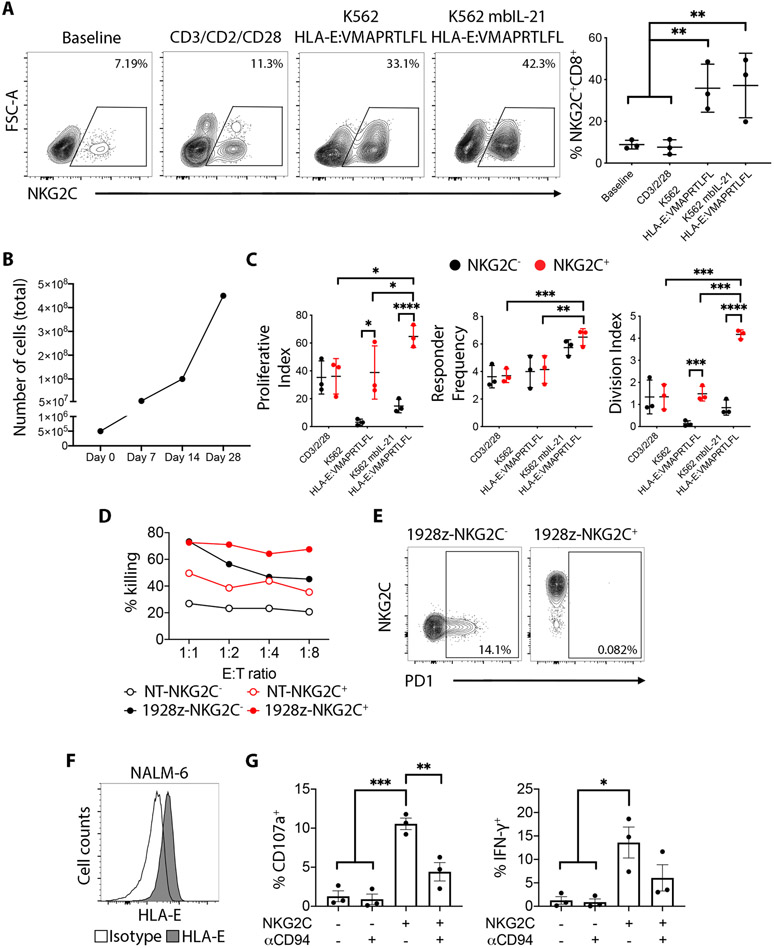
We next examined if HLA-E specifically induces proliferation of NKG2C+CD8+ T cells by co-incubating peripheral blood mononuclear cells (PBMC) with K562 HLA-E:VMAPRTLFL cells. Following one week of stimulation, an increase in the frequency of NKG2C+CD8+ T cells could be observed, indicating preferential expansion of the cell population (Fig. 8A). Co-incubation of NKG2C+CD8+ T cells with the K562 mbIL21 HLA-E:VMAPRTLFL also induced expansion of the NKG2C+CD8+ T cell population (Fig. 8A), 1000-fold after 28 days of stimulation (Fig.8B). Responder frequency and division index were also higher among NKG2C+CD8+ T cells in response to both K562 HLA-E:VMAPRTLFL and K562 mbIL21 HLA-E:VMAPRTLFL, compared to NKG2C−CD8+ T cell counterpart, whereas there was no difference between the two populations following CD3/CD2/CD28 stimulation (Fig. 8C). After a 7-day stimulation with either CD3/CD2/CD28 beads or K562 mbIL21 HLA-E:VMAPRTLFL, NKG2C+CD8+ T cells acquired surface expression of CD45RO (Fig. S6A-B) and maintained higher capacity to degranulate and produce IFN-γ compared to the NKG2C−CD8+ T cell counterpart (Fig. S6C-D). Taken together, our data demonstrate that HLA-E on the target cell can provide an extrinsic signal for selective proliferation.
Fig. 8. NKG2C+CD8+ T cells proliferate in response to HLA-E and display superior tumor killing if transduced with a CD19-CAR.
(A) Left: Representative plots showing total PBMC cells stimulated for 7 days with the indicated stimuli and pre-gated on CD3+CD8+ cells. Right: Cumulative analysis from 3 independent donors. Statistical significance was calculated by one-way ANOVA with Turkey’s multiple comparison test. (B) Total cell counts of pre-sorted NKG2C+CD8+ T cells stimulated with K562 mbIL21 HLA-E:VMAPRTLFL (ratio 1:1) for the indicated number of days. (C) Total CD8+ T cells were CTV labelled and exposed for 7 days to the indicated stimuli. Pre-gated NKG2C+ or NKG2C−CD8+ T cells were then analyzed for proliferation monitoring CTV dilutions. Statistical analysis was calculated using a 2-way ANOVA. (D) NKG2C− and NKG2C+CD8+ T cells from one representative donor were FACS sorted and left untransduced or transduced with a 1928z CD19-targeting CAR construct. Cytotoxicity was assessed using a standard 51Cr assay after 24h of exposure to the CD19+ cell line NALM6. (E) PD-1 surface expression on NKG2C−CD8+ and NKG2C+CD8+ cells transduced with the 1928z CD19-targeting CAR construct. (F) Histogram shows HLA-E surface staining on NALM-6 cells. (G) Cumulative analysis of 3 independent donors showing degranulation (CD107a) and intracellular IFN-γ expression by pre-gated NKG2C−CD8+ T cells and NKG2C+CD8+ T cells against NALM6 in the presence of a CD94 blocking mAb (10 μg/mL). Statistical significance was calculated by one-way ANOVA with Turkey’s multiple comparison test. ****P-value ≤ 0.0001, ***P-value ≤ 0.001, **P-value ≤ 0.01, *P-value ≤ 0.05.
Having demonstrated rapid and extensive expansion upon HLA-E stimulation, NKG2C+CD8+ and NKG2C−CD8+ T cells from one donor were isolated and, after two weeks of expansion using K562 mbIL21 HLA-E:VMAPRTLFL, they were transduced with the CD19-specific CAR construct 1928z(30). When co-cultured with the CD19-expressing target cell line NALM-6 the 1928z-NKG2C+CD8+ cells demonstrated higher killing capacity than their 1928z-NKG2C−CD8+ T cell counterpart, particularly at low effector to target (E:T) ratios (Fig. 8D). In contrast to 1928z-NKG2C−CD8+ T cells, the 1928z-NKG2C+CD8+ T cells did not upregulate PD-1 (Fig. 8E). Basal NALM-6 recognition by non-transduced NKG2C+CD8+ T cells was also higher compared to non-transduced NKG2C−CD8+ T cells, due to activation trough recognition of HLA-E expressed on the NALM-6 targets (Fig. 8F-G). These results illustrate the greater cytotoxicity potential of the NKG2C+CD8+ T cells against a tumor target and its amenability for cell engineering without the limitation of cell exhaustion.
