Abstract
HIV-1 transactivator of transcription (Tat) protein is required for HIV-1 replication, and it has been implicated in the pathogenesis of HIV-1-associated neurocognitive disorder (HAND). HIV-1 Tat can enter cells via receptor-mediated endocytosis where it can reside in endolysosomes; upon its escape from these acidic organelles, HIV-1 Tat can enter the cytosol and nucleus where it activates the HIV-1 LTR promoter. Although it is known that HIV-1 replication is affected by the iron status of people living with HIV-1 (PLWH), very little is known about how iron affects HIV-1 Tat activation of the HIV-1 LTR promoter. Because HIV-1 proteins de-acidify endolysosomes and endolysosome de-acidification affects subcellular levels and actions of iron, we tested the hypothesis that the endolysosome pool of iron is sufficient to affect Tat-induced HIV-1 LTR transactivation. Ferric (Fe3+) and ferrous (Fe2+) iron both restricted Tat-mediated HIV-1 LTR transactivation. Chelation of endolysosome iron with deferoxamine (DFO) and 2–2 bipyridyl, but not chelation of cytosolic iron with deferiprone and deferasirox, significantly enhanced Tat-mediated HIV-1 LTR transactivation. In the presence of iron, HIV-1 Tat increasingly oligomerized and DFO prevented the oligomerization. DFO also reduced protein expression levels of the HIV-1 restriction agent beta-catenin in the cytosol and nucleus. These findings suggest that DFO increases HIV-1 LTR transactivation by increasing levels of the more active dimeric form of Tat relative to the less active oligomerized form of Tat, increasing the escape of dimeric Tat from endolysosomes, and/or reducing beta-catenin protein expression levels. Thus, intracellular iron might play a significant role in regulating HIV-1 replication, and these findings raise cautionary notes for chelation therapies in PLWH.
Keywords: Transactivator of transcription (Tat), HIV-1-associated neurocognitive disorder (HAND), Tat-mediated HIV-1 LTR transactivation, People living with HIV-1 (PLWH), Endolysosomes, Oligomerization, Deferoxamine (DFO)
Introduction
Currently, more than 40 million people worldwide are infected with human immunodeficiency (type-1) virus (HIV-1) (Spudich and Gonzalez-Scarano 2012; Valcour et al. 2012), the cause of acquired immune deficiency syndrome (AIDS). Encouragingly, people living with HIV-1 (PLWH) are able to live almost full lifespans because of effective antiretroviral therapeutics (ART), and HIV-1 infection is now considered to be a chronic managed neuroinflammatory disease. However, PLWH are experiencing almost 50% prevalence rates of HIV-1-associated neurocognitive disorders (HAND) (Heaton et al. 2010; Velasquez et al. 2020). Although ART is able to suppresses HIV-1 levels to below detectable levels in plasma and CSF (Gray et al. 2014; Marban et al. 2016), HIV-1 still resides in reservoirs both in the periphery as well as in the CNS (Churchill et al. 2009; Fois and Brew 2015; Joseph et al. 2015; Thompson et al. 2011). Unfortunately, total eradication of HIV-1 from PLWH remains an extremely challenging task in large part due to the limited access of ART drugs to HIV-1 reservoirs (Chahroudi et al. 2018; Meeker et al. 2014; Saylor et al. 2016; Veenhuis et al. 2019).
HIV-1 replication is controlled by a number of HIV-1 proteins including the transactivator of transcription (Tat) (Arhel and Kirchhoff, 2010; Freed, 2001; Jeang et al. 1999; Liang and Wainberg 2002; Romani et al. 2010). In addition to its importance in regulating HIV-1 long terminal repeat (LTR) transactivation, HIV-1 Tat has been implicated in the pathogenesis of HAND in part because it is highly toxic to neurons, and Tat levels can remain elevated even in the presence of ART-induced suppression of HIV-1 to below detectable limits (Chen et al. 2013; Deshmane et al. 2011; Haughey and Mattson 2002; Hui et al. 2012; Jeang et al. 1999; Johnson et al. 2013; Nath et al. 1996; Sonia et al. 2012).
HIV-1 Tat is composed of two introns; the first has 72 amino acids, and the second has up to 32 amino acids (Frankel et al. 1988). Functionally, HIV-1 Tat is composed of four distinct domains (Clark et al. 2017; Frankel et al. 1988; Jeang et al. 1999; Romani et al. 2010). Of direct relevance to the current study, HIV-1 Tat domain number 2 is a series of amino acids from 22 to 37 (Tat22-37) that contains a number of cysteines, amino acids that bind divalent cations and that are involved in HIV-1 Tat oligomerization (Frankel et al. 1988). HIV-1 Tat is functionally active as a dimer (Jeang et al. 1999) and Tat oligomerization can lessen its biological activity as well as hinder its escape from endolysosomes (Tosi et al. 2000).
HIV-1 Tat is secreted actively from HIV-1 infected or transfected cells and it can be internalized into bystander cells by receptor-mediated endocytosis (Ensoli et al. 1993; Mann and Frankel, 1991; Sonia et al. 2012; Vendeville et al. 2004; Vives 2003). The receptors implicated in HIV-1 Tat endocytosis include low-density lipoprotein receptor-1 (LRP1), heparin sulfate proteoglycan, CD26, and the chemokine receptor CXCR4 (Liu et al. 2000; Tyagi et al. 2001). Following endocytosis, in order for HIV-1 Tat to affect HIV-1 LTR transactivation, it must first escape from endolysosomes into the cytosol and the nucleus where it can impact HIV-1 replication.
Endolysosomes are part of a complicated and dynamic organellar system of physiological significance and pathological relevance (Bright et al. 2005; Chauhan et al. 2014; de Duve 2005; Fredericksen et al. 2002; Grimm et al. 2017; Huotari and Helenius 2011; Khan et al. 2020b; Khan et al. 2021; Luzio et al. 2007; Luzio et al. 2005; Perera and Zoncu 2016; Vijaykumar et al. 2008; Wideman et al. 2014), and they are subject to stress responses (Lakpa et al. 2021). Endolysosomes are acidic organelles whose functions are significantly affected by their luminal pH (Nixon and Cataldo 2006; Christensen et al. 2002; de Duve 2005; Huotari and Helenius 2011; Khan 2020; Khan et al. 2019a; Rivera et al. 2014; Tooze and Dikic 2016; Wideman et al. 2014). Endolysosomes contain high levels of divalent cations including iron and insults including those that affect endolysosome pH can alter the release of these cations from endolysosomes (Halcrow et al. 2019; McGuire et al. 2017; Mindell 2012; Prasad and Rao 2018; Terman and Kurz 2013; Xiong and Zhu 2016; Xu and Ren 2015). Following HIV-1 protein-induced deacidification by Tat and gp120 (Bae et al. 2014; El-Hage et al. 2015; Halcrow et al. 2021; Hui et al. 2012), HIV-1 Tat can enter the cytosol and nucleus where it can activate the HIV-1 LTR promoter (Ensoli et al. 1993; Khan et al. 2018, 2019b; Kolson et al. 1994). Thus, endolysosome deacidification may be an important regulator of HIV-1 replication (Chauhan et al. 2014; Chauhan and Tikoo 2015; Vijaykumar et al. 2008).
Iron status both intracellularly and extracellularly can affect HIV-1 infection and disease progression (Afacan et al. 2002; Banjoko et al. 2012; Doherty 2007; Esan et al. 2013; Khan et al. 2020a; Mancone et al. 2017; Nekhai et al. 2013). Furthermore, HIV-1 infection can affect iron metabolism (Afacan et al. 2002; Banjoko et al. 2012; Kumari et al. 2016); depending on the progression of HIV-1 infection, levels of serum iron are either decreased or increased (Chang et al. 2015; Nekhai et al. 2013). Here, we investigated the role of iron in HIV-1 Tat escape from endolysosomes and subsequent HIV-1 LTR transactivation using a Tat-mediated HIV-1 LTR transactivation reporter assay (Khan et al. 2018, 2019b). Our principal findings are that endolysosome iron can affect the oligomerization of HIV-1 Tat, that iron inhibited Tat-mediated HIV-1 LTR transactivation, and that endolysosome-specific iron chelators (DFO and 2–2 bipyridyl) enhanced Tat-mediated HIV-1 LTR transactivation. Thus, iron can increase levels of HIV-1 Tat oligomers relative to levels of HIV-1 Tat dimers and thereby restrict HIV-1 Tat escape from endolysosomes as well as decrease levels of Tat-induced HIV-1 LTR transactivation.
Material and methods
Cell cultures
U87MG astrocytoma cells were cultured in 1 × DMEM (Invitrogen) supplemented with 10% fetal calf serum and penicillin/streptomycin (Invitrogen) and incubated in a 5% CO2 incubator at 37 °C. Cells were grown to ~ 50% confluence in 35-mm2 dishes as well as in 96-well plates; cells were not used past their tenth passage. U87MG cells were stably transfected with an integrated luciferase gene under the control of an HIV-1 Tat-driven LTR promoter following neomycin antibiotic (Sigma-Aldrich) selection pressure; these cells were provided generously by Dr. Lena Al-Harthi (Rush University, Chicago).
Luciferase reporter assay for Tat-mediated HIV-1 LTR transactivation
U87MG cells were seeded at 30 to 40% confluency (~ 10,000 cells) on 96-well plates 1 day prior to being taken for experimentation. Cells were incubated with 2 μg/ml HIV-1 Tat in the presence of 100 μM chloroquine (Sigma-Aldrich). Three sources of HIV-1 Tat were used: ABL Inc., NIH AIDS program, and Dr. Tory Johnson from Johns Hopkins University. In some experiments, cells were co-treated with FeCl2 (Millipore), FeCl3 (Millipore), ammonium iron citrate (NH4–Fe–citrate, Sigma-Aldrich), iron dextran (Sigma-Aldrich), ZnCl2 (Sigma-Aldrich), MgCl2 (Sigma-Aldrich), CuCl2 (Fisher Scientific), CoCl2 (Sigma-Aldrich), deferoxamine (DFO, Sigma-Aldrich), 2–2 bipyridyl (2–2 BP, Sigma-Aldrich), deferiprone (Sigma-Aldrich), and deferasirox (Advanced Chemblocks Inc.). Luciferase activity assays (Promega) were performed 48 h post-incubation and relative luminescence units were quantified using a fluorometer/luminometer plate reader (Molecular Devices, Spectra MAX GEMINI EM).
Immunoblotting
Cells were harvested and lysed in 1 × RIPA lysis buffer (Thermo Fisher) containing 10 mM NaF, 1 mM Na3VO4, and 1 × protease inhibitor cocktail (Sigma). After centrifugation (14,000 × g for 10 min at 4 °C), supernatants were collected, and protein concentrations were determined with a DC protein assay (Bio-Rad). Proteins (10 μg) were separated by SDS-PAGE (12% gel) and transferred to PVDF membranes using the iBlot 2 dry transfer system (Invitrogen). The membranes were incubated overnight at 4 °C with antibodies against GAPDH (Abcam, Ab181603), histone (Abcam, Ab1791), β-catenin (Abcam, Ab32572), and HIV-1 Tat (Santa Cruz, Sc-65913). Blots were developed with enhanced chemiluminescence, and the density of antibody-positive protein bands was determined using a Li-COR Odyssey Fc Imaging System (LiCor).
