Abstract
p53, a tumor suppressor, inhibits cell proliferation by inducing cellular genes involved in the regulation of the cell cycle. MCG10, a novel cellular p53 target gene, was identified in a cDNA subtraction assay with mRNA isolated from a p53-producing cell line. MCG10 can be induced by wild-type but not mutant p53 and by DNA damage via two potential p53-responsive elements in the promoter of the MCG10 gene. The MCG10 gene contains 10 exons and is located at chromosome 3p21, a region highly susceptible to aberrant chromosomal rearrangements and deletions in human neoplasia. The MCG10 gene locus encodes at least two alternatively spliced transcripts, MCG10 and MCG10as. The MCG10 and MCG10as proteins contain two domains homologous to the heterogeneous nuclear ribonucleoprotein K homology (KH) domain. By generating cell lines that inducibly express either wild-type or mutated forms of MCG10 and MCG10as, we found that MCG10 and MCG10as can suppress cell proliferation by inducing apoptosis and cell cycle arrest in G2-M. In addition, we found that MCG10 and MCG10as, through their KH domains, can bind poly(C) and that their RNA-binding activity is necessary for inducing apoptosis and cell cycle arrest. Furthermore, we found that the level of the poly(C) binding MCG10 protein is increased in cells treated with the DNA-damaging agent camptothecin in a p53-dependent manner. These results suggest that the MCG10 RNA-binding protein is a potential mediator of p53 tumor suppression.
RNA-binding proteins are a large family of proteins with diverse functions which contain one or more RNA-binding domains (RBDs) and other auxiliary domains for protein-protein interaction and subcellular targeting (22, 23, 46, 65, 71, 78). Several ribosomal proteins are RNA-binding proteins, for example, S6, S15, and L11, which are necessary for ribosome assembly and may be a target for translational regulation (23, 82). Several groups of RNA-binding proteins have been shown to play an important role in alternate splicing, RNA editing, and alternate poly(A) site selection. Among these are the abundant heterogeneous nuclear ribonucleoproteins (hnRNPs), which shuttle between the nucleus and cytoplasm (48, 74, 82).
Three major RNA-binding motifs have been found in hnRNPs, that is, the RBD, arginine/glycine-rich box (RGG), and hnRNP K homology (KH) domain. The consensus RBD structure is composed of 90 to 100 amino acids with a βαββαβ secondary structure (9). A majority of hnRNPs, such as A, B, C, D, F, G, and H, contain one or more RBDs, which are necessary for the ability of these hnRNPs in the regulation of splicing, RNA trafficking, and mRNA stability (48, 82). RNA-binding experiments demonstrate that RBD motif proteins can bind RNA with a wide range of affinities and specificities (9). The RGG box is composed of several closely spaced arginine-glycine-glycine repeats with a β-spiral secondary structure (9). Several hnRNPs contain RGG boxes along with RBD or KH motifs. RNA-binding experiments have demonstrated that the RGG box has a relatively weak RNA-binding affinity and specificity (9, 48, 82). However, the RGG box can unstack RNA bases and destabilize RNA secondary structures, which enhances RNA binding for one or more other RNA-binding motifs present in the protein. The KH domain consists of 50 to 70 amino acids with a stable βααββα secondary structure (9, 48, 66, 74, 82). A potential surface for RNA binding is centered on the loop between the first two helices (66). The KH motif proteins have a relatively high binding affinity for dCdT elements and cytosine-rich RNA elements, such as oligo(C) polymer and CU-rich elements (74). Several hnRNPs contain one or more KH domains, for example, hnRNP K and E. The KH motif hnRNPs have been shown to play a role in the regulation of transcriptional activation and repression, mRNA stability, and translational silencing (48, 74, 82). Sam68, a target of the Src tyrosine kinase in mitosis, contains one KH domain (4, 53). Interestingly, a splicing variant, Sam68ΔKH, which lacks the KH domain inhibits cell proliferation and cell cycle transition from G1 to S (4). The fragile X syndrome gene FMR1 encodes an RNA-binding protein with two KH domains (83). Transcriptional silencing of FMR1 or a mutation in the C-terminal KH domain leads to fragile X syndrome (93, 96).
p53, a cellular gatekeeper, plays an important role in the regulation of numerous processes, including cell cycle progression and apoptosis (1, 13, 34, 46, 52), differentiation (2), senescence (52), and tumor surveillance (110). Many studies have shown that p53 transcriptional activity is required to regulate these processes (3, 76, 92, 108, 109). Consistent with this idea, the majority of tumor-derived mutations in p53, which is the most frequently mutated gene in human cancers, occurs in the central, conserved sequence-specific DNA-binding domain, which is necessary for transactivation (34, 46). A number of cellular genes have been found to be induced by p53 (27, 46). They can be classified into three major functional groups: (i) genes whose products are capable of mediating p53-dependent cell cycle arrest (13, 27, 46), (ii) genes whose products are capable of mediating p53-dependent apoptosis (13, 27, 46), and (iii) genes whose products are capable of mediating other p53 activities, such as TAP1, which is involved in tumor surveillance (110), the p48 xeroderma pigmentosa gene which is involved in nucleotide excision repair (37), and the KAI1 gene, involved in suppression of metastasis (56).
Several cellular genes are capable of mediating p53-dependent cell cycle arrest. p21, a well-characterized inhibitor of cyclin-dependent kinase, can induce arrest in G1 (1, 46, 52) and can also induce G2-M arrest in cells that harbor a dysfunctional retinoblastoma (RB) gene (69). G2-M arrest can be induced by 14-3-3ς (12, 35), which inhibits Cdc25C phosphatase activity; GADD45 (95), which is necessary for maintaining genome stability and DNA repair; B99 (88), which is a microtubule-localized protein with G2-phase-specific expression; and B-cell translocation gene 2 (BTG2) (79), whose loss disrupts G2-M arrest when cells are treated with DNA-damaging agents.
Several candidate genes may mediate p53-dependent apoptosis. These are bax (62), KILLER/DR5 (103), phosphatidyl inositol 3-kinase regulatory subunit p85 (105), PAG608 (39, 90), Siah-1 (57, 78), cathepsin D (104), and CD95 (also called Apo-1 or Fas) receptor (5, 64). Nevertheless, it is still not clear whether these p53 targets are necessary or sufficient for inducing apoptosis. Since p53 transcriptional activity is necessary for inducing apoptosis, it is likely that one or more cellular genes must be involved in mediating p53-dependent apoptosis.
In the search for novel cellular target genes responsible for p53 tumor suppression, we performed a cDNA subtraction assay and found one gene, MCG10, that is specifically induced by wild-type but not mutant p53 and by DNA damage. This induction occurs via two potential p53-responsive elements. The MCG10 gene, located at chromosome 3p21 with 10 exons, encodes at least two alternatively spliced transcripts, MCG10 and MCG10as. The MCG10 and MCG10as proteins contain two domains homologous to an hnRNP KH domain. By generating cell lines that inducibly express either wild-type or mutated forms of MCG10 and MCG10as, we found that MCG10 and MCG10as can induce apoptosis and cell cycle arrest in G2-M and that both KH domains are necessary for these activities. We also found that MCG10 and MCG10as are capable of binding to poly(C) and that their RNA-binding activity is necessary for inducing apoptosis and cell cycle arrest. These results suggest that the MCG10 RNA-binding protein is a potential mediator of p53 tumor suppression.
MATERIALS AND METHODS
Cell culture and cell lines.
HCT116, LS174T, and MCF7 cell lines were purchased from the American Type Culture Collection. RKO, HCT116p53−/−, and 80S14 were cultured as described previously (8, 42, 94). RKOE6 and HCT116E6 are derivatives of RKO and HCT116, respectively, that were stably transfected with the E6 gene from human papillomavirus 16 (65). HCT116p53−/− and 80S14 are derivatives of HCT116 in which the genes encoding p53 and p21, respectively, were somatically knocked out (8, 94). The MCF7 cell line, which expresses Tet-VP16 for the generation of tetracycline-inducible cell lines, was purchased from ClonTech (Palo Alto, Calif.). MCF7-p53, an MCF7 derivative that inducibly expresses p53, was generated as previously described (15, 109). p53-3, p53(R249S)-4, and p53(ΔPRD)-5 cell lines, derivatives of H1299 that inducibly express wild-type p53, p53(R249S), and p53(ΔPRD), respectively, were cultured as described previously (15, 108, 109). H1299 cell lines that inducibly express wild-type or mutated forms of MCG10 and MCG10as were generated as previously described (15, 109).
RNA isolation, cDNA subtraction assay, and Northern blot analysis.
Polyadenylated RNA was isolated from p53-3 cells using an mRNA purification kit (Pharmacia, Piscataway, N.J.). Total RNA was isolated from cells using Trizol reagents (Life Technologies, Inc., Gaithersburg, Md.). The cDNA subtraction assay was performed using the ClonTech PCR-Select cDNA subtraction kit according to the manufacturer's instruction (ClonTech). Subtracted cDNA fragments were cloned into pCRII vector (Invitrogen, Carlsbad, Calif.). Northern blot analysis was performed as described previously (14, 109). p21 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes were prepared as described previously (109). The MCG10 probe, a 1.7-kb PstI fragment, was prepared from MCG10 cDNA.
Plasmids and mutagenesis.
The full-length cDNAs for MCG10 and MCG10as were isolated from a cDNA library made with mRNA purified from p53-3 cells and individually cloned into a tetracycline-regulated expression vector, pUHD10-3 (33), between the EcoRI and XbaI sites. Mutant MCG10 and MCG10as cDNA constructs were generated by PCR and used to replace the corresponding regions of wild-type MCG10 or MCG10as in pUHD10-3. To generate MCG10-ΔKH1, the cDNA fragment encoding amino acids 1 to 188 but lacking amino acids 78 to 185 was amplified using the T3 promoter primer as the 5′-end primer and the 3′-end primer GCA GAT CTG ACT GGC AGG GAT GAC. The resulting fragment was used to replace the corresponding region in MCG10 between the EcoRI and BglII sites. To generate MCG10-ΔKH2 and MCG10as-ΔKH2, the cDNA fragment encoding amino acids 278 to 424 but lacking amino acids 281 to 329 of MCG10 was amplified by the 5′-end primer ATC GGG CGC CAT GTC ACC ATC ACT and the 3′-end primer TAG GAT CCG GTC GCT GAG AAT AT. The resulting cDNA fragment was used to replace the corresponding region in MCG10 or MCG10as between the KasI and BamHI sites. To generate MCG10as-KH2− (Ile230Asp), the cDNA fragment encoding amino acids 224 to 369 of MCG10as was amplified by the 5′-end primer CGG GCG CCA GGG CAG CAA GAA CAG CGA G and the 3′-end primer TAG GAT CCG GTC GCT GAG AAT AT. The resulting fragment was used to replace the corresponding region in MCG10as between the NarI and BamHI sites.
Antibody production and Western blot analysis.
To generate anti-MCG10 antibody, a 1,530-bp PstI-NcoI cDNA fragment encoding amino acids 10 to 424 of the MCG10 polypeptide was inserted in frame into the pRSET expression vector (Invitrogen). The His-tagged MCG10 protein was produced in bacteria and purified with Ni-agarose beads. Anti-MCG10 antibody was raised in a rabbit and affinity purified using the His-tagged MCG10 protein (14). For Western blot analysis, cells were collected from culture plates in phosphate-buffered saline (PBS), resuspended in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and boiled for 5 min. Western blot analysis was performed as previously described (109). Antiactin antibody was purchased from Sigma (St. Louis, Mo.).
