Abstract
The pertussis toxin (PT) promoter region is a frequently used target for DNA-based diagnosis of pertussis and parapertussis infections. The reported polymorphism in this region has also allowed discrimination of species in mixtures with several Bordetella species by their specific PCR amplicon restriction patterns. In the present study, we investigated the degree of polymorphism in order to confirm the reliability of the assay. Five different sequence types of the amplified 239- or 249-bp region were found among the 33 Bordetella pertussis, B. parapertussis, and B. bronchiseptica American Type Culture Collection reference strains and patient isolates analyzed. According to the sequences that were obtained and according to the PT promoter sequences already available in the databases, restriction enzyme analysis with TaqI, BglI, and HaeII, which gave four different patterns, can be performed to reliably identify B. pertussis, B. parapertussis, and B. bronchiseptica.
In pertussis, an early diagnosis is of vital importance both for the antibiotic treatment to effectively reduce symptoms and to avoid the spread of this highly contagious disease (9, 40). Both isolation of Bordetella pertussis from nasopharyngeal secretions by culture and serological diagnosis are highly specific methods, but the sensitivity of culture is low (32). Since culture takes 3 to 12 days to complete (24) and paired serum specimens collected 1 month apart are preferred for definitive diagnosis (21), more rapid methods are needed. In several studies, detection of B. pertussis by PCR has been compared to conventional methods, i.e., culture, serology, and clinical diagnosis (7, 10, 19, 27, 35, 37, 44). Different regions in Bordetella sp. DNA are used as targets in diagnostic PCR, such as insertion sequences (8, 19, 22, 26, 33, 45), the adenylate cyclase gene (6), the porin gene (25), and the pertussis toxin (PT) promoter region (22, 36, 41, 42). PT is an important virulence factor for B. pertussis. Both the PT promoter and the structural genes are also present in B. parapertussis and B. bronchiseptica (3). Due to a number of mutations in the PT promoter regions of these two species, PT is not expressed, resulting in milder symptoms with a lower frequency of spasmodic cough compared to the frequency in patients with classical pertussis (15, 30). An advantage to using the PT promoter in pertussis diagnostics is the possibility of amplifying B. pertussis, B. parapertussis, and B. bronchiseptica DNAs in a single PCR with generic primers, with subsequent differentiation of the three species by restriction analysis of the PCR amplicons (36, 37, 41). This enables both a rapid diagnosis and epidemiological studies of the occurrence of the three species when they coexist in the same sample (37). A drawback to using the PT promoter region for diagnosis could be the high degree of polymorphism in this region. In recent years, studies involving genetic and functional analyses of the PT promoter and its regulation (2, 4, 12, 18, 29, 43, 46) have also resulted in publication of several different PT promoter sequences that differ from the B. pertussis sequence first described by Locht and Keith (28) and the B. parapertussis and B. bronchiseptica PT promoter sequences reported by Aricò and Rappuoli (3).
The aim of this study was to analyze in detail the PT promoter regions from a number of strains to investigate if this region can be used for reliable discrimination of the three major Bordetella species causing human infection. Swedish clinical isolates retrieved from 1975 to 1994 as well as American Type Culture Collection (ATCC) type strains of the three species were used for this purpose. In addition, clinical isolates from samples originally negative by a PT promoter PCR directly with nasopharyngeal aspirates but positive by culture (37) were analyzed. Another objective was to investigate if DNA sequence polymorphism could explain the lower diagnostic sensitivity obtained for patients with B. parapertussis infection than for patients with B. pertussis infection in a validation of a nested PT promoter PCR in a pertussis vaccine trial (37).
MATERIALS AND METHODS
Bacterial strains.
The clinical isolates and ATCC strains used in the study are listed in Table 1. All strains were obtained from the Swedish Institute for Infectious Disease Control, where they were cultured on charcoal medium with 40 mg of cephalexin per liter as described by Regan and Lowe (34). ATCC strains were originally cultured separately, aliquoted after harvest, and stored lyophilized, after which they were recultured for quality control. Patient strains were identified at the time of isolation by slide agglutination, biochemical verification, and serotyping as described elsewhere (14, 16, 17) and were stored at −70°C in a sucrose-phosphate-glutamate medium containing 2.5% bovine serum albumin. For bacterial lysates, approximately 10 μl of colony material was suspended in 0.5 ml of distilled water, and the mixture was incubated at 100°C for 10 min and frozen until amplified.
TABLE 1.
