Abstract
Objectives
To find plasma biomarkers prognostic of type 2 diabetes, which could also inform on pancreatic β-cell deregulations or defects in the function of insulin target tissues.
Methods
We conducted a systems biology approach to characterize the plasma lipidomes of C57Bl/6J, DBA/2J, and BALB/cJ mice under different nutritional conditions, as well as their pancreatic islet and liver transcriptomes. We searched for correlations between plasma lipids and tissue gene expression modules.
Results
We identified strong correlation between plasma triacylglycerols (TAGs) and islet gene modules that comprise key regulators of glucose- and lipid-regulated insulin secretion and of the insulin signaling pathway, the two top hits were Gck and Abhd6 for negative and positive correlations, respectively. Correlations were also found between sphingomyelins and islet gene modules that overlapped in part with the gene modules correlated with TAGs. In the liver, the gene module most strongly correlated with plasma TAGs was enriched in mRNAs encoding fatty acid and carnitine transporters as well as multiple enzymes of the β-oxidation pathway. In humans, plasma TAGs also correlated with the expression of several of the same key regulators of insulin secretion and the insulin signaling pathway identified in mice. This cross-species comparative analysis further led to the identification of PITPNC1 as a candidate regulator of glucose-stimulated insulin secretion.
Conclusion
TAGs emerge as biomarkers of a liver-to-β-cell axis that links hepatic β-oxidation to β-cell functional mass and insulin secretion.
Keywords: Triacylglycerols, β-cell function, Systems biology, Type 2 diabetes, PITPNC1, Biomarkers
Abbreviations: TAGs, triacylglycerols; T2D, Type 2 diabetes; GSIS, glucose-stimulated insulin secretion; RC, regular chow; HFD, high-fat diet; PCA, principal component analysis; AUC, area under the curve; SMs, sphingomyelins; CE, cholesteryl esters; PC, phosphatidylcholines; Chol, cholesterol; LPC, lysophosphatidylcholines; PI, phosphatidylinositols; WGCNA, weighted gene co-expression network analysis; KEGG, Kyoto encyclopedia of genes and genomes; GO, Gene Ontology; ER, endoplasmic reticulum; OXPHOS, Oxidative phosphorylation; Lpl, lipoprotein lipase; LDL, low-density lipoproteins; HDL, high-density lipoproteins; VDR, vitamin D receptor
Highlights
-
•
Plasma triacylglycerols correlated with genes controlling β-cell mass and function.
-
•
Plasma triacylglycerols correlated with genes controlling liver β-oxidation.
-
•
In humans, triacylglycerols also correlated with key regulators of insulin secretion.
-
•
Mouse and human data identified PITPNC1 as a candidate regulator of insulin secretion.
-
•
Triacylglycerols are biomarkers of the liver-to-β-cell axis and β-cell function.
1. Introduction
Type 2 diabetes (T2D) is a chronic hyperglycemic condition characterized by reduced glucose-stimulated insulin secretion (GSIS) and increased insulin resistance of insulin target tissues [1,2]. Multiple initiating pathogenic mechanisms can affect any tissue primarily involved in glucose homeostasis, including the pancreatic islet β-cells, liver, muscle, adipose tissue, or the autonomic nervous system [3]. The deregulations in insulin secretion or in insulin action then lead to defects in inter-organ communications causing imbalanced glucose homeostasis, thus T2D. For developing better prevention strategies or treatment options, identification of the primary tissue defects that underlie the appearance of T2D in individual patients will be very useful. The presence of myriads of metabolites in the plasma represents a potential source of information about the function and deregulation of individual tissues [3,4]. Thus, their quantitative measurements could, in principle, inform the deregulations of specific tissue metabolic pathways. In addition, as circulating metabolites can modulate the function of various cell types, their identification could help define modes of interorgan communication that regulate glucose homeostasis.
In our previous study, we initiated a search for circulating biomarkers candidates for the susceptibility to T2D using preclinical models followed by replication of the mouse data in two pre-diabetes cohorts [5]. We first analyzed the correlation between plasma lipids and various glucose homeostasis phenotypes in mice from different genetic backgrounds and fed regular chow or a high-fat diet (HFD) for different periods. This analysis led to the identification of dihydroceramides as candidate biomarkers for T2D development. In human cohorts, we found that the same plasma dihydroceramides were elevated in the plasma of individuals who would develop T2D up to nine years later. Thus, preclinical studies in mice can provide valuable information to identify novel human T2D prognostic biomarkers.
In the present study, we attempted to further exploit this experimental mouse paradigm to investigate whether plasma lipid biomarkers could be identified that predict the function of pancreatic β-cells and whether we could identify tissue metabolic pathways involved in the regulation of the plasma concentrations of these lipids. Thus, we performed a new set of experiments using mice from three different genetic backgrounds fed for different periods with regular chow (RC) or HFD and searched for a correlation between plasma lipids and islets and liver gene co-expression modules. This analysis revealed strong positive and negative correlations between plasma TAGs and islets gene modules that comprise major regulators of insulin secretion such as glucokinase and the KATP channel or mRNAs encoding lipid metabolic enzymes that modulate GSIS. In the liver, we found that plasma TAGs showed a strong correlation with a gene module enriched in fatty acid β-oxidation genes. Together our data indicate that plasma TAGs reflect the activity of a liver-to-β-cell axis that may regulate β-cell function. Similar observations were made in humans linking plasma TAGs and islet genes controlling β-cell mass and function.
2. Methods
2.1. Mouse phenotyping
Eight-week-old male C57Bl/6J, DBA/2J, and BALB/cJ mice were fed ad libitum with a high fat, high sucrose diet (SAFE 235F, with 46% fat expressed in Kcal/kg) or a regular diet (SAFE A04). Oral glucose tolerance tests (OGTT, 2 g/kg from a 30% glucose solution) and insulin tolerance tests (ITT, Novorapid, 0.5 UI/kg) were performed in five-hour fasted mice on days 2, 10, and 30, as described by Cruciani-Guglielmacci et al. [6]. Analysis of the ITTs revealed basal glycemia, using a glucometer (A. Menarini Diagnostics, France), and insulin resistance calculated as the area under the curve of glycemia (AUC; mg/dL∗t) measured at 0, 15, 30, 45, 60, 90, and 120 min after insulin administration. From the OGTTs we also obtained basal insulinemia, using an Ultra-Sensitive Mouse Insulin ELISA Kit (Crystal Chem Inc., #90080); stimulated insulinemia calculated as the AUC of insulinemia (ng/mL∗t) measured at 0, 15, and 90 min after glucose administration, and glucose intolerance calculated as the AUC of the glycemia (mg/dL∗t) measured at 0, 15, 30, 45, 60, 90, and 120 min after glucose administration. The number of mice used in these phenotyping experiments ranged between 185 and 195.
2.2. Quantitative lipidomics analysis
Plasma lipid concentrations were measured by mass spectrometry at the Lipotype shotgun lipidomics platform. Samples processing, lipid extraction, spectra acquisition, data processing, and normalization were as described in Surma et al., 2015 [7]. A principal component analysis (PCA) was performed using the 'prcomp' function in R package.
2.3. RNA-Seq and bioinformatics analysis
Complementary DNA (cDNA) libraries were prepared from RNA isolated from mice tissues using the Illumina TruSeq protocol. RNA-Seq was performed on the Illumina HiSeq platform to generate ∼40Mio 125 nt single-end reads per sample. Reads were mapped and quantified with STAR-2.5.3a software [8] using M.musculus-mm10 as reference genome and GRCm38.83 from ENSEMBL as the reference annotation index. For each sample, quality control included verification of the total number of reads, percent of uniquely mapped reads, number of detected expressed genes, gene body coverage, and cumulative gene diversity. The resulting counts per gene from different samples were integrated to construct a single count matrix for each tissue that was filtered, excluding those genes with <1 count per million with 'edgeR' [9]. We excluded three clear outliers identified by PCA and hierarchical clustering in the islets data set. The count matrix was normalized using the trimmed mean (TMM) normalization method. Differentially expressed genes (DEGs) comparing HF and RC, and the different strains were detected using the Limma package in R [10]. P-values were adjusted for multiple comparisons with the Benjamini-Hochberg procedure, and those genes with adjusted p-value of ≤0.05 were considered as differentially expressed.
Weighted gene correlation network analysis (WGCNA) was performed on the RNA-Seq dataset from all time points, mouse strains, and diets to generate modules of co-expressed genes. Co-expression networks for each tissue were constructed by calculating signed adjacency matrices using a soft-thresholding power of six and a pair-wise Pearson correlation among all genes. A signed topological overlap matrix (TOM) was then calculated from each adjacency matrix, converted to distances, and clustered by hierarchical clustering using average linkage clustering. Modules were identified in the resulting dendrogram by the Dynamic Hybrid tree cut with a cut height of 0.995 and a minimum module size of 20 genes. The correlation between the resulting modules and plasma lipids was calculated in two steps for the liver data set, and the three steps for the islets data set are as follows: first, a PCA was calculated for each module in each data set using only module constituent genes. Second, the Spearman correlation coefficient was calculated between the summarized values of expression of each module (the first principal component or eigenvalue) and plasma lipids. In the islets data set, an intermediate step was added because plasma lipidomics and islet transcriptomics data were derived from different mouse individuals and direct correlation was not possible. Thus, 18 mouse groups were defined by the three strains, two diets, and three-time points of harvesting. Module eigenvalues and plasma lipid concentrations were summarized per mouse group using the mean, which yielded 18 pairs of values to be considered per lipid and module. Student asymptotic p-values were calculated for the given correlations and were adjusted for multiple comparisons using the Benjamini-Hochberg procedure [11].
