Summary
The yeast Saccharomyces cerevisiae uses the pyruvate dehydrogenase‐bypass for acetyl‐CoA biosynthesis. This relatively inefficient pathway limits production potential for acetyl‐CoA‐derived biochemical due to carbon loss and the cost of two high‐energy phosphate bonds per molecule of acetyl‐CoA. Here, we attempted to improve acetyl‐CoA production efficiency by introducing heterologous acetylating aldehyde dehydrogenase and phosphoketolase pathways for acetyl‐CoA synthesis to enhance production of the sesquiterpene trans‐nerolidol. In addition, we introduced auxin‐mediated degradation of the glucose‐dependent repressor Mig1p to allow induced expression of GAL promoters on glucose so that production potential on glucose could be examined. The novel genes that we used to reconstruct the heterologous acetyl‐CoA pathways did not sufficiently complement the loss of endogenous acetyl‐CoA pathways, indicating that superior heterologous enzymes are necessary to establish fully functional synthetic acetyl‐CoA pathways and properly explore their potential for nerolidol synthesis. Notwithstanding this, nerolidol production was improved twofold to a titre of ˜ 900 mg l−1 in flask cultivation using a combination of heterologous acetyl‐CoA pathways and Mig1p degradation. Conditional Mig1p depletion is presented as a valuable strategy to improve the productivities in the strains engineered with GAL promoters‐controlled pathways when growing on glucose.
We attempted to improve acetyl‐CoA production efficiency by introducing heterologous acetylating aldehyde dehydrogenase and phosphoketolase pathways for acetyl‐CoA synthesis to enhance production of the sesquiterpene nerolidol. Conditional Mig1p depletion is presented as a valuable strategy to improve the productivities in the strains engineered with GAL promoters‐controlled pathways when growing on glucose.
Introduction
The commonly used yeast chassis organism Saccharomyces cerevisiae has been intensively engineered for improved production of various chemicals (Stephanopoulos, 2012; Jensen and Keasling, 2015; Nielsen, 2019). It has been shown to be a superior host for the synthesis of terpenoids (a.k.a. isoprenoids), a large group of natural products with a wide range of biological functions and commercial applications (Vickers et al., 2017; Alonso‐Gutierrez et al., 2018). For maximal terpenoid productivities (yield/titre/rate), it is crucial to optimize the metabolic networks, including sugar catabolic (central carbon) metabolism and terpene anabolism (Vickers et al., 2014; Meadows et al., 2016; Aslan et al., 2017; Vickers et al., 2017) as well as regulatory mechanisms to regulate these pathways (Stephanopoulos, 2012; Peng et al., 2015; Meadows et al., 2016; Shen et al., 2020).
In S. cerevisiae, the universal terpene precursors, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), are synthesised through the mevalonate pathway (Fig. 1; Vickers et al., 2017). The flux through this pathway is tightly regulated at both transcriptional and post‐transcriptional levels. In wild type yeast, the pathway synthesises structural terpenoids including sterols, as well as ubiquinones and prenyl groups for protein prenylation (Peng et al., 2017a,2017b). To increase mevalonate pathway flux for high‐level production of heterologous terpenoids, the enzymes from the mevalonate pathway must be deregulated and overexpressed (Bian et al., 2017; Vickers et al., 2017). Redirection of carbon flux to heterologous terpenoid products is achieved by expression of the heterologous terpenoid pathway enzymes and downregulation of enzymes catalysing the synthesis of native terpenoids (Peng et al., 2017a,2017b; Peng et al., 2018a,2018b). This approach targets the terpenoid anabolic pathways and has enabled the production of more than 40 g l−1 sesquiterpene in yeast fed‐batch fermentation (Westfall et al., 2012), but maximal productivities have not been reached (Gruchattka et al., 2013; Meadows et al., 2016).
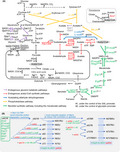
Fig. 1.
Metabolic pathways for the synthesis of acetyl‐CoA and nerolidol in S. cerevisiae (A) and diagram of strain construction processes (B). Acetyl‐CoA synthesis pathways: Lr.xfp, Lactobacillus reuteri fructose 6‐phosphate/xylulose 5‐phosphate phosphoketolase; Mb.pta, Methanosarcina barkeri phosphate acetyl‐transferase; Lr.pduP and Lr.eutE, L. reuteri acetylating aldehyde dehydrogenase; Ald2/3/4/5/6, ADA; and Acs1/2, acetyl‐CoA synthase. Terpene anabolic pathways: Ef.mvaE, Enterococcus faecalis acetoacetyl‐CoA thiolase/HMG‐CoA reductase; Ef.mvaS, E. faecalis HMG‐CoA synthase; HMG2, HMG‐CoA reductase 2; Da.mvaA, Delftia acidovorans NADH‐dependent HMG‐CoA reductase; Erg12, mevalonate kinase; Erg8, phosphomevalonate kinase; Mvd1, mevalonate pyrophosphate decarboxylase; Idi1, IPP:DMAPP isomerase, Erg20, farnesyl pyrophosphate synthetase, AcNES1 Actinidia chinensis trans‐nerolidol synthase. Glycerol metabolic pathway: Gpd1/2, glycerol‐3‐phosphate dehydrogenase; Gpp1/2, glycerol‐3‐phosphate phosphatase; Gut1, glycerol kinase; Gut2, glycerol‐3‐phosphate dehydrogenase; Gcy1, glycerol dehydrogenase; Dak1/2, dihydroxyacetone kinase. Others: Skp1, component of the SCF ubiquitin ligase complex; Os.TIR1, Oryza sativa auxin receptor; Mig1, transcriptional repressor in response to glucose; HMG‐CoA, 3‐hydroxy‐3‐methylglutaryl‐CoA; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; FPP, farnesyl pyrophosphate.
Production of IPP and DMAPP via the mevalonate requires an input of acetyl‐CoA from central carbon metabolism (Fig. 1). Acetyl‐CoA metabolism in S. cerevisiae is compartmentalised to the cytosol, mitochondria and peroxisome (Pronk et al., 1996; Boubekeur et al., 1999; Chen et al., 2012; Fig. 1A). The mevalonate pathway is fed by cytosolic acetyl‐CoA, which is synthesised through the so‐called pyruvate dehydrogenase (PDH) bypass (Boubekeur et al., 1999; Fig. 1). The PDH bypass comprises of three steps of reaction catalysed by the pyruvate decarboxylases Pdc1p, Pdc5 and Pdc6p, cytosolic acetaldehyde dehydrogenase Ald6p, Ald2p and Ald3p or mitochondrial acetaldehyde dehydrogenase Ald4p and Ald5p (Boubekeur et al., 1999; Bakker et al., 2001; Boubekeur et al., 2001) and nuclear/cytosolic acetyla‐CoA synthase Acs2p (Acetate‐CoA ligase) and the cytosolic/peroxisomal acetyl‐CoA synthase Acs1p (Chen et al., 2012). Use of the PDH bypass to synthesize acetyl‐CoA has two key drawbacks: (i) the loss of carbon via CO2 production in the pyruvate decarboxylation step and (ii) the cost of two high‐energy phosphate bonds for acetyl‐CoA ligation (Bogorad et al., 2013; Henard et al., 2015; Meadows et al., 2016). Reducing the flux through the PDH bypass and introducing alternative acetyl‐CoA synthetic reactions increase the theoretical carbon yield of terpenoid production in yeast and decrease the amount of oxygen required for energy production (Meadows et al., 2016).
Alternative acetyl‐CoA synthesis can be engineered with, for example, an NAD+‐preferring acetylating aldehyde dehydrogenase (ADA; Fig. 1A, pduP and eutE, Kozak et al., 2014a,2014b; Meadows et al., 2016), the phosphoketolase pathway (Fig. 1A, xfp and pta, Bogorad et al., 2013; Meadows et al., 2016), cytosolic pyruvate dehydrogenase complex (Kozak et al., 2014a,2014b), pyruvate‐formate lyase (Kozak et al., 2014a,2014b) and ATP citrate lyase (Yu et al., 2018). Combining the ADA and the phosphoketolase pathway, an NADH‐preferring 3‐hydroxy‐3‐methyl‐glutaryl‐coenzyme A (HMG‐CoA) reductase (Fig. 1A, mvaA) can be introduced to oxidize the NADH produced in glycolysis and by NAD+‐dependent ADA. This combination provides a maximum theoretical yield of terpene products from glucose through the mevalonate pathway (Ma et al., 2011; Meadows et al., 2016). In combination with the strategies strengthening terpene anabolic pathway, this led to the production of sesquiterpene farnesene at a titre exceeding 130 g l−1 in an industrial process (Meadows et al., 2016).
Previously, we augmented the mevalonate pathway for improved production of sesquiterpene trans‐nerolidol (Peng et al., 2017a,2017b). We showed that restricting the consumption of acetyl‐CoA for lipogenesis redirected carbon flux towards nerolidol production (Lu et al., 2021). However, we have not introduced heterologous acetyl‐CoA pathways for improved terpene production. Meanwhile, the genes for nerolidol anabolism were overexpressed under the regulation of GAL promoters, in parallel with disruption of the GAL transcription repressor gene GAL80 to enable diaxuie‐inducible expression from GAL promoters (Peng et al., 2017a,2017b; Peng et al., 2018a,2018b; Fig. 1A). In gal80Δ background strain, GAL promoters are repressed by glucose via Mig1p‐mediated repression (Frolova et al., 1999). This is problematic for characterisation of the effects of engineered acetyl‐CoA pathways on nerolidol production when glucose is used as carbon source. This might restrict industrial production in glucose‐pulse fed‐batch cultivation (Westfall et al., 2012). A regulatory mechanism is therefore required for conditional induction of GAL promoters on glucose.
