Abstract
Neonatal calf mortality is a major concern to livestock sector worldwide. Neonatal calf diarrhoea (NCD), an acute severe condition causes morbidity and mortality in calves. Amongst various pathogens involved in NCD, E. coli is considered as one of the major causes. The study was targeted to characterize E. coli isolates from neonatal calves for diarrhoeagenic Escherichia coli (DEC) types (pathotyping), antimicrobial resistance (AMR) profiling and to correlate with epidemiological parameters. From neonates, a total of 113 faecal samples were collected, out of that 308, lactose fermenting colonies were confirmed as E. coli. Pathotypable isolates (12.3%) were represented by STEC (6.1%), EPEC (2.9%), ETEC (1.9%), EAEC (0.9%) and EHEC (0.3%). Occurrence of STEC was more in non-diarrhoeic calves, whereas ETEC was observed more in diarrhoeic calves. EPEC occurrence was observed in both diarrhoeic and non-diarrhoeic calves. Fishers extract test showed no significant association for occurrence of DEC types to type of dairies, health status, species, breed, age and sex of neonatal calves. Two hundred and eighty isolates were tested for antimicrobial susceptibility. The isolates showed maximum resistance towards ampicillin (55.4%) followed by tetracycline (54.3%), while minimum resistance was observed towards meropenem (2.5%). Multidrug resistant E. coli isolates were found to be 139 (49.6%), and Extended-spectrum beta-lactamase (ESBL) producers were 120 (42.9%). DEC pathotypes like STEC, ETEC, EHEC and EAEC that are also multidrug resistant present in neonatal calves have zoonotic potential and hence are of public health significance.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11259-021-09857-5.
Keywords: Neonatal calves, Diarrhoeagenic Escherichia coli (DEC), Antimicrobial susceptibility, Pathotype, India
Introduction
Neonatal calf mortality is one of the most common losses in dairy farming which includes direct loss due to neonate’s death and indirect losses like treatment cost, reduced lifetime, productivity and survivorship (Waltner-Toews et al. 1986; Randhawa et al. 2012). As per Blood and Radostits (1989), there is a 40% reduction in profit if calf mortality is of 20%. Calf mortality in India mostly ranged from 12.5 to 30% (Verma et al. 1980), which may be as high as 81% in rare conditions (Tiwari et al. 2007). Infectious causes lead to neonatal diarrhoea or calf scour, septicaemia and pneumonia (Kochewad et al. 2013). Acute diarrhoea is the main reason behind 75% neonatal calf mortality during pre-weaning period (Muktar et al. 2015).
Neonatal calf diarrhoea (NCD) is considered as a complex multifactorial condition that causes calf morbidity and mortality in early months of life, characterized by onset of secretory diarrhoea and dehydration (Malik et al. 2012). Infectious causes that lead to NCD mainly include E. coli, rotavirus, coronavirus and Cryptosporidium parvum. The reported prevalence of E. coli ranges from 2.6 to 45.1% (Meganck et al. 2015). E. coli is a lactose fermenting Gram negative rod belonging to family Enterobacteriaceae with categorization of diarrhoea causing E. coli into Enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), Enterohemorrhagic E. coli (EHEC), Enteroinvasive E. coli (EIEC), Enteroaggregative E. coli (EAEC), Enterotoxigenic E. coli (ETEC), Diffusely adherent E. coli (DAEC), and Adherent invasive E. coli (AIEC), a new pathotype associated with Crohn’s disease which is not a diarrhoeagenic disease (Croxen et al. 2013; Liu 2015). Non-infectious risk factors that predispose for occurrence and severity of NCD are stress, inappropriate colostrum feeding with reference to quality, quantity and time of feeding and inadequate nutrition to pregnant dam mainly during the third trimester of gestation period. Along with these, poor hygienic condition of shed and nearby surrounding, overcrowding in calving shed and neonate’s calf shed and close association with adult cows and housing of various age groups together contribute towards risk factors associated with NCD. Along with the stated non-infectious factors, other factors like low temperature and/or contaminated incoming air and inadequate ventilation also play role in lowering the defence mechanism in early age of calf (Lance et al. 1992; Tewari 2012; Muktar et al. 2015).
Antimicrobial resistance (AMR) is an important emerging multi-faceted problem worldwide as it is affecting livelihood in terms of livestock morbidity and mortality, causing huge economic losses. In veterinary and human medicine, multidrug resistant E. coli is an emerging issue. E. coli being intrinsically sensitive to most of the antimicrobial classes, but through horizontal gene transfer, it can acquire and donate resistance genes. Gene transfer occurs through mobile genetic elements like plasmids (multidrug-resistant plasmids), transposons and gene cassettes. In India, few studies are reported in neonatal calves with respect to carriage of DEC. The present study was carried out in and around Bareilly, India with an objective to identify carriage of DEC in neonatal calves and to infer significant epidemiological factors. Considering present threat of antimicrobial resistance, E. coli isolates were also screened for antimicrobial susceptibility.
Materials and methods
Sample collection
Calves up to one month age were selected for faecal sample collection, from Bareilly city, Uttar Pradesh, India. Sample collection was carried out from 24 dairy farms in the region, out of which two were organized and 22 were unorganized dairies. In the present study, government or Institute’s dairy farm is considered as organised dairy farm, where standard housing and management practices for keeping the cattle are practiced. The small farms which mushroomed in and around a city without standard housing and management practices of animals are classified as unorganised dairies. A total of 113 neonate’s faecal samples were collected, among that 57 were from diarrhoeic and 56 were from non-diarrhoeic calves. From each calf, approximately 10 g of faecal sample were collected per-rectally and transported on ice for laboratory processing.
Isolation and identification of E. coli
A loop full of faecal material was inoculated in nine ml MacConeky broth (HiMedia, India) and incubated overnight at 37 °C. Enriched culture was further plated on MacConkey agar (HiMedia, India) and incubated at 37 °C for 24 h. Three to four lactose fermenting colonies suspected as E. coli were further plated on selective media, Eosin methylene blue agar (HiMedia) and incubated at 37 °C for 24 h. Colonies producing green metallic sheen were further confirmed by gram staining and IMViC (HiMedia) test for confirmation as E. coli.
Pathotyping of E. coli isolates
Isolates confirmed as E. coli were processed for DEC pathotyping by PCR technique using boiled extracted DNA as template. The multiplex PCR protocol followed for detection of shiga toxin producing E. coli (stx1, stx2 and eae), enterohemorraghic E. coli (eae, stx1, stx2 and hly), enteropathogenic E. coli (eae and bfp), enterotoxigenic E. coli (elt, esta, and estb), enteroaggregative (aggR and pic) and enteroinvasive E. coli (ipaH and invE) (Vidal et al. 2005) is mentioned in Table S1. For enterotoxigenic E. coli detection, an additional multiplex PCR protocol was followed (Johura et al. 2017). The Enterotoxigenic E. coli MTCC 723 (H10407) strain was used as positive control. The primer sequence, gene name, amplicon sizes and references are listed in Table S1 and PCR protocols are represented in Table 1.
