In this work, Liu et al. used metabolic engineering strategies to construct S. cerevisiae strains for high‐level production of tyrosol and salidroside from glucose. Finally, titers of 9.90 ± 0.06 g l−1 of tyrosol and 26.55 ± 0.43 g l−1 of salidroside were achieved in 5 l bioreactors, both are the highest titers reported to date.
![]()
Summary
Tyrosol and its glycosylated product salidroside are important ingredients in pharmaceuticals, nutraceuticals and cosmetics. Despite the ability of Saccharomyces cerevisiae to naturally synthesize tyrosol, high yield from de novo synthesis remains a challenge. Here, we used metabolic engineering strategies to construct S. cerevisiae strains for high‐level production of tyrosol and salidroside from glucose. First, tyrosol production was unlocked from feedback inhibition. Then, transketolase and ribose‐5‐phosphate ketol‐isomerase were overexpressed to balance the supply of precursors. Next, chorismate synthase and chorismate mutase were overexpressed to maximize the aromatic amino acid flux towards tyrosol synthesis. Finally, the competing pathway was knocked out to further direct the carbon flux into tyrosol synthesis. Through a combination of these interventions, tyrosol titres reached 702.30 ± 0.41 mg l−1 in shake flasks, which were approximately 26‐fold greater than that of the WT strain. RrU8GT33 from Rhodiola rosea was also applied to cells and maximized salidroside production from tyrosol in S. cerevisiae. Salidroside titres of 1575.45 ± 19.35 mg l−1 were accomplished in shake flasks. Furthermore, titres of 9.90 ± 0.06 g l−1 of tyrosol and 26.55 ± 0.43 g l−1 of salidroside were achieved in 5 l bioreactors, both are the highest titres reported to date. The synergistic engineering strategies presented in this study could be further applied to increase the production of high value‐added aromatic compounds derived from the aromatic amino acid biosynthesis pathway in S. cerevisiae.
Introduction
Tyrosol and its glycosylated derivative salidroside (8‐O‐β‐D‐glucoside of tyrosol) are widely used in food and medicine industries. For example, tyrosol serves as the precursor for several phenylethanoids (Xue and Yang, 2016; Chung et al., 2017) and anticancer drug candidates (Ahn et al., 2008). Salidroside has anticancer potential (Chen et al., 2016b) and can be used to ameliorate insulin resistance (Zheng et al., 2015) in cardiovascular and cerebrovascular diseases (Tang et al., 2014; Chen et al., 2016a; Zhang et al., 2017a, 2018). Salidroside has also been used in cosmetic industries due to its antioxidant and anti‐photoaging properties (Mao et al., 2015).
Currently, tyrosol and salidroside are mainly extracted from plants, such as olive and Rhodiola roots, or chemically synthesized. Low extraction efficiency (Mao et al., 2007; Miralles et al., 2015) and dwindling plant resources (Galambosi, 2006; Moyo et al., 2015; Chen et al., 2016c) limit the applications of plant‐extraction. Furthermore, chemical synthesis and semi‐synthesis of tyrosol and salidroside require expensive substrates that are often not environmentally friendly (Woodburn and Stuntz, 1950; Troshchenko and Yuodvirshis, 1969; Delépée et al., 2007). Conversely, microbial biosynthesis provides a sustainable and economically feasible platform for producing natural products originally derived from plants (Lee et al., 2012; Borodina and Nielsen, 2014; Paddon and Keasling, 2014). In recent years, E. coil was successfully used to produce tyrosol and salidroside (Satoh et al., 2012; Bai et al., 2014; Chung et al., 2017; Fan et al., 2017; Xue et al., 2017; Liu et al., 2018; Yang et al., 2019). Through these studies, titres of tyrosol achieved using sugar (glucose, or xylose and glucose mixture) as the carbon source, have increased from 69.08 mg l−1 (0.5 mM) to 1.47 g l−1 (Satoh et al., 2012; Liu et al., 2018; Yang et al., 2019). The production of salidroside from sugar increased from 56.90 to 670.58 mg l−1 in shake flasks and to 6.03 g l−1 in a 5 l bioreactor, using a codon‐optimized UDP‐glycosyltransferase synUgt85a1 from Arabidopsis thaliana and adopting a co‐culture strategy (Liu et al., 2018).
Yeasts offer a suitable chassis for high‐value plant molecule production, due to properties such as the availability of versatile genetic engineering tools and their robustness in large‐scale fermentations (Zhang et al., 2017b, 2015; Lian et al., 2018; Xu et al., 2020). Saccharomyces cerevisiae is a GRAS (generally regarded as safe) yeast organism that is highly suitable for food, potentially pharmaceutical compound and cosmetic production (Borodina and Nielsen, 2014; Sewalt et al., 2016). The aromatic amino acid synthesis pathway of S. cerevisiae was successfully engineered to increase the production of aromatic molecules such as reticuline (Trenchard et al., 2015), p‐coumaric acid (Rodriguez et al., 2015), mandelic acid and 4‐hydroxymandelic acid (Suastegui et al., 2017). Therefore, the construction of a high‐titre tyrosol or salidroside‐producing S. cerevisiae strain is highly desirable.
In S. cerevisiae, tyrosol is produced from the shikimate pathway and the l‐tyrosine branch. Erythrose‐4‐phosphate (E4P) and phosphoenolpyruvate (PEP) are derived from the pentose phosphate pathway (PPP) and glycolysis, respectively, and are condensed by DAHP synthase (encoded by ARO4 and ARO3) into 3‐deoxy‐D‐arabino‐heptulosonate‐7‐phosphate (DAHP; Hazelwood et al., 2008; Suastegui and Shao, 2016). Next, pentafunctional enzyme (encoded by ARO1) and chorismate synthase (encoded by ARO2) catalyse chorismate synthesis from DAHP. The conversion of chorismate to prephenate is catalysed by the enzyme chorismate mutase (encoded by ARO7), followed by conversion of prephenate to 4‐hydroxyphenylpyruvate (4‐HPP) by prephenate dehydrogenase (encoded by TYR1). Phenylpyruvate decarboxylase (encoded by ARO10) converts 4‐HPP into 4‐hydroxyphenylacetaldehyde (4‐HPAA). 4‐HPAA is reduced by endogenous alcohol dehydrogenases (ADHs) to tyrosol (Fig. 1). Salidroside is produced via the glycosylation of tyrosol at the 8‐OH group, which is catalysed by a regio‐specific uridine 5’‐diphospho‐glucosyltransferase (UGT; Fan et al., 2017). Previous studies increased tyrosol titres to 927.68 ± 25.26 mg l−1 in shake flasks and 8.37 g l−1 in a bioreactor in a haploid industrial S. cerevisiae strain HLF‐Dα (Jiang et al., 2018; Torrens‐Spence et al., 2018; Guo et al., 2019; Guo et al., 2020). The highest salidroside titres reported for engineered S. cerevisiae are 239.5 mg l−1 in a shake flask and 1.82 g l−1 in a bioreactor (Jiang et al., 2018; Torrens‐Spence et al., 2018; Guo et al., 2020).
Fig. 1.
