Abstract
The relationship between specimen input volume and the frequency of reported human immunodeficiency virus type 1 (HIV-1) RNA copy numbers by nucleic acid amplification technology (the NASBA HIV-1 RNA QT system) was investigated. Results obtained with both clinical specimens and dilution panels indicated that both the absolute number of reported results and the reported HIV-1 RNA copy number were directly proportional to the specimen input volumes evaluated (0.1, 0.5, and 1.0 ml). Conversion of the reported HIV-1 RNA copy numbers to a constant 1.0-ml volume indicated that the numerical relationship among the specimen input volumes and the HIV-1 RNA copy numbers was multiplicative. The HIV-1 RNA copy numbers reported for the 0.5-ml input volume were approximately 5-fold increased over those reported for the 0.1-ml input volume, and those reported for the 1.0-ml input volume were 10-fold increased over those reported for the 0.1-ml input volume. For the specimen input volumes investigated, a common linear range of 264 to 5,400,000 HIV-1 RNA copies was observed. The use of increased specimen input volumes did not result in a loss of assay specificity, as the results reported for specimens from 50 seronegative blood donors were negative at all three specimen input volumes. In conclusion, an increase in the input volume of specimens analyzed by nucleic acid amplification technology can be useful for the enhanced detection of HIV-1 RNA.
The sensitivity of nucleic acid amplification assays to detect and report the analyte target is an important performance characteristic that has a significant impact on clinical utility. In the case of human immunodeficiency virus type 1 (HIV-1), assay sensitivity is particularly relevant, as the use of highly active antiretroviral therapy (HAART) can reduce the amount of RNA in plasma to levels that are often lower than the detection capability of a given assay (2, 8).
Approaches to enhancing the sensitivity of nucleic acid amplification assays for detection of HIV-1 RNA have been directed toward concentration either of the virion contained in the specimen, by ultracentrifugation (7), or of the isolated analyte (F. Simons, M. Sjeps, C. De Laat, M. Cronin, and H. Cuypers, Abstr. 6th Conf. Retrovir. Opportun. Infect., poster 148, 1999). These procedures are followed by standard nucleic acid amplification techniques for reverse transcription-PCR (RT-PCR) and nucleic acid sequence-based amplification (NASBA), respectively. While such approaches may have efficacy, they require specialized equipment and additional technical steps that may not be reproducible or efficient in terms of time.
In this study, we conducted experimentation to ascertain the feasibility of another approach to enhancing the sensitivity of a nucleic acid amplification assay with only a minor modification of the standard isolation methodology used with NASBA technology. We reasoned that the capacity for capture of the HIV-1 RNA analyte on silica particles by following the methodology of Boom et al. (1) would be independent of specimen volume. Thus, we sought to determine whether an increase in the size of the specimen volume used for analysis resulted in greater clinical sensitivity, which is defined in terms of the number of specimens with reported HIV-1 RNA copy results, without loss of specificity. Further, the mathematical relationship between the reported HIV-1 RNA copy number and the volume of the specimen input was determined.
MATERIALS AND METHODS
Nucleic acid amplification assay.
The NASBA HIV-1 RNA QT system (Organon Teknika Corp., Durham, N.C.) was used as the nucleic acid amplification assay per the manufacturer's directions (9). The assay incorporates a stringent nucleic acid isolation procedure based on the release of RNA with detergent and guanidine thiocyanate followed by capture on silica particles (1). The numerical ratio of the specimen input volume to the lysis buffer volume was held constant for the three specimen input volumes studied: 0.1 ml of the specimen and 0.9 ml of NASBA lysis buffer, 0.5 ml of the specimen and 4.5 ml of NASBA lysis buffer, and 1.0 ml of the specimen and 9.0 ml of NASBA lysis buffer. Following the addition of the specimen to the different volumes of NASBA lysis buffer, the specimen was mixed thoroughly by rocking. The subsequent steps of the assay (i.e., isolation, amplification, hybridization, and detection) were performed identically for all specimen input volumes as provided by the manufacturer.
