Abstract
Since 1970 Mycoplasma fermentans has been suspected of being associated with rheumatoid arthritis. However, this association has been difficult to prove, and this has been our goal. The distribution of M. fermentans was studied in the synovial fluid of patients suffering from different arthritides. Samples of synovial fluid were taken from patients with well-defined disease and a clear diagnosis. After removal of the inflammatory cells and hyaluran, they were treated with proteinase K and tested by a single or fully nested PCR with primers directed against part of the two 16S rRNA genes of M. fermentans. The product was sequenced automatically, by using an ALF Express automatic sequencer, to confirm the mycoplasma species and to identify the strain since the two genes were usually found to be polymorphic. This was also true of the type strain, strain PG18. M. fermentans was detected in 23 of 26 (88%) rheumatoid arthritis patients, and four different strains were found. It was also found in 7 of 8 (88%) of the nonrheumatoid inflammatory arthritis patient group, which consisted of one patient with reactive arthritis, one patient with pauciarticular juvenile chronic arthritis, two patients with gout, two patients with ankylosing spondylitis, and two patients with psoriatic arthritis, only one of whom was infected with M. fermentans. It was not detected in any of the 10 osteoarthritis patients. M. fermentans was therefore found to be a variable and very common organism in arthritic patients with inflammatory joint exudates and may well prove to be important in the etiology of the diseases.
Mycoplasmas are proven causes of arthritis in animals, notably, Mycoplasma bovis and M. capricolum in ruminants, M. hyosynoviae in pigs, M. synoviae in poultry, and M. pulmonis and M. arthritidis in rodents (18). Mycoplasmas have been suspected of being associated with arthritis in humans. This is particularly the case for M. fermentans, which was first isolated from rheumatoid arthritis (RA) patients by Williams et al. (20), and an increase in cell-mediated immunity to this organism in RA patients was noted (21). However, others (12) failed to confirm these findings due to the fastidious growth requirements of M. fermentans and poor success with culture. When PCR methods became available, this organism was detected in the saliva samples from 44% of healthy people (4), in the throat, urine, or peripheral blood mononuclear cells of 33% of human immunodeficiency virus-seronegative patients attending a venereal diseases clinic (10), in 11% of the peripheral blood mononuclear cells from human immunodeficiency virus-seronegative subjects (11), and in the synovial fluid of 14% of patients with rheumatoid and other inflammatory arthritides (15, 16), although the organism was present in 40% of the biopsy specimens of the RA patients' synovial lining cells. It has not been reported from osteoarthritis (OA) patients, who were included in the present study as controls. Unfortunately, however, the PCR detection methods for M. fermentans were based on supposedly specific primers (19) which have recently been found to detect M. orale also (5), especially if this organism is present at a high concentration. The work described in this paper was designed to search for and identify M. fermentans in human synovial fluid by a very sensitive, fully nested PCR method and to unequivocally identify the organism by sequencing part of both of its 16S rRNA genes, which proved to be polymorphic. A similar method has been used to identify strains of M. mycoides (14).
MATERIALS AND METHODS
Patients.
Synovial fluid samples were collected from the knees of 26 patients with RA, all of which fulfilled the criteria for diagnosis of the American Rheumatism Association (2). Samples were also collected from 10 patients with OA and from 8 patients with other inflammatory arthritides. The latter included 2 patients suffering from ankylosing spondylitis (AS), 2 patients with gout, 2 patients with psoriatic arthritis (PsA), and one patient each with reactive arthritis (ReA) and pauciarticular juvenile chronic arthritis (JCA). The total number of samples collected from patients with RA was 34, since on five occasions samples were obtained from both the patient's knees, and 2 consecutive samples were obtained from three patients at least 3 months apart. The total number of samples from the eight non-rheumatoid inflammatory arthritis patients was 11, since samples were obtained from both knees for one AS patient and for one ReA patient and two consecutive samples 1 year apart were obtained from the JCA patient. Nonsteroidal anti-inflammatory drugs (NSAIDS) and/or prednisolone were taken by 23 of 26 RA patients and 6 of 8 of the non-rheumatoid inflammatory arthritis patients, whereas only 3 of 10 of the OA patients were taking NSAIDS. Patient information is shown in Table 1.