Table 1.
PCR protocols used in the present study. Reaction conditions employed for pathoyping of E. coli (1), Enterotoxigenic E. coli (ETEC) detection (2), carbapenem (3) and extended spectrum beta-lactamase (ESBL) resistance gene detections (4, 5, 6, 7)
| Sr.no | Initial activation | Denaturation | Annealing | Extension | Elongation | ||
|---|---|---|---|---|---|---|---|
| 1 | E. coli Pathotyping | Temperature | 95 °C | 95 °C | 63 °C | 72 °C | 72 °C |
| Duration | 5 min | 30 s | 1:30 s | 1:30 s | 15 min | ||
| Cycles | 1 | 35 | 35 | 35 | 1 | ||
| 2 | ETEC PCR | Temperature | 94 °C | 94 °C | 57 °C | 72 °C | 72 °C |
| Duration | 4 min | 1 min | 1 min | 1:30 s | 7 min | ||
| Cycles | 1 | 35 | 35 | 35 | 1 | ||
| 3 | Carbapenem Resistance genes (Two multiplex PCR assays) | Temperature | 94 °C | 94 °C | 60 °C | 72 °C | 72 °C |
| Duration | 10 min | 30 s | 40 s | 50 s | 5 min | ||
| Cycles | 1 | 35 | 35 | 35 | 1 | ||
| 4 | AmpC | Temperature | 94 °C | 94 °C | 60 °C | 72 °C | 72 °C |
| Duration | 5 min | 1 min | 1 min | 1 min | 5 min | ||
| Cycles | 1 | 30 | 30 | 30 | 1 | ||
| 5 | TEM | Temperature | 94 °C | 94 °C | 55 °C | 72 °C | 72 °C |
| Duration | 5 min | 1 min | 1 min | 1 min | 7 min | ||
| Cycles | 1 | 35 | 35 | 35 | 1 | ||
| 6 | SHV | Temperature | 94 °C | 94 °C | 60 °C | 72 °C | 72 °C |
| Duration | 5 min | 30 s | 50 s | 50 s | 10 min | ||
| Cycles | 1 | 35 | 35 | 35 | 1 | ||
| 7 | CTX-M | Temperature | 94 °C | 94 °C | 52 °C | 72 °C | 72 °C |
| Duration | 5 min | 25 s | 1 min | 1 min | 6 min | ||
| Cycles | 1 | 30 | 30 | 30 | 1 |
Phenotypic characterization of antimicrobial resistance
Antimicrobial susceptibility testing of isolates was performed against 14 antibiotics (HiMedia) belonging to eight different antimicrobial classes by disk diffusion method (CLSI 2018). The antibiotics selected were ampicillin; AMP (10 μg), ceftazidime; CAZ (30 μg), cefepime; CPM (30 μg), cefotaxime; CTX (30 μg), ciprofloxacin; CIP (5 μg), co-trimoxazole; COT (25 μg), colistin; CST (10 μg), gentamicin; GEN (10 μg), imipenem; IPM (10 μg), meropenem; MRP (10 μg), piperacillin; PI (100 μg), piperacillin-tazobactam; PIT (100/10 μg), sulphadiazine; SZ (100 μg) and tetracycline; TE (30 μg). Phenotypically extended-spectrum beta-lactamase (ESBL) production by E. coli was determined by combined disk diffusion method (CLSI 2018). The third generation cephalosporins used were cefotaxime and ceftazidime along with the ESBL inhibitor clavulanic (CLA) acid.
Genotypic characterization of antimicrobial resistance
E. coli isolates observed to be carbapenem resistant and as ESBL producer by phenotypic approach were processed for antimicrobial resistance gene detection based on PCR. Two standardized multiplex PCR reported for the detection of blaIMP, blaVIM, blaSPM, (Poirel et al. 2011) blaOXA-23 (Paul et al. 2017) and blaOXA-48, blaKPC, blaBIC and blaNDM (Poirel et al. 2011), respectively were employed for the detection of carbapenem resistance genes. The reference strains used as positive controls were blaVIM positive Klebsiella pneumoniae NCTC 13440, blaNDM positive E. coli ATCC 2469 and blaOXA-48 positive K. pneumoniae NCTC 13442. The primer details are listed in Table S1 and PCR protocols are represented in Table 1.
E. coli isolates were subjected to PCR assays for the detection of blaCTXM (Woodford et al. 2006), blaAmpC (Féria et al. 2002), blaTEM (Bhattacharjee et al. 2007), blaSHV (Lob et al. 2015) genes associated with ESBL production. A single multiplex PCR was employed for CTXM family detection, including CTXM-1, CTXM-2, CTXM-8, CTXM-9 and CTXM-25. The reference strains used as positive control include blaSHV positive K. pneumoniae ATCC 700603 and blaTEM positive E. coli ATCC 35218. The primer details are listed in Table S1 and PCR protocols are represented in Table 1.
Statistical analysis
Statistical analysis was performed using SPSS ver. 22.0 (IBM Corp., Armonk, NY) on Windows platform. The association between DEC occurrence, health status, and type of dairy, species, breeds, sex and age group was performed by Fisher’s exact test.
Results
Out of the 113 faecal samples collected from neonatal calves, 35 were from organized dairies and 78 were from unorganized dairies. Among 380 colonies (approximately three per sample) initially selected from MacConkey agar, 308 colonies produced green metallic sheen on EMB agar. These 308 isolates confirmed as E. coli by gram staining and biochemical characterization were subjected to pathotyping. Thirty eight isolates (12.3%) were found to be DEC. Among DEC, STEC: 19 isolates (6.1%) was found to be the most predominant followed by EPEC: nine isolates (2.9%), ETEC: six isolates (1.9%), EAEC: three isolates (0.9%) and EHEC: one isolate (0.3%). The prevalence of DEC pathotypes varied in organized dairies and unorganized dairies and was 10% and 13.5%, respectively. The details of distribution of different DEC are presented in Table 2. Statistically no significant difference (p > 0.05) was observed in occurrence of E. coli pathotypes in different types of dairy, species, breed, sex, health status, and age of neonatal calves.
Table 2.
