Abstract
The scenario of cholera that existed previously changed in 1992 and 1993 with the emergence of toxigenic Vibrio cholerae O139 in India. The genesis of the new serogroup formed the impetus to search for O139 phages in and around the country. A total of five newly isolated phages lytic to V. cholerae O139 strains were used for the development of this phage typing scheme. These phages differed from each other and also differed from the existing O1 phages in their lytic patterns, morphologies, restriction endonuclease digestion profiles, and immunological criteria. With this scheme, 500 V. cholerae O139 strains were evaluated for their phage types, and almost all strains were found to be typeable. The strains clustered into 10 different phage types, of which type 1 (38.2%) was the dominant type, followed by type 2 (22.4%) and type 3 (18%). Additionally, a comparative study of phage types in 1993 and 1994 versus those from 1996 to 1998 for O139 strains showed a higher percentage of phage type 1 (40.5%), followed by type 3 (18.8%) during the period between 1993 and 1994, whereas phage type 2 (32.1%) was the next major type during the period from 1996 to 1998. This scheme comprising five newly isolated phages would be another useful tool in the study of the epidemiology of cholera caused by V. cholerae O139.
Despite more than a century of study, cholera remains an important cause of morbidity and mortality and still presents a devastating global problem. Until 1992, Vibrio cholerae O1 was considered the sole causative agent of the disease cholera. In October 1992 the scenario of cholera that had existed changed because of the emergence of a new etiologic serogroup of cholera, which is now known as O139 Bengal (21). These strains emerged abruptly in the south Indian coastal city of Madras and rapidly spread to other areas of India and neighboring countries where cholera is endemic (15, 16).
Strategies for the prevention and control of an infectious disease like cholera depend on understanding the origin, transmission, and other characteristics associated with the epidemic and its spread. Among the several typing methods, phage typing is one of the important and useful methods for the identification and differentiation of V. cholerae strains. The use of bacteriophages as a method of strain differentiation has contributed greatly to the understanding of the epidemiology of the disease cholera. Basu and Mukerjee (4) proposed a phage typing scheme for V. cholerae O1 biotype E1 Tor which was efficiently used to study the spread of the E1 Tor biotype of V. cholerae O1. A new phage typing scheme was subsequently developed at the National Institute of Cholera and Enteric Diseases for V. cholerae O1 (7, 18). The recent emergence of toxigenic V. cholerae O139 has prompted us to conduct a search for bacteriophages for V. cholerae O139 and to develop an effective phage typing scheme for this organism.
MATERIALS AND METHODS
Bacterial strains.
V. cholerae O139 NPR-4 (6), isolated from a patient in the city of Nagpur, India, in 1993, was used as the host for propagation of V. cholerae O139 phages as it was lysed by all the phages. A total of 500 V. cholerae O139 strains isolated from different areas in India where cholera is endemic between 1993 and 1998 were included in the phage typing study. Additionally, V. cholerae O1 biotype E1 Tor strain MAK 757 (ATCC 51352), 10 E1 Tor strains (ATCC 51610 to ATCC 51619) that had been phage typed (19), and classical biotype strains V. cholerae 154 and 569B were also used in this study. Other enteropathogens included in the present study were Salmonella spp., enteropathogenic Escherichia coli, and Shigella spp. (five strains each).
Isolation and purification of bacteriophages.
In the present study the bacteriophages were isolated from raw sewage water samples collected from different areas in India where cholera is endemic. Samples were collected from selected points where the sewage water connects with the main canal, which receives both domestic and industrial effluents. Apart from V. cholerae O139 phages, V. cholerae O1 biotype E1 Tor typing phage D-10 (ATCC 51352) and classical phage φ149 were also included in the present study.
All the phages isolated were purified by successive single-plaque isolation until homogeneous plaques against the standard propagating strain NPR-4 were obtained. The broth lysis procedure was used to prepare phage lysates. Nutrient broth was inoculated with a 1% (vol/vol) cell suspension from the culture of NPR-4 grown overnight. Cultures were grown at 37°C for 3 to 4 h with continuous shaking in a gyratory shaker. Mid-logarithmic-phase cells were infected with phage at a multiplicity of infection (MOI) of 0.1, and the cells were incubated with shaking at 37°C until complete lysis occurred. A few drops of chloroform containing 1% ethanol was added, and the mixture was centrifuged (10,000 rpm at 4°C) for 20 min. The phage suspension was pelleted at 30,000 rpm for 2 h at 4°C with a Beckman 50.2 Ti rotor. The pellet was suspended in 0.05 M Tris (pH 7.5) with 0.02 M MgCl2 and was maintained at 4°C (9).