Discussion
There is now increasing evidence to challenge the concept that clear phenotypic and functional distinctions exists between the innate and adaptive immune cell populations. It has long been observed that T cells can express the NK-associated KIR and NKG2A receptors(14-16), although their functions remain unclear. A CD8+ T cell subset expressing the NKG2C receptor has previously been observed(31, 32), suggesting that the NKG2C may constitute an alternative T-cell activation pathway(16). The origin of NKG2C+CD8+ T cells, the factors leading to their expansion, and the functional peculiarities of this lymphocyte population, however, have largely been unexplored.
Expanded in the peripheral blood of healthy HCMV-seropositive individuals, these “unconventional” NKG2C+ T cells are already committed to the CD8-single positive stage of T cell development, but have lost expression of the transcription factor BCL11B and have become phenotypically and functionally similar to NK cells. While NKG2C+CD8+ T cells may exist in the peripheral blood of healthy HCMV-seronegative individuals, their frequency notably remains below 2%. Together with the observation that the population expands in the setting of HCMV reactivation following allogeneic hematopoietic cell transplantation, these findings strongly indicate that HCMV infection is the primary stimulus leading to the expansion of the innate-like NKG2C+CD8+ T cells.
Despite the likelihood that both NKG2C+ NK and T lymphocytes can expand upon HCMV infection or reactivation, there is no apparent correlation between expansion of one NKG2C+ lymphocyte population with the other. Furthermore, it is not clear why some individuals exclusively develop NKG2C+NK cells or NKG2C+CD8+ T cells and why some expand both populations or neither. It is evident, however, that NKG2C+CD8+ T cells occur at the highest frequency in HCMV-seropositive individuals homozygous for the NKG2C gene KLRC2, as previously seen for NK cells(33). Accordingly, the frequency of HCMV reactivation is higher among recipients of hematopoietic cell allografts from donors negative for KLRC2(34). While initial expansion of the NKG2C+CD8+ T cells may occur via TCR engagement in response to primary HCMV infection, repeated expansion of the cell population would likely follow lifelong recurrences of HCMV reactivation. Activation via co-expressed NKG2C may provide additional protection from HCMV reactivation and disease.
The T cell response to HCMV is known to be robust and directed against many peptides of the virus(35). Prior studies have primarily focused on pp65-specific T cells, but the presentation of non-pp65 epitopes may stimulate expansion of alternative T cell subpopulations such as NKG2C+ CD8+ T cells. Selective expansion of NKG2C+ CD8+ T cell clones may be related to viral strain and peptide presentation by HLA-E(8), which also presents leader peptides of HLA class I molecules(36). Variation in viral strain has been proposed as a reason for expansion of NKG2C+ NK cells; however, this alone would not explain why one lymphocyte population (NK vs T cell) and not the other would expand in the same individual, when both populations express the same receptor. Specificity of the co-expressed TCR on the NKG2C+CD8+ T cell therefore likely plays a role, a hypothesis supported by the observation that the expanded population of NKG2C+CD8+ T cells is highly oligoclonal. Together with the finding that the small fraction of NKG2C+CD8+ T cells found in HCMV-seronegative individuals is polyclonal, these results support the concept of a strong selective and antigen-driven TCR expansion occurring among NKG2C+ T cell clones in HCMV-infected individuals.
Expression of inhibitory KIR2DL1, KIR2DL2/L3 and KIR3DL1 receptors is evident on NKG2C+CD8+ T cells, consistent with previous reports of KIR expression on terminally differentiated (CD45RA+CD57+CCR7−CD27−CD28−IL7R−) CD8+ T cells(15, 37). NKG2C+ adaptive NK cells preferentially express self-HLA-specific inhibitory KIR. In a previous study on total CD8+ T cells, self- and nonself-specific KIR appear to be randomly distributed (15). In contrast, we found a higher frequency of KIR2DL1+NKG2C+CD8+ T cells in C2/C2 individuals compared to C1/C1 individuals, suggesting a modest self-KIR bias. In our study and in line with previous observations on other T cell subsets(38), the KIR+NKG2C+CD8+ T cell population can be functionally inhibited by cognate HLA class-I molecules expressed on the target cell, indicating that a self-regulating mechanism to control TCR responses exists possibly to avoid cross-reactivity against self-antigens.
Ex vivo phenotype analysis reveals that NKG2C+CD8+ T cells seemingly resemble the TEMRA subset, a classification of memory CD8+ T cells associated with chronic infections, including HCMV(39). We observed, however, that NK cells show the same expression pattern for the markers commonly used to classify TEMRA cells, indicating that this phenotype could be associated with the overall T to NK cell shift rather than a terminally differentiated T cell status. Indeed, after prolonged stimulation, NKG2C+CD8+ T cells can acquire expression of CD45RO. Furthermore, the transcriptional profile of classic TEMRA cells is distinct from that observed in NKG2C+CD8+ T cells, which are more transcriptionally related to NK cells. In the setting of chronic viral infections or malignancies, cellular immunity may be impaired due to the upregulation of cell surface exhaustion molecules, such as PD-1. In our study, absence of PD-1 expression is an exceptional feature of the NKG2C+CD8+ T cell, suggesting a TCR with low affinity and avidity, as has been reported for melanoma-specific CD8+ T cells with low PD-1 expression(40, 41). This characteristic could be critical to ensure long-term proliferation and survival of NKG2C+CD8+ T cells(42).
We unexpectedly found that the transcription factor BCL11B is markedly downregulated in NKG2C+CD8+ T cells. Bcl11b is a zinc-finger transcription factor required for the development of T cells in mice(43) and is first expressed at the CD4−CD8− stage of T cell development. Germline deletion of Bcl11b leads to decreased sensitivity in T cells to Notch signaling and differentiation into NK cells(44). Paradoxically, as NK cells differentiate, BCL11B expression increases, together with several genes associated with T cell signaling (e.g., CD3D, CD3E, CD5, CD6, THEMIS), with the highest BCL11B expression evident in the NKG2C+ “adaptive” subset (45). Thus, T cells can be reprogrammed into NK cells via BCL11B deletion, while NK cells may acquire T cell features via BCL11B upregulation, reinforcing a model that the identities of NK and T cell subpopulations are governed by reciprocal BCL11B-dependent gene programs, at times overlapping.
