In this article we have summarised our findings resulting from analysing the complete genome sequence of Pseudomonas umsongensis GO16 and we have determined experimentally its potential for the upcycling of monomers resulting from PET hydrolysis. Besides a general genomic characterisation, we have identified and validated the set of genes required for terephthalate and ethylene glycol degradation as well as the ability of the strain for producing short and medium‐chain length polyhydroxyalkanoates.
Summary
The throwaway culture related to the single‐use materials such as polyethylene terephthalate (PET) has created a major environmental concern. Recycling of PET waste into biodegradable plastic polyhydroxyalkanoate (PHA) creates an opportunity to improve resource efficiency and contribute to a circular economy. We sequenced the genome of Pseudomonas umsongensis GO16 previously shown to convert PET‐derived terephthalic acid (TA) into PHA and performed an in‐depth genome analysis. GO16 can degrade a range of aromatic substrates in addition to TA, due to the presence of a catabolic plasmid pENK22. The genetic complement required for the degradation of TA via protocatechuate was identified and its functionality was confirmed by transferring the tph operon into Pseudomonas putida KT2440, which is unable to utilize TA naturally. We also identified the genes involved in ethylene glycol (EG) metabolism, the second PET monomer, and validated the capacity of GO16 to use EG as a sole source of carbon and energy. Moreover, GO16 possesses genes for the synthesis of both medium and short chain length PHA and we have demonstrated the capacity of the strain to convert mixed TA and EG into PHA. The metabolic versatility of GO16 highlights the potential of this organism for biotransformations using PET waste as a feedstock.
Introduction
Polyethylene terephthalate (PET) is one of the most commonly used plastic polymers with an annual demand of approximately 33 million tonnes (Geyer et al., 2017). Even though PET bottles are used as an example of successful recycling, globally only 7% of PET produced annually is actually recycled (Forum, 2015) with a large majority of plastic waste still landfilled (Kasper, 2013; B, ; Plastics Europe, 2016). Among the options to reduce PET waste, the possibility of using microorganisms to both degrade and upcycle PET has gained attention (Narancic and O'Connor, 2017; Wei and Zimmermann, 2017; Blank et al., 2019). However, the ability to degrade PET is rare in nature (Wierckx et al., 2018; Salvador et al., 2019). PET belongs to the group of hydrolysable polymers and there are several examples of bacterial hydrolytic enzymes that were shown to break down PET into oligomers and monomers of terephthalate (TA) and ethylene glycol (EG; (Wei and Zimmermann, 2017). While the emergence of ‘plastic eating’ bacteria such as Ideonella sakaiensis (Yoshida et al., 2016) grabbed the attention of scientific and general audiences, there is still uncertainty about the rate of bacterial depolymerization of PET and the efficiency of PET monomer catabolism (Yang et al., 2016). Furthermore, the complete mineralization of PET to CO2 will not encourage circularity (Wierckx et al., 2015). Recycling of PET waste into a material such as a biodegradable plastic like polyhydroxyalkanoate (PHA) creates an opportunity to improve resource efficiency by extending and diversifying the life of the material, thus contributing to a circular economy (European Commission, 2015; Wierckx et al., 2015). It is necessary to optimize the enzymatic hydrolysis of PET but also to develop efficient microbial transformation of PET‐derived monomers TA and EG, arising from enzymatic degradation, into other molecules of value.
The strain Pseudomonas umsongensis GO16 was isolated from soil exposed to PET granules at a PET bottle processing plant (Kenny et al., 2008). The biotechnological potential of this strain was demonstrated by developing a process for the conversion of TA obtained from pyrolysis of PET into a biodegradable polymer, namely medium chain length polyhydroxyalkanoate (mcl‐PHA) (Kenny et al., 2008, 2012). However, the metabolic basis of TA conversion into PHA was not investigated.
Since the genus Pseudomonas was first described in 1894, over 190 species have been identified to date (Peix et al., 2018). The ability of pseudomonads to thrive in soil, sediments, hot springs, extremely cold environments, air, plants, animals and others, is largely due to their tremendous metabolic versatility allowing them to cope with harsh and stressful environmental conditions (Poblete‐Castro et al., 2017; Peix et al., 2018). The adaptability of the species belonging to the Pseudomonas genus to very different lifestyles has inspired the biotechnological use of these organisms as microbial cell factories for the production of chemicals, polymers, as bio‐controlling agents, as well as in bioremediation (Poblete‐Castro et al., 2017).
In this study, we have conducted a genome analysis of P. umsongensis GO16 and identified not only the genes responsible for TA and EG metabolism but also for catabolism of a wide range of aromatics. Moreover, we have identified the set of genes responsible for the synthesis of both short chain length (scl) and mcl‐PHA, and experimentally validated the accumulation of these biopolymers from different substrates including an equimolar mixture of TA and EG.
Results and discussion
Overall genomic organization
The P. umsongensis GO16 genome characteristics are given in Table 1. The genome shows a very high similarity with the previously reported P. umsongensis DSM 16611, shown to degrade a wide range of xenobiotics, such as phenol, trinitrotoluene, xylene, polyaromatic hydrocarbons and petroleum (Furmanczyk et al., 2017). The GC content of these two strains is similar, however P. umsongensis GO16 genome is 650 485 bp longer (Table 1). The strain GO16 contains a 7.3 Mbp chromosome (GenBank: CP044409.1) and an 82 kbp plasmid named pENK22 (GenBank: CP044408.1).
Table 1.
P. umsongensis GO16 genome features and comparison with the genome of P. umsongensis DSM 16611.
| Features | P. umsongensis GO16 | P. umsongensis DSM 16611 |
|---|---|---|
| Length (bp) | 7 269 974 chromosome + 81 914 pENK22 | 6 701 403 |
| GC content (%) | 59.2 | 59.7 |
| CDS | 6867 | 6152 |
| rRNA genes | 6 | 7 |
| tRNA genes | 57 | 62 |
The genome sequence was compared to notable organisms belonging to the same genus and for which a closed genome sequence is available (Fig. 1A). P. umsongensis GO16 possesses the largest genome of all of them, even larger than that of P. protegens Pf‐5 (7 074 893 bp). The phylogenetic analysis conducted with coding sequences of 30 species of the Pseudomonas genus places GO16 as a member of the P. umsongensis species, closely related to P. mandellii and P. frederiksbergensis (Fig. S1). Even though there is a high degree of functional conservation across species (Table S1), P. umsongensis GO16 has a distinct set of genome sections when compared P. umsongensis BS3657, which is the closest species for which a complete genome is available (Fig. 1B). The main differences, in addition to rearrangements, correspond to the insertion of segments likely resulting from horizontal gene transfer events in the chromosome of GO16. This is the case of a prophage (located in coordinates 805 892–830 810 bp); IS6 and IS2 transposons (1 212 038–1 253 316 bp), a region containing conjugative integrative elements, group II introns, IS3, IS5 and IS110 transposons (1 815 827–2 062 259 bp), IS3 transposon (2 661 452–2 775 901 bp); a region containing IS1182, IS110 transposons and an integrative element (4 806 223–4 889 799 bp); and regions with integrative elements (5 642 431–5 745 063 and 7 046 312–7 057 127 bp).
Fig. 1.
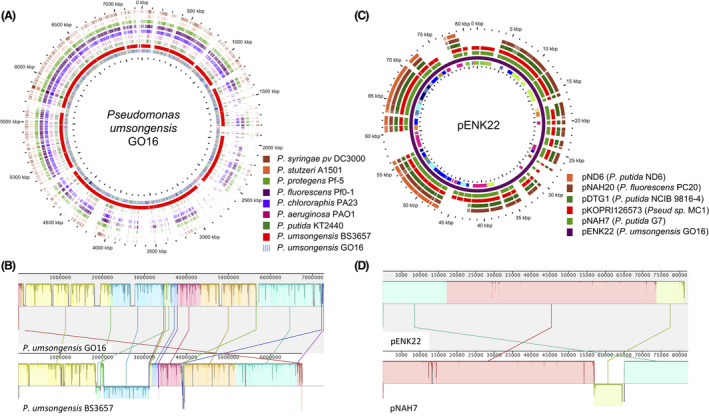
Global analysis of the P. umsongensis GO16 genome.
A. BLAST atlas comparing genes present in strain GO16 (used as a reference) with other representative Pseudomonas strains. The circles represent the genomes of the species shown in the insert starting with Pseudomonas syringae pv tomato DC3000 (outer circle) and finishing with P. umsongensis GO16 shown as the set of clusters of orthologous groups (COGs; inner circle).
B. Genome alignment of strains of P. umsongensis GO16 (upper bar) and BS3657 (lower bar).
C. BLAST atlas comparing present genes in plasmid pENK22 with other representative plasmids of Pseudomonas species containing naphthalene‐catabolic genes (nah).
D. Detailed alignment between pENK22 and the naphthalene degrading plasmid NAH7. Genes, COG (represented by the innermost circle) and blast analysis resulting from the analysis are summarized in Tables S1 and S2.
The plasmid pENK22 of P. umsongensis GO16 has a size of 81 914 kb and shows 99% identity with the 81 kb plasmid from Pseudomonas sp. MC1 (Ahn et al., 2017) and the 82 kb P. putida G7 NAH7 plasmid (Sota et al., 2006) (Fig. 1C). Like those plasmids, pENK22 encodes for a complete pathway for naphthalene mineralization and has a genetic organization almost identical to NAH7 (Sota et al., 2006) with the exception of a gene rearrangement (Fig. 1D; Table S2).
Central metabolism
The central metabolism of P. umsongensis GO16 was reconstructed based on the annotation derived from the genome sequence (Fig. 2; Table S3). Like in most Pseudomonas species, glucose is likely metabolized by the Entner‐Doudoroff pathway (ED), the Embden‐Meyerhof‐Parnas (EMP) pathway and the pentose phosphate pathway (PP), which produce a surplus of reducing power that allows for coping with stressful environments (Nikel et al., 2015). In this metabolism glucose is mainly transformed to gluconate in the periplasm prior to its conversion in 6‐phosphogluconate. Similar to P. putida KT2440, GO16 lacks a cytoplasmic glucose dehydrogenase which could carry out that conversion inside the cell, and a phosphofructokinase that could transform fructose‐6‐phosphate into fructose‐1,6‐bisphosphate following the conventional EMP glycolytic pathway.
Fig. 2.
