Summary
Directed evolution is a powerful method to optimize proteins and metabolic reactions towards user‐defined goals. It usually involves subjecting genes or pathways to iterative rounds of mutagenesis, selection and amplification. While powerful, systematic searches through large sequence‐spaces is a labour‐intensive task, and can be further limited by a priori knowledge about the optimal initial search space, and/or limits in terms of screening throughput. Here, we demonstrate an integrated directed evolution workflow for metabolic pathway enzymes that continuously generate enzyme variants using the recently developed orthogonal replication system, OrthoRep and screens for optimal performance in high‐throughput using a transcription factor‐based biosensor. We demonstrate the strengths of this workflow by evolving a rate‐limiting enzymatic reaction of the biosynthetic pathway for cis,cis‐muconic acid (CCM), a precursor used for bioplastic and coatings, in Saccharomyces cerevisiae. After two weeks of simply iterating between passaging of cells to generate variant enzymes via OrthoRep and high‐throughput sorting of best‐performing variants using a transcription factor‐based biosensor for CCM, we ultimately identified variant enzymes improving CCM titers > 13‐fold compared with reference enzymes. Taken together, the combination of synthetic biology tools as adopted in this study is an efficient approach to debottleneck repetitive workflows associated with directed evolution of metabolic enzymes.
Directed evolution is a powerful method to optimize proteins and metabolic reactions. Here we demonstrate an integrated directed evolution workflow for metabolic pathway enzymes that continuously generates enzyme variants using the recently developed orthogonal replication system, OrthoRep, and screens for optimal performance in high‐throughput using a transcription factor‐based biosensor. Using this workflow we evolve variant enzymes improving CCM titers > 13‐fold compared to reference enzymes by simple passaging of evolving cell populations over the course of 2 weeks.
Introduction
Industrial biotechnology has offered commercialization of environmentally friendly transportation fuels, amino acids and value‐added chemicals by the use of fermentation feedstocks and microbial cell factories (Choi et al., 2019). Yet, industrializing microbial cells for a broad range of applications within manufacturing, health and transportation industries often requires extensive engineering of both the microbial chassis and at the level of scaling up the fermentation processes (Van Dien, 2013; Nielsen and Keasling, 2016). Indeed, in the design of cell factories for fermentation‐based manufacturing of value‐added chemicals and therapeutics, biosynthetic pathways are often composed of enzymes from several different sources, and with enzyme activities and expression levels requiring careful balancing in order to achieve optimal pathway flux (Galanie et al., 2015; Zhang et al., 2020). While such multi‐dimensional optimization can be streamlined using design‐of‐experiment approaches and machine learning algorithms (Jeschek et al., 2016; Xu et al., 2017; Carbonell et al., 2018), the regulatory and cellular complexity of living cells and the constraints in speed, scale, depth and costs of even rational trial‐and‐error engineering approaches challenge the development of microbial cell factories.
As a complementary approach to bottom‐up rational engineering, evolution‐guided cell factory engineering has gained substantial traction over the last decade (Mundhada et al., 2016; Sandberg et al., 2019). Here, the key principles of evolutionary engineering includes targeted or genome‐wide genetic diversification coupled with screening of variant libraries (Packer and Liu, 2015). Numerous metabolic engineering studies have successfully applied directed evolution to improve product and feedstock tolerance and cell factory performance (Caspeta et al., 2014; Park et al., 2014; Mundhada et al., 2016). While powerful, both the generation of large numbers of genetic variants and the development of proper selection regimes, as well as the cloning and transformation procedures associated with directed evolution cycles, are often time‐ and cost‐intensive. To overcome this, in vivo directed evolution uses endogenous or orthogonal cellular machineries to maintain high‐mutation rates without the need for iterative cycles of library cloning and transformation, apart from propagating the evolving population (Esvelt et al., 2011; Ravikumar et al., 2014; Crook et al., 2016). One such system is OrthoRep enabling continuous generation of variant genes of interest expressed from a linear cytoplasmic chromosome that is propagated via an orthogonal error‐prone DNA polymerase (Ravikumar et al., 2014, 2018). With orthogonal in vivo evolution machineries at hand, any trait that can be coupled to growth (e.g. antibiotic resistance, tolerance to cultivation conditions and/or complementation of auxotrophies) enables facile identification of improved target genes without need for direct screening (Esvelt et al., 2011; Ravikumar et al., 2014; García‐García et al., 2020; Rix et al., 2020).
However, for metabolic engineering, the expression of heterologous enzymes and proteins towards biobased production of value‐added chemicals seldom allows direct coupling of production to growth, or other high‐throughput screens, needed to capitalize on the massive diversity generated by in vivo evolution systems (Esvelt and Wang, 2013). Here, the recent development of biosensors based on allosterically regulated transcription factors (aTFs) can provide a complementary technology for coupling enzymatic activity or pathway flux with facile screening of large variant libraries in multiplex through fluorescence‐activated cell sorting (FACS) or growth (Raman et al., 2014; Flachbart et al., 2019). Briefly, such biosensors link binding of small‐molecule ligands to aTFs as input, with changes in expression of reporter genes or actuators as output (Mahr and Frunzke, 2016). Taken together, the coupling of continuous evolution systems with biosensing could allow metabolic engineers to cost‐effectively search for optimal pathway designs.