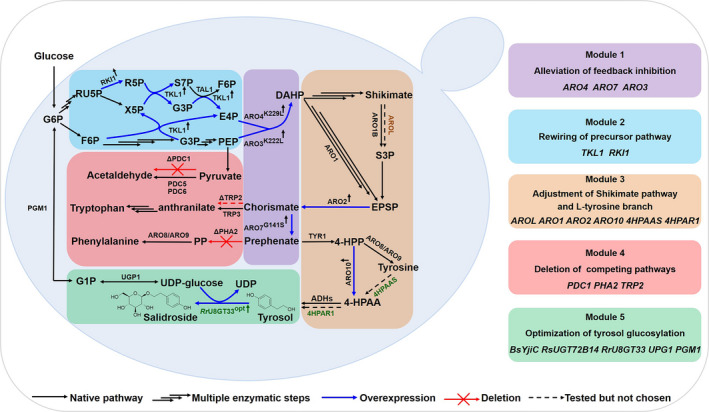
Schematic representation of the modular engineering strategy for high‐level tyrosol and salidroside production in S. cerevisiae. The modular engineering strategy consists of alleviation of feedback inhibition (module 1), rewiring of precursor pathway (module 2), adjustment of shikimate pathway and l‐tyrosine branch (module 3), deletion of competing pathways (module 4) and optimization of tyrosol glycosylation (module 5). Black arrows represent the native pathways in S. cerevisiae; blue bold arrows represent the overexpressed genes in this study; red crosses represent gene deletions; dashed arrows indicate that the reactions were tested but not used in this study and the treble arrows represent multiple enzymatic steps. Native genes of S. cerevisiae are shown in black colour; the gene AROL from E. coil is indicated in orange; the heterologous genes from Rhodiola rosea were shown in green. Metabolite abbreviations: G6P: Glucose 6‐phosphate; RU5P: D‐Ribulose 5‐phosphate; X5P: D‐Xylulose 5‐phosphate; R5P: D‐Ribose 5‐phosphate; S7P: Sedoheptulose 7‐phosphate; G3P: D‐Glyceraldehyde 3‐phosphate; F6P: beta‐D‐Fructose 6‐phosphate; PEP: phosphoenolpyruvate; E4P: D‐Erythrose 4‐phosphate; DAHP: 3‐deoxy‐D‐arabino‐heptulosonate‐7‐phosphate; S3P: shikimate‐3‐phosphate; EPSP: 5‐enolpyruvyl‐3‐shikimate; 4‐HPP: p‐hydroxyphenylpyruvate; PP: phenylpyruvate; 4‐HPAA: 4‐hydroxyphenylacetylaldehyde; G1P: Glucose 1‐phosphate; UDP: Uridine diphosphate; Gene abbreviations: RKI1: ribose‐5‐phosphate ketol‐isomerase1; TKL1: transketolase 1; TAL1: transaldolase 1; ARO3K222L/ARO4K229L : feedback‐insensitive DAHP synthases; ARO1: pentafunctional aromatic enzyme; AROL: shikimate kinase Ⅱ from E. coil; ARO2: chorismate synthase; ARO7G141S : feedback‐insensitive chorismate mutase; PHA2: prephenate dehydratase; TRP2: anthranilate synthase; TRP3: bifunctional anthranilate synthase/indole‐3‐glycerol‐phosphate synthase; PDC1: pyruvate decarboxylase 1; PDC5: pyruvate decarboxylase 5; PDC6: pyruvate decarboxylase 6; TYR1: prephenate dehydrogenase; ARO10: phenylpyruvate decarboxylase; ARO8/9: aromatic amino transferases; ADHs: alcohol dehydrogenases; 4HPAAS: 4‐HPAA synthase from Rhodiola. rosea; 4HPAR1: 4‐HPAA reductase 1 from Rhodiola. rosea; RrU8GT33: tyrosol: UDP‐glucose 8‐O‐glucosyltransferase 33 from Rhodiola. rosea; PGM1: Phosphoglycerate mutase 1; UPG1: UDP‐glucose pyrophosphorylase 1.
In this study, a widely used model organism, S. cerevisiae strain CEN.PK2‐1C was engineered to produce tyrosol and salidroside. In short, four modules in the pathway for tyrosol production were systematically optimized (Fig. 1). Firstly, ARO4K229L and ARO7G141S mutations were introduced, which encoded feedback‐inhibition‐insensitive DAHP synthases and chorismate mutase respectively. Then, RKI1 and TKL1 were overexpressed to tune the flux of the precursor pathway. To direct the carbon flux into tyrosol, the genes of the shikimate pathway and the l‐tyrosine branch from different species were screened, and competing pathways were blocked by deleting PHA2 and PDC1. Finally, another copy of ARO4K229L , ARO3K222L and ARO7G141S was integrated into the 308a locus to enable preferential production of tyrosol. To produce salidroside, three UDP‐glycosyltransferases from diverse species were tested; codon‐optimized RrU8GT33opt was optimal for the glycosylation of tyrosol. Finally, 9.90 ± 0.06 g l−1 tyrosol and 26.55 ± 0.43 g l−1 salidroside were produced in separate 5 l bioreactors, both presenting the highest titres reported to date. Our study paves the way for further industrial production of tyrosol and salidroside.
Results
Unlocking the carbon flux into the aromatic amino acid pathway
Previous studies showed that the mutations of ARO4K229L and ARO7G141S could efficiently relieve feedback inhibition and increase the production of phenylethanol and their para‐hydroxyl analogues in S. cerevisiae (Luttik et al., 2008). To develop a fully integrated and plasmid‐free S. cerevisiae strain for industrial applications, in the present study, the in situ ARO4K229L and ARO7G141S mutations were performed in CEN.PK2‐1C using the PCRCT system (Bao et al., 2015), generating strains LYTY1. After 72 h of fermentation with LYTY1 strains, tyrosol production increased ˜ 3‐fold compared to that of the WT strain, reaching the titre of 105.10 ± 0.66 mg l−1.
Erythrose‐4‐phosphate (E4P) derived from the pentose phosphate pathway (PPP) and phosphoenolpyruvate (PEP) produced from glycolysis are starting precursors for the production of aromatic amino acid‐derived compounds in S. cerevisiae. Based on the catalytic mechanism of the condensation of PEP and E4P to DAHP (Konig et al., 2004), a 1:1 molar ratio of PEP and E4P seems to promote the biosynthesis of aromatic amino acid derivatives. However, in S. cerevisiae, according to the metabolic flux analysis, E4P flux is at least one order of magnitude lower than that of PEP flux (Suastegui et al., 2016). To facilitate the carbon flux afflux in E4P, we first overexpressed RKI1 under the control of the TEF1 promoter in strain LYTY1 to generate strain LYTY1R. Strain LYTY1R produced 132.40 ± 3.52 mg l−1 of tyrosol, increasing 29.78% than that of strain LYTY1. Then, overexpression of TKL1 in LYTY1R led to about 38.10% increase in tyrosol production compared to that of strain LYTY1R. (Fig. 2C).
Fig. 2.
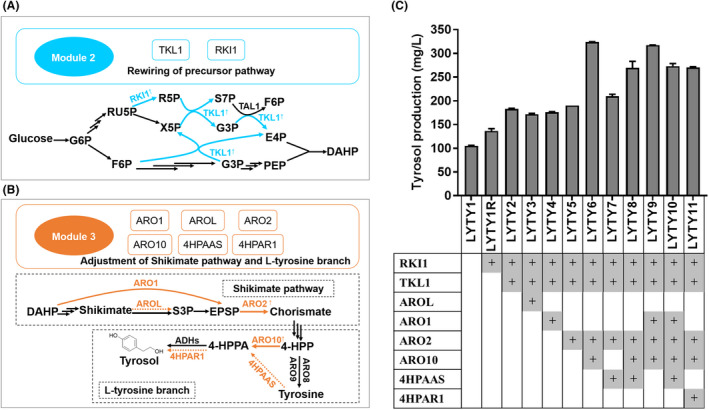
(A) Schematic illustration of rewiring the metabolic pathway for improving supply of E4P. Cyan arrows represent the overexpressed genes. The abbreviations of metabolites and gene details were shown in Fig. 1. (B) Overview of genetic modifications of Module 3. Module 3 consists of Shikimate pathway and l‐tyrosine branch (the dashed boxes). Orange arrows indicate that the actions were tested. See Fig. 1 legend regarding abbreviations of these metabolites. (C) Tyrosol production after engineering of module 2 and module 3. The fermentations were performed at 30 °C in 50 ml YPD medium in shake flasks. Titre of tyrosol was detected after 72 h fermentation. Error bars represent the standard deviation of three biological duplicates.