A plate lysis procedure was used for phage propagation to get a higher titer of phages. A young broth culture of V. cholerae NPR-4 was infected with phage at an MOI of 0.01, and adsorption was allowed to occur at room temperature. The mixture was then plated by the soft agar overlay method and was incubated at 37°C until complete lysis occurred. The soft agar layer was scraped off, suspended in 2 ml of Tris-MgCl2 buffer, and centrifuged at 10,000 rpm for 20 min at 4°C (Beckman J2-21M/E).
The bacteriophages used in the present study were purified by density gradient centrifugation. Three of the phages, phages VE-2, CAL-3, and MS-3, were purified by use of a cesium chloride density (1.3 to 1.7 g/ml) gradient. Phages MAD-5 and S-2 were unstable in cesium chloride; hence, they were purified by a neutral sucrose step gradient (20 to 60%) procedure. An electron microscopic study was performed by the method described by Ghosh et al. (9) to determine the morphologies of the phages.
SDS-polyacrylamide gel electrophoresis (PAGE).
A phage lysate containing 1010 PFU/ml was solubilized in 0.065 M Tris-HCl (pH 6.8) containing 1% sodium dodecyl sulfate (SDS), 1% mercaptoethanol, 15% glycerol, and bromophenol blue. Samples were prepared for electrophoresis by boiling for 3 min prior to loading. Electrophoresis was performed by the standard method (11). The bands were detected by staining the gel with Coomassie brilliant blue.
Preparation and titration of antiphage serum.
Antiphage serum was prepared by immunizing New Zealand White outbred rabbits of either sex (weight, between 1.3 and 1.5 kg) as described previously (14). One milliliter each of the high-titer purified phage lysate was injected once a week. A booster dose was given intravenously after 4 weeks. Test bleeding was performed a week after the last injection. After aseptic separation, the serum was kept in a refrigerator in a sealed ampoule without any preservatives. Preimmune serum from the same rabbit was used as a control for the neutralization test.
The antiphage serum neutralization test was performed by the methodology of Mukerjee (14). For the cross-neutralization test, phage stocks were diluted to 107 PFU/ml, and the antiphage sera were diluted to 1 in 50 in broth. One volume of each of the phage groups was mixed with 99 volumes of serum, and the mixture was kept at 37°C in a water bath for 10 min. An aliquot of the mixture from all the tubes was then diluted 1 in 100 in broth to stop the further action of the serum. A total of 100 μl of these dilutions was then plated by the soft agar overlay method. The immunological cross-reactivities among the five phages were determined by a standard enzyme-linked immunosorbent assay (ELISA) method (10).
Phage characterization.
Adsorption of phage MAD-5 on its host, strain NPR-4, was studied as elaborated by Stent (22) for bacteriophage T4. Exponentially grown V. cholerae O139 NPR-4 (2 × 108 cells/ml) was infected with phage MAD-5 at an MOI of 0.1 at 37°C. After infection, at every 2-min interval over a period of 15 min, the concentration of nonadsorbed phages was determined by diluting the supernatant and plating it before lysis. Thermal and pH inactivation of MAD-5 phage was determined by the procedure described by Mukerjee (14).
The interaction between V. cholerae O139 and phages in a ligated ileal loop of a rabbit was studied by the procedure described by Sarkar et al. (19).
Isolation of phage DNA.
Phage DNA was isolated by the procedure of Sambrook et al. (17). A high-titer phage (1011 to 1012 PFU/ml) stock was treated with phenol-chloroform-isoamyl alcohol, and the DNA that was extracted was precipitated with chilled ethanol. Restriction endonuclease digestion of phage DNA was done by the procedure recommended by the manufacturer (Genei, Bangalore, India).
Phage typing.
The plaque morphologies of the five newly isolated phages were studied on nutrient agar plates by the soft agar overlay method after serial decimal dilution. Initially, 100 V. cholerae strains were included for screening of the phages. The phages were selected on the basis of the sensitivity pattern for development of the phage typing scheme. The routine test dilutions (RTDs) of the phages were determined by the method described by Chattopadhyay et al. (7). High-titer phage stocks were serially diluted, and the dilutions were spotted onto the lawn of standard propagating strain NPR-4. The RTD used for the phage typing study was the highest dilution that just failed to lyse the standard propagating strain clearly or confluently. The phage typing study was performed on the basis of the standard methodology adopted in our laboratory (6, 7, 18). The set of phages used for typing, namely, phages MAD-5, VE-2, CAL-3, S-2, and MS-3, were checked for RTD prior to phage typing of each batch of strains. A batch of V. cholerae O139 strains was streaked onto a nutrient agar plate and the plate was incubated for 18 h at 37°C. On the following morning, a single colony from the nutrient agar plate was inoculated into nutrient broth (2 to 3 ml) and was incubated under stationary conditions for 3 to 4 h. A young broth culture mixed with 3.5 ml of molten soft agar (maintained at 42°C in a water bath) was poured into a nutrient agar plate to prepare a uniform lawn of growth of the bacterial strain on the surface of the agar to provide an adequate substrate for phage action. The plates were then allowed to dry at room temperature, preferably for 20 to 30 min. After the plates were dried, drops of phage lysate at the RTD were applied onto the plates with a Pasteur pipette without touching the agar. The plates were kept at room temperature so that the drops could dry and were then incubated at 37°C for 14 to 18 h. On the following day, the plates were observed to evaluate the degree of lysis. Strong lysis, semiconfluent lysis, or the appearance of at least five plaques were considered positive results. Lesser degrees of lysis or reactions of inhibition were disregarded.
