Abstract
The draft genome sequence of Bacillus subtilis A1, isolated from beach soil, has been shown to produce biofilm. The genome size is 4,215,114 bp with an average G+C content of 43.5%. The genome of Bacillus subtilis A1 has 4413 total genes which include 4166 protein-coding sequences, 126 pseudo genes, 10 rRNA genes with 3 operons (5S, 16S and 23S), 86 tRNA genes and 5 noncoding RNA (ncRNA) genes. The genome contains genes coding for surfactin, fengycin, bacillaene, sublancin 168, bacillibactin, subtilosin A, bacilysin. The whole genome project has been deposited in GenBank under the accession number CP075344.1. The raw data is available at https://www.ncbi.nlm.nih.gov/nuccore/CP075344.1.
Keywords: Genome sequencing, Biofilm, Antimicrobial peptide, Phylogenetic analysis
Specifications Table
| Subject | Microbiology |
| Specific subject area | Genomics |
| Type of data | Figures, Table, genome sequencing data is in FASTA format |
| How data were acquired | Genome was sequenced with Illumina Hiseq-X sequencing at AgriGenome (Kerala, India) |
| Data format | Raw and analysed data sequence |
| Parameters for data collection | The pure culture of Bacillus subtilis A1 was used to isolate genomic DNA. Further genomic DNA was sequenced and analysed. |
| Description of data collection | Genomic DNA extraction was carried out using SDS and ultrasonic lysis and sequenced by Illumina Hiseq X. The adaptor sequences and low quality bases are trimmed using AdapterRemoval-v2 (version 2.3.1). The pre-processed reads are aligned to the reference genome NC_000964.3. The alignment is performed using the BWA MEM Program. Genome annotation was carried out using NCBI Genome Automatic Annotation Pipeline (PGAP) |
| Data source location | Bacillus subtilis A1 was isolated from Beach soil (Midalam, Tamil Nadu) |
| Data accessibility | The sequence data has been deposited at GenBank with the BioProject number: PRJNA729632, BioSample number: SAMN19136531 under the accession number CP075344.1 (https://www.ncbi.nlm.nih.gov/nuccore/CP075344.1). The SRA records could be accessed (https://www.ncbi.nlm.nih.gov/sra/PRJNA729632). |
Value of the Data
-
•
The genome data of strain Bacillus subtilis A-1 helps to identify genes related to biofilm and used for discovering mechanism involved in quorum sensing.
-
•