In this study, we first introduced new heterologous acetyl‐CoA synthesis pathways in yeast with the aim to increase precursor availability for terpene production. To address the problem of a regulatory mechanism for conditional induction of GAL promoters on glucose, we implemented an auxin‐inducible degradation approach (Lu et al., 2021) to deliver auxin‐mediated degradation of the glucose‐dependent transcriptional repressor Mig1p. This provided high‐level expression of heterologous nerolidol pathway genes across the fermentation on different carbon sources (glucose and ethanol). Using this tool, we characterised the impact of engineered acetyl‐CoA synthetic pathways on nerolidol production and sugar metabolism.
Results
Engineered acetyl‐CoA synthetic pathways in nerolidol‐producing strains
We previously engineered trans‐nerolidol production in S. cerevisiae by overexpressing the mevalonate pathway and the genes encoding farnesyl pyrophosphate synthase and nerolidol synthase (terpene anabolic genes; Peng et al., 2017a,2017b; Peng et al., 2018a,2018b). The background mevalonate engineered pathway had one copy of ACS2, Ef‐mvaE, Ef‐mvaS, HMG2K6R , ERG8, MVD1 and IDI1 and two copies of ERG12 overexpressed under the control of the GAL promoters; one copy of ERG8, MVD1 and IDI1 are overexpressed under the control of glucose‐inducible promoters (strain o401R). These modifications delivered titres of ˜ 400 mg l−1 nerolidol in flask cultivation (Peng et al., 2017a,2017b; Peng et al., 2018a,2018b). The auxin‐mediated protein degradation mechanism was previously engineered for investigation of novel metabolic engineering strategies in terpene‐overproducing strains (Lu et al., 2021). To apply this tool into the strains in this work, we introduced the fusion of SKP1 (component of the SCF ubiquitin ligase complex) and Oryza sativa auxin receptor (SKP1‐Os.TIR1; Lu et al., 2021) into o401R to generate a new background strain o501R (Fig. 1B; Table 1; Table S3).
Table 1.
S. cerevisiae strains used in this work.
| Strain | Genotype | Resource/references |
|---|---|---|
| S. cerevisiae | ||
| CEN.PK2‐1C | MATa ura3‐52 trp1‐289 leu2‐3,112 his3Δ1 | Entian and Kötter (1998) |
| CEN.PK113‐5D | MATa ura3‐52 | Entian and Kötter (1998) |
| ILHA series strains | ||
| o401R |
CEN.PK2‐1C derivative; HMG2K6R(−152,‐1)::HIS3‐TEFM1<Ef.mvaS<PGAL1‐PGAL10>ACS2>TACS2‐PGAL2> Ef.mvaE >TEBS1‐PGAL7 pdc5(−31,94)::PGAL2> ERG12>TNAT5‐PTEF2>ERG8>TIDP1‐TPRM9<MVD1<PADH2‐TRPL15A<IDI1<PTEF1‐TRP1 ERG9(1336, 1336)::TURA3‐ PGAL7>MVD1>TPRM9‐PGAL2>ERG12>TNAT5‐TIDP1<ERG8<PGAL10‐PGAL1>IDI1>TRPL15A‐loxP‐ble‐loxP |
Peng et al. (2018a,2018b) |
| o501R |
o401R derivative; ERG9(1336, 1336)::TURA3‐ PGAL7>MVD1>TPRM9‐PGAL2>‐ERG12>TNAT5‐TIDP1<ERG8<PGAL10‐PGAL1>IDI1>TRPL15A‐PACS2>SKP1‐Os.TIR1 |
This work |
| o60 |
o501R derivative; ald6(41, 1053)Δ |
This work |
| o61 |
o501R derivative; ald6(41, 1053):: PADH1 > Lr‐pduP> TPDC1 |
This work |
| o62 |
o501R derivative; ald6(41, 1053):: PADH1 > Lr.pduP> TPDC1 > PSb.GAL2 >Lr. EutE |
This work |
| o63 |
o501R derivative; ald6(41, 1053):: PSk.GAL1 >Da.mvaA>TPGK1 > PADH1 > Lr.pduP> TPDC1 > PGAL2 >Lr. EutE |
This work |
| o73 |
o63 derivative; gpp1(79, 753)::PAg.TEF1>hphMX6>TAg.TEF1‐PENO2> Lr.xfp>TTPI1‐PTDH3>Mb.pta>TFBA1 |
This work |
| N501RU | o501R derivative; [pJT9R] gal80::URA3 | This work |
| N60U | o60 derivative; [pJT9R] gal80::URA3 | This work |
| N61U | o61 derivative; [pJT9R] gal80::URA3 | This work |
| N62U | o62 derivative; [pJT9R] gal80::URA3 | This work |
| N63U | o63 derivative; [pJT9R] gal80::URA3 | This work |
| N73U | o73 derivative; [pJT9R] gal80::URA3 | This work |
| oJ3 | CEN.PK113‐5D derivative; gal80::loxP‐kanMX4‐loxP | Peng et al. (2017a,2017b) |
| o7B | oJ3 derivative; MIG1::CUP1‐AID*‐MIG1 | This work |
| GB5J3 | oJ3 derivative; ura3(1, 704 )::KlURA3‐PGAL1‐yEGFP | Peng et al. (2017a,2017b) |
| GJ38T | oJ3 derivative; ura3(1, 704 )::KlURA3‐PGAL1‐yEGFP‐TPGK1‐PACS2‐SKP1‐OsTIR1 | This work |
| G7B8T | o7B derivative; ura3(1, 704 )::KlURA3‐PGAL1‐yEGFP‐TPGK1‐PACS2‐SKP1 ‐OsTIR1 | This work |
| o57BR |
o501R derivative; MIG1::CUP1‐AID*‐MIG1 |
This work |
| o637B |
o57BR derivative; ald6(41, 1053):: PSk.GAL1 >Da.mvaA>TPGK1 > PADH1 > Lr.pduP> TPDC1 > PGAL2 >Lr. EutE |
This work |
| o737B |
o637B derivative; gpp1(79, 753)::PENO2> Lr.xfp>TTPI1‐PTDH3>Mb.pta>TFBA1 |
This work |
| N57BRU |
o57BR derivative; [pJT9R] gal80::URA3 |
This work |
| N637BU |
o637B derivative; [pJT9R] gal80::URA3 |
This work |
| N737BU |
o737B derivative; [pJT9R] gal80::URA3 |
This work |
| N737B6D1U |
o637B derivative; gpp1(79, 753)::PENO2> Lr.xfp>TTPI1‐PTDH3>Mb.pta>TFBA1 ald4Δ [pJT9R] gal80::URA3 |
This work |
Symbol > or < indicates the direction of open reading frames.
From the new background strain o501R, five strains with a variety of acetyl‐CoA synthesis pathways and further genetic modifications were generated consecutively (Fig. 1B; Table 1). We first disrupted the major cytosolic acetaldehyde dehydrogenase encoding gene ALD6 (strain o60) to reduce acetaldehyde conversion to acetyl‐CoA via the yeast’s inefficient native pathway. The Lactobacillus reuteri ADA gene PduP (Lr.pduP) was then expressed under the control of the ADH1 promoter, resulting in strain o61. PduP expression creates an energy‐efficient short‐cut from acetaldehyde to acetyl‐CoA (Fig. 1). To augment the more efficient pathway, a second ADA encoding gene, Lr. EutE, was expressed under the control of the Saccharomyces bayanus GAL2 promoter (Peng et al., 2018a,2018b; strain o62). Next, a codon‐optimised NADH‐preferring HMG‐CoA reductase gene from Delftia acidovorans (Da.mvaA) was introduced under the control of Saccharomyces kudriavzevii GAL2 promoter, an alternative GAL promoter to avoid untargeted recombination (Peng et al., 2018a,2018b; strain o63). The aim of this step was to balance NADH metabolism by utilising the extra NADH produced by the introduced ADA enzymes, whilst concurrently augmenting mevalonate pathway flux. Finally, a hybrid phosphoketolase pathway, comprising an L. reuteri xylulose‐5‐phosphate/fructose‐6‐phosphate phosphoketolase gene (Lr.xfp) and a codon‐optimised Methanosarcina barkeri phosphate acetyltransferase gene (Mb.pta), was expressed under the control of glycolytic promoters, and GPP1 was disrupted to reduce acetate formation from acetate‐phosphate (Bergman et al., 2016; Meadowset al., 2016), resulting in strain o73. In these background strains o501R (reference), o60 (ald6Δ), o61 (ald6Δ, PADH1‐Lr.pduP), o62 (ald6Δ, PADH1‐Lr.pduP, PSb.GAL2‐Lr.eutE), o63 (ald6Δ, PADH1‐Lr.pduP, PSb.GAL2‐Lr.eutE, PSk.GAL2‐Da.mvaA) and o73 (ald6Δ, gpp1Δ, PADH1‐Lr.pduP, PSb.GAL2‐Lr.eutE, PSk.GAL2‐Da.mvaA, PENO2‐Lr.xfp, PTDH3‐Mb.pta), the plasmid pJT9R carrying GAL‐promoters‐controlled nerolidol synthetic genes was introduced (Peng et al., 2017a,2017b). The transcriptional repressor gene GAL80 was also disrupted in each strain to enable the diauxie‐coupled auto‐induction of the GAL promoters. These engineering steps resulted in the six nerolidol‐producing strains: N501RU (nerolidol reference strain), N60U, N61U, N62U, N63U and N73U respectively.