Here, we combine the power of targeted in vivo mutagenesis using OrthoRep with high‐throughput biosensing for the rapid evolution of rate‐limiting metabolic reactions of the cis,cis‐muconic acid (CCM) pathway (Weber et al., 2012; Curran et al., 2013; Suástegui and Shao, 2016). The 3‐step CCM pathway, consisting a dehydroshikimate dehydratase (AroZ), a multi‐subunit protocatechuic acid (PCA) decarboxylase and a catechol 1,2‐dioxygenase (Fig. 1A) (Weber et al., 2012; Curran et al., 2013), has been extensively studied and optimized to support biobased production of plastics and coatings, following hydrogenation of CCM into adipic acid as a building block for nylon‐6,6 (Weber et al., 2012; Curran et al., 2013; Suastegui et al., 2016; Leavitt et al., 2017; Snoek et al., 2018; Wang et al., 2020). Importantly, for CCM pathway optimization, we have previously engineered the aTF BenM as a CCM biosensor in yeast (Skjoedt et al., 2016), and this has further enabled optimization of yeast as a chassis for CCM production (Snoek et al., 2018; Wang et al., 2020), complemented by additional evolution‐ and machine learning‐guided optimization of the endogenous yeast aromatic amino acid pathway from which CCM is derived (Leavitt et al., 2017; Zhang et al., 2020). However, the build‐up and secretion of the CCM pathway intermediate PCA remain rate‐limiting for high CCM production (Suastegui et al., 2016; Leavitt et al., 2017; Snoek et al., 2018; Wang et al., 2020). This observation is propelled by the fact that PCA accumulation supposedly is not limited to suboptimal catalytic activity or expression of the downstream CCM pathway enzyme, PCA decarboxylase, as PCA accumulation also remains a persistent issue for microbial biosynthesis of other PCA‐derived chemicals and nutraceuticals without decarboxylase requirements (Strucko et al., 2015, 2017; D'Ambrosio et al., 2020). Yet, no attempts for directed evolution of the heterologous enzymes towards increased PCA‐derived production has to our knowledge been performed. Here, we demonstrate rapid evolution of PCA decarboxylase subunits using a simple experimental design enabled by OrthoRep and five biosensor‐assisted selection cycles ultimately yielding > 13‐fold higher CCM production compared with the subunits encoded in the wild‐type PCA decarboxylase complex.
Fig. 1.
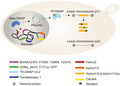
Schematic illustration of the in vivo directed evolution workflow. A. Schematic illustration of the 3‐step cis,cis‐muconic acid pathway, comprising heterologous expression of PaAroZ, KpAroY subunits (B, D, and Ciso), as well as CaCatA and overexpression of Tkl1 (Weber et al., 2012; Curran et al., 2013). B. Schematic illustration of the parental strain (Sc‐105, see Table S5) used for in vivo directed evolution of the cis,cis‐muconic acid pathway enzymes KpAroY. B and KpAroY. Ciso in yeast cells. The strain replicates and expresses the biosensor, all cis,cis‐muconic acid pathway enzymes except KpAroY. B and KpAroY. Ciso, and the variant error‐prone TP‐DNAP (expressed from AR‐Ec633, see Table S4) from the nucleus. All components required for OrthoRep replication and transcription are encoded on p2, whereas, genes encoding KpAroY. B and KpAroY. Ciso are expressed from p1. C. Schematic illustration of the in vivo directed evolution workflow showing the passaging regimes of the parental strain undergoing (i) the five consecutive rounds of OrthoRep coupled with biosensor‐based selection or (ii) fifteen bulk passages to effect drift without biosensor‐based selection.
Results
Parental strain design
In order to efficiently evolve cis,cis‐muconic acid biosynthetic pathway enzymes, we used state‐of‐the‐art orthogonal error‐prone replication and biosensing machineries. For generation of sequence diversity using OrthoRep, we used the highly error‐prone TP‐DNAP1 variant, TP‐DNAP1(L477V, L640Y, I777K, W814N), with mutation‐rate of ~ 1 × 10−5 substitutions per base (Ravikumar et al., 2018), whereas for selection of high‐performing cis,cis‐muconic acid pathway designs, we used the CCM‐binding BenM variant transcription factor, BenM(A230V, F253S, Y286N, Y293H), with a high‐dynamic output range to maximize sorting resolution (Snoek et al., 2019). While the TP‐DNAP1 variant was expressed from a CEN/ARS‐based plasmid, the BenM variant was genomically integrated together with an expression cassette encoding the GFP reporter (Fig. 1A) (Ravikumar et al., 2018; Snoek et al., 2018). With respect to the CCM pathway template, we genomically integrated codon‐optimized AroZ from Podospora anserina (PaAroZ), CatA from Candida albicans (CaCatA), and the gene encoding the D subunit of AroY from Klebsiella pneumoniae (KpAroD) for heterologous expression of the 3‐DHS dehydratase, catechol 1,2‐dioxygenase and D subunit‐mediated PCA decarboxylase reactions respectively (Weber et al., 2012; Curran et al., 2013) (Fig. 1A). In order to evolve the rate‐limiting PCA decarboxylase reaction using OrthoRep, we encoded the codon‐optimized B subunit and the C isoform subunit without oxygen sensitivity of AroY from K. pneumoniae (KpAroY. B and KpAroY. Ciso respectively) (Weber et al., 2012) on the p1 linear plasmid for replication with the error‐prone TP‐DNAP1 variant. Together with overexpression of the transketolase gene (TKL1) to increase the carbon flux into the aromatic amino acid biosynthesis (Luttik et al., 2008; Curran et al., 2013), this parental strain (Sc‐105) was used as a starting point for in vivo directed evolution of CCM production in yeast (Fig. 1B).
Selection of hypermutated CCM biosynthetic pathway enzymes
In order to evolve the parental strain towards higher CCM production, we coupled continuous mutagenesis using OrthoRep with selection using the BenM‐based biosensor output as a proxy for CCM accumulation respectively. Specifically, with the parental starting strain Sc‐105 in place, we cultured cells in Delft minimal medium to allow OrthoRep to generate AroY diversity and selected high CCM‐producing cells by fluorescence‐activated cell sorting (FACS) of 1 mio. events. We iterated culturing and FACS five times, where each sort collected 10 000 events and was followed by a 24‐h recovery cultivation in synthetic complete medium, after which propagated cells were harvested for both glycerol stock and passed for further evolution. For this conservative selection schedule, each such cycle required 3 days. As a control evolution experiment, we cultured the parental starting strain over 15 consecutive 24 h cultivations without FACS selection by bulk passaging 20% of cells between each cultivation. By monitoring population‐level fluorescence changes in this control evolution experiment, we could ensure that no changes in CCM production resulted from simply drifting AroY on OrthoRep. In total, both regimes were conducted over 15 days, totalling five sorted populations and 15 bulk populations (Fig. 1C).
