Abstract
DNA replication in the human β-globin locus is subject to long-distance regulation. In murine and human erythroid cells, the human locus replicates in early S phase from a bidirectional origin located near the β-globin gene. This Hispanic thalassemia deletion removes regulatory sequences located over 52 kb from the origin, resulting in replication of the locus from a different origin, a shift in replication timing to late S phase, adoption of a closed chromatin conformation, and silencing of globin gene expression in murine erythroid cells. The sequences deleted include nuclease-hypersensitive sites 2 to 5 (5′HS2-5) of the locus control region (LCR) plus an additional 27-kb upstream region. We tested a targeted deletion of 5′HS2-5 in the normal chromosomal context of the human β-globin locus to determine the role of these elements in replication origin choice and replication timing. We demonstrate that the 5′HS2-5-deleted locus initiates replication at the appropriate origin and with normal timing in murine erythroid cells, and therefore we conclude that 5′HS2-5 in the classically defined LCR do not control replication in the human β-globin locus. Recent studies also show that targeted deletion of 5′HS2-5 results in a locus that lacks globin gene expression yet retains an open chromatin conformation. Thus, the replication timing of the locus is closely correlated with nuclease sensitivity but not globin gene expression.
The eukaryotic genome is divided into independently regulated domains that initiate DNA replication from defined sequences at specific times during S phase. Control mechanisms exist to specify both the sites of replication initiation (origins) and the temporal order of replication throughout the genome. Although great progress has been made in understanding the control of DNA replication in viruses, bacteria, and yeasts, we are only beginning to understand the control of DNA replication in higher eukaryotes. Over a dozen origins have been mapped in metazoans (reviewed in reference 10), and it appears that DNA replication in these organisms, in contrast to prokaryotes and lower eukaryotes, does not depend only on origin-proximal sequences, but is subject to long-distance regulation as well. The human β-globin locus is one of several loci in which this long-distance regulation has so far been demonstrated; sequences over 50 kb from the origin of replication are necessary for proper replication initiation and replication timing of the locus. Because of the tools available with which to make genetic modifications and perform functional analyses, the human β-globin locus offers an excellent opportunity to dissect possible replication control mechanisms.
A single bidirectional origin, located in the vicinity of the β-globin gene, is used to replicate the human β-globin locus (Fig. 1) (1, 35). Although this origin is used in both erythroid and nonerythroid cells, replication timing follows patterns of gene activity and chromatin structure: in erythroid cells, the globin genes are transcribed, the locus is generally sensitive to DNase I digestion (an “open” chromatin conformation), and the locus replicates early in S phase, while in nonerythroid cells the globin genes are silent, the locus is generally insensitive to nuclease digestion (closed chromatin), and replication occurs late in S phase (11, 16, 19, 23).
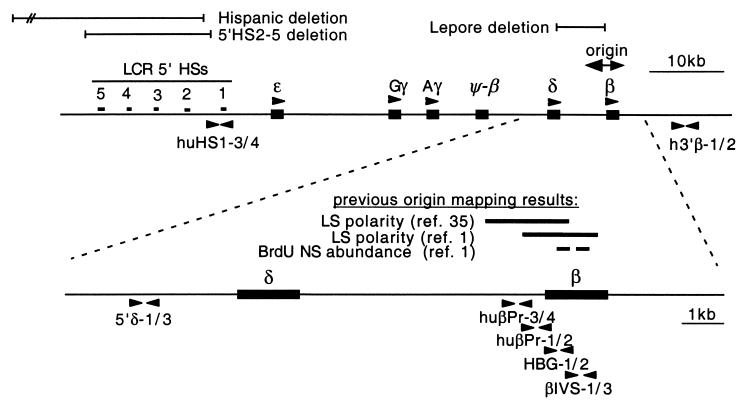
FIG. 1.
Organization of the human β-globin locus. The positions of the globin gene reading frames and LCR DNase I-hypersensitive sites are indicated. The regions removed by the Hispanic, Lepore, and 5′HS2-5 deletions are denoted by lines. The bidirectional origin used to replicate the entire locus is located in the region of the β gene. Results of previous origin-mapping assays in the wild-type locus are summarized in the expanded view of the origin region; leading-strand polarity assays and a nascent strand abundance assay yield results in good agreement. The primer pairs used in this study to assay replication timing and origin activity are indicated by solid arrowheads.
A naturally occurring deletion found in a patient with Hispanic thalassemia results in drastic changes in activity of the human β-globin locus in an erythroid environment. Analysis of the wild-type and Hispanic deletion alleles in a murine erythroid environment (following transfer of these chromosomes from the heterozygous patient into MEL cells) reveals that in contrast to the wild-type hybrid (referred to as N-MEL), in which normal behavior of the human locus is observed, the Hispanic locus (in the hybrid referred to as T-MEL) is transcriptionally silent, adopts a nuclease-resistant chromatin structure, replicates late in S phase, and is replicated unidirectionally from an origin located at least 15 kb downstream of the β gene (1, 19). The Hispanic deletion removes a 40-kb region (located 52 to 92 kb from the origin), including DNase I-hypersensitive sites 2 to 5 (5′HS2-5) upstream of the ɛ-globin gene plus an additional 27 kb of upstream sequence (15, 19). The 5′HSs are powerful regulatory elements that control chromatin structure and transcriptional activity of linked sequences in transient and transgenic assays (22, 45). Consequently, these sites became known collectively as the locus control region (LCR), and it was hypothesized that these HSs were responsible for controlling chromatin structure, expression, and replication of the endogenous β-globin locus.
Here, we analyze replication of a human β-globin locus from which 5′HS2-5 have been deleted by targeted mutagenesis to test the hypothesis that these elements are responsible for controlling replication of the locus. We find that in murine erythroid cells, the 5′HS2-5-deleted locus initiates replication from the normal origin and at the appropriate time during S phase. We conclude that 5′HS2-5 are not responsible for long-distance activation of the normal origin or for early replication timing. Combined with our recent finding that the 5′HS2-5-deleted locus maintains an open chromatin conformation yet lacks expression of the β-globin genes (46, 47), our results also indicate that early replication timing is correlated with an open chromatin structure but not gene transcription in the locus.
MATERIALS AND METHODS
Cell lines and culture conditions.
The hybrids N-MEL and T-MEL contain the wild-type and Hispanic deletion human β-globin loci, respectively, on intact human chromosomes 11 transferred from lymphocytes (from a heterozygous patient) into murine erythroleukemia (MEL) cells, as described previously (19).
The wt-MEL and Δ2-5-MEL hybrids that we analyze here were generated by a series of homologous and site-specific recombinations and chromosome transfers similar to those we have described previously (46), with the addition of an extra transfer of human chromosome 11 from DT40 cells (in which homologous recombination was performed) into ES cells prior to transfer into MEL. This transfer was accomplished by microcell fusion (13) and was found to be necessary for recovery of an intact human chromosome in MEL (thereby avoiding possible position effects), as transfer from DT40 directly into MEL typically results in breakage of the human chromosome and integration of these fragments into the mouse genome genome (46; A. Reik, A. Telling, and M. Groudine, unpublished data). Expression of site-specific recombinases to delete 5′HS2-5 or to recreate a wild-type locus was accomplished as before and was performed in ES cells prior to transfer into MEL for phenotypic analysis. To demonstrate integrity of the transferred chromosomes, fluorescent in situ hybridization (FISH) of wt-MEL and Δ2-5-MEL metaphase chromosomes was performed using a fluorescein isothiocyanate-conjugated human 11-specific chromosome paint (Cambio) a biotinylated human pancentromeric probe (Oncor), and a 15.4-kb ClaI fragment from the human β-globin locus containing the δ- and β-globin genes and the origin region (labeled with digoxygenin by nick translation; Boehringer). Hybridization and detection of these probes were carried out as described previously (47). In addition to FISH, the hygromycin resistance of these clones (conferred by a Hygror cassette inserted at the H-ras locus [13]) and the presence of all non-β-globin loci analyzed (by Southern blot or PCR) on both arms of human chromosome 11, including myoD, PTH, and cbl2, further suggest that no chromosomal breakage or loss has occurred (Results and data not shown). The wt-MEL and Δ2-5-MEL hybrids described here display transcription and chromatin properties similar to those we have previously described for the wild-type and Δ2-5 loci on integrated chromosomal fragments in MEL (46); specifically, these loci are both generally DNase I sensitive, and the Δ2-5 locus (in contrast to the wild-type locus) expresses no detectable β-globin-like mRNA (47; Reik et al., unpublished).
