Abstract
Glycans introduce complexity to the proteins to which they are attached. These modifications vary during the progression of many diseases; thus, they serve as potential biomarkers for disease diagnosis and prognosis. The immense structural diversity of glycans makes glycosylation analysis and quantitation difficult. Fortunately, recent advances in analytical techniques provide the opportunity to quantify even low-abundant glycopeptides and glycans derived from complex biological mixtures, allowing for the identification of glycosylation differences between healthy samples and those derived from disease states. Understanding the strengths and weaknesses of different quantitative glycomics analysis methods is important for selecting the best strategy to analyze glycosylation changes in any given set of clinical samples. To provide guidance towards selecting the proper approach, we discuss four widely used quantitative glycomics analysis platforms, including fluorescence-based analysis of released N-linked glycans and three different varieties of MS-based analysis: LC-MS analysis of glycopeptides, MALDI-TOF MS, and LC-ESI-MS analysis of released N-linked glycans. These methods’ strengths and weaknesses are compared, particularly associated with the figures of merit that are important for clinical biomarker studies, including: the initial sample requirements, the methods’ throughput, sample preparation time, the number of species identified, the methods’ utility for isomer separation and structural characterization, method-related challenges associated with quantitation, repeatability, the expertise required, and the cost for each analysis. This review, therefore, provides unique guidance to researchers who endeavor to undertake a clinical glycomics analysis by offering insights on the available analysis technologies.
1. INTRODUCTION
Protein glycosylation is the most complex post-translational modification, and more than 50% of human proteins are glycosylated. The process of protein glycosylation occurs within the endoplasmic reticulum and Golgi apparatus, and it is controlled by a series of enzymes that modify the carbohydrates that are covalently attached to proteins through certain amino acid residues.1–3 This modification is complex to study, in part, because of the heterogeneous nature of the glycans. Unlike protein biosynthesis, glycan biosynthesis does not rely on an underlying template, and the resultant glycan structures can be very heterogeneous. Both the enzyme availability and the cellular environment can affect the final glycosylation profile.2 In addition, this complexity is further enhanced by the presence of multiple monosaccharide units, which are linked together in a variety of ways to form glycan structures; glycans can have various compositions, and even differently-linked isomers with identical composition, due to variety in linkage and branching.4–5
These heterogeneous glycans (oligosaccharides) attached on proteins play crucial roles in regulating various biological processes such as fertilization,6–7 protein folding and stabilization,8–9 cellular recognition, cellular adhesion,10–11 and immune defense.12 In addition, glycosylation is considered as a critical quality attribute in biotherapeutics production since the glycans attached on proteins greatly affect the safety and the efficacy of protein-based drugs. Thus, a minor change in glycosylation profile of these drugs can lead to serious conditions, such as adverse immune reactions,13–14 rapid clearance,15–16 and loss of therapeutic potency. Furthermore, aberrant glycosylation of various endogenous proteins have been associated with the progression of diseases, such as cancers,17–19 kidney diseases20 among others; thus, glycans may serve as clinical biomarkers for disease diagnosis and prognosis.21 Therefore, deeper understanding of this complex modification, and current glycosylation profiling strategies, is critical, not only to identify sensitive biomarkers, but also to provide information necessary to regulate the glycosylation in biotherapeutics, so the safety and the activity of glycoprotein-based drugs is ensured. While multiple types of glycosylation exist, including lipid glycosylation, glycosaminoglycans, N- and O-linked glycosylation on proteins, this review is focused on N-linked glycosylation of proteins.
1.1. N-linked Glycosylation
The most commonly studied glycosylation type, N-linked glycosylation, occurs when the glycans are attached to the proteins through the amide nitrogen on the side chain of an Asn residue. These glycosylated Asn are usually located within a unique amino acid sequence: Asn-Xxx-Ser/Thr, where Xxx can be any amino acid except proline.1–3, 22 Glc3Man9GlcNAc2 is the common precursor for all the N-linked glycans, as shown in Figure 1. This precursor is attached to the protein during the initial phase of the glycosylation process. The precursor undergoes many enzymatic trimming and monosaccharide addition steps that introduce modifications to the precursor glycan while preserving the tri-mannosyl-pentasaccharide core (Man3GlcNAc2).2–3 These modifications to the precursor glycan result three major types of N-linked glycan structures; they are high-mannose (Man), complex, and hybrid. (See Figure 1.)
Figure 1.
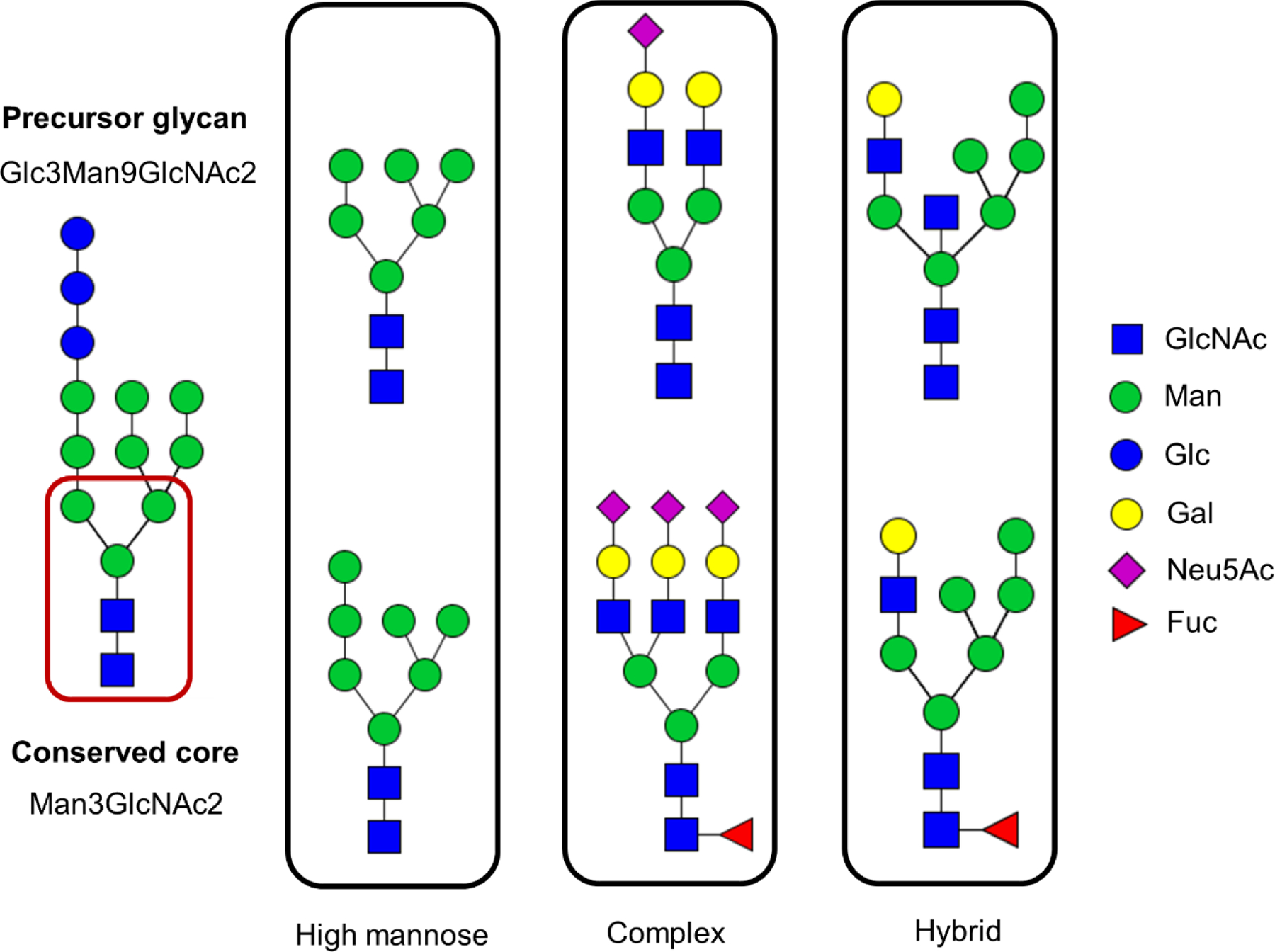
Symbolic representation of different N-linked glycans. High mannose, complex and hybrid are three major N-linked glycan types; all derived from the common precursor Glc3Man9GlcNAc2. These glycans are made of N-acetyl glucosamine (GlcNAc), mannose (Man), glucose (Glc), Galactose (Gal), N-acetyl neuraminic acid (Neu5Ac), and fucose (Fuc).
The high-mannose type glycans are formed by trimming of monosaccharides from the precursor without addition of new monosaccharides, thus leaving only Man residues attached to the core structure. In contrast, complex- type glycans are formed by trimming monosaccharides of the precursor glycan, followed by addition of new sugars to the terminal mannose residues of both arms of the Man3GlcNAc2 core. In complex type structures, GlcNAc is the very first monosaccharide unit directly linked to the terminal mannoses in the core-structure, and it is further extended with additional monosaccharides; the most common pattern involves attachment of galactose (Gal) units and terminal sialic acid (N-acetylneuraminic acid) units. Based on the number of GlcNAc attached to the terminal mannose sugars in the core structure of the complex-type glycans, the number of branches are defined as bi-, tri- and tetra-antennary. In addition, in complex-type glycans, core-fucosylation and/or antennary fucosylation can be observed when the fucose (Fuc) is attached to the innermost GlcNAc of the core structure or the GlcNAc at the non-reducing end. The hybrid-type glycans, the last glycan category, share the characteristic features of both high-mannose and complex-type glycan structures.2–3
1.2. Altered N-linked Glycosylation and Diseases
During the progression of many diseases, alteration of the N-linked glycosylation profile is observed. These alterations may include both upregulation and/or downregulation of glycans, elevated branching, size increase, and modifications to the core-structure.20, 23–26 For instance, during colorectal cancer progression, several glycosylation changes were observed including: differentially expressed serum IgG N-linked glycans, decreased levels of high-mannose structures, reduced core-fucosylation, and less sialylation.23 Decreased levels of fully galactosylated N-linked glycan structures were identified in gastric cancers,24 lung adenocarcinoma tissues,25 and rheumatoid arthritis (RA).27 In addition, significantly decreased levels of Man5 and bi-antennary N-linked glycans, along with elevated levels of branching, antennary fucosylation, and core-fucosylation were observed in the serum glycans of primary epithelial ovarian cancer patients.26 The altered glycans and glycosylation patterns that are unique to certain types of diseases may serve as biomarkers, and discovery of those biomarkers is important, not only to understand disease pathology, but also to perform more selective treatments and disease diagnoses.
In the past few decades, impressive efforts have been made to identify clinically relevant glycan biomarkers for diseases. More than 90 potential N-linked glycan biomarkers have been identified based on previously published studies, including biomarkers for certain types of cancers, such as breast, liver, ovarian, kidney, and pancreatic cancers, as well as for Hepatitis B and C, Alzheimer’s disease, and diabetes.28 This large number of potential glycan-based biomarkers clearly shows the significance of quantitative glycomic studies in discovering selective candidate glycan biomarkers for distinguishing disease states from healthy states and also in disease prognosis, diagnosis and/or treatment. While the number of potential glycan-based biomarkers is impressive, we are unaware of any of these having yet transitioned to clinical diagnostics; undoubtably, part of the reason for this is due to the need for even better analysis technologies that can handle large sample sets. The discovery of unique biomarkers for various diseases is greatly dependent on not only the availability of sensitive and reliable analytical methods but also on careful selection of the most appropriate and cost-effective approach for any clinical glycomics study. Thus, in this review, we compare the performance of four commonly used quantitative glycomics methods to guide the selection of an appropriate analytical strategy for clinical and pre-clinical studies.
2. GENERAL CONSIDERATIONS
2.1. Glycosylation Analysis
Glycosylation analysis is most commonly performed in two ways: glycopeptide-based analysis and glycan analysis.2–3, 29 During glycopeptide-based analysis, the glycans remain attached to the glycosylation site, and therefore, retain information about the protein to which they are attached and the site of attachment. This information increases the specificity of the analysis, but the trade-off is that the analyses are more complex. Site-specific glycosylation analysis is used limitedly in the glycan biomarker discovery field,30 due to the glycopeptides’ lower abundance, lower ionization efficiency, need of method optimization for each glycoprotein,31 and difficulties in data interpretation.32 By contrast, glycan-based analysis, where glycans are released from the glycoproteins and then are analyzed, is useful for obtaining aggregate information about the total glycan pool. One disadvantage of this approach is that a change in glycosylation is hard to interpret: It may be due to changes in glycosyltransferase activity or changes in protein expression of certain glycosylated proteins. While the method provides substantially less specificity, it is frequently employed in clinical glycomics due to the availability of universal and well-established protocols for glycan analysis. One way to balance the strengths of both methods is to perform glycan analysis on a specific protein target that has been purified from the sample. When the glycoprotein of interest can be purified from the matrix, glycan analysis provides information specific to the protein of interest, and well-established methods can be used to facilitate quantitation and analysis.