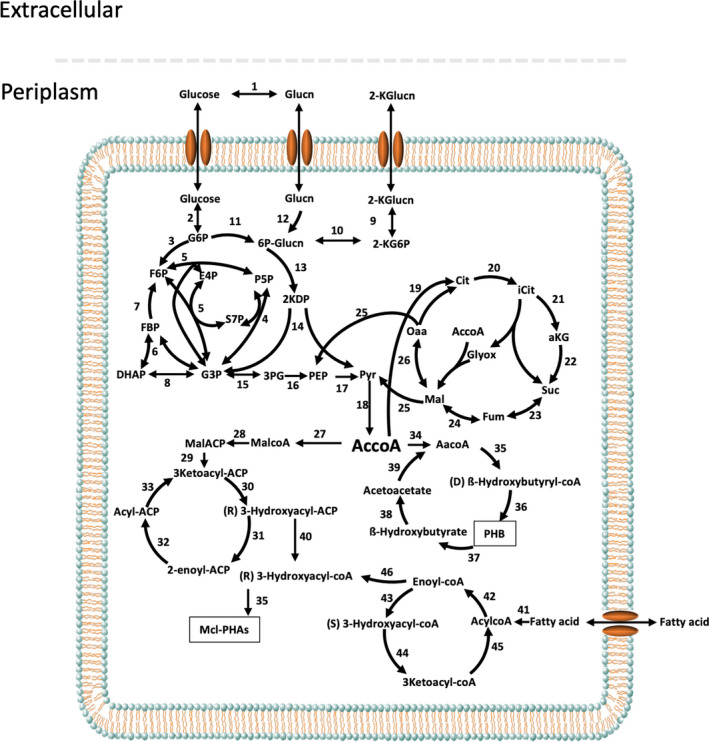
Proposed central metabolism of P. umsogensis GO16. The complete list of the enzymes and isozymes catalyzing each reaction is provided in Table S3. Abbreviations are as follows: Glucn, gluconate; 2‐KGlucn, 2‐keto‐gluconate; 2‐KG6P, 2‐keto‐gluconate‐6P; G6P, glucose‐6‐P; F6P, fructose‐6‐P; FBP, fructose‐1,6‐P2; DHAP, dihydroxyacetone‐P; 6P‐Glucn, 6‐phosphogluconate; 2KDP, 2‐keto‐3‐deoxy‐6‐phosphogluconate; P5P, Pentose‐5‐P; S7P, sedoheptulose‐7‐P; E4P, erythrose‐4‐P; G3P, glyceraldehyde‐3‐P; 3PG, glycerate‐3‐P; PEP, phosphoenolpyruvate; AccoA, acetyl‐coenzyme A; Aacoa, acetoacetyl‐coA; Oaa, oxaloacetate; aKG, α‐ketoglutarate; iCit, Isocitrate; Cit, Citrate; Glyox, Glyoxilate; Suc, Succinate; Fum, Fumarate; Mal, Malate; MalcoA, Malonyl‐coA; MalACP, Malonyl‐ACP; mcl‐PHAs, medium chain length Polyhydroxyalkanoate; PHB, Polyhydroxybutyrate..
According to the functional annotation, the genome of GO16 has a large number of potentially redundant enzymes related to a likely diverse metabolism of lipids. There are a number of activities that could constitute a β‐oxidation pathway including, for example, 23 genes annotated as 3‐oxoacyl‐(acyl‐carrier protein) reductases, 5 genes coding for enoyl‐(acyl‐carrier protein) reductase III, 4 genes coding for 3‐oxoacyl‐(acyl‐carrier‐protein) synthase II, 17 genes for the β‐oxidation enzyme acetyl‐CoA C‐acetyltransferase and 10 for 3‐hydroxyacyl‐CoA dehydrogenases.
Degradation of aromatics
Given that P. umsongensis GO16 was isolated using the aromatic compound TA as the sole carbon and energy source, we analysed the genome for the presence of pathways involved in the mineralization of other aromatics. This species shows remarkable metabolic versatility comparable or even larger than other members of the genus, possibly due to the above average size of the genome (Fig. 3) (Jiménez et al., 2004).
Fig. 3.
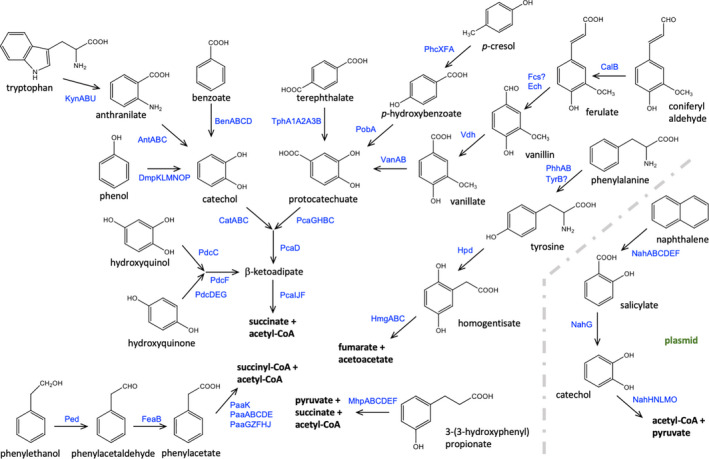
Proposed metabolism of aromatics in P. umsongensis GO16. The diagram shows the range of aromatic substrates metabolized by the bacterium. In blue are shown the required for the conversion of the substrates into central metabolites. The location and homology of the genes encoding for the corresponding enzymes as well as potential transcriptional regulators and transporters is shown in Table S4.
Our analysis identified a set of central pathways likely responsible for the degradation of monoaromatic molecules (Fig. 3 and Table S4). These include the ortho‐ pathway for degradation of catechol (cat), the 3,4‐dioxygenolytic pathway for degradation of protocatechuate (pca) and a pathway for the degradation of hydroxyquinol and hydroquinone (pdc), all of which converge in the β‐ketoadipate pathway that leads to the central metabolism. In addition, we found putative central pathways for the metabolism of homogentisate (hmg), phenylacetate (paa), 3‐hydroxyphenylpropionate (mhp) and the meta‐ cleavage for catechol (xyl) contained in the plasmid pENK22. Notably, the likely pcaGH and pcaIJ genes encoding, respectively, for the protocatechuate 3,4‐dioxygenase and 3‐oxoadipate CoA‐transferase involved in protocatechuate degradation were duplicated in the genome (Table S4).
These pathways are used for funnelling a plethora of aromatic molecules towards central metabolism. We identified genes potentially involved in the degradation of benzoate, tryptophan, anthranilate and phenol leading to catechol; ferulate, vanillin, vanillate, coniferyl aldehyde, p‐hydroxybenzoate (pOHB) and p‐cresol in addition to terephthalate degraded via protocatechuate; phenylalanine and tyrosine metabolized via homogentisate; phenylethanol and phenylacetaldehyde mineralized through phenylacetate. Naphthalene and salicylate are metabolized through the putative meta‐pathway for catechol encoded by the plasmid (Fig. 3 and Table S4). We confirmed experimentally that, in addition to TA, the GO16 strain can grow in 24 h on benzoate, tryptophan, pOHB, vanillate, protocatechuate, phenylacetate, phenylalanine and tyrosine (Fig. S2A and B). The strain exhibited low but detectable growth on the lignin derivatives p‐coumarate and ferulate (Fig. S2C). Likewise, we confirmed growth when in the presence of naphthalene vapours over a period of 4 days (Fig. S2D). The strain was unable to use vanillin, 2‐phenylethanol, anthranilate, p‐cresol, 3‐phenylpropionate and coniferyl aldehyde as sole carbon source in the conditions tested.
The pENK22 plasmid contains two putative operons for the catabolism of naphthalene, the upper pathway encoded by genes nahAa – nahD for the conversion of naphthalene to salicylate, and the lower pathway encoded by genes nahG – nahY for the conversion of salicylate to pyruvate and acetaldehyde via a meta‐cleavage pathway spanning over 26.4 kb (Fig. 3; Table S4). This pathway organization was identified in several bacterial strains capable of aerobic degradation of aromatic compounds (Williams and Sayers, 1994; Sota et al., 2006).
TA and EG metabolism
As already mentioned, TA and EG are PET constituent monomers and can be obtained by pyrolysis or enzymatic hydrolysis of PET (Kenny et al., 2008; Kenny et al., 2012; Wei and Zimmermann, 2017). Genome mining of GO16 revealed the presence of genes for the complete mineralization of both, and the capacity of GO16 to use EG as a sole source of carbon and energy was also validated experimentally.
The operon for the catabolism of TA to protocatechuate (PCA) is 6085 bp long (Fig. 4A; Table S4). The catabolic genes are preceded by a regulator that belongs to the isocitrate lyase regulator‐type (IclR‐type) transcriptional regulators, in general involved in regulation of carbon metabolism, multidrug resistance, quorum sensing, etc. (Molina‐Henares et al., 2006). In Pseudomonas strains IclR‐type regulators are frequently involved in the regulation of the β‐ketoadipate pathway (Molina‐Henares et al., 2006). The regulator is followed by terephthalate 1,2‐dioxygenase subunits α (tphA2) and β (tphA3), a reductase component (tphA1), a dehydrogenase (tphB) and terephthalate transporter (tphK). Similar organization of TA degradation operon was reported for Comamonas testosteroni strains YZW‐D and E6, and Rhodococcus opacus, where TA is degraded to 1,2‐dihydroxy‐3,5‐cyclohexadiene‐1,4‐dicarboxylic acid by the action of terephthalate dioxygenase (TphA2A3A1), followed by the activity of TphB to convert this intermediate into protocatechuate (Sasoh et al., 2006).
Fig. 4.
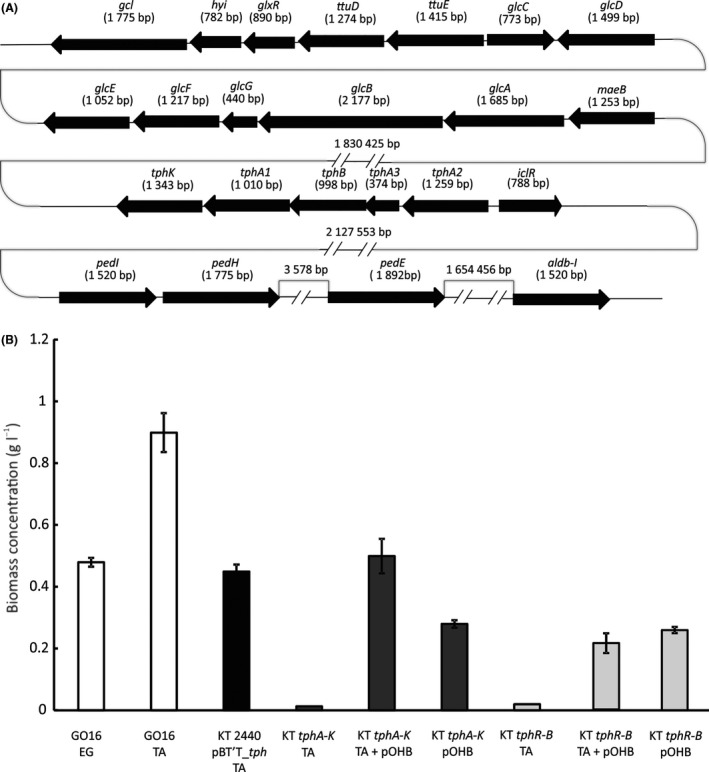
(A) Organization of genes in P. umsongensis GO16 that encode enzymes that enable growth with EG and TA when used as a sole source of carbon and energy. (B) Biomass (CDW g l−1) when P. umsongensis GO16 was grown with EG, TA and growth of P. putida KT2440 pBT’T_tph, KT2440 expressing whole tph operon including the native regulator from P. umsongensis GO16 grown with TA. tphA‐K represents KT2440 expressing the tph operon without the native regulator and this strain was grown with TA, p‐hydroxybenzoate (pOHB) or a mixture of both substrates. tphR‐B represents the growth of KT2440 expressing the tph operon without the transporter tphK grown with TA, pOHB or a mixture of both. The values represent the average of three independent biological replicates and correspond to the biomass accumulated in 48 h.