Growth and nerolidol synthesis in the five strains with modified acetyl‐CoA synthesis pathways and the reference strain N501RU were evaluated in flask cultivations in synthetic mineral medium (YNB) with 20 g l−1 glucose as the sole carbon source. The five acetyl‐CoA pathway engineered strains all had decreased final biomass production (Fig. 2A; P < 0.05) and slightly decreased specific maximum growth rates (Fig. 2D; P < 0.05) compared with the reference strain.
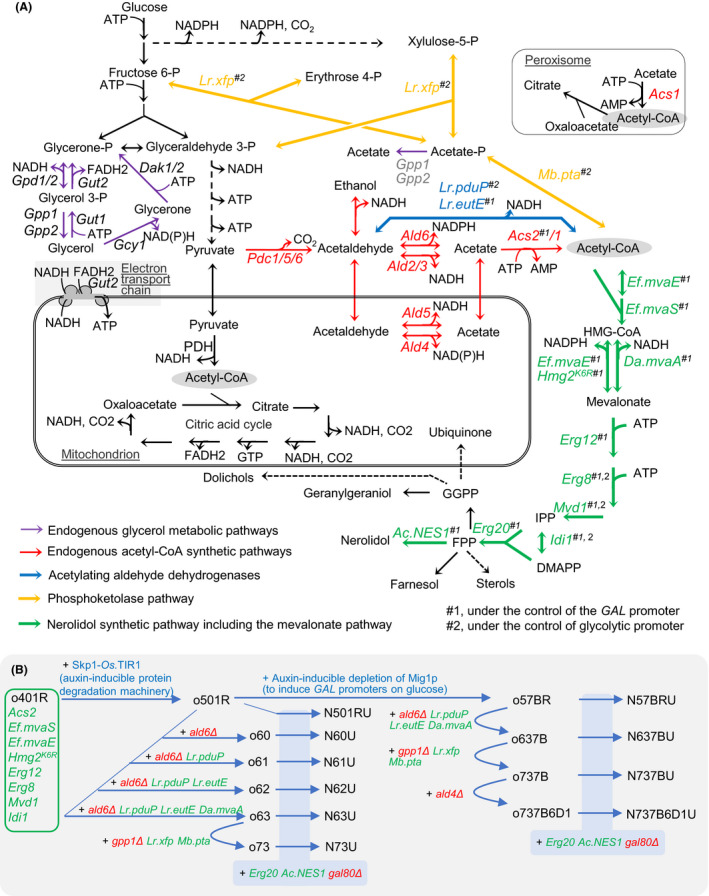
Fig. 2.
Characterising strains with engineered acetyl‐CoA synthesis pathways: (A) growth profiles; (B) nerolidol titre at 72 h and overall specific nerolidol production rate from 0 to 72 h; (C) farnesol and geranylgeraniol titre at 72 h; (D) maximum specific growth rate. Two‐phase flask cultivation on 20 g l−1 glucose was employed. For B, D and E, two‐tailed Welch’s t‐test was used for statistical analysis relative to N501RU: n, P> 0.1; m, P ∈ [0.05, 0.1]; s, P < 0.05. Mean values ± standard deviations are shown (N ≥ 4).
The nerolidol titres at 72 h for strains harbouring heterologous acetyl‐CoA synthesis pathways were slightly increased compared with the reference strains (Fig. 2B); however, those differences were not statistically significant (P > 0.05). Despite this, the overall specific nerolidol production rates were improved significantly by ˜ 70% in strain N73U (ADA pathway + NADH‐preferring HMG‐CoA reductase + PKA pathway), and ˜80% in the strains N62U (ADA pathway) and N63U (ADA pathway + NADH‐preferring HMG‐CoA reductase).
All strains produced small amounts of farnesol and geranylgeraniol (Fig. 2C), which are produced in the presence of excess prenyl phosphate accumulation by the action of non‐specific dephosphorylases (Peng et al., 2017a,2017b). While the farnesol titres in the strain N62U, N63U and N73U were similar to that in the reference, strains N60U and N61U showed an ˜ 50% decrease. Geranylgeraniol titres remained unchanged among all tested strains. Non‐specific prenyl alcohol production levels do not map to the nerolidol synthase‐driven specific production of nerolidol across strains, and it is unclear if these data are biologically relevant with the overall flux through the mevalonate pathway.
Auxin‐mediated Mig1p depletion de‐repressing the GAL promoter on glucose
We used the strong inducible GAL promoters to control the expression of heterologous genes for nerolidol production (Fig. 1A). GAL promoters are regulated by the galactose‐derepressible transcriptional repressor Gal80p and the glucose‐mediated transcriptional repressor Mig1p (Fig. 3A; Nehlin et al., 1991). When GAL80 is disrupted, GAL promoters are auto‐induced in aerobic cultivations in the absence of, or after depletion of, glucose. This diauxie‐inducible expression system has been used to increase the production of various isoprenoids in metabolically engineered yeast by avoiding the metabolic burden caused by high‐level gene expression during the exponential growth phase and strongly driving the production in the post‐exponential growth phase. However, the disadvantage of applying GAL promoters in gal80Δ strains is that it prohibits the characterisation of acetyl‐CoA pathway‐engineered strains on glucose.
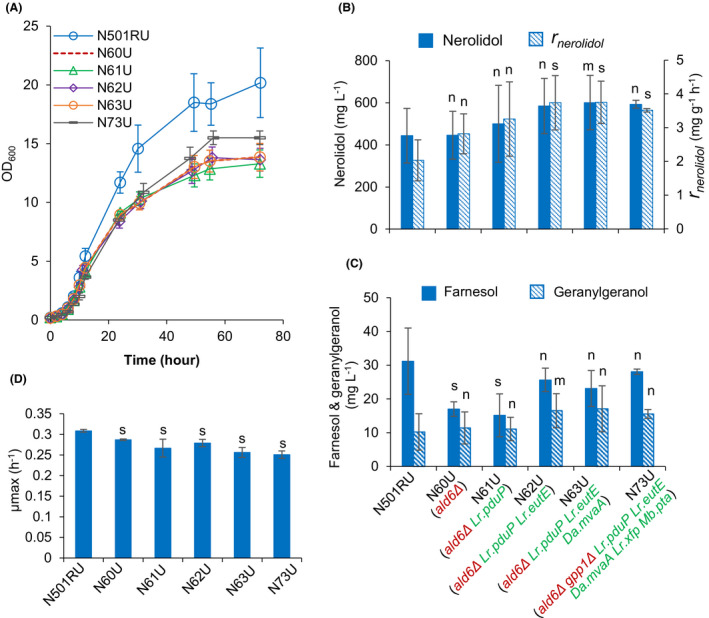
Fig. 3.
Engineering auxin‐mediated Mig1p depletion to induce the GAL1 promoter. Yeast strains include GB5J3 (gal80Δ MIG1 PGAL1‐yEGFP‐TURA3 ), GJ38T (gal80Δ MIG1 PGAL1‐yEGFP‐TPGK1‐PACS1‐SKP1‐OsTIR1‐TURA3 ) and G7B8T (gal80Δ CUP1‐AID*‐MIG1 PGAL1‐yEGFP‐TPGK1‐PACS1‐SKP1‐OsTIR1‐TURA3 ).
A. Schematic representation of the regulation on the GAL1 promoter.
B, C. The growth profile of engineered strains with or without naphthaleneacetic acid (NAA) addition at 1 h.
D, E. GFP fluorescence levels in engineered strains under the conditions with or without NAA addition. NAA was added at 1 h. Mean values ± absolute errors are shown (N = 2).
Double disruption of GAL80 and MIG1 results in constitutive activation of GAL promoters even in the presence of glucose (Nehlin et al., 1991). However, this is problematic if the induction of GAL promoter‐controlled genes, like constitutively overexpressing terpene synthetic pathways (Peng et al., 2017a,2017b; Peng et al., 2018a,2018b), causes metabolic burden/imbalance and slowed growth rates during the glucose phase. To avoid this, we exploited the auxin‐inducible protein degradation mechanism from plants for a conditional depletion of Mig1p (see schematic in Fig. 3A). This protein degradation mechanism has previously been reconstructed in yeast (Morawska and Ulrich, 2013) and consists of two key components: the F‐box protein TIR1 (auxin receptor) and the auxin‐inducible protein degron (AID). The system is powerful but is subject to substantial ‘basal degradation’ in the absence of auxin (Li et al., 2019; Sathyan et al., 2019). Fusion of the endogenous Skp1 protein (an essential component of the ubiquitin ligase complex in the degradation pathway) to TIR1 decreases the basal degradation, decreasing the leakiness and improving the inducibility of the system (Lu et al., 2021).
To test the effect of auxin‐mediated depletion of Mig1p on the expression level of GAL promoters, a Skp1p‐TIR1 fusion (Kanke et al., 2011) was expressed under the control of the constitutive ACS2 promoter. Mig1p was tagged with a mini‐AID tag (AID*; Arabidopsis thaliana IAA17 truncated to amino acids 71–114; Morawska and Ulrich, 2013) fused to the metallothionein protein Cup1p. Fusion of Cup1p to AID* also decreases basal degradation in the absence of auxin (Lu et al., 2021).