The number and duration of the iterative cycles were selected based on (i) the mutation‐rate and copy number of TP‐DNAP1‐4‐2, (ii) the sequence length of the bait genes encoding AroY. B and AroY. Ciso, and (iii) the generation time of S. cerevisiae (Ravikumar et al., 2018). Briefly, with an estimated mutation‐rate of 1 x 10−5 substitutions per base at 10× copies of p1 per cell (Ravikumar et al., 2018), the actual mutation‐rate per cell approximates 1 × 10‐4 substitutions per base. With AroY. B and AroY. Ciso sequences on the p1 plasmid totalling 2.1 kb (Table S1, Fig. S1), we expected 0.21 mutations/replication/generation, or 1 mutation every fifth generation. In addition to this, for the actual cultivations, we assumed that no mutations would occur during the first 24 h recovery following each sort, and that the exponentially growing cells would double every ~ 3 h reaching a max OD600 (optical density measured at a wavelength of 600 nm) of eight following 5–6 doublings in Delft minimal medium before each sort. These experimental choices would on average result in approximately 1 additional mutation accumulated in AroY. B and AroY. Ciso per cell for each additional sort.
Characteristics and validation of selected enzyme variants
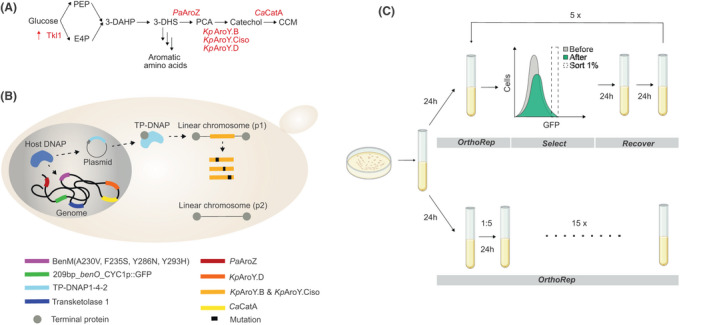
Following the five iterations of OrthoRep coupled with selection using FACS, we compared the population‐level fluorescence distributions from the five sorted populations and the 15 bulk passaged populations (Fig. 2A). Over the course of the evolution experiment the population‐level fluorescence outputs increased following each round of sorting from the first to the fourth sort, reaching a total increase of 2.5‐fold. From the fourth to fifth sorting, no further increase in fluorescence was observed (Fig. 2A). In total, a maximum of 3.9‐fold increase was observed when comparing mean fluorescence outputs following five rounds of sorting versus fifteen rounds of neutral drifting (Fig. 2A, ‘Sort 5’ vs. ‘Bulk 15’). With the interest to inspect fluorescence outputs of cells undergoing drifting vs selection, we isolated 20 colonies obtained from ‘Bulk 15’ and > 300 colonies obtained from “Sort 4 + 5” respectively. Here, the increased fluorescence was confirmed, with an average fluorescence output of colonies from sorted populations > 5‐fold higher than the average fluorescence output of colonies from cells undergoing neutral drifting (Fig. 2B, insert). Importantly, and corroborating the conservative selection regime, more than 96% (309/319) of the colonies from sorted populations (‘Sort4 + 5’) had cells with higher fluorescence than cells from colonies undergoing neutral drifting (‘Bulk 15’) (Fig. 2B, dashed line).
Fig. 2.
Population‐level fluorescence outputs from parental strains expressing evolvable PCA decarboxylase subunits. A. Population‐level fluorescence outputs following 15 passages of cultures of parental strains undergoing neutral drifting (Bulk 1–15) by OrthoRep (OrthoRep – No selection), and following five consecutive iterations (Sort 1–5) of OrthoRep coupled to CCM biosensor‐based selection (OrthoRep + Selection). Bars indicate mean fluorescence intensity (MFI) of 10 000 events. AU: arbitrary units. B. Mean fluorescence intensity of cells from 20 colonies propagated from population ‘Bulk 15’ and 309 colonies propagated from Sort 4 and Sort 5 (‘Sort4–5’). Insert shows box plots from all 20 and 309 mean fluorescence intensities obtained from the 10 000 events measured for each of the ‘Bulk15’ and ‘Sort4 + 5’ populations respectively. Bars indicate mean fluorescence intensity (MFI) of 10 000 events. Error bars represent standard deviation of the mean from three biological replicate samplings. AU: arbitrary units.