While a permease tphC is involved in the facilitated diffusion of TA in C. testosteroni strains, in GO16 the transport of TA is mediated by an MFS transporter of the aromatic acid:H+ symporter (AAHS) family (tphK in Table S4), which shows homology to the p‐hydroxybenzoate transporter pcaK (Salvador et al., 2019) (Fig. 4A; Table S4).
We selected P. putida KT2440 for the recombinant expression of the tph genes identified in GO16. KT2440 has been certified as a safe microorganism and is considered a workhorse for biotransformations (Belda et al., 2016; Volke et al., 2020; Weimer et al., 2020). While P. putida KT2440 is equipped with numerous dioxygenases and can grow with a range of aromatic hydrocarbons, it cannot utilize TA. In fact, out of the four tph catabolic genes, only tphA2 and tphA1 have homologs in the chromosome of KT2440. They show identities of, respectively, 35% and 26% with the subunits BenA and BenC of the benzoate 1,2‐dioxygenase. When the whole tph operon, including the transporter and the regulator, was expressed in P. putida KT2440 on plasmid pBT'T_tph it conferred this organism the ability to use TA as a sole source of carbon and energy (Fig. 4B). The growth rate of the KT2440 pBT’T_tph with TA as a sole source of carbon and energy was 0.3 h−1, which is 1.7‐fold lower compared to glucose as a sole source of carbon and energy (Fig. S3). The final biomass reached by KT2440 pBT’T_tph was 1 g l−1, which is 1.6‐fold lower than the final biomass of GO16 when TA is used as a sole source of carbon and energy (Fig. S3, Table 3). It is worth noting that the whole tph operon, including the transporter tphK and the native regulator iclR (Fig. 4A) were required to allow growth of KT2440 pBT’T_tph with TA as the carbon source (Fig. 4B). A construct lacking the transporter tphK (fragment tphRA2A3BA1) failed to grow on TA as the sole carbon source. Moreover, no growth was observed in TA when the catabolic and transport tphA2A3BK genes cloned into pBT’T were expressed constitutively from the strong promoter Ptac . However, this construct showed increased biomass formation (0.5 g l−1 CDW) when co‐cultured in TA (4.2 g l−1 corresponding to 1.96 gC l−1) and pOHB (0.7 g l−1, providing 0.42 gC l−1) used as an inducer of the pca genes, compared to the culture with pOHB alone (0.28 g l−1 CDW; Fig. 4B) (Kim et al., 2006). These results suggest the tphR can coordinate the expression of the tph genes and the downstream pca pathway required for PCA degradation in P. putida KT2440. Although a specific regulator for the expression of pcaGH in this organism has not been identified (Jimenez et al., 2002), TphR shares a 53% similarity with PcaU, another regulator of the IclR‐family known to control the expression of pcaGH in Acinetobacter sp. ADP1 in response to PCA (Gerischer et al., 1998). These results show that both transport and regulation are specific for TA and cannot be replaced by genes present in the genome of P. putida KT2440.
Table 3.
Comparison of growth characteristics of P. umsongensis GO16 when glucose, TA and EG were used as a sole source of carbon and energy. Three carbon sources were used in amounts to provide 1.96 gC l−1. The specific growth rate was calculated for the exponential phase of growth
| Carbon source | Final biomass (CDW; g l−1) | Specific growth rate (h−1) | Specific rate of C consumption (g l−1 h−1) | Biomass yield (gCDW/gC) |
|---|---|---|---|---|
| Glucose | 1.30 ± 0.06 | 0.22 ± 0.01 | 0.26 ± 0.02 | 0.30 |
| TA | 1.60 ± 0.07 | 0.17 ± 0.01 | 0.18 ± 0.02 | 0.35 |
| EG | 0.40 ± 0.13 | 0.09 ± 0.03 | 0.05 ± 0.02 | 0.15 |
In addition, we observed a transient PCA accumulation in the supernatant of KT2440 pBT’T_tph grown with TA as a sole source of carbon and energy (Fig. S4). These results indicated that the tph genes were functional in KT2440 and produced PCA, however further strain optimization is required to co‐regulate the upper (TA to PCA) and lower (PCA cleavage) pathways.
EG is industrially produced at large volumes and it is used in a range of applications, production of PET being one of them. Aerobic metabolism of EG was described in some Pseudomonas species (Mückschel et al., 2012; Franden et al., 2018; Orellana‐Saez et al., 2019). Functionally redundant periplasmic quinoproteins have been found to catalyse the initial conversion of EG to glycolaldehyde (Mückschel et al., 2012; Wehrmann et al., 2017). In P. putida KT2440 this function is assigned to PedE (PP_2674) and PedH (PP_2679) (Wehrmann et al., 2017). In the next step, catalysed by cytoplasmic aldehyde dehydrogenases PedI (PP_2680) and PP_0545 in KT2440, glycolaldehyde is converted to glycolate, followed by oxidation to glyoxylate by the activity of GlcDEF (Franden et al., 2018). All of the genes encoding the enzymes involved in the oxidation of EG to glyoxylate have been identified in the strain GO16 (Fig. 4A; Table 2).
Table 2.
Comparison of the genes encoding enzymes involved in the sequential oxidation of ethylene glycol (EG) to glyoxylate between P. umsongensis GO16 and P. putida KT2440.
| Gene | Locus | Function | Query cover (%) | Identity AA (%) | Reference gene |
|---|---|---|---|---|---|
| pedI | F6476_19660 | Aldehyde dehydrogenase | 100 | 92 | PP_2680 |
| pedH | F6476_19665 | Quinoprotein ethanol dehydrogenase | 99 | 91 | PP_2679 |
| pedE | F6476_19690 | PQQ dependent dehydrogenase | 100 | 94 | PP_2674 |
| aldB‐I | F6476_27375 | Aldehyde dehydrogenase | 100 | 92 | PP_0545 |
| gcl | F6476_01220 | Carboxylate ligase | 100 | 86 | PP_4297 |
| hyi | F6476_01215 | Hydroxypyruvate isomerase | 100 | 78 | PP_4298 |
| glxR | F6476_01210 | Tartronate reductase | 100 | 86 | PP_4299 |
| ttuD | F6476_01205 | Hydroxypyruvate reductase | 99 | 81 | PP_4300 |
| ttuE | F6476_01200 | Pyruvate kinase | 99 | 79 | PP_4301 |
| glcC | F6476_01190 | Glc operon transcriptional regulator | 98 | 73 | PP_3744 |
| glcD | F6476_01185 | Glycolate oxidase FAD‐linked subunit | 100 | 86 | PP_3745 |
| glcE | F6476_01180 | Glycolate oxidase FAD‐binding subunit | 100 | 72 | PP_3746 |
| glcF | F6476_01175 | Glycolate oxidase Fe‐S subunit | 100 | 77 | PP_3747 |
| glcG | F6476_01170 | Uncharacterized protein | 90 | 73 | PP_3748 |
| glcB | F6476_01165 | Malate synthase | 99 | 63 | PP_0356 |
| glcA | F6476_01160 | Glycolate permease | 100 | 81 | PP_4735 |
| maeB | F6476_01155 | Malate dehydrogenase/malic enzyme | 98 | 71 | PP_5085 |
The growth rate, carbon depletion and biomass yield were compared when GO16 was cultivated with TA, EG or glucose (Table 3, Fig. S5). EG supplied as a sole source of carbon and energy supports the biomass formation in P. umsongensis GO16. In comparison with the growth with TA, EG yields biomass 0.4 g l−1 of cell dry weight (CDW), which is fourfold lower than CDW observed with the corresponding amount of carbon (1.96 gC l−1) of TA (Table 3). When TA was used as a sole source of carbon and energy the specific growth rate was twofold lower compared to the specific growth rate with glucose. However, the final biomass and biomass yield were similar when GO16 was cultivated with glucose or TA as a sole carbon and energy source (Table 3).
The catabolic pathway that allows biomass formation from EG in P. putida JM37 proceeds via Gcl pathway including a glyoxylate carboxyligase (Mückschel et al., 2012). While the wild type KT2440 contains the genetic capacity to form biomass from EG, it can only utilize it as an energy source (Li et al., 2019). It was shown that the repression of the gcl operon is the reason for this, and once this repression is removed, KT2440 can efficiently grow with EG as a sole source of carbon and energy (Li et al., 2019). In GO16 the gcl operon was identified (Table 2). This operon is regulated by a LysR transcriptional regulator (ttdR) and followed by carboxylate ligase (gcl), hydroxypyruvate isomerase (hyi), tartronate semialdehyde reductase (glxR), hydroxypyruvate reductase (ttuD) and pyruvate kinase (ttuE). The gcl operon is followed by the glc operon, consisting of a regulator glcC and genes encoding the subunits of a glycolate oxidase, glcDEF.
It is worth noting that during the cultivation of GO16 with EG as a sole source of carbon and energy we did not observe the formation of EG oxidation products, glycolate, glyoxylate and oxalate, which was the case when P. putida KT2440 was grown in the presence of EG (Li et al., 2019).
PHA metabolism
PHAs are a family of biological polyesters which represented 1.2% of the global bioplastic market in 2019 (European Bioplastics, 2019). PHAs are bacterial carbon and energy storage polyesters usually accumulated as a response to stress (Rehm, 2010). They are grouped into scl‐ polymers, containing (R)‐3‐hydroxyalkanoic acids with four or five carbon atoms with polyhydroxybutyrate (PHB) as a typical example, and mcl‐ polymers of (R)‐3‐hydroxyalkanoic acids containing 6‐12 carbon atoms (Sudesh et al., 2000). With over 150 known PHA monomers, (R)‐3‐hydroxyalkanoic acids, PHAs have highly diverse material properties and therefore a broad range of applications (Rehm, 2010).
P. umsongensis GO16 has a typical mcl‐PHA synthesis genes organization, with a phaC1ZC2D cluster (Table 4). This cluster is well conserved among the mcl‐PHA producing bacteria, and the two PhaCs encoded by it belong to class II PHA synthases typically involved in the synthesis of mcl‐PHA (Chek et al., 2019). We have identified an additional putative PHA synthase that also belongs to class II (Table 4), potentially contributing to mcl‐PHA synthesis in GO16. This synthase is located outside of other PHA metabolism gene clusters, and it has no PHA related genes in its proximity.
Table 4.
