Abstract
The data described in this article presents the toxicity of rotenone and the neuroprotective effect of Yashtimadhu choorna (powder) in an in vitro Parkinson's disease model [1]. Yashtimadhu choorna is prepared from the roots of Glycyrrhiza glabra L., commonly known as licorice/ liquorice. The effects of rotenone and Yashtimadhu was assessed using cellular and molecular assays such as cell cytotoxicity assay, live-dead cell staining assay, cell cycle analysis, and western blotting. Protein-protein interaction was studied using ANAT plug-in in Cytoscape. Rotenone displayed time and dose-dependent toxicity, as evidenced by cell cytotoxicity assay and live-dead cell staining assay. Yashtimadhu showed no toxicity and prevented rotenone-induced toxicity. Rotenone and Yashtimadhu displayed differential control on the cell cycle. The Protein-interaction network showed the proteins interacting with ERK-1/2 and the pathways regulated by these interactions. The pathways regulated were primarily involved in cellular oxidative stress and apoptosis response. The data described here will enable the extent of cellular toxicity as a result of rotenone treatment and the neuroprotection conferred by Yashtimadhu choorna. This will enable understanding and exploring the effect of traditional and complementary medicine and aiding the identification of molecular targets to confer neuroprotection in Parkinson's disease.
Keywords: Neurodegeneration, Complementary and alternative medicine, Ayurveda, Actionable molecules
Specifications Table
| Subject | Biological sciences Neuroscience: Cellular and Molecular |
| Specific subject area | Cell and molecular biology |
| Type of data | Table Image Graph Figure |
| How data were acquired | Cytotoxicity data was acquired from IMR-32 cells treated with rotenone and Yashtimadhu extract (Glycyrrhiza glabra L.) using BMG LABTECH FLUOstar Omega multidetection microplate reader. Cell cycle analysis data was acquired from Guava® easyCyte Flow Cytometer and processed using FCS Express software, version 6. Live-dead assay and ROS assay cell imaging were carried out using ZOE Fluorescent Cell Imager, Bio-Rad, and data processed using ImageJ. Western blotting data were analyzed using ImageJ software. Statistical analysis using GraphPad Prism 5. |
| Data format | Raw and analyzed data |
| Parameters for data collection | Parameters for the data collection were as follows: IMR-32 cells were seeded at different densities based on the cell culture plate; 96-well plate: 5000 cells/well, 12-well plate: 10,000 cells/well, and 6-well plate: 30,000 cells/ well. Cells were differentiated with 10 µM retinoic acid for 9 days and treated with different concentrations of rotenone and Yashtimadhu (Glycyrrhiza glabra L.) aqueous extract. |
| Description of data collection | Treated cells were assayed for cytotoxicity using MTT reagent, live-dead cell staining assay using propidium iodide and HOECHST stains, ROS assay was carried out with cell staining using 2′,7′ –dichlorofluorescin diacetate and HOECHST stains, cell cycle analysis was carried out using propidium iodide staining, and protein expression was assayed using western blotting analysis. Protein interaction analysis using ANAT plug-in of Cytoscape version 3.8. |
| Data source location | Institution: Center for Systems Biology and Molecular Medicine, Yenepoya Research Centre, Yenepoya (Deemed to be University) City/Town/Region: Mangalore, Karnataka Country: India Latitude and longitude for collected samples/data: 12.8113° N, 74.8814° E |
| Data accessibility | With the article |
| Related research article | Karthikkeyan, G., Pervaje, R., Pervaje, S. K., Prasad, T.S.K., Modi, P.K. Prevention of MEK-ERK-1/2 hyper-activation underlines the neuroprotective effect of Glycyrrhiza glabra L. (Yashtimadhu) against rotenone-induced cellular and molecular aberrations. J Ethnopharmacol 274 (2021)114025. |
Value of the Data
-
•
The data described here explains the effect of Yashtimadhu (Glycyrrhiza glabra L.) extract and rotenone on neuronal differentiated IMR-32 cells.
-
•
The data also describes the dose-dependent effect of rotenone and the extent of the resulting cellular damage.
-
•
The data will also be useful for researchers working on the effect of traditional and complementary medicine and for researchers working on Parkinson's disease (PD) model.
-
•
The protein-protein interactions and the pathways regulated by the proteins regulated by Yashtimadu can offer newer insights to understanding its effect in an in vitro PD model.
-
•
The dataset can be used to explore the neuroprotective mechanism of traditional and complementary medicine.
-
•
The dataset can be used to identify molecular targets that can confer neuroprotection in PD.
1. Data Description
The data in this article depicts the effect of rotenone and Yashtimadhu extract (Glycyrrhiza glabra L.) on differentiated IMR-32 cells in the context of neuroprotection conferred by Yashtimadhu in the Parkinson's disease model [1]. Parkinson's disease is the selective loss of dopaminergic neurons, attributable to mitochondrial dysfunction and oxidative stress [2]. Rotenone is a mitochondrial complex-I inhibitor, which leads to the loss of the mitochondrial membrane potential, oxidative stress, and thereby inducing apoptosis [3]. Indian Ayurvedic system classifies plants with neuroprotective properties as Medhya Rasayana, which includes, Yashtimadhu, Glycyrrhiza glabra; Mandukaparni, Centella asiatica and, Guduchi, Tinospora cordifolia [4,5]. Yashtimadhu powder is prepared from licorice root is known for its potential as neuroprotective, memory enhancer, and immunomodulatory effects [5], [6], [7], [8]. The data explored the cellular toxicity of rotenone and neuroprotection by Yashtimadhu.