Luciferase assay.
A 28-bp fragment (5′AGCTTGGTCTTGGCCCAGACTTAGCACA3′) that contains the potential p53-responsive element 1, a 36-bp fragment (5′AGCTTGAACTTAAGACCGAGGCTCTGGACAAGTTGA3′) that contains the potential p53-responsive element 2, and a 27-bp fragment (5′AGCTTGCTCTAGTTCTGGCCATGTTCA3′) that contains the potential p53-responsive element 3 were synthesized and cloned upstream of a minimal c-fos promoter and a firefly luciferase reporter gene (41). The resulting constructs were designated p53RE-1, p53RE-2, and p53RE-3, respectively. To mutate the p53-responsive elements in the MCG10 gene, four nucleotides in p53RE-1 (5′AGCTTGGTaTTtGCCCAGAaTTAtCACA3′) and p53RE-2 (5′AGCTTGAAaTTAAtACCGAGGCTCTGGAaAAtTTGA3′) which are predicted to be critical for p53 binding (shown in lowercase) were replaced. We then generated two reporter vectors, designated m-p53RE-1 and m-p53RE-2, and 2 μg of p53RE-1, m-p53RE-1, p53RE-2, m-p53RE-2, or p53RE-3 was cotransfected into H1299 cells with 1 μg of pcDNA3 control vector or a vector that expresses wild-type or mutant p53. Then 0.1 μg of Renilla luciferase assay vector pRL-CMV (Promega, Madison, Wis.) was also cotransfected as an internal control. The dual luciferase assay was performed according to the manufacturer's instructions (Promega).
EMSA.
The electrophoretic mobility shift assay (EMSA) probes were 28-bp (p53RE-1) and 36-bp (p53RE-2) oligonucleotides containing a potential p53-responsive element in the MCG10 gene. The labeled probe DNA (5 ng) was added to a mixture [20 mM HEPES (pH 7.9), 25 mM KCl, 0.1 mM EDTA, 10% glycerol, 2 mM MgCl2, 2 mM spermidine, 0.5 mM dithiothreitol, 0.025% NP-40, 100 ng of double-stranded poly(dI·dC), and 2 μg of bovine serum albumin containing 20 ng of p53 protein. The p53 protein was expressed in a baculovirus expression system and affinity purified using anti-p53 monoclonal antibody Pab421. The p53-DNA complex was resolved in a 4% polyacrylamide gel. For supershifting the p53-DNA complex, 1 μg of anti-p53 monoclonal antibody Pab1801 was added to the reaction. For competition assays, the unlabeled wild-type RGC (20 and 100 ng) or probe DNA (20 and 100 ng) was added to the reaction.
Growth rate analysis and trypan blue exclusion assay.
To determine the rate of cell growth, cells were seeded at approximately 5 × 104 to 7.5 × 104 cells per 60-mm plate with or without tetracycline (1.0 μg/ml). The medium was replaced every 72 h. At the times indicated, two plates were rinsed with PBS twice to remove dead cells and debris. Live cells on the plates were trypsinized and collected separately. Cells from each plate were counted at least three times using the Coulter cell counter. The average number of cells from two plates was used for growth rate determination. For the trypan blue dye exclusion assay, all cells were collected separately from two plates at the times indicated. The cells were stained with trypan blue (Sigma) for 10 min. The stained (dead) and unstained (live) cells were counted at least three times using a hemocytometer. The percentage of dead cells was used as an index for the degree of apoptosis.
DNA histogram analysis and annexin V staining assay.
Cells were seeded at 2 × 105 per 90-mm plate with or without tetracycline. For DNA histogram analysis, both floating dead cells in the medium and live cells on the plate were collected and fixed with 2 ml of 70% ethanol for at least 30 min. The fixed cells were centrifuged and resuspended in 1 ml of PBS solution containing 50 μg each of RNase A (Sigma) and propidium iodide (Sigma) per ml. The stained cells were analyzed in a fluorescence-activated cell sorter within 4 h. The percentages of cells in the sub-G1, G0-G1, S, and G2-M phases were determined using the ModFit program. For the annexin V staining assay, both dead and live cells were collected and washed twice with cold PBS. The cells were resuspended in 0.1 ml of annexin V binding buffer to a density of 106/ml and stained according to the manufacturer's instructions (Boehringer, Mannheim, Germany).
Mitochondrial membrane potential assay.
To determine whether the cell death mediated by MCG10 goes through the mitochondrial pathway, cells were seeded at approximately 6 × 103 cells/chamber (Fisher Scientific) with or without tetracycline (2 μg/ml) for 3 days. Cells were then rinsed with PBS and stained with ApoAlert Mitochondrial Membrane Sensor reagents according to the manufacturer's instructions (ClonTech). In normal cells, Mitosensor, a cationic dye, is taken up in the mitochondria, where it forms aggregates and exhibits red fluorescence. In apoptotic cells, Mitosensor cannot aggregate in the mitochondria because of altered mitochondrial potentials. As a result, the dye remains in monomeric form in the cytoplasm, where it fluoresces green.
Caspase activity assay.
Cells were seeded at approximately 3 × 105 to 5 × 105 per 90-mm plate with or without tetracycline for 3 days. Cells were then rinsed with cold PBS, and caspase activity was assayed using the caspase 3 or 6 colorimetric protease assay reagent according to the manufacturer's instructions (Chemicon International, Inc.). The percent increase in relative caspase activity was the activity in cells expressing p53 or MCG10 divided by that in control cells.
Ribonucleotide homopolymer binding assay.
The RNA-binding assay was performed as previously described with modifications (84). Briefly, cells were collected, washed two times with cold PBS, and resuspended in 1 ml of RNA-binding buffer (50 mM Tris-HCl [pH 7.4], 100 mM KCl, 2 mM MgCl2, 1 mM EDTA, 0.5% NP-40, 0.5% aprotinin, 2 μg of leupeptin per ml, and 0.5 mM phenylmethylsulfonyl fluoride). Cytoplasmic and nuclear extracts were prepared as previously described (77). For the RNA-binding assay, 0.8 ml of cytoplasmic extracts or nuclear extracts was mixed with 0.2 ml of 5 M NaCl and 5 mg of ribonucleotide homopolymer [poly(A), poly(U), poly(G), or poly(C)]-agarose beads. The mixtures were incubated and rocked at room temperature for 20 min. The beads in the mixture were pelleted and washed three times with RNA-binding buffer. RNA-binding proteins on the beads were resuspended in 2× SDS-PAGE sample buffer, boiled for 8 min, and assayed by Western blot analysis with anti-MCG10 polyclonal antibody.
Nucleotide sequence accession numbers.
The human MCG10 genomic DNA sequence was submitted to GenBank under accession number AF257772. The human MCG10 and MCG10as cDNA sequences were submitted to GenBank under accession numbers AF257770 and AF257771, respectively.
RESULTS
Upregulation of MCG10 by p53.
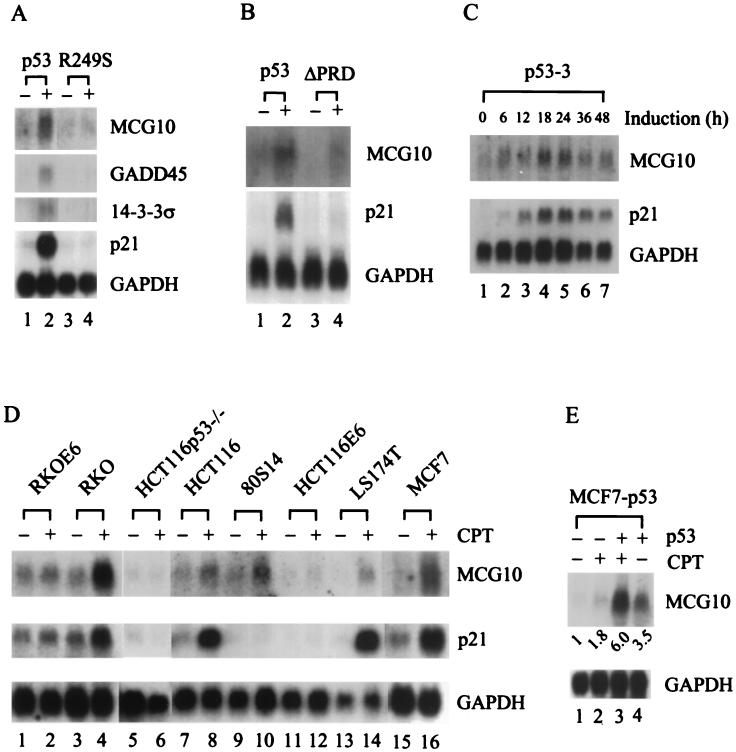
In our ongoing effort to identify novel p53 target genes, the ClonTech PCR-Select cDNA subtraction assay was performed using mRNA isolated from p53-3, a derivative of the H1299 cell line that inducibly expresses p53 under a tetracycline-regulated promoter (15, 109). Several cDNA fragments that may represent genes induced by p53 were isolated. Among these is MCG10, which is a novel gene and encodes a protein with two regions homologous to the KH domain of the hnRNP K protein. To confirm that MCG10 can be induced by p53, Northern blot analysis was performed using MCG10 cDNA as the probe. We found that MCG10 was induced in p53-3 cells when p53 was expressed (Fig. 1A, upper panel, compare lanes 1 and 2). As a control, we tested the expression of three well-defined cellular p53 target genes, p21, GADD45, and 14-3-3ς (28, 35, 42). We found that these genes were also induced by p53 (Fig. 1A, lower panel). The level of induction for MCG10 was higher than that for GADD45 and 14-3-3ς, albeit lower than that for p21. In addition, mutant p53(R249S) was incapable of activating MCG10, p21, GADD45, or 14-3-3ς (Fig. 1A, compare lanes 3 and 4), consistent with the fact that this tumor-derived p53 mutant is defective in transactivation (30). We and others have shown recently that p53(ΔPRD), which lacks the proline-rich domain, is deficient in inducing apoptosis and certain p53 target genes (91, 108). Here we found that p53(ΔPRD) is deficient in inducing MCG10 (Fig. 1B, compare lanes 3 and 4), suggesting that MCG10 is a potential mediator of p53-dependent apoptosis (see more below). Furthermore, we determined the kinetics for p53 induction of MCG10 (Fig. 1C). We found that enhanced expression of MCG10 was detected as early as 6 h after p53 induction and that maximum induction occurred at 18 and 24 h. Induction of p21 showed similar kinetics.
FIG. 1.
