Abstract
To provide reliable molecular markers and effective therapeutic targets for chondrosarcoma and glioma.
Gene Set Enrichment (GSE) 29745 and GSE48420 were downloaded from the Gene Expression Omnibus (GEO) database. Differently expressed genes (DEGs) were identified by the GEO2R. We annotated the function of common DEGs through Digital Audio/Video Interactive Decoder (DAVID) and Metascape. Protein–protein interaction network construction was performed through STRING. Hub genes were identified by the two different algorithms (MCC, EPC). DDX10 and BYSL were key factors in embryo implantation and development, and plays a role in a variety of cancers. The role of the DDX10 and BYSL on the glioma derived from the chondrosarcoma would be explored by the clinical samples.
A total of 1442 DEGs were identified. The variations in DEGs were mainly enriched in vasculature development, cell motion, blood vessel development, cell migration, regulation of cell proliferation, regulation of cell proliferation, wound healing, biological adhesion, growth factor binding, identical pathways in cancer, and p53 signaling pathway. Dead-box helicase 10 (DDX10), Bystin-like (BYSL), and WD repeat domain 12 (WDR12) were identified as the hub genes, and the three hub genes were up-regulated in the chondrosarcoma. Chondrosarcoma patients with high expression levels of DDX10 (Logrank P = .0052; HR (high) = 1.8; n (high) = 131, 50%), and BYSL (P = 6.5e-05; HR (high) = 2.3; n (high) = 131, 50%) had poorer overall survival times than those with low expression levels.
DDX10 and BYSL genes may provide reliable molecular markers and effective therapeutic targets for chondrosarcoma and glioma.
Keywords: BYSL, chondrosarcoma, DDX10, glioma, hub genes
1. Introduction
Chondrosarcoma is a malignant tumor originating from cartilage or cartilaginous connective tissue, with a strong ability to invade locally and cause distant metastasis. And it is the second most common malignant bone tumor after osteosarcoma, accounting for about 30% of malignant bone tumors.[1] Histologically, chondrosarcoma is classified into clear, mucoid, fibrochondrosarcoma, mixed type and clear cell type.[2] The disease progresses slowly in general. Clinical symptoms are related to tumor site, growth mode and degree of nerve compression. In the early stage, symptoms are mostly asymptomatic, while in the later stage, pain and swelling of the affected area may occur. In patients with chondrosarcoma, there may be lumps in some affected areas. There is no obvious pain when pressing the lumps. Redness and fever can be observed around the skin of the affected area. The movement of adjacent joints is limited, and complications such as fractures and varicose veins may occur later. With the gradual enlargement of the mass, it can also produce symptoms of compression, manifested as weakness, weakness, numbness, dysfunction and so on. If not treated in time, complications such as limb paralysis and intestinal obstruction may occur. Presently, even if the five-year survival livability of patients has been improved by curative operation combined with chemotherapy, the overall therapeutic effect is still unsatisfactory.[3] The reason may be chondrosarcoma is not sensitive to radiotherapy and chemotherapy. As a result, targeted therapy may become a new and effective method, and the molecular biological mechanism of occurrence and development of chondrosarcoma are needed by the research of targeted drugs. However, the cause of chondrosarcoma is still unknown, which may be related to genetic factors, chromosomal abnormalities, gene fusion and other factors. Furthermore, there is a new research direction for the pathogenesis of chondrosarcoma with the development of molecular cytogenetics and modern cytogenetics. And the chondrosarcoma might transfer to the brain, and the glioma could occur in the patient. Therefore, it is important to study the molecular mechanism of chondrosarcoma and glioma.
Bioinformatics is a new interdisciplinary subject which combines life science and computer science. It mainly studies the collection, storage, processing, transmission, analysis and interpretation of biological information. Bioinformatics techniques can be used to process and analyze large amounts of complex biological data. Microarray data information analysis has been widely used in the study of tumors and other diseases to explore the genetic correlation among them.[4,5] Microarray analysis technology can simultaneously obtain the expression information of tens of thousands of genes, and then discover the genomic changes related to the occurrence and development of diseases. At present, a large number of studies[6,7] have used bioinformatics technology to analyze differentially expressed genes in tumor progression, and then studied their roles in biological processes, molecular functions and signaling pathways, and clarified the pathogenesis of diseases, so as to provide theoretical basis for early diagnosis and treatment. However, there are a few of studies using the application of bioinformatics technology into the molecular mechanism of chondrosarcoma and glioma.
DDX10 (dead-box helicase 10) is a member of the DDX protein family that encodes Ribonucleic Acid (RNA) helicase. Many studies have found that DDX10 is abnormally expressed in ovarian cell carcinoma,[8] hepatocellular carcinoma,[9] acute myeloid leukemia,[10,11] osteosarcoma[12] and other tumor tissues. Bystin (BYSL) is a 306-amino acid protein encoded in humans by the BYSL gene which is located on the 6p21.1 chromosome. It is conserved across a wide range of eukaryotes.[13] It is also a key factor in embryo implantation and development,[14] and plays a role in a variety of cancers.[15,16] Wang et al established a hepatocellular carcinoma model, and the final result showed that BYSL is essential for the growth of Hepatocellular Carcinoma (HCC) cells in vitro and in vivo. Compared with adjacent non-cancerous tissues, the expression levels of BYSL mRNA and protein in human liver cancer specimens increased significantly.[15] It is suggested that BYSL with IGF-I- and amplified in breast cancer 1(AIB1)-dependent also plays a vital role in the molecular mechanism of the occurrence and development of breast cancer.[17] However, their association with chondrosarcoma has been poorly studied, and its association with chondrosarcoma and glioma remains unclear.
In summary, first of all, the effects of radiotherapy and chemotherapy in the treatment of chondrosarcoma are not satisfactory. Targeted therapy is likely to become a new effective treatment method, but new biomarker is still lacking. Secondly, lots of research have been done on chondrosarcoma, they have not yet fully elucidated the molecular mechanism of its pathogenesis. Finally, many studies have shown that DDX10 and BYSL play vital roles in a variety of malignant tumors, but the evidence for chondrosarcoma and glioma is insufficient.
Therefore, this paper intends to use bioinformatics technology to excavate the core genes between chondrosarcoma and normal tissue and conduct enrichment analysis, pathway analysis and survival analysis. Moreover, the present study aimed to investigate the functional involvement of DDX10 and BYSL genes in chondrosarcoma and its potentially underlying mechanism. Finally, it suggested whether DDX10 (dead-box helicase 10) and BYSL (a key factor in embryo implantation and development) genes could be new targets for the diagnosis and targeted therapy of chondrosarcoma. Use public data to verify the role of core genes in chondrosarcoma. And the role of the DDX10 and BYSL on the glioma derived from the chondrosarcoma would be explored by the experiments.