RESULTS
Bacteriophage.
A total of 79 sewage water samples were collected from different areas in India where cholera is endemic for isolation of V. cholerae O139 phages. Of these, 40 phages sensitive to V. cholerae O139 strains were isolated by using NPR-4 as the host.
Host range.
V. cholerae O1 biotype E1 Tor strain MAK757 (ATCC 51352), 10 E1 Tor strains (strains ATCC 51610 to ATCC 51619) that had been phage typed, V. cholerae O1 Classical biotype strain 569B, and V. cholerae 154 were resistant to these five O139 phages and caused lysis of V. cholerae O139 strains. Strains of common enteropathogens such as Salmonella, Shigella, and enteropathogenic E. coli were not sensitive to these phages, indicating that they were true cholera phages.
Morphology and characteristics of MAD-5.
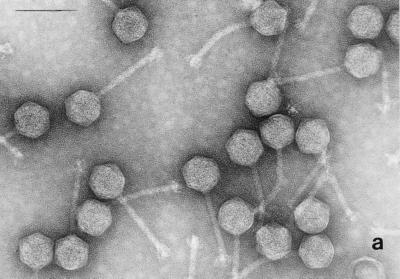
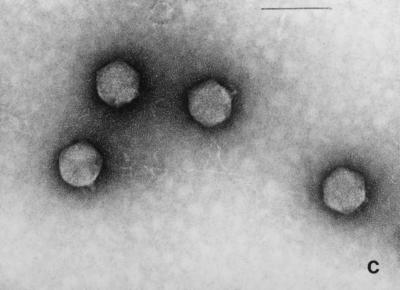
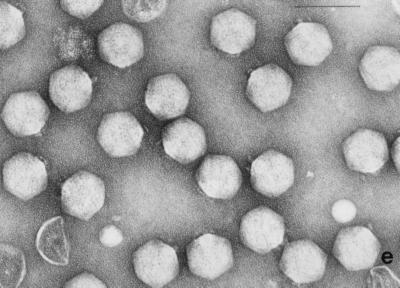
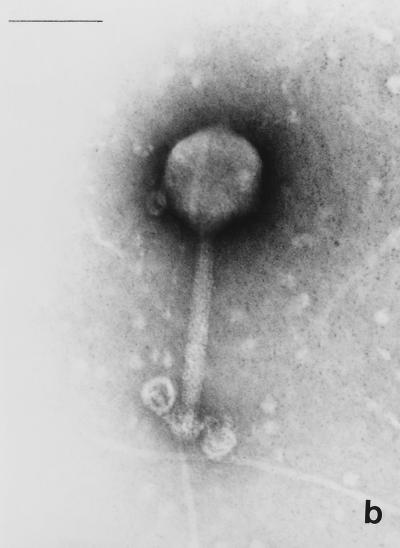
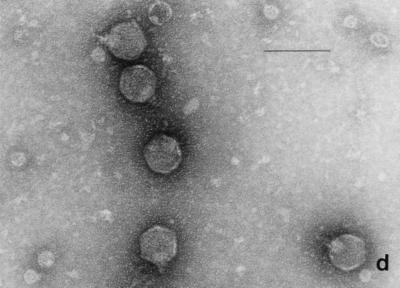
Electron micrographs of the five newly isolated phages are shown in Fig. 1. The diameters of the heads (distance between opposite apices) of MAD-5, VE-2, CAL-3, S-2, and MS-3 were 58 ± 2.7, 112.5 ± 1.8, 67.2 ± 2.9, 60.8 ± 1.8, and 73.9 ± 2.3 nm, respectively, while the lengths of the tails were 141.2 ± 4.8, 204 ± 2.8, 11.5 ± 2.8, 16.5 ± 1.2, and 10.8 ± 1.8, respectively. From the dimensions, the phages could be categorized into two distinct groups. Phages CAL-3, MS-3, and S-2 have hexagonal heads with short noncontractile tails and belong to group C in the classification proposed by Bradley (5). According to the International Committee on Taxonomy of Viruses, they belong to the family Podoviridae (1). Phages MAD-5 and VE-2 have hexagonal heads and long contractile tails and belong to group A or the family Myoviridae.