Upregulation of MCG10 by p53. (A) Wild-type p53 but not mutant p53 induces MCG10. Northern blots were prepared using 10 μg of total RNA isolated from p53-3 or p53(R249S) cells that were uninduced (−) or induced (+) to express wild-type p53 or mutant p53(R249S), respectively. The blots were probed with cDNAs derived from the MCG10, 14-3-3ς, GADD45, p21 and GAPDH genes. (B) The apoptosis-deficient deletion mutant p53(ΔPRD) is incapable of inducing MCG10. A Northern blot was prepared using 10 μg of total RNA isolated from p53-3 or p53(ΔPRD)-5 cells that were uninduced (−) or induced (+) to express wild-type p53 or mutant p53(ΔPRD), respectively. The blot was probed with MCG10 cDNA and then reprobed with both p21 and GAPDH cDNAs. (C) Kinetics of p53 induction of MCG10. A Northern blot was prepared using 10 μg of total RNA isolated from p53-3 cells that were induced for 0, 6, 12, 18, 24, 36, or 48 h. The blot was probed with MCG10 cDNA and then reprobed with both p21 and GAPDH cDNAs. (D) MCG10 is induced by DNA damage in cells that contain an endogenous wild-type p53 gene but not in cells that are functionally p53-null. Northern blots were prepared using 10 μg of total RNA isolated from seven individual cell lines (see text for details) as indicated above the figure, which were untreated (−) or treated (+) with 300 nM camptothecin for 24 h. The blots were probed with cDNAs derived from MCG10, p21, and GAPDH. (E) Exogenous inducible p53 cooperates with endogenous wild-type p53 in MCF7 cells to induce MCG10. A Northern blot was prepared using 10 μg of total RNA isolated from MCF7-p53 cells that were untreated (lane 1), treated with 300 nM camptothecin (CPT) to induce endogenous wild-type p53 (lane 2), induced to express exogenous p53 and treated with 300 nM camptothecin to induce endogenous wild-type p53 (lane 3), or induced to express exogenous p53 (lane 4). The blot was probed with cDNAs derived from MCG10 and GAPDH.
DNA damage stabilizes and activates p53, leading to induction of p53 target genes (32, 46, 52). If MCG10 is a true p53 target, it would be induced by DNA damage in cells that contain an endogenous wild-type p53 gene but not in cells that are p53-null. To this end, we tested eight cell lines using the DNA-damaging agent camptothecin, which is an inhibitor of topoisomerase I and can induce double-strand DNA breaks (68). These cells were treated with camptothecin, and the levels of MCG10 and p21 transcripts were determined by Northern blot analysis (Fig. 1D). We found that both MCG10 and p21 were induced in camptothecin-treated RKO, HCT116, LS174T, and MCF7 cells, which all contain wild-type p53 (Fig. 1D, lanes 3, 4, 7 to 10, and 13 to 16). Although p21 was not expressed in p21-null 80S14 cells (94), MCG10 was still induced by DNA damage (Fig. 1D, lanes 9 and 10), indicating that p53 can induce MCG10 independently of p21. In contrast, MCG10 was not induced in p53-knockout cells (HCT116p53−/−) (Fig. 1D, lanes 5 and 6) or p53-null-like cells (RKOE6 and HCT116E6) (Fig. 1D, lanes 1 and 2 and 11 and 12).
Since exogenous p53 in H1299 cells and endogenous p53 in MCF7 cells are capable of inducing MCG10, we wanted to determine whether MCG10 can be cooperatively induced when both endogenous and exogenous p53s are expressed. To do this, we generated an MCF7 cell line, MCF7-p53, that inducibly expresses HA-tagged p53 under a tetracycline-regulated promoter. We found that MCG10 was induced in MCF7-p53 cells treated with camptothecin to induce endogenous p53 (Fig. 1E, lane 2) or induced to express exogenous HA-tagged p53 (Fig. 1E, lane 4). In contrast, when both endogenous and exogenous p53s were expressed, the level of MCG10 induction (6-fold) was more than additive to that induced by endogenous (1.8-fold) or exogenous (3.5-fold) p53 individually (Fig. 1E, compare lane 3 with lanes 2 and 4).
Identification of two potential p53-responsive elements in the MCG10 gene.
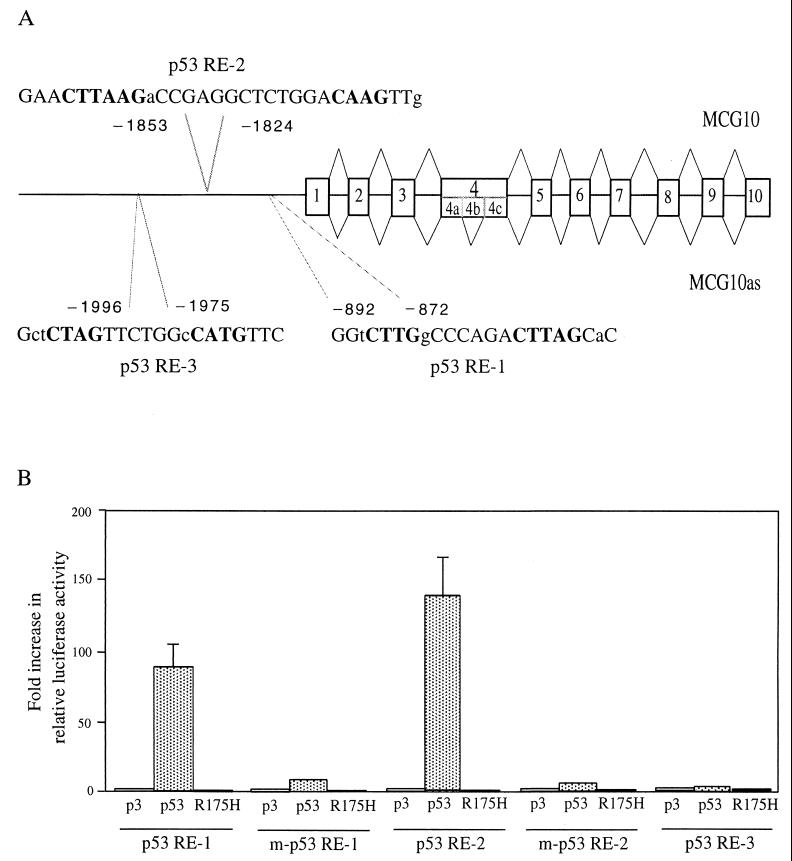
To determine whether MCG10 is a true target of p53, we needed to look for a p53-responsive element in the genomic DNA sequence of the MCG10 gene. To do this, we screened a human bacterial artificial chromosome library and identified a genomic clone containing MCG10. We then sequenced a region of 7,083 nucleotides that spans the entire MCG10 gene locus (Fig. 2A). We found three potential p53-binding sites, p53-responsive elements 1, 2, and 3, located approximately 900, 1,800, and 2,000 nucleotides upstream of the MCG10 transcription start site, respectively (Fig. 2A). All three sequences (p53RE-1, GAA CTTAAG aCC GAGGCTCT GGA CAAG TTg; p53RE-2, GGt CTTG gCC C AGA CTTAG CaC; and p53RE-3, Gct CTAG TTC T GGc CATG TTC) contain mismatches (in lowercase) in the noncritical positions within the consensus p53-binding site. Recently, an 81,512-bp genomic DNA sequence from a P1 artificial chromosome clone that contains the MCG10 gene locus was deposited in GenBank (AC006255). The P1 clone was mapped at chromosome 3p21, a region highly susceptible to aberrant chromosomal rearrangements and deletions in neoplasia (61).
FIG. 2.
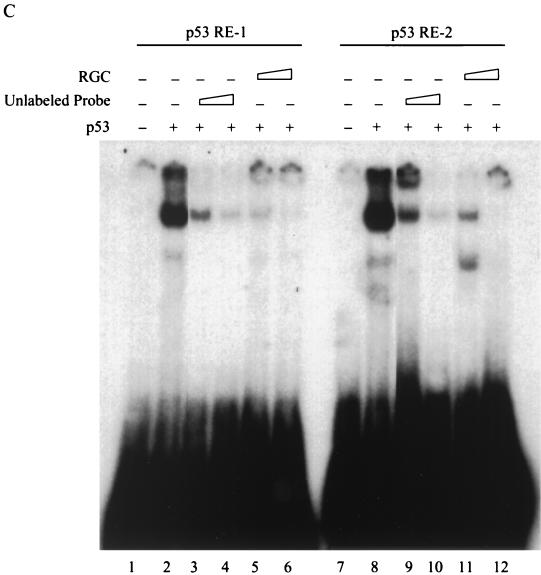
Identification of two p53-responsive elements in the MCG10 gene. (A) Schematic representation of the MCG10 genomic DNA structure. Exons are shown as numbered boxes, and introns are shown as lines. The locations of two potential p53-responsive elements in the promoter of the MCG10 gene are indicated. Bold uppercase letters represent nucleotides predicted to be critical for the consensus p53-responsive element. Lowercase letters represent mismatches. The transcript for MCG10 is drawn above the gene structure, and the transcript for MCG10as is shown below. Exon 4b is not present in the MCG10as transcript. (B) Two of the three potential p53-binding sites but not their mutated forms in the MCG10 gene are responsive to wild-type p53 in vivo. p53RE-1, m-p53RE-1, p53RE-2, m-p53RE-2, or p53RE-3 (2 μg) was cotransfected into H1299 cells with 1 μg of pcDNA3 control vector or a vector that expresses wild-type p53 or mutant p53(R175H). The fold increase in relative luciferase activity is the luciferase activity induced by p53 divided by that induced by pcDNA3. Error bars represent the standard deviations from at least three experiments. (C) p53 binds specifically to both p53RE-1 and p53RE-2 in vitro. The 28- and 36-bp oligonucleotide fragments containing p53RE-1 and p53RE-2, respectively, in the MCG10 gene were labeled with [α-32P]dCTP. The labeled probe DNA (5 ng) was added to a mixture containing 20 ng of p53 protein. The p53-DNA complex was resolved in a 4% polyacrylamide gel. For competition assays, 5- or 20-fold excess unlabeled 28-bp probe DNA (lanes 3 and 4), 36-bp unlabeled probe DNA (lanes 9 and 10), or RGC (lanes 5 and 6 and 11 and 12) was added to the reactions.
To determine whether these binding sites are responsive to p53 in vivo, three fragments that contain these potential p53-responsive elements (see Materials and Methods) were synthesized and cloned upstream of a minimal c-fos promoter and a luciferase reporter gene. The resulting reporter vectors were designated p53RE-1, p53RE-2, and p53RE-3. Each of the reporter vectors was cotransfected into H1299 cells with either pcDNA3 control vector or a vector that expresses wild-type p53 or mutant p53(R175H). The Renilla luciferase assay vector pRL-CMV was also cotransfected as an internal control. We found that the luciferase activity of p53RE-1 and p53RE-2 but not p53RE-3 was markedly increased by wild-type p53 (Fig. 2B). Mutant p53(R175H) was incapable of increasing the luciferase activity of p53RE-1 and p53RE-2 (Fig. 2B). We also replaced four nucleotides in p53RE-1 and p53RE-2 predicted to be critical for p53 binding (see Materials and Methods) and generated two reporters, designated m-p53RE-1 and m-p53RE-2. We found that the luciferase activity for both m-p53RE-1 and m-p53RE-2 was not significantly increased by wild-type p53 or mutant p53(R175H) (Fig. 2B). These results suggest that two of the three potential p53-responsive elements in MCG10 do function in vivo.
To further determine whether p53 binds to the responsive elements in the MCG10 gene, two DNA fragments (28 and 36 bp) that contain p53RE-1 and -2 (see Materials and Methods) were synthesized, 32P-labeled, and used in an EMSA (Fig. 2C). We found that when the purified p53 protein was mixed with these DNA fragments, a complex that presumably contained both p53 and p53RE-1 or -2 was detected (Fig. 2C, lanes 2 and 8). The complex was supershifted with the anti-p53 monoclonal antibody Pab1801 (data not shown). We also used the unlabeled probe DNA and a fragment that contains a wild-type p53-binding site from the ribosomal gene cluster (RGC) (43) as competitors. The unlabeled probe DNA and wild-type RGC competed with the 32P-labeled 28- and 36-bp probe DNA fragments from the MCG10 gene and inhibited the formation of the p53-DNA complex in a dose-dependent manner (Fig. 2C, lanes 3 to 6 and 9 to 12). These results indicate that p53 interacts specifically with both p53RE-1 and -2 in the MCG10 gene.