The functional enrichment analysis was performed using the R library “clusterProfiler” [12]. Pathways from KEGG and GO databases were searched using a hypergeometric test to examine the overrepresentation of the terms within the functional annotation and to determine the p-values using the hypergeometric distribution. P-values were adjusted for multiple comparisons by the Benjamini-Hochberg procedure. Terms with an adjusted p-value ≤0.05 were considered as overrepresented.
2.4. Correlation analysis in partially pancreatectomised patients
The correlation analysis was performed with 60 samples that included T2D, impaired glucose tolerance (IGT), pancreatogenic diabetes (T3cD), and control patients (18, 21, 16, and 4 cases respectively). Pairwise correlations between the pancreatic islets transcriptome and TAGs plasmatic concentrations were performed with Spearman correlation coefficients. For the correlations, Student asymptotic p-values were calculated.
2.5. Cell culture
EndoC-βH1 cells [13] were cultured in 5.6 mM glucose Dulbecco's Modified Eagle's Medium (DMEM, Thermo Fisher Scientific), supplemented with 2% BSA fraction V (Roche-Diagnostics), 50 μM β-mercaptoethanol (Sigma–Aldrich), 5.5 μg/mL transferrin (Sigma–Aldrich), 6.7 ng/mL sodium selenite (Sigma–Aldrich), 10 mM nicotinamide (Calbiochem), 100 μg/mL streptomycin and 100 U/mL penicillin (Thermo Fisher Scientific). Cells were seeded on plates coated with 1.2% Matrigel/3 μg/mL fibronectin (Sigma–Aldrich), and cultured at 37 °C and in 5% CO2.
MIN6B1 cells [14] were cultured in 25 mM glucose DMEM (Thermo Fisher Scientific), supplemented with 15% heat-inactivated fetal bovine serum (Sigma–Aldrich), 71 mM β-mercaptoethanol, and maintained at 37 °C and 5% CO2 (24).
2.6. Small interfering RNA transfection of EndoC-βH1 cells
EndoC-βH1 cells were seeded on 12-well plates and transfected with siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific) 24 h later. The PITPNC1 specific siRNA (siPITPNC1 #1 and #2) sequences were CCACAGACGCACCCGAAUU and CGAUGAAAUUCCAGAGCGC (Microsynth), respectively. Briefly, siPITPNC1 or siCTRL (negative control siRNA, Microysnth) were diluted in OptiMEM (Thermo Fisher Scientific); then Lipofectamine RNAiMAX was added. After 10 min of incubation, the lipid-siRNA complex was added to cells to obtain a final siRNA concentration of 80 nM. The medium was changed 4 h later for fresh culture medium. Cells were used for GSIS experiments 72 h post-transfection.
2.7. RNA isolation and qRT-PCR
Total RNA was extracted from EndoC-βH1 cells and MIN6B1 cells using RNeasy Plus Micro kit (Qiagen) and RNeasy Mini kit (Qiagen), respectively. cDNAs were synthesized using SuperScript II Reverse Transcriptase (Invitrogen), according to the manufacturer's instructions. Expression of target genes was measured by real-time quantitative PCR (qRT-PCR) using the 7500 Fast Real-Time PCR System (Applied Biosystems). qRT-PCR was run in a final volume of 10 μL containing 2 μL of cDNA and 8 μL of Power SYBR Green PCR mix (Applied Biosystems), in the presence of forward and reverse primers (sequences in Key resources table). GusB transcript levels were used to normalize each sample. All the primers were obtained from Microsynth.
2.8. Glucose-stimulated insulin secretion
Forty eight hours after siRNA transfection, EndoC-βH1 cells were starved in 0.5 mM glucose DMEM, supplemented with 2% BSA fraction V, 50 μM β-mercaptoethanol, 5.5 μg/mL transferrin, 6.7 ng/mL sodium selenite, 10 mM nicotinamide, 100 μg/mL streptomycin, and 100 U/mL penicillin. After 24-h of starvation, cells were washed and pre-incubated in Krebs–Ringer bicarbonate HEPES buffer (KRBH1) containing 0.2% BSA fraction V and 0 mM glucose for 1 h. Insulin secretion was measured following a 4-h incubation with KRBH containing 0.2% BSA fraction V and 0.5 mM or 20 mM glucose, in the presence or absence of 45 μM 3-isobutyl-1-methylxanthine (IBMX, Sigma–Aldrich). At the end of the incubation, supernatants were collected; cells lysed for 1 h on ice in TETG buffer: 20 mM Tris HCl pH 8, 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EGTA; supplemented with protease inhibitor cocktail (complete Tablets Mini EDTA-free, Roche). Insulin secretion and cellular insulin content were measured by ELISA according to the manufacturer's protocols using the Human Insulin kit (Mercodia).
2.9. Glucose-stimulated insulin secretion statistics
Data were analyzed using R software (v. 3.6.1) and are presented as the means ± S.E.M. from three independent experiments. Comparisons were performed using two-sample unpaired t-tests, and the p-values adjusted for multiple comparisons. Statistically significant differences are indicated with asterisks (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
3. Results
3.1. Metabolic, lipidomic, and transcriptomic phenotyping
3.1.1. Metabolic phenotyping
To search for potential correlations between plasma lipids and islet genes controlling β-cell function, we exploited the natural diversity of mice with different genetic backgrounds. In a previous study of the differential metabolic adaptation of six different strains of mice [6], we found that different genetic backgrounds directed various insulin secretion capacities and sensitivities to insulin of target tissues, and to different metabolic adaptations to HFD. Here, we decided to work with C57Bl/6J, BALB/cJ, and DBA/2J mice because of their distinct metabolic characteristics [6]. Groups of 8-week-old mice were fed with an RC or an HFD and phenotyped at 2, 10, and 30 days. They were then sacrificed at respectively, 5, 13, and 33 days, for transcriptomic and plasma lipidomic analysis (Figure 1A). These short periods of feeding were selected because we previously showed that important changes in gene expression that predict the long-term (>3 months) establishment of different obesity/diabetes phenotypes occur early after the initiation of the HFD [6,15,16]. In addition, time-series analysis provides additional power to establish meaningful correlations between physiological and omics data.
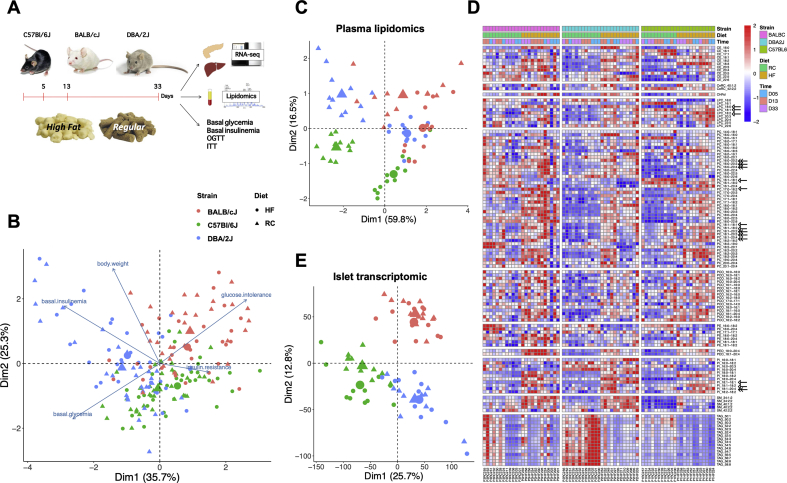
Figure 1.
Metabolic, lipidomic, and transcriptomic phenotyping. A. Experimental scheme. B. Biplot of the principal component analysis (PCA) of the physiological data and of the phenotypic traits from each mouse used in the study. Large circles and triangles represent the centroids of each group. (n = 185 mice). C. Principal component analysis of the lipidomics data. Each symbol represents data from one mouse; n = 71 mice. D. Heatmap of the plasma concentrations of each lipid species in each mouse. E. Principal component analysis of the islets transcriptomic data; n = 69 mice.
Mouse phenotyping included the assessment at days 2, 10, and 30 of body weight, five hours fasted glycemia and insulinemia, stimulated insulinemia (AUC of insulinemia measured during an OGTT), glucose intolerance, and insulin resistance (n = 185–195 mice, Figure S1). PCA of the physiological data revealed clear separation of mice by strain (Figure 1B) and a weak effect of diet, as indicated by the shift in the average position of each strain of mice, represented by large triangles and circles in the biplot of Figure 1B. This biplot also indicates that “glucose intolerance” was the main driver separating BALB/cJ mice and that “basal insulinemia” and “basal glycemia” drove the separation of DBA/2J mice.
3.1.2. Lipidomic analysis
Four mice from each group were sacrificed in the random fed state at days 5, 13, and 33 of RC or HFD feeding. Their quantitative lipidomic measurements were obtained for 71 out of 72 plasma samples. The total amount of lipids extracted from each plasma sample was in the optimal range for lipidomic measurements (5200–12900 pmol; Figure S2A and Suppl. Table 1) [7]. We identified ∼130 lipid molecules from 11 classes of lipids (Table 1). The percentage of total lipids accounted for by each class of lipids (Figure S2B) showed that the major classes were cholesteryl esters (CEs), phosphatidylcholines (PCs), cholesterols (Chol), triacylglycerols (TAGs), lysophosphatidylcholines (LPCs), and phosphatidylinositols (PIs). The other lipid classes formed only small proportions of the total plasma lipids. The percentage of plasma lipids contributed by TAGs showed significant variation across different mouse strains and feeding conditions whereas the contribution of the other lipid class was relatively constant.
Table 1.