Strains used in the study
| Strain or patient isolate | Sequence type | Restriction pattern | Source | Year of isolation |
|---|---|---|---|---|
| B. pertussis | ||||
| KP92\2\92 | a | I | PVTa 92-95 | 1992 |
| KP92\9\92 | a | I | PVT 92-95 | 1992 |
| KP92\12\92 | a | I | PVT 92-95 | 1992 |
| KP92\42\92 | a | I | PVT 92-95 | 1992 |
| KP92\47\92 | a | I | PVT 92-95 | 1992 |
| KP92\357\93 | a | I | PVT 92-95 | 1993 |
| KP92\757\94 | a | I | PVT 92-95 | 1994 |
| KP92\1145\94 | a | I | PVT 92-95 | 1994 |
| KP92\1162\94 | a | I | PVT 92-95 | 1994 |
| R613257 SBL 77-09-29 | a | I | Routine clinical sample | 1977 |
| RA776 SBL 75-10-02 | a | I | Routine clinical sample | 1975 |
| ATCC 9340 | b | I | ATCC | |
| ATCC 9797 | b | I | ATCC | |
| B. parapertussis | ||||
| KP92\11\92 | c | II | PVT 92-95 | 1992 |
| KP92\41\92 | c | II | PVT 92-95 | 1992 |
| KP92\83\93 | c | II | PVT 92-95 | 1993 |
| KP92\106\93 | c | II | PVT 92-95 | 1993 |
| KP86\148\86 | c | II | PVT 86-87 | 1986 |
| KP92\214\93 | c | II | PVT 92-95 | 1993 |
| KP92\218\93 | c | II | PVT 92-95 | 1993 |
| KP86\250\86 | c | II | PVT 86-87 | 1986 |
| KP92\283\93 | c | II | PVT 92-95 | 1993 |
| KP92\308\93 | c | II | PVT 92-95 | 1993 |
| KP92\373\93 | c | II | PVT 92-95 | 1993 |
| KP92\377\93 | c | II | PVT 92-95 | 1993 |
| KP92\437\93 | c | II | PVT 92-95 | 1993 |
| KP92\756\94 | c | II | PVT 92-95 | 1994 |
| KP92\1180\94 | c | II | PVT 92-95 | 1994 |
| ATCC 15237 | c | II | ATCC | |
| ATCC 15311 | c | II | ATCC | |
| B. bronchiseptica | ||||
| ATCC 786 | d | III | ATCC | |
| AB 1254 | d | III | SSb | |
| ATCC 19395 | e | IV | ATCC |
In vitro amplification.
Bacterial lysates were diluted 1:1,000, and 10 μl was used as template in a nested PCR with outer primers STMA10 (5′-CTG CTC AAC CGC CAC ATC AAC GA-3′) and STMA11 (5′-CCA GCC ACG TCA GCC AGC CTG TT-3′) and inner primers STMA17 (5′-biotin-CAG CCC TCC AAC GCG CCA TCC C-3′) and STMA18 (5′-AAT TGT TAT CCG CTC ACA ATT GCC CGA GTG CAA CGC AT-3′) as described previously (41). Negative controls were processed in parallel between every fifth sample (37). The risk for cross contamination was minimized by frequent changing of gloves and by using three separate rooms for mixing of reagents, addition of template, and post-PCR work. Alternatively, 10 μl of bacterial lysate (not diluted) was amplified in a single PCR with primers STMA17 and STMA18. These inner primers encompass a 239-bp region of the PT promoter region (nucleotides 268 to 506 according to Locht and Keith [28]), enabling amplification of B. pertussis, B. parapertussis, and B. bronchiseptica. After amplification, species can be identified by DNA sequencing or restriction enzyme analysis.
Agarose gel electrophoresis.
After amplification, 5 μl of the product obtained by PCR (with primers STMA17 and STMA18) was analyzed on a 3.5% NuSieve agarose gel at 100 V for 5 h in TAE buffer (40 mM Tris-acetate [pH 8.0], 1 mM EDTA). The gel was stained with ethidium bromide (1 μg/ml) for 30 min, destained in water for 10 min, and analyzed under UV light. A 50-bp ladder (Amersham Pharmacia Biotech, Uppsala, Sweden) was used as a marker.
DNA sequence analysis.
Several strategies were applied to determine the sequence of the PCR-amplified PT promoter region as described by Ronaghi et al. (38): briefly, conventional bidirectional Sanger DNA sequencing by either (i) a solid-phase method (23) with fluorescein isothiocyanate-labeled terminators, primers STMA17 and STMA18, and T7 DNA polymerase, (ii) a cycle sequencing protocol with Cy5-labeled terminators, primers STMA17 and STMA18, and ThermoSequenase (Amersham Pharmacia Biotech), or (iii) a cycle sequencing protocol with Cy5-labeled primers STMA31 (5′-Cy5-CCC-TCC AAC GCG CCA TCC C-3′) and STMA32 (5′-Cy5-ATT GCC CGA GTG CAA CGC AT-3′) and ThermoSequenase. The generated Sanger fragments were loaded onto an ALF or ALF Express electrophoretic instrument (Amersham Pharmacia Biotech). To resolve a region containing sequence compressions, pyrosequencing (39) was performed. Two internal primers (primers STMA33 [5′-CGT GTT GGC AAC CGC CAA CGC G-3′] and STMA34 [5′-GGA AGG ATT GAG GGC TTT GTG CGA CG-3′]) were synthesized and used in bidirectional pyrosequencing as described by Ronaghi et al. (39). Briefly, the template (sequencing primer hybridized to the single-stranded DNA [23]) was added to a four-enzyme mixture containing Klenow DNA polymerase, ATP sulfurylase, luciferase, and apyrase. The pyrophosphate released during the nucleotide incorporation is coupled to the ATP generated by sulfurylase and a proportional amount of light is emitted by luciferase, which is detected with a photon detector device. Unincorporated nucleotides are degraded by apyrase, allowing sequential, iterative addition of the four different nucleotides.
Colorimetric restriction analysis of PCR products.