MCG10 gene locus encodes at least two alternatively spliced transcripts for novel KH motif RNA-binding proteins.
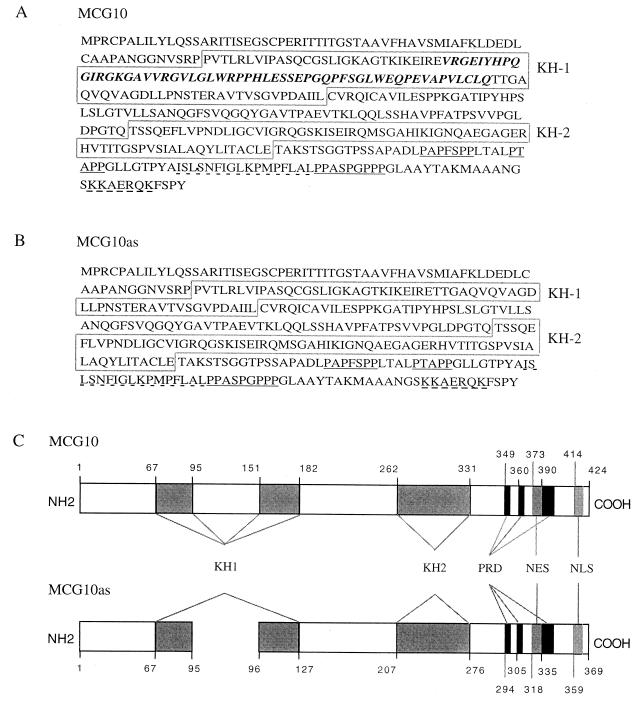
To analyze the activity of the MCG10 gene product, we used the 163-bp cDNA fragment from the cDNA subtraction assay to screen a cDNA library made from mRNA isolated from p53-3 cells. Two cDNA clones (2,623 and 2,458 nucleotides) were identified. When both cDNA sequences were aligned with the 7,083-nucleotide genomic DNA sequence, we found that 10 exons encode the 2,623-nucleotide MCG10 transcript. The 2,458-nucleotide cDNA clone represents an alternatively spliced transcript, MCG10as, which lacks 165 nucleotides within exon 4. We refer to the region not expressed in MCG10as as exon 4b (see Fig. 2A). The MCG10 and MCG10as transcripts encode novel polypeptides of 424 and 369 amino acids, respectively. Each protein contains two KH domains, three proline-rich domains, one potential nuclear export signal, and one potential nuclear localization signal (Fig. 3A to C). A sequence alignment of the KH domains from hnRNP K, FMR1, MCG10, and MCG10as showed that the critical residues in the KH domains of hnRNP K and FMR1 are conserved in those of MCG10 and MCG10as (Fig. 3D). For example, the GXXG motif within the KH domain of hnRNP K (82) and the critical Ile (at residue 304, marked with an asterisk) in the FMR1 KH2 (66) are conserved in the KH domains of MCG10 and MCG10as.
FIG. 3.
(A and B) Deduced amino acid sequences of MCG10 and MCG10as. The N-terminal KH domain (KH1) and C-terminal KH domain (KH2) in MCG10 and MCG10as are boxed. The bold italic letters represent a 55-amino-acid insertion in the N-terminal KH domain of MCG10. Three proline-rich domains (PRD) are underlined. The nuclear export signal (NES) and nuclear localization signal (NLS) are marked by dashes. (C) Schematic representations of MCG10 and MCG10as protein structures. The locations of specific features are indicated by the amino acid number. (D) Sequence alignment of eight KH domains from hnRNP K, FMR1, MCG10, and MCG10as. Numbers on the right indicate positions of the ending amino acids in the KH domain. Highly conserved positions are highlighted in colors. The GXXG motif is shown below the alignment. The critical isoleucine residue for FMR1 KH2 that is mutated in fragile X syndrome is indicated (∗).
MCG10 and MCG10as can induce apoptosis and cell cycle arrest in G2-M.
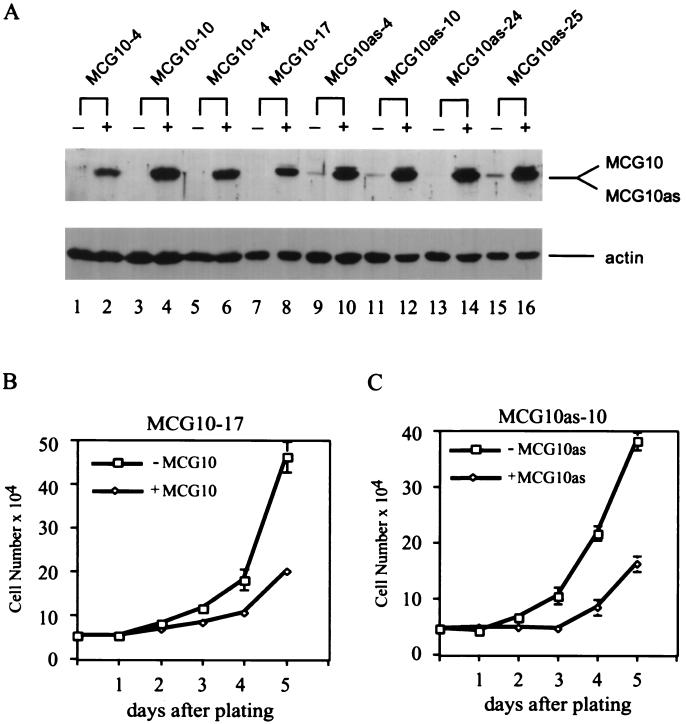
Activation of p53 leads to at least two well-characterized cellular responses, cell cycle arrest and apoptosis (1, 13, 52). Since MCG10 can be induced by p53, we wanted to determine whether MCG10 is capable of mediating p53 tumor suppression. To this end, we generated several cell lines that inducibly express MCG10 and MCG10as under the control of a tetracycline-regulated promoter. The levels of the MCG10 and MCG10as proteins in four representative H1299 cell lines were determined by Western blot analysis with anti-MCG10 antibody (Fig. 4A). A 45-kDa polypeptide was specifically recognized by anti-MCG10 antibody in both MCG10- and MCG10as-producing cells when induced. Interestingly, we found that the apparent molecular masses of MCG10 and MCG10as are nearly identical, although the MCG10 polypeptide is 55 amino acids longer than MCG10as (Fig. 3A and B). When the levels of actin protein were normalized in various cells, we found that MCG10 and MCG10as were expressed at comparable levels. We then measured the growth rates of MCG10-17 and MCG10as-10 cells in the absence and presence of MCG10 and MCG10as over a 5-day period. We found that both MCG10 and MCG10as can suppress cell proliferation (Fig. 4B and C).
FIG. 4.
MCG10 and MCG10as are capable of suppressing cell proliferation. (A) Levels of MCG10, MCG10as, and actin were assayed by Western blot analysis in cell lines that inducibly express MCG10 or MCG10as. Cell extracts were prepared from uninduced cells (−) or cells induced (+) to express MCG10 or MCG10as. The blot was probed with affinity-purified anti-MCG10 polyclonal antibody (upper panel) and then reprobed with antiactin polyclonal antibody (lower panel). (B and C) Growth rates of MCG10-17 and MCG10as-10 cells in the presence (◊) or absence (□) of MCG10 or MCG10as, respectively, were measured as described in Materials and Methods. Error bars represent the standard deviations from at least three experiments.
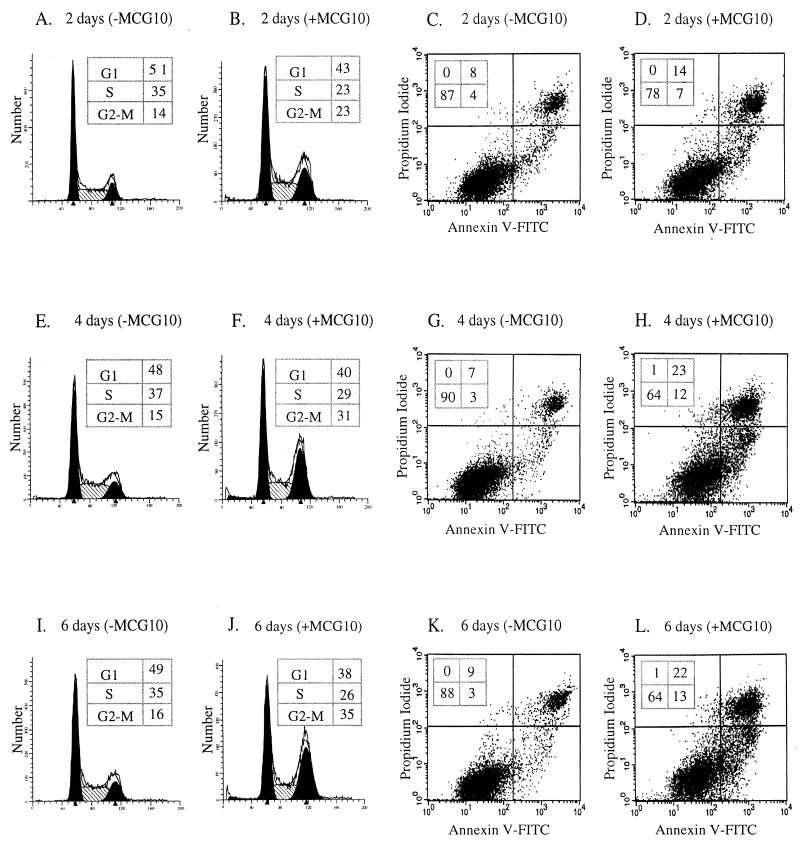
To determine whether the growth suppression by MCG10 and MCG10as is due to cell cycle arrest, apoptosis, or both, we performed DNA histogram analysis. When cells were induced to express MCG10 for 2, 4, and 6 days, we found that the percentage of cells in S phase decreased from 35 to 23% (Fig. 5A and B), 37 to 29% (Fig. 5E and F), and 35 to 26% (Fig. 5I and J), respectively. In contrast, we found that the percentage of cells in G2-M phase increased from 14 to 23% (Fig. 5A and B), 15 to 31% (Fig. 5E and F), and 16 to 35% (Fig. 5I and J), respectively. We also found that the number of cells in G2/M was increased when MCG10 was induced for 1 day (data not shown). The maximum effect was observed between 2 and 4 days following induction of MCG10. This is consistent with the fact that p53-mediated cell cycle arrest occurs within 24 h but remains incomplete till 48 h (15). Furthermore, we found that the ability of MCG10 to induce arrest in G2/M is higher than that of p53 in H1299 cells, although slightly lower than that of GADD45 (95, 108). These results suggest that MCG10 can induce cell cycle arrest in G2-M. However, no substantial increase was detected for cells in sub-G1. Since cells can undergo apoptosis without DNA fragmentation (67, 71, 80), we determined whether MCG10 can induce cell death by the annexin V staining assay. We found that when cells were induced to express MCG10 for 2, 4, and 6 days, the percentage of stained cells (a combination of cells in both the upper right and lower right boxes) was increased from 12 to 21% (Fig. 5C and D), 10 to 35% (Fig. 5G and H), and 12 to 35% (Fig. 5K and L), respectively.
FIG. 5.
MCG10 is capable of inducing cell cycle arrest in G2-M and apoptosis. DNA content was quantitated by propidium iodide staining of fixed cells that were uninduced (−MCG10) or induced (+MCG10) to express MCG10 for 2 days (A and B), 4 days (E and F), and 6 days (I and J). Apoptotic cells were quantitated by propidium iodide-annexin V staining of cells that were uninduced (−MCG10) or induced (+MCG10) to express MCG10 for 2 days (C and D), 4 days (G and H), and 6 days (K and L).
