Summary
Small peptides are a group of natural products with low molecular weights and complex structures. The diverse structures of small peptides endow them with broad bioactivities and suggest their potential therapeutic use in the medical field. The remaining challenge is methods to address the main limitations, namely (i) the low amount of available small peptides from natural sources, and (ii) complex processes required for traditional chemical synthesis. Therefore, harnessing microbial cells as workhorse appears to be a promising approach to synthesize these bioactive peptides. As an emerging engineering technology, synthetic biology aims to create standard, well‐characterized and controllable synthetic systems for the biosynthesis of natural products. In this review, we describe the recent developments in the microbial production of small peptides. More importantly, synthetic biology approaches are considered for the production of small peptides, with an emphasis on chassis cells, the evolution of biosynthetic pathways, strain improvements and fermentation.
Small peptides are a group of bioactive compounds with low molecular weights and complex structures. This review covers recent advances in the microbial production of small peptide, focusing on the development of chassis cells functioning as green factories using synthetic biology approaches.

Introduction
Natural products from plants or microorganisms are fertile sources of therapeutics and lead drugs, which have played a vital role in medicinal applications (Atanasov et al., 2015). Among these applications, antimicrobial agents derived from natural products have developed rapidly since penicillin was first discovered (Singh and Barrett, 2006). The remaining challenge is that new antimicrobial compounds are still urgently needed because drug resistance inevitably occurs with the extensive use of antibiotics. Small peptides are a group of natural products with relatively low molecular weights (< 10 kDa). These bioactive compounds can be classified into two main categories: ribosomally synthesized and post‐translationally modified peptides (RiPPs) and non‐ribosomal peptides (NRPs; Dang and Süssmuth, 2017; Fig. 1). The biosynthesis of RiPPs requires two steps: ribosomal synthesis of the precursor peptide and posttranslational modifications (PTMs; Montalbán‐López et al., 2020). Due to differences in structural features (e.g. lanthionine), RiPPs are divided into different subfamilies, such as lanthipeptides. The structural diversity endows RiPPs with a broad range of bioactivities, including antifungal, antibacterial and antiviral activities (Arnison et al., 2013). NRPs are a diverse family of natural products that are synthesized by large enzyme complexes known as non‐ribosomal peptide synthetases (NRPSs; Podevels et al., 2008). NRPs are structurally diverse secondary metabolites with a variety of biological activities that can be exploited as therapeutic agents, such as the antibiotic daptomycin and the immunosuppressant cyclosporine A (Winn et al., 2016).
Fig. 1.
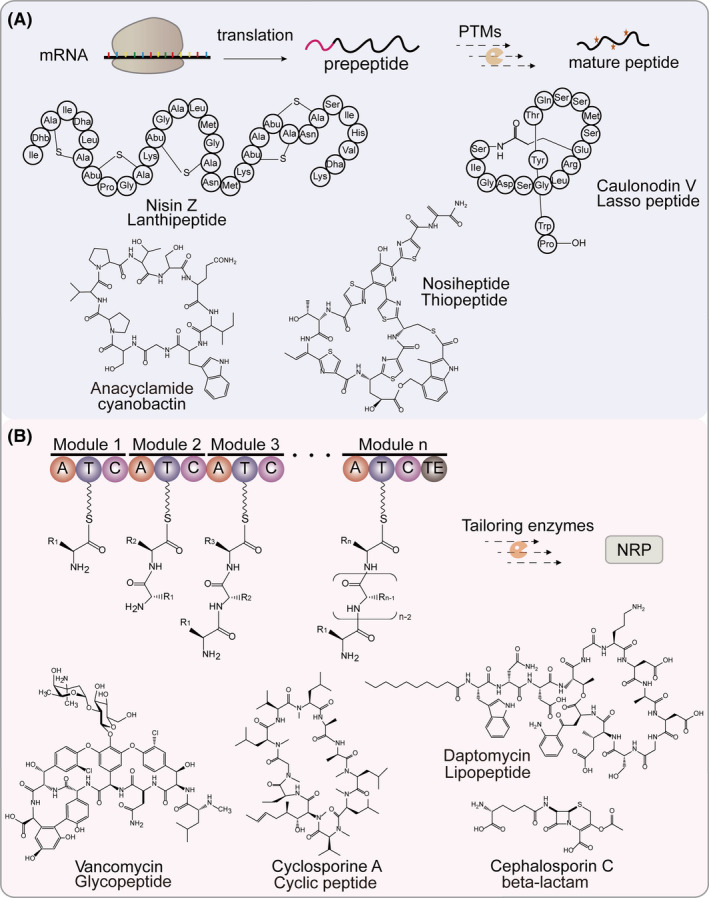
Schematic representation of biosynthetic mechanisms of RiPPs and NRPs.
A. The biosynthesis of RiPPs undergoes the ribosome and post‐translational modification machinery. The precursor peptide usually consists of an N‐terminal leader peptide (for recognition by PTM enzymes and for export) and a core peptide (harbouring various PTM sites). In some cases, the C‐terminal follower peptide serves as a leader peptide or transcriptional factor. Following the completion of PTMs, the leader peptide and follower peptide are removed by proteolysis. Examples of different small peptides belong to the RiPPs family.
B. Model of an NRPS assembly line showing the typical linear. (A)‐adenylation domain, (T)‐thiolation domain, (C)‐condensation domains, (TE)‐thioesterase domain. The module of NRPS may contain additional domains including epimerization (E), N‐methylation (M) and cyclization (Cy) domains. Examples of different small peptides produced by NRPS assembly lines.
In recent decades, bioactive peptides have attracted increasing attention because they may bridge the gap between small molecules and protein drugs. Small peptides have promising prospects in the pharmaceutical industry due to their broad‐spectrum bioactivities, low toxicity and low propensity to induce the development of drug resistance, although further investigations are still needed to unravel their reliability (Barbosa et al., 2015). However, (i) the amount of small peptides available from natural sources is extremely low, and the inefficient isolation and extraction methods increase the difficulty of accessing these peptides. For example, the yield of the lanthipeptide lichenicidin produced by Bacillus licheniformis have just reached 4–6 mg l−1 (Kuthning et al., 2015), which has a huge gap compared with that of extracellular proteins, such as α‐amylase (17.6 g l−1; Niu et al., 2009). Meanwhile, (ii) bioactive peptides with sophisticated structures are particularly difficult to be produced using traditional chemical synthesis, which usually requires multiple reactions to introduce specific functional groups. As exemplified by lactocin S, a 34‐residue lanthipeptide was isolated from Lactobacillus sakei L45, its biosynthesis involved only two enzymatic steps (modification, cleavage), after the prepeptide was synthesized in ribosome (Skaugen et al., 1997). The total synthesis of lactocin S required 71 steps using solid‐supported synthesis (Ross et al., 2010). The intricate construction of cells may not only aid in normal cell metabolism but also serve as an arsenal to synthesize various secondary metabolites. In this regard, microbial production of bioactive peptides in diverse biosystems has been pursued as a potential approach. Although biosynthetic genes are ubiquitous in the currently sequenced genomes and transcriptomes of different microbes, the common feature of the complex biosynthetic mechanism remains unclear. In addition, compared to the other types of molecules, small peptide as a bioactive compound often triggers host immune responses, and its synthesis is thereby subjected to a more complicated regulation (Bartholomae et al., 2017; Yang et al., 2020). The manipulation of biosynthetic pathways encoded by large gene clusters is still a challenge due to the shortage of robust genetic tools and engineering strategy. Therefore, the development of engineered production platforms for the production of small peptides has been substantially hindered in the past few decades.
As a new discipline that combines biology and engineering, synthetic biology endeavours to create a standard, well‐characterized and controllable synthetic system for the production of natural products. The developed chassis cells function as green manufacturing factories for the production of natural products of interest. Furthermore, the biosynthetic pathway encoding target products can be refactored by assembling a series of biological components based on the ‘bottom‐up’ principle, and it can be precisely regulated in microbial cells in a reliable, predictable and standardized manner using robust genetic manipulation toolkits (Slusarczyk et al., 2012). Recently, synthetic biology has been valuable in the development of microbial platforms for the production of various natural products, including terpenoids (Zhang and Hong, 2020), flavonoids (Pandey et al., 2016) and polyketides (Cummings et al., 2014). In this review, attention has been paid to small peptides (3–50 amino acids) from microbial sources that act as antimicrobial substances and can be envisaged as potential therapeutic drugs. We describe the recent developments in the microbial production of small peptides using synthetic biology approaches.
Chassis cells as hosts
Native hosts
Microbes are capable of producing various necessary compounds to maintain growth and numerous unnecessary metabolites that are involved in defending against damage derived from extreme environments, such as limited nutrients. Small peptides are usually isolated from bacteria (e.g. Actinomycetes) or fungi (e.g. Aspergillus niger), and most of these peptides display antimicrobial activity that may contribute to viability by inhibiting the growth of other microorganisms (Dang and Süssmuth, 2017). Native hosts are desirable sources of drug‐like small peptides because of their unique advantages, including the synthetic machinery, precursor supply, transport, self‐resistance, modification and regulatory mechanisms, although a few biosynthetic gene clusters (BGCs) are silent under normal laboratory culture conditions. In addition, the expression of biosynthetic genes in heterologous hosts may fail because of the adaptive mechanism known as the restriction–modification (RM) systems of hosts (Mruk and Kobayashi, 2014). With these advantages, native hosts do not require additional modifications (i.e. the introduction of heterologous biosynthetic pathways) for the de novo synthesis of small peptides, suggesting that the challenge lies in improving peptide productivity. However, several limitations of native hosts should not be overlooked. For instance, most native hosts may not adapt to normal cultivation conditions, leading to slow growth, or may not be amenable to gene manipulations, which hampers the improvement in host production performance through bioengineering strategies.
Heterologous hosts
Regardless of whether metabolic engineering strategies or synthetic biology approaches are used to produce compounds of interest, some ideal host traits are required to balance growth and genetic modification. Therefore, a promising strategy is to develop surrogate hosts for peptide biosynthesis. In general, these heterologous hosts would possess the following collective characteristics: (i) a clear genetic background, (ii) tractable gene manipulation, and (iii) ease of cultivation. Moreover, some traits that are similar to the native hosts might facilitate heterologous expression. For example, Streptomyces are the main sources of secondary metabolites that are synthesized by large‐sized gene clusters, and strains might provide various precursor pools and extensive modification machineries. These ideal traits make them potential chassis cells for the production of natural peptide products through the catalysis of a series of enzymes encoded by multiple genes. A representative example is Streptomyces coelicolor, a well‐known host that is widely used to produce natural products (Gomez‐Escribano and Bibb, 2011). In addition, pathway‐specific biology must be considered because some enzymatic reactions required for peptide biosynthesis rely on specific cellular structures (Hoppert et al., 2001; Meijer et al., 2010). As shown in the study by Gidijala et al. (2009), penicillin production is lower in a yeast strain lacking peroxisomes. The rapid progress in genetic manipulation technologies, including the CRISPR/Cas 9 system, along with the increased availability of synthetic biology tools has promoted the cloning and refactoring of BGCs of natural products in heterologous hosts. The biosynthesis of small peptides has been successfully achieved by harnessing various model hosts, such as Escherichia coli, Bacillus subtilis and Saccharomyces cerevisiae (Table 1). These well‐characterized hosts are beneficial not only for elucidating the catalytic mechanisms of biosynthetic enzymes via protein expression engineering but also for improving productivity based on genetic engineering. Furthermore, on this basis, synthetic chassis cells could be designed through the optimization of these heterologous hosts by using standard elements and rational engineering strategies (Adams, 2016). These synthetic chassis cells could serve as potential microbial cell platforms for large‐scale production of various desired products. Here, chassis cells with specific traits for the biosynthesis of small peptides are proposed (Table 2).
Table 1.
Common strategies used for the biosynthesis of small peptides in heterologous hosts.
| Peptide | Characterization | Native host | Heterologous host | Strategy | Yield | References |
|---|---|---|---|---|---|---|