Clinical specimens.
Specimens were collected from HIV-1-infected patients from the North Shore University Hospital Center for AIDS Research and Treatment (Manhasset, N.Y.) and from the Miriam Hospital (Providence, R.I.) over a 4-month period from July to November 1996. Demographic information concerning the patients' overall health status and medication was obtained from each of the subjects. No exclusionary criteria were established for subject participation in the study. Informed consent was obtained from each subject prior to specimen collection. Peripheral blood was collected by venipuncture from each subject by using EDTA-VACUTAINER tubes (Becton Dickinson and Co., Franklin Lakes, N.J.). Each specimen was processed by standard techniques to obtain plasma, which was used for the subsequent nucleic acid amplification testing. Aliquots of the plasma specimens were added to each of the three volumes of lysis buffer at the volumes indicated above and were stored at −70°C until they were tested. Previous studies performed with the NASBA HIV-1 RNA QT system have indicated that no observable loss of reported HIV-1 RNA occurs following thawing after ultralow-temperature storage in lysis buffer (6). Each specimen was tested three times with the NASBA HIV-1 RNA QT assay, once with each of the three specimen input volumes.
Blood donor specimens.
Whole blood was collected from 50 volunteer donors into EDTA-VACUTAINER tubes. Plasma was obtained by standard techniques. Aliquots of the plasma specimens were added to each of the three volumes of lysis buffer as described above.
Dilution studies. (i) Terminal dilution of clinical specimens.
Ten clinical specimens containing HIV-1 RNA were diluted in lysis buffer to the end point for the 0.1-ml specimen input volume. Additional aliquots of the same specimens at input volumes of 0.5 and 1.0 ml were also tested at each dilution in order to determine the relationship between the specimen input volume and detectability by the NASBA HIV-1 RNA QT system at decreasing concentrations of the analyte. Each specimen of the dilution series was tested three times at each input volume, for a total of nine tests for each dilution. The total number of tests performed was 360.
(ii) Terminal dilution of in vitro culture stock.
Studies were performed with a set of serially diluted specimens derived from HIV-1-infected cell cultures; the virus-infected cell cultures originated from three HIV-1 clinical isolates (11). The concentration of HIV-1 virions from the supernatant of the culture was estimated by electron microscopy and p24 determination at 5 × 108 HIV-1 RNA copies/ml. The stock material was diluted in twofold steps by using a fresh-frozen plasma unit (600 ml containing 55 ml of anticoagulant sodium citrate solution, USP) from a seronegative blood donor as the diluent to create a series of 16 specimens with different concentrations of HIV-1 RNA. The estimated concentrations of HIV-1 RNA in the dilution series ranged from 5.4 × 106 to 1.65 × 102 copies/ml in the dilution series. Each specimen at each dilution was tested six times for each input volume, for a total of 288 tests.
CD4+ cell concentration determination.
CD4+ lymphocyte concentrations in the clinical specimens were determined by standard flow cytometry techniques.
Statistical analysis.
The linear ranges for each specimen input volume of the in vitro culture stock dilutions tested with the NASBA HIV-1 RNA QT system were determined by using linear regression models. The HIV-1 RNA copy number reported for each specimen was regressed on the expected number of input copies, as provided by the AIDS Clinical Trial Group Viral Quality Assurance Laboratory (Rush-Presbyterian-St. Luke's Medical Center, Chicago, Ill.). Since variability in copy number upon repeated testing of the same specimen is directly proportional to the mean, both dependent and independent variables were logarithmically transformed prior to analysis. After transformation, the residual variation in the dependent variable at each level of the independent variable was assumed to be Gaussian with zero mean and constant variance.