SDS-PAGE and native gels for HIV-1 Tat
HIV-1 Tat proteins (7.0 μg/m1) were treated with different preparations of iron at pH 5.5 for 3 h at 37 °C followed by protein separation using 15% SDS-PAGE and Coomassie blue staining. HIV-1 Tat protein mobility shifts used 4–16% Bis–Tris native gels (Thermo Fisher Bis–Tris Native Gel kit).
SDS-PAGE for HIV-1 Tat-treated cells
U87MG cells were incubated with HIV-1 Tat protein (7.0 μg/ml) in the absence or presence of FeCl3, DFO, or a combination of FeCl3 and DFO) for 6 h (pH 5.5, 37 °C). Sample proteins were then separated using 15% SDS-PAGE, and immunoblots were performed for determination of HIV-1 Tat protein mobility shifts and HIV-1 Tat oligomerization.
Effects of extracellular HIV-1 Tat on HIV-1 Tat oligomerization and HIV-1 LTR transactivation
To determine the extent of HIV-1 Tat oligomerization, HIV-1 Tat proteins (7.0 μg/m1) from two separate sources (ABL Inc. and Dr. T. Johnson from Johns Hopkins University) were treated with either 100 or 500 μM DFO for 3 h at room temperature. Aliquots were then added to 4 to 16% Bis–Tris native gels for determination of Tat mobility shifts. To determine the extent of HIV-1 LTR transactivation, U87MG cells expressing HIV-1 LTR were incubated for 4 h with HIV-1 Tat protein (2 μg/m1), CQ (100 μM), and FeCl3 (20 μM) prior to the addition of DFO (20 μM). Cells were then incubated for 48 h in a 5% CO2 incubator at 37 °C prior to being taken for measurement of luciferase activity associated with Tat-mediated HIV-1 LTR transactivation.
Molecular interactions of Fe2+ with HIV-1 Tat
The structure for HIV-1 Tat protein was retrieved from the Protein Data Bank (PDB) using the PDB Id:3MI9. The docking of Fe2+ on the HIV-1 Tat protein was performed using metal ion binding (MIB) prediction models; the docking server (http://bioinfo.cmu.edu.tw/MIB/) used the C-chain of PDB for the structure of HIV-1 Tat (PDB Id: 3MI9) (Lin et al. 2016). Using fragment transformation, comparisons were made between the local structure with known metal ion binding sites and a query protein structure with an unknown binding site. After comparison, all residues were scored, and residues above a set threshold were predicted as Fe2+ binding residues. Illustration of the Tat protein with bound Fe2+ was generated using Chimera v.1.6.2 (Pettersen et al. 2004), and molecular interaction plots of Fe2+ with HIV-1 Tat protein were generated using LigPlot + v.1.4.5 (Laskowski and Swindells 2011).
Generation of U87MG-LTR cells stably expressing HIV-1 Tat-GFP
We generated a cell line stably expressing Tat-GFP by transducing U87MG-LTR cells with lentivirus particles of Tat-GFP encoded lentivector (Origene, CV1011716 subcloned into PS10093) at a multiplicity of infection of 1.0; control cells were transduced with GFP lentivirus particles (Origene, PS100093). Cells were then selected under puromycin pressure (5.0 μg/ml); the molecular mass of the HIV-1 Tat protein was determined by immunoblotting, and levels of Tat-mediated HIV-1 LTR transactivation were measured using the below described Steady Glow Luciferase Assay (Promega).
Effects of intracellular HIV-1 Tat on LTR transactivation
One day prior to being taken for experimentation, 10,000 U87MG-LTR cells stably expressing Tat-GFP were seeded for 24 h in 96-well plates. Cells were then treated for 6 or 24 h with vehicle or 25-μM concentrations of the iron chelators DFO, deferiprone, or 2–2 bipyridyl before being taken for determination of HIV-1 LTR activity using the Steady Glow Luciferase Assay (Promega); HIV-1 LTR activity was measured as relative luminescence units (RLU).
Lysosome leakage assay
U87MG cells were seeded at a density of about 10,000 cells on lysine-coated 35 mm2 culture dishes. After culturing the cells in a 5% CO2 incubator at 37 °C for 24 h, cells were treated for 3 h with the endocytosed dye Alexa 488-conjugated dextran (10 μM, Thermo Fisher) with DMSO, FeCl3 (20 μM), or DFO (20 μM) in the absence or presence of chloroquine (100 μM). Post-incubation, cells were washed 3 times with 1 × PBS, and images were captured by confocal microscopy (Zeiss LSM800). Images were analyzed using ImageJ software (NIH) to determine the intensity of Alexa-488 conjugated dextran fluorescence in endolysosomes and cytosol.
Effects of iron and iron chelators on protein expression levels of β-catenin and β-catenin-mediated TCF/LEF transcription activity
For determination of β-catenin protein levels, U87MG cells were seeded at a density of about 2,000,000 cells on 100-mm2 cell culture dishes. After culturing the cells in a 5% CO2 incubator at 37 °C for 24 h, cells were treated for 36 h with 20 μM concentrations of the iron chelators DFO and DFX or the iron supplements FeCl2, FeCl3, and NH4–Fe–Citrate. Following incubations, cells were harvested and divided into two parts; one part was lysed in 1 × lysis buffer for determination of total protein levels using the Bradford method, and the second part was used to obtain cytosol and nuclear fractions (Thermo Fisher, kit #78,833). Whole-cell lysates, cytosol, and nuclear fractions were used for determination of β-catenin protein levels. For determination of β-catenin-mediated TCF/LEF transcription activity, U87MG cells were transfected with TCF/LEF reporter plasmids (1 μg) (TOPFlash and FOPFlash) using Jet prime reagent (DNA/Jet prime reagent, 1:2 ratio) on 12-well cell culture plates. Twenty-four-hour post-transfection, cells were seeded onto 96-well plates and treated with iron or iron chelators for 24 h followed by determination of luciferase activity (Steady Glow Luciferase assay) as a measure of TCF/LEF transcription activity (Coghlan et al. 2000).
Statistical analysis
All data were presented as means ± standard deviations. Statistical significance between two groups was determined with a Student’s t-test, and statistical significance among multiple groups was determined using a one-way ANOVA plus a Tukey post hoc test. p < 0.05 was accepted to be statistically significant.
Results
Iron decreased Tat-induced HIV-1 LTR transactivation
We first determined the extent to which different forms of iron affected the ability of HIV-1 Tat to promote HIV-1 LTR transactivation. Using a U87MG cell-based reporter assay we determined the effects of FeCl2, FeCl3, ammonium iron citrate (NH4–Fe(III)–citrate), and iron(III)–dextran at concentrations ranging from 0.5 to 100 μM on levels of Tat-mediated HIV-1 LTR transactivation. When co-incubated for 48 h with HIV-1 Tat (2 μg/ml) and chloroquine (CQ, 100 μM), all four forms of iron decreased Tat-mediated HIV-1 LTR transactivation; statistically significant decreases were observed for FeCl2 (Fig. 1A) and FeCl3 (Fig. 1B) starting at 1.0 μM, for NH4–Fe(III)–citrate (Fig. 1C) starting at 2.5 μM, and for iron(III)–dextran (Fig. 1D) starting at 5.0 μM. However, no statistically significant changes in Tat-mediated HIV-1 LTR transactivation were observed when cells were incubated for 24 h with HIV-1 Tat and then incubated for another 24 h with FeCl2, FeCl3, NH4–Fe–citrate, or irondextran at concentrations as high as 50 μM (Fig. 2). These results suggested to us that iron affected HIV-1 Tat and its ability to initiate but not reverse Tat-mediated HIV-1 LTR transactivation.
Fig. 1.
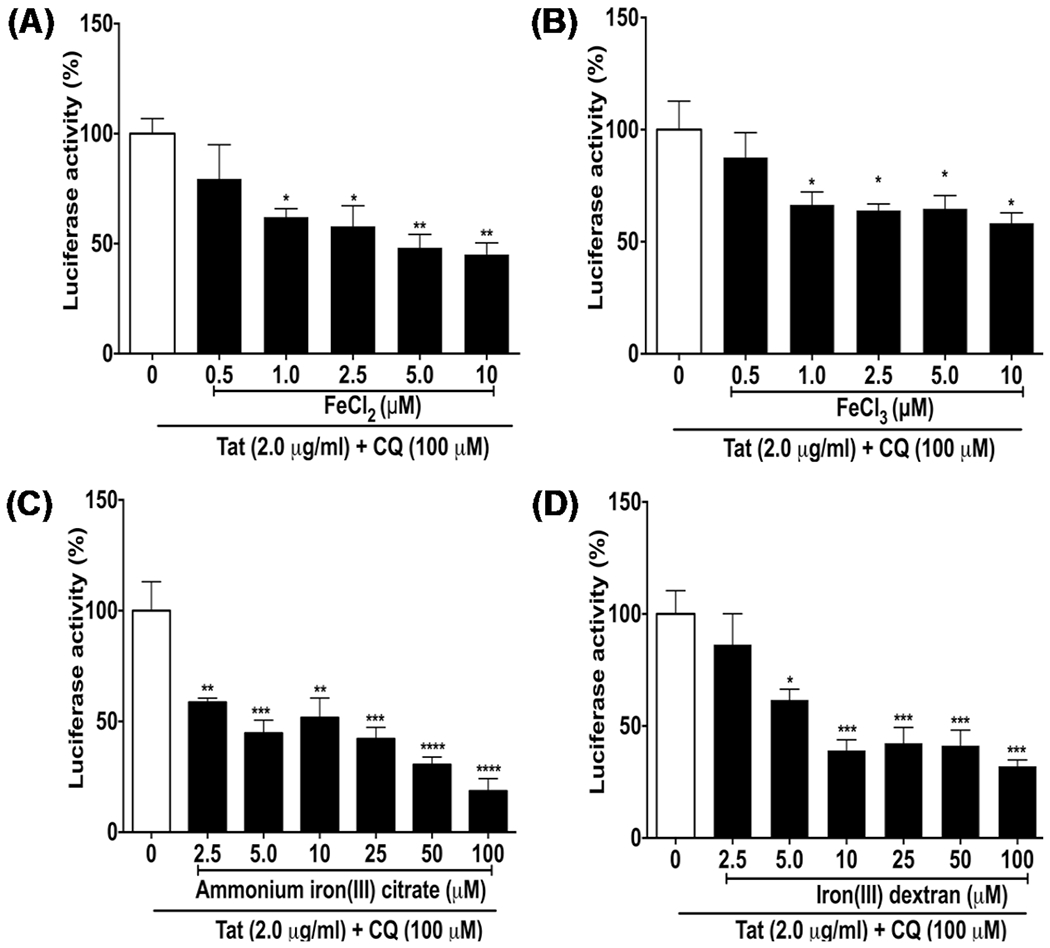
Iron decreased Tat-mediated HIV-1 LTR transactivation. U87MG cells were co-incubated for 48 h with four types of iron A FeCl2, B FeCl3, C ammonium iron(III) citrate, and D iron(III) dextran at concentrations ranging from 0.5 to 100 μM in the absence or presence of Tat protein (2 μg/ml) and chloroquine (CQ, 100 μM). Tat-mediated HIV-1 LTR transactivation measured as luciferase activity (percent of control) was significantly decreased by A FeCl2 starting at 1.0 μM, by B FeCl3 starting at 1.0 μM, by C ammonium iron(III) citrate starting at 2.5 μM, and by D iron(III) dextran starting at 5.0 μM. (n = 3; *p < 0.05; **p < 0.01, ***p < 0.001)
Fig. 2.

