Abstract
Successful pregnancy in placental mammals substantially depends on the establishment of maternal immune tolerance to the semi-allogenic fetus. Disorders in this process are tightly associated with adverse pregnancy outcomes including recurrent miscarriage (RM). However, an in-depth understanding of the systematic and decidual immune environment in RM remains largely lacking. In this study, we utilized single-cell RNA-sequencing (scRNA-seq) to comparably analyze the cellular and molecular signatures of decidual and peripheral leukocytes in normal and unexplained RM pregnancies at the early stage of gestation. Integrative analysis identifies 22 distinct cell clusters in total, and a dramatic difference in leukocyte subsets and molecular properties in RM cases is revealed. Specifically, the cytotoxic properties of CD8+ effector T cells, nature killer (NK), and mucosal-associated invariant T (MAIT) cells in peripheral blood indicates apparently enhanced pro-inflammatory status, and the population proportions and ligand–receptor interactions of the decidual leukocyte subsets demonstrate preferential immune activation in RM patients. The molecular features, spatial distribution, and the developmental trajectories of five decidual NK (dNK) subsets have been elaborately illustrated. In RM patients, a dNK subset that supports embryonic growth is diminished in proportion, while the ratio of another dNK subset with cytotoxic and immune-active signature is significantly increased. Notably, a unique pro-inflammatory CD56+CD16+ dNK subset substantially accumulates in RM decidua. These findings reveal a comprehensive cellular and molecular atlas of decidual and peripheral leukocytes in human early pregnancy and provide an in-depth insight into the immune pathogenesis for early pregnancy loss.
Keywords: Decidual and peripheral leukocytes, Single-cell RNA-sequencing, Recurrent miscarriage, Early pregnancy, Developmental trajectory
Introduction
Reproductive success in placental mammals substantially depends on the establishment of maternal immune tolerance to the semi-allogenic fetus during pregnancy, which means the immune response of mother during pregnancy is dampened against fetally-expressed antigens [1], [2]. Meanwhile, it remains intact against others, such as microbe-specific antigens. At the maternal-fetal interface, the local immune cells are also responsible for the protection against placental infection. Multiple independent lines of evidence from clinical analysis or animal model studies have indicated the tight association of failures in maternal immune adaptation with various adverse pregnancy outcomes, such as recurrent miscarriage (RM) and preeclampsia [3], [4], [5].
At early pregnancy, maternal immune cells populating the uterine mucosa comprise decidual nature killer (dNK) cells (about 50%–70%), macrophages (about 20%), T cells (about 10%–20%), and a small amount of dendritic cells (DC), mast cells, and B cells. A dynamic and well-orchestrated interaction network among extraembryonic placental trophoblast cells and various maternal immune cells at the feto-maternal interface constructs the immune adaptation to pregnancy. For instance, the numerically dominant dNK cells, being characterized by low cytotoxicity and strong cytokine-producing capacity, have multiple functions related to immune tolerance, embryo development, and infection protection. They produce interferon γ (IFNγ) that inhibit inflammatory Th17 cell responses. Fetal trophoblasts expressing human leukocyte antigen-C (HLA-C), HLA-G, and HLA-E are recognized by inhibitory and activating receptors on dNK cells, which reduces cytotoxicity and promotes secretion capability of dNK cells. The cytokines secreted by dNK cells participate in regulation of trophoblast invasion and differentiation, vascular remodeling, and uterine vascularization [6], [7]. The biased production of Th2 cytokines by various decidual leukocytes is vital to the maternal tolerance to fetal cells [8], [9]. In addition, many decidual immune cells, such as various T cell subsets, macrophages, and dNK cells, play roles in modulating immunity to placental infection [10], [11], [12]. A very recent study has revealed the fascinating mechanism by which dNK cells selectively kill bacteria in trophoblasts via transferring granulysin through nanotube connections [13].
Recent paradigm-shifting studies utilizing single-cell RNA-sequencing (scRNA-seq), mass cytometry, and comprehensive data analysis have constructed detailed cellular map and the elaborate cell communication patterns at human decidual-placental interface. Specifically, three major subsets of dNK cells that have distinctive immunomodulatory and chemokine profiles have been identified [14], [15], [16]. However, high-resolution immune landscape of dysregulated decidua in the contexts of pregnancy complications, for example in RM, is still unknown, which greatly limits the exploration of immune-related pathogenesis for RM.
In the present study, we utilized scRNA-seq to comparably analyze the cellular and molecular signatures of decidual and peripheral leukocytes in normal and RM pregnancies at the early stage of gestation. By integrating the gene expression properties, ligand–receptor interactions, and spatial localization patterns, the cell type-specific communications among various leukocyte subsets and the developmental trajectory of dNK cells were constructed. A pathological leukocyte atlas in decidua and peripheral blood from RM patients was systematically illustrated. Our findings reveal a detailed molecular and cellular map of decidual and peripheral leukocytes in human early pregnancy and highlight the integral immune-conflicts that are associated with early pregnancy failure.
Results
Clustering of immune cells from peripheral blood and decidua in normal and RM pregnancies
To characterize the immune cells in RM, we applied scRNA-seq to study CD45+ cells isolated from peripheral blood and decidual tissues of three pairs of normal pregnant women and RM patients with unknown cause (Table 1). Viable CD45+ leukocytes were enriched by fluorescent activated cell sorting (FACS) and subjected to droplet-based scRNA-seq using the 10X Genomics Chromium platform (Figure S1A). Sequencing data from six peripheral blood samples and six decidual tissue samples were obtained from 12 scRNA-seq libraries. Following computational quality control and filtering using the Seurat package [17], [18], the final datasets containing 56,758 high-quality cells were subjected to further analysis (Figure S1B–D).
Table 1.
Clinical characteristics of the pregnant women enrolled in this study
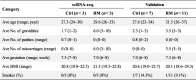 |
Note: Among the seven Ctrl cases in the validation group, decidual tissues from two cases were subjected to immunofluorescent staining as shown in Figure 5, and decidual tissues from five cases were subjected to flow cytometry assay as shown in Figure 7. Ave, average; Ctrl, normal pregnancy; RM, recurrent miscarriage; BMI, body mass index.
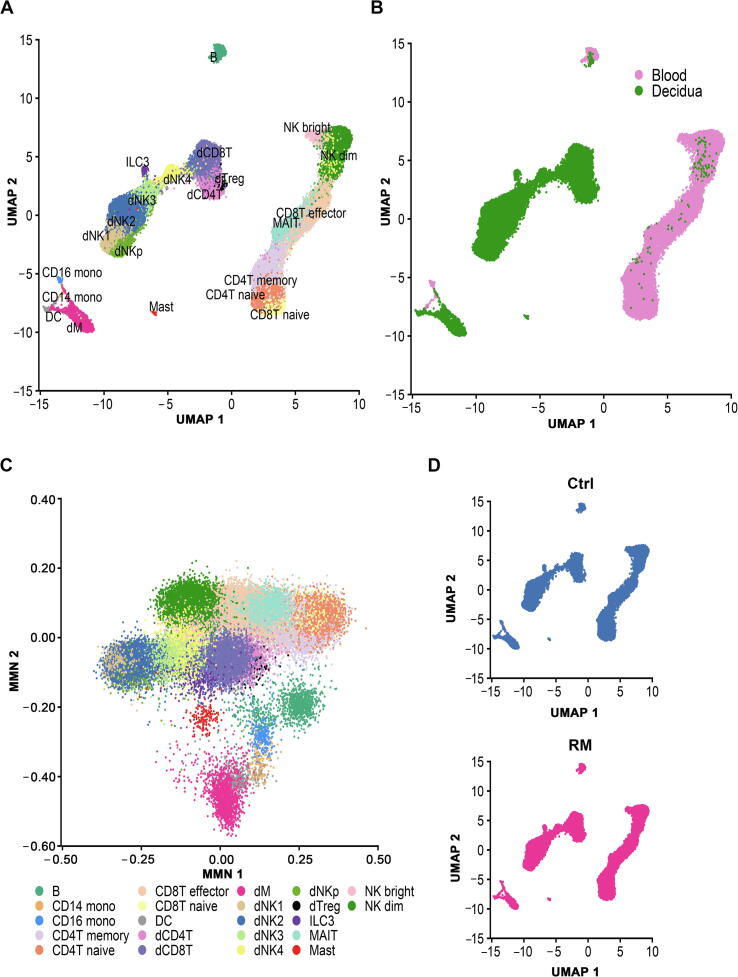
We performed an unsupervised graph-based clustering to analyze scRNA-seq data in Seurat (version 3.0.3). In order to eliminate unreasonable clustering due to batch effects, we integrated the data using the reciprocal principal component analysis (PCA) function of Seurat. Overall, 22 transcriptionally distinct clusters were identified (Figure 1A–C), and were further confirmed using the mutual nearest neighbor (MNN) algorithm (Figure S2) [19]. Cell identity of the 22 clusters was assigned on the basis of known marker genes and literature evidence (Figure 1D) [15], [16].
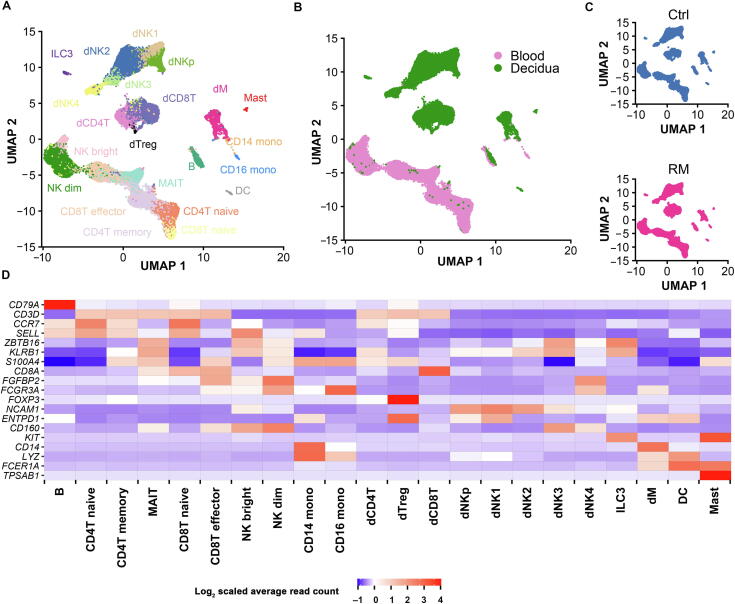
Figure 1.