Lipid abbreviation.
| Abbreviation | Lipid |
|---|---|
| CE | Cholesteryl Esters |
| CeRC | Ceramides |
| CHFol | Cholesterol |
| LPC | Lysophosphatidylcholines |
| PC | Phosphatidylcholine |
| PCO | Ether-linked/plasmalogen lysophosphatidylcholines |
| PE | Phosphatidylethanolamine |
| PEO | Ether-linked/plasmalogen phosphatidylethanolamines |
| PI | Phosphatidylinositol |
| SM | Sphingomyelins |
Note: Lipid species are annotated according to their molecular composition as an abbreviation, sum of the carbon atoms in the hydrocarbon moiety, the sum of the double bonds in the hydrocarbon moiety, and the sum of hydroxyl groups. For example, SM 34:1:2 denotes a sphingomyelin species with a total of 34 carbon atoms, 1 double bond, and 2 hydroxyl groups in the ceramide backbone. In the case of cholesteryl esters, lysophosphatidylcholines, lysophosphatidylethanolamines, and triacylglycerols there are no hydroxyl groups. Phosphatidylcholines, phosphatidylethanolamines, and phosphatidylinositols annotation contain information on the exact identity of their fatty acids. For example, PI18:1-16:0 denotes phosphatidylinositol with C18:1 (oleic) and C16:0 (palmitic) fatty acids. The exact position of fatty acids in relation to a glycerol backbone (sn1 or sn2) cannot be determined.
PCA showed that plasma lipidomics was influenced by both strains and feeding conditions (Figure 1C). A detailed description of the relative concentration of each lipid species in all the plasma samples analyzed is presented in the heat map of Figure 1D. This illustrates that the concentrations of individual lipids were strain- and diet-dependent. In addition, feeding the HFD, which comprises mostly C16:0, C: 18:0, C18:1 and C18:2 fatty acids [17] led to a general tendency to increase the concentrations of lipids containing these fatty acyl side chains (see examples: open arrows in Figure 1D). Nevertheless, several lipid species were also increased that contained one of these C16–C18 fatty acyl side chains and one longer, and more unsaturated fatty acyl side chain (C20:2–C20:4; double arrow in Figure 1D), suggesting that increased activity of various elongases and desaturases also contribute to the changes in lipid species induced by HFD feeding [18]. Figure 1D shows that the plasma concentrations of TAGs were reduced by HFD feeding in the three strains of mice. This observation can be explained by the coordinated induction by HFD feeding of several hepatic β-oxidation genes (See Suppl. Table 2 and further results below).
3.1.3. Islet transcriptomic analysis
Islet transcriptomic data were obtained from islets isolated from 3 to 6 randomly fed mice for each of the 18 experimental groups. A total of 69 islet transcriptomic data sets were used for the subsequent analysis as three samples were rejected after quality control.
PCA showed that the islets transcriptomic was influenced mostly by the mouse genetic backgrounds and to a lower extent by the feeding conditions (Figure 1E). The Venn diagrams of Figure S3A show that differential gene expression between islets from RC- and HFD-fed mice was dependent on strains and period of HFD feeding (Suppl. Table 3). The highest number of differentially regulated genes was found at day 5 of HFD feeding. Islets from C57Bl/6J mice displayed the highest number of differentially expressed genes (354 vs. 113 for DBA/2J mice and 43 for BALB/cJ mice). Analysis of islet genes differentially expressed (adj. p < 0.05) between strains, and considering all feeding conditions, revealed that of the 13,780 genes considered, 9,104 genes were differentially expressed between DBA/2J and C57Bl/6J islets, 8,941 when comparing BALB/cJ with C57Bl/6J islets, and 6,268 when comparing BALB/cJ with DBA/2J islets (Figure S2B and Suppl. Table 4).
Together the above multiomics data sets form the basis for the subsequent, unbiased search for salient correlations between plasma lipids and islet gene expression.
3.2. Correlation between islet gene expression modules and plasma lipids
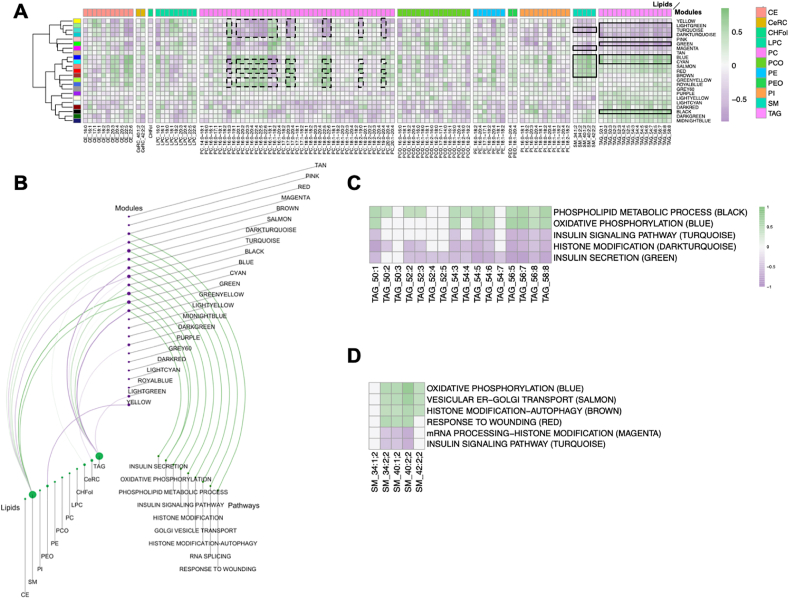
Islet transcriptomic data were subjected to weighted gene co-expression network analysis (WGCNA) [19] to identify groups of genes that were co-regulated across all mouse strains and feeding conditions (gene modules). The Spearman correlation between the summarized value of the expression (eigenvalue) for each of these modules and the concentration of individual lipids was then calculated as described in the Materials and Methods section. These results are presented as a heat map in Figure 2A and the Suppl. Table 5. This shows that some lipid classes correlated homogenously, positively or negatively, with gene modules identified by a color code. For instance, triacylglycerols (TAGs) were negatively correlated (|rho|≥0.4), with the light green, turquoise, dark turquoise, and green modules and positively with the blue, cyan, and black modules (Figure 2A). Sphingomyelins, as a class, also correlated with unique modules (magenta, salmon, red, and brown) and the modules also correlated with TAGs (turquoise, blue, and cyan). In other classes of lipids, only a small number of individual species correlated with gene modules. This was the case for PCs, where only the indicated molecules (dashed lines in Figure 2A) showed correlations (|rho|≥0.2) with some gene modules. Several of these modules were shared with the TAGs (light green, turquoise, dark turquoise, and cyan modules) or SMs (red module). Because TAGs and SMs correlated as classes with islet gene modules, we focused our further analysis on these lipids.
Figure 2.
TAGs and SMs correlation with gene modules and islet functional pathways. A. Heatmap of the lipid-gene module correlations. The color code for the lipid classes is indicated on the right of the Figure. The color scale on the right indicates the value of Spearman's rank correlation coefficient. The seven modules that are most correlated with plasma TAGs and SMs are highlighted with black lines. Dashed black lines indicate the individual PCs correlated with the indicated gene modules. B. Hiveplot displaying the correlation between lipid classes, islet gene modules, and the functional terms enriched in each module. The size of the circles indicates the relative strength of the correlations. Green lines: positive correlations; violet lines: anti-correlations. C. Heatmap of the correlations between individual plasma TAGs and the pathways over-represented in the correlated modules. D. Heatmap of the correlation between plasma SMs and the pathways over-represented in the correlated modules.
3.3. Correlation of TAGs and SMs with islet gene modules and functional pathways
We performed KEGG and Gene Ontology (GO) analysis of the genes included in the islet modules, thus showing the strongest correlations with plasma TAGs and SMs. The results of this analysis are presented in Figure S4 (for TAGs) and S5 (for SMs). The modules that negatively correlated with TAGs were enriched in “insulin secretion” genes (green module), “insulin signaling pathway” genes (turquoise module), and “histone modification” genes (dark turquoise module). The positively correlated modules were enriched for “phospholipid metabolic process” genes (black module) and “oxidative phosphorylation” genes (blue and cyan modules). No overrepresented terms were found among the light green module genes.
The modules that negatively correlated with SMs included not only the same “insulin signaling pathway” genes (turquoise module) but also “mRNA processing/histone modification” (magenta module). The positively correlated modules were enriched in “vesicular ER/Golgi transport” genes (salmon module), “histone modification/autophagy” genes (brown module), and “response to wounding” genes (red module).
The relationship between lipidomic data, islet gene modules, and functional pathways is represented in the hive plot of Figure 2B. This shows that the lipid classes with the highest correlations to gene expression modules were TAGs and SMs and that these correlated with essential β-cell functional pathways, such as insulin secretion, oxidative phosphorylation, phospholipid metabolic processes, and insulin signaling. The heat maps of Figure 2C,D shows the correlation of individual TAGs and SMs with the most important β-cell functional pathways identified. Most individual species showed identical correlations with the islet gene modules.
Collectively, the above data showed that the plasma concentrations of TAGs and SMs, showed a significant correlation with islet gene pathways controlling key β-cell functions. Because of the variability of plasma TAGs concentrations across mouse strains and feeding conditions we next focused on the genes characterizing the functional terms correlated with TAGs.