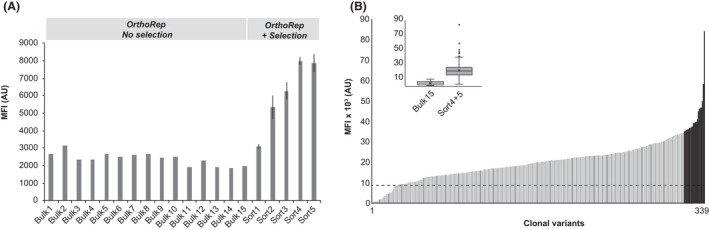
Next, from the > 300 colonies re‐screened using flow cytometry (Fig. 2B), we sequenced the KpAroY. B and KpAroY. Ciso alleles amplified from the p1 plasmid of cells derived from 30 of the highest fluorescent colonies (Fig. 2B, dark bars). Here, we found 11 amino acid changes, two deletion and nine silent mutations (Fig. 3A), with all mutations occurring only once, except the KpAroY. Ciso_666T > del (deletion of single nucleotide following 666T, leading to stop codon at residue 244), occurring twice. Of the missense and non‐sense mutations, four were located in KpAroY. B and nine in KpAroY. Ciso (Fig. 3A, Table S2). To validate that the evolved KpAroY. B and KpAroY. Ciso mutant alleles, and no other potential mutations in the p1 plasmid or nuclear genome, were causal for the observed increase in fluorescence, we cloned individual KpAroY. B and KpAroY. Ciso alleles into the genome of a parental strain (Sc‐78) under the control of the strong constitutive promoters TDH3 and TEF1 respectively (Fig. S2). For each KpAroY. B variant allele, a wild‐type KpAroY. Ciso allele was included in the integration and vice‐versa. The parental strain harboured the PaAroZ, CaCatA and CCM biosensor genes genomically integrated, and fluorescence and metabolites compared with the parental strain expressing wild‐type KpAroY. B and KpAroY. Ciso. A total of 8 KpAroY variants, 2 KpAroY. B and 6 KpAroY. Ciso, were tested and compared with the strain expressing wild‐type KpAroY. B and KpAroY. Ciso. Firstly, we tested fluorescence of the 8 + 1 strains following 24 h of cultivation. Here, we found that only 5 of the 8 variants showed significantly higher fluorescence outputs compared with the wild‐type KpAroY reference strain (P‐value < 0.01), with KpAroY. B_P146T performing the best (10.7×) (Fig. 3B). Next, we analysed the amounts of CCM and PCA of all strains following 72 h of cultivation, as previously shown to be a relevant benchmark time‐point for comparing biosensor fluorescence outputs and CCM titers (Skjoedt et al., 2016). Here, we observed that six out of eight strains showed significantly higher CCM titers compared with the wild‐type KpAroY reference strain (t‐test, P‐value < 0.01), again with KpAroY. B_P146T performing the best with a 13.7‐fold higher CCM titer when expressed together with wild‐type KpAroY. Ciso, compared with expressing wild‐type KpAroY. B and KpAroY. Ciso together (Fig. 3C). Interestingly, when comparing fluorescence and CCM titers, it becomes clear that while the CCM biosensor variant used in this study (BenM(A230V, F253S, Y286N, Y293H)) indeed enables selection of KpAroY. B and KpAroY. Ciso variant alleles yielding higher CCM production, the biosensor operational range cannot discriminate CCM titers higher than approximately 80 mg l−1 under this cultivation regime (Fig. 3B and C). While this is an inherent limitation of the specific BenM variant, it is critical to underscore that this variant was chosen due to its high‐dynamic output range (Snoek et al., 2019), which was deemed necessary for selection in multiplex. Lastly, having verified that CCM product titers were increased when expressing six of the 8 KpAroY variants, we next investigated whether PCA titers for the pathways expressing PCA decarboxylase variants were perturbed. Importantly, PCA titers of no higher than 250–300 mg l−1 are known to cause a significant fitness burden to S. cerevisiae (D'Ambrosio et al., 2020), making it critical to investigate if the observed increases in CCM titers for the six different PCA decarboxylase variants would support catalytic activities to lower PCA below fitness‐burdening PCA levels. Indeed, of the KpAroY variants supporting increased CCM titers, KpAroY. B_P146T also showed significantly reduced PCA titers (77%, P < 0.01) (Fig. 3D), further highlighting this variant as a new and catalytically improved PCA decarboxylase.
Fig. 3.
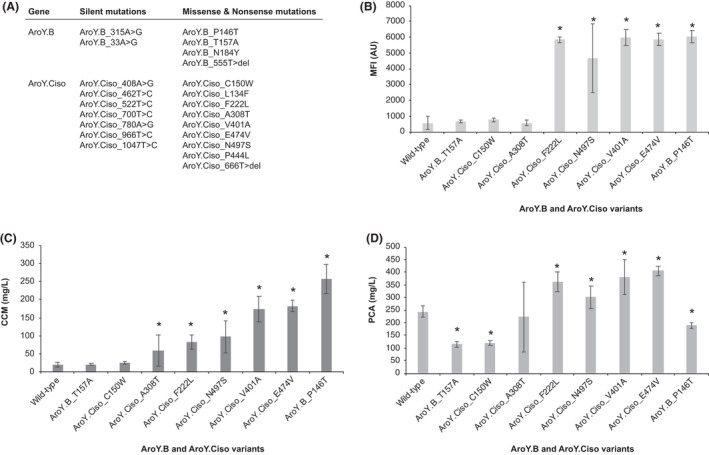
Characterization of PCA decarboxylase subunit variants from OrthoRep‐evolved populations. A. Parent populations from which OrthoRep‐evolved KpAroY. B and KpAroY. Ciso variants derived, and the silent, missense and non‐sense mutations present in them. B. CCM biosensor‐based evaluation of eight evolved PCA decarboxylase subunit variants KpAroY. B and KpAroY. Ciso compared with wild‐type PCA decarboxylase subunits through flow cytometry assays. Mean fluorescence intensity (MFI) following 24 h cultivation for parental strain Sc‐78 integrated with the indicated eight evolved KpAroY. B and KpAroY. Ciso PCA decarboxylase subunit variants or wild‐type PCA decarboxylase subunits (Sc‐194). Data represent means of 4–5 biological replicates, and error bars represent standard deviation of the mean. C. Extracellular cis,cis‐mucinic acid (CCM) concentrations in cultivation broth from the same as in (B) eight evolved PCA decarboxylase subunit variants of KpAroY. B and KpAroY. Ciso compared with wild‐type PCA decarboxylase subunits following 72 h cultivation. Data represent means of 4‐5 biological replicates, and error bars represent standard deviation of the mean (D) Extracellular protocatechuic acid (PCA) concentrations in cultivation broth from the same as in (B) and (C) eight evolved PCA decarboxylase subunit variants KpAroY. B and KpAroY. Ciso compared with wild‐type PCA decarboxylase subunits following 72 h of cultivation. Data represent means of 4–5 biological replicates, and error bars represent standard deviation of the mean.
Discussion
In this study, we demonstrate the successful merger of two synthetic biology tools for the benefit of evolving superior enzymes without the use of labour‐intensive library designs or costly low/semi‐throughput analytical facilities. Importantly, this study show‐cases the evolution of metabolic pathway enzymes without any native growth advantage for the cells, a condition that most previous in vivo directed evolution requires (Esvelt et al., 2011). Furthermore, the merger of OrthoRep and biosensors for directed evolution as demonstrated in this study complements the development of selections associated with growth under (strong) selection pressures (Ravikumar et al., 2018; Zhong et al., 2020).