Hybrid cell lines wt-GM and Δ2-5-GM contain a wild-type and 5′HS2-5-deleted human β-globin locus, respectively, transferred into murine GM979 erythroid cells and have been described previously (46). These hybrids were obtained by direct transfer of human chromosomes from DT40 to GM979 and contain the human β-globin locus centrally located on a large (>12 Mb) fragment of human chromosome 11 integrated into the mouse genome, as determined by FISH analysis (48).
MEL and GM979 hybrids were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% bovine calf serum. DT40 hybrids were cultured in DMEM with 10% bovine calf serum, 10% tryptone phosphate buffer, and 1% chicken serum. MEL, GM, and DT40 hybrids were periodically maintained in hygromycin to select cells containing human chromosome 11. N- and T-MEL hybrids, which do not contain a Hygror marker, were periodically enriched for human chromosome 11 using monoclonal antibody 53/6, as described previously (19).
Replication initiation analysis.
Nascent strands resistant to digestion by lambda exonuclease by virtue of their 5′ RNA primers (21) were prepared by a modification of procedures used to analyze replication in the hamster DHFR locus (36). Rapidly dividing cells (1.7 × 108 to 2.4 × 108) were harvested, washed with phosphate-buffered saline (PBS), and lysed in 5 ml of lysis buffer (0.5% sodium dodecyl sulfate [SDS], 10 mM Tris [pH 7.4], 0.3 M NaCl, 5 mM EDTA). Proteinase K was added to 200 μg/ml, and the lysate was incubated at 55°C overnight. The lysate was diluted to 10 ml with Tris-EDTA (TE) and extracted twice with phenol and once with chloroform. DNA was precipitated with sodium acetate-ethanol and spooled out using a drawn pasteur pipette. The precipitate was rinsed in 70% ethanol and thoroughly resuspended in 4 ml of TE at 4°C. At this point, 0.5 ml from each cell line was set aside for use as a genomic control for the duplex PCR. The remainder was boiled for 3 min, chilled on ice, and loaded onto a single neutral 5 to 30% sucrose gradient prepared in TNE (10 mM Tris [pH 8.0], 1 mM EDTA, 0.3 M NaCl) in a 38.5-ml centrifuge tube. Gradients were centrifuged in a Beckman SW28 rotor for 22 h at 21 krpm and 20°C. Fractions (1.5 ml) were withdrawn from the top of the gradient using a wide-bore pipette tip, and 25 μl of each fraction was run with appropriate size markers on a 0.9% alkaline agarose gel, which was subsequently neutralized and stained with ethidium bromide. Fractions corresponding to 1 to 3 kb were pooled, dialyzed against TE, and precipitated with sodium acetate-ethanol. Pellets were resuspended in 100 μl of TE with 1 mM dithiothreitol and 100 U of RNasin (Promega).
As a control for the completeness of the lambda exonuclease digestion, 600 ng of linearized, calf intestinal phosphatase-treated pBluescript was added to 20 μl of each sample. The DNA mixture was boiled for 5 min, chilled on ice, and phosphorylated with T4 polynucleotide kinase (NEB) in a volume of 40 μl. The reaction was stopped by the addition of 0.2% Sarkosyl and 20 mM EDTA. Proteinase K was added to 0.25 μg/ml, and the reaction was incubated at 55°C for 30 min. The reaction was diluted with 150 μl of TE, extracted once with phenol and once with chloroform, and ethanol-sodium acetate precipitated. The pellet was resuspended in 25 μl of TE; 5 μl was set aside as a negative control, and the remainder was digested with 4 μl of lambda exonuclease (Promega,-4.9 U/μl) in 67 mM glycine-KOH (pH 9.4)–2.5 mM MgCl2–50 μg of bovine serum albumin per ml–25 U of RNasin in a volume of 80 μl overnight at 37°C. Aliquots of both the digested DNA and the undigested control were run on an agarose gel; the Bluescript fragment was used to assess the degree of digestion. If necessary, additional enzyme was added and incubation was continued until complete digestion was achieved, at which point the enzyme was heat inactivated. RNase A (5 μg) was added for 10 min at room temperature, and the nascent strands were size fractionated on a 0.9% low-melting-point agarose-TBE gel. Gel slices corresponding roughly to 0.5 to 0.7 kb (ns1), 0.8 to 1.0 kb (ns2), and 1.0 to 1.9 kb (ns3) were excised, the agarose was digested with β-agarase (NEB), and the nascent strands were isopropanol precipitated and resuspended in 100 μl of TE. Results from each of the size fractions were similar (see Results).
For analysis of nascent strand abundance, duplex PCR (29 cycles: 95°C for 30 s, 60°C for 30 s, 72°C for 30 s) was performed using 2 μl of each nascent strand sample as a template and 100 ng of each primer. PCR products were labeled by including 0.1 μl of [α-32P]dCTP (NEN) in each 25-μl reaction. PCR products were electrophoresed on nondenaturing 5% polyacrylamide gels, and the gels were dried and exposed to both film and phosphorimager screens. Quantification and background correction were performed using ImageQuant software. To correct for differences in the efficiency of the two primer pairs, for each experiment the same PCR premix used for the nascent strands was used to amplify (in duplicate) 1 ng of genomic DNA, which was prepared from each cell line at the time nascent strands were harvested and was sonicated to a size range similar to that of the nascent strands (0.5 to 2 kb). Product ratios from each nascent strand sample were divided by the average product ratio from the corresponding pair of genomic control reactions. For the control reactions shown in Fig. 3, 2 ng of wt-MEL genomic DNA was amplified for 27, 28, 29, and 30 cycles using the conditions above.
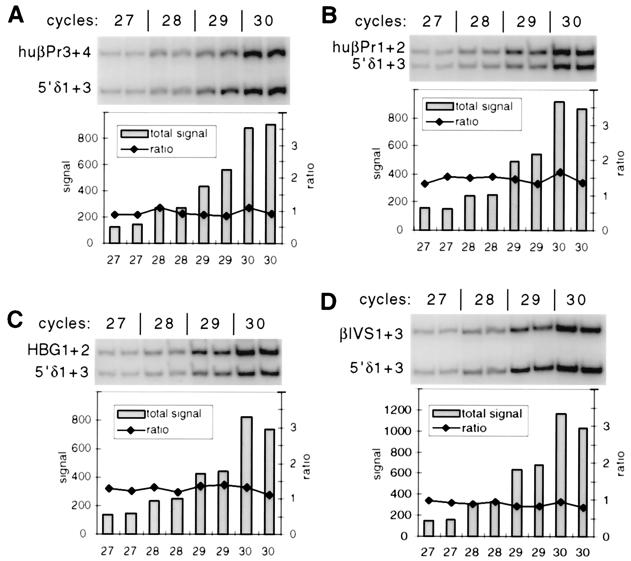
FIG. 3.
Quantitative analysis of control duplex PCR products from the human β-globin locus in MEL hybrids. Amplification products from 2 ng of wt-MEL genomic DNA following 27, 28, 29, and 30 cycles of duplex PCR with primer pair 5′δ1+3 in combination with huβPr3+4 (A), huβPr1+2 (B), HBG1+2 (C), and βIVS1+3 (D) are shown; each reaction was performed in duplicate. Quantitation of these products reveals that in each case, the total amounts of product (bars) increase linearly with cycle number and the product ratios (lines) do not vary significantly between identical reactions or as the cycle number increases. These results demonstrate that each primer is in the linear phase of amplification.
Replication timing analysis.
