Abstract
A broad-range bacterial PCR targeting rRNA genes (rDNAs) was used to directly analyze 536 clinical samples obtained from 459 hospitalized patients during a 4-year study period. The molecular diagnosis based on DNA sequencing of the PCR product was compared to that obtained by bacterial culture. The bacteriological diagnosis was concordant for 447 (83%) specimens. Broad-range rDNA PCR was the only method that yielded an etiologic diagnosis for 11 (2.4%) of 459 patients. Compared to culture and clinical assessment, the sensitivity of the PCR method combined with sequencing was 74.2%, and the specificity was between 98.7 and 99.6%. At present, the described molecular approach proved superior to bacterial culture in two clinical situations: infections caused by bacteria with unusual growth requirements and specimens taken during antimicrobial treatment of the patient.
The phylogenetic classification of microbes based on rRNA gene (rDNA) sequences (4, 34) offers a well-defined framework which can be used to develop molecular tools for microbial identification (1, 29). The rRNA molecules typically consist of highly conserved sequences interspersed with regions of more variable sequences (7, 17). Together with the development of the PCR technique (21), this knowledge has provided an approach for recognition of bacterial pathogens. It is possible to rationally design probes covering a desired phylogenetic group of bacteria, whether this group is a certain division, genus, or species.
When PCR primers are targeted at the most conserved rDNA gene sequences, amplification of the genes of virtually any bacterial species is possible. On the basis of the DNA sequence of the amplification product, the bacterium can then be located in the phylogenetic tree (32). During recent years, this approach has proven its usefulness for the identification of bacterial isolates that are difficult to classify by their phenotypic properties. Even more distinguishing and fascinating has been the application of this method to the identification of rare pathogens with unusual growth requirements and, indeed, of those which can be visualized in tissue but not cultured. The recognition of Bartonella henselae as the cause of bacillary angiomatosis (26) and that of Tropheryma whippelii as the uncultured bacillus associated with Whipple's disease (27, 33) were among the first demonstrations of the power of this method. Subsequently, broad-range bacterial PCR has been used to amplify bacterial genes directly from clinical samples, e.g., biopsy specimens from diseased tissue, with the aim of establishing etiologic diagnoses of various bacterial diseases in a clinical setting. These earlier studies have involved a limited number of specimens and have focused on one defined bacterial disease only, e.g., endocarditis (6, 11), intra-amniotic infection (8, 12), meningitis (16), bacteremia (18), and keratitis (15).
Since the beginning of 1994, our laboratory has applied the PCR technique to the amplification of the 23S or 16S rDNA genes and DNA sequencing of the PCR products for the diagnosis of disease in hospitalized patients. We report here on our 4-year laboratory experience with the use of this molecular approach as a diagnostic tool in a clinical setting and delineate the potential advantages and disadvantages associated with the clinical application of this method.
MATERIALS AND METHODS
Samples.
From the beginning of 1994 through the end of 1997, a total of 536 clinical specimens from 459 patients were analyzed at the Department of Medical Microbiology, Turku University, Turku, Finland, by the broad-range bacterial PCR assay. These included 415 specimens of body fluids, 63 biopsy specimens from tissues, and 58 pus samples from various abscesses (Table 1). All samples were sent from the Turku University Central Hospital. During this study, the PCR assays were requested by the attending medical doctors for clinical purposes.
TABLE 1.
Specimens analyzed by the 23S rDNA PCR and culture as well as the number and percentage of positive results for various sample types
| Specimen | No. (%) of specimens
|
||
|---|---|---|---|
| Analyzed | 23S rDNA PCR positive | Culture positivea | |
| Biopsy | |||
| Bone | 29 | 5 (17) | 4 (14) |
| Heart valve | 8 | 3 (38) | 1 (13) |
| Other or not defined | 26 | 4 (15) | 5 (19) |
| Subtotal | 63 | 12 (19) | 10 (16) |
| Body fluids | |||
| Cerebrospinal fluid | 253 | 15 (6) | 32 (13) |
| Amniotic fluid | 36 | 7 (19) | 4 (11) |
| Synovial fluid | 45 | 6 (13) | 6 (13) |
| Pleural fluid | 58 | 8 (14) | 10 (17) |
| Ascitic fluid | 6 | 2 (33) | 1 (17) |
| Pericardial fluid | 7 | 0 | 1 (14) |
| Bone marrow | 4 | 0 | 0 |
| Other or not defined | 6 | 3 (50) | 2 (33) |
| Subtotal | 415 | 41 (10) | 55 (13) |
| Abscesses | 58 | 20 (35) | 23 (40) |
| Total | 536 | 73 (14) | 88 (16) |
Cultures yielding any bacteria, including those subsequently designated as contamination.
DNA purification.
DNA was extracted from the fresh tissue samples after proteinase K (0.1 mg/ml) digestion (56°C, 2 to 17 h) with two phenol-chloroform-isoamyl alcohol extractions followed by one ether wash as described earlier (11). Synovial, pleural, and amniotic fluids as well as pus samples were concentrated by centrifugation, and organic DNA extraction was performed as described above (12). Extraction of DNA from cells of bone marrow was performed similarly, after gradient centrifugation (Ficoll, Pharmacia, Sweden) and proteinase K treatment. Cerebrospinal fluid and small volumes (0.5 to 1 ml) of pericardial fluid were heated (94°C, 10 min) and treated with proteinase K (16), followed by heat inactivation of the enzyme.
PCR.
The primers used in the 23S and 16S rDNA PCRs have been described earlier (16). To screen for the presence of bacteria in a sample, a PCR with primers MS 37 and MS 38 (which amplify an approximately 850-bp sequence of the 23S rDNA) was performed (16). On the basis of sequence analysis of the 23S rDNA and experimental data, these primers cover several bacterial subdivisions, as described previously (16).