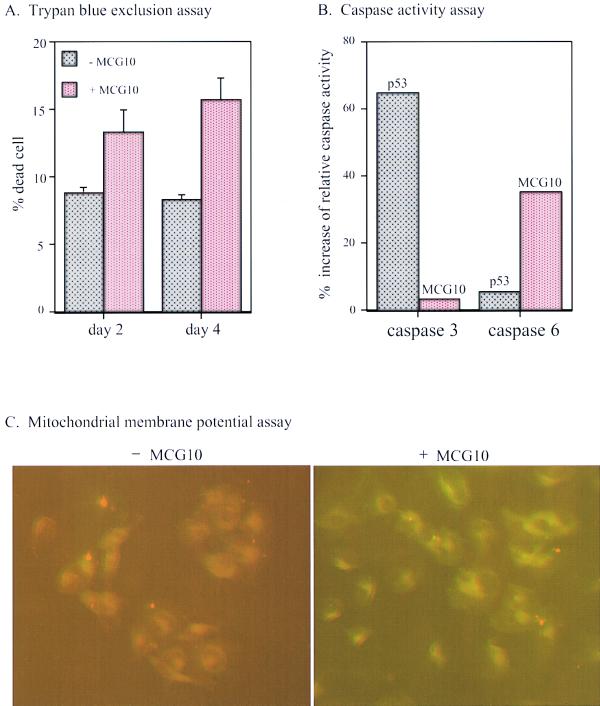
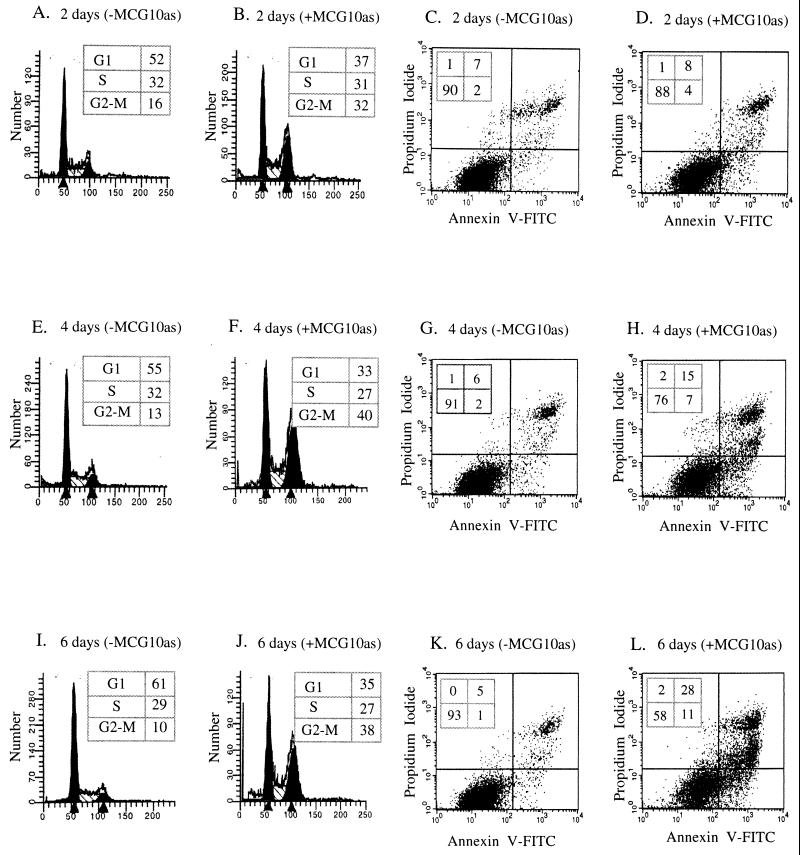
To further demonstrate that MCG10 can induce apoptosis, we performed a trypan blue dye exclusion assay. We found that the percentage of dead (trypan blue stained) cells was significantly increased in cells induced to express MCG10 for 2 and 4 days (Fig. 6A). It is well established that during the apoptotic cascade, several caspases are activated and the mitochondrial membrane potential of apoptotic cells is altered (67). Therefore, we analyzed the activity of caspases 3 and 6 and the mitochondrial membrane potential in cells with and without induction of MCG10. We found that the activity of caspase 6 but not caspase 3 was significantly increased by MCG10 (Fig. 6B). We also found that p53 substantially activated caspase 3 and, to a lesser extent, caspase 6 (Fig. 6B). Furthermore, the mitochondrial membrane was not permeable to Mitosensor, a cationic dye in cells expressing MCG10 (Fig. 6C), or p53 (data not shown), suggesting that the mitochondrial membrane potential is altered. Similar results were obtained for MCG10as-producing cells (Fig. 7). These results suggest that MCG10 can induce apoptosis without causing DNA fragmentation.
FIG. 6.
MCG10 activates caspase 6 and induces apoptosis through the mitochondrial pathway. (A) The percentage of dead cells induced by MCG10 was quantified by trypan blue dye exclusion. Cells were seeded in the presence (+) or absence (−) of MCG10 for 2 or 4 days. Both unstained and trypan blue-stained cells were counted using a hemocytometer. Error bars represent the standard deviations from at least three experiments. (B) Caspase 6 is activated by MCG10. p53-3 or MCG10-17 cells were uninduced or induced to express p53 or MCG10 for 3 days. Cells were then collected and assayed for the activity of caspases 3 and 6 as described in Materials and Methods. (C) The mitochondrial membrane potentials were altered in cells induced to express MCG10. MCG10-17 cells were uninduced (−MCG10) or induced to express MCG10 (+MCG10) for 3 days, stained with Mitosensor, and analyzed by fluorescence microscopy.
FIG. 7.
MCG10as is capable of inducing both cell cycle arrest in G2-M and apoptosis. DNA content was quantitated by propidium iodide staining of fixed cells that were uninduced (−MCG10as) or induced (+MCG10as) to express MCG10as for 2 days (A and B), 4 days (E and F), and 6 days (I and J). Apoptotic cells were quantitated by propidium iodide-annexin V staining of cells that were uninduced (−MCG10as) or induced (+MCG10as) to express MCG10as for 2 days (C and D), 4 days (G and H), and 6 days (K and L).
Role of the KH domain in the activity of MCG10 and MCG10as.
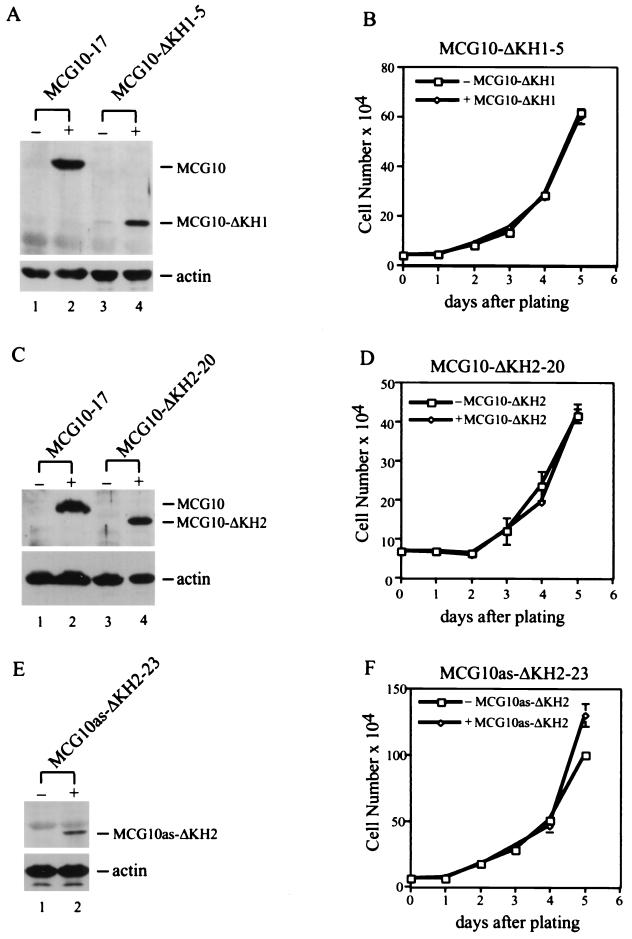
To determine whether the KH domain is necessary for the ability of MCG10 and MCG10as to induce cell cycle arrest and apoptosis, we constructed three KH domain deletion mutants, MCG10-ΔKH1, MCG10-ΔKH2, and MCG10as-ΔKH2. We then generated several cell lines that inducibly express these mutants. Expression of the mutant MCG10 and MCG10as proteins was assayed in Western blots using anti-MCG10 antibody (Fig. 8A, C, and E). Levels of the mutant proteins in MCG10-ΔKH1-5 and MCG10-ΔKH2-20 were fairly comparable to that in MCG10-17 cells (Fig. 8A and C). The level of the mutant protein expressed in MCG10as-ΔKH2-23 cells was relatively low compared to that in MCG10as-10 cells (data not shown). We then measured the growth rates of MCG10-ΔKH1-5, MCG10-ΔKH2-20, and MCG10as-ΔKH2-23 cells in the absence and presence of protein induction over a 5-day period. We found that none of the mutants were capable of suppressing cell proliferation (Fig. 8B, D, and F). Since a single KH domain remains in each mutant, the results suggest that both KH domains are required for the activity of MCG10 and MCG10as.
FIG. 8.
Both KH domains in MCG10 and MCG10as are necessary for inducing cell cycle arrest and apoptosis. (A) Levels of MCG10 and actin in MCG10-17 and MCG10-ΔKH1-5 cell lines were assayed by Western blot analysis. Cell extracts were prepared from uninduced cells (−) or cells induced (+) to express MCG10 or MCG10-ΔKH1. The blot was probed with affinity-purified anti-MCG10 polyclonal antibody (upper panel) and then reprobed with antiactin polyclonal antibody (lower panel). (B) Growth rates of MCG10-ΔKH1-5 cells in the presence (◊) and absence (□) of MCG10-ΔKH1 were measured as described in Materials and Methods. (C) Levels of MCG10 and actin in MCG10-17 and MCG10-ΔKH2-20 cell lines were assayed by Western blot analysis as described for panel A. (D) Growth rates of MCG10-ΔKH2-20 cells in the presence (◊) and absence (□) of MCG10-ΔKH2. (E) Levels of MCG10as-ΔKH2 and actin in the MCG10as-ΔKH2-23 cell line were assayed by Western blot analysis as described for panel A. (F) Growth rates of MCG10as-ΔKH2-23 cells in the presence (◊) and absence (□) of MCG10as-ΔKH2. Error bars represent the standard deviations from at least three experiments.
Both KH domains in MCG10 and MCG10as are necessary for binding RNA.