Pathotype associations to health, farm and animal factors. Occurrence of diarrhoeagenic E. coli (DEC) with different factors like health, type of dairy, species and breed, sex, age group of neonatal calves. Statistically no significant difference (p = 15.68, p > 0.1) was observed in occurrence of E. coli pathotypes with different factors like, health, type of dairy, species and breed, sex, and age group of neonatal calves.
| Factors | No. | DEC (%) | STEC (%) | EPEC (%) | ETEC (%) | EAEC (%) | EHEC (%) | P value | |
|---|---|---|---|---|---|---|---|---|---|
| Health | Non-diarrhoeic | 151 | 17 (11.2) | 13 (8.6) | 01 (0.7) | 02 (1.3) | 01(0.7) | 0 | 0.86NS |
| Diarrhoeic | 157 | 21 (13.4) | 06 (3.8) | 08 (5.1) | 04 (2.5) | 02(1.3) | 01(0.6) | ||
| Type of dairy | Organized dairy | 100 | 10 (10.0) | 08 (8.0) | 02 (2.0) | 0 | 0 | 0 | 0.46NS |
| Unorganized dairy | 208 | 28 (13.5) | 11 (5.3) | 07 (3.4) | 06 (2.9) | 03(1.4) | 01 (0.5) | ||
| Species | Cattle | 176 | 24 (13.6) | 13 (7.4) | 07 (3.4) | 01 (0.6) | 03(1.7) | 0 | 0.49NS |
| Buffalo | 132 | 14 (10.6) | 06 (4.5) | 02 (1.5) | 05 (3.8) | 0 | 01 (0.7) | ||
| Breeds | Cattle Vrindavani | 34 | 03 (8.8) | 03 (8.8) | 0 | 0 | 0 | 0 | 0.60NS |
| Cattle Non-descript | 91 | 12 (13.2) | 02 (2.2) | 07 (7.7) | 0 | 03(3.3) | 0 | ||
| Cattle Crossbred | 51 | 09 (17.6) | 08 (15.7) | 0 | 01(1.9) | 0 | 0 | ||
| Buffalo Murrah | 120 | 13 (10.8) | 06 (5.0) | 02 (1.7) | 05 (4.2) | 0 | 0 | ||
| Buffalo Non- descript | 12 | 01 (8.3) | 0 | 0 | 0 | 0 | 01 (8.3) | ||
| Sex | Male | 162 | 22 (13.6) | 13 (8.0) | 4 (2.5) | 4 (2.5) | 0 | 1 (0.6) | 0.60NS |
| Female | 146 | 16 (10.9) | 06 (4.1) | 05 (3.4) | 02 (1.4) | 03(2.0) | 0 | ||
| Age group | 0-10 days | 67 | 05 (7.5) | 02 (2.9) | 02 (2.9) | 0 | 01(1.5) | 0 | 0.02* |
| 11-20 days | 90 | 19 (21.1) | 10 (11.1) | 05 (5.5) | 02 (2.2) | 02 (2.2) | 0 | ||
| 21-30 days | 151 | 13 (8.6) | 07 (4.6) | 02 (1.3) | 04 (2.6) | 0 | 01(0.7) |
NS- non significant; *- Significant at 95% confidence level
Out of 308, only 280 E. coli isolates were tested for antibiotic sensitivity test (ABST). Percentages of isolates observed to be resistant against various antibiotics are as follows: ampicillin (55.4%), tetracycline (54.3%), co-trimoxazole (44.6%), sulphadiazine (44.3%), cefotaxime (42.1%), ciprofloxacin (35.4%), cefepime (33.2%), ceftazidime (23.9%), imipenem (20.4%), piperacillin (13.2%), piperacillin-tazobactam (5.4%), gentamicin (3.9%), and meropenem (2.5%). In polymyxin group none of the E. coli isolates were resistant to colistin. Forty five isolates (16.1%) were resistant to one antibiotic, 20 (7.1%) to two antibiotics, 29 (10.4%) to three antibiotics, 34 (12.1%) to four antibiotics, 27 (9.6%) to five antibiotics, 17 (6.0%) to six antibiotics, 24 (8.6%) to seven antibiotics, 25 (8.9%) to eight antibiotics, four (1.4%) to nine antibiotics, two (0.7%) to ten antibiotics and two (0.7%) to more than ten antibiotics. Isolates sensitive to all antibiotics were 51(18.2%) out of 280. Multidrug resistant (MDR) E. coli isolates were found to be 139 (49.6%), which are resistant to at least three antimicrobial classes. The occurrence of antimicrobial resistance with respect to type of dairies, health status and species of neonatal calves are depicted in Table 3. Antimicrobial resistance pattern shown in diarrhoeagenic E. coli is depicted in Table 4.
Table 3.
Antimicrobial resistance pattern among E. coli isolates. Antimicrobial resistance pattern with respect to type of dairy, health status and species of neonatal calves
| Antibiotics | Total Number of Resistance E. cole isolates | |||||
|---|---|---|---|---|---|---|
| Type of Dairy | Health Status of neonate | Species of neonatal calves | ||||
| Organized Dairies (%) | Unorganized Dairies (%) | Non-diarrhoeic (%) | Diarrhoeic (%) | Cattle (%) | Buffalo (%) | |
| AMP | 63 (64.3) | 92 (50.5) | 72 (52.2) | 83 (58.4) | 80 (54.0) | 75 (56.8) |
| CAZ | 26 (26.5) | 41 (22.5) | 38 (27.5) | 29 (20.4) | 34 (22.9) | 33 (25) |
| CTX | 44 (44.9) | 74 (40.7) | 60 (43.5) | 58 (40.8) | 64 (43.2) | 54 (40.9) |
| CIP | 42 (42.8) | 57 (31.3) | 44 (31.9) | 55(38.7) | 55 (37.2) | 44 (33.3) |
| COT | 61 (62.2) | 64 (35.2) | 61 (44.2) | 64 (45.1) | 61 (41.2) | 64 (48.5) |
| CPM | 20 (20.4) | 73 (40.1) | 46 (33.3) | 47 (33.1) | 49 (33.1) | 44 (33.3) |
| GEN | 08 (8.2) | 03 (1.6) | 07 (5.1) | 04 (2.8) | 07 (4.7) | 04 (3.0) |
| IPM | 28 (28.6) | 29 (15.9) | 32 (23.2) | 25 (17.6) | 25 (16.9) | 32 (24.2) |
| MRP | 03 (3.1) | 04 (2.2) | 06 (4.3) | 01 (0.7) | 06 (4.0) | 01 (0.8) |
| PI | 07 (7.1) | 30 (16.5) | 20 (14.5) | 17 (11.9) | 14 (9.5) | 23 (17.4) |
| PIT | 01 (1.0) | 14 (7.7) | 11 (7.9) | 04 (2.8) | 10 (6.7) | 05 (3.8) |
| SZ | 63 (64.3) | 61 (33.5) | 61 (44.2) | 63 (44.4) | 60 (40.5) | 64 (48.5) |
| TE | 54 (55.1) | 98 (53.8) | 63 (45.6) | 89 (62.7) | 78 (52.7) | 74 (56.1) |
Table 4.
