Summary
Yarrowia lipolytica is widely used as a microbial producer of lipids and lipid derivatives. Here, we exploited this yeast’s potential to generate aromatic amino acids by developing chassis strains optimized for the production of phenylalanine, tyrosine and tryptophan. We engineered the shikimate pathway to overexpress a combination of Y. lipolytica and heterologous feedback‐insensitive enzyme variants. Our best chassis strain displayed high levels of de novo Ehrlich metabolite production (up to 0.14 g l−1 in minimal growth medium), which represented a 93‐fold increase compared to the wild‐type strain (0.0015 g l−1). Production was further boosted to 0.48 g l−1 when glycerol, a low‐cost carbon source, was used, concomitantly to high secretion of phenylalanine precursor (1 g l−1). Among these metabolites, 2‐phenylethanol is of particular interest due to its rose‐like flavour. We also established a production pathway for generating protodeoxyviolaceinic acid, a dye derived from tryptophan, in a chassis strain optimized for chorismate, the precursor of tryptophan.
We have thus demonstrated that Y. lipolytica can serve as a platform for the sustainable de novo bio‐production of high‐value aromatic compounds, and we have greatly improved our understanding of the potential feedback‐based regulation of the shikimate pathway in this yeast.
Yarrowia lipolytica chassis strains optimized for the de novo production of aromatic amino acids have been generated by engineering the shikimate pathway and deciphering its feedback regulations. The chassis were used to produce high‐value aromatic compounds i.e. Ehrlich metabolites and protodeoxyviolaceinic acid.
Introduction
Yarrowia lipolytica is an oleaginous yeast that is widely used as an industrial chassis to produce lipids and lipid derivatives because the species naturally accumulates high levels of these compounds (Ledesma‐Amaro and Nicaud, 2016a). As an industrial host, Y. lipolytica presents several advantages. For example, a vast repertoire of synthetic biology tools have been developed for this microbe (Larroude et al., 2018, 2019). It naturally grows in low‐cost substrates such as glycerol or molasses (Ledesma‐Amaro and Nicaud, 2016b) and, once engineered, can also grow in xylose, raw starch, inulin and even cellobiose or cellulose (Zhao et al., 2010; Wei et al., 2014; Guo et al., 2015; Ledesma‐Amaro et al., 2015, 2016). It is therefore a highly promising microbial factory for sustainably producing biobased molecules. In addition to lipids, Y. lipolytica can also generate aromatic compounds such as 2‐phenylethanol (2‐PE), via the Ehrlich pathway (Celińska et al., 2019, 2013, 2015) or flavonoids when engineered (e.g. naringenin or taxifolin; Lv et al., 2019). These compounds are of particular interest to the cosmetics, food and healthcare industries. The aromatic amino acids (AAA) biosynthetic pathway is a major source of biobased aromatic compounds. It provides key precursors for synthetizing chemicals with potential applications in human health and nutrition, including flavonoids, stilbenes and alkaloids (Koopman et al., 2012; Gottardi et al., 2017; Suástegui and Shao, 2016). Consequently, this metabolic pathway could be an excellent target for engineering efforts that seek to transform Y. lipolytica into a cell factory for aromatic compounds of commercial value.
The AAA pathway is present in many microorganisms and plants and is made up of three main parts: (i) the shikimic acid pathway, (ii) the L‐tryptophan (TRP) branch and (iii) the l‐tyrosine (TYR) and l‐phenylalanine (PHE) branches (Fig. 1).
Fig. 1.
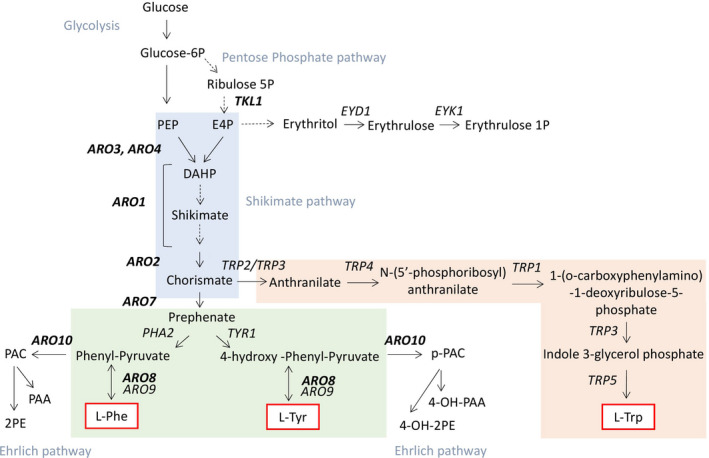
Schematic representation of the AAA pathway separated into three parts: (1) the shikimate pathway (in grey), (2) the L‐TRP branch (in orange) and (3) the L‐TYR and L‐PHE branches (in green). Also shown is precursor availability through the glycolysis and pentose phosphate pathways as well as catabolism through the Ehrlich pathway and the erythritol pathway. The genes involved in the pathways are indicated next to the arrows representing the reactions. Dotted arrows represent multistep pathways, and bidirectional arrows indicate reversible reactions. PAC: phenyl acetaldehyde; 2PE: 2‐phenylethanol; 4OH2PE: 2‐(4‐hydroxyphenyl)ethanol; PAA: 2‐phenylacetate; and 4OHPAA: 2‐(4‐hydroxyphenyl)acetate.
The shikimate pathway produces chorismate via a set of seven enzyme‐catalysed reactions. In Saccharomyces cerevisiae, this pathway is encoded by four genes. The first committed step in the shikimate pathway is the condensation of erythrose 4‐phosphate (E4P) and phosphoenolpyruvate (PEP), a reaction that is catalysed by deoxy‐d‐arabino‐heptulosonate‐7‐phosphate (DAHP) synthase. This reaction can be performed by two isoenzymes – Aro3 and Aro4. DAHP is then converted into chorismate via the successive action of Aro1 and Aro2. In yeasts, Aro1 is a multifunctional enzyme that catalyses five reactions and that is encoded by the ARO1 gene. Chorismate lies at the pathway’s first branching point: one branch leads to TRP, and the other leads to PHE and TYR. The biosynthesis of TRP involves five steps. The first step is the conversion of chorismate to anthranilate by anthranilate synthase (Trp2) and glutamine amidotransferase (Trp3). Anthranilate is then successively transformed into TRP by the enzymes Trp4, Trp1, Trp3 and Trp5. In contrast, only two steps are needed to create TYR and PHE from chorismate. Chorismate mutase, encoded by ARO7, produces prephenate (PPA), which is then directed to become either TYR or PHE. Prephenate dehydrogenase, encoded by TYR1, transforms PPA into p‐hydroxyphenylpyruvate (pOHPPY), while prephenate dehydratase, encoded by PHA2, transforms PPA into 2‐oxo acid phenylpyruvate (PPY). pOHPPY and PPY can be transaminated to generate TYR or PHE, respectively, via the AAA transaminases I (Aro8) or II (Aro9) (Braus, 1991; Iraqui et al., 1998). Alternatively, pOHPPY and PPY can be decarboxylated to form a fusel aldehyde by the action of Aro10, as part of the catabolic Ehrlich pathway. The fusel aldehyde can either be reduced to generate a fusel alcohol or oxidized to generate a fusel acid, depending on the redox status of the cell (Hazelwood et al., 2008).
There are several reasons why Y. lipolytica presents advantages for AAA derivative production. First, when glycerol concentrations are high, the yeast increases its production of erythritol, whose precursor is also E4P (Carly et al., 2017b). This phenomenon shows the organism’s potential to produce high levels of E4P under certain conditions. The catabolism of erythritol was recently deciphered (Carly et al., 2017a, 2018), and the process can be engineered to promote E4P accumulation. Second, malonyl‐CoA is readily accessible in Y. lipolytica. This compound is a precursor for several molecules derived from AAAs, which means that this yeast has strong potential to serve as a chassis in the production of AAA derivatives.
The metabolic pathway leading to AAA production in Y. lipolytica was relatively unexplored until very recently. Therefore, it is important to take a closer look at this pathway if we wish to develop strains of industrial interest. More specifically, the objective should be increasing the available pool of AAAs.
The regulatory dynamics of the shikimate pathway remain poorly described in Y. lipolytica. However, in S. cerevisiae, feedback inhibition regulates the pathway’s first step: the action of Aro4 and Aro3 is inhibited by the presence of TYR and PHE respectively. Additionally, at the first branching point, Aro7 is inhibited by TYR and activated by TRP (Braus, 1991). A quantitative study examining the effects of feedback inhibition on AAA biosynthesis in S. cerevisiae found that introducing feedback‐insensitive alleles of Aro4 and Aro7 resulted in a 4.5‐fold increase of the flux through the AAA biosynthetic pathway (Luttik et al., 2008). Another study showed that the overexpression of a feedback‐resistant variant of Aro3 led to an enhanced flux through the shikimate pathway (Brückner et al., 2018). Deciphering how this pathway is regulated in Y. lipolytica will facilitate efforts to engineer it.