2. Material and methods
2.1. Dataset
We downloaded tow gene profiles, GSE29745 and GSE48420, from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GSE29745 includes 6 chondrosarcoma samples and 2 normal cartilage samples, while the GSE48420 includes 3 chondrosarcoma samples and 3 normal cartilage samples.
2.2. DEGs identification
We applied GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r), an online tool based on GEO query and limma R packages, to identify DEGs between esophagus cancer and normal group. The cut-off criteria were that P-value < .05 and a log (FC) > 1 or log (FC) < -1.
Log (FC) is the logarithmic value of fold change (FC), meaning “difference multiples”, and is an important indicator in difference analysis. When log (FC)> 1, it means that the gene is up-regulated relative to the control group; when log (FC) < -1 means that the gene is relative to the control group down.
2.3. DEGs annotation
DAVID (https://david.ncifcrf.gov/home.jsp) and Metascape (http://metascape.org/gp/inde x.html) are two powerful annotation tools which can perform the biological process (BP), cellular component (CC), molecular function (MF) and KEGG analysis on genes. We annotated the function of common DEGs through DAVID and Metascape.
2.4. Protein–protein interaction network construction
The Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org), can convert DEGs into expressed proteins and construct PPI network. We got PPI network of common DEGs through STRING, and visualized it by Cytoscape (version 3.8.0, it is available at the official Cytoscape app store: https://cytoscape.org/).
2.5. Hub genes identification and expression
Molecular Complex Detection tool (MCODE) (version 1.6.1), an open plug-in of Cytoscape, was performed to identify significant module from PPI network. The criteria were that the maximum depth = 100, MCODE scores > 5, cut-off = 2, k-score = 2, and node score cut-off = 0.2. Besides, we also applied cytoHub to screened out hub genes from key module of MCODE and ranked them by two different algorithms, MCC, EPC. And the expression of hub genes was visualized by the R language.
2.5.1. K-score
The larger the value, the smaller the cluster. The degree of nodes in the subnet must be greater than k.
2.5.2. Node score cut-off
Affects the size of the cluster, the smaller the value, the smaller the cluster obtained.
2.6. Conventional treatment
Surgery is the best choice for chondrosarcoma. First of all, the choice of anesthesia is different depending on the tumor growth site of the patient. Epidural anesthesia or even general anesthesia are used. The position also needs to be changed according to the changes in the tumor growth position, such as supine or prone position, etc. When it comes to approach and exposure, pathological changes, resection, fixation or placement of prosthesis, and reconstruction, adopted surgical procedures and ideas commonly used in China.[18]
2.7. RT-qPCR assay
A total of 10 participates were recruited, including 5 control individuals and 5 patients with glioma derived from the chondrosarcoma. After surgery, 5 glioma tumor samples from patients with glioma derived from the chondrosarcoma and 5 control brain samples from control individuals were obtained. The research conformed to the Declaration of Helsinki and was authorized by the Human Ethics and Research Ethics Committees of the Cangzhou Central Hospital. The informed consents were obtained from all participates.
Total RNA was extracted 10 samples by the RNAiso Plus (Trizol) kit (Thermofisher, Massachusetts, America), and reverse transcribed to cDNA. RT-qPCR was performed using a Light Cycler 4800 System with specific primers for DDX10 and BYSL. Table 1 presents the primer sequences used in the experiments. The RQ values (2−ΔΔCt, where Ct is the threshold cycle) of each sample were calculated, and are presented as fold change in gene expression relative to the control group. GAPDH was used as an endogenous control.
Table 1.
Primers and their sequences for RT-PCR analysis.
| Primer | Sequence (5′–3′) |
| GAPDH-hF | GAAAGGTCCCAGTGAACGGAT |
| GAPDH-hR | CTTTCTCAGCCAAGACCGTG |
| DDX10-hF | TGCTAATTAGCAAACGAA |
| DDX10-hR | CGATTAATTGAGGGTGAG |
| BYSL-hF | ATCGAACCAGTTACTCCG |
| BYSL-hR | GCCCATCTGACTGTACCCT |
3. Results
3.1. DEGs
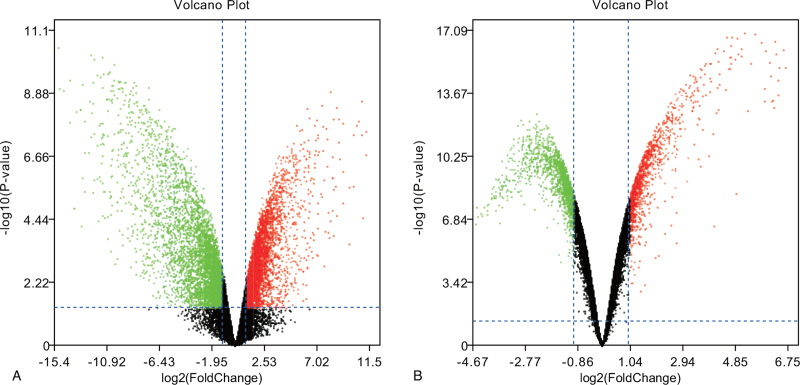
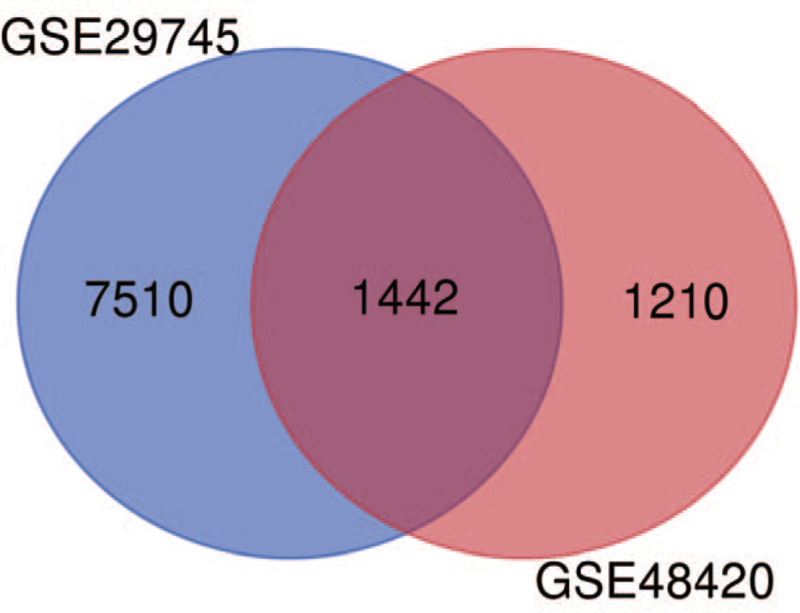
Two volcano plots present the DEGs in the GSE29745 and GSE48420 (Fig. 1A, B). The Venn diagram revealed 1442 DEGs shared by the two datasets (Fig. 2).