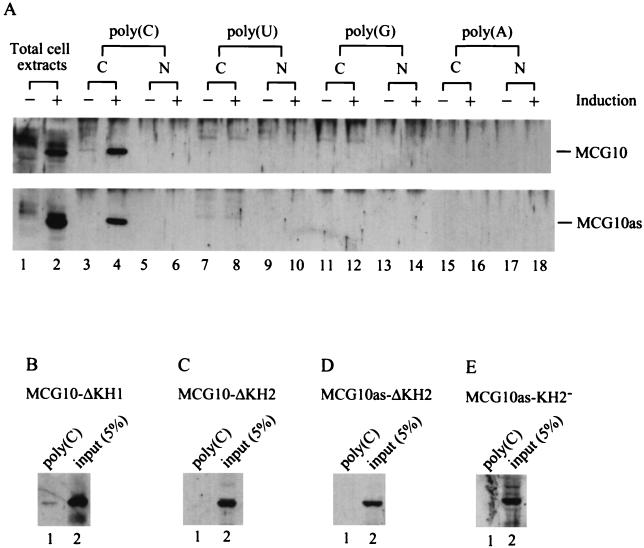
MCG10 and MCG10as each contain two KH domains. Since KH domains are known to bind RNA, we wanted to determine whether the KH domains in MCG10 and MCG10as also bind to RNA. To do this, poly(C)-, poly(G)-, poly(U)-, or poly(A)-agarose beads were added to cytoplasmic or nuclear extracts purified from uninduced cells or cells induced to express MCG10 or MCG10as. Proteins that specifically bound to the homopolymer beads were isolated, and the MCG10 and MCG10as proteins were identified by Western blot analysis. We found that MCG10 and MCG10as can bind to poly(C) but not to poly(A), poly(U), or poly(G) (Fig. 9A). This is consistent with the RNA-binding specificity for the KH domain (48, 74, 82). We did not detect any MCG10 and MCG10as in the nuclear extracts, suggesting that these proteins are predominantly located in the cytoplasm.
FIG. 9.
KH domain in MCG10 and MCG10as is capable of and necessary for binding poly(C). (A) MCG10 and MCG10as can bind to poly(C) but not to poly(U), poly(G), or poly(A). Total cell extracts run in lanes 1 and 2 were prepared from MCG10-17 and MCG10as-10 cells that were uninduced (−) and induced (+) to express MCG10 (upper panel) or MCG10as (lower panel). Cytoplasmic extracts (C) and nuclear extracts (N) were prepared from uninduced cells (−) or cells induced (+) to express MCG10 or MCG10as and mixed with poly(C)-, poly(U)-, poly(G)-, or poly(A)-agarose beads. Proteins bound to the beads were isolated and assayed by Western blot analysis using anti-MCG10 antibody. (B) The KH1 domain in MCG10 is necessary for binding poly(C). Cytoplasmic extracts were prepared from cells induced to express MCG10-ΔKH1, and the RNA-binding assay was performed as described for panel A. (C and D) The KH2 domain in MCG10 and MCG10as is necessary for binding poly(C). Cytoplasmic extracts were prepared from cells induced to express MCG10-ΔKH2 or MCG10as-ΔKH2, and the RNA-binding assay was performed. (E) A point mutation (Ile230Asp) in the KH2 domain abrogates the ability of MCG10as to bind to poly(C). Cytoplasmic extracts were prepared from cells induced to express MCG10as-KH2−, and the RNA-binding assay was performed.
To determine whether the KH domain deletion mutants that are defective in suppressing cell proliferation are also inert in binding RNA, the poly(C) RNA-binding assay was performed using cytoplasmic extracts from cells expressing MCG10-ΔKH1, MCG10-ΔKH2, and MCG10as-ΔKH2. We found that MCG10-ΔKH2 and MCG10as-ΔKH2 were incapable of binding poly(C) (Fig. 9C and D), whereas MCG10-ΔKH1 bound poly(C) extremely weakly (Fig. 9B). It has been reported that a missense mutation from Ile to Asp at residue 304 in KH2 of FMR1 abrogates its RNA-binding activity (66). To determine whether such a mutation would affect the RNA-binding activity of MCG10as, we generated a cell line that inducibly expresses the analogous mutant, designated MCG10as-KH2−. We found that, like the FMR1 mutant, MCG10as-KH2− was defective in binding RNA (Fig. 9E).
Poly(C)-binding MCG10 protein level is increased in cells following DNA damage in a p53-dependent manner.
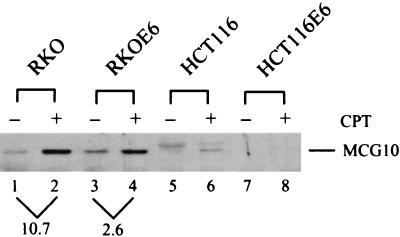
We have shown above that the MCG10 gene is induced by p53 and DNA damage (Fig. 1). To determine whether the level of MCG10 protein is increased in cells following a genotoxic stress, cytoplasmic cell extracts were prepared from RKO, RKOE6, HCT116, and HCT116E6 cells that were untreated or treated with 300 nM camptothecin for 24 h. MCG10 was isolated using the poly(C) beads and assayed by Western blot analysis with anti-MCG10 antibody. We found that the level of MCG10 protein was increased nearly 11-fold in RKO cells (Fig. 10, compare lanes 1 and 2), but only 2.6-fold in RKOE6 cells that are functionally p53-null when treated with camptothecin (Fig. 10, compare lanes 3 and 4). In addition, MCG10 was detected in HCT116 cells only when treated with camptothecin, but not in HCT116E6 cells, which are functionally p53-null (Fig. 10, lanes 5 to 8). A nonspecific protein that migrated slightly slower than MCG10 was detected in HCT116 cells (lanes 5 and 6). These results are consistent with the data obtained by Northern blot analysis (Fig. 1C) that induction of MCG10 by DNA damage is p53 dependent. It should be noted that, although MCG10 mRNA is not induced by DNA damage in RKOE6 cells (Fig. 1C, lanes 1 and 2), the level of poly(C)-binding MCG10 protein is increased, albeit to a lesser extent than in RKO cells. This suggests that MCG10 can be regulated posttranscriptionally by DNA damage in a p53-independent manner.
FIG. 10.
Level of the poly(C)-binding MCG10 protein is increased in cells treated with DNA-damaging agent camptothecin in a p53-dependent manner. Cytoplasmic extracts were prepared from RKO, RKOE6, HCT116, and HCT116E6 cells that were untreated (−) or treated (+) with camptothecin (CPT). The RNA-binding assay was performed as described in the legend to Fig. 9A.
DISCUSSION
RNA-binding proteins have diverse functions in the regulation of gene expression. This is the first report, to our knowledge, that a KH motif RNA-binding protein is regulated by p53 and that it serves as a mediator in inducing apoptosis and cell cycle arrest in G2-M. We have demonstrated that deletion of either of the KH domains or a point mutation in the C-terminal KH domain of MCG10as abrogates or severely diminishes the activity of MCG10 and MCG10as in binding RNA. As a result, the MCG10 and MCG10as mutants defective in RNA binding are also defective in inducing apoptosis and cell cycle arrest. These results indicate that, like other RNA-binding proteins, the RNA-binding activity is critical for the function of MCG10 and MCG10as. Interestingly, a 55-amino-acid insertion in the N-terminal KH domain does not interfere with the RNA-binding activity of MCG10.
Previously, we and others have shown that p53 cellular target genes are differentially regulated by p73 (21, 50, 107). We found that the MCG10 gene is among the group that is not induced by p73, further supporting the idea that the p73 signaling pathway is different from that for p53 (13). It should be mentioned that, like other p53 target genes, the MCG10 gene is induced by DNA damage in a p53-dependent manner (Fig. 1). DNA-damaging agents can induce a number of DNA-binding proteins by both transcriptional and posttranscriptional mechanisms, such as p53 (47), c-jun (106), and c-fos (29). However, the role of RNA-binding proteins in response to genotoxic stresses is mostly unexplored. A18 hnRNP, which contains one each of the RBD and RGG RNA-binding motifs, can be induced in response to UV-induced DNA damage (81). Nevertheless, it is still not clear what the physiological function of the A18 hnRNP protein is and whether DNA damage induction of the A18 hnRNP gene is p53 dependent. In addition, up to 13 DNA damage-inducible proteins were found to be capable of binding to a viral RNA probe consisting of the trans-activation-responsive element of human immunodeficiency virus type 1 and to a G+C-rich RNA probe (11). Since the genes encoding these RNA-binding proteins have not been characterized, it is not clear whether any of these genes can be regulated by p53.
How does the MCG10 protein mediate p53-dependent apoptosis and cell cycle arrest in G2-M? Based on the activities conferred by the KH domain in other proteins, it is likely that MCG10 may regulate expression of genes responsible for the control of the cell cycle by both transcriptional and posttranscriptional mechanisms. For example, by binding to the CT-rich repeat elements in the promoter of c-myc, hnRNP K enhances transcriptional initiation, possibly by promoting remodeling of chromatin architecture to facilitate interactions between transcription factors (59, 60, 87). In contrast, by binding to the CT-rich element adjacent to the Sp1-responsive element (E2) in the promoter of the neuronal nicotinic acetylcholine receptor β4 subunit (nACH β4) gene, hnRNP K may directly block Sp1 binding to E2, leading to transcriptional repression of the nACH β4 gene (26). In addition, hnRNP K and E can bind to a CU-rich repetitive element in the 3′ untranslated region (3′-UTR) of erythroid 15-lipooxygenase (LOX) mRNA and block 80S ribosome complex assembly on LOX RNA, leading to translational silencing of the LOX gene (73). In contrast, by binding to a CU-rich RNA element in the 3′-UTR of α-globin mRNA, hnRNP E can stabilize α-globin mRNA, leading to enhanced expression of the α-globin gene (45, 97). Interestingly, five GADD mRNAs, including GADD45, which is a cellular target of p53 and whose product can mediate cell cycle arrest in G2-M (95), are stabilized in hamster cells when treated with DNA-damaging agents (40). However, it is still not clear whether DNA damage-induced stabilization of these GADD mRNAs is p53 dependent. It will be interesting to determine whether MCG10 can regulate these GADD genes.
Tumorigenesis involves multistep sequential alterations of genetic materials. One of the early outcomes of this process is immortalization of cells, leading to an unlimited replicative life span. Recent studies have shown that overexpression of telomerase, whose activity can be regulated by p53 (16, 49), immortalizes cells, suggesting that the length of the telomere is critical for a limited replicative life span (19). Telomerase is a specialized reverse transcriptase that synthesizes a DNA sequence using an RNA template (19, 54). The RNA template is usually 100 to 200 nucleotides long and contains several repeats of C-rich elements. Interestingly, loss of heterozygosity (LOH) at 3p21, the mapped location of MCG10, is associated with an increased telomerase activity in head, neck, and renal carcinomas (55, 58). Since MCG10 is a potent poly(C)-binding protein, it is possible that, by binding to the C-rich repeats in the RNA template, MCG10 and MCG10as can sequester the RNA template and inhibit telomere synthesis, thereby suppressing cell proliferation.
In addition to the RNA-binding motifs, hnRNPs often contain other auxiliary domains, most notably the proline-rich PXXP motif (P represents proline, whereas X is any amino acid). PXXP residues can form a left-handed polyproline type II helix, which creates a binding site for Src homology 3 (SH3) domains (18). The proline-rich domains in hnRNP K and Sam68 have been shown to interact with several protooncogene products, including Src (85, 98), Fyn (98), Lyn (98), and Vav (10, 36). In addition, upon interaction with Src, hnRNP K and Sam68 can be phosphorylated at tyrosine residues by Src tyrosine kinase (85, 89). These results support a hypothesis that extracellular signals can be received by a membrane-associated tyrosine kinase, such as Src, which transmits the signal to an RNA-binding protein, such as hnRNP K and Sam68. The RNA-binding protein would then regulate the expression of genes that control cellular responses to various extracellular signals. MCG10 and MCG10as contain three proline-rich domains at their carboxyl termini. Therefore, future studies are needed to determine with what protein MCG10 and MCG10as interact and what the physiological response is, if indeed an interaction occurs.