Optimization of the shikimate pathway and the L‐tyrosine branch
In S. cerevisiae, tyrosol is derived from the shikimate pathway and the subsequent l‐tyrosine branch, as shown in Fig. 2B. In the shikimate pathway, a single pentafunctional enzyme (encoded by ARO1) catalyses the formation of 5‐enolpyruvyl‐3‐shikimate phosphate (EPSP) from DAHP. Then, the conversion of chorismate from EPSP is catalysed by the Aro2. In contrast to S. cerevisiae, the formation of EPSP from DAHP is catalysed by five monofunctional enzymes in E. coil. Overexpression of the shikimate kinase Ⅱ gene AROL from E. coil or endogenous ARO1 from S. cerevisiae was regarded as a useful strategy to produce high level of shikimate derivatives (Rodriguez et al., 2015). However, by integrating AROL or ARO1 into the 1622b locus (high expression levels were observed when gene cassettes were integrated in this locus; Reider Apel et al., 2017) of strain LYTY2, the titre of tyrosol was marginally decreased (Fig. 2C). These results suggested that methods used to optimize the conversion of DAHP to EPSP did not lead to increased tyrosol production in S. cerevisiae. Next, ARO2 was overexpressed under the control of the PGK1 promoter (resulting in strain LYTY5), and the titre of tyrosol was slightly greater than that of strain LYTY2.
Tyrosol biosynthesis diverges from the conversion of chorismate in both yeasts and plants. In S. cerevisiae, chorismate is first converted into prephenate by Aro7p, then prephenate can be transformed into 4‐HPP by Tyr1p. Next, 4‐HPP is decarboxylated to 4‐HPAA by Aro10p, and alcohol dehydrogenases finally converts 4‐HPPA into tyrosol (Fig. 1). In plants, chorismate is converted into tyrosine over several steps, and then, tyrosine can be directly converted into 4‐HPPA by 4‐HPAA synthase (encoded by 4HPAAS). 4‐HPAA reductase (encoded by 4HPAR1 and 4HPAR2) finally converts 4‐HPPA into tyrosol (Schenck and Maeda, 2018; Torrens‐Spence et al., 2018). To direct flux towards tyrosol, we first overexpressed the endogenous gene ARO10 and the codon‐optimized gene 4HPAASopt from R. rosea in strain LYTY5, resulting in strains LYTY6 and LYTY7 respectively. The yields of strains LYTY6 and LYTY7 reached 323.90 ± 0.41 and 209.70 ± 2.88 mg l−1, which were approximately 70% and 10% greater compared to that of strain LYTY5. Due to these positive effects, both genes (ARO10 and 4HPAASopt ) were overexpressed in strain LYTY5, generating strain LYTY8, which led to the accumulation of 42% more tyrosol than that of the parental LYTY5 but 16% less than that of LYTY6 (Fig. 2C), demonstrating that Aro10p can efficiently convert 4‐HPP into 4‐HPAA. Furthermore, ARO1 overexpression in strains LYTY6 and LYTY8 did not lead to increase in tyrosol production (Fig. 2C). Finally, the overexpression of the plant‐derived gene 4HPAR1, which showed high catalytic activity and specificity for the reduction of 4‐HPAA to tyrosol (Torrens‐Spence et al., 2018), resulted in decrease in tyrosol production (Fig. 2C). These results indicated that the conversion of DAHP to EPSP and the reduction of 4‐HPAA to tyrosol may not be limiting steps in the tyrosol synthetic pathway, whereas the conversion of 4‐HPP to 4‐HPAA was a rate‐limiting step.
Deletion of competing pathways
Pyruvate decarboxylases (encoded by PDC1, PDC5 and PDC6) catalyse the decarboxylation of pyruvate to acetaldehyde, which is a competing pathway for the accumulation of PEP and E4P. Disruption of PDC1 resulted in ˜ 30% lower total pyruvate decarboxylase activity and increased the production of aromatic compound vanillin in S. cerevisiae (Brochado et al., 2010). We disrupted PDC1 in strain LYTY6, generating strain LYTY12, with a yield of 338.72 ± 0.1 mg l−1. As chorismate serves as a precursor to the biosynthesis of l‐tryptophan, l‐phenylalanine and l‐tyrosine, TRP2 and PHA2 were knocked out separately in strain LYTY12 (Fig. 1), yielding strains LYTY13 and LYTY14 respectively. Strain LYTY14 accumulated 522.39 ± 0.99 mg l−1 tyrosol, which was 54.2% greater than that of strain LYTY12. However, unexpectedly, tyrosol titre was 19.7% less in strain LYTY13 than that of strain LYTY12 (Fig. 3), which could be due to significant growth deficiency caused by the TRP2 deletion (Fig. S1).
Fig. 3.
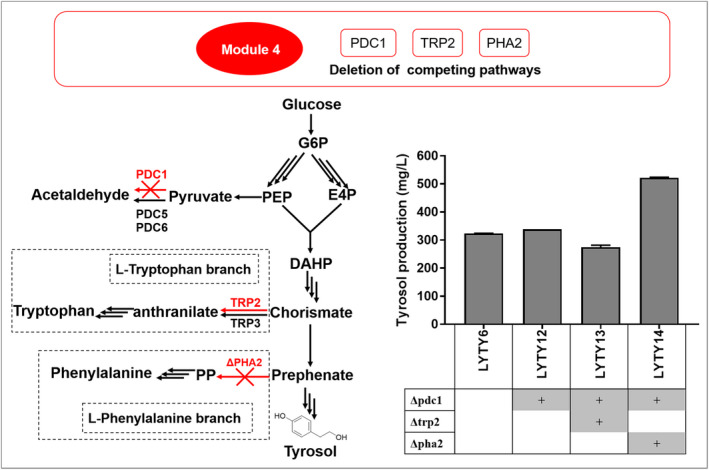
Effect of PDC1, PHA2 and TRP2 deletion on tyrosol production. The left schematic represents the competing pathways. The right pane represents the tyrosol production of the modified strains. Titre of tyrosol was produced after 72 h fermentation in 50 ml YPD medium. Error bars represented the standard deviation from three replicates.
Optimization of tyrosol glycosylation for salidroside production
According to previous studies, salidroside and tyrosol production are positively correlated (Jiang et al., 2018; Liu et al., 2018); therefore, the key aim in this study was to improve the salidroside yield through increasing tyrosol yield. Previous studies have also shown that the addition of a copy of ARO4K229L/ARO7G141S/ARO3K222L under a strongconstitutive promoter led to increased aromatic compounds production (Rodriguez et al., 2015; Reifenrath and Boles, 2018). To further increase tyrosol production, a copy of ARO4K229L/ARO7G141S or ARO4K229L/ARO7G141S/ARO3K222L with strong constitutive promoters (ARO4K229L under TDH3 promoter, ARO7G141S under TEF2 promoter and ARO3K222L under HSP26 promoter) was introduced into LYTY14. However, no viable clones were obtained using this approach. On the other hand, by integrating ARO4K229L/ARO7G141S or ARO4K229L/ARO7G141S/ARO3K222L with native promoters into 308a locus (Reider Apel et al., 2017) of LYTY14, the tyrosol titres of 653.79 ± 0.06 mg l−1 (strain LYTY15) or 702.30 ± 0.41 mg l−1 (strain LYTY16) were obtained, respectively, after 72 h of shake flask cultivation in YPD medium.