Separate data sets were formed from the data for each specimen volume input level, and consequently each data set represented a different range of the x variable. For each data set, a test of linearity was performed by partitioning regression analysis residual variation into two components: one representing error due to lack of fit to a linear model and the other representing pure error (3). The ratio of expected mean squares of these sources of error follows an F distribution. For each range, an F value estimating this ratio was computed, and the probability (P) of obtaining an F value greater than the observed value under a null hypothesis of no lack of fit to a linear model was calculated. A P value lower than 0.15 was interpreted as evidence of nonlinearity.
RESULTS
Clinical specimens from HIV-1-infected individuals.
Of the 102 specimens evaluated with the three investigational input volumes, HIV-1 RNA was reported by the NASBA HIV-1 RNA QT system for the majority (66 of 102; 65%) with all three input volumes. For 15 specimens (15%), no HIV-1 RNA was reported by the NASBA HIV-1 RNA QT system with any of the three input volumes tested; of these, 14 (93%) were from patients receiving antiretroviral therapy. For another 13 specimens (13%), HIV-1 RNA was reported by the NASBA HIV-1 RNA QT system with two input volumes, 0.5 and 1.0 ml. For eight specimens (8%), HIV-1 RNA was reported by the NASBA HIV-1 RNA QT system only with the highest input volume, 1.0 ml. The numbers of specimens for which HIV-1 RNA was reported were similar among the three groups stratified by CD4+ counts for each of the input volumes (Table 1).
TABLE 1.
Clinical specimen input volume versus frequency of reported HIV-1 RNAa
| CD4+ count | No. of specimens for which HIV-1 RNA was reported/no. tested (% positive) with the following input volume:
|
||
|---|---|---|---|
| 0.1 ml | 0.5 ml | 1.0 ml | |
| 500–1,716 | 11/18 (61) | 13/18 (72) | 16/18 (89) |
| 200–499 | 21/31 (68) | 24/31 (77) | 27/31 (87) |
| <200 | 34/53 (64) | 42/53 (79) | 44/53 (83) |
| Total | 66/102 (65) | 79/102 (77) | 87/102 (85) |
The clinical specimens were stratified by CD4+ counts. HIV-1 RNA copy numbers were obtained with the NASBA HIV-1 RNA QT system.
The rate of HIV-1 RNA copy number reporting with the NASBA HIV-1 RNA QT system was proportionately related to the specimen input volumes when all specimens were considered cumulatively. The highest detection rate for HIV-1 RNA was found with the 1.0-ml specimen input volume (87 specimens; 85%). The next greatest number of specimens for which HIV-1 RNA was reported was obtained with the 0.5-ml input volume (79 specimens; 77%). A specimen input volume of 0.1 ml resulted in the lowest rate of HIV-1 RNA detection (66 specimens; 65%). These results indicated a significant increase with the use of larger specimen input volumes compared to the 0.1-ml volume (a 15% increase with the 0.5-ml input volume and a 21% increase with the 1.0-ml input volume).
The HIV-1 RNA copy number reported by the NASBA assay was also consistent as a function of specimen input volume. For each clinical specimen tested, the highest HIV-1 RNA copy number was reported with the 1.0-ml input volume (mean = 169,430) and the lowest copy number was reported with the 0.1-ml input volume (mean = 19,909). The HIV-1 RNA copy numbers reported with the 0.5-ml input volumes were intermediate between those with the 0.1-ml input volume and those with the 1.0-ml input volume (mean = 85,299) for each clinical specimen. For the clinical specimens tested, the HIV-1 RNA copy numbers observed were in relatively good agreement with the HIV-1 RNA copy numbers expected based on the interval relationship of the three specimen input volumes. The mean HIV-1 RNA copy number for the 0.5-ml input volume was 4.3 times that for the 0.1-ml input volume, and that for the 1.0-ml input volume was 8.5 times that for the 0.1-ml input volume. The mean HIV-1 RNA copy number for the 0.5-ml input volume was 50% of the mean HIV-1 RNA copy number obtained with the 1.0-ml input volume. In no case was HIV-1 RNA reported with the smallest specimen input volume and not reported with the two larger input volumes.
Whole-blood donor specimens.