Comparison of the whole genome sequencing data of the strain Bacillus subtilis A-1 with that of other Bacillus subtilis strains might provide wide knowledge on the enzymes and its production mechanism.
-
•
It also helps to understand the genetic structure and production of secondary metabolites especially non-ribosomal peptides which might provide pharmaceutical implications.
-
•
Comparative genomics might provide knowledge on identifying genes with respect to metal resistance since the bacterium is isolated from metal rich environment.
1. Data Description
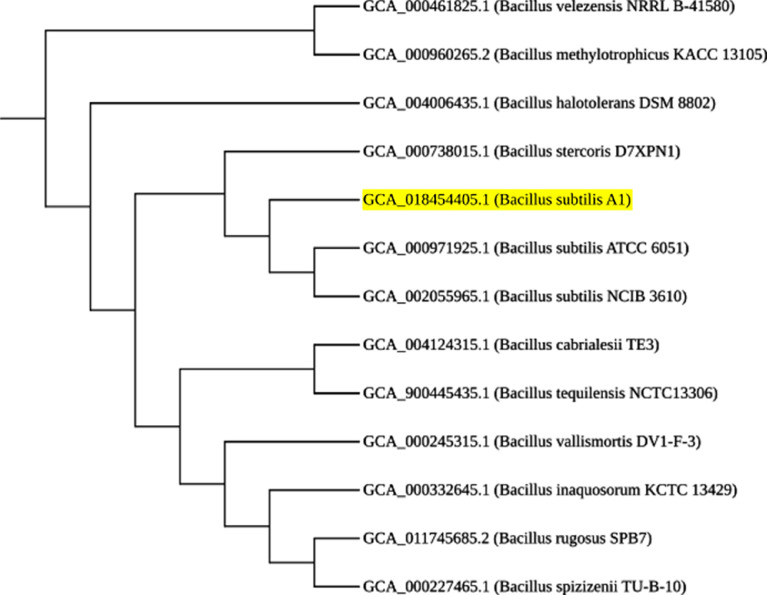
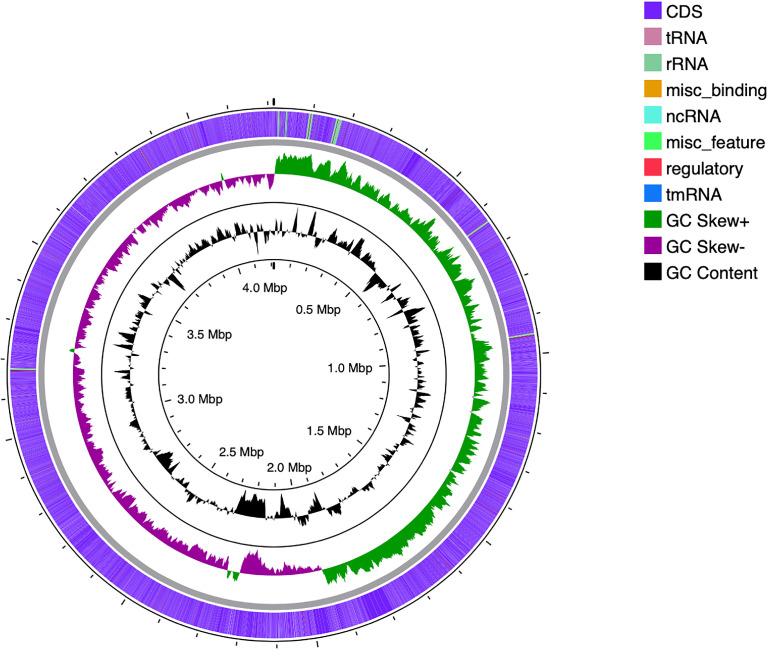
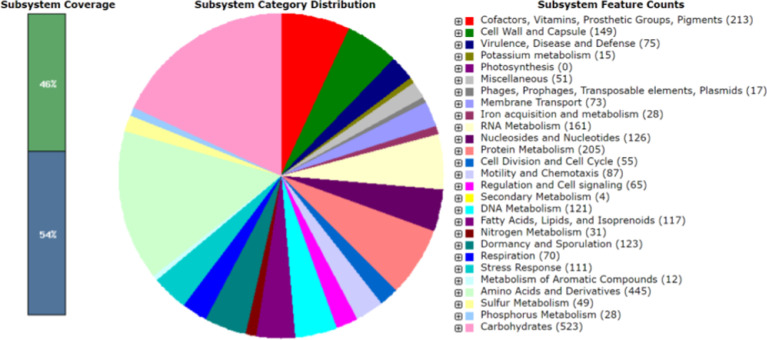
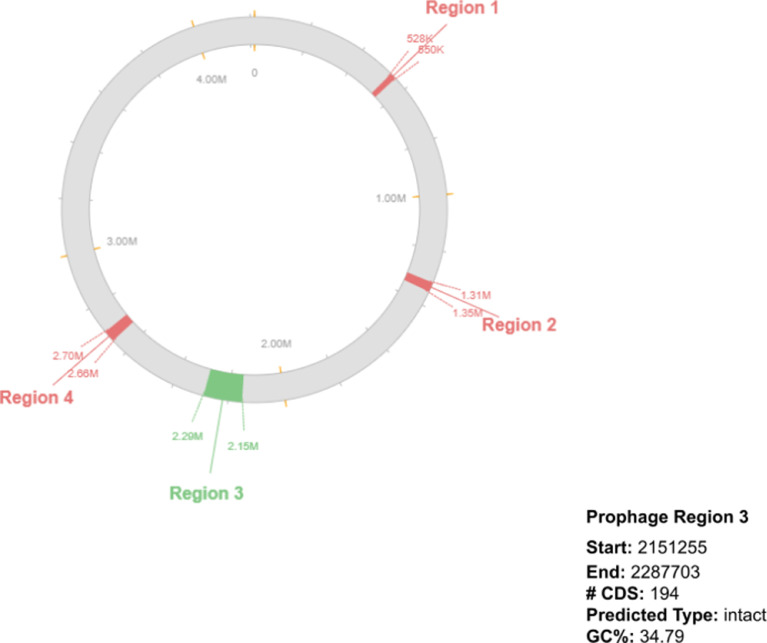
Bacillus subtilis is a Gram positive, catalase positive, non-pathogenic bacteria and is one of the most commonly studied bacterial species and used for producing several proteins and the organism is considered generally as safe (GRAS). It is a rod-shaped bacterium also called hay or grass bacillus, and capable of forming stress resistant endospores. They form biofilms and are sensitive to most antibiotics [1]. Bacillus subtilis also yields a variety of secondary metabolites that can be advantageous for commercial extraction and use. These include lipopeptides (surfactin, fengycin, iturin etc.), and RiPPs (lanthipeptides, thiopeptides etc.) among others. These metabolites have antimicrobial features that enhance its function as a biocontrol agent. Specifically, fengycin and surfactin are antibacterial and antifungal agents. Additionally, lanthipeptides and sactipeptides are commonly used as antibiotics [2]. In this study, the organism was isolated from beach soil and characterized using 16s rRNA sequencing and the sequence was submitted with MT361322.1. Fig. 1 represents the phylogenetic analysis of 16s sequences of isolates showing close relativity with the strain Bacillus subtilis. The whole genome sequencing of the isolate Bacillus subtilis A1 showing the evolutionary history was inferred and revealed close proximity to the strains of Bacillus subtilis (Fig. 2). Whole genome sequencing analysis along with genome annotation was performed and the assembled sequence was submitted in Gen bank with the accession id CP075344.1. The genome annotation features are provided in Table 1. The draft genome contains single contig with 4215114 bp. The genome of Bacillus subtilis A1 has 4413 total genes which include 4166 protein-coding sequences, 126 