Antimicrobial resistance among different pathotypes of diarrhoeagenic E. coli (DEC). Antimicrobial resistance observed among isolates belonging to various DEC pathotypes are represented in both number of isolates and in percentage
| Antibiotics | STEC (%) | EPEC (%) | ETEC (%) | EAEC (%) |
|---|---|---|---|---|
| AMP | 06 (31.6) | 05(55.5) | 00 (0) | 03 (100) |
| CAZ | 01 (5.3) | 01 (11.1) | 00 (0) | 02 (66.7) |
| CTX | 05 (26.3) | 03 (33.3) | 00 (0) | 02 (66.7) |
| CIP | 01 (5.3) | 01 (11.1) | 00 (0) | 03 (100) |
| COT | 09 (47.4) | 03 (33.3) | 00 (0) | 03 (100) |
| CPM | 00 (0) | 01 (11.1) | 00 (0) | 02 (66.7) |
| IPM | 01 (5.3) | 00 (0) | 02 (33.3) | 00 (0) |
| MRP | 00 (0) | 00 (0) | 00 (0) | 01 (33.3) |
| PI | 00 (0) | 00 (0) | 00 (0) | 02 (66.7) |
| PIT | 00 (0) | 00 (0) | 00 (0) | 02 (66.7) |
| SZ | 09 (47.4) | 03 (33.3) | 00 (0) | 03 (100) |
| TE | 12 (63.2) | 04 (44.4) | 05 (83.3) | 03 (100) |
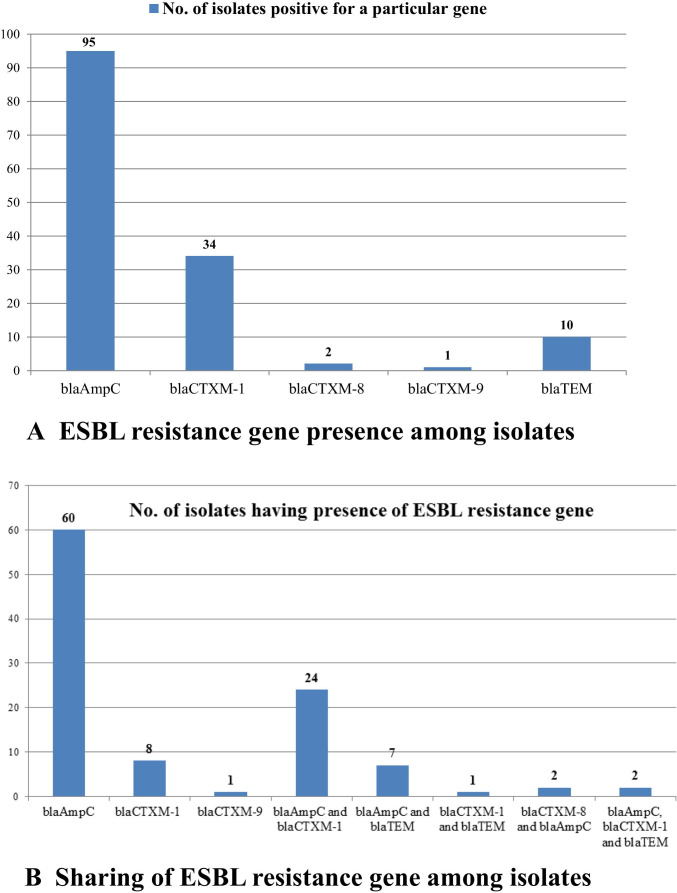
Altogether 120 (42.9%) E. coli isolates were found as positive for ESBL production by combined disk diffusion method. Based on cefotaxime and cefotaxime plus clavulanic acid resistance pattern, 80 (28.6%) E. coli isolates were found to be ESBL producers. Based on ceftazidime and ceftazidime plus clavulanic acid antibiotic resistance pattern, 88 (31.4%) E. coli isolates were found to be ESBL producers. Fifty three isolates (18.9%) were resistant to carbapenem drug phenotypically. Whereas, none of the isolates with phenotypic carbapenem resistance showed the presence of any of the carbapenem resistance genes (blaIMP, blaVIM, blaSPM, blaOXA-23, blaOXA-48, blaKPC, blaBIC and blaNDM) screened in the study. Among 120 phenotypic ESBL producers, 105 (87.5%) isolates were genotypically positive for ESBL, while 15 (12.5%) isolates were genotypically negative. Among the six genes, blaAmpC 90 (75%) was observed maximum, followed by blaCTXM-1 34 (28.3%) and blaTEM 10 (8.3%). Graphical representation of ESBL resistance gene presence in E. coli isolates is depicted in Fig. 1A. ESBL resistance genes shared among E. coli isolates were found as, presence of both blaCTXM-1 and blaAmpC were 24 (20.0%), blaAmpC and blaTEM were seven (5.8%), blaCTXM-8 and blaAmpC were two (1.7%), blaCTXM-1 and blaTEM was one (0.8%) and for three genes blaCTXM-1, blaAmpC, and blaTEM were two (1.7%). E. coli positive for only blaAmpC, blaCTXM-1, blaCTXM-9 alone were 60 (50%), 08(6.7%) and one (0.8%), respectively. Graphical representation of sharing of ESBL resistance gene among E. coli isolates is depicted in Fig. 1B.
Fig. 1.
ESBL resistance gene presence in E. coli isolates. ESBL resistance genes (six) and number of E. coli isolates are shown in X and Y axis respectively. Figure 1A represents ESBL resistance gene presence and Fig. 1B represents sharing of ESBL resistance genes
Discussion
In the study, DEC was characterized from both organized and unorganised dairies. In organized dairies, colostrum feeding in calves within eight hours after birth and thereafter milk feeding two times in a day was practiced. In case of unorganized dairies, insufficient colostrum feeding 8 h after birth was observed and suckling of dam milk was not regularly practised. Considering housing facility, organized dairies were having concrete flooring with separate area for calves. In unorganized dairies, muddy flooring with limited or absence of segregation in housing of adult and calves, healthy and diseased animals was observed. As per livestock schedule, deworming and vaccinations were carried out at the third and fourth month, respectively in organised farm. Deworming and vaccination was not practiced in calves in unorganized dairies. This indicates organized and unorganized diaries differed with respect to feeding, housing and management practices followed.