To determine whether an increase in specimen input volume might affect the specificity of the assay, specimens from 50 HIV-1-seronegative volunteer whole-blood donors were evaluated with each of the three input volumes. No HIV-1 RNA copies were reported from any of the 50 specimens evaluated at any of the three input volumes.
Dilution studies.
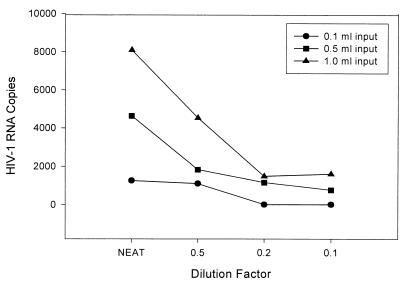
Dilutions of both clinical specimens from HIV-1-infected individuals and a characterized stock were analyzed to determine the relative sensitivities of the three input volumes. This empirical approach with the stock material also enabled estimation of the linear range for each specimen input volume. For each of 10 clinical specimens diluted to the end point of detection by the assay, the reporting of HIV-1 RNA reached the assay threshold first with the 0.1-ml input volume. Further dilutions were necessary to reach the end point for detection with both the 0.5- and the 1.0-ml input volume. The relationship between the dilution required to attain the end point and the input volume was directly proportional, as exemplified in Fig. 1. The mean HIV-1 RNA copy number reported at each dilution was proportional to the input volume (Table 2). As with the undiluted clinical specimens tested with different input volumes, there was no specimen for which HIV-1 RNA was reported at the 0.1-ml input volume and not at the larger input volumes.
FIG. 1.
Relationship of specimen input volume to HIV-1 RNA copy number reported by the NASBA HIV-1 RNA QT system for a representative clinical specimen that was terminally diluted.
TABLE 2.
Relationship between reported HIV-1 RNA copy numbers in terminally diluted clinical specimens and specimen input volume
| Input vol (ml) | No. (%) of diluted and undiluted specimensa:
|
Mean copy no. for specimens at the indicated dilutionb
|
|||||
|---|---|---|---|---|---|---|---|
| Below lower limit of detection | With HIV-1 RNA detected | Undiluted | Dilution 1 | Dilution 2 | Dilution 3 | Σ | |
| 0.1 | 29 (24) | 91 (76) | 28,040 | 3,810 | 1,001 | 413 | 33,264 |
| 0.5 | 21 (17) | 99 (83) | 130,193 | 15,193 | 2,478 | 1,017 | 149,601 |
| 1.0 | 7 (6) | 113 (94) | 221,509 | 30,441 | 4,976 | 1,501 | 479,936 |
Each of 10 specimens from HIV-1-infected individuals was tested undiluted and at three serial dilutions with the NASBA HIV-1 RNA QT system. Three tests were performed on the specimens at each dilution per input volume.
The dilution series for the clinical specimens were as follows. Five specimens were tested undiluted and at dilutions of 1:2, 1:5, and 1:10. Two specimens were tested undiluted and at dilutions of 1:5, 1:10, and 1:50. Three specimens were tested undiluted and at dilutions of 1:10, 1:100, and 1:10,000.
