Abstract
The astrocyte is a central glial cell and plays a critical role in the architecture and activity of neuronal circuits and brain functions through forming a tripartite synapse with neurons. Emerging evidence suggests that dysfunction of tripartite synaptic connections contributes to a variety of psychiatric and neurodevelopmental disorders. Furthermore, recent advancements with transcriptome profiling, cell biological and physiological approaches have provided new insights into the molecular mechanisms into how astrocytes control synaptogenesis in the brain. In addition to these findings, we have recently developed in vivo cell-surface proximity-dependent biotinylation (BioID) approaches, TurboID-surface and Split-TurboID, to comprehensively understand the molecular composition between astrocytes and neuronal synapses. These proteomic approaches have discovered a novel molecular framework for understanding the tripartite synaptic cleft that arbitrates neuronal circuit formation and function. Here, this short review highlights novel in vivo cell-surface BioID approaches and recent advances in this rapidly evolving field, emphasizing how astrocytes regulate excitatory and inhibitory synapse formation in vitro and in vivo.
Keywords: Split-TurboID, BioID, Astrocyte, Synapse, Neuron, Tripartite synapse, Synaptomics
1. Introduction
Astrocytes are the most abundant glial cells in the central nervous system (CNS) and extend thousands of fine processes that structurally and functionally associate with neuronal synapses to form “tripartite synapses”. Most neuronal synapses (approximately 50-75%) in the cortex and hippocampus are contacted by astrocytes and form tripartite synapses (Lanjakornsiripan et al., 2018; Ventura and Harris, 1999). Astrocytes have traditionally been considered to have key roles in metabolic homeostasis and synaptic transmission through ionic balance and neurotransmitter clearance as supporting cells (Araque et al., 2014). Interestingly, in addition to this traditional concept of astrocytes, recent evidence has discovered that astrocytes also tightly control individual local synaptic development and circuit connectivity in the brain (Allen and Eroglu, 2017; Baldwin and Eroglu, 2017; Stogsdill et al., 2017; Takano et al., 2020). This new conceptual framing of the tripartite synapse has emerged as a rapidly expanding field. It is one of the most exciting topics in cellular neuroscience that is changing our understandings of brain circuitry formation and function. Furthermore, new evidence from genomic and physiological studies reveals that dysfunction of astrocyte-synaptic interactions may contribute to psychiatric and neurodevelopmental disorders (Baldwin and Eroglu, 2017; Stogsdill and Eroglu, 2017; Yu et al., 2020). Despite the vital role of astrocytes in synaptic development and physiology, deciphering the molecular composition of tripartite synaptic connections that drive these processes remains a significant challenge.
In recent years several synapse-specific proteomics profiling techniques, such as a cell sorting of growth cones and synaptosomes, imaging-based approaches, and affinity purification, have been established for analyzing synaptic molecular networks in vivo (Apostolo et al., 2020; Cizeron et al., 2020; Li et al., 2020b; Loh et al., 2016; Micheva et al., 2010; Poulopoulos et al., 2019; Zhu et al., 2018). However, despite these advances, it has been technically difficult to profile the molecular composition at the cell-type-specific cell-cell contacts such as tripartite synaptic sites in vivo. Recently, we developed novel in vivo cell-surface proximity-dependent biotinylation (BioID) approaches, TurboID-surface, and Split-TurboID, and demonstrated they are highly successful for discovering the molecular network of tripartite synaptic connections (Takano et al., 2020). Here, we highlight the application of these novel proteomic approaches and the current understating of how astrocytes control synapse formation and function in vivo.
Cell-surface TurboID-based proteome in vivo
2-1. Proximity-based labeling via BioID and APEX
Spatially-restricted, proximity-dependent biotinylation (BioID and APEX) is a powerful chemico-genetic approach that enables the identification of specific intracellular and extracellular proteomes as they exist in situ (Branon et al., 2018; Kim et al., 2016; Roux et al., 2012; Spence et al., 2019; Uezu et al., 2016). The BioID approach first utilized an Escherichia coli-derived mutant biotin ligase (BirA*-R118G), which generates reactive biotin (biotinoyl-5’- AMP) and has an enhanced off-rate so that biotin covalently attaches to exposed lysine residues of any neighboring protein (approximately 10 nm) (Kim et al., 2016; Roux et al., 2012). More recently, TurboID, a directed-evolution mutant of BirA, was developed that exhibits higher enzymatic activity for proximity-dependent labeling than BirA*-R118G (Branon et al., 2018). For BioID, expression constructs encoding a bait protein fused to BirA*-R118G or TurboID are delivered into the cells in vitro or in vivo, and they are treated with biotin, which is taken up into cells, to label nearby proteins (Fig. 1). The biotinylated proteins are purified by affinity-isolation using streptavidin-coupled beads and then subjected to liquid chromatography-tandem mass spectrometry (LC/MS/MS) to discover the local proteomes (Fig. 1). BioID has high spatial resolution and can label and identify insoluble proteins, membrane-associated proteins, and weak and/or transient protein-protein interactions (Kim et al., 2016; Roux et al., 2012). Indeed, two examples of this approach from our laboratory, gephyrin-BirA that is an inhibitory synaptic organizer, and Wrp-BirA that is a Rac-GAP localized to nascent dendritic spines, both identified a large number of proteins from these synaptic sites in the brain that were previously difficult to dissect by traditional biochemistry methods such as affinity purification and subcellular fractionation method (Spence et al., 2019; Uezu et al., 2016).
Fig. 1.