We find that in humans, downregulation of BCL11B occurs naturally in the NKG2C+CD8+ T cells, and indeed is a marker for a general NK reprogramming of a mature T cell, as supported by the transcriptional upregulation of genes classically associated with “NK cell identity” and “NK cell cytotoxicity” and the simultaneous downregulation of genes associated with “T cell identity”. In support of the active role BCL11B plays in the NK reprogramming of the T cell population, transcriptome comparison with a murine dataset where Bcl11b was deleted in young adult thymocytes at the DN2 stage of T cell development(22) reveals that the majority of genes observed to be differentially expressed in naturally occurring human NKG2C+CD8+ T cells are modulated in the same direction in the Bcl11b-knockout murine cells. Strikingly, BCL11B deletion in canonical mature NKG2C−CD8+ T cells gives rise to CD45RA+CCR7−CD56+NKG2C+DAP12+ cells, providing further evidence that loss of BCL11B is directly responsible for the overall reprogramming of NKG2C+CD8+ T cells into NK-like cells. Of note, NKG2A+CD8+ T cells also expand upon BCL11B deletion. This is somewhat paradoxical, given the fact that stimulation with HLA-E should suppress the expansion of NKG2A+ cells for which HLA-E represents an inhibitory signal. Others have proposed that NK cells do not downregulate the expression of CD94/NKG2A receptors upon ligation(46), but rather that NKG2A is constantly recycled at a slow rate, whereupon HLA-E serially triggers its expression to maintain an inhibitory signal(47). A similar mechanism of sustained NKG2A surface expression may therefore take place in T cells, as supported by our data. Absence of BCL11B is likely responsible for the resistance to PD-1 upregulation observed in NKG2C+CD8+ T cells. In our transcriptome analysis, PDCD1 was found to be controlled by BCL11B both in the human and murine datasets. Moreover, NK cells, which express BCL11B at similarly low levels as NKG2C+CD8+ T cells, have minimal to no PD-1 expression(48), further supporting a role for BCL11B in regulating the similarities between the two lymphocyte subsets.
TCR analysis indicates that these reprogrammed T cells have undergone significant clonal expansion in HCMV-seropositive individuals, suggesting that a strong response to a pathogenic challenge occurs in vivo. Whether reprogramming of the CD8+ T cell occurs after TCR engagement and expansion of the T cell clone or it is an intrinsic characteristic of a T cell subset that during development is diverted from a “conventional” CD8+ T cell program, remains unclear. As our data suggest, the presence of a small fraction of CD8+ T cells displaying NKG2C surface expression in HCMV-seronegative individuals supports the second hypothesis. The finding that NKG2C activation of CD8+ T cells can trigger cytotoxic granule release and IFN-γ production in the absence of CD3 stimulation reveals a TCR-independent pathway for activation. Moreover, NKG2C signaling complements TCR signaling in activation of the T cell, revealing two non-overlapping pathways that serve to enhance each other. NKG2C+CD8+ T cells from multiple individuals were able to mount a robust and efficient cytotoxic response against HLA-E:VMAPRTLFL-expressing targets and HCMV-infected targets, which are known to upregulate HLA-E upon HCMV infection(49). While the HLA-G-derived leader sequence VMAPRTLFL is an optimal ligand for CD94:NKG2C receptors(26, 50), we found that the TRBV-14 TCR expressed by NKG2C+CD8+ T cells is not specific for the self-peptide VMAPRTLFL, but rather for a non-self HLA-E-presented peptide. Thus, NKG2C+CD8+ T cells exhibit an HLA-E-restricted TCR that cooperates with NKG2C for target cell recognition. Although HLA-E expression on healthy tissue is relatively low compared to classical HLA-class I molecules, it is frequently overexpressed in many types of tumors(51), suggesting a potent immunosurveillance role via HLA-E for NKG2C+CD8+ T cells.
Previous reports have elucidated a clinical benefit to a population of polycytotoxic T cells expressing NKG2C in leprosy and tuberculosis(18, 52), suggesting that NKG2C could be used as a biomarker to monitor protective CD8+ T cell responses to intracellular pathogens. Beyond this, however, the innate capacity of the NKG2C+CD8+ T cell population to respond to virally infected cells and tumor cells, combined with a capacity for ex vivo proliferation and resistance to PD-1 upregulation, reveal a potential utility as a cellular immunotherapy, either unmodified or as a cellular vehicle for modification with a chimeric antigen receptor. Indeed, we found that 1928z-NKG2C+CD8+ T cells were superior effector cells compared to 1928z-NKG2C−CD8+ T cells from the same donor.
Finally, our findings have important implications for the development and differentiation of T and NK cell lineages. Downregulation of BCL11B transcription in NKG2C+CD8+ T cells indicates that a lymphocyte already committed to the T cell lineage can still be diverted toward an innate lymphoid fate, gaining innate capacity to respond through NKG2C without sacrificing any ability to signal via the native TCR. What molecular events trigger this diversion late in T cell development remains unknown but raises the possibility of reprogramming T cells to adopt NK-like features for immune advantage by targeting BCL11B.
Materials and Methods
Study design
The goal of this study was to characterize the human CD8+ T cell population that expresses NKG2C. To this end, we relied on phenotypic analysis of healthy human volunteer donors, cell-based functional assays and RNA-sequencing. Validation of BCL11B expression was performed by flow cytometry and assessment of BCL11B function used in vitro transduction KO experiments. Replicates and statistical tests are provided in the figure legends. All data points were included and no outliers were excluded. Researchers were not blinded as to the source of the blood samples and HCMV serostatus.
Cell sources and preparation
Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll (GE Healthcare) centrifugation. Additional PBMC were isolated from buffy coats obtained from healthy volunteer donors via the New York Blood Center (NYBC). The MSKCC Institutional Review Board (IRB) waived the need for additional research consent for anonymous NYBC samples. PBMC were cryopreserved in fetal bovine serum with 10% DMSO. DNA was extracted from whole blood (Qiagen) and KLRC2 copy number was determined by PCR(53) for 130 individuals. Human cytomegalovirus (HCMV) serostatus was provided by NYBC. A total of 59 adult patients who underwent double umbilical cord blood (DUCB) hematopoietic cell transplantation (HCT) at Memorial Sloan Kettering Cancer Center (MSKCC) between 2006 and 2017 were included in the study, 30 patients with and 29 patients without HCMV reactivation. Patients provided informed written consent for research, and studies were approved by the MSKCC IRB.