Comparison of the genes encoding proteins involved in the synthesis and degradation of PHA present in P. umosongensis GO16 and those of related organisms. Gene codes starting with PP_, P or WP_ correspond, respectively, to P. putida KT2440, Pseudomonas oleovorans or Pseudomonas extremoaustralis.
| Gene | Locus | Function | Query cover (%) | Identity AA (%) | Reference gene |
|---|---|---|---|---|---|
| phaC1 | F6476_32385 | Poly(3‐hydroxyalkanoate) synthase 1 | 100 | 82 | PP_5003 |
| phaZ1 | F6476_32380 | Poly(3‐hydroxyalkanoate) depolymerase | 99 | 91 | PP_5004 |
| phaC2 | F6476_32375 | Poly(3‐hydroxyalkanoate) synthase 2 | 100 | 74 | PP_5005 |
| phaD | F6476_32370 | TetR family transcriptional regulator | 99 | 77 | PP_5006 |
| GA2 | F6476_32365 | Granule associated protein | 65 | 66 | PP_5007 |
| GA1 | F6476_32360 | Granule associated protein | 100 | 61 | PP_5008 |
| GA3 | F6476_32350 | Putative granule associated protein | 98 | 57 | PP_5010 |
| PHA synthase | F6476_07555 | Poly(3‐hydroxyalkanoate) synthase | 94 | 49 | P26494 |
| phaZ2 | F6476_23055 | PHB depolymerase/α/β hydrolase | 88 | 37 | P26495 |
| phaC3 | F6476_22230 | Class I poly(3‐hydroxyalkanoate) synthase | 99 | 73 | WP_042946539 |
| phaA | F6476_22225 | Acetyl‐CoA acetyltransferase | 100 | 82 | WP_010563427 |
| phaB | F6476_22220 | Acetoacetyl‐CoA reductase | 98 | 80 | WP_010563428 |
| GA4 | F6476_22205 | Granule associated protein | 97 | 66 | WP_003464225 |
| phbF | F6476_22200 | PHA synthesis repressor | 98 | 69 | WP_010563433 |
Furthermore, the P. umsongensis GO16 chromosome encodes a scl‐PHA synthesis pathway (Fig. 5A; Table 4). While the organization of the genes encoding a class I PHA synthase (phaC3), acetyl‐CoA acetyltransferase (β‐ketothiolase; phaA), acetoacetyl‐CoA reductase (phaB) is the same as in a scl‐PHA model organism Cupriavidus necator H16, in this model organism the phaCAB operon is followed by the phaR encoding the PHA synthesis repressor (Pohlmann et al., 2006). The phaCAB genes of GO16 are followed by two genes encoding hypothetical proteins, and genes encoding a phasin family protein and PhaR repressor (Table 4). One of the two hypothetical proteins, positioned immediately after the acetoacetyl‐CoA reductase, shows 99% identity with AraC transcriptional regulators found in Pseudomonas species (Gallegos et al., 1997). The second hypothetical protein from this cluster contains a DUF3141 domain, also found in PHA synthase of class III involved in scl‐PHA synthesis (Batista et al., 2016; Chek et al., 2019).
Fig. 5.
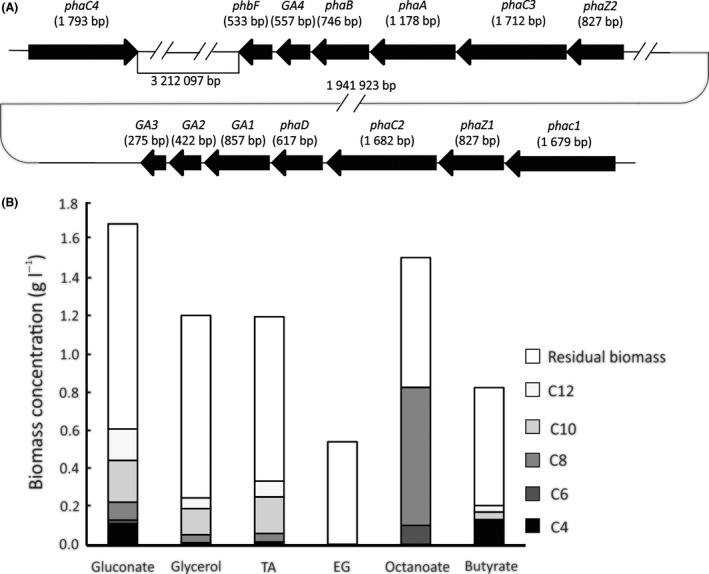
(A) Organization of the PHA metabolism genes in P. umsongensis GO16. (B) PHA accumulation and monomer composition (mol%) from carbon unrelated (sodium gluconate, glycerol, TA and EG) and carbon related substrates (sodium octanoate, sodium butyrate). The values represent the average of three independent biological replicates and correspond to the biomass accumulated in 48 h.
While mcl‐PHA accumulation is widely reported in pseudomonads, scl‐PHA production is not a common characteristic of Pseudomonas species (Diard et al., 2002). P. oleovorans group including P. oleovorans, P. pseudoalcaligenes, Pseudomonas sp., as well as recently described P. extremoaustralis and Pseudomonas sp. MPC6 are examples of Pseudomonas species that have the capacity to accumulate scl‐PHA when grown with sodium octanoate (Diard et al., 2002; Lopez et al., 2009; Catone et al., 2014; Orellana‐Saez et al., 2019). In P. extremoaustralis, the scl‐PHA, polyhydroxybutyrate (PHB) cluster phaRBAC is located in a genomic island of 32.3 kb containing 28 ORFs (Ayub et al., 2007). We have also identified an integrase 3 ORFs upstream of the scl‐PHA synthase, as well as three transposases (4806223‐4807659, 4818701‐4820131, 4837609‐4837980), suggesting that scl‐PHA gene cluster could have been acquired by horizontal gene transfer.
We investigated the ability of GO16 to form mcl‐PHA and scl‐PHA using a variety of growth substrates. GO16 accumulated mcl‐PHA when grown with gluconate, TA, glycerol, octanoic acid, but no PHA accumulation was observed when EG was used as a sole source of carbon and energy (Fig. 5B). Typically, when PHA monomer‐unrelated substrates were used, C10 was the major monomer detected with approximately 50 mol% (0.13–0.20 g l−1). We observed a higher fraction of C12, up to 30 mol% (0.05–0.15 g l−1) in mcl‐PHA accumulated by GO16 in comparison with mcl‐PHA accumulated by P. putida KT2440, which is also a model organism for mcl‐PHA accumulation (Sohn et al., 2010; Davis et al., 2013). In addition to typical mcl‐PHA monomers, C4 monomer was identified when butyrate (61 mol%; 0.11 g l−1), gluconate (18 mol%; 0.10 g l−1) or TA (1 mol%; 0.01 g l−1) were used as the substrates. When butyrate was used, C4 was predominant monomer, with C8 (3 mol%; 0.007 g l−1), C10 (19 mol%; 0.036 g l−1) and C12 (17 mol%; 0.032 g l−1) also present (Fig. 5B).
Besides the natural capabilities of GO16, we also tested the accumulation of PHA in KT2440 containing the pBT’T_tph plasmid when grown with TA as a sole source of carbon and energy. The biomass accumulated under nitrogen limitation was 0.36 g l−1 CDW, and 17% CDW (0.0612 g l−1) was PHA. The major detected monomer was C10 (75 mol%), followed by C8 (17 mol%) and C12 (8 mol%). The biomass achieved by KT2440 pBT’T_tph was threefold lower compared to GO16 grown under the same conditions, and total PHA amount was 4.9‐fold lower. These results show the potential to use the tph genes of GO16 for the upcycling of PET monomers in other bacterial species.
We assessed the potential of GO16 for the upcycling of PET by analysing the growth and PHA production of P. umsongensis GO16 using equimolar mixture of TA and EG as carbon sources. Under PHA non‐accumulating conditions, TA and EG were depleted and a final CDW of 3.6 g l−1 was achieved, resulting in a total yield of 0.74 gCDW gC −1. GO16 showed preferential utilization of TA over EG with utilization of EG beginning after complete degradation of TA (Fig. 6). After TA was completely depleted, a lag period of approximately 5 h was observed prior to commencement of EG metabolism. TA contributed 3.2 g l−1 CDW, with total TA utilization occurring within 10 h, resulting in a yield of 0.38 gCDW gTA −1 or 0.82 gCDW gC −1. The consumption of EG contributed 0.4 g l−1 CDW, resulting in a yield of 0.2 gCDW gEG −1 or 0.42 gCDW gC −1. A maximum specific growth rate (μ) of 0.37 h−1 was observed when utilizing TA, and a 15‐fold lower maximum specific growth rate (μ) of 0.024 h−1 was recorded during EG utilization compared to TA. Total depletion of EG occurred within 8 h since the start of EG consumption.
Fig. 6.
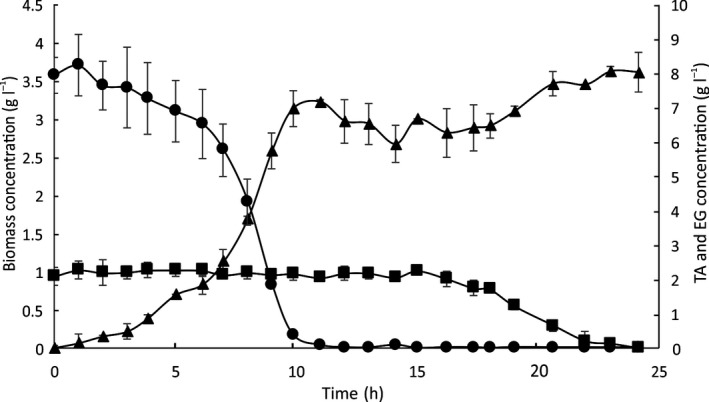
Growth and substrate consumption of P. umsongensis GO16 on minimal salts medium (MSMfull) supplemented with an equimolar synthetic mixture of TA and EG to a final concentration of 40 mM (8.4 g l−1 TA, 2.48 g l−1 EG), mimicking enzymatically hydrolysed PET, in a 5 l bioreactor with a 3 l working volume at 30°C. The figure shows biomass accumulation (CDW; g l−1; ▲), TA utilization (g l−1; ●) and EG utilization (g l−1; ■). Error bars represent the standard deviation of three biological replicates.
Under PHA accumulating conditions, a final CDW of 1.5 g l−1 was achieved (Fig. 7), a twofold decrease compared to cultivation of P. umsongensis GO16 under non‐limiting conditions. Nitrogen was completely exhausted by 8 h, leading to the onset of PHA accumulation. A total PHA content of 0.13 g l−1 (9% CDW) was achieved. A consumption rate of 0.97 g (l h)−1 TA was observed after nitrogen depletion (Fig. 7) with complete TA utilization occurring within 12 h. No EG consumption was observed, similar to what we have observed in shake flask experiments when EG was used as a sole source of carbon and energy under PHA accumulating conditions. The medium chain length PHA produced by Pseudomonas sp. GO16 from the equimolar synthetic mixture of TA and EG consisted of C10 (53 mol%), C8 (25 mol%) and C12 (24 mol%). These results indicate that upcycling of hydrolysed PET is possible mainly at the expense of TA, which is preferentially used over EG by P. umsongensis GO16.