TABLE 1.
Patient details
| Disease | Patient no. | Mean age (yr) | Female:male ratio | Mean disease duration (yr) | Disease duration (range [yr]) |
|---|---|---|---|---|---|
| RA | 26 | 59.7 | 1.6:1 | 9.8 | 1–39 |
| OA | 10 | 65.6 | 2.3:1 | 5.2 | 0.33–14 |
| AS | 2 | 44.5 | Male | 3.9 | 1.8–6 |
| JCA | 1 | 25 | Female | 23 | |
| ReA | 1 | 28 | Female | 2.6 | |
| Gout | 2 | 49.5 | Male | 0.13 | 0–0.25 |
| PsA | 2 | 48 | Male | 23 | 22–24 |
Synovial fluid collection.
The synovial fluid samples were obtained by sterile aspiration from the knees of patients attending the rheumatology clinic when aspiration was indicated as part of routine clinical practice. Consent for evaluation of the fluid was obtained from the patients. The samples were handled aseptically and were centrifuged at 500 × g for 30 min at 4°C to remove the cells. Aliquots (usually 1 ml) were stored at −70°C. It was important that these samples not be thawed and refrozen.
Materials.
The suppliers of the apparatus and chemicals are listed below. Hyaluronidase and proteinase K were obtained from Roche Diagnostics Ltd., Lewes, United Kingdom. The primers, including the biotin-labelled primer with the hexamethylene spacer, came from Oswel DNA Service, University of Southampton, Boldrewood, United Kingdom. The Alf Express automatic sequencer, Cy5-labelled sequencing primers, Taq polymerase, and the solid-phase sequencing plastics and chemicals came from Amersham Pharmacia Biotech Ltd., Little Chalfont, United Kingdom. The thermal cycler and Taq Gold came from Perkin-Elmer Applied Biosystems Ltd., Warrington, United Kingdom, and Takara Ex Taq came from BioWhittaker UK Ltd., Wokingham, United Kingdom. The Tris-borate sequencing buffer came from National Diagnostics, Hull, United Kingdom, and the Finnpipettes from Life Sciences International, Basingstoke, United Kingdom.
Preparation of samples for PCR.
Synovial fluid samples were prepared in batches of 16 or 32 and normally contained two blanks with 1 ml of Tris-buffered saline. A 10-mg/ml solution of hyaluronidase in pyrogen-free water was freshly prepared and was sterilized by filtration. The viscosity of the 1-ml synovial fluid samples was lowered by adding 20 μl of hyaluronidase solution per ml and allowing the mixture to stand for 0.5 h at room temperature. The samples were then centrifuged at 18,000 × g for 0.5 h at 20°C, and the small pellet that was obtained was washed twice with sterile pyrogen-free saline that had been buffered at pH 8.0 with 10 mM Tris-HCl (TBS). For the blanks, hyaluronidase was added to 1 ml of TBS, and the tubes were treated as described above for the other samples.
A fresh 10-mg/ml solution of DNA-free proteinase K was prepared. The sample buffer contained 10 μl of 1 M Tris-HCl (pH 8.0), 12 μl of proteinase K solution, and 0.978 ml of pyrogen-free water. Then, 50 μl of filter-sterilized 9% Tween 20 was added, and 0.1 ml of this mixture was added to the samples and the blanks. These were heated at 37°C for 30 min and 60°C for 1 h, and then the proteinase K activity was destroyed by heating the samples at 96°C for 10 min. These samples were stored frozen at −20°C.
Primers.
The PCR primers were based on those described by Hopert et al. (7), but only those which fit the published 16S rRNA gene sequence for M. fermentans exactly were used. The primers were purified by high-performance liquid chromatography, and their sequences are shown in Table 2.
TABLE 2.