Salidroside is produced by the glycosylation of tyrosol at its 8‐OH group, which is catalysed by regio‐specific uridine 5'‐diphospho‐glucosyltransferases (UGTs). The RsUGT72B14 and RrU8GT33 from Rhodiola sachalinensis and Rhodiola rosea, respectively, showed high catalytic efficiencies for salidroside synthesis (Yu et al., 2011; Torrens‐Spence et al., 2018). The glycosyltransferase BsYjiC from Bacillus subtilis 168 performs robust glycosylation on a variety of glycosides (Dai et al., 2017). Here, codon‐optimized RsUGT72B14opt , RrU8GT33opt and BsYjiCopt under the control of a strong constitutive TEF1 promoter were expressed in strain LYTY16, showing more than 20‐fold higher transcription levels compared to that of the housekeeping gene ALG9 (Fig. S2B). Strain SAL3 expressing RrU8GT33opt produced the highest salidroside titre of 1575.45 ± 19.35 mg l−1 and presented a conversion rate of approximately 95% (Fig. 4). UDP‐glucose is a substrate for tyrosol glycosylation. A previous study showed that overexpression of PGM1 and UGP1 together can increase the UDP‐glucose accumulation (Zhuang et al., 2017). Thus, PGM1 and UPG1 genes were overexpressed in strain SAL3 under HSP26 and RAL3 promoters, respectively, yielding strain SAL4. The transcription levels of PGM1 and UPG1 genes in strain SAL4 are 1.5‐fold higher than those of the WT strain (Fig. S2). However, this strategy led to a salidroside titre of 1560.34 ± 5.89 mg l−1, which was not significantly different to that of strain SAL3. To our knowledge, this is the highest titre of salidroside reported for shake flask cultures.
Fig. 4.
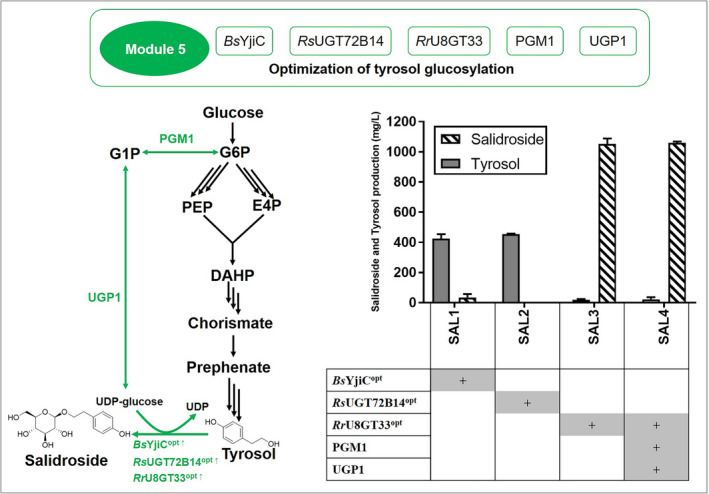
Production of salidroside (striated bars) and tyrosol (grey bars) after overexpression of different UGTs and native PGM1 and UPG 1. The left schematic represents the biosynthesis of salidroside through optimization of tyrosol glycosylation. BsYjiC from Bacillus subtilis 168, RsUGT72B14 from Rhodiola sachalinensis and RrU8GT33 from Rhodiola rosea were codon optimized for salidroside synthesis. The right pane represents the salidroside and tyrosol production of the modified strains. The strains were grown in 50 ml YPD medium in shaker flasks at 220 rpm, 30°C. Titres of tyrosol and salidroside were detected after 72 h fermentation. Standard deviations are based on three biological duplicates.
Production of Tyrosol and Salidroside in Fed‐Batch Fermentation
Until this stage, strain LYTY16 had produced the highest tyrosol titres; therefore, it was used to further promote tyrosol production in a 5‐L fed‐batch bioreactor. Similarly, strain SAL3 was used for salidroside production by fed‐batch fermentation. After the initial glucose was consumed, a carbon restriction strategy was applied to glucose supplementation. The glucose concentration was controlled at 0.5–5 g l−1 by adjusting the feeding rate. As shown in Fig. 5A, the cell density (OD600) of strain LYTY16 reached approximately 93.4 (the biomass reached about 36.05 g l−1) after 168 h cultivation, and the tyrosol titre consistently increased and finally reached 9.90 ± 0.06 g l−1. The maximal biomass reached about 42.32 g l−1 (OD600 ≈ 109.6) of strain SAL3 was achieved after 168 h cultivation and the production of salidroside reached up to 26.55 ± 0.43 g l−1. In addition, 2.12 ± 0.06 g l−1 tyrosol was produced during the fermentation of strain SAL3 after 168 h (Fig. 5B).
Fig. 5.
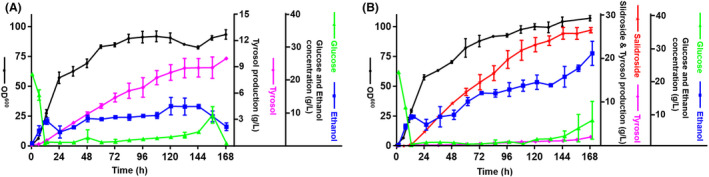
Fed‐batch fermentation for tyrosol and salidroside production. (A) Tyrosol production in 5‐l fed‐batch bioreactors using engineered strain LYTY16. (B) Salidroside production (red line) of strain SAL3 in 5‐l fed‐batch bioreactors. The accumulation of low tyrosol titre (purple line) was observed during fed‐batch fermentation. Purple lines indicated tyrosol production, red lines indicated salidroside production, black line showed cell densities (OD600), blue lines represented the ethanol production and green lines represented concentration of glucose. The error bars are represented by the standard deviation from two biological replicates.
Discussion
Previous studies on tyrosol production in S. cerevisiae have mainly focused on the introduction of heterologous pathways, such as plant‐derived pathways and bacteria‐derived pathways (Jiang et al., 2018; Guo et al., 2019; Guo et al., 2020). Particularly, Guo (Guo et al., 2020) recently increased tyrosol titre ˜ 15‐fold compared to that of the WT strain in a haploid industrial strain S. cerevisiae strain HLF‐Dα through introduction of the Xfpk‐based pathway, disruption of endogenous genes (PDC1, PHA2 and TRP3) and introduction of heterogenous genes (PcAAS and EcTyrAM53I/A354V ). In our work, the tyrosol‐producing capability of S. cerevisiae itself was highly inspired. All modifications in our final tyrosol‐producing strain LYTY16 were based on endogenous genes, containing overexpression of endogenous genes TKL1, RKI1, ARO2, ARO10, ARO4K229L , ARO7G141S and ARO3K222L and disruption of PDC1 and PHA2. As shown in Fig. S2A, the transcription levels of these overexpressed genes are at least 2‐fold higher compared to those of the WT strain. Finally, tyrosol titres of 702.30 ± 0.41 mg l−1 were achieved in an engineered S. cerevisiae strain, which were 26‐fold higher than those of the WT strain. With bioprocess engineering, the final strain LYTY16 produced 9.90 ± 0.06 g l−1 tyrosol, which highlights this as a promising platform for further industrial applications.
In our work, in suit mutation of Aro4pK229L and Aro7pG141S in S. cerevisiae led to 3‐fold increase in tyrosol production. In combination with rewiring the glycolysis pathway and pentose phosphate pathway through overexpression of Tki1p and Rki1p, the titre of tyrosol was 7‐fold compared to that of the WT strain (Fig. 2A). To further increase the tyrosol production and to simplify the fermentation conditions, constitutive promoters were given priority rather than inducible promoters. At first time, we tried several times to overexpress the ARO4K229L /ARO7G141S or ARO4K229L /ARO7G141S /ARO3K222L under the control of strong constitutive promoters (ARO4K229L under TDH3 promoter, ARO7G141S under TEF2 promoter and ARO3K222L under HSP26 promoter) in the engineered strain LYTY14. Unfortunately, no viable clones were obtained. A possible explanation for this might be that these manipulations down‐regulated some transporter genes. Finally, another copy of feedback‐inhibition‐insensitive DAHP synthase and chorismate mutase (Aro4pK229L/Aro7pG141S, or Aro4pK229L/Aro7pG141S/Aro3pK222L) was integrated into LYTY14, resulting in about 30% increase in tyrosol production. These results were consistent with previous reports, where overexpression of Aro4pK229L, Aro7pG141S and Aro3pK222L unlocked the production of aromatic amino acid derivates (Luttik et al., 2008; Bruckner et al., 2018).