FIG. 1.
Electron micrograph of five newly isolated V. cholerae O139 phages. (a) MAD-5; (b) VE-2; (c) CAL-3; (d) S-2; (e) MS-3. Bars, 100 nm.
One of the phages (MAD-5) was characterized by its physicochemical parameters. Phage MAD-5 showed a typical biphasic pattern of adsorption on its host, strain NPR-4. Adsorption of phage MAD-5 was dependent on the divalent cation Mg2+, but with replacement of Mg2+ by Ca2+, Na+, or K+, the adsorption was almost identical in nature to that with Mg2+. This phage was found to be stable at pHs ranging from 4 to 11. Below pH 4 and above pH 11, phage MAD-5 was found to be unstable. Like serology, morphology, and host range pattern, the thermal death point is one of the parameters used to identify a phage. Phage MAD-5 was found to be most stable at 4°C, and its thermal death point ranged between 67 and 70°C.
Protein profile.
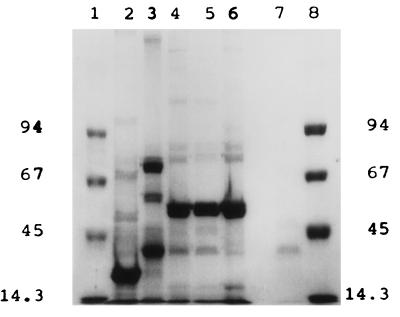
The protein profiles of the five V. cholerae O139 phages are shown in Fig. 2. SDS-PAGE pattern analysis of the CAL-3 and MS-3 virions showed that they had more or less similar profiles but that the profiles differed from those exhibited by MAD-5, VE-2, and S-2, which were again different among themselves. The newly isolated phages showed distinctly different protein profiles compared to the profile of O1 biotype E1 Tor typing phage D-10 (ATCC 51352).
FIG. 2.
SDS-PAGE patterns of structural proteins of five newly isolated V. cholerae O139 phages stained with Coomassie brilliant blue. Lanes 1 and 8, molecular size marker (in kilobases); lane 2, MAD-5; lane 3, VE-2; lane 4, CAL-3; lane 5, S-2; lane 6, MS-3; lane 7, D-10.
Immunological analysis.
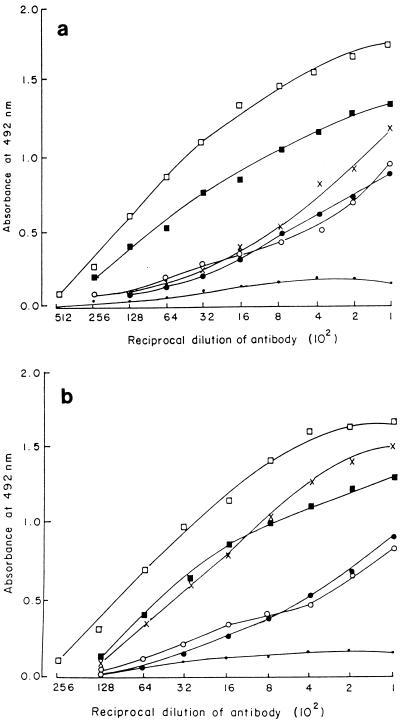
The five phages were tested to determine their relatedness in terms of inhibition of infectivity by using antiphage sera raised against the individual phages. Antisera against phages MAD-5, CAL-3, and MS-3 inhibited these three phages but did not inhibit VE-2 and S-2, while antisera against VE-2 and S-2 inhibited these two phages but did not inhibit MAD-5, CAL-3, and MS-3. Thus, on the basis of the inhibition of infectivity of the phage particles (cross-neutralization test) they could be categorized into two groups, with phages MAD-5, CAL-3, and MS-3 belonging to one group and phages VE-2 and S-2 belonging to another group. Analysis of the results of ELISA (Fig. 3) revealed that the five phages cross-reacted immunologically but showed no immunological relatedness to O1 typing phage D-10 (ATCC 51352).
FIG. 3.
(a) Determination of antigenic relatedness among the five newly isolated V. cholerae O139 phages by ELISA. Anti-MAD-5 phage antiserum was used as the antibody, and five V. cholerae O139 phages along with E1 Tor phage D-10 were used as antigens (□, MAD-5; ×, VE-2; ■, CAL-3; ●, S-2; ○, MS-3; •, D-10). (b) Determination of antigenic relatedness among the five newly isolated V. cholerae O139 phages by ELISA. Anti-VE-2 antisera was used as the antibody, and five V. cholerae O139 phages along with E1 Tor phage D-10 were used as antigens (□, VE-2; ×, MAD-5; ●, CAL-3; ■, S-2; ○, MS-3; •, D-10).