Landscape of immune cells from peripheral blood and decidual tissues at early pregnancy
A. UMAP plot of scRNA-seq data to show the 22 leukocyte clusters in peripheral blood and decidual tissues at early pregnancy from both Ctrl and RM patients. B. UMAP plot showing cell populations from peripheral blood and decidual tissue from both Ctrl and RM patients. C. UMAP visualization of cell clustering in Ctrl (top) and RM (bottom) pregnancies. D. Cell type annotation of the 22 leukocyte clusters based on the expression of their canonical marker genes. Ctrl, normal; RM, recurrent miscarriage; B, peripheral B cell; CD4T naive, peripheral CD4+ naive T cell; CD8T naive, peripheral CD8+ naive T cell; MAIT, peripheral mucosal-associated invariant T cell; CD4T memory, peripheral CD4+ memory T cell; CD8T effector, CD8+ T effector cell; NK dim, peripheral CD56dimCD16+ NK cell; NK bright, peripheral CD56bright CD16− NK cell; CD14 mono, peripheral CD14+ monocyte; CD16 mono, peripheral CD16+ monocyte; DC, dendritic cell; dNK, decidual natural killer cell; dCD4, decidual CD4+ T cell; dCD8, decidual CD8+ T cell; dTreg, decidual regulatory T cell; dM, decidual macrophage; ILC3, group 3 innate lymphoid cell.
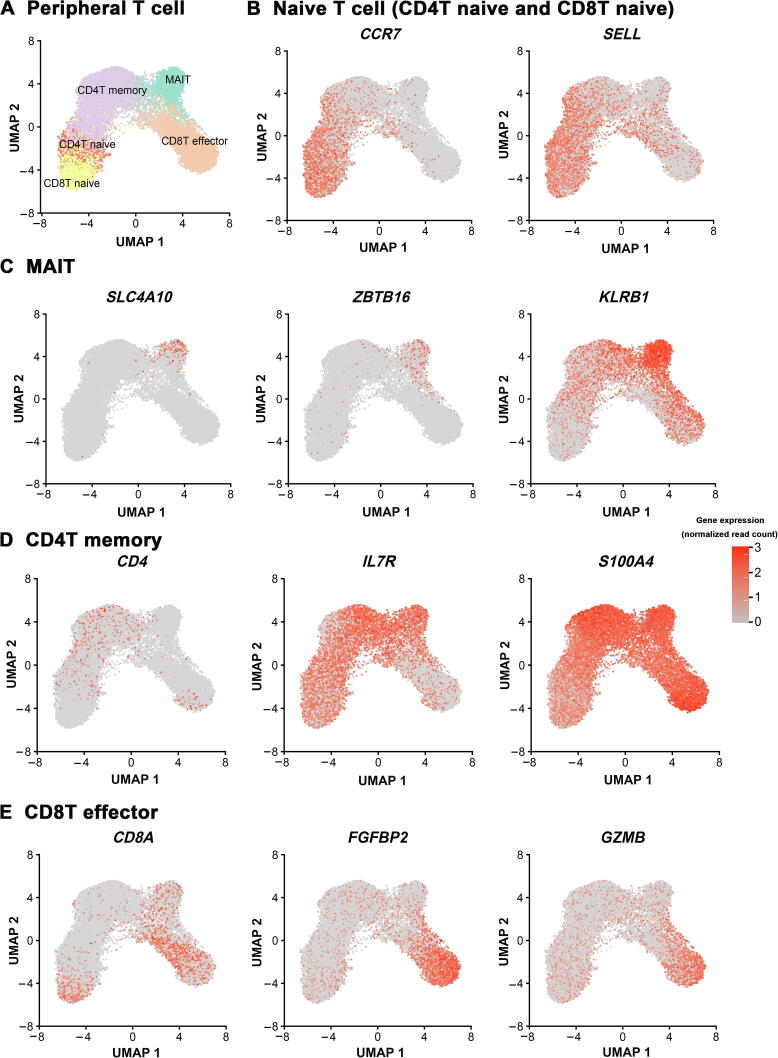
In peripheral blood, T cells were identified and grouped into five major clusters, including two subsets of naive T cells (CD4T naive and CD8T naive) expressing marker gene CCR7, mucosal-associated invariant T (MAIT) cells specifically expressing ZBTB16, CD4+ memory T cells (CD4T memory) annotated based on S100A4 expression, and CD8+ effector T cells (CD8T effector) that express FCGR3A and FGFBP2 (Figure 1D and Figure S3). Peripheral NK cells were clustered as CD56dimCD16+ NK cells (NK dim) and CD56brightCD16− NK cells (NK bright). Monocytes include CD14+ subset and CD16+ subset. In addition, a low abundance of B cells (identified based on CD79A expression) and DCs (annotated based on LYZ and FCER1A expression) were captured in peripheral blood.
In decidual tissues, the most abundant population appeared as dNK cells, which were sub-grouped into five clusters, named as dNKp, dNK1, dNK2, dNK3, and dNK4. The dNK4 subset is positive for expression of FCGR3A/CD16, which was uniquely accumulated in the decidual tissues from RM patients. Decidual T cells were clustered as CD4+ (dCD4T), CD8+ (dCD8T), and a relatively small number of FOXP3 regulatory T cells (dTreg). Other decidual leukocyte clusters include KIT-expressing group 3 innate lymphoid cells (ILC3), CD14-expressing decidual macrophage (dM) cells, FCER1A-expressing DCs, and TPSAB1-marked mast cells (Figure 1A and D).
Cellular and molecular characteristics of the peripheral leukocytes indicate a systematic pro-inflammatory feature in RM patients
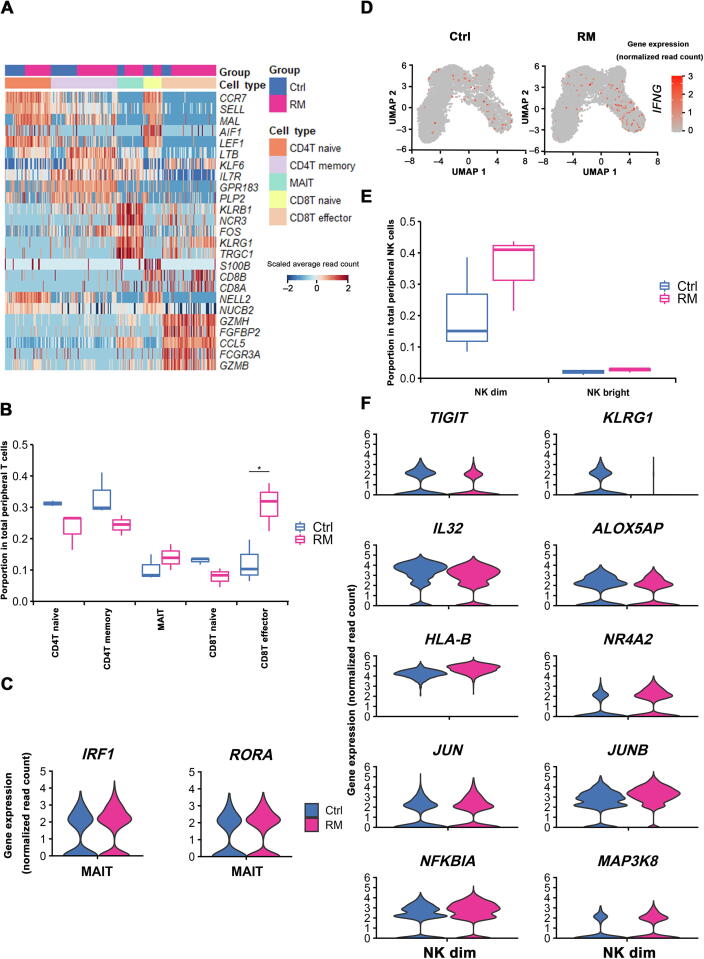
Given T cells are present predominantly in peripheral leukocytes, we therefore first analyzed the properties of T cell clusters in RM and normal pregnancies. Gene expression profiles of the five peripheral T cell subsets were indistinguishable between RM and normal pregnancies (Figure 2A). However, proportion of each T cell cluster exhibited evident alterations in RM patients, with reduced CD4T naive, CD8T naive, and CD4T memory populations, but augmented CD8T effector and MAIT populations (Figure 2B). In addition, transcription of genes encoding inflammatory factors, such as IRF1 and RORA in MAIT cells and IFNG in CD8T effector cells, was remarkably increased in RM cases (Figure 2C and D).
Figure 2.
Characteristics of the peripheral blood leukocyte clusters indicate the maternal immune inflammatory status in RM patients
A. Heatmap of the top 5 genes expressed in peripheral blood T cell subsets. Subject group and cell type information is indicated above the plot. B. Box plots depicting proportion of various peripheral T cell subtypes in Ctrl and RM patients. C. Violin plots showing the differential expression (absolute log2 FC > 0.5 and Bonferroni-adjusted P < 0.05) of typical marker genes of MAIT subset in Ctrl and RM patients using the method of non-parametric two-sided Wilcoxon rank sum test in Seurat. D. UMAP visualization of the log-transformed, normalized IFNG expression in CD8T effector subset in Ctrl (left) and RM (right) patients. High expression is shown in red, and low expression in gray. E. Box plots displaying proportion of peripheral NK cell subsets in Ctrl and RM patients. F. Violin plots showing the differential expression (absolute log2 FC > 0.5 and Bonferroni-adjusted P < 0.05) of anti-inflammation genes including TIGIT, KLRG1, IL32, and ALOX5AP, and pro-inflammation genes including HLA-B, NR4A2, JUN, JUNB, NFKBIA, and MAP3K8 in peripheral NK dim subset from Ctrl and RM patients, using method of the non-parametric two-sided Wilcoxon rank sum test in Seurat. FC, fold change.
The other significant change in peripheral leukocytes from RM patients was the markedly increased proportion of NK dim cells (Figure 2E). Furthermore, in NK dim cells from RM cases, an evidently decreased expression of immunosuppressive genes such as TIGIT, KLRG1, IL32, and ALOX5AP was identified, whereas increased expression of pro-inflammation genes, including HLA-B, NR4A2, JUN, JUNB, NFKBIA, and MAP3K8, was found (Figure 2F). Taken together, properties of the peripheral leukocytes, including aforementioned cell composition shift and specific gene expression pattern in certain cell clusters, strongly indicate an overall enhanced systematic pro-inflammatory immune feature of RM patients.