Identification of bacterial species by restriction enzyme analysis was performed as described previously (41). Briefly, biotinylated PCR products containing a lac operator sequence, which is introduced as a handle in the nonbiotinylated inner primer (STMA18), was immobilized onto streptavidin-coated super paramagnetic beads (Dynal AS, Oslo, Norway). The magnetic beads with the captured PCR product were divided into three aliquots and were separately treated with TaqI (which cleaves only the amplified sequences originating from B. pertussis and B. bronchiseptica), BglI (which cleaves only amplicons originating from B. parapertussis), and HaeII (which cleaves B. parapertussis and B. bronchiseptica amplicons). The beads were then incubated with the DNA-binding LacI–β-galactosidase fusion protein, which specifically recognizes the lac operator sequence. After washing, substrate was added, the change in absorbance was measured in an enzyme-linked immunosorbent assay plate reader, and species were identified on the basis of the colorimetric restriction pattern; i.e., only noncleaved amplicons yielded a colorimetric response.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF157332 to AF157364).
RESULTS
PCR amplification and agarose gel analysis.
To evaluate the degree of polymorphism in the PT promoter region, a part of this region encompassing nucleotides 268 to 506 was amplified with primers STMA17 and STMA18 (28). These generic primers amplify the promoter regions from B. pertussis, B. parapertussis, and B. bronchiseptica, which can subsequently be identified by restriction enzyme analyses due to sequence variations in the three species (3, 36, 41).
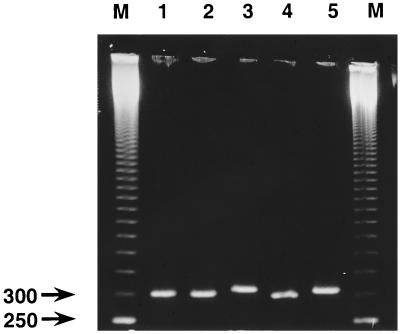
All strains in the study (Table 1) were amplified by the PCR, including those in the culture-positive nasopharyngeal samples which were originally negative by PCR when amplification directly with clinical nasopharyngeal aspirate samples was used (37). When the PCR products were analyzed by the highly resolving agarose gel electrophoresis, two different fragment lengths of the PCR products were observed; these differed by approximately 10 bp in length (Fig. 1). Amplification of all B. pertussis strains resulted in a shorter fragment (approximately 300 bp), which is in accordance with the length of the fragment expected with primers STMA17 and STMA18 by using the sequence published by Locht and Keith (28). Amplification of all B. parapertussis strains resulted in slightly longer PCR amplicons. The majority of the published B. parapertussis PT promoter sequences are 5 to 10 bp longer than the corresponding B. pertussis sequence (3, 18). Surprisingly, fragments of both lengths were obtained when amplifying the PT promoter region from B. bronchiseptica. Strain ATCC 19395 had a longer PCR fragment, while the two strains ATCC 786 and AB 1254 had shorter PCR amplicons. Figure 1 shows the resulting PCR products from two B. pertussis strains (patient isolate KP92\2\92 and type strain ATCC 9797), one B. parapertussis strain (ATCC 15311), and two B. bronchiseptica strains (ATCC 786 and ATCC 19395) and demonstrates the two different fragment sizes.
FIG. 1.
Results of agarose gel electrophoresis demonstrating the two different fragment sizes obtained with PCR primers STMA17 and STMA18. Lane 1, amplified product of B. pertussis patient isolate KP92\2\92; lane 2, B. pertussis ATCC 9797; lane 3, B. parapertussis ATCC 15311; lane 4, B. bronchiseptica ATCC 786; lane 5, B. bronchiseptica 19395; lane M, a 50-bp ladder.
DNA sequencing.
In order to elucidate the reason for the length polymorphisms, the variations at the nucleotide level were investigated. All strains investigated in this study (Table 1) were sequenced by conventional Sanger DNA sequencing by one or more strategies. These included (i) a solid-phase method with fluorescein isothiocyanate-labeled terminators, primers STMA17 and STMA18, and T7 DNA polymerase, (ii) a cycle sequencing protocol with Cy5-labeled terminators, primers STMA17 and STMA18, and ThermoSequenase, or (iii) cycle sequencing with Cy5-labeled primers STMA31 and STMA32 and ThermoSequenase. However, a short region of the sequence remained difficult to resolve (this was especially pronounced in all the B. parapertussis strains and in B. bronchiseptica ATCC 19395) due to sequence compressions as a result of strong secondary structures. The same sequencing pattern in this short region (nucleotides 48 to 71 in Fig. 2) appeared in several strains. Therefore, we used an alternative, non-gel-based sequencing method, pyrosequencing (39), to resolve the sequence in this region for strains B. pertussis KP92\2\92, a patient isolate, and ATCC 9797, B. parapertussis ATCC 15311, and B. bronchiseptica ATCC 786 and ATCC 19395. Briefly, internal primers were designed to hybridize close to the region comprising the sequence compressions, and pyrosequencing was successfully performed with both strands (38).
FIG. 2.