Cell cytotoxicity assay was used to determine the treatment concentrations of rotenone and Yashtimadhu extract. Yashtimadhu aqueous extract and rotenone dissolved in DMSO were used to test their effects on IMR-32 cells. Upon treatment with different concentrations of rotenone for 24 h and 48 h, the cells displayed a time- and dose-dependent toxicity. The absorbance and the percentage of cell death estimated with respect to control are shown in Tables 1 and 2.
Table 1.
Cell cytotoxicity analysis with rotenone treatment for 24 h.
| Sample | Replicate | 560 nm | 650 nm | Background subtraction | Blank subtraction | % cell viability | Average % cell viability |
|---|---|---|---|---|---|---|---|
| Blank | Rep_1 | 0.036 | 0.031 | 0.005 | |||
| Rep_2 | 0.034 | 0.032 | 0.002 | 0 | NA | NA | |
| Rep_3 | 0.042 | 0.039 | 0.003 | ||||
| Ctrl | Rep_1 | 0.93 | 0.163 | 0.767 | 0.762 | 100.00 | 100.00 |
| Rep_2 | 0.988 | 0.19 | 0.798 | 0.796 | 100.00 | ||
| Rep_3 | 0.921 | 0.174 | 0.747 | 0.744 | 100.00 | ||
| DMSO | Rep_1 | 1.008 | 0.205 | 0.803 | 0.798 | 104.72 | 96.69 |
| Rep_2 | 0.892 | 0.169 | 0.723 | 0.721 | 90.58 | ||
| Rep_3 | 0.832 | 0.124 | 0.708 | 0.705 | 94.76 | ||
| 0.25 nM | Rep_1 | 0.92 | 0.219 | 0.701 | 0.696 | 91.34 | 87.50 |
| Rep_2 | 0.883 | 0.167 | 0.716 | 0.714 | 89.70 | ||
| Rep_3 | 0.734 | 0.125 | 0.609 | 0.606 | 81.45 | ||
| 0.5 nM | Rep_1 | 0.792 | 0.109 | 0.683 | 0.678 | 88.98 | 88.78 |
| Rep_2 | 0.829 | 0.148 | 0.681 | 0.679 | 85.30 | ||
| Rep_3 | 0.801 | 0.113 | 0.688 | 0.685 | 92.07 | ||
| 1 nM | Rep_1 | 0.66 | 0.089 | 0.571 | 0.566 | 74.28 | 80.50 |
| Rep_2 | 0.674 | 0.091 | 0.583 | 0.581 | 72.99 | ||
| Rep_3 | 0.821 | 0.117 | 0.704 | 0.701 | 94.22 | ||
| 10 nM | Rep_1 | 0.623 | 0.097 | 0.526 | 0.521 | 68.37 | 77.23 |
| Rep_2 | 0.694 | 0.097 | 0.597 | 0.595 | 74.75 | ||
| Rep_3 | 0.815 | 0.153 | 0.662 | 0.659 | 88.58 | ||
| 100 nM | Rep_1 | 0.608 | 0.101 | 0.507 | 0.502 | 65.88 | 67.77 |
| Rep_2 | 0.675 | 0.145 | 0.53 | 0.528 | 66.33 | ||
| Rep_3 | 0.621 | 0.089 | 0.532 | 0.529 | 71.10 | ||
| 200 nM | Rep_1 | 0.616 | 0.166 | 0.45 | 0.445 | 58.40 | 60.18 |
| Rep_2 | 0.527 | 0.109 | 0.418 | 0.416 | 52.26 | ||
| Rep_3 | 0.696 | 0.173 | 0.523 | 0.52 | 69.89 | ||
| 500 nM | Rep_1 | 0.564 | 0.117 | 0.447 | 0.442 | 58.01 | 57.71 |
| Rep_2 | 0.552 | 0.1 | 0.452 | 0.45 | 56.53 | ||
| Rep_3 | 0.538 | 0.099 | 0.439 | 0.436 | 58.60 | ||
| 1 µM | Rep_1 | 0.723 | 0.219 | 0.504 | 0.499 | 65.49 | 59.26 |
| Rep_2 | 0.551 | 0.129 | 0.422 | 0.42 | 52.76 | ||
| Rep_3 | 0.573 | 0.127 | 0.446 | 0.443 | 59.54 | ||
| 10 µM | Rep_1 | 0.54 | 0.118 | 0.422 | 0.417 | 54.72 | 54.41 |
| Rep_2 | 0.507 | 0.082 | 0.425 | 0.423 | 53.14 | ||
| Rep_3 | 0.522 | 0.107 | 0.415 | 0.412 | 55.38 |
Table 2.
Cell cytotoxicity analysis with rotenone treatment for 48 h.