Primers used in the studya
| Primer type | Primer name | 5′ sequence | Primer or primer specificity | 3′ sequence |
|---|---|---|---|---|
| Outer | 9A | CGCCTGAGTAGTACGTTCGC | 3A | GCGGTGTGTACAAGACCCGA |
| Inner | *8A | TGGTGCATGGTTGTCGTCAG | 5B | GAACGTATTCACCGTAGCGTA |
| Sequencing | ||||
| SEQ1 | Cy5-GACCCGAGAACGTATTCACC | General | ||
| SEQ2 | Cy5-TAGCGTACGTGATCTACGAT | M. fermentans only |
The asterisk for primer *8A indicates that the primer was biotinylated at its 5′ end. The biotin interfered with the binding of primer *8A, but this problem was overcome with a hexaethylene glycol spacer. The sequencing primers were labelled with the fluorescent dye Cy5 and were designed to bind to the nonbiotinylated end of the PCR product. The sequences obtained were reversed and complementary to the coding strand.
PCR.
The PCR mixture (50 μl) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% (wt/vol) gelatin (Perkin-Elmer Taq Gold buffer diluted 10 times), the nucleotides dATP, dCTP, dGTP, and dTTP, each at a concentration of 200 μM, each primer at a concentration of 200 nM, 1 U of Taq polymerase, and 5 μl of sample. When a nested PCR was to be performed, the first round used Taq polymerase from Pharmacia or, in some cases, TaKaRa Ex Taq, which contains a proofreading enzyme which increases the sensitivity of the reaction two- to threefold. In this case the buffer and deoxynucleoside triphosphate solutions supplied by the manufacturer were used. It was important to keep the pH at 8.3 for the biotinylated primer used in the second round, so highly specific Taq Gold was used. The primer 8A recommended by Hopert et al. (7) caused problems when it was biotinylated, probably because the primer binds close to a loop on the DNA. However, primer 8A worked well when the biotin was attached with a hexaethylene glycol spacer. The PCR was carried out in 0.2-ml tubes, and the reaction mixture was overlaid with 30 μl of sterile mineral oil. The thermal cycler was a Perkin-Elmer GeneAMP PCR system 9600.
The sample was initially dissociated by heating at 95°C for 3 min (Taq Gold for 15 min) and was then thermally cycled 50 times at 95°C for 35 s, 58°C for 60 s, and 72°C for 35 s, finally being left to extend at 72°C for 7 min before being cooled to 4°C. The cycling conditions were the same for both sets of primers, and 50 rather than 30 cycles were used to obtain as strong a PCR band as possible for sequencing. The relatively long annealing period was found to produce the best results. An 8-μl sample of the final reaction products was run on a 3% agarose gel containing 0.5 μg of ethidium bromide ml−1, and the gel was photographed under a UV light. Those samples showing a clear PCR band were retained for sequencing. Initially, primers *8A and 3A were used with Taq Gold under the same PCR conditions described above. This gave a single-stage PCR and, with the Cy5-labelled sequencing primer SEQ1, allowed the type strain PG18 to be partially sequenced and identified a missing base (N = A), which was used in SEQ2. Samples from 12 of 32 of the inflammatory arthritis patients gave bands which could be sequenced by the single-stage PCR. However, some bands which were too weak to sequence were found, so the nested PCR was used, together with the Cy5-labelled primer SEQ2. The single-stage PCR and sequencing primer SEQ1 were aimed at human mycoplasmas in general and were not specific for M. fermentans, although this was the only organism detected by the single-stage PCR. The nested PCR used primers 9A and 3A as the outer primer pair and primers *8A and 5B as the inner primer pair, with SEQ2 as the sequencing primer. These were specific only for M. fermentans.
Sequencing.