The five different sequences encompassing the PT promoter region (the 239 or 249 nucleotides upstream of the start codon of the PT subunit 1 gene amplified with flanking primers) found among the 33 ATCC reference strains and patient isolates sequenced. The three restriction sites used for species identification are marked with boxes. Restriction enzyme recognition sequences are underlined. The nucleotide positions in sequence b (B. pertussis ATCC 9797 and ATCC 9340) that differ from those in sequence a (B. pertussis patient isolates) are marked in boldface. The nucleotide positions in sequence e (B. bronchiseptica ATCC 19395) that differ from those in sequence d (B. bronchiseptica ATCC 786 and AB 1254) are also marked in boldface. Sequence c is the sequence common to all of the B. parapertussis strains.
When comparing all sequences we could identify five different consensus sequences in this region (sequences a to e in Fig. 2 and Table 1). Two different sequences of the same length (239 bp) were found among the B. pertussis strains sequenced. All clinical isolates showed the same sequence (sequence a in Fig. 2), which differed at eight positions compared to the sequences of two ATCC type strains (ATCC 9797 and ATCC 9340) (marked in boldface in sequence b, Fig. 2). When a database search was performed with an advanced BLASTN algorithm (version 2.0.4), several sequences identical to sequence a in Fig. 2 were found (GenBank accession nos. AJ006157, M14378, X16347, AJ007363, M13223, AJ006155, and AJ006153), indicating that this pattern is not specific for Swedish isolates. One perfect match to sequence b was also found in the databases (accession no. AJ006159 [unpublished; submitted in May 1998]). All B. parapertussis strains sequenced (clinical isolates as well as ATCC strains) had exactly the same sequence (sequence c in Fig. 2). However, this sequence is 10 bp longer than the B. pertussis sequences and differ from the two B. pertussis sequences at 23 and 17 positions, respectively. The sequence most similar to the B. parapertussis sequence, as determined by database searches (sequence c; 10 bp shorter and two different nucleotides), is the sequence published by Aricò and Rappuoli (3) (GenBank accession no. M16493). Among the B. bronchiseptica strains, two different sequences were found. Two of the strains (ATCC 786 and AB 1254) (sequence d in Fig. 2) are of the same length as the B. pertussis sequences (239 bp) but differ from the a and b sequences at 18 and 14 positions, respectively. When performing a database search with B. bronchiseptica sequence d (Fig. 2) as the query sequence, the most similar sequence found (differing at five nucleotide positions) was the B. bronchiseptica sequence published by Aricò and Rappuoli (3) (GenBank accession no. M16492). The sequence of the third B. bronchiseptica strain sequenced (ATCC 19395) differs remarkably from the other two B. bronchiseptica sequences in that it contains the 10-bp insertion present in B. parapertussis and has nine nucleotide substitutions (marked in boldface in sequence e, Fig. 2) compared to the sequences of the other B. bronchiseptica strains. The sequence most similar to B. bronchiseptica ATCC 19395 (sequence e in Fig. 2) is, surprisingly, a B. parapertussis sequence (GenBank accession no. M16493) (3). The sequence of B. bronchiseptica ATCC 19395 differs from the database sequence at six nucleotide positions and contains the same 10-bp insertion that we found in B. parapertussis. Strain ATCC 19395 was identified at the Culture Collection, University of Gothenburg, Gothenburg, Sweden, as B. bronchiseptica by use of 103 classical biochemical features and numerical analysis (data not shown; E. Falsen, personal communication).
Colorimetric restriction analysis.
The polymorphic positions in the PT promoter region of B. pertussis, B. parapertussis, and B. bronchiseptica has previously been shown to be amenable for species identification on the basis of restriction analysis (36, 41). After alignment of the observed sequences in this study (Fig. 2), we focused on the restriction enzyme recognition sequences used for species differentiation. In both B. pertussis sequences, a recognition sequence for TaqI is present and there are no BglI or HaeII restriction sites. The B. parapertussis sequence (identical for all strains in the study) contains, as expected, a BglI and an HaeII recognition sequence but no TaqI site. In both B. bronchiseptica sequence types, the TaqI site is intact. B. bronchiseptica ATCC 786 and AB 1254 (sequence d in Fig. 2) also contain the HaeII site and lack the BglI site, as expected. However, B. bronchiseptica ATCC 19395 contains the BglI recognition sequence, in addition to the TaqI site. The HaeII site, present in the other B. bronchiseptica strains, is disturbed by a G-to-A substitution (sequence e in Fig. 2).