While this study was a successful demonstration of continuous hypermutation of target genes applied for metabolic enzyme evolution, more prospecting and evolution‐guided engineering of the CCM pathway is still warranted. For biobased CCM production, such efforts should not be limited to improving the suboptimal catalytic activity or expression of pathway enzymes downstream of PCA, i.e. PCA decarboxylase and catechol 1,2‐dioxygenase. One strategy to consider further is the need to limit the secretion or passive diffusion, of PCA across the cellular membrane as is often observed in yeast engineered to produce PCA‐derived chemicals and nutraceuticals (Hansen et al., 2009; Weber et al., 2012; Curran et al., 2013; Suastegui et al., 2016; Leavitt et al., 2017). With the evolved PCA decarboxylase subunits identified from this study, a logical next step could be to move from in vivo directed evolution of predefined target genes towards genome‐wide adaptive laboratory evolution. For such purposes, transferring the biosensor read‐out from fluorescence to growth would be beneficial from a technical and scalability point of view (Zhong et al., 2020). For such a purpose, the coupling of CCM to growth using synthetic control circuits founded on BenM, or the recently developed vanillin biosensor, could be useful for genome‐wide searches of nucleotide polymorphisms and chromosomal re‐arrangements limiting PCA efflux and/or further boosting PCA metabolic flux respectively (Ambri et al., 2020; D'Ambrosio et al., 2020).
Another aspect to consider is to diversify the selection criteria beyond the stringent one used in this study (Fig. 1C). Specifically, tuning the selection strength during continuous evolution regimes has previously been demonstrated to enable mutational drifting and adaptation of robust proteins (Bershtein et al., 2008; Steinberg and Ostermeier, 2016; Zhong et al., 2020). With the coupling of OrthoRep to FACS‐compatible screens as demonstrated in this study, such tuning should be possible to implement and further explored, in order not to outpace the rate of adaption using the conservative and stringent cut‐off for selection as applied in this study. Ultimately, this could expand both the robustness and catalytic activity of further evolved PCA decarboxylases, but also increase the relatively low number of mutations observed per evolved PCA decarboxylase variant. Furthermore, toggled selection regimes of neutral drifting interrupted by selection (Rix et al., 2020; Zhong et al., 2020), may also increase the hit‐rate of the continuous evolution, and limit the false‐discovery rate observed in this study (> 9/22, > 0.45) (Fig. 3). Extending from this, it is also worth considering the use of biosensor variants with operational ranges > 100 mg l−1 CCM (Fig. 3B and C) or tuning of cultivation time (Skjoedt et al. 2016; Snoek et al. 2019), to increase the hit‐rate of PCA decarboxylase variants with even higher catalytic activity.
In summary, we consider that our study serves as a first demonstration of rapid evolution of metabolic enzymes without any direct fitness advantage using continuous hypermutation, and furthermore moves forward the engineering of S. cerevisiae, and potentially other microbial chassis, for the industrial production of CCM as a precursor for further hydrogenation into adipic acid and nylon‐6,6 for the bioplastics industry.
Experimental procedures
Cloning
Plasmid pEDJ366 carries a URA3 targeting gRNA and was made by inverse amplification of pCfB3050 (Jessop‐Fabre et al., 2016) with oligos EDJ382 and EDJ383 followed by T4 DNA ligation (NEB). Plasmid p1237 (Skjoedt et al., 2016) was modified for SpHIS5::TRP1 by inverse amplification with oligos EDJ386 and EDJ387, and assembled with CEN. PK2‐1C genomic TRP1 amplified with EDJ388 and EDJ389 by T4 DNA ligation (pEDJ371). pCfB2764 was amplified with EDJ432 and EDJ433 and assembled with gEDJ12 obtained with primers EDJ434 and EDJ435 to give plasmid pEDJ515. gEZ475 was amplified by EDJ390 and EDJ391 and assembled with ZZ‐Ec475 (Zhong et al., 2020) after restriction digest by XhoI and XbaI (pMB10). The p1 (Ravikumar et al., 2014) integration cassette (Table S1) that contains wild‐type KpAroY. B and KpAroY. Ciso was made by USER assembly (Jensen et al., 2014) with pMB10 as backbone vector and KpAroY. B and KpAroY. Ciso amplified from p1241 (Skjoedt et al., 2016) with primer pairs EDJ413 and EDJ414, and EDJ415 and EDJ416 respectively (pMB11). All oligonucleotides used in this study can be found in Table S3.
Media
One litre of mineral medium (Delft) with 2% glucose (Verduyn et al., 1992) contained 75 ml (NH4)2SO4 (100 g l−1), 120 ml KH2PO4 (120 g l−1), 10 ml MgSO4, 7H2O (50 g l−1), 2 ml trace metals, 1 ml vitamins and 20 g glucose. 1 l of trace metals contain 4.5 g CaCl2·2H2O, 4.5 g ZnSO4·7H2O, 3 g FeSO4·7H2O, 1 g H3BO3, 1 g MnCl2·4H2O, 0.4 g Na2MoO4·2H2O, 0.3 g CoCl2·6H2O, 0.1 g CuSO4·5H2O, 0.1 g KI and 15 g EDTA. 1 l of vitamins contain 50 mg biotin, 200 mg p‐aminobenzoic acid, 1 g nicotinic acid, 1 g Ca‐pantotenate, 1 g pyridoxine HCl, 1 g thiamine HCl and 25 g myo‐Inositol. Synthetic complete dropout media were bought from Sigma‐Aldrich (St. Louis, MO, USA).