An ALF Express automated sequencer was used. It was developed particularly for the sequencing of human DNA and readily detects gene polymorphisms. It was used with the solid-phase sequencing kit supplied by the manufacturer. The biotinylated PCR product was removed from the reaction mixture by allowing it to adhere to four streptavidin-coated plastic combs. The Cy5-labelled sequencing primer was allowed to bind to the PCR product, and the sequencing reaction was run with T7 polymerase and the dideoxynucleoside triphosphate mixtures provided by the manufacturer. The reaction products were dissociated from the combs with the iodoacetamide mixture provided by the manufacturer and were electrophoresed down a 6% polyacrylamide gel with 0.089 M Tris-borate buffer (pH 8.3) with 2 mM EDTA, obtained as a 10× concentrate. The dye-labelled products were visualized with a laser, and the sequence was shown on a computer screen. This sequence, when reversed and complemented, was compared to the prokaryotic DNA sequences on the PCGENE or EMBL databases to identify the organism.
Precautions against contamination.
As the mycoplasmal contents of the synovial fluid samples were extremely small, they were both difficult to measure and easy to contaminate. It was essential to do the DNA extraction and PCR procedures under sterile conditions since the primers used might react to some extent with other bacteria and contaminate the PCR band. If this happened, the sequencing primer was normally blocked so no sequence was obtained, even if a band had been observed.
Pyrogen-free saline and water for injection were used in the isolation of the sample. The Eppendorf tubes and 0.2-ml PCR tubes were sterilized in an autoclave, and the other plastics and pipette filter tips were purchased in radiation-sterilized condition. Given that the PCR bands were sequenced, there was no danger that a band due to a bacterial contaminant would be identified as a mycoplasma. New Gilson and Finnpipettes were acquired and were used solely with the filter tips for PCR. Disposable plastic trays were used to prepare PCRs.
The batch preparation of mixed samples and blanks and the preparation of more than one sample from each individual in different batches were again precautions against contamination, although sometimes not enough material was available for more than one test with a sample. The sequencing of the PCR band identified the organism, and since identifiably different M. fermentans strains were discovered, it was often possible to check for contamination by sequencing.
Nucleotide sequence accession number.
The partial sequence of the second gene in strain PG18 and in most of the clinical samples was deposited in GenBank, under the name MF16SRRNA2 and with accession no. AF031374.
RESULTS
M. fermentans was found in 31 of 34 samples from the RA patients, 9 of 11 samples from the non-rheumatoid inflammatory arthritis patients, and in none of 10 samples from the OA patients. If more than one sample was obtained from one patient the same strain was always found in both knees or in two consecutive samples. An exception was for the first JCA patient sample, which was negative and which may have been affected by a bacterial contaminant, as only a poor-quality PCR band which could not be sequenced was obtained. The second sample from this patient was positive. Table 3 shows the distribution of M. fermentans among the patient groups. It was confined to patients with inflammatory arthritis but not to patients with RA alone and was not found in the 10 patients with OA.
TABLE 3.
Detection of M. fermentans in synovial fluid samples
| Disease | No. of patients positive for M. fermentans/total no. of patients (%) |
|---|---|
| RA | 23/26 (88) |
| OA | 0/10 (0) |
| JCA | 1/1 (100) |
| AS | 2/2 (100) |
| Gout | 2/2 (100) |
| ReA | 1/1 (100) |
| PsA | 1/2 (50) |
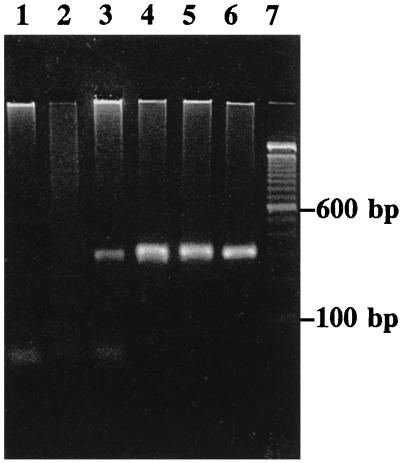
Figure 1 is a photograph of an agarose gel stained with ethidium bromide. It shows the second-round PCR product obtained from some RA, AS, gout, and OA patients with the primer pair *8A-5B. Bands can be seen only for samples from the patients with inflammatory arthritis and not for samples from the patients with OA.
FIG. 1.