Most p53 target genes can mediate one defined p53 activity. For example, p21 is necessary for mediating G1 arrest (7, 20), 14-3-3ς mediates G2-M arrest (35), and Bax possibly mediates apoptosis (62). Interestingly, MCG10 and MCG10as can mediate two p53 activities, that is, apoptosis and cell cycle arrest in G2-M. This may not be surprising. Since the mechanism by which MCG10 and MCG10as may function as a potential p53 mediator is their ability to regulate gene expression and/or to interact with one or more signaling proteins responsible for the control of the cell cycle, multiple pathways could be regulated. It should be noted that MCG10 and MCG10as are potent in inducing apoptosis, but unlike wild-type p53, they do so without inducing significant cellular DNA fragmentation. Since the RNA-binding activity is necessary for apoptosis, it is likely that one or more cellular genes whose products can lead to DNA breakdown are not regulated by MCG10 and MCG10as. Indeed, caspase 3 is not significantly activated by MCG10 (Fig. 6B). Caspase 3 is the primary effector enzyme that proteolytically inactivates DFF45 (DNA fragmentation factor 45) (also called ICAD [inhibitor of caspase-activated DNase]) and releases active DFF40 (also called CAD [caspase-activated DNase]), leading to internucleosomal DNA cleavage (102).
Is MCG10 a tumor suppressor?
p53 is a bona fide tumor suppressor because it fulfills the “classical features” of a tumor suppressor (17). The ability of MCG10 to inhibit the growth of transformed cells fulfills one of the criteria for a tumor suppressor. Second, the MCG10 gene maps to chromosome 3p21, a region highly susceptible to aberrant chromosomal rearrangements and deletions (61). LOH at 3p21 has been found in many types of human cancers, such as breast carcinomas, small and non-small cell lung carcinomas, uterine and cervical carcinomas, renal cell carcinomas, head, neck, and oral squamous cell carcinomas, ovarian cancers, and pancreatic islet cell tumors (6, 24, 25, 31, 70, 72, 75, 100, 101). Homozygous deletions of 3p21 are also found in several lung tumors and lung cancer cell lines (86). In esophageal carcinomas, LOH at 3p21 is an early event, preceding loss of RB and p53 functions (63). In addition, LOH in a region syntenic with 3p21 is also found in many types of mouse cancers (22, 70). When scid mouse tumors, which are induced by human chromosome 3-mouse microcell hybrids, were used to screen for a common eliminated region, one was often found at 3p21 (38, 44), suggesting that loss of a tumor suppressor gene may be necessary for microcell hybrids to induce tumors in scid mice. The human mismatch repair gene (hMLH) also maps to 3p21, and loss of hMLH function is associated with microsatellite instability at one or more loci (51). However, only a subset (less than 30%) of non-small cell lung carcinomas contain LOH at 3p21 with microsatellite instability (99), suggesting that, in non-small cell lung carcinomas without microsatellite instability, LOH at 3p21 probably involves another tumor suppressor gene(s). Therefore, future studies are needed to determine whether MCG10 LOH occurs in these tumors and whether loss of MCG10 contributes to tumorigenesis.
ACKNOWLEDGMENTS
We thank Jason Paik for technical help and Rhea Markowitz for critical reading of the manuscript.
This work is supported in part by National Cancer Institute grant CA 76069 and the Department of Defense Army Breast Cancer Program DAMD17-97-1-7019.
REFERENCES
- 1.Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Almog N, Rotter V. Involvement of p53 in cell differentiation and development. Biochim Biophys Acta. 1997;1333:F1–F27. doi: 10.1016/s0304-419x(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 3.Attardi L D, Lowe S W, Brugarolas J, Jacks T. Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. EMBO J. 1996;15:3693–3701. [PMC free article] [PubMed] [Google Scholar]
- 4.Barlat I, Maurier F, Duchesne M, Guitard E, Tocque B, Schweighoffer F. A role for Sam68 in cell cycle progression antagonized by a spliced variant within the KH domain. J Biol Chem. 1997;272:3129–3132. doi: 10.1074/jbc.272.6.3129. [DOI] [PubMed] [Google Scholar]
- 5.Bennett M, Macdonald K, Chan S W, Luzio J P, Simari R, Weissberg P. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 1998;282:290–293. doi: 10.1126/science.282.5387.290. [DOI] [PubMed] [Google Scholar]
- 6.Braga E, Pugacheva E, Bazov I, Ermilova V, Kazubskaya T, Mazurenko N, Kisseljov F, Liu J, Garkavtseva R, Zabarovsky E, Kisselev L. Comparative allelotyping of the short arm of human chromosome 3 in epithelial tumors of four different types. FEBS Lett. 1999;454:215–219. doi: 10.1016/s0014-5793(99)00807-8. [DOI] [PubMed] [Google Scholar]
- 7.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 8.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 9.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 10.Bustelo X R, Suen K L, Michael W M, Dreyfuss G, Barbacid M. Association of the vav proto-oncogene product with poly(rC)-specific RNA-binding proteins. Mol Cell Biol. 1995;15:1324–1332. doi: 10.1128/mcb.15.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrier F, Gatignol A, Hollander M C, Jeang K T, Fornace A J., Jr Induction of RNA-binding proteins in mammalian cells by DNA-damaging agents. Proc Natl Acad Sci USA. 1994;91:1554–1558. doi: 10.1073/pnas.91.4.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan T A, Hermeking H, Lengauer C, Kinzler K W, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 13.Chen X. The p53 family: same response, different signals? Mol Med Today. 1999;5:387–392. doi: 10.1016/s1357-4310(99)01545-2. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Bargonetti J, Prives C. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 1995;55:4257–4263. [PubMed] [Google Scholar]
- 15.Chen X, Ko L J, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 16.Chin L, Artandi S E, Shen Q, Tam A, Lee S L, Gottlieb G J, Greider C W, DePinho R A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 17.Clurman B, Groudine M. Tumour-suppressor genes. Killer in search of a motive? Nature. 1997;389:122–123. doi: 10.1038/38116. [DOI] [PubMed] [Google Scholar]
- 18.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 19.Colgin L M, Reddel R R. Telomere maintenance mechanisms and cellular immortalization. Curr Opin Genet Dev. 1999;9:97–103. doi: 10.1016/s0959-437x(99)80014-8. . (Erratum, 9:247.) [DOI] [PubMed] [Google Scholar]
- 20.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 21.Di Como C J, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietrich W F, Radany E H, Smith J S, Bishop J M, Hanahan D, Lander E S. Genome-wide search for loss of heterozygosity in transgenic mouse tumors reveals candidate tumor suppressor genes on chromosomes 9 and 16. Proc Natl Acad Sci USA. 1994;91:9451–9455. doi: 10.1073/pnas.91.20.9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 24.Driouch K, Briffod M, Bieche I, Champeme M H, Lidereau R. Location of several putative genes possibly involved in human breast cancer progression. Cancer Res. 1998;58:2081–2086. [PubMed] [Google Scholar]
- 25.Druck T, Kastury K, Hadaczek P, Podolski J, Toloczko A, Sikorski A, Ohta M, LaForgia S, Lasota J, McCue P, et al. Loss of heterozygosity at the familial RCC t(3;8) locus in most clear cell renal carcinomas. Cancer Res. 1995;55:5348–5353. [PubMed] [Google Scholar]
- 26.Du Q, Melnikova I N, Gardner P D. Differential effects of heterogeneous nuclear ribonucleoprotein K on Sp1- and Sp3-mediated transcriptional activation of a neuronal nicotinic acetylcholine receptor promoter. J Biol Chem. 1998;273:19877–19883. doi: 10.1074/jbc.273.31.19877. [DOI] [PubMed] [Google Scholar]
- 27.el-Deiry W S. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 28.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 29.Elkeles A, Juven-Gershon T, Israeli D, Wilder S, Zalcenstein A, Oren M. The c-fos proto-oncogene is a target for transactivation by the p53 tumor suppressor. Mol Cell Biol. 1999;19:2594–2600. doi: 10.1128/mcb.19.4.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedlander P, Haupt Y, Prives C, Oren M. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol Cell Biol. 1996;16:4961–4971. doi: 10.1128/mcb.16.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fullwood P, Marchini S, Rader J S, Martinez A, Macartney D, Broggini M, Morelli C, Barbanti-Brodano G, Maher E R, Latif F. Detailed genetic and physical mapping of tumor suppressor loci on chromosome 3p in ovarian cancer. Cancer Res. 1999;59:4662–4667. [PubMed] [Google Scholar]
- 32.Giaccia A J, Kastan M B. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 33.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottlieb T M, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 35.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 36.Hobert O, Jallal B, Schlessinger J, Ullrich A. Novel signaling pathway suggested by SH3 domain-mediated p95vav/heterogeneous ribonucleoprotein K interaction. J Biol Chem. 1994;269:20225–20228. [PubMed] [Google Scholar]
- 37.Hwang B J, Ford J M, Hanawalt P C, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imreh S, Kost-Alimova M, Kholodnyuk I, Yang Y, Szeles A, Kiss H, Liu Y, Foster K, Zabarovsky E, Stanbridge E, Klein G. Differential elimination of 3p and retention of 3q segments in human/mouse microcell hybrids during tumor growth. Genes Chromosomes Cancer. 1997;20:224–233. [PubMed] [Google Scholar]
- 39.Israeli D, Tessler E, Haupt Y, Elkeles A, Wilder S, Amson R, Telerman A, Oren M. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackman J, Alamo I, Jr, Fornace A J., Jr Genotoxic stress confers preferential and coordinate messenger RNA stability on the five gadd genes. Cancer Res. 1994;54:5656–5662. [PubMed] [Google Scholar]
- 41.Johansen F E, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 43.Kern S E, Kinzler K W, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 44.Kholodnyuk I, Kost-Alimova M, Kashuba V, Gizatulin R, Szeles A, Stanbridge E J, Zabarovsky E R, Klein G, Imreh S. A 3p21.3 region is preferentially eliminated from human chromosome 3/mouse microcell hybrids during tumor growth in SCID mice. Genes Chromosomes Cancer. 1997;18:200–211. [PubMed] [Google Scholar]
- 45.Kiledjian M, Wang X, Liebhaber S A. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 47.Ko L J, Shieh S Y, Chen X, Jayaraman L, Tamai K, Taya Y, Prives C, Pan Z Q. p53 is phosphorylated by CDK7-cyclin H in a p36MAT1-dependent manner. Mol Cell Biol. 1997;17:7220–7229. doi: 10.1128/mcb.17.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krecic A M, Swanson M S. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 49.Kusumoto M, Ogawa T, Mizumoto K, Ueno H, Niiyama H, Sato N, Nakamura M, Tanaka M. Adenovirus-mediated p53 gene transduction inhibits telomerase activity independent of its effects on cell cycle arrest and apoptosis in human pancreatic cancer cells. Clin Cancer Res. 1999;5:2140–2147. [PubMed] [Google Scholar]
- 50.Lee C W, La Thangue N B. Promoter specificity and stability control of the p53-related protein p73. Oncogene. 1999;18:4171–4181. doi: 10.1038/sj.onc.1202793. [DOI] [PubMed] [Google Scholar]