Figure 1.
Two volcano plots present the DEGs. (A) The DEGs in the GSE29745. (B) The DEGs in the GSE48420.
Figure 2.
3.2. DEGs annotation
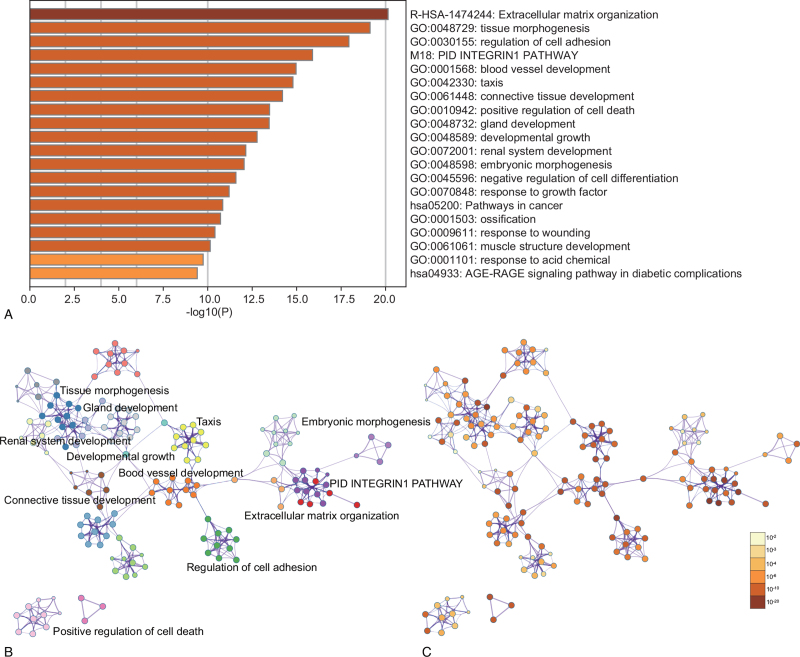
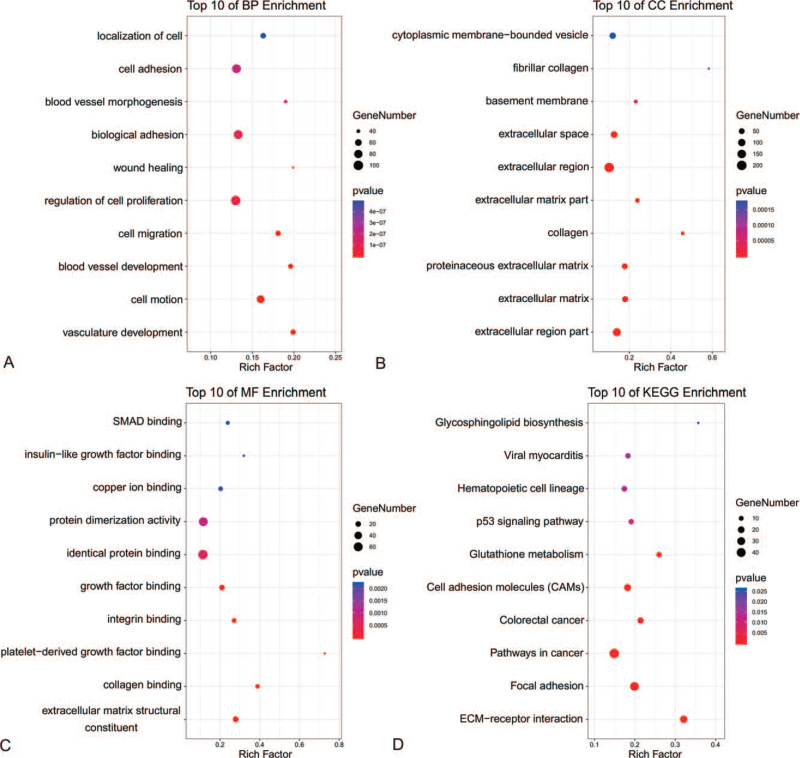
Enrichment analysis by Metascape was displayed in Figure 3. The DEGs related with biological process (BP), cellular component (CC), molecular function (MF), and KEGG enrichment analysis were displayed in bubble diagrams separately (Fig. 4). The variations in DEGs linked with BP were mainly enriched in vasculature development, cell motion, blood vessel development, cell migration, regulation of cell proliferation, regulation of cell proliferation, wound healing, biological adhesion, blood vessel morphogenesis, cell adhesion, and localization of cell (Fig. 4A). The variations in DEGs linked with CC were mainly enriched in extracellular region part, extracellular matrix, proteinaceous extracellular matrix, collagen, extracellular matrix part, extracellular region, extracellular space, basement membrane, fibrillar collagen, and cytoplasmic membrane-bounded vesicle (Fig. 4B). The variations in DEGs linked with MF were mainly enriched in extracellular matrix structural constituent, collagen binding, platelet-derived growth factor binding, integrin binding, growth factor binding, identical protein binding, protein dimerization activity, copper ion binding, insulin-like growth factor binding, and drosophila mothers against decapentaplegic (SMAD)binding (Fig. 4C). The variations in DEGs linked with Kyoto Encyclopedia of Genes and Genomes (KEGG) were mainly enriched in Extracellular Matrix (ECM)-receptor interaction, Focal adhesion, Pathways in cancer, Colorectal cancer, Cell adhesion molecules, Glutathione metabolism, p53 signaling pathway, Hematopoietic cell lineage, Viral myocarditis, and Glycosphingolipid biosynthesis (Fig. 4D).
Figure 3.
Enrichment analysis for the DEGs by Metascape. (A) Heatmap of enriched terms across input differently expressed gene lists, colored by P-values, via the Metascape. (B) Network of enriched terms colored by cluster identity, where nodes that share the same cluster identity are typically close to each other. (C) Network of enriched terms colored by P-value, where terms containing more genes tend to have a more significant P-value.
Figure 4.
The functional annotation for the DEGs based on the DAVID. (A) BP, (B) CC, (C) MF, and (D) KEGG.