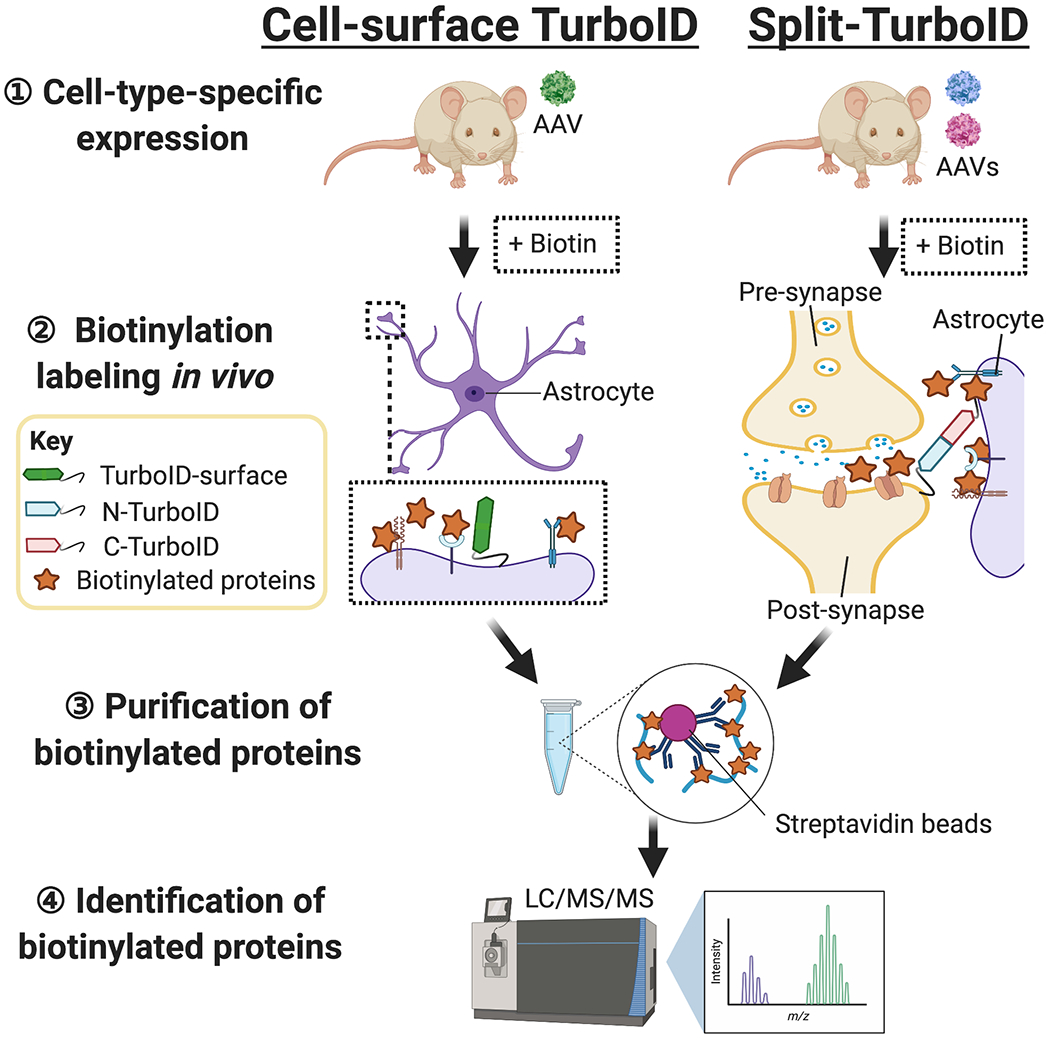
Outline of cell-surface TurboID and Split-TurboID-based proteomics in vivo. 1) Cell-surface TurboID or Split-TurboID is delivered to the brain using cell-type-specific AAVs. 2) After the expression of TurboID probes, mice are given subcutaneous injections of biotin (24 mg/kg) for 7 days to induce proximity-dependent biotinylation labeling at cell-type-specific cell-surface or cell-cell contact sites in vivo. 3) The biotinylated proteins are purified by affinity-isolation using streptavidin-coupled beads. 4) The bound biotinylated proteins are subjected to liquid chromatography-tandem mass spectrometry (LC/MS/MS) to discover the molecular networks.
APEX is an ascorbate peroxidase that catalyzes the oxidation of biotin-phenol to the biotin-phenoxyl radical in the presence of H2O2. APEX-based labeling has some advantages to BirA-based approaches, including the ability to label the proteins by biotinylation within 1 minute in living cells (Loh et al., 2016; Martell et al., 2016). Furthermore, a recent study demonstrates that APEX can be applied to transcriptome profiling through direct proximity labeling of endogenous RNA within specific cellular compartments of the living cells (Fazal et al., 2019; Han et al., 2020). The APEX peroxidase also catalyzes the polymerization and local deposition of diaminobenzidine (DAB) and enables to recruit of electron-dense osmium that is easily visualized by electron microscopy. Thus, APEX is useful for proteomic analysis and spatiotemporally high-resolution transcriptome profiling and imaging in vitro. However, the application of APEX in vivo has limited because it requires H2O2, which can be toxic and is less amenable to labeling reactions in tissue.
2-2. Astrocyte-specific cell-surface TurboID in vivo
Barres and colleagues initially discovered that treatment with astrocytic-conditioned media (ACM) or neuronal and astrocyte co-cultures promote the maturation and functional formation of excitatory synapses (Pfrieger and Barres, 1997; Ullian et al., 2001). In addition to the excitatory synapse, these conditions also induce inhibitory synapse formation in vitro, suggesting that astrocytes control both excitatory and inhibitory synapse formation and function through the astrocytic-secreted and/or cell-adhesion molecules (Allen and Eroglu, 2017; Baldwin and Eroglu, 2017; Elmariah et al., 2005; Stogsdill and Eroglu, 2017; Takano et al., 2020). Interestingly, astrocytic morphogenesis is also tightly linked to synaptogenesis during brain development (Sakers and Eroglu, 2019; Stogsdill et al., 2017). These findings imply that bidirectional signalings between astrocytes and neurons play a critical role in proper brain development and function. However, because cell-surface proteins are typically low abundance with high hydrophobicity and heterogeneity (Kuhlmann et al., 2018; Li et al., 2019), the identification of the molecular composition at astrocytic cell-surface and tripartite synaptic contacts in vivo has been significantly limited.
Proximity-dependent cell-surface labeling of cells can reveal their extracellular molecular landscape and also provide a roadmap to investigate mechanisms of how astrocytes control synaptic connectivity and function in the brain. Recently, horseradish peroxidase (HRP)-based cell-surface proteomic profiling has been achieved for neuronal synapse in vitro and ex vivo (Cijsouw et al., 2018; Li et al., 2020b; Loh et al., 2016). For example, HRP-fused with the known synaptic cleft proteins such as LRRTM1, LRRTM2, Siltrk3, or Neuroligin2 (NL2) has been used to specifically dissect the excitatory and inhibitory synaptic cleft proteomes in cultured neurons (Cijsouw et al., 2018; Loh et al., 2016). These HRP-based cell-surface proteomics at excitatory and inhibitory synaptic clefts identified 199 glutamatergic and 42 GABAergic synaptic cleft proteins in vitro and found that a novel synaptic cleft protein Mdga2 controls inhibitory synapse formation through the postsynaptic recruitment of NL2 (Loh et al., 2016). In addition, HRP-fused with the known excitatory cell adhesion molecule SynCAM1 identified several excitatory synaptic cleft proteins including, Receptor-type tyrosine-protein phosphatase zeta (R-PTP-zeta) (Cijsouw et al., 2018). Furthermore, cell-type-specific expression of HRP fused to the N-terminal extracellular domain CD2 (HRP-CD2) was recently used to identify novel cell surface molecules of Drosophila olfactory projection neurons ex vivo (Li et al., 2020b).