Flow cytometry
PBMC (2 × 105 cells per well) were stained for 20 minutes with LIVE/DEAD™ Fixable Aqua - Dead Cell Stain Kit (405 nm, Invitrogen) followed by 30 minutes staining in FACS buffer (PBS with 0.5% BSA and 2mM EDTA) at room temperature with the following antibodies: CD3 (UCHT1, BD Biosciences), CD56 (N901, Beckman Coulter), CD4 (VIT4, Miltenyi Biotec), CD8 (RPA-T8, Biolegend), TCR-γδ (B1, Biolegend), TCR-Vδ1 (REA173, Miltenyi Biotec), KIR2DL1/S1 (EB6B, Beckman Coulter), KIR2DL2/L3/S2 (CH-L, BD Biosciences), KIR3DL1 (DX9, BD Biosciences), NKG2C (REA205, Miltenyi Biotec), NKG2A (Z199, Beckman Coulter), PD-1 (EH12.2H7, Biolegend). FcεRIγ (rabbit polyclonal, Millipore Sigma) was detected by intracellular staining using the FIX & PERM™ Cell Permeabilization Kit (Thermo Fisher).
For BCL11B staining, 1 × 106 cells per well previously stained for extracellular markers as indicated above and were fixed and permeabilized using BD Phosflow kit (BD Bioscience). After two washes, intracellular staining was performed using BCL11B antibody (Abcam) or a Rat IgG2a Isotype control (Abcam) in FACS buffer.
TCR repertoire studies were performed using a IOTest® Beta Mark TCR Vβ Repertoire Kit (Beckman Coulter). For T cell differentiation analysis, 2 × 105 PBMC were stained with the following antibodies: CD3 (UCHT1, BD Biosciences), CD8 (RPA-T8, Biolegend), CD4 (VIT4, Miltenyi Biotec), CD45RO (UCHL1, Biolegend), CD45RA (HI100, Biolegend), CD27 (M-T271, Biolegend), CD28 (CD28.2, Biolegend), CD197 (G043H7, Biolegend), CD95 (DX2, Biolegend), CD127 (A019D5, Biolegend), CD57 (HNK-1, Biolegend), CD62L (DREG-56, Biolegend).
The following tetrameric peptide/HLA class I tetramer complexes from MBL® Interational Corporation were used in this study: HCMV pp65 NLVPMVATV/HLA A*0201 and HCMV pp65 TPRVTGGGAM/HLA B*0702. The APC-labelled HLA-E tetramers were provided by the NIH Tetramer Core Facility at Emory University.
For HLA-E surface detection on tumor cell lines, clone 3D12 (Biolegend) was used. All FACS analyses were performed on an LSR Fortessa (BD Biosciences) and analyzed using FlowJo software (10.6.2).
Cell lines and culture
Human acute myelogenous leukemia (AML) cell lines KG1, HL-60, THP-1, MOLM-13, U-937, SET-2 and the erythroleukemia cell line K562 expressing HLA-E with the HLA-G*01 leader sequence peptide (VMAPRTLFL) were cultured in RPMI supplemented with 10% FCS and 1% Penicillin-Streptomycin. All the AML cell lines were stably transduced with a GFP-firefly luciferase fusion protein as described elsewhere(54).
The 721.221 B lymphoblastoid cell line and 721.221 transfected with cDNA encoding HLA-Cw3 or HLA-Cw4 (kindly provided by Dr. Peter Parham, Stanford University, Stanford, CA) were cultured in RPMI supplemented with 10% FCS and 1% Penicillin-Streptomycin.
The HLA-E*01:01 gene with the signal peptide exchanged for that of HLA-G*01 (MVVMAPRTLFLLLSGALTLTETWA) was codon-optimized for human translation and synthesized in pUC57 (Genscript). Site-directed mutagenesis to convert HLA-E*01:01 for HLA-E*01:03 (R128G) was performed using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) with the forward primer 5’-GGGACCAGACGGGAGATTCCTG-3’ and reverse primer 5’-AGCTCGCATCCGTGCATC-3’ (IDT). The HLA-E*01:03-HLA-G*01 gene was then cloned in the 3rd generation lentivector pERRL by restriction digest cloning with XbaI and SalI (NEB); pERRL was derived from pRRLSIN.cPPT.PGK-GFP.WPRE (a gift from Didier Trono, Addgene #12252), exchanging the human PGK promoter for the human EF1α promoter and inserting an XbaI restriction site in the 5’ MCS. Lentivirus was produced as previously described(55); briefly, 293T cells (ATCC) were transiently transfected with 15μg pERRL, 18μg pRSV-Rev (a gift from Didier Trono, Addgene #12253), 18μg pMDLg/pRRE (a gift from Didier Trono, Addgene #12251), and 7μg pCI-VSVG (a gift from Gary Nolan, Addgene #1733) using Lipofectamine 2000 (Invitrogen), supernatants were collected at 24 and 48hr post-transfection and concentrated by centrifugation at ≥30,000g for ≥6hr. Viral pellets were combined and resuspended in RPMI complete media and used to transduce parental K562 cells or K562 Clone 9 mbIL21 cells (provided by Dean Lee, Nationwide Children’s Hospital, OH). Transduced K562 cells were sorted by FACS for uniform high expression of HLA-E, while a high-expressing clone of transduced K562 Clone 9 mbIL21 was isolated by limiting dilution and grown out.
Human primary fibroblasts cultures were obtained by outgrowth of cells from explanted skin biopsies(56). Briefly, after dissection, the dermis was minced into 1mm3 pieces and 8 fragments were plated on the bottom of a 75 cm2 culture dish. Explants were let air dry for 15 minutes and then 8 mL of DMEM supplemented with 20% FCS was added. After 2 weeks cells were dissociated with trypsin-EDTA. Human fibroblasts were infected with the HCMV TB40/E strain derivative TB40-BACKL7-SE-EGFP as previously described(27). The percentage of infected cells was assessed by flow cytometry (EGFP) prior to functional experiments.
For T cell proliferation analysis, 2 x 105 CellTrace™ Violet (Invitrogen) dye-labelled PBMC were cultured in 50U/mL IL-2 (Proleukin, Prometheus Laboratories) and stimulated with CD3/CD2/CD28 beads (Miltenyi Biotec) at a bead-to-cell ratio of 1:2 or irradiated K562 HLA-E:VMAPRTLFL at a ratio of 1:1 for 7 days, replenishing the IL-2 and irradiated K562 HLA-E:VMAPRTLFL 4 days post-stimulation. Responder frequency, proliferation index, and division index were calculated as previously described(57).
For CAR-T experiments, NKG2C+ and NKG2C−CD8+ T cells were first FACS-sorted and stimulated with CD3/CD2/CD28 beads (Miltenyi Biotec) at a bead-to-cell ratio of 1:2 for 48h in the presence of 100U/ml IL2. After 48h, beads were magnetically removed and cells were transduced as previously described(58). Prior to functional experiments, cells were expanded for 2 weeks with K562 mbIL21 HLA-E:VMAPRTLFL at a ratio of 1:1, in the presence of 100U/ml IL2.