Second-round PCR products produced by the synovial fluid samples which were treated with primer pair *8A-5B. Lanes 1 and 2, samples from OA patients; lane 3, sample from a gout patient infected with M. fermentans; lane 4, samples from an AS patient infected with M. fermentans; lanes 5 and 6, samples from RA patients infected with M. fermentans; lane 7, molecular size ladder.
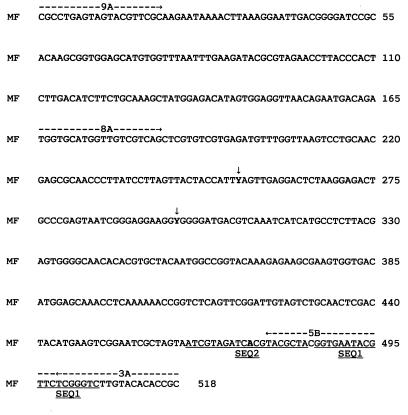
Figure 2 shows the coding strands of part of the 16S rRNA gene from M. fermentans. It was discovered that for the type strain of M. fermentans, strain PG18, and for most of the strains found in the clinical samples, the two 16S rRNA genes of M. fermentans were not identical. The ambiguities occurred at positions 253 and 298 in the M. fermentans sequence shown, where the base was T on one gene and C on the other and is described by the letter Y in the sequence. For strain PG18 and most of the strains from patients, position 253 was heterogenous (Y) but the base at position 298 was T on both genes and was Y for only one strain from an RA patient. PCR products from strains from two RA patients, the PsA patient, and strain “incognitus” homogeneously showed a T base at position 253, and strains from four RA patients homogeneously showed a C base at position 253. Unusual sequences were resequenced for confirmation. Strains from two RA patients were either C or Y, as the (repeated) sequences obtained were poor and ambiguous. This could have been an experimental defect, or perhaps two M. fermentans strains were present. One other base, described as N on the original sequence, was identified and is underlined and in boldface type in Fig. 2. It is part of SEQ2.
FIG. 2.
Sequence of part of the 16S rRNA gene of M. fermentans showing the positions of the primers and the ambiguities in the sequence. The positions of the outer and inner primers are marked with dotted lines and arrows, and the sequencing primers are underlined. MF, edited part of M. fermentans sequence MFRRNAF. Y (arrows) is C or T.
DISCUSSION
M. fermentans was restricted to the joints of patients with inflammatory arthritis and was not detected in any of the patients with OA. Two of three of the RA patients negative for M. fermentans were not taking anti-inflammatory drugs and had clinically quiescent disease. The third patient was taking NSAIDS, and this patient, as well as one of the other M. fermentans-free patients, had only recently developed the disease. A recent publication by O'Dell et al. (13) has shown that treatment of recent-onset RA patients with minocycline, to which M. fermentans is susceptible (6), produces a long-term decrease in the severity of their symptoms.
Comparison with previous studies.
The results of tests for the detection of M. fermentans confirm those of Williams et al. (20) and Schaeverbecke et al. (15, 16), except that 88% of the synovial fluid samples from our RA patients contained M. fermentans, whereas 14% of the samples reported by Schaeverbeke et al. (15, 16) contained M. fermentans, and both of our patients with gout were infected with M. fermentans. Four different strains of M. fermentans were found, and different strains have been reported to have been cultivated from arthritis patients (17).
There are a number of reasons for the greater sensitivity of the PCR procedures reported from this work. M. fermentans has been reported to associate with B cells rather than neutrophils in the peripheral blood (3), so it was possible to remove the irrelevant, largely neutrophilic infiltrate from the synovial fluid, allowing the small amount of residual precipitate to be treated with proteinase K, which is a 10-fold more sensitive method of DNA preparation than ethanol precipitation from phenol-chloroform (1). This was followed by the use of a fully nested PCR, which is about 100-fold more sensitive than a single or seminested PCR. However, other measures contributed to the result, such as the use of Taq Gold and the treatment of the sample, particularly the specific removal of hyaluran with hyaluronidase, which left any mycoplasmas behind.