2.2. Sample Preparation
Figure 2. shows multiple ways of generating glycans or glycopeptides from complex biological samples for glycosylation analysis. Of these sample preparation steps, glycoprotein purification at the crude protein mixture level is performed especially when targeted quantitation of a specific protein’s glycosylation profile is necessary; for example, IgG is affinity purified with protein A or G, prior to the quantitation of IgG N-linked glycans associated with cancer in serum samples.23–24 On the other hand, glycans can be released directly from the non-enriched biological samples when the total glycome pool of a biological matrix is quantified;33 however, the method yields limited specificity. In glycopeptide analysis, proteolysis at crude mixture level is performed when the targeted glycoprotein is in high abundance;30, 34 but the proteolysis on purified glycoprotein(s)32, 35–36 is used more frequently, as it improves the sensitivity of the analysis. Once these glycans or glycopeptides are generated, further purification can be performed by solid phase extraction-based methods (SPE) with porous graphitized carbon (PGC) 33, 37 and hydrophilic interaction liquid chromatography (HILIC),32, 38–39 or by using specific lectins for glycopeptides.40
Figure 2.
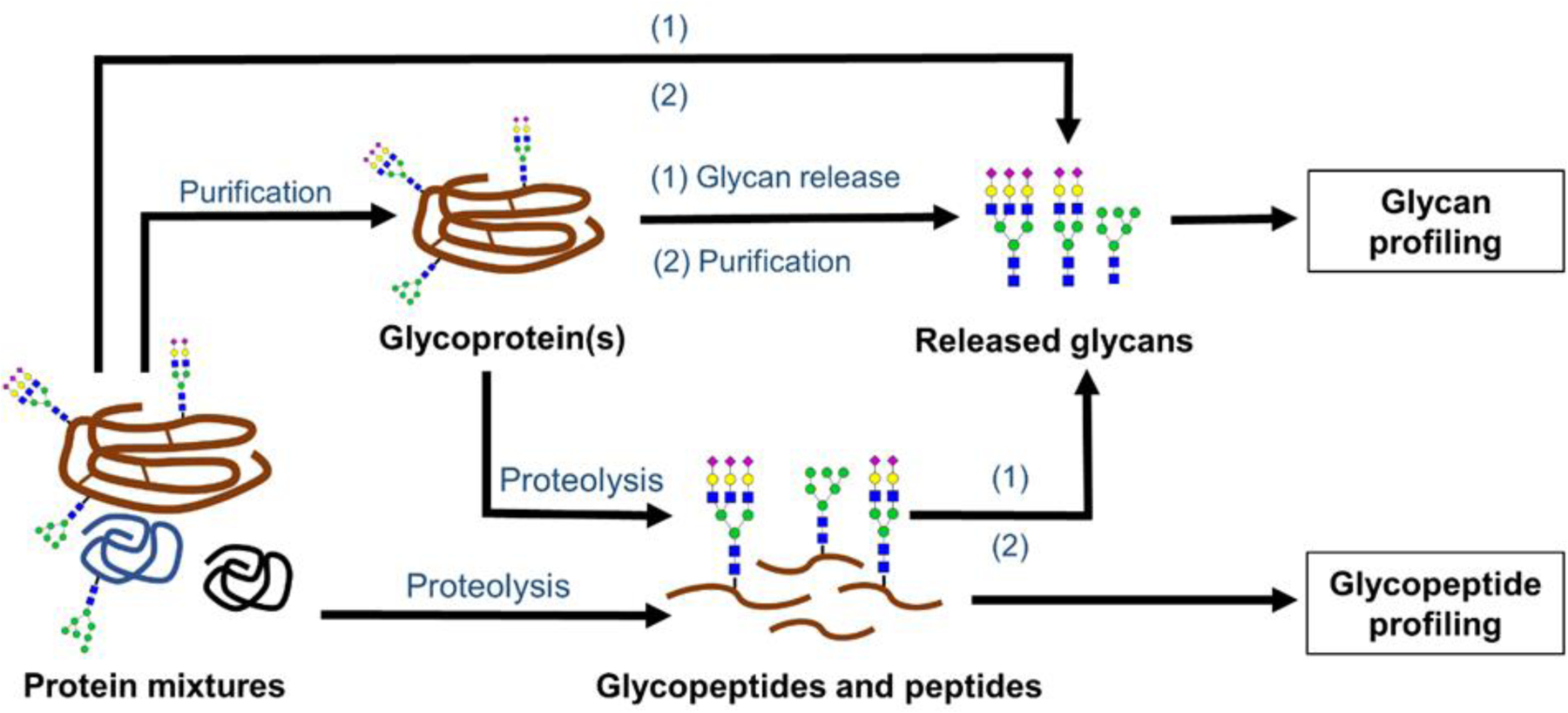
Different sample preparation routes for generating glycans and glycopeptides from complex biological samples for quantitative glycosylation analysis. In this workflow, for glycan profiling: glycans are released from purified glycoprotein(s), or directly from the crude biological mixture. Alternatively, glycans can be released from glycopeptides. For glycopeptide profiling: glycopeptides that are generated from either the purified glycoprotein(s) or directly from complex biological mixtures (for high abundant glycoproteins) are subjected to proteolysis; then, the resulting mixture of glycopeptides and peptides are subjected to quantitative analysis.
2.3. Quantitation
Glycan abundances from healthy patients versus those of a disease state are compared by either absolute4, 37 or relative quantitation;4, 17, 37 relative quantitation is the more common choice, since absolute quantitation usually requires glycan standards that are not readily available.41 In relative quantitation, the proportion of glycans present in the two sample types (healthy versus disease state) is reported by dividing an individual glycan abundance by the total glycan abundance,42–43 or by the abundance of the most intense glycan peak,31, 44 or by a peak among the major signals.31 While these methods do not report exact glycan concentrations, the ratios are useful for allowing the identification of under- or over-expressed glycans between healthy versus disease groups, which is the ultimate goal of the analysis.
3. QUANTITATIVE STRATEGIES IN CLINICAL GLYCOMICS
3.1. MASS SPECTROMETRY (MS)-BASED APPROACHES
MS-based approaches are widely used in clinical and pre-clinical glycomic studies. This choice is preferred by many researchers because the method is sensitive, and it can be used to differentiate species with unique masses. Structural information can also be obtained through MS/MS and MSn experiments.45–49 These benefits, especially when coupled with separation and enrichment techniques, facilitate the identification and quantitation of glycans originating from complex biological matrices. Matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) and electrospray ionization mass spectrometry (ESI-MS) are the most common ionization techniques for glycan analysis, and the latter may be done in conjunction with widely used separation techniques, such as liquid chromatography and capillary electrophoresis, while the former requires offline separation.
3.1.1. MALDI-TOF MS Analysis of Released N-linked Glycans
MALDI-TOF MS is widely applied in quantitative clinical glycomics. It is a simple, sensitive, high-throughput method.20, 23, 33, 37, 50–51 The most common sample preparation procedure for glycan analysis by MALDI-TOF MS is the workflow appearing at the top of Figure 2: the desired glycoprotein of interest is isolated and purified from the complex biological matrix; next, release of N-linked glycans from glycoproteins via PNGase F treatment typically follows. The glycans are subsequently purified and labeled20, 23, 33, 37, 51 for MALDI-TOF MS analysis.
Figure 3 (A) shows more detail about the MALDI-MS specific sample preparation steps. Once the glycans are purified, labeling of released N-linked glycans is usually performed to enhance the ionization efficiency of glycans, to improve the sensitivity of the analysis, and sometimes to allow simultaneous detection of both neutral and acidic N-linked glycans.23, 51 MALDI-TOF MS analysis of permethylated N-linked glycans20, 33, 37, 50 has been performed in many clinical glycomics studies. In addition, derivatization of sialic acid via methyl esterification43 or ethyl esterification38 is another useful labelling method. Once the labeled glycans are purified again, they are prepared for MALDI-TOF MS analysis. While permethylation has been the go-to labeling method in the field, many researchers are currently taking advantage of the esterification method pioneered by Wuhrer and colleagues, due to its dual advantages of stabilizing the sialic acids while also providing linkage information.
Figure 3.
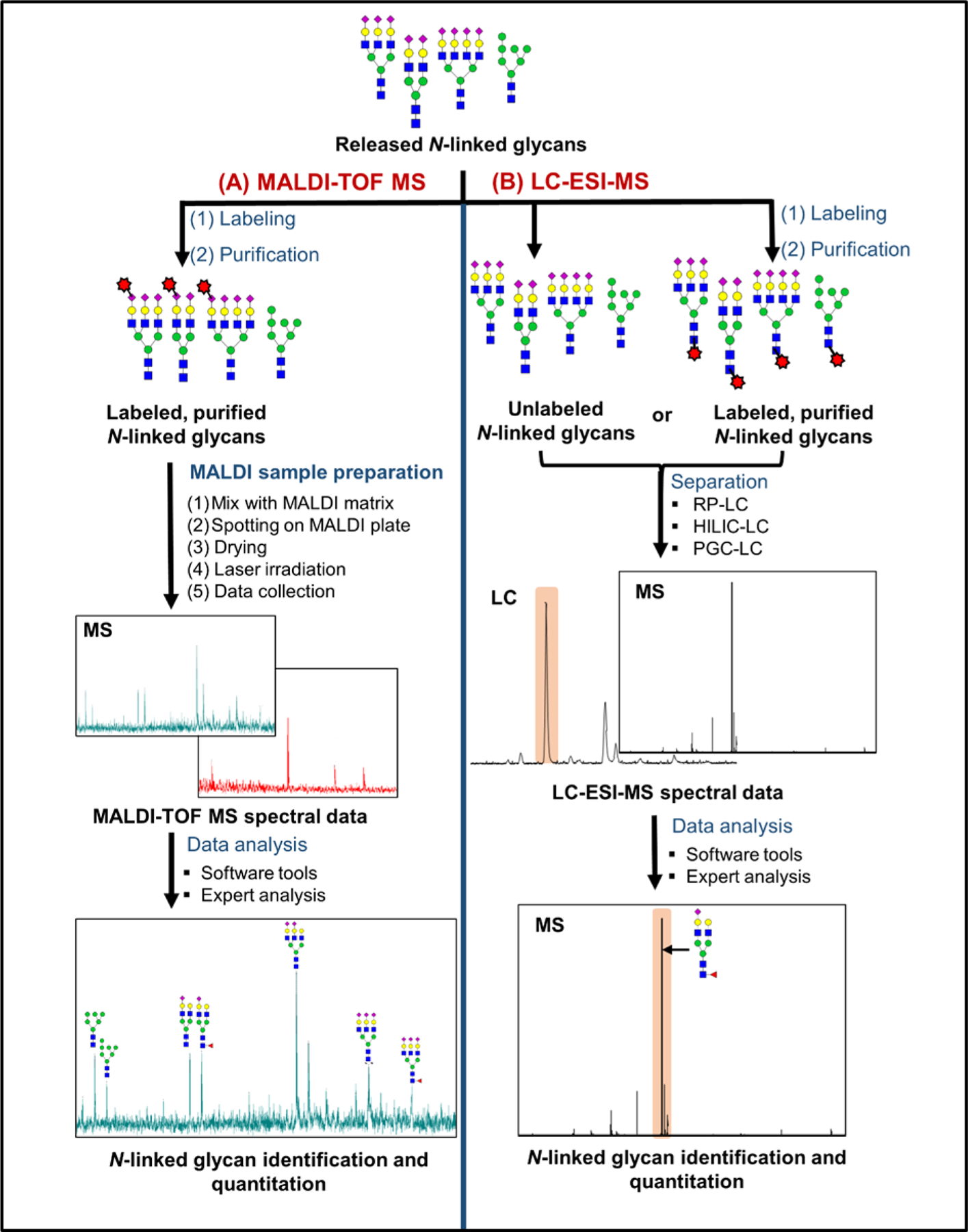
General workflows for Released N-linked glycan quantitation by MALDI-TOF MS (A) and by LC-ESI-MS (B). In MALDI-TOF MS analysis, labeled N-linked glycans are mixed with a MALDI-matrix and irradiated with laser shots to collect MS data (A). In LC-ESI-MS, labeled or unlabeled N-linked glycans are separated by using a liquid chromatography method, followed by ESI-MS analysis (B). During LC-MS analysis of labeled glycans, the label is most commonly installed at the reducing end. By contrast, a common labeling strategy for MALDI-MS involves derivatizing sialic acids, which are at the nonreducing end.
Another reason to label glycans prior to MALDI-MS analysis is to incorporate a stable isotopic label for quantitative purposes. One attractive strategy is dual labeling, which enables simultaneous quantification of neutral and acidic glycans.51–53 This approach involves incorporation of isotopic species at both ends of the glycan. Notably, this method provides added value through stabilization of sialic acid moieties, simplified sialic acid counting, and enhanced ionization.51, 54 Li and colleagues implemented this strategy in their mdSUGAR tag, using reductive amination and periodate oxidation to derivatize the reducing end and sialic acids, respectively, which introduces a larger mass difference and renders this method amenable to lower resolution instruments.55 Isotopic PFBHA and isotopic methylamine was used in a similar fashion; this dual labeling strategy also enables glycan enrichment via fluorous SPE.54, 56 More traditional methods include enzymatic 18O labeling,57 isotopic permethylation,58 and metabolic labeling.59 Sample-matrix preparation greatly affects the quality of the resultant MS spectra, as matrix plays a vital role in promoting solid phase analytes into the gaseous phase; DHB is the most common matrix for glycan analysis by MALDI-MS, although other choices are available and sometimes prove to be worth exploring. Prior to MALDI-TOF MS analysis, the labeled glycans are mixed in a 1:1 ratio with a matrix, including 2,5-dihydroxy-benzoic acid (2,5-DHB),38, 60 Super DHB,20 4-chloro-α-cyanocinnamic acid (Cl-CCA),60 2,4,6-trihydroxyacetophenone (THAP),60 or 9-Aminoacridine (9-AA),61 followed by spotting the aliquots of the mixed sample solutions onto a MALDI plate. Then multiple laser shots are applied on each sample spot to ionize the samples, followed by the MS analysis.20, 23, 33, 37, 51 During the analysis, reflectron-positive20, 37–38, 62–63 and negative,60 as well as the linear-positive and negative ion modes60 are used. Among them, positive-ion mode is more commonly used due to higher ionization efficiency and higher S/N ratio reported for labeled glycans.29 However, negative ion mode is also used to detect acidic glycans with improved detection sensitivity.60
3.1.2. LC-ESI-MS Analysis of Released N-linked Glycans
ESI-MS is also used in quantitative glycomics studies. A general LC-ESI-MS workflow of glycan quantitation includes: glycan release, purification, labeling, followed by glycan separation, and ESI-MS quantitation. Similar to MALDI-TOF MS, in LC-ESI-MS-based glycan quantitation, glycan release is commonly performed at the crude mixture level for total glycome quantitation, followed by enrichment of released glycans using SPE,4, 25 but it can also be performed after enriching for a target protein.
