Abstract
Objective:
To train a model to predict vasopressor use in ICU patients with sepsis, and optimize external performance across hospital systems using domain adaptation, a transfer learning approach.
Design:
Observational cohort study
Setting:
Two academic medical centers from January 2014 to June 2017.
Patients:
Data were analyzed from 14,512 patients (9,423 at the development site, 5,089 at the validation site) who were admitted to an intensive care unit (ICU) and met Center for Medicare and Medicaid Services (CMS) definition of severe sepsis either before or during the ICU stay. Patients were excluded if they never developed sepsis, if the ICU length of stay was less than 8 hours or more than 20 days, or if they developed shock up to the first 4 hours of ICU admission.
Measurements and Main Results:
Forty retrospectively collected features from the electronic medical records (EMR) of adult ICU patients at the development site (4 hospitals) were used as inputs for a neural network Weibull-Cox survival model to derive a prediction tool for future need of vasopressors. Domain adaptation updated parameters to optimize model performance in the validation site (2 hospitals), a different healthcare system over 2,000 miles away. The cohorts at both sites were randomly split into training and testing sets (80% and 20%, respectively). When applied to the test set in the development site, the model predicted vasopressor use 4 to 24 hours in advance with an area under the receiver operator characteristic curve [AUCroc], specificity, and positive predictive value (PPV) ranging from 0.80-0.81, 56.2-61.8% and 5.6-12.1% respectively. Domain adaptation improved performance of the model to predict vasopressor use within 4 hours at the validation site (AUCroc 0.81 [CI 0.80-0.81] from 0.77 [CI 0.76-0.77], p<0.01; specificity 59.7% [CI 58.9-62.5%] from 49.9% [CI 49.5-50.7%], p<0.01; PPV 8.9% [CI 8.5-9.4%] from 7.3 [7.1-7.4%], p<0.01).
Conclusions:
Domain adaptation improved performance of a model predicting sepsis-associated vasopressor use during external validation.
Keywords: sepsis, septic shock, intensive care units, clinical decision support, artificial intelligence, disease progression
INTRODUCTION
Sepsis, life-threatening organ dysfunction caused by a dysregulated host response to infection, can lead to poor outcomes, especially among those who experience disease progression. Patients with sepsis who develop septic shock have a higher mortality than those without progression.[1-4] Patients with hospital-acquired sepsis and those who progress to septic shock are among the groups with the highest mortality,[5] providing a potential opportunity to improve outcomes through early recognition and management.
While a few data-driven sepsis prediction models have been externally validated,[6, 7] there is an expected degradation in model performance when those models are tested on unseen data. Some of that diminished performance may be due to different incidences of sepsis or sequelae like septic shock; lower incidences of the outcome of interest at validating sites may lead to worse model performance.[8] Other factors such as differences in provider antibiotic prescribing practices and lactate measurement could also play a role. Newer analytics tools could improve generalizability of models that predict sepsis and its sequelae.
Domain adaptation is a machine learning approach that can fine-tune data-driven models to improve performance characteristics in a different context.[9, 10] Domain adaptation has been adopted by the medical community to repurpose models once used in other domains in which training data is scarce. For instance, online photo recognition software has been employed as a “base model” to help machine learning algorithms more quickly and accurately detect diabetic retinopathy from retinal optic imaging, and improve pneumonia detection from chest radiographs in pediatric patients.[9] More recently, domain adaptation has been used to test the ability of a model to predict mortality in patients across different types of ICUs- medical and surgical.[11] In this context, domain adaptation can adjust the weights given to predictor variables of a model that was trained on one dataset (source data) to apply to a different dataset (target data). The goal would be to give priority to the combinations of model inputs that optimize performance in the target dataset.
Domain adaptation can potentially improve external validity of models that predict sepsis and its progression,[12] but such an approach has not been applied to risk prediction in the ICU. The objectives of this study are: (1) to derive a machine learning-based model that predicts future vasopressor use among sepsis patients; and (2) to determine the impact of domain adaptation on model generalizability.