Restriction endonuclease digestion profile.
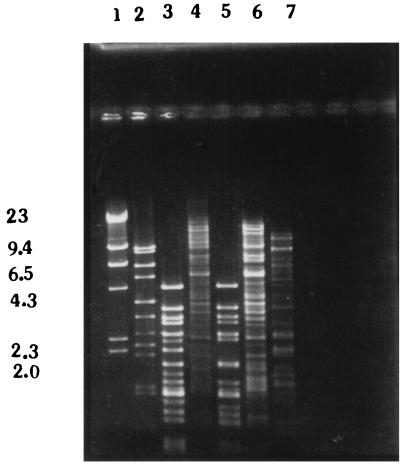
DNAs from the five phages were extracted and digested with the restriction endonuclease HindIII. The restriction endonuclease digestion patterns (Fig. 4) of the DNAs of the five phages showed polymorphism, indicating that they were different from each other as well as from E1 Tor phage D-10 (ATCC 51352). The restriction digestion patterns showed that phages VE-2 and S-2 and phages CAL-3 and MS-3 were similar but not identical.
FIG. 4.
Restriction endonuclease (HindIII) digestion patterns of DNA isolated from five newly isolated V. cholerae O139 phages. Lane 1, lambda phage DNA digested with HindIII; lane 2, MAD-5; lane 3, VE-2; lane 4, CAL-3; lane 5, S-2; lane 6, MS-3; lane 7, D-10. Numbers on the left are in kilobases.
Phage typing.
A total of 500 V. cholerae O139 strains isolated from different areas in India where cholera is endemic were used for phage typing with the set of typing phages. It was observed that almost all the strains were typeable and clustered into 10 distinct phage types (Table 1). Only four strains could not be typed. Phage type 1 (38.2%) was the major phage type, followed by phage type 2 (22.4%) and phage type 3 (18%) (Table 2). Strains belonging to phage type 1 were widely distributed and were found in all 12 areas where the O139 strains were isolated. It was interesting that O139 strains from Madras exhibited the maximum number of phage types (8 of 10 types). Incidentally, the outbreak of V. cholerae O139 originated in this southern city of India (16).
TABLE 1.
Phage typing patterns observed with the five V. cholerae O139 phages
| Phage type | Lytic patterna
|
||||
|---|---|---|---|---|---|
| MAD-5 | VE-2 | CAL-3 | S-2 | MS-3 | |
| 1 | + | + | + | + | + |
| 2 | + | − | + | + | + |
| 3 | + | − | + | − | + |
| 4 | + | + | + | − | + |
| 5 | + | + | − | + | + |
| 6 | − | + | + | + | + |
| 7 | − | + | − | + | − |
| 8 | − | + | − | − | − |
| 9 | − | − | − | + | − |
| 10 | − | − | − | − | − |
+, more than 20 plaques, semiconfluent lysis, confluent lysis (with or without secondary growth), i.e., strong lytic reaction; −, no plaques; also +, sensitive; −, resistant.
TABLE 2.
Distribution of phage types of V. cholerae O139 in different cities in India
| City | No. of strains tested | No. of strains isolated during 1992–1993 and 1996–1998
|
No. (%) of strains with the following phage typing pattern:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Period | No. of strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Madras | 77 | 1993–1994 | 77 | 38 | 6 | 21 | 5 | 1 | 3 | 2 | 1 | ||
| 1996–1998 | NAa | ||||||||||||
| Madurai | 52 | 1993–1994 | 52 | 16 | 11 | 8 | 12 | 2 | 2 | 1 | |||
| 1996–1998 | NA | ||||||||||||
| Calcutta | 129 | 1993–1994 | 41 | 14 | 15 | 4 | 5 | 3 | |||||
| 1996–1998 | 88 | 33 | 21 | 14 | 19 | 1 | |||||||
| Nagpur | 104 | 1993–1994 | 47 | 26 | 7 | 7 | 1 | 3 | 2 | 1 | |||
| 1996–1998 | 57 | 8 | 26 | 19 | 4 | ||||||||
| Pune | 21 | 1993–1994 | 21 | 2 | 2 | 14 | 2 | 1 | |||||
| 1996–1998 | NA | ||||||||||||
| Vellore | 19 | 1993–1994 | 19 | 7 | 4 | 3 | 4 | 1 | |||||
| 1996–1998 | NA | ||||||||||||
| Aurangabad | 31 | 1993–1994 | 31 | 9 | 4 | 3 | 8 | 2 | 4 | 1 | |||
| 1996–1998 | NA | ||||||||||||
| Ahmedabad | 16 | 1993–1994 | 16 | 11 | 4 | 1 | |||||||
| 1996–1998 | NA | ||||||||||||
| Sevagram | 17 | 1993–1994 | NA | ||||||||||
| 1996–1998 | 17 | 9 | 4 | 2 | 2 | ||||||||
| Ambajugai | 12 | 1993–1994 | NA | ||||||||||
| 1996–1998 | 12 | 5 | 5 | 2 | |||||||||
| Alleppy | 12 | 1993–1994 | NA | ||||||||||
| 1996–1998 | 12 | 12 | |||||||||||
| Pondicherry | 10 | 1993–1994 | NA | ||||||||||
| 1996–1998 | 10 | 1 | 7 | 2 | |||||||||
| Total no. of strains tested | 500 | 500 | 191 (38.2) | 112 (22.4) | 90 (18) | 61 (12.2) | 6 (1.2) | 9 (1.8) | 12 (2.4) | 6 (1.2) | 9 (1.8) | 4 (0.8) | |
NA, not available.