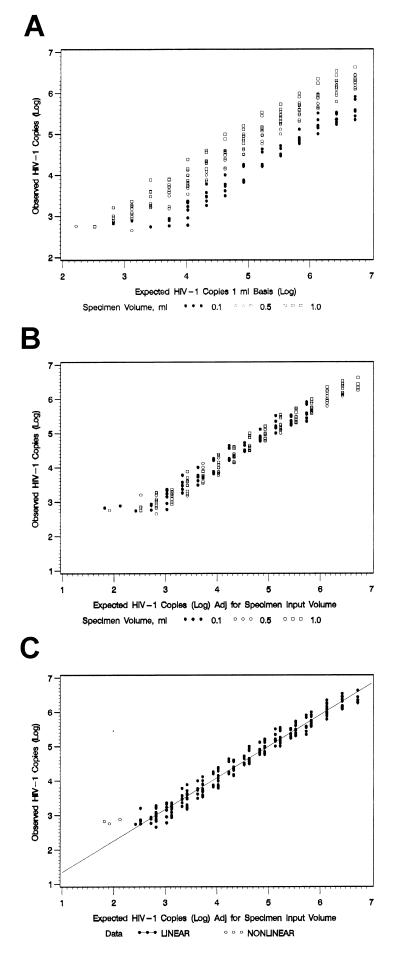
Detection of HIV-1 RNA following dilution of the in vitro HIV-1 stock followed the same general pattern as that observed for the clinical specimens. For the stock material, the threshold of detection by the NASBA HIV-1 RNA QT system was attained first in the dilution series with the 0.1-ml specimen input volume and last with the 1.0-ml specimen input volume. The threshold of detection for specimens with the 0.5-ml input volume was at dilutions intermediate between those with the highest and lowest input volumes (Table 3). Subsequent conversion of these data to a constant 1.0-ml input volume and logarithmic transformation of the data indicated no detectable difference in the reported number of HIV-1 RNA copies as a function of input volume (Figures 2A and B). The reported HIV-1 copy numbers were found to be described as a linear function across a defined range of HIV-1 RNA copy numbers from 264 to 5,400,000 for all three specimen input volumes, and no evidence on a log-log scale of a departure from linearity was observed (P = 0.44). In this linear range of HIV-1 RNA copy numbers, there was no significant difference between the intercepts (P = 0.44) or slopes (P = 0.57) describing the results with the three specimen input volumes. As a result, a common slope and intercept could be used to describe the relationship [log y = 0.419 + 0.914 · log x] (Fig. 2C). Thus, these results indicate that the number of copies reported was directly proportional to the number of copies expected per milliliter multiplied by the specimen input volume.
TABLE 3.
Relationship of reported HIV-1 RNA copy numbers in a terminally diluted HIV-1 stock and specimen input volumes
| Estimated HIV-1 RNA concn in specimen (no. of copies/ml) | Result at an input vol of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.1 ml
|
0.5 ml
|
1.0 ml
|
|||||||
| No. reported/totala (%) | Mean copy no.b | SD | No. reported/total (%) | Mean copy no. | SD | No. reported/total (%) | Mean copy no. | SD | |
| 5.4 × 105 | 6/6 (100) | 450,000 | 234,776 | 6/6 (100) | 1,716,667 | 673,548 | 6/6 (100) | 2,850,000 | 1,087,658 |
| 2.7 × 106 | 6/6 (100) | 240,000 | 62,610 | 6/6 (100) | 1,145,000 | 420,844 | 6/6 (100) | 2,033,333 | 823,812 |
| 1.35 × 106 | 6/6 (100) | 181,666 | 78,081 | 6/6 (100) | 593,333 | 217,868 | 6/6 (100) | 1,075,000 | 585,892 |
| 6.75 × 105 | 6/6 (100) | 79,333 | 26,242 | 6/6 (100) | 336,667 | 133,666 | 6/6 (100) | 595,000 | 241,060 |
| 3.37 × 105 | 6/6 (100) | 42,166 | 9,766 | 6/6 (100) | 205,000 | 98,539 | 6/6 (100) | 298,333 | 129,525 |
| 1.69 × 105 | 6/6 (100) | 24,333 | 11,978 | 6/6 (100) | 98,667 | 35,607 | 6/6 (100) | 190,000 | 92,880 |
| 8.44 × 104 | 6/6 (100) | 12,016 | 5,485 | 6/6 (100) | 51,500 | 23,279 | 6/6 (100) | 103,166 | 37,650 |
| 4.22 × 104 | 6/6 (100) | 5,416 | 2,453 | 6/6 (100) | 26,000 | 11,314 | 6/6 (100) | 52,333 | 28,268 |
| 2.11 × 104 | 6/6 (100) | 3,283 | 1,480 | 6/6 (100) | 12,933 | 6,199 | 6/6 (100) | 27,166 | 10,154 |