MATERIALS AND METHODS
Key reporting metrics for prediction models were adapted from the TRIPOD checklist[13] and guidelines published by Leisman and colleagues,[14] and are summarized below:
Study Population
Training of the Deep Artificial Intelligence Sepsis Expert (AISE2) algorithm was performed on a subset of 52,799 adult ICU patients (aged 18 years or over) from four hospitals within the Emory Healthcare system (the development site), and validated on a subset of 19,923 adult ICU patients from two hospitals within the University of California San Diego (UCSD) system (the validation site). Data was extracted from January 2014 to June 2017. This investigation was approved by the Institutional Review Boards of both sites (Emory IRB #110675; UCSD IRB #191098), with a waiver of informed consent. Data were included if patients developed sepsis prior to, or during their ICU stay. The time of sepsis, or t0, was the last of all events that define severe sepsis according to the Centers for Medicare and Medicaid Services (CMS).[15] (See Appendix A.) Suspicion of infection was defined as the administration of antibiotics plus acquisition of body fluid cultures. Patients were excluded if they never developed sepsis prior to or during the ICU stay, if their length of ICU stay was less than 8 hours or more than 20 days, or if they needed vasopressors prior to and including the first four hours of ICU admission.
Sepsis was alternately defined according to the Third International Consensus Definitions for Sepsis (Sepsis-3)[16] to identify any changes in the prediction of vasopressor use by AISE2.
Predictors (Features), and Data Sources
A total of 40 commonly-available and easily obtained input variables were included from electronic medical record (EMR) data for model development, as previously outlined.[17] (Supplemental Table 1) Data from the Cerner Powerchart medical record system (Cerner, Kansas City, MO) were extracted through a clinical data warehouse (MicroStrategy, Tysons Corner, VA). Data from the Epic medical record system (Epic Systems Corporation, Verona, Wisconsin) was retrieved by accessing Clarity, Epic’s main data archiving and reporting environment.
Outcome
The outcome of interest was the first recorded timestamp of vasopressor use in ICU patients with preexisting sepsis, which was used as a surrogate for septic shock. Drugs classified as vasopressors include norepinephrine, vasopressin, epinephrine, dopamine, and phenylephrine. The main objective of the algorithm was to predict vasopressor administration within a 4-hour prediction window.
Model Development and Domain Adaptation
Data for all 40 variables were placed into hourly bins. Carry-forward or mean imputation approaches were used to impute missing values if there was at least one value or no prior value during the ICU stay, respectively. (See Appendix A for further details on feature pre-processing.) Data were randomly split into training and testing sets (80% and 20%) for each hospital system. AISE2, composed of neural network and Weibull-Cox layers, were trained and tested at the development site.
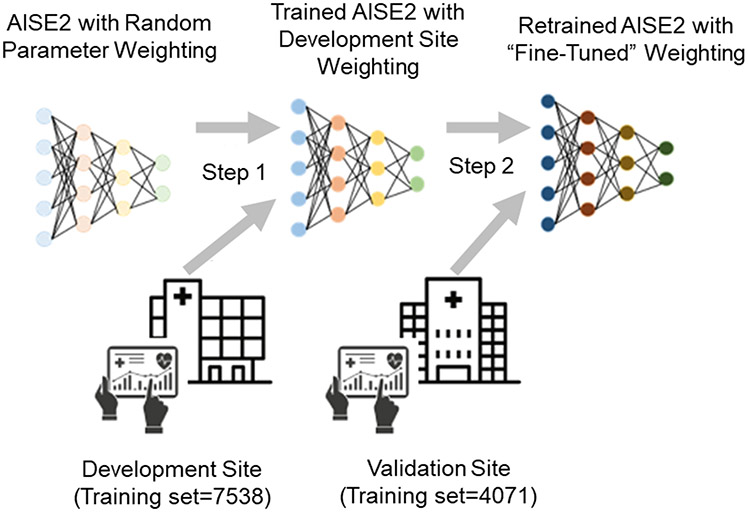
The model weights learned at the development site were used as a starting point for model retraining and optimization on the training set at a remote validation site in a process called domain adaptation. (See Figure 1 and Appendix A.) The newly-tuned model was then tested on the validation site’s test set.
Figure 1: Schematic diagram of domain adaptation*.
*-The connections between each node in the AISE2 model are initially assigned random “weights” to determine model output. The model was trained on a sample of data from the development site (the training set), which updated the model weights (step 1). Before the model was applied to a remote external site, domain adaptation was used to refine the model weights using a sample of new training data from the remote (validation) site (step 2). Therefore, the model was customized for prediction at the validation site.
