Summary
The large diversity of neuron types provides the means by which cortical circuits perform complex operations. Neuron can be described by biophysical and molecular characteristics, afferent inputs, neuron targets. To quantify, visualize, and standardize these features, we developed the open-source Matlab-based framework, CellExplorer. It consists of three components: a processing module, a flexible data structure, and a powerful graphical interface. The processing module calculates standardized physiological metrics, performs neuron type classification, finds putative monosynaptic connections and saves it to a standardized yet flexible machine-readable format. The graphical interface makes it possible to explore the computed features at the speed of a mouse click. The framework allows users to process, curate and relate their data to a growing public collection of neurons. CellExplorer can link genetically identified cell types to physiological properties of neurons collected across laboratories, and potentially lead to interlaboratory standards of single cell metrics.
Discovering novel mechanisms in brain circuits requires high-resolution monitoring the constituent neurons and understanding the nature of their interactions. Large-scale extracellular electrophysiology aims to establish the relationship between neuronal firing and behavioral or cognitive variables in order to provide insights about the computational role of neurons and neuronal assemblies (Barlow, 1972; Buzsáki, 2004; Steinmetz et al., 2019). Exploiting the power of correlations between neuronal firing and behavioral variables requires multi-level characterization of single neurons and their interactions. Simultaneous recordings from large numbers of neurons, preferably identified by optogenetic and other methods, make it possible to build an extensive list of neuron features and their assigned ‘cell type’ properties (Fig. 1). Identification and manipulation of different neuron types in the behaving animal is a prerequisite for deciphering their role in circuit dynamics and behavior. Yet, currently, a gap exists between neuron classification schemes based on molecular and physiological methods (Gouwens et al., 2019; Jia et al., 2019; Kepecs and Fishell, 2014; Klausberger and Somogyi, 2008; McBain and Fisahn, 2001; Roux and Buzsáki, 2015; Rudy et al., 2011), largely because of vast differences in processing data across laboratories and even within the same laboratory. Ideally, the acquired data must be findable, accessible, interoperable, and reusable (FAIR; Wilkinson et al., 2016). This requires an agreed platform for data and metadata curation that allows to share datasets across laboratories for cross-examination and building ‘big data’ from experiments collected in multiple labs. Sharing easily interpretable data will facilitate neuroscientists to effectively and transparently communicate about their experiments (Sejnowski et al., 2014; Teeters et al., 2015; Bouchard et al., 2016; Martone et al., 2020).
Figure 1: Experimental paradigm-independent characterization of single neurons.
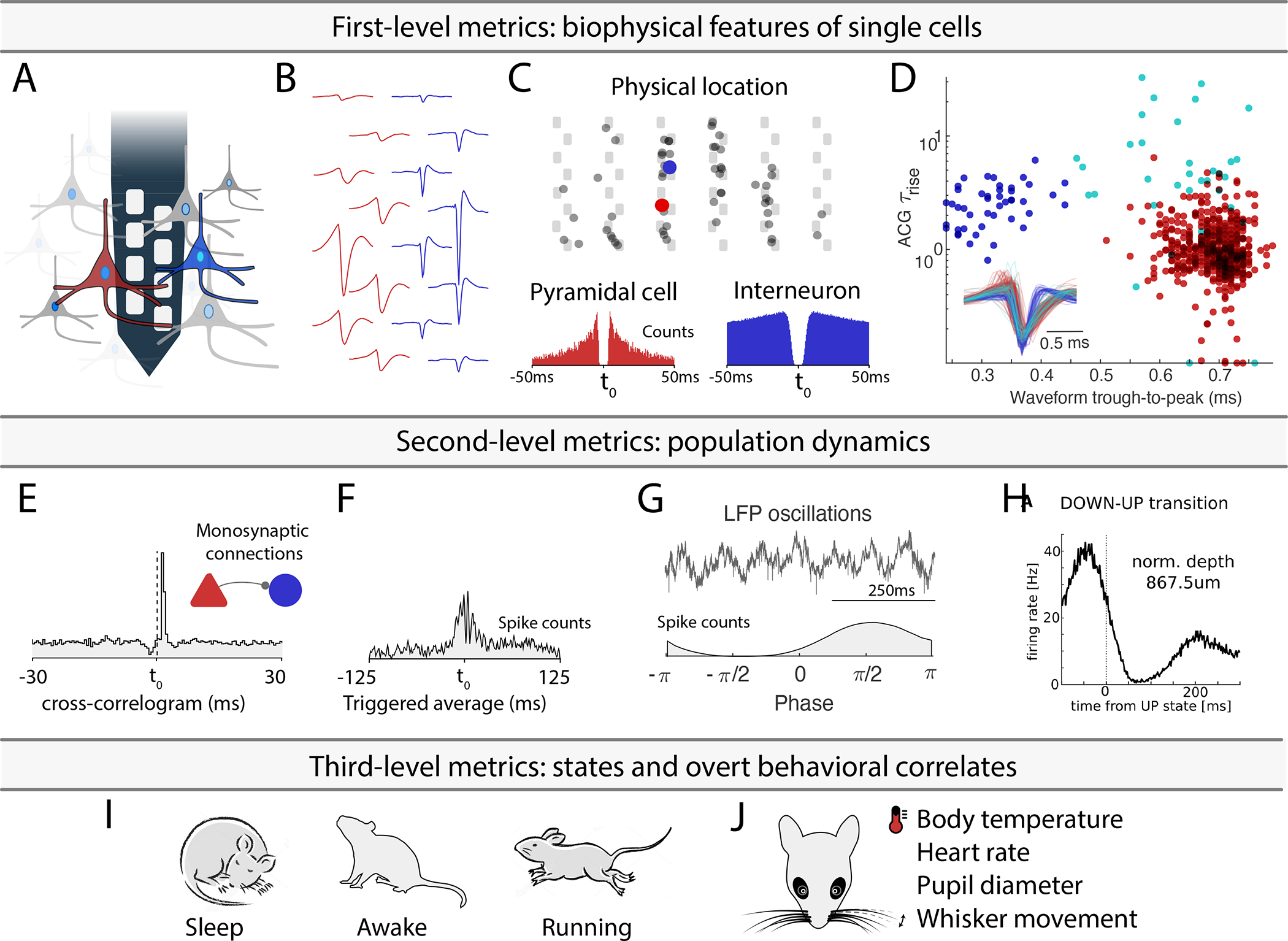
A. Using high-density silicon probes or multiple tetrodes (shown is a single shank with 8 recording sites), dozens to hundreds of neurons can be recorded simultaneously. B. Spikes of putative single neurons are extracted from the recorded traces and assigned to individual neurons through spike sorting algorithms. C. Their relative position determined through trilateration (the top panel shows neurons projected on a silicon probe with 6 shanks and a staggered electrode layout). Autocorrelograms (ACGs; lower two panels) are used to characterize the neurons (a bursting pyramidal cell with a wide waveform in red; a fast spiking interneuron with