Strains
Strain CEN. PK2‐1C (EUROSCARF) expressing pRS414‐TEF1p‐Cas9‐CYC1t (DiCarlo et al., 2013) was transformed with gRNA plasmid pEDJ366 and URA3‐KO‐90‐mer to completely remove ura3‐52 (Sc‐62). CCM‐producing strain Sc‐78 was made essentially as previously described (Skjoedt et al., 2016) with minor modifications; NotI treated plasmids p1237 and pEDJ371 were sequentially integrated in Sc‐62 to give Sc‐67 and Sc‐68, followed by integration of similarly treated pCfB2553 and pEDJ515. Sc‐78 was transformed with AR‐Ec633 (Ravikumar et al., 2018) and saved as Sc‐79 from histidine dropout medium. F102‐2 was transformed with ScaI treated pMB11 (Sc‐93), and resulting transformants were picked from uracil dropout medium. Sc‐93 was protoplast fused with Sc‐79 as previously described (Ravikumar et al., 2014) and selected in synthetic complete histidine and uracil dropout medium to give Sc‐105. Screened variants and wild‐type KpAroY. B and KpAroY. Ciso (Sc‐194) were synthesized as gene blocks by IDT and integrated with overlapping homology into Easyclone site XII‐5 (Jensen et al., 2014) in strain Sc‐78 with Cas9 and pCfB3050. Oligos ORP3‐10 were used for amplifying individual parts from gene blocks or genomic DNA as indicated in Table S3 prior to reverse engineering of variant enzymes, and ORP1 with ORP2 and ORP11 with ORP12 for amplification of terminators with chr:XII‐5 homology from pCfB2909 (Jessop‐Fabre et al., 2016).
All plasmids and strains designed and constructed in this study can be found in Tables S4 and S5.
Flow cytometer analysis
Strains were inoculated into appropriate synthetic complete dropout media (Sigma‐Aldrich) and incubated for 48 h in 96 deep‐well plates at 30 °C with shaking. All cultures were then diluted 10‐fold in fresh synthetic complete dropout media and incubated as before for 24 or 72 h as indicated. All cultures were diluted 5‐fold in a total volume of 150 μl with 1× phosphate buffer saline (PBS) from Life Technologies immediately before analysis. The BD LSRFortessa™ from BD Biosciences was used for analysing 10 000 single events per culture with a blue laser at 488 nm and settings FITC: 450, FSC: 150 and SSC: 250. FlowLogic (Invai Technologies, Victoria, Australia) was used for processing of collected data.
Mutation analysis
KpAroY. B and KpAroY. Ciso amplicons, derived with primers p1F and p1R from the p1 plasmid, were Sanger sequenced on individual clones. Three primers were used for sequencing to cover both genes (p1F, p1S and p1R). The sequencing trace files were analysed with non‐evolved p1 plasmid as a reference in mutation surveyor using default settings (SoftGenetics) (Minton et al., 2011). Poor‐quality sequences at the beginning and the end of the trace files were trimmed, and all called mutations were manually verified. All mutant sequences are listed in Table S2.
Fluorescence‐assisted cell sorting
Parental strain Sc‐105 was streaked on synthetic complete histidine and uracil dropout plates and incubated at 30 °C for 3 days; a single colony was grown in 5 ml of fresh Delft minimal medium for 24 h at 30 °C with shaking. Pre‐inoculo culture was obtained by diluting individual cell culture to OD of 0.2 in a total volume of 5 ml fresh Delft minimal medium and grown for an additional day at 30 °C with shaking. The culture was diluted again to OD of 2 in a total volume of 5 ml fresh Delft minimal medium and grown for another day at 30 °C with shaking. After 24 h, 1 ml of the culture was named ‘Bulk 1’ and saved as glycerol stock at −80 °C; 1 ml was diluted in a total volume of 5 ml fresh Delft minimal medium to continue the ‘Bulk’ cell culture and grown at 30 °C with shaking, and the remaining of the culture was diluted to OD of 0.5 in a total volume of 1 ml phosphate‐buffered saline (PBS) in sterile tubes to arrest cell growth for flow cytometry acquisition. Using a SH800S cell sorter (Sony Biotechnology), 1 million events were analysed with a blue laser at 488 nm, and 10 000 cells of the top 2% fluorescent output were sorted into 2 ml of fresh synthetic complete medium and grown for 24 h at 30 °C with shaking. The sorted population was then spun down and re‐suspended in 2 mL of fresh Delft minimal medium for additional 24 h of recovery at 30 °C with shaking. Finally, 1 ml of the culture was named ‘Sort 1’ and saved as glycerol stock at −80 °C; the remaining population was diluted to OD of 2 in a total volume of 5 ml Delft minimal to be grown again for flow cytometry acquisition and additional sorting, totalling five ‘Sort’ samples over the course of the 2 weeks experiment (Fig. 1B). The ‘Bulk’ culture was re‐incolutated daily into a total volume of 5 ml fresh Delft minimal medium totalling to 15 ‘Bulk’ samples. Since the ‘Bulk’ samples were not subjected to fluorescence enrichment they represent the neutrally drifted strains as a control for the sorted populations. The experiment has been done in triplicates, and 14 ml Falcon™ Round‐Bottom Polypropylene Tubes (Thermo Fisher Scientific, Waltham, MA, USA) were used for every cultivation.
HPLC
Three replicates from each individual test strain were inoculated in 200 μl mineral medium (pH 4.5) supplemented with 20 mg l−1 Histidine and Uracil in 96 deep‐well plates with air‐penetrable lids (EnzyScreen) and incubated for 24 h at 30 °C with shaking (250 rpm). 50 μl of the O/N culture was transferred into 450 μL fresh mineral medium and incubated for 72 h with the same conditions as described above. Cultures were centrifuged at 3500 rpm, and supernatants were diluted 10‐fold into mQ water before analysis on an Aminex HPX‐87H ion exclusion column. Samples were analysed for 45 min each at 60 °C and with a flow rate of 0.6 ml min−1 of 1 mM H2SO4. Quantifications of PCA and CCM were performed by comparison with the spectrum of standards ranging from 16 to 160 mg l−1.
Conflict of interest
JDK has a financial interest in Amyris, Lygos, Demetrix, Maple Bio, Napigen, Ansa Biotechnologies, Berkeley Yeast, Apertor Pharmaceuticals and Zero Acre Farms. The authors declare that they have no other competing interests.
Author contributions
EDJ, MKJ, CCL and JDK conceived the study. EDJ, FA, MBB and AAJ conducted all experimental work related to strain designs, constructions and characterizations. EDJ, FA and MBB performed all data analysis. EDJ and MKJ wrote the manuscript. All authors reviewed and approved the manuscript.
Supporting information
Table S1. p1 integration cassette sequence from pMB11.
Table S2. List of all sequences relevant to this study.
Table S3. List of oligonucleotides used in this study.
Table S4. List of plasmids used in this study.