3.3.1. Islet modules negatively correlates with TAGs
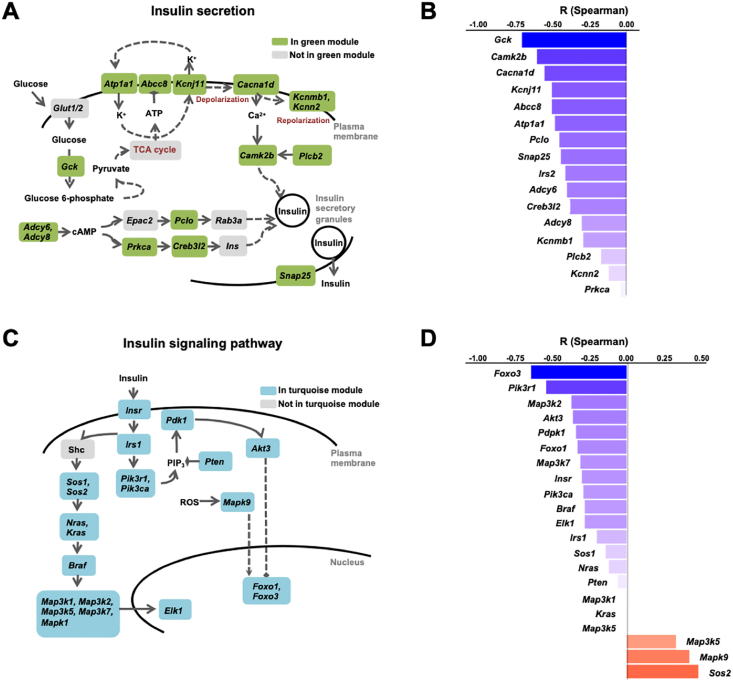
Analysis of the “insulin secretion” term showed that it included a large number of mRNAs encoding key regulators of GSIS (Figure 3A). The highest correlation with plasma TAGs was found for the Gck mRNA (r = −0.70; p = 6.52E-09), which encodes the rate-controlling enzyme in GSIS [20] (Figure 3B). Other highly correlated mRNAs encoded the subunits of the KATP channel (Abcc8 and KcnJ11), the alpha subunit of the Na+/K+ ATPase (Atp1a1), the voltage-gated Ca++ channel subunit alpha-1 (Cacna1d). They also included regulators of the gluco-incretin signaling pathway, which depends on cyclic adenosine monophosphate (cAMP) production (Adcy6, Adcy8, Prkca, Creb3l2, Pclo) and other regulators of insulin granule exocytosis (Camk2b, Snap25).
Figure 3.
Plasma TAGs anti-correlated with islet insulin secretion and insulin signaling pathways. A. Scheme of the insulin secretion pathway with, in green, the genes of the “insulin secretion” term present in the islet green module. B. Spearman correlation coefficients between the expression of the “insulin secretion” genes and plasma TAGs. C. Scheme of the insulin signaling pathway with, in blue, the genes of the “insulin signaling pathway” term present in the turquoise module. D. Spearman correlation coefficients between the expression of the “insulin signaling pathway” genes and plasma TAGs.
The top correlated “signaling” terms (turquoise module, Figure S4B) included “MAPK signaling”, “mTOR signaling”, and “Foxo signaling”. The complete list of genes forming these terms and their correlation with TAGs are presented in Suppl. Table 6. The mRNAs most highly correlated to plasma TAGs encoded most of the components of the insulin receptor signaling pathway, Insr, Irs1, Pi3kca, Pik3r1, Pten, Pdpk1, Akt3, Foxo1, Foxo3, Mapk1, Mapk9, Sos2, Braf, Nras, and Kras (Figure 3C). The correlation of each of these genes with TAGs is presented in Figure 3D.
The dark turquoise module contained only 29 genes; a significant association of these genes with the “histone modification” term was identified and eight mRNAs supported this association (Suppl. Table 7). These were the acetylated epigenetic mark readers (Brd1, Brd3); a component of a histone acetyltransferase complex (Ep400); a histone methyltransferase (Setd1b) and demethylase (Phf2), and Hcfc1 and Arid1a, which are part of chromatin remodeling complexes.
3.3.2. Islet modules positively correlated with TAGs
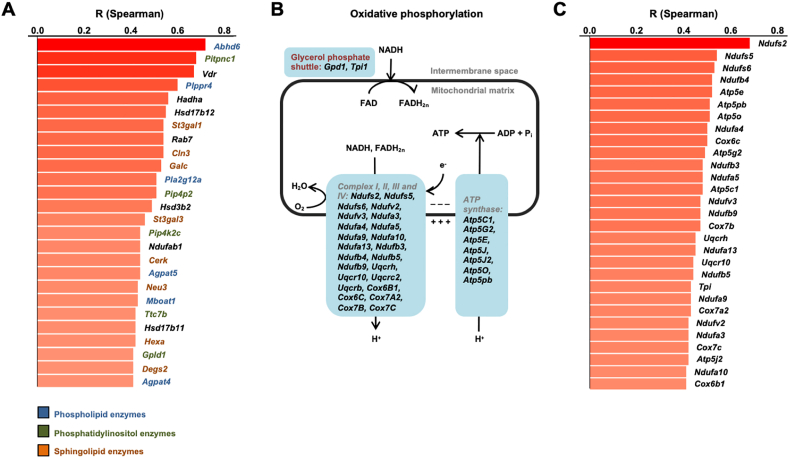
The GO analysis of the positively correlated black module showed enrichment in “phospholipid metabolic process” and other lipid metabolism-related terms (Figure S4D). These included genes controlling the metabolism of phospholipids (Plppr4, Pla2g12a, Mboat1, Agpat5, Agpat4), of phosphatidyl-inositols (Pitpnc1, Pip4p2, Pip4kc2, Ttc7b, Gpld1), and of sphingolipids (Cln3, Galc, Cerk, Degs2, St3gal1, Neu3, HexA), with each lipid class playing a modulatory role in insulin secretion [[21], [22], [23], [24]] (Figure 4A). The highest positive correlation was with Abhd6 (r = 0.72, and p = 8.49E-10), encoding monoacylglycerol lipase alpha/beta hydrolase Domain 6, an endocannabinoid degrading enzyme that negatively controls insulin secretion [25].
Figure 4.
Plasma TAGs correlate with islet lipid signaling and oxidative phosphorylation pathways. A. Genes of the “lipid signaling” term of the black module and their Spearman correlation coefficients with plasma TAGs (shown only for correlations with adjusted p-values <0.05 and |rho|≥0.4). These genes encoded enzymes contributing to the phospholipids (blue), phosphatidylinositol (green), and sphingolipids (orange) metabolic pathways. B. Scheme of the oxidative phosphorylation pathway with, in blue boxes, the genes of the “oxidative phosphorylation” term of the blue and cyan modules. C. Spearman correlation coefficients between expression of the “oxidative phosphorylation” genes and plasma TAGs.
The blue and cyan modules were enriched in terms (“Thermogenesis”, “Huntington”, “Alzheimer”, “Parkinson”, and “Oxidative phosphorylation”), which primarily comprised oxidative phosphorylation mRNAs (Figures S4E,F). The scheme of Figure 4B shows that most of these genes belong to the OXPHOS chain up to the ATP synthase step and Figure 4C shows the correlation of individual mRNAs with plasma TAGs.
Overall, the above data show that plasma TAGs were negatively correlated with the GSIS pathway and with the insulin signaling pathway, which controls β-cell proliferation and functional mass [26]. Contrastively, plasma TAGs were positively correlated with lipid signaling pathways, notably with Abhd6, a negative regulator of insulin secretion.
3.4. Plasma TAGs correlate with the liver β-oxidation pathway
The liver is the major tissue involved in the biosynthesis and secretion of TAGs. In addition, a liver-to-β-cell axis, which controls β-cell insulin secretion capacity, has previously been reported to involve various liver-derived signals [[27], [28], [29]]. We, therefore, investigated whether a hepatic metabolic pathway could be identified that could link plasma TAGs and β-cell function. We, thus, obtained RNA-Seq data from 72 mouse livers (Figure 1A). The scatter plot of Figure S6A shows the separation of the transcriptomic profiles by strain and, in part, by diet, particularly in BALB/cJ mice. These data were then used for WGCN analysis and for identification of gene co-expression modules.
The heat map of Figure S6B shows a correlation between the plasma lipids and liver gene modules. Four modules showed strong negative correlations with plasma TAGs and were enriched in specific GO terms: “fatty acid degradation” (cyan), “Golgi vesicle transport” (bisque4), “response to endoplasmic stress” (light yellow); no pathway enrichments were found in the saddle brown module.
Four modules, enriched in specific GO terms showed a positive correlation with TAGs: “sterol biosynthesis process” (dark orange), “cell cycle phase transition” (red), “lipid transport” (brown 4), and “regulation of sterol biosynthetic process” (black). The correlation between individual TAGs and these GO terms is presented in Figure S6C.
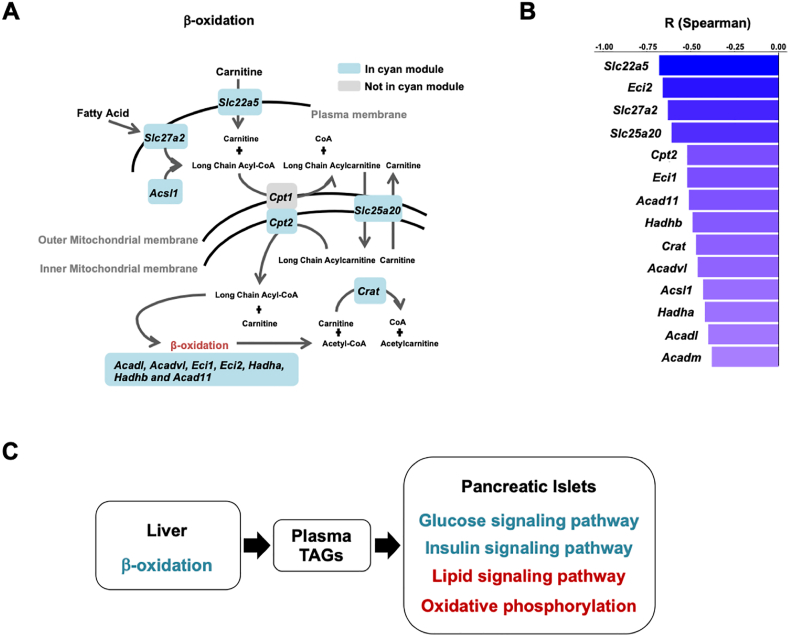
Among the negatively correlated modules, only the “fatty acid degradation” cyan module contained metabolism-related genes. These included a large number of β-oxidation mRNAs (Acadl, Acadvl, Eci1, Eci2, Hadha, Hadhb and Acad11) and two mRNAs encoding a plasma membrane (Slc22a5) and a mitochondrial (Slc25a20) carnitine transporter. The position of the encoded proteins in the overall fatty acid uptake and catabolism pathway is described in the scheme of Figure 5A. The individual correlations between these mRNAs with plasma TAGs are listed in Figure 5B. The expression of these mRNAs was increased by HFD in the livers of the three mouse strains, as mentioned above (Suppl. Table 2).