Fig. 7.
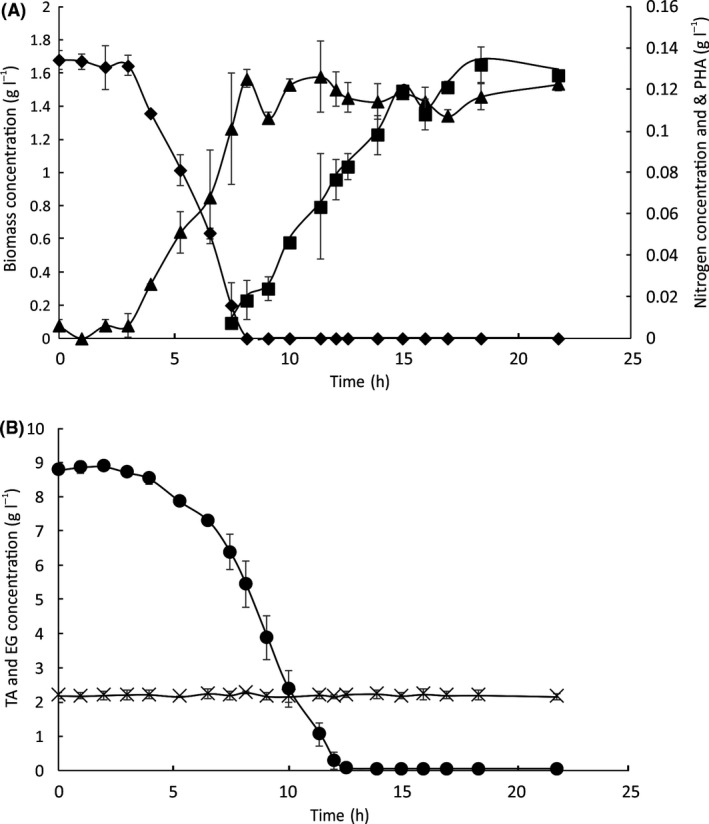
Growth and PHA accumulation of P. umsongensis GO16 on minimal salts media (MSMlim) supplemented with an equimolar synthetic mixture of TA and EG to a final concentration of 40 mM (8.4 g l−1 TA, 2.48 g l−1 EG), mimicking enzymatically hydrolysed PET, in a 5 l bioreactor with a 3 l working volume at 30°C. The figure shows (A) biomass accumulation (CDW; g l−1; ▲), nitrogen utilization (g l−1; ♦) and PHA accumulation (g l−1; ■) (B) TA (●) and EG (×) utilization. Error bars represent the standard deviation of three biological replicates.
Conclusions
In this work we have analysed the complete genome of P. putida GO16, a promising tool for the upcycling of PET hydrolysis monomers TA and EG (Tiso et al., 2020). The central metabolism of GO16 shows similarities with other species of Pseudomonas. For instance, glucose is most likely metabolized via the periplasmic conversion to gluconate and its complete metabolism combines the ED, EMP and PP pathways. This allows for the production of a larger surplus of reducing equivalents compared to the canonical glycolysis and it is linked to a better fitness in stressful environments including demanding bioconversions such as those required for the degradation of aromatics (Chavarria et al., 2013). Related to this, P. umsongensis GO16 is capable of degrading a wide diversity of aromatics such as naphthalene, due to the presence of pENK22, and most notably TA, which is uncommon in Pseudomonas species. The presence of this pathway in GO16 enables the synthesis of different molecules of interest derived from the hydrolysis of PET. These include other aromatic molecules (Kim et al., 2019), but also the products of the cleavage of the aromatic ring and posterior modifications, some of which containing two or more functional groups and can be used as building blocks for synthesizing different types of polymers (Johnson et al., 2019). One example is adipic acid which could be obtained by the recombinant expression of a protocatechuate decarboxylase transforming the protocatechuate obtained from TA into catechol (Johnson et al., 2016). Catechol can then be cleaved by the chromosomally encoded catechol‐1,2‐dioxygenase to render the adipic acid precursor cis,cis‐muconate.
Another interesting feature of P. umsongensis GO16 is the high abundance of genes related to the metabolism of lipids including several involved in different stages of β‐oxidation. Also related to the metabolism of lipids, GO16 harbours genes for the synthesis of both scl‐ and mcl‐PHA. C4 monomer was dominant when butyrate was used as a substrate. However, the other tested substrates drove biosynthesis of predominantly mcl‐PHA monomers. It is worth highlighting that the proportion of longer acids (e.g. C12) is enriched in GO16 compared to other Pseudomonas species.
As a result of the complete sequencing and genome analysis of P. umosongensis GO16, we have been able to identify some key properties of this organism. In particular, we have identified some aspects of its metabolism that make it a versatile workhorse for biotransformation using PET and its degradation products as feedstocks that could facilitate the development of bio‐based recycling strategies for this critical polymer with the potential for its conversion to not only one but two biodegradable polymers (scl‐PHA and mcl‐PHA).
Experimental procedures
Bacterial strain, medium and growth conditions
P. umsongensis GO16 (accession number NCIMB 41538, NCIMB Aberdeen, Scotland, UK) was inoculated from glycerol stock onto mineral salts medium (MSM) solidified with 1.5% agar supplemented with 4.4 g l−1 (20 mM) disodium terephthalate (TA; Sigma‐Aldrich, UK). MSM contained 9 g l−1 Na2HPO4·12H2O, 1.5 g l−1 KH2PO4 and 1 g l−1 (MSMfull) or 0.25 g l−1 (MSMlim) NH4Cl. Prior to inoculation MSM was supplemented with MgSO4 (200 mg ml−1) and trace elements (per litre: 4 g ZnSO4∙7H2O; 1 g MnCl2∙4H2O; 0.2 g Na2B4O7∙10H2O; 0.3 g NiCl2∙6H2O; 1 g Na2MoO4∙2H2O; 1 g CuCl∙2H2O; 7.6 g FeSO4∙7H2O).
To test the growth of GO16 with the volatile aromatic hydrocarbons naphthalene, a single colony was cultured overnight in glucose and then diluted 1/100 when transferred into a flask with 50 ml of M9 minimal medium supplemented with vitamins and trace elements (Harwood and Cutting, 1990). Naphthalene was supplemented via vapour phase from an open eppendorf tube containing crystals of the substrate (10 mg) and suspended above the culture. Flasks were incubated in an orbital shaker at 30°C and 200 rpm for 4 days monitoring the absorbance at 600 nm periodically. Growth was confirmed by comparison with control flasks without substrate. For testing the growth using other soluble aromatic substrates, cultures of 200 µl of M9 supplemented with 5 mM of the aromatic molecule (all from Sigma‐Aldrich, UK) used as a carbon source, were incubated in a Clariostar microplate reader (BMG Labtech, Germany) at 30°C by measuring the optical density of the cultures at 600 nm.
For the growth and PHA accumulation analysis, a single colony was inoculated into 3 ml of MSMFull supplemented with various carbon sources (sodium gluconate, TA, EG, sodium butyrate, sodium octanoate, glycerol) in the amount corresponding to 1.96 gC l−1, and incubated for 18 h at 200 rpm and 30°C. The seed culture (1 ml) was inoculated into 250 ml Erlenmeyer flasks containing 50 ml MSMlim media supplemented with the corresponding carbon source and incubated for 48 h at 200 rpm and 30°C. The cells were harvested by centrifugation at 5000 g for 10 min at 4°C (Benchtop 5430R centrifuge; Eppendorf, Germany) and washed with 10 ml of phosphate buffer (50 mM, pH 7). The pellets were frozen at −80°C.
For bioreactor experiments, the pre‐inoculum was prepared by inoculating 250 ml Erlenmeyer flasks containing 50 ml MSMfull with 1 ml of seed culture supplemented with 20 mM TA and 20 mM EG. Flasks were incubated in a shake incubator (New Brunswick Scientific, Innova 44; USA) for 18 h at 200 rpm and 30°C. Optical density readings (OD540) of the seed culture was taken prior to bioreactor inoculation using a spectrophotometer (Spectrophotometer 6300, Jenway; UK) at 540 nm.
Batch fermentation experiments were carried out in a 5 l Biostat B bioreactor (Sartorious, Germany), containing 3 l of MSMfull or MSMlim broth supplemented with 40 mM TA and EG. The bioreactor was set up with 5% (v/v) inoculum of cells, with an OD540 of 3.5 ± 0.49. Air was supplied at a constant rate of 3 l min−1 (1 VVM) throughout the fermentation and dissolved oxygen (DO) was maintained at a minimum of 20%, via a control loop. Impeller speed was set to a minimum of 500 rpm and maximum of 1500 rpm. Temperature was maintained constant at 30°C. The pH of the culture was maintained at a value of pH 7 by the automatic addition of 20% (v/v) NH4OH or 15% (v/v) H2SO4. DO and rpm were monitored online by BioPAT MFCS/win fermentation data acquisition software (Sartorious; Germany). 2 ml samples were taken in duplicate at hourly intervals for CDW, carbon, nitrogen and PHA analysis.
To analyse the dynamics of growth and carbon depletion, P. umsongensis GO16 and P. putida KT2440_tph were grown in 50 ml MSMfull with glucose, TA or EG as a sole source of carbon and energy. The carbon and energy source was supplemented in an amount to provide 1.96 gC l−1. The cultures were incubated for 48 h at 200 rpm and 30°C, and samples were withdrawn at 6 h intervals. OD was measured at absorbance 540 nm (JENWAY 6300 spectrophotometer; Cole‐Parmer, Staffordshire, UK) and 2 ml of supernatants were kept for carbon consumption analysis. The cells were harvested by centrifugation, freeze dried using a Labconco® (Fisher Scientific) freeze‐dryer for CDW determination. The supernatant was retained, filtered and analysed by HPLC for carbon depletion.
For the purpose of the genomic DNA isolation, Lysogeny broth (LB; Sigma‐Aldrich, Ireland) medium was used for the cultivation of P. umsongensis GO16. A single colony of P. umsongensis GO16 was inoculated into 4 ml of LB and cultivated for 16 h at 200 rpm and 30°C. The cells were harvested by centrifugation at 16 000 g, 5 min, 4°C (Benchtop 5430R centrifuge; Eppendorf, Hamburg, Germany).
Genomic DNA extraction
The total DNA was extracted using Blood and Cell culture DNA midi kit (QIAGEN, Germany) according to manufacturer’s instructions for the lysis of bacteria. The quality and concentration of isolated DNA were verified by Qubit® Fluorometer (Thermo Fisher Scientific, Ireland) according to the manufacturer’s instructions.
Genome sequencing and analysis
The genome of P. umsongensis GO16 was sequenced by BaseClear BV (Leiden, NL). Using Illumina HiSeq platform and PacBio RSII platform a 300 bp paired‐end library and 10 kb library respectively were prepared. This was followed by a de novo assembly of the reads and an automated gap closure using GapFiller version 1.10.