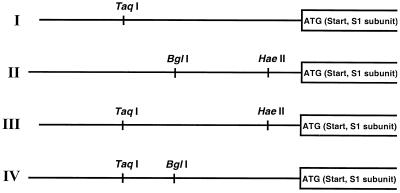
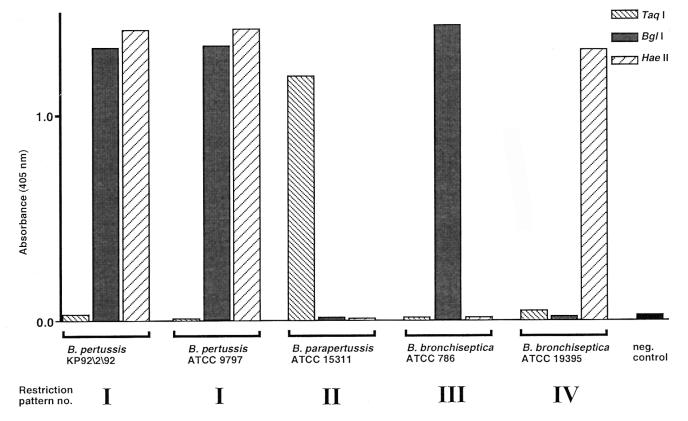
Altogether, five different sequence types for the PT promoter region and four different restriction patterns (Fig. 3 and Table 1) were found among the 33 strains analyzed. To confirm the restriction sites found by sequencing, we performed the previously described (41) colorimetric restriction analysis with the PCR products. Briefly, biotinylated PCR products containing a lac operator sequence were immobilized onto streptavidin-coated super paramagnetic beads and were separately treated with TaqI, BglI, and HaeII. After washing and binding of a DNA binding fusion protein (LacI–β-galactosidase) that specifically recognizes the lac operator sequence, substrate was added and the change in absorbance resulting from successful cleavage of the PCR amplicon was measured. Figure 4 shows the results for the five sequence types (sequences a to e in Fig. 2) represented by B. pertussis KP92\2\92, a patient isolate, and ATCC 9797, B. parapertussis ATCC 15311, and B. bronchiseptica ATCC 786 and ATCC 19395. The results demonstrate that for the strains sequenced in this study, discrimination between the three different Bordetella spp. is possible by use of TaqI, BglI, and HaeII. In addition, the two different B. bronchiseptica sequence types can be separated.
FIG. 3.
The four different restriction patterns resulting from the five different sequence types in Fig. 2 obtained with restriction enzymes TaqI, BglI, and HaeII.
FIG. 4.
Results from colorimetric restriction enzyme analysis of Bordetella strains representing the five different PT promoter sequence types in Fig. 2 to confirm the four restriction patterns in Fig. 3. The negative control corresponds to a PCR-negative sample that was immobilized onto magnetic beads and mixed with fusion protein and substrate (see text for details).
DISCUSSION
Several PT promoter sequences that differ from the sequences originally published (3, 28) have been reported since Aricò et al. (2) demonstrated sequence heterogeneity within the same species. The PT promoter and its genes have been used both to try to solve the classification problems of the Bordetella species (11) and to construct phylogenetic trees to elucidate the evolutionary relationships in the genus Bordetella (2, 11). Since the sequence encompassing the PT promoter region is also widely used for pertussis diagnosis by performing PCR (22, 36, 41, 42), subsequent detection by hybridization (5, 22), and restriction enzyme analysis (36, 41) of this region, detailed knowledge about sequence polymorphism is of great importance. For example, nucleotide variations in the primer annealing region have been suggested as a source of false-negative PCR results (31).
We decided to undertake sequence analysis of the promoter region used by us to detect and distinguish B. pertussis, B. parapertussis, and B. bronchiseptica strains (41) with PCR primers encompassing nucleotides 268 to 506, as described by Locht and Keith (28), to further investigate whether restriction analysis can be used to reliably identify the three species. From the results that we obtained we found that the two different B. pertussis sequences generated the same restriction pattern (restriction pattern I in Fig. 3) and that all B. parapertussis strains had the same sequence in this region (restriction pattern II). Two different sequences were found among the B. bronchiseptica strains analyzed, resulting in restriction patterns III and IV. When the sequence of B. bronchiseptica ATCC 19395 (sequence e in Fig. 2 and restriction pattern IV) was compared to the Bordetella sequences deposited in public databases, the most similar sequence was, surprisingly, a B. parapertussis sequence. However, B. bronchiseptica ATCC 19395 was analyzed further by use of 103 classical biochemical features and numerical analysis and was classified as a B. bronchiseptica strain.
When analyzing the 33 ATCC type strains and clinical isolates, we experienced technical difficulties in determining the nucleotide sequence in the promoter region (nucleotides 48 to 71) of all 17 B. parapertussis strains and of B. bronchiseptica ATCC 19395 due to sequence compressions in a palindromic region (38). Consequently, there is uncertainty whether variations in earlier published sequences are due to problems associated with conventional DNA sequencing methodology or reflect a natural nucleotide variation. However, by retrieving the 12 sequences for this region that have been deposited in the databases and that match in whole or in part the region that we sequenced, we found that all sequences contain the expected restriction enzyme recognition sequences. All nine B. pertussis sequences in the database contain restriction sites, according to restriction pattern I in Fig. 3. The two retrieved B. parapertussis sequences contain restriction sites for BglI and HaeII (restriction pattern II) and the B. bronchiseptica sequence contains restriction sites for TaqI and HaeII (restriction pattern III). Thus, according to the PT promoter sequences available today, restriction analysis of amplicons from the promoter region with TaqI, BglI, and HaeII results in identification of the three species. In addition, the results indicate that the two different B. bronchiseptica sequence variants can be distinguished. Alternatively, the five different sequence types could be identified by designing a sequencing primer (which hybridizes to nucleotides 113 to 90 in Fig. 2) and performing pyrosequencing to detect at least 32 bases that could distinguish the three species.