It was reported that the overexpression of ARO1 or AROL gene could improve the production of shikimate pathway‐derived products in S. cerevisiae. For example, p‐coumaric acid production increased ˜ 50% compared to that of a control strain when ARO1 or AROL were overexpressed (Rodriguez et al., 2015); para‐hydroxybenzoic acid production also increased ˜ 13% compared to that of the control strain due to AROL overexpression (Averesch et al., 2017). However, when the ARO1 or AROL gene was overexpressed in the engineered strain LYTY2 in this study, the transcription level of ARO1 was more than 20‐fold compared to that of the WT strain (Fig. S2A) and the transcription level of AROL was 3.5‐fold higher than that of the housekeeping gene ALG9 (Fig. S2B), but the production of tyrosol decreased slightly (Fig. 2C). It seems that the step from DAHP to EPSP may not be a rate‐limiting step of the tyrosol biosynthetic pathway in S. cerevisiae.
Early engineering of the l‐tyrosine branch for tyrosol production in S. cerevisiae was performed via a combination with the plant‐derived pathway (Jiang et al., 2018; Guo et al., 2019; Guo et al., 2020). In this work, the effect of overexpressing endogenous genes and plant‐derived genes on tyrosol production was compared. As a result, the overexpression of endogenous ARO10 led to higher titres of tyrosol than overexpressing codon‐optimized 4HPAASopt from Rhodiola rosea or co‐overexpression of ARO10 and 4HPAASopt (Fig. 2). We hypothesized that this result might be attributed to the shortage of l‐tyrosine after the combinational overexpression strategy had been conducted. This also indicates that the conversion from 4‐HPP to 4‐HPAA in S. cerevisiae was a rate‐limiting step in tyrosol biosynthesis.
The deletion of PDC1 led to an approximate 5% increase in the tyrosol titre in strains LYTY6, whereas the additional deletion PHA2 increased above 50% the titre of tyrosol (Fig. 4). Although the deletion of PHA2 was thought to introduce phenylalanine auxotrophy in S. cerevisiae (Urrestarazu et al., 1998), our pha2 disrupting strains did not result in growth deficiency by culturing in YPD medium. On the contrary, although supplementing enough extra tryptophan (20 mg l−1) to the fermentation medium, deletion of TRP2 resulted in an obvious growth defect and low tyrosol production (Fig. S1).
Recent studies (Jiang et al., 2018; Guo et al., 2020) showed that glucosyltransferase UGT85a1 from Arabidopsis thaliana was used to transform tyrosol to salidroside in S. cerevisiae. Nevertheless, in those studies, less than 25% of tyrosol was glycosylated into salidroside through expressing UGT85a1. We screened several reported UGTs, and discovered the best performer, RrU8GT33 from Rhodiola rosea. This strain, achieved 95% and 80% conversion of tyrosol into salidroside in shake flasks and fed‐batch fermentation respectively. Finally, in 5‐l fed‐batch bioreactors, salidroside over‐producing strain SAL3 produced 26.55 ± 0.43 g l−1 salidroside, which to our knowledge is the highest level of salidroside production via microbial biosynthesis.
Conclusions
In summary, this study demonstrated that high titres of tyrosol and salidroside can be produced in a widely used model organism S. cerevisiae CEN.PK2‐1C and the multi‐modular engineering strategy increased the tyrosol titre ˜ 26‐fold over the initial strain CEN.PK2‐1C. Two best performing strains LYTY16 and SAL3 produced tyrosol and salidroside at titres of 702.30 ± 0.41 and 1575.45 ± 19.35 mg l−1, respectively, in shake flasks. The strains have the capacity to produce tyrosol and salidroside at titres of 9.90 ± 0.06 and 26.55 ± 0.43 g l−1, respectively, in 5 l fed‐batch bioreactors. To our knowledge, these are the highest levels of tyrosol or salidroside production to date in microbial cell factories.
Experimental procedures
Strains and plasmids
Saccharomyces cerevisiae strain CEN.PK2‐1C was used for genetic manipulations. All engineered strains used in this work are listed in Table 1. E. coil strains DH5α and DH10β were used for sub‐cloning. Plasmids used in this study are listed in Table S1.
Table 1.
Strains used in this study.
| Strains | Characteristics |
|---|---|
| BY4741α | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| CEN.PK2‐1C | MAT a ura3‐52 his3‐Δ1 leu2‐3_112 trp1‐289, MAL2‐8c SUC2 |
| LYTY1 | CEN.PK2‐1C ARO4K229L ARO7G141S |
| LYTY1R | LYTY1 ZWF1 :: P TEF1 ‐RKI1‐T PGK1 TRP1 |
| LYTY2 | LYTY1 ZWF1 :: P HXT7 ‐TKL1‐T ADH1 ‐P TEF1 ‐RKI1‐T PGK1 TRP1 |
| LYTY3 | LYTY2 1622b :: P ENO2 ‐AROL‐T TEF1 |
| LYTY4 | LYTY2 URA3 :: P ENO2 ‐ARO1‐T TEF1 |
| LYTY5 | LYTY2 URA3 :: P PGK1 ‐ARO2‐T GPD |
| LYTY6 | LYTY2 URA3 :: P PGK1 ‐ARO2‐T GPD ‐P TEF1 ‐ARO10‐T PGK1 |
| LYTY7 | LYTY2 URA3 :: P PGK1 ‐ARO2‐T GPD ‐P TEF1 ‐4HPAASopt ‐T PGK1 |
| LYTY8 | LYTY2 URA3 :: P PGK1 ‐ARO2‐T GPD ‐P CCW12 ‐ARO10‐T ADH1 ‐P TEF1 ‐4HPAASopt ‐T PGK1 |
| LYTY9 | LYTY2 URA3 :: P PGK1 ‐ARO2‐T GPD ‐P CCW12 ‐ARO10‐T ADH1 ‐ P ENO2 ‐ARO1‐T TEF1 |
| LYTY10 | LYTY2 URA3 :: P PGK1 ‐ARO2‐T GPD ‐P CCW12 ‐ARO10‐T ADH1 ‐P TEF1 ‐4HPAASopt ‐T PGK1 ‐P ENO2 ‐ARO1‐T TEF1 |
| LYTY11 | LYTY6 1622b :: P TPI ‐4HPAR1opt ‐T PGK1 |
| LYTY12 | LYTY6 Δpdc1 |
| LYTY13 | LYTY6 Δpdc1 Δtrp2 |
| LYTY14 | LYTY6 Δpdc1 Δpha2 |
| LYTY15 | LYTY6 Δpdc1 Δpha2 308a :: P ARO4 ‐ARO4K229L ‐T ARO4 ‐P ARO7 ‐ARO7G141S ‐T ARO7 |
| LYTY16 | LYTY6 Δpdc1 Δpha2 308a :: P ARO4 ‐ARO4K229L ‐T ARO4 ‐P ARO7 ‐ARO7G141S ‐T ARO7 ‐P ARO3 ‐ARO3K222L ‐T ARO3 |
| SAL1 | LYTY16 LEU2 :: P TEF1 ‐YjiCopt ‐T PGK1 |
| SAL2 | LYTY16 LEU2 :: P TEF1 ‐UGT72B14opt ‐T PGK1 |
| SAL3 | LYTY16 LEU2 :: P TEF1 ‐RrU8GT33opt ‐T PGK1 |
| SAL4 | LYTY16 LEU2 :: P TEF1 ‐RrU8GT33opt ‐T PGK1 ‐P HSP26 ‐PGM1‐T TDH2 ‐P RAL3 ‐UPG1‐T ENO2 |
All native genes (TKL1, RKI1, ARO1, ARO2, ARO10, ARO3, ARO4, ARO7, UPG1, PGM1) were amplified from CEN.PK2‐1C genomic DNA. The AROL gene was amplified from the genomic DNA of E. coil DH5α. Codon‐optimized RrU8GT33, BsYjiC, RsUGT72B14, 4HPAR1 and 4HPAAS were synthesized by GENEWIZ, China. All promoters and terminators were amplified from S. cerevisiae BY4741 genomic DNA except for ARO4‐promoter, ARO4‐terminator, ARO3‐promoter, ARO3‐terminater, ARO7‐promoter and ARO7‐terminator, which were amplified from CEN.PK2‐1C genomic DNA. All primers used in this study are listed in Table S2.