Cellular atlas of the decidual leukocytes demonstrates compromised immune cell interactions in RM pregnancy
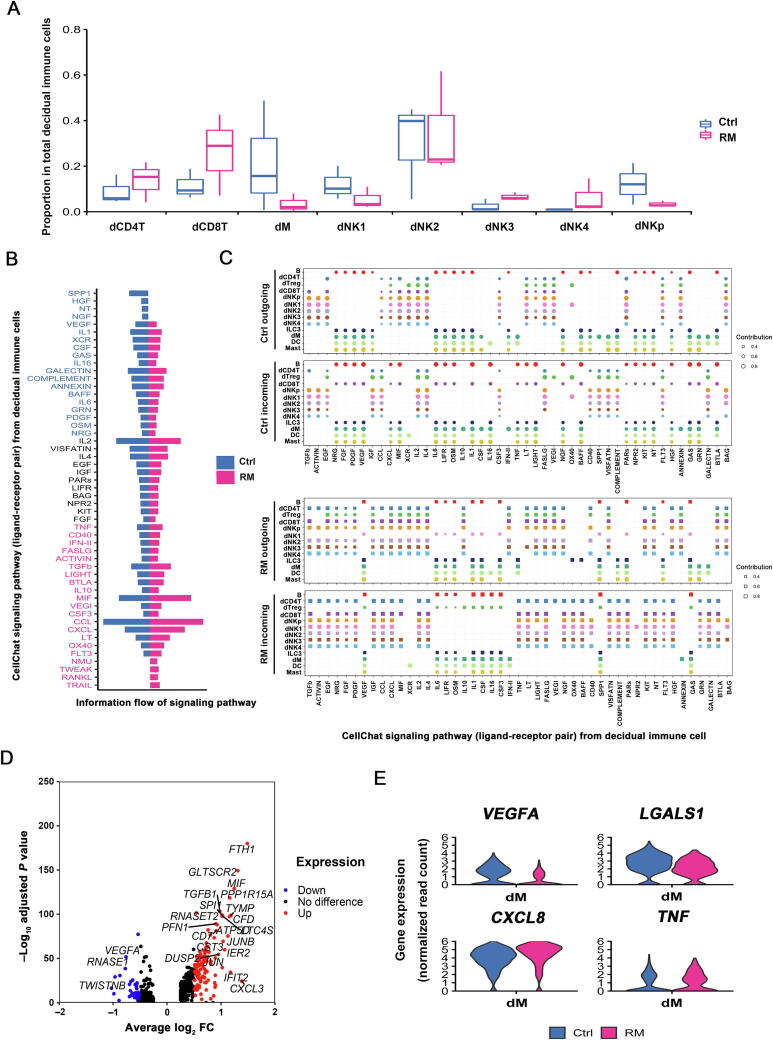
Increasing evidence has demonstrated compromised immune cell differentiation or function in the decidua from RM patients. However, a global profile of the cellular changes remains largely undefined. Our analysis of cell subset proportion in decidua clearly revealed an increased portion of dCD8T, dNK3, and dNK4 populations, and lowered ratio of dNKp, dNK1, and dM cells in decidual tissues from RM patients (Figure 3A). To systematically investigate the cell–cell communications among these cell subsets, we analyzed the expression levels of immune cell revenant ligand–receptor interacting pairs within cell types using CellChat [20]. Intriguingly, the status of signaling activation was different between RM and normal pregnancies. As shown, decidual leukocytes in RM patients exhibited more active signaling pathways of IFN-II, MIF, CCL, RANKL, and TRAIL, whereas repression in signal transduction of VEGF, NGF, HGF, IL1, and CSF pathways (Figure 3B). Using pattern recognition analysis, we further investigated the detailed changes in the outgoing signaling (levels of ligands) and incoming signaling (levels of receptors) across these pathways (Figure 3C). For instance, increased expression of cytokines in TNF pathway in dCD8T, ILC3, and dM cells in decidual tissues from RM patients might account for the accumulation of dNK4 or dCD4T cells that had higher expression of the corresponding receptors for these cytokines. Cytokine production and responses to the cytokines in the CCL pathway were more active in decidual T and dNK cells of RM patients (Figure 3C). Previous studies have demonstrated that protein expression of C-C motif chemokine ligand 3 (CCL3), CCL4, and CCL5 in different immune cells reflects the activation of the immune micro-environment at the maternal-fetal interface [21]. The pattern of molecular interactions among cell populations via specific ligand–receptor complexes in RM leukocytes provides clues to reveal potential mechanisms of compromised immune environment in the decidua from RM patients.
Figure 3.
Characteristics of decidual leukocyte subpopulations demonstrate an immune activation status at the maternal-fetal interface of RM pregnancy
A. Box plots displaying proportion of decidual immune cell subsets in Ctrl and RM patients. B. Differences in overall information flow of the significant signaling pathways between Ctrl and RM decidua. All significant signaling pathways collected by CellChat were ranked based on their differences in overall information flow within the inferred networks between Ctrl and RM. Signaling pathways colored in blue are more enriched in Ctrl, pathways colored in black are equally enriched in Ctrl and RM, and pathways colored in red are more enriched in RM. C. Dot plots showing the alteration in outgoing (ligand) or incoming (receptor) signaling pathways in decidual leukocyte subsets from Ctrl (top) and RM (bottom) patients. The dot size is proportional to the contribution score, which is calculated from pattern recognition analysis. Higher contribution score suggests that the signaling pathway is more enriched in the corresponding cell subset. D. Volcano plot showing the differentially expressed genes (absolute log2 FC > 0.5 and Bonferroni-adjusted P < 0.05) in decidual macrophages between Ctrl and RM patients, using the method of non-parametric two-sided Wilcoxon rank sum test in Seurat. E. Violin plots illustrating the differential expression of inflammation-associated genes in decidual macrophages in Ctrl and RM patients.
dM cells in RM pregnancy preferentially display pro-inflammatory feature
Alteration in global gene expression profiles in the leukocyte subset from RM patients was typically observed in dM cells. As shown in the volcano plot, using absolute log2 fold change (FC) > 0.5 and Bonferroni-adjusted P < 0.05 as cutoffs, 96 upregulated genes and 27 downregulated genes were identified in dM cells from RM patients relative to its normal counterpart (Figure 3D).
dM cells mediate multifunctional immunoregulation, such as antigen presentation, phagocytosis, and recruitment of other immune cells [22], [23]. The increased expression of CXCL8, TNF, and IFIT2 in dM cells from RM patients indicates impaired immune protection. Moreover, we also observed decreases in the expression of angiogenic factor gene VEGFA and immunosuppressive molecule-coding gene LAGLAS1, but increases in the expression of pro-inflammatory factor-coding genes CXCL8 and TNF in dM cells from RM patients (Figure 3E). These data indicate dM cells in RM patients may fail to modulate uterine vessel remodeling, but they are preferentially pro-inflammatory instead.
Molecular aspects and developmental trajectories of human dNK subtypes
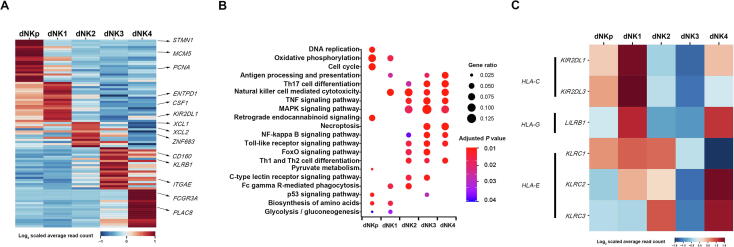
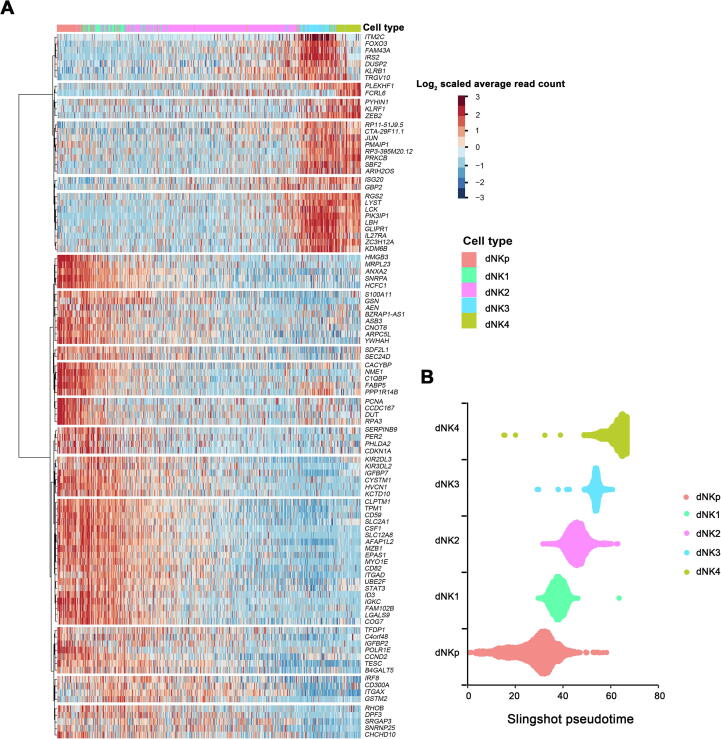
Consistent with the findings in previous literature [24], [25], dNK cells account for more than 70% of total decidual leukocytes in our dataset. Characteristics of the five dNK subpopulations were further illustrated by the expression of top 20 marker genes selected for each subset using method of non-parametric two-sided Wilcoxon rank sum test in Seurat (Figure 4A) and KEGG pathway analysis (Figure 4B). As shown, dNKp cells highly expressed MCM5, STMN1, and PCNA, indicating their manifest proliferative capacity. dNK1 cells strongly expressed ENTPD1/CD39, CSF1, and KIR2DL1, and showed active oxidative phosphorylation and glycolysis activities. The defining markers for dNK2 cells included XCL1, XCL2, and ZNF683. dNK3 cells were defined by ITGAE/ CD103, CD160, and KLRB1, and exhibited certain cytotoxic and immune-activated property. FCGR3A/CD16, and PLAC8 were enriched in dNK4 cells, which indicates an active cytotoxicity status of these cells.
Figure 4.
Molecular annotation of human dNK subtypes
A. Heatmap showing the relative expression of the top marker genes defining the 5 dNK subsets. The top 20 genes for each subset are defined using the method of non-parametric two-sided Wilcoxon rank sum test in Seurat, and typical marker genes for each subset are indicated om the right. B. KEGG enrichment analysis using differential genes (log2 FC > 0.5 and Bonferroni-adjusted P < 0.05) from the five dNK subsets to illustrate the functional signature of these cells. The differential expression analyses is performed using the method of non-parametric two-sided Wilcoxon rank sum test in Seurat. C. Heatmap showing the expression of genes encoding receptors for HLA antigens in dNK subsets. HLA, human leukocyte antigen.