The bacterial DNA present in a 23S rDNA PCR-positive sample was identified by sequencing the 16S and/or 23S rDNA. The 23S rDNA PCR assay was used for the initial screening of the samples because of its higher sensitivity compared to that of the previously described 16S rDNA PCR method. 16S rDNA was preferably used for sequencing, since (in 1997) our database contained 6,800 bacterial 16S rDNA sequences (3,400 of which were complete) but only 204 bacterial 23S rDNA sequences (131 of which were complete).
Analytic sensitivity of the assay.
When serial dilutions of Escherichia coli ATCC 25922 (American Type Culture Collection [ATCC], Manassas, Va.) cells were used as the template, the analytical sensitivity of the 23S rDNA PCR was about 10 CFU/reaction, and this was also the case in the presence of 50,000 Ficoll-isolated human mononuclear blood cells per reaction when the cells were mixed with the bacteria prior to DNA isolation. However, on the basis of the intensity of the band it was estimated that at least 100 CFU would be needed for a successful sequencing reaction. When Streptococcus pyogenes ATCC 8184 was used as a target, the sensitivity of the 23S rDNA PCR assay was 2,000 CFU/reaction.
Precautions to avoid false-positive and false-negative PCR results.
Precautions taken to avoid laboratory contamination included physical separation of the pre-PCR areas used for sample preparation, DNA extraction, and preparation of the reaction mixtures from the post-PCR areas (22). The staff working in the post-PCR areas did not participate in handling of the samples or PCR reagents before amplification. Biosafety hoods and positive-displacement pipettes or barrier tips were used when handling specimens and reagents to avoid carryover and amplicon contamination.
Two negative controls were included in each PCR series: a sample preparation control with sterile water that went through the same DNA extraction steps as the clinical samples and a reagent control with sterile water as a template. As a positive control, 50 ng of DNA from Neisseria meningitidis group B strain 10026 (National Collection of Type Cultures, London, United Kingdom) was used. To avoid amplification of possible bacterial DNA contaminants in the reagents, reaction mixtures were irradiated with UV light for 3 to 3.5 min before addition of the target DNA.
Since April 1997, all samples have also been examined for inhibition of PCR and/or degradation of DNA by amplifying a 429-bp fragment of the human growth hormone gene (24) in a separate reaction by using the same parameters used for the 23S rDNA PCR. When tested by serial dilution experiments with human mononuclear cells, the detection limit of this PCR was about 1,000 cells/reaction. All human samples that were originally PCR negative were further analyzed for inhibition by spiking 90 ng of human DNA (about 10-fold the minimal amount required) in the reaction tube before retesting. Samples found to be strongly inhibitory were excluded from further analyses.
Sequencing.
The sequencing reactions were performed as described earlier, either manually (11) or semiautomatedly (16). The practice of sequencing of the amplified PCR product varied during the study period from sequencing of selected samples after a special request by the clinician in 1994 to routine sequencing of all PCR-positive samples in 1997.
Comparative sequence analyses and databases.
The 16S rDNA sequences that were obtained were compared with sequences in a database made up of sequences from GenBank (2), EMBL (30), and the ribosomal database project (19) by using an in-house algorithm (16). At the beginning of the study period in 1994 the database contained about 3,000 bacterial sequences, and at the end of the study period in 1997 it contained about 6,800 bacterial sequences. For comparison of the 23S rDNA sequences, the FastA program (25) was used. The interpretation of the sequencing results was based on the guidelines given by Stackebrandt and Goebel (28). Below, the best matches and sequence homologies are reported according to the original results; that is, the sequences that were originally analyzed were not reanalyzed by comparison with the sequences in the expanded database that was available at the end of the study. Most often the result of the sequence analysis was reported at the genus level. Some bacterial 16S rDNA or 23S rDNA sequences in the database were distinct enough to allow identification to the species level (6).
Conventional microbiological methods.
In addition to being referred for PCR analysis, the clinical samples were also sent to the microbiology laboratory of the Turku University Central Hospital for bacterial or mycobacterial cultures by conventional methods. The routine culture methods for most samples included aerobic culture on blood agar and chocolate agar plates and, when appropriate, anaerobic culture on a fastidious anaerobic agar plate. Prior to cultivation, liquid samples were concentrated by centrifugation and biopsy specimens were homogenized in brain heart infusion broth (Gibco BRL, Life Technologies, Paisley, Scotland). Identification of the colonies isolated was based on routine microbiological methods (10). Gram staining was performed with the specimen if requested by the attending clinician. Detection of mycobacteria was based on routine culture methods with the use of the BACTEC 460 TB Hood system (Becton Dickinson Diagnostic Instrument Systems, Cockeysville, Md.), followed by identification with AccuProbe (GenProbe, San Diego, Calif.).
Analysis of the microbiological methods.
An infectious diseases specialist (P.K. or O.R.) examined the hospital records of all patients with positive PCR and/or culture results with the aim of evaluating the clinical importance of these findings.
Statistics.
Statistical analyses were performed by Fisher's exact test with the use of the SAS System for Windows (release 6.12; SAS Institute Inc., Cary, N.C.).
RESULTS
PCR and sequencing.
Of all 536 samples analyzed, 73 (14%) were positive by the 23S rDNA PCR (Table 1), and sequencing was attempted for 59 of them. The microbe was recognized to the genus level in 45 (76%) samples, among which the recognition was based on the 16S rDNA sequence for 35 samples and on the 23S rDNA sequence for 10 samples. In one sample from which only the 23S rDNA sequence was available, no matching sequence was found in the database. The sequence was later found to be identical to that of the Propionibacterium acnes strain isolated by bacterial culture from the same sample. In six samples, the presence of several species was suggested on the basis of the typical electropherogram produced by the sequencer. In seven samples, sequencing was not considered possible even after reamplification or the reaction did not produce an interpretable sequence.
Comparison of molecular methods and bacterial culture.