Variations in phage typing patterns of V. cholerae O139 strains isolated in 1993 and 1994 and from 1996 to 1998.
Of the 500 strains of V. cholerae O139 examined, 304 strains were isolated during the period between 1993 and 1994, while 196 were isolated from 1996 to 1998 when a resurgence of V. cholerae O139 was observed in Calcutta (3, 13). A comparative analysis of the phage typing patterns of the O139 strains isolated in 1993 and 1994 versus those of the O139 strains isolated from 1996 to 1998 showed that during both periods phage types 1, 2, 3, and 4 were the major phage types (Table 2). Phage type 1 was found as the predominant type in the strains recovered during both periods. However, a higher percentage of phage type 1 (40.46%) O139 strains was isolated in 1993 and 1994 than from 1996 to 1998 (34.69%). Strains were available from Calcutta and Nagpur during both periods. Strains obtained from Calcutta during both periods were divided into five different phage types. Although strains isolated in Nagpur in 1993 and 1994 were of six different types, phage types 6 and 7 were missing from O139 strains recovered in Nagpur from 1996 to 1998. For all other cities, only strains isolated in 1993 and 1994 or from 1996 to 1998 were available. It was observed that strains of all 10 phage types were detected among the O139 strains recovered in 1993 and 1994, but strains of only 5 phage types were detected among the O139 strains isolated from 1996 to 1998. It was also observed that phage type 3 was the second major type among the O139 strains recovered in 1993 and 1994, whereas phage type 2 was found as the second major type among the O139 isolates recovered from 1996 to 1998.
DISCUSSION
Phages can be isolated from the environment in which host bacterial strains generally survive and can be found in sewage, feces, soil, and water (2). In the present study, a total of 40 bacteriophages were isolated from 79 sewage water samples collected from different areas in India where cholera is endemic. A set of five phages were selected from the total of 40 isolates. The lytic patterns of these phages were found to vary among themselves as well as from those of the existing O1 E1 Tor and classical phages. Strains of V. cholerae O1 E1 Tor and classical biotypes were resistant to these phages. These five phages were characterized on the basis of electron microscopy and immunological and physicochemical parameters, and their utility for use in the development of a phage typing scheme for V. cholerae O139 was also evaluated.
The plaques formed by a particular phage are one of the important parameters for characterization of the phage. In this study, the diameters of the plaques formed by the five phages differed. According to the International Committee on Taxonomy of Viruses, phages MAD-5 and VE-2 belong to the family Myoviridae and phages CAL-3, S-2, and MS-3 belong to the family Podoviridae. The structural proteins of the five phages selected for the present study showed marked differences from those of the E1 Tor O1 phages, as revealed by SDS-PAGE (Fig. 2). The restriction endonuclease digestion profiles of the five phages were different from each other as well as from that of E1 Tor O1 phage D-10 (ATCC 51352), indicating heterogeneity in the DNAs of O139 phages.