DNA manipulations
All native or synthetic genes were first cloned to helper plasmids and then assembled to integrated plasmids (PRS404, PRS405 and PRS406) or PUC19 plasmid to generate integration‐cassette plasmids and donor plasmids respectively. First, vector backbone PRS423 was linearized using restriction enzymes EcoRI‐HF and BamHI‐HF (New England Biolabs, US). Adjacent promoters and terminators with the restriction site PmeI were cloned into the linearized PRS423 vector, which generated helper plasmids (Table S1). Then, amplified gene fragments were assembled to helper plasmids (digested by PmeI), yielding gene‐cassette plasmids. Integrated plasmids and donor plasmids were constructed with Gibson assembly by adding corresponding gene‐cassette fragments. The pCas plasmid (Ryan et al., 2014) was modified by introducing two BsaI sites between HDV ribozyme and sgRNA to generate the pCas‐BsaI plasmid. All Cas9‐gRNA plasmids were constructed using the Golden Gate assembly method (Engler et al., 2009).
Strains construction
All integrated plasmids harbouring gene‐overexpression cassette were integrated into chromosomal loci. To integrate plasmid pTYR1 into ZWF1 loci, pTYR1 was linearized using the restriction enzyme BamHI‐HF. One microgram of the purified linearized fragment was transformed into yeast using the LiAc/ssDNA/PEG method (Gietz and Schiestl, 2007), and then, yeast were plated on a SC‐TRP plate. The integration of other fragments was similar to the integration of linearized pTYR1 except transformants were plated on SC‐LEU or SC‐URA for selection. Gene‐overexpression cassettes of AROL, 4HPAR1opt , ARO4K229L/ARO7G141S and ARO4K229L/ARO7G141S/ARO3K222L were integrated into specific S. cerevisiae chromosomal loci (308a locus or 1622b locus) using a Cas9‐based toolkit (Reider Apel et al., 2017). Clones were screened using PCR on yeast colonies with KOD FX PCR mix (Toyobo, Osaka, Japan), which were then verified by DNA sequencing. Deletion of PHA2, PDC1and TRP2 was performed using the pCas system (Ryan et al., 2014; Table S2). The specific guide RNA sequences were designed using the CHOPCHOP web tool (http://crispor.tefor.net; Labun et al., 2016). All gRNA sequences used in this study are listed in Table S3.
Cultivations and fermentations
Yeast strains were picked from pre‐cultured plates and cultured in 2 ml YPD medium (20 g l−1 Peptone, bacteriological (Sangon Biotech﹟A100636), 10 g l−1 yeast extract and 20 g l−1 dextrose) at 30°C, shaking at 220 rpm for 18 h. Portions of pre‐cultured mixture were then transferred into 50 ml YPD medium in 250 ml shake flasks until the OD600 value reached approximately 0.2. The strains were cultured at 30°C, shaking at 220 rpm for 72 h. To monitor cell growth, samples fermented for various lengths of time were measured at 600 nm using a spectrophotometer. Samples of 500 μl were collected after 72 h and centrifuged at 5000 rpm for 5 min. The supernatant was stored at −20°C until analysis.
Fed‐batch fermentation for tyrosol and salidroside production
Strain LYTY16 was used to produce tyrosol and strain SAL3 was used to produce salidroside. Seed cultures of 250 ml were transferred into 5 l bioreactors (BaiLun, China) containing 2.25 l fed‐batch medium (20 g l−1 peptone, 20 g l−1 yeast extract and 20 g l−1 glucose) when the OD600 of seed cultures reached 5. Fermentation was performed at 30°C; pH was maintained at 5.7 by automated addition of 5 M NaOH. Air flow was set at 2.5 vvm (air volume/working volume min−1) and the dissolved oxygen (dO2) concentration was controlled above 40% saturation by agitation cascade (400–600 rpm). Glucose solution (600 g l−1) was fed periodically into the fermentation system to maintain the glucose concentration under 5.0 g l−1. To maintain the glucose concentration under 5.0 g l−1, the glucose concentration was monitored every 4 h. After the initial glucose was depleted, the glucose was added to the fermenter at a rate of 3 g l−1 h−1 (initiated at 9.5 h). The supplementary rate of glucose was adjusted to ˜0.6 g l−1 h−1 at 32 h and this rate was maintained to the end of the fermentation. Yeast extract (125 ml, 500 g l−1) was added into the bioreactor at 8, 16, 24 and 32 h time points. Cell density, glucose and ethanol concentration were constantly monitored during the fermentation process. One millilitre supernatant of fermentation cultures was stored every four hours at −20°C until analysis.
Analytical methods
Samples were analysed using an Agilent HPLC 1260 series instrument equipped with a Zorbax SB‐C18 column (Agilent, 5 μm, 4.6 mm × 250 mm), which was maintained at 30°C. Mobile phase A was 0.05% formic acid in water and solvent D was acetonitrile; the flow rate was 1 ml min−1. The gradient profile was as follows: 10% D (0–20 min), then 5% D from 20 min to 25 min, then a linear gradient from 5% D to 95% D (25–35 min), then hold 95% D for 5 min, finally return to 10% D for 10 min. Tyrosol and salidroside absorbance was detected at 224 nm. Tyrosol (Solarbio, China) and salidroside (Solarbio, China) standards were added to culture medium and used to generate standard curves for metabolite quantification. The R2 coefficient for the calibration curve was higher than 0.999. At least three biological replicates were analysed for each strain.
Conflict of interest
The authors have declared the competing financial interests in the form of a pending patent application.
Author contributions
Huayi Liu, Wenhai Xiao and Yunzi Luo designed the experiments. Huayi Liu, Yujuan Tian, Yi Zhou, Yeyi Kan and Tingting Wu performed the experiments. Huayi Liu and Yunzi Luo wrote the manuscript.
Supporting information
Fig. S1. (A) Deletion of TRP2 decreased the cell growth rate of the engineered strain LYTY12. (B). The tyrosol production after adding 20 mg l−1 extra tryptophan to the fermentation medium in strain LYTY13. The strains were cultured in shake flasks at 220 rpm, 30°C with YPD medium, or YPD medium with additional 20 mg l−1 tryptophan. The cell densities were monitored with the spectrophotometer at 600 nm wavelength. Titers of tyrosol were detected after 72 h fermentation. Error bars represented the standard deviation from three replicates.
Fig. S2. (A) The transcription levels of endogenous genes compared to those of the WT strains. (B) The transcription levels of heterogenous genes compared to that of the housekeeping gene ALG9. Strains were cultured for 24 h before RNA extraction. The transcription levels of those genes were measured with RT‐qPCR, and the housekeeping gene ALG9 was selected as the reference. The expression level of ALG9 was set as 1. Error bars represent the standard deviation of three biological replicates.
Table S1. Plasmids used in this study.
Table S2. Primers used in this study.
Table S3. List of the gRNA sequences used for gene deletions, mutations, and insertions via the CRISPR system.
Microbial Biotechnology (2021) 14(6), 2605–2616
Funding information
We gratefully acknowledge the financial supports from the National Key R&D Program of China (Grant No. 2018YFA0903300), and the Natural Science Foundation of Tianjin City (Grant No. 19JCYBJC24200).