Sequencing of P. umsongensis GO16 total DNA (BaseClear BV, Leiden, The Netherlands) yielded 5 contigs. Specific primers corresponding to the ends of these contigs were designed and used to amplify fragments from the total DNA using Q5® High‐Fidelity DNA Polymerase (New England BioLabs, Ipswich, MA, USA). The amplified products were cloned into pGEM®‐T Easy vector (Promega, Madison, WI, USA) and sequenced (GATC Biotech, Ebersberg, Germany). The resulting sequences were assembled using DNASTAR® Lasergene® Genomics Suite Software (Thermo Fisher Scientific, Waltham, MA, USA). These additional end sequences and the 5 assembled contigs were orientated, overlapped and ordered relative to the genome of the closely related P. umsongensis BS3657 strain. Each sequence was mapped to the BS3657 strain using ‘blastn’, part of the Basic Local Alignment Search Tool (blast) suite of software (Altschul et al., 1990). The relative genomic coordinates for each sequence’s location were then extracted and used to merge all contigs into one larger chromosome. The phylogenetic analysis was conducted by an alignment‐free genome‐wide comparison of coding regions using a composite vector approach. The algorithm compared the amino acid counts of all predicted proteins and was implemented with the online tool cvtree3 (Qi et al., 2004; Zuo and Hao, 2015). Blast atlases were generated by gview java package software (https://server.gview.ca; Petkau et al., 2010) by carrying out genome‐wide tblastx searches between GO16 and each of the representative Pseudomonas genomes. The same approach was taken for comparisons between plasmid pENK22 and other plasmids containing naphthalene‐catabolic genes. Regions on each genome reporting a BLAST hit above the threshold cut‐off (80% identity, minimum HSP length of 100 bp, and expect value of 1e−10) were considered a valid match and represented in the figures. Genome alignments were generated using Progressive Mauve (Darling et al., 2004). BLAST searches were performed at the NCBI suite (https://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al., 1990) with the ‘blastp’ algorithm using either specific databases (e.g. P. putida KT2440) or the Swissprot database when running untargeted searches. BLAST searches against the genome of P. umsongensis GO16 were carried out locally with the Genome Workbench suite of NCBI (https://www.ncbi.nlm.nih.gov/tools/gbench/).
Generation of P. putida KT2440 pBT’T_tph, pBT’T_tphA2A3BA1K and pBT’T_tphRA2A3BA1
Based on the available genome sequence of P. umsongensis GO16, the tph operon (1 982 772‐ 1 988 856 in CP044409.1; 6128 bp) was synthesized (TwistBioscience, UK) and cloned into pBT’T (Koopman et al., 2010) vector using NEBuilder® HiFi DNA Assembly Master Mix (NEB, UK). To allow Gibson assembly of the insert and the vector, the tph operon was amplified using the following primers RF 5′‐CAG TGA GCC CCA TCC CAA CAT CAA AGC ATA‐3′ and GR 5′‐CCG ACG TCG CAT GCT CCT CTA GA‐3′, while pBT’T was amplified using V1F 5′‐TCT AGA GGA GCA TGC GAC GTC GG‐3′ and V1rR 5′‐TAT GCT TTG ATG TTG GGA TGG GGC TCA CTG‐3′. 100 ng of vector and twofold molar excess of insert (351.4 ng of insert RT) were incubated with NEBuilder® HiFi DNA Assembly Master Mix as recommended by the manufacturer. Gibson reaction (10 µl) was incubated in ice 1 h and transferred by heat shock into E. coli DH5α chemically competent cells. Prior to plating on selective media, cells were allowed to recover at 37°C for 2 h. The pBT’T_tph construct was verified by sequencing (Eurofins, Ireland) transformed into P. putida KT2440 (Choi et al., 2006) and the positive transformants were selected on MSMfull with TA as the sole carbon and energy source and confirmed by colony PCR using the primers DHD_FWD 5′‐CTA TCG CGC AGC CAT GGA TCT ATG A‐3′ and DHD_REV 5′‐TGG ACC TTG GTG GTA ATG ACC TTG CG‐3′, which amplify the region of the tph operon between tphA2 and tphA1 giving a product of 1809 bp. The stability of the plasmid after growth on TA was verified by miniprep (Qiagen, UK) followed by digestion with SphI, and PCR of the backbone and the tph genes with primers V1F and V1rR and RT 5′‐CAG TGA GCC CCA TCC CAA CAT CAA AGC ATA‐3′ and GR respectively (see Fig. S6 for details). The growth was tested in 50 ml MSMlim as described above. The wild type P. putida KT2440 was used as a negative control, as this strain does not grow with TA as a sole source of carbon and energy.
The tphA2A3BA1K fragment (5160 bp) was synthesized (TwistBioscience, UK) and cloned into pBT’T (Koopman et al., 2010) vector using NEBuilder® HiFi DNA Assembly Master Mix (NEB, UK). The insert was amplified using the following primers A2‐K F 5′‐CAG GAG GTC AAC AAT GAA CAT CAT TAC TGA‐3′ and A2‐K R 5′‐ACG TCG CAT GCT CCT CTA GAT TAA AGC GTG‐3′, while pBT’T was amplified using V2F 5′‐CAC GCT TTA ATC TAG AGG AGC ATG CGA CGT‐3′ and V2R 5′‐TCA GTA ATG ATG TTC ATT GTT GAC CTC CTG‐3′. The ligation and transformation were performed as described above.
The insert pcaRtphA2A3B1A1 (4715 bp) that includes the native regulator, but excludes the transporter was synthesized by TwistBioscience (UK) and cloned into pBT’T (Koopman et al., 2010) vector using NEBuilder® HiFi DNA Assembly Master Mix (NEB, UK). The primers used for Gibson assembly were RF 5′‐CAG TGA GCC CCA TCC CAA CAT CAA AGC ATA‐3′ and A2R 5′‐GCA TGC TCC TCT AGA CTA TGA AGG CGG CAG‐3′ for the insert, and V3F 5′‐CTG CCG CCT TCA TAG TCT AGA GGA GCA TGC‐3′ and V1rR 5′‐TAT GCT TTG ATG TTG GGA TGG GGC TCA CTG‐3′ for the vector. The ligation and transformation were performed as described above.
Nutrient analysis
For the analysis of TA consumption, the supernatant collected during cultivation had to be diluted so that the concentration of TA in the final preparation did not exceed 0.63 g l−1. A 1100series HPLC (Agilent, USA) equipped with a C18 ODS Hypersil column (125 × 3 mm, particle size 5 μm; Thermo Scientific, USA) was used, and samples were isocratically eluted using 0.4% formic acid at a flow rate of 1 ml min−1 and read on a UV‐vis detector at 230 nm. The TA retention time under the above conditions was 4.2 min. The PCA retention time under the above conditions was 3.2 min.
EG depletion was monitored using an Aminex HPX‐87H ion exclusion column (300 mm × 7.8 mm, particle size 9 μm; Bio‐rad). The column was maintained at 40°C and samples were isocratically eluted using 0.014 N H2SO4 at a flow rate of 0.55 ml min−1 and read on a refractive index detector (RID). The EG retention time under the above conditions was 23 min.
Glucose consumption was determined from supernatant by HPLC (i‐series, Shimadzu, Kyoto, Japan) using an Aminex HPX‐87H Column (300 × 7.8 mm, particle size 9 µm particle size; Bio‐rad, UK). The samples were eluted with 0.014N H2SO4 at a flow rate of 0.55 ml min−1.
PHA extraction and content determination
The polymer content was assayed by subjecting the lyophilized cells to acidic methanolysis as previously described (Lageveen et al., 1988). The PHA monomers’ methylesters were assayed by GC using a Hewlett‐Packard 6890N chromatograph equipped with a HP‐Innowax capillary column (30 m × 0.25 mm, 0.50μm film thickness; Agilent Technologies) and a flame ionization detector (FID), using the temperature programme previously described (Lageveen et al., 1988). Total PHA content was determined as a percentage of CDW.
Conflict of interests
Author Shane Kenny is employed by the company Bioplastech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
TN, SK, KO and JJ designed the study and supervised the research. TN, NB, SK, JU, UA and MS conducted experimental work. TN, MS, GH, HW and JJ performed the bioinformatic analyses. All authors contributed to writing the manuscript.
Supporting information
Table S1. Blast atlas of P. umsongensis GO16 chromosomal genes. The table shows the results of the tblastx analysis comparing the annotated features present in the chromosome of P. umsongenis GO16 against the genomes of representative Pseudomonas species (see the experimental procedures section for details on the computational analysis).
Table S2. Blast atlas of the plasmid pENK22. The table shows the results of the tblastx analysis comparing the annotated features present in the plasmid pENK22 against representative similar plasmids (see the experimental procedures section for details on the computational analysis).
Table S3. Blast comparison of P. umsongensis GO16 putative genes belonging to the central metabolism. The analysis was conducted using the tblastx feature of the blast suite at NCBI (see the experimental procedures section for details on the computational analysis).
Table S4. Blast comparison of P. umsongensis GO16 putitave genes taking part in the metabolism of aromatics. The analysis was conducted using the blastp feature of the blast suite and the genome workbench tool at NCBI using known enzymes for comparison (see the experimental procedures section for details).
Fig. S1. Phylogenetic analysis of different members of the Pseudomonas genus. Genome‐wide comparison between P. umsongensis GO16 and notable Pseudomonas species was conducted using a composite vector approach (see the experimental procedures section in the main text for details).
Fig. S2. Growth profile of P. umsongensis GO16 in different soluble aromatics as the sole carbon source. In panels A to C cells were cultured in the microplate reader using the indicated aromatic substrates at a final concentration of 5 mM. Panel D shows bacterial growth in flasks in the presence of naphthalene vapours as the sole carbon source (see methods for details). Results correspond to the mean and standard deviation of three biological replicates.
Fig. S3. Growth profile and substrate consumption of P. putida KT2440 expressing the tph genes from P. umsongensis GO16. P. putida KT2440 was transformed with the plasmid pBT’T_tph (KT_tph) or the empty control pBT’T (WT in the plot) for comparison. Only KT_tph was able to use TA as the sole carbon source for growth (shown in red; solid line for growth and dashed line for substrate consumption). Expression of the tph genes (green lines) did not affect growth in glucose compare to the control (blue lines). Results correspond to the mean and standard deviation of three biological replicates.
Fig. S4. Terephthalic acid (TA) depletion and protocatechuate (PCA) accumulation in the supernatant of P. putida KT_tph grown with TA as a sole source of carbon and energy. (A) Kinetics of PCA accumulation and TA consumption when the strain was cultivated in MSM medium without nitrogen limitation (full nitrogen), or under polyhydroxyalkanoate (PHA) accumulating conditions (limited nitrogen) determined by HPLC‐UV. (B) After 12 hours of incubation in TA and N limited conditions the culture exhibits a characteristic purple colour corresponding to PCA accumulation in the supernatant (left flask). The culture with full nitrogen that does not accumulate PCA is shown for comparison (right flask). (C) Chromatograms of TA and PCA determination. The upper panel represents a supernatant after 12 hours of culturing in N limited conditions. Mid and lower panels represent, respectively, a standard of 0.075 g L‐1 of PCA and a standard with a mixture of 0.08 g L‐1 of PCA and 0.11 g L‐1 of TA.