In this study, we could not find any sequence differences correlated to year of isolation (1975 to 1994) or to whether the original aspirate sample was positive by culture but negative by PCR. Nor did we find any nucleotide variation in B. parapertussis that explains the lower diagnostic sensitivity obtained for B. parapertussis in a recent study (37). We suggest that these problems are more associated with processing of the clinical samples prior to PCR than with polymorphisms in the amplified PT region. However, since only the internal regions encompassed by the PCR primers were sequenced, the possibility that nucleotide variations are present in the primer annealing regions cannot be disregarded. Furthermore, this study indicates the internal consensus sequences that can be used to design new generic primers. A high degree of PT promoter sequence homology among the clinical B. pertussis isolates as well as clinical B. parapertussis isolates was observed in this study, although the number of sequenced isolates is limited (n = 11 and n = 17 isolates with identical sequences, respectively). The sequence differences between the two B. pertussis ATCC strains and the clinical isolates are in agreement with previous findings which suggested that type strain 18323 (ATCC 9797) might not be a typical B. pertussis strain (2).
Recent reports emphasize the need to consider B. parapertussis and B. bronchiseptica in the diagnosis of Bordetella infections. B. bronchiseptica might be of increased importance in the diagnosis of disease in humans in the future, since B. bronchiseptica was recently identified in two vaccinated children with cough illnesses (43). The sequence of the PT promoter region in these strains is identical to sequence e (Fig. 2) in this study except for the absence of the 10-nucleotide insertion. Several studies demonstrate the importance of B. parapertussis, which also causes a whooping cough-like disease in vaccinated populations (15, 30). Hausman et al. (18) have analyzed three B. parapertussis strains isolated from unvaccinated children exhibiting antibodies to PT. According to those investigators, the possibility of a mixed infection was investigated, but no such infection was demonstrated. The sequences of those isolates are identical to all (n = 17) B. parapertussis sequences in the present study (sequence c). In a recent study estimating the prevalence of B. pertussis and B. parapertussis in a highly immunized population, B. parapertussis was found to be the cause of one-third of laboratory-confirmed Bordetella infections (20). In pertussis vaccine trials, the use of PCR methods that differentiate pertussis and parapertussis infections is highly recommended (31). Detection and species identification with common PCR primers and subsequent restriction analysis can therefore be used as complements to biochemical and serological methods, which in some cases produce inconclusive results (42). However, further sequence analysis of patient isolates for which biochemical and serological reactions and clinical history contradict is recommended to discover any alternative PT promoter sequences in possible intermediate or variant Bordetella strains.
ACKNOWLEDGMENTS
This work was supported by Dynal AS.
We thank Lena Lindberg for technical assistance, Enevold Falsen for complementary biochemical testing of bacterial strains, and Hans Hallander for critical comments.
REFERENCES
- 1.Ad Hoc Group for the Study of Pertussis Vaccines. Placebo-controlled trial of two acellular pertussis vaccines in Sweden—protective efficacy and adverse events. Lancet. 1988;i:955–960. . (Erratum i:1238.) [PubMed] [Google Scholar]
- 2.Aricò B, Gross R, Smida J, Rappuoli R. Evolutionary relationships in the genus Bordetella. Mol Microbiol. 1987;1:301–308. doi: 10.1111/j.1365-2958.1987.tb01936.x. [DOI] [PubMed] [Google Scholar]
- 3.Aricò B, Rappuoli R. Bordetella parapertussis and Bordetella bronchiseptica contain transcriptionally silent pertussis toxin genes. J Bacteriol. 1987;169:2847–2853. doi: 10.1128/jb.169.6.2847-2853.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher P E, Stibitz S. Synergistic binding of RNA polymerase and BvgA phosphate to the pertussis toxin promoter of Bordetella pertussis. J Bacteriol. 1995;177:6486–6491. doi: 10.1128/jb.177.22.6486-6491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck G E. Detection of Bordetella pertussis by rapid-cycle PCR and colorimetric microwell hybridization. J Clin Microbiol. 1996;34:1355–1358. doi: 10.1128/jcm.34.6.1355-1358.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas E, Coote J G, Parton R, McPheat W. Identification of Bordetella pertussis in nasopharyngeal swabs by PCR amplification of a region of the adenylate cyclase gene. J Med Microbiol. 1993;38:140–144. doi: 10.1099/00222615-38-2-140. [DOI] [PubMed] [Google Scholar]