Figure 5.
Plasma TAGs anti-correlate with the liver ß-oxidation pathway. A. Scheme of the liver fatty acid and carnitine uptake and β-oxidation pathways with, in blue, the genes present in the liver “fatty acid degradation term” of the cyan module. B. Spearman correlations between expression of the fatty acid metabolism-related genes of (A) and plasma TAGs. C. Plasma TAGs link liver β-oxidation to β-cell pathways controlling insulin secretion and functional mass. Pathways in blue are anti-correlated with plasma TAGs, those in red are positively correlated with TAGs.
Among the positively correlated modules, the dark orange module was enriched with mRNAs encoding enzymes involved in cholesterol biosynthesis (Figure S7). The brown4 module included 4 mRNAs in the GO term “lipid transport” (Aqp8, Lipc, Cyp2j5, Atp8b4). The red module was enriched in biological processes related to cell cycle regulation (Suppl. Table 8). The black module also contained five mRNAs involved in cholesterol biosynthesis (Srebpf1, Abcg1, Erlin1, Erlin2, Scp2).
Together, the above data showed a striking inverse correlation between liver β-oxidation and plasma TAGs concentrations, which conforms with the established relationship between these two parameters [30]. It further suggests a link between hepatic β-oxidation, plasma TAGs, and β-cell function (Figure 5C).
3.5. Correlation between plasma TAGs and islet transcripts in humans
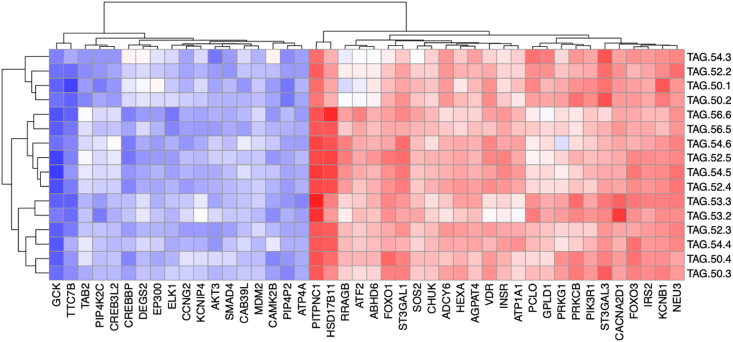
We next assessed whether similar correlations between plasma TAGs and key functional β-cell genes could be observed in humans. To this end, we analyzed RNA-Seq data generated from laser capture micro-dissected islets and quantitative plasma lipidomic data obtained from a cohort of 60 partially pancreatectomized patients [31].
For correlation analysis, we selected the aforementioned human orthologues of the mouse islet mRNAs that encode components of the GSIS pathway, lipid metabolic enzymes, and proteins of the insulin signaling pathway. Out of the 86 considered mouse genes, 85 had human orthologs that were expressed in the human islets data set. Figure 6 shows the heatmap of the correlation between plasma TAGs and 44 of the most highly correlated human islet mRNAs. Strikingly, as in mice, the top negative correlation was found for GCK. The correlation was also found with CREBP and KCNB1, important regulators of the insulin signaling pathway. Strong correlations were also found for several of the lipid metabolic enzymes. The strongest correlations were found for PITPNC1 (Pitpnc1 was the gene with the second highest correlation to TAGs in the list of Figure 4A, related to lipid signaling), for ABHD6, and for HSD17B11. Key elements of the insulin signaling pathway showed significant correlation with TAGs: IRS2, PI3KR1, AKT3, FOXO1, FOXO3 (see Figure 6B).
Figure 6.
Plasma TAGs also correlate with key ß-cell transcripts in humans. Heatmap of the Spearman correlation coefficients between individual TAGs and human islet mRNAs. The human islet mRNAs analyzed were homologous of the mRNAs showing the highest correlation with plasma TAGs in mice. The human data were from paired islet transcriptomic and plasma lipidomic analysis generated from biosamples obtained from partially pancreatectomized patients.
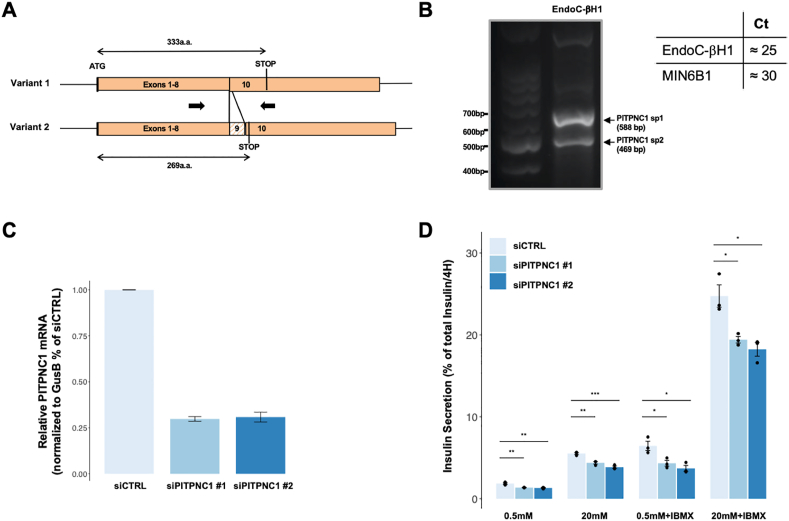
3.6. PITPNC1 as a novel regulator of insulin secretion
Because islet PITPNC1, which codes for a phosphatidyl-inositol transfer protein [32], showed a strong correlation with TAGs both in mice and humans, we performed preliminary experiments to assess whether this gene could be a so-far uncharacterized regulator of insulin secretion. We first measured its expression in the mouse MIN6B1 and the human EndoC-βH1 insulin cell lines. MIN6B1 cells have an almost undetectable level of the Pitpnc1 mRNA (Ct ∼30). In contrast, the two splice variants of PITPNC1 [32] were robustly expressed in EndoC-βH1 cells (Ct ∼25; Figure 7A,B). We, therefore, investigated whether silencing PITPNC1 in EndoC-βH1 would impact GSIS. Figure 7C shows that transfecting the cells with two distinct siRNAs, which target both splice variants, reduced PITPNC1 expression by ∼60–70%. This led to a significant reduction in insulin secretion in basal conditions as well as in the presence of 20 mM glucose or of 20 mM glucose and IBMX, a phosphodiesterase inhibitor (Figure 7D).
Figure 7.
PITPNC1 as a novel regulator of insulin secretion. Analysis of a regulatory role of PITPNC1 in glucose-stimulated insulin secretion. A.PITPNC1 is expressed as two alternative splicing variants. Variant 2 includes exon 9, which leads to the presence of an in-frame stop codon at the indicated position. The location of the PCR primers is indicated by arrows. B. EndoC-bH1 cells express both PTPNC1 variants (left) at a relatively high level as compared to the expression of Pitpnc1 mRNA in MIN6B1 cells (right). C. Shows the efficacy of PITPNC1 silencing in EndoC-bH1 cells transfected with a CTRL or two different PITPNC1 siRNAs. D. EndoC-bH1 cells were transfected with the indicated siRNAs and exposed 48 h later to 0.5 or 20 mM glucose for 4 h, in the presence or absence of IBMX. Insulin secretion is expressed as a percent of total insulin. The data are the means ± S.E.M. from three independent experiments. Statistically significant differences are indicated with asterisks (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
4. Discussion
In the present study, we aimed to determine whether plasma lipids could be the indicators of β-cell function and whether the tissues and pathways controlling the production of these lipids could be identified. We found that plasma TAGs, as a class, correlate strongly with islet gene co-expression modules. The most correlated modules are enriched in mRNAs encoding regulators of GSIS, enzymes producing various lipids that modulate insulin secretion, and transducers of the insulin signaling pathway. In the liver, the highest correlation was with the β-oxidation pathway. In humans, analysis of paired plasma lipid and islet transcriptomic data revealed a similar pattern of correlation between plasma TAGs and key regulators of insulin secretion as found in mice. This cross-species correlation analysis led to the identification of PITPNC1 as a new candidate regulator of insulin secretion. Thus, plasma TAGs emerge as biomarkers of β-cell function both in mice and humans and as potential effectors of a liver-β-cell axis.
It is usually considered that T2D appears when pancreatic β-cells can no longer compensate for the insulin resistance of the liver, adipose tissue, and muscle [1,33]. However, whether the primary defects lie in insulin secretion or in insulin action may vary among patients. Assessing insulin sensitivity and appearance of insulin resistance or determining the insulin secretion capacity of an individual during the progression of pre-diabetes or T2D is a complex process that relies on glucose or insulin tolerance tests or on insulinemic/glycemic clamp techniques. Therefore, finding prognostic plasma biomarkers for T2D, which could also give information about primary defects in pancreatic β-cells and/or in insulin target tissues would pave the way for improved preventive or therapeutic options adapted to individual patients.