A proxy construct was included to examine GAL promoter activity in the presence of Mig1p engineering. This construct consisted of a GAL1 promoter fused to a yeast enhanced green fluorescent protein (yEGFP), allowing GFP fluorescence to report on GAL1 promoter activity in response to various perturbations. In our previous works, we used the URA3 terminator to control yEGFP transcription termination (strain GB5J3; Peng et al., 2017a,2017b). In the new construct, the PGK1 terminator (Loison et al., 1989) was fused to downstream of yEGFP gene for cloning SKP1‐TIR1 expression cassette into a single plasmid (pJAIDB58T; Table 2 and Supplementary Table 2). With yEGFP fused to the PGK1 terminator, GFP fluorescence was ˜threefold higher than that under the control of the URA3 promoter (Fig. 3D). Terminators are known to have a significant effect on expression strength (Curran et al., 2013); in this case, the PGK1 terminator provides a higher expression level than the URA3 terminator.
Table 2.
Plasmids used in this work.
| Plasmid | Features | References |
|---|---|---|
| pRS424 | E. coli/S. cerevisiae shuttle plasmid; 2 μ, TRP1 | Christianson et al. (1992) |
| pRS425 | E. coli/S. cerevisiae shuttle plasmid; 2 μ, LEU2 | Christianson et al. (1992) |
| pJT9R | pRS425: TRPL3<ERG20<PGAL1‐PGAL2>Ac.NES1>TRPL41B | Peng et al. (2017a,2017b) |
| pIR3DH8 | Plasmid used to disrupt GAL80 with URA3 marker | This work |
| pML104 | E. coli/S. cerevisiae shuttle plasmid; 2 μ, URA3, PTDH3‐CRSIPR/Cas9‐TADH1 , PSNR52‐guideRNA‐swaI‐TSUP4 | Laughery et al. (2015) |
| pJCble | pML104; URA3 marker was replaced with ble marker (Goldstein and McCusker, 1999) | This work |
| pIALD2 | pRS424: ALD6 (−125, 40)‐ PADH1 > Lr.pduP> TPDC1 ‐ PSb.GAL2 >Lr. EutE> ALD6 (1054, 1749) | This work |
| pIALD2S | pRS424: ALD6 (−125, 40)> PADH1 > Lr.pduP> TPDC1 ‐ALD6 (1054, 1749) | This work |
| pIALD2E | pRS424: ALD(−125, 40)‐ ALD6(1054, 1749) | This work |
| pIALD2HMGr | pRS424: ALD6(−125, 40)> PSk.GAL1 >Da.mvaA>TPGK1 ‐PADH1 > Lr.pduP> TPDC1 ‐PGAL2 >Lr. EutE‐ALD6(1054, 1749) | This work |
| pIPKA2 | pRS424: GPP1(−220,80)> PENO2 >Lr.xfp>TTPI1 ‐PTDH3 >Mb.pta>TFBA1 ‐GPP1(754, 1400) | This work |
| pIPKAH | pRS424: GPP1(−220,80)> PAg.TEF1>hphMX6(Goldstein and McCusker, 1999)>TAg.TEF1‐PENO2> Lr.xfp>TTPI1‐PTDH3>Mb.pta>TFBA1 ‐GPP1(754, 1400) | This work |
| pJT9R | pRS425: TRPL3<ERG20<PGAL1‐PGAL2>Ac.NES1>TRPL41B | Peng et al. (2017a,2017b) |
| pILGB5A | Yeast integration plasmid; PURA3‐KlURA3‐TAgTEF1‐PGAL2‐yEGFP‐TURA3 | Peng et al. (2015) |
| pJAIDB58T | pILGB5A; PURA3‐KlURA3‐TAgTEF1‐PGAL2‐yEGFP‐ TPGK1‐PACS2‐SKP1 ‐OsTIR1‐TURA3 | This work |
Symbol > or < indicates the direction of the open reading frame.
Further engineering steps delivered two strains. The control strain GJ38T had the gal80 disruption, the PGAL1‐yEGFP‐TPGK1 expression cassette and constitutive expression of SKP1‐OsTIR1. In addition to the modifications found in GJ38T, strain G7B8T had the chromosomal MIG1 fused with the AID*‐CUP1 fusion. Comparison between these strains allowed specific examination of the effect of Mig1p degradation on GAL promoter activity.
Growth and GFP fluorescence of GJ38T (MIG1) and G7B8T (CUP1‐AID*‐MIG1) were examined in flask cultivations using MES‐buffered mineral medium with 20 g l−1 glucose as the carbon source (Fig. 3). Glucose is expected to be depleted after 12 h and then yeast started to use ethanol as the carbon source (Peng et al., 2015; Peng et al., 2017a,2017b). Both strains showed similar growth patterns during the exponential growth phase of the culture in the presence or absence of synthetic auxin analog NAA. However, the strain with the AID*‐tagged MIG1 (G7B8T; Fig. 3C) grew faster and produced more biomass during the post‐exponential growth phase compared with the untagged strain (GJ38T; Fig. 3B). This occurred in both the presence of NAA (added at 1 h cultivation) and in the absence of NAA. The effect was more marked in the presence of NAA, although very high variability of growth rate was also observed in these strains. The increased growth in the absence of NAA is presumably due to residual leaky degradation of Mig1p (Lu et al., 2021). NAA had no effect in the absence of the AID* tag (strain expressing only SKP1‐OsTIR1; Fig. 3b). The increased biomass production in the presence of the Mig1p degradation system may be caused by lifting Mig1p‐mediated repression on respiratory metabolisms (Bonander et al., 2008; Fendt and Sauer, 2010).
Fluorescence patterns from the GAL1 promoter reporter construct were very similar between the two strains in the absence of NAA, with low level fluorescence during the exponential (glucose) phase of the culture and a gradual increase after 12 h when glucose was depleted (Peng et al., 2015), over the remainder of the 72 h cultivation (Fig. 3D). Addition of NAA resulted in a dramatic change in fluorescence in the AID*‐tagged MIG1 strain, with a sharp increase in fluorescence during the exponential (glucose) phase of the culture, peaking at 12 h at the same level that the other strains/conditions reached after 72 h, and remaining at this level over the rest of the culture (Fig. 3E). No effect on fluorescence was seen in the wild type MIG1 strain expressing only the SKP1‐OsTIR1 fusion. These data confirm effective modification of the GAL1 promoter behaviour in the presence of an AID*‐tagged MIG1, presumably as the result of degradation of the Mig1 protein and consequent release of glucose‐mediated repression of the GAL1 promoter.
Auxin‐mediated depletion of Mig1p improves nerolidol production in acetyl‐CoA pathway‐engineered strains
Next, we tested the combined effect of GAL promoter induction on glucose and the synthetic acetyl‐CoA pathways for nerolidol production. We used strain o501R (mevalonate pathway augmented; auxin‐inducible protein degradation machinery gene SKP1‐TIR1 expressed under the control of the ACS2 promoter; Fig. 1B; Table 1), as a background strain. To introduce auxin‐mediated induction of GAL promoters into strain o501R, Cup1p‐AID* was tagged to the N‐terminus of Mig1p (strain o57BR). Three nerolidol‐producing strains were built from strain o57BR: N57BRU, with auxin‐mediated degradation of Mig1p and nerolidol production (reference strain); N637BU, with auxin‐mediated degradation of Mig1p, an ADA pathway (ald6Δ, PADH1‐Lr.pduP, PSb.GAL2‐Lr. EutE) and nerolidol production; and N737BU, with auxin‐mediated degradation of Mig1p, an ADA pathway and a phosphoketolase pathway (ald6Δ, gpp1Δ, PADH1‐Lr.pduP, PSb.GAL2‐Lr. EutE, PENO2‐Lr.xfp and PTDH3‐Mb.pta), and nerolidol production. These three strains were characterised under the induction of the GAL promoter controlled genes by adding NAA during the pre‐cultivation and again to the main culture immediately after inoculation.
Compared with the strains with wild‐type Mig1p (Fig. 2A and D; N501RU, N63U, N73U), the Mig1p depletion strains N57BRU, N637BU and N737BU all showed decreased exponential growth in the presence of NAA (Fig. 4A and B). Strain N57BRU had a µmax of ˜ 0.23 h−1 (Fig. 4B; with NAA addition) compared with its reference N501RU, which had a of µmax ˜ 0.31 h−1 (Fig. 2D). Strain N57BRU also accumulated less biomass than N501RU during the post‐exponential phase (Figs 2A and 4A). These support the idea that Mig1p depletion or GAL promoter induction on glucose results in a metabolic burden or imbalance. Strain N637BU (ADA pathway + nerolidol production) showed growth arrest during the ethanol consumption phase (Fig. 4A), suggesting a significant extra metabolic burden of additional nerolidol production in the presence of acetyl‐CoA metabolism modifications in this strain. Consistent with the observation for strain N73U (ADA pathway and phosphoketolase pathway; MIG1‐wildtype; Fig. 2C and E), strain N737BU (ADA pathway and phosphoketolase pathway; Mig1p depleted + nerolidol production) showed slower exponential growth rate compared with the other strains (Fig. 4B), and superior post‐exponential growth (Fig. 4A). Despite the decreased growth rate relative to N73U, no negative effect on the biomass yield was observed for N737BU (Figs 2A and 4A).