Exponentially growing cells (1 × 107 to 3 × 107) were cultured in the presence of 50 μM bromodeoxyuridine (BrdU) for 60 min. Cells were washed two times with cold PBS, resuspended in 2.5 ml of cold PBS, fixed by the addition of 7.5 ml of cold ethanol, and stored overnight at −20°C. For sorting, cells were resuspended in 5 ml of resuspension buffer (40 mM Tris [pH 7.4], 0.8% NaCl, 21 mM MgCl2), which was then supplemented with 0.05% NP-40, propidium iodide, (50 μg/ml), and RNase A (0.5 mg/ml). Cells were allowed to stain in the dark for 30 min prior to sorting. Equal numbers of cells from each cell cycle compartment (typically 6,000 to 8,000) were sorted (using a Becton Dickinson FACS Vantage sorter) into microcentrifuge tubes containing lysis buffer (1 M NaCl, 10 mM EDTA, 50 mM Tris [pH 8.0], 0.5% SDS, 0.2 mg of proteinase K per ml, 0.25 mg of denatured, sheared salmon sperm DNA per ml); lysates were incubated at 50°C for 2 h and then stored at −20°C. Lysates were extracted once with phenol-chloroform and once with chloroform. DNA was precipitated and resuspended in 460 μl of TE with 40 μl of 5-mg of denatured, sheared salmon sperm DNA per ml. DNA was sonicated to an average size of 0.8 kb, heat denatured, and cooled on ice. Then, 50 μl of 10 immunoprecipitation buffer (1 ml of 1 M sodium phosphate [pH 7.0], 1.4 M NaCl, 0.5% Triton X-100) and 40 μl of mouse anti-BrdU DNA monoclonal antibody (25 μg/ml; Becton Dickinson) was added to each tube. After 20 min of constant rocking at room temperature, 8.5 μl of rabbit anti-mouse immunoglobulin G (IgG) (2.1 mg/ml; Sigma) was added, and incubation continued for an additional 20 min at room temperature with rocking. DNA-protein complexes were pelleted by microcentrifuging for 5 min at 4°C. After washing with 750 μl of 1 X IP buffer, pellets were resuspended in 200 μl of digestion buffer (50 mM Tris [pH 8.0], 10 mM EDTA, 0.5% SDS, 250 μg of proteinase K per ml). Digestion was allowed to proceed overnight at 37°C and then for 1 h at 50°C after the addition of 100 μl of fresh buffer. Then 4 μl of Escherichia coli tRNA (5 mg/ml) was added to each tube, extractions were performed as above. DNA was precipitated with sodium acetate and ethanol, briefly dried, and resuspended in TE at a concentration of 250 cell equivalents/μl.
PCR (23 cycles) was performed using 2 μl (500 cell equivalents) of each nascent strand sample as a template. Southern blots were prepared and probed with radiolabeled probes synthesized by random priming the equivalent PCR product amplified separately from either MEL or K562 genomic DNA. Blots were washed and exposed to a phosphorimager cassette and to film. In each experiment, genomic DNA from both MEL and K562 were included as controls for the strength and specificity of the PCR; all primers were species specific and yielded a single primary product.
Primers.
The locations, predicted product sizes, and sequences (5′→3′) of the primers are listed below. The locations refer to the more distant primer of the pair. Amplification of MEL (mouse) and K562 (human) genomic DNA was performed to verify that each primer pair yields a single, species-specific product. For origin-mapping experiments, we designed origin-proximal primers to yield a product slightly larger (by 30 to 69 nucleotides) than the origin-distal primer pairs to avoid any artificial enrichment of the origin sequences due to preferential amplification of a smaller product.
Primers 5′Ey3 and 5′Ey4 (5′Ey3+4): 1.1 kb upstream of mouse Ey globin start codon; 376-bp product; 5′Ey3, GCACATGGATGCAGTTAAACAC; 5′Ey4, GAGTGACAGTGTAGAGAAGATG. Primers mAmyl1+2: 0.6 kb upstream of mouse pancreatic amylase 2.1y start codon; 370-bp product; mAmyl1, AGCACTGAGGATTCAGTCTATG; mAmyl2, CCCGTACAAGGAGAATTACAAC. Primers Fra11B1+2: at the human cbl2 locus on chromosome 11q; 297-bp product; Fra11B1, CGGAACTGATACTGACAC; Fra11B2, CTACTCAGAGCATTGCAC. Primers huHS1−3+4: near 5′HS1 in the human LCR; 355-bp product; huHS1-3, CCTGCAAGTTATCTGGTCAC; huHS1-4, CTGGGCAGCGTCAGAAACTG. Primers 5′δ1+3: 2.5 kb upstream of the human δ-globin start codon (9.9 kb upstream of the human β-globin start codon); 350-bp product; 5′δ1, GTAACCAGATCTCCCAATGTG; 5′δ3, ATATGTGGATCTGGAGCTCAG. Primers βIVS1+3: 0.9 kb downstream of the human β-globin start codon; 419-bp product; βIVS1, TACCCTGATTTGGTCAATATGTG; βIVS3, GGAAGGGGAGAAGTAACAGGG. Primers HBG1+2: 0.4 kb downstream of the human β-globin start codon; 395-bp product; HBG1, CCTGAGGAGAAGTCTGCCGT; HBG2, CAGTGCAGCTCACTCAGTGT. Primers huβPr1+2: 0.4 kb upstream of the human β-globin start codon; 380-bp product; huβPr1, TGCTTACCAAGCTGTGATTCC; huβPr2, AACGGCAGACTTCTCCTCAGG. Primers huβPr3+4: 0.7 kb upstream of the human β-globin start codon; 407-bp product; huβPr3, TAAGTAACTAATGCACAGAGCAC; huβPr4, TGGAATCACAGCTTGGTAAGC. Primers h3′β1+2: 11.1 kb downstream of the human β-globin start codon; 367-bp product; h3′β1, AGTTCATGTCCTTTGTAGGGAC; h3′β2, GCTCTACGGATGTGTGAGATC. Primers m3′βmaj1+2: 1.8 kb downstream of the mouse β-major globin start codon; 369-bp product; m3′βmaj1, CAGTAAGCCACAGATCCTATTG; m3′βmaj2, CCCATAGTGACTATTGACTGTG. Primers mLCR1+2: near 5′HS2 in the mouse LCR; 356-bp product; mLCR1, TGTAATCTAGCACAGAACCTTAG; mLCR2, TCTTTCATGGATGGTAACTAGTG.
RESULTS
Replication initiation occurs at the appropriate origin in the absence of 5′HS2-5.
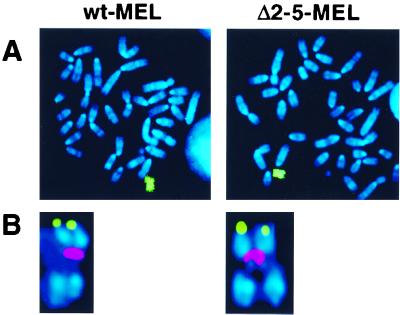
The Hispanic deletion removes 5′HS2-5 of the LCR and additional upstream sequences, resulting in inactivation of the normal origin in the locus located over 50 kb away (Fig. 1). To determine whether 5′HS2-5 are the cis-acting elements responsible for long-distance replication control, we examined replication initiation in murine erythroleukemia (MEL) cell hybrids carrying a human chromosome 11 from which 5′HS2-5 had been deleted (Δ2-5-MEL). For comparison, we examined MEL hybrids with a human chromosome 11 carrying a wild-type human β-globin locus (wt-MEL) or the Hispanic deletion locus (T-MEL). The wt-MEL and Δ2-5-MEL lines were generated by a series of modifications and chromosome transfers similar to those we have described previously (46), with an additional transfer step to facilitate the recovery of intact human chromosomes in these hybrids. FISH of wt-MEL and Δ2-5-MEL metaphase chromosomes reveals that in both hybrids, the human β-globin locus is in its normal position on an intact chromosome 11 (Fig. 2).
FIG. 2.
Cytogenetic analysis of wt-MEL and Δ2-5-MEL hybrids. Metaphase chromosomes from each cell line were hybridized with (A) a human chromosome 11-specific paint (green) or (B) a human β-globin probe (green) in combination with a human centromeric probe (red). Chromosomes were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) (blue). Both cell lines contain a single intact human chromosome 11 with the human β-globin locus in its normal position near the telomere of 11p.
To determine whether the normal β-globin origin of replication is active in these hybrids, we employed a PCR-based nascent strand abundance assay. Short, RNA-primed nascent strands that originate at a site of replication initiation were isolated using a combination of size fractionation (to exclude unligated lagging strands and long nascent strands) and removal of contaminating broken genomic DNA by lambda exonuclease digestion (which digests DNA lacking a 5′ RNA primer) (21). This nascent strand purification strategy has been used recently to map origins of replication in the hamster DHFR locus with high resolution and in good agreement with results from BrdU-labeled nascent strands (36).
For analysis of the nascent strands, we developed a quantitative duplex PCR assay. PCR is performed simultaneously with two pairs of primers (one located near the origin of replication and another located some distance away), and comparison of the amount of product from each primer pair is used to determine the relative abundance of the corresponding template sequences in the nascent strands. Primer pairs with similar amplification efficiency were chosen for duplex PCR. However, to accurately correct for subtle differences in primer efficiency, for each experiment the same PCR premix used to amplify nascent strands is used in parallel to amplify genomic DNA (prepared from the same culture used for the nascent strand preparation), and the product ratios obtained from nascent strands are normalized to ratios obtained from genomic DNA. Because of the importance of this normalization, duplicate genomic control reactions were used in the following analyses, and nascent strand ratios were normalized to the average of these two genomic ratios; however, variation between identical genomic reactions is low (see below).