E. coli are a normal commensal of human and animal intestine whereas DEC is pathogenic by producing several virulence factors (Picco et al. 2015). In the present study, among diarrhoeagenic E. coli (DEC) pathotypes, STEC (6.1%) was found to be the most prevalent followed by EPEC (2.9%), ETEC (1.9%), and EAEC (0.9%) in faecal samples of neonatal calves. A similar observation with a higher prevalence of STEC (30.7%) followed by ETEC (12.7%), and EPEC (2.7%) was reported by Awad et al. (2020). In 2019, study conducted by Coura et al. reported the presence of STEC in diarrhoeic calves as 23.8% followed by Necrotoxic E. coli (NTEC) (19.0%), EPEC (3.6%), EHEC (3.6%) and ETEC (2.4%). In case of non-diarrhoeic calves, presence of NTEC as high as 42% followed by STEC (34%) and EHEC (2%) was reported (Coura et al. 2019).
Several studies reported that cattle are the reservoir for EHEC, and calves are the carrier of DEC pathotypes of public health importance (Suleiman et al. 2020; Cengiz and Adiguzel 2020). Intimin gene (eaeA) observed in EPEC were found to be more pathogenic in humans than in calves and are involved in zoonotic transmission (Thiry et al. 2017). Similarly, Janke et al. (1989) and Moxley and Smith (2010) reported that EPEC and STEC are less commonly associated with diarrhoea in cattle as they lack receptors in vascular endothelium that enables binding of STEC/EHEC leading to systemic disease. Hence, with the help of immunomodulation and intestinal colonization they survive and multiply in intestine. In the present study, isolates belonging to DEC did not showed significant association to diarrhoeic calves. Similar observations were made based on meta-analysis study data where STEC, EHEC and EPEC presence were reported in both diarrhoeic and healthy calves with some studies also reports their presence more in healthy calves (Kolenda et al. 2015).
Among DEC, ETEC is considered the predominant pathotype associated with NCD (Kolenda et al. 2015). In non-diarrhoeic calves, predominant pathotype was STEC followed by ETEC whereas in diarrhoeic calves, predominant pathotype was EPEC followed by STEC (Table 2). The ETEC isolation rate was low (1.9%) as compared to earlier study from Kashmir, India reporting 8 % E. coli isolates from diarrhoeic calves as ETEC (Wani et al. 2013). Also, the ETEC isolates recovered in the study from non-diarrhoeic calf had a past history of diarrhoea indicative of its possible role in neonatal diarrhoea. It has been reported that recovered and sub-clinically infected calves shed ETEC for months after recovery (Acres 1985). The lower prevalence of various pathotypes and especially ETEC in diarrhoeic samples in present study could also be due to the presence of other pathogens associated with diarrhoea and has not been ruled out in the study. Another possible reason could be due to the under detection of toxin genes which are mostly encoded in plasmids (Acres 1985) that are lost during repeated sub culturing of isolates (Sengupta 2011; Tazzyman and Bonhoeffer 2014).
The prevalence of DEC pathotypes was 10.0% and 13.5% in organized and unorganized dairies, respectively (Table 2). This indicates slightly more DEC is associated with unorganized dairies. Unorganized dairies from where samples were collected had muddy or wet flooring, improper shelter facilities, poor and small housing in comparison to herd size, no segregation in housing of adult and calves, as well as healthy and diseased animals. The dairy owners lack animal husbandry management knowledge, and dairy was operated by a family member in leisure time. These non-infectious factors may contribute to a higher prevalence of diarrhoeagenic E. coli as Yeshiwas and Fentahun (2017) found the occurrence of E. coli more in case of a muddy or wet floor. Recent study indicated significant association of climatic and management conditions on O157:H7 shedding, while the study indicated limited role of age or sex for shedding in Egyptian sheep (Kamel et al. 2015).
Neonatal calves of age group zero to ten days harboured STEC, EPEC and EAEC with 2.9%, 2.9% and 1.5% prevalence, respectively. The presence of DEC in this age group may be due to the alkaline pH of abomasum, truancy/absence of competing microflora and sluggish motility of intestine favouring their growth (Smith 1965). Also, bacterial fimbrial binding receptor is more expressed during younger age in calves favouring binding of pathogenic bacteria (Acres 1985; Dubreuil et al. 2016). In the present study, a higher presence of pathotype STEC (8.0%) and ETEC (2.5%) were observed in male neonatal calves. Male calves are considered being of no economic importance in dairy farms, and thus, are insufficiently fed with colostrum at initial hours after birth (Murugan et al. 2019). During the sample collection, it has been observed that diarrhoeic male calves were not provided with proper treatment in unorganised dairies. A significant association of DEC to age group 10 to 20 days was observed in this study (Table 2). The increased DEC colonization could be probably associated with stoppage or reduced milk feeding to calves after few weeks.
Among all antibiotics used for AMR study, the highest resistance was observed towards ampicillin (55.4%) followed by tetracycline (54.3%) (Table 4). In several countries, including India, ampicillin, tetracycline, and sulphonamide are the most commonly used antibiotics to treat animals, their misuse or overuse could be related to its high resistance levels in E. coli (Chantziaras et al. 2014; Maciel et al. 2019). Studies conducted on E. coli isolates exhibited 75% (Gharieb et al. 2019), 83% (Wani et al. 2013) and 87% (Gupta et al. 2017) resistance for ampicillin. Several studies also reported resistance toward ampicillin as 100% in E. coli isolates from calves (Donaldson et al. 2006; Mailk et al. 2013; Pandey et al. 2015; Mohammed et al. 2019). With respect to tetracycline resistance, the resistance level indicated in this study (54.3%) also corroborated earlier studies mostly ranging from 57% (Hang et al. 2019) to 63% (Srivani et al. 2017; Maciel et al. 2019). Resistance to tetracycline to even 100% has been reported in E. coli isolates (Gupta et al. 2017). The possible reason for acquiring E. coli ampicillin and tetracycline resistance in neonatal calves may be from their dams or surroundings or due to the transfer of resistant genes from one ecosystem to another (Johnson et al. 2007; Singh 2011).
ESBL resistance was observed as 42.9% in E. coli isolates. In the present study, resistance towards cephalosporins were highest for cefotaxime (42.1%) followed by cefepime (33.2%), and ceftazidime (23.9%). Wani et al. (2013) reported 83% resistance in E. coli isolates from calf to cephalosporins. Similar high resistance for ceftazidime (98%) and cefotaxime (91.8%) was reported by Srivani et al. (2017). Even 100% resistance to cefotaxime and 91.6% resistance to ceftazidime were reported by Batabyal et al. (2018). The result thus indicates the resistance pattern of cephalosporin in E. coli isolates from calf was lesser than earlier studies from India. Recent study in neonatal calves less than 2 weeks old in Germany revealed ESBL-producing enterobacteria on all farms and 96.5% of the calves investigated shed ESBL-positive bacteria (Waade et al. 2021). The lower resistance could be due to involvement of more strains from unorganized dairies that are less exposed to antibiotics in comparison to organized dairies.