Pseudo genes, 10 rRNA genes with 3 operons (5S, 16S and 23S), 86 tRNA genes and 5 noncoding RNA (ncRNA) genes. A genomic circular map is provided in Fig. 3. The taxonomic position of the strain A1 was determined by multilocus sequence typing (MLST), using internal fragments of seven genes including purH, glpF, pycA, ilvD, rpoD, tpiA and pta. The RAST server identified the genome sequence of size 4,215,114 to have 4553 features, comprising 4437 coding sequences and 116 RNAs (5S RNA - 10, LSU rRNA - 10, SSU rRNA - 10, tRNA - 86). The sequence has GC content of 43.5% and 476 subsystems which is represented in Fig. 4. The presence of prophage sequences in the Bacillus genome A1 was analyzed and identified four prophage regions, of which 1 region was intact, 3 regions were incomplete (Fig. 5). Intact regions of prophages were located between positions 2151255 and 2287703 bp of length 136.4 Kb with total proteins of 194 is highlighted in Fig. 5. The strain A1 codes for major facilitator superfamily (MFS) antibiotic efflux pump and it codes for virulence factors mph (k) coding for spiramycin, telithromycin resistance, gene aadk coding for streptomycin and a gene tet (L) coding for doxycycline and tetracycline. No plasmids were found in the isolated genome. The genome contains genes responsible for the production of several bioactive secondary metabolites. The organism codes for surfactin, Bacillaen, fengycin, Sublancin 168, Bacillibactin, Subtilosin A and Bacilysin. The details of the secondary metabolites are provided in Table 2. The variant data was annotated using snpEff (supplementary file 1).
Fig. 1.
Phylogenetic relationship of 16s sequence of Bacillus subtilis A1.
Fig. 2.
Phylogenetic relationship of whole genome sequencing from Bacillus subtilis A1.
Table 1.
General genomic feature of Bacillus subtilis A1.
| Attributes | Value |
|---|---|
| Genome size (bp) | 4,215,114 |
| Genes (total) | 4413 |
| CDSs (total) | 4292 |
| Genes (coding) | 4166 |
| Genes (RNA) | 121 |
| rRNAs | 10 |
| tRNAs | 86 |
| ncRNAs | 5 |
| Pseudo Genes (total) | 126 |
Fig. 3.
Genomic organisation of Bacillus subtilis A1- Midalam.
Fig. 4.
The whole genome sequence of the strain Bacillus subtilis A1 was annotated using the Rapid Annotation System Technology (RAST) server. The pie chart demonstrates the subsystem category distribution.
Fig. 5.
Prophage regions of Bacillus subtilis A1.
Table 2.