Figure 3 (B) represents the general workflow for LC-ESI-MS analysis. Once the glycans are purified, analysis can be performed on labeled, 33, 48, 64–66 unlabeled,4, 24 or chemically reduced25, 67 glycans. Glycan labeling is performed to improve the sample throughput, by allowing for multiple, differentially labeled samples to be analyzed together, and to enhance the ionization efficiency of the glycans. 68 Isobaric tags or tandem mass tags (TMT)48, 64–66 that have identical masses, but with various heavy isotopes distributed within the structure, are commonly used in labeling experiments. AminoxyTMT,48, 66 is one such tag that allows simultaneous labeling of glycans derived from multiple samples, resulting in a single chromatographic peak at the full MS level for various glycans, yielding sample-specific reporter ions at the MS/MS level for comparative N-linked glycan quantitation. Alternatively, stable isotopic labeling of glycans where small mass differences to the glycans derived from multiple samples are introduced through isotopically labeled reducing-end labeling agents, or isotopic permethylation, are used to quantify glycans at the MS1 level.64 For example, 8-plex quantitative glycan analysis of multiple breast cancer cell lines was feasible through isotopic permethylation performed by using isotopically labeled iodomethane during the permethylation reaction.64 Furthermore, metabolic isotope labeling, where cells from different samples are labeled isotopically, is also used in quantitative glycomics analyses to reliably quantify glycans from different samples while minimizing potential sample preparation bias.33 At the end of the appropriate glycan labeling step, the samples are purified from the labeling agent and/or biological matrix molecules, and they are analyzed by MS. Labeling agent removal is not trivial, and different labeling agents have different optimal clean-up procedures.
Glycan separation prior to MS detection facilitates the enrichment of various glycan structures derived from complex biological samples, while allowing sensitive detection of multiple glycans by MS. Liquid chromatography (LC) is used frequently, due to its ability to resolve complex mixtures, its compatibility with MS methods, and its capacity for facilitating automation. As compared to traditional LC-ESI-MS or MALDI-TOF MS, nano-LC-MS is used in many studies, as it significantly improves the detection sensitivity.4 Nano-LC columns packed with C1864, 68–69 or PGC-bonded stationary phases66 are frequently used to separate permethylated N-linked glycans; PGC is also regularly used for non-permethylated glycans, while HILIC columns are used to separate more hydrophilic glycans, which are often labeled prior to analysis.48 Alternatively, by incorporating a microfluidic chip to the nano-LC workflow, a greater retention time reproducibility, better separation, and high sensitivity for the glycans can be achieved.4, 24 For example, PGC / nano-LC chip-based separation has been used in many quantitative glycomic studies, as PGC is capable of separating glycans by their polarity, size, and 3D structure, while exhibiting good isomer separation capacity.4, 24–25, 67 Once the glycans are effectively separated, they are detected with MS for quantitation.
Electrospray ionization (ESI) is a commonly used ionization technique in quantitative glycomics studies because it generates glycan ions without the loss of labile groups; thus, it provides complete composition information. In many studies where ESI-MS is used, the instrument is operated in a data dependent mode,65–66, 69 acquiring full MS scans in an Orbitrap,65, 69 for example, followed by MS/MS scans of the most intense ions. Also, in some studies, targeted quantitation is performed using a triple quadrupole mass spectrometer operated in multiple reaction monitoring (MRM) mode.69 Once the LC-MS data are acquired, they are analyzed by using software tools or by combining both expert analysis and automated tools prior to the N-linked glycan quantitation.
3.1.3. LC-MS Analysis of Glycopeptides
LC-MS analysis of glycopeptides is another method used in biomarker discovery; this approach provides glycosylation site-specific information. However, the method is challenging because it involves determination of both an unknown peptide and an unknown glycan. The general workflow for glycopeptide analysis using LC-MS includes: isolation of desired glycoprotein(s) from the biological matrices, glycoprotein denaturation, reduction and alkylation, all prior to the enzymatic digestion; then separation of enriched or non-enriched glycopeptides is achieved, usually by HPLC, followed by MS analysis.
In this workflow, isolation of glycoprotein(s) at either the crude mixture level32 or at glycopeptide level35–36 is important due to the glycopeptides’ low abundance as compared to the non-glycosylated peptides. Of these enrichment methods, lectin-based enrichment, where carbohydrate-binding proteins or lectins bind to specific carbohydrate moieties via affinity binding, can be implemented for both glycoprotein level70 and glyopeptide level enrichment.40, 46, 71 For example, Aleuria aurantia lectin (AAL) 70 and Lens culinaris Agglutinin (LCA)46 are two lectins that bind to fucosylated carbohydrate motifs, and Sambucus nigra lectin (SNA) is another lectin that binds to carbohydrates that contain sialic acids.70 One obvious drawback of using lectins to enrich the glycans is that the lectins will differentially enrich certain glycoforms, so this purification strategy can be quite deleterious if the goal of the analysis is to capture all glycoforms present at their natural abundance. Alternatively, unique antibodies that identify specific epitopes present on protein backbones are also used to isolate specific glycoproteins from complex biological samples.32, 72 Anti-IgA72 and antihuman Haptoglobin (Hp)32 are two such antibodies that have been used to isolate IgA and haptoglobin, derived from cancer-associated serum samples. Additionally, protein G or protein A-based isolation of IgG72–73 and glycopeptide level enrichment with sepharose beads73 are also reported. However, when the targeted glycoprotein is in high abundance, such as immunoglobulin G in serum, enrichment steps at either the crude mixture level or glycopeptide level can be avoided, while performing proteolytic digestion directly on crude biological samples, as shown in Figure 2.30, 34
To generate glycopeptides, the (enriched) samples are subjected to proteolytic digestion. Most commonly, site-specific proteases, such as trypsin,30, 74 are usedThese proteases produce peptides that can be predicted in advance, and the number of different peptides generated per glycosylation site is very limited; often a single unique peptide will be generated per glycosylation site. Some researchers, however, are concerned that specific proteases limit the number of glycopeptides identified when the resultant glycopeptides are multiply glycosylated or miscleaved. They support the use of non-specific proteases, which may be better in the specific cases of multiply glycosylated and difficult-to-digest glycoproteins. This strategy, either on its own, or in combination with specific proteases,32, 34, 41 may provide more complete glycopeptide identification in some cases.34 After protease digestion, the resulting glycopeptide mixture, is subjected to purification/enrichment35, 71, 74 or not.30, 34 Most commonly, enrichment is not part of the workflow prior to LC-MS, but sometimes it proves to be beneficial. After sample preparation is complete, the sample can be analyzed by LC-MS, as shown in Figure 4.
Figure 4.
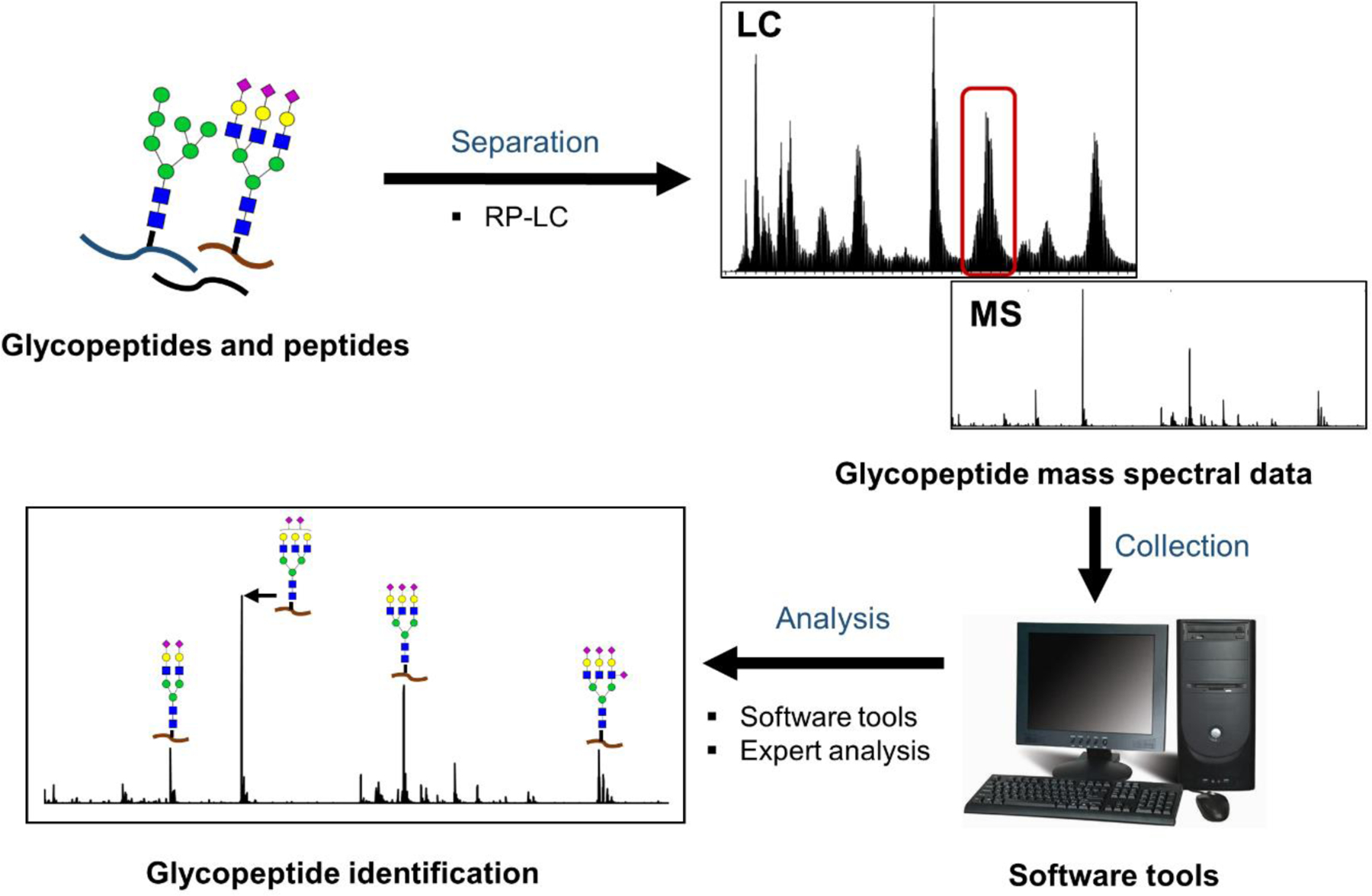
General workflow for LC-MS analysis of glycopeptides. In this workflow, enriched or non-enriched mixtures of glycopeptides generated at purified glycoproteins level or at crude mixture level are further separated followed by the mass spectrometry detection and analysis for glycopeptide identification and quantification.
Glycopeptide separation prior to MS detection is important because it permits enrichment of glycopeptides from peptide counterparts that co-exist in the mixture; those can reduce the ionization of glycopeptides if they co-elute. LC is the method of choice for glycopeptide separation owing to its MS compatibility, glycopeptide resolving capacity, and ability to be automated.3 Reverse phase (RPLC) columns with C18- bonded phases are the most popular in glycopeptide separation;30, 36, 40–41, 74–80 these columns separate glycopeptides predominantly based on the interactions between the peptide backbone and the hydrophobic stationary phase, although the presence of sialic acids also can impact the retention time of the glycopeptides, albeit to a lesser degree.
Once the glycopeptides are separated, they are detected using MS. Multiple reaction monitoring (MRM) mode, a targeted mass spectrometry approach, is frequently used for quantifying glycopeptides due to its high sensitivity and selectivity.30, 34, 71, 81 Alternatively, untargeted approaches are possible; in these cases, glycopeptides can be quantified by their high-resolution ESI-MS signal82–83 or by using data-dependent LC-MS/MS.70 ESI with positive ionization mode is frequently used, but in some cases, negative ion mode is also performed to enhance the ionization of sialylated glycopeptides.40 Finally, the data analysis is performed by using either software tools30, 34 or by combining both software tools and manual verification.32
3.2. SPECTROSCOPY- BASED APPROACHES
3.2.1. LC/CE-Fluorescence Profiling of Released N-linked Glycans
Another widely used method in quantitative clinical glycomics studies is HPLC with fluorescence detection. In this case, labeled glycans are analyzed. As with previously discussed methods, this one also requires isolation and purification of glycoprotein(s) of interest from complex biological samples, prior to sample preparation; see Figure 2. Then the isolated glycoprotein(s), which are either denatured84 or non-denatured20 are subjected to enzymatic glycan release with PNGase F enzyme, followed by fluorescent labeling and glycan purification.20, 85–86 Some common fluorescent labeling reagents include 2-aminobenzoic acid (2-AA)38 and 2-aminobenzamide (2-AB).20, 43, 85–86 Once the glycans are fluorescently labeled, the excess labeling reagent is removed using SPE,31, 43–44, 84 paper chromatography,85, 87 or size-exclusion chromatography88 and the labeled glycans are separated by HPLC. Options include HILIC-LC (hydrophilic interaction liquid chromatography),43–44 HPAEC (high pH anion exchange chromatography),20, 86 and NP-HPLC (normal phase high performance liquid chromatography)42, with the HILIC-LC method dominating the field. The separated, derivatized glycans are quantified based on their fluorescence signal. See Figure 5 for a representative workflow for this method.