It has been well known that dNK cells possess various receptors for recognition of HLA-C, HLA-G, and HLA-E molecules in placental trophoblast cells, which primarily mediate dNK–trophoblast communications [6]. We found that dNK1 cells expressed high level of KIR2DL3 and KIR2DL1, which encode inhibitory killer cell immunoglobulin like receptor (KIR) for HLA-C, and LILRB1, which encodes the high-affinity receptor for HLA-G. dNK1 and dNK2 cells also moderately expressed KLRC2/NKG2C, KLRC3/NKG2E, and KLRC1/NKG2A, which are receptors for HLA-E. dNK3 cells possessed much lower gene expression of all these receptors. Interestingly, high levels of LILRB1, KLRC2/NKG2C, and KLRC3/NKG2E were observed in dNK4 cells (Figure 4C). Interestingly, LILRB1 has been indicated as a marker gene of 'memory' NK cells [7]. The high expression of LILRB1 in RM-enriched dNK4 cells may indicate the participation of the CD16+ dNK4 cells in the repeated adverse pregnancy outcomes of RM patients.
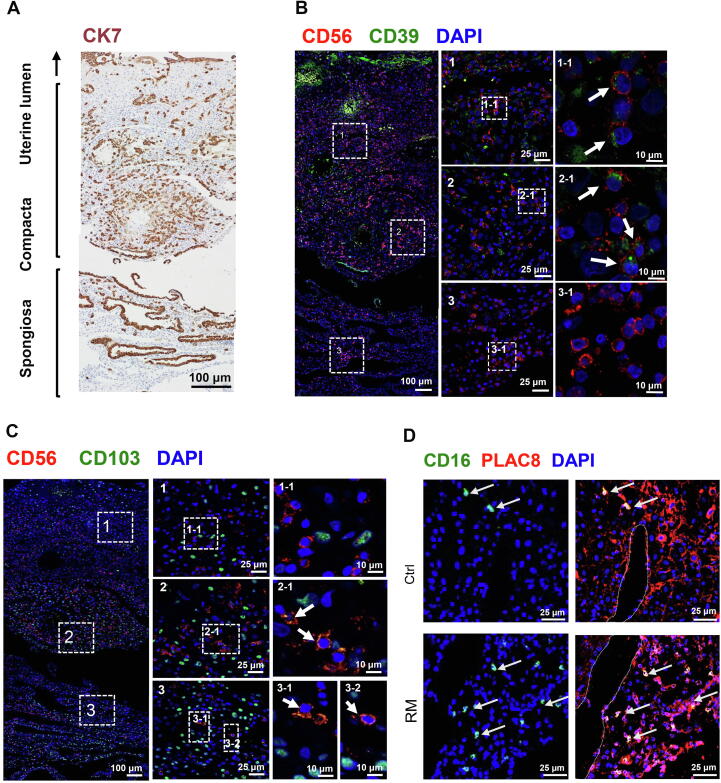
To determine whether the dNK subtypes exhibit cell type-dependent localization pattern at the feto-maternal interface, we performed immunofluorescent staining for CD56, CD39, CD103, CD16, and PLAC8 on serial sections of decidual tissues. Immunohistochemistry for cytokeratin 7 (CK7) was used to locate trophoblast and uterine gland epithelium in the sections (Figure 5A). CD39+ dNK1 cells were more abundant in decidual compacta where trophoblasts infiltrate, while CD103+ dNK3 cells were mainly distributed in distal compacta through spongiosa, but less in proximal compacta (Figure 5A–C). A very small number of CD16-expressing dNK4 cells could be found in decidual compacta, usually near uterine spiral artery (Figure 5D). These results suggest the potential of dNK1 cells in the recognition and protection of trophoblasts, and of dNK3 cells in maintaining cytotoxic and immune-active environment in the decidua.
Figure 5.
Spatial distribution of dNK subsets at the feto-maternal interface at early pregnancy
A. A representative immunohistochemistry image for CK7 (brown), labeling trophoblasts and uterine gland epithelium in a paraffin section of the feto-maternal interface at gestational week 7. Spongiosa, decidual spongiosa; Compacta, decidual compacta. B. Representative immunofluorescent images of sections adjacent to the section in panel A, showing the distribution of dNK1 cells (CD56+CD39+; white arrows) at the feto-maternal interface. C. Representative immunofluorescent images of sections adjacent to the section in panel A, showing the distribution of dNK3 cells (CD56+CD103+; white arrows) at the feto-maternal interface. In both panels, boxed areas from different regions of the feto-maternal interface on the left (scale bar, 100 μm) are zoomed in and shown in the middle (scale bar, 25 μm), and boxed areas in the middle are further zoomed in and shown on the right (scale bar, 10 μm). D. Representative immunofluorescent images showing the distribution of dNK4 cells (CD16+ PLAC8+; white arrows) in decidual compacta from Ctrl (top) and RM (bottom) pregnancies. Dotted lines indicate the position of uterine blood vessels. Scale bar, 25 μm.
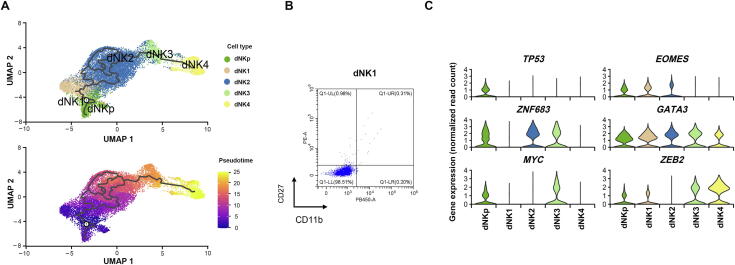
Interestingly, apart from dNK4 cells, the other four dNK subsets co-expressed the tissue-resident markers ITGA1/CD49A, indicating their uterine resident feature. By far, the developmental route of uterine resident dNK cells remains unclear. Therefore, we further analyzed the developmental trajectory of dNK cells by carrying out pseudotime analysis on our scRNA-seq data. As shown, the trajectory of the root cells in dNK1 sequentially passed through dNK2 cells with a complex series of branches and reunions, and further through dNK3 to dNK4 toward the end of trajectory (Figure 6A). Similar results were obtained using Slingshot [26] and general additive model (GAM) presenting top 100 genes whose expression changed in a continuous manner over pseudotime of dNK development (Figure S4). Furthermore, flow cytometry analysis showed dNK1 cells as CD27−CD11b− (Figure 6B), indicating their immature developmental status according to previous studies [27]. dNK1 cells emerged at the early developmental stage with high expression of classical transcription factor gene EOMES that marks immature NK cells. These cells tend to differentiate toward dNK2 and finally dNK3 cells, as partially indicated by the expression of ZNF683, MYC, and ZEB2 that mark mature NK cells (Figure 6C). Distance between dNK3 and dNK4 cells indicated the probably alternative origin of CD49a−CD56+CD16+ dNK4 cells (Figure 6A).
Figure 6.
Developmental trajectories of human dNK subtypes
A. Monocle3 analysis displaying the developmental trajectories of dNK subsets (top), and an additional representation of trajectory over calculated pseudotime (bottom). B. Flow cytometry analysis demonstrating the low expression of CD27 and CD11b in dNK1 cells. C. Violin plots illustrating the expression of classical transcription factor genes in the 5 dNK subsets.
Altered cell proportion and transcription patterns of dNK subsets in the decidua of RM pregnancy
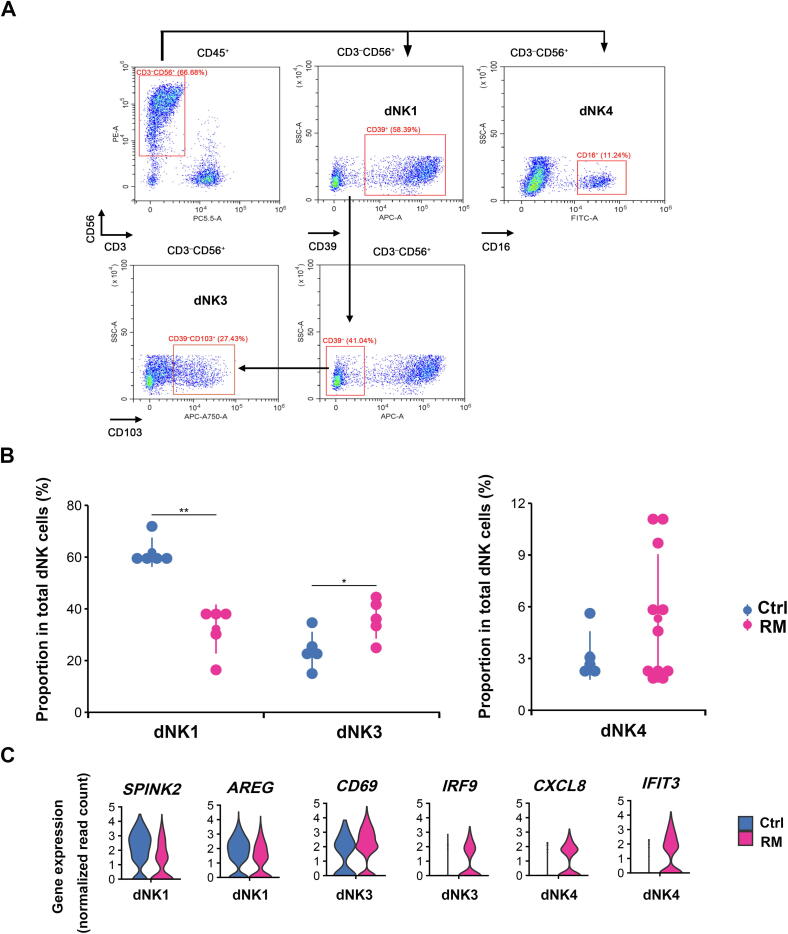
To verify the feature of dNK subset proportion in decidual tissues from RM patients as identified by scRNA-seq in Figure 3A, we analyzed dNK cell population in decidual tissues by flow cytometry in a larger sample cohort of normal (n = 5) and RM (n = 5 for dNK1 and dNK3; n = 11 for dNK4) subjects (Table 1). We sorted cells by using CD56 (expressed by all dNK cells) and CD3 (negative sorting marker), combined with markers for dNK1 (CD39), dNK3 (CD103), and dNK4 (CD16) that were identified from our scRNA-seq data (Figure 7A). As shown, percentage of dNK1 cells was uniformly and significantly reduced by around 50% in RM patients (unpaired Student’s t-test, P < 0.01), whereas percentage of dNK3 cells was significantly increased by approximately 1-fold (unpaired Student’s t-test, P < 0.05) in RM patients, compared to the corresponding normal pregnant controls (Figure 7B). Percentage of dNK4 cells was rather low in normal decidua, accounting for < 4% of total dNK cells. However, in approximately half of the enrolled RM cases, dNK4 cell population increased to about 12% of total NK cells (Figure 7B). Consistently, immunofluorescent staining for CD16 in decidual tissues from RM patients showed evidently more dNK4 cells in decidua compacta niche close to blood vessels (Figure 5D).