a narrow waveform in blue). D. Neuron-type classification based on first-order biophysical parameters, such as spike waveform width (trough-to-peak) and the temporal scale of the rising phase of the ACGs (τrise). Optogenetic and other direct identification methods can further ground units to neuron types. E. Interactions between neurons are characterized by their cross-correlograms and monosynaptic connections (determined via spike transmission probabilities). F. Event related histogram. G. Relating spikes to LFP patterns. H. Relating spikes to brain state changes. I-J. Spike pattern correlations with brain states and overt behaviors. Only a few possible examples are shown. See also Supplementary Table 1.
Properties of neurons can be described at multiple levels of complexity. The first level is a description of their biophysical characteristics. This level includes waveform features (Fig. 1B), their position relative to the recording sites, and other units and metrics related to firing patterns: interspike interval statistics, autocorrelograms, and derived metrics (Fig. 1C). These first level features can be used for a first-order separation of single neurons into putative major classes, typically excitatory and inhibitory cells (Fig. 1D). Single neuron properties can be related to genetically identified neuron classes with optogenetics and other more direct methods like juxtacellular and intracellular recordings (Boyden et al., 2005; Klausberger and Somogyi, 2008; Rudy et al., 2011; Buzsáki et al., 2015; Roux and Buzsáki, 2015; Lima et al., 2009). Antidromic and unit-LFP coupling techniques provide further assignment of single neurons to cortical regions, layers, and target projections (Bishop et al., 1962; Zhang et al., 2013; Ciocchi et al., 2015; Senzai et al., 2019; Shamash et al., 2018). The second level relates properties of single neurons to other neurons. Examples include cross-correlations and putative monosynaptic connections between excitatory and inhibitory neurons derived from spike transmission probabilities (Fig. 1E), pairwise and population synchrony, relationship to multiple oscillatory and irregular local field potentials (LFP; e.g., rhythmic patterns, up-down transitions in cortex). These metrics can be expanded for specific brain regions and questions. The third level metrics of single-unit activity include the relationship between its firing patterns and brain states (e.g. non-REM, REM, awake; Fig. 1H) and overt behavioral correlates (Fig. 1I). These include, but not restricted to, arousal states (sleep states and waking), spontaneous motor patterns, movement pattern changes, locomotion speed, head turns, whisker movements, respiration, heart rate, body temperature, pupil diameter and other autonomic parameters (McGinley et al., 2015; Stringer et al., 2019).
These three levels provide generic features of neuronal activity common to all experimental paradigms in the same species and, therefore, are communicable across different experiments and laboratories, leading to joint data bases and standardized metrics across different laboratories. Next, these three-level paradigm-independent features can be contrasted to and compared with experiment-unique manipulations and higher-level correlates of spiking activity. Because these latter correlations are often paradigm-specific and differ across laboratories, the three-level analysis can safeguard against mistakenly assigning cognitive and other roles of neuronal spiking when spike pattern changes can be explained by measurable overt behavioral correlates. Yet, even if all of the above information is available separately, factoring out critical variables and their combinations is possible only when the multitudes of single neuron characteristics can be compared flexibly.