The use of an immunological technique is an important approach to the examination of a phage particle and its components. The antiserum neutralization test was performed with the phages and their homologous antisera, and marked inhibition of the infectivities of the phages were observed. The cross-neutralization test with the antiserum of a particular phage with the five groups of phages showed that three of the five phages, phages MAD-5, CAL-3, and MS-3, were related and differed from phages VE-2 and S-2, which were related to each other. Thus, the five phages fall into two groups in terms of inhibition of infectivity. To correlate with immunological analysis, the antiphage serum of one of the three related phages (anti-MAD-5 phage antisera) and that of one of the two other related phages (anti-VE-2 phage antisera) were further tested. The interaction of the phage protein as the antigen with anti-MAD-5 phage antisera and anti-VE-2 phage antisera by ELISA proves that all of the O139 phages cross-react antigenically. Antisera raised against phage MAD-5 can probably recognize the proteins of CAL-3 and MS-3, including the tail protein (receptor recognition site), which is responsible for phage adsorption and which thus inactivates lytic propagation, whereas anti-MAD-5 phage antisera can recognize proteins of phages VE-2 and S-2 other than their receptor recognition sites and thus could not inhibit lytic propagation by inhibiting their infectivities. Similarly, anti-VE-2 phage antibody can recognize proteins other than the receptor recognition sites of phages MAD-5, CAL-3, and MS-3. Interestingly, O139 phages do not cross-react with O1 E1 Tor phages. It should be mentioned that V. cholerae O1 biotype E1 Tor phages have no prophylactic value against V. cholerae infection (19). Likewise, V. cholerae O139 phages were also tested in a ligated rabbit ileal loop, and it was found that O139 phages are also unable to lyse the host cell inside the intestine. It is presumed that cholera phages have no prophylactic value against V. cholerae infection.
All of the five phages were used at the RTD to develop the phage typing scheme. Five hundred V. cholerae O139 strains were differentiated into 10 distinct phage types. Most of these strains grouped into phage type 1 (38.2%), followed by phage type 2 (22.4%) and phage type 3 (18.0%). Eight of the phage types were present among the O139 strains received from Madras, where the O139 serogroup originated and rapidly spread to other areas in India where cholera is endemic. The phage sensitivities of the V. cholerae O139 strains received from different places in India showed variable patterns. Phage types 5 and 6 were present only in strains isolated from Ahmedabad, Madurai, and Calcutta. This variation in the distribution of phage types in different areas indicates the presence of different clones of V. cholerae O139 strains. Molecular studies have shown the existence of different ribotypes and CTX (cholera toxin genetic element) genotypes of V. cholerae O139 (3, 8, 20). In this study, it was interesting that all 10 phage types were detected among the O139 strains isolated in 1992 and 1993, while only 5 phage types were detected among those isolated from 1996 to 1998. A comparative study of the phage types of the O139 strains isolated in 1992 and 1993 and from 1996 to 1998 showed that during both periods phage type 1 was the predominant type. However, the percentage of the O139 strains that were isolated in 1992 and 1993 and that belonged to phage type 1 (40.46%) was much higher than the percentage of O139 strains of that type recovered from 1996 to 1998 (34.69%). Again, molecular studies have shown substantial changes in the organization of the CTX phage module of O139 strains isolated from 1996 to 1998 compared to the organization of the CTX phage module of O139 strains isolated in 1993 and 1994 (3, 20).
Phages absorb to specific receptor sites on the bacterial cell wall. In gram-negative bacteria the receptors have been identified as protein and lipopolysaccharide components of the outer membrane layer surrounding the peptidoglycan. A particular phage or group of phages will absorb to a specific site, and different phages will absorb to different sites. Thus, on the surface of a given bacterial cell a variety of different receptors are present, each type being present in a large number of copies. Phage typing methods can test large numbers of strains rapidly. It is a cost-effective and a simple laboratory method and does not require sophisticated instrumentation. Besides, it has been shown that the presence of Vibrio phages in sewage water is a potential tool for the prediction of cholera outbreaks (12). The most important finding of our study was the almost complete typeabilities of the O139 strains with the set of five phages, and the 500 O139 strains could be differentiated into 10 distinct phage types. The V. cholerae O139-specific phages isolated and characterized in the present study were unique in their ability to identify and discriminate V. cholerae O139 strains. The proposed phage typing scheme consists of a set of five newly isolated phages, and the phage typing scheme developed by using the five phages would be useful in the study of the epidemiology of cholera caused by V. cholerae O139.