Synthetic biology tools have sped up pathway assembly and expression in heterologous hosts. However, to simplify and accelerate the development of microbial factories, it is crucial to construct chassis strains that efficiently produce generic precursors for numerous compounds of interest. Such an investment will also help reduce the costs of future development efforts.
Here, we constructed and characterized Y. lipolytica chassis strains that were optimized for AAA production via the overexpression of the AAA pathway. We achieved the latter by combining Y. lipolytica and S. cerevisiae genes, including those that encode deregulated enzyme variants and by deciphering feedback regulations of Aro4 and Aro7 in Y. lipolytica. Using Ehrlich metabolites derived from PHE and TYR as readouts, we demonstrate that metabolite production was greatly improved. Our best chassis strain had a production titre that was 93‐fold higher than that of the wild type. We were able to double this titre by using glycerol as a carbon source. The latter is known to favour erythritol production in Y. lipolytica, and erythritol is just one step away from the E4P precursor (Carly et al., 2017b, 2018; Carly and Fickers, 2018). We also created and validated a specific chassis strain for producing a TRP derivative by introducing a heterologous pathway that generated protodeoxyviolaceinic acid (PVA). PVA is a green dye that is a precursor of violacein, a violet dye with antibacterial activity.
Results
Construction of a chassis strain for Ehrlich metabolites
To construct a Y. lipolytica chassis strain for AAA production, we first overexpressed genes from the shikimate pathway, taking advantage of the recently developed Golden Gate cloning strategy (Larroude et al., 2019). This tool allows multiple genes to be overexpressed simultaneously, thus reducing transformation number and limiting the side‐effects that arise from multiple cycles of gene integration and marker rescue (Fickers et al., 2003; Guo et al., 2018).
All the ORFs coding for enzymes in the AAA pathway in Y. lipolytica were identified via BLAST: information for the Y. lipolytica genome found in the GRYC database (http://gryc.inra.fr/) and the KEGG database (https://www.genome.jp/kegg/) was validated with amino acid sequences of well‐characterized proteins in the AAA pathway in S. cerevisiae. The list of the Y. lipolytica genes of relevance for the AAA pathway and their homologies with S. cerevisiae genes are presented in Table S1.
To evaluate whether the pathway had been successfully engineered, we first attempted to measure AAA levels in the supernatant using HPLC. However, no AAA were detectable for the wild‐type strain (data not shown). We therefore used the levels of Ehrlich metabolites in the supernatant as a readout. These metabolites were 2‐(4‐hydroxyphenyl)ethanol (2‐4OHPE), 4‐hydroxyphenylacetic acid (4OHPAA), phenylethanol (2‐PE) and phenylacetic acid (PAA). They are derived from PHE and TYR and are secreted and detectable in S. cerevisiae (Luttik et al., 2008). In addition to serving as a readout for pathway engineering, they are themselves of industrial interest. In particular, 2‐PE is a higher alcohol with a rose flavour that is widely used in cosmetics and food production. Ehrlich metabolites have previously been found in the supernatant of some Y. lipolytica wild‐type strains, but not in all of them (Celińska et al., 2013). Follow‐up proteomics research identified proteins involved in the degradation of PHE via the Ehrlich pathway in Y. lipolytica. These identified proteins were solely detected in situations where PHE had been added to the medium (Celińska et al., 2015). This work revealed that YlAro10 plays an important role in PHE degradation and that YlAro8 is constitutively expressed. The overexpression of the two genes encoding these proteins resulted in a dramatic increase (50%) in 2‐PE production in media supplemented with 2 g l−1 of PHE (Celińska et al., 2019). In our study, Ehrlich metabolites occurred at a very low level (1.5 mg l−1) in the wild‐type strain in minimal medium without amino acid supplementation (Fig. 2 – strain W29). However, when YlARO8 and YlARO10 were overexpressed, the level of Ehrlich metabolites climbed (5 mg l−1) even though no precursors (i.e. PHE or TYR) had been added to the medium (Fig. 2 – strain JMY7891). This strain was therefore a good starting point for further modifications to the shikimate pathway and was used as a tool to develop a chassis strain for AAA production.
Fig. 2.
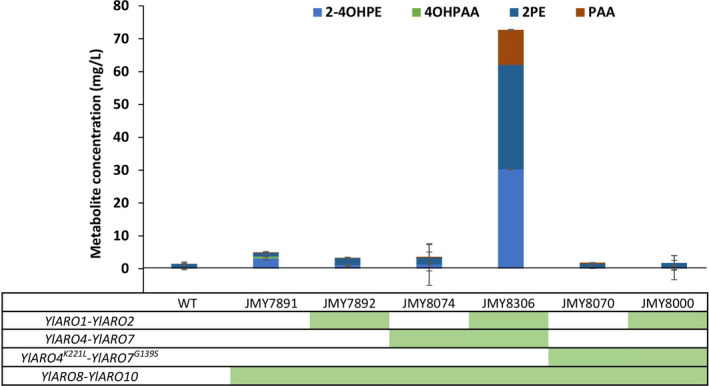
The series of engineered strains and their production of Ehrlich metabolites (in mg l−1) when cultured for 5 days in YNB medium containing 10 g l−1 of glucose. The strains’ genotypes are indicated by the green boxes located below the strains’ names. The mutated forms of Aro4 (YlAro4K221L) and Aro7 (YlAro7G139S) were equivalent to the feedback‐inhibited forms of the enzymes in S. cerevisiae (ScAro4K229L and ScAro7G141S). WT: wild‐type strain; 2‐4OHPE: 2‐(4‐hydroxyphenyl) ethanol; 4OHPAA: 4‐hydroxyphenylacetic acid; 2PE: phenylethanol; and PAA: phenylacetic acid.
The next step was to sequentially overexpress pairs of genes encoding shikimate pathway enzymes in this specific background. First, we overexpressed YlARO1‐YlARO2 or YlARO4‐YlARO7. Neither situation resulted in significant impacts on Ehrlich metabolite production (Fig. 2 – strains JMY7892 and JMY8074 respectively). However, when the four genes were overexpressed simultaneously (YlARO1‐YlARO2 and YlARO4‐YlARO7), we saw a considerable increase in production (up to 70 mg l−1; Fig. 2 – strain JMY8306).
In S. cerevisiae, two reactions in the shikimate pathway are known to be strongly regulated by feedback inhibition: (i) the pathway’s first committed step, which is catalysed by two isoforms of DAHP synthase (Aro4 and Aro3) and (ii) the reaction catalysed by chorismate mutase (Aro7). In S. cerevisiae, a single residue substitution can interfere with these feedback inhibitions. The expression of feedback‐insensitive forms of two of these enzymes (ScAro4K229L and ScAro7G141S) significantly increased flux through the AAA pathway (Luttik et al., 2008; Reifenrath et al., 2018). The amino acid sequences of YlAro4 and YlAro7 were compared to those of their homologues in S. cerevisiae. The sequences were well conserved (identity = 72% and 55% respectively), as were the amino acids involved in feedback regulation (Fig. S1). Consequently, we introduced mutations into these amino acids via PCR with the goal of expressing deregulated versions of the Y. lipolytica enzymes. Thus, following the approach used to create the feedback‐insensitive S. cerevisiae mutant, we converted the lysine at position 221 in the Aro4 of Y. lipolytica (which corresponds to the lysine at position 229 in the Aro4 of S. cerevisiae) into leucine (K221L), generating the YlAro4K221L variant. In S. cerevisiae, several point mutations were used to deregulate the enzyme Aro7 – the best characterized are G141S and T226I (Schnappauf et al., 1998; Krappmann et al., 2000; Luttik et al., 2008). The G141S mutation abolishes the effects of both TYR and TRP, while the T226I mutation abolishes TYR‐mediated inhibition but preserves TRP‐mediated stimulation. However, during our comparison of protein sequences, we found that only the equivalent of the G141 position was conserved in Y. lipolytica. As a result, we introduced a mutation to change glycine into serine at this site, which resulted in a Aro7 Y. lipolytica enzyme with the mutation G139S, generating the YlAro7G139S variant.
An expression cassette containing both mutated genes YlARO4K221L and YlARO7G139S was introduced into Y. lipolytica either overexpressing YlARO1 and YlARO2 or not (Fig. 2 – strains 8000 and 8070 respectively). Despite these efforts, neither of these mutant strains displayed Ehrlich metabolite levels that differed significantly from those of the parent strain.
Regulation of YlAro4 and YlAro7 in Y. lipolytica
Since we saw significantly improved AAA flux in the strain expressing the YlARO4‐YlARO7 cassette but not in the strains expressing the cassette containing the mutated YlARO4K221L and YlARO7G139S genes, we wanted to verify that the enzyme variants had been correctly expressed. All the genes – YlARO1, YlARO2, YlARO4K221L and YlARO7G139S – were tested by qRT‐PCR and were properly overexpressed in the strain JMY8000. Their expression levels were 20‐ to 70‐fold higher than those in the wild‐type strain (Fig. S2), which indicated that our results were not due to defects in gene overexpression.