Fig. 4.
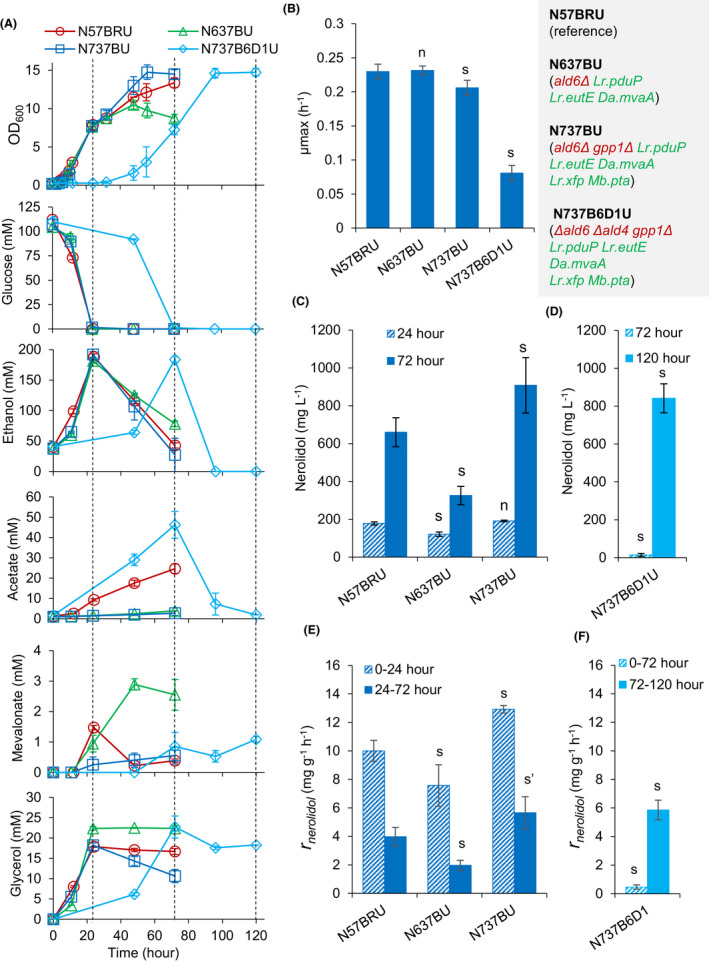
Characterising the strains with engineered acetyl‐CoA synthesis pathways and auxin‐mediated induction of GAL promoters: (A) growth (N = 4) and metabolic profiles (N = 2); (B) maximum specific growth rate (N = 4); (C and D), nerolidol titre (N = 4); (E and F), specific nerolidol production rate (N = 4). Two‐phase flask cultivation on 20 g l−1 glucose was employed, and NAA dissolved in ethanol was added in preculture and at the beginning of the cultivation. Dashed lines in A indicate 24, 72 and 120 h. For C–F, two‐tailed Welch’s t‐test was used for statistical analysis relative to N57BRU: n, P > 0.1; m, P ∈ [.05, 0.1]; s and s′ (calculated from log‐transformed data), P < 0.05. Mean values ± standard deviations are shown.
Nerolidol production in the strains N57BRU, N637BU and N737BU was evaluated by measuring the titres at 24 and 72 h and calculating the specific nerolidol production rates for the period from 0 to 24 h and from 24 to 72 h. These two periods approximately resemble the growth phases on glucose and ethanol respectively (Fig. 4A). At 24 h, i.e., on glucose, strain N57BRU produced ˜ 180 mg l−1 nerolidol (Fig. 4C). This is much higher than the 24 h titre (˜ 40 mg l−1) in a very similar strain previously engineered with GAL‐promoter‐controlled nerolidol synthetic pathway but in the absence of MIG1 depletion (strain N391DA; Peng et al., 2017a). The final nerolidol titre of ˜ 660 mg l−1 at 72 h of N57BRU (Fig. 4C) was significantly higher than titres achieved with strain N391DA 392 mg l−1 (Peng et al., 2017a,2017b) and N501RA (444 mg l−1; Fig. 2B; two‐tailed Welch’s t‐test, P = 0.04), which are comparable to each other. Nerolidol production in N637BU (ADA pathway) was only slightly lower at 24 h than strain N57BRU (reference strain), but significantly lower at 72 h (about half; Fig. 4C). This is consistent with the growth arrest phenotype during the ethanol phase (Fig. 4A), which suggests a major metabolic perturbation in this strain. However, strain N737BU (ADA pathway + phosphoketolase pathway) accumulated ˜ 910 mg l−1 nerolidol at 72 h, an improvement of ˜40% compared with N57BRU (Fig. 4C). For all three strains, the specific nerolidol production rate on glucose was more than twofold higher than that on ethanol (Fig. 4E).
Other extracellular metabolites acetate, mevalonate and glycerol were also measured (Fig. 4A). Compared with the reference N57BRU, acetate production was reduced in the acetyl‐CoA engineered strains. Mevalonate was secreted by all three strains (Fig. 4A), indicating an imbalance of pathway enzyme levels or an energy limitation of the ATP‐dependent downstream steps. In strain N57BRU, mevalonate peaked at 24 h and was consumed in the following ethanol growth phase. In contrast, strain N637BU (ADA pathway) continued to produce mevalonate into the ethanol phase. Production peaked at 48 h (concomitant with growth arrest) and the mevalonate was not re‐assimilated. Strain N737BU (ADA pathway + phosphoketolase pathway) produced only minor amounts of mevalonate. All strains exhibited similarly high glycerol formation during the glucose phase (Fig. 4A). Consistent with the reported deficiency of the parental S. cerevisiae CEN.PK strain in glycerol utilisation (Swinnen et al., 2013; Ho et al., 2017), strains N57BRU and N637BU did not consume glycerol during the ethanol phase (Fig. 4A). Interestingly, strain N737BU re‐assimilated glycerol during the ethanol phase.
These showed that Mig1p depletion improved nerolidol production when glucose was used as carbon source, overall nerolidol production in the reference strain without heterologous acetyl‐CoA pathways and in the strain with both ADA pathway and phosphoketolase pathway. In the strain with ADA pathway only, Mig1p depletion caused negative effects on growth and nerolidol production. It was also shown that engineered acetyl‐CoA pathways improved nerolidol production and causes perturbations on carbohydrate metabolism.
Disruption of ALD4 and ACS2 in strains with engineered acetyl‐CoA synthetic pathways
To further force carbon flux through the engineered acetyl‐CoA synthetic pathways, we disrupted additional endogenous genes involved in acetyl‐CoA synthesis. We targeted ALD4, which encodes a mitochondrial ADA, and ACS2, which encodes the major acetyl‐CoA synthase (Fig. 1). ALD4 and ACS2 were disrupted separately in the background strains o60 (ald6Δ), o61 (ald6Δ, PADH1‐Lr.pdu) and o73 (ald6Δ, PADH1‐Lr.pduP, PSb.GAL2‐Lr.eutE, PSk.GAL2‐Da.mvaA, PENO2‐Lr.xfp, PTDH3‐Mb.pta). The heterologous acetyl‐CoA synthesis pathways in strains o61 (ADA) and o73 (ADA + phosphoketolase pathway) are theoretically able to complement the deficiency caused by double disruption of ALD6 and ALD4 or double disruption of ALD6 and ACS2. However, in contrast to the respective o60 derivatives without complementary genes for acetyl‐CoA synthesis, the ald4Δ derivatives and the acs2Δ derivatives of o61 and o73 did not show a restored growth on the synthetic mineral salt agar with glucose as the carbon source (Fig. S1). Supplementing acetate restored the growth deficiency caused by double disruption of ALD6 and ALD4 (Fig. S1). Acetyl‐CoA synthesis through acetylating ADA (Kozak et al., 2014a,2014b; Kozak et al., 2016; Meadows et al., 2016) and the phosphoketolase pathway (Meadows et al., 2016) have previously been shown to complement growth defects caused by the removal of endogenous reactions essential for acetyl‐CoA synthesis and to improve sesquiterpene production. These presumably indicate the novel gene combinations used here did not provide sufficient acetyl‐CoA in the presence of the disrupted endogenous acetyl‐CoA synthesis genes (ald4Δ ald6Δ; or ald6Δ acs2Δ).
Despite the apparent insufficiency in heterologous acetyl‐coA synthesis, we were interested in the impact of ALD4 disruption on nerolidol production to confirm the phenotype for nerolidol production in the strain with major acetaldehyde dehydrogenases disrupted. We deleted this gene in strain N737BU (ADA + phosphoketolase pathways) and characterised the resulting strain N737B6D1U in the presence of NAA for GAL promoter induction (Fig. 4). Consistent with that double disruption of ALD4 and ALD6 caused growth deficiency (Fig. S1), the ald4Δ mutant showed extremely slow growth on glucose, with a maximum specific growth rate of ˜ 0.08 h−1 (Fig. 4A and B). Glucose utilisation was similarly slow and was accompanied by high production of acetate and glycerol (Fig. 4A).