Fig. S5. CDW (blue lines) and substrate consumption (red lines) of P. umsongensis GO16 growing on TA (upper), EG (mid) and glucose (lower panel). All cultures contained 1.96 gC L‐1. Plots show the mean and standard deviation of three biological replicates.
Fig. S6. Analysis of the stability of the plasmid pBT’T_tph in P. putida KT2440. The plasmid was purified from KT2440 grown on TA as the sole carbon source using a standard miniprep protocol. The plasmid preparation was digested with SphI (lane sphI) rendering the expected fragments of 6.3 and 3.8 kb. The plasmid was also used as a template for PCR reactions with oligonucleotides V1F and V1rR annealing on the backbone and producing a 3.9 kb DNA fragment (lane pBT’T), and with oligonucleotides RT and GR that render a 6.1 kb DNA product (lane RT). Size in bp of the molecular weight markers is shown for comparison.
Acknowledgements
This work was supported by EU Horizon 2020 research and innovation programme grant number 633962 for the project P4SB and number 863922 for the project MIX‐UP. TN and KO’C are supported by Science Foundation Ireland research centre grant number 16/RC/3889. JJ and MS acknowledge the support received from the Biotechnology and Biological Sciences Research Council (BBSRC) (grants BB/M009769/1 and BB/T011289/1). UA is the recipient of a PhD studentship from the Petroleum Technology Development Fund of Nigeria. TN and GH are funded by an Ad Astra Fellowship at UCD. Dr Federico Cerrone is acknowledged for his help with analysing PHA monomer composition. Dr Helen King, Dr Mandy Hughes and Anita Sicilia are acknowledged for their technical support.
Microbial Biotechnology (2021) 14(6), 2463–2480
To the loving memory of Dr Shane Kenny. A wonderful human being and a true example for all.
Funding informationThis work was supported by EU Horizon 2020 research and innovation programme grant number 633962 for the project P4SB and number 863922 for the project MIX‐UP. TN and KO’C are supported by Science Foundation Ireland research centre grant number 16/RC/3889. JJ and MS acknowledge the support received from the Biotechnology and Biological Sciences Research Council (BBSRC) (grants BB/M009769/1 and BB/T011289/1) H2020: 870294. UA is the recipient of a PhD studentship from the Petroleum Technology Development Fund of Nigeria. TN and GH are funded by an Ad Astra Fellowship at UCD.
Contributor Information
Kevin E. O'Connor, Email: kevin.oconnor@ucd.ie.
José I. Jiménez, Email: j.jimenez@imperial.ac.uk.
References
- Ahn, E. , Choi, K.Y. , Kang, B.S. , Zylstra, G.J. , Kim, D. , and Kim, E. (2017) Salicylate degradation by a cold‐adapted Pseudomonas sp. Ann Microbiol 67: 417–424. [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. , and Lipman, D.J. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Ayub, N.D. , Pettinari, M.J. , Mendez, B.S. , and Lopez, N.I. (2007) The polyhydroxyalkanoate genes of a stress resistant Antarctic Pseudomonas are situated within a genomic island. Plasmid 58: 240–248. [DOI] [PubMed] [Google Scholar]
- Batista, M.B. , Müller‐Santos, M. , Pedrosa, F.O. , and de Souza, E.M. (2016) Potentiality of Herbaspirillum seropedicae as a platform for bioplastic production. In Microbial Models: From Environmental to Industrial Sustainability. Microorganisms for Sustainability, vol. 1. Singapore: Springer, pp. 23–39. [Google Scholar]
- Belda, E. , van Heck, R.G.A. , Lopez‐Sanchez, M.J. , Cruveiller, S. , Barbe, V. , Fraser, C. , et al. (2016) The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ Microbiol 18: 3403–3424. [DOI] [PubMed] [Google Scholar]
- Blank, L.M. , Narancic, T. , Mampel, J. , Tiso, T. , and O'Connor, K. (2019) Biotechnological upcycling of plastic waste and other non‐conventional feedstocks in a circular economy. Curr Opin Biotechnol 62: 212–219. [DOI] [PubMed] [Google Scholar]
- Catone, M.V. , Ruiz, J.A. , Castellanos, M. , Segura, D. , Espin, G. , and Lopez, N.I. (2014) High polyhydroxybutyrate production in Pseudomonas extremaustralis is associated with differential expression of horizontally acquired and core genome polyhydroxyalkanoate synthase genes. PLoS One 9: e98873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria, M. , Nikel, P.I. , Perez‐Pantoja, D. , and Lorenzo, V. (2013) The Entner‐Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ Microbiol 15: 1772–1785. [DOI] [PubMed] [Google Scholar]
- Chek, M.F. , Hiroe, A. , Hakoshima, T. , Sudesh, K. , and Taguchi, S. (2019) PHA synthase (PhaC): interpreting the functions of bioplastic‐producing enzyme from a structural perspective. Appl Microbiol Biot 103: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Choi, K.H. , Kumar, A. , and Schweizer, H.P. (2006) A 10‐min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Meth 64: 391–397. [DOI] [PubMed] [Google Scholar]
- Darling, A.C.E. , Mau, B. , Blattner, F.R. , and Perna, N.T. (2004) Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R. , Kataria, R. , Cerrone, F. , Woods, T. , Kenny, S. , O'Donovan, A. , et al. (2013) Conversion of grass biomass into fermentable sugars and its utilization for medium chain length polyhydroxyalkanoate (mcl‐PHA) production by Pseudomonas strains. Biores Technol 150: 202–209. [DOI] [PubMed] [Google Scholar]
- Diard, S. , Carlier, J.‐P. , Ageron, E. , Grimont, P.A.D. , Langlois, V. , Guérin, P. , and Bouvet, O.M.M. (2002) Accumulation of poly(3‐hydroxybutyrate) from octanoate in different Pseudomonas belonging to the rRNA homology group I. Syst Appl Microbiol 25: 183–188. [DOI] [PubMed] [Google Scholar]
- European Bioplastics (2019) Bioplastics market development update 2019. European Bioplastics. [Google Scholar]
- European Commission (2015) Directive of the European Parliament and of The Council amending Directive 2008/98/EC on waste. [Google Scholar]
- Forum, W.E. (2015) Project MainStream – a global collaboration to accelerate the transition towards the circular economy. Status Update. [Google Scholar]
- Franden, M.A. , Jayakody, L.N. , Li, W.J. , Wagner, N.J. , Cleveland, N.S. , Michener, W.E. , et al. (2018) Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab Eng 48: 197–207. [DOI] [PubMed] [Google Scholar]
- Furmanczyk, E.M. , Kaminski, M.A. , Dziembowski, A. , Lipinski, L. , and Sobczak, A. (2017) Draft genome sequence of the type strain Pseudomonas umsongensis DSM 16611. Genome Announc 5: e01038‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos, M.T. , Schleif, R. , Bairoch, A. , Hofmann, K. , and Ramos, J.L. (1997) AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerischer, U. , Segura, A. , and Ornston, L.N. (1998) PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter . J Bacteriol 180: 1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, R. , Jambeck, J.R. , and Law, K.L. (2017) Production, use, and fate of all plastics ever made. Sci Adv 3: e1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood, C.R. , and Cutting, S.M. (1990) Chemically defined growth media and supplements. In Molecular biological Methods for Bacillus. Harwood, C.R. , and Cutting, S.M. (eds.). Chichester, UK: Wiley, p. 548. [Google Scholar]
- Jimenez, J.I. , Minambres, B. , Garcia, J.L. , and Diaz, E. (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4: 824–841. [DOI] [PubMed] [Google Scholar]
- Jiménez, J.I. , Miñambres, B. , García, J.L. , and Díaz, E. (2004) Genomic Insights in the Metabolism of Aromatic Compounds in Pseudomonas. In Pseudomonas. Ramos, J.L. (ed). Springer, Boston, MA: Springer. [Google Scholar]
- Johnson, C.W. , Salvachúa, D. , Khanna, P. , Smith, H. , Peterson, D.J. , and Beckham, G.T. (2016) Enhancing muconic acid production from glucose and lignin‐derived aromatic compounds via increased protocatechuate decarboxylase activity. Metabolic Eng Commun 3: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C.W. , Salvachúa, D. , Rorrer, N.A. , Black, B.A. , Vardon, D.R. , St. John, P.C. , et al. (2019) Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule 3: 1523–1537. [Google Scholar]
- Kasper, M. (2013) Energy from Waste can Help Curb Greenhouse Gas Emissions. Center for American Progress. [Google Scholar]
- Kenny, S.T. , Runic, J.N. , Kaminsky, W. , Woods, T. , Babu, R.P. , Keely, C.M. , et al. (2008) Up‐cycling of PET (polyethylene terephthalate) to the biodegradable plastic PHA (polyhydroxyalkanoate). Environ Sci Technol 42: 7696–7701. [DOI] [PubMed] [Google Scholar]
- Kenny, S. , Runic, J.N. , Kaminsky, W. , Woods, T. , Babu, R. , and Oconnor, K.E. (2012) Development of a bioprocess to convert PET derived terephthalic acid and biodiesel derived glycerol to medium chain length polyhydroxyalkanoate. Appl Microbiol Biotechnol 95: 623–633. [DOI] [PubMed] [Google Scholar]
- Kim, Y.H. , Cho, K. , Yun, S.H. , Kim, J.Y. , Kwon, K.H. , Yoo, J.S. , and Kim, S.I. (2006) Analysis of aromatic catabolic pathways in Pseudomonas putida KT 2440 using a combined proteomic approach: 2‐DE/MS and cleavable isotope‐coded affinity tag analysis. Proteomics 6: 1301–1318. [DOI] [PubMed] [Google Scholar]
- Kim, H.T. , Kim, J.K. , Cha, H.G. , Kang, M.J. , Lee, H.S. , Khang, T.U. , et al. (2019) Biological valorization of poly(ethylene terephthalate) monomers for upcycling waste PET. ACS Sustain Chem Eng 7: 19396–19406. [Google Scholar]
- Koopman, F. , Wierckx, N. , de Winde, J.H. , and Ruijssenaars, H.J. (2010) Identification and characterization of the furfural and 5‐(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. Proc Natl Acad Sci USA 107: 4919–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageveen, R.G. , Huisman, G.W. , Preusting, H. , Ketelaar, P. , Eggink, G. , and Witholt, B. (1988) Formation of polyesters by Pseudomonas oleovorans: Effect of substrates on formation and composition of Poly‐(R)‐3‐Hydroxyalkanoates and Poly‐(R)‐3‐Hydroxyalkenoates. Appl Environ Microbiol 54: 2924–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.J. , Jayakody, L.N. , Franden, M.A. , Wehrmann, M. , Daun, T. , Hauer, B. , et al. (2019) Laboratory evolution reveals the metabolic and regulatory basis of ethylene glycol metabolism by Pseudomonas putida KT2440. Environ Microbiol 21: 3669–3682. [DOI] [PubMed] [Google Scholar]
- Lopez, N.I. , Pettinari, M.J. , Stackebrandt, E. , Tribelli, P.M. , Potter, M. , Steinbuchel, A. , and Mendez, B.S. (2009) Pseudomonas extremaustralis sp nov., a poly(3‐hydroxybutyrate) producer isolated from an Antarctic environment. Curr Microbiol 59: 514–519. [DOI] [PubMed] [Google Scholar]