For analysis of the human β-globin locus, we chose four primer pairs in the vicinity of the origin and three primer pairs located away from the origin (Fig. 1). To establish appropriate reaction conditions for these primers and to demonstrate the reliability of this quantitative approach, a series of control reactions were performed (Fig. 3). Genomic DNA was amplified for 27, 28, 29, and 30 cycles with each of the four origin-proximal primer pairs in duplex with one of the distant primer pairs (5′δ1+3). Quantitation of the products reveals that as cycle number increases, there is a linear increase in the amount of products with no significant change in the product ratio. These results demonstrate that under these reaction conditions, both primer pairs in each duplex PCR are in the linear phase of amplification and that the product ratio remains constant over a large range of product amounts. For analysis of nascent strands (below), 29 cycles was chosen as a satisfactory balance between signal strength and maintaining the reactions in the linear phase.
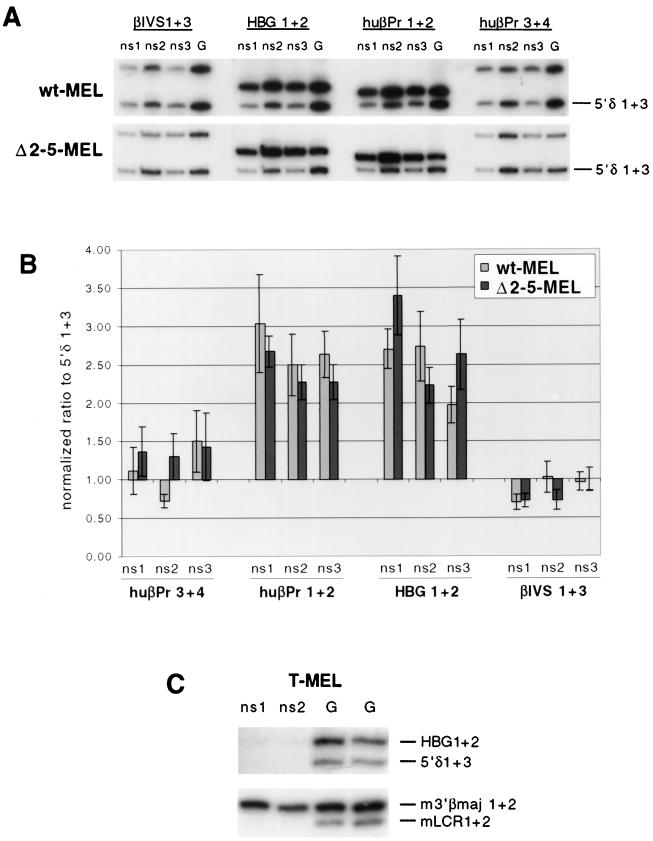
Typical duplex PCR results from wt-MEL and Δ2-5-MEL nascent strands are shown in Fig. 4A. Compared to genomic controls, both wt-MEL and Δ2-5-MEL nascent strands show a clear enrichment of huβPr1+2 and HBG1+2 sequences relative to 5′δ1+3. This enrichment is consistent in all three nascent strand size fractions (ns1 to ns3; see Materials and Methods). In contrast, no significant enrichment of huβPr3+4 or βIVS1+3 sequences is seen in either cell line. Three independent PCR analyses were performed, and each yielded similar results. Quantitative analysis of these results (Fig. 4B) reveals a similar pattern of enrichment in wt-MEL and Δ2-5-MEL: the primer pairs huβPr1+2 and HBG1+2 (located centrally within the previously identified origin region) are enriched 2.0- to 3.0-fold in wt-MEL and 2.2- to 3.4-fold in Δ2-5-MEL, while the flanking primer pairs (huβPr3+4 and βIVS1+3) show little or no enrichment (≤1.5-fold) in either cell line. A second independent nascent strand preparation from each cell line yielded similar results (data not shown).
FIG. 4.
DNA replication initiation in wild-type and mutant human β-globin loci in MEL hybrids. (A) Representative duplex PCR results from lambda exonuclease-resistant nascent strands prepared from wt-MEL and Δ2-5-MEL. Short nascent strands (ns1 to ns3; see Materials and Methods) and genomic DNA (G) from each cell line were used as templates for duplex PCR with one primer pair in the origin region in combination with a primer pair distant from the origin (5′δ1+3). Compared to genomic DNA, nascent strands from both cell lines are enriched for HBG1+2 and huβPr1+2 sequences, the primer pairs located centrally in the initiation region depicted in Fig. 1. Little or no enrichment is seen with flanking primer sets huβPr3+4 and βIVS1+3. (B) Quantification of duplex PCR analysis of wt-MEL and Δ2-5-MEL nascent strands. Three independent PCR analyses were performed; the mean value and standard error of the mean are plotted. A value above 1.0 represents enrichment of the origin-proximal sequences. The wild-type and Δ2-5 loci yield the same pattern: huβPr1+2 and HBG1+2 products are enriched, while products from adjacent primers show little or no enrichment. An independent nascent strand preparation from each hybrid yielded similar results. (C) Duplex PCR analysis of T-MEL (Hispanic deletion) nascent strands. Primers used are from the human (upper panel) and mouse (lower panel) β-globin loci. Primers in the human locus yield almost no products from nascent strands, despite strong amplification of genomic DNA with the same PCR premix; the signal strength from the nascent strands was not high enough above background to allow a ratio to be reliably determined. This PCR was repeated with the same result. In contrast, primers in the mouse locus yield comparable amounts of products from the nascent strands and genomic DNA, despite the fact that half the amount of nascent strand template was used for the mouse PCR as was used for the human PCR. Note the enrichment of 3′βmajor1+2 relative to LCR1+2 sequences, consistent with the location of the β-major primers within the initiation zone in the mouse locus (see text).
As controls, we verified that duplex PCR with two pairs of primers located outside the origin region (hu3′β1+2, located 11 kb downstream of the origin, and 5′δ1+3) yields no significant enrichment of either product (data not shown). In addition, we find that enrichment of origin-proximal sequences is not dependent on the use of 5′δ1+3 as the distant primer pair: duplex PCR with HBG1+2 and the distant primer pair huHS1−3+4 (located near LCR 5′HS1, 49 kb upstream of the origin region) yielded a similar degree of enrichment of the origin sequences (data not shown).
These results demonstrate replication initiation activity at the normal origin in both wt-MEL and Δ2-5-MEL. To ensure that this assay is capable of identifying whether the origin is nonfunctional, we examined the Hispanic locus in T-MEL, in which deletion of the LCR and upstream sequences results in inactivation of the normal origin. In contrast to results in wt-MEL and Δ2-5-MEL, duplex PCR using T-MEL nascent strands as a template reveals greatly reduced abundance of human β-globin sequences (Fig. 4C). This PCR was repeated with the same result; in neither case was the signal strength high enough above background to allow reliable quantitation. This lack of amplification from nascent strands is not due to loss of the human chromosome from the T-MEL hybrid, since T-MEL genomic DNA (prepared at the same time that nascent strands were harvested) yields strong signals with both human primers. To demonstrate that the T-MEL preparation contains origin-proximal nascent strands, we performed duplex PCR with primers in the endogenous mouse β-globin locus. The mouse β-globin locus is characterized by replication initiation throughout a broad region that includes the adult β-globin genes but not the LCR (M. Aladjem, L. Rodewald, D. Cimbora, E. Epner, M. Groudine, and G. Wahl, unpublished data; C. Schildkraut, personal communication). Duplex PCR with murine origin-proximal primers m3′βmaj1+2 (within the initiation zone) and origin-distal primers mLCR1+2 (outside the initiation zone) yields abundant signal from nascent strands (similar in intensity to the control genomic DNA) and an enrichment of m3′βmaj1+2 relative to mLCR1+2, as expected (Fig. 4C). This result demonstrates that the T-MEL nascent strand preparation is enriched for origin-proximal sequences, and therefore we infer that the lack of amplification of human β-globin sequences reflects the inactivity of the normal human β-globin origin in the Hispanic deletion locus.
Taken together, these results demonstrate the utility of the lambda exonuclease nascent strand assay in determining origin activity in the human β-globin locus and allow us to conclude that the normal origin of replication in the human β-globin locus is active in both wt-MEL and Δ2-5-MEL despite the absence of LCR 5′HS2-5 in the latter.
Early replication of the 5′HS2-5-deleted locus in erythroid cells.