Model outputs and explainability
A risk score and relevance scores were reported at every time bin. The risk score is the probability that at least one vasopressor would be initiated within the prediction window. The explainability of AISE2 was derived from the relevance scores of each variable, which represent the amount that the risk score reacted to the new feature values. (See Appendix B for further details on AISE2 and its explainability.)
Statistical methods
Model performance was assessed using three metrics- the area under receiver operating characteristic curves (AUCroc), specificity, and positive predictive value (PPV)- of AISE2 classification over 4 hours. Those performance metrics were compared to performance using prediction windows of 6, 8, 12, and 24-hours. Sensitivity was set at 85% to optimize specificity without creating an unreasonably high threshold for alerts. Main results were bootstrapped to report confidence intervals and minimize overfitting. Calibration curves assessed the ability of the model to predict sepsis-associated vasopressor use at different levels of risk, as determined by AISE2.
Comparisons to AISE2 performance
The ability of AISE2 to predict future vasopressor use, as measured by the AUCroc, was compared to that of two other tools: (1) a Weibull-Cox proportional hazards model; and (2) serial SOFA assessment. (See Appendix A for further details on model derivation.)
Software
MATLAB v2020a (Natick, MA, USA) was used for all pre-processing and analyses.
RESULTS
Demographics and characteristics in the cohort defined by CMS severe sepsis definition
After applying all exclusion criteria, 1324 (14.0%) out of 9,423 septic patients were started on vasopressors in the ICU in the development site, and 1,046 (20.5%) out of 5,089 septic patients needed vasopressors in the validation site. (Supplemental Table 2 & Supplemental Figure 1). The vasopressor group in the validation site had slightly lower SOFA scores than that of the development site (5.2 vs 5.8), but longer ICU length of stay (228.7 vs 169.2 hours), longer median times from ICU admission to vasopressors (33 vs 25 hours) and from sepsis to vasopressors (36.3 vs 23.2 hours). (Supplemental Table 2) The validation site had a slightly higher inpatient mortality in the vasopressor group compared to the development site (42.4% vs 38.1%), but more patients in the vasopressor group at the development site were sent to hospice (14.4% vs 1.2%).
Feature missingness
Features were missing to varying degrees depending on the baseline frequency of routine measurement. (Supplemental Table 1) Laboratory data were missing a higher percentage of time compared to vital sign data. Patterns in feature missingness were similar in the development and validation cohorts, except there were higher percentages of missing data for systolic, diastolic and mean blood pressures in the validation cohort. (See Appendix C for further details on missingness and imputation.)
AISE2 ability to predict septic-associated vasopressor use
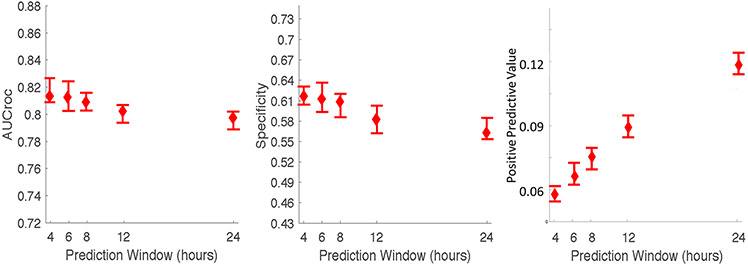
The AUCroc, specificity and PPV of the AISE2 algorithm to forecast sepsis-associated vasopressor use with 85% sensitivity in the test set of the development site within a 4-hour prediction window were 0.81 [CI 0.81-0.83], 61.8% [CI 60.6-63.2%] and 5.6% [CI 5.3-6.2%], respectively. (Figure 2) The AUCroc at the development site decreased slightly for longer prediction windows up to 24 hours, the lowest value being 0.80 [CI 0.79-0.80] at 24 hours. (Figure 2) The specificity also decreased slightly from 4 to 24 hours (61.8% [CI 60.6-63.2%] to 56.2% [CI 55.4-58.2%]), but the PPV doubled from 5.6% [CI 5.3-6.2%] to 12.1% [CI 11.4-12.6%]. Model performance was similar in the cohort defined by sepsis 3 criteria. (See Supplemental Figure 2 and Appendix C for details.)