Functional assays
CD107a mobilization and IFN-γ production were used to determine CD8+ T cell activation. Frozen PBMC samples were thawed and rested overnight in RPMI complete media with 200 U/mL IL-2 in a 37°C incubator with 5% CO2. PBMC (2 × 105 cells per well) were incubated in 96-V bottom plates with target cells at a 5:1 ratio in the presence of anti-CD107a antibody (BD Biosciences). After 2 hours of co-culture, 55.5 μg/mL of Brefeldin A (MP Biomedicals) was added to the cells. After additional 4 hours of co-culture, cells were washed, fixed/permeabilized and stained with anti-IFNγ antibody (BD Biosciences).
For plate-bound Ab stimulation assays, 96-well flat-bottom plates were pre-coated overnight at 4°C with the following antibodies (10μg/mL): anti-NKG2C (134522, R&D systems), anti-NKG2D (1D11, Biolegend), anti-DNAM1 (DX11, BD Biosciences) or MICA-Fc (1300-MA, R&D systems). 2 × 105 cells were added to the wells in complete medium containing anti-CD107a antibody. After 2h of incubation, Brefeldin A was added and incubation followed for 4 additional hours at 37°C. Afterwards, cells were washed, fixed/permeabilized and stained with anti-IFN-γ antibody. In blocking experiments, the following antibodies (10μg/mL) were added at the beginning of the co-culture: anti-CD94 (HP-3B1, Santa Cruz Biotechnology), anti-NKG2D (1D11, Biolegend) or anti-DNAM1 (DX11, BD Biosciences) and the procedure continued as described above. The anti-CD94 antibody was first digested to purify F(ab´)2 fragments using the Pierce™ F(ab')2 Preparation Kit (Thermo Fisher Scientific).
For HCMV antigen-specific stimulation, T cells were stimulated with 1μg/mL of the indicated ORFs (JPT), containing 15 amino acid peptides spanning the complete amino acid sequence of the indicated protein antigen. Following O/N peptide stimulation cells were stained with anti-IFN-γ antibody.
RNA-seq processing and analysis
Paired-end reads were trimmed for adaptors and removed of low quality reads using Trimmomatic (v0.38)(59). Transcript quantification was based on the hg38 UCSC Known Gene models and performed using the quasi-mapping-based mode of Salmon (v.0.13.1)(60), correcting for potential GC bias. Counts were summarized to the gene level using tximport (v1.10.1)(61). For those samples that were sequenced across two runs, summarized reads determined by tximport were summed, and the means of average transcript length offsets calculated for each run was used for downstream differential analyses executed by DESeq2 (v1.22.2)(62). Genes were considered differentially expressed (DE) if they showed a false discovery rate (FDR)-adjusted P-value < 0.05. Gene set analysis was performed with GOseq (v.1.34.1)(63), using either DE genes higher in NKG2C+ CD8+ T cells or DE genes higher in NKG2C− NKG2A− (DN) CD8+ T cells, and those showing an absolute log2 fold change > 1. Gene sets were retrieved from the MSigDB database (v.3.0)(64, 65). FDR-corrected P-values were calculated from P-values calculated by GOseq, and only gene ontologies passing a threshold of P < 0.05 were considered. Up to the top 10 gene sets ranked on p-value are shown. For transcription factor assignment, a list of curated human genes associated with transcription factors were obtained from the Animal TFDB (v3.0)(66) and gene IDs were matched to DE gene IDs. To determine potential BCL11B-dependent genes, DE genes that were defined to be either repressed (upregulated in Bcl11b-deficiency) or activated (downregulated in Bcl11b-deficiency) were retrieved from a previously published study on mouse thymocytes(22). These gene names were converted into human orthologs using the NCBI gene IDs from the HCOP database(67), in addition to matching gene symbols between orthologs. Hypergeometric tests were calculated on the overlap between BCL11B-dependent genes and DE genes (NKG2C+ versus DN) using all the sufficiently expressed genes (18,943) that remained after independent filtering performed by DESeq2 as the total population size. The NK cell signature was defined as all DE genes that were expressed higher (log2 fold change > 0) in NKG2C− NKG2A− NK cells compared to NKG2C−NKG2A− CD8+ T cells. To derive the TEMRA signature, FASTQ files downloaded from the Gene Expression Omnibus (accession number GSE80306)(24) and processed using the same software and methods described above, except for single-end reads. The TEMRA signature was defined as DE genes that were expressed higher (log2 fold change > 0) in TEMRA T cells compared to naïve T cells. All heatmaps were generated using ComplexHeatmap (v1.99.7)(68).
Gene targeting
For BCL11B locus targeting, NKG2A−NKG2C−KIRs−CD8+ T cells were FACS-sorted and maintained in RPMI complete media supplemented with 200U/ml IL-2 O/N at a density of 106 cells per ml. The next day, cells were transfected by electrotransfer of Cas9 protein and gRNA using a Nucleofector II Amaxa system (Lonza). 5x106 cells were mixed with 10 μg TrueCut Cas9 protein (Thermo Fisher Scientific) for each sgRNA and either 2 μl (200 pmol) of a control (TrueGuide synthetic sgRNA, Thermo Fisher Scientific) or 2 μl (200 pmol) each of three different BCL11B sgRNA guides (TrueGuide synthetic sgRNA, Thermo Fisher Scientific) in a 1ml cuvette. The target DNA sequences of the BCL11B sgRNA guides were: 5’-CGCCATCCTCGAAGAAGACG-3’, 5’-GTTCATTTGACACTGGCCAC-3’, 5’ACTTGGATCCCGATCTCCAC-3’. Following electroporation, cells were diluted in RPMI complete medium supplemented with 100U/ml IL-2 and incubated at 37 °C, 5% CO2. 48h after electrotransfer cells were stimulated with CD3/CD2/CD28 beads (Miltenyi Biotec) at a bead-to-cell ratio of 1:2 and, after two weeks, with irradiated K562 mbIL21 HLA-E:VMAPRTLFL at a ratio of 1:1, replenishing the IL-2 every 3 to 4 days and irradiated K562 every 7 days.
For TRAC locus targeting, total T lymphocytes were purified using the EasySep™ Human T Cell Enrichment Kit (STEMCELL Technologies) and activated with CD3/CD2/CD28 beads (Miltenyi Biotec) at a bead-to-cell ratio of 1:2. 48 hours after initiating T cell activation, beads were magnetically removed and 2x106 cells were transfected by electrotransfer of Cas9 ptotein and gRNA using a 4D Nucleofector X Unit system (Lonza), as previously described(69).