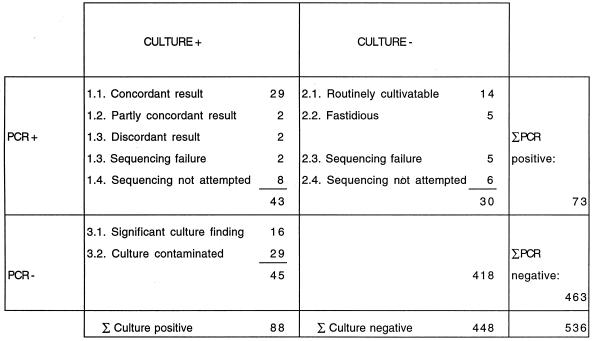
An overall presentation of the results obtained by PCR and sequencing compared to the results of routine culture after resolution of the clinical importance of the findings is shown in Fig. 1.
FIG. 1.
Overall results obtained by broad-range PCR and sequencing compared to those obtained by culture for 536 clinical samples. +, positive result; −, negative result. Σ, sum.
Of the 43 PCR-positive and culture-positive samples, sequencing was not attempted for 8 and was not successful for 2 samples (Fig. 1). A detailed description of the remaining 33 samples is given in Table 2. Classification of the results for two samples as partly concordant was based on recognition of an identical bacterial species by both sequencing and culture and an additional species by one of the methods. Such samples included one amniotic fluid sample, which yielded Capnocytophaga sp. by both methods but also Fusobacterium sp. by culture, and one pus sample, which yielded only Staphylococcus aureus by culture, although the low level of sequence homology of 86.3% suggested the presence of other bacteria in addition to staphylococci. The two samples classified as having discordant results on the basis of the identification of different bacterial species by these two methods included one amniotic fluid sample, which contained Bacteroides ureolyticus on the basis of sequencing but which yielded a Fusobacterium sp. by culture, and one pleural fluid sample, from which Bacillus, Staphylococcus epidermidis, and non-group A beta-hemolytic streptococci were grown but from which only a Haemophilus sp. was sequenced. No definite conclusions about the contribution of these microbes to the clinical disease in these two patients can be made. Since the patient with pleuropneumonia was an alcoholic, the Haemophilus sp. was a probable causative agent, but a mixed infection is also possible. The isolate identified as a Fusobacterium sp. in the clinical microbiology laboratory on the basis of phenotypic properties and resistance to brilliant green and vancomycin but susceptibility to kanamycin by disc tests is no longer available for retesting in order to verify the species identification.
TABLE 2.
Description of 33 clinical specimens which were positive by broad-range bacterial PCR assay and bacterial culture and clinical data regarding the results of blood cultures, administration of antimicrobial therapy, and the final clinical diagnosis for the patient
| Specimen no. | Specimen | PCR result | Sequencing resulta | % Homologyb | Sequence length (bp)b | Culture resultc |
|---|---|---|---|---|---|---|
| 1 | Lung biopsy | + | Staphylococcus sp. | 100 | 300 | Staphylococcus epidermidis (++) |
| 2 | CSFh | + | Streptococcus (S. pneumoniae) | 97.8 | 269 | S. pneumoniae (+++) |
| 3 | CSF | + | Streptococcus (S. pneumoniae) | 100 | 230 | S. pneumoniae (+++) |
| 4 | CSF | + | Neisseria (N. meningitidis) | 99.1 | 228 | N. meningitidis B (+++) |
| 5 | CSF | + | Neisseria (N. meningitidis) | 98.7i | 287 | N. meningitidis B (+) |
| 6 | CSF | + | Neisseria (N. meningitidis) | 99.7i | 287 | N. meningitidis B (+++) |
| 7 | CSF | + | Neisseria (N. meningitidis) | 100 | 230 | N. meningitidis C (+++) |
| 8 | CSF | + | Neisseria (N. meningitidis) | 98.3i | 287 | N. meningitidis B (++) |
| 9 | CSF | + | E. coli | 99.0i | 300 | E. coli (++) |
| 10 | CSF | + | Enterobacterial sp.l | 90.2i | 246 | E. coli |
| 11 | Ascites | + | Enterococcus sp. | 99.0 | 471 | Enterococcus faecium (++) |
| 12 | Pleural fluid | + | Streptococcus sp. | 99.7 | 319 | S. pyogenes (+) |
| 13 | Pleural fluid | + | Pseudomonas (P. aeruginosa) | 98.6 | 217 | P. aeruginosa (++) |
| 14 | Amniotic fluid | + | Streptococcus (S. sanguis) | 100 | 260 | Streptococcus, viridans group (+++) |
| 15 | Synovial fluid | + | Staphylococcus sp. | 100 | 279 | S. aureus (+++) |
| 16 | Synovial fluid | + | Staphylococcus sp. | 99.7 | 285 | S. aureus (+) |
| 17 | Synovial fluid | + | Neisseria sp. | 98.1 | 422 | N. meningitidis B (++) |
| 18 | Synovial fluid | + | Salmonella (S. enteritidis) | 100i | 292 | S. enteritidis (++) |
| 19 | Brain abscess | + | Streptococcus sp. | 95.8 | 484 | Streptococcus equinus (+++) |
| 20 | Brain abscess | + | Streptococcus sp. | 96.5i | 285 | Streptococcus, viridans group (+) |
| 21 | Brain abscess | + | Streptococcus (S. milleri group) | 99.6 | 458 | Streptococcus, viridans group (+++) |
| 22 | Brain abscess | + | Peptostreptococcus sp. | 95.7 | 243 | Anaerobic gram-positive cocci (+++) |
| 23 | Brain abscess | + | Propionibacterium acnes | 99.7 | 299 | P. acnes (+++) |
| 24 | Brain abscess | + | Several species | Bacteroides gracilis (+), Peptostreptococcus sp. (+), S. intermedius (++) | ||
| 25 | Gall bladder abscess | + | P. aeruginosa | 100 | 240 | P. aeruginosa (+++) |
| 26 | Paravertebral abscess | + | Several species | S. epidermidis (+); Streptococcus, group C (+); Streptococcus, nonhemolytic (+) | ||
| 27 | Pus from periost | + | Staphylococcus sp. | 99.6 | 239 | S. aureus (+++) |
| 28 | Pus from soft tissue | + | Staphylococcus sp. | 100 | 513 | S. aureus (++) |