- Molina‐Henares, A.J. , Krell, T. , Guazzaroni, M.E. , Segura, A. , and Ramos, J.L. (2006) Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol Rev 30: 157–186. [DOI] [PubMed] [Google Scholar]
- Mückschel, B. , Simon, O. , Klebensberger, J. , Graf, N. , Rosche, B. , Altenbuchner, J. , et al. (2012) Ethylene glycol metabolism by Pseudomonas putida . Appl Environ Microbiol 78: 8531–8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narancic, T. , and O'Connor, K.E. (2017) Microbial biotechnology addressing the plastic waste disaster. Microb Biotechnol 10: 1232–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel, P.I. , Chavarria, M. , Fuhrer, T. , Sauer, U. , and de Lorenzo, V. (2015) Pseudomonas putida KT2440 strain metabolizes glucose through accle formed by enzymes of the Entner‐Doudoroff. Embden‐Meyerhof‐Parnas, and pentose phosphate pathways. J Biol Chem 290: 25920–25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana‐Saez, M. , Pacheco, N. , Costa, J.I. , Mendez, K.N. , Miossec, M.J. , Meneses, C. , et al. (2019) In‐depth genomic and phenotypic characterization of the antarctic psychrotolerant strain Pseudomonas sp. MPC6 reveals unique metabolic features, plasticity, and biotechnological potential. Front Microbiol 10: 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peix, A. , Ramirez‐Bahena, M.H. , and Velazquez, E. (2018) The current status on the taxonomy of Pseudomonas revisited: an update. Infect Genet Evol 57: 106–116. [DOI] [PubMed] [Google Scholar]
- Petkau, A. , Stuart‐Edwards, M. , Stothard, P. , and Van Domselaar, G. (2010) Interactive microbial genome visualization with GView. Bioinformatics 26: 3125–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plastics Europe (2016) Plastics – the Facts 2016. [Google Scholar]
- Poblete‐Castro, I. , Borrero‐de Acuña, J.M. , Nikel, P.I. , Kohlstedt, M. , and Wittmann, C. (2017) Host organism: Pseudomonas putida . In Industrial Biotechnology: Microorganisms. Wittmann, C. , and Liao, J.C. (eds.). Weinheim: Wiley‐VCH, pp. 299–326. [Google Scholar]
- Pohlmann, A. , Fricke, W.F. , Reinecke, F. , Kusian, B. , Liesegang, H. , Cramm, R. , et al. (2006) Genome sequence of the bioplastic‐producing “Knallgas” bacterium Ralstonia eutropha H16. Nat Biotechnol 24: 1257–1262. [DOI] [PubMed] [Google Scholar]
- Qi, J. , Wang, B. , and Hao, B.I. (2004) Whole proteome prokaryote phylogeny without sequence alignment: a K‐string composition approach. J Mol Evol 58: 1–11. [DOI] [PubMed] [Google Scholar]
- Rehm, B.H.A. (2010) Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol 8: 578–592. [DOI] [PubMed] [Google Scholar]
- Salvador, M. , Abdulmutalib, U. , Gonzalez, J. , Kim, J. , Smith, A.A. , Faulon, J.L. , et al. (2019) Microbial genes for a circular and sustainable bio‐PET economy. Genes‐Basel 10: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasoh, M. , Masai, E. , Ishibashi, S. , Hara, H. , Kamimura, N. , Miyauchi, K. , and Fukuda, M. (2006) Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl Environ Microb 72: 1825–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, S.B. , Kim, T.Y. , Park, J.M. , and Lee, S.Y. (2010) In silico genome‐scale metabolic analysis of Pseudomonas putida KT2440 for polyhydroxyalkanoate synthesis, degradation of aromatics and anaerobic survival. Biotechnol J 5: 739–750. [DOI] [PubMed] [Google Scholar]
- Sota, M. , Yano, H. , Ono, A. , Miyazaki, R. , Ishii, H. , Genka, H. , et al. (2006) Genomic and functional analysis of the IncP‐9 naphthalene‐catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J Bacteriol 188: 4057–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudesh, K. , Abe, H. , and Doi, Y. (2000) Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog Polym Sci 25: 1503–1555. [Google Scholar]
- Tiso, T. , Narancic, T. , Wei, R. , Pollet, E. , Beagan, N. , Schröder, K. , et al. (2020) Bio‐upcycling of polyethylene terephthalate. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Volke, D.C. , Calero, P. , and Nikel, P.I. (2020) Pseudomonas putida. Trends Microbiol 28: 512–513. [DOI] [PubMed] [Google Scholar]
- Wehrmann, M. , Billard, P. , Martin‐Meriadec, A. , Zegeye, A. , and Klebensberger, J. (2017) Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone‐dependent alcohol dehydrogenases in Pseudomonas putida KT2440. MBio 8: e00570–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, R. , and Zimmermann, W. (2017) Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb Biotechnol 10: 1302–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer, A. , Kohlstedt, M. , Volke, D.C. , Nikel, P.I. , and Wittmann, C. (2020) Industrial biotechnology of Pseudomonas putida: advances and prospects. Appl Microbiol Biotechnol 104: 7745–7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierckx, N. , Narancic, T. , Eberlein, C. , Wei, R. , Drzyzga, O. , Magnin, A. , et al. (2018) Plastic biodegradation: challenges and opportunities. In Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids: Biodegradation and Bioremediation. Steffan, R. (ed). Cham: Springer International Publishing, pp. 1–29. [Google Scholar]
- Wierckx, N. , Prieto, M.A. , Pomposiello, P. , de Lorenzo, V. , O'Connor, K. , and Blank, L.M. (2015) Plastic waste as a novel substrate for industrial biotechnology. Microb Biotechnol 8: 900–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, P.A. , and Sayers, J.R. (1994) The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas . Biodegradation 5: 195–217. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Yang, J. , and Jiang, L. (2016) Comment on “A bacterium that degrades and assimilates poly(ethylene terephthalate)”. Science 353: 759. [DOI] [PubMed] [Google Scholar]
- Yoshida, S. , Hiraga, K. , Takehana, T. , Taniguchi, I. , Yamaji, H. , Maeda, Y. , et al. (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351: 1196–1199. [DOI] [PubMed] [Google Scholar]
- Zuo, G.H. , and Hao, B.L. (2015) CVTree3 web server for whole‐genome‐based and alignment‐free prokaryotic phylogeny and taxonomy. Genom Proteom Bioinf 13: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Blast atlas of P. umsongensis GO16 chromosomal genes. The table shows the results of the tblastx analysis comparing the annotated features present in the chromosome of P. umsongenis GO16 against the genomes of representative Pseudomonas species (see the experimental procedures section for details on the computational analysis).
Table S2. Blast atlas of the plasmid pENK22. The table shows the results of the tblastx analysis comparing the annotated features present in the plasmid pENK22 against representative similar plasmids (see the experimental procedures section for details on the computational analysis).
Table S3. Blast comparison of P. umsongensis GO16 putative genes belonging to the central metabolism. The analysis was conducted using the tblastx feature of the blast suite at NCBI (see the experimental procedures section for details on the computational analysis).
Table S4. Blast comparison of P. umsongensis GO16 putitave genes taking part in the metabolism of aromatics. The analysis was conducted using the blastp feature of the blast suite and the genome workbench tool at NCBI using known enzymes for comparison (see the experimental procedures section for details).
Fig. S1. Phylogenetic analysis of different members of the Pseudomonas genus. Genome‐wide comparison between P. umsongensis GO16 and notable Pseudomonas species was conducted using a composite vector approach (see the experimental procedures section in the main text for details).
Fig. S2. Growth profile of P. umsongensis GO16 in different soluble aromatics as the sole carbon source. In panels A to C cells were cultured in the microplate reader using the indicated aromatic substrates at a final concentration of 5 mM. Panel D shows bacterial growth in flasks in the presence of naphthalene vapours as the sole carbon source (see methods for details). Results correspond to the mean and standard deviation of three biological replicates.
Fig. S3. Growth profile and substrate consumption of P. putida KT2440 expressing the tph genes from P. umsongensis GO16. P. putida KT2440 was transformed with the plasmid pBT’T_tph (KT_tph) or the empty control pBT’T (WT in the plot) for comparison. Only KT_tph was able to use TA as the sole carbon source for growth (shown in red; solid line for growth and dashed line for substrate consumption). Expression of the tph genes (green lines) did not affect growth in glucose compare to the control (blue lines). Results correspond to the mean and standard deviation of three biological replicates.
Fig. S4. Terephthalic acid (TA) depletion and protocatechuate (PCA) accumulation in the supernatant of P. putida KT_tph grown with TA as a sole source of carbon and energy. (A) Kinetics of PCA accumulation and TA consumption when the strain was cultivated in MSM medium without nitrogen limitation (full nitrogen), or under polyhydroxyalkanoate (PHA) accumulating conditions (limited nitrogen) determined by HPLC‐UV. (B) After 12 hours of incubation in TA and N limited conditions the culture exhibits a characteristic purple colour corresponding to PCA accumulation in the supernatant (left flask). The culture with full nitrogen that does not accumulate PCA is shown for comparison (right flask). (C) Chromatograms of TA and PCA determination. The upper panel represents a supernatant after 12 hours of culturing in N limited conditions. Mid and lower panels represent, respectively, a standard of 0.075 g L‐1 of PCA and a standard with a mixture of 0.08 g L‐1 of PCA and 0.11 g L‐1 of TA.
Fig. S5. CDW (blue lines) and substrate consumption (red lines) of P. umsongensis GO16 growing on TA (upper), EG (mid) and glucose (lower panel). All cultures contained 1.96 gC L‐1. Plots show the mean and standard deviation of three biological replicates.
Fig. S6. Analysis of the stability of the plasmid pBT’T_tph in P. putida KT2440. The plasmid was purified from KT2440 grown on TA as the sole carbon source using a standard miniprep protocol. The plasmid preparation was digested with SphI (lane sphI) rendering the expected fragments of 6.3 and 3.8 kb. The plasmid was also used as a template for PCR reactions with oligonucleotides V1F and V1rR annealing on the backbone and producing a 3.9 kb DNA fragment (lane pBT’T), and with oligonucleotides RT and GR that render a 6.1 kb DNA product (lane RT). Size in bp of the molecular weight markers is shown for comparison.