The sequencing of the mycoplasmal PCR product not only confirmed the identity of the mycoplasma but also showed the presence of different strains. M. fermentans has an unusual rRNA gene organization, with two sets of rRNA genes and with the two 16S-23S rRNA gene clusters being arranged in a tail-to-tail orientation (8). The sequencing method used in this work was originally developed to sequence the human HLA genes. The whole PCR product is sequenced, so heterozygotes (or polymorphism) can be detected. The sequences of both of the two 16S rRNA genes of M. fermentans were therefore determined, and if a lack of identity occurred they were usually not identical at only one or rarely two bases.
M. fermentans was found in the synovial fluid of nearly all the inflammatory arthritis patients. It has not previously been detected in gout patients, and this discrepancy is attributed to the greater sensitivity of the methods described in this work. In patients with gout it is possible either that the organism arrived in the joint subsequent to the inflammation or even that it initiated the sodium urate crystal formation by a biochemical reaction or by acting as a nucleus for crystal formation. RA patients have significantly more mycoplasmal rRNA in their synovial fluid than the patients with other arthritides (9), so they may have a particular difficulty dealing with these organisms, or special strains may be found in RA patients alone. M. fermentans may well contribute to the inflammation and chronicity of RA.
ACKNOWLEDGMENTS
We thank David Pitcher for providing the mycoplasmal DNAs with the primers were tested and Angus Dalgleish for allowing us to use his automated sequencer.
This work was supported by the St. George's Hospital Special Trustees.
REFERENCES
- 1.Abele-Horn M, Busch U, Nitschko H, Jacobs E, Bax R, Pfaff F, Schaffer B, Heesemann J. Molecular approaches to diagnosis of pulmonary diseases due to Mycoplasma pneumoniae. J Clin Microbiol. 1998;36:548–551. doi: 10.1128/jcm.36.2.548-551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnett F C, Edworthy S M, Bloch D A, McShane D J, Fries J F, Cooper N S, Healey L A, Kaplan S R, Liang M H, Luthra H S, Medsger T A, Jr, Mitchell D M, Neustadt D H, Pinals R S, Schaller J G, Sharp J T, Wilder R L, Hunder G G. The American Rheumatism Association 1987. Revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 3.Cheek R F, Olsak I, Madoff S, Preffer F I. In vitro detection of Mycoplasma fermentans binding to B-lymphocytes in fresh peripheral blood using flow cytometry. Cytometry. 1997;28:90–95. doi: 10.1002/(sici)1097-0320(19970501)28:1<90::aid-cyto11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 4.Chingbingyong M I, Hughes C V. Detection of Mycoplasma fermentans in human saliva with a polymerase chain reaction-based assay. Arch Oral Biol. 1996;41:311–314. doi: 10.1016/0003-9969(96)84556-0. [DOI] [PubMed] [Google Scholar]
- 5.Ditty S E, Connolly M A, Li B J, Lo S-C. Mycoplasma orale has a sequence similar to the insertion-like sequence of M. fermentans. Mol Cell Probes. 1999;13:183–189. doi: 10.1006/mcpr.1999.0233. [DOI] [PubMed] [Google Scholar]
- 6.Hannan P C T. Comparative susceptibilities of various AIDS-associated and human urogenital tract mycoplasmas and strains of Mycoplasma pneumoniae to 10 classes of antimicrobial agent in vitro. J Med Microbiol. 1998;47:1115–1122. doi: 10.1099/00222615-47-12-1115. [DOI] [PubMed] [Google Scholar]
- 7.Hopert A, Uphoff C C, Wirth M, Hauser H, Drexler H G. Specificity and sensitivity of polymerase chain reaction (PCR) in comparison with other methods for the detection of mycoplasma contamination in cell lines. J Immunol Methods. 1993;164:91–100. doi: 10.1016/0022-1759(93)90279-g. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Robertson J A, Stemke G W. An unusual rRNA gene organization in Mycoplasma fermentans (incognitus strain) Can J Microbiol. 1995;41:424–427. doi: 10.1139/m95-056. [DOI] [PubMed] [Google Scholar]