Short-term quantitative cytotoxicity assay
The short-term cytotoxicity of NKG2C− or NKG2C+CD8+ T cells was determined by a standard luciferase-based killing assay. 5x103 target tumor cells expressing firefly luciferase were co-cultured with FACS-sorted NKG2C− or NKG2C+CD8+ T cells at different effector-to-target ratios in triplicate in white-walled 96-well plates (Corning) in a total volume of 200 ul of cell media. Target cells alone were plated at the same cell density to determine the maximal luciferase expression as a reference (“max signal”), and 16 h later, 75 ng of D-Luciferin (Gold Biotechnology) dissolved in 50 ul of PBS was added to each well. Emitted luminescence of each sample (“sample signal”) was detected in a Spark plate reader (Tecan) and quantified using the SparkControl software (Tecan). Percent lysis was determined as (1 – (“sample signal” / “max signal”)) x 100.
Statistical analysis
Paired t-test was applied for the comparison between groups with observations from the same donor cells. Unpaired t-test was used to compare independent groups. ANOVA with multiple comparisons was used to analyze groups of more than two. All tests are indicated in each individual figure legend. *P ≤0.05, **P ≤0.01, ***P ≤0.001 and ****P ≤0.0001 were used as significant p values. The analysis was performed using Prism 8 software (GraphPad).
Supplementary Material
Data file S1. Raw data file (Excel file).
Fig. S1. NKG2C+CD8+ T cells do not expand with age and are not specific for the most immunogenic HCMV peptides.
Fig. S2. T cell differentiation markers analysis on HCMV-seronegative donors and NKG2C+CD8+ T cell comparison with NK cells.
Fig. S3. NKG2C+CD8+ T cells are transcriptionally distinct from TEMRA cells.
Fig. S4. NKG2C+CD8+ T cells showed the narrowest TCR Vβ chain usage and those from HCMV-seronegative donors are polyclonal for their TCR Vβ chain repertoire.
Fig. S5. Functional characterization of NKG2C+CD8+ T cells.
Fig. S6. Phenotype and function of NKG2C+CD8+ T cells after one week expansion with CD3/CD2/CD28 beads or K562 mbIl21 HLA-E:VMAPRTLFL.
Table S1. Characteristics of DUCB-HCT patients.
Table S2. Single-cell sequencing of the productive α and β TCR pairs expressed by NKG2C+CD8+ T cells.
Table S3. HLA class I genotypes of effector and target cells.
Acknowledgments:
The authors would like to thank Dr. Ming Li (Memorial Sloan Kettering Cancer Center) and Dr. Kattria van der Ploeg for the helpful discussions. The authors are also thankful to Dr. Karlo Perica (Memorial Sloan Kettering Cancer Center) for the assistance with TCR knock-out experiments, and Dr. Sanam Shahid (Memorial Sloan Kettering Cancer Center) for providing patients clinical data.
Funding:
The following funding was used to support these studies: NIH U01 AI069197 (RS, JBL, MKP, KCH), R01 HL155741 (RS, JBL, MKP, KCH), R01 AI150999 (RS, JBL, MKP, KCH) and MSKCC Center for Experimental Therapeutics Big Bets program (RS, MKP, KCH); NIH/NCI Cancer Center Support Grant P30 CA008748 (RS, JLB, MKP, KCH). CML was supported by the Cancer Research Institute as a Cancer Research Institute-Carson Family Fellow.
Footnotes
Competing interests: MKP and KCH are inventors on a provisional patent for the design and use of HLA-E:peptide chimeric molecules (US provisional patent application number: 63/173,966). RS, KCH, MKP and JBL are inventors on a provisional patent for manufacture and use of NKG2C+CD8+ T cells (US provisional patent application number: 63/188,435). KT is presently an employee of Daiichi Sankyo Co., Ltd. AFD is a co-inventor of intellectual property related to CD371 CAR T-cell technology and field of use specific for allogeneic cell therapies, licensed by MSK to Caribou Biosciences. Caribou is a private biotechnology company that develops CRISPR technologies and allogenic cell therapies for oncology.
Data and material availability: Data generated in this study have been deposited in The Gene Expression Omnibus (accession number GSE179981).
References
- 1.Picarda G, Benedict CA, Cytomegalovirus: Shape-Shifting the Immune System. J Immunol 200, 3881–3889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowd J, Aiello A, Alley D, Socioeconomic disparities in the seroprevalence of cytomegalovirus infection in the US population: NHANES III. Epidemiol Infect 137, 58–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falk C, Mach M, Schendel D, Weiss E, Hilgert I, Hahn G, NK cell activity during human cytomegalovirus infection is dominated by US2-11-mediated HLA class I down-regulation. J Immunol 169, 3257–3266 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Klenerman P, Oxenius A, T cell responses to cytomegalovirus. Nat Rev Immunol 16, 367–377 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Sylwester A, Mitchell B, Edgar J, Taormina C, Pelte C, Ruchti F, Sleath P, Grabstein K, Hosken N, Kern F, Nelson J, Picker L, Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 202, 673–685 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun J, Beilke J, Bezman N, Lanier L, Homeostatic proliferation generates long-lived natural killer cells that respond against viral infection. J Exp Med 208, 357–368 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prod'homme V, Tomasec P, Cunningham C, Lemberg MK, Stanton RJ, McSharry BP, Wang EC, Cuff S, Martoglio B, Davison AJ, Braud VM, Wilkinson GW, Human cytomegalovirus UL40 signal peptide regulates cell surface expression of the NK cell ligands HLA-E and gpUL18. J Immunol 188, 2794–2804 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammer Q, Rückert T, Borst E, Dunst J, Haubner A, Durek P, Heinrich F, Gasparoni G, Babic M, Tomic A, Pietra G, Nienen M, Blau I, Hofmann J, Na I, Prinz I, Koenecke C, Hemmati P, Babel N, Arnold R, Walter J, Thurley K, Mashreghi M, Messerle M, Romagnani C, Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol 19, 453–463 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Rölle A, Pollmann J, Ewen E, Le V, Halenius A, Hengel H, Cerwenka A, IL-12-producing monocytes and HLA-E control HCMV-driven NKG2C+ NK cell expansion. J Clin Invest 124, 5305–5316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M, Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104, 3664–3671 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes T, Han H, Chiang S, Foley B, Mattsson K, Larsson S, Schaffer M, Malmberg K-J, Ljunggren H-G, Miller J, Bryceson Y, Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott J, Kamimura Y, Lanier L, Kim S, Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42, 431–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon C, Zajac A, Jamieson A, Corral L, Hammer G, Ahmed R, Raulet D, Viral and bacterial infections induce expression of multiple NK cell receptors in responding CD8(+) T