Figure 7.
Alterations of dNK subsets in the decidua of RM pregnancy
A. Representative flow cytometry plots showing the strategy to sort dNK cells in human decidual tissues at gestational weeks 6–8, including dNK1 (CD3−CD56+CD39+), dNK3 (CD3−CD56+CD39−CD103+), and dNK4 (CD3−CD56+CD16+). B. Quantification of cell population of dNK1 and dNK3 (left panel), as well as dNK4 (right panel) subsets in decidual tissues from Ctrl (n = 5) and RM patients (n = 5 in left panel, and n = 11 in right panel). Significant difference between the two groups is analyzed using unpaired t-test. *, P < 0.05; **, P < 0.01. C. Violin plots illustrating the expression of typical differential genes (absolute log2 FC > 0.5 and Bonferroni-adjusted P < 0.05) in dNK1, dNK3, and dNK4 cells from Ctrl and RM patients. The differential expression analysis is performed using the method of non-parametric two-sided Wilcoxon rank sum test in Seurat.
Comparison of the differential gene expression in dNK subsets between RM and normal pregnancies demonstrated increased expression of inflammation-related genes such as CD69 and IRF9 in dNK3 cells, CXCL8 and IFIT3 in dNK4 cells, as well as downregulation of anti-inflammation genes including SPINK2 and AREG in dNK1 cells from RM patients (Figure 7C).
Discussion
Cellular and molecular mechanisms accounting for maternal immune tolerance to semi-allogenic fetus in placental mammals have been a fascinating scientific question. Increasing evidence demonstrates the significance of the complicated and precisely controlled cell–cell communications within the feto-maternal interface in concert with the well-coordinated systematic immune adaptations along gestation [1], [2], [28], [29], [30]. To our knowledge, our study is the first comprehensive single-cell transcriptomics atlas of the decidual and peripheral immune cells in human RM pregnancy at early gestation. We integrate complex information of immune cells, including cell composition, functional status, and developmental trajectory, predict cellular interactions among various subsets, and illustrate an integral framework of the compromised immune environment in RM pregnancy.
The immuno-mechanism for RM, especially those without fetal chromosomal or congenital abnormalities or other known pathological causes, has been largely debatable. Studies focusing on limited cell types may result in contradictive observations or misleading interpretations. To date, strategies for early diagnosis or intervention of RM have been lacking, making it hard to reduce the threat to the patients [31]. Here by taking advantage of single-cell sequencing technology, we profile the overall pathological change in the properties of leukocyte subsets in peripheral blood, which may pave the way to pre-symptomatic diagnosis of this disordered pregnancy. The significantly enhanced differentiation of CD8T naive cells to CD8T effector cells, and a preferential increase in number of NK dim cells, together with the upregulated expression of inflammation-related genes, demonstrate a systematic pro-inflammatory status in RM patients. These results are consistent with previous reports [32], [33]. An interesting finding is the increased frequency of MAIT cells in the peripheral blood of RM patients. As a non-conventional T cell subset, MAIT cells are mainly activated by exposure to microbes, while they can also be turned on by inflammatory stimuli in the absence of T-cell receptor-mediated antigen recognition. There is evidence suggesting the involvement of MAIT cells in a broad range of infectious and non-infectious diseases [34]. Studies in liver disorders suggest that MAIT cells may play a protective role against bacterial infections in a normal liver, but might be detrimental, with over-inflammation, in liver diseases [35]. Notably, MAIT cells express several cytokine receptors, including IL-1R, IL-7R, IL-12R, IL-15R, IL-18R, and IL-23R, and respond to the stimulations of multiple cytokines [34]. Our sequencing data showed the evidently enhanced expression of IRF1 and RORA in MAIT cells of periphery blood from RM patients, indicating their highly immune-activated property. By far, evidence supporting the role of MAIT cells in pregnant condition has been very limited, and our study for the first time indicates the association of MAIT activation with the maternal inflammation condition that may be detrimental to fetal survival. However, whether the change of MAIT activation is the cause or effect of miscarriage remains to be established.
At the maternal-fetal interface, decidual NK cells constitute the largest population of decidual leukocytes at early pregnancy. Unlike their peripheral counterparts, dNK cells are less cytotoxic but actively secret a vast array of factors and cytokines. They have been found to play roles in many processes, including facilitating the remodeling of uterine spiral arteries, promoting trophoblast invasion and fetal growth, regulating T cell differentiation, and increasing the availability of maternal blood at the implantation site [2]. In unexplained infertile patients, substantially fewer uterine NK cells were observed when compared to fertile controls [36]. However, discrepant results have been reported regarding the change of dNK cell number in RM patients [31], [37]. Our study clearly demonstrates the abnormal properties of dNK subsets in decidua from RM patients, including subset composition and gene expression, which suggest impaired regulation of dNK subset development in these patients.
The highly active dNK1 subset is numerically dominant dNK cells and demonstrates the typical dNK functions mentioned above. The CD11b− CD27− property of dNK1 cells manifests their immature characteristic and the potential to differentiate [27]. Specific expression of pre-b-cell leukemia homeobox 1 (PBX1) was found in CD27− CD11b− dNK cells, and a recent study has demonstrated the association of decreased PBX1 expression or PBX1G21S mutation with unexplained RM [38], further suggesting the crucial role of this dNK subset in pregnancy maintenance. Interestingly, a recently identified pregnancy trained dNK (PT dNK) subset share similar features with dNK1 [7]. This subset has been speculated to be enriched in decidua and boost decidua receptivity in subsequent pregnancy, therefore it may be responsible for the “memory” of reproductive outcomes in next pregnancies. Thus, the strikingly lowered frequency of this subset in RM patients may partially explain why failed pregnancies repeatedly occur in these patients. In addition to the decreased proportion, the altered gene expression in dNK1 cells from RM decidua further indicates their diminished immune-protective capability.
The development trajectory of dNK subsets reveals that dNK3 cells are relatively mature and immune-activated. They are likely to maintain certain levels of cytotoxic and immune-active environment in decidua, which may contribute to the appropriate degree of trophoblast invasion. However, the portion of dNK3 cells in decidua from RM patients rises to more than one-fold of that from their normal counterparts. Besides, they exert enhanced production of pro-inflammatory cytokines, which may render the decidual environment harmful to the fetal cells and other maternal immune cells, such as T cells and macrophages [39], [40], [41], [42], [43].
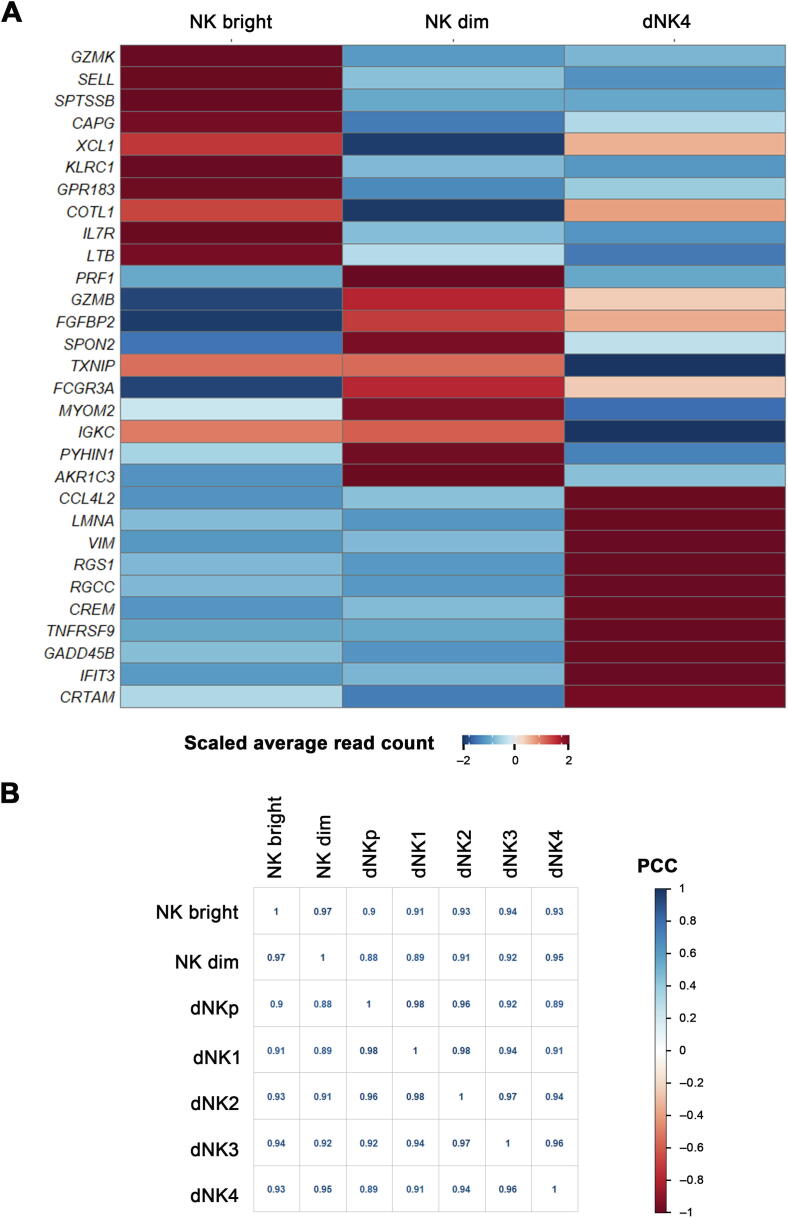
Another notable dNK subset in our study is the newly identified CD56+CD16+ dNK4 subset, which is largely enriched in RM decidua. Being different from all the other four subsets, dNK4 cells do not express the tissue-resident marker CD49a. Consistently, the developmental trajectory shows the separation of dNK4 from the maturation route of other dNK subsets. It is therefore likely that this subset may origin from other sources. Currently there are three hypotheses for the origin of dNK cells, i.e., (1) recruitment of peripheral blood NK cells to decidua [44], (2) maturation of uterine-resident NK cells in response to IL15 or progesterone [45], and (3) direct differentiation from hematopoietic precursors in the decidua upon stimulation of specific decidual factors [46]. Considering the common CD49a−CD16+ feature of dNK4 and peripheral NK cells, we compared the gene expression pattern of dNK4 and pNK (NK dim and NK bright) cells and found obvious difference in gene expression patterns among these three NK subsets. Analysis of the correlation between dNK4 and other NK subsets based on the overall transcription level revealed a closer transcription pattern between dNK3 and dNK4 (Figure S5). Thus, it is likely that peripheral NK dim cells are recruited to the decidua and further educated toward dNK4 by some decidual factors. Many studies have demonstrated the crucial role of IFN-γ, CCL3/MIP-1, CXCL10/IP-10, CXCL12/SDF-1 in enrolling pNK to decidua [47], [48], [49]. Moreover, an in vitro study reveals that human peripheral CD16+ NK cells can be converted to a dNK-like phenotype upon the stimulation of hypoxia, TGFβ, and the demethylating agent 5-aza-2′-deoxycytidine [50]. Here our data reveal the evident increase in INFG in dCD8T, as well as CXCL16, CCL3, and TGFB in dM cells of decidua from RM patients, which may be responsible for enrolling and educating excessive pNK cells to decidua compartment. The much higher expression of several pro-inflammatory factors in dNK4 cells from RM patients also indicates their enhanced cytotoxicity in these patients. An interesting previous study demonstrated the extravagant enrichment of CD16+ NK cells in endometrium of RM patients during their pre-pregnancy period [51]. Taking into consideration the strong expression of LILRB1 in dNK4 cells, which indicates their “memory” of pregnancy outcomes, we propose that patients with more abundant dNK4 cells may suffer from a greater chance of pregnancy failure in their subsequent pregnancies.