Genotypic resistance was in correlation with phenotypic ESBL resistance but there was no correlation between phenotypic carbapenem resistance and genotypic resistance as none of the targeted carbapenem resistance genes were detected (Fig. 1). This possibly indicates the isolates either harbouring novel resistance genes or are with substantial genetic variations hindering successful PCR amplification. A similar study has been reported with less congruence within phenotypic and genotypic antibiotic resistance in E. coli isolates of calf origin, and revealed the presence of blaVIM carbapenemase gene in only 1 out of 81 phenotypically positive for carbapenem resistant isolates (Murugan et al. 2019). Another study also reported only 26 Gram negative bacilli isolates to be genotypically positive for carbapenemase gene while by disc diffusion test 111 isolates were phenotypically resistant. Among 26 isolates, presence of carbapenem gene distribution was blaNDM in 16, blaVIM in 8 and blaOXA48 in 2 isolates (Codjoe et al. 2019). Based on genome sequence, high genetic diversity was observed in genes conferring resistance to seven different antibiotic classes among E. coli strains isolated from veal calves (Kim et al. 2021). For food animal’s treatment there is a restriction for the use of carbapenem drug, so carbapenem resistance observed in livestock population may be either from natural environment or through human contact (Murugan et al. 2019).
In the present study, higher antimicrobial resistance was observed in organized dairies (Table 3) in comparison to unorganized dairies towards all antibiotics, except piperacillin and piperacillin - tazobactam. Another study carried out in piglets of organised farms showed similar kind of observation with ESBL occurrence of 64% (VinodhKumar et al. 2019). In unorganized diaries, antibiotic usage history was not known, whereas, the most commonly used antibiotics in organized farms were fluroquinolone, beta lactam and cephalosporins group. Resistance correlating usage in farms were observed for some antibiotics especially for ampicillin (beta lactam) showing 64.3% resistance, ciprofloxacin (quinolone) showing 42.8% resistance in case of organized dairies and cefotaxime (cephalosporins) showing 44.9% resistance (Table 3). Other possible reason may be the role of concrete flooring in organized diaries carrying more resistant isolates than the muddy or earthen floor, as concrete flooring led to ineffective cleaning and washing with mild detergent (Murugan et al. 2019).
Conclusion
The neonatal period of calf life, is considered to be the most critical period, as majority of mortality occurs due to diarrhoea, septicaemia, and pneumonia. Among bacterial causes, E. coli is one of the most important cause for NCD. Among 308 E. coli isolates from 24 farms, 12.3% represented DEC. Occurrence of STEC was highest in non-diarrhoeic calves whereas ETEC occurrence predominated in diarrhoeic calves. The prevalence of DEC pathotypes was lower in organized diaries, indicative of the importance of good farm management and animal husbandry practices followed there. Multi drug resistant isolates were found to be 49.6% among all isolates. This indicates the importance of E. coli infections in calves as well as its public health significance.
Supplementary Information
(DOCX 24 kb)
Acknowledgments
Financial support received under All India Network Programme on Neonatal Mortality in Farm Animals, Indian Council of Agricultural Research (ICAR), New Delhi is gratefully acknowledged.
Code availability
Not Applicable.
Abbreviations
- NCD
Neonatal calf diarrhoea
- DEC
Diarrhoeagenic E. coli
- EPEC
Enteropathogenic E. coli
- STEC
Shiga toxin-producing E. coli
- EHEC
Enterohemorrhagic E. coli
- EAEC
Enteroaggregative E. coli
- ETEC
Enterotoxigenic E. coli
- AMR
Antimicrobial resistance
- ESBL
Extended-spectrum beta-lactamase
Author’s contributions
DK, DS and PT conceptualized and designed the study, DK, ML, ATV and MP carried out sample collection and isolation. DK, VOR and LC carried out statistical analysis. ABV and SI carried out Antimicrobial Susceptibility Testing. PT, DS, PC and BS supervised the study. DK,VOR, DS and PT wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
All India Network Programme on Neonatal Mortality in Farm Animals, Indian Council of Agricultural Research (ICAR), New Delhi.
Declarations
Conflict of interest
Authors declare no conflict of interests.
Ethics approval
Not Applicable as animal experiments was not conducted for this research. All sample collections were made after approval and as per the guidelines of the Institutional Animal Ethics Committee, ICAR-IVRI.
Consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
D. K. Sinha, Email: sinhaivri@rediffmail.com
Prasad Thomas, Email: prasadthomas99@gmail.com.