Several previous studies led to the identification of prognostic biomarkers for T2D or which can help T2D patients’ stratification [34] and to the identification of islet gene modules characteristic of diabetic β-cell deregulations [31,[35], [36], [37]]. Plasma metabolomic biomarkers include various classes of hydrophilic and lipophilic molecules [34,[38], [39], [40], [41], [42], [43], [44], [45]]. In a previous study, we identified dihydroceramide as prognostic biomarker candidates for T2D in mice and humans [5]. Plasma TAGs showed a correlation in mice in our previous study with fasting insulinemia and glucose intolerance. Here, we exploited the vast diversity of metabolic traits of mice with different genetic backgrounds to search, in an unbiased manner, for salient correlations between plasma lipids and tissue gene expression modules.
We found that plasma TAGs and SMs displayed high correlations with islet gene modules that regulate critical β-cell physiological functions. The highest correlations with plasma TAGs were found for mRNAs that encoded key regulators of insulin secretion. These included the negative correlations with Gck and the subunits of the KATP channel, Abcc8, and Kcnj11, and the positive correlation with Abhd6, a negative regulator of insulin secretion [25]. Thus, during the initial period of HFD feeding, when plasma TAGs are reduced, increased expression of the Gck gene and the KATP channel and lower levels of Abhd6 represent a mechanism for β-cell compensation and insulin hypersecretion.
A negative correlation between plasma TAGs and a large number of genes involved in the insulin signaling pathway was also documented. The insulin/IGF1R signaling pathway in β-cells participates in the control of β-cell proliferation, differentiation, and protection against apoptosis [26,[46], [47], [48], [49], [50]]. Thus, we suggest a strong association between the activity of this signaling pathway and GSIS in our studied mouse models. Contrastively, no correlation between plasma TAGs and transducers or modulators of the insulin/IGF1R signaling pathway was established in the liver. This suggests that the insulin signaling pathway may be differentially regulated in β-cells and the liver. Transcriptional control of the expression of the various signaling components may predominate in the pancreas whereas phosphorylation events play a major role in modulating the insulin signaling cascade in the liver [51].
Correlation between SMs and islet gene modules overlapped, in part, with that between gene modules and plasma TAGs but also included other modules, characterized by “vesicular ER/Golgi transport”, “histone modification/autophagy” or “response to wounding” genes. These were less directly related to the processes of acute GSIS and control of β-cell mass and function; therefore, we focused on the modules correlated with plasma TAGs.
High levels of plasma TAGs can result from increased liver de novo lipogenesis, increased hepatic uptake of fatty acids originating from the adipose tissue or the ingested food, or reduced hepatic β-oxidation [30]. Additionally, we found that plasma TAGs displayed a strong inverse correlation only with a liver gene module enriched in β-oxidation genes (Acadl, Acadvl, Eci1, Eci2, Hadha, Hadhb, and Acad11). This suggests that in our experimental model increased hepatic fatty acid catabolism is the main regulator of plasma TAGs concentrations. This is consistent, for instance, with the observation that fenofibrate-induced hepatic β-oxidation reduces hypertriglyceridemia [30,52]. The same gene co-expression module also included genes for the plasma membrane (Slc22a5) and mitochondrial membrane (Slc25a20) carnitine transporters. Carnitine is required for transporting fatty acids into mitochondria by carnitine palmitoyl-transferase for their subsequent β-oxidation. The appearance of two carnitine transporters in our correlation analysis indicates that they regulate the availability of carnitine for free fatty acid degradation. Carnitine nutritional supplementation can reduce plasma TAGs levels in rodents and humans [[53], [54], [55]].
Together, our lipidomic–transcriptomic correlation analysis suggests the existence of a liver-to-β-cell axis whereby hepatic β-oxidation, by regulating plasma TAGs levels, may influence the expression of key regulators of GSIS. Although the mechanistic link between TAGs and β-cell insulin secretion capacity remains poorly understood, it is well known that exposure of islets to high glucose, high free fatty acids, or a combination of both lead to increased basal insulin secretion [[56], [57], [58]] and defects in β-cell function. β-cells express the enzyme lipoprotein lipase, which releases free fatty acids from circulating TAGs for cellular uptake [59,60]. Free fatty acids are required to maintain insulin secretion capacity in the fasted state [61]. However, in the long term, elevated concentrations of free fatty acids increase basal- and reduce glucose-stimulated insulin secretion [[62], [63], [64]]. We found that the lipoprotein lipase (Lpl) mRNA, which is expressed in β-cells and is required for normal GSIS [59], is expressed at a similar level in all islet preparations (not shown). Thus, differential free fatty acid uptake may primarily be determined by the circulating levels of TAGs. The mentioned detrimental effects of free fatty acids on β-cell function may explain why elevated plasma TAGs can be causal biomarkers of β-cell dysfunction and T2D susceptibility [65,66]. An alternative or complementary explanation is that plasma TAGs are secreted by the liver as part of lipoprotein particles; studies have shown that low-density lipoproteins (LDL) have a detrimental effect on GSIS, whereas high-density lipoproteins (HDL) protect the β-cell functional mass [67]. The fact that higher circulating LDL/HDL ratios may increase diabetes susceptibility is in agreement with the observation that single nucleotide polymorphisms leading to reduced expression of LPL in humans are associated with an increased risk of T2D development [68].
Our study further unveiled similar correlations between plasma TAGs and islet genes involved in glucose and lipid regulation of insulin secretion and insulin signaling in mice and humans. This is the case for ABHD6, a negative regulator of insulin secretion [25], or the vitamin D receptor (VDR), which has also been suggested to control the glucose competence of β-cells [69,70]. But this cross-species analysis also identified several highly correlated genes whose function in regulating insulin secretion remains undefined. Examples include PITPNC1, the mRNA that showed the second highest correlation with TAGs in mice and the highest correlation in humans. PITPNC1 encodes a phosphatidyl-inositol transfer protein, which associates with the Golgi complex through its binding to resident PI4P and which recruits Rab1b to enhance protein secretion [71,72]. Single nucleotide polymorphism in PITPNC1 has also been linked to an increased risk of T2D [73]. Other potentially interesting mRNAs are HSD17D11 and HSD17B12 genes, which encode, respectively, a short-chain fatty acid dehydrogenase/reductase and an enzyme that converts estrone into estradiol and which also has a fatty acid elongase activity. Further work is, however, needed to determine the exact mechanism underlying the regulation of insulin secretion by PITPNC1.
Collectively, our data show that plasma TAGs display high correlations with islet gene co-expression modules encoding key regulators of β-cell function and strong correlation, in the liver, with a module that contains mRNAs encoding fatty acid transporters, carnitine transporters, and β-oxidation genes. These correlations indicate that lower hepatic β-oxidation activity leads to increased plasma TAGs, which then may negatively impact the expression of genes controlling GSIS. Therefore, TAGs can be considered not only as biomarkers of type 2 diabetes [74] but also as indicators of the correlation that exists between liver fatty acid degradation and β-cell pathways that control these cells’ integrity and secretion capacity. Finally, this research provides a rationale for reducing plasma TAGs, for instance through carnitine supplementation or by fenofibrate-like molecule treatments, to prevent type 2 diabetes development.
Author contributions
B.T., C.M., M.I., and C.C.G. conceived the experiments. C.C.-G., J.L., J.D. and N.K. performed the mouse phenotyping and collected and processed the samples. C.R. and K.S. performed the lipidomic analysis. A.R.S.-A. performed the analysis and multi-omics integration in the pre-clinical model and the human cohort of PPP. L.W. and F.M. performed lipidomics quality controls and phenotypic measures quality controls and statistical tests. C.R. performed the experimental validation in the mouse cell lines. M.S., M.B., J.W., M.D., and M.I. contributed human transcriptomic data. B.T. and A.R.S.-A. wrote the manuscript with contributions from M.I. who also supervised the bioinformatics analysis. B.T. coordinated and managed the research project with support from C.M. and M.I. All authors have critically read the manuscript and approved it for publication.
Data availability
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (59) and are accessible through GEO Series accession number GSE164673 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164673). Pancreatic islets expression data SubSeries are available with the accession number GSE140369 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140369) and liver expression with accession number GSE164672 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164672).
Funding
This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 115881 (RHAPSODY). This Joint Undertaking receives support from the European Union's Horizon 2020 Research and Innovation Program and EFPIA. This work is also supported by the Swiss State Secretariat for Education, Research and Innovation (SERI) under contract number 16.0097. The opinions expressed and arguments employed herein do not necessarily reflect the official views of these funding bodies. B.T. also received support from a Swiss National Science Foundation Grant (310030_1824969).
Acknowledgements
We wish to thank Wanda Dolci, Xavier Pascal Berney, and the Genomic Technologies Facility of the University of Lausanne for their technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101355.
Contributor Information
Ana Rodríguez Sánchez-Archidona, Email: ana.rodriguez.1@unil.ch.
Céline Cruciani-Guglielmacci, Email: cruciani@univ-paris-diderot.fr.
Clara Roujeau, Email: clara.roujeau@sciprom.ch.
Leonore Wigger, Email: leonore.wigger@sib.swiss.
Justine Lallement, Email: justine.lallement@univ-paris-diderot.fr.
Jessica Denom, Email: jessica.denom@univ-paris-diderot.fr.
Marko Barovic, Email: marko.barovic@helmholtz-muenchen.de.
Nadim Kassis, Email: nadim.kassis@univ-paris-diderot.fr.
Florence Mehl, Email: florence.mehl@sib.swiss.
Jurgen Weitz, Email: juergen.weitz@uniklinikum-dresden.de.