|
RiPP, antibacterial agent (e.g. against Clostridia) | Ruminococcus gnavus | E. coli |
|
|
Ongey et al. (2018) |

|
RiPP, antibacterial agent (e.g. against B. subtilis) | Streptococcus bovis | E. coli |
|
|
Xiao et al. (2004); Lin et al. (2011) |

|
RiPP, antibacterial agent (e.g. against Caldibacillus sp.) | Aeribacillus pallidus | E. coli |
|
|
Kaunietis et al. (2019) |

|
RiPP, antibacterial agent (e.g. against Listeria Monocytogenes), commercialization | Lactococcus lactis | B. subtilis |
|
__ | Yuksel and Hansen(2007) |

|
RiPP, antibacterial agent (e.g. against Staphylococcus aureus) | Bacillus sp. HIL Y‐85,54728 | B. amyloliquefaciens |
|
__ | Barbosa et al. (2015); Herzner et al. (2011) |

|
RiPP, enzyme inhibitor (e.g. protein tyrosine phosphatase1B) | Streptomyces collinus | S. coelicolor |
|
|
Iftime et al. (2015) |


|
RiPP, antibacterial agent (e.g. against L. monocytogenes) | B. licheniformis | E. coli |
|
|
Begley et al. (2009); Kuthning et al. (2015) |

|
RiPP, inhibit cell invasion | Streptomyces leeuwenhoekii | E. coli |
|
|
Martin‐Gómez et al. (2018) |

|
RiPP | Thermobifida fusca | E. coli |
|
__ | Koos and Link (2019) |

|
RiPP, antibacterial agent (e.g. against S. aureus) | Planobispora rosea | S. coelicolor |
|
|
Flinspach et al. (2014) |

|
RiPP, antibacterial agent (e.g. against S. aureus) | Streptomyces | S. lividans |
|
|
Young and Walsh (2011) |

|
RiPP, telomerase inhibitor | Streptomyces anulatus | S. avermitilis |
|
|
Amagai et al. (2017) |

|
NRP, antibiotic, commercialization | Penicillium chrysogenum | S. cerevisiae |
|
|
Awan et al. (2017) |

|
NRP, pigment | Streptomyces. lavendulae | S. cerevisiae |
|
|
Wehrs et al. (2018) |

|
NRP, antibiotic, anthelmintic, cytotoxic, et al. commercialization | Beauveria bassiana | Aspergillus niger |
|
|
Boecker et al. (2018) |

|
NRP, antitumor | Streptomyces lasaliensis | E. coli |
|
|
Watanabe et al. (2006) |