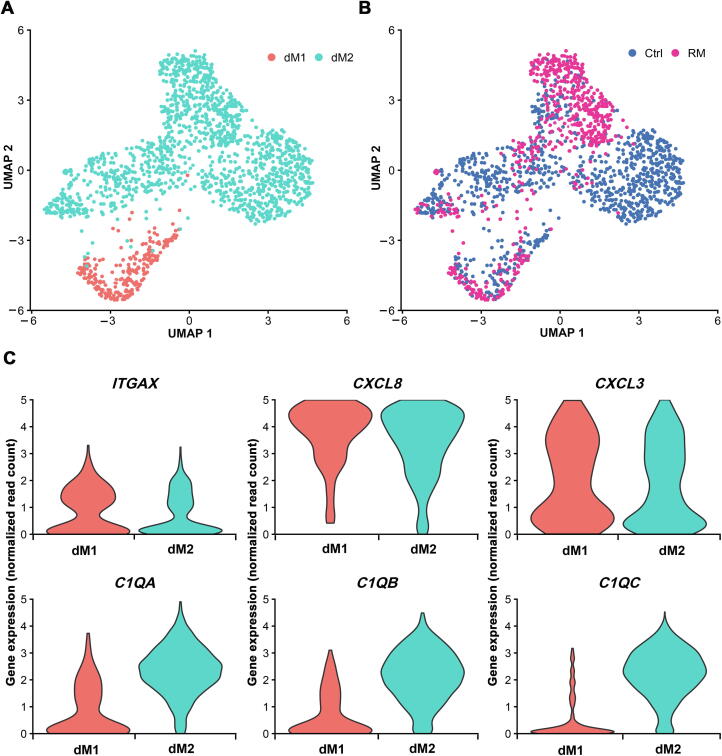
Decidual macrophages account for around 20% of leukocytes at the feto-maternal interface, and they have many diverse functions during pregnancy. In this study, a relatively large number of differentially expressed genes were identified in decidual macrophages from RM patients, which potentially suggest their functional abnormalities. For instance, the obviously repressed expression of VEGFA in dM cells from RM patients indicates the link to impaired remodeling of the spiral arteries and angiogenesis. The upregulated genes include those encoding immunoinflammatory factors and the relevant signaling molecules, such as CXCL8, TNF, IFIT2, JUN, and JUNB, predicting the diminishment of the anti-inflammatory capacity. Furthermore, the increased expression of CXCL8 in dM cells and enhanced IFNG expression in dNK4 of RM patients are likely highly correlated, since it has been demonstrated that CXCL8 (IL-8) from dM cells enhances the production of IFN-γ in dNK cells [52], [53]. In addition, our analysis of the ligand–receptor signaling pathways indicates the potential of dM cells in recruiting dCD8T or dNK4 through CXCL16-CXCR6 or TNF-TNFRSF1B interactions. Although decidual macrophages are believed to exist predominantly in a regulatory/homeostatic M2-like phenotype, while less in pro-inflammatory M1 phenotype during pregnancy [1], we did not find an alteration in the proportion of dM1 and dM2 cells in RM patients (Figure S6). This is probably due to the relatively small amount of captured dM cells for sequencing.
In general, our study comprehensively illustrates the compromised immune response in periphery and feto-maternal interface of RM patients. The findings generate a data-driven hypothesis about immune-related pathogenesis for recurrent miscarriage. Further functional studies using appropriate in vitro or in vivo models are necessary to eventually clarify the immune causes of RM, which may provide new insight into strategies for intervention of RM.
Materials and methods
Sample collection
Clinical samples of anti-coagulant peripheral blood and decidual tissues from normal (n = 10) or RM (n = 14) pregnancies at gestational weeks 6–8 were obtained upon therapeutic termination of pregnancy at Peking University Third Hospital, Beijing, China. The decidual tissues were immersed in iced RPMI-1640 medium and the blood samples were kept on ice. All samples were subjected to cell isolation or fixation within 1 h following the surgery.
RM was defined according to the criteria of Practice Committee of the American Society for Reproductive Medicine. In brief, these patients had history of two or more failed pregnancies with unknown cause [54]. Women who manifest endocrine disorder, fetal chromosomal or congenital abnormalities, uterine anatomical disorders, renal disease, or pregnancies conceived by fertility treatment were excluded from this study. In addition, to avoid the influence of secondary inflammation by a prolonged in utero fetal death as far as possible, the enrolled RM patients had undergone careful medical care every week, with ultrasound monitor every two weeks. Within 2–3 days of the fetal heartbeat ceasing, the patients took an induced abortion. Therefore, all the RM cases examined in this study were early missed abortion cases (at a very early stage of abortion), but not inevitable abortion cases (at a late stage of abortion). Moreover, we excluded miscarriage cases with abnormal fetal chromosomal karyotypes. The clinical characteristics of the enrolled pregnant women were summarized in Table 1.
Cell isolation and purification
Freshly collected human decidual tissues were trimmed into 1-mm3 piece by GentelMACS Dissociator (Catalog No. 130-093-235, Miltenyi Biotec, Bergisch Gladbach, Germany) and digested twice for 30 min each at 37 °C with 1.0 mg/ml type IV collagenase (Catalog No. 9001121, Gibco, Grand Island, NY) and 10 U/ml type I DNase (Catalog No. DN25, Sigma, St Louis, MO). The cell suspensions were filtered through 60 mesh and 200 mesh sieves and were collected by centrifuging at 1000 rpm for 10 min. The resuspended cells were subjected to lymphocyte enrichment using Ficoll-Paque Plus (Catalog No. 17-1440-02, GE Healthcare, Buckinghamshire, UK). The decidual leukocytes were further purified by FACS with 7-aminoactinomycin D (7-AAD; Catalog No. 420404, BioLegend, San Diego, CA) and FITC-labeled anti-CD45 antibody (Catalog No. 11-0459-42, eBioscience, San Diego, CA).
Freshly collected peripheral blood samples were diluted 1:1 with PBS and subjected to lymphocyte enrichment using Ficoll-Paque. The peripheral blood leukocytes were further purified by FACS with 7-AAD and FITC-labeled anti-CD45 antibody.
Freshly purified decidual or peripheral leukocytes were immediately subjected to scRNA-seq as described below.
Generation of single-cell library and transcriptome sequencing
The purified leukocytes from three pairs of normal and RM cases were separately loaded on Chromium Single Cell Controller (10X Genomics, Pleasanton, CA) using the Chromium Single Cell 3’ kit v2 to capture 5000–8000 cells per sample. Libraries were sequenced on an Illumina NovaSeq 6000 with a read length of 26 bp for read 1 (cell barcode and UMI), 8 bp i7 index read (sample barcode), and 98 bp for read 2 (actual RNA read). Reads were first sequenced in the rapid run mode, allowing for fine-tuning sample ratios in the following high-output run. Combining the data from both flow cells yielded approximately > 40,000 reads per cell.
scRNA-seq data processing and analysis
The raw sequencing reads were processed using Cell Ranger (version 2.0.1) [55]. The reference index was built using the GRCh38 (Ensembl 93) human reference genome assembly. Cells with < 600 detected genes or with the total mitochondrial gene expression > 5% were removed. Then we converted the obtained matrix into a Seurat object for downstream analysis. All Seurat objects for individual samples were merged into one combined object. According to the integration method reciprocal PCA provided by Seurat, we first performed standard normalization and variable feature selection on each individual sample. Next, we selected features for downstream integration, and ran PCA on each individual sample in the combined object. Given our data include RM patients and normal pregnant women, we chose data from one RM patient and one normal pregnant woman as the reference and then used the function FindIntegrationAnchors provided by Seurat to integrate the samples of 6 individuals. Downstream analyses, including normalization, shared nearest neighbor graph-based clustering, differential expression analysis, and visualization, were performed using the standard workflow provided by Seurat (version 3.0.3). Differentially expressed genes (log2 FC > 0.5 and Bonferroni-adjusted P < 0.05) from the comparison of the five dNK subsets were subjected to KEGG analysis using R package clusterProfiler (version 3.12.0) to illustrate the functional signature of the 5 dNK subsets. The differential expression analyses were performed using the method of non-parametric two-sided Wilcoxon rank sum test in Seurat. [56].
