Abstract
Human pluripotent stem cell (hPSC)-derived progenies are immature versions of cells, presenting a potential limitation to the accurate modelling of diseases associated with maturity or age. Hence, it is important to characterise how closely cells used in culture resemble their native counterparts. In order to select appropriate time points of retinal pigment epithelium (RPE) cultures that reflect native counterparts, we characterised the transcriptomic profiles of the hPSC-derived RPE cells from 1- and 12-month cultures. We differentiated the human embryonic stem cell line H9 into RPE cells, performed single-cell RNA-sequencing of a total of 16,576 cells to assess the molecular changes of the RPE cells across these two culture time points. Our results indicate the stability of the RPE transcriptomic signature, with no evidence of an epithelial–mesenchymal transition, and with the maturing populations of the RPE observed with time in culture. Assessment of Gene Ontology pathways revealed that as the cultures age, RPE cells upregulate expression of genes involved in metal binding and antioxidant functions. This might reflect an increased ability to handle oxidative stress as cells mature. Comparison with native human RPE data confirms a maturing transcriptional profile of RPE cells in culture. These results suggest that long-term in vitro culture of RPE cells allows the modelling of specific phenotypes observed in native mature tissues. Our work highlights the transcriptional landscape of hPSC-derived RPE cells as they age in culture, which provides a reference for native and patient samples to be benchmarked against.
Keywords: Human embryonic stem cell, Human pluripotent stem cell, Retinal pigment epithelium, Single-cell RNA sequencing, Ageing
Introduction
The retinal pigment epithelium (RPE) is a monolayer of post-mitotic, pigmented polarised cells that is key to the health and function of photoreceptors and underlying vasculature. In particular, the RPE protects the retina against photo-oxidation and phagocytoses photoreceptor outer segments. The RPE is also essential to the immune privilege of the eye, as it physically contributes to the blood–retina barrier and also expresses molecules repressing the migration of immune cells into the retina [1]. In the human retina, ageing is associated with vision decline and delayed dark adaptation, both of which are direct consequences of tissue stress and retinal damage [2]. It is hypothesised that over time, oxidative stress leads to the death of retinal neurons, a decrease in the number of RPE cells, an accumulation of the toxic waste lipofuscin within the RPE, and an accumulation of basal toxic deposits called drusen underneath the RPE [2]. Together, these events contribute to a loss of homeostasis and low-grade inflammation within the retina [2]. Although it is clear that the RPE is key to the health of the retina, the precise molecular mechanisms underlying its ageing are not well understood.
Human pluripotent stem cells (hPSCs) have the ability to propagate indefinitely in vitro and give rise to any cell types in the body, including cells that form the retina. Various protocols have been described to differentiate hPSCs into RPE cells [3], [4], [5], [6], [7], [8]. RPE cells are generally assayed after a few weeks of differentiation, at which stage they demonstrate similarity to their human native counterparts, in terms of morphology/expression of key proteins, functions, and expression profiles, however with a profile closer to a foetal stage than adult stage [4], [9], [10]. Interestingly, the transcriptome profile of hPSC-derived RPE cells as they age in culture is unknown. To date, most RNA-sequencing (RNA-seq) studies of RPE cells have been performed on bulk samples. Yet, the ability to sequence individual cells provides a powerful tool to precisely uncover potential heterogeneity in cell population, especially as these cells develop and mature in vitro. Here, we used single-cell RNA-seq (scRNA-seq) of hPSC-derived RPE cells maintained in culture for 1 month or 12 months to assess the impact of time on the RPE transcriptome and whether genetic hallmarks of maturation can be observed over time. A short-time differentiation (1 month) was chosen as it represents a time point routinely used in in vitro assays of RPE [3]. A prolonged time course of differentiation (12 months) was chosen as its characterization could be subsequently used for comparison with other retinal cell differentiation methods, in particular of retinal organoids and photoreceptors, for which differentiation and relative maturity are obtained after prolonged time in culture and would thus be present at that later time point [11], [12], [13], [14].
Results
scRNA-seq profiles the transcriptomes of 16,576 cells
The human embryonic stem cell (hESC) line H9 was differentiated to RPE cells following the protocol described in the Materials and methods section. To generate a transcriptional map of the RPE cells reflecting time in culture, RPE cells from the same culture and original passaging were isolated after 1 month or 12 months of differentiation, dissociated to single cells, and processed to generate libraries for scRNA-seq analysis (Figure 1A). The capture of single-cell library from the 1-month-old culture detected 12,873 cells at the mean read depth of 40,499 reads per cell, while the capture from the 12-month-old culture detected 4850 cells at the mean read depth of 114,503 reads per cell (Table S1). Both datasets were subjected to cell-specific quality control, where 510 cells and 637 cells were removed from the 1-month-old and 12-month-old culture datasets, respectively. The remaining 16,576 cells were retained for further analysis.
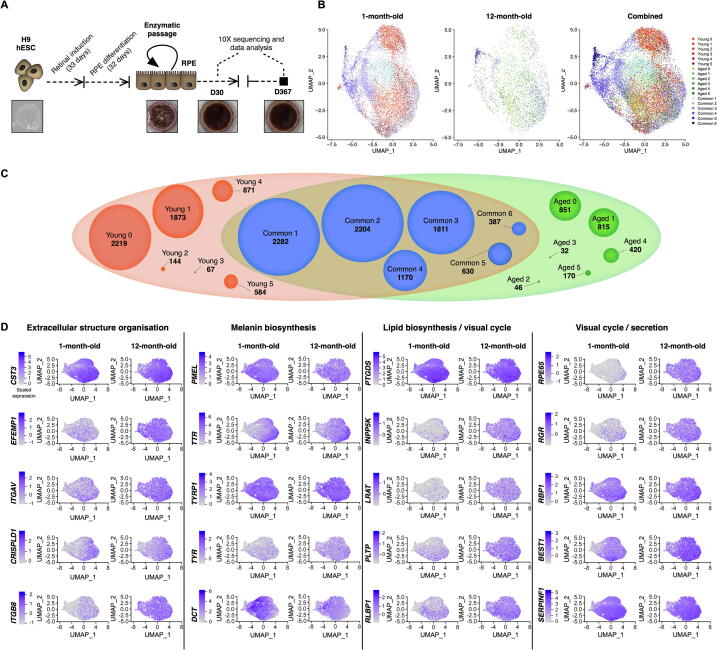
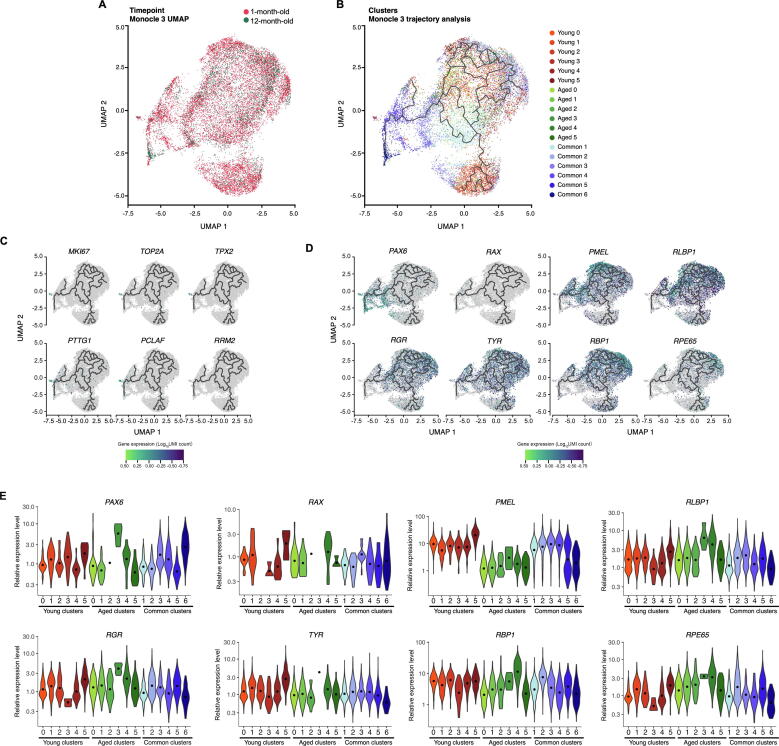
Figure 1.
scRNA-seq transcriptome profiling of hPSC-derived RPE cells reveals 18 subpopulations
A. Schematic representations of the experimental flow. B. UMAP of single-cell expression profile from 16,576 cells, clustered into 18 subpopulations, split by condition (1-month-old and 12-month-old) and combined. C. Cluster grouping represented by a Venn diagram, identifying 18 subpopulations, showing Young (red), Aged (green), and their common subpopulations (blue). Number of cells for each subpopulation is indicated in bold below the subpopulation name. D. UMAP of canonical RPE markers in 1-month-old and 12-month-old cultures, organised by cellular functions: extracellular structure organisation (CST3, EFEMP1, ITGAV, CRISPLD1, and ITGB8); melanin biosynthesis (PMEL, TTR, TYRP1, TYR, and DCT), lipid biosynthesis (PTGDS and INPP5K), visual cycle (LRAT, PLTP, RLBP1, RPE65, RGR, RBP1, and BEST1), as well as secretion (SERPINF1). Levels of gene expression per cell (percentage expressed) are shown with colour gradients. scRNA-seq, single-cell RNA sequencing; hPSC, human pluripotent stem cell; RPE, retinal pigment epithelium; UMAP, Uniform Manifold Approximation and Projection for Dimension Reduction.
We compared variations in the transcriptomic profiles between the 1-month-old and 12-month-old samples (Figure 1B and C) to identify potential changes in phenotypes upon ageing of RPE cells in vitro, by analysing differential expression. A range of RPE markers were observed as conserved between both time points (Figure 1D). In particular, canonical RPE markers [15] associated with extracellular structure organisation (CST3, EFEMP1, ITGAV, CRISPLD1, and ITGB8), melanin biosynthesis (PMEL, TTR, TYRP1, TYR, and DCT), lipid biosynthesis (PTGDS and INPP5K), visual cycle (LRAT, PLTP, ABHD2/RLBP1, RPE65, RGR, RBP1, and BEST1), and secretion (SERPINF1) were expressed at both time points (Figure 1D).
Clustering analysis highlights 12 subpopulations of RPE cells
Clustering analysis was performed independently for samples obtained at the two time points and identified 12 subpopulations (clusters) in each sample (Figure 1C; Table S2). After integrating with anchors identified with a method described previously [16], MetaNeighbor was used to match common subpopulations across both samples [17], denoted as “Common”. Clusters unique to 1-month-old and 12-month-old samples are denoted as “Young” and “Aged”, respectively (Figure 1C). In total, 18 subpopulations were identified, including six common subpopulations (Common 1–6; 8484 cells; Table S2), six subpopulations exclusive to the 1-month-old dataset (Young 0–5; 5758 cells; Table S2), and six subpopulations exclusive to the 12-month-old dataset (Aged 0–5; 2334 cells; Table S2). Cell counts per cluster (Figure 1C; Table S2) and the top conserved markers for each distinct cluster (Figure 2A; Tables S3–S5) were identified. Clusters were visualised using the Uniform Manifold Approximation and Projection (UMAP) plots (Figure 1B). In total, 3070 cells were considered singletons. These cells had fewer connections with similar cells (neighbours) relative to the rest of the cell population and could not be assigned to a subpopulation. Cluster 0 in the 1-month-old dataset (Young 0) comprises 2219 cells, accounting for 18% of all 1-month-old cells, whereas Cluster 0 in the 12-month-old dataset (Aged 0) comprises 851 cells, accounting for 20% of all 12-month-old cells (Table S2). There were more singleton cells in the Young population as we captured more cells from this group. We assessed the expression profile of genes characteristic of progenitors (BMP7 and SOX4) and canonical RPE genes [15] across all subpopulations, both in terms of frequency and intensity of expression (Figure 2B; Tables S3–S5). These genes are linked to lipid biosynthesis (INPP5K and PTGDS), visual cycle (LRAT, RGR, PLTP, RLBP1/CRALBP, RBP1/CRBP1, BEST1, and RPE65), melanin biosynthesis (DCT, TYRP1, TYR, TTR, and PMEL); secretion (ENPP2, VEGFA, and SERPINF1), phagocytic activity (GULP1), and extracellular structure organisation (ITGAV, CRISPLD1, CST3, and EFEMP1). Most canonical RPE genes were expressed across most populations, although many were found at lower levels in the 1-month-old cells, confirming the purity of the RPE cell cultures over time (Figure 2B). As these transcripts are associated with stages of RPE maturity, our data indicate that all subpopulations are of RPE lineage, potentially at various stages of differentiation and maturation.