| 29 | Pus from soft tissue | + | Staphylococcus sp. | 99.6 | 260 | S. aureus (+++) |
| 30 | Amniotic fluidi | + | Capniocytophaga (C. sputigena) | 99.7 | 319 | Capnocytophaga sputigena (+++), Fusobacterium sp. (++) |
| 31 | Pus from spinal abscessj | + | Staphylococcus sp., probably also other species | 86.3 | 164 | S. aureus (++) |
| 32 | Pleural fluidk | + | Haemophilus sp. | 93.0 | 199 | Bacillus sp.; S. epidermidis; Streptococcus, beta-hemolytic, non-A |
| 33 | Amniotic fluidk | + | Bacteroides ureolyticus | 98.9 | 273 | Fusobacterium sp. (+++) |
| Gram stain resultd | Blood culture resulte | Antimicrobial therapyf | Final diagnosis |
|---|---|---|---|
| − | NDg | Yes | Pneumonia in a compromised host |
| GPC (+++) | S. pneumoniae | No | Bacterial meningitis |
| GPC (+) | S. pneumoniae | No | Bacterial meningitis |
| GNC (+) | − | No | Bacterial meningitis |
| − | − | No | Bacterial meningitis |
| GNC (+) | N. meningitidis B | No | Bacterial meningitis |
| GNC (+) | − | No | Bacterial meningitis |
| GNC (+) | − | No | Bacterial meningitis |
| GPC (+) | E. coli | No | Bacterial meningitis |
| ND | E. coli | No | Bacterial meningitis |
| − | − | Yes | Mesenterial thrombosis and intra-abdominal infection |
| GPC (+++) | − | No | Pneumonia and pleuritis |
| − | ND | Yes | Hospital-acquired pneumonia |
| ND | S. mitis | Yes (2 h) | Intra-amniotic infection |
| GPC (+++) | − | No | Septic arthritis |
| − | S. aureus | Yes (4 days) | Septic arthritis |
| − | ND | No | Septic coxitis |
| − | Salmonella sp. | No | Septic arthritis |
| GPC (++) | − | Yes (14 h) | Brain abscess |
| − | − | No | Brain abscess |
| GPC (+++) | − | No | Brain abscess |
| GPC (+++) | ND | Yes (12 h) | Brain abscess |
| − | − | No | Postoperative brain abscess |
| GPC (+++) | − | No | Brain abscess |
| ND | − | Yes | Recurrent cholecystitis |
| ND | ND | Yes | Postoperative spondylitis |
| GPC (+++) | ND | Yes (5 days) | Osteomyelitis |
| GPC (++) | S. aureus (−3 days) | Yes (3 days) | Septicemia |
| ND | ND | No | Subcutaneous abscess |
| ND | ND | No | Intra-amniotic infection |
| ND | S. aureus (−8 days) | Yes (8 days) | Sepsis, spondylitis, and spinal abscess |
| ND | ND | Yes (1 day) | Pneumonia and empyema |
| ND | ND | No | Intra-amniotic infection |
The sequencing result is reported according to the principles reported in the text.
The percent homology and the length of the overlapping sequence for the best match are presented.
Symbols in parentheses are relative number of colonies on the primary culture plate: +, few; ++, moderate; +++, many.
GPC, gram-positive cocci; GNC, gram-negative cocci. Symbols in parentheses are relative number of cells seen in the stain: +, few; ++, moderate; +++, many; −, negative result.
Unless otherwise indicated, the blood for culture was taken on the same day (±2 days) that the specimen for the PCR assay and culture was taken. −, negative result.
The durations of antimicrobial therapy effective against the microbe(s) recognized in the specimen are given in parentheses. Patients 1, 11, 13, 25, and 26 received ineffective therapy.
ND, the test was not done.
CSF, cerebrospinal fluid.
Recognition was based on the 23S rDNA sequence. Preferably, the 16S rDNA sequence was used.
Specimens had partly concordant results.
Specimens had discordant results.
A species belonging to the family Enterobacteriaceae.
Among the 30 PCR-positive and culture-negative specimens, sequencing gave an interpretable result for 18 specimens, suggested the presence of several species in 1 specimen, was not attempted for 6 specimens, and was not successful for five specimens (Fig. 1). A description of the 19 specimens is given in Table 3. Thirteen of the bacterial species sequenced had normal growth requirements and five were fastidious. For six patients, blood culture yielded the same bacterial species or genus as identified in the patient's biopsy or body fluid specimen by sequencing, with the median point for a positive blood culture result being 22 days (range, 2 h to 30 days) before the sample for PCR and culture was taken. For 11 patients (2.4% of the 459 patients), PCR combined with sequencing was the only method that yielded evidence of the etiological agent of the infection, as blood culture either remained negative or was not done. On the basis of a clinical evaluation, the enterobacterial DNA recognized in a lung biopsy specimen of a leukemia patient with aspergillosis was designated as probable PCR contamination, while the remaining findings were considered clinically significant. The etiologic role of the Prevotella sp. (most likely Prevotella oris) DNA detected in the pleural fluid of patient 13 was supported by the detection of the same DNA sequence from pulmonary abscess material, which was, erroneously, not cultured. These findings are in agreement with the severe immunosuppression of this patient due to heart transplantation. In patient 5, the recognition of several bacterial species in a lung biopsy specimen after bone marrow transplantation was considered suggestive of a polymicrobial infection in an immunocompromised host.
TABLE 3.