3.3. Protein-protein interaction network and hub genes
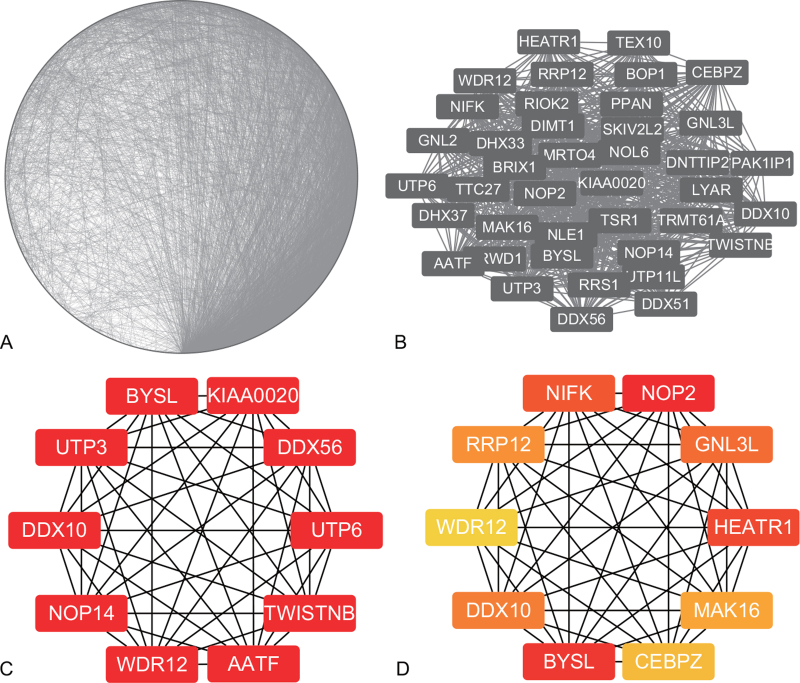
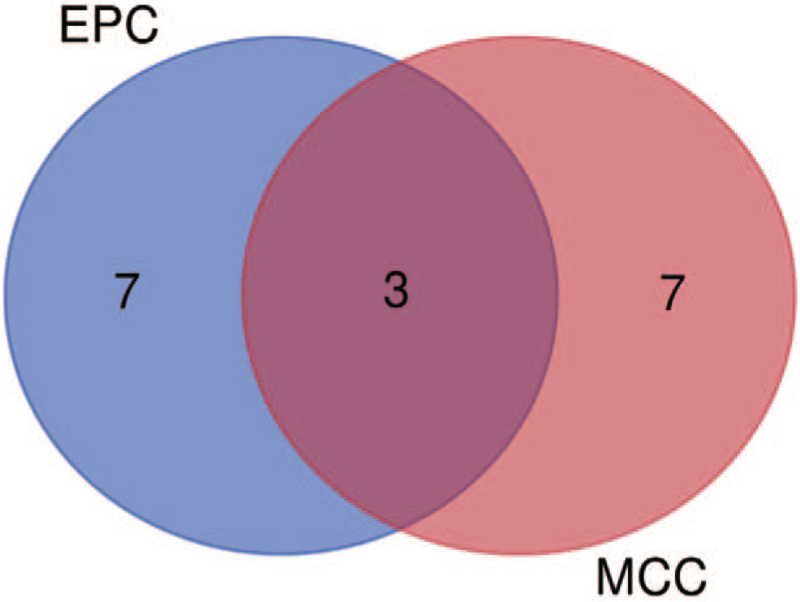
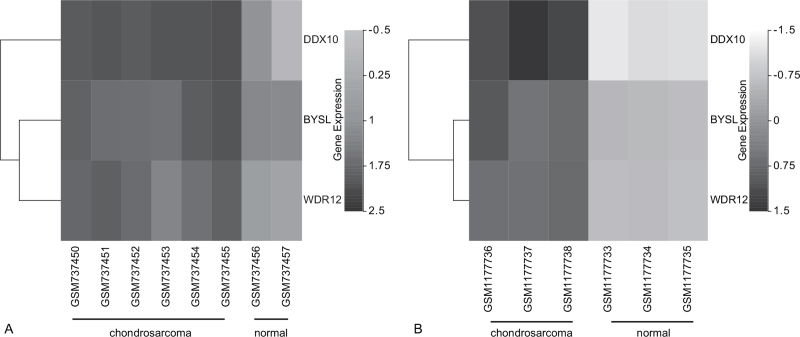
PPI was displayed in Figure 5A. The key module of MCODE analysis was shown (Fig. 5B). The top 10 genes screened by cytoHubb in two algorithms were shown (Fig. 5C, D), and Venn diagram figured out three mutual genes between the algorithms, which included DDX10, BYSL, and WDR12. (Fig. 6). Two heat maps showed the expressions of the hub genes in GSE29745 (Fig. 7A) and GSE48420 (Fig. 7B). And the three hub genes were up-regulated in the chondrosarcoma compared with the normal tissues.
Figure 5.
The PPI network and hub genes. (A) PPI network. (B) The key module of MCODE analysis. (C) The top 10 genes screened by cytoHubb in MCC. (D) The top 10 genes screened by cytoHubb in EPC.
Figure 6.
Venn diagram figured out three mutual genes between the algorithms, which included DDX10, BYSL, and WDR12.
Figure 7.
The expression analysis for the hub genes. (A) The heat map showed the expressions of the hub genes in GSE29745. (B) The heat map showed the expressions of the hub genes in GSE48420.
3.4. Overall survival analysis based on the expression of hub genes
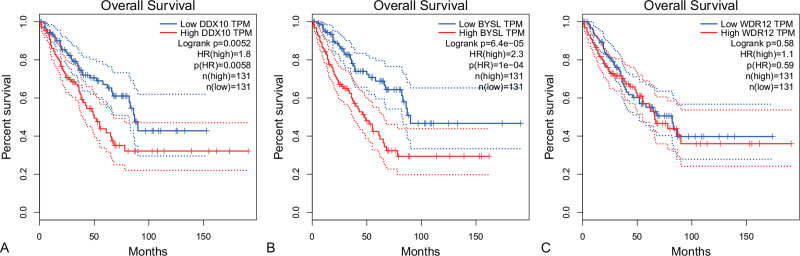
Chondrosarcoma patients with high expression levels of DDX10 had poorer overall survival times than those with low expression levels (P = .0052; HR (high) = 1.8; n (high) = 131, 50%; Fig. 8A). Chondrosarcoma patients with high expression levels of BYSL also had poorer overall survival times than those with low expression levels (P = 6.5e−05; HR (high) = 2.3; n (high) = 131, 50%; Fig. 8B). However, there was no statistically significant effect on overall survival associated with the expression of WDR12 (P = .58; Fig. 8C).
Figure 8.
The overall survival analysis. (A) DDX10, (B) BYSL, (C) WDR12.