Pseudotime trajectory analysis
Developmental trajectories were inferred with the Monocle3 (version 0.2.3.0) [57]. Corrected principal component (PC) values for the merged RM or normal pregnancy datasets (PC values were integrated by reciprocal PCA provided by Seurat) were used as input for the UMAP dimension reduction based on construction of pseudotime trajectories. The pseudotime analysis was validated using Slingshot (version 1.4.0) R package [26]. After running Slingshot, genes with altered expression over the development course of dNK cells were identified, and GAM was used to regress each gene on the pseudotime variable.
Cell communication analysis
A systematic analysis of cell communication was based on the network analysis and pattern recognition approaches provided by CellChat (version 0.0.1) R package [20]. We used the standard workflow to predict major signaling inputs and outputs of cells and how these cells and signals coordinate for functions. Subsequently, we classified signaling pathways and depicted conserved and context-specific pathways between RM and normal pregnancies.
Immunostaining analysis
Human decidual tissues were briefly fixed in 4% paraformaldehyde (PFA) and embedded in O.C.T. Compound (Catalog No. 4583, Sakura Finetek, Torrance, CA). The frozen sections at 10 μm were further fixed in 4% PFA and treated with 0.1% triton, and subjected to the incubation with specific antibodies against NCAM1/CD56 (Catalog No. ab75813, Abcam, Cambridge, MA), CK7 (Catalog No. ab181598, Abcam), CD39 (Catalog No. 14211-1-AP, Proteintech, Wuhan, China), or CD103 (Catalog No. 350227, BioLegend). Binding of the antibody was visualized using FITC-conjugated or TRITC-conjugated secondary antibody (Catalog No. ZF-0311 or ZF-0313, ZSGB-BIO, Beijing, China), and cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; Catalog No. 28718-90-3, Sigma). Immunofluorescent staining was examined using Zeiss LSM780 confocal system (Carl Zeiss, Jena, Germany) and processed with ZEN 2012 software (Carl Zeiss). Immunohistochemical staining for CK in decidua was performed by using antibody against CK7 (Catalog No. ab181598, Abcam) and HRP-conjugated second antibody (Catalog No. PV-6001, ZSGB-BIO) followed by recovery of substrate diaminodbenzidine (DAB) (Catalog No. ZLI-9019, ZSGB-BIO). The imagines were recorded on a light microscope with charge-coupled device (CCD) (Olympus, Tokyo, Japan).
Flow cytometry assay
Flow cytometry assay for dNK cells was carried out in CytoFLEX (Beckman Coulter, Miami, FL) using the following antibodies: PE-labeled anti-CD56 (Catalog No. 362508, BioLegend), PerCP-Cy5.5-labeled anti-CD3 (Catalog No. 300328, BioLegend), APC-labeled ani-CD39 (Catalog No. 328209, BioLegend), APC-Cy7-labeled anti-CD103 (Catalog No. 350227, BioLegend), FITC-labeled anti-CD16 (Catalog No. 302206, BioLegend), PE-labeled anti-CD27 (Catalog No. 356405, BioLegend), and Pacific Blue-labeled anti CD11b (Catalog No. 301316, BioLegend) according to the manufacturer’s instructions. Data were analyzed using CytExpert (Beckman Coulter).
Statistical analysis
Comparison of cell proportions between normal and RM pregnancies was analyzed with GraphPad Prism version 7.00 (GraphPad Software, San Diego, CA). Data were shown as mean ± SEM and comparison was carried out by unpaired Student’s t-test. Differences with P < 0.05 were considered significant.
Ethical statement
The collection of human samples was approved by the Local Ethical Committees in Peking University Third Hospital. The written informed consent was obtained from the participating subjects.
Data availability
The raw sequencing data reported in this paper have been deposited in the Genome Sequence Archive [58] in the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: HRA000237 with BioProject: PRJCA003061), and are publicly accessible at http://bigd.big.ac.cn/gsa-human and https://ngdc.cncb.ac.cn/bioproject/.
CRediT author statement
Feiyang Wang: Conceptualization, Software, Writing - review & editing. Wentong Jia: Validation, Data curation. Mengjie Fan: Resources, Data curation. Xuan Shao: Validation, Data curation. Zhilang Li: Data curation. Yongjie Liu: Software, Methodology. Yeling Ma: Investigation, Writing - review & editing. Yu-Xia Li: Methodology. Rong Li: Resources, Methodology. Qiang Tu: Software, Methodology, Writing - review & editing. Yan-Ling Wang: Conceptualization, Supervision, Writing - review & editing, Funding acquisition. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Key R&D Program of China (Grant Nos. 2018YFC1004100, 2017YFC1001404, 2016YFC1000401, and 2016YFC1000200) and the National Natural Science Foundation of China (Grant Nos. 81730040 and 81490740). We are grateful to Prof. Bin Cao at Xiamen University and Mr. Zhenghui Zhao at Institute of Zoology, Chinese Academy of Sciences for their critical comments to this study. The technical support from Shiwen Li, Xia Yang, and Qing Meng for confocal analysis and FACS is appreciated. We acknowledge all the enrolled patients for their contribution to the study.
Handled by Fuchou Tang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2020.11.002.
Contributor Information
Rong Li, Email: roseli001@sina.com.
Qiang Tu, Email: qtu@genetics.ac.cn.
Yan-Ling Wang, Email: wangyl@ioz.ac.cn.
Supplementary material
The following are the Supplementary data to this article:
Supplementary Figure S1.
Gating strategy and quality control for droplet scRNA-seq A. Flow sorting of living leukocytes from peripheral blood and decidual tissue using CD45 antibody and 7-AAD. B. Total number of cells that met the requirement of quality control and subjected to droplet scRNA-seq. C. Workflow depicting the dissociation and sorting of CD45+ leukocytes from peripheral blood and decidual to generate scRNA transcriptome profile. D. Quality evaluation on sequencing data including the total number of reads per cell (left panel) and total number of genes per cell (right panel) and in peripheral blood and decidua. 7-AAD, 7-aminoactinomycin D.
Supplementary Figure S2.
MNN algorithm to confirm the elimination of batch effect of the data A. Data from the 6 samples were integrated with MNN and visualized using UMAP. B. Distribution of leukocytes from peripheral blood and decidual tissues after MNN integration. C. MNN 1 and MNN 2 of the integral analysis of decidual and peripheral blood leukocytes from the droplet-based datasets. D. Distribution of leukocytes from Ctrl and RM patients after MNN integration. MNN, mutual nearest neighbor.
Supplementary Figure S3.
Identification of peripheral blood T cell subpopulations by specific marker genes A. UMAP visualization of peripheral blood T cells. Feature plot of naive T cells (B), MAIT cells (C), CD4T memory cells (D), and CD8T effector cells (E) with expression of their specific marker genes.
Supplementary Figure S4.
Analysis of dNK pseudotime trajectory using Slingshot A. Heatmap showing the top 100 variable genes in dNK cells that change in a continuous manner over pseudotime. The top 100 variable were selected by using GAM to calculate a dynamic changes over pseudotime in dNK development. B. Slingshot map pseudotime analysis of dNK cells. GAM, general additive model.
Supplementary Figure S5.
Global gene expression analysis in CD16+ NK cells including dNK4 and peripheral NK bright and NK dim subsets A. Heatmap showing the top 10 differentially expressed genes (log2FC > 0.5 and Bonferroni-adjusted P < 0.05) in three NK subsets by using the method of non-parametric two-sided Wilcoxon rank sum test in Seurat. B. Correlation analyses between dNK4 and other NK subsets based on the overall transcription level. PCC, Pearson correlation coefficient.
Supplementary Figure S6.
Identification of decidual macrophage subpopulations A. UMAP showing two subpopulations of decidual macrophage. B. UMAP showing decidual macrophage subpopulations in Ctrl (blue) and RM (red) patients. C. Characterization of the two decidual macrophage subpopulations using marker genes.