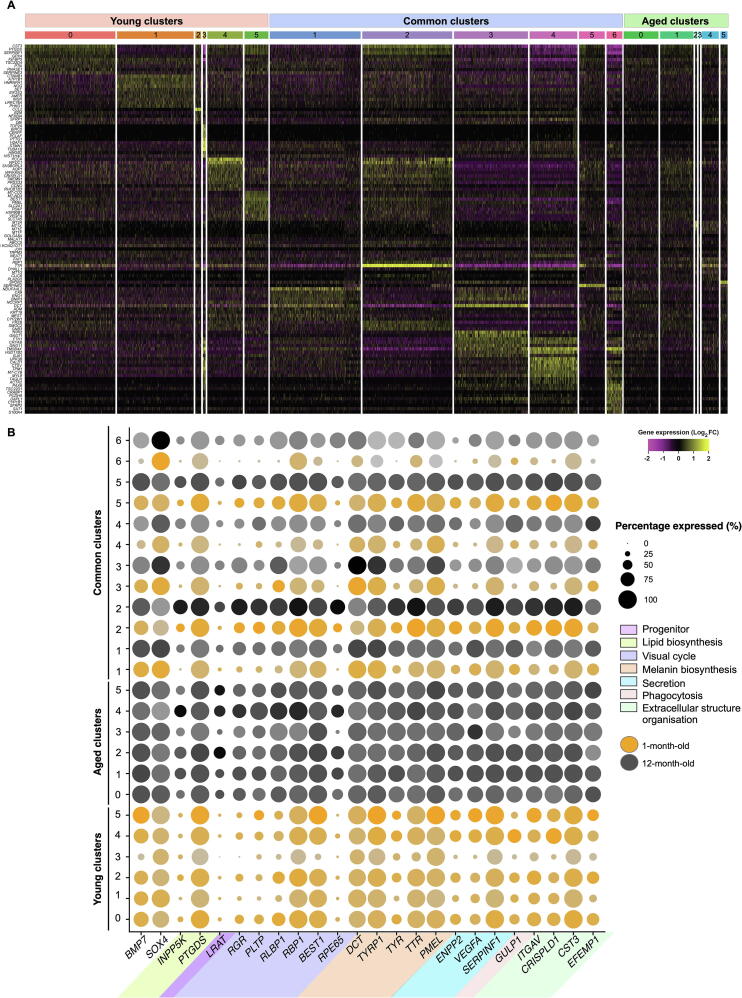
Figure 2.
Characterisation of hPSC-derived RPE populations
A. Heatmap showing the most conserved markers (gene symbols are indicated on the left side) in all individual cells in each of the 18 subpopulations (indicated on top, with colours matching those of subpopulations shown in Figure 1C). Gene expression levels were scaled and presented as average of Log2-transformed FC. B. Dotplot representation of single-cell expression profile from 1-month-old and 12-month-old cells for selected gene markers, representative of progenitor cells, or RPE with genes linked to RPE functions. Populations arising from 1-month-old cultures are represented in orange and those from 12-month-old cultures in black. Levels of gene expression per cell are shown with colour gradients, and frequencies of cells expressing the respective gene (percentage expressed) are shown with size of dots. RPE markers are coloured according to their cellular functions. FC, fold change.
Most cells share common transcriptomic profiles suggestive of maturing RPE cells
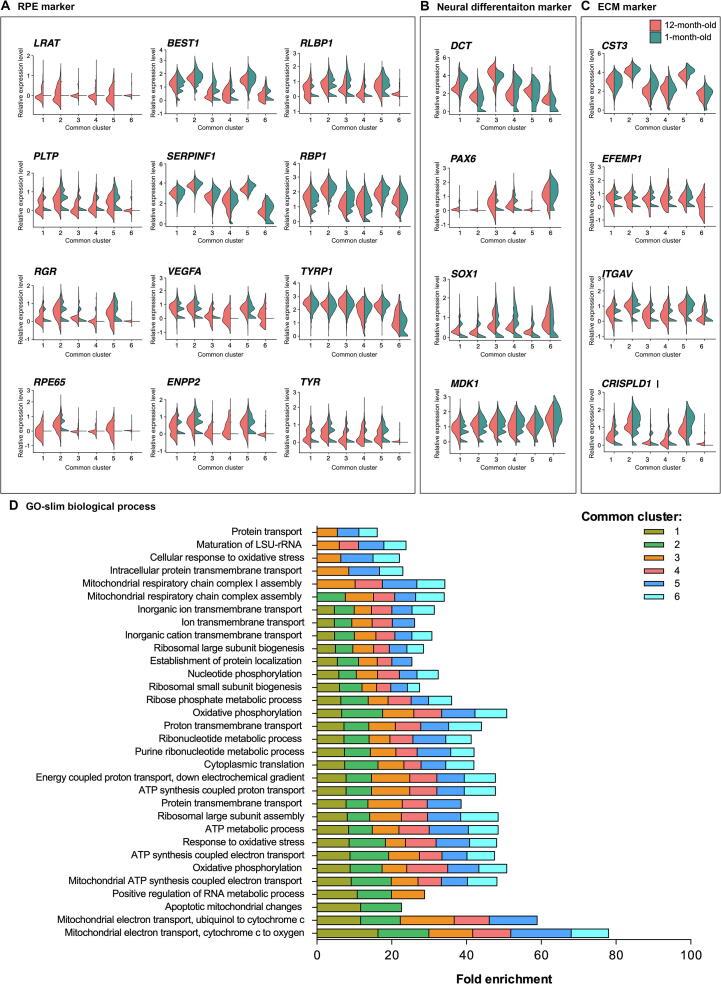
We then performed differential gene expression analysis and pathway enrichment analysis to characterise the molecular signature of these subpopulations. Of the cells examined, more than half (8484 cells) were clustered into six Common subpopulations that intersect the 1-month-old and 12-month-old cell cultures (Figure 1B and C), indicating a large shared transcriptional profile between the two conditions. Some commonalities and differences were observed between the Common samples arising from the 1-month-old and 12-month-old cultures. A range of RPE markers was observed as conserved between samples collected at both time points (Figure 2B). In particular, RPE markers associated with melanin biosynthesis (MITF, PMEL, TTR, TYR, TYRP1, and DCT), extracellular structure organisation (EFEMP1, CST3, CRISPLD1, ITGAV, and ITGB8), secretion (SERPINF1 and VEGFA), visual cycle (RPE65, BEST1, RBP1, RLBP1, PLTP, RGR, and LRAT), tight junctions (TJP1), phagocytic activity (GULP1), and lipid biosynthesis (PTGDS, CYP27A1, INPP5K, PLA2G16, and PLCE1) were conserved in all or some of the subpopulations (Table S3). Expression of some genes related to RPE maturity did not appear to differ between the 1-month-old and 12-month-old RPE cells. These include RBP1, TYRP1, and SERPINF1, demonstrating that some genes encoding proteins necessary for retinoid-cycle binding, melanin biosynthesis and secretory are expressed in early RPE development (Figure 2B). More heterogeneity was observed in the expression of RPE markers RGR, PLTP, RLBP1, BEST1, ENPP2, VEGFA, and TYR in the 1-month-old RPE cells relative to the 12-month-old RPE cells, in which expression of these genes was generally high and stable (Figures 1D and 2B). Expression of LRAT and RPE65 was predominantly observed in the 12-month-old RPE cells, with the exception of low expression of RPE65 in the 1-month-old sample for the Common 2 subpopulation. This suggests that the catabolic machinery converting all-trans-retinol into all-trans-retinyl ester (LRAT) and 11-cis-retinol (RPE65) for phototransduction is expressed at low levels in the 1-month-old RPE samples but becomes more comprehensive as cells age in culture (Figure 2C and D). Variations in the pattern of gene expression were observed between cells identified from the 1-month-old or 12-month-old cultures within each subpopulation (Figure 3A–C). Genes involved in neural differentiation, including DCT, PAX6, SOX1, and MDK, exhibited notable differences in expression between the 1-month-old and 12-month-old samples (Figure 3B). Similar patterns were also observed in genes involved in the extracellular matrix (ECM) formation and maintenance, including CST3, EFEMP1, ITGAV, and CRISPLD1 (Figure 3C), which were more heterogeneous in the 1-month-old samples, suggesting transitional changes in ECM markers during early RPE differentiation. Examples of genes characteristics of RPE, neural differentiation, and ECM are illustrated in Figure 3A–C, respectively.
Figure 3.
Expression patterns of selected conserved markers and GO pathway in the hPSC-derived RPE cells
Expression values are measured as normalised UMI counts. A. Violin plot of selected conserved markers in each Common subpopulation characteristic of the RPE. B. Violin plot of selected conserved markers in each Common subpopulation characteristic of the neural differentiation. C. Violin plot of selected conserved markers in each Common subpopulation characteristic of the ECM. The plots describe the distribution and relative expression of each transcript in each common subpopulation, with separation of cells belonging to the 1-month-old (blue) and 12-month-old (brown) cultures. D. PANTHER GO-slim (biological process) pathways associated with each of the Common subpopulations (Common 1–6; colour-coded) identified via over-representation analysis. Association is measured by fold enrichment, that is calculated from the number of genes observed, divided by the expected number of genes to be present by chance. UMI, unique molecular identifier; ECM, extracellular matrix.
The common subpopulation 1 (Common 1, 2282 cells) was characterised by 891 conserved markers identified (P < 0.74) (Table S3). The most highly conserved markers include NDUFA4L2 (a gene associated with the macula retina [18]), CA9 (zinc metalloenzyme gene), as well as DCT, PMEL, MITF, TYR, and TYRP1 (genes involved in pigment/melanin biosynthesis). This subpopulation also expressed genes involved in early retinal development including of the RPE and eye morphogenesis (SOX4, EFEMP1, BMP7, VIM, GJA1, and PTN) and in the retinoid cycle (RPE65 and RLBP1). In addition, 79 ribosomal genes (32 RPS and 47 RPL) and 11 mitochondrially-encoded genes were identified. Expression of ribosomal genes and mitochondrially-encoded genes has been correlated with development and maturation [19], including of the retina [20], [21]. This is supported by the GO analysis showing an overrepresentation of pathways involved in mitochondrial and ribosomal functions; protein biogenesis, transport, assembly, and function; as well as ATP biosynthesis and metabolism (Figure 3D; Table S3). Hence, together with the presence of RPE markers, the data indicate this subpopulation comprises a highly metabolically active maturing RPE phenotype.
The subpopulation Common 2 (2204 cells) identified 1077 conserved markers. The 15 most conserved markers in this subpopulation were all known RPE markers. Many of the expressed markers are involved in the generation of RPE cells or in maturation and homeostasis of these cells. For instance, cystatin C encoded by CST3 is abundantly produced by RPE cells [22] and its secretion diminishes with age [23]. DCT is expressed in the developing retina [15] and is important for melanin production and RPE homeostasis [24], [25]. Its downregulation is associated with mature native RPE [26]. This demonstrates that this subpopulation is a mature functional RPE population.
The subpopulation Common 3 (1811 cells) identified 1226 conserved markers. The most conserved markers in this subpopulation were mostly known to be expressed in the RPE (DCT, TTR, CST3, AQP1, FTH1 [27], and BEST1), with some markers, such as TFPI2, known to promote survival and maintenance of RPE cells [28]. The markers were similar to those of Common 2. Other markers identified are not necessarily RPE-specific, e.g., GNGT1 that encodes a protein found in rod outer segments, suggesting that cells are not yet fully committed. Together, these markers indicate cellular functions suggestive of functional and maturing RPE cells.
The subpopulation Common 4 (1170 cells) identified 1421 conserved markers. Many of the most conserved markers of this subpopulation are not specifically linked to the RPE. For instance, TMSB4X is linked to the cytoplasmic sequestering of NF-κB but has not yet been reported to be associated with molecular events in RPE cells. Many other genes are associated with the cytoskeleton, such as TAGLN, TNNC1, CALD1, MYLK, TPM1, ACTA2, and MYL9. On the other hand, there are many markers known to be expressed by the RPE cells, including TTR, BEST1, CST3, CSTV [29], CRYAB, SERPINF1, PMEL, VEGFA, RBP1, RLBP1, TYR, and TYRP1, supporting the RPE identity of this subpopulation. The presence of genes associated with early differentiation, such as IGFBP5 (downregulated relative to other clusters, as observed upon RPE differentiation [30]) and CRB2 [31], and genes involved in RPE polarity [32], is indicative of an early stage of RPE maturity and of a differentiating RPE population.
The subpopulation Common 5 (630 cells; 907 conserved markers) was characterised by expression of conserved RPE markers (SERPINE2 [33], SFRP5 [34]). Expression of many genes (AQP1, CST3, PTGDS, SERPINF1, BEST1, and SMOC2) was downregulated when compared to that in other populations. Expression patterns of other genes indicate an immaturity/ differentiation or proliferation of cells, such as GAP43, DAAM1, CD44 [35], and DUSP4. Together, these data describe a subpopulation comprised of cells in early differentiation to RPE.
The subpopulation Common 6 (387 cells; 1664 conserved markers) was characterised by markers associated with retinal cell types other than RPE [36] (such as SPP1, CPODXL, STAC2, and PCDH9) or found at low levels in the RPE (such as CPAMD8 and SFRP2 [37]). Only the marker CRABP1 was highly conserved with expression upregulated, whilst expression of other RPE markers (SERPINF1, BEST1, RLBP1, and RPE65) was downregulated. These data thus suggest a subpopulation of immature cells.
Finally, the analysis of the PANTHER GO slim biological processes conserved within the Common subpopulations identified pathways that are predominantly involved in mitochondrial, metabolic, and ribosomal processes, as well as purine biosynthesis, nucleotide metabolism, protein biogenesis, localisation, and transport (Figure 3D; Table S3). Altogether, these data demonstrate that the population common to cultures at both time points is heterogeneous, with subpopulations representing different stages of RPE cell differentiation.