Post-treatment with iron did not reverse Tat-mediated increases in HIV-1 LTR transactivation: U87MG cells were incubated for 24 h with HIV-1 Tat protein (2 μg/ml) and CQ (100 μM) prior to the addition of and incubation for another 24 h with four types of iron A FeCl2, B FeCl3, C ammonium iron(III) citrate, and D iron(III) dextran at concentrations ranging from 10 to 50 μM. Post-treatment with iron did not significantly affect the ability of HIV-1 Tat to increase HIV-1 LTR transactivation. (n = 3; p > 0.05)
Tat-mediated HIV-1 LTR transactivation was not significantly affected by ZnCl2, CuCl2, MgCl2, and CoCl2
Because endolysosomes contain high levels of divalent cations in addition to iron, we next determined the extent to which other divalent cations might affect Tat-mediated HIV-1 LTR transactivation; co-incubation of cells for 48 h with HIV-1 Tat (2 μg/ml) and chloroquine (CQ, 100 μM) with ZnCl2, CuCl2, MgCl2, and CoCl2 at concentrations ranging from 0.5 to 100 μM did not produce any statistically significant differences in Tat-mediated HIV-1 LTR transactivation (Fig. 3). Thus, of the divalent cations tested, only iron had a restrictive action on Tat-mediated HIV-1 LTR transactivation. Furthermore, incubations with ZnCl2, CuCl2, MgCl2, and CoCl2 at the various concentrations used were for 4 h. Under those conditions, we did not observe any statistically significant changes in endolysosome pH. Although we did not measure cell life and death, the cells were inspected under light microscopy and appeared healthy (data not shown).
Fig. 3.
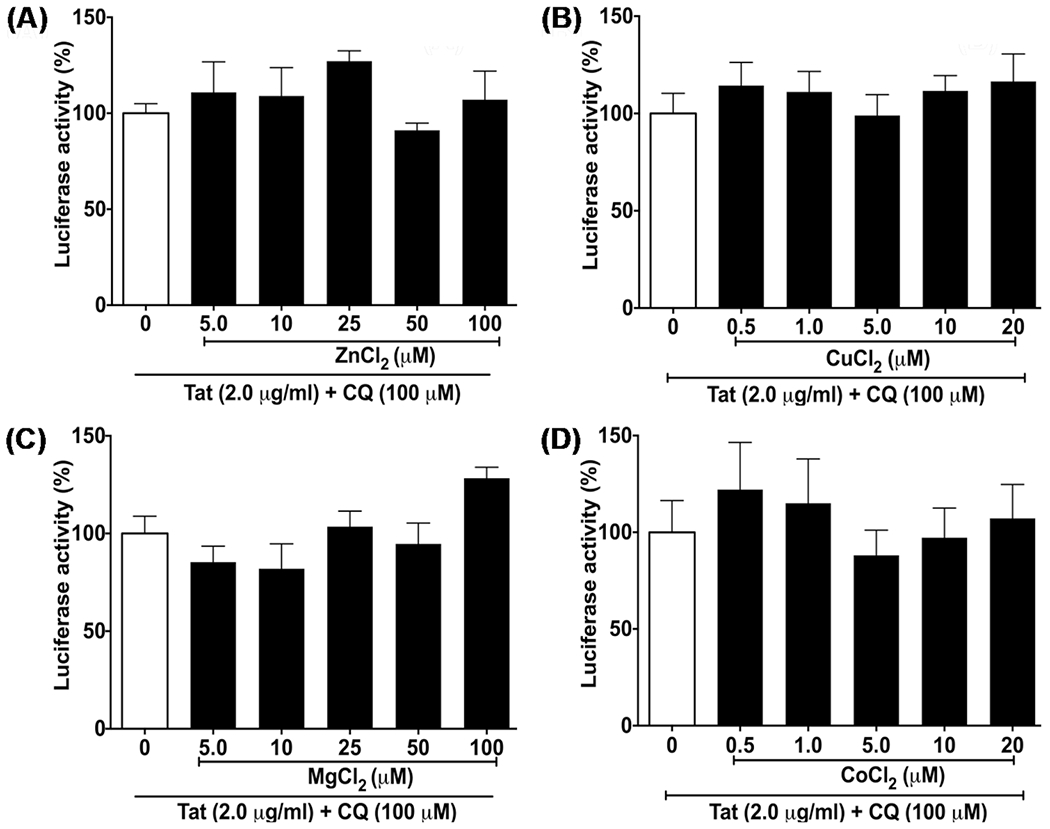
ZnCl2, CuCl2, MgCl2, and CoCl2 did not significantly affect Tat-mediated HIV-1 LTR transactivation. U87MG cells were co-incubated for 48 h with A ZnCl2, B CuCl2, C MgCl2, and D CoCl2 at concentrations ranging from 0.5 to 100 μM in the absence or presence of HIV-1 Tat protein (2 μg/ml) and chloroquine (CQ, 100 μM). Tat-mediated HIV-1 LTR luciferase activity (percent of control) was not significantly affected by pretreatment of cells with ZnCl2, CuCl2, MgCl2, or CoCl2. (n = 3; p > 0.05)
Endolysosome-specific iron chelators enhanced Tat-mediated HIV-1 LTR transactivation
Because the actions of HIV-1 Tat on LTR transactivation are mediated intracellularly and because iron is known to accumulate in subcellular organelles and the cytosol, we next determined the extent to which chelation of iron in endolysosomes and/or cytosol affected HIV-1 LTR transactivation induced by exogenously applied HIV-1 Tat. Deferoxamine (DFO) is endocytosed and chelates iron in endolysosomes (Castino et al. 2011; De Domenico et al. 2009; Doulias et al. 2003; Espósito et al. 2002; Glickstein et al. 2005; Kurz et al. 2006; Lloyd et al. 1991) and 2–2-bipyridyl (2–2-BP) enters cells by diffusion and chelates iron both in the cytosol and in endolysosomes (Espósito et al. 2002; Fernández et al. 2016). DFO and 2–2-BP both caused statistically significant increases in Tat protein (2 μg/ml) plus chloroquine (CQ, 100 μM)-mediated HIV-1 LTR transactivation concentration-dependently; statistically significant increases with DFO started at 1.0 μM (p < 0.05) and with 2–2-BP started at 50 μM (Fig. 4A). In contrast, two cytosol-specific iron chelators, deferiprone and deferasirox, (Espósito et al. 2002; FISCHER et al. 2005; Glickstein et al. 2005; Hider and Zhou 2005; Mobarra et al. 2016) at concentrations ranging from 2.5 to 100 μM had no statistically significant effects on Tat-mediated HIV-1 LTR transactivation (Fig. 4B). Thus, it appears that endolysosome iron and not cytosolic iron regulates Tat-mediated HIV-1 LTR transactivation.
Fig. 4.

Endolysosome but not cytosolic iron chelators enhanced Tat-mediated HIV-1 LTR transactivation. A U87MG cells stably transfected with a luciferase gene under the control of the Tat responsive HIV-1 LTR promoter were incubated for 48 h with the endolysosome-specific iron chelators deferoxamine (DFO) or 2–2 bipyridyl at concentrations ranging from 0.25 to 100 μM and HIV-1 Tat protein (2 μg/ml) in the presence of chloroquine (CQ, 100 μM). Luciferase activity (percent of control) was significantly increased by DFO starting at concentrations of 1.0 μM (left panel) and by 2–2 bipyridyl starting at concentrations of 50 μM (right panel). B No statistically significant effects on Tat-mediated HIV-1 LTR transactivation were observed when U87MG cells were incubated for 48 h with the cytosolic iron chelators deferiprone or deferasirox at concentrations ranging from 2.5 to 100 μM in the presence of Tat protein (2 μg/ml) and CQ (100 μM). (n = 3; *p < 0.05; **p < 0.01, ***p < 0.001, ****p < 0.0001)
Molecular interactions of Fe2+ with the cysteine-rich domain HIV-1 Tat
Some divalent cations bind to and interact with the cysteine-rich helical domain of HIV-1 Tat thereby causing protein oligomerization (Frankel et al. 1988; Tosi et al. 2000). Here, we modeled the amino acid sites at which Fe2+ binds to the cysteine-rich helical domain of HIV-1 Tat (Tat22-37). Molecular interaction modeling predicted that Fe2+ interacted at two sites of Tat22-37; both involve the cysteine-rich helical region of the protein (Fig. 5A). More specifically, 7 residues (Cys-22, Cys-25, Cys-27, Cys-30, His-33, Cys-34, Cys-37) of HIV-1 Tat protein interacted with the two Fe2+ cations. At one site, Fe2+ formed three interaction bonds with sulfhydryl (SH) groups on Cys-25, Cys-27, and Cys-30 (Fig. 5B). At the other site, Fe2+ formed four interaction bonds with SH groups on Cys-22, Cys-34, and Cys-37 and the N-atom of the histidine imidazole ring on His-33 (Fig. 5B).
Fig. 5.