REFERENCES
- 1.Ackerman H W, Furniss A L, Kasatiya S S, Lee J V, Mbiguino A, Newman F S, Takeya K, View J F. Morphology of Vibrio cholerae typing phages. Ann Virol (Inst Pasteur) 1983;134E:387–404. [Google Scholar]
- 2.Almeida R J, Cameron D N, Cook W L, Wachsmuth I K. Vibriophage VCA-3 as an epidemic strain marker for U.S. Gulf coast Vibrio cholerae. J Clin Microbiol. 1992;30:300–304. doi: 10.1128/jcm.30.2.300-304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basu A, Mukhopadhyay A K, Sharma C, Jyot J, Gupta N, Ghosh A, Bhattacharya S K, Takeda Y, Faruque A S G, Albert M S, Nair G B. Heterogeneity in the organization of the CTX genetic element in the strains of Vibrio cholerae O139 Bengal isolated from Calcutta, India and Dhaka, Bangladesh and its possible link to the dissimilar incidence of O139 cholera in the two locus. Microb Pathog. 1998;24:175–183. doi: 10.1006/mpat.1997.0186. [DOI] [PubMed] [Google Scholar]
- 4.Basu S, Mukerjee S. Bacteriophage typing of Vibrio ElTor. Experientia. 1968;24:299–300. doi: 10.1007/BF02152832. [DOI] [PubMed] [Google Scholar]
- 5.Bradley D E. Ultrastructure of bacteriophages and bacteriocins. Bacteriol Rev. 1967;31:230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti A K, Ghosh A N, Sarkar B L. Isolation of Vibrio cholerae O139 phages to develop a phage typing scheme. Ind J Med Res. 1997;105:254–257. [PubMed] [Google Scholar]
- 7.Chattopadhyay D J, Sarkar B L, Ansari M Q, Chakrabarti B K, Roy M K, Ghosh A N, Pal S C. New phage typing scheme for Vibrio cholerae O1 biotype E1 Tor strains. J Clin Microbiol. 1993;31:1579–1585. doi: 10.1128/jcm.31.6.1579-1585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque S M, Alin A R, Albert M J, Islam K M, Mekalanos J J. Induction of lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh A N, Ansari M Q, Dutta G C. Isolation and morphological characterization of E1 Tor cholera phages. J Gen Virol. 1989;70:2241–2243. doi: 10.1099/0022-1317-70-8-2241. [DOI] [PubMed] [Google Scholar]
- 10.Hornbeck P. Antibodies. In: Colligan J E, Kruisbeck A, Margulies D A, Shevach E M, Storber W, editors. Current protocols in immunology. New York, N.Y: John Willey & Sons, Inc.; 1994. pp. 212–220. [Google Scholar]
- 11.Laemmli U K. Clevage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Madico G, Checkley W, Gilman R H, Bravo N, Cabrera L, Calderon M, Ceballos A. Active surveillance for Vibrio cholerae O1 and vibriophages in sewage water as a potential tool to predict cholera outbreak. J Clin Microbiol. 1996;34:2968–2972. doi: 10.1128/jcm.34.12.2968-2972.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra R, Basu A, Dutta D, Nair G B, Takeda Y. Resurgence of Vibrio cholerae O139 Bengal with altered antibiogram in Calcutta, India. Lancet. 1996;348:1181. doi: 10.1016/s0140-6736(05)65326-3. [DOI] [PubMed] [Google Scholar]
- 14.Mukerjee S. Principles and practice of typing Vibrio cholerae. Methods Microbiol. 1978;12:51–115. [Google Scholar]
- 15.Nair G B, Shimada T, Kurazono H, Okuda J, Pal A, Karasawa T, Mihara T, Uesaka Y, Shirai H, Garg S, Saha P K, Mukhopadhyay A K, Ohashi T, Tada J, Nakayama T, Fukushima S, Takeda T, Takeda Y. Characterization of phenotypic, serological, and toxigenic traits of Vibrio cholerae O139 Bengal. J Clin Microbiol. 1994;32:2775–2779. doi: 10.1128/jcm.32.11.2775-2779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazono H, Pal A, Takeda Y. Emergence of novel strain of Vibrio cholerae with epidemic potential in Southern and Eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Manniatis T. Bacteriophage λ growth, purification and DNA extraction. In: Ford N, Nolan C, Ferguson M, editors. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 2.60–2.81. [Google Scholar]
- 18.Sarkar B L, De S P, Saha M R, Niyogi S K, Roy M K. Validity of new phage typing scheme against Vibrio cholerae O1 biotype E1 Tor strains. Ind J Med Res. 1994;99:159–161. [PubMed] [Google Scholar]
- 19.Sarkar B L, Chakrabarti A K, Koley H, Chakrabarti M K, De S P. Biological activity and interaction of Vibrio cholerae bacteriophages in rabbit ileal loop. Ind J Med Res. 1996;104:139–141. [PubMed] [Google Scholar]
- 20.Sharma C, Maiti S, Mukhopadhyay A K, Basu A, Basu I, Nair G B, Mukhopadhyay R, Das B, Kar S, Ghosh R K, Ghosh A. Unique organization of the CTX genetic element in Vibrio cholerae O139 strains which reemerged in Calcutta, India, in September 1996. J Clin Microbiol. 1997;35:2248–3350. doi: 10.1128/jcm.35.12.3348-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada T, Nair G B, Deb B C, Albert M J, Sack R B, Takeda Y. Outbreaks of Vibrio cholerae non O1 in India and Bangladesh. Lancet. 1993;341:1347. doi: 10.1016/0140-6736(93)90855-b. [DOI] [PubMed] [Google Scholar]
- 22.Stent G S. Molecular biology of bacterial viruses. San Francisco, Calif: W. H. Freeman & Co.; 1963. pp. 94–96. [Google Scholar]