To examine the role of 5′HS2-5 in replication timing, we adapted an assay used previously to study replication timing of several human loci (25). This assay determines the relative abundance of specific sequences in nascent DNA synthesized during different windows of the cell cycle. An exponentially growing culture is briefly labeled with BrdU, and cells are sorted by fluorescence-activated cell sorting (FACS) into different compartments of the cell cycle on the basis of DNA content. DNA is prepared from equal numbers of cells in each cell cycle compartment, and BrdU-labeled (nascent) DNA is purified by immunoprecipitation with an anti-BrdU–DNA monoclonal antibody. The immunoprecipitate is then used as a template for PCR, using primers specific for either the human β-globin locus or for various control loci. PCR products are analyzed by Southern blot hybridization to enhance signal strength and specificity.
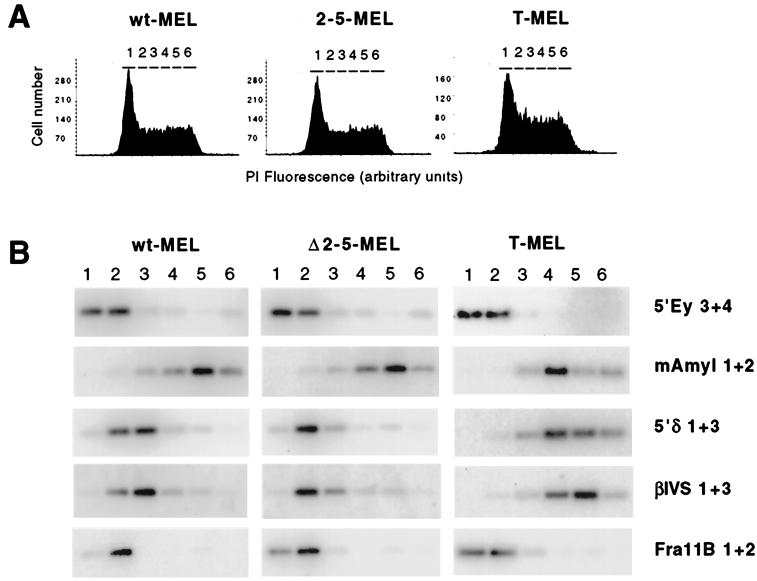
This replication timing assay was applied to the MEL hybrids described above (wt-MEL, Δ2-5-MEL, and the Hispanic deletion in T-MEL). The cell cycle profile of each culture and the sort gates corresponding to G1, S1 to S4, and G2 are shown in Fig. 5. To assay timing of the human β-globin locus, primers upstream of the δ-globin gene and in the first intron of the β-globin gene were used. Sequences in this region of the locus have been used previously to analyze replication timing of the human β-globin locus in MEL hybrids and were found to accurately reflect the replication timing of a >185-kb domain (19). As controls, we used primers to other mouse and human loci with known replication timing. For early-replicating controls, we used the endogenous mouse β-globin locus (18, 26) and a fragile site at the human cbl2 proto-oncogene locus on the long arm of human chromosome 11, designated Fra11B (32, 57). For a late-replicating control, we used the endogenous mouse pancreatic amylase 2.1y gene (59), which is late-replicating in all erythroid cells examined (12, 19, 26). We find that the human β-globin locus in wt-MEL is replicated early in S phase, concurrently with Fra11B and the mouse β-globin locus, and significantly earlier than the amylase locus (Fig. 5). Analysis of the human β-globin locus in N-MEL (which contains the wild-type allele from the heterozygous Hispanic patient) yielded a similar early pattern of replication (data not shown). Analysis of Δ2-5-MEL reveals early replication timing of the human β-globin locus (Fig. 5). Analysis of a second Δ2-5-MEL clone yielded similar results (data not shown). Subtle differences are apparent between wt-MEL and Δ2-5-MEL: in wt-MEL, the peak of human β-globin replication occurs in fractions 2 and 3, while in Δ2-5-MEL the peak is predominantly in fraction 2. However, because these differences are also evident in the early controls (the 5′Ey and Fra11B peaks are shifted slightly earlier in Δ2-5-MEL than in wt-MEL), we conclude that this is due to small differences in the separation of cell cycle compartments during FACS; minor sort-to-sort variations such as this have been reported with this assay (25, 58). In contrast to the early-replicating behavior of the human locus in wt-MEL and Δ2-5-MEL, the Hispanic deletion locus in T-MEL is late replicating (Fig. 5), as we have previously shown with an independent assay based on centrifugal elutriation (19).
FIG. 5.
Replication timing of wild-type and mutant human β-globin loci in MEL hybrids. (A) Histogram of propidium iodide (PI) staining intensity (DNA content) of cells sorted for timing analysis. The gates used to sort cells into compartments, corresponding approximately to G1, S1 to S4, and G2, are labeled 1 to 6, respectively. (B) PCR and Southern analysis of replication timing in β-globin and control loci. Analysis was performed as described in the text, using primers for the human β-globin locus (5′δ1+3 and βIVS1+3), early-replicating control loci (endogenous murine β-globin [5′Ey3+4] and human cbl2 [Fra11B1+2]), and a late-replicating control locus (murine amylase, mAmyl1+2). In wt-MEL, the human β-globin locus replicates early (concurrently with murine β-globin and human Fra11B and earlier than amylase). In Δ2-5-MEL, the human β-globin locus replicates early, with a temporal profile similar to that seen in wt-MEL. Minor differences in timing between the human β-globin loci in wt-MEL and Δ2-5-MEL are also seen in the early controls and thus do not represent a difference in replication timing (see text). Analysis of a second independent Δ2-5-MEL clone yielded similar results. In contrast to wt-MEL and Δ2-5-MEL, the Hispanic deletion locus in T-MEL is late replicating.
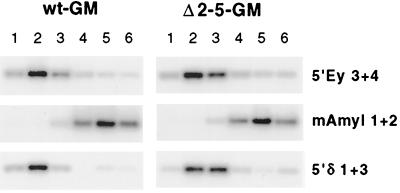
To determine whether the observed early replication timing of the 5′HS2-5-deleted locus is specific for a particular erythroid environment, we analyzed timing of the wild-type and Δ2-5 human loci in a second mouse erythroid cell line (GM979). In contrast to the MEL line used above, in which only the adult globin genes are expressed, GM979 hybrids express embryonic, fetal, and adult human globin genes (5, 46, 61); as in MEL, we have shown that expression of these globin genes is dependent on the presence of 5′HS2-5 (46, 47). The GM979 hybrids were derived by direct transfer of the human chromosome from DT40 rather than through the ES cell intermediate used in generating the wt-MEL and Δ2-5-MEL hybrids (see Materials and Methods) (46). Thus, the GM979 hybrids allow us to test for a possible influence of either chromosomal history or the trans-acting environment of the cell on replication timing. Analysis of GM979 hybrids with the FACS-BrdU assay (Fig. 6) reveals early replication of both the wild-type and Δ2-5 human loci relative to the early (mouse globin) and late (amylase) controls.
FIG. 6.
Replication timing of wild-type and Δ2-5 loci in GM979 hybrids. Replication timing was assayed as in the MEL hybrids (see legend to Fig. 5). In both wt-GM and Δ2-5-GM, the human β-globin locus replicates early, relative to the early and late controls.
Based on these results, we conclude that the human β-globin locus is early replicating in erythroid cells despite the absence of LCR 5′HS2-5 and that this early replication is not dependent on trans-acting factors specific to a particular erythroid cell line.
5′HS2-5 are not required for late replication of the human locus in nonerythroid cells.
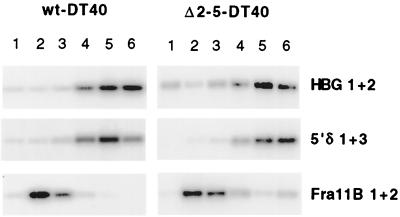
In nonerythroid cells, the wild-type human β-globin locus is replicated from the same origin as in erythroid cells (1, 35), but replication occurs late in S phase, correlating with the closed chromatin and transcriptional inactivity of the locus in this environment (11, 16). Little is known about the control of late replication of this locus in nonerythroid environments; one possibility is that the LCR HSs are required to maintain the locus in its late-replicating state. To test this, we examined replication timing of the wild-type and 5′HS2-5-deleted loci in the avian pre-B-cell line DT40. DT40 hybrids containing the wild-type human β-globin locus were generated by transfer of an intact human chromosome 11 from N-MEL; deletion of 5′HS2-5 was accomplished by a combination of homologous and site-specific recombinations (see Materials and Methods) (46). The normal origin of replication in the human β-globin locus is active in DT40, indicating that origin function is conserved (1).
Analysis of these hybrids with the FACS-BrdU replication timing assay (Fig. 7) reveals that the wild-type locus is late replicating relative to the early-replicating Fra11B control, as expected for a nonerythroid environment. Furthermore, deletion of 5′HS2-5 does not result in early replication timing of the locus. We conclude that 5′HS2-5 do not play an active role in maintenance of the late-replicating state of the β-globin locus in a nonerythroid environment.