Figure 2: Septic-associated vasopressor use prediction in the test set of the development site at different prediction windows*.
*Confidence intervals for the AUCroc, specificity and PPV of each prediction window were calculated using bootstrapped resampling
Among the features that contributed most to algorithm performance were vital signs (e.g., systolic blood pressure, respiratory rate, and heart rate), laboratory data (e.g., white blood cell count, lactate, and glucose), demographic information (e.g., gender), and other miscellaneous hospital-specific information (e.g., the patient’s assigned ICU). (Supplemental Table 4) Of these, seven of the top ten features were based on vital signs. The top three important features at both sites were systolic blood pressure, respiratory rate, and heart rate, despite site differences in systolic blood pressure and respiratory rate missingness. (Supplemental Table 1)
Impact of domain adaptation
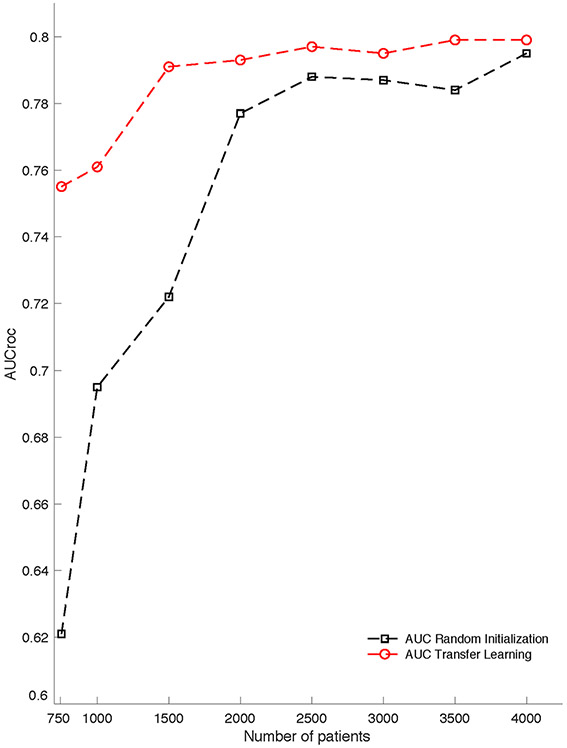
All performance characteristics for the model improved at the validation site with domain adaptation, approximating performance in the development site (AUCroc 0.77 [CI 0.76-0.78] to 0.81 [CI 0.80-0.81]). (Table 1) To achieve comparable performance, a model that was only trained and tested on the validation site (without involvement of a development site) would require a much larger proportion of training data than the amount needed from a model applying domain adaptation. (Figure 3) The following metrics also improved with domain adaptation: specificity (49.9% [CI 49.5-50.7%] to 59.7% [CI 58.9-62.5%]), PPV (7.3% [CI 7.1-7.4%] to 8.9 [CI 8.5-9.4%]), and the workup detection index (13.7 [CI 13.5-14.1] to 11.2 [CI 10.6-11.8]). (Table 1) With domain adaptation, all measured performance metrics of AISE2 on the validation cohort were at least comparable to those measured in the development cohort. Based on the workup detection indices, domain adaptation would generate an average of 2.5 fewer false alerts for every true alert at the validation site, compared to the model without retraining.
Table 1:
Model performance at the validation site using 4-hour prediction windows, with and without retraininga
| No retraining | Domain adaptationb | |
|---|---|---|
| AUCroc | 0.77 [0.76, 0.78] | 0.81 [0.80, 0.81] % |
| Specificityc | 49.9 [49.5 50.7] % | 59.7 [58.9 62.5] % |
| PPVc,d | 7.3 [7.1, 7.4] % | 8.9 [8.5, 9.4] % |
| Accuracy | 51.5 [51.0, 52.2] % | 60.8 [60.0, 63.4] % |
| Workup detection index | 13.7 [13.5, 14.1] | 11.2 [10.6, 11.8] |
AUCroc=Area under the receiver operator curve; PPV=positive predictive value
All performance metrics are reported for a prediction window of four hours. Confidence intervals are also reported, after bootstrapping. False negative labels were defined as instances in which patients are assigned a risk score <0.35, but they required vasopressors within 4 hours. False positive labels were instances in which patients were assigned a risk score of >0.35, but they did not require vasopressors within 4 hours. The p-values were <0.01 for all comparisons between each performance metric associated with retraining and no retraining.