Marius Distler, Email: marius.distler@uniklinikum-dresden.de.
Christian Klose, Email: klose@lipotype.com.
Kai Simons, Email: simons@lipotype.com.
Mark Ibberson, Email: mark.ibberson@sib.swiss.
Michele Solimena, Email: michele.solimena@tu-dresden.de.
Christophe Magnan, Email: christophe.magnan@univ-paris-diderot.fr.
Bernard Thorens, Email: bernard.thorens@unil.ch.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Nolan C.J., Damm P., Prentki M. Type 2 diabetes across generations: from pathophysiology to prevention and management. Lancet. 2011;378(9786):169–181. doi: 10.1016/S0140-6736(11)60614-4. [DOI] [PubMed] [Google Scholar]
- 2.Prentki M., Nolan C.J. Islet beta-cell failure in type 2 diabetes. Journal of Clinical Investigation. 2006;116(7):1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorens B., Rodriguez A., Cruciani-Guglielmacci C., Wigger L., Ibberson M., Magnan C. Use of preclinical models to identify markers of type 2 diabetes susceptibility and novel regulators of insulin secretion - a step towards precision medicine. Molecular Metabolism. 2019;27S(Suppl):S147–S154. doi: 10.1016/j.molmet.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauling L., Robinson A.B., Teranishi R., Cary P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proceedings of the National Academy of Sciences of the United States of America. 1971;68(10):2374–2376. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wigger L., Cruciani-Guglielmacci C., Nicolas A., Denom J., Fernandez N., Fumeron F., et al. Plasma dihydroceramides are diabetes susceptibility biomarker candidates in mice and humans. Cell Reports. 2017;18(9):2269–2279. doi: 10.1016/j.celrep.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Cruciani-Guglielmacci C., Bellini L., Denom J., Oshima M., Fernandez N., Normandie-Levi P., et al. Molecular phenotyping of multiple mouse strains under metabolic challenge uncovers a role for Elovl2 in glucose-induced insulin secretion. Molecular Metabolism. 2017;6(4):340–351. doi: 10.1016/j.molmet.2017.1001.1009. eCollection 2017 Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surma M.A., Herzog R., Vasilj A., Klose C., Christinat N., Morin-Rivron D., et al. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. European Journal of Lipid Science and Technology. 2015;117(10):1540–1549. doi: 10.1002/ejlt.201500145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochberg Y., Benjamini Y. More powerful procedures for multiple significance testing. Statistics in Medicine. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 12.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P., et al. A genetically engineered human pancreatic beta-cell line exhibiting glucose-inducible insulin secretion. Journal of Clinical Investigation. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y., et al. Establishment of a pancreatic beta-cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127(1):126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 15.Hall D., Poussin C., Velagapudi V.R., Empsen C., Joffraud M., Beckmann J.S., et al. Peroxisomal and microsomal lipid pathways associated with resistance to hepatic steatosis and reduced pro-inflammatory state. Journal of Biological Chemistry. 2010;285(40):31011–31023. doi: 10.1074/jbc.M110.127159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poussin C., Ibberson M., Hall D., Ding J., Soto J., Abel E.D., et al. Oxidative phosphorylation flexibility in the liver of mice resistant to high-fat diet-induced hepatic steatosis. Diabetes. 2011;60(9):2216–2224. doi: 10.2337/db11-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding J., Loizides-Mangold U., Rando G., Zoete V., Michielin O., Reddy J.K., et al. The peroxisomal enzyme L-PBE is required to prevent the dietary toxicity of medium-chain fatty acids. Cell Reports. 2013;5(1):248–258. doi: 10.1016/j.celrep.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 18.Guillou H., Zadravec D., Martin P.G., Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Progress in Lipid Research. 2010;49(2):186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-1189-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matschinsky F.M., Glaser B., Magnuson M.A. Pancreatic beta-cell glucokinase: closing the gap between theoretical concepts and experimental realities. Diabetes. 1998;47(3):307–315. doi: 10.2337/diabetes.47.3.307. [DOI] [PubMed] [Google Scholar]
- 21.Tengholm A., Gylfe E. Oscillatory control of insulin secretion. Molecular and Cellular Endocrinology. 2009;297(1–2):58–72. doi: 10.1016/j.mce.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Lang J. PIPs and pools in insulin secretion. Trends in Endocrinology and Metabolism. 2003;14(7):297–299. doi: 10.1016/s1043-2760(03)00136-x. [DOI] [PubMed] [Google Scholar]
- 23.Prentki M., Matschinsky F.M., Madiraju S.R. Metabolic signaling in fuel-induced insulin secretion. Cell Metabolism. 2013;18(2):162–185. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Boslem E., Meikle P.J., Biden T.J. Roles of ceramide and sphingolipids in pancreatic β-cell function and dysfunction. Islets. 2012;4(3):177–187. doi: 10.4161/isl.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao S., Mugabo Y., Iglesias J., Xie L., Delghingaro-Augusto V., Lussier R., et al. α/β-Hydrolase domain-6-accessible monoacylglycerol controls glucose-stimulated insulin secretion. Cell Metabolism. 2014;19(6):993–1007. doi: 10.1016/j.cmet.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Kulkarni R.N. New insights into the roles of insulin/IGF-I in the development and maintenance of beta-cell mass. Reviews in Endocrine & Metabolic Disorders. 2005;6(3):199–210. doi: 10.1007/s11154-005-3051-y. [DOI] [PubMed] [Google Scholar]
- 27.Seyer P., Vallois D., Poitry-Yamate C., Schutz F., Metref S., Tarussio D., et al. Hepatic glucose sensing is required to preserve beta cell glucose competence. Journal of Clinical Investigation. 2013;123(4):1662–1676. doi: 10.1172/JCI65538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imai J., Katagiri H., Yamada T., Ishigaki Y., Suzuki T., Kudo H., et al. Regulation of pancreatic beta cell mass by neuronal signals from the liver. Science. 2008;322(5905):1250–1254. doi: 10.1126/science.1163971. [DOI] [PubMed] [Google Scholar]
- 29.El Ouaamari A., Kawamori D., Dirice E., Liew C.W., Shadrach J.L., Hu J., et al. Liver-derived systemic factors drive beta cell hyperplasia in insulin-resistant states. Cell Reports. 2013;3(2):401–410. doi: 10.1016/j.celrep.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postic C., Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. Journal of Clinical Investigation. 2008;118(3):829–838. doi: 10.1172/JCI34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wigger L., Barovic M., Brunner A.D., Marzetta F., Schöniger E., Mehl F., et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nature Metabolism. 2021;3(7):1017–1031. doi: 10.1038/s42255-021-00420-9. [DOI] [PubMed] [Google Scholar]
- 32.Garner K., Hunt A.N., Koster G., Somerharju P., Groves E., Li M., et al. Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. Journal of Biological Chemistry. 2012;287(38):32263–32276. doi: 10.1074/jbc.M112.375840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weir G.C., Gaglia J., Bonner-Weir S. Inadequate β-cell mass is essential for the pathogenesis of type 2 diabetes. The Lancet Diabetes & Endocrinology. 2020;8(3):249–256. doi: 10.1016/S2213-8587(20)30022-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbasi A., Sahlqvist A.S., Lotta L., Brosnan J.M., Vollenweider P., Giabbanelli P., et al. A systematic review of biomarkers and risk of incident type 2 diabetes: an overview of epidemiological, prediction and aetiological research literature. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0163721. eCollection 0162016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller M.P., Choi Y., Wang P., Davis D.B., Rabaglia M.E., Oler A.T., et al. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Research. 2008;18(5):706–716. doi: 10.1101/gr.074914.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solimena M., Schulte A.M., Marselli L., Ehehalt F., Richter D., Kleeberg M., et al. Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia. 2018;61(3):641–657. doi: 10.1007/s00125-00017-04500-00123. Epub 02017 Nov 00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L., Pan Z., Yang S., Shan W., Yang Y. Identification of key gene pathways and coexpression networks of islets in human type 2 diabetes. Diabetes, Metabolic Syndrome and Obesity. 2018;11:553–563. doi: 10.2147/DMSO.S178894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T.J., Larson M.G., Vasan R.S., Cheng S., Rhee E.P., McCabe E., et al. Metabolite profiles and the risk of developing diabetes. Nature Medicine. 2011;17(4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T.J., Ngo D., Psychogios N., Dejam A., Larson M.G., Vasan R.S., et al. 2-Aminoadipic acid is a biomarker for diabetes risk. Journal of Clinical Investigation. 2013;123(10):4309–4317. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang-Sattler R., Yu Z., Herder C., Messias A.C., Floegel A., He Y., et al. Novel biomarkers for pre-diabetes identified by metabolomics. Molecular Systems Biology. 2012;8:615. doi: 10.1038/msb.2012.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wurtz P., Tiainen M., Makinen V.P., Kangas A.J., Soininen P., Saltevo J., et al. Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care. 2012;35(8):1749–1756. doi: 10.2337/dc11-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menni C., Fauman E., Erte I., Perry J.R., Kastenmuller G., Shin S.Y., et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62(12):4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrannini E., Natali A., Camastra S., Nannipieri M., Mari A., Adam K.P., et al. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62(5):1730–1737. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molnos S., Wahl S., Haid M., Eekhoff E.M.W., Pool R., Floegel A., et al. Metabolite ratios as potential biomarkers for type 2 diabetes: a DIRECT study. Diabetologia. 2018;61(1):117–129. doi: 10.1007/s00125-017-4436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mousa A., Naderpoor N., Mellett N., Wilson K., Plebanski M., Meikle P.J., et al. Lipidomic profiling reveals early-stage metabolic dysfunction in overweight or obese humans. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2019;1864(3):335–343. doi: 10.1016/j.bbalip.2018.1012.1014. Epub 2018 Dec 1023. [DOI] [PubMed] [Google Scholar]