3.5. Results of RT-qPCR analysis
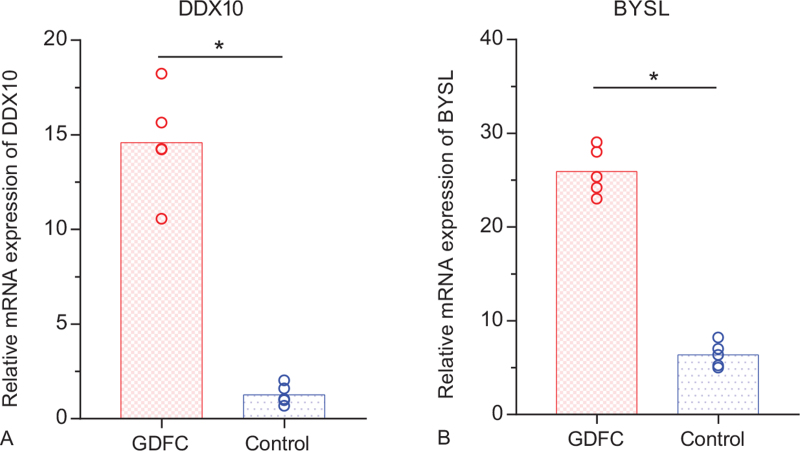
According to the above expression analysis, DDX10 and BYSL were markedly up-regulated in glioma derived from the chondrosarcoma. As presented in Figure 9, the relative expression levels of DDX10 and BYSL were significantly higher in glioma derived from the chondrosarcoma, compared with the control groups. The result demonstrated that DDX10 and BYSL might be considered as biomarkers for glioma and chondrosarcoma.
Figure 9.
Relative expression of DDX10 and BYSL by RT-qPCR analysis. ∗P < .05, compared with controls. GDFC: glioma derived from the chondrosarcoma.
4. Discussion
Chondrosarcoma is a cartilaginous cap-shaped bony protrusion, usually on the surface of a long tubular bone. Because chondrosarcomas are often insensitive to chemotherapy and radiation, surgery is the preferred treatment for chondrosarcomas.[19] Extensive resection is recommended to prevent recurrence and metastasis, however, this can lead to dysfunction in patients. Moreover, when the tumor is growing rapidly or has already spread widely, the effect of extensive resection is very limited. Tumor location and insensitivity to chemotherapy and radiotherapy lead to poor quality of life and prognosis.[20] The limitations of radiotherapy, chemotherapy and surgery for chondrosarcoma and glioma necessitate the search for new treatments.
4.1. DDX10 gene and BYSL gene were highly expressed in chondrosarcoma and glioma
Molecular targeted therapy works by targeting specific molecular targets involved in tumor formation and development. Research on targeted drugs is extremely important. There are many important signaling pathways involved in the development of chondrosarcoma tumors, including Hedgehog (Hh), Parathyroid hormone-related peptide (PTHrP), Insulin-like Growth Factor (IGF), emergency room (ER), histoplasma tissue inhibitory factor (HIF)-1 and Nicotinamide Adenine Dinucleotide (NAD) pathways.[21] Molecular targeted therapy may be considered as an alternative treatment strategy in the future. In this study, bioinformatics technology was used to analyze chondrosarcoma and normal tissues, and core genes were screened. It was found that DDX10 gene and BYSL gene were highly expressed in chondrosarcoma, and when the two genes were highly expressed, the prognosis of the patients was poor.
DDX10 may be involved in ribosome assembly. Nup98-ddx10 fused transcripts may contribute to the development of secondary hematologic malignancies caused by DNA topoisomerase II inhibitors through abnormal nucleoplasmic transport and/or altered ribosome assembly. C-myc-induced apoptosis in colonic epithelial cells can be either p53-dependent or independent.[22,23] BYSL empty embryos survive and develop in the preimplantation stage. Overall gene expression analysis in the delayed implantation model revealed differentially expressed genes between dormant and activated blastocysts. BYSL is one of the estrogen-activated genes.[24] BYSL, an adhesion molecule in mammalian preimplantation embryos, is critical in the processing of ribosomal preRNA (rRNA), and its depletion leads to the accumulation of 18S rRNA precursors.[25] Loss of BYSL leads to deletion of zygotic ribosomes and failure to initiate zygotic translation, which in turn affects protein synthesis important for cell division and differentiation. In fact, BYSL silences had a slight or insignificant effect on the mRNA expression of EndoA and Cdx2, but their protein expression levels were significantly reduced.[14] BYSL is highly expressed in embryonic stem cells and certain types of adult stem cells, but its expression is decreased and its differentiation ability is decreased. Global gene expression profiles obtained by large-scale cDNA sequencing showed that BYSL was expressed not only in embryos, including preembryonic and postembryonic, but also in fetal tissues and some types of adult tissues, suggesting that BYSL plays an important role in a broader cell spectrum. In fact, BYSL mRNA levels in ES cells did not change significantly at least for a short period of time after suppression of LIF or forced suppression of OCT / 4-induced differentiation.[14]
4.2. The functional involvement and underlying mechanism of DDX10 in chondrosarcoma and glioma
DDX10 (dead-box helicase 10) is a member of the DDX protein family that encodes RNA helicase. Members of this family were first found to be involved in embryo formation, spermatogenesis and cell division and growth, and many members of this family are closely related to tumorigenesis and development. DDX10 plays an important role in biological processes such as ribosome synthesis, cell proliferation and apoptosis by activating the production of various transcription factors. Several studies have found that DDX10 plays a key role in the proliferation, apoptosis, cell cycle and metabolism of tumor cells. Gai et al[26] found that DDX10 promotes proliferation of ovarian cancer cells through activation of the protein kinase B/nuclear factor κB (Akt/NF-κB) signaling pathway. Luizon et al[9] found that DDX10 expression was significantly up-regulated in liver cancer cells, and DDX10 promoted the proliferation of liver cancer cells through the Wnt/β-catenin signaling pathway. In addition, when DDX10 levels were inhibited, the expression levels of β-catenin molecules regulating downstream target gene transcription factors in the nucleus were also decreased. These findings suggest that DDX10 can promote β-catenin expression and entry into the nucleus to some extent, ultimately promoting cell proliferation and growth.[27] In acute myeloid leukemia, the Recombinant Nucleoporin (NUP) 98 gene is rearranged and leads to fusion protein expression. The most common one is NUP98-DDX10 fusion protein. A portion of NUP98 fuses with a portion of DDX10. Yassin et al[11] found that NUP98-DDX10 significantly increased the proliferation and self-renewal of human primary CD34+ cells, while NUP98-DDX10 affected the differentiation of CD34+ cells into erythrocytes and medullary cells. Shi et al[28] found that the expression level of RNA helicase DDX10 was significantly increased in patients with osteosarcoma, and DDX10 gene inhibited the proliferation of osteosarcoma cell MG63 by inhibiting the MAPK signaling pathway. The high expression of DDX10 gene may be related to the poor prognosis of patients with osteosarcoma. There have been no reports on DDX10 gene and chondrosarcoma. In this study, it was found that DDX10 gene was highly expressed in chondrosarcoma, and the patients with high expression had a poor prognosis. We hypothesized that DDX10 may be involved in tumor genesis and development through the following mechanisms:
-
1.
direct regulation of cell proliferation and cell cycle;
-
2.