References
- Acres SD. Enterotoxigenic Escherichia coli infections in newborn calves: a review. J Dairy Sci. 1985;68(1):229–256. doi: 10.3168/jds.S0022-0302(85)80814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antikainen J, Tarkka E, Haukka K, et al. New 16-plex PCR method for rapid detection of diarrheagenic Escherichia coli directly from stool samples. Eur J Clin Microbiol Infect Dis. 2009;28(8):899–908. doi: 10.1007/s10096-009-0720-x. [DOI] [PubMed] [Google Scholar]
- Awad WS, El-Sayed, AA, Mohammed FF et al (2020) Molecular characterization of pathogenic Escherichia coli isolated from diarrheic and in-contact cattle and buffalo calves. Trop Anim Health Pro 1-13. 10.1007/s11250-020-02343-1 [DOI] [PMC free article] [PubMed]
- Batabyal K, Banerjee A, Pal S, et al. Detection, characterization, and antibiogram of extended spectrum beta-lactamase Escherichia coli isolated from bovine milk samples in West Bengal. India. Vet World. 2018;11(10):1423. doi: 10.14202/vetworld.2018.1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee A, Sen MR, Anupurba S, et al. Detection of OXA-2 group extended-spectrum-β-lactamase-producing clinical isolates of Escherichia coli from India. J Antimicrob Chemother. 2007;60(3):703–704. doi: 10.1093/jac/dkm267. [DOI] [PubMed] [Google Scholar]
- Blood DC, Radostits OM (1989) Veterinary Medicine. 7th ed. ESBL. Oxford. P1502
- Brandal LT, Lindstedt BA, AAS L, et al. Octaplex PCR and fluorescence-based capillary electrophoresis for identification of human diarrheagenic Escherichia coli and Shigella spp. J Microbiol Methods. 2007;68(2):331–341. doi: 10.1016/j.mimet.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Cengiz S, Adiguzel MC (2020) Determination of virulence factors and antimicrobial resistance of E. coli isolated from calf diarrhea, part of eastern Turkey. Vet Fak Derg. 10.33988/auvfd.640990
- Chantziaras I, Boyen F, Callens B, et al. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals :report on seven countries. J Antimicrob Chemothery. 2014;69(3):827–834. doi: 10.1093/jac/dkt443. [DOI] [PubMed] [Google Scholar]
- CLSI (2018) Performance standards for antimicrobial susceptibility testing, 28th edn. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne
- Croxen MA, Law RJ, Scholz R et al (2013) Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26 (4), 822-880. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed]
- Codjoe FS, Donkor ES, Smith TJ, et al. Phenotypic and genotypic characterization of carbapenem-resistant gram-negative bacilli pathogens from hospitals in Ghana. Microb Drug Resist. 2019;25(10):1449–1457. doi: 10.1089/mdr.2018.0278. [DOI] [PubMed] [Google Scholar]
- Coura FM, Diniz SDA, Silva MX et al (2019) Virulence factors and phylotyping of Escherichia coli isolated from non-diarrheic and diarrheic water buffalo calves. Ciênc. Rural 49(5). 10.1590/0103-8478cr20180998
- Donaldson SC, Straley BA, Hegde NV, et al. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl Environ Microbiol. 2006;72(6):3940–3948. doi: 10.1128/AEM.02770-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil JD, Isaacson RE, Schifferli DM (2016) Animal enterotoxigenic Escherichia coli. Eco Sal Plus 7(1). 10.1128/2Fecosalplus.ESP-0006-2016 [DOI] [PMC free article] [PubMed]
- Féria C, Ferreira E, Correia JD, et al. Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J Antimicrob Chemother. 2002;49(1):77–85. doi: 10.1093/jac/49.1.77. [DOI] [PubMed] [Google Scholar]
- Furrer B, Candrian U, Lüthy J. Detection and identification of E. coli producing heat-labile enterotoxin type I by enzymatic amplification of a specific DNA fragment. Lett Appl Microbiol. 1990;10(1):31–34. doi: 10.1111/j.1472-765X.1990.tb00088.x. [DOI] [PubMed] [Google Scholar]
- Gharieb R, Fawzi E, Elsohaby I. Antibiogram, virulotyping and genetic diversity of Escherichia coli and Salmonella serovars isolated from diarrheic calves and calf handlers. Comp Immunol Microbiol Infect Dis. 2019;67:101367. doi: 10.1016/j.cimid.2019.101367. [DOI] [PubMed] [Google Scholar]
- Gupta MD, Islam M, Sen A, et al. Prevalence and antibiotic susceptibility pattern of Escherichia coli in cattle on Bathan and intensive rearing system. Microbes and Health. 2017;6(1):1–4. doi: 10.3329/mh.v6i1.34062. [DOI] [Google Scholar]
- Hang BPT, Wredle E, Börjesson S, et al. High level of multi drug-resistant Escherichia coli in young dairy calves in southern Vietnam. Trop Anim Health Pro. 2019;51(6):1405–1411. doi: 10.1007/s11250-019-01820-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke BH, Francis DH, Collins JE, et al. Attaching and effacing Escherichia coli infections in calves, pigs, lambs, and dogs. J Vet Diagn. 1989;1(1):6–11. doi: 10.1177/2F104063878900100104. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Sannes MR, Croy C et al (2007) Antimicrobial drug– resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002–2004. Emerg Infect Dis 13(6):838. 10.3201/2Feid1306.061576 [DOI] [PMC free article] [PubMed]
- Johura FT, Parveen R, Islam A, et al. Occurrence of hybrid Escherichia coli strains carrying Shiga toxin and heat-stable toxin in livestock of Bangladesh. Public Health Front. 2017;4:287. doi: 10.3389/fpubh.2016.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel M, El-Hassan DGA, El-Sayed A. Epidemiological studies on Escherichia coli O157: H7 in Egyptian sheep. Trop Anim Health Prod. 2015;47(6):1161–1167. doi: 10.1007/s11250-015-0843-2. [DOI] [PubMed] [Google Scholar]
- Kim SW, Van Kessel JAS, Haley BJ (2021) Genome sequences of antibiotic-resistant Escherichia coli isolated from veal calves in the United States. J Glob Antimicrob Resist. 10.1016/j.jgar.2021.04.024 [DOI] [PubMed]
- Kochewad SA, Singh JP, Patil VM, et al. Calf mortality. Indian Farming. 2013;62(10):23–26. [Google Scholar]
- Kolenda R, Burdukiewicz M, Schierack P. A systematic review and meta-analysis of the epidemiology of pathogenic Escherichia coli of calves and the role of calves as reservoirs for human pathogenic E coli. Front Cell Infect Mi. 2015;5:23. doi: 10.3389/fcimb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuralayanapalya SP, Patil SS, Hamsapriya S et al (2019) Prevalence of extended- spectrum beta- lactamase producing bacteria from animal origin: a systematic review and meta-analysis report from India. PloSone 14(9). 10.1371/journal.pone.0221771 [DOI] [PMC free article] [PubMed]
- Lance SE, Miller GY, Hancock DD, et al. Salmonella infections in neonatal dairy calves. J Am Vet Med Assoc. 1992;201(6):864–868. [PubMed] [Google Scholar]
- Liu D (2015) Diarrhoeagenic Escherichia coli. In Molecular Medical Microbiology 1133- 1146