FIG. 7.
Replication timing of wild-type and Δ2-5 loci in DT40 hybrids. Replication timing assays were performed as in MEL and GM979 hybrids. In DT40, both the wild-type and Δ2-5 human β-globin loci (HBG1+2 and 5′δ1+3) are late replicating relative to the early-replicating Fra11B control.
DISCUSSION
The Hispanic deletion removes DNase-hypersensitive sites 2 to 5 of the LCR and additional upstream sequences and identifies this region as necessary in cis for proper replication initiation and timing in the human β-globin locus. We now show that the deleted HSs, despite their requirement for globin gene transcription, are not responsible for origin choice or early replication of the locus in an erythroid environment. We conclude that early replication timing correlates with an open chromatin structure rather than globin gene expression, while origin choice correlates with neither of these properties.
Long-distance control of DNA replication.
According to the replicon model proposed 35 years ago (31), bacterial DNA replication is controlled by proteins (termed initiators) which bind to cis-acting genetic elements (replicators) necessary for initiation of DNA synthesis. This model remains valid not only for bacteria and simple eukaryotes but also for the complex genomes of higher organisms. While the identification of initiator proteins in metazoans has been a fruitful area of research due to the conservation of these proteins through evolution, identification of replicators in higher eukaryotes has been more problematic. The human β-globin locus provided an early insight into the nature of a mammalian replicator: a naturally occurring deletion (found in a patient with hemoglobin Lepore syndrome) removes the primary site of replication initiation in the locus (Fig. 1), and as a result, replication initiates from a point within or upstream of the LCR (35). Thus, in mammals, as in lower eukaryotes and prokaryotes, origins are located within regions genetically identified as replicators. However, analysis of the Hispanic thalassemia deletion (Fig. 1) identified a second region, located over 50 kb away from the origin, that is also necessary for origin function (1). This was the first demonstration in mammals of the control of replication initiation by a distant sequence element; since then, a second example has been found in the hamster DHFR locus, in which sequences located >10 kb from the primary origin are required for normal initiation (33). These examples of long-distance replication control are reminiscent of the Drosophila chorion gene cluster, where proper replication depends on the amplification control element located 1.5 kb from the origin (42). In all of these cases, the DNA sequence elements that influence origin activity from a distance meet the classical definition of a replicator (a cis-acting genetic element necessary for initiation of DNA replication [52]), demonstrating that in metazoans, not all replicators are origins of replication.
Given the powerful functions of 5′HS2-5 in transgenic and transient assays (22, 45), it was reasonable to hypothesize that the loss of these 5′HSs was responsible for the Hispanic deletion phenotypes. Surprisingly, targeted deletion of 5′HS2-5 (leaving the upstream sequences intact) results in a locus that lacks globin gene expression but retains general DNase I sensitivity (46), indicating that 5′HS2-5 are necessary for activity of the globin gene promoters but not for open chromatin structure of the locus. The results presented here demonstrate that neither replication initiation at the normal origin nor early replication timing of the locus in erythroid cells is dependent on 5′HS2-5. One explanation for the different replication phenotypes of the Hispanic and 5′HS2-5 deletions is that an element(s) located upstream of 5′HS5, either alone or in combination with 5′HS2-5, is responsible for normal replication of the locus. These elements, if they exist, may correspond to the erythroid-specific DNase hypersensitive sites that we have recently identified between 5′HS5 and the upstream endpoint of the Hispanic deletion (7). Alternatively, it is possible that the different cellular histories of the Δ2-5 and Hispanic deletion chromosomes (Table 1) affect their mutant phenotypes, perhaps by a mechanism involving epigenetic modification of the chromosome. One argument against such a possibility is that despite different chromosomal histories, the transferred wild-type β-globin loci in wt-MEL and N-MEL and the endogenous locus in human erythroid cells all exhibit the same transcription, chromatin, and replication properties (see Table 1 and references therein). An alternative possibility is that passage of the ΔHS2-5 chromosome through the germ line is necessary for full manifestation of its mutant phenotype, as we and others have discussed (17, 27, 46). However, we have recently demonstrated that in the murine β-globin locus, deletion of the LCR yields similar transcription and chromatin phenotypes whether the analyses are performed in a heterologous environment (human erythroid cells in culture) or in mice after passage of the mutation through the germ line (3, 17). Based on these observations, the most reasonable explanation for the different replication phenotypes of ΔHS2-5 and the Hispanic deletion is the existence of replication control elements upstream of 5′HS5 that are removed by the Hispanic deletion but are still present in the ΔHS2-5 locus.
TABLE 1.
Chromatin structure, globin gene expression, and replication behavior of wild-type and mutant human β-globin loci in erythroid and nonerythroid cell typesa
| Genotype of human locus | Cell type used for analysis | Chromosomal history (transfers) | Chromatin structure | Globin transcription | Replication timing | Normal origin | Reference(s) |
|---|---|---|---|---|---|---|---|
| WT | Hum eryth | None | Open | Expressed | Early | Active | 11, 16, 23, 35 |
| WT (N-MEL) | Mur eryth | Lymph>eryth | Open | Expressed | Early | Active | 1, 19, 47; this work |
| WT (wt-MEL) | Mur eryth | Lymph>eryth>lymph>ES>eryth | Open | Expressed | Early | Active | 46, 47; this work |
| Hispanic deletion | Mur eryth | Lymph>eryth | Closed | Silent | Late | Inactive | 1, 19, 47; this work |
| ΔHS2-5 | Mur eryth | Lymph>eryth>lymph>ES>eryth | Open | Silent | Early | Active | 46, 47; this work |
| WT | Hum lymph | None | Closed | Silent | Late | Active | 11, 16, 35, 60 |
| WT | Avian lymph | Lymph>eryth>lymph | —b | — | Late | — | This work |
| ΔHS2-5 | Avian lymph | Lymph>eryth>lymph | — | — | Late | — | This work |
WT, wild type; hum, human; eryth, erythroid; mur, murine; lymph, lymphoid; ES, embryonic stem cells.
—, not determined.
How might such elements control activity of the origin, located over 50 kb away? The mechanism does not simply involve maintaining an open chromatin structure or transcriptional activity, since the normal origin is functional in nonerythroid cells, in which the locus is DNase resistant and transcriptionally silent (Table 1 and see below) (35). It is possible, however, that some other aspect of chromatin structure affects origin function; recently, for example, we have demonstrated that in erythroid cells, the Hispanic locus is characterized by hypoacetylation of histones H3 and H4, while these histones are hyperacetylated in the wild-type and Δ2-5 loci (47). Alternatively, elements within the Hispanic deletion endpoints may influence origin choice directly, by recruiting proteins that interact with origin sequences (e.g., origin recognition complex [ORC] and Purα) and assisting in the assembly of an active initiation complex. In addition, the replication control elements could control origin choice by localizing the locus to a nuclear compartment (such as the nuclear matrix or scaffold) in which activity of the normal origin is favored; consistent with this model, we find that while the Hispanic deletion locus is localized near pericentromeric heterochromatin in interphase erythroid nuclei, the wild-type and ΔHS2-5 loci are localized away from this heterochromatic compartment (see below) (47).
Analysis of the human β-globin origin of replication.
Two independent analyses of leading-strand polarity demonstrated that the entire locus (>135 kb) is replicated by forks that emanate from a ∼2.5-kb region at the β-globin promoter and gene; a nascent strand abundance assay (which analyzed BrdU-labeled nascent strands) confirmed initiation activity in this region (Fig. 1) (1, 35). Our results, obtained with lambda exonuclease-resistant nascent strands, demonstrate replication initiation within a ∼0.8-kb region roughly centered on the β-globin gene transcription start site. We cannot exclude, based on these results, the possibility that other initiation sites exist in the vicinity of the β-globin gene or elsewhere in the locus. However, the fact that the initiation site that we have identified is centrally located within the region delineated by the leading-strand analyses suggests that this is the primary site from which the entire locus is replicated.