Regulate the expression of key molecules of tumor cells, such as P53, cellular-myelocytomatosis viral oncogene (C-MYC), mitogen-activated protein kinase (MAPK), etc., or regulate microRNA to shear and modify mRNA at the transcription and translation levels, thus affecting the expression of key genes.
Mdm-2 is a negative regulator of p53 protein by promoting proteomic degradation. As a regulatory ring, p53 is up-regulated by Mdm2.[29] Alternative Reading Frame (Arf) also plays a role in this regulatory mechanism because it inhibits Mdm2 and C-myC function. By increasing the expression of Arf, c-myC protein plays an important role in the regulation of p53, which ultimately leads to p53-dependent apoptosis. Crosstalk between C-myC and p53 is essential in inducing pro-survival or pro-death responses to apoptotic stimuli. Both c-myC overexpression and the disruption of normal p53 activity are highly associated with human cancer. Therefore, it is reasonable to assume that developing drugs that target both pathways at the same time could provide better outcomes for cancer treatment.[30] Cancer cells often become resistant to SUBSTANCES that damage DNA during chemotherapy, especially when the tumor cells are treated with high concentrations of the drug. One of the potential mechanisms of chemotherapeutic resistance is that cancer cells manage to promote c-MYC expression after exposure to chemotherapeutic drugs, suggesting that elevated C-MyC expression may be at least partially responsible for cancer chemotherapeutic resistance.[31,32] Here, we found that INZ(c) inhibits c-MYC expression through the miRNA pathway, offering a potential drug to inhibit or delay cancer cell resistance during chemotherapy. Further research into the effects of INZ(c) on the growth of drug-resistant cancer cells will provide more information on how INZ(c) can promote the resensitivity of drug-resistant cancer cells to chemotherapeutic drugs. Knockout of C-MyC expression can stimulate the expression of P53, suggesting that c-MyC inhibition of P53 expression occurs at the transcriptional level. Notably, c-MYC was predicted as a potential transcription factor binding to the P53 promoter through the online database described above. C-myc inhibits the transcription of P53 by binding to the P53 promoter and reducing its activity. In addition, in NASopharyngeal carcinoma cells with c-MyC overexpression, p53 transduction not only reduced cell growth and EdU staining, but also inhibited EGFR/PI3K/AKT/ C-Myc signal transduction and up-regulated the expression of Mir-133A-3p. Inhibition of EGFR decreased the PI3K/AKT/ C-MyC signaling pathway, and increased the expression of P53 and Mir-133A-3p.[33] Under normal circumstances, DDX10 may be transported to and play a role in the nucleus. The widespread expression and putative nuclear localization of DDX10 are consistent with the possible role of ribosome assembly in rRNA processing.[34]
4.3. The functional involvement and underlying mechanism of BYSL in chondrosarcoma and glioma.
BYSL (Bystin-like) gene can encode nucleolar protein and participate in early embryo implantation and the 40S subunit biosynthesis process of ribosome, which plays an important role in the maturation process of pre-ribosomal RNA.[35] Wang et al found that BYSL plays an important role in the rapid growth of liver cancer cells. BYSL mediates the formation of nucleolus-derived foci (NDF) and prenucleolar bodies (PNBs), which are ultimately involved in nucleolar assembly during cell division. Inhibition of BYSL-mediated nuclear formation in mitotic cells may be an effective means of destroying tumor genesis.[15] Kasugai et al found that BYSL gene was overexpressed in diffuse large B cell lymphoma. In addition, the expression of BYSL is up-regulated in various cancer tissues, such as breast cancer,[17] diffuse large B cell lymphoma[36] and gastric adenocarcinoma.[37] BYSL has been shown to inhibit the proliferation of cancer cells in vitro.[38] BYSL is a direct downstream target of c-MYC proto-oncogene, suggesting a close relationship with cancer. Taken together, these results suggest that upregulation of BYSL may be a common feature of human cancers and is essential for tumor formation. In this study, it was found that BYSL gene was highly expressed in chondrosarcoma, and the patients with high expression had a poor prognosis. BYSL is an important promoter of multiple tumor progression and may be a candidate target for the treatment of chondrosarcoma and glioma.
The high expression of BYSL in glioma tissues promoted the growth and survival of tumor cells.[39] Mild chemical hypoxia can also induce significant BYSL expression in vitro. These increases were not accompanied by an increase in GFAP expression. In experimental animal models of brain injury, BYSL was elevated 5 weeks after injury.[40] It is suggested that BYSL expression is increased during il-β transformation in astrocytes in vitro, which may be involved in the activation and differentiation of astrocytes. BYSL expression appears to be more prominent in ischemic attacks (e.g., fatal bleeding) and to a moderate extent in hypoxic attacks (e.g., CO poisoning), but the likely duration of attacks plays an important role here. BYSL is highly expressed in liver cancer, ovarian cancer tissue and prostate cancer cells near peripheral nerves. Here, we find that BYSL-IR is up-regulated in GBM. These results suggest that the high expression of BYSL may be common in different types of tumors.[41] BYSL is a highly evolutionarily conserved gene from yeast to humans. It encodes the human protein bestin. In yeast and humans, BYSL is involved in 18S pre-rRNA processing and 40S ribosome subunit biogenesis. Repositioning and reactivation of rRNA preprocessing mechanisms are critical for nucleolus assembly. Wang et al. proposed that down-regulation of BYSL may impair nucleogenesis and thereby inhibit the growth of hepatocellular carcinoma cells. The dynamic assembly of nucleoli is an important event during mitotic withdrawal of eukaryotic cells. Our data showed that BYSL silencing led to glioma cell cycle arrest at G1 phase, which was further confirmed by down-regulation of Cyclin D1 and CDKs (CDK4 and CDK6) and up-regulation of CDKIs (P21 and P27). Therefore, the down-regulation of BYSL may impair the processing of pre-RRNA and the assembly of nucleoli, thus hindering the mitotic process and proliferation of glioma cells.