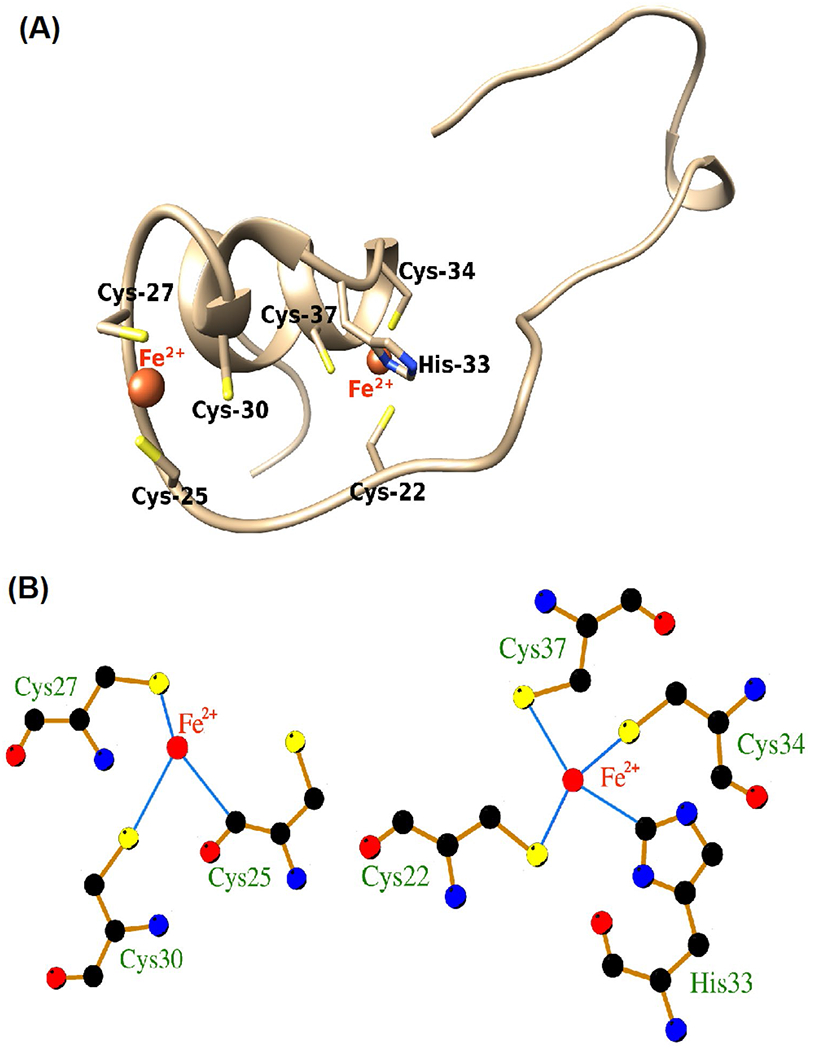
Computer modeling of iron-binding sites on HIV-1 Tat protein. Computer modeling indicated that the cysteine-rich domain of HIV-1 Tat protein (Cys-22, Cys-25, Cys-27, Cys-30, His-33, Cys-34, Cys-37) is capable of interacting with ferrous (Fe2+) and ferric (Fe3+) iron. A Molecular interaction modeling predicted that Fe2+ interacted at two sites of Tat22-37; both involve the cysteine-rich helical region of the protein involving 7 amino acid residues (Cys-22, Cys-25, Cys-27, Cys-30, His-33, Cys-34, Cys-37). B At one site, Fe2+ formed three interaction bonds with sulfhydryl (–SH) groups on Cys-25, Cys-27, and Cys-30. At other sites, Fe2+ formed four interaction bonds with –SH groups on Cys-22, Cys-34, and Cys-37 and the N-atom of the histidine imidazole ring on His-33. Illustrated are Fe2+ and Fe3+ (red), hydrogen bonds (green), bond lengths, and coordinate bonds (thin blue lines) (http://bioinfo.cmu.edu.tw/MIB/)
Iron increased and zinc decreased HIV-1 Tat oligomerization
We next determined the effects of iron, and for comparison zinc, on the molecular mass of HIV-1 Tat using Bis–Tris native gel and SDS-PAGE immunoblots. HIV-1 Tat protein incubated with 20 μM FeCl2, FeCl3, or ZnCl2 and then subjected to electrophoretic separation using 4 to 16% Bis–Tris native gels shifted the HIV-1 Tat-positive band to a higher molecular mass with FeCl3 and less so FeCl2; ZnCl2 did not affect the molecular mass of HIV-1 Tat (Fig. 6A). Following immunoblot separation of HIV-1 Tat protein on 15% SDS-PAGE gels, HIV-1 Tat-positive bands were observed both at molecular masses of about 14 and 24 kDa (Fig. 6B). The density of staining expressed as a ratio of oligomer (24 kDa)/dimer (14 kDa) was increased significantly (p < 0.001) with FeCl3 and FeCl2 but was decreased significantly (p < 0.05) by ZnCl2 (Fig. 6C). Thus, iron increased, and zinc decreased levels of the less-active oligomeric form of HIV-1 Tat.
Fig. 6.

Iron increases and zinc decreases HIV-1 Tat oligomerization. A HIV-1 Tat protein (7.0 μg/ml) was incubated at 37 °C for 3 h with 20 μM concentrations (pH 5.5) of FeCl2, FeCl3, and ZnCl2 followed by gel electrophoresis separation using 4–16% Bis–Tris native gels. FeCl3 and less so FeCl2 increased the molecular mass of HIV-1 Tat; ZnCl2 had no observable effect. B, C Tat protein (7.0 μg/ml) was incubated at 37 °C for 3 h with 20 μM concentrations (pH 5.5) of FeCl2, FeCl3, and ZnCl2 followed by gel electrophoresis separation using 15% SDS-PAGE and immunoblotting. FeCl2 and FeCl3 increased significantly (p < 0.001) and ZnCl2 decreased significantly (p < 0.05) the ratio of HIV-1 Tat oligomer to dimer. Thus, iron increased, and zinc decreased levels of the less-active oligomeric form of HIV-1 Tat
Deferoxamine prevented and reversed FeCl3-induced HIV-1 Tat oligomerization and increased Tat-mediated HIV-1 LTR transactivation
First, we determined if the endocytosed iron chelator deferoxamine (DFO) could prevent and/or reverse HIV-1 Tat oligomerization. U87MG cells expressing HIV-1 Tat were incubated for 3 h at 37 °C with either aqueous buffer (control), DMSO vehicle, DFO (20 μM), FeCl3 (20 μM), or FeCl3 (20 μM) plus DFO (20 μM); samples were then immunoblotted for HIV-1 Tat using 15% SDS-PAGE. FeCl3 increased significantly (p < 0.001) and DFO decreased significantly (p < 0.01) levels of oligomerized HIV-1 Tat; DFO decreased significantly (p < 0.05) FeCl3-induced HIV-1 Tat oligomerization (Fig. 7A). Next, we determined whether DFO could reverse HIV-1 Tat oligomerization. HIV-1 Tat proteins from two separate sources (JHU and ABL Inc.) were treated with DFO at concentrations of 100 or 500 μM for 3 h at room temperature and then processed on 4 to 16% Bis–Tris native gels. Both forms of HIV-1 Tat were heavily oligomerized and DFO reversed significantly (p < 0.001) levels of oligomeric relative to dimeric HIV-1 Tat (Fig. 7B). To confirm that changes in levels of oligomerized HIV-1 Tat were functionally relevant, we next determined the extent to which FeCl3 and DFO affected Tat-mediated HIV-1 LTR transactivation. Incubation of U87MG cells with FeCl3 (20 μM) decreased significantly (p < 0.05) HIV-1 LTR transactivation, DFO (20 μM) increased significantly (p < 0.05) HIV-1 LTR activation, and co-incubation of FeCl3 with DFO blocked significantly (p < 0.01) FeCl3-induced decreases in HIV-1 LTR activation (Fig. 7C). The ability of DFO to increase Tat-mediated HIV-1 LTR transactivation was concentration-dependent, and significant increases in Tat-mediated HIV-1 LTR transactivation were observed starting at 1.0 μM DFO (Fig. 7D).
Fig. 7.
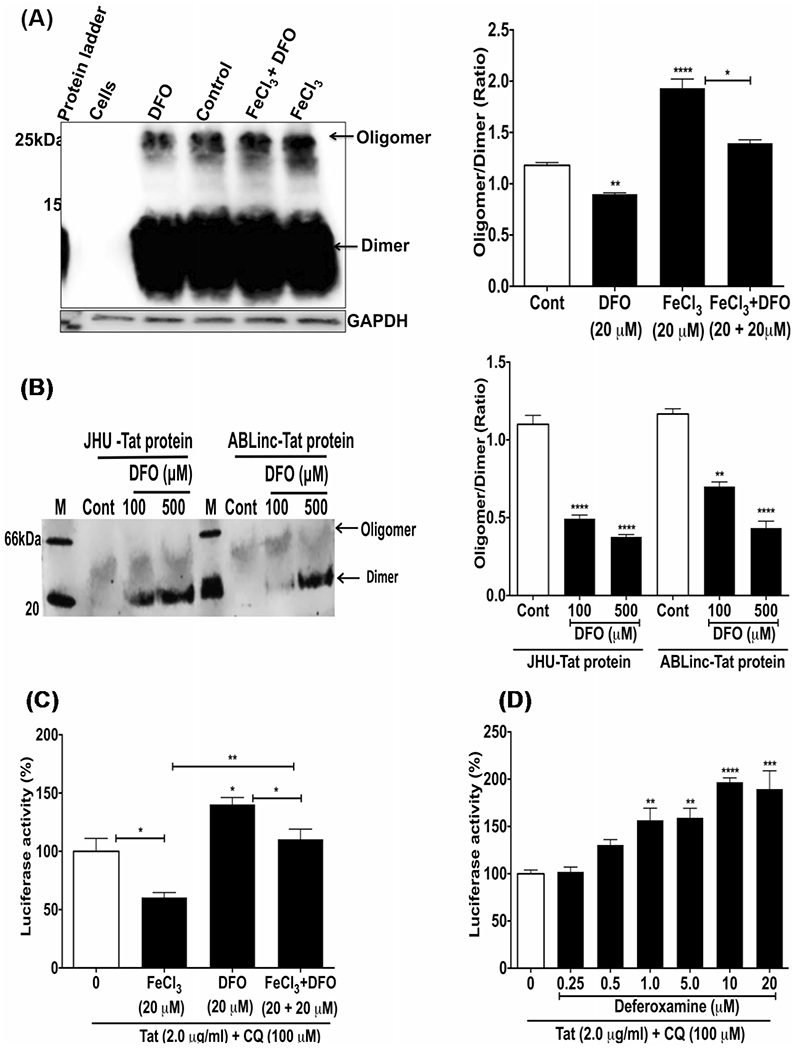
Deferoxamine (DFO) prevented and reversed FeCl3-induced HIV-1 Tat oligomerization, and increased Tat-mediated HIV-1 LTR transactivation. A Tat protein (7.0 μg/ml) was incubated for 6 h at 37 °C with U87MG cells alone and in combination with 20 μM FeCl3, 20 μM DFO, and 20 μM FeCl3 plus 20 μM DFO. Immunoblots for HIV-1 Tat using 15% SDS-PAGE showed that DFO decreased significantly (p < 0.01) Tat oligomerization, that FeCl3 increased significantly (p < 0.001) Tat oligomerization, and that DFO blocked significantly (p < 0.05) FeCl3-induced Tat oligomerization. B HIV-1 Tat from two sources (JHU and ABL) contained oligomerized Tat, but DFO was able to reverse the oligomerization when Tat protein (7.0 μg/ml) was incubated with DFO (100 and 500 μM) for 3 h at room temperature, and samples were separated using 4–16% Bis–Tris native gels (Thermo Fisher). C U87MG cells were incubated with HIV-1 Tat (2 μg/ml) and CQ (100 μM) for 4 h prior to 48 h incubation with FeCl3 (20 μM), DFO (20 μM), or FeCl3 (20 μM) plus DFO (20 μM). Tat-mediated HIV-1 LTR transactivation was decreased significantly (p < 0.05) by FeCl3, was increased significantly (p < 0.05) by DFO, and DFO blocked significantly (p < 0.05) FeCl3-induced decreases in Tat-mediated HIV-1 LTR transactivation. D U87MG cells were incubated with HIV-1 Tat protein (2 μg/ml) for 4 h in the presence of CQ (100 μM) prior to the addition of DFO (0.25 to 20 μM) and incubation for an additional 24 h. DFO significantly increased Tat-mediated HIV-1 LTR transactivation starting at 1.0 μM. (n = 3; *p < 0.05, **p < 0.01, ****p < 0.0001)
FeCl3 and DFO did not affect lysosome leakage and did not affect chloroquine-induced lysosome leakage
We reported previously that CQ increased lysosome leakage as well as promoted the escape of HIV-1 Tat from endolysosomes (Khan et al. 2020c). Therefore, it was important to discount the possibility that FeCl3 and/or DFO influence Tat-mediated HIV-1 LTR transactivation because of effects on lysosome leakage. Using U87MG reporter cells preloaded with Alexa-488 conjugated dextran, CQ (100 μM), but not FeCl3 (20 μM) or DFO (20 μM), significantly decreased (p < 0.001) endolysosome levels of Alexa-488 fluorescence and increased (p < 0.05) cytosol levels of Alexa-488 fluorescence (Fig. 8A). Additionally, neither FeCl3 (20 μM) nor DFO (20 μM) significantly affected the ability of CQ (100 μM) to alter lysosome leakage (Fig. 8B).