- 9.Johnson S M, Bruckner F E, Collins D A. Mycoplasmal RNA and 5′-nucleotidase in synovial fluid from arthritis patients. Int Org Mycoplasmol Lett. 1996;4:140. [Google Scholar]
- 10.Katseni V L, Gilroy C B, Ryait B K, Ariyoshi K, Bieniasz P D, Weber J N, Taylor-Robinson D. Mycoplasma fermentans in individuals seropositive and seronegative for HIV-1. Lancet. 1993;341:271–273. doi: 10.1016/0140-6736(93)92617-3. [DOI] [PubMed] [Google Scholar]
- 11.Kovacic R, Launey V, Tuppin P, Lafeuillade A, Feuillie V, Montagnier L, Grau O. Search for the presence of six Mycoplasma species in peripheral blood mononuclear cells of subjects seropositive and seronegative for human immunodeficiency virus. J Clin Microbiol. 1996;34:1808–1810. doi: 10.1128/jcm.34.7.1808-1810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mårdh P A, Nilsson F J, Bjelle A. Mycoplasma and bacteria in synovial fluid from patients with arthritis. Ann Rheum Dis. 1973;32:319–325. doi: 10.1136/ard.32.4.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Dell, J. R., G. Paulsen, C. E. Haire, K. Blakely, W. Palmer, S. Wees, P. J. Eckhoff, L. W. Klassen, M. Churchill, D. Doud, A. Weaver, and G. F. Moore. Treatment of early seropositive rheumatoid arthritis with minocycline. Arthritis Rheum. 42:1691–1695. [DOI] [PubMed]
- 14.Pettersson B, Leitner T, Ronaghi M, Bolske G, Uhler M, Johansson K-E. Phylogeny of the Mycoplasma mycoides cluster as determined by sequence analysis of the 16S rRNA genes from the two rRNA operons. J Bacteriol. 1996;178:4131–4142. doi: 10.1128/jb.178.14.4131-4142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaeverbeke T, Gilroy C B, Bébéar C, Dehais J, Taylor-Robinson D. Mycoplasma fermentans in joints of patients with rheumatoid arthritis and other joint disorders. Lancet. 1996;347:1418. doi: 10.1016/s0140-6736(96)91065-x. [DOI] [PubMed] [Google Scholar]
- 16.Schaeverbeke T, Gilroy C B, Bébéar C, Dehais J, Taylor-Robinson D. Mycoplasma fermentans, but not M. penetrans, detected by PCR assays in synovium from patients with rheumatoid arthritis and other rheumatic disorders. J Clin Pathol. 1996;49:824–828. doi: 10.1136/jcp.49.10.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaeverbeke T, Clerc M, Lequen L, Charron A, Bébéar C, de Barbeyrac B, Bannwarth B, Dehais J, Bébéar C. Genotypic characterization of seven strains of Mycoplasma fermentans isolated from synovial fluids of patients with arthritis. J Clin Microbiol. 1998;36:1226–1231. doi: 10.1128/jcm.36.5.1226-1231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simecka J W, Davis J K, Davidson M K, Ross S E, Städtlander C T, Cassell G H. Mycoplasma diseases of animals. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C.: American Society for Microbiology; 1992. pp. 391–415. [Google Scholar]
- 19.Wang R Y, Hu W S, Dawson M S, Shih J W, Lo S C. Selective detection of Mycoplasma fermentans by polymerase chain reaction and by using a nucleotide insertion sequence within the insertion sequence-like element. J Clin Microbiol. 1992;30:245–248. doi: 10.1128/jcm.30.1.245-248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams M H, Brostoff J, Roitt I M. Possible role of Mycoplasma fermentans in the pathogenesis of rheumatoid arthritis. Lancet. 1970;ii:277–280. doi: 10.1016/s0140-6736(70)91328-0. [DOI] [PubMed] [Google Scholar]
- 21.Williams M H, Bruckner F E. Immunological reactivity to Mycoplasma fermentans in patients with rheumatoid arthritis. Preliminary communication. Ann Rheum Dis. 1971;30:271–273. doi: 10.1136/ard.30.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]