Only ˜ 14 mg l−1 nerolidol (Fig. 4D) and < 1 mM mevalonate (Fig. 4A) were detected after 72 h (extended glucose phase). After glucose depletion, all the ethanol and 85% of the accumulated acetate were consumed within 24 h (Fig. 4A). The ethanol consumption rate was much faster than that of strains N57BRU, N637BU and N737BU. Like strain N737BU, N737B6D1 U also re‐utilized glycerol after glucose depletion. Interestingly, the nerolidol titre at 120 h was ˜ 840 mg l−1, ˜ 28% higher than that in the reference strain N57BRU at 72 h (a comparable time relative to the physiological stage of the cultures) and similar to N737BU at 72 h. While the specific nerolidol production rate on glucose was very reduced, it was much higher than the reference strain N57BRU during the ethanol phase (Fig. 4E and F). These results indicate that the heterologous acetyl‐CoA pathways engineered in this work were not active enough for acetyl‐CoA production to support cell growth, despite the improved overall nerolidol production.
Discussion
Heterologous acetyl‐CoA pathways have been reconstructed previously in S. cerevisiae and have improved terpenoid production (Meadows et al., 2016; Vickers et al., 2017; Zhang et al., 2017). In this work, heterologous genes encoding ADA and phosphoketolase pathways were introduced in sesquiterpene nerolidol‐producing yeasts, attempting to construct carbon/energy‐efficient acetyl‐CoA synthesis pathways to improve isoprenoid (nerolidol) production. In addition to the acetyl‐CoA pathway engineering, we deployed an auxin‐mediated Mig1p depletion mechanism for GAL promoter induction on glucose, which improved nerolidol production on the glucose growth phase and overall nerolidol production. Combination of Mig1p depletion and engineered acetyl‐CoA synthetic pathway further improved nerolidol production and allowed us observe metabolic perturbation in these strains.
Previously, expressing various ADA genes in an ald2/3/4/5/6Δ background strain resulted in a fast‐growing strain with a maximum specific growth rate of 0.27 h−1 (Kozak et al., 2014a,2014b). Similar results were observed by expressing the ADA (EutE) from E. coli, in an acs2Δ background strain (Meadows et al., 2016). Expressing xfp from Leuconostoc mesenteroides and pta from Clostridium kluyveri enabled growth of an acs2Δ ald6Δ background strain on glucose (Meadows et al., 2016). However, while we observed enhanced specific nerolidol production in the strains with engineered acetyl‐CoA synthetic pathways in this work, they still exhibited growth defects when endogenous acetyl‐CoA pathways were inactivated. As the genes we used were from different species than the previous studies, this result underlines the importance of identifying optimal heterologous genes for efficient engineering.
Notwithstanding the deleterious physiological effects, the engineered heterologous pathways did deliver some of the targeted metabolic perturbation effects that we were aiming to achieve – including improved nerolidol production. Consistent with previous studies (Remize et al., 2000; Medina et al., 2010), acetate production was reduced in the ald6Δ strain (Fig. 4A; N637BU). While ADA pathway expression in combination with ALD6 disruption (strain Ν63U) improved nerolidol production, negative effects were seen in the Mig1p‐depleted strain N637BU, including decreased biomass accumulation on ethanol, increased mevalonate and glycerol accumulation, and decreased nerolidol productivities. Mevalonate accumulation may indicate the presence of increased pathway flux and production/consumption imbalance. Additional modifications, i.e., disrupting GPP1 and expression of phosphoketolase pathway genes Lr.xfp and Mb.pta (strain N737BU), alleviated the negative effects seen in strain N637BU and delivered almost 1 g l−1 nerolidol. Various phosphoketolases, including ones from Lactobacillus, have been expressed in S. cerevisiae previously (Sonderegger et al., 2004; Bergman et al., 2016) and expressing phosphoketolase has been shown to increase respiratory capacity (Bergman et al., 2019). This may contribute to improved nerolidol production in the strain harbouring phosphoketolase pathway. Further deletion of ALD4 in this strain caused acetate accumulation on glucose growth phase (Strain N737B6D1 U; Fig. 4). Acetate accumulation was not shown previously in an ald4Δ ald6Δ strain (Remize et al., 2000). We presume that the presence of the engineered heterologous acetyl‐CoA synthetic pathways in this strain results in an imbalance which favours acetate production. However, more biochemical analysis would be required to understand these perturbation effects.
We applied auxin‐inducible protein degradation mechanism to investigate a new strategy allowing induction of GAL promoters on glucose. Previously, we showed that depletion of hexokinase 2 (Hxk2p), which interacts with Snf1p complex and coordinates Mig1p repression on GAL promoters (Ahuatzi et al., 2007), led to induction of GAL promoters on glucose and improved nerolidol production. Similarly, depleting Mig1p also delivered GAL promoter induction (Fig. 3) and improved nerolidol production (Fig. 4). This indicate that auxin‐inducible protein degradation is applicable to control depletion of a transcription factor for transcriptional regulation and that inactivating Mig1p can be adopted as a strategy to induce GAL‐promoter‐controlled pathway for improved production on glucose.
In summary, production of target sesquiterpene nerolidol was improved through engineering of acetyl‐CoA synthetic pathway and depletion of glucose‐dependent repressor Mig1p. The genes we introduced to construct ADA and phosphokinases pathways did not deliver enough acetyl‐CoA to support yeast growth but did cause improved production. It therefore is important to select optimal genes for construction of functional heterologous acetyl‐CoA pathways before applying them in metabolic engineering. This, on the other hand, shows that untargeted metabolic perturbation in metabolic engineering may cause improved production of target product. Depleting Mig1p was validated to induce GAL promoters and improve nerolidol production from engineered pathways under control of GAL promoters. This can be a general regulatory strategy for metabolic engineering in the yeast where GAL promoters are used to control pathway genes. However, instead of adding auxin in culture to trigger Mig1p depletion, dynamic mechanisms like quorum sensing (Lu et al., 2021) may be further exploited to control its depletion for application in industrial fed‐batch production.
Experiment procedures
Plasmid and strain construction
Strains and plasmids used in this study are listed in Tables 1 and 2. Primers and PCR products used in this study are listed in Table S1. Plasmid construction processes are described in Table S2. Strain construction processes are described in Fig. 1B and Table S3. Genes Lr.pduP, Lr.eutE and Lr.xfp were amplified from Lactobacillus reuteri genomic DNA, which was isolated from Blackmores Digestive Bio BalanceTM pills (purchased at Chemist Warehouse, Australia). The codon‐optimised gene sequences of Delftia acidovorans mvaA (Genbank: M24015.1), Methanosarcina barkeri pta (Genbank: AKB52167), Oryza sativa TIR1 (Dharmasiri et al., 2005; Nishimura et al., 2009) and the minimal, auxin‐inducible degron AID* (Morawska and Ulrich, 2013), were synthesized by Integrated DNA Technologies (USA) as gBlocks® Gene Fragments.
Two‐phase flask cultivation
Two‐phase flask cultivation was used to characterize the nerolidol‐producing strains (Peng et al., 2017a,2017b). Yeast cells were recovered from glycerol stocks by plating on yeast nitrogen base (YNB; with ammonia sulfate without amino acids) agar with 200 g l−1 glucose. YNB medium with 20 g l−1 glucose and 100 mM 2‐(N‐morpholino) ethanesulfonic acid (MES, Sigma‐Aldrich#M8250)‐ammonia buffer (pH 6) was used in preculture and flask cultivations. MES buffer was used to alleviate the alleviation of medium (Peng et al., 2015). Yeast cells were precultured to exponential phase in 15 ml MES‐buffered YNB‐glucose medium in a 50 ml Erlenmeyer flask (optical density at 600 nm from 0.8 to 5). Two‐phase flask cultivation was initiated by inoculating precultured cells to an OD600 of 0.2 in 23 ml MES‐buffered YNB‐glucose medium in a 250 ml Erlenmeyer flask; 2 ml dodecane were added to extract isoprenoid products. Flask cultivation was performed at 30°C and 200 rpm. A 500 mM ethanolic stock solution of the auxin analog NAA was prepared and added to the culture to a final concentration of 1 mM at indicated time points. For all cultivations, about 3 ml culture was sampled in the first 12 h for growth curve measurement. The dodecane phase and aqueous cell suspension were sampled for metabolite analysis and stored at −80°C. OD600 was measured using a SHIMAZU UV‐2450 UV–visible spectrophotometer or a WPA CO8000 cell density metre.
Metabolite analysis
Nerolidol, farnesol and geranylgeraniol in dodecane samples were analysed using a reverse‐phase high‐performance liquid chromatography method (Peng et al., 2017a,2017b). If necessary, dodecane samples were diluted using dodecane to enable nerolidol concentration in the analytic range. Dodecane samples were diluted in a 40‐fold volume of ethanol. Ethanol‐diluted dodecane samples of 20 μl were injected into a Zorbax Extend C18 column (4.6 × 150 mm, 3.5 µm; Agilent, Santa Clara, CA, USA; part number: 763953‐902) with a guard column (SecurityGuard Gemini C18; Phenomenex, Lane Cove, NSW, Australia; part number: AJO‐7597). Analytes were eluted at 35°C at 0.9 ml min−1 using the mixture of solvent A (high purity water, 18.2 kΩ) and solvent B (45% acetonitrile, 45% methanol and 10% water), with a linear gradient of 5%–100% solvent B from 0 to 24 min, then 100% from 24 to 30 min and finally 5% from 30.1 to 35 min. Analytes of interest were monitored using a diode array detector (Agilent, Santa Clara, CA, USA; DAD SL, G1315C) at 202 nm wavelength. Analytical standards of trans,trans‐farnesol (96% purity; Sigma‐Aldrich, North Ryde, NSW, Australia; #277541), trans‐nerolidol (93.7% purity; Sigma‐Aldrich, North Ryde, NSW, Australia; #04610590) and geranylgeraniol (85% purity; Sigma‐Aldrich, North Ryde, NSW, Australia; #G3278), were used to prepare the standard curve for quantification.