The Young subpopulations are characterised by immature and differentiating cells
Around one third of all cells (5625 cells) were clustered into six Young subpopulations (Table S4). Common RPE markers were conserved within several Young subpopulations (Figure 2A and B; Table S4). These genes were associated with lipid biosynthesis (PTGDS), visual cycle (RGR, RLBP1, RBP1, and BEST1), melanin biosynthesis (DCT, TYRP1, TYR, TTR, and PMEL), phagocytic activity (GULP1), secretion (ENPP2, VEGFA, and SERPINF1), and extracellular structure organisation (ITGAV, CRISPLD1, CST3, EFEMP1, and ITGB8).
The subpopulation Young 0 (2219 cells; 20 conserved markers) comprises singleton cells that expressed SERPINF1, CST3, DCT, TSC22D4, IGFBP5, and RNASE1. Expression of most of these genes has been reported in the retina (Courtesy of Human Protein Atlas, www.proteinatlas.org) [36] and in the human RPE (IGFBP5 [30]). PANTHER GO biological process analysis indicates involvement of these genes in regulation of neuroblast proliferation, indicative of progenitor cells (Table S4).
The subpopulation Young 1 (1873 cells; 58 conserved markers) was characterised by the expression of CTNNB1, NOG, ATP1B1, GSTP1, CD63, and HNRNPH1. All these genes play fundamental roles in the homeostasis and functions of the RPE. ATP1B1 encodes an apical Na+/K+ ATPase, whose expression reduces with age and in age-related macular degeneration (AMD) [38]. Defects in CTNNB1 are linked to abnormalities in RPE development and pigmentation [39], whereas GSTP1 is a survival factor for RPE cells, whose expression increases as cells mature [40]; CD63 is a late endosome/exosome marker known to be released by RPE cells [41]; and HNRNPH1 levels are associated with improved survival of RPE cells in culture [42]. Hence, expression of these markers indicates functional RPE cells. It is interesting to note that known canonical RPE markers, such as SERPINF1, RLBP1, TTR, PMEL, and CRYAB, were expressed at lower levels in this subpopulation than in all other subpopulations. GO biological process analysis highlights that this population is highly metabolically active (Table S4). These data are suggestive of an earlier stage of maturation of the RPE cells.
The subpopulation Young 2 (144 cells; 16 conserved markers) was characterised by the specific upregulation of genes including CCL2, SFRP1, and B2M, downregulation of CTSV and TMSB4X, as well as expression of DCT, which is known to be expressed during RPE development [43], indicative of an immature RPE population.
The subpopulation Young 3 (67 cells; 369 conserved markers) was characterised by the expression of genes such as TOP2A, PCLAF, PTTG1, ANLN, MKI67, RRM2, TPX2, and PBK. Although all genes are found to be expressed in the retina [36], none are associated with a specific RPE signature. However, these genes are associated with cell proliferation (TOP2A, PCLAF, PTTG1, MKI67, RRM2, TPX2, and PBK) and cellular rearrangements (ANLN), which have been described as characteristics of immature RPE cells [44]. In particular, ANLN is reported to promote maturity of intercellular adhesions (tight junctions and adherens junctions) in epithelial cells [45]. TOP2A is associated with retinal development and proliferation [46], which, combined with expression of PCLAF, PTTG1, and MKI67, suggests a proliferating cell population. Low expression of RPE markers (RBP1, ENPP2, CRABP1, and HNRNPH1) further demonstrates the RPE identity of the developing retinal cell subpopulation. Altogether, this expression profile specifies an immature differentiating cell population.
The subpopulation Young 4 (871 cells; 49 conserved markers) was characterised by expression of genes that are not traditionally associated with the RPE identity. The expression of CRYAB (downregulated), CRX, FTH1, TFPI2 (known to promote survival and maintenance of RPE cells [28]), and DCT (expressed in the native RPE) suggests a differentiation to RPE, yet the presence of the photoreceptor-specific GNGT1 expression could also indicate an early differentiation step where cells are not yet fully committed.
Interestingly, the subpopulation Young 5 (584 cells; 99 conserved markers) displayed a signature comprising mitochondrial and ribosomal transcripts with 9 mitochondrial genes (MT-) and 14 ribosomal genes (RPS- or RPL-). These genes are ubiquitous and have not been specifically correlated to the retina or the RPE, however they are known to facilitate fundamental biological processes, including electron transfer, energy provision, ribosome biogenesis, and protein synthesis. PANTHER GO-slim biological process and GO biological process analyses confirmed that the conserved pathways within this subpopulation are mostly related to the mitochondrial energetic metabolism and nucleotide metabolism (Table S4). This subpopulation also expressed RPE markers, such as BEST1, VEGFA, ENPP2, TIMP3, and TYRP1, as well as genes involved in early retinal development including of the RPE and eye morphogenesis (SOX11, PMEL, EFEMP1, BMP7, VIM, GJA1, and PTN) [15], [47]. Hence the presence of high level of expression of mitochondrial genes and ribosomal genes in RPE is likely significative of highly active cells with high levels of protein synthesis, implying that this is a maturing RPE population.
Taken together, our results demonstrate that all Young subpopulations are immature cells developing to RPE cells.
The Aged subpopulations are characterised by higher maturity of RPE cells
Less than 10% of all cells (2334 cells) are clustered into the “Aged” category, which comprises six subpopulations (Table S5). All six identified “Aged” subpopulations were subjected to the same analysis as the “Young” and “Common” subpopulations. However, a statistical overrepresentation test returned no positive results for subpopulations Aged 0, 1, and 5, most likely owing to the low number of conserved genes identified. Only a few common RPE markers associated with lipid biosynthesis (INPP5K), visual cycle (RLBP1), melanin biosynthesis (TTR and DCT), secretion (SERPINF1 and VEGFA), and extracellular structure organisation (CST3 and CRISPLD1) were conserved in some of the “Aged” subpopulations (Figure 2; Table S5).
The subpopulation Aged 0 (851 cells; 6 conserved markers) is composed of singleton cells. These cells expressed CRISPLD1, PCCA, WFDC1, TTR, SH3BGRL3, and TMSB4X, but at lower average levels per cell than those in all other subpopulations. Some of these genes have been found to be expressed in the RPE (CRISPLD1, WFDC1 [48], TTR, and TMSB4X [43]) and are associated with late RPE development (CRISPLD1 and TTR [15]). Some other genes have a wider expression pattern (PCCA and SH3BGRL3) and encode for proteins involved in more universal cellular events. For instance, mitochondrial protein PCCA plays roles in death/survival, SH3BGRL3 is involved in oxidoreduction, whilst TMSB4X encodes proteins of the cytoskeleton. Likewise, the subpopulation Aged 1 (815 cells; 12 conserved markers) presented more cells expressing HSD17B2, TPM1, MYL9, NDUFA4L2, BNIP3, CALD1, TTR, DCT, MT-CYB, FTH1, CRYAB, and TMSB4X but at lower average levels per cell than in all other subpopulations. Nine out of the twelve genes are known to be expressed in the RPE (TTR, TMSB4X [43], CALD1 [49], DCT [43], HSD17B2 [50], NDUFA4L2 [15], BNIP3 [27], FTH1 [27], and CRYAB). Some of these markers are associated with late RPE development (TTR [15]), whilst others are associated with a geographic localisation of cells within the retina (HSD17B2 [51] and NDUFA4L2 [18]). BNIP3 and NDUFA4L2 encode for mitochondrial proteins playing roles in death/survival and electron transport, respectively, whilst TPM1, MYL9, TMSB4X, and CALD1 encode proteins of the cytoskeleton. A similar pattern of expression was observed in the subpopulation Aged 5 (170 cells; 6 conserved markers) with PCCA, WFDC1, SERPINE2, TMSB4X, and TTR being expressed in more cells with lower average levels than all other subpopulations. In addition, SPON2, which encodes ECM proteins important for cell adhesion, was expressed in more cells but at higher levels. The close similarities of these three Aged subpopulations point to a late RPE phenotype, with higher levels of maturation, towards regionalisation of cells.
Similarly, the subpopulations Aged 2 (46 cells; 22 conserved markers), Aged 3 (32 cells; 121 conserved markers), and Aged 4 (420 cells; 34 conserved markers) showed close similarities in terms of gene expression profiles. The presence of known RPE markers, such as DCT (Aged 2, downregulated), CALD1 (Aged 2, downregulated), TTR (Aged 3, downregulated; Aged 4), SOX9 (Aged 3, downregulated), RBP1 (Aged 3, downregulated; Aged 4), SERPINF1 (Aged 3, downregulated), VEGFA (Aged 4, downregulated), TMSB4X (downregulated in all three subpopulations), and CRYAB (downregulated in Aged 2 and Aged 3) confirms their RPE identity.
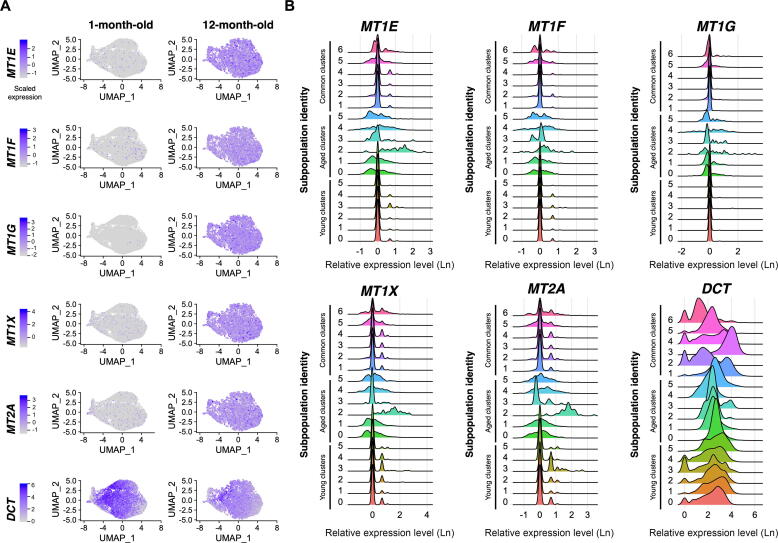
PANTHER GO-slim biological process (Aged 2) and PANTHER GO biological process analyses (Aged 2, 3, and 4) were performed. Subpopulations Aged 2 and 4 were similar, with high significance in pathways associated with response to metal ions (particularly cadmium, copper, iron, and zinc); response to stress, chemicals, and toxins; and neural crest fate commitment. Among them, neural crest fate commitment is the most significantly identified biological process in Aged 3, with a 92.9-fold enrichment. Interestingly, GO analysis indicates that nuclear genes encoding metallothioneins involved in metal binding (MT1E, MT1F, MT1G, DCT, MT2A), as well as in oxidoreduction (DCT), were significantly differentially expressed between the “Aged” subpopulations compared to all others, with an overall increased expression per cell as cells age (Table S5). Further analysis of the metallothioneins MT1E, MT1F, MT1G, MT1X, and MT2A across all subpopulations at the two time points confirms a large increase in their mRNA expression in the 12-month-old culture when compared to the 1-month-old cells (Figure 4A and B). On the other hand, expression of DCT was reduced in the 12-month-old culture compared to the 1-month-old cells, which is consistent with a mature RPE profile [26] (Figure 4A and B). Altogether, these data demonstrate that the RPE cells of these “Aged” subpopulations are increasing their handling of metals and antioxidant abilities, which likely reflects a further maturation of the RPE cells.
Figure 4.
Aged RPE subpopulations likely increase their handling of metals and antioxidant abilities
A. Feature plots of expression profiles of DCT and genes encoding key metallothioneins (including MT1E, MT1F, MT1G, MT1X, and MT2A) across the 1-month-old and 12-month-old cells. The intensity of gene expression is indicated by colour gradient. B. Ridge plots of expression profiles (measured in natural log-normalised UMI counts) of DCT and genes encoding key metallothioneins and across all subpopulations. Different colours are used for each subpopulation for ease of reading.
RPE cells collected from different time points share common developmental trajectories
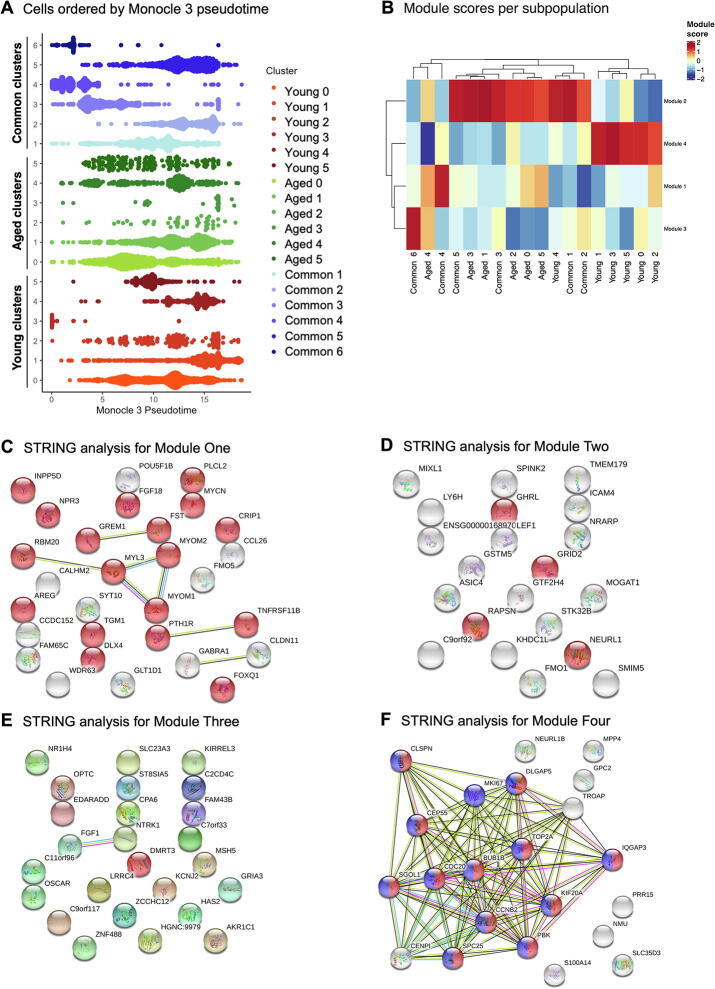
We investigated the pseudo-temporal transition of 1-month-old cells to 12-month-old cells using trajectory analysis methods to identify trajectories originating from proliferative cells in the subpopulation Young 3. Monocle 3 [52] identified a complex, branched development trajectory that included cells from both time points (Figure 5A and B). Interestingly, the pseudo-temporal ordering of cells across the trajectory did not correspond to time points (Figure S3A). To determine the nature of the trajectories, we used Moran’s I test to identify 158 genes that were significantly associated with pseudotime (Table S6). These genes formed four co-expression modules that were further analysed with STRING (Table S6; Figure S3B–F). As illustrated in Figure S3, Module 1 revealed genes involved in development (red) and Module 4 revealed genes involved in cell cycle (blue) and mitosis (red). The other two modules did not show clear biological process associations but genes were associated with biological process for neural development. The differential expression of the residual gene trajectory for the four modules confirms differences among various clusters. In particular, all Young subpopulations except Young 4 showed the highest expression levels of genes of the proliferative Module 4 (Figure S3B), confirming their immature proliferative nature.