We then tested whether differences in the regulation of YlAro4 and YlAro7 activity provided an explanation. At present, we know little about the feedback regulation of Aro enzymes in Y. lipolytica. However, in S. cerevisiae, it was found that ScAro4 is repressed by TYR, while ScAro7 is repressed by TYR and activated by TRP (Braus, 1991). We therefore tested whether wild‐type and mutated YlAro4 and YlAro7 experienced feedback regulation mediated by TYR or TRP and determined their level of activity. For this purpose, each enzyme was cloned and expressed individually (strains JMY8016, JMY8018, JMY8020 and JMY8022 – Table 1). We discovered that TYR repressed the activity of YlAro4 when it is expressed under its own promoter (wild‐type strain JMY195 – Fig. 3A). The same inhibition was observed in strain where YlARO4 is overexpressed under pTEF promoter (strain JMY8016 – Fig. 3A). As for S. cerevisiae, the overexpression of mutated version of YlAro4K221L did not show inhibition by TYR in vitro (JMY8018 – Fig. 3A). In contrast, TYR repressed the activity of YlAro7 when it is expressed under its own promoter while TRP activated it (wild‐type strain JMY195‐Fig. 3B) as it was reported for S. cerevisiae (Braus, 1991). When YlARO7 is overexpressed, regulation seems to be lost in vitro (strain JMY8020‐Fig. 3B). On the other hand, the mutated version of YlAro7G139S behaved like the wild‐type version in the presence of TYR or TRP (strain JMY8022 – Fig. 3B). This result suggests that the mutated version of YlAro7G139S is not feedback‐insensitive.
Table 1.
Strains used in this study
| Y. lipolytica strains | ||||
|---|---|---|---|---|
| Name | Genotype | Auxotrophy | Reference | |
| JMY195, Po1d | MATA ura3‐302 leu2‐270 xpr2‐322 | U‐L‐ | Barth and Gaillardin 1996 | |
| JMY 2101 | JMY195 + LEU2 | U‐ | Dulermo et al. 2017 | |
| JMY7891 | Po1d + NAT‐YlARO8‐YlARO10 | U‐L‐ | This study | |
| JMY7892 | Po1d + URA3‐YlARO1‐YlARO2 + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8074 | Po1d + URA3‐YlARO4‐YlARO7 + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8070 | Po1d + URA3‐YlARO4K221L‐YlARO7G139S + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY7902 | Po1d + LEU‐ScARO4K229L ‐ScARO7T226I + NAT‐YlARO8‐YlARO10 | U‐ | This study | |
| JMY8306 | Po1d + URA3‐YlARO1‐YlARO2 + YlARO4‐YlARO7 + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8000 | Po1d + LEU2‐YlARO1‐YlARO2 + URA3‐YlARO4K221L‐YlARO7G139S + NAT‐YlARO8‐YlARO10 | Prototroph | This study | |
| JMY7903 | Po1d + URA3‐YlARO1‐YlARO2 + LEU‐ScARO4K229L‐ScARO7T226I + NAT‐YlARO8‐YlARO10 | Prototroph | This study | |
| JMY8109 | Po1d + URA3‐YlARO1‐YlARO2 + ScARO4K229L‐ScARO7T226I + NAT‐ScARO3K222L + HPH‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8032 | Po1d + URA3‐YlARO1‐YlARO2 + ScARO4K229L‐ScARO7T226I + LEU2‐YlTKL + NAT‐YlARO8‐YlARO10 | Prototroph | This study | |
| JMY8054 | Po1d URA3‐YlARO1‐YlARO2 + ScARO4K229L‐ScARO7T226I + ScARO3K222L + LEU2‐YlTKL + NAT‐YlARO8‐YlARO10 | Prototroph | This study | |
| JMY7906 | Po1d + URA3‐ScARO4K229L‐ScARO3K222L + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8002 | Po1d + URA3‐YlARO1‐YlARO2 + LEU2‐ ScARO4K229L‐ScARO3K222L + NAT‐YlARO8‐YlARO10 | Prototroph | This study | |
| JMY8175 | Po1d + URA3‐YlARO1‐YlARO2 + ScARO4K229L‐ScARO7T226I + NAT‐ScARO3K222L | L‐ | This study | |
| JMY8073 | Po1d + URA3‐YlARO4 + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8077 | Po1d + URA3‐YlARO4K221L + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8079 | Po1d + URA3‐YlARO7 + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8081 | Po1d + URA3‐YlARO7G139S + NAT‐YlARO8‐YlARO10 | L‐ | This study | |
| JMY8354 | Po1d + URA3‐YlARO1‐YlARO2 + LEU‐YlARO4 + NAT‐YlARO8‐YlARO10 | Prototroph | This study | |
| JMY8356 | Po1d + URA3‐YlARO1‐YlARO2 + LEU2‐YlARO4K221L + NAT‐YlARO8‐YlARO10 | Prototroph | This study | |
| JMY8358 | Po1d + URA3‐YlARO1‐YlARO2 + LEU2‐YlARO7 + NAT‐YlARO8Yl‐ARO10 | Prototroph | This study | |
| JMY8360 | Po1d + URA3‐YlARO1‐YlARO2 + LEU2‐YlARO7G139S + NAT‐YlARO8‐YlARO10 | Prototroph | This study | |
| JMY7739 | Po1d + CvVioA‐VioB‐VioE; ura3‐302 leu2‐270 xpr2‐322 | L‐ U‐ | Kholany et al. (2019) | |
| JMY7751 | JMY7739 + URA3‐YlARO1‐YlARO2 | Prototroph | This study | |
| JMY7793 | JMY7739 + URA3‐YlARO1‐YlARO2 + LEU2‐ScARO4K229L‐ScARO7T226I | Prototroph | This study | |
| JMY7795 | JMY7739 + URA3‐YlARO1‐YlARO2 + LEU2‐ScARO4K229L‐ScARO3K222L | Prototroph | This study | |
| JMY8016 | JMY195 + URA3‐YlARO4 | L‐ | This study | |
| JMY8018 | JMY195 + URA3‐YlARO4K221L | L‐ | This study | |
| JMY8020 | JMY195 + URA3‐YlARO7 | L‐ | This study | |
| JMY8022 | JMY195 + URA3‐YlARO7G139S | L‐ | This study | |
Fig. 3.
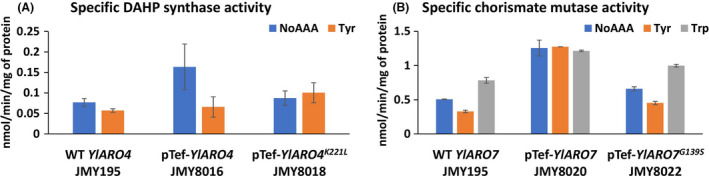
Specific enzymatic activity of (A) Aro4 and (B) Aro7 in Y. lipolytica measured in the presence of TYR or TRP and compared to the control (i.e. no amino acid supplementation). Error bars correspond to standard deviation of two replicates.
Because Ehrlich metabolite production was unaffected in the strains co‐expressing YlAro4K221L and YlAro7G139S variants compared to the wild‐type strain, it seems likely that the mutation we introduced had a deleterious effect in terms of level of activity.
Therefore, the wild‐type and mutant enzyme variants were expressed separately in a strain overexpressing the two cassettes YlARO1‐YlARO2 and YlARO8‐YlARO10 (Fig. 4) in order to determine the effect on Ehrlich metabolite production. A similar level of improvement in Ehrlich metabolite production was observed when YlARO4 was expressed alone and when there was co‐expression of YlARO4‐YlARO7. Compared to the wild‐type form of the enzyme, YlARO4 K221L improved the production by 50% (Fig. 4), suggesting that this particular mutation may have made the enzyme insensitive to feedback regulation as suggested by the enzymatic tests.
Fig. 4.
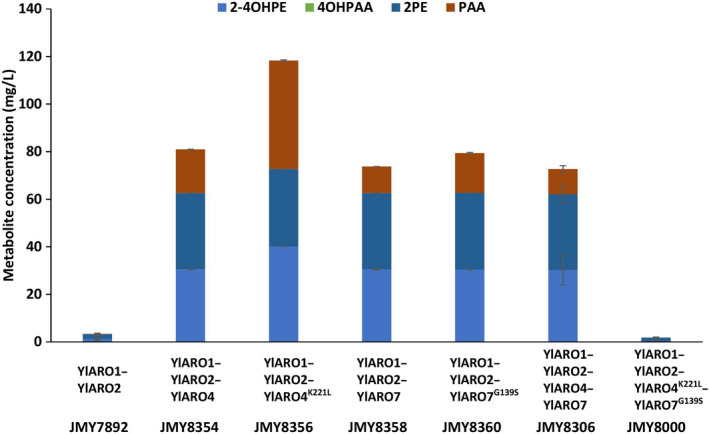
Enzymatic activity of YlAro4, YlAro7, YlAro4K221L and Aro7 YlAro7G139S in Y. lipolytica as quantified via Ehrlich metabolite production (in mg l−1). All the strains also overexpressed YlARO8‐YlARO10. The mutated forms of Aro4 (YlAro4K221L) and Aro7 (YlAro7G139S) were equivalent to the feedback‐inhibited forms of the enzymes in S. cerevisiae (ScAro4K229L and ScAro7G141S). 2‐4OHPE: 2‐(4‐hydroxyphenyl)ethanol; 4OHPAA: 4‐hydroxyphenylacetic acid; 2PE: phenylethanol; and PAA: phenylacetic acid.