GFP fluorescence assay
GFP fluorescence was monitored in cells cultivated aerobically in 20 ml MES‐buffered YNB‐glucose medium in 100 ml flasks. NAA was added to the culture to a final concentration of 1 mM as indicated. Samples were taken at indicated time points; the GFP fluorescence in single cells was analysed immediately after sampling using a flow cytometer (BD Accuri™ C6; BD Biosciences, Franklin Lakes, NJ, USA). Cultures were diluted after 12 h with 10 volumes of water before flow cytometry analysis. GFP fluorescence was excited by a 488 nm laser and monitored through a 530/20 nm band‐pass filter (FL1.A); 10 000 events were counted per sample. The GFP fluorescence was expressed as percentage of the average background auto‐fluorescence from exponentially growing cells of the GFP‐negative reference strain GH4J3.
Physiological feature calculation
The maximum specific growth rates (μmax) in flask cultivations were determined from the slope of the linear regression of the natural logarithm of the OD600 values versus time curve during the exponential growth phase. The biomass concentration was estimated using the OD600‐biomass correlation of 0.23 g l−1 of cell dry weight per unit of OD600 (Peng et al., 2012). The specific nerolidol production rate (rnerolidol , mg g−1 biomass h−1) was calculated by dividing the difference in the nerolidol titre (mg l−1) with the integral of biomass in defined time (g l−1 h−1).
Funding Information
BP and this project were supported by a CSIRO Synthetic Biology Future Science Fellowship and the University of Queensland. IHF was supported by an Indonesia Endowment Fund for Education scholarship.
Conflict of interests
The authors declare that they have no competing interests.
Author contributions
BP and CEV conceived the study. IHF and BP performed experiments. MP performed the metabolite analyses. BP and IHF drafted the manuscript, and BEE, GD, and CEV revised the manuscript. All authors contributed to the result analysis and the discussion of the research. All authors read and approved the final manuscript.
Supporting information
Figure S1. Testing growth complimentary effects of heterologous acetyl‐CoA synthetic genes in the strains with double disruption of ALD6 and ALD4 (A) or in the strains with double disruption of ALD6 and ACS2 (B). The strains in a are: strain o60 (ald6Δ ALD4), strain o60 with ALD4 disruption (ald6Δ, ald4Δ), strain o61 with ALD4 disruption (ald6Δ, ald4Δ, Lr.pduP), and strain o73 with ALD4 disruption (ald6Δ, ald4Δ, Lr.pduP, Lr.xfp, Mb.pta); and were growth at 30 °C for 2 days. Strains in b are: strain o60 (ald6Δ ACS2), strain o60 with ACS2 disruption (ald6Δ, acs2Δ), strain o61 with ACS2 disruption (ald6Δ, acs2Δ, Lr.pduP), and strain o73 with ACS2 disruption (ald6Δ, acs2Δ, Lr.pduP, Lr.xfp, Mb.pta); and were growth at 30 °C for 3 days. ACS2 was disrupted by transforming PCR fragment #19 (Table S1).
Table S1. List of primers and PCR fragments used in this work. PXXX and TXXX indicate promoter and terminator sequence of gene XXX, respectively; red coloured sequences indicate a part that complementary to the DNA template; restriction enzyme sites used in cloning are shown in bold; 20‐mer CRISPR/Cas9 guide sequence was underlined in bold.
Table S2. Plasmid construction.
Table S3. Strain construction. DNA fragments refer to Supplementary Table S1, and plasmids refer to Table 2.
Acknowledgements
BP and this project were supported by a CSIRO Synthetic Biology Future Science Fellowship and the University of Queensland. IHF was supported by an Indonesia Endowment Fund for Education scholarship.
Microb. Biotechnol. (2021) 14(6), 2627–2642
Contributor Information
Claudia E. Vickers, Email: Claudia.vickers@csiro.au.
Bingyin Peng, Email: b.peng@uq.edu.au.
References
- Ahuatzi, D. , Riera, A. , Pelaez, R. , Herrero, P. , and Moreno, F. (2007) Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J Biol Chem 282: 4485–4493. [DOI] [PubMed] [Google Scholar]
- Alonso‐Gutierrez, J. , Koma, D. , Hu, Q. , Yang, Y. , Chan, L.J.G. , Petzold, C.J. , et al. (2018) Toward industrial production of isoprenoids in Escherichia coli: Lessons learned from CRISPR‐Cas9 based optimization of a chromosomally integrated mevalonate pathway. Biotechnol Bioeng 115: 1000–1013. [DOI] [PubMed] [Google Scholar]
- Aslan, S. , Noor, E. , and Bar‐Even, A. (2017) Holistic bioengineering: rewiring central metabolism for enhanced bioproduction. Biochem J 474: 3935–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, B.M. , Overkamp, K.M. , van Maris, A.J. , Kotter, P. , Luttik, M.A. , van Dijken, J.P. , and Pronk, J.T. (2001) Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae . FEMS Microbiol Rev 25: 15–37. [DOI] [PubMed] [Google Scholar]
- Bergman, A. , Hellgren, J. , Moritz, T. , Siewers, V. , Nielsen, J. , and Chen, Y. (2019) Heterologous phosphoketolase expression redirects flux towards acetate, perturbs sugar phosphate pools and increases respiratory demand in Saccharomyces cerevisiae . Microb Cell Fact 18: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, A. , Siewers, V. , Nielsen, J. , and Chen, Y. (2016) Functional expression and evaluation of heterologous phosphoketolases in Saccharomyces cerevisiae . AMB Express 6: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, G. , Deng, Z. , and Liu, T. (2017) Strategies for terpenoid overproduction and new terpenoid discovery. Curr Opin Biotechnol 48: 234–241. [DOI] [PubMed] [Google Scholar]
- Bogorad, I.W. , Lin, T.S. , and Liao, J.C. (2013) Synthetic non‐oxidative glycolysis enables complete carbon conservation. Nature 502: 693–697. [DOI] [PubMed] [Google Scholar]
- Bonander, N. , Ferndahl, C. , Mostad, P. , Wilks, M.D.B. , Chang, C. , Showe, L. , et al. (2008) Transcriptome analysis of a respiratory Saccharomyces cerevisiae strain suggests the expression of its phenotype is glucose insensitive and predominantly controlled by Hap4, Cat8 and Mig1. BMC Genom 9: 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubekeur, S. , Bunoust, O. , Camougrand, N. , Castroviejo, M. , Rigoulet, M. , and Guerin, B. (1999) A mitochondrial pyruvate dehydrogenase bypass in the yeast Saccharomyces cerevisiae . J Biol Chem 274: 21044–21048. [DOI] [PubMed] [Google Scholar]
- Boubekeur, S. , Camougrand, N. , Bunoust, O. , Rigoulet, M. , and Guerin, B. (2001) Participation of acetaldehyde dehydrogenases in ethanol and pyruvate metabolism of the yeast Saccharomyces cerevisiae . Eur J Biochemistry/FEBS 268: 5057–5065. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Siewers, V. , and Nielsen, J. (2012) Profiling of cytosolic and peroxisomal acetyl‐CoA metabolism in Saccharomyces cerevisiae . PLoS One 7: e42475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T.W. , Sikorski, R.S. , Dante, M. , Shero, J.H. , and Hieter, P. (1992) Multifunctional yeast high‐copy‐number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Curran, K.A. , Karim, A.S. , Gupta, A. , and Alper, H.S. (2013) Use of expression‐enhancing terminators in Saccharomyces cerevisiae to increase mRNA half‐life and improve gene expression control for metabolic engineering applications. Metab Eng 19: 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri, N. , Dharmasiri, S. , and Estelle, M. (2005) The F‐box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Entian, K.‐D. , and Kötter, P. (1998) 23 yeast mutant and plasmid collections. Methods Microbiol 26: 431–449. [Google Scholar]
- Fendt, S. M. , and Sauer, U. (2010) Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst Biol 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova, E. , Johnston, M. , and Majors, J. (1999) Binding of the glucose‐dependent Mig1p repressor to the GAL1 and GAL4 promoters in vivo: regulation by glucose and chromatin structure. Nucleic Acids Res 27: 1350–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A.L. , and McCusker, J.H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae . Yeast (Chichester, England) 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gruchattka, E. , Hadicke, O. , Klamt, S. , Schutz, V. , and Kayser, O. (2013) In silico profiling of Escherichia coli and Saccharomyces cerevisiae as terpenoid factories. Microb Cell Fact 12: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henard, C.A. , Freed, E.F. , and Guarnieri, M.T. (2015) Phosphoketolase pathway engineering for carbon‐efficient biocatalysis. Curr Opin Biotechnol 36: 183–188. [DOI] [PubMed] [Google Scholar]
- Ho, P.W. , Swinnen, S. , Duitama, J. , and Nevoigt, E. (2017) The sole introduction of two single‐point mutations establishes glycerol utilization in Saccharomyces cerevisiae CEN.PK derivatives. Biotechnol Biofuels 10: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, M.K. , and Keasling, J.D. (2015) Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res 15: 1–10. [DOI] [PubMed] [Google Scholar]