Table S5. List of strains used in this study.
Fig. S1. Graphical overview of p1 sequence after pMB11 insert integration.
Fig. S2. Graphical overview of wild‐type genome integration cassette for reverse engineering with homology regions shown.
Fig. S3. Graphical overview of identified mutations in sequenced p1 amplicons.
Acknowledgements
This work was supported by the Novo Nordisk Foundation. Authors would also like to thank Mette Christensen and Lars Schrübbers for technical advice related to HPLC.
Microb. Biotechnol. (2021) 14(6), 2617–2626
Funding information
This work was supported by the Novo Nordisk Foundation.
References
- Ambri, F. , D'Ambrosio, V. , Di Blasi, R. , Maury, J. , Jacobsen, S.A.B. , McCloskey, D. , et al. (2020) High‐resolution scanning of optimal biosensor reporter promoters in yeast. ACS Synth Biol 9: 218–226. [DOI] [PubMed] [Google Scholar]
- Bershtein, S. , Goldin, K. , and Tawfik, D.S. (2008) Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol 379: 1029–1044. [DOI] [PubMed] [Google Scholar]
- Carbonell, P. , Jervis, A.J. , Robinson, C.J. , Yan, C. , Dunstan, M. , Swainston, N. , et al. (2018) An automated Design‐Build‐Test‐Learn pipeline for enhanced microbial production of fine chemicals. Commun Biol 1: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspeta, L. , Chen, Y. , Ghiaci, P. , Feizi, A. , Buskov, S. , Hallström, B.M. , et al. (2014) Biofuels. Altered sterol composition renders yeast thermotolerant. Science 346: 75–78. [DOI] [PubMed] [Google Scholar]
- Choi, K.R. , Jang, W.D. , Yang, D. , Cho, J.S. , Park, D. , and Lee, S.Y. (2019) Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol 37: 817–837. [DOI] [PubMed] [Google Scholar]
- Crook, N. , Abatemarco, J. , Sun, J. , Wagner, J.M. , Schmitz, A. , and Alper, H.S. (2016) In vivo continuous evolution of genes and pathways in yeast. Nat Commun 7: 13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, K.A. , Leavitt, J.M. , Karim, A.S. , and Alper, H.S. (2013) Metabolic engineering of muconic acid production in Saccharomyces cerevisiae . Metab Eng 15: 55–66. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio, V. , Dore, E. , Di Blasi, R. , van den Broek, M. , Sudarsan, S. , Horst, J.T. , et al. (2020) Regulatory control circuits for stabilizing long‐term anabolic product formation in yeast. Metab Eng 61: 369–380. [DOI] [PubMed] [Google Scholar]
- DiCarlo, J.E. , Norville, J.E. , Mali, P. , Rios, X. , Aach, J. , and Church, G.M. (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR‐Cas systems. Nucleic Acids Res 41: 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt, K.M. , Carlson, J.C. , and Liu, D.R. (2011) A system for the continuous directed evolution of biomolecules. Nature 472: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt, K.M. , and Wang, H.H. (2013) Genome‐scale engineering for systems and synthetic biology. Mol Syst Biol 9: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachbart, L.K. , Sokolowsky, S. , and Marienhagen, J. (2019) Displaced by deceivers: prevention of biosensor cross‐talk is pivotal for successful biosensor‐based high‐throughput screening campaigns. ACS Synth Biol 8: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie, S. , Thodey, K. , Trenchard, I.J. , Filsinger Interrante, M. , and Smolke, C.D. (2015) Complete biosynthesis of opioids in yeast. Science 349: 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐García, J.D. , Joshi, J. , Patterson, J.A. , Trujillo‐Rodriguez, L. , Reisch, C.R. , Javanpour, A.A. , et al. (2020) Potential for applying continuous directed evolution to plant enzymes: an exploratory study. Life 10: 179. doi: 10.3390/life10090179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, E.H. , Møller, B.L. , Kock, G.R. , Bünner, C.M. , Kristensen, C. , Jensen, O.R. , et al. (2009) De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker's yeast (Saccharomyces cerevisiae). Appl Environ Microbiol 75: 2765–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, N.B. , Strucko, T. , Kildegaard, K.R. , David, F. , Maury, J. , Mortensen, U.H. , et al. (2014) EasyClone: method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae . FEMS Yeast Res 14: 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschek, M. , Gerngross, D. , and Panke, S. (2016) Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort. Nat Commun 7: 11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop‐Fabre, M.M. , Jakočiūnas, T. , Stovicek, V. , Dai, Z. , Jensen, M.K. , Keasling, J.D. , and Borodina, I. (2016) EasyClone‐MarkerFree: a vector toolkit for marker‐less integration of genes into Saccharomyces cerevisiae via CRISPR‐Cas9. Biotechnol J 11: 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt, J.M. , Wagner, J.M. , Tu, C.C. , Tong, A. , Liu, Y. , and Alper, H.S. (2017) Biosensor‐enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae . Biotechnol J 12: 1600687. 10.1002/biot.201600687. [DOI] [PubMed] [Google Scholar]
- Luttik, M.A.H. , Vuralhan, Z. , Suir, E. , Braus, G.H. , Pronk, J.T. , and Daran, J.M. (2008) Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab Eng 10: 141–153. [DOI] [PubMed] [Google Scholar]
- Mahr, R. , and Frunzke, J. (2016) Transcription factor‐based biosensors in biotechnology: current state and future prospects. Appl Microbiol Biotechnol 100: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton, J.A.L. , Flanagan, S.E. , and Ellard, S. (2011) Mutation surveyor: Software for DNA sequence analysis. In PCR Mutation Detection Protocols. Theophilus, B.D.M. , and Rapley, R. (eds). Totowa, NJ: Humana Press, pp. 143–153. [DOI] [PubMed] [Google Scholar]
- Mundhada, H. , Miguel, J.S. , Schneider, K. , Koza, A. , Christensen, H.B. , Klein, T. , et al. (2016) Increased production of L‐serine in Escherichia coli through adaptive laboratory evolution. Metab Eng 39: 141–150. [DOI] [PubMed] [Google Scholar]