- 7.Erlandsson A, Bäckman A, Törnqvist E, Olcén P. PCR assay or culture for diagnosis of Bordetella pertussis in the routine diagnostic laboratory? J Infect. 1997;35:221–224. doi: 10.1016/s0163-4453(97)92738-9. [DOI] [PubMed] [Google Scholar]
- 8.Glare E M, Paton J C, Premier R R, Lawrence A J, Nisbet I T. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol. 1990;28:1982–1987. doi: 10.1128/jcm.28.9.1982-1987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granström G, Sterner G, Nord C E, Granström M. Use of erythromycin to prevent pertussis in newborns of mothers with pertussis. J Infect Dis. 1987;155:1210–1214. doi: 10.1093/infdis/155.6.1210. [DOI] [PubMed] [Google Scholar]
- 10.Grimprel E, Begue P, Anjak I, Betsou F, Guiso N. Comparison of polymerase chain reaction, culture, and Western immunoblot serology for diagnosis of Bordetella pertussis infection. J Clin Microbiol. 1993;31:2745–2750. doi: 10.1128/jcm.31.10.2745-2750.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross R, Aricò B, Rappuoli R. Genetics of pertussis toxin. Mol Microbiol. 1989;3:119–124. doi: 10.1111/j.1365-2958.1989.tb00111.x. [DOI] [PubMed] [Google Scholar]
- 12.Gross R, Carbonetti N H, Rossi R, Rappuoli R. Functional analysis of the pertussis toxin promoter. Res Microbiol. 1992;143:671–681. doi: 10.1016/0923-2508(92)90062-s. [DOI] [PubMed] [Google Scholar]
- 13.Gustafsson L, Hallander H O, Olin P, Reizenstein E, Storsaeter J. A controlled trial of a two-component acellular, a five-component acellular, and a whole-cell pertussis vaccine. N Engl J Med. 1996;334:349–355. doi: 10.1056/NEJM199602083340602. [DOI] [PubMed] [Google Scholar]
- 14.Gustavsson L, Hallander H O, Olin P, Reizenstein E, Storsaeter J. Efficacy trial of acellular pertussis vaccines. Technical report trial 1. Stockholm, Sweden: Swedish Institute for Infectious Disease Control; 1995. [Google Scholar]
- 15.Hallander H O, Gnarpe J, Gnarpe H, Olin P. Bordetella pertussis, Bordetella parapertussis, Mycoplasma pnuemoniae, Chlamydia pneumoniae and persistent cough in children. Scand J Infect Dis. 1999;31:281–286. doi: 10.1080/00365549950163581. [DOI] [PubMed] [Google Scholar]
- 16.Hallander H O, Reizenstein E, Renemar B, Rasmuson G, Mardin L, Olin P. Comparison of nasopharyngeal aspirates with swabs for culture of Bordetella pertussis. J Clin Microbiol. 1993;31:50–52. doi: 10.1128/jcm.31.1.50-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallander H O, Storsaeter J, Möllby R. Evaluation of serology and nasopharyngeal cultures for diagnosis of pertussis in a vaccine efficacy trial. J Infect Dis. 1991;163:1046–1054. doi: 10.1093/infdis/163.5.1046. [DOI] [PubMed] [Google Scholar]
- 18.Hausman S Z, Cherry J D, Heininger U, Wirsing von Konig C H, Burns D L. Analysis of proteins encoded by the ptx and ptl genes of Bordetella bronchiseptica and Bordetella parapertussis. Infect Immun. 1996;64:4020–4026. doi: 10.1128/iai.64.10.4020-4026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Q, Mertsola J, Soini H, Skurnik M, Ruuskanen O, Viljanen M K. Comparison of polymerase chain reaction with culture and enzyme immunoassay for diagnosis of pertussis. J Clin Microbiol. 1993;31:642–645. doi: 10.1128/jcm.31.3.642-645.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Q, Viljanen M K, Arvilommi H, Aittanen B, Mertsola J. Whooping cough caused by Bordetella pertussis and Bordetella parapertussis in an immunized population. JAMA. 1998;280:635–637. doi: 10.1001/jama.280.7.635. [DOI] [PubMed] [Google Scholar]
- 21.Hoppe J E. Update of epidemiology, diagnosis, and treatment of pertussis. Eur J Clin Microbiol Infect Dis. 1996;15:189–193. doi: 10.1007/BF01591352. . (Editorial.) [DOI] [PubMed] [Google Scholar]
- 22.Houard S, Hackel C, Herzog A, Bollen A. Specific identification of Bordetella pertussis by the polymerase chain reaction. Res Microbiol. 1989;140:477–487. doi: 10.1016/0923-2508(89)90069-7. [DOI] [PubMed] [Google Scholar]
- 23.Hultman T, Ståhl S, Hornes E, Uhlén M. Direct solid phase sequencing of genomic and plasmid DNA using magnetic beads as solid support. Nucleic Acids Res. 1989;17:4937–4946. doi: 10.1093/nar/17.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katzko G, Hofmeister M, Church D. Extended incubation of culture plates improves recovery of Bordetella spp. J Clin Microbiol. 1996;34:1563–1564. doi: 10.1128/jcm.34.6.1563-1564.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Jansen D L, Finn T M, Halperin S A, Kasina A, O'Connor S P, Aoyama T, Manclark C R, Brennan M J. Identification of Bordetella pertussis infection by shared-primer PCR. J Clin Microbiol. 1994;32:783–789. doi: 10.1128/jcm.32.3.783-789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichtinghagen R, Diedrich-Glaubitz R, von Horsten B. Identification of Bordetella pertussis in nasopharyngeal swabs using the polymerase chain reaction: evaluation of detection methods. Eur J Clin Chem Clin Biochem. 1994;32:161–167. doi: 10.1515/cclm.1994.32.3.161. [DOI] [PubMed] [Google Scholar]