|
NRP, antibiotic, anthelmintic, cytotoxic, et al. commercialization | Filamentous fungi (e.g. Fusarium) | B. subtilis |
|
|
Zobel et al. (2015); Boecker et al. (2018) |
Table 2.
The proposed chassis cells for the production of small peptides.
| Chassis cell characterization | Advantage | Engineering strategy | References |
|---|---|---|---|
| Genome reduction |
|
|
Gao et al. (2010) |
| Increasing resistance |
|
|
Yu et al. (2018); Hu and Ochi (2001); Huo et al. (2012); Zhang et al. (2016); Huo et al. (2012); Qi et al. (2017); Dörr et al. (2016); Cao et al. (2018) |
| Secretion remodelling |
|
|
Kuipers et al. (2004); Li et al. (2015); Kuipers et al. (2006) |
Artificial evolution of biosynthetic pathways
Pathway design
Identification of biosynthetic pathways
Small peptides are synthesized by specific biosynthetic pathways composed of various enzymes encoded by the corresponding gene clusters within the chromosome of native hosts. The identification of biosynthetic pathways is crucial to improve our understanding of synthetic machinery, for the discovery of new compounds, and for heterologous expression (Amagai et al., 2017; Kawahara et al., 2018; Shi et al., 2019). The plummeting cost of genome sequencing has facilitated the increase in the number of sequenced bacterial genomes, which provide a wealth of data for investigating potential BGCs encoding biosynthetic pathways (Vallenet et al., 2009). Due to advances in bioinformatics techniques coupled with the interest in peptide biosynthetic logics, many bioinformatics tools have been developed that enable researchers to mine BGCs of peptide natural products by matching the biosynthetic genes to the most similar candidates through a search of known databases, such as the antiSMASH database (Blin, Medema, et al., 2017). These computational tools are dedicated to the identification and characterization of the BGCs of peptides and other natural products in different species (Fig. 2a), including bacteria (Blin, Wolf, et al., 2017), fungi (Khaldi et al., 2010) and plants (Kautsar et al., 2017). Some of the tools based on specific algorithms have the ability to predict chemical structures based on detected biosynthetic genes. For instance, PRISM 3 is an available web server that predicts the chemical structures of natural peptide products based on homology to known BGCs or biosynthetic enzymes by modelling the structure scaffold as a chemical graph (Skinnider et al., 2017). Recently, mass spectrometry (MS)‐based genome mining has been employed to identify the BGCs of secondary metabolites. Pep2Path, an automated mining tool based on advanced MS detection equipment, precisely links natural peptide products with the likely BGCs according to their conserved biosynthetic logics (Medema et al., 2014). Pep2Path consists of RiPP2Path and NRP2Path, both of which are able to convert a detected mass shift into a possible amino acid sequence tag that can be further used to match the candidate gene clusters. Unlike RiPPquest, an algorithm that is dedicated to identifying the prepeptide‐encoding genes of RiPPs (Mohimani et al., 2014), RiPP2Path is suitable for all RiPPs. Additional genome mining techniques for natural peptide products have previously been reviewed in detail elsewhere (Boddy, 2014; Hetrick and van der Donk, 2017). All of these user‐friendly tools not only accelerate the discovery of BGCs for small peptides and other secondary metabolites but also provide avenues for the production of natural products by expressing BGCs in heterologous hosts.
Fig. 2.
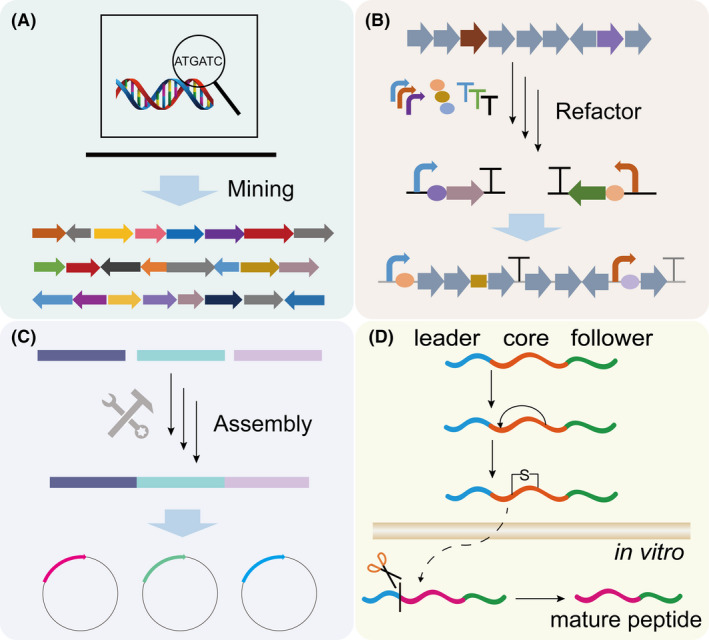
Evolution of artificial pathways.
A. Mining BGCs, the BGCs encoding the biosynthesis of small peptides are identified based on advanced computational tools.
B. Refactor BGCs, including the remove of native regulation elements, fine‐tuning of gene expression using diverse synthetic biology tools.
C. Assembly BGCs, the fusion of different genetic fragments based on various assembly strategies, and further clone the BGCs into suitable vectors.
D. Scheme of semi‐in vitro biosynthesis. The prepeptide of RiPP is synthetized and modified within the cell, and the mature peptide can subsequently be released by removing the leader peptide through the in vitro cleavage system.
Refactoring of biosynthetic pathways
The gene clusters responsible for the biosynthesis of small peptides are composed of multiple operons. The native BGCs may be subjected to endogenous cellular regulation, which will affect the levels of gene expression within the biosynthetic pathway (Shao et al., 2013). A controllable biological process that is based on the rewiring of BGCs with well‐characterized DNA parts is required to circumvent this limitation (Larroude et al., 2018). This process might be facilitated by refactoring, in which native biosynthetic pathways are reassembled using genetic parts (Smanski et al., 2016; Fig. 2b). During the process of refactoring, non‐coding genes and native regulatory elements within the BGC are removed. Furthermore, the uncharacterized regulatory elements within the coding DNA sequences (CDSs) will be identified and deleted by randomly assembling coding genes in silico. The remaining coding genes are obtained using DNA synthesis techniques. By refactoring the BGCs, the expression of genes is controlled using diverse biological parts, such as the promoter, RBS, terminator (Khalil and Collins, 2010). Expanded parts, such as insulators, are used to construct synthetic pathways because of their ability to increase the reliability of the functions of each part and to predict the behaviour of synthetic circuits in different genetic contexts (Lou et al., 2012). The BGCs that are equipped with synthetic regulatory elements are theoretically relatively orthogonal, which would allow natural regulatory networks to be bypassed and force the expression of the controlled genes (Myronovskyi and Luzhetskyy, 2016). Another advantage is the potential application of high‐throughput techniques to the refactored BGCs for the optimization of biosynthetic pathways. For example, synthetic promoter libraries have been established to refactor the biosynthetic pathway for the production of desired products based on high‐throughput screening (Siegl et al., 2013). Different promoters that vary in strength enable the optimization of BGC expression through a rational combinatorial strategy (Awan et al., 2017). MacPherson et al. constructed a series of short synthetic terminators by randomly linking terminators with a consensus sequence. These synthetic short terminators avoided repetitive sequences and improved the stability of recombinant synthetic circuits and the assembly of biosynthetic pathways into S. cerevisiae (MacPherson and Saka, 2017). Additionally, refactoring permits the creation of new structures by swapping genetic modules encoding diverse modification enzymes. This strategy is particularly important for natural products with similarly biosynthetic mechanisms. For instance, NRPSs are usually composed of multiple modules that are responsible for integrating diverse monomers into the peptide backbone, which facilitates the production of new chemical structures through the modification of biosynthetic modules (Nguyen et al., 2006). The state‐of‐the‐art structures are generated through the combination of different biosynthetic modules derived from the BGCs of non‐ribosomal peptides (Zobel et al., 2016). Furthermore, hybrid enzymes that are capable of synthesizing new compounds have been generated by the fusion of NRPS with other megaenzymes, such as polyketide synthases (PKSs; Zhang et al., 2020).
Pathway construction
In general, two strategies have been developed for the construction of biosynthetic pathways. The first is de novo DNA synthesis. The availability of sequenced genomes along with advanced gene mining tools has allowed biologists to access the BGCs of natural products. Furthermore, the cost of DNA synthesis has decreased, which has facilitated the construction of biosynthetic pathways through commercial synthesis. Researchers were able to perform de novo synthesis of codon‐optimized large fragments that closely match the codon usage of heterologous hosts. The other strategy for pathway construction is DNA assembly (Fig. 2c). Many assembly methods are available and have been reviewed, including Gibson assembly, ligase cycling reaction, scarless stitching and Golden Gate assembly (Kosuri and Church, 2014). When the BGCs are obtained, robust vectors are necessary to carry them. Common tools include cosmids and fosmids, which have been used to clone and express the large gene clusters of small peptides (Caetano et al., 2011; Shi et al., 2019). The expression of the intact gene clusters based on multiplasmid systems is also an effective strategy for the manipulation of large gene clusters (Watanabe et al., 2006). In addition, bacterial artificial chromosome (BAC) vectors have been applied to express all BGCs of secondary metabolites (Liu et al., 2009). BACs containing the ϕC31 att/int and oriT functions have been used to integrate large gene clusters into the chromosome of heterologous hosts (Baltz, 2012). This feature has been employed to construct small peptide‐producing strains by inserting the BGCs into the chromosomes of the heterologous hosts (Alexander et al., 2010). Advances in next‐generation gene sequencing technologies known as ‘plug‐and‐play’ approaches are required to express orphan pathways from sequenced microorganisms for the biosynthesis of natural products of interest, which enable the construction of novel vectors that are capable of directly cloning, refactoring, and expressing large gene clusters for the design, construction, and discovery of novel secondary metabolites (Yamanaka et al., 2014). Recently, synthetic vectors have been developed due to their abilities to construct artificial biosynthetic pathways and control the expression of genes by integrating various biological parts (Pandey et al., 2016). As a versatile platform for manipulating biosynthetic pathways involved in multiple genes, the ePathBrick vector system supports the modular assembly of pathway genes using multiple compatible plasmids simultaneously and allows the fine‐tuning of gene expression by controlling transcription signals (Xu et al., 2012).
For the biosynthesis of RiPPs synthesized via the ribosomal machinery, specific structures (e.g. lanthionine) are essential for the bioactivity of RiPPs, which are formed by modifying enzymes that are capable of recognizing conserved sequences of precursor peptides. The in vivo PTMs will not be obstructed when functional tags are inserted into the N‐terminus of the leader peptide (Huo and Donk, 2016). Based on this mechanism, semi‐in vitro biosynthesis (SIVB) might be an alternative strategy for producing RiPPs using the artificial prepeptide chain (Fig. 2d). The modified prepeptides are produced by expressing the precursor peptide in the form of a fusion protein with an N‐terminal tag, followed by the activation of the modified prepeptide in vitro through the cleavage of the leader peptide by a specific endonuclease (Lin et al., 2011). The modified prepeptide may provide the desired properties following the introduction of various functional tags, such as His tags (Zambaldo et al., 2017).
Pathway optimization