- Lob SH, Kazmierczak KM, Badal RE, et al. Trends in susceptibility of Escherichia coli from intra-abdominal infections to ertapenem and comparators in the United States according to data from the SMART program, 2009 to 2013. Antimicrob Agents Chemother. 2015;59(6):3606–3610. doi: 10.1128/AAC.05186-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lortie LA, Dubreuil JD, Harel JOSEE. Characterization of Escherichia coli strains producing heat-stable enterotoxin b (STb) isolated from humans with diarrhea. J Clinic Microbiol. 1991;29(3):656–659. doi: 10.1128/jcm.29.3.656-659.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciel JF, Matter LB, Tasca C, et al. Characterization of intestinal Escherichia coli isolated from calves with diarrhea due to rotavirus and coronavirus. J Med Microbiol. 2019;68(3):417–423. doi: 10.1099/jmm.0.000937. [DOI] [PubMed] [Google Scholar]
- Mailk S, Kumar A, Verma AK, et al. Incidence and drug resistance pattern of collibacillosis in cattle and buffalo calves in Western Uttar Pradesh in India. J Anim Health Prod. 2013;1(1):15–19. [Google Scholar]
- Malik S, Verma AK, Kumar A, et al. Incidence of calf diarrhea in cattle and buffalo calves in Uttar Pradesh. India. Asian J Anim Vet Adv. 2012;7:1049–1054. doi: 10.3923/ajava.2012.1049.1054. [DOI] [Google Scholar]
- Meganck V, Hoflack G, Piepers S, et al. Evaluation of a protocol to reduce the incidence of neonatal calf diarrhoea on dairy herds. Prev Vet Med. 2015;118(1):64–70. doi: 10.1016/j.prevetmed.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed SAEM, Marouf SAEM, Erfana AM, et al. Risk factors associated with E. coli causing neonatal calf diarrhea. Saudi J Biol Sci. 2019;26(5):1084–1088. doi: 10.1016/j.sjbs.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxley RA, Smith DR. Attaching-effacing Escherichia coli infections in cattle. Veterinary Clinics: Food Animal Practice. 2010;26(1):29–56. doi: 10.1016/j.cvfa.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muktar Y, Mamo G, Tesfaye B, et al. A review on major bacterial causes of calf diarrhea and its diagnostic method. J Vet Med Anim Health. 2015;7(5):173–185. doi: 10.5897/JVMAH2014.0351. [DOI] [Google Scholar]
- Müller D, Greune L, Heusipp G, et al. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol. 2007;73(10):3380. doi: 10.1128/2FAEM.02855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan MS, Sinha DK, Kumar OV et al (2019) Epidemiology of carbapenem-resistant Escherichia coli and first report of blaVIM carbapenemases gene in calves from India. Epidemiol Infect 147. 10.1017/s0950268819000463 [DOI] [PMC free article] [PubMed]
- Ngeleka M, Pritchard J, Appleyard G, et al. Isolation and association of Escherichia coli AIDA-I/STb, rather than EAST1 pathotype, with diarrhea in piglets and antibiotic sensitivity of isolates. J Vet Diagn Invest. 2003;15(3):242–252. doi: 10.1177/2F104063870301500305. [DOI] [PubMed] [Google Scholar]
- Pandey A, Joshi N, Joshi KR, et al. Virulence attributes and antibiotic resistance pattern of E. coli isolated from human and animals. Asian J Anim Vet Adv. 2015;11:67–72. doi: 10.3923/ajava.2016.67.72. [DOI] [Google Scholar]
- Paul D, Ingti B, Bhattacharjee D, et al. An unusual occurrence of plasmid-mediated blaOXA-23 carbapenemase in clinical isolates of Escherichia coli from India. Int J Antimicrob Agents. 2017;49(5):642–645. doi: 10.1016/j.ijantimicag.2017.01.012. [DOI] [PubMed] [Google Scholar]
- Picco NY, Alustiza FE, Bellingeri RV, et al. Molecular screening of pathogenic Escherichia coli strains isolated from dairy neonatal calves in Cordoba province, Argentina. Rev Argent Microbiol. 2015;47(2):95–102. doi: 10.1016/j.ram.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Randhawa SS, Randhawa SS, Zahid UN, et al. Drug combination therapy in control of cryptosporidiosis in Ludhiana district of Punjab. J Parasit Dis. 2012;36(2):269–272. doi: 10.1007/s12639-012-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M, Austin S (2011) Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect Immun 79(7):2502- 2509. 10.1128/IAI.00127-11 [DOI] [PMC free article] [PubMed]
- Singh BR (2011) Drug resistant bacteria in the environment: In: Environmental Security, SR Garg, HD, Department of Veterinary Public Health,Veterinary College, LLRU Vet. and Animal Sciences, Hissar, IBDC Publishers, Lucknow, India. pp.313- 326
- Smith HW. The development of the flora of the alimentary tract in young animals. J Pathol. 1965;90:495–513. doi: 10.1002/path.1700900218. [DOI] [PubMed] [Google Scholar]
- Srivani M, Reddy YN, Subramanyam KV et al (2017) Prevalence and antimicrobial resistance pattern of Shigatoxigenic Escherichia coli in diarrheic buffalo calves. Vet World 10(7):774. 10.14202/2Fvetworld.2017.774-778 [DOI] [PMC free article] [PubMed]
- Suleiman JM, Saed OAS, AL-Kubaisi SMA et al (2020) Molecular characterization of Eae and Stxs genes for E. coli O157:H7 isolates from calves. Medico legal update 20(1):788-791. 10.37506/mlu.v20i1.464
- Tazzyman SJ, Bonhoeffer S. Why there are no essential genes on plasmids. Mol Biol Evol. 2014;32(12):3079–3088. doi: 10.1093/molbev/msu293. [DOI] [PubMed] [Google Scholar]
- Tewari A (2012) Neonatal calf Diarrhoea. Indian Dairyman p54
- Thiry D, Saulmont M, Takaki S, et al. Enteropathogenic Escherichia coli O80:H2 in young calves with diarrhea, Belgium. Emerg Infect Dis. 2017;23(12):2093. doi: 10.3201/2Feid2312.170450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari R, Sharma M, Singh B. Buffalo calf health care in commercial dairy farms: a field study in Uttar Pradesh (India) Livest Res Rural Dev. 2007;19(3):8. [Google Scholar]
- Verma GS, Sadana DK, Basu SB et al (1980) Studies on mortality in buffalo calves. Indian J Dairy Sci
- Vidal M, Kruger E, Durán C, et al. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clinic Microbiol. 2005;43(10):5362–5365. doi: 10.1128/JCM.43.10.5362-5365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinodh Kumar OR, Singh BR, Sinha DK et al (2019) Risk factor analysis, antimicrobial resistance and pathotyping of Escherichia coli associated with pre-and post-weaning piglet diarrhoea in organised farms, India. Epidemiol Infect 147. 10.1017/S0950268819000591 [DOI] [PMC free article] [PubMed]
- Waade J, Seibt U, Honscha W, et al. Multidrug-resistant enterobacteria in newborn dairy calves in Germany. PloS one. 2021;16(3):e0248291. doi: 10.1371/journal.pone.0248291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltner-Toews D, Martin SW, Meek AH. An epidemiological study of selected calf pathogens on Holstein dairy farms in south western Ontario. Can J Vet Res. 1986;50(3):307. [PMC free article] [PubMed] [Google Scholar]
- Wani SA, Hussain I, Beg SA et al (2013) Diarrhoeagenic Escherichia coli and salmonellae in calves and lambs in Kashmir :absence, prevalence and antibiogram. Revscitech Offint Epiz 32:1-7. 10.20506/rst.32.2.2213 [DOI] [PubMed]
- Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2006;57(1):154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- Yeshiwas T, Fentahun WM (2017) The prevalence of E. coli from diarrheic calves and their antibiotic sensitivity test in selected dairy farms of Debre Zeit, Ethiopia. Adv Biotech & Micro 6(1):555680
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 24 kb)