Description of 19 clinical specimens which were positive by broad-range bacterial PCR assay and negative by culture as well as clinical data regarding the results of blood cultures, administration of antimicrobial therapy, and the final clinical diagnosis for the patient
| Specimen no. | Specimen | PCR result | Sequencing resulta | % Homologyb | Sequence length (bp)b | Culture result | Gram staining result |
|---|---|---|---|---|---|---|---|
| 1 | Bone biopsy | + | Staphylococcus sp., probably also other species | 94.7 | 216 | − | − |
| 2 | Bone biopsy | + | Staphylococcus sp. | 100 | 284 | − | NDe |
| 3 | Bone biopsy | + | Enterobacterial sp.f | 96.3g | 297 | − | − |
| 4 | Lung biopsy | + | Enterobacterial sp.f | 92.9g | 266 | − | − |
| 5g | Lung biopsy | + | Several species | − | ND | ||
| 6 | Muscle biopsy | + | Bacteroides sp. | 98.5 | 262 | − | ND |
| 7 | Heart valve | + | Abiotrophia defectiva | 100 | 495 | − | − |
| 8 | Heart valve | + | Bartonella (B. quintana) | 99.8 | 1,406 | − | − |
| 9 | CSFh | + | Streptococcus (S. pneumoniae) | 100 | 360 | − | GPC+i |
| 10 | CSF | + | Neisseria (N. meningitidis) | 100 | 230 | − | − |
| 11 | CSF | + | Neisseria (N. meningitidis) | 100 | 237 | − | ND |
| 12 | Ascitic fluid | + | Bacteroides fragilis | 99.6 | 239 | − | − |
| 13 | Pleural fluid | + | Prevotella (P. oris) | 100 | 310 | − | − |
| 14 | Amniotic fluid | + | Ureaplasma urealyticum | 100 | 300 | − | ND |
| 15 | Amniotic fluid | + | U. urealyticum, probably also other species | 88.1 | 208 | − | ND |
| 16 | Puncture from Douglas' pouch | + | U. urealyticum | 100 | 468 | − | − |
| 17 | Brain abscess | + | Staphylococcus sp. | 100 | 300 | − | − |
| 18 | Pus from empyema | + | Streptococcus sp. | 100 | 343 | − | ND |
| 19 | Pus from soft tissue | + | Bartonella sp. | 96.7g | 271 | − | − |
| Blood culture resultc | Antimicrobial therapyd | Final diagnosis |
|---|---|---|
| S. aureus (15 days) | Yes (15 days) | Sepsis, spondylitis and spinal abscess |
| ND | Yes (10 days) | Osteomyelitis |
| Klebsiella pneumoniae (30 days) | Yes (30 days) | Spondylitis and paravertebral abscess |
| ND | Yes (>30 days) | Aspergillosis, probable PCR contamination |
| Lactobacillus sp., coagulase-negative Staphylococcus | Yes | Hospital-acquired pneumonia in a compromised host |
| ND | Yes (>30 days) | Intramuscular abscess |
| Abiotrophia defectiva (28 days) | Yes (28 days) | Infective endocarditis |
| − | Yes (5 days) | Infective endocarditis |
| S. pneumoniae (2 days) | Yes (2 days) | Bacterial meningitis |
| − | Yes (7 hours) | Bacterial meningitis |
| N. meningitidis B (2 hours) | Yes (2 hours) | Bacterial meningitis |
| − | Yes (17 days) | Spontaneous peritonitis |
| ND | Yes (11 days) | Lung abscess and pleural empyema in a compromised host |
| ND | Yes | Intra-amniotic infection |
| ND | Yes | Intra-amniotic infection |
| − | Yes | Mesenterial thrombosis and intra-abdominal infection |
| − | Yes (5 days) | Postoperative brain abscess |
| S. pneumoniae (30 days) | Yes (30 days) | Pneumonia and pleural empyema |
| ND | Yes (7 days) | Cat-scratch disease |
The sequencing result is reported according to the principles described in the text.
The percent homology and the length of the overlapping sequence for the best match are presented.
The times of a positive blood culture result before the specimen for the PCR assay and culture was taken are given in parentheses. The blood samples that were negative by culture were taken on the same day (±2 days.)
The durations of antimicrobial therapy usually effective against the microbe recognized by sequencing are given in parentheses. Patient 5 received ceftazidime, and patients 14 to 16 received ineffective therapy.
ND, the test was not done.
A species belonging to the family Enterobacteriaceae.
Recognition was based on the 23S rDNA sequence. Preferably, the 16S rDNA sequence was used. Data for patient 5 were excluded from the final analysis of sensitivity and specificity.
CSF, cerebrospinal fluid.
GPC+, few cells of gram-positive cocci.
Of the 46 PCR-negative and culture-positive samples, the culture result was designated as contamination for 29 samples (Fig. 1). This decision was made on the basis of (i) scarce growth of coagulase-negative staphylococci and/or gram-positive rods such as Corynebacterium sp., Propionibacterium sp., or Bacillus sp. in the primary culture plate or (ii) detection of one or more of the aforementioned species or other species known to have low levels of pathogenicity, such as an Enterococcus sp. or pseudomonas-like rods, in one of three enrichment cultures, often after a prolonged culture period. In all patients, the clinical presentation of the disease and the immunologic status of the patient were verified before classification of the culture finding as contamination. Among the 16 PCR-negative specimens yielding significant growth (Table 4), the microbe recovered was a gram-positive organism for 11 specimens and belonged to the mycobacteria for 4 specimens. Only once was a gram-negative bacterium missed by the bacterial PCR methodology. The distribution of gram-positive and gram-negative bacteria in PCR-negative versus PCR-positive samples differed significantly (P = 0.0001; Fisher's exact test), indicating that gram-positive bacteria were not detected as efficiently as the gram-negative ones. In 6 of the 16 samples, the number of bacteria was sufficient to be visualized by routine staining procedures. The possibility of PCR inhibition in these 16 samples was excluded by successful amplification of the human growth hormone gene.