More recently, TurboID-based cell surface proteomic profiling (TurboID-surface) has been engineered for deciphering the molecular composition of the cell-type-specific surface proteomes in vivo (Fig. 1 left) (Takano et al., 2020). TurboID-conjugated with a glycosylphosphatidylinositol (GPI) anchored reconstitution-activated proteins highlight intercellular connections (GRAPHIC) was delivered to cortical astrocytes using cell-type selective adeno-associated virus (AAV) to label and purify proteins for LC/MS/MS of the astrocytic surface proteome in vivo (Kinoshita et al., 2019; Takano et al., 2020). Super-resolution Stimulated Emission Depletion (STED) microscopy showed that almost half of the proteins labeled by astrocytic TurboID-surface are localized at the peri-synaptic sites in vivo (Takano et al., 2020). Using label-free quantitative LC/MS/MS analysis after affinity isolation by streptavidin-coupled beads (Fig. 1 left), a large number of proteins (~3, 000 proteins) are identified. These proteins are further analyzed for those with significant enrichment in astrocytic TurboID-surface the over control group (soluble TurboID) as described previously (Courtland et al., 2021; Spence et al., 2019; Uezu et al., 2016). Astrocytic TurboID-surface revealed 178 extracellular proteins (fold-change>1.5 and p<0.05), including 58 known synaptic proteins based on the synaptic SYNGO database (Koopmans et al., 2019; Takano et al., 2020). These findings suggest that cell-surface proximity-dependent labeling using TurboID and HRP are both robust approaches to interrogate the cell-surface proteomes of genetically defined cell types in vitro and in vivo. This genetic access to cell-type derived surface compartments now enables the proteomic interrogation of sites previously not readily accessible to biochemical studies (Li et al., 2020a, b; Takano et al., 2020).
Split-TurboID enables the labeling and molecular profiling of specific cell-type interfaces
Deciphering the extracellular adhesion codes of cell-type-specific connections in the brain remains a major challenge. Recently, surface HRP or TurboID fragment complementation approaches, split HRP (sHRP) and Split-TurboID, have been engineered to enable the visualization of synapses (sHRP) or the proteomic mapping of the specific cell-type interfaces such as synaptic clefts and astrocyte-neuron contacts (Split-TurboID) (Takano et al., 2020; Martell et al., 2016). These techniques, sHRP and Split-TurboID, divide the proximity-based enzyme into N- and C-terminal fragments such that they can reconstitute the functional enzyme by fragment complementation at specific cell-cell contact sites when brought into proximity of each other (Fig. 1 right). For example, biotin-conjugated synaptic cleft proteins based on sHRP reconstitution at synapses can be used to visualize the specific synapses between amacrine cells and retinal ganglion cells (RGCs) in vivo, indicating that this technique is helpful for mapping neuronal connections in the brain (Martell et al., 2016). Split-TurboID (also Split-BirA) has also been utilized for investigating protein-protein complexes and intracellular membrane-membrane contacts such as ER-mitochondria interactions (Cho et al., 2020; Martell et al., 2016; Schopp et al., 2017).
Split-TurboID at the surface of cells is a newly developed and valuable tool for discovering the molecules at the specific cell-type interfaces such as astrocyte-synapse contacts in vivo (Takano et al., 2020). Split-TurboID splits TurboID into N-terminal (N TurboID) and C-terminal (C TurboID) halves and expresses them on the surface of cells using GPI-anchor (Fig. 1 right). Each Split-TurboID fragment is expressed in vivo in either neurons or astrocytes using AAVs with cell-type-specific promoters. Where astrocytes ensheath neuronal synapses N-TurboID and C-TurboID reconstitute functional TurboID at the tripartite synaptic clefts (Fig. 1 right). Thus, Split-TurboID enables molecular profiling at specific cell-type interfaces in vivo, which is a significant advance over other approaches (Fig. 1). Indeed, STED microscopy reveals that Split-TurboID highly labels the proteins between astrocytes at excitatory or inhibitory synapses in the cerebral cortex (Takano et al., 2020). Using LC/MS/MS analysis after affinity isolation by streptavidin (Fig. 1 right), a large number of proteins (~3, 000 proteins) are identified. These proteins are further analyzed for those with significant enrichment in Split-TurboID over the control group (soluble TurboID) as described previously (Courtland et al., 2021; Spence et al., 2019; Uezu et al., 2016). Split-TurboID discovered 173 proteins (fold-change> 1.5 and p<0.05), including 63 known synaptic proteins identified in the synaptic biology SYNGO database. These included synaptic proteins such as Neuroligin/Neurexin (Neuroligin-3, Neurexin I), calcium channel auxiliary subunits that also regulate glutamate receptor trafficking (Cacna2d3, Ccicng2-3), AMPA receptors (Gria2-3), and known inhibitory synaptic proteins such as GABAA receptors (Gabra1, Gabra4, Gabrb2, Gabrg2) (Koopmans et al., 2019; Takano et al., 2020). Interestingly, over half of the proteins identified by Split-TurboID are unique and have not been previously reported (Takano et al., 2020). Thus, Split-TurboID provided a new molecular framework for understanding tripartite synaptic connections and how these contacts control synapse formation and function in the brain.
Overview of the molecular mechanisms by which astrocytes control synaptic connectivity
4-1. Astrocytic molecules control excitatory synapse in vivo
Several recent studies have highlighted new molecular mechanisms of astrocyte-neuron communication to generate excitatory synapses (Allen and Eroglu, 2017; Baldwin and Eroglu, 2017; Stogsdill and Eroglu, 2017; Takano et al., 2020). Remarkably, some of these studies have described a role for astrocytes in directing the formation of specific types of synaptic connections to build different circuits. Based on our tripartite synaptic cleft proteomes, using astrocytic TurboID-surface and Split-TurboID, several extracellular proteins likely play synaptogenic roles at astrocyte contact sites were identified. These included Hevin (SPARC-like 1), Cacna2d3 (α2δ-3), Semaphorin 7A (Sema 7A), Plexin A4, Erythropoietin producing hepatocellular receptor tyrosine kinase B2 (EphB2), Neurexin I (NRX1), and Neuroligin 3 (NL3); all crucial regulators of astrocyte-neuron signaling communication for excitatory synapse formation and function in vitro and in vivo (Allen and Eroglu, 2017; Baldwin and Eroglu, 2017; Stogsdill and Eroglu, 2017; Takano et al., 2020). For example, secreted protein Hevin, and its homolog protein SPARC, are highly expressed in astrocytes of the cortex during the synaptogenesis (Kucukdereli et al., 2011; Risher et al., 2014). Purified Hevin from astrocyte conditioned media (ACM) promotes synapse formation, resulting in structurally normal but postsynaptically silent excitatory synapses in cultured retinal ganglion cells (RGCs) (Kucukdereli et al., 2011). Mechanistically, the astrocytic secreted Hevin binds to presynaptic NRX1α and postsynaptic NL1B (Fig. 2) (Kucukdereli et al., 2011; Singh et al., 2016). In contrast, SPARC doesn’t enhance synapse formation but antagonizes Hevin-induced synapse formation (Kucukdereli et al., 2011). Consistently, Hevin knockout in mouse impairs synapse formation in the thalamocortical circuit that results from the morphological immature dendritic spine (Kucukdereli et al., 2011; Risher et al., 2014).