- 46.Okada T., Liew C.W., Hu J., Hinault C., Michael M.D., Krtzfeldt J., et al. Insulin receptors in beta-cells are critical for islet compensatory growth response to insulin resistance. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(21):8977–8982. doi: 10.1073/pnas.0608703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueki K., Okada T., Hu J., Liew C.W., Assmann A., Dahlgren G.M., et al. Total insulin and IGF-I resistance in pancreatic beta cells causes overt diabetes. Nature Genetics. 2006;38(5):583–588. doi: 10.1038/ng1787. [DOI] [PubMed] [Google Scholar]
- 48.Cornu M., Modi H., Kawamori D., Kulkarni R.N., Joffraud M., Thorens B. Glucagon-like peptide-1 increases beta-cell glucose competence and proliferation by translational induction of insulin-like growth factor-1 receptor expression. Journal of Biological Chemistry. 2010;285(14):10538–10545. doi: 10.1074/jbc.M109.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Modi H., Jacovetti C., Tarussio D., Metref S., Madsen O.D., Zhang F.P., et al. Autocrine action of IGF2 regulates adult beta-cell mass and function. Diabetes. 2015;64(12):4148–4157. doi: 10.2337/db14-1735. [DOI] [PubMed] [Google Scholar]
- 50.Ansarullah, Jain C., Far F.F., Homberg S., Wiβmiller K., von Hahn F.G., et al. Inceptor counteracts insulin signalling in β-cells to control glycaemia. Nature. 2021;590(7845):326–331. doi: 10.1038/s41586-021-03225-8. [DOI] [PubMed] [Google Scholar]
- 51.Saltiel A.R. Insulin signaling in health and disease. Journal of Clinical Investigation. 2021;131(1) doi: 10.1172/JCI142241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fruchart J.C., Duriez P. Mode of action of fibrates in the regulation of triglyceride and HDL-cholesterol metabolism. Drugs Today (Barc) 2006;42(1):39–64. doi: 10.1358/dot.2006.42.1.963528. [DOI] [PubMed] [Google Scholar]
- 53.Askarpour M., Hadi A., Symonds M.E., Miraghajani M., Omid S., Sheikhi A., et al. Efficacy of l-carnitine supplementation for management of blood lipids: a systematic review and dose-response meta-analysis of randomized controlled trials. Nutrition, Metabolism, and Cardiovascular Diseases. 2019;29(11):1151–1167. doi: 10.1016/j.numecd.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 54.Couturier A., Ringseis R., Mooren F.C., Krüger K., Most E., Eder K. Carnitine supplementation to obese Zucker rats prevents obesity-induced type II to type I muscle fiber transition and favors an oxidative phenotype of skeletal muscle. Nutrition and Metabolism. 2013;10:48. doi: 10.1186/1743-7075-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malaguarnera M., Gargante M.P., Russo C., Antic T., Vacante M., Malaguarnera M., et al. L-carnitine supplementation to diet: a new tool in treatment of nonalcoholic steatohepatitis--a randomized and controlled clinical trial. American Journal of Gastroenterology. 2010;105(6):1338–1345. doi: 10.1038/ajg.2009.719. [DOI] [PubMed] [Google Scholar]
- 56.Lee Y., Hirose H., Ohneda M., Johnson J.H., McGarry J.D., Unger R.H. b-cell lipotoxicity in the pathogenesis of non-insulin-dependent diabetes mellitus of obese rats: impairment in adipocytes b-cell relationship. Proceedings of the National Academy of Sciences USA. 1994;91:10878–10882. doi: 10.1073/pnas.91.23.10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ling Z., Pipeleers D.G. Prolonged exposure of human beta cells to elevated glucose levels results in sustained cellular activation leading to a loss of glucose regulation. Journal of Clinical Investigation. 1996;98(12):2805–2812. doi: 10.1172/JCI119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marselli L., Piron A., Suleiman M., Colli M.L., Yi X., Khamis A., et al. Persistent or transient human β cell dysfunction induced by metabolic stress: specific signatures and shared gene expression with type 2 diabetes. Cell Reports. 2020;33(9):108466. doi: 10.1016/j.celrep.2020.108466. [DOI] [PubMed] [Google Scholar]
- 59.Pappan K.L., Pan Z., Kwon G., Marshall C.A., Coleman T., Goldberg I.J., et al. Pancreatic beta-cell lipoprotein lipase independently regulates islet glucose metabolism and normal insulin secretion. Journal of Biological Chemistry. 2005;280(10):9023–9029. doi: 10.1074/jbc.M409706200. Epub 409702005 Jan 409706206. [DOI] [PubMed] [Google Scholar]
- 60.Marshall B.A., Tordjman K., Host H.H., Ensor N.J., Kwon G., Marshall C.A., et al. Relative hypoglycemia and hyperinsulinemia in mice with heterozygous lipoprotein lipase (LPL) deficiency. Islet LPL regulates insulin secretion. Journal of Biological Chemistry. 1999;274(39):27426–27432. doi: 10.1074/jbc.27274.27439.27426. [DOI] [PubMed] [Google Scholar]
- 61.Stein D.T., Esser V., Stevenson B.E., Lane K.E., Whiteside J.H., Daniels M.B., et al. Essentiality of circulating fatty acids for glucose-stimulated insulin secretion in the fasted rat. Journal of Clinical Investigation. 1996;97(12):2728–2735. doi: 10.1172/JCI118727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee Y., Hirose H., Ohneda M., Johnson J.H., McGarry J.D., Unger R.H. Beta-cell lipotoxicity in the pathogenesis of non-insulin-dependent diabetes mellitus of obese rats: impairment in adipocyte-beta-cell relationships. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(23):10878–10882. doi: 10.1073/pnas.91.23.10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y.-P., Grill V.E. Long-term exposure of rat pancreatic islets to fatty acids inhibits glucose-induced insulin secretion and biosynthesis through a glucose fatty acid cycle. Journal of Clinical Investigation. 1994;93:870–876. doi: 10.1172/JCI117042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sako Y., Grill V.E. A 48-hour lipid infusion in the rat time-dependently inhibits glucose-induced insulin secretion and B cell oxidation through a process likely coupled to fatty acid oxidation. Endocrinology. 1990;127:1580–1589. doi: 10.1210/endo-127-4-1580. [DOI] [PubMed] [Google Scholar]
- 65.Taskinen M.R., Borén J. New insights into the pathophysiology of dyslipidemia in type 2 diabetes. Atherosclerosis. 2015;239(2):483–495. doi: 10.1016/j.atherosclerosis.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 66.Hiukka A., Fruchart-Najib J., Leinonen E., Hilden H., Fruchart J.-C., Taskinen M.-R. Alterations of lipids and apolipoprotein CIII in very low-density lipoprotein subspecies in type 2 diabetes. Diabetologia. 2005;48(6):1207–1215. doi: 10.1007/s00125-005-1753-z. [DOI] [PubMed] [Google Scholar]
- 67.Rütti S., Ehses J.A., Sibler R.A., Prazak R., Rohrer L., Georgopoulos S., et al. Low- and high-density lipoproteins modulate function, apoptosis, and proliferation of primary human and murine pancreatic beta-cells. Endocrinology. 2009;150(10):4521–4530. doi: 10.1210/en.2009-0252. [DOI] [PubMed] [Google Scholar]
- 68.Liu D.J., Peloso G.M., Yu H., Butterworth A.S., Wang X., Mahajan A., et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nature Genetics. 2017;49(12):1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bornstedt M.E., Gjerlaugsen N., Pepaj M., Bredahl M.K.L., Thorsby P.M. Vitamin D increases glucose stimulated insulin secretion from insulin producing beta cells (INS1E) International Journal of Endocrinology and Metabolism. 2019;17(1) doi: 10.5812/ijem.74255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kjalarsdottir L., Tersey S.A., Vishwanath M., Chuang J.C., Posner B.A., Mirmira R.G., et al. 1,25-Dihydroxyvitamin D(3) enhances glucose-stimulated insulin secretion in mouse and human islets: a role for transcriptional regulation of voltage-gated calcium channels by the vitamin D receptor. The Journal of Steroid Biochemistry and Molecular Biology. 2019;185:17–26. doi: 10.1016/j.jsbmb.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 71.Halberg N., Sengelaub C.A., Navrazhina K., Molina H., Uryu K., Tavazoie S.F. PITPNC1 recruits RAB1B to the Golgi network to drive malignant secretion. Cancer Cell. 2016;29(3):339–353. doi: 10.1016/j.ccell.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waugh M.G. The Great Escape: how phosphatidylinositol 4-kinases and PI4P promote vesicle exit from the Golgi (and drive cancer) Biochemical Journal. 2019;476(16):2321–2346. doi: 10.1042/BCJ20180622. [DOI] [PubMed] [Google Scholar]
- 73.Greenawalt D.M., Sieberts S.K., Cornelis M.C., Girman C.J., Zhong H., Yang X., et al. Integrating genetic association, genetics of gene expression, and single nucleotide polymorphism set analysis to identify susceptibility Loci for type 2 diabetes mellitus. American Journal of Epidemiology. 2012;176(5):423–430. doi: 10.1093/aje/kws123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slieker R.C., Donnelly L.A., Fitipaldi H., Bouland G.A., Giordano G.N., Åkerlund M., et al. Diabetes; 2021. Distinct molecular signatures of clinical clusters in people with type 2 diabetes: an IMIRHAPSODY study. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (59) and are accessible through GEO Series accession number GSE164673 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164673). Pancreatic islets expression data SubSeries are available with the accession number GSE140369 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140369) and liver expression with accession number GSE164672 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164672).