Figure 5.
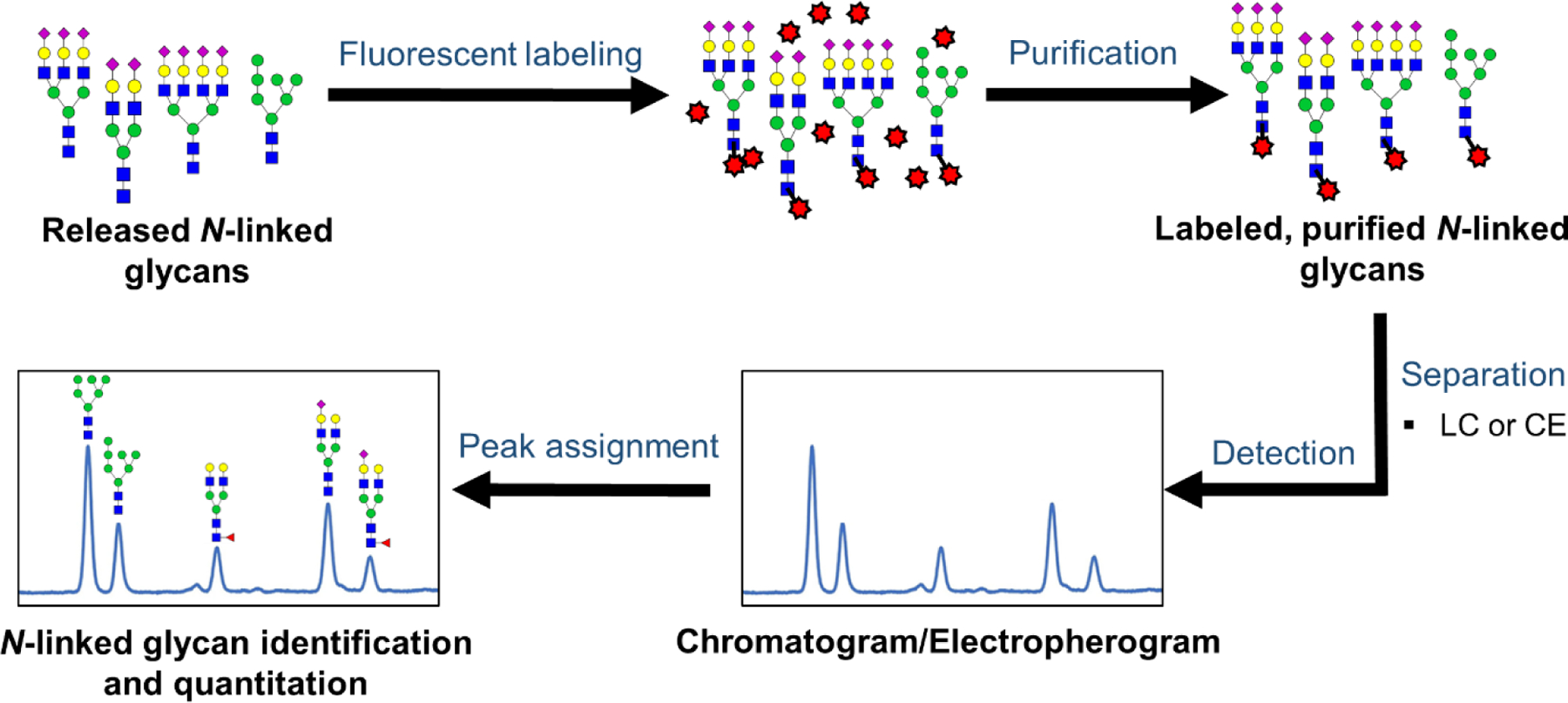
Workflow for the analysis of released N-linked glycans via liquid chromatography (LC)/ capillary electrophoresis (CE)-fluorescence detection. In this workflow, released N-linked glycans are labeled at the reducing end with a suitable fluorescence labeling reagent, purified, and then are separated by using LC or CE. Finally, the resultant peaks of chromatograms or electropherograms are assigned by using established data bases and/or follow-up experiments followed by N-linked glycans quantitation.
Capillary electrophoresis (CE) separation paired with fluorescence detection is also used to profile N-linked glycans in clinical glycomics studies, as the method is high-throughput and readily adaptable to microfluidic devices.63, 88–89 The sample preparation for CE and HPLC are similar; however, 8-aminopyrene-1,3,6-trisulfonic acid (APTS),20, 31, 43, 85, 90 and 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS)91 labels are often used; these labels carry three negative charges from sulfonic acids. The charges on the fluorophores increase the electrophoretic separation.92 However, when the glycans carry negatively-charged sialic acid groups, cleavage of the sialic acids89 or neutralization via chemical modifications such as methylamidation63 is typically performed before the APTS labeling step. This modification yields glycans all bearing the same charge state, thus, allowing glycans’ electrophoretic migrations to be based on their hydrodynamic volumes, resulting in increased migration times for sialylated glycans while preserving the efficiency of separation.63 Once the glycans are labeled, they are separated by using various CE modes, such as conventional CE63, 88 or capillary gel electrophoresis (CGE);43, 85 then they are detected by fluorescence.
Peak assignment for fluorescence-based methods is typically accomplished with the use of a glucose unit index (GUI), in which calibration of the system with a standard dextran ladder allows for the normalization of glycans’ retention/migration times by converting them to glucose units (GU).85, 93–96 The GU value, determined based on the number and linkage types of its constituent monosaccharides,94 is used to determine the elution/migration positions of individual glycans. A database of normal-phase HPLC elution positions for over 350 2-AB labeled glycans based on GU values was previously established in an effort to automate the process of glycan identification.94 However, full structural characterization of glycans requires additional experiments such as exoglycosidase digestion and/or complementary studies with tandem-MS.90, 97–98
4. PERFORMANCE COMPARISON
As described in the previous section, various glycomic analysis and quantitation strategies have been developed; each of these methods are currently used in glycomics-based biomarker studies. However, each of these methods has differences in their workflows and unique advantages and disadvantages. Thus, one must carefully consider when to use a particular quantitative strategy for a biomarker study. To assist in this selection process, we compared the performance of the four quantitative strategies described above with respect to various key figures of merit. Table 1 summarizes the methods’ performance.
Table 1.
Performance Comparison of Four Glycomics Analysis Platforms.
| MALDI-TOF MS of Glycans | LC-ESI-MS of Glycans | Fluorescence Detection of Glycans | LC-MS of Glycopeptides |
|---|---|---|---|
| 4.1 Initial Sample Amounts | |||
| Low µg to ≤ 50 µg | Low µg to ≤ 50 µg | ~20 µg to a few hundred µg | Low µg to ≤ 50 µg |
| 4.2 Sample Throughput | |||
| Highest throughput: 96–384 samples per run | Mid- to low- throughput; limited by online separation, can be improved with multiplexing agents | Mid- to low-throughput; When CE-LIF is multiplexed, provides the second-best throughput | Mid- to low- throughput; limited by online separation |
| 4.3 Sample Preparation Time | |||
| Fastest: 96 samples in <4 hours. | In principle, equal to that of fluorescence detection. | Second-fastest: 96 samples in <8 hours. | Longest sample prep time. |
| 4.4 Number of Structures Identified | |||
| Worst MS method but better than LC-fluorescence. | >>100 glycans; lacks site-specific information | Low performance | Up to 30,000 glyccopeptides. Both glycan and attachment site information |
| 4.5 Isomer Separation and Structural Characterization Ability | |||
| No isomer separation; but, sialic acid linkages can be identified with derivatization. Intermediate performance in structural characterization; tandem capabilities are needed for structural elucidation. |
Superior isomer separation Superior performance in structural characterization; but no site-specific information is available |
Low isomer separation Low performance in structural characterization; complementary methods are needed to obtain structural information |
Intermediate isomer separation Intermediate structural characterization; site-specific information is available |
| 4.6 Differences in Quantitative Data Generation | |||
| Ionization differences hinder quantification of different glycans within a sample | Ionization differences hinder quantification of different glycans within a sample | Glycans within a sample and among different samples can be quantified easily | Ionization differences hinder quantification of different glycans within a sample |
| 4.7 Method Precision | |||
| Sufficient repeatability | Sufficient repeatability | Highest repeatability (<10% CV) | Sufficient repeatability |
| 4.8 Required Expertise | |||
| Mid-level technical expertise required | Highest degree of expertise required | Least expertise required | Highest degree of expertise required |
| 4.9 Cost for Instrumentation and Per Sample | |||
| Intermediate cost | High cost | Lowest cost | High cost |
4.1. Sample Size
In complex biological mixtures, the glycoprotein(s) of interest are usually present at low abundance compared to the non-glycosylated proteins. Therefore, the detection of these low abundant glycans/glycopeptides depends on not only the method sensitivity, but also the initial sample amount used for the analysis. Table 1. shows the comparison of methods in terms of typically used initial glycoprotein amounts for the quantitative glycomics analysis.
Generally, MS-based methods are highly sensitive; they can be implemented with lower microgram quantities of glycoproteins to provide reliable quantitative glycomics data. When LC-MS-based analysis of glycopeptides/glycans is implemented, higher quantitation sensitivity is achieved by using different MS-based strategies, such as targeted multiple reaction monitoring (MRM)30, 34, 68 or carefully designed data-dependent scan sequences that selectively target glycopeptides;32, 73 these methods permit lower initial sample amounts. For examples, two studies performed by Hong et al.30, 34 used MRM-mode to quantify immunoglobulin glycopeptides generated from approximately 24 µg of IgG,30, 34 5 µg of IgA,34 and 3 µg of IgM,34 derived from 2 µL of un-enriched serum samples; the glycoprotein amounts indicated here are approximate values calculated based on these glycoproteins’ average plasma concentrations reported in the literature.99 In addition, MS/MS techniques, such as LC-ESI-MS/MS73 and LC-EThcD MS/MS32 were used to quantify glycopeptides of IgG (~24 µg),73 and haptoglobin (3 µg)32 derived from serum samples of pancreatic cancer and liver cirrhosis patients, respectively. LC-MS analysis of released N-linked glycans is also performed at lower microgram levels;24, 68 one study quantified more than 55 permethylated serum N-linked glycans, by using approximately 0.1 µg of total glycoproteins derived from 0.005 µL of enriched-serum sample injected on column;68 yet readers should be aware that the amount of glycoprotein initially subjected to sample preparation for these studies was more than 100 times greater than the reported injection amounts. Similar to LC-MS analysis methods, MALDI-TOF MS analysis also uses initial glycoprotein amounts ranging from 0.5 – 30 µg, derived from up to 5 µL of biological samples for N-linked glycan quantitation.20, 23, 37, 100 The study performed by Gao and coworkers100 quantified more than 50 TMPP-Ac-labeled N-linked glycan structures derived from approximately 0.5 µg of serum glycoproteins per MALDI spot; however, the starting quantity, prior to sample preparation, was 20 times higher.
Compared to all MS-based methods, fluorescence-based methods typically require comparatively higher initial glycoprotein amounts, ranging from approximately twenty to a few hundred micrograms.84, 88
4.2. Sample Throughput
High-throughput methods that are capable of analyzing glycomic profiles of several thousands of biological samples are necessary to perform large-scale clinical studies.42 The throughput of MALDI-TOF MS is superior to all other glyco-analytical techniques, and it is followed by CE-LIF and HPLC-FLD.39, 43, 85, 90 See Table 1 for an abbreviated comparison.
To assess the throughput of profiling human plasma IgG N-glycosylation of 1201 individuals, four platforms were compared: MALDI-TOF MS, LC-ESI-MS, and two spectroscopic approaches.85 MS analysis was performed on purified tryptic glycopeptides, while non-mass spectrometric methods, UPLC-HILIC-FLR and multiplex capillary gel electrophoresis-laser induced fluorescence detection (xCGE-LIF), were performed on 2-AB and APTS labeled, released N-linked glycans, respectively.85 MALDI-TOF MS proved to be far superior, while LC-ESI-MS was the slowest. In another example, HILIC-UHPLC-FLD, xCGE-LIF and MALDI-TOF MS approaches were compared for identifying the serum N-glycome changes associated with rheumatoid arthritis and pregnancy. Again, MALDI-TOF MS sample throughput outperformed the spectroscopic methods.43, 85 These studies showed that 96–384 samples could be analyzed by MALDI within a single run, providing the measurement of a sample at a sub-minute time scale.85, 101
Apart from MALDI-TOF MS, CGE-LIF, when multiplexed, proves to be the method with the second-best throughput. It allows for the analysis of thousands of samples within a day.43, 85 The typical run time for either the CGE-LIF or HPLC-FLD is approximately in 40 – 60 min range, but once CGE is multiplexed with up to 96 capillaries in parallel, the required analysis time per sample can be reduced to the low minute scale.43 As compared with CGE-LIF, the throughput of conventional CE-LIF is lower as it lacks multiplexing ability, but the typical run time is generally lower than both CGE-LIF and UPLC-FLR.