Fig. 2.
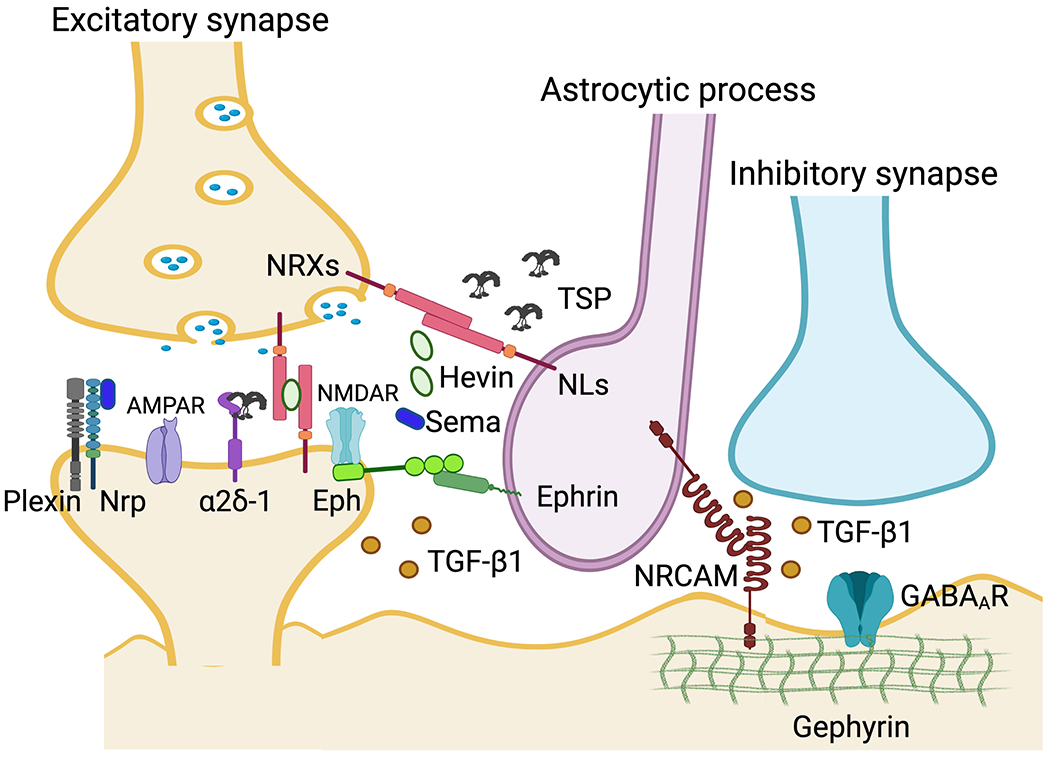
Overview of the signal networks that astrocytes-regulated excitatory and inhibitory synapse formation. Astrocytic secreted proteins thrombospondins (TSP) interacts with voltage-dependent calcium channel subunit α2δ-1 (α2δ-1), thereby leads to excitatory synapse formation. Astrocytic Neuroligin 2 (NL2) controls synaptogenesis through neuronal neurexins (NRXs) during development. The NL2-NRXs complex also regulates astrocytic morphogenesis. Astrocytic secreted protein Hevin promotes synapse formation through its interactions with NRX1α and NL1B. Semaphorin 3A (Sema 3A)/Plexin A4/Neuroplins (Nrps) signaling between astrocyte and neuron in the cortex may be involved in excitatory synaptic connection and function. Eph receptors (EphA and EphB) and its ligand ephrin lead to contact-dependent astrocyte-neuron communication to generate synapse. Astrocyte-secreted TGF-β1 induces excitatory synapse formation. Astrocytic cell adhesion protein NRCAM interacts with neuronal NRCAM that is coupled to gephyrin at inhibitory postsynapse, thereby leads to inhibitory synapse formation in vivo.
Cacna2d3/α2δ-3 (Calcium channel, voltage-dependent, alpha 2/delta subunit 3) is an L-type calcium channel voltage-dependent subunit. Aberrant α2δ subunit expression has been implicated in the pathogenesis of several syndromes and diseases, such as chronic neuropathic pain, autism spectrum disorder, and epilepsy (Geisler et al., 2015; Vergult et al., 2015; Zamponi et al., 2015). Among of α2δ subunits, α2δ-1 and α2δ-3 are highly expressed in the cerebral cortex and hippocampus (Cole et al., 2005; Klugbauer et al., 1999; Schlick et al., 2010). α2δ-1 promotes excitatory synapse formation in vivo through the interaction with astrocytic secreted factor thrombospondin (TSP) (Fig. 2) (Eroglu et al., 2009). α2δ-1 is also a receptor for Gabapentin, which is a drug used to treat epilepsy and neuropathic pain. Gabapentin antagonizes the interaction of α2δ-1 with TSP and inhibits excitatory synapse formation (Eroglu et al., 2009). It has recently been reported that the expression of α2δ-3 increases the excitatory and inhibitory synaptic density and facilitates spontaneous GABA release in cultured neurons (Bikbaev et al., 2020). Interestingly, α2δ-3 knockout mice exhibit anxiety-like behavior and auditory deficits (Landmann et al., 2019). These findings demonstrated that α2δ-1 and α2δ-3 might have distinct roles in the formation of specific synaptic connections in vivo, and further studies are needed to elucidate that how astrocytes regulate α2δ-3 for generating excitatory and inhibitory synapses in vivo.