The challenge for the production of natural products is developing a method to control the biosynthetic flux in synthetic biosystems. In synthetic biology, the regulation of biosynthetic pathway has been achieved at the transcriptional and translational levels. The expression of genes within the biosynthetic pathway is precisely controlled using various genetic parts (Seo et al., 2013). A common strategy for the biosynthesis of small peptides is to increase the expression of the genes related to biosynthesis by inserting strong promoters in front of the operons (Qiu et al., 2014; Iftime et al., 2015; Ji et al., 2015; Ni et al., 2017; Wang et al., 2018). The constitutive promoter ermE*, which is derived from the native promoter of the erythromycin‐resistance gene, is widely used to drive gene or gene cluster expression for the production of small peptides (Thykaer et al., 2010; Flinspach et al., 2014; Iftime et al., 2015). The ability to control the levels of gene expression via inducible promoters is an alternative temporal strategy for coordinating cell growth and product formation (Li et al., 2018a). A representative example is the T7 promoter, which has been used to increase the expression of biosynthetic genes in E. coli for the heterologous production of peptides (Weiz et al., 2011; Kuthning et al., 2015). The translational regulation of the biosynthetic pathway has been achieved by modulating the efficiency of protein synthesis (Thiel et al., 2018). For instance, Martin‐Gómez and colleagues substituted native RBS sequences with E. coli‐optimized RBS sequences in front of the biosynthetic genes cptB1 and cptB2 within the BGC of the lasso peptide chaxapeptin, resulting in chaxapeptin production in E. coli (Martin‐Gómez et al., 2018).
Although the overexpression of biosynthesis‐related genes might efficiently increase productivity in most cases, a prerequisite for pathway optimization is the balanced expression of genes in operons (Pfleger et al., 2006). The challenge is that each gene might be subjected to complex regulations in the cell and the optimized expression levels of gene clusters are unknown, which might further hamper the increased production of the molecules of interest. This problem might be circumvented through the refactoring of BGCs based on the libraries of genetic parts (Ji et al., 2018). The use of diverse regulatory parts in a combinatorial manner may exert positive synergistic effects on the optimization of biosynthetic pathways (Kosuri et al., 2013). Meanwhile, genetically encoded biosensors coupled with high‐throughput fluorescence‐activated cell sorting (FACS) have provided a method to screen or select desired metabolites at the single‐cell level (Hossain et al., 2020). A practical application is that cells producing high levels of the desired metabolites are selected via a biosensor‐based high‐throughput screen without knowing the expression level of each gene within the biosynthetic pathway (Wang et al., 2019). The dynamic regulation of the biosynthetic pathway is necessary for the optimal production of secondary metabolites, while avoiding the excess accumulation of intermediates, especially toxic compounds (Dahl et al., 2013; Xu et al., 2014). The well‐characterized dynamic behaviours of synthetic circuits from libraries of synthetic parts will guide the design of synthetic pathways using computational methods (Rodrigo et al., 2011). As exemplified in the biosynthesis of penicillin in yeast, Awan and coworkers optimized the production of the non‐ribosomal peptide penicillin by constructing and testing hundreds of different combinations of penicillin pathway genes with different promoters with varying strengths and expression dynamics (Awan et al., 2017). Recently, a random rational strategy was applied to engineer the gene cluster of bottromycin for the construction of overproducing strains. The cluster was refactored with a combination of various promoters with unknown strengths, which allowed researchers to construct a library of bot clusters that would enable the ideal expression levels to be screened in an appropriate host. Furthermore, this strategy allows other genetic regulatory parts (e.g. RBSs) to be used to refactor the BGCs of interest and does not require detailed knowledge of the biosynthesis in the initial stage (Horbal et al., 2018).
Strain improvement
Activation of positive regulators
The improvement of strain performance can be achieved within the bio‐based peptide‐producing system through the use of pathway engineering strategies, such as transcription regulation (Fig. 3). The biosynthesis of RiPPs is regulated by complex regulatory mechanisms that are commonly mediated by specific pathway regulators (Bartholomae et al., 2017). For example, the biosynthesis of the class I lanthipeptide nisin is regulated by quorum sensing, which is mediated by the two‐component system nisRK, the specific regulatory genes within the gene cluster (Lubelski et al., 2008). Specific pathway regulators control the transcription of biosynthetic genes within the gene clusters by binding to the promoter regions of operons (Zhang et al., 2014). The overexpression of these positive regulators increases the transcription of structural genes within gene clusters and improves productivity (Li et al., 2018b). In addition to pathway‐specific regulators, global regulators also exert positive effects on the biosynthesis of RiPPs. An example is microcins, a class of RiPPs with a low molecular weight that are produced by E. coli and related Enterobacteria (Severinov et al., 2007). The transcription of the microcin C51 operon is regulated by RNA polymerase sigma S factor (σS) and the global transcriptional regulator CRP that interacts with cyclic AMP in the cyclic complex CRP‐cAMP. The production of microcin C51 is markedly decreased or abolished upon the mutation of σS, CRP or cAMP (Fomenko et al., 2001).
Fig. 3.
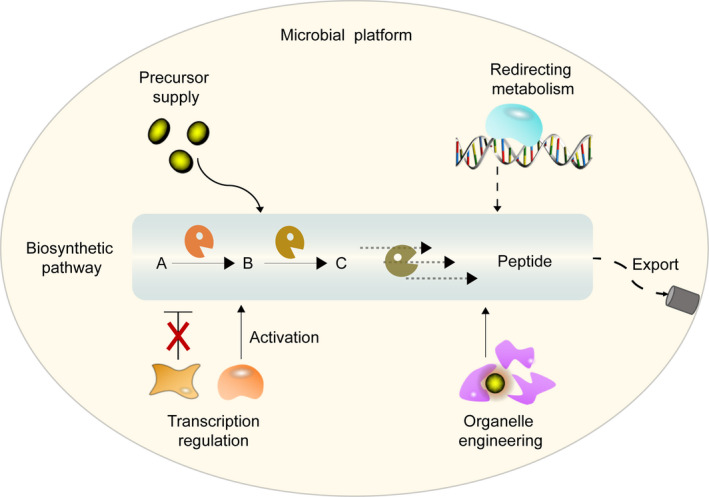
Metabolic engineering strategies for strain improvement.
The activation of positive regulators of specific pathways is also essential to increase biosynthetic gene transcription and increase production in the biosynthesis of NRPs. The gene cluster responsible for the synthesis of the glycopeptide teicoplanin is regulated by two positive regulators, tei15* and tei16*, which are StrR‐ and LuxR‐type transcriptional regulators, respectively. The expression of tei15* and tei16* under the aac(3)IV promoter in Actinoplanes teichomyceticus increases the production of teicoplanin (Horbal et al., 2014). Daptomycin is a cyclic lipopeptide produced by Streptomyces roseosporus that may represent a clinical therapeutic agent for the treatment of skin infections (Baltz, 2010). Yuan and coworkers confirmed that DepR1, a TetR family transcriptional regulator, is a positive regulator of daptomycin biosynthesis. DepR1 might mediate the positive feedback regulation of daptomycin production by directly binding to its own promoters. The overexpression of DepR1 results in the improved production of daptomycin and a shorter fermentation period (Yuan et al., 2016). Unlike the action of DepR1, the TetR family transcriptional regulator AtrA regulates the production of daptomycin by activating the A‐factor signalling pathway (Mao et al., 2015). Additionally, specific molecules also serve as positive regulators to promote biosynthetic gene transcription by mediating signal transduction. An example is the biosynthesis of the non‐ribosomal peptide surfactin, which is produced by B. subtilis through a process by a cell density‐responsive mechanism. The specific extracellular signalling peptides comX and phrC induce the expression of quorum‐responsive genes (srfA). The overexpression of comX and phrC increases the yield of surfactin (Jung et al., 2012).
Disruption of negative regulators
The negative regulators will downregulate the expression of biosynthetic genes by binding to the promoter of biosynthetic genes, leading to a decrease in the production of target products (Reverchon et al., 2002). The removal of these negative regulators could relieve the transcriptional repression of the BGC and contribute to peptide synthesis (Choi et al., 2009). For example, the ArsR family transcriptional regulator DepR2 represses the expression of the daptomycin gene cluster by interacting with the dptEp promoter. The deletion of depR2 caused an approximately 2.5‐fold increase in the production of daptomycin (Mao et al., 2017). The biosynthesis of peptide antibiotics is related to the endogenous metabolite states of the cell, such as sporulation (Marahier et al., 1993). AbrB is a global regulator that controls the expression of genes involved in the transition state between the postexponential phase and stationary phase and sporulation in B. subtilis (Strauch et al., 1989). Moreover, AbrB plays an important role in the production of antibiotics and degradative enzymes and many other physiological processes in B. subtilis (Phillips and Strauch, 2002). AbrB‐encoding genes have been identified in various bacterial species, including archaea (http://mbgd.genome.ad.jp/; Chumsakul et al., 2011). In a previous study, researchers introduced the BGC of polymyxin into B. subtilis and successfully observed product generation. However, quantitative real‐time PCR results indicated that the transcription level of the lipopeptide antibiotic polymyxin biosynthetic gene pmxA was reduced in the presence of AbrB. Transcription was significantly recovered upon the deletion of the abrB gene encoding the AbrB regulator, resulting in the efficient production of polymyxin (Park et al., 2012). The unexpected repressive effects of the AbrB regulator on the expression of biosynthetic genes have also been observed in the biosynthesis of RiPPs in B. subtilis. For example, the biosynthesis of the lantibiotic subtilin was repressed by the transcriptional regulator AbrB, which caused a low level of expression of biosynthetic proteins during the exponential growth phase. The production of subtilin was substantially increased by deleting of the abrB gene in B. subtilis ATCC 6633 (Stein et al., 2002). Additionally, global regulators that are involved in primary metabolism may exert negative effects on the biosynthesis of secondary metabolites. For example, the two‐component system PhoR‐PhoP, which is related to phosphorus metabolism, was proven to suppress antibiotic synthesis in Streptomyces lividans and the deletion of phoP or phoR‐phoP relieved this repression (Sola‐Landa et al., 2003). Deletion of the phoP gene in B. licheniformis also increased the yield of bacitracin, a NRP produced by B. subtilis and B. licheniformis (Cai et al., 2019).
Precursor supply
For the biosynthesis of RiPPs, the building blocks are usually proteinogenic amino acids and unnatural amino acids that are incorporated into the peptide backbone through amber‐suppression technology (Himes et al., 2016). In addition to proteinogenic/non‐proteinogenic amino acids (Hubbard et al., 2000), some specific moieties may be incorporated into the non‐ribosomal peptide backbones, such as fatty acids (Wu et al., 2019). Here, the emphasis is placed on the precursors supplied for the biosynthesis of NRPs (Fig. 3). The supply of precursors is increased through rational genetic engineering to perturb metabolic fluxes, including (i) the overexpression of enzymes involved in the precursor synthesis pathway (Thykaer et al., 2010), (ii) the disruption of competing pathways or by‐product pathways (Lee et al., 2015), and (iii) the manipulation of transcriptional regulators related to primary metabolism (Cai et al., 2019). Recently, a promising synthetic biology approach was reported to increase the precursor supply through the redesign of a biosynthetic pathway by replacing native enzymes with heterologous proteins from unrelated pathways. Hydroxyphenylglycine (HPG), the precursor for the calcium‐dependent antibiotic of S. coelicolor, was synthesized efficiently using this method (Diez et al., 2015).
Redirecting metabolism