Domain adaptation involved fine-tuning; training started from the model coefficient values derived at the development site, with 20 iterations of gradient descent to optimize model performance
Sensitivity was held constant at 85%
A label was deemed a true positive if and only if vasopressor use occurred within 4 hours, regardless of vasopressor use thereafter
Figure 3: Contrasting the effect of training vs retraining (“domain adaptation”) on AISE2 performance in the validation site*.
*-Domain adaptation achieved superior performance using a relatively small training set at the target institution. The line with square points represents a model trained from scratch (randomly initialized weights) using 750, 1000, …, 4000 patients at the target institution (Hospital System B). The line with circular points represents a model fine-tuned on the target institution data using 750, 1000, …, 4000 patients used a model trained from a separate institution (Hospital System A). (All other hyperparameters were kept constant.) The domain adaptation approach achieved similar performance regardless of the size of retraining data from the target population
Through the addition of domain adaptation, the AUCroc of AISE2 was higher than a Weibull-Cox model without a neural network component. Without model retraining, the AISE2 AUCroc was higher than that of serial SOFA assessment at the validation site (0.73 vs 0.67, respectively), but not higher than that of a Weibull-Cox-only model (0.73 vs 0.73). The AISE2 AUCroc was higher than that of serial SOFA assessment and the Weibull-Cox-only model in the validation site after domain adaptation (AUCroc 0.81 vs 0.67 and 0.76, respectively). (Supplemental Figure 3)
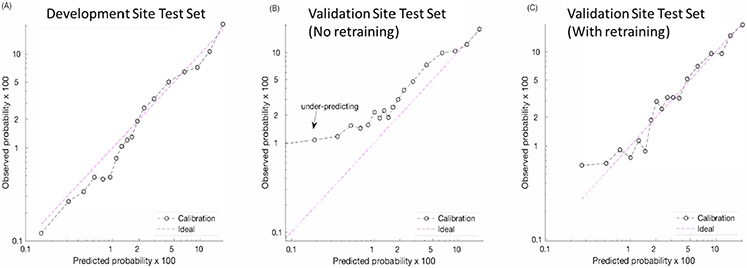
Domain adaptation improved model calibration. The AISE2 model was well-calibrated across all risk scores in the development cohort, (Figure 4, panel A) but in the absence of retraining it underestimated risk for those with very low probability (<10%) of requiring vasopressors in the validation cohort. (Figure 4, panel B) Domain adaptation corrected the risk prediction in that group, making it more closely approximate the calibration seen in the development cohort. (Figure 4, panel C).
Figure 4: Calibration plots*.
*-Observations were binned into quantiles; each point represented an average of the observations in each quantile. Log-log scales were used for better visualization. The left-most plot shows the calibration plot of the model on the testing set of the development site. Due to the higher incidence of sepsis-associated vasopressor use at the validation site, the original model under-predicted the probability of that outcome at lower ranges of predicted risk (center plot). The fine-tuned model using domain adaptation (right-most plot) had better discrimination and calibration properties.
DISCUSSION
We demonstrated that an explainable, data-driven algorithm predicted the need for vasopressors within four hours among septic patients with an AUCroc of 0.81 [CI 0.81-0.83], using time-variant EMR data. Most of the top ten important features used to create the risk scores were vital signs. The algorithm maintained an AUCroc and specificity of at least 0.80 and 55%, respectively, regardless of the prediction window (up to 24 hours), or the definition of sepsis used for patient inclusion. Performance of the model was externally updated and validated at a separate site using domain adaptation.