UPLC-FLR and LC-ESI-MS show medium throughput;85 the throughput is limited by the front-end gradient time. For example, one study quantified total plasma N-linked glycan profiles obtained from 2396 individuals by using an HPLC-FLD. The reported total analysis time was 106 min per sample.44
While LC-ESI-MS/MS analysis of glycans is one of the slowest methods, sample throughput can be improved by using multiplexing agents, such as tandem mass tags (TMT), isobaric labels,66 and stable isotopic labels.64 Introducing multiplexing agents is useful not only for improving the reliability of the quantitation, but also for increasing the number of samples that can be analyzed within a single LC-MS run, while lowering the analysis time per sample.66 Recently, sixplex AminoxyTMT mass tags were used by Merchef and co-workers66 to reliably quantify serum N-linked glycans derived from individuals with various esophageal diseases. They quantified 44 glycans after labeling them with TMT sixplex reagents, followed by glycan separation on a PGC column and analysis with nano-LC-ESI-MS/MS.66 In addition, a recent study performed by Li and coworkers102 presented mass-defect-based, duplex-dimethyl pyrimidinyl ornithine (DiPyrO) tags with a mass difference of 45.3 mDa at the MS1 level; these tags were used to quantify N-glycome profile differences of human serum samples derived from cancer patients before and after chemotherapy; the study permitted quantification of 36 glycans, presented at relatively high abundance in the control samples as compared to the samples collected after chemotherapy.102
4.3. Sample Preparation Time
Sample preparation is required for all these methods because of the complexity of the sample matrix and the heterogeneity within the sample. Typically, on a glycoprotein, a variety of glycans can be attached to either a single glycosylation site or to multiple glycosylation sites found on the peptide backbone. This heterogeneity results many different protein glycoforms, which are usually in low abundance compared to the non-glycosylated proteins present in the biological matrix.2–3 Therefore, efficient glycoprotein purification and separation at the crude-mixture level or the glycan/glycopeptide level is vital in glycomics analysis, as any contaminant present in the sample can affect the detection sensitivity, reproducibility, and relative glycan quantitation.84 Therefore, many improvements in glycoprotein purification, sample preparation, including release of N-linked glycans, glycan enrichment, and labeling, have been reported in the literature; these methods are described in sections 3.1.1, 3.1.2, 3.1.3 and 3.2.1. However, the complexity of these multiple glycan/glycopeptide processing steps directly affects the total sample preparation time of the analysis, making it difficult to perform large-scale clinical studies on disease-related glycan biomarkers.88
The throughput of preparing samples for HPLC-FLD, CE-LIF and MALDI-TOF MS analysis of released N-linked glycans is quite similar.43 All of these methods include glycoprotein enrichment, enzymatic N-linked glycan release performed overnight, glycan derivatization, and detection of purified glycans. However, the sample preparation throughput of MALDI-TOF MS currently surpasses the non-MS based methods. This is well-evidenced by two studies performed by Shubhakar et al.50 and Bladergroen et al.;103 they have presented high-throughput, clinically-feasible, automated sample preparation for MALDI-TOF MS analysis; these automated protocols allowed 96 clinical samples to be processed and detected within about 7 h for permethylated samples 50 and 3.5 h for samples with sialic acids esterified.103 During these studies, the sample preparation workflow was expedited through introducing robotic liquid handling systems, which significantly reduced the sample preparation time for the analysis. On the other hand, automation of non-MS-based sample preparation, for example HPLC-FLD, has been also reported; one of these studies reduced the sample processing time for 96 2-AB labeled samples from 72 h87 to 22 h84 by introducing an efficient way to conduct glycan release and derivatization in an automated fashion. Another study done by the same group96 reported processing of 100 samples within approximately 14 hours, by using the same method, but with some modifications that permitted whole serum glycan analysis.
Compared to MALDI-TOF MS and fluorescence-based methods, less effort has been directed at speeding up sample preparation for LC-ESI-MS of released glycans and LC-MS of glycopeptides. In principle, sample preparation for LC-ESI-MS analysis of glycans would be approximately equivalent to that of MALDI-TOF MS, since the same types of analytes are studied. In contrast to analyzing released glycans, LC-MS analysis of glycopeptides requires different sample preparation steps that must be done in advance: glycoprotein enrichment, denaturation, reduction, alkylation, and enzymatic digestion. The sample preparation time consumes about 3 hours prior to the enzymatic digestion; enzymatic digestion is typically performed overnight.32 The time allotted to the LC-MS/MS analysis can vary from 20 min34, 81 to a few hours,41 which reduces the analytical throughput of the method.
4.4. Number of Structures Identified
Identification of many unique glycan/glycopeptide species present in biological samples is important in biomarker research: the more structures quantified, the greater the likelihood that researchers will be able to identify glycans whose abundance changes with the disease sate.
Among different MS-based methods, LC-MS analysis of glycopeptides is capable of identifying the highest number of analytes per analysis. Glycopeptides have diversity in both the peptide and glycan portions, resulting in a larger number of possible glycopeptides being present than if the glycans alone were analyzed. One recent study quantified more than 600 glycopeptides across over 50 serum glycoproteins by implementing a dynamic multiple-reaction monitoring (dMRM) method optimized in a UHPLC-QqQ; the study permitted quantitation of sialylated and fucosylated glycans, in addition to low abundant high mannose-type glycans.41 In another study, Kazuhiro et al.40 identified more than 30 000 AAL-affinity-enriched glycopeptides derived from serum samples of hepatocellular carcinoma (HCC) patients, chronic hepatitis patients, and healthy controls via LC-TOF-MS while allowing the identification of multifucosylated glycans of alpha-1-acid glycoprotein (AGP), as candidate HCC biomarkers.
LC-ESI-MS analysis of released N-linked glycans, provides the second-best coverage of glycosylated analytes. Among many studies where a higher number of glycan identifications were reported, PGC-chip-based separation was used. Song et al.67 performed an analysis on reduced serum N-linked glycans and identified more than 170 N-linked glycan structures, including complete (50) and partially elucidated (100) structures that were included in a representative library for serum. Moreover, in another study, out of 115 glycan structures identified, 29 were altered in lung adenocarcinoma tissue samples as compared to the non-malignant tissues.25
Among all MS-based methods, MALDI-TOF MS shows the lowest coverage of unique glycans. However, compared to fluorescence-based methods, MALDI-TOF MS is capable of assigning more glycan compositions, and it provides good separation for more complex tri- and tetra-antennary glycan structures.43
In fluorescence-based methods, the number of unique species detected is limited43, 63, 104 and the assignment of each individual analyte peak requires prior knowledge about the retention time or the migration time of the species being analyzed. Table 1 summarizes the different methods’ capacities.
4.5. Isomer Separation and Structural Characterization Ability
Glycomics analysis is complicated because of the structural diversity introduced by different glycan compositions, linkages, and branching patterns.4–5 Accurate identification of numerous glycan compositions, and in-depth structural characterization of different glycans or glycopeptides structures, including isomers, is particularly important when the goal is to identify the glycan signatures that change during disease progression. As indicated in Table 1, LC-MS-based methods are thus far the most informative for structural assignment of glycans/glycopeptides; however, both tandem MS techniques and optimized separation strategies are required to perform isomer separation and characterization.5
Considerable research has been invested into achieving isomeric separation for released glycans, followed by characterization by tandem mass spectrometry. Porous graphitized carbon (PGC),4, 105–106 hydrophilic-interaction liquid chromatography (HILIC),107 and reversed-phase (RP)-LC69 are potential choices for the stationary phases for isomer separation, while tandem MS techniques, such as collision induced dissociation (CID)67, 69, 105–106 and higher energy collision dissociation (HCD) 97–98 are main choices for glycans’ structural characterization. Two recent studies performed by Yehia Mechref and coworkers,105–106 used a PGC-nLC-MS/MS method performed at higher temperature (75 oC), to effectively separate and characterize permethylated glycans derived from multiple cancer cell lines. This study allowed efficient separation of glycan structures including many different glycan isomers, such as monosaccharide positional isomers (core- or branched-fucose and α3- or α6-branched galactose) and linkage isomers; these structures were then effectively identified by using specific diagnostic fragment ions or by comparing their intensity distributions observed in the resulting tandem MS spectra to that of the standards. Overall, these studies allowed identification of more than 100 glycan isomer structures derived from less than 50 glycan compositions. Apart from the frequently used PGC-nLC-MS/MS, RP-nLC-MS/MS is also used for glycan isomer separation and characterization; in one example, permethylated N-linked glycans from HCC patients were characterized, and 82 potential isomeric glycans from 52 glycan compositions were identified.69 However, use of RP-LC for N-linked glycan isomer separation is limited due to the poor resolution observed for permethylated isomeric glycans, and the poor retention observed for hydrophilic glycans.105 HILIC is also used to separate N-linked glycan isomers, for an example, linkage isomers of ProA-labeled sialylated glycans.107
Isomer separation at the glycopeptide level is also important, as it permits the quantitation of site-specific isomeric glycan alterations. Generally, RP-LC is the method of choice for glycopeptide separation; it successfully separates multiple glycopetides with different peptide backbones, but it poorly resolves the glycan isomers that have a common peptide backbone.108–109 Therefore, RP-LC has been used limitedly in glycopeptides’ isomer-specific studies; one such example is the study performed by Yuan et al.;47 they used RP-nLC-MRM-MS to quantify linkage-specific fucosylation differences of N-linked glycopeptides of seven plasma-derived glycoproteins of liver cirrhosis patients. These authors used outer arm fucosylation-specific fragment ion(s) in the tandem MS spectra for targeted transitions; they found that increased outer arm fucosylation is related to the progression of disease.
Instead of reverse phase chromatography, HILIC separation obtained significant attention in recent years because of its glycopeptide isomer separation ability.108–109 Two recent studies performed by Huang et al.108 and van der Burgt et al.109 used HILIC-LC in combined with targeted MS approaches to separate glycopeptide isomers of human IgG108 and prostate-specific antigen (PSA);109 both of these studies allowed differentiation of linkage-specific sialylated glycopeptides and also resolved galactose position of G1F glycan of IgG glycopeptides.108
While glycopeptide-based analysis provides site-specific information, it typically provides poor isomer separation ability and limited glycan-specific structural information compared to LC-ESI-MS analysis of released N-linked glycans. Therefore, if the goal of the study is to obtain comprehensive structural information of various glycans, the best choice would be LC-ESI-MS of released glycans, which enables effective separation and in-detail characterization of glycan structures including many different isomers.
As compared to LC-MS-based methods discussed in this review, MALDI-TOF MS lacks the ability to separate glycan isomers, as the method is not supported by front-end glycan separation. As a result, it typically provides glycans’ compositional assignments but not isomer-specific information.43 However, MALDI-TOF MS by itself permits sialic acid linkage differentiation when the sialic acids are subjected to linkage-specific derivatization prior to the analysis.38, 43, 110 For an example, Reiding et al.38 identified 77 plasma N-linked glycan compositions belonging to 108 glycan structures, after subjecting sialic acid α−2,6 and α- 2,3 linkages to ethyl esterification and lactonization, respectively. In another study, MALDI-TOF-MS was used to identify differences in sialylation linkages of ethyl esterified serum N-linked glycans derived from the samples of normal pregnant individuals and those with rheumatoid arthritis.43 Moreover, though MALDI-TOF-MS is useful for assigning different glycan compositions, structural elucidation of those compositions requires additional tandem capabilities.
Many studies have compared the number of glycans detected byf MALDI-TOF MS vs. non-MS-based approaches to demonstrate that other methods are generally more appropriate for resolving isomers. Among various methods discussed in this review, CE-LIF and UPLC-FLR methods also allow for effective separation of N-linked glycans while permitting branch-specific information and separation of various isomers.43, 85, 90 HILIC-FLR and CGE-LIF methods are able to distinguish between the 3-arm and 6-arm galactosylation differences in addition to the fucose position (core- or branched-) of fucosylated glycans.43, 85 By contrast, during these studies, MALDI-TOF MS analysis was not able to provide isomer-specific information for these glycans.
Fluorescence is not a method that is well-suited to provide structural information about the glycans in a sample, as it primarily is used for quantitation. When using fluorescence to quantify glycans, other methods or tools need to be paired with it if information about the glycan is needed. For example, well-characterized glycan standards can be used to match retention times in LC-fluorescence analyses, or additional follow-up MS experiments44, 63, 92 could be done, or sequential enzymatic digestion can be used 39, 42–43, 92 to obtain structural information. These additional methods, which need to be done in conjunction with fluorescence-based quantification, introduce limitations when one of the researchers’ goals is to identify the structure(s) of the glycan(s) that interest them.