In contrast, while Ehrlich metabolite production displayed similar improvements regardless of whether YlARO7 was expressed alone or in tandem (i.e. YlARO4‐YlARO7) in the same genetic background, production did not differ between the wild type and the mutated form YlAro7G139S, indicating that the enzyme was probably not deregulated by the mutation (Fig. 4), in accordance to what we observed with the enzymatic tests. The fact that Ehrlich metabolites production dropped dramatically when the two mutated enzyme variants were co‐expressed underscore a putative antagonism effect in this particular genetic background.
These results suggested that shikimate pathway regulatory differences likely exist between Y. lipolytica and S. cerevisiae. We therefore evaluated the effects of overexpressing feedback‐insensitive S. cerevisiae enzyme variants as part of our chassis strain construction process.
Deregulation of the shikimate pathway by overexpressing S. cerevisiae feedback insensitivity
Our attempts to deregulate Aro4 and Aro7 in Y. lipolytica had an unexpected impact on Ehrlich metabolite production. Moreover, the feedback regulation results revealed some differences with those observed in S. cerevisiae, which implies that we cannot completely extrapolate the behaviour of Y. lipolytica enzymes from that of S. cerevisiae enzymes. Therefore, in an effort to enhance the flux through the shikimate pathway, codon‐optimized deregulated variants of S. cerevisiae genes were expressed in Y. lipolytica (ScARO4K229L and ScARO7T226I ). The result was a higher level of Ehrlich metabolite production than in the strain overexpressing Y. lipolytica genes (Fig. 5 – strain JMY7903).
Fig. 5.
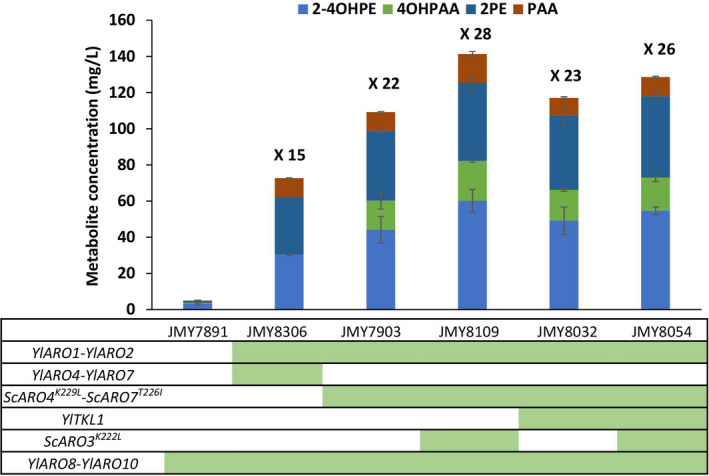
Ehrlich metabolites production by the engineered strains (in mg l−1). The strains’ genotypes are indicated by the green boxes located below the strains’ names. Yl: Y. lipolytica; Sc: S. cerevisiae; 2‐4OHPE: 2‐(4‐hydroxyphenyl)ethanol; 4OHPAA: 4‐hydroxyphenylacetic acid; 2PE: phenylethanol; and PAA: phenylacetic acid. Up above the bars are the estimates of fold improvement relative to the control strain (JMY7891).
To improve strain performance even more, we evaluated the effect of overexpressing two additional enzymes: (i) the deregulated version of Aro3 of S. cerevisiae (ScAro3K222L), hypothesized to be more effective than its isoenzyme Aro4 (Hartmann et al., 2003; Brückner et al., 2018) and (ii) Tkl1 of Y. lipolytica, with the aim of optimizing the availability of the precursor erythrose‐4‐phosphate (E4P). The expression of ScAro3K222L had a positive impact on Ehrlich metabolite production (Fig. 5 – strain JMY8109); the expression of YlTkl1 had no effect (Fig. 5 – strain JMY8032). When both were co‐expressed, YlTkl1 seemed to slightly reduce the improvement brought about by ScAro3K222L (Fig. 5 – strain 8054), likely because Tkl1 can catalyse the opposite reaction (Curran et al., 2013).
Among all the strains constructed, the JMY8109 strain – bearing YlARO1, YlARO2, ScARO3K222L , ScARO4K229L , ScARO7T226I , YlARO8 and YlARO10 – was the one to produce the largest amounts of Ehrlich metabolites (i.e. 28‐fold increase compared to strain JMY7891). JMY8109 produced over 140 mg l−1 of total Ehrlich metabolites, while this was just 5 mg l−1 for the strain that overexpressed YlARO8 and YlARO10 in the wild‐type background (JMY7891), demonstrating that the flux through the shikimate pathway was significantly increased in the former strain (Fig. 5).
To better characterize our best chassis strain (JMY8109), we compared the expression levels of all the overexpressed genes to the expression levels in the wild‐type strain. Fold change was calculated for YlARO1 and YlARO2, for which endogenous forms were available. Both genes were overexpressed by a factor of more than 50 (Fig. 6A). However, when looking at the relative expression level (2−ΔCT), we can see that, for the wild type, the expression level of YlARO2 is 24 times lower than that of YlARO1. A similar trend was observed in the chassis strain (Fig. 6B), which makes sense given that both genes displayed the same overexpression factor (Fig. 6A). All the heterologous genes overexpressed in our chassis strain had relative expression levels that were higher than that of ACT1, which indicated that the entire pathway was strongly overexpressed.
Fig. 6.
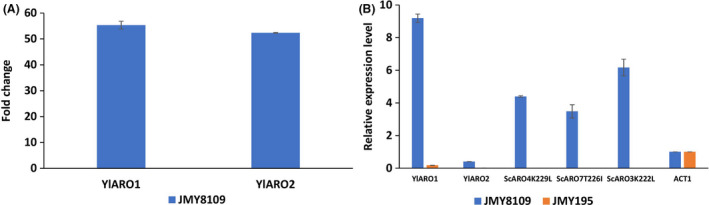
Gene expression in the chassis strain. Results from a qRT‐PCR analysis comparing the chassis strain and the wild‐type strain are shown.
A. Increase in the expression of endogenous genes in the chassis strain versus the wild‐type strain.
B. Expression levels for overexpressed genes versus ACT1. Sc: S. cerevisiae; Yl: Y. lipolytica; ACT1: gene encoding actin. Error bars correspond to standard deviation of two replicates. JMY195: MATA ura3‐302 leu2‐270 xpr2‐322. JMY8109: Po1d + URA3‐YlARO1‐YlARO2 + ScARO4K229L‐ScARO7T226I + NAT‐ScARO3K222L + HPH‐YlARO8‐YlARO10.
Overall, strain JMY8175, which was equivalent to strain JMY8109 except that it did not overexpress YlARO8‐YlARO10 and thus avoided AAA catabolism, could serve as a good chassis strain if the objective were to produce aromatic derivative compounds other than Ehrlich metabolites.
Improving Ehrlich metabolite production via culture conditions
After we built and validated a robust and efficient chassis strain (JMY8109) for producing Ehrlich metabolites, we tuned culture conditions to generate further improvements. In particular, we changed the glucose concentration; we kept the minimal medium as the medium background for the sake of comparison. E4P is a precursor of both AAAs and erythritol, and previous research has shown that using glycerol as a carbon source can boost erythritol production (Carly et al., 2017b). Therefore, we also tested glycerol utilization compared to glucose utilization. In this experiment, we compared the results for our best chassis strain (JMY8109), the strain that overexpressed TKL1 instead of ScARO3K222L (JMY8032) and the strain that overexpressed both TKL1 and ScARO3K222L (JMY8054), given that the Tkl1 enzyme is involved in E4P synthesis and could improve production when glycerol serves as the carbon source.
Increasing the glucose concentration from 10 g l−1 to 40 g l−1 greatly enhanced the production of Ehrlich metabolites reaching around 0.3 g l−1 for the 3 strains (Fig. 7A). Using glycerol (concentration: 40 g l−1) yielded an even greater improvement from 0.48 g l−1 for JMY8109 to 0.59 g l−1 for JMY8054. As growth rates were similar, this increased titre corresponds to an increase in production rate (corresponding to 70 mg g−1 CDW for the strain JMY8109 in glycerol 40 g l−1). Growth on glycerol also triggered the accumulation of AAA in the supernatant in relatively high amount by the 3 strains, in particular PHE, reaching up to 1 g l−1 in JMY8109 (Fig. 7B). AAA accumulation was not observed in at 10 g l−1 glucose and was only detectable at 40 g l−1 glucose for the strain JMY8109 (Fig. 7B). Even if growth in glycerol shows a slightly less improvement in Ehrlich metabolites production for our chassis strain compared to JMY8054, it produced in this condition a high amount of PHE, which is one of the precursors of the Ehrlich pathway. Thus, it indicated a bottleneck in the Ehrlich metabolite production reached by this strain and confirmed its high potential for PHE derivative products.
Fig. 7.