Semaphorins (Semas) and their receptors, Plexins, form a protein complex with Neuroplins (Nrps) that are known to regulate many different developmental steps, including axon and dendrite outgrowth, neuronal migration, and synapse formation (Takano et al., 2019; Takano et al., 2015). In the spinal cord, Sema3A is highly expressed in astrocytes, and this astrocytic Sema3A controls proper motor neuron and sensory neurocircuit organization (Molofsky et al., 2014). The deletion of astrocytic Sema3A decreases excitatory and increases inhibitory inputs in vivo, indicating that astrocytic Sema3A regulates the motor neuron firing properties (Molofsky et al., 2014). Consistent with these data, the Sema3A receptor Plexin A4 was detected by our tripartite synaptic cleft proteome from the cerebral cortex (Takano et al., 2020). Sema3A-Plexin A4 signaling between astrocytes and neurons may be involved in excitatory synaptic connection and function in the brain (Fig. 2). In addition to Sema3A, Sema7A is highly expressed in astrocytes and is involved in the formation of glial scars after spinal cord injury (Eixarch et al., 2013; Kopp et al., 2010). In olfactory sensory neurons, the deletion of Sema7A perturbs activity-dependent synapse formation (Inoue et al., 2018). However, the role of astrocytic Sema7A in modulating synaptic connections and their functions remains to be determined.
Eph receptors (EphA and EphB) and their ligand ephrin have a critical role in contact-dependent astrocyte-neuron communication to generate synapses (Carmona et al., 2009; Nguyen et al., 2020; Nishida and Okabe, 2007). Ephrin-A3 is expressed in hippocampal neurons and ephrin-A3 is particularly enriched on fine astrocytic processes (Carmona et al., 2009). Ephrin-A3 knockout mice have abnormal dendritic spines and impaired the acquisition of contextual fear memory, suggesting astrocytic ephrin-A3 controls proper spine formation important for behavior in vivo (Carmona et al., 2009). Interestingly, ephrin-A3 reverse signaling to astrocytes from neuronal synapses regulates the glutamate uptake through the glutamate transporters GLAST (EAAT1) and GLT1 (EAAT2) (Carmona et al., 2009). These results indicate that astrocytes and neurons utilize bidirectional signaling through EphA4 and ephrin-A3 at the tripartite synaptic sites. More recently, it was reported that astrocyte-specific ephrin-B1 knockout mice exhibit an increase of excitatory synapses and elevated excitation in CA1 pyramidal neurons that is responsible for imbalanced excitatory and inhibitory synaptic activity and associated with impaired sociability. These data indicate that astrocytic ephrin-B1 negatively regulates excitatory synapse formation and function in vivo (Koeppen et al., 2018; Nguyen et al., 2020).
Recent cell-type-specific RNAseq reveals that the NL1-3 family is highly expressed in astrocytes in addition to neurons (Sakers and Eroglu, 2019; Stogsdill et al., 2017). The individual knockdown of astrocytic NL1-3 impaired astrocytic morphogenesis in vitro and in vivo (Stogsdill et al., 2017). Interestingly, the knockdown of neuronal NRX1/2 also prevents astrocytic morphogenesis, indicating that astrocyte-neuron communication through the trans-synaptic interactions of astrocytic NLs with neuronal NRXs is essential for astrocytic development in vivo (Scheiffele et al., 2000; Stogsdill et al., 2017). Notably, each astrocytic NL isoform might have different roles for brain development. For example, the loss of astrocytic NL2 prevented astrocytic morphogenesis at both early and late developmental stages, while the loss of astrocytic NL1 and NL3 impaired the astrocytic complexity of only early or late developmental stages, respectively (Stogsdill et al., 2017). Furthermore, the astrocyte-specific NL2 knockout mice display an impairment of excitatory synapse formation and function in the cortex (Stogsdill et al., 2017). Thus, astrocytic NL and neuronal NRX complexes play a critical role in bidirectional signaling between astrocyte and neuronal synapses that are responsible for the astrocytic morphogenesis and excitatory synapse formation during development.
4-2. Astrocyte control inhibitory synapse in vivo
In addition to excitatory synapse formation and function, the astrocyte has a critical role in establishing GABAergic inhibitory synapses (Elmariah et al., 2005). However, the molecular mechanism by which astrocytes control inhibitory synapse formation and function has remained less understood than their excitatory counterparts and it is an ongoing research field. It has been reported that astrocytic-derived transforming growth factor beta 1 (TGF-β1) is involved in inhibitory synapse formation and function (Diniz et al., 2014). TGF-β1 induces inhibitory synapse formation through the Ca2+/calmodulin-dependent kinase II (CaMKII)-mediated clustering of NL2 at the inhibitory postsynaptic terminal in vitro and in vivo (Diniz et al., 2014). Interestingly, astrocytic TGF-β1 also induces excitatory synapse formation that involves neuronal activity and secretion of the NMDA co-agonist D-serine (Diniz et al., 2014). However, the molecular mechanisms that modulate or signal astrocytic TGF-β1 release remain unclear.
Both synaptic proteomes, using astrocyte-neuron Split-TurboID and inhibitory postsynaptic proteome using gephyrin-BirA, identified the neuronal cell adhesion molecular (NRCAM), and our later Split-TurboID study demonstrated it is a key regulator of astrocytic morphogenesis and inhibitory synaptogenesis (Takano et al., 2020; Uezu et al., 2016). Multiple human genetic analyses indicate that the NRCAM gene may be associated with autism spectrum disorders (Bonora et al., 2005; Marui et al., 2009; Sakurai et al., 2006), yet most studies have focused on its role in neurons and have reported that NRCAM controls spine formation and axon guidance (Demyanenko et al., 2014; Mohan et al., 2019). Despite this, the expression level of NRCAM in astrocytes is higher than that of neurons, and the endogenous NRCAM is highly enriched at the astrocytic plasma membrane in vivo (Takano et al., 2020; Zhang et al., 2014; Zhang et al., 2016). Interestingly, the deletion of astrocytic NRCAM stimulates astrocytic expansion of territory and infiltration during brain development, indicating that NRCAM is a unique negative regulator of astrocytic outgrowth (Takano et al., 2020). Notably, the regulation of astrocytic morphogenesis by NRCAM appears to require homophilic binding to neuronal NRCAM. What might be the astrocyte signaling pathway downstream of NRCAM? It is reported that neuronal NRCAM limits spine development through the interaction with the Npn-2/PlexinA3 complex that stimulates Semaphorin signaling pathway (Demyanenko et al., 2014; Mohan et al., 2019). This signaling pathway suppresses small GTPase Rap1 and Rac1 activity that is required for the clustering of a cell adhesion molecule β1 integrin and cytoskeletal reorganization (Mohan et al., 2019; Takano et al., 2019; Takano et al., 2015). Thus, the astrocytic NRCAM may activate the Npn-2/Rap1/Rac1 pathway through neuronal NRCAM to establish a large number of astrocytic peri-synaptic processes. Further exploration of the extracellular and intracellular signaling pathways regulating astrocytic morphogenesis is a crucial issue in the brain developmental research field.