References
- Ahn, E.‐Y. , Jiang, Y. , Zhang, Y. , Son, E. , You, S. , Kang, S.‐W. , et al. (2008) Cytotoxicity of p‐tyrosol and its derivatives may correlate with the inhibition of DNA replication initiation. Oncol Rep 19: 527–534. [PubMed] [Google Scholar]
- Averesch, N.J.H. , Prima, A. , and Kromer, J.O. (2017) Enhanced production of para‐hydroxybenzoic acid by genetically engineered Saccharomyces cerevisiae . Bioprocess Biosyst Eng 40: 1283–1289. [DOI] [PubMed] [Google Scholar]
- Bai, Y. , Bi, H. , Zhuang, Y. , Liu, C. , Cai, T. , Liu, X. , et al. (2014) Production of salidroside in metabolically engineered Escherichia coli . Sci Rep 4: 6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, Z. , Xiao, H. , Liang, J. , Zhang, L. , Xiong, X. , Sun, N. , et al. (2015) Homology‐integrated CRISPR‐Cas (HI‐CRISPR) system for one‐step multigene disruption in Saccharomyces cerevisiae . ACS Synth Biol 4: 585–594. [DOI] [PubMed] [Google Scholar]
- Borodina, I. , and Nielsen, J. (2014) Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol J 9: 609–620. [DOI] [PubMed] [Google Scholar]
- Brochado, A.R. , Matos, C. , Moller, B.L. , Hansen, J. , Mortensen, U.H. , and Patil, K.R. (2010) Improved vanillin production in baker's yeast through in silico design. Microb Cell Fact 9: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner, C. , Oreb, M. , Kunze, G. , Boles, E. , and Tripp, J. (2018) An expanded enzyme toolbox for production of cis, cis‐muconic acid and other shikimate pathway derivatives in Saccharomyces cerevisiae. FEMS Yeast Research 18. 10.1093/femsyr/foy017 [DOI] [PubMed] [Google Scholar]
- Chen, T. , Ma, Z. , Zhu, L. , Jiang, W. , Wei, T. , Zhou, R. , et al. (2016a) Suppressing receptor‐interacting protein 140: a new sight for salidroside to treat cerebral Ischemia. Mol Neurobiol 53: 6240–6250. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Wu, Y. , Yang, T. , Wei, M. , Wang, Y. , Deng, X. , et al. (2016b) Salidroside alleviates cachexia symptoms in mouse models of cancer cachexia via activating mTOR signalling. J Cachexia Sarcopenia Muscle 7: 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S.L. , Yu, H. , Luo, H.M. , Wu, Q. , Li, C.F. , and Steinmetz, A. (2016c) Conservation and sustainable use of medicinal plants: problems, progress, and prospects. Chin Med 11: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, D. , Kim, S.Y. , and Ahn, J.H. (2017) Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli . Sci Rep 7: 2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, L. , Li, J. , Yao, P. , Zhu, Y. , Men, Y. , Zeng, Y. , et al. (2017) Exploiting the aglycon promiscuity of glycosyltransferase Bs‐YjiC from Bacillus subtilis and its application in synthesis of glycosides. J Biotechnol 248: 69–76. [DOI] [PubMed] [Google Scholar]
- Delépée, R. , Berteina‐Raboin, S. , Lafosse, M. , Lamy, C. , Darnault, S. , Renimel, I. , et al. (2007) Synthesis, purification, and activity of salidroside. J Liq Chromatogr Relat Technol 30: 2069–2080. [Google Scholar]
- Engler, C. , Gruetzner, R. , Kandzia, R. , and Marillonnet, S. (2009) Golden gate shuffling: a one‐pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, B. , Chen, T. , Zhang, S. , Wu, B. , and He, B. (2017) Mining of efficient microbial UDP‐glycosyltransferases by motif evolution cross plant kingdom for application in biosynthesis of salidroside. Sci Rep 7: 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galambosi, B. (2006) Demand and availability of Rhodiola rosea raw material. In Medicinal and aromatic plants: agricultural, commercial, ecological, legal, pharmacological and social aspects. Bogers, R.J. , Craker, L.E. , and Lange, D. (eds). Dordrecht, Netherlands: Springer Netherlands, pp. 223–236. [Google Scholar]
- Gietz, R.D. , and Schiestl, R.H. (2007) High‐efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2: 31–34. [DOI] [PubMed] [Google Scholar]
- Guo, W. , Huang, Q. , Feng, Y. , Tan, T. , Niu, S. , Hou, S. , et al. (2020) Rewiring central carbon metabolism for tyrosol and salidroside production in Saccharomyces cerevisiae. Biotechnology and Bioengineering 117: 2410–2419. 10.1002/bit.27370. [DOI] [PubMed] [Google Scholar]
- Guo, W. , Huang, Q. , Liu, H. , Hou, S. , Niu, S. , Jiang, Y. , et al. (2019) Rational engineering of chorismate‐related pathways in Saccharomyces cerevisiae for improving tyrosol production. Front Bioeng Biotechnol 7: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelwood, L.A. , Daran, J.M. , van Maris, A.J. , Pronk, J.T. , and Dickinson, J.R. (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74: 2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. , Yin, H. , Wang, S. , Zhuang, Y. , Liu, S. , Liu, T. , and Ma, Y. (2018) Metabolic engineering of Saccharomyces cerevisiae for high‐level production of salidroside from glucose. J Agric Food Chem 66: 4431–4438. [DOI] [PubMed] [Google Scholar]
- Konig, V. , Pfeil, A. , Braus, G.H. , and Schneider, T.R. (2004) Substrate and metal complexes of 3‐deoxy‐D‐arabino‐heptulosonate‐7‐phosphate synthase from Saccharomyces cerevisiae provide new insights into the catalytic mechanism. J Mol Biol 337: 675–690. [DOI] [PubMed] [Google Scholar]
- Labun, K. , Montague, T.G. , Gagnon, J.A. , Thyme, S.B. , and Valen, E. (2016) CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 44: W272–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.W. , Na, D. , Park, J.M. , Lee, J. , Choi, S. , and Lee, S.Y. (2012) Systems metabolic engineering of microorganisms for natural and non‐natural chemicals. Nat Chem Biol 8: 536–546. [DOI] [PubMed] [Google Scholar]
- Lian, J. , Mishra, S. , and Zhao, H. (2018) Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications. Metab Eng 50: 85–108. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Li, X.B. , Jiang, J. , Liu, Z.N. , Qiao, B. , Li, F.F. , et al. (2018) Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab Eng 47: 243–253. [DOI] [PubMed] [Google Scholar]
- Luttik, M.A. , Vuralhan, Z. , Suir, E. , Braus, G.H. , Pronk, J.T. , and Daran, J.M. (2008) Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab Eng 10: 141–153. [DOI] [PubMed] [Google Scholar]
- Mao, Y. , Li, Y. , and Yao, N. (2007) Simultaneous determination of salidroside and tyrosol in extracts of Rhodiola L. by microwave assisted extraction and high‐performance liquid chromatography. J Pharm Biomed Anal 45: 510–515. [DOI] [PubMed] [Google Scholar]
- Mao, G.X. , Xing, W.M. , Wen, X.L. , Jia, B.B. , Yang, Z.X. , Wang, Y.Z. , et al. (2015) Salidroside protects against premature senescence induced by ultraviolet B irradiation in human dermal fibroblasts. Int J Cosmet Sci 37: 321–328. [DOI] [PubMed] [Google Scholar]
- Miralles, P. , Chisvert, A. , and Salvador, A. (2015) Determination of hydroxytyrosol and tyrosol by liquid chromatography for the quality control of cosmetic products based on olive extracts. J Pharm Biomed Anal 102: 157–161. [DOI] [PubMed] [Google Scholar]
- Moyo, M. , Aremu, A.O. , and Van Staden, J. (2015) Medicinal plants: an invaluable, dwindling resource in sub‐Saharan Africa. J Ethnopharmacol 174: 595–606. [DOI] [PubMed] [Google Scholar]
- Paddon, C.J. , and Keasling, J.D. (2014) Semi‐synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol 12: 355–367. [DOI] [PubMed] [Google Scholar]