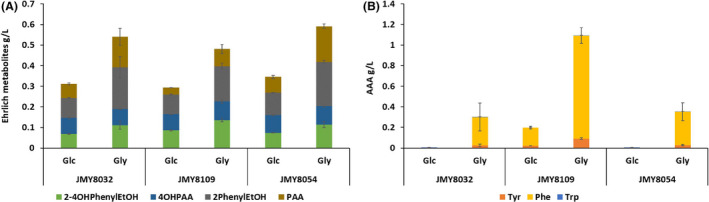
Effect of carbon source on Ehrlich metabolite production after 5 days of growth. Three AAA‐engineered strains overexpressing ARO1‐ARO2‐scARO4K229L‐scARO7T226I were tested using different concentrations and types of carbon sources. Strain JMY8032 overexpressed YlTKL1, strain JMY8109 overexpressed the deregulated form of ScARO3 (ScARO3K222L ) and strain JMY8054 overexpressed YlTKL1 and ScARO3K222L . The graph shows the total amounts of Ehrlich metabolites (A) and the total amounts of PHE, TYR and TRP (B) produced by each strain under each set of conditions. The three strains overexpressed YlARO8 and YlARO10. YNB: minimal yeast nitrogen base medium; Glc40: 40 g l−1 glucose; Gly40: 40 g l−1 glycerol; 2‐4OHPE: 2‐(4‐hydroxyphenyl)ethanol; 4OHPAA: 4‐hydroxyphenylacetic acid; 2PE: phenylethanol; and PAA: phenylacetic acid.
Chassis for tryptophan‐derived products
While the pathway presented here is dedicated to PHE‐ and TYR‐derived products, an intermediate chassis could be constructed for TRP‐derived products. TRP is a precursor for many alkaloids of commercial value (Ramani et al., 2013) as well as for various pigments (Huccetogullari et al., 2019). Chorismate lies at the branching point between TRP synthesis and PHE/TYR synthesis. Therefore, overexpressing the shikimate pathway up to chorismate should generate a chassis suitable for TRP derivative products.
As a proof of concept, we attempted to produce PVA, a green pigment derived from TRP. We used a strain overexpressing three genes (VioA, VioB and VioE), allowing the conversion of TRP to PVA (Fig. 8A), and we introduced different AAA pathway modifications. Thus, the VioABE cassette was expressed into (i) a wild‐type strain; (ii) a strain overexpressing YlARO1 and YlARO2; (iii) a strain overexpressing the genes up to the chorismate branching point (YlARO1, YlARO2, ScARO4K229L and ScARO3K222L ; Fig. 8B); and (iv) a strain overexpressing the overall shikimate pathway up to the production of prephenate, after the chorismate branching point (YlARO1, YlARO2, ScARO4K229L and ScARO7T226I ; Fig. 8C).
Fig. 8.
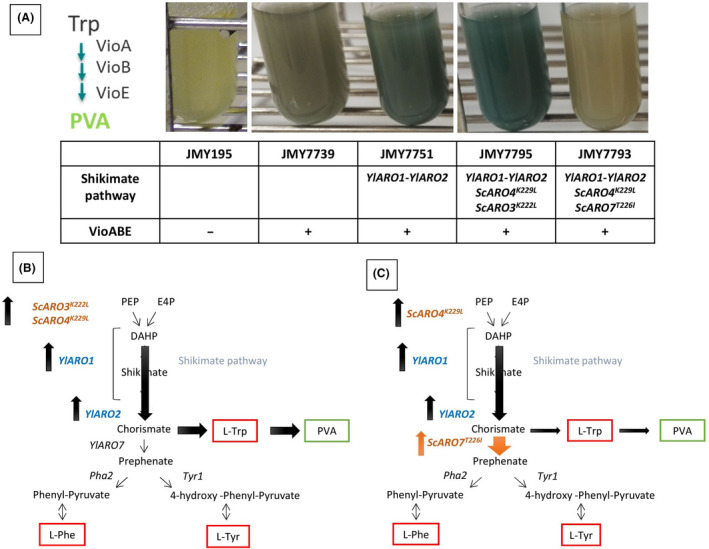
Depiction of the PVA pathway, which serves as an intracellular reporter system for the shikimate pathway.
A. Enzymes from the PVA pathway that were overexpressed in Y. lipolytica. (A) Strains expressing the PVA pathway in different genetic backgrounds. The strains’ names are indicated as well as the shikimate pathway overexpressed variant genes and the overexpression of the PVA pathway.
B. Schematic representation of the shikimate pathway and how the flux was modified when genes upstream from the chorismate branching point were overexpressed. This approach can increase the flux to TRP and consequently the production of PVA (strain JMY7795).
C. Schematic representation of the shikimate pathway and how the flux was modified when genes downstream from the chorismate branching point were overexpressed. This approach decreases the TRP that is available for PVA production, resulting in much lighter green cultures (strain JMY7793). The bidirectional arrows indicate reversible reactions.
The wild‐type strain overexpressing the PVA pathway (JMY7739) was light green in colour. The strain overexpressing YlARO1 and YlARO2 (JMY7751) was dark green. The strain overexpressing the shikimate pathway up to the chorismate branching point (JMY7795) was very dark green. These results show that TRP availability increased PVA production (Fig. 8A).
In contrast, chorismate availability declined when scARO7T226I was overexpressed (JMY7793), given that the flux was pushed towards PHE and TYR (as demonstrated in the results for our previous chassis strain). The strain was only a very faint green, even lighter in colour than the strain with the wild‐type background (Fig. 8A), reflecting the substantial reduction in the flux towards PVA. To verify that the differences in colour intensity were not due to inconsistent expression of the AAA modification cassettes, inverse construction was performed: the cassette expressing the PVA pathway was introduced into the relevant pre‐existing chassis strains. Similar results were obtained (data not shown). Thus, strain JMY7795 appears to be a good chassis for producing TRP‐derived products like violacein or alkaloids.
Discussion
Using a Golden Gate multiple gene cloning approach, we created two chassis strains for producing AAA derivatives. We achieved this task by combining the expression of Y. lipolytica and S. cerevisiae genes to produce strains with a higher flux through the shikimate pathway; these genes included variants encoding feedback‐inhibited versions of S. cerevisiae enzymes. Overall, the strain JMY8109 – which overexpressed YlARO1, YlARO2, ScARO3K222L , ScARO4K229L , ScARO7T226I , YlARO8 and YlARO10 – was the best chassis for PHE‐ and TYR‐derived products, as shown by the successful production of Ehrlich metabolites. The latter are valuable products, especially 2‐PE, which is a flavouring and fragrance compound. The strain JMY7795 – which overexpressed YlARO1, YlARO2, ScARO3K222L and ScARO4K229L – was the best chassis for TRP‐derived products, as shown by the successful production of PVA, a green dye that is a violacein precursor.
The AAA biosynthetic pathway has been only very recently exploited in Y. lipolytica, and few studies have reported the production of AAAs or derived metabolite products in this yeast. Carreira and colleagues used bioconversion and TYR supplementation (1.5 mM [0.27 g l−1]) to generate the TYR derivatives pOH‐PE and pOH‐PAA, achieving levels of 0.5 mM (0.07 g l−1) and 0.25 mM (0.038 g l−1) respectively (Carreira et al., 2001). These values are largely similar to those obtained with our chassis strain in this study with de novo synthesis in minimal medium, without AAA feeding. Celińska and colleagues produced 2 g l−1 of PE by adding 7 g l−1 of PHE to the medium once the culture had reached its stationary phase, meaning that all the PHE added was used for biotransformation rather than for growth (Celińska et al., 2013). Very recently, Gu and colleagues achieved 2.7 g l−1 of PE by bioconversion, adding 4 g l−1 of PHE to the medium with a Y. lipolytica strain that was engineered to overexpress the Ehrlich pathway, to improve precursors availability and to block competing pathways (Gu et al., 2020b). Overall, most studies in which yeast or bacteria have been engineered for 2‐PE production have achieved titres in a g l−1 range with PHE supplementation, and these titre levels declined around 10‐fold when there was no supplementation (Wang et al., 2019). Therefore, optimizing the shikimate pathway using a synthetic biology approach is essential for producing target molecules from standard carbon sources, as described here.
Similarly, our TRP chassis strain resulted in PVA production in the absence of precursors supplementation, while violacein is usually generated using TRP‐supplemented media. For example, Wong and colleagues produced violacein in Y. lipolytica with a CSM medium containing TRP (Wong et al., 2017), whereas Kholany and colleagues utilized a minimal or rich medium containing TRP (25 mg l−1) (Kholany et al., 2019).
Very recently, several works have been published involving engineering of the shikimate pathway to a different extent to produce AAA derivatives. Researchers successfully optimized both the shikimate pathway and the availability of DAHP synthase substrates in S. cerevisiae and achieved 2‐PE production levels of 12.7 mM (1.55 g l−1) (Hassing et al., 2019). In Y. lipolytica, by overexpressing endogenous and heterologous genes, including feedback‐insensitive enzymes, and deleting competing pathways, Gu and colleagues achieved 2.4 g l−1 of de novo 2‐PE among other derivatives (Gu et al., 2020a). Lv and colleagues achieved 0.25 g l−1 naringenin in flask conditions (Lv et al., 2019), and Palmer and colleagues produced 0.9 g l−1 naringenin in bioreactor fermentation conditions (Palmer et al., 2020) in Y. lipolytica. Finally, Sáez‐Sáez and colleagues reached 12.4 g l−1 resveratrol synthesis in bioreactor fermentation conditions with a Y. lipolytica strain optimized for the shikimate pathway and expressing multiple copies of resveratrol pathway (Sáez‐Sáez et al., 2020).