- Kanke, M. , Nishimura, K. , Kanemaki, M. , Kakimoto, T. , Takahashi, T.S. , Nakagawa, T. , and Masukata, H. (2011) Auxin‐inducible protein depletion system in fission yeast. Bmc Cell Biol 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, B.U. , van Rossum, H.M. , Benjamin, K.R. , Wu, L. , Daran, J.M. , Pronk, J.T. , and van Maris, A.J. (2014a) Replacement of the Saccharomyces cerevisiae acetyl‐CoA synthetases by alternative pathways for cytosolic acetyl‐CoA synthesis. Metab Eng 21: 46–59. [DOI] [PubMed] [Google Scholar]
- Kozak, B.U. , van Rossum, H.M. , Niemeijer, M.S. , van Dijk, M. , Benjamin, K. , Wu, L. , et al. (2016) Replacement of the initial steps of ethanol metabolism in Saccharomyces cerevisiae by ATP‐independent acetylating acetaldehyde dehydrogenase. FEMS Yeast Res 16: fow006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, B.U. , van Rossum, H.M. , Luttik, M.A.H. , Akeroyd, M. , Benjamin, K.R. , Wu, L. , et al. (2014b) Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae . mBio 5: e01696–e1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughery, M.F. , Hunter, T. , Brown, A. , Hoopes, J. , Ostbye, T. , Shumaker, T. , and Wyrick, J.J. (2015) New vectors for simple and streamlined CRISPR‐Cas9 genome editing in Saccharomyces cerevisiae . Yeast (Chichester, England) 32: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Prasanna, X. , Salo, V.T. , Vattulainen, I. , and Ikonen, E. (2019) An efficient auxin‐inducible degron system with low basal degradation in human cells. Nat Methods 16: 866–869. [DOI] [PubMed] [Google Scholar]
- Loison, G. , Vidal, A. , Findeli, A. , Roitsch, C. , Balloul, J.M. , and Lemoine, Y. (1989) High level of expression of a protective antigen of schistosomes in Saccharomyces cerevisiae . Yeast (Chichester, England) 5: 497–507. [DOI] [PubMed] [Google Scholar]
- Lu, Z. , Peng, B. , Ebert, B.E. , Dumsday, G. , and Vickers, C.E. (2021) Auxin‐mediated protein depletion for metabolic engineering in terpene‐producing yeast. Nat Commun 12: 1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, S.M. , Garcia, D.E. , Redding‐Johanson, A.M. , Friedland, G.D. , Chan, R. , Batth, T.S. , et al. (2011) Optimization of a heterologous mevalonate pathway through the use of variant HMG‐CoA reductases. Metab Eng 13: 588–597. [DOI] [PubMed] [Google Scholar]
- Meadows, A.L. , Hawkins, K.M. , Tsegaye, Y. , Antipov, E. , Kim, Y. , Raetz, L. , et al. (2016) Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 537: 694–697. [DOI] [PubMed] [Google Scholar]
- Medina, V.G. , Almering, M.J.H. , van Maris, A.J.A. , and Pronk, J.T. (2010) Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microbiol 76: 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska, M. , and Ulrich, H.D. (2013) An expanded tool kit for the auxin‐inducible degron system in budding yeast. Yeast (Chichester, England) 30: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin, J.O. , Carlberg, M. , and Ronne, H. (1991) Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J 10: 3373–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J. (2019) Yeast systems biology: model organism and cell factory. Biotechnol J 14: e1800421. [DOI] [PubMed] [Google Scholar]
- Nishimura, K. , Fukagawa, T. , Takisawa, H. , Kakimoto, T. , and Kanemaki, M. (2009) An auxin‐based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922. [DOI] [PubMed] [Google Scholar]
- Peng, B. , Nielsen, L.K. , Kampranis, S.C. , and Vickers, C.E. (2018) Engineered protein degradation of farnesyl pyrophosphate synthase is an effective regulatory mechanism to increase monoterpene production in Saccharomyces cerevisiae . Metab Eng 47: 83–93. [DOI] [PubMed] [Google Scholar]
- Peng, B. , Plan, M.R. , Carpenter, A. , Nielsen, L.K. , and Vickers, C.E. (2017a) Coupling gene regulatory patterns to bioprocess conditions to optimize synthetic metabolic modules for improved sesquiterpene production in yeast. Biotechnol Biofuels 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, B. , Plan, M.R. , Chrysanthopoulos, P. , Hodson, M.P. , Nielsen, L.K. , and Vickers, C.E. (2017b) A squalene synthase protein degradation method for improved sesquiterpene production in Saccharomyces cerevisiae . Metab Eng 39: 209–219. [DOI] [PubMed] [Google Scholar]
- Peng, B. , Shen, Y. , Li, X. , Chen, X. , Hou, J. , and Bao, X. (2012) Improvement of xylose fermentation in respiratory‐deficient xylose‐fermenting Saccharomyces cerevisiae . Metab Eng 14: 9–18. [DOI] [PubMed] [Google Scholar]
- Peng, B. , Williams, T. , Henry, M. , Nielsen, L. , and Vickers, C. (2015) Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microb Cell Fact 14: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, B. , Wood, R.J. , Nielsen, L.K. , and Vickers, C.E. (2018) An expanded heterologous GAL promoter collection for diauxie‐inducible expression in Saccharomyces cerevisiae . ACS Synth Biol 7: 748–751. [DOI] [PubMed] [Google Scholar]
- Pronk, J.T. , Yde Steensma, H. , and Van Dijken, J.P. (1996) Pyruvate metabolism in Saccharomyces cerevisiae . Yeast (Chichester, England) 12: 1607–1633. [DOI] [PubMed] [Google Scholar]
- Remize, F. , Andrieu, E. , and Dequin, S. (2000) Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl Environ Microbiol 66: 3151–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan, K.M. , McKenna, B.D. , Anderson, W.D. , Duarte, F.M. , Core, L. , and Guertin, M.J. (2019) An improved auxin‐inducible degron system preserves native protein levels and enables rapid and specific protein depletion. Genes Dev 33: 1441–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, B. , Zhou, P. , Jiao, X. , Yao, Z. , Ye, L. , and Yu, H. (2020) Fermentative production of Vitamin E tocotrienols in Saccharomyces cerevisiae under cold‐shock‐triggered temperature control. Nat Commun 11: 5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderegger, M. , Schumperli, M. , and Sauer, U. (2004) Metabolic engineering of a phosphoketolase pathway for pentose catabolism in Saccharomyces cerevisiae . Appl Environ Microbiol 70: 2892–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos, G. (2012) Synthetic biology and metabolic engineering. ACS Synth Biol 1: 514–525. [DOI] [PubMed] [Google Scholar]
- Swinnen, S. , Klein, M. , Carrillo, M. , McInnes, J. , Nguyen, H.T.T. , and Nevoigt, E. (2013) Re‐evaluation of glycerol utilization in Saccharomyces cerevisiae: characterization of an isolate that grows on glycerol without supporting supplements. Biotechnol Biofuels 6: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, C.E. , Bongers, M. , Liu, Q. , Delatte, T. , and Bouwmeester, H. (2014) Metabolic engineering of volatile isoprenoids in plants and microbes. Plant Cell Environ 37: 1753–1775. [DOI] [PubMed] [Google Scholar]
- Vickers, C.E. , Williams, T.C. , Peng, B. , and Cherry, J. (2017) Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr Opin Chem Biol 40: 47–56. [DOI] [PubMed] [Google Scholar]
- Westfall, P.j. , Pitera, D.j. , Lenihan, J.r. , Eng, D. , Woolard, F.x. , Regentin, R. , et al. (2012) Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA 109: E111–E118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T. , Zhou, Y.J.J. , Huang, M.T. , Liu, Q.L. , Pereira, R. , David, F. , and Nielsen, J. (2018) Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell 174: 1549–1558.e14. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Nielsen, J. , and Liu, Z. (2017) Engineering yeast metabolism for production of terpenoids for use as perfume ingredients, pharmaceuticals and biofuels. FEMS Yeast Res 17: fox080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Testing growth complimentary effects of heterologous acetyl‐CoA synthetic genes in the strains with double disruption of ALD6 and ALD4 (A) or in the strains with double disruption of ALD6 and ACS2 (B). The strains in a are: strain o60 (ald6Δ ALD4), strain o60 with ALD4 disruption (ald6Δ, ald4Δ), strain o61 with ALD4 disruption (ald6Δ, ald4Δ, Lr.pduP), and strain o73 with ALD4 disruption (ald6Δ, ald4Δ, Lr.pduP, Lr.xfp, Mb.pta); and were growth at 30 °C for 2 days. Strains in b are: strain o60 (ald6Δ ACS2), strain o60 with ACS2 disruption (ald6Δ, acs2Δ), strain o61 with ACS2 disruption (ald6Δ, acs2Δ, Lr.pduP), and strain o73 with ACS2 disruption (ald6Δ, acs2Δ, Lr.pduP, Lr.xfp, Mb.pta); and were growth at 30 °C for 3 days. ACS2 was disrupted by transforming PCR fragment #19 (Table S1).
Table S1. List of primers and PCR fragments used in this work. PXXX and TXXX indicate promoter and terminator sequence of gene XXX, respectively; red coloured sequences indicate a part that complementary to the DNA template; restriction enzyme sites used in cloning are shown in bold; 20‐mer CRISPR/Cas9 guide sequence was underlined in bold.
Table S2. Plasmid construction.
Table S3. Strain construction. DNA fragments refer to Supplementary Table S1, and plasmids refer to Table 2.