Figure 5.
Only a few cells retain a proliferative profile
Highly dimensional expression data were reduced into two dimensions using UMAP. Plot axes (UMAP1 and UMAP2) represent coordinates in the resulting 2D space. A. UMAP of single cell expression profile split by conditions (1-month-old and 12-month-old). B. UMAP of single cell expression profile for trajectory analysis using Monocle 3. C. UMAP of single cell expression profile for markers associated with proliferation (MKI67, TOP2A, PCLAF, RRM2, TPX2, and PTTG1) across all cells. Expression levels measured as Log10 normalised UMI counts are represented by colour intensity. Lines represent differentiation trajectories as calculated by Monocle3. D. Pseudotime analysis of early retinal markers (PAX6 and RAX) and RPE genes (PMEL, RLBP1, RGR, TYR, RBP1, and RPE65) across the 1-month-old and 12-month-old cells. Expression levels measured as Log10 normalised UMI counts are represented by colour intensity. Lines represent differentiation trajectories as calculated by Monocle3. E. Violin plots of normalised UMI counts of early retinal markers and RPE genes across all subpopulations showing variations in expression across cluster groups.
Differentiated RPE cells are post-mitotic and fully committed to the RPE lineage, in large part due to their cell–cell contact inhibition [53], [54]. The expression of the proliferation marker MKI67 and of other genes associated with proliferation (TOP2A, TPX2, PTTG1, PCLAF, and RRM2) was examined to assess whether some progenitor populations remain with proliferative potentials, and whether these vary over time in culture. Our data indicate that only a small portion of cells remained proliferative over time in culture. In particular, MKI67 was almost uniquely expressed in the immature subpopulation Young 3, together with other cell proliferation markers (TOP2A, TPX2, PTTG1, PCLAF, and RRM2; Figure 5C; Table S4). Very few cells of the 12-month-old population were found to express markers associated with proliferation (Figure 5C). This confirms that only a small subpopulation of progenitor/immature cells present early in culture is proliferative and these cells disappear with time in culture.
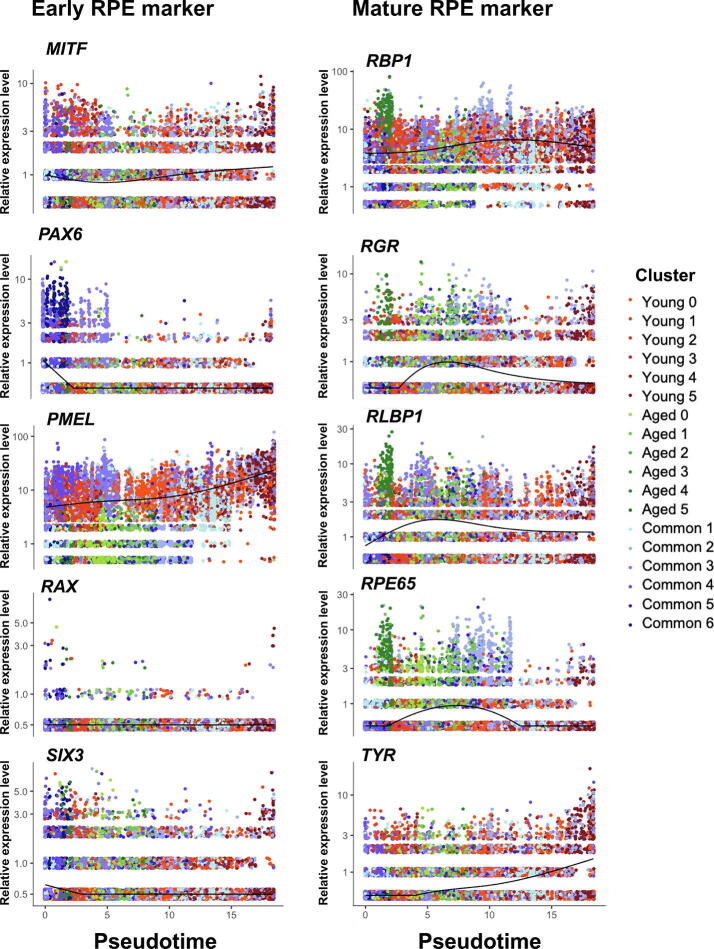
Pseudotime analyses of early retinal markers (PAX6 and RAX) and RPE genes (PMEL, RLPB1, RGR, TYR, RBP1, and RPE65) were performed in all subpopulations to measure gene expression trajectories (Figure 5D and E; Figure S4). The early retinal markers PAX and RAX, were expressed in similar manners across subpopulations, with minor variations between samples, with the exception of the subpopulation Aged 3, which showed higher levels of PAX6 and absence of expression of RAX (Figure 5D and E; Figure S4). The pigmentation marker TYR was consistently expressed across all populations, whilst expression of PMEL was downregulated in all aged subpopulations and in subpopulations Common 5 and 6 (Figure 5D and E; Figure S4). Similarly, comparable expression levels of RGR, RLBP1, and RBP1 were observed across all subpopulations, with the exception of subpopulations Aged 3 and 4, which showed higher levels of expression (Figure 5D and E; Figure S4). These variations in gene expression are likely indicative of RPE cell maturation in culture.
RPE subpopulations contribute to paracrine signalling
RPE cells can modulate immune responses and differentiation of other retinal cell types, in part via paracrine signalling [1]. Such modulation can be detected in vitro through examining expression of specific secretion factors and receptors known to play roles in immune or developmental events. For instance, in the retina, the chemokine CCL2 is implicated in monocyte infiltration following damage [55] and its secretion by RPE cells contributes to the regulation of the immunological response to inflammation [56]. CCL2 was found to be faintly expressed in cells at both time points (Figure 6A). CCL2 was identified as a marker of the immature subpopulation Young 2 (Table S2) and was also expressed, albeit at lower levels, across other “Aged” and “Common” subpopulations (Figure 6A). Unsurprisingly, CCR2, which encodes the cognate receptor of CCL2, was not expressed in any subpopulations, as it is most likely that CCL2 targets monocytes but not endogenous cells of the retina (data not shown). CCL2 was more uniformly expressed in the 12-month-old sample, suggesting more homogeneity in its expression as cells mature.
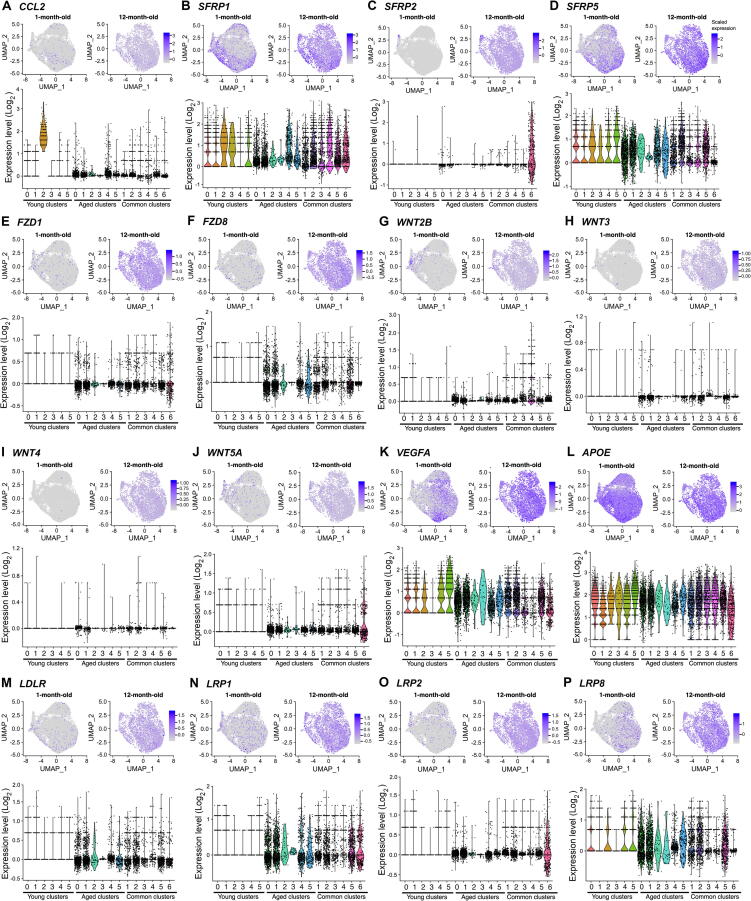
Figure 6.
RPE subpopulations contribute to paracrine signalling
UMAP of single-cell expression profile of markers for 1-month-old and 12-month-old cultures, with associated violin plots, representing Log2 UMI counts across all populations for CCL2 (A); SFRPs, FZDs, and WNTS (B); VEGFA (C); APOE, LDLR, and LRPs (D). In the UMAPs, the intensity of gene expression is indicated by colour gradient. UMAP of single-cell expression profile of markers for 1-month-old and 12-month-old cultures, with associated violin plots across all populations for CCL2 (A), SFRP1 (B), SFRP2 (C), SFRP5 (D), FZD1 (E), FZD8 (F), WNT2B (G), WNT3 (H), WNT4 (I), WNT5A (J), VEGFA (K), APOE (L), LDLR (M), LRP1 (N), LRP2 (O), LRP8 (P). In the UMAP plots, the intensity of gene expression is indicated by colour gradient. In the violin plots, different colours are used for each subpopulation for ease of reading.
Secreted frizzled-related proteins (SFRPs) are secreted ligands for receptors Frizzled (FZD1–10), which are also receptors for WNT proteins (WNTs). Therefore, SFRPs are considered WNT antagonists and can modulate WNT signalling. Cells in culture from both time points expressed genes involved in the WNT signalling pathways, in particular SFRP 1, 2, and 5; FZD 1 and 8; as well as WNT 2B, 3, 4, and 5A, with more expression homogeneity in the 12-month-old samples for many of them (Figure 6B–J). SFRP1 was expressed across all subpopulations with varied levels and was identified as a marker of the immature subpopulation Young 2 (Figure 6B; Table S4). SFRP2 was identified as a marker of the immature subpopulation Common 6 where it was mainly expressed, whilst SFRP5 was identified as a marker of the immature subpopulation Common 5 and expressed across most subpopulations (Figure 6C and D; Table S3). It is interesting to note that expression of SFRP1 and SFRP5 was both downregulated in the subpopulation Aged 3 (relative to other “Aged” subpopulations), reflective of a dynamic expression pattern of SFRP genes by RPE cells in culture (Figure 6C and D; Table S3). Interestingly, 1-month-old RPE cells showed low expression levels of FZD1, FZD8, WNT2B, WNT3, WNT4, and WNT5A with expression of all other FZD and WNT genes undetectable (Figure 6E–J). The expression of all detectible SFRP, FZD, and WNT genes was higher in the 12-month-old RPE cells (Figure 6B–J). This dynamic profile of expression of molecules involved in WNT signalling indicates that RPE cells can signal via the WNT pathway. This correlates with the known role of WNT signalling in retinal development including the RPE [34], [57].
Pigment epithelium-derived factor (PEDF) is another factor key to various retinal cell differentiation, maturation, and survival, including RPE and photoreceptors [58]. It is encoded by SERPINF1, which is highly expressed by RPE cells and was found as a marker of subpopulations within the Young, Common, and Aged subpopulations (Figure 1D; Tables S3–S5). However, the PNPLA2, the gene encoding PEDF receptor, was not found to be expressed in the RPE cells, indicating that PEDF secreted from RPE largely acts in a paracrine manner. Likewise, VEGFA, the gene encoding pleiotropic factor VEGFα [59], was also identified as a marker of subpopulations within the three populations, but no expression of the gene encoding VEGF receptor (VEGFR1–3) was detected in the samples, again indicating that the RPE-secreted VEGF likely acts on neighbouring cells to maintain homeostasis (Figure 6K; Tables S3–S5). These examples of paracrine molecules secreted by RPE subpopulations illustrate the important role played by RPE cells in the shaping of a developing retina. These cells may release bioactive factors to regulate events such as cell fate, differentiation, polarity, and maturity, and contribute to the regulation of the immune environment of the retina.