AISE2 could be clinically useful for a few reasons. First, we only included patients who had sepsis (infection with organ dysfunction) to maximize our incidence of vasopressor administration and refine our model’s performance. This contrasts with other algorithms that predict septic shock.[18-22] Patients with sepsis are at highest risk for disease progression; they may require an intensified search for source control, or more aggressive efforts to mitigate organ dysfunction. Second, we used the CMS definition of severe sepsis as our inclusion criteria since it is the standard upon which sepsis core measure (SEP-1) benchmarking is based.[15] The algorithm maintained an AUCroc and specificity of at least 0.80 and 55%, respectively, regardless of the prediction window (up to 24 hours), or the definition of sepsis used for patient inclusion. The AUCroc and specificity at the 24-hour prediction horizon were comparable to similar models.[23]
We employed domain adaptation to improve generalizability of our predictive algorithm. Domain adaptation, the process by which learning a new task is improved through knowledge transfer from a related task,[24] allowed us to optimize prediction of vasopressor use among septic patients at a remote location from the development site. Under normal circumstances (without retraining), the model parameters (weights) for the development site would be applied to the validation site. Instead, we reserved data from the validation site to retrain AISE2, beginning with feature weights it learned from the development site. (Figure 1) There was a small expected decrease in all metrics of performance during external validation without retraining. By contrast, AUCroc, specificity and PPV based on model performance at the validation site after applying domain adaptation were similar to performance metrics taken from the development site. (Figure 2 & Table 1)
The context in which domain adaptation was applied made it appropriately suited for the task. Sepsis patients are a heterogenous group and are subject to varying inflammatory host responses to infection, and varying degrees of organ dysfunction. While the top three most important features remained the same in the development and validation sites, there were some differences with ranking of other features. White blood cell count was ranked higher in the development site than the validation site, suggesting a difference in the inflammatory milieu. (Supplemental Table 4) Furthermore, patients who did not require vasopressors had a slightly lower SOFA score in the validation site compared with the development site. (Supplemental Table 2) Our two cohorts also exhibited some differences in missingness patterns. Systolic blood pressure, the most important feature predicting vasopressor use at both sites, was missing much more in the validation cohort compared to the development cohort (37.5 and 7.5%, respectively), likely due to differences in blood pressure documentation protocols. This probably contributed to the degradation in model performance at the validation site prior to retraining. In spite of this, domain adaptation allowed us to optimize the model’s performance to account for differences in data collection at the validation site.
Our use of domain adaptation is a novel application of this machine learning approach, especially in the early prediction of critical illness. Others have attempted to improve generalizability of predictive algorithms by combining data from the development or source site with that of the target site during model retraining.[6, 25] However, this is an impractical approach due to patient privacy and data ownership concerns. The domain adaptation of AISE2 would tailor its parameters to each new site by adjusting model weights, without transfer of data. Shickel and colleagues [26] showed that domain adaptation increased performance of a model using EMR data to predict hospital discharge. However, this was in the context of a pilot study demonstrating the added benefit of multimodal sensor data in outcome prediction compared to EMR data alone. Unlike our domain adaptation approach, the goal was not to improve generalizability of the same model at a different site, on different patients under different conditions. Our approach is the first to use domain adaptation during external validation of a model that predicts evolving critical illness in an ICU population.
Domain adaptation may also have practical implications for the deployment of our algorithm, particularly in a community hospital environment with a paucity of training data. Though we used 80% of data from the validation site for retraining, the model was able to maintain an AUCroc of 0.8 after retraining with as little as 30% of the total validation cohort (1500 patients). (Figure 3) A model trained on that number of patients at the validation site without prior training at a development site (i.e., “de novo” training) would have had an AUCroc of 0.72. Thus, domain adaptation would allow us to quickly deploy this model in settings that might not otherwise be able to accommodate due to limited data resources.
Despite its strengths, there were some limitations in this study. Our definition for sepsis used blood culture acquisition and antibiotic administration as a proxy for suspicion of infection, concurrent with evidence of organ failure. This approach may have included patients who did not develop sepsis and may have excluded true sepsis cases, but similar methods have been applied to sepsis studies using retrospective databases.[27] The outcome of interest was the initiation of vasopressor support, which was used as a surrogate for septic shock. It was not possible to predict septic shock as defined by sepsis-3[28] due to inconsistent lactate measurement near the time of vasopressor administration, and unreliable assessment of fluid administration. Still, the mortality of those who needed vasopressors was at least about 40% in the derivation and validation cohorts, and at least three-fold higher than the mortality in those who did not require vasopressors (Supplemental Table 2). Death and transfer to hospice was a competing risk to vasopressor use, but predicting vasopressor administration in these patients would still be relevant. Goals of care might be different if vasopressor support is preventable. Our dataset did not include information on the specific cause of sepsis. However, we included a mix of medical, surgical, and subspecialty ICUs, which should have provided some variation in infectious causes. EMR data may have been biased since many features were not missing at random; providers order more tests and check vital signs more frequently based on clinical concern. However, the model needed to be trained on this kind of “real world” data to be most effective in clinical practice. The reported PPVs were fairly low for AISE2, since an alert was only considered a true positive if first vasopressor use occurred within the predefined prediction window. The definition of “true” alerts could be refined to improve the PPV during real-time implementation, yet still retain clinical meaning. (See Appendix D for more details.)