| Sample | Replicate | 560 nm | 650 nm | Background subtraction | Blank subtraction | % cell viability | Average % cell viability |
|---|---|---|---|---|---|---|---|
| Blank | Rep_1 | 0.038 | 0.03 | 0.008 | |||
| Rep_2 | 0.037 | 0.029 | 0.008 | 0 | NA | NA | |
| Rep_3 | 0.032 | 0.03 | 0.002 | ||||
| Ctrl | Rep_1 | 0.658 | 0.15 | 0.508 | 0.5 | 100.00 | 100.00 |
| Rep_2 | 0.682 | 0.108 | 0.574 | 0.566 | 100.00 | ||
| Rep_3 | 0.722 | 0.13 | 0.592 | 0.59 | 100.00 | ||
| DMSO | Rep_1 | 0.605 | 0.117 | 0.488 | 0.48 | 96.00 | 105.47 |
| Rep_2 | 0.742 | 0.111 | 0.631 | 0.623 | 110.07 | ||
| Rep_3 | 0.762 | 0.109 | 0.653 | 0.651 | 110.34 | ||
| 0.25 nM | Rep_1 | 0.58 | 0.106 | 0.474 | 0.466 | 93.20 | 84.90 |
| Rep_2 | 0.534 | 0.08 | 0.454 | 0.446 | 78.80 | ||
| Rep_3 | 0.579 | 0.089 | 0.49 | 0.488 | 82.71 | ||
| 0.5 nM | Rep_1 | 0.505 | 0.082 | 0.423 | 0.415 | 83.00 | 73.33 |
| Rep_2 | 0.473 | 0.08 | 0.393 | 0.385 | 68.02 | ||
| Rep_3 | 0.494 | 0.085 | 0.409 | 0.407 | 68.98 | ||
| 1 nM | Rep_1 | 0.511 | 0.089 | 0.422 | 0.414 | 82.80 | 66.66 |
| Rep_2 | 0.423 | 0.077 | 0.346 | 0.338 | 59.72 | ||
| Rep_3 | 0.424 | 0.083 | 0.341 | 0.339 | 57.46 | ||
| 10 nM | Rep_1 | 0.377 | 0.07 | 0.307 | 0.299 | 59.80 | 62.78 |
| Rep_2 | 0.448 | 0.076 | 0.372 | 0.364 | 64.31 | ||
| Rep_3 | 0.457 | 0.076 | 0.381 | 0.379 | 64.24 | ||
| 100 nM | Rep_1 | 0.34 | 0.082 | 0.258 | 0.25 | 50.00 | 45.88 |
| Rep_2 | 0.353 | 0.085 | 0.268 | 0.26 | 45.94 | ||
| Rep_3 | 0.346 | 0.098 | 0.248 | 0.246 | 41.69 | ||
| 200 nM | Rep_1 | 0.308 | 0.087 | 0.221 | 0.213 | 42.60 | 38.03 |
| Rep_2 | 0.289 | 0.073 | 0.216 | 0.208 | 36.75 | ||
| Rep_3 | 0.271 | 0.064 | 0.207 | 0.205 | 34.75 | ||
| 500 nM | Rep_1 | 0.237 | 0.076 | 0.161 | 0.153 | 30.60 | 30.73 |
| Rep_2 | 0.225 | 0.067 | 0.158 | 0.15 | 26.50 | ||
| Rep_3 | 0.331 | 0.122 | 0.209 | 0.207 | 35.08 | ||
| 1 µM | Rep_1 | 0.244 | 0.075 | 0.169 | 0.161 | 32.20 | 27.58 |
| Rep_2 | 0.236 | 0.081 | 0.155 | 0.147 | 25.97 | ||
| Rep_3 | 0.212 | 0.065 | 0.147 | 0.145 | 24.58 | ||
| 10 µM | Rep_1 | 0.223 | 0.073 | 0.15 | 0.142 | 28.40 | 25.10 |
| Rep_2 | 0.209 | 0.068 | 0.141 | 0.133 | 23.50 | ||
| Rep_3 | 0.214 | 0.074 | 0.14 | 0.138 | 23.39 |
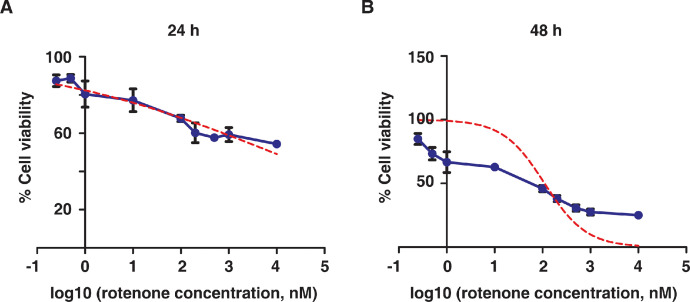
Non-linear curve fitting with the dose-response with respect to rotenone concentration was used for calculating the IC50, using the GraphPad Prism software. The IC50 concentration of rotenone was found to be 8.1 µM and 110 nM for 24 h and 48 h, respectively (Fig. 1).
Fig. 1.
The Dose-response curve of cell viability with respect to treatment with different concentrations of rotenone. (A) Rotenone treatment for 24 h, (B) Rotenone treatment for 48 h. The red-line indicates the normalized dose-response curve-fit, and the blue-line represents the observed cell viability with the respective concentrations.
The treatment of the IMR-32 cells with Yashtimadhu extract did not show any toxicity, and the absorbance observed using the cytotoxicity assay and the cell viability for the different concentrations of Yashtimadhu, are given in Table 3.
Table 3.