APOE is a conserved marker of various RPE subpopulations
Apolipoproteins E (APOE) are proteins involved in lipid metabolism including cholesterol and are also regulated with complement activation in the RPE [60]. APOEs interact with the low-density lipoprotein (LDL) receptors (LDLRs) and very-low-density lipoprotein receptors (VLDLRs). APOE was highly expressed in cells across both time points in culture (Figure 6L). APOE was identified as a conserved marker of the subpopulations Young 1 and 5, as well as all Common subpopulations (Figure 6L; Tables S3 and S4). The levels of APOE expression per cell was however different between subpopulations, with an upregulation in Young 5, and a downregulation in Young 1 as well as in Common 1, 2, 3, and 6 (Figure 6L). Interestingly, within the subpopulation Common 4, cells arising from the 1-month-old culture expressed lower levels of APOE than cells arising from 1-month-old culture of all other Common subpopulations, whereas the opposite was observed for cells from 12-month-old culture (Table S3). The opposite pattern was observed in the subpopulation Common 5 (Table S3). Expression of LDLR, LRP1, LRP2, and LRP8, the genes encoding APOE receptors, was detected in both cell cultures, with increased levels in the 12-month-old samples when compared to the 1-month-old culture (Figure 6M–P). Interestingly, expression of these receptor genes was basically absent from all Young subpopulations, but present at higher levels in the Aged subpopulations (Figure 6M–P). These data illustrate that the expression of APOE and associated receptor genes is dynamic within RPE subpopulations and with time in culture.
The complement pathway is not modulated with time in culture
RPE cells express many complement components in various retinal diseases, inflammation, and/or ageing [61]. C1s, C1r, and C1QBP, the genes encoding complement regulators that form the C1 complex, were conserved markers in a few Young and Common subpopulations (Tables S3 and S4). Other genes associated with the complement response (such as CFH, CFB, CFHR1, CFHR3, and C3) were not identified as markers of any subpopulation (Tables S3–S5). This suggests that the complement components are not modulated with time in culture.
RPE cells do not undergo epithelial mesenchymal transition
It is interesting to note that no markers of epithelial mesenchymal transition (such as SNAI1, SNAI2, ZEB1, TWIST1, and GSC) were characterised in any examined subpopulation (Tables S3–S5). These data demonstrate the stability of the cell culture over time, with no evidence of a transition to a mesenchymal phenotype.
RPE subpopulations express native RPE markers with different patterns
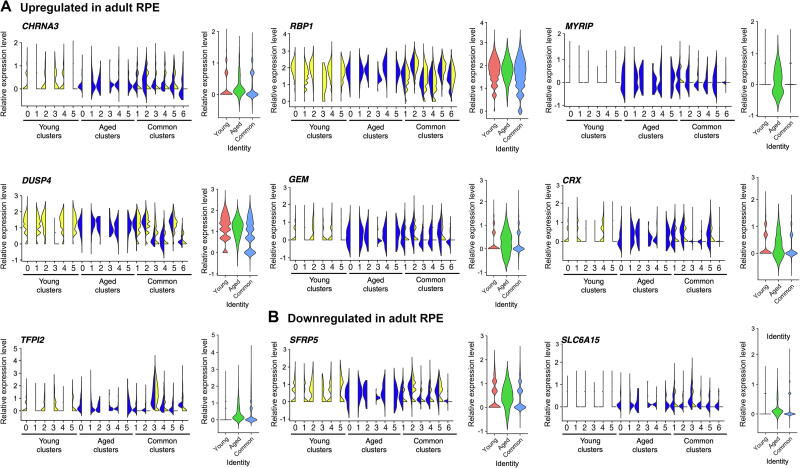
To assess correspondence of the RPE cells with native counterparts, we compared the hPSC-derived RPE signature to that of foetal native RPE cells, which was examined using scRNA-seq previously [15]. As observed in foetal native tissue, most subpopulations of hPSC-derived RPE cells expressed SERPINF1, BEST1, TYR, TTR, and RPE65, and the more immature subpopulations also expressed MKI67 and DCT (Figure 7A and B). Genes commonly expressed in native RPE cells were enriched in 12-month-old subpopulations and in Aged subpopulations (such as RPE65, LRAT, PLTP, RGR, RLBP1, LRAT, INPP5K, ITGB8, EFEMP1, ITGAV, GULP1, and VEGFA), whilst other genes, including TYRP1, were found at similar expression levels in 1-month-old and 12-month-old cultures, as well as in Young, Common, and Aged populations (Figure 7C–H). In addition, expression of some genes, such as PMEL, PTGDS, CST3, and CRISPLD1, was significantly lower in 12-month-old cultures than that in 1-month-old cultures (Figure 7D–F). Some common RPE genes are expressed in the adult native RPE at much higher levels than in foetal RPE [26]. These include TTR, RPE65, BEST1, CHRNA3, RBP1, MYRIP, TFPI2, PTGDS, SERPINF1, DUSP4, GEM, and CRX. Similarly, downregulation of DCT, SFRP5, TYRP1, and SLC6A15 is associated with mature native RPE [26]. We thus compared the expression profiles of these genes in the RPE cultures over time, in order to assess the maturity of the cultured cells [26] (Figure 8). CHRNA3, MYRIP, GEM, CRX, and TFPI2 — whose expression is upregulated in the adult native RPE— were more highly expressed in the Aged population (Figure 8A). However, PTGDS, RBP1, and DUSP4, which are genes also highly expressed in the adult RPE, were not found at higher levels in the Aged or 12-month-old cultures than in the Young population or 1-month-old culture (Figures 7E and 8A). Expression of DCT, PMEL, TYRP1, SFRP5, and SLC6A15 was generally downregulated in the Aged subpopulations and in the 12-month-old Common subpopulations, when compared to the 1-month-old Common subpopulations (Figures 7B and D; 8B).
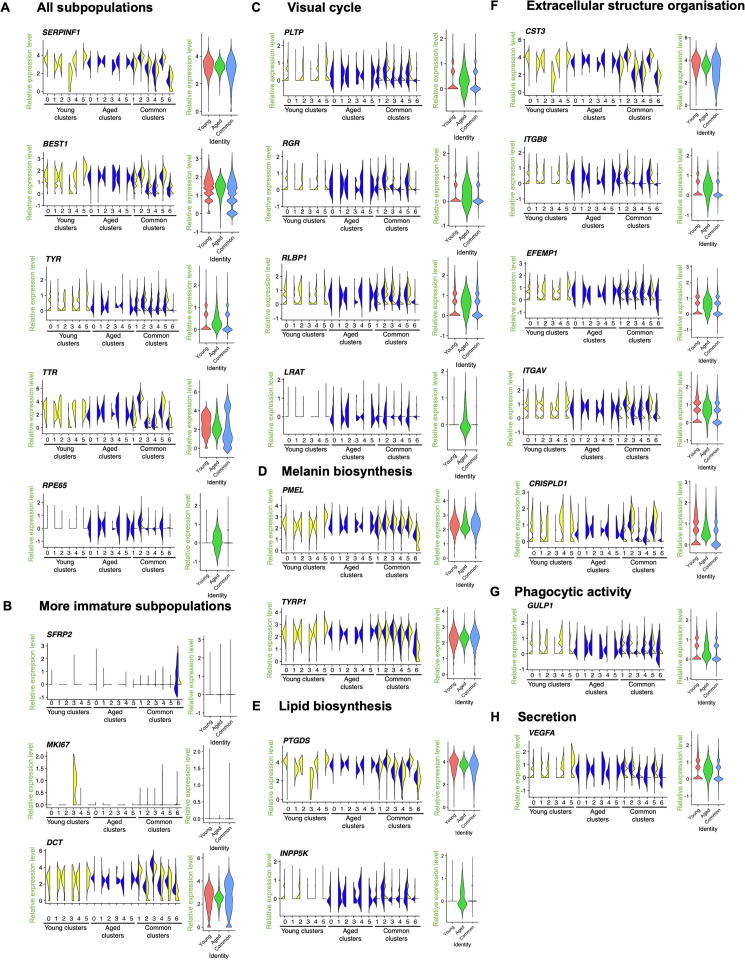
Figure 7.
hPSC-derived RPE subpopulations express native RPE markers with different patterns
Violin plots of selected markers representative of native RPE cells (obtained from [28]) across all subpopulations and in the three main populations “Young”, “Aged”, “Common”, represented in different colours. Subpopulations arising from the 1-month-old culture are indicated in yellow; subpopulations arising from the 12-monht-old culture are indicated in blue. A. Genes found in all subpopulations (SERPINF1, BEST1, TYR, TTR, and RPE65). B. Genes found in more immature subpopulations (SFRP2, MKI67, and DCT). C. Genes with varied expression associated with visual cycle (PLTP, RGR, RLBP1, and LRAT). D. Genes with varied expression associated with melanin biosynthesis (PMEL and TYRP1). E. Genes with varied expression associated with lipid biosynthesis (PTGDS and INPP5K). F. Genes with varied expression associated with extracellular structure organisation (CST3, ITGB8, EFEMP1, ITGAV, and CRISPLD1). G. Genes with varied expression associated with phagocytic activity (GULP1). H. Genes with varied expression associated with secretion (VEGFA). The plots describe the distribution and relative expression of each gene in the subpopulations, measured as normalised UMI counts.
Figure 8.
Some hPSC-derived RPE subpopulations acquire a gene expression profile closer to adult native RPE with time in culture
Violin plots of selected markers representative of native adult RPE cells (obtained from [28]) across all subpopulations and in the three main populations “Young”, “Aged”, “Common”, represented in different colours. Subpopulations arising from the 1-month-old culture are indicated in yellow; subpopulations arising from the 12-monht-old culture are indicated in blue. A. Genes with upregulated expression in adult RPE (CHRNA3, RBP1, MYRIP, DUSP4, GEM, CRX, and TFPI2). B. Genes with downregulated expression in adult RPE (SFRP5 and SLC6A15). The plots describe the distribution and relative expression of each gene in the subpopulations, measured as normalised UMI counts.
Discussion
Here, we provide a dynamic profile of the transcriptome of hPSC-derived RPE cells over 12 months. Our data confirm expression of marker genes of RPE homeostasis and functions in hESC-derived RPE [15], [26], [62], [63] and provide novel information on the timing of expression of these markers. At both early and late time points, we observed that hPSC-derived RPE cells expressed genes associated with lipid metabolism, secretion, visual cycle, melanin synthesis, phagocytic activity, metal binding, and oxidoreductase activity. Based on expression pattern of genes associated with RPE maturity levels and on PANTHER GO analyses, the 18 subpopulations identified regroup into populations from immature and progenitor cells, to maturing RPE cells, and functionally mature RPE cells (based on genes involved in RPE functions). Some subpopulations comprised highly metabolically active cells. The pseudo-temporal analysis could not identify trajectories matching time points, which is likely due to the experimental design of including two time points only. Beyond the scope of this study but of interest, including intermediate culture time points could provide more information on the pseudo-temporal ordering of cells alongside the course of culture.
An essential function of the RPE is photoprotection of the retina, which is accomplished through different mechanisms. These include absorbing radiation, binding and sequestering redox-active metals such as iron, as well as scavenging free radicals and reactive oxygen species [64]. Metallothioneins are metal-binding proteins that are protective against oxidative stress. Compared to the 12-month-old RPE cells, the 1-month-old RPE cells express fewer transcripts for the metallothioneins MT1E, MT1F, MT1G, MT2A, and MT1X, as well as higher levels of DCT. These data suggest a variation in the handling of metals and in the antioxidant abilities of RPE cells, which could be reflective of either a necessity to handle more oxidative stress in an in vitro ageing environment or a maturation of RPE cells towards a more mature and protective phenotype [64]. The assessment of variations in the expression of other genes between the two time points indicates a maturation profile of cells rather than an increased stress. Indeed, DCT is known to be expressed in human retinal progenitor cells [15] and its expression is regulated by the early RPE marker MITF. It is thus not surprising that as RPE cells mature, MITF expression reduces [65] and subsequently DCT expression reduces, as is observed in human foetal retina [15]. Similarly, CTNNB1 regulates MITF and OTX2 expression and subsequently RPE differentiation [39]. Finally, SOX11 is known to be expressed in early retinal progenitor and early in differentiating RPE cells [15], [47], hence its downregulation as cell culture ages further supports a maturation of RPE cells in culture. The subpopulations Young 0 and Young 1 are characterised by the expression of CST3, which encodes the cysteine proteinase inhibitor cystatin. Interestingly, this protein is known to decrease in native RPE cells with ageing [23]. Its presence in the cell population further strengthens the implication of a maturing RPE population over time. The high expression levels of mitochondrial and ribosomal genes in some cell populations likely indicate that cells are metabolically and transcriptionally active, necessitating energy and ribosomal activities for protein synthesis, respectively. It could also suggest that ribosomes potentially contribute to extra-ribosomal functions in the RPE cells, such as cell development and maturation [19], as already reported for melanocyte development [66], retinal development [20], [21], and retinal degeneration [67]. Of note, the absence of markers of epithelial mesenchymal transition highlights the robustness and ability of RPE cells to maintain their phenotype in vitro despite of prolonged culture time.