Engineering cell microbial factories for bioproduction requires to overcome metabolic burden which may place hidden constraints on host productivity (Wu et al., 2016). Progress in systems biology and the development of various omics techniques (e.g. metabolomics), researchers are able to analyse the metabolic levels of biosynthetic pathways from a global perspective (Wang et al., 2020). One example is the establishment of metabolic network models for in silico analyses of metabolic flux (Ma et al., 2018). To date, genome‐scale metabolic network model (GMM) reconstruction has been completed in various strains, such as S. coelicolor (Borodina et al., 2005). The metabolic network model is able to assess the metabolic capabilities and investigate flux bottlenecks through computational simulations (Bum Kim et al., 2004). As exemplified in the production of daptomycin, the metabolic flux of each reconstituted strain was compared with the wild‐type strain, and three potential targets (zwf2, dptI, and dptJ) related to the production of daptomycin were identified. The overexpression of these three target genes individually increased the production of daptomycin. Furthermore, compared with the parental strain, the strain coexpressing these three genes exhibited a 34.4% increase in daptomycin yield, suggesting that the enzymes encoded by these three genes exert a synergistic effect on daptomycin biosynthesis (Huang et al., 2012). Advances in gene editing tools and DNA synthesis enable multiplex genome engineering of industrial chassis organisms for optimize bioproduction (Barbieri et al., 2017; Fig. 3). Multiplexed automated genome engineering (MAGE) permits access to a library of mutants with diverse genotypes by introducing oligonucleotides, including insertions, deletions and mismatches (Auxillos et al., 2019). The combination of this technique with suitable screening approaches could be utilized to investigate target strains that possess optimized in vivo metabolic fluxes for target products. Recently, the CRISPRi‐based programmable circuit was designed to regulate the metabolic pathway, completing dynamic and self‐regulatory dual control of metabolic flux without external inducers (Wu et al., 2020).
Organelle engineering
Subcellular organelles play vital roles in the biosynthesis of secondary metabolites based on the spatial distribution of enzymes in eukaryotes (Keller, 2015; Hammer and Avalos, 2017). As an example, peroxisomes are nearly ubiquitous single‐membrane organelles involved in the biosynthesis of fungal metabolites (Stehlik et al., 2014). The biosynthesis of penicillin and related cephalosporins requires specific enzymatic reactions within the peroxisomes (Bartoszewska et al., 2011). This suggests that subcellular compartmentalization of enzymes involved in secondary metabolites biosynthesis may have a promising engineering potential (Fig. 3). Herr et al. tested the peroxisome targeting strategy for the localization of main enzymes of penicillin biosynthesis in A. nidulans. A peroxisomal targeting signal 1 (PTS1) tag was incorporated into the non‐ribosomal peptide synthase AcvA, which led to a 3.2‐fold increase in penicillin production. Moreover, increased peroxisomes through overexpression of pexK also led to improvement in penicillin production (Herr and Fischer, 2014). The biosynthesis of RiPPs is main involved in the ribosome translation with the incorporation of 20 canonical amino acids into the peptide chain. The non‐canonical amino acids (ncAAs) as building blocks are available through the interference of translation process via ribosome, which paves the way to expand structural diversity of ribosomal peptides (Budisa, 2013). For example, Tianero et al. (2012) revealed that the specific ncAA can be incorporated into the precursor peptide of cyanobactin using an orthogonal tRNA/aminoacyl‐tRNA synthetase (tRNA/aaRS) pair in response to the nonsense or frameshift mutation. Recently, it was demonstrated that ribosome engineering exerted tremendous effects on the production of secondary metabolites (Lopatniuk et al., 2019). Several results showed that ribosomal protein S12 was related to drug resistance of producer and mutations in the ribosomal protein S12 efficiently increased small peptide production (Huo et al., 2012; Wang et al., 2014).
Fine‐tuning fermentation
Fermentation is an important part of bio‐based production using microbial hosts as cell factories. The common strategies including medium optimization (Dang et al., 2019), fed‐bath fermentation (Costas Malvido et al., 2016), and control of parameters such as dissolved oxygen (DO; Guez et al., 2008) have been employed to improve productivity of small peptide‐producing strains. However, the maximum potential is difficult to exploit and achieve the highest productivity in the scaled fermentation process. One reason is that traditional methods for controlling fermentation mainly rely on empirical knowledge based on the metabolic states of the wild‐type strain, which poses a challenge to accurately regulate cellular metabolic states for high productivity. The artificial chassis cells designed for specific purposes (e.g. for the production of small peptides) have a relatively clear genetic background, and intracellular interference signals from unwanted pathways have been minimally modified, which are different from the situation in traditional hosts. Based on these traits, media with defined nutrients can be designed to meet the requirements of growth and product generation based on the biosynthetic pathways embedded in chassis cells. Meanwhile, novel fermentation strategies based on intelligent online detection platforms with high precision and high efficiency have been employed to rationally control the fermentation process at both the extracellular and intracellular levels (Fig. 4). The key real‐time parameters of a fermentation process have been calculated using online physiological data and used to guide the optimization of the fermentation process (Lu et al., 2016). Moreover, multiple component models of a fermentation process and the adaptive fuzzy control algorithm have been established using near‐infrared spectroscopy (NIS; Landgrebe et al., 2010). With the advent of omics techniques, fermentation process control has become a more systematic and scientific method. For example, the advanced UPLC/Q‐TOF‐MS online detection technique enabled the construction of a global chemome that was used to analyse the main components of fermentation broths and target products (Paglia et al., 2012). Additionally, intracellular signals that reflect the metabolic states of chassis cells have also been accurately monitored and evaluated using next‐generation analysis tools, such as Oxford Nanopore Technologies (ONT; Garalde et al., 2018). With the identification of intracellular signals, researchers have been able to quantitatively analyse and control key parameters (e.g. temperature, DO and pH), which usually exert marked effects on the fermentation process and control the metabolic states of chassis cells, via the online feedback system.
Fig. 4.
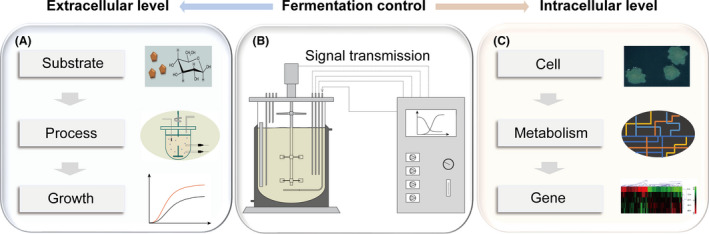
Schematic illustration of fermentation process control based on the extracellular and intracellular levels.
A. Control of the fermentation process at the extracellular level. The fermentation process is affected by changes in various extracellular factors, including substrate and process parameters (e.g. DO), as well as cell growth (e.g. biomass).
B. Fermentation control system.
C. The physiological state of cells can be monitored and regulated by controlling the fermentation process at the intracellular level, which includes controlling the cell morphology and cell metabolism, as well as gene expression.
Concluding remarks
Small peptides, a group of bioactive compounds with low molecular weights and complex structures, have become an area of focus because of their therapeutic potential (Daliri et al., 2018). This review provides a good example of gaining insights into peptide compounds by a combination of chemical and cell biological approaches. Given the complex structures of bioactive peptides, the design and development of cell factories to synthesize these peptides might be a promising approach to bypass the complex chemical synthesis. Microbial production, over the past decade, has made it possible to obtain small peptides from natural isolation to laboratory preparation by engineering the biosynthetic pathway using advanced synthetic biology toolkits and emerging strategies. A part of sophisticated structures has also been realized through the enzymatic reactions within synthetic chassis cells, which further contribute to solving the availability and complexity issue of small peptides. The remained challenge is how to achieve large‐scale production because the yield of small peptide cannot meet market requirements currently. Additionally, novel peptides and their analogs that might possess attractive pharmacological properties are required to be exploited in response to the risk of drug resistance. Recent researches showed that these new compounds can be synthesized by rational biosynthesis approaches, such as combinatorial biosynthesis (Yan et al., 2018). More efforts are still needed to elucidate the biosynthetic mechanisms of small peptides and explore synthetic approaches for accessing to new chemical structures. These state‐of‐the‐art structures will further enrich the library of therapeutic candidates.
Conflict of interest
The authors declare no competing interests.
Acknowledgements
This review was supported by the National Key Research & Development Program of China (2018YFA0900504, 2020YFA0907700, 2018YFA0900300 and 2016YFD0401404), the National Natural Foundation of China (31401674), the National First‐Class Discipline Program of Light Industry Technology and Engineering (LITE2018‐22), and the Top‐notch Academic Programs Project of Jiangsu Higher Education Institutions.
Microb. Biotechnol. (2021) 14(6), 2257–2278
Funding information This review was supported by the National Key Research & Development Program of China (2018YFA0900504, 2020YFA0907700, 2018YFA0900300 and 2016YFD0401404), the National Natural Foundation of China (31401674), the National First‐Class Discipline Program of Light Industry Technology and Engineering (LITE2018‐22), and the Top‐notch Academic Programs Project of Jiangsu Higher Education Institutions.
Contributor Information
Youran Li, Email: liyouran@jiangnan.edu.cn.
Guiyang Shi, Email: gyshi@jiangnan.edu.cn.
References
- Adams, B.L. (2016) The next generation of synthetic biology chassis: moving synthetic biology from the laboratory to the field. ACS Synth Biol 5: 1328–1330. [DOI] [PubMed] [Google Scholar]
- Alexander, D.C. , Rock, J. , He, X. , Brian, P. , Miao, V. , and Baltz, R.H. (2010) Development of a genetic system for combinatorial biosynthesis of lipopeptides in Streptomyces fradiae and heterologous expression of the a54145 biosynthesis gene cluster. Appl Environ Microbiol 76: 6877–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amagai, K. , Ikeda, H. , Hashimoto, J. , Kozone, I. , Izumikawa, M. , Kudo, F. , et al. (2017) Identification of a gene cluster for telomestatin biosynthesis and heterologous expression using a specific promoter in a clean host. Sci Rep 7: 2–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnison, P.G. , Bibb, M.J. , Bierbaum, G. , Bowers, A.A. , Bugni, T.S. , Bulaj, G. , et al. (2013) Ribosomally synthesized and post‐translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30: 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasov, A.G. , Waltenberger, B. , Pferschy‐Wenzig, E.‐M. , Linder, T. , Wawrosch, C. , Uhrin, P. , et al. (2015) Discovery and resupply of pharmacologically active plant‐derived natural products: a review. Biotechnol Adv 33: 1582–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auxillos, J.Y. , Garcia‐Ruiz, E. , Jones, S. , Li, T. , Jiang, S. , Dai, J. , and Cai, Y. (2019) Multiplex genome engineering for optimizing bioproduction in Saccharomyces cerevisiae . Biochemistry 58: 1492–1500. [DOI] [PubMed] [Google Scholar]