Cell cytotoxicity assay with Yashtimadhu extract treatment for 48 h.
| Sample | Replicate | 560 nm | 630 nm | Background subtraction | Blank subtraction | % cell viability | Average % cell viability |
|---|---|---|---|---|---|---|---|
| Blank | Rep_1 | 0.046 | 0.0325 | 0.0135 | |||
| Rep_2 | 0.044 | 0.031 | 0.013 | 0 | NA | NA | |
| Rep_3 | 0.046 | 0.0335 | 0.0125 | ||||
| Control | Rep_1 | 0.736 | 0.1225 | 0.6135 | 0.6 | 100.00 | 100.00 |
| Rep_2 | 0.7575 | 0.1235 | 0.634 | 0.621 | 100.00 | ||
| Rep_3 | 0.605 | 0.1005 | 0.5045 | 0.492 | 100.00 | ||
| 50 µg/ml | Rep_1 | 0.737 | 0.1275 | 0.6095 | 0.596 | 99.33 | 98.13 |
| Rep_2 | 0.7775 | 0.13 | 0.6475 | 0.6345 | 102.17 | ||
| Rep_3 | 0.592 | 0.1225 | 0.4695 | 0.457 | 92.89 | ||
| 100 µg/ml | Rep_1 | 0.762 | 0.1175 | 0.6445 | 0.631 | 105.17 | 98.65 |
| Rep_2 | 0.807 | 0.145 | 0.662 | 0.649 | 104.51 | ||
| Rep_3 | 0.5295 | 0.0925 | 0.437 | 0.4245 | 86.28 | ||
| 200 µg/ml | Rep_1 | 0.665 | 0.1155 | 0.5495 | 0.536 | 89.33 | 99.18 |
| Rep_2 | 0.82 | 0.149 | 0.671 | 0.658 | 105.96 | ||
| Rep_3 | 0.622 | 0.1065 | 0.5155 | 0.503 | 102.24 | ||
| 500 µg/ml | Rep_1 | 0.7235 | 0.152 | 0.5715 | 0.558 | 93.00 | 98.76 |
| Rep_2 | 0.7655 | 0.1565 | 0.609 | 0.596 | 95.97 | ||
| Rep_3 | 0.6855 | 0.145 | 0.5405 | 0.528 | 107.32 | ||
| 1000 µg/ml | Rep_1 | 0.598 | 0.1225 | 0.4755 | 0.462 | 77.00 | 78.98 |
| Rep_2 | 0.635 | 0.1235 | 0.5115 | 0.4985 | 80.27 | ||
| Rep_3 | 0.514 | 0.1095 | 0.4045 | 0.392 | 79.67 | ||
| 1500 µg/ml | Rep_1 | 0.722 | 0.1435 | 0.5785 | 0.565 | 94.17 | 84.31 |
| Rep_2 | 0.56 | 0.1145 | 0.4455 | 0.4325 | 69.65 | ||
| Rep_3 | 0.549 | 0.098 | 0.451 | 0.4385 | 89.13 |
The cells were also co-treated with Yashtimadhu extract at 200 µg/ml, and two different concentrations of rotenone, 50 nM, and 100 nM. The co-treatment with Yashtimadhu showed lesser toxicity in comparison with the treatment of respective rotenone concentrations (Tables 4 and 5).
Table 4.
Cell cytotoxicity assay with 50 nM rotenone and 200 µg/ml Yashtimadhu extract co-treatment for 48 h.
| Sample | Replicate | 560 nm | 630 nm | Background subtraction | Blank subtraction | % cell viability | Average % cell viability |
|---|---|---|---|---|---|---|---|
| Blank | Rep_1 | 0.0427 | 0.0303 | 0.0123 | |||
| Rep_2 | 0.0383 | 0.0303 | 0.0080 | 0 | NA | NA | |
| Rep_3 | 0.0383 | 0.0303 | 0.0080 | ||||
| Control | Rep_1 | 1.0713 | 0.2233 | 0.8480 | 0.8357 | 100.00 | 100.00 |
| Rep_2 | 1.1007 | 0.1610 | 0.9397 | 0.9317 | 100.00 | ||
| Rep_3 | 1.1053 | 0.1633 | 0.9420 | 0.9340 | 100.00 | ||
| Rotenone 50 nM | Rep_1 | 0.8130 | 0.1050 | 0.7080 | 0.6957 | 83.25 | 76.48 |
| Rep_2 | 0.8000 | 0.1053 | 0.6947 | 0.6867 | 73.70 | ||
| Rep_3 | 0.7993 | 0.1143 | 0.6850 | 0.6770 | 72.48 | ||
| Rotenone 50 nM + Yashtimadhu 200 µg/ml | Rep_1 | 0.9323 | 0.1803 | 0.7520 | 0.7397 | 88.51 | 86.08 |
| Rep_2 | 0.9297 | 0.1570 | 0.7727 | 0.7647 | 82.08 | ||
| Rep_3 | 0.9800 | 0.1533 | 0.8267 | 0.8187 | 87.65 | ||
| Yashtimadhu 200 µg/ml | Rep_1 | 1.0863 | 0.1717 | 0.9147 | 0.9023 | 107.98 | 103.09 |
| Rep_2 | 1.0157 | 0.1450 | 0.8707 | 0.8627 | 92.59 | ||
| Rep_3 | 1.1733 | 0.1500 | 1.0233 | 1.0153 | 108.71 |
Table 5.
Cell cytotoxicity assay with 100 nM rotenone and 200 µg/ml Yashtimadhu extract co-treatment for 48 h.