The expression analysis of ligands and receptors involved in retinal development and homeostasis demonstrates a dynamic profile of gene expression of hPSC-derived RPE cells. This highlights the importance of the RPE in the development and homeostasis of the whole retina. For instance, WNT signalling is important to the early stages of RPE differentiation [39], [68]. Some SFRPs and WNTs are expressed in RPE cell subpopulations and the genes encoding FZD receptors are very lowly expressed in most subpopulations. This suggests that the ligand expression could be directed to paracrine signalling, playing roles in the development and homeostasis of other neighbouring retinal cells. For instance, SFRP 1, 2, and 5, found to be expressed in the hPSC-derived RPE cells, are associated with retinal development [34], [57], and known to promote the differentiation of retinal ganglion cells and photoreceptors [69], and axon guidance [70]. Similarly, high expression levels of PEDF/SERPINF1 and of VEGF were found across many subpopulations, yet genes encoding their respective receptors, PNPLA2 and VEGFR, were not expressed in the cells, suggestive of a paracrine signalling mechanism between RPE and neighbouring cells as well. This is in accordance with the role of these growth factors in the biology and survival of retinal cells, including photoreceptors [58], [71] and retinal ganglion cells [72], or beyond the retina in neighbouring endothelial cells. Interestingly, APOE was also expressed in cells across both time points in culture, with variations observed between subpopulations. In the RPE, APOE is involved in lipid metabolism including cholesterol and drusen content [60]. APOE also plays a striking role in melanogenesis, regulating the formation of functional premelanosome protein (PMEL) amyloid fibrils in RPE cells [73]. Hence, the variations observed in its expression levels and that of its receptors could possibly be indicators of melanogenesis within RPE subpopulations. Altogether, the expression analysis of genes encoding ligands and receptors in RPE cells also hints at the possible value of co-culturing RPE cells with retinal organoid cultures to further support retinal differentiation and cell maturation, and to improve the in vitro modelling of retinal biology.
Our analyses also reveal that cells in culture can develop a transcriptomic profile more closely related to the adult native RPE with higher expression levels of some RPE genes and lower expression levels of others, as observed in their native counterpart. Altogether, these data thus strongly suggest that as hPSC-derived RPE cells mature with time in culture, they acquire characteristics more closely resembling those of an adult RPE profile.
Conclusion
The novel insight into the underlying genetic architecture of hPSC-derived RPE cells at short and long durations in culture conditions reveals a gradual differentiation and maturation process, as well as a stable RPE phenotype over time. Most cells with a clear RPE signature are found in the Common subpopulations, indicating that RPE cells are present from an early time point in culture and maintain this identity with time. The clustering analysis also reveals that whilst some subpopulations express more genes associated with retinal and RPE biology, other RPE subpopulations demonstrate increased expression in mitochondrial and/or ribosomal genes. Altogether, these data suggest that hPSC-derived RPE cells develop their characteristic signatures early during the differentiation process and continue to mature over time in culture. Our analysis also warrants the use of hPSC-derived RPE cells for modelling of RPE biology at early and later differentiation timings.
Materials and methods
Cell culture and differentiation of hESCs to RPE cells
The hESC line H9 (Wicell) was maintained on vitronectin-coated plates using StemFlex (Catalogue No. A3349401, ThermoFisher Scientific, Waltham, MA), with medium changed every second day [74]. Cells were differentiated into RPE cells as previously described [3] with the following modifications. Briefly, hESCs were maintained in culture until reaching 70%–80% confluency, at which stage StemFlex was replaced with Complete E6 (Catalogue No. 05946, Stem Cell Technologies, Vancouver, Canada) supplemented with N2 (Catalogue No. 17502048, ThermoFisher Scientific) to induce retinal differentiation, with media changes 3 times/week for 33 days. On Day 33, medium was replaced with RPEM containing α-MEM (Catalogue No. 12571071, ThermoFisher Scientific), 5% foetal bovine serum (Catalogue No. 26140079, Thermo-Fisher Scientific), non-essential amino acids (Catalogue No. 11140050, ThermoFisher Scientific), penicillin- streptomycin- glutamine (Catalogue No. 10378016, ThermoFisher Scientific), N1 (Catalogue No. N6530, Sigma-Aldrich, St Louis, MO), and taurine-hydrocortisone-triiodo-thyronin (in-house) to promote RPE differentiation, with medium changed every second day. Cells were cultured for 32 days, a time point at which maximal pigmentation is routinely observed. Cells were harvested with an 8-min exposure to 0.25% trypsin-EDTA (Catalogue No. 25200056, ThermoFisher Scientific) and inactivated with RPEM. Non-RPE contaminants (visible as unpigmented cells) were manually removed from the culture, which begin shedding off the culture plate after ~2 min. Cells were seeded at a density of 75,000 cells/cm2 onto growth factor-reduced Matrigel-coated tissue culture plates (Corning Matrigel hESC-qualified Matrix, Catalogue No. 354277, In vitro Technologies, Melbourne, Australia). Media was changed every second day, with the first sample of cells harvested after 30 days (D30) and the second sample harvested on day 367 (D367) for scRNA-seq analysis (Figure 1A).
RPE cell harvest and single-cell preparation
RPE cells were dissociated to single cells using 0.25% trypsin-EDTA for 8 min and inactivated with RPEM. Cells were centrifuged at 300 g for 1 min to pellet and resuspended in a small volume of RPEM containing 0.1% v/v propidium iodide (PI, Sigma-Aldrich) to exclude non-viable cells. Single cell suspensions were passed through a 35-µm filter prior to sorting. A minimum of 60,000 live cells (PI-negative) were sorted on a BD FACSAria IIu (100 µm, 20 psi; BD-Biosciences, San Jose, CA) into culture medium. Cells were centrifuged at 300 g for 5 min and resuspended in PBS containing 0.04% BSA to a concentration of ~800–1000 cells/µl. Approximately 17,400 cells were loaded onto a 10X chip for a target recovery of 10,000 cells. Cultures at the two time points were captured separately to prepare two separate 10X reactions.
Generation of single-cell gel beads in emulsion and sequencing libraries
To generate single-cell gel beads in emulsion, single cell suspensions were loaded onto 10X Genomics Single Cell 3′ Chips together with the reverse transcription master mix following the manufacturer's protocol for the Chromium Single Cell 3′ v2 Library (PN-120233, 10X Genomics, Pleasanton, CA). For each sample, sequencing libraries were generated with unique sample indices (SI), assessed by gel electrophoresis (Agilent D1000 ScreenTape Assay, Santa Clara, CA) and quantified with qPCR (Illumina KAPA Library Quantification Kit, Roche, Pleasanton, CA). Following pooling and normalisation to 4 nM, libraries were denatured and diluted to 1.6 pM for loading onto the sequencer. Libraries were sequenced on an Illumina NextSeq 500 (NextSeq Control Software v2.2.0/Real Time Analysis v2.4.11) using NextSeq 500/550 High Output Kit v2.5 (150 Cycles) (Catalogue No. 20024907, Illumina, San Diego, CA) as follows: 26 bp (Read 1), 8 bp (i7 Index), and 98 bp (Read 2).
Preprocessing, mapping, and quantification of scRNA-seq data
We used the cellranger mkfastq and cellranger count pipelines from the Cell Ranger Single Cell Software Suite (version 3.0.2) by 10x Genomics (http://10xgenomics.com) for initial quality control, sample demultiplexing, mapping, and quantification of raw sequencing data. The cellranger count pipeline was run with the following argument: “--expect-cells = 10,000”, and reads were mapped to the Homo sapiens reference genome (GRCh38, Annotation: Gencode v29). Filtered count matrices were then used for downstream analyses in R.
Quality control and normalisation
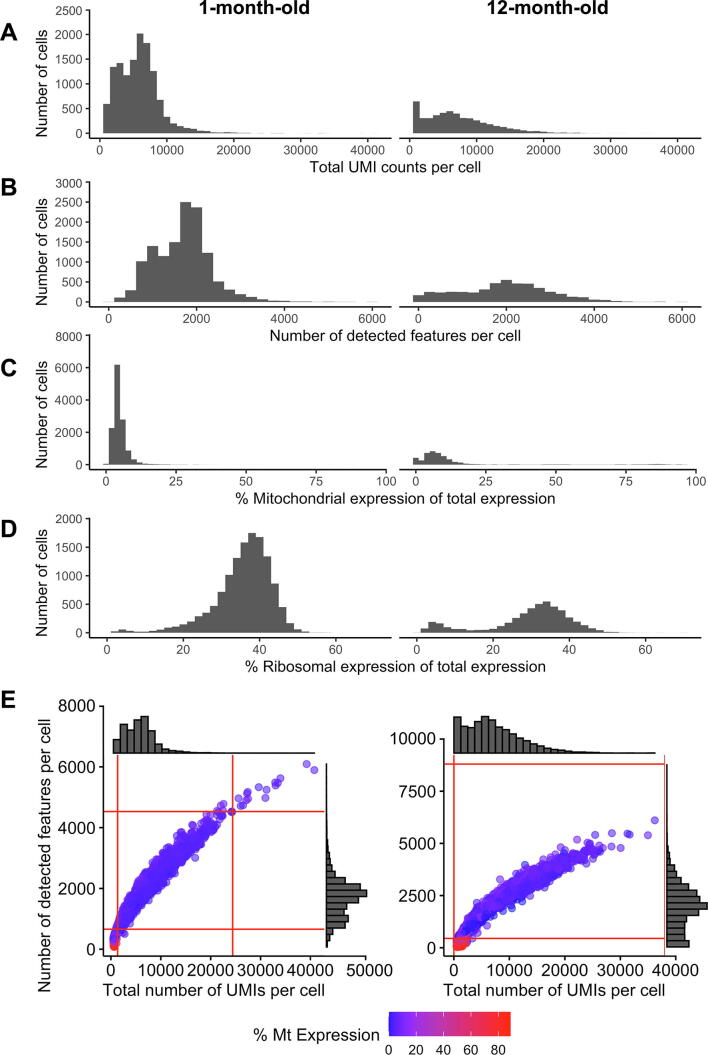
Using Seurat v3.1.3 [16], data from the two culture time points underwent quality control and normalisation separately. The following values were calculated for each cell: total number of unique molecular identifiers (UMIs), number of detected genes, as well as proportion of mitochondrial and ribosomal transcripts relative to total expression. Cells were removed from subsequent analysis if the total UMI count of the cell exceeded the threefold median absolute deviation (MAD) across all cells in the sample. Manual thresholds were derived from outlier peaks in the distributions of the number of detected genes, and fraction of mitochondrial and ribosomal transcripts to total expression (Figure S1). Cells were removed from the 1-month time point sample if the number of detected genes exceeded the lower and upper limits of 220 and 5000, respectively, while cells from the 12-month time point sample were removed if the number of detected genes was lower than 220. Cells from both time points were removed if mitochondrial transcripts accounted for more than 25% of total expression, and/or ribosomal transcripts accounted for more than 60% of total expression (Figure S1A–D; Table S1). The confounding effect of these mitochondrial and ribosomal transcript QC metrics in remaining cells was regressed out during cell–cell normalisation, using the SCTransform function from Seurat [75] (Table S7).
Integration, dimensionality reduction, and clustering
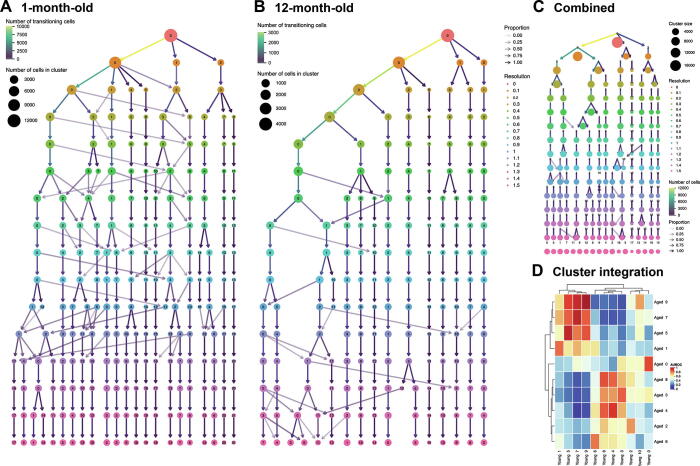
Data dimensionality was reduced with principal component analysis (PCA). Subsequently, the 30 most statistically significant PCs were reduced to two dimensions using the Uniform Manifold Approximation and Projection (UMAP) and used to construct a shared nearest neighbour (SNN) graph for each cell. The Louvain method for community detection was then used to identify clusters in each dataset with resolutions ranging from 0 to 1.5. The results for all resolutions were plotted using clustree [76], which showed the stabilisation of cell population identities at the resolution of 0.6 in the 1-month culture and 0.7 in the 12-month culture (Figure S2A–C). Datasets at both time points were combined into one dataset for comparative analysis with the integration workflow from Seurat [77]. This workflow used canonical correlation analysis (CCA) to identify 22,023 anchors based on 3000 most variable genes. The anchors were then used to align both datasets. To integrate the clusters across both time points, the unsupervised version of MetaNeighbor [17] was used to evaluate the similarities between the 1-month clusters and 12-month clusters. Cluster pairs that were reciprocally top hits and received a mean area under the receiver operating characteristic (AUROC) score greater than 0.8 were merged into one cluster (Figure S2D).