CONCLUSIONS
We have retrospectively validated the use of a model to predict future vasopressor use among ICU patients with sepsis using commonly available clinical data from two hospital systems with different commercial EMR systems. Model retraining through domain adaptation allowed our model to achieve comparable performance between the source development site and a remote target validation site, despite differences in data sample frequency and potential differences in sepsis case mix. The model preserved its performance despite using small amounts of retraining data. This would accommodate model evaluation at a range of sites, including remote community hospitals and other health systems with limited data.
Supplementary Material
Conflicts of interest and sources of funding:
Dr. Holder is supported by the National Institute of General Medical Sciences of the National Institutes of Health (K23GM37182). Dr. Nemati is funded by the National Institutes of Health (#K01ES025445) and the Gordon and Betty Moore Foundation (#GBMF9052). Dr. Wardi is supported by the National Foundation of Emergency Medicine and funding from the Gordon and Betty Moore Foundation (#GBMF9052). He has received speakers’ fees from Thermo-Fisher and consulting fees from General Electric. "Dr. Buchman’s institution received funding from the Henry M. Jackson Foundation for his role as site director in Surgical Critical Care Institute, www.sc2i.org, funded through the Department of Defense’s Health Program – Joint Program Committee 6/Combat Casualty Care (USUHS HT9404-13-1-0032 and HU0001- 15-2-0001) and from Society of Critical Care Medicine for his role as Editor-in-Chief of Critical Care Medicine. Dr. Buchman is a 0.5 FTE under an IPA between Emory University and HHS/ASPR/BARDA. Funders played no role in study design, collection of data, analysis, reporting or interpretation of results.
Copyright Form Disclosure: Drs. Holder and Nemati’s institutions received funding from the National Institutes of Health (NIH). Dr. Holder received funding from the Gordon and Betty Moore Foundation, Thermo-Fisher, and General Electric, and he received support for article research from the NIH. Dr. Wardi’s institution received funding from the National Foundation of Emergency Medicine, he received funding from Thermo-Fisher and Roche/GE, and he received grants from the Gordon and Betty Moore Foundation. Dr. Buchman’s institution received funding from the Henry M. Jackson Foundation and the Society of Critical Care Medicine (SCCM), and he received funding from the Department of Defense and from SCCM. Dr. Nemati received support for article research from the NIH. Dr. Shashikumar has disclosed that he has no potential conflicts of interest.