| Sample | Replicate | 560 nm | 630 nm | Background subtraction | Blank subtraction | % cell viability | Average % cell viability |
|---|---|---|---|---|---|---|---|
| Blank | Rep_1 | 0.0427 | 0.0303 | 0.0123 | |||
| Rep_2 | 0.0564 | 0.0455 | 0.0109 | 0 | NA | NA | |
| Rep_3 | 0.0557 | 0.0439 | 0.0118 | ||||
| Control | Rep_1 | 1.0943 | 0.2040 | 0.8903 | 0.8780 | 100.00 | 100.00 |
| Rep_2 | 2.2680 | 0.2322 | 2.0358 | 2.0249 | 100.00 | ||
| Rep_3 | 2.3037 | 0.2362 | 2.0675 | 2.0557 | 100.00 | ||
| Rotenone 100nM | Rep_1 | 0.6525 | 0.1000 | 0.5525 | 0.5402 | 61.52 | 53.93 |
| Rep_2 | 1.1915 | 0.1546 | 1.0369 | 1.0260 | 50.67 | ||
| Rep_3 | 1.1709 | 0.1396 | 1.0314 | 1.0196 | 49.60 | ||
| Rotenone 100 nM + Yashtimadhu 200 µg/ml | Rep_1 | 0.8633 | 0.1557 | 0.7077 | 0.6953 | 79.20 | 74.76 |
| Rep_2 | 1.8263 | 0.1956 | 1.6307 | 1.6198 | 79.99 | ||
| Rep_3 | 1.5189 | 0.1692 | 1.3497 | 1.3379 | 65.08 | ||
| Yashtimadhu 200 µg/ml | Rep_1 | 1.0833 | 0.1617 | 0.9217 | 0.9093 | 103.57 | 109.89 |
| Rep_2 | 2.5736 | 0.2595 | 2.3141 | 2.3032 | 113.74 | ||
| Rep_3 | 2.5775 | 0.2560 | 2.3216 | 2.3098 | 112.36 |
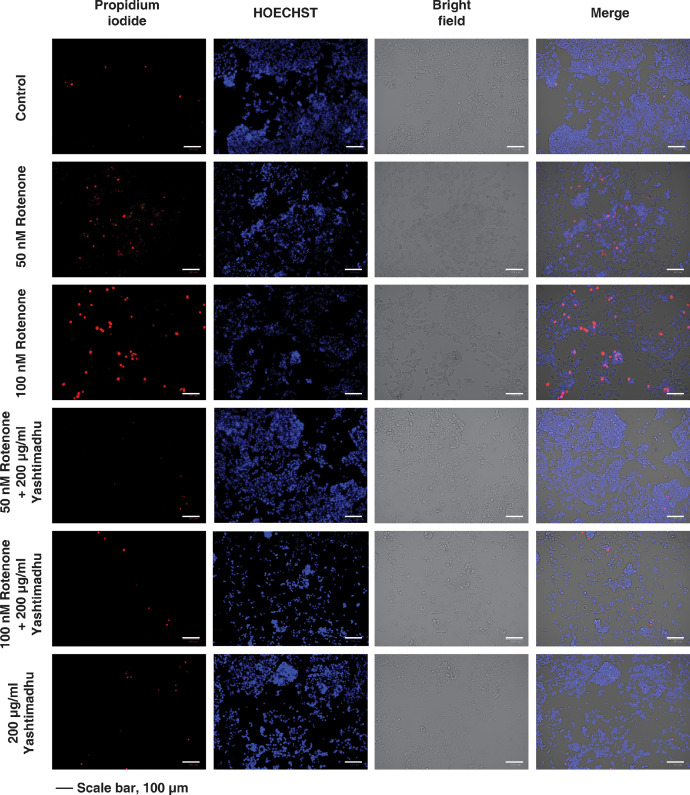
In addition to the cell cytotoxicity assay, a live-dead staining assay was also carried out on the differentiated IMR-32 cells to evaluate the effect of rotenone at 50 nM and 100 nM concentrations and the protective effect of Yashtimadhu extract at 200 µg/ml concentration (Fig. 2).
Fig. 2.
Live/dead cell staining assay using propidium iodide (red) to stain dead cells and HOECHST nuclear stain as a counterstain. Cells were treated with different concentrations of rotenone, 50 nM and 100 nM, and 200 µg/ml Yashtimadhu extract. The 50 nM and 100 nM rotenone treatment resulted in a significant (p < 0.05) reduction in live cells compared to control, and the rotenone+Yashtimadhu co-treatment resulted in a significant (p < 0.05) restoration of the live cells compared to the rotenone alone treatment groups.
The differentiated IMR-32 cells were treated with 100 nM rotenone and 200 µg/ml Yashtimadhu extract to analyze the effect of the treatments on cell cycle progression. The undifferentiated cells were also subject to cell cycle analysis. The scatter plots from the treatments are given in Fig. 3, and the percentage of cells in each of the cell cycle phases, such as G0/G1 (Population of cells in Go and G1 phases of cell cycle), S (population of cells in Synthesis phase), and G2/M (Population of cells in G2 and mitotic phases of cell cycle), is given in Table 6 and the raw FCS files are provided as supplementary files.
Fig. 3.
Scatter plot of cell cycle analysis of undifferentiated cells, and the differentiated cells treated with rotenone, rotenone + Yashtimadhu co-treatment and Yashtimadhu alone, which was assessed after 48 h of treatment.
Table 6.