- Awan, A.R. , Blount, B.A. , Bell, D.J. , Shaw, W.M. , Ho, J.C.H. , McKiernan, R.M. , and Ellis, T. (2017) Biosynthesis of the antibiotic nonribosomal peptide penicillin in Baker’s yeast. Nat Commun 8: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz, R.H. (2010) Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot (Tokyo) 63: 506–511. [DOI] [PubMed] [Google Scholar]
- Baltz, R.H. (2012) Streptomyces temperate bacteriophage integration systems for stable genetic engineering of actinomycetes (and other organisms). J Ind Microbiol Biotechnol 39: 661–672. [DOI] [PubMed] [Google Scholar]
- Barbieri, E.M. , Muir, P. , Akhuetie‐Oni, B.O. , Yellman, C.M. , and Isaacs, F.J. (2017) Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell 171: 1453–1467.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa, J. , Caetano, T. , and Mendo, S. (2015) Class I and class II lanthipeptides produced by Bacillus spp. J Nat Prod 78: 2850–2866. [DOI] [PubMed] [Google Scholar]
- Bartholomae, M. , Buivydas, A. , Viel, J.H. , Montalbán‐López, M. , and Kuipers, O.P. (2017) Major gene‐regulatory mechanisms operating in ribosomally synthesized and post‐translationally modified peptide (RiPP) biosynthesis. Mol Microbiol 106: 186–206. [DOI] [PubMed] [Google Scholar]
- Bartoszewska, M. , Opaliński, Ł. , Veenhuis, M. , and van der Klei, I.J. (2011) The significance of peroxisomes in secondary metabolite biosynthesis in filamentous fungi. Biotechnol Lett 33: 1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley, M. , Cotter, P.D. , Hill, C. , and Ross, R.P. (2009) Identification of a novel two‐peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl Environ Microbiol 75: 5451–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin, K. , Medema, M.H. , Kottmann, R. , Lee, S.Y. , and Weber, T. (2017) The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res 45: D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin, K. , Wolf, T. , Chevrette, M.G. , Lu, X. , Schwalen, C.J. , Kautsar, S.A. , et al. (2017) AntiSMASH 4.0 ‐ improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45: W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, C.N. (2014) Bioinformatics tools for genome mining of polyketide and non‐ribosomal peptides. J Ind Microbiol Biotechnol 41: 443–450. [DOI] [PubMed] [Google Scholar]
- Boecker, S. , Grätz, S. , Kerwat, D. , Adam, L. , Schirmer, D. , Richter, L. , et al. (2018) Aspergillus niger is a superior expression host for the production of bioactive fungal cyclodepsipeptides. Fungal Biol Biotechnol 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodina, I. , Krabben, P. , and Nielsen, J. (2005) Genome‐scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res 3: 820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisa, N. (2013) Expanded genetic code for the engineering of ribosomally synthetized and post‐translationally modified peptide natural products (RiPPs). Curr Opin Biotechnol 24: 591–598. [DOI] [PubMed] [Google Scholar]
- Bum Kim, H. , Smith, C.P. , Micklefield, J. , and Mavituna, F. (2004) Metabolic flux analysis for calcium dependent antibiotic (CDA) production in Streptomyces coelicolor . Metab Eng 6: 313–325. [DOI] [PubMed] [Google Scholar]
- Caetano, T. , Krawczyk, J.M. , Mösker, E. , Süssmuth, R.D. , and Mendo, S. (2011) Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli . Chem Biol 18: 90–100. [DOI] [PubMed] [Google Scholar]
- Cai, D. , Zhu, J. , Zhu, S. , Lu, Y. , Zhang, B. , Lu, K. , et al. (2019) Metabolic engineering of main transcription factors in carbon, nitrogen, and phosphorus metabolisms for enhanced production of bacitracin in Bacillus licheniformis . ACS Synth Biol 8: 866–875. [DOI] [PubMed] [Google Scholar]
- Cao, L. , Liang, D. , Hao, P. , Song, Q. , Xue, E. , Caiyin, Q. , et al. (2018) The increase of O‐acetylation and N‐deacetylation in cell wall promotes acid resistance and nisin production through improving cell wall integrity in Lactococcus lactis . J Ind Microbiol Biotechnol 45: 813–825. [DOI] [PubMed] [Google Scholar]
- Choi, S.K. , Park, S.Y. , Kim, R. , Kim, S. Bin , Lee, C.H. , Kim, J.F. , and Park, S.H. (2009) Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis . J Bacteriol 191: 3350–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumsakul, O. , Takahashi, H. , Oshima, T. , Hishimoto, T. , Kanaya, S. , Ogasawara, N. , and Ishikawa, S. (2011) Genome‐wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res 39: 414–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas Malvido, M. , Alonso González, E. , and Pérez Guerra, N. (2016) Nisin production in realkalized fed‐batch cultures in whey with feeding with lactose‐ or glucose‐containing substrates. Appl Microbiol Biotechnol 100: 7899–7908. [DOI] [PubMed] [Google Scholar]
- Cummings, M. , Breitling, R. , and Takano, E. (2014) Steps towards the synthetic biology of polyketide biosynthesis. FEMS Microbiol Lett 351: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, R.H. , Zhang, F. , Alonso‐Gutierrez, J. , Baidoo, E. , Batth, T.S. , Redding‐Johanson, A.M. , et al. (2013) Engineering dynamic pathway regulation using stress‐response promoters. Nat Biotechnol 31: 1039–1046. [DOI] [PubMed] [Google Scholar]
- Daliri, E.B.M. , Lee, B.H. , and Oh, D.H. (2018) Current trends and perspectives of bioactive peptides. Crit Rev Food Sci Nutr 58: 2273–2284. [DOI] [PubMed] [Google Scholar]
- Dang, T. , and Süssmuth, R.D. (2017) Bioactive peptide natural products as lead structures for medicinal use. Acc Chem Res 50: 1566–1576. [DOI] [PubMed] [Google Scholar]
- Dang, Y. , Zhao, F. , Liu, X. , Fan, X. , Huang, R. , Gao, W. , et al. (2019) Enhanced production of antifungal lipopeptide iturin A by Bacillus amyloliquefaciens LL3 through metabolic engineering and culture conditions optimization. Microb Cell Fact 18: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez, V. , Loznik, M. , Taylor, S. , Winn, M. , Rattray, N.J.W. , Podmore, H. , et al. (2015) Functional exchangeability of oxidase and dehydrogenase reactions in the biosynthesis of hydroxyphenylglycine, a nonribosomal peptide building block. ACS Synth Biol 4: 796–807. [DOI] [PubMed] [Google Scholar]
- Dörr, T. , Alvarez, L. , Delgado, F. , Davis, B.M. , Cava, F. , and Waldor, M.K. (2016) A cell wall damage response mediated by a sensor kinase/response regulator pair enables beta‐lactam tolerance. Proc Natl Acad Sci USA 113: 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinspach, K. , Kapitzke, C. , Tocchetti, A. , Sosio, M. , and Apel, A.K. (2014) Heterologous expression of the thiopeptide antibiotic GE2270 from Planobispora rosea ATCC 53733 in Streptomyces coelicolor requires deletion of ribosomal genes from the expression construct. PLoS One 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomenko, D. , Veselovskii, A. , and Khmel, I. (2001) Regulation of microcin C51 operon expression: the role of global regulators of transcription. Res Microbiol 152: 469–479. [DOI] [PubMed] [Google Scholar]
- Gao, H. , Zhuo, Y. , Ashforth, E. , and Zhang, L. (2010) Engineering of a genome‐reduced host: practical application of synthetic biology in the overproduction of desired secondary metabolites. Protein Cell 1: 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde, D.R. , Snell, E.A. , Jachimowicz, D. , Sipos, B. , Lloyd, J.H. , Bruce, M. , et al. (2018) Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15: 201–206. [DOI] [PubMed] [Google Scholar]
- Gidijala, L. , Kiel, J.A.K.W. , Douma, R.D. , Seifar, R.M. , van Gulik, W.M. , Bovenberg, R.A.L. , et al. (2009) An engineered yeast efficiently secreting penicillin. PLoS One 4: e8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Escribano, J.P. , and Bibb, M.J. (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guez, J.S. , Müller, C.H. , Danze, P.M. , Büchs, J. , and Jacques, P. (2008) Respiration activity monitoring system (RAMOS), an efficient tool to study the influence of the oxygen transfer rate on the synthesis of lipopeptide by Bacillus subtilis ATCC6633. J Biotechnol 134: 121–126. [DOI] [PubMed] [Google Scholar]
- Hammer, S.K. , and Avalos, J.L. (2017) Harnessing yeast organelles for metabolic engineering. Nat Chem Biol 13: 823–832. [DOI] [PubMed] [Google Scholar]
- Herr, A. , and Fischer, R. (2014) Improvement of Aspergillus nidulans penicillin production by targeting AcvA to peroxisomes. Metab Eng 25: 131–139. [DOI] [PubMed] [Google Scholar]
- Herzner, A.M. , Dischinger, J. , Szekat, C. , Josten, M. , Schmitz, S. , Yakéléba, A. , et al. (2011) Expression of the lantibiotic mersacidin in Bacillus amyloliquefaciens FZB42. PLoS One 6: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick, K.J. , and van der Donk, W.A. (2017) Ribosomally synthesized and post‐translationally modified peptide natural product discovery in the genomic era. Curr Opin Chem Biol 38: 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes, P.M. , Allen, S.E. , Hwang, S. , and Bowers, A.A. (2016) Production of sactipeptides in Escherichia coli: probing the substrate promiscuity of subtilosin A biosynthesis. ACS Chem Biol 11: 1737–1744. [DOI] [PubMed] [Google Scholar]
- Hoppert, M. , Gentzsch, C. , and Schörgendorfer, K. (2001) Structure and localization of cyclosporin synthetase, the key enzyme of cyclosporin biosynthesis in Tolypocladium inflatum . Arch Microbiol 176: 285–293. [DOI] [PubMed] [Google Scholar]
- Horbal, L. , Kobylyanskyy, A. , Truman, A.W. , Zaburranyi, N. , Ostash, B. , Luzhetskyy, A. , et al. (2014) The pathway‐specific regulatory genes, tei15* and tei16*, are the master switches of teicoplanin production in Actinoplanes teichomyceticus . Appl Microbiol Biotechnol 98: 9295–9309. [DOI] [PubMed] [Google Scholar]
- Horbal, L. , Marques, F. , Nadmid, S. , Mendes, M.V. , and Luzhetskyy, A. (2018) Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab Eng 49: 299–315. [DOI] [PubMed] [Google Scholar]
- Hossain, G.S. , Saini, M. , Miyake, R. , Ling, H. , and Chang, M.W. (2020) Genetic biosensor design for natural product biosynthesis in microorganisms. Trends Biotechnol 38: 797–810. [DOI] [PubMed] [Google Scholar]
- Hu, H. , and Ochi, K. (2001) Novel approach for improving the productivity of antibiotic‐producing strains by inducing combined resistant mutations. Appl Environ Microbiol 67: 1885–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. , Wen, J. , Wang, G. , Yu, G. , Jia, X. , and Chen, Y. (2012) In silico aided metabolic engineering of Streptomyces roseosporus for daptomycin yield improvement. Appl Microbiol Biotechnol 94: 637–649. [DOI] [PubMed] [Google Scholar]
- Hubbard, B.K. , Thomas, M.G. , and Walsh, C.T. (2000) Biosynthesis of L‐p‐hydroxyphenylglycine, a non‐proteinogenic amino acid constituent of peptide antibiotics. Chem Biol 7: 931–942. [DOI] [PubMed] [Google Scholar]
- Huo, L. , Rachid, S. , Stadler, M. , Wenzel, S.C. , and Müller, R. (2012) Synthetic biotechnology to study and engineer ribosomal bottromycin biosynthesis. Chem Biol 19: 1278–1287. [DOI] [PubMed] [Google Scholar]
- Huo, L. , and Van Der Donk, W.A. (2016) Discovery and characterization of bicereucin, an unusual D‐amino acid‐containing mixed two‐component lantibiotic. J Am Chem Soc 138: 8–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftime, D. , Jasyk, M. , Kulik, A. , Imhoff, J.F. , Stegmann, E. , Wohlleben, W. , et al. (2015) Streptocollin, a type IV lanthipeptide produced by Streptomyces collinus Tü 365. ChemBioChem 16: 2615–2623. [DOI] [PubMed] [Google Scholar]
- Ji, C.H. , Kim, J.P. , and Kang, H.S. (2018) Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters. ACS Synth Biol 7: 1946–1955. [DOI] [PubMed] [Google Scholar]