4.6. Differences in Quantitative Data Generation
MS-based approaches used in glycomic quantitation are more complex than LC/ CE-fluorescence-based methods. In MS-based methods, the glycan response (peak abundances) are affected by both structural composition of the glycans as well as the co-eluting interferences that suppress the ionization of glycans or glycopeptides (in LC-MS based approaches). Therefore, the absolute signal of underivatized glycans or glycopeptides does not directly correlate to the glycan’s abundance in the sample. Relative quantitation, or comparing ratios of glycans within a sample, is a more reliable quantitative method. Some labeling agents correct for these differences in ionization efficiency by dramatically improving the ionization of all the glycans to which they attach. In these cases, the intensity of the MS signals of the derivatized glycans are comparable to the fluorescence intensity of the same derivatized glycans.111 It should be noted, however, that heavily sialylated or sulfated glycans may not ionize equivalently to neutral glycans, even with these derivatizing agents attached. In LC/CE-fluorescence methods, glycan labeling is stoichiometric and is not affected by the nature of the glycan type or composition. In these methods, the fluorescent dye is attached only to the reducing end of the glycan, and none of the structural differences of N-linked glycans are found at this end.42 Therefore, researchers typically assume that all the labeled N-linked glycans fluoresce with a similar quantum yield, while allowing reliable quantitation of glycan peak areas in the same sample and between samples.42, 85 This aspect makes LC/CE-fluorescence based methods, or MS methods that rely on the same derivatizing agents, preferable if the research study requires that the relative quantities of glycans within a sample be measured accurately.
4.7. Method Precision
Method precision is an important factor that needs to be considered during quantitative clinical glycomics studies where many sample sets are being analyzed. When the method is highly reproducible, it permits improved sensitivity, thus, allowing for differentiation of minor changes occurring in multiple samples.34
Many studies show that the repeatability of LC-FLD analysis of released N-linked glycans is superior to all other analytical methods.43–44, 84 Typically, HPLC-FLD yields lower than 10% coefficient of variation (CV), especially for major glycan peaks of the sample,43–44, 84 and even a lower CV value (1.6%) was reported for the 10 most abundant N-linked glycans derived from plasma samples of RA patients.43 CE-LIF analysis of glycans is also highly reproducible, but it is second to the LC-FLD method.31, 43
In MALDI-TOF MS, the repeatability is affected by not only the variation of the analyte ionization but also the spot-to-spot variation of the laser pulse. Therefore, compared to the non-MS based methods, MALDI-TOF MS analysis reported the least precise glycan quantitation data in many studies;43, 103 these studies showed that the precision of the analysis can be improved when quantifying glycan-derived traits instead of individual glycan peaks. Using traits, instead of peaks, can improve quantitative studies of non-MALDI-based methods as well.
Similar to MALDI-TOF MS, other MS-based methods also show lower repeatability as a result of both LC run-to-run variation and the ionization differences that occur during the MS analysis.66 Throughout the literature, the use of MS-based methods with sufficient repeatability (with Coefficient of Variation, CV, of <15%) for the quantitative clinical glycomics analysis have been reported for both LC-MS analysis of glycopeptides and released N-linked glycans. Lebrilla and colleagues34, 41 and Shih et al.73 reported a less than 15% intra-day and inter-day repeatability for serum glycopeptide quantitation. Similarly, for LC-ESI-MS N-linked glycan quantitation, sufficient repeatability was reported in many studies with lower run-to-run variation and over multiple sample preparations.66–67
4.8. Required Expertise
MS-based methods typically require higher expertise compared to non-MS based methods; both the operation of the mass spectrometer and the more complex data analysis require significant experience.43, 85 Among the MS-based methods, MALDI-TOF MS is the most straightforward, but for LC-ESI-MS analysis of both glycopeptides and released N-linked glycans, the required expertise is very high; researchers not only have to have a solid command of mass spectrometry, but also HPLC separation. Additionally, ESI data is often more complicated to analyze, especially if it is from glycopeptides. In contrast to MS-based methods, the primary skill necessary to perform UPLC-FLR and CE-LIF methods is expertise in separations. While these methods also require training for sample preparation and instrument handling, well-established glycan preparation protocols are available; a straight-forward detection method and well-established data bases also simplify data analysis for fluorescence-based methods. 43, 84–85
4.9. Cost for Instrumentation and per Sample
Typically, the instrumentation cost for high-resolution LC-ESI-MS is higher than MALDI-TOF MS, followed by the UPLC-FLR, and the cost for the CE-LIF instrumentation is the lowest. In terms of costs per sample, when the analysis is performed in high-throughput mode, both CE-LIF and MALDI-TOF MS provide low costs per sample, as they are the highest-throughput methods. In contrast, UPLC-FLD can be rather expensive, due to the low throughput of the method, and LC-ESI-MS provides the highest cost per sample as a result of the cost associated with the instrumentation as well the low throughput of the method.85
5. CONCLUSIONS
Recent advances in quantitative glycomics analytical methods, along with efficient glycoprotein purification, glycan labeling, and separation, allow successful application of these analyses in quantitative clinical assays. These different techniques exhibit their own strengths, while showing their utility in quantifying glycosylation differences of challenging sets of clinical samples. Among the methods described in this review, MALDI-TOF MS provides the highest sample throughput; LC-ESI-MS of released glycans is the best method for isomer separation, and LC/CE-fluorescence permits superior repeatability and simplicity; finally, LC-MS analysis of glycopeptides, the most complicated method, allows for the largest number of species to be detected.
There is no “one best method” that can be applied to quantify protein glycosylation of any given set of clinical samples, and the selection of an appropriate method depends on the specifics of the experiment. Therefore, Table 1 and the additional information included in the review provide guidance for selecting the best approach for a variety of circumstances.
ACKNOWLEDGEMENTS
The authors acknowledge funding from the National Institute of Health through grant R35GM130354 to H.D.
Biographies

Milani Wijeweera Patabandige received her Ph.D. from the Department of Chemistry at the University of Kansas in 2020. During her graduate studies, she developed liquid chromatography-mass spectrometry (LC-MS)-based methods for glycopeptide and glycan analysis. Currently, she works as a post-doctoral fellow at the Center for Devices and Radiological Health of the U.S. Food and Drug Administration in Maryland developing efficient, solvent-less extraction approaches to detect volatile extractables derived from polymeric medical devices by using GC-MS.

Leah Danielle Pfeifer is currently a third-year graduate student pursuing a Ph.D. in chemistry at the University of Kansas. She holds a Bachelor of Science Degree in Chemistry (2018) from Creighton University in Omaha, Nebraska. Her research involves quantifying lipid changes in different biological states using mass spectrometry data and machine learning.

Hanna T. Nguyen received her B. S. degree in Chemistry from Creighton University and subsequently worked for a few years at an agricultural/environmental lab and a pharmaceutical company before coming to the University of Kansas. She is currently a graduate research assistant in the Department of Chemistry. Her research is in bioanalytical chemistry, focusing on glycomic analysis using fluorescence detection.

Heather Desaire received her BA degree in Chemistry from Grinnell College, Grinnell IA (1997), and her Ph.D. from the University of California Berkeley (2001). After a short stint in industry, she began her academic career at the University of Kansas in 2002. Her research focuses on the intersection of mass spectrometry, glycobiology, and data science.
Footnotes
DISCLOSURE STATEMENT
The authors declare no competing interests.
LITERATURE CITED
- 1.Cho BG; Veillon L; Mechref Y, N-glycan profile of cerebrospinal fluids from Alzheimer’s Disease patients using LC-MS. J Proteome Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalpathado DS; Desaire H, Glycopeptide analysis by mass spectrometry. The Analyst 2008, 133 (6), 731–8. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Z; Desaire H, Carbohydrates on proteins: site-specific glycosylation analysis by mass spectrometry. Annual Review of Analytical Chemistry 2015, 8, 463–483. [DOI] [PubMed] [Google Scholar]
- 4.Hua S; An HJ; Ozcan S; Ro GS; Soares S; DeVere-White R; Lebrilla CB, Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. The Analyst 2011, 136 (18), 3663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veillon L; Huang Y; Peng W; Dong X; Cho BG; Mechref Y, Characterization of isomeric glycan structures by LC-MS/MS. Electrophoresis 2017, 38 (17), 2100–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahuja KK, Carbohydrate determinants involved in mammalian fertilization. The American journal of anatomy 1985, 174 (3), 207–23. [DOI] [PubMed] [Google Scholar]
- 7.Gupta SK; Bhandari B; Shrestha A; Biswal BK; Palaniappan C; Malhotra SS; Gupta N, Mammalian zona pellucida glycoproteins: structure and function during fertilization. Cell and tissue research 2012, 349 (3), 665–78. [DOI] [PubMed] [Google Scholar]
- 8.Glozman R; Okiyoneda T; Mulvihill CM; Rini JM; Barriere H; Lukacs GL, N-glycans are direct determinants of CFTR folding and stability in secretory and endocytic membrane traffic. The Journal of cell biology 2009, 184 (6), 847–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson SR; Culyba EK; Hsu TL; Wong CH; Kelly JW; Powers ET, The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proceedings of the National Academy of Sciences of the United States of America 2009, 106 (9), 3131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hang Q; Isaji T; Hou S; Wang Y; Fukuda T; Gu J, A Key Regulator of Cell Adhesion: Identification and Characterization of Important N-Glycosylation Sites on Integrin alpha5 for Cell Migration. Molecular and cellular biology 2017, 37 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsiao CT; Cheng HW; Huang CM; Li HR; Ou MH; Huang JR; Khoo KH; Yu HW; Chen YQ; Wang YK; Chiou A; Kuo JC, Fibronectin in cell adhesion and migration via N-glycosylation. Oncotarget 2017, 8 (41), 70653–70668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zabczynska M; Pochec E, [The role of protein glycosylation in immune system]. Postepy biochemii 2015, 61 (2), 129–37. [PubMed] [Google Scholar]
- 13.Bosques CJ; Collins BE; Meador JW 3rd; Sarvaiya H; Murphy JL; Dellorusso G; Bulik DA; Hsu IH; Washburn N; Sipsey SF; Myette JR; Raman R; Shriver Z; Sasisekharan R; Venkataraman G, Chinese hamster ovary cells can produce galactose-alpha-1,3-galactose antigens on proteins. Nature biotechnology 2010, 28 (11), 1153–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shubhakar A; Reiding KR; Gardner RA; Spencer DI; Fernandes DL; Wuhrer M, High-Throughput Analysis and Automation for Glycomics Studies. Chromatographia 2015, 78 (5–6), 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alessandri L; Ouellette D; Acquah A; Rieser M; Leblond D; Saltarelli M; Radziejewski C; Fujimori T; Correia I, Increased serum clearance of oligomannose species present on a human IgG1 molecule. mAbs 2012, 4 (4), 509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetze AM; Liu YD; Zhang Z; Shah B; Lee E; Bondarenko PV; Flynn GC, High-mannose glycans on the Fc region of therapeutic IgG antibodies increase serum clearance in humans. Glycobiology 2011, 21 (7), 949–59. [DOI] [PubMed] [Google Scholar]
- 17.Ju L; Wang Y; Xie Q; Xu X; Li Y; Chen Z; Li Y, Elevated level of serum glycoprotein bifucosylation and prognostic value in Chinese breast cancer. Glycobiology 2016, 26 (5), 460–71. [DOI] [PubMed] [Google Scholar]