Cluster characterization and analysis
Network analysis was performed on differentially expressed genes (DEGs) using Reactome functional interaction analysis [78], [79]. Differential expression (DE) analysis was performed using the FindMarkers function based on the likelihood ratio test adapted for single-cell gene expression [80]. GO analysis [81], [82] was performed using a PANTHER overrepresentation test [Fisher exact test, false discovery rate (FDR) < 0.05] against the Homo sapiens genome (PANTHER version 14.1 released 2019-03-12). For some clusters, insufficient gene markers are available for GO analysis (Aged Clusters 0, 1, and 5). Canonical RPE markers, gene expression profiles, and their associated GO terms specific to each cluster are provided in Tables S3–S5.
Trajectory analysis
Trajectory analysis was performed with Monocle 3 v0.2.4 [52]. Harmonised Pearson residuals produced by the integration step underwent dimensionality reduction with UMAP, and the resulting projection was used to initialize trajectory inference. The node closest to cells expressing proliferative markers was selected as the root of the trajectory, and pseudotime values were calculated. Gene expression dynamics across the trajectory was characterised with Moran I’s test, which was applied via the “graph_test” function using the following arguments: neighbor_graph = “principal_graph”, reduction_method = “UMAP”, and expression_family = “quasiposson”. DEGs with FDR ≤ 0.05 were clustered into co-expression modules using the “find_gene_modules” function, and the resulting protein interactions were characterised with STRING.
Ethical statement
The experimental work was approved by the Human Research Ethics Committee of the University of Melbourne (1545484), with the requirements of the National Health and Medical Research Council (NHMRC) of Australia in accordance with the Declaration of Helsinki.
Code availability
Code and usage notes are available at: https://github.com/powellgenomicslab/RPE_scRNA_AgedStudy. This repository consists of code used to process raw sequencing data in FASTQ format to cell-gene expression tables via the Cell Ranger pipeline, and code used to perform the following analyses: quality control, normalisation, dimensionality reduction, clustering, differential expression, and integration.
Data availability
Sequencing data are available at ArrayExpress (ArrayExpress: E-MTAB-8511). Files are raw FASTQ files, and a tab separated matrix of UMIs per gene for each cell passing quality control filtering. BAM files can be generated using the supplied repository to process the FASTQ files via Cell Ranger.
CRediT author statement
Grace E. Lidgerwood: Conceptualization, Data curation, Formal analysis, Visualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing - original draft, Writing - review & editing. Anne Senabouth: Conceptualization, Data curation, Formal analysis, Visualization, Methodology, Investigation, Software, Writing - original draft, Writing - review & editing. Casey J.A. Smith-Anttila: Conceptualization, Methodology, Investigation, Writing - review & editing. Vikkitharan Gnanasambandapillai: Conceptualization, Methodology, Investigation, Writing - review & editing. Dominik C. Kaczorowski: Conceptualization, Methodology, Investigation, Writing - review & editing. Daniela Amann-Zalcenstein: Conceptualization, Methodology, Investigation, Writing - review & editing. Erica L. Fletcher: Conceptualization, Methodology, Funding acquisition, Writing - review & editing. Shalin H. Naik: Conceptualization, Methodology, Funding acquisition, Writing - review & editing. Alex W. Hewitt: Conceptualization, Funding acquisition, Methodology, Resources, Writing - review & editing, Supervision. Joseph E. Powell: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Software, Writing - original draft, Writing - review & editing, Supervision. Alice Pébay: Conceptualization, Formal analysis, Visualization, Funding acquisition, Methodology, Project administration, Resources, Writing - original draft, Writing - review & editing, Supervision. All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work was supported by a National Health and Medical Research Council (NHMRC-Australia) Practitioner Fellowship (awarded to AWH), Career Development Fellowship (awarded to JEP), and Senior Research Fellowship (Grant No. 1154389, awarded to AP), an Australian Research Council Future Fellowship (Grant No. FT140100047, awarded to AP), NHMRC project grants (Grant Nos. 1138253 awarded to ELF and AP, as well as 1062820 and 1124812 awarded to SHN), a NHMRC synergy grant (Grant No. 1181010 awarded to ELF and AP), grants from the Macular Disease Foundation Australia (awarded to AP, JEP, and AWH), the Jack Brockhoff Foundation (awarded to GEL), the DHB Foundation (awarded to GEL and AP), the Ophthalmic Research Institute of Australia (awarded to AP and AWH), Stem Cells Australia – the Australian Research Council Special Research Initiative in Stem Cell Science (awarded to SHN, AWH, JEP, and AP), the TMG Family Fund (awarded to AP and GEL), a donation from Ms Jacqueline Pascual, the University of Melbourne, and the Operational Infrastructure Support from the Victorian Government, Australia.
Handled by Fuchou Tang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2020.08.002.
Contributor Information
Grace E. Lidgerwood, Email: grace.lidgerwood@unimelb.edu.au.
Alice Pébay, Email: apebay@unimelb.edu.au.
Supplementary material
The following are the Supplementary data to this article:
Supplementary figure S1.
Filtering of cells based on quality control metrics. A. Total number of UMIs per cell. B. Total number of features per cell. C. Percentage of mitochondrial gene expression relative to total expression. D. Percentage of ribosomal gene expression relative to total expression. E. Relationship between total UMIs, detected genes, and percentage of mitochondrial gene expression. Histograms for total UMIs and detected features are located on X and Y margins of scatter plot, respectively. Scatter plot represents the relationships between the two metrics and are coloured by percentage of mitochondrial expression. Plots for 1-month-old culture and 12-month-old culture are shown on the left and right, respectively. UMI, unique molecular identifier; MAD, median absolute deviation.
Supplementary figure S2.
Characterisation of stable cell subpopulations. Graph-based clustering was performed at different resolutions ranging from 0 to 1.5, in increments of 0.1 for 1-month-old (A), 12-month-old (B), and combined (C) datasets using clustree. Regions of stability are represented by minimal branching. Values within circles are arbitrary cluster identifiers and arrows indicate the proportion of cells moving from one group to another (transitioning cells, colour coded). D. Integration of clusters identified in 1-month-old and 12-month-old cells via MetaNeighbor. Heatmap represents degree of similarity of clusters as AUROC values. AUROC, area under receiver operating characteristic curve.
Supplementary figure S3.
Residual trajectory genes modules analyses measured with Monocle 3. A. Monocle 3 pseudotime represented in beeswarm plots. Different colours are used for each subpopulation for ease of reading. B. Heatmap of module scores for each subpopulation. Modules were identified via Monocle 3 gene trajectory analysis, and module scores were calculated via differential expression of genes from each module. C. STRING analysis of genes from Table S6 for Module 1. Genes involved in regulation of development are shown in red. D. STRING analysis of genes from Table S6 for Module 2. Genes involved in regulation of synapse organisation are shown in red. E. STRING analysis of genes from Table S6 for Module 3. No relevant biological processes was revealed. F. STRING analysis of genes from Table S6 for Module 4. Genes involved in regulation of cell cycle and mitosis are shown in blue and red, respectively.
Supplementary figure S4.
Gene trajectory by pseudotime analyses of early and mature RPE markers. Gene expression (measured as normalised UMI count) of early (MITF, PAX6, PMEL, RAX, and SIX3) and mature RPE markers (RBP1, RGR, RLBP1, RPE65, and TYR) using Monocle 3, with each subpopulation coded in colour as indicated. Trend of expression is represented by a black line within each panel.
Quality control parameters.
Cluster analysis with number of cells per subpopulation.
Conserved cluster markers, PANTHER Gene Ontology pathways and differential expression for each subpopulation within the “Common” population.
Conserved cluster markers and Gene Ontology pathways for each subpopulation within the “Young” population.
Conserved cluster markers and Gene Ontology pathways for each subpopulation within the “Aged” population.
Monocle3 residual trajectory.
Details of cell filtration criteria.
References
- 1.Perez V.L., Saeed A.M., Tan Y., Urbieta M., Cruz-Guilloty F. The eye: a window to the soul of the immune system. J Autoimmun. 2013;45:7–14. doi: 10.1016/j.jaut.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Xu H., Chen M., Forrester J.V. Para-inflammation in the aging retina. Prog Retin Eye Res. 2009;28:348–368. doi: 10.1016/j.preteyeres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Lidgerwood G.E., Morris A.J., Conquest A., Daniszewski M., Rooney L.A., Lim S.Y., et al. Role of lysophosphatidic acid in the retinal pigment epithelium and photoreceptors. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:750–761. doi: 10.1016/j.bbalip.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Lidgerwood G.E., Lim S.Y., Crombie D.E., Ali R., Gill K.P., Hernández D., et al. Defined medium conditions for the induction and expansion of human pluripotent stem cell-derived retinal pigment epithelium. Stem Cell Rev Rep. 2016;12:179–188. doi: 10.1007/s12015-015-9636-2. [DOI] [PubMed] [Google Scholar]
- 5.Kamao H., Mandai M., Okamoto S., Sakai N., Suga A., Sugita S., et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osakada F., Jin Z.B., Hirami Y., Ikeda H., Danjyo T., Watanabe K., et al. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci. 2009;122:3169–3179. doi: 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- 7.Buchholz D.E., Pennington B.O., Croze R.H., Hinman C.R., Coffey P.J., Clegg D.O. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl Med. 2013;2:384–393. doi: 10.5966/sctm.2012-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foltz L.P., Clegg D.O. Rapid, directed differentiation of retinal pigment epithelial cells from human embryonic or induced pluripotent stem cells. J Vis Exp. 2017:56274. doi: 10.3791/56274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennis A., Jacobs J.G., Catsburg L.A.E., ten Brink J.B., Koster C., Schlingemann R.O., et al. Stem cell derived retinal pigment epithelium: the role of pigmentation as maturation marker and gene expression profile comparison with human endogenous retinal pigment epithelium. Stem Cell Rev Rep. 2017;13:659–669. doi: 10.1007/s12015-017-9754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokkinaki M., Sahibzada N., Golestaneh N. Human induced pluripotent stem-derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells. 2011;29:825–835. doi: 10.1002/stem.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lidgerwood G.E., Hewitt A.W., Pébay A. Human pluripotent stem cells for the modelling of diseases of the retina and optic nerve: toward a retina in a dish. Curr Opin Pharmacol. 2019;48:114–119. doi: 10.1016/j.coph.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Llonch S., Carido M., Ader M. Organoid technology for retinal repair. Dev Biol. 2018;433:132–143. doi: 10.1016/j.ydbio.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 13.Gagliardi G., Ben M'Barek K., Goureau O. Photoreceptor cell replacement in macular degeneration and retinitis pigmentosa: a pluripotent stem cell-based approach. Prog Retin Eye Res. 2019;71:1–25. doi: 10.1016/j.preteyeres.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Sridhar A., Hoshino A., Finkbeiner C.R., Chitsazan A., Dai L., Haugan A.K., et al. Single-cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures. Cell Rep. 2020;30 doi: 10.1016/j.celrep.2020.01.007. 1644–59.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y., Wang X., Hu B., Mao Y., Chen Y., Yan L., et al. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 2019;17:e3000365. doi: 10.1371/journal.pbio.3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177 doi: 10.1016/j.cell.2019.05.031. 1888–902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crow M., Paul A., Ballouz S., Huang Z.J., Gillis J. Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat Commun. 2018;9:884. doi: 10.1038/s41467-018-03282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarau I.S., Berlin A., Curcio C.A., Ach T. The cytoskeleton of the retinal pigment epithelium: from normal aging to age-related macular degeneration. Int J Mol Sci. 2019;20:3578. doi: 10.3390/ijms20143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X., Liao W.J., Liao J.M., Liao P., Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7:92. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barkić M., Crnomarković S., Grabusić K., Bogetić I., Panić L., Tamarut S., et al. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol Cell Biol. 2009;29:2489–2504. doi: 10.1128/MCB.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watkins-Chow D.E., Cooke J., Pidsley R., Edwards A., Slotkin R., Leeds K.E., et al. Mutation of the diamond-blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet. 2013;9:e1003094. doi: 10.1371/journal.pgen.1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paraoan L., Grierson I., Maden B.E. Analysis of expressed sequence tags of retinal pigment epithelium: cystatin C is an abundant transcript. Int J Biochem Cell Biol. 2000;32:417–426. doi: 10.1016/s1357-2725(99)00143-0. [DOI] [PubMed] [Google Scholar]
- 23.Kay P., Yang Y.C., Hiscott P., Gray D., Maminishkis A., Paraoan L. Age-related changes of cystatin C expression and polarized secretion by retinal pigment epithelium: potential age-related macular degeneration links. Invest Ophthalmol Vis Sci. 2014;55:926–934. doi: 10.1167/iovs.13-13239. [DOI] [PubMed] [Google Scholar]