- Ji, S. , Li, W. , Baloch, A.R. , Wang, M. , and Cao, B. (2015) Improved production of sublancin via introduction of three characteristic promoters into operon clusters responsible for this novel distinct glycopeptide biosynthesis. Microb Cell Fact 14: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J. , Yu, K.O. , Ramzi, A.B. , Choe, S.H. , Kim, S.W. , and Han, S.O. (2012) Improvement of surfactin production in Bacillus subtilis using synthetic wastewater by overexpression of specific extracellular signaling peptides, comX and phrC. Biotechnol Bioeng 109: 2349–2356. [DOI] [PubMed] [Google Scholar]
- Kaunietis, A. , Buivydas, A. , Čitavičius, D.J. , and Kuipers, O.P. (2019) Heterologous biosynthesis and characterization of a glycocin from a thermophilic bacterium. Nat Commun 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar, S.A. , Suarez Duran, H.G. , Blin, K. , Osbourn, A. , and Medema, M.H. (2017) PlantiSMASH: Automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res 45: W55–W63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara, T. , Izumikawa, M. , Kozone, I. , Hashimoto, J. , Kagaya, N. , Koiwai, H. , et al. (2018) Neothioviridamide, a polythioamide compound produced by heterologous expression of a Streptomyces sp. cryptic RiPP biosynthetic gene cluster. J Nat Prod 81: 264–269. [DOI] [PubMed] [Google Scholar]
- Keller, N.P. (2015) Translating biosynthetic gene clusters into fungal armor and weaponry. Nat Chem Biol 11: 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi, N. , Seifuddin, F.T. , Turner, G. , Haft, D. , Nierman, W.C. , Wolfe, K.H. , and Fedorova, N.D. (2010) SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47: 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, A.S. , and Collins, J.J. (2010) Synthetic biology: applications come of age. Nat Rev Genet 11: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koos, J.D. , and Link, A.J. (2019) Heterologous and in vitro reconstitution of fuscanodin, a lasso peptide from Thermobifida fusca . J Am Chem Soc 141: 928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri, S. , and Church, G.M. (2014) Large‐scale de novo DNA synthesis: technologies and applications. Nat Methods 11: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri, S. , Goodman, D.B. , Cambray, G. , Mutalik, V.K. , Gao, Y. , Arkin, A.P. , et al. (2013) Composability of regulatory sequences controlling transcription and translation in Escherichia coli . Proc Natl Acad Sci USA 110: 14024–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers, A. , de Boef, E. , Rink, R. , Fekken, S. , Kluskens, L.D. , Driessen, A.J.M. , et al. (2004) NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with non‐lantibiotic peptides. J Biol Chem 279: 22176–22182. [DOI] [PubMed] [Google Scholar]
- Kuipers, A. , Wierenga, J. , Rink, R. , Kluskens, L.D. , Driessen, A.J.M. , Kuipers, O.P. , and Moll, G.N. (2006) Sec‐mediated transport of posttranslationally dehydrated peptides in Lactococcus lactis . Appl Environ Microbiol 72: 7626–7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuthning, A. , Mösker, E. , and Süssmuth, R.D. (2015) Engineering the heterologous expression of lanthipeptides in Escherichia coli by multigene assembly. Appl Microbiol Biotechnol 99: 6351–6361. [DOI] [PubMed] [Google Scholar]
- Landgrebe, D. , Haake, C. , Höpfner, T. , Beutel, S. , Hitzmann, B. , Scheper, T. , et al. (2010) On‐line infrared spectroscopy for bioprocess monitoring. Appl Microbiol Biotechnol 88: 11–22. [DOI] [PubMed] [Google Scholar]
- Larroude, M. , Rossignol, T. , Nicaud, J.M. , and Ledesma‐Amaro, R. (2018) Synthetic biology tools for engineering Yarrowia lipolytica . Biotechnol Adv 36: 2150–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.W. , Ng, B.G. , and Kim, B.S. (2015) Increased valinomycin production in mutants of Streptomyces sp. M10 defective in bafilomycin biosynthesis and branched‐chain α‐keto acid dehydrogenase complex expression. J Ind Microbiol Biotechnol 42: 1507–1517. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Jin, K. , Zhang, L. , Ding, Z. , Gu, Z. , and Shi, G. (2018a) Development of an inducible secretory expression system in Bacillus licheniformis based on an engineered xylose operon. J Agric Food Chem 66: 9456–9464. [DOI] [PubMed] [Google Scholar]
- Li, J. , Li, Y. , Niu, G. , Guo, H. , Qiu, Y. , Lin, Z. , et al. (2018b) NosP‐regulated nosiheptide production responds to both peptidyl and small‐molecule ligands derived from the precursor peptide. Cell Chem Biol 25: 143–153. [DOI] [PubMed] [Google Scholar]
- Li, Xu , Yang, H. , Zhang, D. , Li, X. , Yu, H. , and Shen, Z. (2015) Overexpression of specific proton motive force‐dependent transporters facilitate the export of surfactin in Bacillus subtilis . J Ind Microbiol Biotechnol 42: 93–103. [DOI] [PubMed] [Google Scholar]
- Lin, Y. , Teng, K. , Huan, L. , and Zhong, J. (2011) Dissection of the bridging pattern of bovicin HJ50, a lantibiotic containing a characteristic disulfide bridge. Microbiol Res 166: 146–154. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Jiang, H. , Haltli, B. , Kulowski, K. , Muszynska, E. , Feng, X. , et al. (2009) Rapid cloning and heterologous expression of the meridamycin biosynthetic gene cluster using a versatile Escherichia coli‐Streptomyces artificial chromosome vector, pSBAC. J Nat Prod 72: 389–395. [DOI] [PubMed] [Google Scholar]
- Lopatniuk, M. , Myronovskyi, M. , Nottebrock, A. , Busche, T. , Kalinowski, J. , Ostash, B. , et al. (2019) Effect of “ribosome engineering” on the transcription level and production of S. albus indigenous secondary metabolites. Appl Microbiol Biotechnol 103: 7097–7110. [DOI] [PubMed] [Google Scholar]
- Lou, C. , Stanton, B. , Chen, Y.J. , Munsky, B. , and Voigt, C.A. (2012) Ribozyme‐based insulator parts buffer synthetic circuits from genetic context. Nat Biotechnol 30: 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, F. , Wang, Z. , Zhao, W. , Chu, J. , and Zhuang, Y. (2016) A simple novel approach for real‐time monitoring of sodium gluconate production by on‐line physiological parameters in batch fermentation by Aspergillus niger . Bioresour Technol 202: 133–141. [DOI] [PubMed] [Google Scholar]
- Lubelski, J. , Rink, R. , Khusainov, R. , Moll, G.N. , and Kuipers, O.P. (2008) Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci 65: 455–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z. , Ye, C. , Deng, W. , Xu, M. , Wang, Q. , Liu, G. , et al. (2018) Reconstruction and analysis of a genome‐scale metabolic model of Ganoderma lucidum for improved extracellular polysaccharide production. Front Microbiol 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson, M. , and Saka, Y. (2017) Short synthetic terminators for assembly of transcription units in vitro and stable chromosomal integration in yeast S. Cerevisiae . ACS Synth Biol 6: 130–138. [DOI] [PubMed] [Google Scholar]
- Mao, X.M. , Luo, S. , and Li, Y.Q. (2017) Negative regulation of daptomycin production by DepR2, an ArsR‐family transcriptional factor. J Ind Microbiol Biotechnol 44: 1653–1658. [DOI] [PubMed] [Google Scholar]
- Mao, X.M. , Luo, S. , Zhou, R.C. , Wang, F. , Yu, P. , Sun, N. , et al. (2015) Transcriptional regulation of the daptomycin gene cluster in Streptomyces roseosporus by an autoregulator, AtrA. J Biol Chem 290: 7992–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahier, M.A. , Nakano, M.M. , and Zuber, P. (1993) Regulation of peptide antibiotic production in Bacillus . Mol Microbiol 7: 631–636. [DOI] [PubMed] [Google Scholar]
- Martin‐Gómez, H. , Linne, U. , Albericio, F. , Tulla‐Puche, J. , and Hegemann, J.D. (2018) Investigation of the biosynthesis of the lasso peptide chaxapeptin using an E. coli‐based production system. J Nat Prod 81: 2050–2056. [DOI] [PubMed] [Google Scholar]
- Medema, M.H. , Paalvast, Y. , Nguyen, D.D. , Melnik, A. , Dorrestein, P.C. , Takano, E. , and Breitling, R. (2014) Pep2Path: Automated mass spectrometry‐guided genome mining of peptidic natural products. PLoS Comput Biol 10: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, W.H. , Gidijala, L. , Fekken, S. , Kiel, J.A.K.W. , van den Berg, M.A. , Lascaris, R. , et al. (2010) Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum . Appl Environ Microbiol 76: 5702–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani, H. , Kersten, R.D. , Liu, W.‐T. , Wang, M. , Purvine, S.O. , Wu, S. , et al. (2014) Automated genome mining of ribosomal peptide natural products. ACS Chem Biol 9: 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalbán‐López, M. , Scott, T.A. , Ramesh, S. , Rahman, I.R. , van Heel, A.J. , Viel, J.H. , et al. (2020) New developments in RiPP discovery, enzymology and engineering. Nat Prod Rep. 10.1039/d0np00027b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk, I. , and Kobayashi, I. (2014) To be or not to be: regulation of restriction‐modification systems and other toxin‐antitoxin systems. Nucleic Acids Res 42: 70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myronovskyi, M. , and Luzhetskyy, A. (2016) Native and engineered promoters in natural product discovery. Nat Prod Rep 33: 1006–1019. [DOI] [PubMed] [Google Scholar]
- Nguyen, K. t , Ritz, D. , Gu, J‐q , Alexander, D. , Chu, M. , Miao, V. , et al. (2006) Combinatorial biosynthesis of novel antibiotics related to daptomycin. Proc Natl Acad Sci USA 103: 17462–17467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Z.‐J. , Zhang, X.‐y. , Liu, F. , Wang, M. , Hao, R.‐h. , Ling, P.‐x. , and Zhu, X.‐Q. (2017) Effect of co‐overexpression of nisin key genes on nisin production improvement in Lactococcus lactis LS01. Probiotics Antimicrob Proteins 9: 204–212. [DOI] [PubMed] [Google Scholar]
- Niu, D. , Zuo, Z. , Shi, G.Y. , and Wang, Z.X. (2009) High yield recombinant thermostable α‐amylase production using an improved Bacillus licheniformis system. Microb Cell Fact 8: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongey, E.L. , Giessmann, R.T. , Fons, M. , Rappsilber, J. , Adrian, L. , and Neubauer, P. (2018) Heterologous biosynthesis, modifications and structural characterization of ruminococcin‐A, a lanthipeptide from the gut bacterium Ruminococcus gnavus E1, in Escherichia coli . Front Microbiol 9: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglia, G. , Hrafnsdóttir, S. , Magnúsdóttir, M. , Fleming, R.M.T. , Thorlacius, S. , Palsson, B. , and Thiele, I. (2012) Monitoring metabolites consumption and secretion in cultured cells using ultra‐performance liquid chromatography quadrupole‐time of flight mass spectrometry (UPLC‐Q‐ToF‐MS). Anal Bioanal Chem 402: 1183–1198. [DOI] [PubMed] [Google Scholar]
- Pandey, R.P. , Parajuli, P. , Koffas, M.A.G. , and Sohng, J.K. (2016) Microbial production of natural and non‐natural flavonoids: Pathway engineering, directed evolution and systems/synthetic biology. Biotechnol Adv 34: 634–662. [DOI] [PubMed] [Google Scholar]
- Park, S.‐Y. , Choi, S.‐K. , Kim, J. , Oh, T.‐K. , and Park, S.‐H. (2012) Efficient production of polymyxin in the surrogate host Bacillus subtilis by introducing a foreign ectB gene and disrupting the abrB gene. Appl Environ Microbiol 78: 4194–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger, B.F. , Pitera, D.J. , Smolke, C.D. , and Keasling, J.D. (2006) Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol 24: 1027–1032. [DOI] [PubMed] [Google Scholar]
- Phillips, Z.E.V. , and Strauch, M.A. (2002) Bacillus subtilis sporulation and stationary phase gene expression. Cell Mol Life Sci 59: 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podevels, A.M. , Felnagle, E.A. , McMahon, M.D. , Berti, A.D. , Chan, Y.A. , Thomas, M.G. , and Jackson, E.E. (2008) Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm 5: 191–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, J. , Caiyin, Q. , Wu, H. , Tian, K. , Wang, B. , Li, Y. , and Qiao, J. (2017) The novel sRNA s015 improves nisin yield by increasing acid tolerance of Lactococcus lactis F44. Appl Microbiol Biotechnol 101: 6483–6493. [DOI] [PubMed] [Google Scholar]