TABLE 4.
Description of 16 clinical specimens which were negative by broad-range bacterial PCR assay and positive by culture as well as clinical data regarding the results of blood cultures, administration of antimicrobial therapy, and the final clinical diagnosis for the patient
| Specimen no. | Specimen | PCR result | Culture findinga | Gram staining resultb | Blood culture resultc | Antimicrobials therapyd | Final diagnosis |
|---|---|---|---|---|---|---|---|
| 1 | CSFe | − | Streptococcus pneumoniae (+) | − | S. pneumoniae | No | Bacterial meningitis |
| 2 | CSF | − | S. pneumoniae (+) | − | − | No | Bacterial meningitis |
| 3 | CSF | − | Streptococcus mitis (+++) | GPC (+) | NDf | Yes | Bacterial meningitis |
| 4 | CSF | − | Streptococcus agalactiae (+) | GPC (+) | S. agalactiae | No | Bacterial meningitis |
| 5ag | CSF | − | P. acnes (+++) | GPR (++) | P. acnes | No | Cerebrospinal shunt infection |
| 5bg | CSF | P. acnes (+++) | ND | ||||
| 6 | CSF | − | Listeria monocytogenes (+) | − | L. monocytogenes | No | Bacterial meningitis |
| 7 | CSF | − | Mycobacterium tuberculosis | − | − | No | Tuberculous vasculitis |
| 8 | Pleural fluid | − | S. aureus (+) | ND | − | Yes (6 days) | Pneumonia and pleural empyema |
| 9 | Pleural fluid | − | Streptococcus pyogenes (+++) | GPC (++) | − | Yes (1 day) | Pneumonia and pleural empyema |
| 10 | Pus from empyema | − | S. aureus (+++) | GPC (+) | − | Yes (3 days) | Postoperative subdural empyema |
| 11 | Pus from middle ear | − | S. aureus (+++) | ND | ND | No | Chronic otitis media |
| 12 | Pus from spinal abscess | − | P. aeruginosa (++) | − | P. aeruginosa | Yes | Postoperative spondylodiscitis and spinal abscess |
| 13 | Pus from spinal abscess | − | M. tuberculosis | − | − | Yes | Tuberculous spondylitis |
| 14 | Pus from knee joint | − | M. tuberculosis | AFB (+) | − | No | Miliary tuberculosis with bone involvement |
| 15 | Pus from soft tissue | − | Mycobacterium avium | − | − | No | M. avium abscess |
Symbols in parentheses are relative number of colonies on the primary culture plate; +, few; ++, moderate; +++, many.
For patients 13 and 14, auramine-rhodamine stain. GPC, gram-positive cocci; GPR, gram-positive rods; AFB, acid-fast bacilli. Symbols in parentheses are relative number of cells seen in the strain: +, few; ++, moderate; +++, many.
On the same day (±2 days) as the specimen for the PCR assay and culture was taken.
The durations that antimicrobial therapy was effective against the microbe recognized in the specimen are given in parentheses. Patients 3, 12, and 13 received ineffective therapy.
CSF, cerebrospinal fluid.
ND, the test was not done.
Two successive cerebrospinal fluid samples, designated samples 5a and 5b, were obtained from patient 5.
Effect of antimicrobial treatment on recovery of microbes.
A tendency for a specimen to be culture negative but PCR positive was observed if the patient was receiving antimicrobial treatment at the time that the specimen was taken. A comparison of the antimicrobial status (effective antimicrobial agents versus no antimicrobial treatment) of the patients for whom results are presented in Tables 2 and 3 showed that if the patient was receiving effective antimicrobial treatment, the infective agent was significantly more likely to be detected by rRNA amplification only (Table 3) than by both conventional and molecular methods (Table 2) (P = 0.000016; Fisher's exact test). Data for the specimens that were taken during ineffective antimicrobial treatment and those that yielded fastidious microbes were excluded from the statistical analysis.
Sensitivities and specificities of the methods.
For evaluation of sensitivity and specificity, samples from groups 1.1, 2.1, and 2.2 in Fig. 1 were designated true PCR positive, samples from group 3.1 were designated false PCR negative, and samples from groups 3.2 and 4 were designated true PCR negative. The sample from patient 4 (Table 2) as well as a second sample with evident PCR contamination found in the clinical evaluation (a synovial fluid sample belonging to group 2.3) was designated as false PCR positive. Data for the remaining samples were excluded from the analysis because of the difficulty in judging the significance of the finding in the absence of sequence data.
As resolved by the culture result and clinical evaluation as described above, the sensitivity of the PCR assay combined with sequencing was 74.2%, and the specificity was 99.6%, while the positive and negative predictive values of this method were 95.8 and 96.5%, respectively. If all five samples with sequencing failures in group 2.3 were designated contaminations, the specificity of the method would be 98.7%, with a positive predictive value of 88.5%. The performance of the PCR with various sample types is shown in Table 5.
TABLE 5.
Performance of broad-range bacterial PCR compared to culture results and clinical evaluation for various sample typesa
| Sample type | No. of samples | Sensitivity (%) | Specificity (%) | PPVb (%) | NPVc (%) |
|---|---|---|---|---|---|
| Tissue biopsy specimen | 59 | 100 | 98.1 | 87.5 | 100 |
| Cerebrospinal fluid | 250 | 63.2 | 99.6 | 92.3 | 97.1 |
| Amniotic fluid | 33 | 100 | 100 | 100 | 100 |
| Pleural fluid | 54 | 60.0 | 98.0 | 75.0 | 96.0 |
| Synovial fluid | 46 | 100 | 95.1 | 71.4 | 100 |
| Pus | 50 | 68.4 | 96.8 | 92.9 | 83.3 |
| All samples | 515 | 74.2 | 98.7 | 88.5 | 96.6 |
Sequencing failures for culture-negative samples were designated PCR contaminations.