List of Antimicrobial Peptides in Bacillus subtilis A1.
| Type | From | To | Most similar known cluster | Similarity(%) |
|---|---|---|---|---|
| Ranthipeptide, sactipeptide | 204175 | 226248 | Sporulation killing factor | 100 |
| NRPS | 358303 | 421744 | Surfactin | 82 |
| TransAT-PKS, PKS-like, T3PKS, transAT-PKS-like, NRPS | 1763763 | 1878521 | Bacillaene | 100 |
| NRPS, betalactone | 1935448 | 2017660 | Fengycin | 100 |
| Glycocin | 2259521 | 2279691 | Sublancin 168 | 100 |
| NRPS | 3260519 | 3310260 | Bacillibactin | 100 |
| Sactipeptide | 3826058 | 3847669 | Subtilosin A | 100 |
| Other | 3850668 | 3892086 | Bacilysin | 100 |
2. Experimental Design, Materials and Methods
2.1. Phylogenetic analysis
The organism was isolated from beach soil and characterized by PCR using 16s rRNA sequencing and whole genome sequencing of the isolates. The evolutionary history was inferred using TYGS tool [3] to reveal a close proximity to other strains.
2.2. Extraction of DNA and whole genome sequencing
A single colony of Bacillus spp was cultivated in Luria- Bertani medium at 37 ° C overnight. The bacterial cells were lysed using ultrasonic [4] and pelleted using a centrifuge for the extraction of DNA and sequenced at AgriGenome Labs, Kochi, Kerala. Sequencing was performed using Illumina HiSeq X. The library preparation was carried out using NEBNext Ultra DNA Library Prep kit. The reads are generated R1=3,175,214 and R2= 3,175,214 with a mean length of 150 bp. The raw reads (.fastq) of the sequenced genome were subjected to pre-processing. Low quality reads were trimmed using Adapter Removal v-2. The sample genome was compared to the reference strain 168 (lab strain of Bacillus subtilis), using BWA MEM program. Once genome assembly was complete, the aligned reads were sorted and duplicates were removed using sambamba (v-0.8.0). Bcftools was utilized to assess Single Nucleotide Polymorphisms (SNPs) and Indels. The taxonomic position of strain A1 was determined by the MLST database (PubMLST; http://pubmlst.org/bsubtilis/) [5]. Genes were predicted by NCBI Prokaryotic Genome annotation pipeline (PGAP) [6]. Annotation was performed with RAST [7] using the RASTtk scheme [8]. Functional analysis was carried out using the tools available in SEED portal [9] with phaster [10]. Antibiotic resistance was determined using CARD [11]. The genome was screened for the presence of plasmids using PlasmidFinder 2.1 [12]. Several bioactive secondary metabolites were revealed by antiSMASH [13].
Ethics Approval
CSP/20/FEB/84/97
CRediT Author Statement
Sneha Pramod: Writing – original draft, Formal analysis; Rhea Thomas Thommana: Writing – original draft, Software, Formal analysis; Harini Kulanthaivelu Kanagam: Writing – original draft, Software, Formal analysis; Ashmita Suresh Kumar: Writing – original draft, Formal analysis; Santha Kalaikumari S: Visualization, Writing – review & editing; Elavarashi Elangovan: Writing – review & editing, Supervision; Kumar Perumal: Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships, which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgement
The authors thank Sri Ramachandra Institute of Higher Education and Research for the GATE - Young Faculty Research Grant.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2021.107552.
Appendix. Supplementary materials
Supplementary file 1. Variant data of Bacillus subtilis A1
References
- 1.Earl A.M., Losick R., Kolter R. Ecology and genomics of Bacillus subtilis. Trends. Microbiol. 2008;16:269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westers L., Westers H., Quax W.J. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim. Biophys. Acta. 2004;1694:299–310. doi: 10.1016/j.bbamcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Meier-Kolthoff J.P., Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Z., Sun J., Lv A., Sung Y., Sun X., Shi H., Hu X., Wang A., Xing K. A modified method for genomic DNA extraction from the fish intestinal microflora. AMB Express. 2018;8:52. doi: 10.1186/s13568-018-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jolley K.A., Bray J.E., Maiden M. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz R.K., Bartels D., Best A.A. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brettin T., Davis J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., Shukla M., Thomason J.A., Stevens R., Vonstein V., Wattam A.R., Xia F. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5 doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., Vonstein V., Wattam A.R., Xia F., Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic. Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.V, Cheng A.A., Liu S., Min S.Y., et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zankari C.E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., Møller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blin K., Wolf T., Chevrette M.G., Lu X., Schwalen C.J., Kautsar S.A., Suarez Duran H.G., de Los Santos E., Kim H.U., Nave M., Dickschat J.S., et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1. Variant data of Bacillus subtilis A1