- 51.Lengauer C, Kinzler K W, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 52.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 53.Lin Q, Taylor S J, Shalloway D. Specificity and determinants of Sam68 RNA binding: implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 54.Lingner J, Cech T R. Telomerase and chromosome end maintenance. Curr Opin Genet Dev. 1998;8:226–232. doi: 10.1016/s0959-437x(98)80145-7. [DOI] [PubMed] [Google Scholar]
- 55.Loughran O, Clark L J, Bond J, Baker A, Berry I J, Edington K G, Ly I S, Simmons R, Haw R, Black D M, Newbold R F, Parkinson E K. Evidence for the inactivation of multiple replicative lifespan genes in immortal human squamous cell carcinoma keratinocytes. Oncogene. 1997;14:1955–1964. doi: 10.1038/sj.onc.1201028. [DOI] [PubMed] [Google Scholar]
- 56.Mashimo T, Watabe M, Hirota S, Hosobe S, Miura K, Tegtmeyer P J, Rinker-Shaeffer C W, Watabe K. The expression of the KAI1 gene, a tumor metastasis suppressor, is directly activated by p53. Proc Natl Acad Sci USA. 1998;95:11307–11311. doi: 10.1073/pnas.95.19.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuzawa S, Takayama S, Froesch B A, Zapata J M, Reed J C. p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J. 1998;17:2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mehle C, Lindblom A, Ljungberg B, Stenling R, Roos G. Loss of heterozygosity at chromosome 3p correlates with telomerase activity in renal cell carcinoma. Int J Oncol. 1998;13:289–295. doi: 10.3892/ijo.13.2.289. [DOI] [PubMed] [Google Scholar]
- 59.Michelotti E F, Michelotti G A, Aronsohn A I, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michelotti G A, Michelotti E F, Pullner A, Duncan R C, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitelman F, Mertens F, Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet. 1997;15(Spec. No.):417–474. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 62.Miyashita T, Krajewski S, Krajewska M, Wang H G, Lin H K, Liebermann D A, Hoffman B, Reed J C. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 63.Mori T, Yanagisawa A, Kato Y, Miura K, Nishihira T, Mori S, Nakamura Y. Accumulation of genetic alterations during esophageal carcinogenesis. Hum Mol Genet. 1994;3:1969–1971. doi: 10.1093/hmg/3.11.1969. [DOI] [PubMed] [Google Scholar]
- 64.Muller M, Wilder S, Bannasch D, Israeli D, Lehlbach K, Li-Weber M, Friedman S L, Galle P R, Stremmel W, Oren M, Krammer P H. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med. 1998;188:2033–2045. doi: 10.1084/jem.188.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munger K, Phelps W C, Bubb V, Howley P M, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Musco G, Stier G, Joseph C, Castiglione Morelli M A, Nilges M, Gibson T J, Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 67.Nagata S. Apoptotic DNA fragmentation. Exp Cell Res. 2000;256:12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- 68.Nelson W G, Kastan M B. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niculescu A B, 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed S I. Effects of p21 (Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nikiforova M N, Nikiforov Y E, Biddinger P, Gnepp D R, Grosembacher L A, Wajchenberg B L, Fagin J A, Cohen R M. Frequent loss of heterozygosity at chromosome 3p14.2-3p21 in human pancreatic islet cell tumours. Clin Endocrinol (Oxford) 1999;51:27–33. doi: 10.1046/j.1365-2265.1999.00785.x. [DOI] [PubMed] [Google Scholar]
- 71.Oberhammer F, Wilson J W, Dive C, Morris I D, Hickman J A, Wakeling A E, Walker P R, Sikorska M. Apoptotic death in epithelial cells: cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 1993;12:3679–3684. doi: 10.1002/j.1460-2075.1993.tb06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogasawara S, Maesawa C, Tamura G, Satodate R. Frequent microsatellite alterations on chromosome 3p in esophageal squamous cell carcinoma. Cancer Res. 1995;55:891–894. [PubMed] [Google Scholar]
- 73.Ostareck D H, Ostareck-Lederer A, Wilm M, Thiele B J, Mann M, Hentze M W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 74.Ostareck-Lederer A, Ostareck D H, Hentze M W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 75.Partridge M, Emilion G, Langdon J D. LOH at 3p correlates with a poor survival in oral squamous cell carcinoma. Br J Cancer. 1996;73:366–371. doi: 10.1038/bjc.1996.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pietenpol J A, Tokino T, Thiagalingam S, el-Deiry W S, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pinol-Roma S, Choi Y D, Matunis M J, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. . (Erratum, 2:190.) [DOI] [PubMed] [Google Scholar]
- 78.Roperch J P, Lethrone F, Prieur S, Piouffre L, Israeli D, Tuynder M, Nemani M, Pasturaud P, Gendron M C, Dausset J, Oren M, Amson R B, Telerman A. SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21 (Waf1) Proc Natl Acad Sci USA. 1999;96:8070–8073. doi: 10.1073/pnas.96.14.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rouault J P, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, Shaw P, Berger R, Samarut J, Magaud J P, Ozturk M, Samarut C, Puisieux A. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 80.Sakahira H, Enari M, Ohsawa Y, Uchiyama Y, Nagata S. Apoptotic nuclear morphological change without DNA fragmentation. Curr Biol. 1999;9:543–546. doi: 10.1016/s0960-9822(99)80240-1. [DOI] [PubMed] [Google Scholar]
- 81.Sheikh M S, Carrier F, Papathanasiou M A, Hollander M C, Zhan Q, Yu K, Fornace A J., Jr Identification of several human homologs of hamster DNA damage-inducible transcripts: cloning and characterization of a novel UV-inducible cDNA that codes for a putative RNA-binding protein. J Biol Chem. 1997;272:26720–26726. doi: 10.1074/jbc.272.42.26720. [DOI] [PubMed] [Google Scholar]
- 82.Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- 83.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 84.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taylor S J, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 86.Todd S, Franklin W A, Varella-Garcia M, Kennedy T, Hilliker C E, Jr, Hahner L, Anderson M, Wiest J S, Drabkin H A, Gemmill R M. Homozygous deletions of human chromosome 3p in lung tumors. Cancer Res. 1997;57:1344–1352. [PubMed] [Google Scholar]
- 87.Tomonaga T, Michelotti G A, Libutti D, Uy A, Sauer B, Levens D. Unrestraining genetic processes with a protein-DNA hinge. Mol Cell. 1998;1:759–764. doi: 10.1016/s1097-2765(00)80075-1. [DOI] [PubMed] [Google Scholar]
- 88.Utrera R, Collavin L, Lazarevic D, Delia D, Schneider C. A novel p53-inducible gene coding for a microtubule-localized protein with G2-phase-specific expression. EMBO J. 1998;17:5015–5025. doi: 10.1093/emboj/17.17.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Seuningen I, Ostrowski J, Bustelo X R, Sleath P R, Bomsztyk K. The K protein domain that recruits the interleukin 1-responsive K protein kinase lies adjacent to a cluster of c-Src and Vav SH3-binding sites: implications that K protein acts as a docking platform. J Biol Chem. 1995;270:26976–26985. doi: 10.1074/jbc.270.45.26976. [DOI] [PubMed] [Google Scholar]
- 90.Varmeh-Ziaie S, Okan I, Wang Y, Magnusson K P, Warthoe P, Strauss M, Wiman K G. Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene. 1997;15:2699–2704. doi: 10.1038/sj.onc.1201454. [DOI] [PubMed] [Google Scholar]
- 91.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Venot C, Maratrat M, Sierra V, Conseiller E, Debussche L. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene. 1999;18:2405–2410. doi: 10.1038/sj.onc.1202539. [DOI] [PubMed] [Google Scholar]
- 93.Verkerk A J, deVries B B, Niermeijer M F, Fu Y H, Nelson D L, Warren S T, Majoor-Krakauer D F, Halley D J, Oostra B A. Intragenic probe used for diagnostics in fragile X families. Am J Med Genet. 1992;43:192–196. doi: 10.1002/ajmg.1320430132. [DOI] [PubMed] [Google Scholar]
- 94.Waldman T, Lengauer C, Kinzler K W, Vogelstein B. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature. 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 95.Wang X W, Zhan Q, Coursen J D, Khan M A, Kontny H U, Yu L, Hollander M C, O'Connor P M, Fornace A J, Jr, Harris C C. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Warren S T, Nelson D L. Advances in molecular analysis of fragile X syndrome. JAMA. 1994;271:536–542. [PubMed] [Google Scholar]
- 97.Weiss I M, Liebhaber S A. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weng Z, Thomas S M, Rickles R J, Taylor J A, Brauer A W, Seidel-Dugan C, Michael W M, Dreyfuss G, Brugge J S. Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol Cell Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wieland I, Ammermuller T, Bohm M, Totzeck B, Rajewsky M F. Microsatellite instability and loss of heterozygosity at the hMLH1 locus on chromosome 3p21 occur in a subset of nonsmall cell lung carcinomas. Oncol Res. 1996;8:1–5. [PubMed] [Google Scholar]
- 100.Wistuba I I, Behrens C, Milchgrub S, Bryant D, Hung J, Minna J D, Gazdar A F. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene. 1999;18:643–650. doi: 10.1038/sj.onc.1202349. [DOI] [PubMed] [Google Scholar]
- 101.Wistuba I I, Montellano F D, Milchgrub S, Virmani A K, Behrens C, Chen H, Ahmadian M, Nowak J A, Muller C, Minna J D, Gazdar A F. Deletions of chromosome 3p are frequent and early events in the pathogenesis of uterine cervical carcinoma. Cancer Res. 1997;57:3154–3158. [PubMed] [Google Scholar]
- 102.Wolf B B, Schuler M, Echeverri F, Green D R. Caspase-3 is the primary activator of apoptotic DNA fragmentation via DNA fragmentation factor-45/inhibitor of caspase-activated DNase inactivation. J Biol Chem. 1999;274:30651–30656. doi: 10.1074/jbc.274.43.30651. [DOI] [PubMed] [Google Scholar]
- 103.Wu G S, Burns T F, McDonald E R, 3rd, Jiang W, Meng R, Krantz I D, Kao G, Gan D D, Zhou J Y, Muschel R, Hamilton S R, Spinner N B, Markowitz S, Wu G, El-Deiry W S. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 104.Wu G S, Saftig P, Peters C, El-Deiry W S. Potential role for cathepsin D in p53-dependent tumor suppression and chemosensitivity. Oncogene. 1998;16:2177–2183. doi: 10.1038/sj.onc.1201755. [DOI] [PubMed] [Google Scholar]
- 105.Yin Y, Terauchi Y, Solomon G G, Aizawa S, Rangarajan P N, Yazaki Y, Kadowaki T, Barrett J C. Involvement of p85 in p53-dependent apoptotic response to oxidative stress. Nature. 1998;391:707–710. doi: 10.1038/35648. [DOI] [PubMed] [Google Scholar]
- 106.Zhan Q, Bae I, Kastan M B, Fornace A J., Jr The p53-dependent gamma-ray response of GADD45. Cancer Res. 1994;54:2755–2760. [PubMed] [Google Scholar]
- 107.Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58:5061–5065. [PubMed] [Google Scholar]
- 108.Zhu J, Jiang J, Zhou W, Zhu K, Chen X. Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene. 1999;18:2149–2155. doi: 10.1038/sj.onc.1202533. [DOI] [PubMed] [Google Scholar]
- 109.Zhu J, Zhou W, Jiang J, Chen X. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem. 1998;273:13030–13036. doi: 10.1074/jbc.273.21.13030. [DOI] [PubMed] [Google Scholar]
- 110.Zhu, K., W. J., J. Zhu, J. Jiang, J. Shou, and X. Chen. 1999. p53 induces TAP1 and enhances the transport of MHC class I peptides. Oncogene 18:7740–7747. [DOI] [PubMed]