For our Y. lipolytica chassis strain, changing the carbon source markedly improved the production of Ehrlich metabolites. When glucose was replaced by glycerol, a low‐cost carbon source, Ehrlich metabolite production reached a level of 0.48 g l−1 without the need for any further optimization in our chassis strain. Thus, this result lays the foundation for process improvement via a well‐reasoned fermentation strategy. In addition, using glycerol as the carbon source greatly increases TYR and PHE detectable in the medium, the Ehrlich metabolites pathway overexpression reaching probably its limits in this condition. Therefore, the pool of AAA is even greater in our chassis to what can be measured indirectly with the Ehrlich metabolites.
Our chassis strains can therefore be used as tools for producing a wide range of AAA derivatives by introducing heterologous pathways for biobased aromatic compounds. Such molecules like naringenin, resveratrol or even violacein have recently been successfully produced in Y. lipolytica, showing that the expression of these heterologous pathways is efficient in this yeast (Lv et al., 2019; Kholany et al., 2019; Palmer et al., 2020; Sáez‐Sáez et al., 2020; Gu et al., 2020a).
Sáez‐Sáez and colleagues observed that glucose performed better than glycerol for the production of resveratrol (Sáez‐Sáez et al., 2020). Our strain produced more 2‐PE and PHE in glycerol. This discrepancy might come from the heterologous pathway introduced. Thus, carbon source may be reconsidered depending on the target product.
The chassis strain constructed in this work could be further improved by a better deciphering of shikimate pathway metabolic regulations in Y. lipolytica. The effect of TRP and TYR on the enzymatic activity of YlAro4 and YlAro7 enzymes was analysed in vitro during this study. TYR repressed YlAro4 and this repression was lost in the mutated version of the enzyme (YlAro4K221L). In addition, this feedback‐insensitive enzyme contributed to increase the titres of Ehrlich metabolites produced when expressed in vivo. These results are in accordance with two very recent works done in Y. lipolytica, in which the same YlAro4K221L feedback‐insensitive variant was constructed, allowing an increase on the final product titres of naringenin and resveratrol (Palmer et al., 2020; Sáez‐Sáez et al., 2020). YlAro7 is repressed by TYR and activated by TRP, which is in accordance with S. cerevisiae data (Luttik et al., 2008). The mutated version of YlAro7 (YlAro7G139S) behaves like the wild‐type enzyme in vitro, suggesting that the feedback regulation was not lost by this mutation. However, this result is in disagreement with a very recent work where the same mutation was done in YlARO7 and enable an increase on the production of resveratrol, even when YlAro7G139S was expressed together with YlAro4K221L (Sáez‐Sáez et al., 2020), while in our conditions, co‐expression of the mutated version has a deleterious effect on the Ehrlich metabolite productions.
Finally, in Y. lipolytica, the deregulated version of ScAro3 seemed to be more efficient than the deregulated version of ScAro4. Compared to Aro4, Aro3 appears to have a greater affinity for E4P and PEP (Hartmann et al., 2003). Consequently, Aro3 may be the more effective enzyme, given its lower Km values for E4P and PEP and its higher Ki value for its specific inhibitor (Hartmann et al., 2003; Brückner et al., 2018).
Even though there are still many unknowns regarding the regulation of the shikimate pathway in Y. lipolytica, our results suggest the process differs between Y. lipolytica and S. cerevisiae, at least in the case of Aro7.
The recent and fast growing interest in aromatic compounds production by Y. lipolytica encourages us to push forward our chassis strains. The next step in this research will be to improve the fitness of our chassis strains, as the overexpression of the shikimate pathway has negative impacts on growth. Such effects probably result from 2‐PE toxicity, which has been reported in similar AAA‐optimized chassis strains in S. cerevisiae (Hassing et al., 2019).
Experimental procedures
Strains and media
The E. coli strain DH5α was used for cloning and plasmid propagation. Cells were grown at 37°C in Luria‐Bertani medium (10 g l−1 tryptone, 5 g l−1 yeast extract and 10 g l−1 NaCl) containing the appropriate antibiotic for plasmid selection (either 100 μg l−1 ampicillin or 50 μg l−1 kanamycin).
The Y. lipolytica strains used in this study were derived from the Po1d strain, which was itself derived from the wild‐type W29 strain (ATCC20460) strain; they are listed in Table 1. The strains were grown at 28 °C in rich YPD medium (10 g l−1 glucose), containing 10 g l−1 peptone and 10 g l−1 yeast extract, or in minimal YNB medium (10 g l−1 glucose), containing 1.7 g l−1 yeast nitrogen base, 5 g l−1 NH4Cl and 50mM phosphate buffer (pH 6.8). YNBglc40 contained 40 g l−1 glucose, and YNBgly40 contained 40 g l−1 glycerol. We added uracil at 100 mg l−1 (YNBura) and leucine at 700 mg l−1 (YNBleu) to supplement the auxotrophic requirements of the strains. When needed, antibiotics were added for strain selection: hygromycin B at 250 mg l−1 (YNBhygro) or nourseothricin at 400 mg l−1 (YNBnour).
Solid media for E. coli and Y. lipolytica were prepared by adding 15 g l−1 agar to liquid media.
Culture conditions
Precultures were inoculated into tubes containing 5 ml of YPD medium and cultured overnight (28°C, 180 rpm). The precultures were then centrifuged, washed twice with sterile distilled water and used to inoculate the culture at an OD600 of 0.05. Strains were grown in 50 ml of YNB media in 500‐ml baffled flasks kept at 28 °C and 180 rpm. We took 1‐ml samples after 2 and 5 days of culture. OD600 was measured for each sample, and after centrifugation, the supernatant was used in HPLC analysis. For each strain, two replicates were used. Optical density was measured at 600 nm using a spectrophotometer.
Plasmid and strain construction
The Y. lipolytica genes used to construct the overexpression cassettes were amplified via PCR from the W29 strain. The sequences were analysed in silico and, if present, internal BsaI recognition sites were eliminated via site‐directed mutagenesis using PCR. The 4nt overhangs needed for Golden Gate Assembly were also added using PCR (Larroude et al., 2019). Punctual mutations to create the deregulated forms of YlAro4K221L and YlAro7G139S were also carried out via site‐directed mutagenesis using PCR. All the genes and variants from S. cerevisiae were codon‐optimized and synthesized by TWIST Bioscience or Genecust.
The overexpression cassettes (listed in Table S2) were constructed using a Golden Gate Assembly technique described elsewhere (Celińska et al., 2017; Larroude et al., 2019) and were used to transform Y. lipolytica via the lithium acetate method (Le Dall et al., 1994). These cassettes allowed random integration into the genome. Selection marker removal was performed using the LoxP‐Cre system, whose use in Y. lipolytica has been previously described (Fickers et al., 2003). Mutants were selected on YNB, YNBleu, YNBura, YNBhygro or YNBnour media, depending on their genotype, and were then verified using colony PCR.
Restriction enzymes were purchased from New England Biolabs (Ipswich, MA, USA). PCR amplification was performed using Q5 High‐Fidelity DNA Polymerase (New England Biolabs) or GoTaq DNA polymerases (Promega, Charbonnières‐les‐Bains, France). When needed, PCR fragments were purified with a NucleoSpin® Gel and PCR Clean‐up Kit (Macherey‐Nagel, Duren, Germany). All the reactions were carried out in accordance with the manufacturer’s instructions. Transformation of chemically competent E. coli cells was performed using the heat shock protocol. Plasmids were isolated from E. coli with the NucleoSpin® Plasmid EasyPure Kit (Macherey‐Nagel).
All the primers used in this study are listed in Table S3.
Analytical methods
Metabolite measurements
Ehrlich metabolites (phenyl acetic acid, phenyl ethanol, p‐OH‐phenylethanol and p‐OH‐phenylacetic acid) were identified via HPLC using an Agilent Zorbax Eclipse Plus C18 Column (4.6 × 100; 3.5 microns) operating at 40°C. The eluent was a gradient of acetonitrile and 20 mM KH2PO4 (pH 2) with 1% acetonitrile. The flow rate was 0.8 ml·min−1; it increased from 0 to 10% acetonitrile in 6 min and then climbed to 40% acetonitrile by 23 min. From 23 min to 27 min, the flow was 99% KH2PO4. Metabolites were measured at 214 nm using a diode array detector (DAD).
Phenylethanol, phenylacetic acid, p‐OH‐phenylethanol and p‐OH‐phenylacetic acid standards were obtained from Sigma‐Aldrich (France). Solutions of 10 mM were created for each compound (in their specific solvents as per manufacturer’s instructions) and then diluted to final concentrations of 0.1, 0.2, 0.5, 1 and 2 mM for HPLC calibration.