References
- 1.Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. 2013;31:387–411. doi: 10.1146/annurev-immunol-032712-100003. [DOI] [PubMed] [Google Scholar]
- 2.Ander S.E., Diamond M.S., Coyne C.B. Immune responses at the maternal-fetal interface. Sci Immunol. 2019;4:eaat6114. doi: 10.1126/sciimmunol.aat6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher S.J. Why is placentation abnormal in preeclampsia? Am J Obstet Gynecol. 2015;213:S115–S122. doi: 10.1016/j.ajog.2015.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kheshtchin N., Gharagozloo M., Andalib A., Ghahiri A., Maracy M.R., Rezaei A. The expression of Th1- and Th2-related chemokine receptors in women with recurrent miscarriage: the impact of lymphocyte immunotherapy. Am J Reprod Immunol. 2010;64:104–112. doi: 10.1111/j.1600-0897.2010.00829.x. [DOI] [PubMed] [Google Scholar]
- 5.Deshmukh H., Way S.S. Immunological basis for recurrent fetal loss and pregnancy complications. Annu Rev Pathol. 2019;14:185–210. doi: 10.1146/annurev-pathmechdis-012418-012743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parham P., Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamliel M., Goldman-Wohl D., Isaacson B., Gur C., Stein N., Yamin R., et al. Trained memory of human uterine NK cells enhances their function in subsequent pregnancies. Immunity. 2018;48:951–962.e5. doi: 10.1016/j.immuni.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Wang W.J., Hao C.F., Yi L., Yin G.J., Bao S.H., Qiu L.H., et al. Increased prevalence of T helper 17 (Th17) cells in peripheral blood and decidua in unexplained recurrent spontaneous abortion patients. J Reprod Immunol. 2010;84:164–170. doi: 10.1016/j.jri.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Yang H., Qiu L., Chen G., Ye Z., Lu C., Lin Q. Proportional change of CD4+CD25+ regulatory T cells in decidua and peripheral blood in unexplained recurrent spontaneous abortion patients. Fertil Steril. 2008;89:656–661. doi: 10.1016/j.fertnstert.2007.03.037. [DOI] [PubMed] [Google Scholar]
- 10.Svensson-Arvelund J., Mehta R.B., Lindau R., Mirrasekhian E., Rodriguez-Martinez H., Berg G., et al. The human fetal placenta promotes tolerance against the semiallogeneic fetus by inducing regulatory T cells and homeostatic M2 macrophages. J Immunol. 2015;194:1534–1544. doi: 10.4049/jimmunol.1401536. [DOI] [PubMed] [Google Scholar]
- 11.Van der Zwan A., Bi K., Norwitz E.R., Crespo A.C., Claas F.H.J., Strominger J.L., et al. Mixed signature of activation and dysfunction allows human decidual CD8+ T cells to provide both tolerance and immunity. Proc Natl Acad Sci U S A. 2018;115:385–390. doi: 10.1073/pnas.1713957115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salvany-Celades M., van der Zwan A., Benner M., Setrajcic-Dragos V., Bougleux Gomes H.A., Iyer V., et al. Three types of functional regulatory T cells control T cell responses at the human maternal-fetal interface. Cell Rep. 2019;27:2537–2547.e5. doi: 10.1016/j.celrep.2019.04.109. [DOI] [PubMed] [Google Scholar]
- 13.Crespo A.C., Mulik S., Dotiwala F., Ansara J.A., Sen Santara S., Ingersoll K., et al. Decidual NK cells transfer granulysin to selectively kill bacteria in trophoblasts. Cell. 2020;182:1125–1139.e18. doi: 10.1016/j.cell.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huhn O., Ivarsson M.A., Gardner L., Hollinshead M., Stinchcombe J.C., Chen P., et al. Distinctive phenotypes and functions of innate lymphoid cells in human decidua during early pregnancy. Nat Commun. 2020;11:381. doi: 10.1038/s41467-019-14123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suryawanshi H., Morozov P., Straus A., Sahasrabudhe N., Max K.E.A., Garzia A., et al. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv. 2018;4:eaau4788. doi: 10.1126/sciadv.aau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Myung P., Plikus M.V., et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du M.R., Wang S.C., Li D.J. The integrative roles of chemokines at the maternal-fetal interface in early pregnancy. Cell Mol Immunol. 2014;11:438–448. doi: 10.1038/cmi.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varol C., Mildner A., Jung S. Macrophages: development and tissue specialization. Annu Rev Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- 23.Gentek R., Molawi K., Sieweke M.H. Tissue macrophage identity and self-renewal. Immunol Rev. 2014;262:56–73. doi: 10.1111/imr.12224. [DOI] [PubMed] [Google Scholar]
- 24.King A., Balendran N., Wooding P., Carter N.P., Loke Y.W. CD3– leukocytes present in the human uterus during early placentation: phenotypic and morphologic characterization of the CD56+ population. Dev Immunol. 1991;1:169–190. doi: 10.1155/1991/83493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulmer J.N., Morrison L., Longfellow M., Ritson A., Pace D. Granulated lymphocytes in human endometrium: histochemical and immunohistochemical studies. Hum Reprod. 1991;6:791–798. doi: 10.1093/oxfordjournals.humrep.a137430. [DOI] [PubMed] [Google Scholar]
- 26.Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu B., Wang F., Sun R., Ling B., Tian Z., Wei H. CD11b and CD27 reflect distinct population and functional specialization in human natural killer cells. Immunology. 2011;133:350–359. doi: 10.1111/j.1365-2567.2011.03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arck P.C., Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat Med. 2013;19:548–556. doi: 10.1038/nm.3160. [DOI] [PubMed] [Google Scholar]
- 29.Nancy P., Tagliani E., Tay C.S., Asp P., Levy D.E., Erlebacher A. Chemokine gene silencing in decidual stromal cells limits T cell access to the maternal-fetal interface. Science. 2012;336:1317–1321. doi: 10.1126/science.1220030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna J., Goldman-Wohl D., Hamani Y., Avraham I., Greenfield C., Natanson-Yaron S., et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006;12:1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 31.Seshadri S., Sunkara S.K. Natural killer cells in female infertility and recurrent miscarriage: a systematic review and meta-analysis. Hum Reprod Update. 2014;20:429–438. doi: 10.1093/humupd/dmt056. [DOI] [PubMed] [Google Scholar]
- 32.Ebina Y., Nishino Y., Deguchi M., Maesawa Y., Nakashima Y., Yamada H. Natural killer cell activity in women with recurrent miscarriage: etiology and pregnancy outcome. J Reprod Immunol. 2017;120:42–47. doi: 10.1016/j.jri.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Kuon R.J., Vomstein K., Weber M., Müller F., Seitz C., Wallwiener S., et al. The “killer cell story” in recurrent miscarriage: association between activated peripheral lymphocytes and uterine natural killer cells. J Reprod Immunol. 2017;119:9–14. doi: 10.1016/j.jri.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Godfrey D.I., Koay H.F., McCluskey J., Gherardin N.A. The biology and functional importance of MAIT cells. Nat Immunol. 2019;20:1110–1128. doi: 10.1038/s41590-019-0444-8. [DOI] [PubMed] [Google Scholar]
- 35.Bertrand L., Lehuen A. MAIT cells in metabolic diseases. Mol Metab. 2019;27:S114–S121. doi: 10.1016/j.molmet.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klentzeris L.D., Bulmer J.N., Warren M.A., Morrison L., Li T.C., Cooke I.D. Lymphoid-tissue in the endometrium of women with unexplained infertility- morphometric and immunohistochemical aspects. Hum Reprod. 1994;9:646–652. doi: 10.1093/oxfordjournals.humrep.a138564. [DOI] [PubMed] [Google Scholar]
- 37.King K., Smith S., Chapman M., Sacks G. Detailed analysis of peripheral blood natural killer (NK) cells in women with recurrent miscarriage. Hum Reprod. 2010;25:52–58. doi: 10.1093/humrep/dep349. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y., Fu B., Xu X., Zhang J., Tong X., Wang Y., et al. PBX1 expression in uterine natural killer cells drives fetal growth. Sci Transl Med. 2020;12:eaax1798. doi: 10.1126/scitranslmed.aax1798. [DOI] [PubMed] [Google Scholar]
- 39.Yang F., Zheng Q., Jin L. Dynamic function and composition changes of immune cells during normal and pathological pregnancy at the maternal-fetal interface. Front Immunol. 2019;10:2317. doi: 10.3389/fimmu.2019.02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L., Li G., Cao G., Zhu Y., Du M., Zhao Y., et al. dNK cells facilitate the interaction between trophoblastic and endothelial cells via VEGF-C and HGF. Immunol Cell Biol. 2017;95:695–704. doi: 10.1038/icb.2017.45. [DOI] [PubMed] [Google Scholar]
- 41.Sotnikova N., Voronin D., Antsiferova Y., Bukina E. Interaction of decidual CD56+ NK with trophoblast cells during normal pregnancy and recurrent spontaneous abortion at early term of gestation. Scand J Immunol. 2014;80:198–208. doi: 10.1111/sji.12196. [DOI] [PubMed] [Google Scholar]
- 42.Fridman W.H., Pages F., Sautes-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 43.Fu B., Li X., Sun R., Tong X., Ling B., Tian Z., et al. Natural killer cells promote immune tolerance by regulating inflammatory TH17 cells at the human maternal-fetal interface. Proc Natl Acad Sci U S A. 2013;110:E231–E240. doi: 10.1073/pnas.1206322110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlino C., Stabile H., Morrone S., Bulla R., Soriani A., Agostinis C., et al. Recruitment of circulating NK cells through decidual tissues: a possible mechanism controlling NK cell accumulation in the uterus during early pregnancy. Blood. 2008;111:3108–3115. doi: 10.1182/blood-2007-08-105965. [DOI] [PubMed] [Google Scholar]
- 45.Manaster I., Mizrahi S., Goldman-Wohl D., Sela H.Y., Stern-Ginossar N., Lankry D., et al. Endometrial NK cells are special immature cells that await pregnancy. J Immunol. 2008;181:1869–1876. doi: 10.4049/jimmunol.181.3.1869. [DOI] [PubMed] [Google Scholar]
- 46.Vacca P., Vitale C., Montaldo E., Conte R., Cantoni C., Fulcheri E., et al. CD34+ hematopoietic precursors are present in human decidua and differentiate into natural killer cells upon interaction with stromal cells. Proc Natl Acad Sci U S A. 2011;108:2402–2407. doi: 10.1073/pnas.1016257108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanna J., Wald O., Goldman-Wohl D., Prus D., Markel G., Gazit R., et al. CXCL12 expression by invasive trophoblasts induces the specific migration of CD16– human natural killer cells. Blood. 2003;102:1569–1577. doi: 10.1182/blood-2003-02-0517. [DOI] [PubMed] [Google Scholar]
- 48.Wu X., Jin L.P., Yuan M.M., Zhu Y., Wang M.Y., Li D.J. Human first-trimester trophoblast cells recruit CD56brightCD16– NK cells into decidua by way of expressing and secreting of CXCL12/stromal cell-derived factor 1. J Immunol. 2005;175:61–68. doi: 10.4049/jimmunol.175.1.61. [DOI] [PubMed] [Google Scholar]
- 49.Wallace A.E., Host A.J., Whitley G.S., Cartwright J.E. Decidual natural killer cell interactions with trophoblasts are impaired in pregnancies at increased risk of preeclampsia. Am J Pathol. 2013;183:1853–1861. doi: 10.1016/j.ajpath.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Dunk C., Croy A.B., Lye S.J. To serve and to protect: the role of decidual innate immune cells on human pregnancy. Cell Tissue Res. 2016;363:249–265. doi: 10.1007/s00441-015-2315-4. [DOI] [PubMed] [Google Scholar]
- 51.Lachapelle M.H., Miron P., Hemmings R., Roy D.C. Endometrial T, B, and NK cells in patients with recurrent spontaneous abortion. Altered profile and pregnancy outcome. J Immunol. 1996;156:4027–4034. [PubMed] [Google Scholar]
- 52.Baratin M., Roetynck S., Lepolard C., Falk C., Sawadogo S., Uematsu S., et al. Natural killer cell and macrophage cooperation in MyD88-dependent innate responses to Plasmodium falciparum. Proc Natl Acad Sci U S A. 2005;102:14747–14752. doi: 10.1073/pnas.0507355102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dalbeth N., Gundle R., Davies R.J., Lee Y.C., McMichael A.J., Callan M.F. CD56bright NK cells are enriched at inflammatory sites and can engage with monocytes in a reciprocal program of activation. J Immunol. 2004;173:6418–6426. doi: 10.4049/jimmunol.173.10.6418. [DOI] [PubMed] [Google Scholar]
- 54.Practice Committee of the American Society for Reproductive M Definitions of infertility and recurrent pregnancy loss: a committee opinion. Fertil Steril. 2013;99:63. doi: 10.1016/j.fertnstert.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 55.Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., et al. GSA: Genome Sequence Archive. Genomics Proteomics Bioinformatics. 2017;15:14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequencing data reported in this paper have been deposited in the Genome Sequence Archive [58] in the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: HRA000237 with BioProject: PRJCA003061), and are publicly accessible at http://bigd.big.ac.cn/gsa-human and https://ngdc.cncb.ac.cn/bioproject/.