Table showing the percentage of cells in different phases of cell cycle assessed after 48 h treatment.
| Sample | Replicate | % G0/G1 | % S | % G2/M | Average % G0/G1 | Average % S | Average % G2/M |
|---|---|---|---|---|---|---|---|
| Un-differentiated cells | Rep_1 | 52.59 | 32.01 | 15.39 | 56.93 | 28.95 | 14.13 |
| Rep_2 | 61.26 | 25.88 | 12.86 | ||||
| Rep_3 | 62.86 | 25.88 | 12.86 | ||||
| Control | Rep_1 | 68.89 | 24.53 | 6.58 | 56.13 | 38.84 | 5.03 |
| Rep_2 | 50.13 | 46.21 | 3.66 | ||||
| Rep_3 | 49.36 | 45.78 | 4.86 | ||||
| Rotenone 100nM | Rep_1 | 44.94 | 27.40 | 27.66 | 40.28 | 21.24 | 38.49 |
| Rep_2 | 46.65 | 19.12 | 34.23 | ||||
| Rep_3 | 29.24 | 17.19 | 53.57 | ||||
| Rotenone 100 nM + Yashtimadhu200 µg/ml | Rep_1 | 60.89 | 28.79 | 10.33 | 57.72 | 36.50 | 5.79 |
| Rep_2 | 55.76 | 37.20 | 7.04 | ||||
| Rep_3 | 56.50 | 43.50 | 0.00 | ||||
| Yashtimadhu 200 µg/ml | Rep_1 | 52.54 | 46.44 | 1.03 | 53.39 | 45.08 | 1.53 |
| Rep_2 | 62.94 | 33.50 | 3.56 | ||||
| Rep_3 | 44.70 | 55.30 | 0.00 |
1.1. Protein expression and interaction
Rotenone showed the IC50 at 100 nM therefore, the 100 nM rotenone concentration was used to assess the neuroprotective effect of Yashtimadhu.
The expression of proteins was analyzed using western blotting and their fold change of expression was calculated with respect to untreated control cells. The fold change values are presented as a heat map with the values in the boxes representing each replicate from the respective groups, Fig. 4A. The interaction network of these proteins is given in Fig. 4B, and the significance of their interaction is given in Table 7.
Fig. 4.
Protein expression and Protein-protein interaction map. (A) Heat map giving the protein expression across the replicates and groups,which were significantly (p < 0.05) altered between rotenone vs control, all of the proteins were significantly restored by Yashtimadhu (p < 0.05) co-treatment, for 48 h treatment time, (B) Protein-protein interaction. Kinase ERK-1/2 is displayed in green, and the proteins studied for their expression in orange, while the intermediary interaction nodes are given in blue, which were not studied for their expression. Legends: C: Control, R: 100 nM rotenone, R+Y: Rotenone 100 nM + Yashtimadhu 200 µg/ml, Y: Yashtimadhu 200 µg/ml.
Table 7.
Pathways regulated by the protein-protein interaction that were altered with rotenone and Yashtimadhu, after 48 h treatment.
| Interaction nodes | Confidence | Gene Ontology annotation | p-value | Corrected p-value | KEGG annotation | p-value | Corrected p-value |
|---|---|---|---|---|---|---|---|
| ERK1/2, PARP1, CASP3 | 0.96 | Cellular response to chemical stress | 3.0E-06 | 1.8E-03 | Apoptosis | 5E-06 | 4.7E-04 |
| ERK1, TP53, PHB, PDHA1 | 0.84 | Regulation of protein stability | 1.3E-05 | 3.7E-03 | Central carbon metabolism in cancer | 6.6E-07 | 2.0E-04 |
| ERK2, PARP1, ERK1, TP53, PHB, PDHA1 | 0.78 | Cellular response to oxidative stress | 3.2E-07 | 4.1E-04 | Central carbon metabolism in cancer | 2.8E-08 | 3.5E-05 |
| ERK2, PARP1, ERK1, TP53, PHB, HSPD1 | 0.78 | Response to oxidative stress | 1.7E-08 | 1.3E-04 | Apoptosis | 4.1E-07 | 1.5E-04 |
| ERK1/2, PARP1, PCNA | 0.96 | Cellular response to oxidative stress | 1.8E-06 | 1.3E-03 | Base excision repair | 5E-05 | 2.0E-03 |
| ERK1, PARP1, PCNA | 0.96 | Cellular response to oxidative stress | 1.8E-06 | 1.3E-03 | Base excision repair | 5E-05 | 2.0E-03 |
| ERK1/2, PARP1 | 0.96 | Cellular response to metal ion | 9.5E-05 | 1.3E-02 | Apoptosis | 2.9E-04 | 6.1E-03 |
2. Experimental Design, Materials and Methods
2.1. Materials
Propidium iodide, rotenone, retinoic acid, HOECHST-33342, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium-bromide), 2′,7′-Dichlorofluorescein diacetate (DCFDA), and collagen were procured from Sigma-Aldrich, St. Louis, USA. DMEM high glucose media, fetal bovine serum (FBS), and antibiotic/antimycotic solution, Gibco, Thermo Fisher Scientific USA. Antibodies were procured from Cell Signaling Technology, Danvers, USA, and Sigma-Aldrich, St. Louis, USA. Clarity ECL Substrate and Nitrocellulose membrane, Bio-Rad Laboratories, California, USA.
2.2. Yashtimadhu procurement, authentication, and extract preparation
Yashtimadhu choorna (Lot No.64), prepared from the roots of Glycyrrhiza glabra L. was procured from SDP Remedies and Research Centre, Puttur, Kaarnataka, India, a GMP-certified Ayurvedic product manufacturer (http://sdpayurveda.com/products/choorna/yastimadhu-choorna/). A specimen of it is maintained at the SDP Remedies and Research Centre (Identifier No. SDP/YM/001-2017). The industrial process includes the following steps; the roots of G. glabra L. were washed and dried under shade-net, followed by vacuum-drum drying. Dried roots were pulverized and sieved to yield the fine powder, giving a yield of about 90%.
For cell culture, Yashtimadhu root powder was suspended in MilliQ-water (0.1 g/mL) and incubated overnight at room temperature, with continuous rotation. The mixture was centrifuged at 5000 rpm for 10 min, twice. Aqueous supernatant was carefully aspirated into a new tube and dried using SpeedVac (Savant, Thermo Fisher Scientific, USA). The yield from the aqueous extraction of 1g of Yashtimadhu powder was found to be 54% w/w. The dried extract was dissolved in serum-free media for cell culture treatment.