Culture supernatants were treated with 1% (vol/vol) trifluoroacetic acid for at least one hour at 4 °C and were then centrifuged and filtered to 0.22 µm before HPLC analysis.
Enzyme assays
Chorismate mutase (EC 5.4.99.5) and DAHP synthase (EC 2.5.1.54) were assayed as described in Luttik et al. (2008). To measure chorismate mutase activity, 100 µl cell extracts was incubated with 1 ml of incubation mix, containing 100 µl of Tris–HCl (pH 7.6; 50 mM), 10 µl of dithiothreitol (DTT, 1 mM), 10 µl of EDTA (0.1 mM) and H2O to reach the final volume at 30°C. To test Aro7 inhibition and activation, either 0.5 mM of TYR or 0.5 mM of TRP was added to the incubation mix. At t = 0 min, 1 mM (final concentration) of barium chorismate was added. Over a 5 min period, a 100 µl sample was taken every minute and added to 100 µl of 1 M HCl. These samples were incubated at 30°C for 10 min, after which 800 µl of 1 M NaOH was added. Absorbance was measured at 320 nm, and an extinction coefficient of 13.165 mM−1 cm−1 was used to calculate the phenylpyruvate concentration.
DAHP synthase activity was measured as follows: the incubation mix of 0.9 ml contained 100 µl of phosphate buffer (100 mM), 100 µl of phosphoenolpyruvate (PEP; 0.5 mM) and 100 µl of erythrose‐4‐phosphate (E4P, 1 mM); it was completed with H2O. To check for the presence of allosteric control in Aro4, 0.5 mM of TYR was added to the incubation mix. The reaction was initiated with the addition of 100 µl of cell extract. At 2, 3, 4 and 5 min, 200 µl of the reaction mixture was transferred to an Eppendorf tube containing 80 µl of a 10% w/v trichloroacetic acid solution. The resulting mixture was centrifuged for 5 min in an Eppendorf tabletop centrifuge to remove any proteins. Then, 125 µl of the supernatants was transferred to clean Eppendorf tubes, and 125 µl of periodic acid (25 mM in 0.075 M H2SO4) was added to each tube. After a 45 min incubation period at room temperature, 250 µl of 2% (w/v) sodium arsenite in 0.5 M HCl was added to the mixture to destroy excess periodate (2 min at room temperature). Then, 1 ml of thiobarbituric acid (0.3%, w/v, pH 2.0) was added to the tubes, which were subsequently placed in a boiling‐water bath for 5 min. The mixture was cooled in a water bath for 10 min at 40 °C, and the pink colour that resulted was measured at 549 nm in a spectrophotometer. The extinction coefficient of DAHP at 549 nm was 13.6 mM−1 cm−1.
qRT‐PCR
Strains were grown in 10 ml of YPD in 100 ml baffled Erlenmeyer flasks kept at 28°C and 180 rpm. Inoculation was carried out at 0.2 OD using an overnight preculture. Cells were harvested 6 h post‐inoculation, frozen in liquid nitrogen and stored at −80°C.
Total RNA was extracted from the cells using the RNeasy Mini Kit (Qiagen, Hilden, Germany), and 1.5 µg of each sample was treated with DNase (TURBO DNA‐Free Kit). RNA quantities were measured at 260 nm using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Villebon sur Yvette, France). cDNA synthesis and qPCR were carried out in a single step using a Luna Universal One‐Step RT‐qPCR Kit (New England Biolabs, Ipswich, MA, USA) in accordance with the manufacturer’s instructions.
PCR reactions were performed using specific primers that targeted the 3′ ends of the genes (Table S3). A constitutive gene, ACT1 (YALI0D08272g), was utilized as the reference. The following programme was used: 98°C for 3 min, followed by 40 cycles at 98°C for 15 s, 58°C for 30 s and 72°C for 30 s. Expression levels were then quantified using the ΔCT and ΔΔCT methods (Schmittgen and Livak, 2008). All experiments were done in duplicates.
Supporting information
Fig. S1. Comparison of amino acid sequences of S. cerevisiae and Y. lipolytica Aro4 and Aro7 enzymes. Sequences of Y. lipolytica YlAro4 and YlAro7 enzymes blasted against the equivalent S. cerevisiae enzymes. In yellow are highlighted the positions usually mutated in S. cerevisiae to construct the feedback insensitive forms of the enzymes. In green are highlighted the conserved equivalent positions for Y. lipolytica. In Aro7 S. cerevisiae sequence can be observed also blue marks that correspond to other sites that were also proposed as involved in regulation of the enzyme.
Fig. S2. ARO genes relative expression levels in wild‐type and the strain overexpressing YlARO1, YlARO2, YlARO4K221L and YlARO7G139S .
Table S1. List of Y. lipolytica genes involved in the AAA pathway. The blast results of S. cerevisiae orthologous proteins against Y. lipolytica genome are indicated.
Table S2. List of plasmids used in this study.
Table S3. Primers used in this study.
Acknowledgements
This project received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 720824. The authors declare that they have no competing interests. We would like to thank Jessica Pearce for her language editing services.
Microbial Biotechnology (2021) 14(6), 2420–2434
Funding informationThis project received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 720824.
References
- Barth, G. & Gaillardin, C. (1996) Yarrowia lipolytica. Wolf, K. , Breuning, K. D. & Barth, J. , (eds). Non‐conventional Yeasts in Biotechnology. Berlin, Germany: Springer‐Verlag, (pp. 313–388). [Google Scholar]
- Braus, G.H. (1991) Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol Rev 55: 349–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner, C. , Oreb, M. , Kunze, G. , Boles, E. , and Tripp, J. (2018) An expanded enzyme toolbox for production of cis, cis‐muconic acid and other shikimate pathway derivatives in Saccharomyces cerevisiae . FEMS Yeast Res 18:foy017. [DOI] [PubMed] [Google Scholar]
- Carly, F. , and Fickers, P. (2018) Erythritol production by yeasts: a snapshot of current knowledge. Yeast 35: 455–463. [DOI] [PubMed] [Google Scholar]
- Carly, F. , Gamboa‐Melendez, H. , Vandermies, M. , Damblon, C. , Nicaud, J.‐M. , and Fickers, P. (2017a) Identification and characterization of EYK1, a key gene for erythritol catabolism in Yarrowia lipolytica . Appl Microbiol Biotechnol 101: 1–10. [DOI] [PubMed] [Google Scholar]
- Carly, F. , Steels, S. , Telek, S. , Vandermies, M. , Nicaud, J.M. , and Fickers, P. (2018) Identification and characterization of EYD1, encoding an erythritol dehydrogenase in Yarrowia lipolytica and its application to bioconvert erythritol into erythrulose. Bioresour Technol 247: 963–969. [DOI] [PubMed] [Google Scholar]
- Carly, F. , Vandermies, M. , Telek, S. , Steels, S. , Thomas, S. , Nicaud, J.‐M. , and Fickers, P. (2017b) Enhancing erythritol productivity in Yarrowia lipolytica using metabolic engineering. Metab Eng 42: 19–24. [DOI] [PubMed] [Google Scholar]
- Carreira, A. , Ferreira, L.M. , and Loureiro, V. (2001) Brown pigments produced by Yarrowia lipolytica result from extracellular accumulation of homogentisic acid. Appl Environ Microbiol 67: 3463–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celińska, E. , Borkowska, M. , Białas, W. , Kubiak, M. , Korpys, P. , Archacka, M. , et al. (2019) Genetic engineering of Ehrlich pathway modulates production of higher alcohols in engineered Yarrowia lipolytica . FEMS Yeast Res 19: foy122. [DOI] [PubMed] [Google Scholar]
- Celińska, E. , Kubiak, P. , Białas, W. , Dziadas, M. , and Grajek, W. (2013) Yarrowia lipolytica: the novel and promising 2‐phenylethanol producer. J Ind Microbiol Biotechnol 40: 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celińska, E. , Ledesma‐Amaro, R. , Larroude, M. , Rossignol, T. , Pauthenier, C. , and Nicaud, J.‐M. (2017) Golden Gate Assembly system dedicated to complex pathway manipulation in Yarrowia lipolytica . Microb Biotechnol 10: 12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celińska, E. , Olkowicz, M. , and Grajek, W. (2015) L‐Phenylalanine catabolism and 2‐phenylethanol synthesis in Yarrowia lipolytica‐mapping molecular identities through whole‐proteome quantitative mass spectrometry analysis. FEMS Yeast Res 15: fov041. [DOI] [PubMed] [Google Scholar]
- Curran, K.A. , Karim, A.S. , Gupta, A. , and Alper, H.S. (2013) Use of expression‐enhancing terminators in Saccharomyces cerevisiae to increase mRNA half‐life and improve gene expression control for metabolic engineering applications. Metab Eng 19: 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulermo, R. , Brunel, F. , Dulermo, T. , Ledesma‐Amaro, R. , Vion, J. , Trassaert, M. , et al. (2017) Using a vector pool containing variable‐strength promoters to optimize protein production in Yarrowia lipolytica. Microb Cell Fact 16: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickers, P. , Le Dall, M.T. , Gaillardin, C. , Thonart, P. , and Nicaud, J.M. (2003) New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica . J Microbiol Methods 55: 727–737. [DOI] [PubMed] [Google Scholar]