The enrichments that we observe with our assay at the human β-globin origin are modest compared to a previous quantitative analysis of this origin, which revealed as much as ∼100-fold enrichment of origin-proximal sequences (2). This previous study analyzed an 8-kb fragment containing the human β-globin origin, inserted at an ectopic site in nonerythroid simian cells. At this integration site, the human β-globin origin fragment was found to be early replicating (in contrast to the late-replicating behavior of the endogenous locus in human nonerythroid cells) (11, 16), suggesting that the ectopic integration site and/or transgene sequences have a dominant effect on activity of this origin fragment. Technical differences (competitive PCR versus duplex PCR, BrdU-labeled versus lambda exonuclease-resistant nascent strands) further complicate direct comparison of numerical enrichments. Application of quantitative assays to other metazoan origins (chicken, hamster, and human) has yielded intermediate enrichment values (e.g., 8- to 11-fold) (36, 38, 44), suggesting that endogenous origins may not typically exhibit the very high enrichments seen for the transgenic human β-globin origin. However, although differences in techniques and basic experimental design are likely explanations for variations in the observed degree of enrichment, bona fide differences in origin behavior may also be responsible. Frequency of initiation from a given origin within a population, broad versus narrow initiation “zones,” and abortive initiations (without elongation) are possible factors that may affect apparent enrichment. Future studies using assays that can distinguish between these possibilities will be helpful in understanding the behavior and regulation of higher eukaryotic origins.
Origin choice does not correlate with transcription or chromatin structure.
There is abundant evidence in a number of systems that origin activity can be influenced by chromatin structure and nearby transcriptional activity. For example, in viruses and yeasts, transcription factors have been shown to stimulate origin activity (reviewed in references 43 and 55) and transcription factor binding sites contribute to autonomously replicating sequence (ARS) activity in mammalian cells (29, 53). Activity of an ARS in Saccharomyces cerevisiae is dependent on being devoid of nucleosomes (49), and chromatin structure has an effect on the specification of origins in an in vitro replication system based on Xenopus egg extracts (37).
Despite these suggestions, we and others have found that neither globin gene expression nor an open chromatin structure is a requirement for initiation from the normal origin in the human β-globin locus (Table 1). Although a requirement for active transcription can be ruled out, we cannot rule out the possibility that a transcription factor(s) may be bound to the inactive β-globin promoter in the context of the 5′HS2-5-deleted locus and that this binding may have an effect on origin function. Such factors need not be erythroid specific (e.g., Sp1 binding sites exist in the β-globin promoter [51]) and their binding may be responsible for activation of this origin in nonerythroid environments as well. Particularly intriguing is the presence of a binding site for the Purα protein 45 bp upstream of the β transcription start site; Purα is involved in both transcription and replication of viral DNA in mammalian cells and colocalizes with sites of cellular DNA synthesis (4, 9, 30).
Replication timing correlates with chromatin structure but not transcription of the locus.
A variety of observations suggest a link between transcription and replication timing. Constitutively active housekeeping genes, for example, are typically early replicating, while most tissue-specific loci are early replicating in cells in which they are expressed and late replicating in cells in which they are not (26; reviewed in references 28 and 48). In addition, significant changes in both replication timing and transcriptional activity occur during X chromosome inactivation in mammals (54, 56), immunoglobulin heavy-chain rearrangement in B cells (6, 8), and in chromosomal translocations, including those that lead to human leukemias (14, 24, 34). Despite these possibilities, our results demonstrate that the transcriptionally inactive ΔHS2-5 locus in MEL and GM979 erythroid cells is early replicating. Thus, we conclude that in the human β-globin locus, globin gene expression is not necessary for early replication in an erythroid environment and that early replication is not sufficient to achieve globin gene expression.
Our results do, however, suggest a relationship between chromatin structure and replication timing: wild-type and ΔHS2-5 loci are early replicating and in an open conformation in erythroid cells, while the Hispanic deletion locus in erythroid cells and the wild-type locus in nonerythroid cells are closed and late replicating (Table 1). Recent studies of the human α-globin locus on chromosome 16 also reached the conclusion that early replication timing correlates with an open chromatin structure (50). Thus, despite fundamental differences in the regulation and chromosomal context of the α- and β-globin loci, in both cases a correlation is seen between replication timing and chromatin structure.
An open chromatin structure could be either a cause or an effect of early replication timing of the locus. For example, an open chromatin structure may promote early replication by allowing the rapid binding of replication proteins to the locus at the onset of S phase. On the other hand, passage of a replication fork (and the associated disruption of chromatin structure) provides an opportunity for chromatin-opening factors to gain access to the locus; if such factors are limiting in the nucleus, early-replicating sequences may be at an advantage for binding of these factors and therefore may be more likely to adopt an open conformation. Alternatively, despite the correlation between them, early replication and open chromatin structure may be completely independent properties. A final possibility is that both open chromatin structure and early replication timing are the result of some common upstream regulatory event, such as sequestration of the locus in a particular nuclear compartment conducive to early replication and open chromatin. Our recent finding that the Hispanic locus is localized near centromeric heterochromatin in interphase nuclei while the wild-type and ΔHS2-5 loci are not (47) makes such a model attractive and is in agreement with our earlier observations regarding nuclear localization of active and inactive HS2-containing transgenes in human K562 erythroid cells (20). Although it has been demonstrated that early- and late-replicating sequences occupy distinct positions in the nucleus (39–41), the relationship between these replication timing compartments and pericentromeric heterochromatin remains to be determined. Clearly, understanding the role that nuclear compartmentalization plays in controlling gene activity, chromatin structure, and replication timing will be an important avenue for future research.
ACKNOWLEDGMENTS
This work was supported by a fellowship from the American Cancer Society to D.M.C., a fellowship from the Deutsche Forschungsgemeinschaft to D.S., a fellowship from the Leukemia Research Foundation to A.R., and NCI grant CA54337 and NIH grants HL57620 and DK44746 to M.G.
We thank Jim Roberts and Ross Hardison for critical reading of the manuscript; R. Scott Hansen and Mirit Aladjem for helpful discussions; and Urszula Maliszewski, Agnes Telling, and the FHCRC Flow Cytometry and Image Analysis Facilities for technical assistance.
REFERENCES
- 1.Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier R E, Wahl G M, Epner E M. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem M I, Rodewald L W, Kolman J L, Wahl G M. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 3.Bender M, Bulger M, Close J, Groudine M. Globin gene switching and DNaseI sensitivity of the endogenous β globin locus in mice do not require the locus control region. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 4.Bergemann A D, Johnson E M. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol. 1992;12:1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown B A, Padgett R W, Hardies S C, Hutchison C A, Edgell M H. Beta-globin transcript found in induced murine erythroleukemia cells is homologous to the beta h0 and beta h1 genes. Proc Natl Acad Sci USA. 1982;79:2753–2757. doi: 10.1073/pnas.79.9.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown E H, Iqbal M A, Stuart S, Hatton K S, Valinsky J, Schildkraut C L. Rate of replication of the murine immunoglobulin heavy-chain locus: evidence that the region is part of a single replicon. Mol Cell Biol. 1987;7:450–457. doi: 10.1128/mcb.7.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulger M, van Doorninck J H, Saitoh N, Telling A, Farrell C, Bender M A, Felsenfeld G, Axel R, Groudine M. Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calza R E, Eckhardt L A, DelGiudice T, Schildkraut C L. Changes in gene position are accompanied by a change in time of replication. Cell. 1984;36:689–696. doi: 10.1016/0092-8674(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 9.Chang C F, Gallia G L, Muralidharan V, Chen N N, Zoltick P, Johnson E, Khalili K. Evidence that replication of human neurotropic JC virus DNA in glial cells is regulated by the sequence-specific single-stranded DNA-binding protein Pur alpha. J Virol. 1996;70:4150–4156. doi: 10.1128/jvi.70.6.4150-4156.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePamphilis M L. Origins of DNA replication. In: DePamphilis M, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 48–86. [Google Scholar]