PPV, positive predictive value.
NPV, negative predictive value.
DISCUSSION
We assess in this study the usefulness and practicability of molecular methods in a clinical setting on the basis of an analysis of 536 patient samples examined by a broad-range bacterial PCR assay. Our results show that the molecular findings were in good agreement with those obtained by conventional microbiological methods. The results were identical for 447 (83%) samples, including 418 culture-negative and PCR-negative samples and 29 culture-positive samples from which significant pathogens were sequenced, as defined by clinical assessment.
An important finding was that for 17 samples, the PCR assay combined with sequencing succeeded in recognizing an etiologically significant bacterial pathogen that did not grow in conventional bacterial culture, although the species identified was routinely cultivatable from 12 samples. Clinical evaluation of these culture-negative patients revealed that all of them had been receiving antimicrobial treatment at the time that the specimen for PCR and culture was taken. This finding supports our previous hypothesis that in patients receiving empiric antimicrobial treatment, molecular methods might be superior to bacterial culture in providing the etiologic diagnosis (16). In the present study, this hypothesis could be statistically verified.
Another advantage of bacterial PCR is connected to its broad-range nature: rare or unexpected pathogens can be found. Patient 8 (Table 3) was the first patient in Finland to receive a diagnosis of bartonella endocarditis; until then, bartonella infections had been unknown in our country (11). Both Bartonella and the other fastidious pathogen found by molecular methods in the present study, Ureaplasma urealyticum, can be cultured with relative ease, with the precondition that they are regarded as potential etiologic agents of infection and specific growth conditions are provided. Considering the numerous unknown rDNA sequences found in environmental samples (9), it is noteworthy that no novel bacterial species were detected in this study with a large set of clinical material.
A comparison to culture results revealed difficulties in detecting gram-positive species and mycobacteria by PCR. On the basis of sequence analysis, there is no systematic discrimination of gram-positive species (with the exception of the genus Bifidobacterium) with the 16S rDNA-specific primers (14). Also, the 23S rDNA-specific primers have successfully amplified several gram-positive bacteria, including the genera most often missed in the present study, Streptococcus and Staphylococcus, as well as mycobacteria. Thus, the problems are due to difficulties in breaking the cell walls of these organisms during sample preparation, resulting in a failure in the DNA extraction process. Microscopically, an abundance of S. pyogenes cells remains intact after boiling and incubation of the sample with proteinase K (14). Therefore, finding a more effective method of breaking the cell walls would be of vital importance but may be difficult, since the addition of enzymes to the reaction tubes inevitably increases the risk of contamination with bacterial DNA in the reagents. According to our preliminary experiments, mechanical disruption of bacteria could be a more optimal solution.
In any PCR laboratory, strenuous efforts should be taken to avoid contamination of the reagents and samples by amplicons and high-copy-number DNA in other samples. The risk of contamination is still higher for broad-range bacterial PCR than for specific assays, since traces of environmental bacterial fragments from reagents, vessels, etc., will be amplified. Thus, the goal is to determine a level at which the sensitivity of the PCR assay is sufficient for finding the possible pathogens in the samples while simultaneously avoiding the detection of occasional bacterial DNA in the reagents and/or the environment, leading to false-positive results. In the present material, PCR contaminations appeared to be less of a problem than expected, because in comparison to culture results, only one sample with unequivocal contamination and one sample with probable contamination were detected.
Application of rRNA phylogeny to the identification of bacteria, whether the rDNA is amplified directly from clinical samples or is isolated from bacteria in pure cultures, always allows a given (single) sequence to be located somewhere in the bacterial phylogeny (13). Thus, the identity of no isolate, even a novel one, remains totally unknown. At present, a 16S rDNA-based system for identification of bacterial strains is also available commercially (MicroSeq; Applied Biosystems, Foster City, Calif.). However, phylogenic identification based only on ribosomal sequences does have its limitations. Several examples show that information on rRNA sequences (16S or 23S) is not in agreement with DNA-DNA relatedness or the phenotypic properties of a bacterial species. Clearly separated species can have completely identical (within the limits of sequencing errors) 16S rRNA molecule sequences (5, 20, 28), but, on the other hand, intrastrain variations in the 16S rRNA sequences are common (3). In general, it is obvious that the 16S rRNA (or 23S rRNA) identity does not guarantee the species identity (5, 28). This is especially true if only partial 16S or 23S rRNA sequences are used for species identification. In every case, the decision must be made individually on the basis of similarity values obtained from the sequence comparisons, with careful consideration of the quality of the database.
The clinical shortcomings related to the use of broad-range bacterial PCR and sequencing are also of concern. Often only the bacterial genus can be determined, while the probable species remains undefined. This may cause difficulties, for example, regarding various staphylococcal species, as the therapeutic guidelines for S. aureus and coagulase-negative staphylococcal infections are different. In addition, the antimicrobial susceptibility of the causative agent remains unknown. Polymicrobial infections are also problematic due to the inability of the current procedure to identify microorganisms when several are present in the same specimen. Furthermore, sequence homology to species commonly present in water or the environment (31) but of little pathogenic potential should always raise concern about contamination and warrants a careful clinical evaluation.
On account of financial and professional requirements, the performance of the kind of a molecular assay described here is restricted to only a few highly specialized laboratories. The median turnaround time for PCR was 2 days from the time of arrival of the sample at the laboratory, and the results of the sequence analysis were obtained from 2 to 14 days after retrieval of the positive PCR result. Therefore, the approach reported here is rather slow and is too labor-intensive to be used in clinical microbiology. However, the development of automated DNA amplification techniques together with rapid and affordable sequence detection systems will contribute to the usefulness of the direct molecular approach in the near future. The introduction of novel high-density DNA microarrays may also revolutionize pathogen detection based on molecular techniques (23).