- Reider Apel, A. , d'Espaux, L. , Wehrs, M. , Sachs, D. , Li, R.A. , Tong, G.J. , et al. (2017) A Cas9‐based toolkit to program gene expression in Saccharomyces cerevisiae . Nucleic Acids Res 45: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenrath, M. , and Boles, E. (2018) Engineering of hydroxymandelate synthases and the aromatic amino acid pathway enables de novo biosynthesis of mandelic and 4‐hydroxymandelic acid with Saccharomyces cerevisiae . Metab Eng 45: 246–254. [DOI] [PubMed] [Google Scholar]
- Rodriguez, A. , Kildegaard, K.R. , Li, M. , Borodina, I. , and Nielsen, J. (2015) Establishment of a yeast platform strain for production of p‐coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab Eng 31: 181–188. [DOI] [PubMed] [Google Scholar]
- Ryan, O.W. , Skerker, J.M. , Maurer, M.J. , Li, X. , Tsai, J.C. , Poddar, S. , et al. (2014) Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife 3: e03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, Y. , Tajima, K. , Munekata, M. , Keasling, J.D. , and Lee, T.S. (2012) Engineering of a tyrosol‐producing pathway, utilizing simple sugar and the central metabolic tyrosine, in Escherichia coli . J Agric Food Chem 60: 979–984. [DOI] [PubMed] [Google Scholar]
- Schenck, C.A. , and Maeda, H.A. (2018) Tyrosine biosynthesis, metabolism, and catabolism in plants. Phytochemistry 149: 82–102. [DOI] [PubMed] [Google Scholar]
- Sewalt, V. , Shanahan, D. , Gregg, L. , La Marta, J. , and Carrillo, R. (2016) The generally recognized as safe (GRAS) process for industrial microbial enzymes. Ind Biotechnol 12: 295–302. [Google Scholar]
- Suastegui, M. , Guo, W. , Feng, X. , and Shao, Z. (2016) Investigating strain dependency in the production of aromatic compounds in Saccharomyces cerevisiae . Biotechnol Bioeng 113: 2676–2685. [DOI] [PubMed] [Google Scholar]
- Suastegui, M. , and Shao, Z. (2016) Yeast factories for the production of aromatic compounds: from building blocks to plant secondary metabolites. J Ind Microbiol Biotechnol 43: 1611–1624. [DOI] [PubMed] [Google Scholar]
- Suastegui, M. , Yu Ng, C. , Chowdhury, A. , Sun, W. , Cao, M. , House, E. , et al. (2017) Multilevel engineering of the upstream module of aromatic amino acid biosynthesis in Saccharomyces cerevisiae for high production of polymer and drug precursors. Metab Eng 42: 134–144. [DOI] [PubMed] [Google Scholar]
- Tang, Y. , Vater, C. , Jacobi, A. , Liebers, C. , Zou, X. , and Stiehler, M. (2014) Salidroside exerts angiogenic and cytoprotective effects on human bone marrow‐derived endothelial progenitor cells via Akt/mTOR/p70S6K and MAPK signalling pathways. Br J Pharmacol 171: 2440–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrens‐Spence, M.P. , Pluskal, T. , Li, F.S. , Carballo, V. , and Weng, J.K. (2018) Complete pathway elucidation and heterologous reconstitution of Rhodiola salidroside biosynthesis. Mol Plant 11: 205–217. [DOI] [PubMed] [Google Scholar]
- Trenchard, I.J. , Siddiqui, M.S. , Thodey, K. , and Smolke, C.D. (2015) De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab Eng 31: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troshchenko, A.T. , and Yuodvirshis, A.M. (1969) Synthesis of glycosides of 2‐(p‐hydroxyphenyl)ethanol(tyrosol). Chem Nat Compd 5: 217–220. [Google Scholar]
- Urrestarazu, A. , Vissers, S. , Iraqui, I. , and Grenson, M. (1998) Phenylalanine‐ and tyrosine‐auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol Gen Genet 257: 230–237. [DOI] [PubMed] [Google Scholar]
- Woodburn, H.M. , and Stuntz, C.F. (1950) The synthesis of β‐(3‐amino‐4‐hydroxyphenyl)‐ethanol; 3‐aminotyrosol. J Am Chem Soc 72: 1361–1364. [Google Scholar]
- Xu, K. , Qin, L. , Bai, W. , Wang, X. , Li, F. , Ren, S. , et al. (2020) Multilevel Defense System (MDS) relieves multiple stresses for economically boosting ethanol production of industrial Saccharomyces cerevisiae . ACS Energy Lett 5: 572–582. [Google Scholar]
- Xue, Y. , Chen, X. , Yang, C. , Chang, J. , Shen, W. , and Fan, Y. (2017) Engineering Eschericha coli for enhanced tyrosol production. J Agric Food Chem 65: 4708–4714. [DOI] [PubMed] [Google Scholar]
- Xue, Z. , and Yang, B. (2016) Phenylethanoid glycosides: research advances in their phytochemistry, pharmacological activity and pharmacokinetics. Molecules 21: 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Xue, Y. , Yang, C. , Shen, W. , Fan, Y. , and Chen, X. (2019) Modular engineering of tyrosol production in Escherichia coli . J Agric Food Chem 67: 3900–3908. [DOI] [PubMed] [Google Scholar]
- Yu, H.S. , Ma, L.Q. , Zhang, J.X. , Shi, G.L. , Hu, Y.H. , and Wang, Y.N. (2011) Characterization of glycosyltransferases responsible for salidroside biosynthesis in Rhodiola sachalinensis . Phytochemistry 72: 862–870. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Du, Q. , Yang, Y. , Wang, J. , Liu, Y. , Zhao, Z. , et al. (2018) Salidroside alleviates ischemic brain injury in mice with ischemic stroke through regulating BDNK mediated PI3K/Akt pathway. Biochem Pharmacol 156: 99–108. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Kasim, V. , Xie, Y.D. , Huang, C. , Sisjayawan, J. , Dwi Ariyanti, A. , et al. (2017a) Inhibition of PHD3 by salidroside promotes neovascularization through cell‐cell communications mediated by muscle‐secreted angiogenic factors. Sci Rep 7: 43935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Zhang, K. , Mehmood, M.A. , Zhao, Z.K. , Bai, F. , and Zhao, X. (2017b) Deletion of acetate transporter gene ADY2 improved tolerance of Saccharomyces cerevisiae against multiple stresses and enhanced ethanol production in the presence of acetic acid. Bioresour Technol 245: 1461–1468. [DOI] [PubMed] [Google Scholar]
- Zhang, M.M. , Zhao, X.Q. , Cheng, C. , and Bai, F.W. (2015) Improved growth and ethanol fermentation of Saccharomyces cerevisiae in the presence of acetic acid by overexpression of SET5 and PPR1. Biotechnol J 10: 1903–1911. [DOI] [PubMed] [Google Scholar]
- Zheng, T. , Yang, X. , Wu, D. , Xing, S. , Bian, F. , Li, W. , et al. (2015) Salidroside ameliorates insulin resistance through activation of a mitochondria‐associated AMPK/PI3K/Akt/GSK3beta pathway. Br J Pharmacol 172: 3284–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, Y. , Yang, G.Y. , Chen, X. , Liu, Q. , Zhang, X. , Deng, Z. , and Feng, Y. (2017) Biosynthesis of plant‐derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme. Metab Eng 42: 25–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) Deletion of TRP2 decreased the cell growth rate of the engineered strain LYTY12. (B). The tyrosol production after adding 20 mg l−1 extra tryptophan to the fermentation medium in strain LYTY13. The strains were cultured in shake flasks at 220 rpm, 30°C with YPD medium, or YPD medium with additional 20 mg l−1 tryptophan. The cell densities were monitored with the spectrophotometer at 600 nm wavelength. Titers of tyrosol were detected after 72 h fermentation. Error bars represented the standard deviation from three replicates.
Fig. S2. (A) The transcription levels of endogenous genes compared to those of the WT strains. (B) The transcription levels of heterogenous genes compared to that of the housekeeping gene ALG9. Strains were cultured for 24 h before RNA extraction. The transcription levels of those genes were measured with RT‐qPCR, and the housekeeping gene ALG9 was selected as the reference. The expression level of ALG9 was set as 1. Error bars represent the standard deviation of three biological replicates.
Table S1. Plasmids used in this study.
Table S2. Primers used in this study.
Table S3. List of the gRNA sequences used for gene deletions, mutations, and insertions via the CRISPR system.