- 11.Dhar V, Mager D, Iqbal A, Schildkraut C L. The coordinate replication of the human beta-globin gene domain reflects its transcriptional activity and nuclease hypersensitivity. Mol Cell Biol. 1988;8:4958–4965. doi: 10.1128/mcb.8.11.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhar V, Skoultchi A I, Schildkraut C L. Activation and repression of a beta-globin gene in cell hybrids is accompanied by a shift in its temporal replication. Mol Cell Biol. 1989;9:3524–3532. doi: 10.1128/mcb.9.8.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieken E S, Epner E M, Fiering S, Fournier R E, Groudine M. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nat Genet. 1996;12:174–182. doi: 10.1038/ng0296-174. [DOI] [PubMed] [Google Scholar]
- 14.Disteche C M, Eicher E M, Latt S A. Late replication in an X-autosome translocation in the mouse: correlation with genetic inactivation and evidence for selective effects during embryogenesis. Proc Natl Acad Sci USA. 1979;76:5234–5238. doi: 10.1073/pnas.76.10.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driscoll M C, Dobkin C S, Alter B P. Gamma delta beta-thalassemia due to a de novo mutation deleting the 5′ beta-globin gene activation-region hypersensitive sites. Proc Natl Acad Sci USA. 1989;86:7470–7474. doi: 10.1073/pnas.86.19.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epner E, Forrester W C, Groudine M. Asynchronous DNA replication within the human beta-globin gene locus. Proc Natl Acad Sci USA. 1988;85:8081–8085. doi: 10.1073/pnas.85.21.8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epner E, Reik A, Cimbora D, Telling A, Bender M A, Fiering S, Enver T, Martin D I, Kennedy M, Keller G, Groudine M. The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 18.Epner E, Rifkind R A, Marks P A. Replication of alpha and beta globin DNA sequences occurs during early S phase in murine erythroleukemia cells. Proc Natl Acad Sci USA. 1981;78:3058–3062. doi: 10.1073/pnas.78.5.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 20.Francastel C, Walters M C, Groudine M, Martin D I K. A functional enhancer supresses silencing of a transgene and prevents its localization close to centromeric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- 21.Gerbi S A, Bielinsky A K. Replication initiation point mapping. Methods. 1997;13:271–280. doi: 10.1006/meth.1997.0526. [DOI] [PubMed] [Google Scholar]
- 22.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 23.Groudine M, Kohwi-Shigematsu T, Gelinas R, Stamatoyannopoulos G, Papayannopoulou T. Human fetal to adult hemoglobin switching: changes in chromatin structure of the beta-globin gene locus. Proc Natl Acad Sci USA. 1983;80:7551–7555. doi: 10.1073/pnas.80.24.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guinta D R, Tso J Y, Narayanswami S, Hamkalo B A, Korn L J. Early replication and expression of oocyte-type 5S RNA genes in a Xenopus somatic cell line carrying a translocation. Proc Natl Acad Sci USA. 1986;83:5150–5154. doi: 10.1073/pnas.83.14.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen R S, Canfield T K, Lamb M M, Gartler S M, Laird C D. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 26.Hatton K S, Dhar V, Brown E H, Iqbal M A, Stuart S, Didamo V T, Schildkraut C L. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgs D R. Do LCRs open chromatin domains? Cell. 1998;95:299–302. doi: 10.1016/s0092-8674(00)81761-4. [DOI] [PubMed] [Google Scholar]
- 28.Holmquist G P. Role of replication time in the control of tissue-specific gene expression. Am J Hum Genet. 1987;40:151–173. [PMC free article] [PubMed] [Google Scholar]
- 29.Iguchi-Ariga S M, Ogawa N, Ariga H. Identification of the initiation region of DNA replication in the murine immunoglobulin heavy chain gene and possible function of the octamer motif as a putative DNA replication origin in mammalian cells. Biochim Biophys Acta. 1993;1172:73–81. doi: 10.1016/0167-4781(93)90271-e. [DOI] [PubMed] [Google Scholar]
- 30.Itoh H, Wortman M J, Kanovsky M, Uson R R, Gordon R E, Alfano N, Johnson E M. Alterations in Pur(alpha) levels and intracellular localization in the CV-1 cell cycle. Cell Growth Differ. 1998;9:651–665. [PubMed] [Google Scholar]
- 31.Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp Quant Biol. 1964;28:329–348. [Google Scholar]
- 32.Jones C, Penny L, Mattina T, Yu S, Baker E, Voullaire L, Langdon W Y, Sutherland G R, Richards R I, Tunnacliffe A. Association of a chromosome deletion syndrome with a fragile site within the proto-oncogene CBL2. Nature. 1995;376:145–149. doi: 10.1038/376145a0. [DOI] [PubMed] [Google Scholar]
- 33.Kalejta R F, Li X, Mesner L D, Dijkwel P A, Lin H B, Hamlin J L. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol Cell. 1998;2:797–806. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- 34.Karube T, Watanabe S. Analysis of the chromosomal DNA replication pattern using the bromodeoxyuridine labeling method. Cancer Res. 1988;48:219–222. [PubMed] [Google Scholar]
- 35.Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human beta-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T, Rein T, DePamphilis M L. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol Cell Biol. 1998;18:3266–3277. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawlis S J, Keezer S M, Wu J R, Gilbert D M. Chromosome architecture can dictate site-specific initiation of DNA replication in Xenopus egg extracts. J Cell Biol. 1996;135:1207–1218. doi: 10.1083/jcb.135.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malott M, Leffak M. Activity of the c-myc replicator at an ectopic chromosomal location. Mol Cell Biol. 1999;19:5685–5695. doi: 10.1128/mcb.19.8.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manders E M, Stap J, Brakenhoff G J, van Driel R, Aten J A. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 40.Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Keefe R T, Henderson S C, Spector D L. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orr-Weaver T L, Johnston C G, Spradling A C. The role of ACE3 in Drosophila chorion gene amplification. EMBO J. 1989;8:4153–4162. doi: 10.1002/j.1460-2075.1989.tb08600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pederson D, Heintz N. Transcription factors and DNA replication. R. G. Austin, Tex: Landes; 1994. [Google Scholar]
- 44.Phi-van L, Stratling W H. An origin of bidirectional DNA replication is located within a CpG island at the 3′ end of the chicken lysozyme gene. Nucleic Acids Res. 1999;27:3009–3017. doi: 10.1093/nar/27.15.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raich N, Enver T, Nakamoto B, Josephson B, Papayannopoulou T, Stamatoyannopoulos G. Autonomous developmental control of human embryonic globin gene switching in transgenic mice. Science. 1990;250:1147–1149. doi: 10.1126/science.2251502. [DOI] [PubMed] [Google Scholar]
- 46.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biology. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schübeler D, Francastel C, Cimbora D M, Reik A, Martin D I K, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 48.Simon I, Cedar H. Temporal order of DNA replication. In: DePamphilis M, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 387–408. [Google Scholar]
- 49.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 50.Smith Z E, Higgs D R. The pattern of replication at a human telomeric region (16p13.3): its relationship to chromosome structure and gene expression. Hum Mol Genet. 1999;8:1373–1386. doi: 10.1093/hmg/8.8.1373. [DOI] [PubMed] [Google Scholar]
- 51.Stamatoyannopoulos G, Nienhuis A W. Hemoglobin switching. In: Stamatoyannopoulos G, Nienhuis A W, Majerus P W, Varmus H, editors. The molecular basis of blood disease. W. B. Philadelphia, Pa: Saunders; 1994. pp. 107–155. [Google Scholar]
- 52.Stillman B. DNA replication: replicator renaissance. Nature. 1993;366:506–507. doi: 10.1038/366506a0. [DOI] [PubMed] [Google Scholar]
- 53.Taira T, Iguchi-Ariga S M, Ariga H. A novel DNA replication origin identified in the human heat shock protein 70 gene promoter. Mol Cell Biol. 1994;14:6386–6397. doi: 10.1128/mcb.14.9.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takagi N. Differentiation of X chromosomes in early female mouse embryos. Exp Cell Res. 1974;86:127–135. doi: 10.1016/0014-4827(74)90657-0. [DOI] [PubMed] [Google Scholar]
- 55.van der Vliet P. Roles of transcription factors in DNA replication. In: DePamphilis M, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 87–118. [Google Scholar]
- 56.Wahrman J, Richler C, Neufeld E, Friedmann A. The origin of multiple sex chromosomes in the gerbil Gerbillus gerbillus (Rodentia: Gerbillinae) Cytogenet Cell Genet. 1983;35:161–180. doi: 10.1159/000131863. [DOI] [PubMed] [Google Scholar]
- 57.Widrow R J. Ph.D. thesis. Seattle: University of Washington; 1999. [Google Scholar]
- 58.Widrow R J, Hansen R S, Kawame H, Gartler S M, Laird C D. Very late DNA replication in the human cell cycle. Proc Natl Acad Sci USA. 1998;95:11246–11250. doi: 10.1073/pnas.95.19.11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiebauer K, Gumucio D L, Jones J M, Caldwell R M, Hartle H T, Meisler M H. A 78-kilobase region of mouse chromosome 3 contains salivary and pancreatic amylase genes and a pseudogene. Proc Natl Acad Sci USA. 1985;82:5446–5449. doi: 10.1073/pnas.82.16.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yagi M, Gelinas R, Elder J T, Peretz M, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Chromatin structure and developmental expression of the human alpha-globin cluster. Mol Cell Biol. 1986;6:1108–1116. doi: 10.1128/mcb.6.4.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zitnik G, Hines P, Stamatoyannopoulos G, Papayannopoulou T. Murine erythroleukemia cell line GM979 contains factors that can activate silent chromosomal human gamma-globin genes. Proc Natl Acad Sci USA. 1991;88:2530–2534. doi: 10.1073/pnas.88.6.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]