- 18.Kamiyama T; Yokoo H; Furukawa J; Kurogochi M; Togashi T; Miura N; Nakanishi K; Kamachi H; Kakisaka T; Tsuruga Y; Fujiyoshi M; Taketomi A; Nishimura S; Todo S, Identification of novel serum biomarkers of hepatocellular carcinoma using glycomic analysis. Hepatology (Baltimore, Md.) 2013, 57 (6), 2314–25. [DOI] [PubMed] [Google Scholar]
- 19.Kyselova Z; Mechref Y; Kang P; Goetz JA; Dobrolecki LE; Sledge GW; Schnaper L; Hickey RJ; Malkas LH; Novotny MV, Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clinical chemistry 2008, 54 (7), 1166–75. [DOI] [PubMed] [Google Scholar]
- 20.Argade S; Chen T; Shaw T; Berecz Z; Shi W; Choudhury B; Parsons CL; Sur RL, An evaluation of Tamm-Horsfall protein glycans in kidney stone formers using novel techniques. Urolithiasis 2015, 43 (4), 303–12. [DOI] [PubMed] [Google Scholar]
- 21.Chen J; Li X; Edmondson A; Meyers GD; Izumi K; Ackermann AM; Morava E; Ficicioglu C; Bennett MJ; He M, Increased Clinical Sensitivity and Specificity of Plasma Protein N-Glycan Profiling for Diagnosing Congenital Disorders of Glycosylation by Use of Flow Injection-Electrospray Ionization-Quadrupole Time-of-Flight Mass Spectrometry. Clinical chemistry 2019, 65 (5), 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong X; Huang Y; Cho BG; Zhong J; Gautam S; Peng W; Williamson SD; Banazadeh A; Torres-Ulloa KY; Mechref Y, Advances in mass spectrometry-based glycomics. Electrophoresis 2018, 39 (24), 3063–3081. [DOI] [PubMed] [Google Scholar]
- 23.Liu S; Cheng L; Fu Y; Liu BF; Liu X, Characterization of IgG N-glycome profile in colorectal cancer progression by MALDI-TOF-MS. Journal of proteomics 2018, 181, 225–237. [DOI] [PubMed] [Google Scholar]
- 24.Ruhaak LR; Barkauskas DA; Torres J; Cooke CL; Wu LD; Stroble C; Ozcan S; Williams CC; Camorlinga M; Rocke DM; Lebrilla CB; Solnick JV, The Serum Immunoglobulin G Glycosylation Signature of Gastric Cancer. EuPA open proteomics 2015, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruhaak LR; Taylor SL; Stroble C; Nguyen UT; Parker EA; Song T; Lebrilla CB; Rom WN; Pass H; Kim K; Kelly K; Miyamoto S, Differential N-Glycosylation Patterns in Lung Adenocarcinoma Tissue. J Proteome Res 2015, 14 (11), 4538–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwedler C; Kaup M; Weiz S; Hoppe M; Braicu EI; Sehouli J; Hoppe B; Tauber R; Berger M; Blanchard V, Identification of 34 N-glycan isomers in human serum by capillary electrophoresis coupled with laser-induced fluorescence allows improving glycan biomarker discovery. Analytical and bioanalytical chemistry 2014, 406 (28), 7185–93. [DOI] [PubMed] [Google Scholar]
- 27.Su Z; Xie Q; Wang Y; Li Y, Abberant Immunoglobulin G Glycosylation in Rheumatoid Arthritis by LTQ-ESI-MS. International journal of molecular sciences 2020, 21 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng W; Zhao J; Dong X; Banazadeh A; Huang Y; Hussien A; Mechref Y, Clinical application of quantitative glycomics. Expert review of proteomics 2018, 15 (12), 1007–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H; Zhang N; Wan D; Cui M; Liu Z; Liu S, Mass spectrometry-based analysis of glycoproteins and its clinical applications in cancer biomarker discovery. Clinical proteomics 2014, 11 (1), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong Q; Lebrilla CB; Miyamoto S; Ruhaak LR, Absolute quantitation of immunoglobulin G and its glycoforms using multiple reaction monitoring. Anal Chem 2013, 85 (18), 8585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruhaak LR; Koeleman CA; Uh HW; Stam JC; van Heemst D; Maier AB; Houwing-Duistermaat JJ; Hensbergen PJ; Slagboom PE; Deelder AM; Wuhrer M, Targeted biomarker discovery by high throughput glycosylation profiling of human plasma alpha1-antitrypsin and immunoglobulin A. PloS one 2013, 8 (9), e73082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J; Chen Z; Zhang J; An M; Wu J; Yu Q; Skilton SJ; Bern M; Ilker Sen K; Li L; Lubman DM, Differential Quantitative Determination of Site-Specific Intact N-Glycopeptides in Serum Haptoglobin between Hepatocellular Carcinoma and Cirrhosis Using LC-EThcD-MS/MS. J Proteome Res 2019, 18 (1), 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X; Wang Y; Qian Y; Wu X; Zhang Z; Liu X; Zhao R; Zhou L; Ruan Y; Xu J; Liu H; Ren S; Xu C; Gu J, Discovery of specific metastasis-related N-glycan alterations in epithelial ovarian cancer based on quantitative glycomics. PloS one 2014, 9 (2), e87978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong Q; Ruhaak LR; Stroble C; Parker E; Huang J; Maverakis E; Lebrilla CB, A Method for Comprehensive Glycosite-Mapping and Direct Quantitation of Serum Glycoproteins. J Proteome Res 2015, 14 (12), 5179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kammeijer GSM; Nouta J; de la Rosette J; de Reijke TM; Wuhrer M, An In-Depth Glycosylation Assay for Urinary Prostate-Specific Antigen. Anal Chem 2018, 90 (7), 4414–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen IH; Aguilar HA; Paez Paez JS; Wu X; Pan L; Wendt MK; Iliuk AB; Zhang Y; Tao WA, Analytical Pipeline for Discovery and Verification of Glycoproteins from Plasma-Derived Extracellular Vesicles as Breast Cancer Biomarkers. Anal Chem 2018, 90 (10), 6307–6313. [DOI] [PubMed] [Google Scholar]
- 37.Jeong HJ; Kim YG; Yang YH; Kim BG, High-throughput quantitative analysis of total N-glycans by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem 2012, 84 (7), 3453–60. [DOI] [PubMed] [Google Scholar]
- 38.Reiding KR; Blank D; Kuijper DM; Deelder AM; Wuhrer M, High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal Chem 2014, 86 (12), 5784–93. [DOI] [PubMed] [Google Scholar]
- 39.Reiding KR; Ruhaak LR; Uh HW; El Bouhaddani S; van den Akker EB; Plomp R; McDonnell LA; Houwing-Duistermaat JJ; Slagboom PE; Beekman M; Wuhrer M, Human Plasma N-glycosylation as Analyzed by Matrix-Assisted Laser Desorption/Ionization-Fourier Transform Ion Cyclotron Resonance-MS Associates with Markers of Inflammation and Metabolic Health. Molecular & cellular proteomics : MCP 2017, 16 (2), 228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanabe K; Kitagawa K; Kojima N; Iijima S, Multifucosylated Alpha-1-acid Glycoprotein as a Novel Marker for Hepatocellular Carcinoma. J Proteome Res 2016, 15 (9), 2935–44. [DOI] [PubMed] [Google Scholar]
- 41.Li Q; Kailemia MJ; Merleev AA; Xu G; Serie D; Danan LM; Haj FG; Maverakis E; Lebrilla CB, Site-Specific Glycosylation Quantitation of 50 Serum Glycoproteins Enhanced by Predictive Glycopeptidomics for Improved Disease Biomarker Discovery. Anal Chem 2019, 91 (8), 5433–5445. [DOI] [PubMed] [Google Scholar]
- 42.Saldova R; Reuben JM; Abd Hamid UM; Rudd PM; Cristofanilli M, Levels of specific serum N-glycans identify breast cancer patients with higher circulating tumor cell counts. Annals of oncology : official journal of the European Society for Medical Oncology 2011, 22 (5), 1113–9. [DOI] [PubMed] [Google Scholar]
- 43.Reiding KR; Bondt A; Hennig R; Gardner RA; O’Flaherty R; Trbojevic-Akmacic I; Shubhakar A; Hazes JMW; Reichl U; Fernandes DL; Pucic-Bakovic M; Rapp E; Spencer DIR; Dolhain R; Rudd PM; Lauc G; Wuhrer M, High-throughput Serum N-Glycomics: Method Comparison and Application to Study Rheumatoid Arthritis and Pregnancy-associated Changes. Molecular & cellular proteomics : MCP 2019, 18 (1), 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruhaak LR; Uh HW; Beekman M; Hokke CH; Westendorp RG; Houwing-Duistermaat J; Wuhrer M; Deelder AM; Slagboom PE, Plasma protein N-glycan profiles are associated with calendar age, familial longevity and health. J Proteome Res 2011, 10 (4), 1667–74. [DOI] [PubMed] [Google Scholar]
- 45.Wang H; Chen X; Zhang X; Zhang W; Li Y; Yin H; Shao H; Chen G, Comparative Assessment of Glycosylation of a Recombinant Human FSH and a Highly Purified FSH Extracted from Human Urine. J Proteome Res 2016, 15 (3), 923–32. [DOI] [PubMed] [Google Scholar]
- 46.Tan Z; Yin H; Nie S; Lin Z; Zhu J; Ruffin MT; Anderson MA; Simeone DM; Lubman DM, Large-scale identification of core-fucosylated glycopeptide sites in pancreatic cancer serum using mass spectrometry. J Proteome Res 2015, 14 (4), 1968–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan W; Wei R; Goldman R; Sanda M, Optimized Fragmentation for Quantitative Analysis of Fucosylated N-Glycoproteins by LC-MS-MRM. Anal Chem 2019, 91 (14), 9206–9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen B; Zhong X; Feng Y; Snovida S; Xu M; Rogers J; Li L, Targeted MultiNotch MS(3) Approach for Relative Quantification of N-Glycans Using Multiplexed Carbonyl-Reactive Isobaric Tags. Anal Chem 2018, 90 (2), 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng WF; Liu MQ; Zhang Y; Wu JQ; Fang P; Peng C; Nie A; Yan G; Cao W; Liu C; Chi H; Sun RX; Wong CC; He SM; Yang P, pGlyco: a pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3. Scientific reports 2016, 6, 25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shubhakar A; Kozak RP; Reiding KR; Royle L; Spencer DI; Fernandes DL; Wuhrer M, Automated High-Throughput Permethylation for Glycosylation Analysis of Biologics Using MALDI-TOF-MS. Anal Chem 2016, 88 (17), 8562–9. [DOI] [PubMed] [Google Scholar]
- 51.Wei L; Cai Y; Yang L; Zhang Y; Lu H, Duplex Stable Isotope Labeling (DuSIL) for Simultaneous Quantitation and Distinction of Sialylated and Neutral N-Glycans by MALDI-MS. Analytical Chemistry 2018, 90 (17), 10442–10449. [DOI] [PubMed] [Google Scholar]
- 52.Zhou H; Warren PG; Froehlich JW; Lee RS, Dual Modifications Strategy to Quantify Neutral and Sialylated N-Glycans Simultaneously by MALDI-MS. Analytical Chemistry 2014, 86 (13), 6277–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C; Wu Y; Zhang L; Liu BF; Lin Y; Liu X, Relative quantitation of neutral and sialylated N-glycans using stable isotopic labeled d0/d5-benzoyl chloride by MALDI-MS. Analytica chimica acta 2018, 1002, 50–61. [DOI] [PubMed] [Google Scholar]
- 54.Wang L; Yang L; Zhang Y; Lu H, Dual isotopic labeling combined with fluorous solid-phase extraction for simultaneous discovery of neutral/sialylated N-glycans as biomarkers for gastric cancer. Analytica chimica acta 2020, 1104, 87–94. [DOI] [PubMed] [Google Scholar]
- 55.Feng Y; Li M; Lin Y; Chen B; Li L, Multiplex Quantitative Glycomics Enabled by Periodate Oxidation and Triplex Mass Defect Isobaric Multiplex Reagents for Carbonyl-Containing Compound Tags. Analytical Chemistry 2019, 91 (18), 11932–11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang L; Du X; Peng Y; Cai Y; Wei L; Zhang Y; Lu H, Integrated Pipeline of Isotopic Labeling and Selective Enriching for Quantitative Analysis of N-Glycome by Mass Spectrometry. Analytical Chemistry 2019, 91 (2), 1486–1493. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W; Wang H; Tang H; Yang P, Endoglycosidase-Mediated Incorporation of 18O into Glycans for Relative Glycan Quantitation. Analytical Chemistry 2011, 83 (12), 4975–4981. [DOI] [PubMed] [Google Scholar]
- 58.Alvarez-Manilla G; Warren NL; Abney T; Atwood J III; Azadi P; York WS; Pierce M; Orlando R, Tools for glycomics: relative quantitation of glycans by isotopic permethylation using 13CH3I. Glycobiology 2007, 17 (7), 677–687. [DOI] [PubMed] [Google Scholar]
- 59.Orlando R; Lim J-M; Atwood JA; Angel PM; Fang M; Aoki K; Alvarez-Manilla G; Moremen KW; York WS; Tiemeyer M; Pierce M; Dalton S; Wells L, IDAWG: Metabolic incorporation of stable isotope labels for quantitative glycomics of cultured cells. J Proteome Res 2009, 8 (8), 3816–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selman MH; Hoffmann M; Zauner G; McDonnell LA; Balog CI; Rapp E; Deelder AM; Wuhrer M, MALDI-TOF-MS analysis of sialylated glycans and glycopeptides using 4-chloro-alpha-cyanocinnamic acid matrix. Proteomics 2012, 12 (9), 1337–48. [DOI] [PubMed] [Google Scholar]
- 61.Smargiasso N; De Pauw E, Optimization of matrix conditions for the control of MALDI in-source decay of permethylated glycans. Anal Chem 2010, 82 (22), 9248–53. [DOI] [PubMed] [Google Scholar]
- 62.Miura Y; Hato M; Shinohara Y; Kuramoto H; Furukawa J; Kurogochi M; Shimaoka H; Tada M; Nakanishi K; Ozaki M; Todo S; Nishimura S, BlotGlycoABCTM, an integrated glycoblotting technique for rapid and large scale clinical glycomics. Molecular & cellular proteomics : MCP 2008, 7 (2), 370–7. [DOI] [PubMed] [Google Scholar]