Chondrosarcomas are a group of heterogeneous primary bone cancers characterized by hyaline chondroid tumor tissue. They are the second most common primary bone malignancy. The vast majority of chondrosarcomas are routine chondrosarcomas, and most routine chondrosarcomas are low to intermediate tumors (grade 1 or 2) with clinical indolent manifestations and low metastatic potential. Recurrence is a poor predictor of prognosis because conventional chondrosarcomas are resistant to both radiation and chemotherapy. Recent discoveries in the biology, genetics, and epigenetics of conventional chondrosarcomas have greatly advanced our understanding of the pathobiology of these tumors and provided insights into potential therapeutic targets.[19] In the early stages, chondrosarcoma is still considered an intermediate type of tumor and rarely metastases. Unfortunately, late stages show marked resistance to chemotherapy and radiation, and metastasis often occurs.[20]
An elevated level of DDX10 is linked to diseases such as cancer or neurodegeneration. DDX10 is significantly overexpressed in osteosarcoma cancer patients. DDX10 overexpression predicts worse prognosis in osteosarcoma and its deletion prohibits cell activities modulated by MAPK pathway. Importantly, αSyn interacts with Dbp4 and DDX10 to isolate nucleolar proteins into the cytoplasm in yeast and human cells. This effect was significant in human cells, where DDX10 and αSyn formed large cytoplasmic inclusion bodies, but was less significant in yeast cells, where slight BiFC complementary signaling was observed in cytoplasm close to the plasma membrane, similar to αSyn inclusion bodies. Although the interaction between αSyn and Dbp4 in the cytoplasm of yeast cells has been clearly demonstrated by different research methods, it is not obvious whether Dbp4 and αSyn form inclusion bodies similar to human cells. In chromosomal translocations associated with myeloid malignancies, at least one rearrangement gene is usually a transcription factor. NUP98-DDX10 may be a novel fusion gene associated with myeloid malignancy. A special structural feature of DDX10 is the presence of two acidic regions and charged amino acids at the C-terminal. DDX10 may be a transcriptional activator because the acidic motif can act as a transcriptional activation domain like VP16 and GAL4.50. However, the normal functional information of nucleoporin and the putative function of DDX10 suggest that fusion genes may interfere with protein synthesis during normal myeloid differentiation during hematopoietic processes through abnormal cytoplasmic transport or altered ribosome assembly.[34] The high expression of DDX10 in tumor tissues is related to the proliferation, apoptosis and cell cycle of tumor cells.[42]
4.4. Limitation of the study
Despite the rigorous bioinformatics analysis in this paper, there are still some deficiencies. In this study, no animal experiments of gene overexpression or knockout were conducted to further verify its function. Therefore, in the future research, we should carry out in-depth exploration in this aspect. Furthermore, to verify our conclusions, a large number of clinical trials are also needed.
In conclusion, DDX10 and BYSL genes may provide new ideas and evidence for the diagnosis and targeted therapy of chondrosarcoma and glioma.
Acknowledgments
None.
Author contributions
Conceptualization: Qiang Zhang.
Data curation: Rugang Zhao.
Formal analysis: Xuemin Quan, Changsong Zhao.
Funding acquisition: Zhengrong Gao, Qiang Zhang.
Investigation: Yao Zhang.
Methodology: Rugang Zhao
Software: Jingjing Wang
Supervision: Qiang Zhang.
Validation: Xuemin Quan
Visualization: Xuemin Quan
Writing – original draft: Xuemin Quan.
Writing – review & editing: Qiang Zhang.
Footnotes
Abbreviations: AIB1 = amplified in breast cancer 1, BP = biological process, BYSL = Bystin-like, CC = cellular component, CD34 = Recombinant Cluster Of Differentiation, C-MYC = cellular-myelocytomatosis viral oncogene, DAVID = Digital Audio/Video Interactive Decoder, DDX10 = dead-box helicase 10, DEGs = Differently expressed genes, ECM = Extracellular Matrix, EPC = Endothelial Progenitor Cell, ER = emergency room, FC = fibrillar center, GDFC = glioma derived from the chondrosarcoma, GEO = Gene Expression Omnibus, GSE = Gene Set Enrichment, HCC = Hepatocellular Carcinoma, Hh = Hedgehog, HIF = histoplasma tissue inhibitory factor, IGF = Insulin-like Growth Factor, KEGG = Kyoto Encyclopedia of Genes and Genomes, MAPK = mitogen-activated protein kinase, MCC = Merkel Cell Carcinoma, MCODE = Molecular Complex Detection tool, MF = molecular function, MG63 = human osteosarcoma cells, NAD = Nicotinamide Adenine Dinucleotide, NDF = nucleolus-derived foci, NUP98 = Recombinant Nucleoporin98, PNBs = prenucleolar bodies, PPI = protein-protein interaction, PTHrP = Parathyroid hormone-related peptide, RNA = Ribonucleic Acid, SMAD = drosophila mothers against decapentaplegic, WDR12 = WD repeat domain 12.
How to cite this article: Quan X, Zhao C, Gao Z, Zhang Y, Zhao R, Wang J, Zhang Q. DDX10 and BYSL as the potential targets of chondrosarcoma and glioma. Medicine. 2021;100:46(e27669).
Ethical review and patient consent: The research confirmed to the Declaration of Helsinki and was authorized by the Human Ethics and Research Ethics Committees of the Cangzhou Central Hospital. Written informed consent was obtained from all patients and their families.
The authors have no funding to disclose.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Karpik M, Reszeć J. Low grade chondrosarcoma—epidemiology, diagnosis. Treatment Ortop Traumatol Rehabil 2018;20:65–70. [DOI] [PubMed] [Google Scholar]
- [2].Amer KM. 0000-0002-9114-1890 AO, Munn M, Congiusta D, Abraham JA, Basu MA. Survival and prognosis of chondrosarcoma subtypes: SEER database analysis. J Orthop Res 2020;38:311–9. [DOI] [PubMed] [Google Scholar]
- [3].Lozano Martínez GA, Llauger Rosselló J. [Secondary chondrosarcoma: radiopathological correlation]. Radiologia 2015;57:344–59. [DOI] [PubMed] [Google Scholar]
- [4].Zhou Y, Liepe J, Sheng X, Stumpf MP, Barnes C. GPU accelerated biochemical network simulation. Bioinformatics 2011;27:874–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nobile MS, Cazzaniga P, Tangherloni A, Besozzi D. Graphics processing units in bioinformatics, computational biology and systems biology. Brief Bioinform 2017;18:870–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res 2018;24:1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Qiu Z, Li H, Zhang Z, et al. A pharmacogenomic landscape in human liver cancers. Cancer Cell 2019;36:179–93.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer 2017;17:332. [DOI] [PubMed] [Google Scholar]
- [9].Luizon MR, Eckalbar WL, Wang Y, et al. Genomic characterization of metformin hepatic response. PLoS Genet 2016;12:e1006449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baril C, Gavory G, Bidla G, Knævelsrud H, Sauvageau G, Therrien M. Human NUP98-HOXA9 promotes hyperplastic growth of hematopoietic tissues in drosophila. Dev Biol 2017;421:16–26. [DOI] [PubMed] [Google Scholar]