cells. J Immunol 169, 1444–1452 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Mingari M, Schiavetti F, Ponte M, Vitale C, Maggi E, Romagnani S, Demarest J, Pantaleo G, Fauci A, Moretta L, Human CD8+ T lymphocyte subsets that express HLA class I-specific inhibitory receptors represent oligoclonally or monoclonally expanded cell populations. Proceedings of the National Academy of Sciences 93, 12433–12438 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Björkström N, Béziat V, Cichocki F, Liu L, Levine J, Larsson S, Koup R, Anderson S, Ljunggren H, Malmberg K, CD8 T cells express randomly selected KIRs with distinct specificities compared with NK cells. Blood 120, 3455–3465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guma M, Busch L, Salazar-Fontana L, Bellosillo B, Morte C, Garcıa P, Lopez-Botet M, The CD94/NKG2C killer lectin-like receptor constitutes an alternative activation pathway for a subset of CD8+ T cells. Eur J Immunol 35, 2071–2080 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Sullivan LC, Nguyen THO, Harpur CM, Stankovic S, Kanagarajah AR, Koutsakos M, Saunders PM, Cai Z, Gray JA, Widjaja JML, Lin J, Pietra G, Mingari MC, Moretta L, Samir J, Luciani F, Westall GP, Malmberg KJ, Kedzierska K, Brooks AG, Natural killer cell receptors regulate responses of HLA-E-restricted T cells. Sci Immunol 6, (2021). [DOI] [PubMed] [Google Scholar]
- 18.Balin S, Pellegrini M, Klechevsky E, Won S, Weiss D, Choi A, Hakimian J, Lu J, Ochoa M, Bloom B, Lanier L, Stenger S, Modlin R, Human antimicrobial cytotoxic T lymphocytes, defined by NK receptors and antimicrobial proteins, kill intracellular bacteria. Sci Immunol 3, eaat7668, DOI: 76 10.1126/sciimmunol.aat7668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wills MR, Carmichael AJ, Mynard K, Jin X, Weekes MP, Plachter B, Sissons JG, The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol 70, 7569–7579 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaughlin-Taylor E, Pande H, Forman SJ, Tanamachi B, Li CR, Zaia JA, Greenberg PD, Riddell SR, Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol 43, 103–110 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Duraiswamy J, Ibegbu CC, Masopust D, Miller JD, Araki K, Doho GH, Tata P, Gupta S, Zilliox MJ, Nakaya HI, Pulendran B, Haining WN, Freeman GJ, Ahmed R, Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J Immunol 186, 4200–4212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosokawa H, Romero-Wolf M, Yui M, Ungerbäck J, Quiloan M, Matsumoto M, Nakayama K, Tanaka T, Rothenberg E, Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nature 19, 1427–1440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Leid M, Rothenberg EV, An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329, 89–93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulko V, Davies JS, Martinez C, Lanteri MC, Busch MP, Diamond MS, Knox K, Bush EC, Sims PA, Sinari S, Billheimer D, Haddad EK, Murray KO, Wertheimer AM, Nikolich-Zugich J, Human memory T cells with a naive phenotype accumulate with aging and respond to persistent viruses. Nat Immunol 17, 966–975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng R, Spolski R, Finkelstein SE, Oh S, Kovanen PE, Hinrichs CS, Pise-Masison CA, Radonovich MF, Brady JN, Restifo NP, Berzofsky JA, Leonard WJ, Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J Exp Med 201, 139–148 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llano M, Lee N, Navarro F, Garcia P, Albar JP, Geraghty DE, Lopez-Botet M, HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur J Immunol 28, 2854–2863 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Sampaio KL, Weyell A, Subramanian N, Wu Z, Sinzger C, A TB40/E-derived human cytomegalovirus genome with an intact US-gene region and a self-excisable BAC cassette for immunological research. Biotechniques 63, 205–214 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Mahe E, Pugh T, Kamel-Reid S, T cell clonality assessment: past, present and future. J Clin Pathol 71, 195–200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE, HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol 160, 4951–4960 (1998). [PubMed] [Google Scholar]
- 30.Zhao Z, Condomines M, van der Stegen SJC, Perna F, Kloss CC, Gunset G, Plotkin J, Sadelain M, Structural Design of Engineered Costimulation Determines Tumor Rejection Kinetics and Persistence of CAR T Cells. Cancer Cell 28, 415–428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arlettaz L, Villard J, de Rham C, Degermann S, Chapuis B, Huard B, Roosnek E, Activating CD94:NKG2C and inhibitory CD94:NKG2A receptors are expressed by distinct subsets of committed CD8+ TCR alphabeta lymphocytes. Eur J Immunol 34, 3456–3464 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Meresse B, Curran SA, Ciszewski C, Orbelyan G, Setty M, Bhagat G, Lee L, Tretiakova M, Semrad C, Kistner E, Winchester RJ, Braud V, Lanier LL, Geraghty DE, Green PH, Guandalini S, Jabri B, Reprogramming of CTLs into natural killer-like cells in celiac disease. J Exp Med 203, 1343–1355 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manser AR, Scherenschlich N, Thons C, Hengel H, Timm J, Uhrberg M, KIR Polymorphism Modulates the Size of the Adaptive NK Cell Pool in Human Cytomegalovirus-Infected Individuals. J Immunol 203, 2301–2309 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Cao K, Marin D, Sekine T, Rondon G, Zhao W, Smith NT, Daher M, Wang Q, Li L, Saliba RM, Pingali R, Popat U, Hosing C, Olson A, Oran B, Basar R, Mehta RS, Champlin R, Shpall EJ, Rezvani K, Donor NKG2C Copy Number: An Independent Predictor for CMV Reactivation After Double Cord Blood Transplantation. Front Immunol 9, 2444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson SE, Sedikides GX, Okecha G, Wills MR, Generation, maintenance and tissue distribution of T cell responses to human cytomegalovirus in lytic and latent infection. Med Microbiol Immunol 208, 375–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braud VM, Allan DS, O'Callaghan A, Soderstrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Anfossi N, Doisne JM, Peyrat MA, Ugolini S, Bonnaud O, Bossy D, Pitard V, Merville P, Moreau JF, Delfraissy JF, Dechanet-Merville J, Bonneville M, Venet A, Vivier E, Coordinated expression of Ig-like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J Immunol 173, 7223–7229 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Mingari MC, Vitale C, Cambiaggi A, Schiavetti F, Melioli G, Ferrini S, Poggi A, Cytolytic T lymphocytes displaying natural killer (NK)-like activity: expression of NK-related functional receptors for HLA class I molecules (p58 and CD94) and inhibitory effect on the TCR-mediated target cell lysis or lymphokine production. Int Immunol 7, 697–703 