2.3. Cell culture and treatment
IMR32 cells (ATCC® CCL-127™) were procured from National Centre for Cell Science, Pune, India. The cells were cultured in DMEM-high glucose medium supplemented with 10% FBS and antibiotic/antimycotic solution and maintained at 37°C with 5% CO2. Differentiation of IMR-32 cells was carried with DMEM-high glucose medium supplemented with 10 µM retinoic acid and 2% FBS for nine days. The expression of tyrosine hydroxylase was used to assess differentiation. Post-differentiation, the cells were treated with the following; 50 nM or 100 nM rotenone (dissolved in DMSO), 200 µg/mL Yashtimadhu extract, and co-treatment of rotenone and Yashtimadhu, for 48 h, while untreated cells were taken as control.
2.4. Cell cytotoxicity assay with rotenone and Yashtimadhu
IMR32 cells were seeded at 5000 cells/well, in a 96-well plate, and subject to treatment with different concentrations of rotenone (0.25 nM–10 µM) for 24h and 48h, Yashtimadhu (50–1500 µg/mL) for 48h and, co-treatment of Yashtimadhu and rotenone for 48h. MTT-dye was incubated for 3 -4 h; formazan crystals were dissolved using 50:50 ethanol: DMSO and read at 560 nm and 650 nm. Cell cytotoxicity is expressed as percentage cell viability with respect to untreated control. Cell cytotoxicity assay was conducted as three technical replicates and three independent biological replicates.
2.5. Staining of live and dead cells
Differentiated and treated cells (rotenone, Yashtimadhu, and rotenone + Yashtimadhu), were stained with 20 µg/mL of propidium iodide (PI), for dead cells and counter-stain, Hoechst-33342 (5 µg/mL) [9]. Cells were imaged using ZOE™ Fluorescent Cell Imager, BioRad Laboratories, California, USA. ImageJ software [10] was used to calculate the ratio of PI to Hoechst-33342 stained cells. The percentage of cell death was calculated with respect to control and later converted to fold change. The live-dead cell cycle was performed as two technical replicates and three independent biological replicates.
2.6. Analysis of cell cycle
Treated cells were washed with 1X PBS, trypsinized and further washed twice with 1X PBS and re-suspended in hypotonic buffer (2 µg/mL PI, 1 mg/mL trisodium citrate, 0.1% Triton-X 100 and, 100 µg/mL RNase), incubated in the dark for 30 min. The red fluorescence was measured using Guava® easyCyte Flow Cytometer, EMD Millipore, Massachusetts, USA. Cell cycle data analysis was carried out with FCS Express (version-6). The cell cycle data was performed as independent three biological replicates.
2.7. Immunoblotting
Harvested cells were lysed in buffer [4% sodium dodecyl sulfate in 50 mM triethylammonium bicarbonate, with sodium orthovanadate (1mM), sodium pyrophosphatase (2.5 mM), and, beta-glycerophosphate (1 mM)]. Following proteins were assessed, the primary antibody along with dilution factors are mentioned in brackets; cleaved-caspase-9 (1:1000), cleaved caspase-3 (1:1000), cleaved-Poly-ADP-ribose polymerase (PARP, 1:1000), BCL2-associated-X protein (BAX, 1:1000), BCL2-antagonist/killer-1 (BAK1, 1:1000), pyruvate dehydrogenase-E1 alpha-1 (PDHA1, 1:1000), cytochrome-C (1:1000), superoxide dismutase-1 (SOD1, 1:1000), heat-shock protein-60 (HSP60, 1:1000), phosphorylated extracellular-signal-regulated kinase-1/2 (ERK-1/2, 1:1000), total ERK1-/2 (1:1000) were procured from Cell Signaling Technology, USA and, HRP-conjugated β-actin (1:50000) was used a loading-control. Immunoblotting analysis of the above-mentioned proteins were carried out as three independent biological replicates.
Protein concentration estimated with BCA assay and immunoblotting was carried out as described previously [11]. Briefly, equal amounts of proteins from the four conditions were loaded onto SDS-PAGE and resolved. The resolution of proteins was carried out with SDS-PAGE, and the proteins were transferred onto a nitrocellulose membrane. Upon completion of the transfer, the membranes were blocked with 5% skimmed milk for 1h. Membranes were incubated in primary antibodies, which were diluted in 3% bovine serum albumin (BSA) overnight. Washing of membranes was carried out with 1X PBST (phosphate-buffered saline with 0.5% Tween-20) and further incubated with 1:5000 dilution HRP-conjugated secondary antibody in 3 % skimmed milk for 2h. The bands were imaged and developed with Enhanced Chemiluminescence reagent, BioRad, and captured onto X-ray films, Carestream. Densitometry analysis of band intensity was performed using ImageJ, normalized with loading-control, and fold-change calculated with respect to control. A heat map depicting the protein expression was prepared using Morpheus.
2.8. Protein-protein interaction
The proteins assessed with western blotting were further analyzed for their interaction network using ANAT (Advanced Network Analysis Tool) plug-in [12], in Cytoscape, version 3.8. The anchored network was built, and the significance of the interactions was obtained using the Gene Ontology (GO) and KEGG pathways using the ANAT tool, using the default settings for carrying out the protein-protein interaction network.