- 24.Guyonneau L., Murisier F., Rossier A., Moulin A., Beermann F. Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Mol Cell Biol. 2004;24:3396–3403. doi: 10.1128/MCB.24.8.3396-3403.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda K., Yokoyama S., Yasumoto K.I., Saito H., Udono T., Takahashi K., et al. OTX2 regulates expression of DOPAchrome tautomerase in human retinal pigment epithelium. Biochem Biophys Res Commun. 2003;300:908–914. doi: 10.1016/s0006-291x(02)02934-0. [DOI] [PubMed] [Google Scholar]
- 26.Strunnikova N.V., Maminishkis A., Barb J.J., Wang F., Zhi C., Sergeev Y., et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum Mol Genet. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strunnikova N., Zhang C., Teichberg D., Cousins S.W., Baffi J., Becker K.G., et al. Survival of retinal pigment epithelium after exposure to prolonged oxidative injury: a detailed gene expression and cellular analysis. Invest Ophthalmol Vis Sci. 2004;45:3767–3777. doi: 10.1167/iovs.04-0311. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka Y., Utsumi J., Matsui M., Sudo T., Nakamura N., Mutoh M., et al. Purification, molecular cloning, and expression of a novel growth-promoting factor for retinal pigment epithelial cells, REF-1/TFPI-2. Invest Ophthalmol Vis Sci. 2004;45:245–252. doi: 10.1167/iovs.03-0230. [DOI] [PubMed] [Google Scholar]
- 29.Alizadeh P., Smit-McBride Z., Oltjen S.L., Hjelmeland L.M. Regulation of cysteine cathepsin expression by oxidative stress in the retinal pigment epithelium/choroid of the mouse. Exp Eye Res. 2006;83:679–687. doi: 10.1016/j.exer.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuel W., Kutty R.K., Vijayasarathy C., Pascual I., Duncan T., Redmond T.M. Decreased expression of insulin-like growth factor binding protein-5 during N-(4-hydroxyphenyl)retinamide-induced neuronal differentiation of ARPE-19 human retinal pigment epithelial cells: regulation by CCAAT/enhancer-binding protein. J Cell Physiol. 2010;224:827–836. doi: 10.1002/jcp.22191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinn P.M., Buck T.M., Mulder A.A., Ohonin C., Alves C.H., Vos R.M., et al. Human iPSC-derived retinas recapitulate the fetal CRB1 CRB2 complex formation and demonstrate that photoreceptors and Müller glia are targets of AAV5. Stem Cell Rep. 2019;12:906–919. doi: 10.1016/j.stemcr.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paniagua A.E., Herranz-Martín S., Jimeno D., Jimeno Á.M., López-Benito S., Carlos Arévalo J., et al. CRB2 completes a fully expressed Crumbs complex in the retinal pigment epithelium. Sci Rep. 2015;5:14504. doi: 10.1038/srep14504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winokur P.N., Subramanian P., Bullock J.L., Arocas V., Becerra S.P. Comparison of two neurotrophic serpins reveals a small fragment with cell survival activity. Mol Vis. 2017;23:372–384. [PMC free article] [PubMed] [Google Scholar]
- 34.Chang J.T., Esumi N., Moore K., Li Y., Zhang S., Chew C., et al. Cloning and characterization of a secreted frizzled-related protein that is expressed by the retinal pigment epithelium. Hum Mol Genet. 1999;8:575–583. doi: 10.1093/hmg/8.4.575. [DOI] [PubMed] [Google Scholar]
- 35.Liu N.P., Roberts W.L., Hale L.P., Levesque M.C., Patel D.D., Lu C.L., et al. Expression of CD44 and variant isoforms in cultured human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1997;38:2027–2037. [PubMed] [Google Scholar]
- 36.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 37.Bennis A., Ten Brink J.B., Moerland P.D., Heine V.M., Bergen A.A. Comparative gene expression study and pathway analysis of the human iris- and the retinal pigment epithelium. PLoS One. 2017;12:e0182983. doi: 10.1371/journal.pone.0182983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao L., Liu J., Pu J., Milne G., Chen M., Xu H., et al. Polarized retinal pigment epithelium generates electrical signals that diminish with age and regulate retinal pathology. J Cell Mol Med. 2018;22:5552–5564. doi: 10.1111/jcmm.13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westenskow P., Piccolo S., Fuhrmann S. Beta-catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development. 2009;136:2505–2510. doi: 10.1242/dev.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee W.H., Joshi P., Wen R. Glutathione S-transferase pi isoform (GSTP1) expression in murine retina increases with developmental maturity. Adv Exp Med Biol. 2014;801:23–30. doi: 10.1007/978-1-4614-3209-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang A.L., Lukas T.J., Yuan M., Du N., Tso M.O., Neufeld A.H. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS One. 2009;4:e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasovic L., Utheim T.P., Reppe S., Khan A.Z., Jackson C.J., Thiede B., et al. Improvement of storage medium for cultured human retinal pigment epithelial cells using factorial design. Sci Rep. 2018;8:5688. doi: 10.1038/s41598-018-24121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai H., Shin M.C., Tezel T.H., Kaplan H.J., Del Priore L.V. Use of iris pigment epithelium to replace retinal pigment epithelium in age-related macular degeneration: a gene expression analysis. Arch Ophthalmol. 2006;124:1276–1285. doi: 10.1001/archopht.124.9.1276. [DOI] [PubMed] [Google Scholar]
- 44.Alge C.S., Suppmann S., Priglinger S.G., Neubauer A.S., May C.A., Hauck S., et al. Comparative proteome analysis of native differentiated and cultured dedifferentiated human RPE cells. Invest Ophthalmol Vis Sci. 2003;44:3629–3641. doi: 10.1167/iovs.02-1225. [DOI] [PubMed] [Google Scholar]
- 45.Wang D., Chadha G.K., Feygin A., Ivanov A.I. F-actin binding protein, anillin, regulates integrity of intercellular junctions in human epithelial cells. Cell Mol Life Sci. 2015;72:3185–3200. doi: 10.1007/s00018-015-1890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Hao H., Tzatzalos E., Lin R.-K., Doh S., Liu L.F., et al. Topoisomerase IIbeta is required for proper retinal development and survival of postmitotic cells. Biol Open. 2014;3:172–184. doi: 10.1242/bio.20146767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Usui A., Mochizuki Y., Iida A., Miyauchi E., Satoh S., Sock E., et al. The early retinal progenitor-expressed gene Sox11 regulates the timing of the differentiation of retinal cells. Development. 2013;140:740–750. doi: 10.1242/dev.090274. [DOI] [PubMed] [Google Scholar]
- 48.van Soest S.S., de Wit G.M.J., Essing A.H.W., ten Brink J.B., Kamphuis W., de Jong P.T.V.M., et al. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch's membrane. Mol Vis. 2007;13:1608–1617. [PubMed] [Google Scholar]
- 49.Tian J., Ishibashi K., Honda S., Boylan S.A., Hjelmeland L.M., Handa J.T. The expression of native and cultured human retinal pigment epithelial cells grown in different culture conditions. Br J Ophthalmol. 2005;89:1510–1517. doi: 10.1136/bjo.2005.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitmore S.S., Wagner A.H., DeLuca A.P., Drack A.V., Stone E.M., Tucker B.A., et al. Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp Eye Res. 2014;129:93–106. doi: 10.1016/j.exer.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai H., Fields M.A., Hoshino R., Del Priore L.V. Effects of aging and anatomic location on gene expression in human retina. Front Aging Neurosci. 2012;4:8. doi: 10.3389/fnagi.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stern J., Temple S. Retinal pigment epithelial cell proliferation. Exp Biol Med (Maywood) 2015;240:1079–1086. doi: 10.1177/1535370215587530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu T.G. Transplantation of human embryonic stem cell-derived retinal pigment epithelial cells (MA09-hRPE) in macular degeneration. NPJ Regen Med. 2019;4:19. doi: 10.1038/s41536-019-0081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen M, Forrester JV, Xu H. Dysregulation in retinal para-inflammation and age-related retinal degeneration in CCL2 or CCR2 deficient mice. PLoS One 2011;6:e22818. [DOI] [PMC free article] [PubMed]
- 56.Holtkamp G.M., De Vos A.F., Peek R., Kijlsta A. Analysis of the secretion pattern of monocyte chemotactic protein-1 (MCP-1) and transforming growth factor-beta 2 (TGF-beta2) by human retinal pigment epithelial cells. Clin Exp Immunol. 1999;118:35–40. doi: 10.1046/j.1365-2249.1999.01016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bovolenta P., Esteve P., Ruiz J.M., Cisneros E., Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121:737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 58.He X., Cheng R., Benyajati S., Ma J.X. PEDF and its roles in physiological and pathological conditions: implication in diabetic and hypoxia-induced angiogenic diseases. Clin Sci. 2015;128:805–823. doi: 10.1042/CS20130463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Penn J.S., Madan A., Caldwell R.B., Bartoli M., Caldwell R.W., Hartnett M.E. Vascular endothelial growth factor in eye disease. Prog Retin Eye Res. 2008;27:331–371. doi: 10.1016/j.preteyeres.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang P., Skiba N.P., Tewkesbury G.M., Treboschi V.M., Baciu P., Jaffe G.J. Complement-mediated regulation of Apolipoprotein E in cultured human RPE cells. Invest Ophthalmol Vis Sci. 2017;58:3073–3085. doi: 10.1167/iovs.16-20083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toomey C.B., Johnson L.V., Bowes Rickman C. Complement factor H in AMD: bridging genetic associations and pathobiology. Prog Retin Eye Res. 2018;62:38–57. doi: 10.1016/j.preteyeres.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao J.L., Yu J., Huang K., Hu J., Diemer T., Ma Z., et al. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum Mol Genet. 2010;19:4229–4238. doi: 10.1093/hmg/ddq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.May-Simera H.L., Wan Q., Jha B.S., Hartford J., Khristov V., Dejene R., et al. Primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells. Cell Rep. 2018;22:189–205. doi: 10.1016/j.celrep.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodríguez-Menéndez S., García M., Fernández B., Álvarez L., Fernández-Vega-Cueto A., Coca-Prados M., et al. The Zinc-metallothionein redox system reduces oxidative stress in retinal pigment epithelial cells. Nutrients. 2018;10:1874. doi: 10.3390/nu10121874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Capowski E.E., Simonett J.M., Clark E.M., Wright L.S., Howden S.E., Wallace K.A., et al. Loss of MITF expression during human embryonic stem cell differentiation disrupts retinal pigment epithelium development and optic vesicle cell proliferation. Hum Mol Genet. 2014;23:6332–6344. doi: 10.1093/hmg/ddu351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGowan K.A., Li J.Z., Park C.Y., Beaudry V., Tabor H.K., Sabnis A.J., et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grewal R., Stepczynski J., Kelln R., Erickson T., Darrow R., Barsalou L., et al. Coordinated changes in classes of ribosomal protein gene expression is associated with light-induced retinal degeneration. Invest Ophthalmol Vis Sci. 2004;45:3885–3895. doi: 10.1167/iovs.04-0358. [DOI] [PubMed] [Google Scholar]
- 68.Leach L.L., Buchholz D.E., Nadar V.P., Lowenstein S.E., Clegg D.O. Canonical/β-catenin Wnt pathway activation improves retinal pigmented epithelium derivation from human embryonic stem cells. Invest Ophthalmol Vis Sci. 2015;56:1002–1013. doi: 10.1167/iovs.14-15835. [DOI] [PubMed] [Google Scholar]
- 69.Esteve P., Trousse F., Rodríguez J., Bovolenta P. SFRP1 modulates retina cell differentiation through a beta-catenin-independent mechanism. J Cell Sci. 2003;116:2471–2481. doi: 10.1242/jcs.00452. [DOI] [PubMed] [Google Scholar]
- 70.Rodriguez J., Esteve P., Weinl C., Ruiz J.M., Fermin Y., Trousse F., et al. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005;8:1301–1309. doi: 10.1038/nn1547. [DOI] [PubMed] [Google Scholar]
- 71.Saint-Geniez M., Maharaj A.S.R., Walshe T.E., Tucker B.A., Sekiyama E., Kurihara T., et al. Endogenous VEGF is required for visual function: evidence for a survival role on Müller cells and photoreceptors. PLoS One. 2008;3:e3554. doi: 10.1371/journal.pone.0003554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishijima K., Ng Y.S., Zhong L., Bradley J., Schubert W., Jo N., et al. Vascular endothelial growth factor-A is a survival factor for retinal neurons and a critical neuroprotectant during the adaptive response to ischemic injury. Am J Pathol. 2007;171:53–67. doi: 10.2353/ajpath.2007.061237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van-Niel G., Bergam P., Di Cicco A., Hurbain I., Lo Cicero A., Dingli F., et al. Apolipoprotein E regulates amyloid formation within endosomes of pigment cells. Cell Rep. 2015;13:43–51. doi: 10.1016/j.celrep.2015.08.057. [DOI] [PubMed] [Google Scholar]
- 74.Daniszewski M., Nguyen Q., Chy H.S., Singh V., Crombie D.E., Kulkarni T., et al. Single-cell profiling identifies key pathways expressed by iPSCs cultured in different commercial media. iScience. 2018;7:30–39. doi: 10.1016/j.isci.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zappia L, Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience 2018;7:giy083. [DOI] [PMC free article] [PubMed]
- 77.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu G., Feng X., Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDavid A., Finak G., Chattopadyay P.K., Dominguez M., Lamoreaux L., Ma S.S., et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.The Gene Ontology Consortium The Gene Ontology resource: 20 years and still going strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality control parameters.
Cluster analysis with number of cells per subpopulation.
Conserved cluster markers, PANTHER Gene Ontology pathways and differential expression for each subpopulation within the “Common” population.
Conserved cluster markers and Gene Ontology pathways for each subpopulation within the “Young” population.
Conserved cluster markers and Gene Ontology pathways for each subpopulation within the “Aged” population.
Monocle3 residual trajectory.
Details of cell filtration criteria.
Data Availability Statement
Sequencing data are available at ArrayExpress (ArrayExpress: E-MTAB-8511). Files are raw FASTQ files, and a tab separated matrix of UMIs per gene for each cell passing quality control filtering. BAM files can be generated using the supplied repository to process the FASTQ files via Cell Ranger.