Fig. 8.
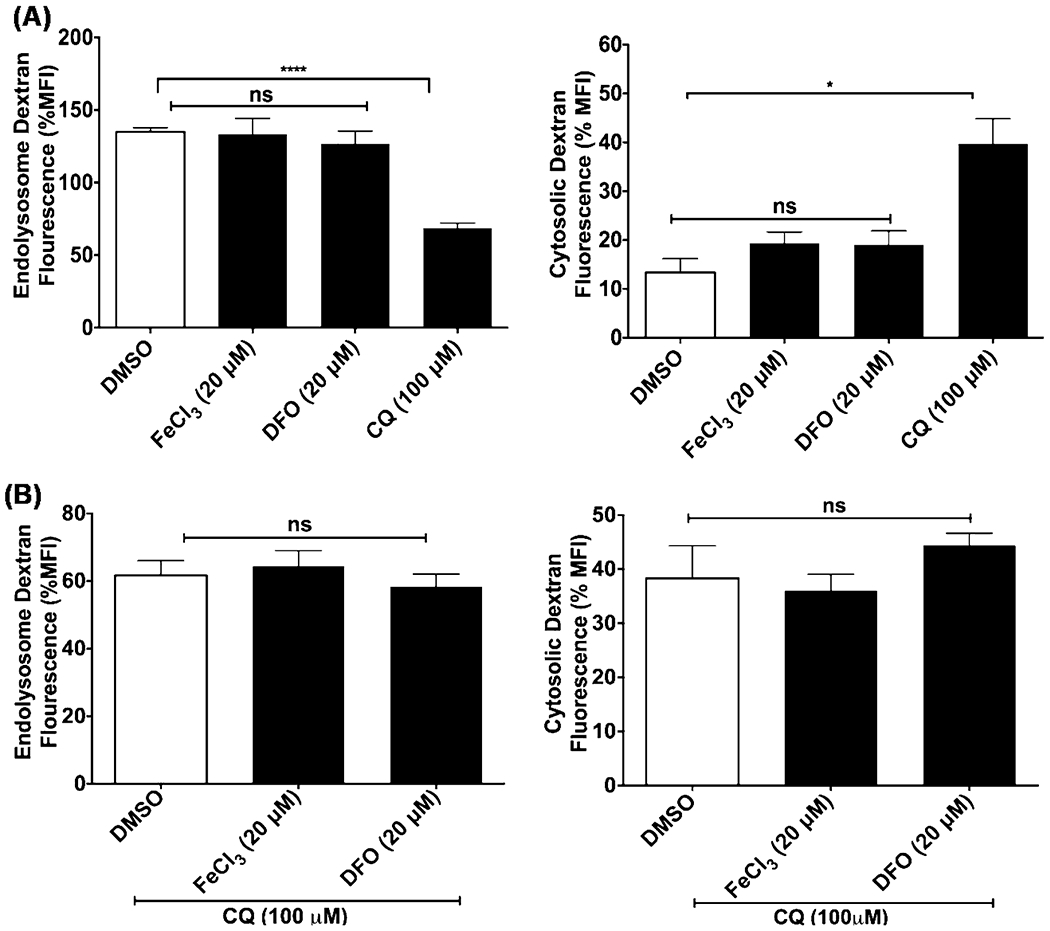
FeCl3 and DFO did not affect lysosome leakage and did not affect chloroquine-induced lysosome leakage. U87MG cells were incubated for 4 h at 37 °C with the endocytosed dye Alexa-488 conjugated dextran (10 μM) prior to incubating the cells for 3 h with A CQ (100 μM), FeCl3 (20 μM), and DFO (20 μM) or B CQ (100 μM) in the absence or presence of FeCl3 (20 μM) and DFO (20 μM). Images were captured by confocal microscopy (Zeiss LSM800), and data were analyzed using ImageJ to determine Alexa-488 conjugated dextran fluorescence intensity in endolysosomes and cytosol. CQ significantly decreased (p < 0.0001) levels of Alexa-488 fluorescence in endolysosomes and increased (p < 0.05) levels of fluorescence in the cytosol (Fig. 8A). FeCl3 and DFO did not significantly affect leakage of Alexa-488 conjugated dextran nor did it affect significantly the lysosomotropic actions of CQ. (n = 3; *p < 0.05; ****p < 0.001)
Effects of iron and iron-chelators on protein expression levels of β-catenin and β-catenin-mediated TCF/LEF transcription activity
β-Catenin is a restriction factor for HIV-1 LTR transactivation and HIV-1 replication. Therefore, it was important to determine possible effects of iron and iron chelators on protein expression levels of β-catenin as well as β-catenin-mediated TCF/LEF transcription activity. Protein expression levels of β-catenin were decreased significantly (p < 0.01) by the iron chelators deferoxamine (DFO, 20 μM) and deferasirox (DFX, 20 μM) but were not changed significantly by 20 μM FeCl2, NH4–Fe–citrate, and FeCl3 in whole-cell extracts (Fig. 9A, D), cytosol (Fig. 9B, E), and nuclear extracts (Fig. 9C, F). β-Catenin-mediated TCF/LEF transcription activity was decreased significantly by the endolysosome iron chelator DFO (20 μM, p < 0.0001) and the cytosolic iron chelator deferasirox (DFX) (20 μM, p < 0.001) but was not changed significantly by 20 μM FeCl2, NH4–Fe–citrate and FeCl3 (Fig. 9G).
Fig. 9.

Effects of iron and iron chelators on protein expression levels of the HIV-1 restriction factor β-catenin and β-catenin-mediated TCF/LEF transcription activity. U87MG cells were treated with the iron chelators deferoxamine (DFO, 20 μM) and deferasirox (DFX, 20 μM) or 20 μM FeCl2, FeCl3, and NH4–Fe–citrate for 36 h prior to harvesting cells for immunoblotting of protein expression levels of the HIV-1 restriction factor β-catenin in whole-cell extracts (A), cytosol (B), and nuclear fractions (C). In all three preparations, protein expression levels of β-catenin were significantly (p < 0.01) reduced by the iron chelators DFO and DFX in whole-cell extracts (D), cytosol (E), and nuclear fractions (F) but was not changed significantly by 20 μM FeCl2, NH4–Fe–citrate and FeCl3. G β-Catenin-mediated TCF/LEF transcription activity was decreased significantly by the endolysosome iron chelator DFO (20 μM, p < 0.0001) and the endolysosome and cytosol iron chelator DFX (20 μM, p < 0.001) but was not changed significantly by 20 μM FeCl2, NH4–Fe–citrate, and FeCl3
Effects of iron and iron chelators on HIV-1 LTR transactivation in U87MG cells stably transduced with HIV-1 Tat-GFP
To determine effects of iron and iron chelators on HIV-1 transactivation induced by intracellularly produced HIV-1 Tat, we first generated a stable HIV-1 Tat-GFP cell line. U87MG-LTR cells were transduced with a Tat-GFP lentivirus plasmid at an MOI of 1.0 followed by puromycin selection pressure (5.0 μg/ml). Using confocal microscopy, stably transduced cells exhibited robust staining for HIV-1 Tat that co-distributed in DAPI-positive nuclei (Fig. 10A). Western blot analysis of HIV-1 Tat showed high levels of Tat-GFP in the stably transduced cells (Fig. 10B). Levels of HIV-1 LTR transactivation were significantly (p < 0.001) higher in stably transduced Tat-GFP cells than in control-GFP U87MG-LTR cells (Fig. 10C). Intracellular Tat expression-mediated HIV-1 LTR transactivation was not affected significantly when stably transduced cells were incubated for 24 h with 20 μM FeCl2, FeCl3, NH4–Fe–citrate, or iron dextran (Fig. 10D). Intracellular Tat expression-mediated HIV-1 LTR transactivation was significantly (p < 0.001) increased when stably transduced cells were incubated for 24 h with 25 μM of the iron chelators deferoxamine (DFO), 2–2 bipyridyl (2–2-BP), and deferiprone (DPO) (Fig. 10E).
Fig. 10.

Effects of iron and iron chelators on HIV-1 LTR transactivation in U87MG cells stably transduced with HIV-1 Tat-GFP. To determine effects of iron and iron chelators on HIV-1 transactivation induced by intracellularly produced HIV-1 Tat, we first generated a stable HIV-1 Tat-GFP cell line. U87MG-LTR cells were transduced with a Tat-GFP lentivirus plasmid at an MOI of 1.0 followed by puromycin selection pressure (5.0 μg/ml). A Using confocal microscopy, stably transduced cells exhibited robust staining for HIV-1 Tat that co-distributed in DAPI-positive nuclei. B Western blot analysis of HIV-1 Tat showed high levels of Tat-GFP in the stably transduced cells. C Levels of HIV-1 LTR transactivation were significantly (p < 0.001) higher in stably transduced Tat-GFP cells than in control-GFP U87MG-LTR cells (n = 3, ***p < 0.001). D Intracellular Tat expression-mediated HIV-1 LTR transactivation was not affected significantly when stably transduced cells were incubated for 24 h with 20 μM FeCl2, FeCl3, NH4–Fe–citrate or iron dextran. E Intracellular Tat expression-mediated HIV-1 LTR transactivation was significantly (p < 0.001) increased when stably transduced cells were incubated for 24 h with 25 μM of the iron chelators deferoxamine (DFO), 2–2 bipyridyl (2–2-BP), and deferiprone (DPO). (n = 3, ***p < 0.001)
Discussion
HIV-1 Tat is endocytosed into cells, is secreted actively from HIV-1 infected cells, and once up-taken it impacts HIV-1 replication (Ensoli et al. 1993; Mann and Frankel 1991; Sonia et al. 2012; Vendeville et al. 2004; Vives 2003). HIV-1 Tat is composed of two introns (Frankel et al. 1988), and functionally it is composed of four distinct domains (Clark et al. 2017; Frankel et al. 1988; Jeang et al. 1999; Romani et al. 2010). Of relevance to the current work, domain 2 (Tat22-37) is a cysteine-rich region that binds divalent cations and once these cations are bound to HIV-1 Tat they induce structural changes including Tat oligomerization (Frankel et al. 1988). Functionally, HIV-1 Tat is most active as a dimer (Jeang et al. 1999) and Tat oligomerization decreases its biological activity as well as its ability to escape endolysosomes (Tosi et al. 2000) (Fig. 11).