2.9. Statistical analysis
The data from the independent biological replicates were used for the statistical testing. GraphPad Prism 5 was used for the statistical analysis, using One-way ANOVA (Analysis of Variance) with Bonferroni-corrections to identify the significant pairs. The ANAT tool gives the p-value of enrichment, which was used for statistical significance. A p-value of ≤ 0.05 was considered statically significant.
Ethics Statement
The data described here did not involve the use of human or animal studies.
CRediT Author Statement
Gayathree Karthikkeyan: Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Visualization; Ashwini Prabhu: Methodology, Investigation, Resources; Ravishankar Pervaje: Conceptualization, Resources; Sameera Krishna Pervaje: Formal analysis; Prashant Kumar Modi: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition; Thottethodi Subrahmanya Keshava Prasad: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
The work was partially funded by Karnataka Biotechnology and Information Technology Services, Government of Karnataka, for support under the Biotechnology Skill Enhancement Programme in Multiomics Technology (BiSEP GO ITD 02MDA2017), and Yenepoya (Deemed to be University) Seed grant (YU/Seed grant/077-2019). The authors also thank Dr. Harkrishna Panaje, SDP Remedies and Research Centre for authenticating the specimen and providing the details of Yashtimadhu choorna. The authors thank Dr. Pushkar Sharma, Staff Scientist, National Institute of Immunology, India, for providing key antibodies used in this study. GK was a recipient of Senior Research Fellowship from Council of Scientific & Industrial Research (CSIR), Government of India (2014-2019), and is currently a recipient of KSTePs DST-Ph.D. Fellowship from Department of Science and Technology-Karnataka Science and Technology Promotion Society, Government of Karnataka (2020-2021).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2021.107535.
Contributor Information
Prashant Kumar Modi, Email: prashantmodi@yenepoya.edu.in.
Thottethodi Subrahmanya Keshava Prasad, Email: keshav@yenepoya.edu.in.
Appendix. Supplementary materials
References
- 1.Karthikkeyan G., Pervaje R., Pervaje S.K., Prasad T.S.K., Modi P.K. Prevention of MEK-ERK-1/2 hyper-activation underlines the neuroprotective effect of Glycyrrhiza glabra L. (Yashtimadhu) against rotenone-induced cellular and molecular aberrations. J. Ethnopharmacol. 2021;274 doi: 10.1016/j.jep.2021.114025. [DOI] [PubMed] [Google Scholar]
- 2.Poewe W., Seppi K., Tanner C.M., Halliday G.M., Brundin P., Volkmann J., et al. Parkinson disease. Nat. Rev. Dis. Primers. 2017;3:17013. doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 3.Condello S., Curro M., Ferlazzo N., Caccamo D., Satriano J., Ientile R. Agmatine effects on mitochondrial membrane potential and NF-kappaB activation protect against rotenone-induced cell damage in human neuronal-like SH-SY5Y cells. J. Neurochem. 2011;116(1):67–75. doi: 10.1111/j.1471-4159.2010.07085.x. [DOI] [PubMed] [Google Scholar]
- 4.Deolankar S.C., Modi P.K., Subbannayya Y., Pervaje R., Prasad TSK. Molecular targets from traditional medicines for neuroprotection in human neurodegenerative diseases. OMICS. 2020;24(7):394–403. doi: 10.1089/omi.2020.0033. [DOI] [PubMed] [Google Scholar]
- 5.Sarokte A.S., Rao M.V. Effects of Medhya Rasayana and Yogic practices in improvement of short-term memory among school-going children. Ayu. 2013;34(4):383–389. doi: 10.4103/0974-8520.127720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheshagiri S., Patel K.S., Rajagopala S. Randomized placebo-controlled clinical study on enhancement of Medha (intelligence quotient) in school going children with Yahstimadhu granules. Ayu. 2015;36(1):56–62. doi: 10.4103/0974-8520.169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karthikkeyan G., Najar M.A., Pervaje R., Pervaje S.K., Modi P.K., Prasad T.S.K. Identification of molecular network associated with neuroprotective effects of Yashtimadhu (Glycyrrhiza glabra L.) by quantitative proteomics of rotenone-induced Parkinson's disease model. ACS Omega. 2020;5(41):26611–26625. doi: 10.1021/acsomega.0c03420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karthikkeyan G., Pervaje R., Subbannayya Y., Patil A.H., Modi P.K., Prasad T.S.K. Plant omics: metabolomics and network pharmacology of Liquorice, Indian Ayurvedic medicine Yashtimadhu. OMICS. 2020;24(12):743–755. doi: 10.1089/omi.2020.0156. [DOI] [PubMed] [Google Scholar]
- 9.Bose B., Kapoor S., Sen U., Nihad As M., Chaudhury D., Shenoy P.S. Assessment of oxidative damage in the primary mouse ocular surface cells/stem cells in response to ultraviolet-C (UV-C) damage. J. V. Exp. JoVE. 2020;(156) doi: 10.3791/59924. [DOI] [PubMed] [Google Scholar]
- 10.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modi P.K., Komaravelli N., Singh N., Sharma P. Interplay between MEK-ERK signaling, cyclin D1, and cyclin-dependent kinase 5 regulates cell cycle reentry and apoptosis of neurons. Mol. Biol. Cell. 2012;23(18):3722–3730. doi: 10.1091/mbc.E12-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yosef N., Zalckvar E., Rubinstein A.D., Homilius M., Atias N., Vardi L., et al. ANAT: a tool for constructing and analyzing functional protein networks. Sci. Signal. 2011;4(196):l1. doi: 10.1126/scisignal.2001935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.