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Bottcher JP, Beyer M, Meissner F, Abdullah Z, Sander J, Hochst B, Eickhoff S, Rieckmann JC, Russo C, Bauer T, Flecken T, Giesen D, Engel D, Jung S, Busch DH, Protzer U, Thimme R, Mann M, Kurts C, Schultze JL, Kastenmuller W, Knolle PA, Functional classification of memory CD8(+) T cells by CX3CR1 expression. Nat Commun 6, 8306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon S, Vignard V, Florenceau L, Dreno B, Khammari A, Lang F, Labarriere N, PD-1 expression conditions T cell avidity within an antigen-specific repertoire. Oncoimmunology 5, e1104448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, Trebska-McGowan K, Wunderlich JR, Yang JC, Rosenberg SA, Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 22, 433–438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez RJ, Evavold BD, Lower Affinity T Cells are Critical Components and Active Participants of the Immune Response. Front Immunol 6, 468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R, Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol 4, 533–539 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee S, Lu D, Campos L, Goulding D, Ng B, Dougan G, Huntly B, Gottgens B, Jenkins N, Copeland N, Colucci F, L. P P, Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329, 85–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmes TD, Pandey RV, Helm EY, Schlums H, Han H, Campbell TM, Drashansky TT, Chiang S, Wu CY, Tao C, Shoukier M, Tolosa E, Von Hardenberg S, Sun M, Klemann C, Marsh RA, Lau CM, Lin Y, Sun JC, Mansson R, Cichocki F, Avram D, Bryceson YT, The transcription factor Bcl11b promotes both canonical and adaptive NK cell differentiation. Sci Immunol 6, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huard B, Karlsson L, Triebel F, KIR down-regulation on NK cells is associated with down-regulation of activating receptors and NK cell inactivation. Eur J Immunol 31, 1728–1735 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Borrego F, Kabat J, Sanni TB, Coligan JE, NK cell CD94/NKG2A inhibitory receptors are internalized and recycle independently of inhibitory signaling processes. J Immunol 169, 6102–6111 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Judge SJ, Dunai C, Aguilar EG, Vick SC, Sturgill IR, Khuat LT, Stoffel KM, Van Dyke J, Longo DL, Darrow MA, Anderson SK, Blazar BR, Monjazeb AM, Serody JS, Canter RJ, Murphy WJ, Minimal PD-1 expression in mouse and human NK cells under diverse conditions. J Clin Invest 130, 3051–3068 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomasec P, Braud V, Rickards C, Powell M, McSharry B, Gadola S, Cerundolo V, Borysiewicz L, McMichael A, Wilkinson G, Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Vales-Gomez M, Reyburn HT, Erskine RA, Lopez-Botet M, Strominger JL, Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J 18, 4250–4260 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, Blery M, Bonnafous C, Gauthier L, Morel A, Rossi B, Remark R, Breso V, Bonnet E, Habif G, Guia S, Lalanne AI, Hoffmann C, Lantz O, Fayette J, Boyer-Chammard A, Zerbib R, Dodion P, Ghadially H, Jure-Kunkel M, Morel Y, Herbst R, Narni-Mancinelli E, Cohen RB, Vivier E, Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 175, 1731–1743 e1713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Busch M, Herzmann C, Kallert S, Zimmermann A, Hofer C, Mayer D, Zenk SF, Muche R, Lange C, Bloom BR, Modlin RL, Stenger S, Network TB, Lipoarabinomannan-Responsive Polycytotoxic T Cells Are Associated with Protection in Human Tuberculosis. Am J Respir Crit Care Med 194, 345–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hikami K, Tsuchiya N, Yabe T, Tokunaga K, Variations of human killer cell lectin-like receptors: common occurrence of NKG2-C deletion in the general population. Genes Immun 4, 160–167 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Pegram HJ, Purdon TJ, van Leeuwen DG, Curran KJ, Giralt SA, Barker JN, Brentjens RJ, IL-12-secreting CD19-targeted cord blood-derived T cells for the immunotherapy of B-cell acute lymphoblastic leukemia. Leukemia 29, 415–422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parry RV, Rumbley CA, Vandenberghe LH, June CH, Riley JL, CD28 and inducible costimulatory protein Src homology 2 binding domains show distinct regulation of phosphatidylinositol 3-kinase, Bcl-xL, and IL-2 expression in primary human CD4 T lymphocytes. J Immunol 171, 166–174 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Rittie L, Fisher GJ, Isolation and culture of skin fibroblasts. Methods Mol Med 117, 83–98 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Lyons AB, Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J Immunol Methods 243, 147–154 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Smith EL, Harrington K, Staehr M, Masakayan R, Jones J, Long TJ, Ng KY, Ghoddusi M, Purdon TJ, Wang X, Do T, Pham MT, Brown JM, De Larrea CF, Olson E, Peguero E, Wang P, Liu H, Xu Y, Garrett-Thomson SC, Almo SC, Wendel HG, Riviere I, Liu C, Sather B, Brentjens RJ, GPRC5D is a target for the immunotherapy of multiple myeloma with rationally designed CAR T cells. Sci Transl Med 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soneson C, Love MI, Robinson MD, Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young MD, Wakefield MJ, Smyth GK, Oshlack A, Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP, Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY, AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res 47, D33–D38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, Bruford EA, Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res 45, D619–D625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu Z, Eils R, Schlesner M, Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, Sadelain M, Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1. Raw data file (Excel file).
Fig. S1. NKG2C+CD8+ T cells do not expand with age and are not specific for the most immunogenic HCMV peptides.
Fig. S2. T cell differentiation markers analysis on HCMV-seronegative donors and NKG2C+CD8+ T cell comparison with NK cells.
Fig. S3. NKG2C+CD8+ T cells are transcriptionally distinct from TEMRA cells.
Fig. S4. NKG2C+CD8+ T cells showed the narrowest TCR Vβ chain usage and those from HCMV-seronegative donors are polyclonal for their TCR Vβ chain repertoire.
Fig. S5. Functional characterization of NKG2C+CD8+ T cells.
Fig. S6. Phenotype and function of NKG2C+CD8+ T cells after one week expansion with CD3/CD2/CD28 beads or K562 mbIl21 HLA-E:VMAPRTLFL.
Table S1. Characteristics of DUCB-HCT patients.
Table S2. Single-cell sequencing of the productive α and β TCR pairs expressed by NKG2C+CD8+ T cells.
Table S3. HLA class I genotypes of effector and target cells.