Fig. 11.

Effects of iron and the iron chelators on Tat-mediated HIV-1 LTR transactivation. Endolysosome iron (FeCl2 and FeCl3) can bind to HIV-1 Tat, induce HIV-1 Tat oligomerization, and thereby decrease its biological activity as measured by Tat-mediated HIV-1 LTR transactivation. Endolysosome-specific iron chelators deferoxamine (DFO) and deferasirox (DFX) can maintain HIV-1 Tat in its more active dimeric form that enhances Tat-mediated HIV-1 LTR transactivation by mechanisms that include decreasing levels of labile iron in endolysosomes and overcoming the actions of the HIV-1 restriction factor β-catenin
Here, we showed (1) that ferric (Fe3+) and ferrous (Fe2+) iron both restricted Tat-mediated HIV-1 LTR transactivation, (2) that in the presence of iron HIV-1 Tat increasingly oligomerized and had a decreased ability to enhance Tat-mediated HIV-1 LTR transactivation, (3) that chelation of endolysosome iron with deferoxamine and 2–2 bipyridyl, but not chelation of cytosolic iron with deferiprone and deferasirox, significantly enhanced Tat-mediated HIV-1 LTR transactivation, (4) that deferoxamine was able to both prevent and reverse HIV-1 Tat oligomerization and thereby increased the effectiveness of Tat to enhance HIV-1 LTR transactivation, and (5) that deferoxamine reduced protein expression levels of the HIV-1 restriction factor beta-catenin in the cytosol and nucleus. These findings demonstrate that reducing levels of endolysosome iron through chelation strategies can increase the levels and actions of the more active dimeric form of HIV-1 Tat relative to its less active oligomerized form. This has potentially important implications both for PLWH as well as experimentalists studying the actions and effects of HIV-1 Tat.
Extracellular levels of iron affect HIV-1 infection and disease progression (Afacan et al. 2002; Banjoko et al. 2012; Doherty 2007; Esan et al. 2013; Khan et al. 2020a; Mancone et al. 2017; Nekhai et al. 2013). Conversely, HIV-1 infection affects iron metabolism (Afacan et al. 2002; Banjoko et al. 2012; Kumari et al. 2016); depending on the progression of HIV-1 infection, levels of serum iron are either decreased or increased (Chang et al. 2015; Nekhai et al. 2013). Increased levels of iron can lead to the generation of reactive oxygen species (ROS) in part through Fenton reactions and increased levels of ROS can increase HIV-1 replication (Couret and Chang 2016; Ivanov et al. 2016; Kanti Das et al. 2015) and metabolic imbalances that can exploit iron (Afacan et al. 2002; Banjoko et al. 2012; Kumari et al. 2016). Moreover, the neurotoxic actions of the HIV-1 proteins gp120 and Tat are mediated at least in part through increased levels of ROS (El-Amine et al. 2018; Louboutin et al. 2014, 2007; Louboutin and Strayer 2014; Nath et al. 2000; Ronaldson and Bendayan 2008). Thus, there is a complicated relationship between iron levels and HIV-1.
Intracellularly, iron may also affect HIV-1 replication. Endolysosomes are acidic organelles that contain high micromolar levels of divalent cations including Ca2+, Fe2+, and Zn2+. When de-acidified, as can occur with a variety of insults, these cations are released from endolysosomes (Halcrow et al. 2019; McGuire et al. 2017; Mindell, 2012; Prasad and Rao 2018; Terman and Kurz 2013; Xiong and Zhu 2016; Xu and Ren 2015), and iron once released can increase levels of iron and ROS in the cytosol and in mitochondria. This is of relevance to our findings because Tat and gp120 de-acidify endolysosomes (El-Hage et al. 2015; Hui et al. 2012), and when deacidified, HIV-1 Tat can escape into the cytosol and nucleus where it activates the HIV-1 LTR promoter (Ensoli et al. 1993; Khan et al. 2018, 2019b; Kolson et al. 1994). Thus, endolysosome deacidification may be an important regulator of HIV-1 replication (Chauhan et al. 2014; Chauhan and Tikoo 2015; Vijaykumar et al. 2008).
Cations released from endolysosomes might impact HIV-1 in various ways. Iron (Fe2+) supramolecular helicates interfered with interactions between HIV-1 Tat and TAR RNA and affected HIV-1 replication (Slice et al. 1992). Ammonium iron citrate negatively regulated viral infections by blocking endolysosome escape processes (Wang et al. 2018). Zn2+ interacted with the cysteine-rich domain of Tat protein and induced Tat oligomerization (Frankel et al. 1988; Song et al. 2003). However, here, we did not find that ZnCl2, MgCl2, CuCl2, and CoCl2 could alter extracellular Tat-mediated HIV-1 LTR transactivation. Because iron was found in our molecular docking studies to bind to the same amino acids as does zinc (Cys-22, Cys-25, Cys-27, Cys-30, His-33, Cys-34, Cys-37), iron-induced Tat oligomerization might affect HIV-1 transactivation by virtue of its ability to either restrict the escape of Tat from endolysosomes or by increasing levels of ROS.
When iron was co-incubated with exogenously applied HIV-1 Tat, we observed lower levels of the active dimeric forms of HIV-1 Tat as well as lower levels of Tat-mediated LTR transactivation; opposite results were observed with endolysosome-specific iron chelators. However, when iron was applied to cells after incubation with HIV-1 Tat, no effects on Tat-mediated LTR transactivation were observed. But, when endolysosome-specific iron chelators were applied to cells after incubation with HIV-1 Tat, enhanced Tat-mediated HIV-1 LTR transactivation was still observed. Thus, chelation of endolysosome iron appears to be an important regulator of the biological actions of HIV-1 Tat whether exogenously applied or produced from cells stably transfected to produce HIV-1 Tat. This may be due to the ability of iron chelators to reduce the expression of anti-HIV-1 factor β-catenin and TCF/LEF transcription activity in U87MG cells (Coombs et al. 2012; Kamihara et al. 2016; Song et al. 2011).
Our results also have implications for experimentalists interested in the biological actions of HIV-1 Tat. There are relatively few sources from which HIV-1 Tat can be obtained. All sources we have tested contain to varying degrees dimeric and oligomeric HIV-1 Tat; HIV-1 Tat is far more active as a dimer than it is as an oligomer. This might help explain great discrepancies in the literature about concentration/effect relationships. Importantly, we have shown that endolysosome-specific chelators of iron can reverse iron-dependent oligomerization and increase Tat-mediated HIV-1 LTR transactivation. Thus, it is possible that people producing HIV-1 Tat might create a more potent source of Tat if they prevented iron-induced HIV-1 Tat oligomerization.
In summary, endolysosome iron might restrict HIV-1 transactivation by promoting HIV-1 Tat oligomerization—a less active form of Tat. Iron might also decrease levels of the more active dimeric form of HIV-1 Tat proteins in the laboratory because of iron-induced oligomerization of HIV-1 Tat. On the other hand, endolysosome-specific iron chelators can increase levels of HIV-1 transactivation by decreasing HIV-1 Tat oligomerization and by reducing β-catenin protein expression. Thus, endolysosome-specific iron chelators might be used with caution in PLWH whereas experimentalists might find benefit by using iron chelators because of their ability to increase levels of the more active dimeric forms of HIV-1 Tat.
Funding
This study was funded by the National Institute of General Medical Sciences (P30GM100329, U54GM115458), the National Institute of Mental Health (R01MH100972, R01MH105329, R01MH119000), the National Institute of Neurological Diseases and Stroke (2R01NS065957), and the National Institute of Drug Abuse (2R01DA032444).
Footnotes
Ethical approval (animals) Although the University of North Dakota Animal Care and Use Committee is adherent with the Guide for the Care and Use of Laboratory Animals (NIH publication number 80–23), no animal studies were conducted as part of this work.
Ethical approval (humans) This work did not use human participants and did not use any materials of human origin.
Conflict of interest The authors declare no competing interests.