- Nielsen, J. , and Keasling, J.D. (2016) Engineering cellular metabolism. Cell 164: 1185–1197. [DOI] [PubMed] [Google Scholar]
- Packer, M.S. , and Liu, D.R. (2015) Methods for the directed evolution of proteins. Nat Rev Genet 16: 379–394. [DOI] [PubMed] [Google Scholar]
- Park, S.H. , Kim, H.U. , Kim, T.Y. , Park, J.S. , Kim, S.‐S. , and Lee, S.Y. (2014) Metabolic engineering of Corynebacterium glutamicum for L‐arginine production. Nat Commun 5: 4618. [DOI] [PubMed] [Google Scholar]
- Raman, S. , Rogers, J.K. , Taylor, N.D. , and Church, G.M. (2014) Evolution‐guided optimization of biosynthetic pathways. Proc Natl Acad Sci USA 111: 201409523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar, A. , Arrieta, A. , and Liu, C.C. (2014) An orthogonal DNA replication system in yeast. Nat Chem Biol 10: 175–177. [DOI] [PubMed] [Google Scholar]
- Ravikumar, A. , Arzumanyan, G.A. , Obadi, M.K.A. , Javanpour, A.A. , and Liu, C.C. (2018) Scalable, continuous evolution of genes at mutation rates above genomic error thresholds. Cell 175: 1946–1957.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rix, G. , Watkins‐Dulaney, E.J. , Almhjell, P.J. , Boville, C.E. , Arnold, F.H. , and Liu, C.C. (2020) Scalable continuous evolution for the generation of diverse enzyme variants encompassing promiscuous activities. Nat Commun 11: 5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg, T.E. , Salazar, M.J. , Weng, L.L. , Palsson, B.O. , and Feist, A.M. (2019) The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng 56: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjoedt, M.L. , Snoek, T. , Kildegaard, K.R. , Arsovska, D. , Eichenberger, M. , Goedecke, T.J. , et al. (2016) Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat Chem Biol 12: 951–958. [DOI] [PubMed] [Google Scholar]
- Snoek, T. , Chaberski, E.K. , Ambri, F. , Kol, S. , Bjørn, S.P. , Pang, B. , et al. (2019) Evolution‐guided engineering of small‐molecule biosensors. Nucleic Acids Res 48: e3. 10.1093/nar/gkz954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek, T. , Romero‐Suarez, D. , Zhang, J. , Ambri, F. , Skjoedt, M.L. , Sudarsan, S. , et al. (2018) An orthogonal and pH‐tunable sensor‐selector for muconic acid biosynthesis in yeast. ACS Synth Biol 7: 995–1003. [DOI] [PubMed] [Google Scholar]
- Steinberg, B. , and Ostermeier, M. (2016) Environmental changes bridge evolutionary valleys. Sci Adv 2: e1500921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strucko, T. , Buron, L.D. , Jarczynska, Z.D. , Nødvig, C.S. , Mølgaard, L. , Halkier, B.A. , and Mortensen, U.H. (2017) CASCADE, a platform for controlled gene amplification for high, tunable and selection‐free gene expression in yeast. Sci Rep 7: 41431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strucko, T. , Magdenoska, O. , and Mortensen, U.H. (2015) Benchmarking two commonly used Saccharomyces cerevisiae strains for heterologous vanillin‐β‐glucoside production. Metab Eng Commun 2: 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suastegui, M. , Matthiesen, J.E. , Carraher, J.M. , Hernandez, N. , Rodriguez Quiroz, N. , Okerlund, A. , et al. (2016) Combining metabolic engineering and electrocatalysis: application to the production of polyamides from sugar. Angew Chem Int Ed Engl 128: 2414–2419. [DOI] [PubMed] [Google Scholar]
- Suástegui, M. , and Shao, Z. (2016) Yeast factories for the production of aromatic compounds: from building blocks to plant secondary metabolites. J Ind Microbiol Biotechnol 43: 1611–1624. [DOI] [PubMed] [Google Scholar]
- Van Dien, S. (2013) From the first drop to the first truckload: commercialization of microbial processes for renewable chemicals. Curr Opin Biotechnol 24: 1061–1068. [DOI] [PubMed] [Google Scholar]
- Verduyn, C. , Postma, E. , Scheffers, W.A. , and Van Dijken, J.P. (1992) Effect of benzoic acid on metabolic fluxes in yeasts: a continuous‐culture study on the regulation of respiration and alcoholic fermentation. Yeast 8: 501–517. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Øzmerih, S. , Guerreiro, R. , Meireles, A.C. , Carolas, A. , Milne, N. , et al. (2020) Improvement of cis, cis‐muconic acid production in Saccharomyces cerevisiae through biosensor‐aided genome engineering. ACS Synth Biol 9: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, C. , Brückner, C. , Weinreb, S. , Lehr, C. , Essl, C. , and Boles, E. (2012) Biosynthesis of cis, cis‐muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae . Appl Environ Microbiol 78: 8421–8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. , Rizzoni, E.A. , Sul, S.‐Y. , and Stephanopoulos, G. (2017) Improving metabolic pathway efficiency by statistical model‐based multivariate regulatory metabolic engineering. ACS Synth Biol 6: 148–158. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Petersen, S.D. , Radivojevic, T. , Ramirez, A. , Pérez‐Manríquez, A. , Abeliuk, E. , et al. (2020) Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism. Nat Commun 11: 4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z. , Wong, B.G. , Ravikumar, A. , Arzumanyan, G.A. , Khalil, A.S. , and Liu, C.C. (2020) Automated continuous evolution of proteins in vivo. ACS Synth Biol 9: 1270–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. p1 integration cassette sequence from pMB11.
Table S2. List of all sequences relevant to this study.
Table S3. List of oligonucleotides used in this study.
Table S4. List of plasmids used in this study.
Table S5. List of strains used in this study.
Fig. S1. Graphical overview of p1 sequence after pMB11 insert integration.
Fig. S2. Graphical overview of wild‐type genome integration cassette for reverse engineering with homology regions shown.
Fig. S3. Graphical overview of identified mutations in sequenced p1 amplicons.