NRCAM plays a critical role in inhibitory synapse formation and function in the cerebral cortex (Fig. 2) (Takano et al., 2020). NRCAM-expressed in HEK293 cells induce GABAergic synapses but not glutamatergic synapses of the cultured neurons (Takano et al., 2020). In the cerebral cortex, the depletion of astrocytic NRCAM significantly impairs astrocyte interactions with inhibitory synapses and thereby leads to a decrease in the number of inhibitory synapses, but not excitatory synapses. This loss of NRCAM also results in significant inhibitory synaptic transmission deficits, with minor effects on glutamatergic responses. Mechanistically, astrocytic NRCAM transcellular binds to neuronal NRCAM that is coupled to gephyrin at inhibitory postsynapses (Fig. 2). Thus, astrocytes directly control inhibitory synapse formation and function through the NRCAM-dependent astrocyte-neuron contact (Fig. 2) (Takano et al., 2020). Together, astrocytes control both excitatory and inhibitory synaptic connectivity and establish proper neurocircuit formation and synaptic function in the brain. Furthermore, our novel TurboID-surface- and Split-TurboID-based proteomic profiling establishes a framework to fully elucidate the molecular mechanisms underlying tripartite synaptic formation and function in vivo (Takano et al., 2020).
Perspectives
Since the cell-surface proteins are typically present at low abundance and have high hydrophobicity and heterogeneity (Kuhlmann et al., 2018; Li et al., 2019), dissecting the molecular composition of specific cell-cell connections from the brain remains a major challenge in the neuroscience field. The development of novel in vivo cell-surface BioID approaches, TurboID-surface and Split-TurboID (Fig. 1), has provided the first insights into the specific extracellular molecular landscape and cell-surface interactome of the tripartite synapse in vivo (Fig. 2) (Takano et al., 2020). However, TurboID-surface and Split-TurboID may have a few potential limitations. The enzyme activity of all BirA variants requires ATP, which is only found at low amounts extracellularly in vivo. In addition, these approaches need the administration of exogenous biotin for 7 days to obtain enough biotinylation labeling (Spence et al., 2019; Uezu et al., 2016). Thus, a further challenge is the need to improve the temporal resolution of these approaches. Nevertheless, these approaches revealed a large number of uncharacterized molecules at the tripartite synaptic cleft in the brain. Interestingly, emerging evidence shows that astrocytes are highly heterogeneous in the brain and may control specific types of synaptic connections to establish different neurocircuits (Allen and Eroglu, 2017; Baldwin and Eroglu, 2017; Sakers and Eroglu, 2019; Stogsdill and Eroglu, 2017). Furthermore, recent genome-wide and imaging studies demonstrate that many proteins that are tightly linked to neurological diseases such as autism and schizophrenia are highly expressed in astrocytes in addition to neurons (Foo et al., 2011; Srinivasan et al., 2016; Zhang et al., 2014; Zhang et al., 2016). However, the majority of studies on these proteins view their neuronal loss of function as the primary cause of synaptic abnormalities seen in neurological diseases. Yet clearly, astrocytes play key roles in organizing synaptogenesis and physiology. An important issue for the field is how defects in the astrocytic compartment of tripartite synapses may cause neurological diseases that result from synaptic disassembly and dysfunction. Our tripartite synaptic cleft proteome provides insight into how astrocytes control neurocircuit connectivity and brain functions and how astrocyte-neuron signaling could be altered in neurological diseases. Future challenges in astrocyte biology will entail exploring these issues using advances in imaging technology, genetic models, and innovative experimental approaches in vivo (Yu et al., 2020). It can be expected that these cutting-edge approaches to study tripartite synaptic connectivity will provide new paradigms for how neurocircuits develop in normal and maladaptive states, and shed light on therapeutic strategies for neurological diseases.
Highlights.
Cell-type-specific proximity-based labeling TurboID-surface enables to map and identify the cell surface proteins from brain tissue.
A novel proteomically approach Split-TurboID enables the mapping and molecular profiling at specific cell-type interfaces in vivo.
Astrocytes control excitatory and inhibitory synapse formation and function in vivo.
Acknowledgments
This work was supported by Brain initiative RO1DA047258 from NIH (S.H.S), Kahn Neurotechnology Award (S.H.S), The Japan Society for the Promotion of Science (21H02582) (T.T), Nakajima Foundation (T.T), and The Uehara Memorial Foundation (T.T).
Footnotes
Conflict of Interest Statement
The authors declare no competing financial interests.
References
- Allen NJ, and Eroglu C (2017). Cell Biology of Astrocyte–Synapse Interactions. Neuron 96; 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolo N, Smukowski SN, Vanderlinden J, Condomitti G, Rybakin V, Ten Bos J, Trobiani L, Portegies S, Vennekens KM, Gounko NV, et al. (2020). Synapse type–specific proteomic dissection identifies IgSF8 as a hippocampal CA3 microcircuit organizer. Nature communications 11, 5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araque A, Carmignoto G, Haydon PG, Oliet SH, Robitaille R, and Volterra A (2014). Gliotransmitters travel in time and space. Neuron 81, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin KT, and Eroglu C (2017). Molecular mechanisms of astrocyte–induced synaptogenesis. Current opinion in neurobiology 45, 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikbaev A, Ciuraszkiewicz-Wojciech A, Heck J, Klatt O, Freund R, Mitlohner J, Enrile Lacalle S, Sun M, Repetto D, Frischknecht R, et al. (2020). Auxiliary alpha2delta1 and alpha2delta3 Subunits of Calcium Channels Drive Excitatory and Inhibitory Neuronal Network Development. The Journal of neuroscience : the official journal of the Society for Neuroscience 40, 4824–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora E, Lamb JA, Barnby G, Sykes N, Moberly T, Beyer KS, Klauck SM, Poustka F, Bacchelli E, Blasi F, et al. (2005). Mutation screening and association analysis of six candidate genes for autism on chromosome 7q. Eur J Hum Genet 13, 198–207. [DOI] [PubMed] [Google Scholar]
- Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, and Ting AY (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36,880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona MA, Murai KK, Wang L, Roberts AJ, and Pasquale EB (2009). Glial ephrin–A3 regulates hippocampal dendritic spine morphology and glutamate transport. Proceedings of the National Academy of Sciences of the United States of America 106 12524–12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KF, Branon TC, Rajeev S, Svinkina T, Udeshi ND, Thoudam T, Kwak C, Rhee H-W, Lee I-K, Carr SA, et al. (2020). Split–TurboID enables contact–dependent proximity labeling in cells bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cijsouw T, Ramsey AM, Lam TT, Carbone BE, Blanpied TA, and Biederer T (2018). Mapping the Proteome of the Synaptic Cleft through Proximity Labeling Reveals New Cleft Proteins. Proteomes 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizeron M, Qiu Z, Koniaris B, Gokhale R, Komiyama NH, Fransen E, and Grant SGN (2020). A brainwide atlas of synapses across the mouse life span. Science (New York, NY) 369, 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RL, Lechner SM, Williams ME, Prodanovich P, Bleicher L, Varney MA, and Gu G (2005). Differential distribution of voltage–gated calcium channel alpha–2 delta (alpha2delta) subunit mRNA–containing cells in the rat central nervous system and the dorsal root ganglia. J Comp Neurol 491, 246–269. [DOI] [PubMed] [Google Scholar]
- Courtland JL, Bradshaw TW, Waitt G, Soderblom EJ, Ho T, Rajab A, Vancini R, Kim IH, and Soderling SH (2021). Genetic disruption of WASHC4 drives endo–lysosomal dysfunction and cognitive–movement impairments in mice and humans. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko GP, Mohan V, Zhang X, Brennaman LH, Dharbal KE, Tran TS, Manis PB, and Maness PF (2014). Neural cell adhesion molecule NrCAM regulates Semaphorin 3F–induced dendritic spine remodeling. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 11274–11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz LP, Matias IC, Garcia MN, and Gomes FC (2014). Astrocytic control of neural circuit formation: highlights on TGF–beta signaling. Neurochem Int 78, 18–27. [DOI] [PubMed] [Google Scholar]
- Eixarch H, Gutierrez-Franco A, Montalban X, and Espejo C (2013). Semaphorins 3A and 7A: potential immune and neuroregenerative targets in multiple sclerosis. Trends Mol Med 19, 157–164. [DOI] [PubMed] [Google Scholar]
- Elmariah SB, Oh EJ, Hughes EG, and Balice-Gordon RJ (2005). Astrocytes regulate inhibitory synapse formation via Trk–mediated modulation of postsynaptic GABAA receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 3638–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu C, Allen NJ, Susman MW, O’Rourke NA, Park CY, Ozkan E, Chakraborty C, Mulinyawe SB, Annis DS, Huberman AD, et al. (2009). Gabapentin receptor alpha2delta–1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell 139, 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, and Ting AY (2019). Atlas of Subcellular RNA Localization Revealed by APEX–Seq. Cell 178, 473–490 e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC, Allen NJ, Bushong EA, Ventura PB, Chung WS, Zhou L, Cahoy JD, Daneman R, Zong H, Ellisman MH, et al. (2011). Development of a method for the purification and culture of rodent astrocytes. Neuron 71, 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Schopf CL, and Obermair GJ (2015). Emerging evidence for specific neuronal functions of auxiliary calcium channel alpha(2)delta subunits. Gen Physiol Biophys 34, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Zhao BS, Myers SA, Carr SA, He C, and Ting AY (2020). RNA-protein interaction mapping via MS2- or Cas13–based APEX targeting. Proceedings of the National Academy of Sciences of the United States of America 117, 22068–22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, Nishizumi H, Naritsuka H, Kiyonari H, and Sakano H (2018). Sema7A/PlxnCI signaling triggers activity–dependent olfactory synapse formation. Nature communications 9, 1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, and Roux KJ (2016). An improved smaller biotin ligase for BioID proximity labeling. Molecular biology of the cell 27, 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Huang AJY, McHugh TJ, Suzuki SC, Masai L, Kim IH, Soderling SH, Miyawaki A, and Shimogori T (2019). Genetically Encoded Fluorescent Indicator GRAPHIC Delineates Intercellular Connections. iScience 15, 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugbauer N, Lacinova L, Marais E, Hobom M, and Hofmann F (1999). Molecular diversity of the calcium channel alpha2delta subunit. The Journal of neuroscience : the official journal of the Society for Neuroscience 19, 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen J, Nguyen AQ, Nikolakopoulou AM, Garcia M, Hanna S, Woodruff S, Figueroa Z, Obenaus A, and Ethell IM (2018). Functional Consequences of Synapse Remodeling Following Astrocyte–Specific Regulation of Ephrin–B1 in the Adult Hippocampus. The Journal of neuroscience : the official journal of the Society for Neuroscience 38, 5710–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans F, van Nierop P, Andres–Alonso M, Byrnes A, Cijsouw T, Coba MP, Cornelisse LN, Farrell RJ, Goldschmidt HL, Howrigan DP, et al. (2019). SynGO: An Evidence–Based, Expert–Curated Knowledge Base for the Synapse. Neuron 103, 217–234 e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp MA, Brommer B, Gatzemeier N, Schwab JM, and Pruss H (2010). Spinal cord injury induces differential expression of the profibrotic semaphorin 7A in the developing and mature glial scar. Glia 58, 1748–1756. [DOI] [PubMed] [Google Scholar]
- Kucukdereli H, Allen NJ, Lee AT, Feng A, Ozlu MI, Conatser LM, Chakraborty C, Workman G, Weaver M, Sage EH, et al. (2011). Control of excitatory CNS synaptogenesis by astrocyte–secreted proteins Hevin and SPARC. Proceedings of the National Academy of Sciences of the United States of America 108, E440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann L, Cummins E, Samudio I, and Kislinger T (2018). Cell–surface proteomics for the identification of novel therapeutic targets in cancer. Expert Rev Proteomics 15, 259–275. [DOI] [PubMed] [Google Scholar]
- Landmann J, Richter F, Classen J, Richter A, Penninger JM, and Bechmann I (2019). Behavioral phenotyping of calcium channel (CACN) subunit alpha2delta3 knockout mice: Consequences of sensory cross–modal activation. Behav Brain Res 364, 393–402. [DOI] [PubMed] [Google Scholar]
- Lanjakornsiripan D, Pior BJ, Kawaguchi D, Furutachi S, Tahara T, Katsuyama Y, Suzuki Y, Fukazawa Y, and Gotoh Y (2018). Layer–specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nature communications 9, 1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Han S, Li H, Udeshi ND, Svinkina T, Mani DR, Xu C, Guajardo R, Xie Q, Li T, et al. (2020a). Cell–Surface Proteomic Profiling in the Fly Brain Uncovers Wiring Regulators. Cell 180, 373–386 e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Han S, Li H, Udeshi ND, Svinkina T, Mani DR, Xu C, Guajardo R, Xie Q, Li T, et al. (2020b). Cell–Surface Proteomic Profiling in the Fly Brain Uncovers Wiring Regulators. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Y, Mao J, Yao Y, Wang K, Qiao Q, Fang Z, and Ye M (2019). Sensitive profiling of cell surface proteome by using an optimized biotinylation method. J Proteomics 196, 33–41. [DOI] [PubMed] [Google Scholar]
- Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, et al. (2016). Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell 166, 1295–1307 e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Yamagata M, Deerinck TJ, Phan S, Kwa CG, Ellisman MH, Sanes JR, and Ting AY (2016). A split horseradish peroxidase for the detection of intercellular protein–protein interactions and sensitive visualization of synapses. Nat Biotechnol 34, 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marui T, Funatogawa I, Koishi S, Yamamoto K, Matsumoto H, Hashimoto O, Nanba E, Nishida H, Sugiyama T, Kasai K, et al. (2009). Association of the neuronal cell adhesion molecule (NRCAM) gene variants with autism. Int J Neuropsychopharmacol 12, 1–10. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Busse B, Weiler NC, O’Rourke N, and Smith SJ (2010). Single–synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron 68, 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan V, Sullivan CS, Guo J, Wade SD, Majumder S, Agarwal A, Anton ES, Temple BS, and Maness PF (2019). Temporal Regulation of Dendritic Spines Through NrCAM–Semaphorin3F Receptor Signaling in Developing Cortical Pyramidal Neurons. Cerebral cortex (New York, NY : 1991) 29, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Kelley KW, Tsai HH, Redmond SA, Chang SM, Madireddy L, Chan JR, Baranzini SE, Ullian EM, and Rowitch DH (2014). Astrocyte–encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AQ, Sutley S, Koeppen J, Mina K, Woodruff S, Hanna S, Vengala A, Hickmott PW, Obenaus A, and Ethell IM (2020). Astrocytic Ephrin–B1 Controls Excitatory–Inhibitory Balance in Developing Hippocampus. The Journal of neuroscience : the official journal of the Society for Neuroscience 40, 6854–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, and Okabe S (2007). Direct astrocytic contacts regulate local maturation of dendritic spines. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrieger FW, and Barres BA (1997). Synaptic efficacy enhanced by glial cells in vitro. Science (New York, NY) 277, 1684–1687. [DOI] [PubMed] [Google Scholar]
- Poulopoulos A, Murphy AJ, Ozkan A, Davis P, Hatch J, Kirchner R, and Macklis JD (2019). Subcellular transcriptomes and proteomes of developing axon projections in the cerebral cortex. Nature 565, 356–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risher WC, Patel S, Kim IH, Uezu A, Bhagat S, Wilton DK, Pilaz LJ, Singh Alvarado J, Calhan OY, Silver DL, et al. (2014). Astrocytes refine cortical connectivity at dendritic spines. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, and Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of cell biology 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakers K, and Eroglu C (2019). Control of neural development and function by glial neuroligins. Current opinion in neurobiology 57, 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Ramoz N, Reichert JG, Corwin TE, Kryzak L, Smith CJ, Silverman JM, Hollander E, and Buxbaum JD (2006). Association analysis of the NrCAM gene in autism and in subsets of families with severe obsessive–compulsive or self–stimulatory behaviors. Psychiatr Genet 16, 251–257. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, and Serafini T (2000). Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669. [DOI] [PubMed] [Google Scholar]
- Schlick B, Flucher BE, and Obermair GJ (2010). Voltage–activated calcium channel expression profiles in mouse brain and cultured hippocampal neurons. Neuroscience 167, 786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopp IM, Amaya Ramirez CC, Debeljak J, Kreibich E, Skribbe M, Wild K, and Bethune J (2017). Split–BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nature communications 8, 15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Stogsdill JA, Pulimood NS, Dingsdale H, Kim YH, Pilaz LJ, Kim IH, Manhaes AC, Rodrigues WS Jr., Pamukcu A, et al. (2016). Astrocytes Assemble Thalamocortical Synapses by Bridging NRX1alpha and NL1 via Hevin. Cell 164, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence EF, Dube S, Uezu A, Locke M, Soderblom EJ, and Soderling SH (2019). In vivo proximity proteomics of nascent synapses reveals a novel regulator of cytoskeleton–mediated synaptic maturation. Nature communications 10, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, and Khakh BS (2016). New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo. Neuron 92, 1181–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogsdill JA, and Eroglu C (2017). The interplay between neurons and glia in synapse development and plasticity. Current opinion in neurobiology 42, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, Ejikeme T, Ji RR, and Eroglu C (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Funahashi Y, and Kaibuchi K (2019). Neuronal Polarity: Positive and Negative Feedback Signals. Front Cell Dev Biol 7, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Wallace JT, Baldwin KT, Purkey AM, Uezu A, Courtland JL, Soderblom EJ, Shimogori T, Maness PF, Eroglu C, et al. (2020). Chemico–genetic discovery of astrocytic control of inhibition in vivo. Nature 588, 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Xu C, Funahashi Y, Namba T, and Kaibuchi K (2015). Neuronal polarization. Development (Cambridge, England) 142, 2088–2093. [DOI] [PubMed] [Google Scholar]
- Uezu A, Kanak DJ, Bradshaw TW, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, and Soderling SH (2016). Identification of an elaborate complex mediating postsynaptic inhibition. Science (New York, NY) 353, 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, and Barres BA (2001). Control of synapse number by glia. Science (New York, NY) 291, 657–661. [DOI] [PubMed] [Google Scholar]
- Ventura R, and Harris KM (1999). Three–dimensional relationships between hippocampal synapses and astrocytes. The Journal of neuroscience : the official journal of the Society for Neuroscience 19, 6897–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergult S, Dheedene A, Meurs A, Faes F, Isidor B, Janssens S, Gautier A, Le Caignec C, and Menten B (2015). Genomic aberrations of the CACNA2D1 gene in three patients with epilepsy and intellectual disability. Eur J Hum Genet 23, 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Nagai J, and Khakh BS (2020). Improved tools to study astrocytes. Nature reviews Neuroscience 21, 121–138. [DOI] [PubMed] [Google Scholar]
- Zamponi GW, Striessnig J, Koschak A, and Dolphin AC (2015). The Physiology, Pathology, and Pharmacology of Voltage–Gated Calcium Channels and Their Future Therapeutic Potential. Pharmacol Rev 67, 821–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA–sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, et al. (2016). Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Cizeron M, Qiu Z, Benavides-Piccione R, Kopanitsa MV, Skene NG, Koniaris B, DeFelipe J, Fransen E, Komiyama NH, et al. (2018). Architecture of the Mouse Brain Synaptome. Neuron 99, 781–799 e710. [DOI] [PMC free article] [PubMed] [Google Scholar]