References
- Afacan YE, Hasan MS, Omene JA (2002) Iron deficiency anemia in HIV infection: immunologic and virologic response. J Natl Med Assoc 94:73–77 [PMC free article] [PubMed] [Google Scholar]
- Arhel N, Kirchhoff F (2010) Host proteins involved in HIV infection: new therapeutic targets. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1802:313–321 [DOI] [PubMed] [Google Scholar]
- Bae M, Patel N, Xu H, Lee M, Tominaga-Yamanaka K, Nath A, Geiger J, Gorospe M, Mattson MP, Haughey NJ (2014) Activation of TRPML1 clears intraneuronal Aβ in preclinical models of HIV infection. J Neurosci 34:11485–11503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjoko SO, Oseni FA, Togun RA, Onayemi O, Emma-Okon BO, Fakunle JB (2012) Iron status in HIV-1 infection: implications in disease pathology. BMC Clin Pathol 12:26–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright NA, Gratian MJ, Luzio JP (2005) Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol 15:360–365 [DOI] [PubMed] [Google Scholar]
- Castino R, Fiorentino I, Cagnin M, Giovia A, Isidoro C (2011) Chelation of lysosomal iron protects dopaminergic SH-SY5Y neuroblastoma cells from hydrogen peroxide toxicity by precluding autophagy and Akt dephosphorylation. Toxicological Sciences : an Official Journal of the Society of Toxicology 123:523–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahroudi A, Wagner TA, Persaud D (2018) CNS persistence of HIV-1 in children: the untapped reservoir. Curr HIV/AIDS Rep 15:382–387 [DOI] [PubMed] [Google Scholar]
- Chang HC, Bayeva M, Taiwo B, Palella FJ Jr, Hope TJ, Ardehali H (2015) Short communication: high cellular iron levels are associated with increased HIV infection and replication. AIDS Res Hum Retroviruses 31:305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A, Mehla R, Vijayakumar TS, Handy I (2014) Endocytosis-mediated HIV-1 entry and its significance in the elusive behavior of the virus in astrocytes. Virology 456–457:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A, Tikoo A (2015) The enigma of the clandestine association between chloroquine and HIV-1 infection. HIV Med 16:585–590 [DOI] [PubMed] [Google Scholar]
- Chen X, Hui L, Geiger NH, Haughey NJ, Geiger JD (2013) Endolysosome involvement in HIV-1 transactivator protein-induced neuronal amyloid beta production. Neurobiol Aging 34:2370–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen KA, Myers JT, Swanson JA (2002) pH-dependent regulation of lysosomal calcium in macrophages. J Cell Sci 115:599–607 [DOI] [PubMed] [Google Scholar]
- Churchill MJ, Wesselingh SL, Cowley D, Pardo CA, McArthur JC, Brew BJ, Gorry PR (2009) Extensive astrocyte infection is prominent in human immunodeficiency virus–associated dementia. Ann Neurol 66:253–258 [DOI] [PubMed] [Google Scholar]
- Clark E, Nava B, Caputi M (2017) Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget 8:27569–27581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan MP, Culbert AA, Cross DA, Corcoran SL, Yates JW, Pearce NJ, Rausch OL, Murphy GJ, Carter PS, Roxbee Cox L, Mills D, Brown MJ, Haigh D, Ward RW, Smith DG, Murray KJ, Reith AD, Holder JC (2000) Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem Biol 7:793–803 [DOI] [PubMed] [Google Scholar]
- Coombs GS, Schmitt AA, Canning CA, Alok A, Low ICC, Banerjee N, Kaur S, Utomo V, Jones CM, Pervaiz S, Toone EJ, Virshup DM (2012) Modulation of Wnt/β-catenin signaling and proliferation by a ferrous iron chelator with therapeutic efficacy in genetically engineered mouse models of cancer. Oncogene 31:213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couret J, Chang TL (2016) Reactive oxygen species in HIV infection. EC Microbiology 3:597–604 [PMC free article] [PubMed] [Google Scholar]
- De Domenico I, Ward DM, Kaplan J (2009) Specific iron chelators determine the route of ferritin degradation. Blood 114:4546–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C (2005) The lysosome turns fifty. Nat Cell Biol 7:847–849 [DOI] [PubMed] [Google Scholar]
- Deshmane SL, Mukerjee R, Fan S, Sawaya BE (2011). High-performance capillary electrophoresis for determining HIV-1 Tat protein in neurons. PLoS One 6:e16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty CP (2007) Host-pathogen interactions: the role of iron. J Nutr 137:1341–1344 [DOI] [PubMed] [Google Scholar]
- Doulias P-T, Christoforidis S, Brunk UT, Galaris D (2003) Endosomal and lysosomal effects of desferrioxamine: protection of HeLa cells from hydrogen peroxide-induced DNA damage and induction of cell-cycle arrest. Free Radical Biol Med 35:719–728 [DOI] [PubMed] [Google Scholar]
- El-Amine R, Germini D, Zakharova VV, Tsfasman T, Sheval EV, Louzada RAN, Dupuy C, Bilhou-Nabera C, Hamade A, Najjar F, Oksenhendler E, Lipinski M, Chernyak BV, Vassetzky YS (2018) HIV-1 Tat protein induces DNA damage in human peripheral blood B-lymphocytes via mitochondrial ROS production. Redox Biol 15:97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N, Rodriguez M, Dever SM, Masvekar RR, Gewirtz DA, Shacka JJ (2015) HIV-1 and morphine regulation of autophagy in microglia: limited interactions in the context of HIV-1 Infection and Opioid Abuse. J Virol 89:1024–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P, Gallo RC (1993) Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol 67:277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esan MO, van Hensbroek MB, Nkhoma E, Musicha C, White SA, ter Kuile FO, Phiri KS (2013) Iron supplementation in HIV-infected Malawian children with anemia: a double-blind, randomized, controlled trial. Clin Infect Dis 57:1626–1634 [DOI] [PubMed] [Google Scholar]
- Espósito BP, Epsztejn S, Breuer W, Cabantchik ZI (2002) A review of fluorescence methods for assessing labile iron in cells and biological fluids. Anal Biochem 304:1–18 [DOI] [PubMed] [Google Scholar]
- Fernández B, Fdez E, Gómez-Suaga P, Gil F, Molina-Villalba I, Ferrer I, Patel S, Churchill GC, Hilfiker S (2016) Iron overload causes endolysosomal deficits modulated by NAADP-regulated 2-pore channels and RAB7A. Autophagy 12:1487–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Piga A, Harmatz P, NIELSEN P, (2005) Monitoring longterm efficacy of iron chelation treatment with biomagnetic liver susceptometry. Ann N Y Acad Sci 1054:350–357 [DOI] [PubMed] [Google Scholar]
- Fois AF, Brew BJ (2015) The potential of the CNS as a reservoir for HIV-1 infection: implications for HIV eradication. Curr HIV/AIDS Rep 12:299–303 [DOI] [PubMed] [Google Scholar]
- Frankel A, Bredt D, Pabo C (1988) Tat protein from human immunodeficiency virus forms a metal-linked dimer. Science 240:70–73 [DOI] [PubMed] [Google Scholar]
- Fredericksen BL, Wei BL, Yao J, Luo T, Garcia JV (2002) Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J Virol 76:11440–11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO (2001) HIV-1 replication. Somat Cell Mol Genet 26:13–33 [DOI] [PubMed] [Google Scholar]
- Glickstein H, El Ben R, Shvartsman M, Cabantchik ZI (2005) Intracellular labile iron pools as direct targets of iron chelators: a fluorescence study of chelator action in living cells. Blood 106:3242–3250 [DOI] [PubMed] [Google Scholar]
- Gray LR, Roche M, Flynn JK, Wesselingh SL, Gorry PR, Churchill MJ (2014) Is the central nervous system a reservoir of HIV-1? Curr Opin HIV AIDS 9:552–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Chen C-C, Wahl-Schott C, Biel M (2017) Two-pore channels: catalyzers of endolysosomal transport and function. Front Pharmacol 8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halcrow P, Khan N, Datta G, Ohm JE, Chen X, Geiger JD (2019) Importance of measuring endolysosome, cytosolic, and extracellular pH in understanding the pathogenesis of and possible treatments for glioblastoma multiforme. Cancer Rep 2(6). 10.1002/cnr2.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halcrow PW, Lakpa KL, Khan N, Afghah Z, Miller N, Datta G, Chen X, Geiger JD (2021) HIV-1 gp120-induced endolysosome deacidification leads to efflux of endolysosome iron, and increases in mitochondrial iron and reactive oxygen species. J Neuroimmune Pharmacol 10.1007/s11481-021-09995-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughey NJ, Mattson MP (2002) Calcium dysregulation and neuronal apoptosis by the HIV-1 proteins Tat and gp120. J Acquir Immune Defic Syndr 31(Suppl 2):S55–61 [DOI] [PubMed] [Google Scholar]
- Heaton R, Clifford D, Franklin D, Woods S, Ake C, Vaida F, Ellis R, Letendre S, Marcotte T, Atkinson J (2010) HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75:2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hider RC, Zhou T (2005) The design of orally active iron chelators. Ann N Y Acad Sci 1054:141–154 [DOI] [PubMed] [Google Scholar]
- Hui L, Chen X, Haughey NJ, Geiger JD (2012) Role of endolysosomes in HIV-1 Tat-induced neurotoxicity. ASN Neuro 4:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J, Helenius A (2011) Endosome maturation. EMBO J 30:3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Valuev-Elliston VT, Ivanova ON, Kochetkov SN, Starodubova ES, Bartosch B, Isaguliants MG (2016) Oxidative stress during HIV infection: mechanisms and consequences. Oxid Med Cell Longev 2016:8910396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang KT, Xiao H, Rich EA (1999) Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem 274:28837–28840 [DOI] [PubMed] [Google Scholar]
- Johnson TP, Patel K, Johnson KR, Maric D, Calabresi PA, Hasbun R, Nath A (2013) Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. Proc Natl Acad Sci U S A 110:13588–13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R (2015) HIV-1 target cells in the CNS. J Neurovirol 21:276–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamihara Y, Takada K, Sato T, Kawano Y, Murase K, Arihara Y, Kikuchi S, Hayasaka N, Usami M, Iyama S, Miyanishi K, Sato Y, Kobune M, Kato J (2016) The iron chelator deferasirox induces apoptosis by targeting oncogenic Pyk2/β-catenin signaling in human multiple myeloma. Oncotarget 7:64330–64341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanti Das T, Wati MR, Fatima-Shad K (2015) Oxidative stress gated by Fenton and Haber Weiss reactions and its association with Alzheimer’s disease. Arch Neurosci 2:e60038 [Google Scholar]
- Khan N (2020) Possible protective role of 17β-estradiol against COVID-19. J Allergy Infect Dis 1:38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Chen X, Geiger JD (2020a) Role of divalent cations in HIV-1 replication and pathogenicity. Viruses 12:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Chen X, Geiger JD (2020b) Role of endolysosomes in severe acute respiratory syndrome coronavirus-2 infection and coronavirus disease 2019 pathogenesis: implications for potential treatments. Front Pharmacol 11:595888. 10.3389/fphar.2020.595888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Chen X, Geiger JD (2021) Possible therapeutic use of natural compounds against COVID-19. J Cell Signal 2:63–79 [PMC free article] [PubMed] [Google Scholar]