REFERENCES
- 1.Leon AL, Hoyos NA, Barrera LI, et al. : Clinical course of sepsis, severe sepsis, and septic shock in a cohort of infected patients from ten Colombian hospitals. BMC Infect Dis 2013; 13:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glickman SW, Cairns CB, Otero RM, et al. : Disease progression in hemodynamically stable patients presenting to the emergency department with sepsis. Acad Emerg Med 2010; 17:383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberti C, Brun-Buisson C, Chevret S, et al. : Systemic inflammatory response and progression to severe sepsis in critically ill infected patients. Am J Respir Crit Care Med 2005; 171:461–8. [DOI] [PubMed] [Google Scholar]
- 4.Wardi G, Wali AR, Villar J, et al. : Unexpected intensive care transfer of admitted patients with severe sepsis. J Intensive Care 2017; 5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones SL, Ashton CM, Kiehne LB, et al. : Outcomes and Resource Use of Sepsis-associated Stays by Presence on Admission, Severity, and Hospital Type. Med Care 2016; 54:303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao Q, Jay M, Hoffman JL, et al. : Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open 2018; 8:e017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faisal M, Scally A, Richardson D, et al. : Development and External Validation of an Automated Computer-Aided Risk Score for Predicting Sepsis in Emergency Medical Admissions Using the Patient's First Electronically Recorded Vital Signs and Blood Test Results. Crit Care Med 2018; 46:612–618. [DOI] [PubMed] [Google Scholar]
- 8.Vergouwe Y, Steyerberg EW, Eijkemans MJ, et al. : Substantial effective sample sizes were required for external validation studies of predictive logistic regression models. J Clin Epidemiol 2005; 58:475–83. [DOI] [PubMed] [Google Scholar]
- 9.Kermany DS, Goldbaum M, Cai W, et al. : Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018; 172:1122–1131 e9. [DOI] [PubMed] [Google Scholar]
- 10.Redko I, Habrard A, Morvant E, et al. : Advances in Domain Adaptation Theory: 1st Edition. London, U.K., ISTE Press - Elsevier, 2019 [Google Scholar]
- 11.Karmakar C, Saha B, Palaniswami M, et al. : Multi-task transfer learning for in-hospital-death prediction of ICU patients. Annu Int Conf IEEE Eng Med Biol Soc 2016; 2016:3321–3324. [DOI] [PubMed] [Google Scholar]
- 12.Wardi G, Carlile M, Holder A, et al. : Predicting Progression to Septic Shock in the Emergency Department Using an Externally Generalizable Machine-Learning Algorithm. Ann Emerg Med 2021; 77:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins GS, Reitsma JB, Altman DG, et al. : Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD). Ann Intern Med 2015; 162:735–6. [DOI] [PubMed] [Google Scholar]
- 14.Leisman DE, Harhay MO, Lederer DJ, et al. : Development and Reporting of Prediction Models: Guidance for Authors From Editors of Respiratory, Sleep, and Critical Care Journals. Crit Care Med 2020; 48:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalantari A, Mallemat H Weingart SD: Sepsis Definitions: The Search for Gold and What CMS Got Wrong. West J Emerg Med 2017; 18:951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer M, Deutschman CS, Seymour CW, et al. : The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyna MA, Josef CS, Jeter R, et al. : Early Prediction of Sepsis From Clinical Data: The PhysioNet/Computing in Cardiology Challenge 2019. Crit Care Med 2020; 48:210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry KE, Hager DN, Pronovost PJ, et al. : A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med 2015; 7:299ra122. [DOI] [PubMed] [Google Scholar]
- 19.Calvert J, Mao Q, Rogers AJ, et al. : A computational approach to mortality prediction of alcohol use disorder inpatients. Comput Biol Med 2016; 75:74–9. [DOI] [PubMed] [Google Scholar]
- 20.Ghalati PF, Samal SS, Bhat JS, et al. : Critical Transitions in Intensive Care Units: A Sepsis Case Study. Sci Rep 2019; 9:12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiel SW, Rosini JM, Shannon W, et al. : Early prediction of septic shock in hospitalized patients. J Hosp Med 2010; 5:19–25. [DOI] [PubMed] [Google Scholar]
- 22.Fagerstrom J, Bang M, Wilhelms D, et al. : LiSep LSTM: A Machine Learning Algorithm for Early Detection of Septic Shock. Sci Rep 2019; 9:15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yee CR, Narain NR, Akmaev VR, et al. : A Data-Driven Approach to Predicting Septic Shock in the Intensive Care Unit. Biomed Inform Insights 2019; 11:1178222619885147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torrey LJ Shavlik: Transfer Learning. In: Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques. Soria Olivas E, et al. (Eds). Hershey, PA, IGI Global, 2010, pp. 892. [Google Scholar]
- 25.Desautels T, Das R, Calvert J, et al. : Prediction of early unplanned intensive care unit readmission in a UK tertiary care hospital: a cross-sectional machine learning approach. BMJ Open 2017; 7:e017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shickel B, Davoudi A, Ozrazgat-Baslanti T, et al. : Deep Multi-Modal Transfer Learning for Augmented Patient Acuity Assessment in the Intelligent ICU. Front Digit Health 2021; 3: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seymour CW, Liu VX, Iwashyna TJ, et al. : Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315:762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shankar-Hari M, Phillips GS, Levy ML, et al. : Developing a New Definition and Assessing New Clinical Criteria for Septic Shock: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315:775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.