- Qiu, Y. , Xiao, F. , Wei, X. , Wen, Z. , and Chen, S. (2014) Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl Microbiol Biotechnol 98: 8895–8903. [DOI] [PubMed] [Google Scholar]
- Reverchon, S. , Rouanet, C. , Expert, D. , and Nasser, W. (2002) Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J Bacteriol 184: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo, G. , Carrera, J. , and Jaramillo, A. (2011) Computational design of synthetic regulatory networks from a genetic library to characterize the designability of dynamical behaviors. Nucleic Acids Res 39: e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, A.C. , Liu, H. , Pattabiraman, V.R. , and Vederas, J.C. (2010) Synthesis of the lantibiotic lactocin S using peptide cyclizations on solid phase. J Am Chem Soc 132: 462–463. [DOI] [PubMed] [Google Scholar]
- Seo, S.W. , Yang, J. , Min, B.E. , Jang, S. , Lim, J.H. , Lim, H.G. , et al. (2013) Synthetic biology: tools to design microbes for the production of chemicals and fuels. Biotechnol Adv 31: 811–817. [DOI] [PubMed] [Google Scholar]
- Severinov, K. , Semenova, E. , Kazakov, A. , Kazakov, T. , and Gelfand, M.S. (2007) Low‐molecular‐weight post‐translationally modified microcins. Mol Microbiol 65: 1380–1394. [DOI] [PubMed] [Google Scholar]
- Shao, Z. , Rao, G. , Li, C. , Abil, Z. , Luo, Y. , and Zhao, H. (2013) Refactoring the silent spectinabilin gene cluster using a plug‐and‐play scaffold. ACS Synth Biol 2: 662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Zeng, Y.J. , Zhang, B. , Shao, F.L. , Chen, Y.C. , Xu, X. , et al. (2019) Comparative genome mining and heterologous expression of an orphan NRPS gene cluster direct the production of ashimides. Chem Sci 10: 3042–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl, T. , Tokovenko, B. , Myronovskyi, M. , and Luzhetskyy, A. (2013) Design, construction and characterisation of a synthetic promoter library for fine‐tuned gene expression in actinomycetes. Metab Eng 19: 98–106. [DOI] [PubMed] [Google Scholar]
- Singh, S.B. , and Barrett, J.F. (2006) Empirical antibacterial drug discovery ‐ Foundation in natural products. Biochem Pharmacol 71: 1006–1015. [DOI] [PubMed] [Google Scholar]
- Skaugen, M. , Abildgaard, C.I.M. , and Nes, I.F. (1997) Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol Gen Genet 253: 674–686. [DOI] [PubMed] [Google Scholar]
- Skinnider, M.A. , Merwin, N.J. , Johnston, C.W. , and Magarvey, N.A. (2017) PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res 45: W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarczyk, A.L. , Lin, A. , and Weiss, R. (2012) Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet 13: 406–420. [DOI] [PubMed] [Google Scholar]
- Smanski, M.J. , Zhou, H. , Claesen, J. , Shen, B. , Fischbach, M.A. , and Voigt, C.A. (2016) Synthetic biology to access and expand nature’s chemical diversity. Nat Rev Microbiol 14: 135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola‐Landa, A. , Moura, R.S. , and Martin, J.F. (2003) The two‐component PhoR‐PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans . Proc Natl Acad Sci USA 100: 6133–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlik, T. , Sandrock, B. , Ast, J. , and Freitag, J. (2014) Fungal peroxisomes as biosynthetic organelles. Curr Opin Microbiol 22: 8–14. [DOI] [PubMed] [Google Scholar]
- Stein, T. , Borchert, S. , Kiesau, P. , Heinzmann, S. , Klöss, S. , Klein, C. , et al. (2002) Dual control of subtilin biosynthesis and immunity in Bacillus subtilis . Mol Microbiol 44: 403–416. [DOI] [PubMed] [Google Scholar]
- Strauch, M.A. , Spiegelman, G.B. , Perego, M. , Johnson, W.C. , Burbulys, D. , and Hoch, J.A. (1989) The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J 8: 1615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, K. , Mulaku, E. , Dandapani, H. , Nagy, C. , Aro, E.M. , and Kallio, P. (2018) Translation efficiency of heterologous proteins is significantly affected by the genetic context of RBS sequences in engineered cyanobacterium Synechocystis sp. PCC 6803. Microb Cell Fact 17: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thykaer, J. , Nielsen, J. , Wohlleben, W. , Weber, T. , Gutknecht, M. , Lantz, A.E. , and Stegmann, E. (2010) Increased glycopeptide production after overexpression of shikimate pathway genes being part of the balhimycin biosynthetic gene cluster. Metab Eng 12: 455–461. [DOI] [PubMed] [Google Scholar]
- Tianero, M.D.B. , Donia, M.S. , Young, T.S. , Schultz, P.G. , and Schmidt, E.W. (2012) Ribosomal route to small‐molecule diversity. J Am Chem Soc 134: 418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet, D. , Engelen, S. , Mornico, D. , Cruveiller, S. , Fleury, L. , Lajus, A. , et al. (2009) MicroScope: a platform for microbial genome annotation and comparative genomics. Database 2009: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Cress, B.F. , Yang, Z. , Hordines, J.C. , Zhao, S. , Jung, G.Y. , et al. (2019) Design and characterization of biosensors for the screening of modular assembled naringenin biosynthetic library in Saccharomyces cerevisiae . ACS Synth Biol 8: 2121–2130. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Li, S. , Li, Z. , Zhang, J. , Fan, K. , Tan, G. , et al. (2020) Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces . Nat Biotechnol 38: 76–83. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Yu, H. , Wang, M. , Yang, H. , and Shen, Z. (2018) Enhanced biosynthesis and characterization of surfactin isoforms with engineered Bacillus subtilis through promoter replacement and Vitreoscilla hemoglobin co‐expression. Process Biochem 70: 36–44. [Google Scholar]
- Wang, Q. , Zhang, D. , Li, Y. , Zhang, F. , Wang, C. , and Liang, X. (2014) Genome shuffling and ribosome engineering of Streptomyces actuosus for high‐yield nosiheptide production. Appl Biochem Biotechnol 173: 1553–1563. [DOI] [PubMed] [Google Scholar]
- Watanabe, K. , Hotta, K. , Praseuth, A.P. , Koketsu, K. , Migita, A. , Boddy, C.N. , et al. (2006) Total biosynthesis of antitumor nonribosomal peptides in Escherichia coli . Nat Chem Biol 2: 423–428. [DOI] [PubMed] [Google Scholar]
- Wehrs, M. , Prahl, J.‐P. , Moon, J. , Li, Y. , Tanjore, D. , Keasling, J.D. , et al. (2018) Production efficiency of the bacterial non‐ribosomal peptide indigoidine relies on the respiratory metabolic state in S. cerevisiae . Microb Cell Fact 17: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiz, A.R. , Ishida, K. , Makower, K. , Ziemert, N. , Hertweck, C. , and Dittmann, E. (2011) Leader peptide and a membrane protein scaffold guide the biosynthesis of the tricyclic peptide microviridin. Chem Biol 18: 1413–1421. [DOI] [PubMed] [Google Scholar]
- Winn, M. , Fyans, J.K. , Zhuo, Y. , and Micklefield, J. (2016) Recent advances in engineering nonribosomal peptide assembly lines. Nat Prod Rep 33: 317–347. [DOI] [PubMed] [Google Scholar]
- Wu, Y. , Chen, T. , Liu, Y. , Tian, R. , Lv, X. , Li, J. , et al. (2020) Design of a programmable biosensor‐CRISPRi genetic circuits for dynamic and autonomous dual‐control of metabolic flux in Bacillus subtilis . Nucleic Acids Res 48: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G. , Yan, Q. , Jones, J.A. , Tang, Y.J. , Fong, S.S. , and Koffas, M.A.G. (2016) Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol 34: 652–664. [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Zhi, Y. , and Xu, Y. (2019) Systematically engineering the biosynthesis of a green biosurfactant surfactin by Bacillus subtilis 168. Metab Eng 52: 87–97. [DOI] [PubMed] [Google Scholar]
- Xiao, H. , Chen, X. , Chen, M. , Tang, S. , Zhao, X. , and Huan, L. (2004) Bovicin HJ50, a novel lantibiotic produced by Streptococcus bovis HJ50. Microbiology 150: 103–108. [DOI] [PubMed] [Google Scholar]
- Xu, P. , Li, L. , Zhang, F. , Stephanopoulos, G. , and Koffas, M. (2014) Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA 111: 11299–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. , Vansiri, A. , Bhan, N. , and Koffas, M.A.G. (2012) EPathBrick: A synthetic biology platform for engineering metabolic pathways in E. coli . ACS Synth Biol 1: 256–266. [DOI] [PubMed] [Google Scholar]
- Yamanaka, K. , Reynolds, K.A. , Kersten, R.D. , Ryan, K.S. , Gonzalez, D.J. , Nizet, V. , et al. (2014) Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci USA 111: 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, F. , Burgard, C. , Popoff, A. , Zaburannyi, N. , Zipf, G. , Maier, J. , et al. (2018) Synthetic biology approaches and combinatorial biosynthesis towards heterologous lipopeptide production. Chem Sci 9: 7510–7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R. , Lei, S. , Xu, X. , Jin, H. , Sun, H. , Zhao, X. , et al. (2020) Key elements and regulation strategies of NRPSs for biosynthesis of lipopeptides by Bacillus . Appl Microbiol Biotechnol 104: 8077–8087. [DOI] [PubMed] [Google Scholar]
- Young, T.S. , and Walsh, C.T. (2011) Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans . Proc Natl Acad Sci USA 108: 13053–13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Hui, M. , Li, R. , Chen, L. , Tian, H. , and Wang, L. (2018) Enhancement of daptomycin production by the method of combining ribosome engineering and genome shuffling in Streptomyces roseosporus . Appl Biochem Microbiol 54: 611–615. [Google Scholar]
- Yuan, P. , Zhou, R. , Chen, X. , Luo, S. , Wang, F. , Mao, X. , and Li, Y. (2016) DepR1, a TetR family transcriptional regulator, positively regulates daptomycin production in an industrial producer, Streptomyces roseosporus SW0702. Appl Environ Microbiol 82: 1898–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuksel, S. , and Hansen, J.N. (2007) Transfer of nisin gene cluster from Lactococcus lactis ATCC 11454 into the chromosome of Bacillus subtilis 168. Appl Microbiol Biotechnol 74: 640–649. [DOI] [PubMed] [Google Scholar]
- Zambaldo, C. , Luo, X. , Mehta, A.P. , and Schultz, P.G. (2017) Recombinant macrocyclic lanthipeptides incorporating non‐canonical amino acids. J Am Chem Soc 139: 11646–11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Caiyin, Q. , Feng, W. , Zhao, X. , Qiao, B. , Zhao, G. , and Qiao, J. (2016) Enhance nisin yield via improving acid‐tolerant capability of Lactococcus lactis F44. Sci Rep 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , and Hong, K. (2020) Production of terpenoids by synthetic biology approaches. Front Bioeng Biotechnol 8: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.M. , Wang, H.H. , Liu, X. , Hu, C.H. , and Zou, Y. (2020) Heterologous and engineered biosynthesis of nematocidal polyketide‐nonribosomal peptide hybrid macrolactone from extreme thermophilic fungi. J Am Chem Soc 142: 1957–1965. [DOI] [PubMed] [Google Scholar]
- Zhang, P. , Wu, H. , Chen, X.L. , Deng, Z. , Bai, L. , and Pang, X. (2014) Regulation of the biosynthesis of thiopeptide antibiotic cyclothiazomycin by the transcriptional regulator SHJG8833 in Streptomyces hygroscopicus 5008. Microbiology 160: 1379–1392. [DOI] [PubMed] [Google Scholar]
- Zobel, S. , Boecker, S. , Kulke, D. , Heimbach, D. , Meyer, V. , and Süssmuth, R.D. (2016) Reprogramming the biosynthesis of cyclodepsipeptide synthetases to obtain new enniatins and beauvericins. ChemBioChem 17: 283–287. [DOI] [PubMed] [Google Scholar]
- Zobel, S. , Kumpfmüller, J. , Süssmuth, R.D. , and Schweder, T. (2015) Bacillus subtilis as heterologous host for the secretory production of the non‐ribosomal cyclodepsipeptide enniatin. Appl Microbiol Biotechnol 99: 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]