- 63.Mitra I; Snyder CM; Zhou X; Campos MI; Alley WR Jr.; Novotny MV; Jacobson SC, Structural Characterization of Serum N-Glycans by Methylamidation, Fluorescent Labeling, and Analysis by Microchip Electrophoresis. Anal Chem 2016, 88 (18), 8965–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong X; Peng W; Yu CY; Zhou S; Donohoo KB; Tang H; Mechref Y, 8-plex LC-MS/MS Analysis of Permethylated N-Glycans Achieved by Using Stable Isotopic Iodomethane. Anal Chem 2019, 91 (18), 11794–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang S; Wang M; Chen L; Yin B; Song G; Turko IV; Phinney KW; Betenbaugh MJ; Zhang H; Li S, QUANTITY: An Isobaric Tag for Quantitative Glycomics. Scientific reports 2015, 5, 17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou S; Hu Y; Veillon L; Snovida SI; Rogers JC; Saba J; Mechref Y, Quantitative LC-MS/MS Glycomic Analysis of Biological Samples Using AminoxyTMT. Anal Chem 2016, 88 (15), 7515–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song T; Aldredge D; Lebrilla CB, A Method for In-Depth Structural Annotation of Human Serum Glycans That Yields Biological Variations. Anal Chem 2015, 87 (15), 7754–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou S; Hu Y; DeSantos-Garcia JL; Mechref Y, Quantitation of permethylated N-glycans through multiple-reaction monitoring (MRM) LC-MS/MS. Journal of the American Society for Mass Spectrometry 2015, 26 (4), 596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai TH; Wang M; Di Poto C; Hu Y; Zhou S; Zhao Y; Varghese RS; Luo Y; Tadesse MG; Ziada DH; Desai CS; Shetty K; Mechref Y; Ressom HW, LC-MS profiling of N-Glycans derived from human serum samples for biomarker discovery in hepatocellular carcinoma. J Proteome Res 2014, 13 (11), 4859–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song E; Zhu R; Hammoud ZT; Mechref Y, LC-MS/MS quantitation of esophagus disease blood serum glycoproteins by enrichment with hydrazide chemistry and lectin affinity chromatography. J Proteome Res 2014, 13 (11), 4808–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim JY; Oh D; Kim SK; Kang D; Moon MH, Isotope-coded carbamidomethylation for quantification of N-glycoproteins with online microbore hollow fiber enzyme reactor-nanoflow liquid chromatography-tandem mass spectrometry. Anal Chem 2014, 86 (15), 7650–7. [DOI] [PubMed] [Google Scholar]
- 72.Miyamoto S; Stroble CD; Taylor S; Hong Q; Lebrilla CB; Leiserowitz GS; Kim K; Ruhaak LR, Multiple Reaction Monitoring for the Quantitation of Serum Protein Glycosylation Profiles: Application to Ovarian Cancer. J Proteome Res 2018, 17 (1), 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shih HC; Chang MC; Chen CH; Tsai IL; Wang SY; Kuo YP; Chen CH; Chang YT, High accuracy differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma by immunoglobulin G glycosylation. Clinical proteomics 2019, 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia X; Chen J; Sun S; Yang W; Yang S; Shah P; Hoti N; Veltri B; Zhang H, Detection of aggressive prostate cancer associated glycoproteins in urine using glycoproteomics and mass spectrometry. Proteomics 2016, 16 (23), 2989–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lundstrom SL; Yang H; Lyutvinskiy Y; Rutishauser D; Herukka SK; Soininen H; Zubarev RA, Blood plasma IgG Fc glycans are significantly altered in Alzheimer’s disease and progressive mild cognitive impairment. Journal of Alzheimer’s disease : JAD 2014, 38 (3), 567–79. [DOI] [PubMed] [Google Scholar]
- 76.Go EP; Cupo A; Ringe R; Pugach P; Moore JP; Desaire H, Native Conformation and Canonical Disulfide Bond Formation Are Interlinked Properties of HIV-1 Env Glycoproteins. Journal of Virology 2016, 90 (6), 2884–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Go EP; Ding H; Zhang S; Ringe RP; Nicely N; Hua D; Steinbock RT; Golabek M; Alin J; Alam SM, Glycosylation Benchmark Profile for HIV-1 Envelope Glycoprotein Production Based on Eleven Env Trimers. Journal of Virology 2017, 91 (9), e02428–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Go EP; Herschhorn A; Gu C; Castillo-Menendez L; Zhang S; Mao Y; Chen H; Ding H; Wakefield JK; Hua D; Liao H-X; Kappes JC; Sodroski J; Desaire H, Comparative Analysis of the Glycosylation Profiles of Membrane-Anchored HIV-1 Envelope Glycoprotein Trimers and Soluble gp140. Journal of Virology 2015, 89 (16), 8245–8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu W; Su X; Zhu Z; Go EP; Desaire H, GlycoPep MassList: software to generate massive inclusion lists for glycopeptide analyses. Analytical and bioanalytical chemistry 2017, 409 (2), 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lakbub JC; Su X; Zhu Z; Patabandige MW; Hua D; Go EP; Desaire H, Two New Tools for Glycopeptide Analysis Researchers: A Glycopeptide Decoy Generator and a Large Data Set of Assigned CID Spectra of Glycopeptides. J Proteome Res 2017, 16 (8), 3002–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruhaak LR; Kim K; Stroble C; Taylor SL; Hong Q; Miyamoto S; Lebrilla CB; Leiserowitz G, Protein-Specific Differential Glycosylation of Immunoglobulins in Serum of Ovarian Cancer Patients. J Proteome Res 2016, 15 (3), 1002–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rebecchi KR; Wenke JL; Go EP; Desaire H, Label-free quantitation: a new glycoproteomics approach. Journal of the American Society for Mass Spectrometry 2009, 20 (6), 1048–59. [DOI] [PubMed] [Google Scholar]
- 83.Hua D; Patabandige MW; Go EP; Desaire H, The Aristotle Classifier: Using the Whole Glycomic Profile To Indicate a Disease State. Anal Chem 2019, 91 (17), 11070–11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stockmann H; Adamczyk B; Hayes J; Rudd PM, Automated, high-throughput IgG-antibody glycoprofiling platform. Anal Chem 2013, 85 (18), 8841–9. [DOI] [PubMed] [Google Scholar]
- 85.Huffman JE; Pucic-Bakovic M; Klaric L; Hennig R; Selman MH; Vuckovic F; Novokmet M; Kristic J; Borowiak M; Muth T; Polasek O; Razdorov G; Gornik O; Plomp R; Theodoratou E; Wright AF; Rudan I; Hayward C; Campbell H; Deelder AM; Reichl U; Aulchenko YS; Rapp E; Wuhrer M; Lauc G, Comparative performance of four methods for high-throughput glycosylation analysis of immunoglobulin G in genetic and epidemiological research. Molecular & cellular proteomics : MCP 2014, 13 (6), 1598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Argade SP; Vanichsarn C; Chenoweth M; Parsons CL, Abnormal glycosylation of Tamm-Horsfall protein in patients with interstitial cystitis. BJU international 2009, 103 (8), 1085–9. [DOI] [PubMed] [Google Scholar]
- 87.Royle L; Campbell MP; Radcliffe CM; White DM; Harvey DJ; Abrahams JL; Kim YG; Henry GW; Shadick NA; Weinblatt ME; Lee DM; Rudd PM; Dwek RA, HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Analytical biochemistry 2008, 376 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- 88.Vanderschaeghe D; Meuris L; Raes T; Grootaert H; Van Hecke A; Verhelst X; Van de Velde F; Lapauw B; Van Vlierberghe H; Callewaert N, Endoglycosidase S Enables a Highly Simplified Clinical Chemistry Procedure for Direct Assessment of Serum IgG Undergalactosylation in Chronic Inflammatory Disease. Molecular & cellular proteomics : MCP 2018, 17 (12), 2508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vanderschaeghe D; Szekrenyes A; Wenz C; Gassmann M; Naik N; Bynum M; Yin H; Delanghe J; Guttman A; Callewaert N, High-throughput profiling of the serum N-glycome on capillary electrophoresis microfluidics systems: toward clinical implementation of GlycoHepatoTest. Anal Chem 2010, 82 (17), 7408–15. [DOI] [PubMed] [Google Scholar]
- 90.Reusch D; Haberger M; Maier B; Maier M; Kloseck R; Zimmermann B; Hook M; Szabo Z; Tep S; Wegstein J; Alt N; Bulau P; Wuhrer M, Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles--part 1: separation-based methods. mAbs 2015, 7 (1), 167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reusch D; Haberger M; Falck D; Peter B; Maier B; Gassner J; Hook M; Wagner K; Bonnington L; Bulau P; Wuhrer M, Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles-Part 2: Mass spectrometric methods. mAbs 2015, 7 (4), 732–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szabo Z; Guttman A; Rejtar T; Karger BL, Improved sample preparation method for glycan analysis of glycoproteins by CE-LIF and CE-MS. Electrophoresis 2010, 31 (8), 1389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gautam S; Peng W; Cho BG; Huang Y; Banazadeh A; Yu A; Dong X; Mechref Y, Glucose unit index (GUI) of permethylated glycans for effective identification of glycans and glycan isomers. The Analyst 2020, 145 (20), 6656–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Campbell MP; Royle L; Radcliffe CM; Dwek RA; Rudd PM, GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics (Oxford, England) 2008, 24 (9), 1214–6. [DOI] [PubMed] [Google Scholar]
- 95.Adamczyk B; Tharmalingam-Jaikaran T; Schomberg M; Szekrényes Á; Kelly RM; Karlsson NG; Guttman A; Rudd PM, Comparison of separation techniques for the elucidation of IgG N-glycans pooled from healthy mammalian species. Carbohydrate Research 2014, 389, 174–185. [DOI] [PubMed] [Google Scholar]
- 96.Stockmann H; O’Flaherty R; Adamczyk B; Saldova R; Rudd PM, Automated, high-throughput serum glycoprofiling platform. Integrative biology : quantitative biosciences from nano to macro 2015, 7 (9), 1026–32. [DOI] [PubMed] [Google Scholar]
- 97.Saldova R; Asadi Shehni A; Haakensen VD; Steinfeld I; Hilliard M; Kifer I; Helland Å; Yakhini Z; Børresen-Dale A-L; Rudd PM, Association of N-Glycosylation with Breast Carcinoma and Systemic Features Using High-Resolution Quantitative UPLC. J Proteome Res 2014, 13 (5), 2314–2327. [DOI] [PubMed] [Google Scholar]
- 98.Stumpo KA; Reinhold VN, The N-glycome of human plasma. J Proteome Res 2010, 9 (9), 4823–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clerc F; Reiding KR; Jansen BC; Kammeijer GS; Bondt A; Wuhrer M, Human plasma protein N-glycosylation. Glycoconjugate journal 2016, 33 (3), 309–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao W; Li H; Liu Y; Liu Y; Feng X; Liu BF; Liu X, Rapid and sensitive analysis of N-glycans by MALDI-MS using permanent charge derivatization and methylamidation. Talanta 2016, 161, 554–559. [DOI] [PubMed] [Google Scholar]
- 101.Parsons CL; Stein P; Zupkas P; Chenoweth M; Argade SP; Proctor JG; Datta A; Trotter RN, Defective Tamm-Horsfall protein in patients with interstitial cystitis. The Journal of urology 2007, 178 (6), 2665–70. [DOI] [PubMed] [Google Scholar]
- 102.Chen B; Feng Y; Frost DC; Zhong X; Buchberger AR; Johnson J; Xu M; Kim M; Puccetti D; Diamond C; Ikonomidou C; Li L, Quantitative Glycomic Analysis by Mass-Defect-Based Dimethyl Pyrimidinyl Ornithine (DiPyrO) Tags and High-Resolution Mass Spectrometry. Anal Chem 2018, 90 (13), 7817–7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bladergroen MR; Reiding KR; Hipgrave Ederveen AL; Vreeker GC; Clerc F; Holst S; Bondt A; Wuhrer M; van der Burgt YE, Automation of High-Throughput Mass Spectrometry-Based Plasma N-Glycome Analysis with Linkage-Specific Sialic Acid Esterification. J Proteome Res 2015, 14 (9), 4080–6. [DOI] [PubMed] [Google Scholar]
- 104.Lauc G; Huffman JE; Pucic M; Zgaga L; Adamczyk B; Muzinic A; Novokmet M; Polasek O; Gornik O; Kristic J; Keser T; Vitart V; Scheijen B; Uh HW; Molokhia M; Patrick AL; McKeigue P; Kolcic I; Lukic IK; Swann O; van Leeuwen FN; Ruhaak LR; Houwing-Duistermaat JJ; Slagboom PE; Beekman M; de Craen AJ; Deelder AM; Zeng Q; Wang W; Hastie ND; Gyllensten U; Wilson JF; Wuhrer M; Wright AF; Rudd PM; Hayward C; Aulchenko Y; Campbell H; Rudan I, Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS genetics 2013, 9 (1), e1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou S; Huang Y; Dong X; Peng W; Veillon L; Kitagawa DAS; Aquino AJA; Mechref Y, Isomeric Separation of Permethylated Glycans by Porous Graphitic Carbon (PGC)-LC-MS/MS at High Temperatures. Anal Chem 2017, 89 (12), 6590–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peng W; Goli M; Mirzaei P; Mechref Y, Revealing the Biological Attributes of N-Glycan Isomers in Breast Cancer Brain Metastasis Using Porous Graphitic Carbon (PGC) Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS). J Proteome Res 2019, 18 (10), 3731–3740. [DOI] [PubMed] [Google Scholar]
- 107.Tao S; Huang Y; Boyes BE; Orlando R, Liquid chromatography-selected reaction monitoring (LC-SRM) approach for the separation and quantitation of sialylated N-glycans linkage isomers. Anal Chem 2014, 86 (21), 10584–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang Y; Nie Y; Boyes B; Orlando R, Resolving Isomeric Glycopeptide Glycoforms with Hydrophilic Interaction Chromatography (HILIC). Journal of biomolecular techniques : JBT 2016, 27 (3), 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van der Burgt YEM; Siliakus KM; Cobbaert CM; Ruhaak LR, HILIC-MRM-MS for Linkage-Specific Separation of Sialylated Glycopeptides to Quantify Prostate-Specific Antigen Proteoforms. J Proteome Res 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li H; Gao W; Feng X; Liu BF; Liu X, MALDI-MS analysis of sialylated N-glycan linkage isomers using solid-phase two step derivatization method. Analytica chimica acta 2016, 924, 77–85. [DOI] [PubMed] [Google Scholar]
- 111.Keser T; Pavić T; Lauc G; Gornik O, Comparison of 2-Aminobenzamide, Procainamide and RapiFluor-MS as Derivatizing Agents for High-Throughput HILIC-UPLC-FLR-MS N-glycan Analysis. Frontiers in Chemistry 2018, 6 (324). [DOI] [PMC free article] [PubMed] [Google Scholar]