- Gottardi, M. , Reifenrath, M. , Boles, E. , and Tripp, J. (2017) Pathway engineering for the production of heterologous aromatic chemicals and their derivatives in Saccharomyces cerevisiae: bioconversion from glucose. FEMS Yeast Res 17: fox035. [DOI] [PubMed] [Google Scholar]
- Gu, Y. , Ma, J. , Zhu, Y. , Ding, X. , and Xu, P. (2020a) Engineering Yarrowia lipolytica as a chassis for de novo synthesis of five aromatic‐derived natural products and chemicals. ACS Synth Biol 9: 2096–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y. , Ma, J. , Zhu, Y. , and Xu, P. (2020b) Refactoring Ehrlich Pathway for high‐yield 2‐phenylethanol production in Yarrowia lipolytica . ACS Synth Biol 9: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z. , Duquesne, S. , Bozonnet, S. , Cioci, G. , Nicaud, J.‐M. , Marty, A. , and O'Donohue, M.J. (2015) Development of cellobiose‐degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnol Biofuels 8: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z. , Robin, J. , Duquesne, S. , O’Donohue, M.J. , Marty, A. , and Bordes, F. (2018) Developing cellulolytic Yarrowia lipolytica as a platform for the production of valuable products in consolidated bioprocessing of cellulose. Biotechnol Biofuels 11: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, M. , Schneider, T.R. , Pfeil, A. , Heinrich, G. , Lipscomb, W.N. , and Braus, G.H. (2003) Evolution of feedback‐inhibited beta /alpha barrel isoenzymes by gene duplication and a single mutation. Proc Natl Acad Sci USA 100: 862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassing, E.J. , de Groot, P.A. , Marquenie, V.R. , Pronk, J.T. , and Daran, J.M.G. (2019) Connecting central carbon and aromatic amino acid metabolisms to improve de novo 2‐phenylethanol production in Saccharomyces cerevisiae . Metab Eng 56: 165–180. [DOI] [PubMed] [Google Scholar]
- Hazelwood, L.A. , Daran, J.M. , Van Maris, A.J.A. , Pronk, J.T. , and Dickinson, J.R. (2008) The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74: 2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huccetogullari, D. , Luo, Z.W. , and Lee, S.Y. (2019) Metabolic engineering of microorganisms for production of aromatic compounds. Microb Cell Fact 18: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui, I. , Vissers, S. , Cartiaux, M. , and Urrestarazu, A. (1998) Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol Gen Genet 257: 238–248. [DOI] [PubMed] [Google Scholar]
- Kholany, M. , Trébulle, P. , Martins, M. , Ventura, S.P. , Nicaud, J.‐M. , and Coutinho, J.A. (2019) Extraction and purification of violacein from Yarrowia lipolytica cells using aqueous solutions of surfactants. J Chem Technol Biotechnol 95:1126–1134. [Google Scholar]
- Koopman, F. , Beekwilder, J. , Crimi, B. , van Houwelingen, A. , Hall, R.D. , Bosch, D. , et al. (2012) De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae . Microb Cell Fact 11: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann, S. , Lipscomb, W.N. , and Braus, G.H. (2000) Coevolution of transcriptional and allosteric regulation at the chorismate metabolic branch point of Saccharomyces cerevisiae . Proc Natl Acad Sci USA 97: 13585–13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroude, M. , Park, Y.K. , Soudier, P. , Kubiak, M. , Nicaud, J.M. , and Rossignol, T. (2019) A modular Golden Gate toolkit for Yarrowia lipolytica synthetic biology. Microb Biotechnol 12:1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroude, M. , Rossignol, T. , Nicaud, J.‐M. , and Ledesma‐Amaro, R. (2018) Synthetic biology tools for engineering Yarrowia lipolytica . Biotechnol Adv 36: 2150–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dall, M.T. , Nicaud, J.M. , and Gaillardin, C. (1994) Multiple‐copy integration in the yeast Yarrowia lipolytica . Curr Genet 26: 38–44. [DOI] [PubMed] [Google Scholar]
- Ledesma‐Amaro, R. , Dulermo, T. , and Nicaud, J.M. (2015) Engineering Yarrowia lipolytica to produce biodiesel from raw starch. Biotechnol Biofuels 8: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma‐Amaro, R. , Lazar, Z. , Rakicka, M. , Guo, Z. , Fouchard, F. , Le Coq, A.‐M. , and Nicaud, J.‐M. (2016) Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab Eng 38: 115–124. [DOI] [PubMed] [Google Scholar]
- Ledesma‐Amaro, R. , and Nicaud, J.‐M. (2016a) Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog Lipid Res 61: 40–50. [DOI] [PubMed] [Google Scholar]
- Ledesma‐Amaro, R. , and Nicaud, J.‐M. (2016b) Metabolic engineering for expanding the substrate range of Yarrowia lipolytica . Trends Biotechnol 34: 798–809. [DOI] [PubMed] [Google Scholar]
- Luttik, M.A.H. , Vuralhan, Z. , Suir, E. , Braus, G.H. , Pronk, J.T. , and Daran, J.M. (2008) Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab Eng 10: 141–153. [DOI] [PubMed] [Google Scholar]
- Lv, Y. , Marsafari, M. , Koffas, M.A.G. , Zhou, J. , and Xu, P. (2019) Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis. ACS Synth Biol 8: 2514–2523. [DOI] [PubMed] [Google Scholar]
- Palmer, C.M. , Miller, K.K. , Nguyen, A. , and Alper, H.S. (2020) Engineering 4‐coumaroyl‐CoA derived polyketide production in Yarrowia lipolytica through a β‐oxidation mediated strategy. Metab Eng 57: 174–181. [DOI] [PubMed] [Google Scholar]
- Ramani, S. , Patil, N. , Nimbalkar, S. , and Jayabaskaran, C. (2013) Alkaloids derived from tryptophan: terpenoid indole alkaloids. In Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes. Berlin, Heidelberg: Springer, pp. 575–604. [Google Scholar]
- Reifenrath, M. , Bauer, M. , Oreb, M. , and Boles, E. (2018) Bacterial bifunctional chorismate mutase‐prephenate dehydratase PheA increases flux into the yeast phenylalanine pathway and improves mandelic acid production. Metab Eng Commun 7: e00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez‐Sáez, J. , Wang, G. , Marella, E.R. , Sudarsan, S. , Pastor, M.C. , and Borodina, I. (2020) Engineering the oleaginous yeast Yarrowia lipolytica for high‐level resveratrol production. Metab Eng 62: 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T.D. , and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schnappauf, G. , Lipscomb, W.N. , and Braus, G.H. (1998) Separation of inhibition and activation of the allosteric yeast chorismate mutase. Proc Natl Acad Sci USA 95: 2868–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suástegui, M. , and Shao, Z. (2016) Yeast factories for the production of aromatic compounds: from building blocks to plant secondary metabolites. J Ind Microbiol Biotechnol 43: 1611–1624. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Zhang, H. , Lu, X. , Zong, H. , and Zhuge, B. (2019) Advances in 2‐phenylethanol production from engineered microorganisms. Biotechnol Adv 37: 403–409. [DOI] [PubMed] [Google Scholar]
- Wei, H. , Wang, W. , Alahuhta, M. , Vander Wall, T. , Baker, J.O. , Taylor, L.E. , et al. (2014) Engineering towards a complete heterologous cellulase secretome in Yarrowia lipolytica reveals its potential for consolidated bioprocessing. Biotechnol Biofuels 7: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, L. , Engel, J. , Jin, E. , Holdridge, B. , and Xu, P. (2017) YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica . Metab Eng Commun 5: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C.‐H. , Cui, W. , Liu, X.‐Y. , Chi, Z.‐M. , and Madzak, C. (2010) Expression of inulinase gene in the oleaginous yeast Yarrowia lipolytica and single cell oil production from inulin‐containing materials. Metab Eng 12: 510–517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Comparison of amino acid sequences of S. cerevisiae and Y. lipolytica Aro4 and Aro7 enzymes. Sequences of Y. lipolytica YlAro4 and YlAro7 enzymes blasted against the equivalent S. cerevisiae enzymes. In yellow are highlighted the positions usually mutated in S. cerevisiae to construct the feedback insensitive forms of the enzymes. In green are highlighted the conserved equivalent positions for Y. lipolytica. In Aro7 S. cerevisiae sequence can be observed also blue marks that correspond to other sites that were also proposed as involved in regulation of the enzyme.
Fig. S2. ARO genes relative expression levels in wild‐type and the strain overexpressing YlARO1, YlARO2, YlARO4K221L and YlARO7G139S .
Table S1. List of Y. lipolytica genes involved in the AAA pathway. The blast results of S. cerevisiae orthologous proteins against Y. lipolytica genome are indicated.
Table S2. List of plasmids used in this study.
Table S3. Primers used in this study.