| 1.05 × 104 | 6/6 (100) | 1,500 | 666 | 6/6 (100) | 7,000 | 3,686 | 6/6 (100) | 10,650 | 6,858 |
| 5.23 × 103 | 3/6 (50) | 743 | 148 | 6/6 (100) | 3,117 | 1,395 | 6/6 (100) | 5,900 | 1,577 |
| 2.64 × 103 | 1/6 (17) | <LL | 6/6 (100) | 1,767 | 197 | 6/6 (100) | 2,778 | 2,168 | |
| 1.32 × 103 | 1/6 (17) | <LL | 6/6 (100) | 1,290 | 588 | 6/6 (100) | 1,423 | 532 | |
| 6.6 × 102 | 1/6 (17) | <LL | 3/6 (50) | 1,003 | 517 | 6/6 (100) | 946 | 348 | |
| 3.3 × 102 | 0/6 (0) | <LL | 0/6 (0) | <LL | 3/6 (17) | 506 | 7 | ||
| 1.7 × 102 | 0/6 (0) | <LL | 1/6 (17) | 570 | 0/6 (0) | <LL | |||
Number of tests in which HIV-1 RNA was detected/total number of tests.
Mean from all six determinations, including results below the lower limit of 400 HIV-1 RNA copies (<LL) obtained with a given concentration.
FIG. 2.
Number of HIV-1 RNA copies observed with the NASBA HIV-1 RNA QT system versus the number of copies expected based on an estimated input of 1 ml (A) or adjusted for specimen input volume (B). (C) Observed linear range. The input specimens were derived by dilution of a well-characterized HIV-1-infected cell lysate provided by the Viral Quality Assurance Laboratory.
DISCUSSION
The results of the present study demonstrated a constant proportional relationship between the specimen input volume and the number of HIV-1 RNA copies reported following amplification with the NASBA HIV-1 RNA QT system. This relationship was observed both with authentic clinical specimens from HIV-1-infected individuals and with specimens derived from an in vitro HIV-1 stock. Conversion of the latter data to reflect input on a copy number basis indicated no statistical difference between the three specimen volumes with respect to the relationship of input copy to copy number of reported HIV-1 RNA. The consistently observed relationship between specimen input volume and HIV-1 RNA copy number should be extrapolatable to other specimen input volumes if the reported HIV-1 RNA copy number is within the defined linear range of the assay (264 to 5,400,000 copies). This extrapolation was reported in a previous study using a modified version of the NASBA HIV-1 RNA QT assay with specimen input volumes of 0.2 and 1.0 ml (5). In addition, by increasing the volume of the specimen used for analysis with this assay, an increase in the number of specimens with HIV-1 RNA copies reported (clinical sensitivity) was achieved.
Efficacious utilization of viral load testing is predicated upon the ability to obtain an accurate report of the HIV-1 RNA copy number in a given patient specimen. The ability of a nucleic acid amplification assay for determination of HIV-1 RNA to report copy numbers accurately under circumstances where the concentration of HIV-1 RNA is diminished, for example, following administration of HAART, is especially important clinically. Clearly, detection of low levels of HIV-1 RNA is essential for clinical management of patients receiving anti-HIV-1 therapeutic regimens (see, for example, reference 5).