- Khan N, Datta G, Geiger JD, Chen X (2018) Apolipoprotein E isoform dependently affects Tat-mediated HIV-1 LTR transactivation. J Neuroinflammation 15:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Halcrow PW, Lakpa KL, Afghah Z, Miller NM, Dowdy SF, Geiger JD, Chen X (2020c) Two-pore channels regulate Tat endolysosome escape and Tat-mediated HIV-1 LTR transactivation. Faseb j 34:4147–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Haughey NJ, Nath A, Geiger JD (2019a) Involvement of organelles and inter-organellar signaling in the pathogenesis of HIV-1 associated neurocognitive disorder and Alzheimer’s disease. Brain Res 1722:146389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Lakpa KL, Halcrow PW, Afghah Z, Miller NM, Geiger JD, Chen X (2019b) BK channels regulate extracellular Tat-mediated HIV-1 LTR transactivation. Sci Rep 9:12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolson DL, Collman R, Hrin R, Balliet JW, Laughlin M, McGann KA, Debouck C, Gonzalez-Scarano F (1994) Human immunodeficiency virus type 1 Tat activity in human neuronal cells: uptake and trans-activation. J Gen Virol 75(Pt 8):1927–1934 [DOI] [PubMed] [Google Scholar]
- Kumari N, Ammosova T, Diaz S, Lin X, Niu X, Ivanov A, Jerebtsova M, Dhawan S, Oneal P, Nekhai S (2016) Increased iron export by ferroportin induces restriction of HIV-1 infection in sickle cell disease. Blood Adv 1:170–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Gustafsson B, Brunk UT (2006) Intralysosomal iron chelation protects against oxidative stress-induced cellular damage. FEBS J 273:3106–3117 [DOI] [PubMed] [Google Scholar]
- Lakpa KL, Khan N, Afghah Z, Chen X, Geiger JD (2021) Lysosomal stress response (LSR): physiological importance and pathological relevance. J Neuroimmune Pharmacol. 10.1007/s11481-021-09990-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786 [DOI] [PubMed] [Google Scholar]
- Liang C, Wainberg M (2002) The role of Tat in HIV-1 replication: an activator and/or a suppressor? AIDS Rev 4:41–49 [PubMed] [Google Scholar]
- Lin YF, Cheng CW, Shih CS, Hwang JK, Yu CS, Lu CH (2016) MIB: metal ion-binding site prediction and docking server. J Chem Inf Model 56:2287–2291 [DOI] [PubMed] [Google Scholar]
- Liu Y, Jones M, Hingtgen CM, Bu G, Laribee N, Tanzi RE, Moir RD, Nath A, He JJ (2000) Uptake of HIV-1 tat protein mediated by low-density lipoprotein receptor-related protein disrupts the neuronal metabolic balance of the receptor ligands. Nat Med 6:1380–1387 [DOI] [PubMed] [Google Scholar]
- Lloyd JB, Cable H, Rice-Evans C (1991) Evidence that desferrioxamine cannot enter cells by passive diffusion. Biochem Pharmacol 41:1361–1363 [DOI] [PubMed] [Google Scholar]
- Louboutin J-P, Agrawal L, Reyes BAS, Van Bockstaele EJ, Strayer DS (2014) Oxidative stress is associated with neuroinflammation in animal models of HIV-1 Tat neurotoxicity. Antioxidants (basel, Switzerland) 3:414–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louboutin J-P, Strayer D (2014. Role of oxidative stress in HIV-1-associated neurocognitive disorder and protection by gene delivery of antioxidant enzymes. In: Antioxidants (Basel, Switzerland), pp 770–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louboutin JP, Agrawal L, Reyes BAS, Van Bockstaele EJ, Strayer DS (2007) Protecting neurons from HIV-1 gp120-induced oxidant stress using both localized intracerebral and generalized intraventricular administration of antioxidant enzymes delivered by SV40-derived vectors. Gene Ther 14:1650–1661 [DOI] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA (2007) Lysosomes: fusion and function. Nat Rev Mol Cell Biol 8:622–632 [DOI] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Gray SR, Gratian MJ, Piper RC, Bright NA (2005) Membrane traffic to and from lysosomes. Biochem Soc Symp 72:77–86 [DOI] [PubMed] [Google Scholar]
- Mancone C, Grimaldi A, Refolo G, Abbate I, Rozera G, Benelli D, Fimia GM, Barnaba V, Tripodi M, Piacentini M, Ciccosanti F (2017) Iron overload down-regulates the expression of the HIV-1 Rev cofactor eIF5A in infected T lymphocytes. Proteome Science 15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DA, Frankel AD (1991) Endocytosis and targeting of exogenous HIV-1 Tat protein. EMBO J 10:1733–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marban C, Forouzanfar F, Ait-Ammar A, Fahmi F, El Mekdad H, Daouad F, Rohr O, Schwartz C (2016) Targeting the brain reservoirs: toward an HIV cure. Front Immunol 7:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire C, Stransky L, Cotter K, Forgac M (2017) Regulation of V-ATPase Activity Front Biosci (landmark Ed) 22:609–622 [DOI] [PubMed] [Google Scholar]
- Meeker RB, Asahchop E, Power C (2014) The brain and HAART: collaborative and combative connections. Curr Opin HIV AIDS 9:579–584 [DOI] [PubMed] [Google Scholar]
- Mindell JA (2012) Lysosomal acidification mechanisms. Annu Rev Physiol 74:69–86 [DOI] [PubMed] [Google Scholar]
- Mobarra N, Shanaki M, Ehteram H, Nasiri H, Sahmani M, Saeidi M, Goudarzi M, Pourkarim H, Azad M (2016) A review on iron chelators in treatment of iron overload syndromes. International Journal of Hematology-Oncology and Stem Cell Research 10:239–247 [PMC free article] [PubMed] [Google Scholar]
- Nath A, Haughey NJ, Jones M, Anderson C, Bell JE, Geiger JD (2000) Synergistic neurotoxicity by human immunodeficiency virus proteins Tat and gp120: protection by memantine. Ann Neurol 47:186–194 [PubMed] [Google Scholar]
- Nath A, Psooy K, Martin C, Knudsen B, Magnuson DS, Haughey N, Geiger JD (1996) Identification of a human immunodeficiency virus type 1 Tat epitope that is neuroexcitatory and neurotoxic. J Virol 70:1475–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekhai S, Kumari N, Dhawan S (2013) Role of cellular iron and oxygen in the regulation of HIV-1 infection. Future Virol 8:301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Cataldo AM (2006) Lysosomal system pathways: genes to neurodegeneration in Alzheimer’s disease. 9(3):277–89 [DOI] [PubMed] [Google Scholar]
- Perera RM, Zoncu R (2016) The lysosome as a regulatory hub. Annu Rev Cell Dev Biol 32:223–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- Prasad H, Rao R (2018) Histone deacetylase-mediated regulation of endolysosomal pH. J Biol Chem 293:6721–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera LE, Colon K, Cantres-Rosario YM, Zenon FM, Melendez LM (2014) Macrophage derived cystatin B/cathepsin B in HIV replication and neuropathogenesis. Curr HIV Res 12:111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani B, Engelbrecht S, Glashoff RH (2010) Functions of Tat: the versatile protein of human immunodeficiency virus type 1. J Gen Virol 91:1–12 [DOI] [PubMed] [Google Scholar]
- Ronaldson PT, Bendayan R (2008) HIV-1 viral envelope glycoprotein gp120 produces oxidative stress and regulates the functional expression of multidrug resistance protein-1 (Mrp1) in glial cells. J Neurochem 106:1298–1313 [DOI] [PubMed] [Google Scholar]
- Saylor D, Dickens AM, Sacktor N, Haughey N, Slusher B, Pletnikov M, Mankowski JL, Brown A, Volsky DJ, McArthur JC (2016) HIV-associated neurocognitive disorder–pathogenesis and prospects for treatment. Nat Rev Neurol 12:234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slice LW, Codner E, Antelman D, Holly M, Wegrzynski B, Wang J, Toome V, Hsu MC, Nalin CM (1992) Characterization of recombinant HIV-1 Tat and its interaction with TAR RNA. Biochemistry 31:12062–12068 [DOI] [PubMed] [Google Scholar]
- Song L, Nath A, Geiger JD, Moore A, Hochman S (2003) Human immunodeficiency virus type 1 Tat protein directly activates neuronal N-methyl-D-aspartate receptors at an allosteric zinc-sensitive site. J Neurovirol 9:399–403 [DOI] [PubMed] [Google Scholar]
- Song S, Christova T, Perusini S, Alizadeh S, Bao R-Y, Miller BW, Hurren R, Jitkova Y, Gronda M, Isaac M, Joseph B, Subramaniam R, Aman A, Chau A, Hogge DE, Weir SJ, Kasper J, Schimmer AD, Al-awar R, Wrana JL, Attisano L (2011) Wnt inhibitor screen reveals iron dependence of β-catenin signaling in cancers. Can Res 71:7628–7639 [DOI] [PubMed] [Google Scholar]
- Sonia M, Albert D, Gilbert B, Isabelle R, Catherine D, Herve T-D, Malika M, Herve M, Catherine T, Corinne B, Pascale P, Francoise D-G, Andreas S, Philippe B, Stephen AS, Grant RC, Erwann PL (2012) Antiretroviral therapy does not block the secretion of the human immunodeficiency virus Tat protein. Infectious Disorders - Drug Targets 12:81–86 [DOI] [PubMed] [Google Scholar]
- Spudich S, Gonzalez-Scarano F (2012) HIV-1-related central nervous system disease: current issues in pathogenesis, diagnosis, and treatment. Cold Spring Harb Perspect Med 2:a007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A, Kurz T (2013) Lysosomal iron, iron chelation, and cell death. Antioxid Redox Signal 18:888–898 [DOI] [PubMed] [Google Scholar]
- Thompson KA, Cherry CL, Bell JE, McLean CA (2011) Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. Am J Pathol 179:1623–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Dikic I (2016) Autophagy captures the nobel prize. Cell 167:1433–1435 [DOI] [PubMed] [Google Scholar]
- Tosi G, Meazza R, De Lerma BA, D’Agostino A, Mazza S, Corradin G, Albini A, Noonan DM, Ferrini S, Accolla RS (2000) Highly stable oligomerization forms of HIV-1 Tat detected by monoclonal antibodies and requirement of monomeric forms for the transactivating function on the HIV-1 LTR. Eur J Immunol 30:1120–1126 [DOI] [PubMed] [Google Scholar]
- Tyagi M, Rusnati M, Presta M, Giacca M (2001) Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J Biol Chem 276:3254–3261 [DOI] [PubMed] [Google Scholar]
- Valcour V, Chalermchai T, Sailasuta N, Marovich M, Lerdlum S, Suttichom D, Suwanwela NC, Jagodzinski L, Michael N, Spudich S, van Griensven F, de Souza M, Kim J, Ananworanich J, Group RSS (2012) Central nervous system viral invasion and inflammation during acute HIV infection. J Infect Dis 206:275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenhuis RT, Clements JE, Gama L (2019) HIV eradication strategies: implications for the central nervous system. Curr HIV/AIDS Rep 16:96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S, Prevedel L, Valdebenito S, Gorska AM, Golovko M, Khan N, Geiger J, Eugenin EA (2020) Circulating levels of ATP is a biomarker of HIV cognitive impairment. EBioMedicine 51:102503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A, Rayne F, Bonhoure A, Bettache N, Montcourrier P, Beaumelle B (2004) HIV-1 Tat enters T cells using coated pits before translocating from acidified endosomes and eliciting biological responses. Mol Biol Cell 15:2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykumar T, Nath A, Chauhan A (2008) Chloroquine mediated molecular tuning of astrocytes for enhanced permissiveness to HIV infection. Virology 381:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives E (2003) Cellular uptake [correction of utake] of the Tat peptide: an endocytosis mechanism following ionic interactions. J Mol Recognit 16:265–271 [DOI] [PubMed] [Google Scholar]
- Wang H, Li Z, Niu J, Xu Y, Ma L, Lu A, Wang X, Qian Z, Huang Z, Jin X, Leng Q, Wang J, Zhong J, Sun B, Meng G (2018) Antiviral effects of ferric ammonium citrate. Cell Discov 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideman JG, Leung KF, Field MC, Dacks JB (2014) The cell biology of the endocytic system from an evolutionary perspective. Cold Spring Harb Perspect Biol 6:a016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Zhu MX (2016) Regulation of lysosomal ion homeostasis by channels and transporters. Science China Life Sciences 59:777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Ren D (2015) Lysosomal physiology. Annu Rev Physiol 77:57–80 [DOI] [PMC free article] [PubMed] [Google Scholar]