- 27.Lind-Brandberg L, Welinder-Olsson C, Lagergård T, Taranger J, Trollfors B, Zackrisson G. Evaluation of PCR for diagnosis of Bordetella pertussis and Bordetella parapertussis infections. J Clin Microbiol. 1998;36:679–683. doi: 10.1128/jcm.36.3.679-683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locht C, Keith J M. Pertussis toxin gene: nucleotide sequence and genetic organization. Science. 1986;232:1258–1264. doi: 10.1126/science.3704651. [DOI] [PubMed] [Google Scholar]
- 29.Marques R R, Carbonetti N H. Genetic analysis of pertussis toxin promoter activation in Bordetella pertussis. Mol Microbiol. 1997;24:1215–1224. doi: 10.1046/j.1365-2958.1997.4371792.x. [DOI] [PubMed] [Google Scholar]
- 30.Mastrantonio P, Stefanelli P, Giuliano M, Herrera Rojas Y, Ciofi degli Atti M, Anemona A, Tozzi A E. Bordetella parapertussis infection in children: epidemiology, clinical symptoms, and molecular characteristics of isolates. J Clin Microbiol. 1998;36:999–1002. doi: 10.1128/jcm.36.4.999-1002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meade B D, Bollen A. Recommendations for use of the polymerase chain reaction in the diagnosis of Bordetella pertussis infections. J Med Microbiol. 1994;41:51–55. doi: 10.1099/00222615-41-1-51. [DOI] [PubMed] [Google Scholar]
- 32.Muller F M, Hoppe J E, Wirsing von Konig C H. Laboratory diagnosis of pertussis: state of the art in 1997. J Clin Microbiol. 1997;35:2435–2443. doi: 10.1128/jcm.35.10.2435-2443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olcén P, Bäckman A, Johansson B, Esbjorner E, Törnqvist E, Bygraves J, McPheat W L. Amplification of DNA by the polymerase chain reaction for the efficient diagnosis of pertussis. Scand J Infect Dis. 1992;24:339–345. doi: 10.3109/00365549209061340. [DOI] [PubMed] [Google Scholar]
- 34.Regan J, Lowe F. Enrichment medium for the isolation of Bordetella. J Clin Microbiol. 1977;6:303–309. doi: 10.1128/jcm.6.3.303-309.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reizenstein E. Diagnostic polymerase chain reaction. Dev Biol Stand. 1997;89:247–254. [PubMed] [Google Scholar]
- 36.Reizenstein E, Johansson B, Mardin L, Abens J, Möllby R, Hallander H O. Diagnostic evaluation of polymerase chain reaction discriminative for Bordetella pertussis, B. parapertussis, and B. bronchiseptica. Diagn Microbiol Infect Dis. 1993;17:185–191. doi: 10.1016/0732-8893(93)90094-n. [DOI] [PubMed] [Google Scholar]
- 37.Reizenstein E, Lindberg L, Möllby R, Hallander H O. Validation of nested Bordetella PCR in pertussis vaccine trial. J Clin Microbiol. 1996;34:810–815. doi: 10.1128/jcm.34.4.810-815.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronaghi M, Nygren M, Lundeberg J, Nyrén P. Analyses of secondary structures in DNA by pyrosequencing. Anal Biochem. 1999;267:65–71. doi: 10.1006/abio.1998.2978. [DOI] [PubMed] [Google Scholar]
- 39.Ronaghi M, Uhlén M, Nyrén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363. doi: 10.1126/science.281.5375.363. , 365. [DOI] [PubMed] [Google Scholar]
- 40.Sprauer M A, Cochi S L, Zell E R, Sutter R W, Mullen J R, Englender S J, Patriarca P A. Prevention of secondary transmission of pertussis in households with early use of erythromycin. Am J Dis Child. 1992;146:177–181. doi: 10.1001/archpedi.1992.02160140043018. [DOI] [PubMed] [Google Scholar]
- 41.Stark M, Reizenstein E, Uhlén M, Lundeberg J. Immunomagnetic separation and solid-phase detection of Bordetella pertussis. J Clin Microbiol. 1996;34:778–784. doi: 10.1128/jcm.34.4.778-784.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stefanelli P, Giuliano M, Bottone M, Spigaglia P, Mastrantonio P. Polymerase chain reaction for the identification of Bordetella pertussis and Bordetella parapertussis. Diagn Microbiol Infect Dis. 1996;24:197–200. doi: 10.1016/0732-8893(96)00064-8. [DOI] [PubMed] [Google Scholar]
- 43.Stefanelli P, Mastrantonio P, Hausman S Z, Giuliano M, Burns D L. Molecular characterization of two Bordetella bronchiseptica strains isolated from children with coughs. J Clin Microbiol. 1997;35:1550–1555. doi: 10.1128/jcm.35.6.1550-1555.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Zee A, Agterberg C, Peeters M, Mooi F, Schellekens J. A clinical validation of Bordetella pertussis and Bordetella parapertussis polymerase chain reaction: comparison with culture and serology using samples from patients with suspected whooping cough from a highly immunized population. J Infect Dis. 1996;174:89–96. doi: 10.1093/infdis/174.1.89. [DOI] [PubMed] [Google Scholar]
- 45.van der Zee A, Agterberg C, Peeters M, Schellekens J, Mooi F R. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J Clin Microbiol. 1993;31:2134–2140. doi: 10.1128/jcm.31.8.2134-2140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zu T, Goyard S, Rappuoli R, Scarlato V. DNA binding of the Bordetella pertussis H1 homolog alters in vitro DNA flexibility. J Bacteriol. 1996;178:2982–2985. doi: 10.1128/jb.178.10.2982-2985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]