The difficulty of detecting low analyte concentrations with standard HIV-1 viral load assay configurations has led to the development of assay refinements to improve detection sensitivity. Concentration of the isolated HIV-1 RNA or of virions from the specimen forms the conceptual basis for such approaches. In the present study, our approach utilized the flexibility of the Boom nucleic acid isolation procedure to increase the specimen input volume, with a concomitant proportional increase in the NASBA lysis buffer volume, and thereby increase the concentration of the HIV-1 RNA analyte available for amplification. In effect, capture of the analyte by the silica matrix used for the Boom nucleic acid isolation procedure is analogous to physical concentration of HIV-1 virions by ultracentrifugation (10). The approach described in this study may be generally more useful as a means for improving the sensitivity of HIV-1 RNA detection, since the probability of HIV-1 RNA detection increases with large specimen volumes. This approach may reduce the necessity of performing a second ultrasensitive test procedure if the initial viral load test result is below the detection limit (4). Further, since the analysis of greater specimen volumes can be accomplished with a minor modification of the standard NASBA isolation procedure, this approach may be more efficient in terms of time than the reported ultrasensitive procedures used in conjunction with HIV-1 RNA viral load testing.
In conclusion, the use of specimen volumes increased as much as 10-fold over the normal 0.1-ml input for the NASBA HIV-1 RNA QT assay resulted in an increased frequency of HIV-1 RNA reporting. The HIV-1 RNA copy numbers reported by the NASBA HIV-1 RNA QT system were found to be directly proportional to the specimen input volume. The observed increase in clinical sensitivity can be attributed to the ability of the Boom nucleic acid isolation procedure to efficiently capture HIV-1 RNA from incremental specimen input volumes.
ACKNOWLEDGMENTS
Organon Teknika Corporation, the manufacturer of the NASBA test kits, provided funding for this work. C. C. Ginocchio was funded in part by the Jane and Dayton Brown and Dayton Brown, Jr., Molecular Diagnostics Laboratory.
REFERENCES
- 1.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Report of the NIH panel to define principles of therapy of HIV infection and guidelines for the use of anti-retroviral agents in HIV-infected adults and adolescents. Morbid Mortal Weekly Rep. 1998;47(RR-5):1–82. [PubMed] [Google Scholar]
- 3.Draper N, Smith H. Applied regression analysis. New York, N.Y: John Wiley & Sons; 1981. [Google Scholar]
- 4.Erali M, Hillyard D R. Evaluation of the ultrasensitive Roche Amplicor HIV-1 Monitor assay for quantitation of human immunodeficiency virus type 1 RNA. J Clin Microbiol. 1999;37:792–795. doi: 10.1128/jcm.37.3.792-795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginocchio C C, Tetali S, Washburn D, Zhang F, Kaplan M H. Comparison of levels of human immunodeficiency virus type 1 RNA in plasma as measured by the NucliSens nucleic acid sequence-based amplification and Quantiplex branched-DNA assays. J Clin Microbiol. 1999;37:1210–1212. doi: 10.1128/jcm.37.4.1210-1212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginocchio C C, Wang X P, Kaplan M H, Mulligan G, Witt D, Romano J W, Cronin M, Carroll R. Effects of specimen collection, processing, and storage conditions on stability of human immunodeficiency virus type 1 RNA levels in plasma. J Clin Microbiol. 1997;35:2886–2893. doi: 10.1128/jcm.35.11.2886-2893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder J, Resnick R, Saget B, Scheibel S, Herman S, Payne H, Harrigan R, Kwok S. A rapid and simple method for extracting human immunodeficiency virus type 1 RNA from plasma: enhanced sensitivity. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natarajan V, Bosche M, Metcalf J A, Ward D J, Lane H C, Kovacs J A. HIV-1 replication in patients with undetectable plasma virus receiving HAART. Lancet. 1999;353:119–120. doi: 10.1016/s0140-6736(05)76156-0. [DOI] [PubMed] [Google Scholar]
- 9.Van Gemen B, van Beuningen R, Nabbe A, van Strijp D, Juriaans S, Lens P, Kievits T. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescence (ECL) labeled probes. J Virol Methods. 1994;49:157–168. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 10.Winters M A, Tan L B, Katzenstein D A, Merigan T C. Biological variation and quality control of plasma human immunodeficiency virus type 1 RNA quantitation by reverse transcriptase polymerase chain reaction. J Clin Microbiol. 1993;31:2960–2966. doi: 10.1128/jcm.31.11.2960-2966.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Tood J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trial Group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]