In conclusion, we report here on a comparison of direct broad-range PCR and sequencing and routine culture in bacteriological diagnostics for 536 samples obtained from 459 hospitalized patients. Although not currently competitive with conventional methods, the direct molecular approach appears to be a valuable complementary technique for the diagnosis of infectious diseases. At present, broad-range bacterial PCR can be recommended for samples obtained during antimicrobial treatment and/or by a demanding clinical procedure, especially if routine cultures remain negative. The findings presented here may be used as a basis for the development of faster and less labor-intensive molecular methodologies for the direct identification of bacterial pathogens.
ACKNOWLEDGMENTS
Pentti Huovinen is acknowledged for critical reading of the manuscript. We thank Päivi Sandberg and Tuula Närä for computational assistance. Tiina Haarala, Kirsi Sundholm, Merja Mikkola, and Anne Peippo are acknowledged for excellent technical work.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson D, Boguski M S, Lipman D J, Ostell J, Oullette B F F, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton R A, Sutton G, Hinkle P S, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 4.Fox G E, Stackebrandt E, Hespell R B, Gibson J, Maniloff J, Dyer T A, Wolfe R S, Balch W E, Tanner R S, Magrum L J, Zablen L B, Blakemore R, Gupta R, Bonen L, Lewis B J, Stahl D A, Luehrsen K R, Chen K N, Woese C R. The phylogeny of procaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 5.Fox G E, Wisotzkey J D, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 6.Goldenberger D, Künzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35:2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutel R R, Weiser B, Woese C R, Noller H F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acids Res. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 8.Hitti J, Riley D E, Krohn M A, Hillier S L, Agnew K J, Krieger J N, Eschenbach D A. Broad-spectrum bacterial rDNA polymerase chain reaction assay for detecting amniotic fluid infection among women in premature labor. Clin Infect Dis. 1997;24:1228–1232. doi: 10.1086/513669. [DOI] [PubMed] [Google Scholar]
- 9.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isenberg H D, editor. Clinical microbiology procedures handbook. Washington, D.C.: American Society for Microbiology; 1992. [Google Scholar]
- 11.Jalava J, Kotilainen P, Nikkari S, Skurnik M, Vänttinen E, Lehtonen O-P, Eerola E, Toivanen P. Use of the polymerase chain reaction and DNA sequencing for detection of Bartonella quintana in the aortic valve of a patient with culture-negative infective endocarditis. Clin Infect Dis. 1995;21:891–896. doi: 10.1093/clinids/21.4.891. [DOI] [PubMed] [Google Scholar]
- 12.Jalava J, Mäntymaa M-L, Ekblad U, Toivanen P, Skurnik M, Lassila O, Alanen A. Bacterial 16S rDNA polymerase chain reaction in the detection of intra-amniotic infection. Br J Obstet Gynecol. 1996;103:664–669. doi: 10.1111/j.1471-0528.1996.tb09835.x. [DOI] [PubMed] [Google Scholar]
- 13.Jalava J, Eerola E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int J Syst Bacteriol. 1999;49:1375–1379. doi: 10.1099/00207713-49-4-1375. [DOI] [PubMed] [Google Scholar]
- 14.Jalava J. Molecular detection and identification of bacteria based on PCR and rRNA phylogeny. Ph.D. thesis. Turku, Finland: University of Turku; 1999. [Google Scholar]
- 15.Knox C M, Cevellos V, Dean D. 16S ribosomal DNA typing for identification of pathogens in patients with bacterial keratitis. J Clin Microbiol. 1998;36:3492–3496. doi: 10.1128/jcm.36.12.3492-3496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotilainen P, Jalava J, Meurman O, Lehtonen O-P, Rintala E, Seppälä O-P, Eerola E, Nikkari S. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J Clin Microbiol. 1998;36:2205–2209. doi: 10.1128/jcm.36.8.2205-2209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley B E, Linton C J, Bennett D M C, Jalal H, Foot A B M, Millar M R. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1998;17:247–253. doi: 10.1007/BF01699981. [DOI] [PubMed] [Google Scholar]
- 19.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C W. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Murcia A J, Benlloch S, Collins M D. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int J Syst Bacteriol. 1992;42:412–421. doi: 10.1099/00207713-42-3-412. [DOI] [PubMed] [Google Scholar]
- 21.Mullis K B, Falcoona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 22.Nikkari S, Merilahti-Palo R, Saario R, Söderström K-O, Granfors K, Skurnik M, Toivanen P. Yersinia-triggered reactive arthritis. Use of polymerase chain reaction and immunocytochemical staining in the detection of bacterial components from synovial specimens. Arthritis Rheum. 1992;35:682–687. doi: 10.1002/art.1780350613. [DOI] [PubMed] [Google Scholar]
- 23.Nikkari S, Relman D A. Molecular approaches for identification of infectious agents in Wegener's granulomatosis and other vasculitides. Curr Opin Rheumatol. 1999;11:11–16. doi: 10.1097/00002281-199901000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Olerup O. HLA-B27 typing by a group-specific PCR amplification. Tissue Antigens. 1994;43:253–256. doi: 10.1111/j.1399-0039.1994.tb02334.x. [DOI] [PubMed] [Google Scholar]
- 25.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tomkins L S. The agent of bacillary angiomatosis—an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 27.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 28.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 29.Stahl D A, Kane M D. Methods of microbial identification, tracking and monitoring of function. Curr Opin Biotechnol. 1992;3:244–252. [Google Scholar]
- 30.Stoesser G, Tuli M A, Lopez R, Sterk P. The EMBL nucleotide sequence database. Nucleic Acids Res. 1999;27:18–24. doi: 10.1093/nar/27.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanner M A, Goebel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson K H, Blitchington R, Frothingam R, Wilson J A P. Phylogeny of the Whipple's-disease-associated bacterium. Lancet. 1991;338:474–475. doi: 10.1016/0140-6736(91)90545-z. [DOI] [PubMed] [Google Scholar]
- 34.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]