- [11].Yassin ER, Abdul-Nabi AM, Takeda A, Yaseen NR. Effects of the NUP98-DDX10 oncogene on primary human CD34+ cells: role of a conserved helicase motif. Leukemia 2010;24:1001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jia F, Zhang Z, Zhang X. MicroRNA-338-3p inhibits tumor growth and metastasis in osteosarcoma cells by targeting RUNX2/CDK4 and inhibition of MAPK pathway. J Cell Biochem 2019;120:6420–30. [DOI] [PubMed] [Google Scholar]
- [13].Olczak M, Chutorański D, Kwiatkowska M, Samojłowicz D, Tarka S, Wierzba-Bobrowicz T. Bystin (BYSL) as a possible marker of severe hypoxic-ischemic changes in neuropathological examination of forensic cases. Forensic Sci Med Pathol 2018;14:26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Adachi K, Soeta-Saneyoshi C, Sagara H, Iwakura Y. Crucial role of Bysl in mammalian preimplantation development as an integral factor for 40S ribosome biogenesis. Mol Cell Biol 2007;27:2202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang H, Xiao W, Zhou Q, et al. Bystin-like protein is upregulated in hepatocellular carcinoma and required for nucleologenesis in cancer cell proliferation. Cell Res 2009;19:1150–64. [DOI] [PubMed] [Google Scholar]
- [16].Azzato EM, Driver KE, Lesueur F, et al. Effects of common germline genetic variation in cell cycle control genes on breast cancer survival: results from a population-based cohort. Breast Cancer Res 2008;10:R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ochnik AM, Peterson MS, Avdulov SV, Oh AS, Bitterman PB, Yee D. Amplified in breast cancer regulates transcription and translation in breast cancer cells. Neoplasia 2016;18:100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li D, Weng JC, Zhang GJ, et al. Proposed treatment paradigm for intracranial chondrosarcomas based on multidisciplinary coordination. World Neurosurg 2018;109:e517–30. [DOI] [PubMed] [Google Scholar]
- [19].Chow WA. Chondrosarcoma: biology, genetics, and epigenetics. F1000Res 2018;07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Boehme KA, Schleicher SB, Traub F, Rolauffs B. Chondrosarcoma: a rare misfortune in aging human cartilage? the role of stem and progenitor cells in proliferation, malignant degeneration and therapeutic resistance. Int J Mol Sci 2018;19(1.): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nazeri E, Gouran SM, Majidzadeh-A K, Esmaeili R. Chondrosarcoma: An overview of clinical behavior, molecular mechanisms mediated drug resistance and potential therapeutic targets. Crit Rev Oncol Hematol 2018;131:102–9. [DOI] [PubMed] [Google Scholar]
- [22].Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol 2006;16:275–87. [DOI] [PubMed] [Google Scholar]
- [23].Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene 2003;22:9007–21. [DOI] [PubMed] [Google Scholar]
- [24].Hamatani T, Daikoku T, Wang H, et al. Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc Natl Acad Sci U S A 2004;101:10326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li Y, Zhang F, Cong Y, Zhao Y. Identification of potential genes and miRNAs associated with sepsis based on microarray analysis. Mol Med Rep 2018;17:6227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gai M, Bo Q, Qi L. Epigenetic down-regulated DDX10 promotes cell proliferation through Akt/NF-(B pathway in ovarian cancer. Biochem Biophys Res Commun 2016 1000-;469:05. [DOI] [PubMed] [Google Scholar]
- [27].Wen JL, Wen XF, Li RB, et al. UBE3C promotes growth and metastasis of renal cell carcinoma via activating Wnt/(-catenin pathway. PLoS One 2015;10:e0115622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shi JH, Hao YJ. DDX10 overexpression predicts worse prognosis in osteosarcoma and its deletion prohibits cell activities modulated by MAPK pathway. Biochem Biophys Res Commun 2019;510:525–9. [DOI] [PubMed] [Google Scholar]
- [29].Shi D, Gu W. Dual roles of MDM2 in the regulation of p53: ubiquitination dependent andubiquitination independent mechanisms of MDM2 repression of p53 activity. Genes Cancer 2012;3:240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jung JH, Liao JM, Zhang Q, et al. Inauhzin(c) inactivates c-Myc independently of p53. Cancer Biol Ther 2015;16:412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Niimi S, Nakagawa K, Yokota J, et al. Resistance to anticancer drugs in NIH3T3 cells transfected with c-myc and/or c-H-ras genes. Br J Cancer 1991;63:237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sklar MD, Prochownik EV. Modulation of cis-platinum resistance in Friend erythroleukemia cells by c-myc. Cancer Res 1991;51:2118–23. [PubMed] [Google Scholar]
- [33].Liang Z, Liu Z, Cheng C, et al. VPS33B interacts with NESG1 to modulate EGFR/PI3K/AKT/c-Myc/P53/miR-133a-3p signaling and induce 5-fluorouracil sensitivity in nasopharyngeal carcinoma. Cell Death Dis 2019;10:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Arai Y, Hosoda F, Kobayashi H, et al. The inv(11)(p15q22) chromosome translocation of de novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood 1997;89:3936–44. [PubMed] [Google Scholar]
- [35].Harada O, Suga T, Suzuki T, et al. The role of trophinin, an adhesion molecule unique to human trophoblasts, in progression of colorectal cancer. Int J Cancer 2007 1072-;121:08. [DOI] [PubMed] [Google Scholar]
- [36].Kasugai Y, Tagawa H, Kameoka Y, Morishima Y, Nakamura S, Seto M. Identification of CCND3 and BYSL as candidate targets for the 6p21 amplification in diffuse large B-cell lymphoma. Clin Cancer Res 2005;11:8265–72. [DOI] [PubMed] [Google Scholar]
- [37].Tsukamoto Y, Uchida T, Karnan S, et al. Genome-wide analysis of DNA copy number alterations and gene expression in gastric cancer. J Pathol 2008;216:471–82. [DOI] [PubMed] [Google Scholar]
- [38].Miyoshi M, Okajima T, Matsuda T, Fukuda MN, Nadano D. Bystin in human cancer cells: intracellular localization and function in ribosome biogenesis. Biochem J 2007;404:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gao S, Sha Z, Zhou J, et al. BYSL contributes to tumor growth by cooperating with the mTORC2 complex in gliomas. Cancer Biol Med 2021;18:88–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sheng J, Yang S, Xu L, et al. Bystin as a novel marker for reactive astrocytes in the adult rat brain following injury. Eur J Neurosci 2004;20:873–84. [DOI] [PubMed] [Google Scholar]
- [41].Sha Z, Zhou J, Wu Y, et al. BYSL promotes glioblastoma cell migration, invasion, and mesenchymal transition through the GSK-3β/β-catenin signaling pathway. Front Oncol 2020;10:565225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu C, Tang J, Duan X, Du Y, Wang X, Cui Y. DDX10 promotes human lung carcinoma proliferation by U3 small nucleolar ribonucleoprotein IMP4. Thorac Cancer 2021;12:1873–80. [DOI] [PMC free article] [PubMed] [Google Scholar]