The above workflow is similar across many physiological experiments. In the analysis of various features, an often-asked question is how one particular feature of unit firing relates to the many other metrics calculated by the experimenter. This is typically done by identifying some unexpected firing patterns in a set of neurons and independently analyze features one by one to find common features or exclude potential artifactual explanations. Whether testing a specific hypothesis or mining the ever-growing number of publicly available datasets, this process can be advanced by user-friendly processing pipelines, standardization of data formats, and highly flexible visualization methods. To provide the needed flexibility and to facilitate new ways of data mining neurophysiological data, we developed the open-source framework, CellExplorer, to characterize and classify single neuron features from multi-site extracellular recordings. CellExplorer consists of a pipeline for extracting and calculating physiological features, a flexible data format, and a powerful graphical interface that allows for fast manual curation and feature exploration. We demonstrate its utility through multiple examples and explain its user-friendly operation though detailed tutorials and video illustrations.
RESULTS
The CellExplorer architecture and operation consist of three main parts: a processing module for feature extraction, a graphical interface for manual curation and exploration, and a standardized yet flexible data structure (Fig 2). A step-by-step tutorial is available in the supplementary section, and more tutorials are available online (Suppl. Video 1). Flow charts are available in Suppl. Figure 1. The first step in running the pipeline is defining the data input.
Figure 2. Three-component framework.
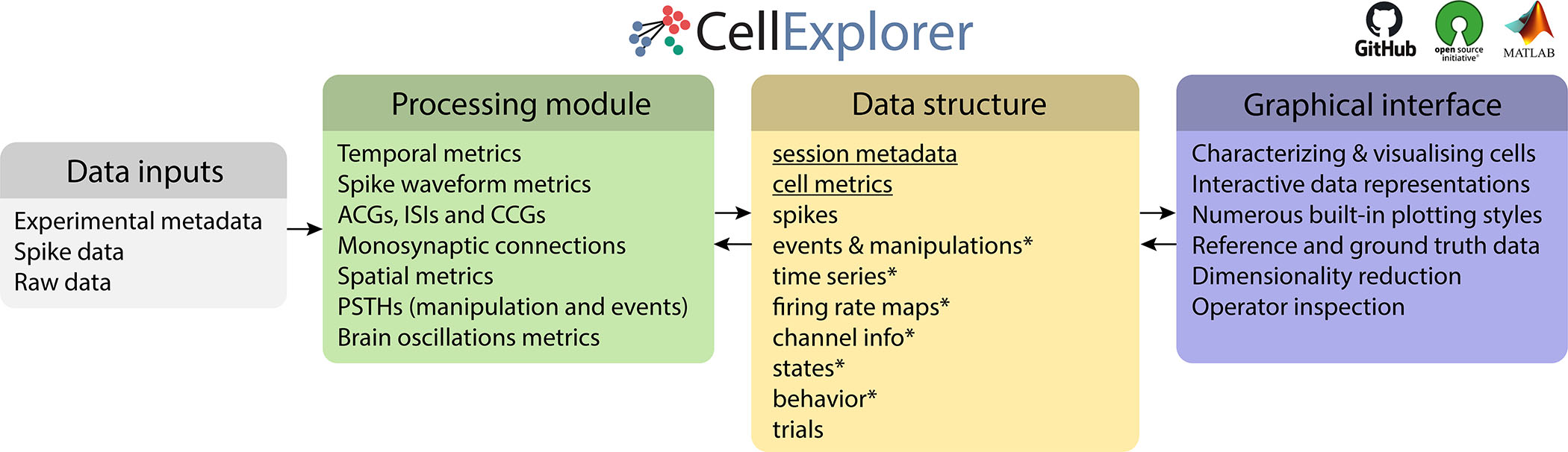
A single extensive processing module (green); Standardized yet flexible data structure (yellow); and a graphical interface (purple). Data inputs are compatible with most existing spike sorting algorithms (grey). The data structure joins the Processing module with the Graphical interface (* signifies data containers). CellExplorer is open-source, built in MATLAB, and available on GitHub. See also Supplementary Figure 1, 2, 3 and Supplementary Table 2.
Data Input
When running the pipeline, relevant metadata describing the spike format, raw data, and experimental metadata must be defined (Fig. 2). All experimental metadata (session-level) are handled in a single MATLAB structure, with an optional graphical interface for inspection and manual entry (Suppl. Figure 2). The platform supports several spike sorting data formats, including Neurosuite, Phy, KiloSort, SpyKING Circus, Wave_Clus, MClust, AllenSDK, NWB, ALF, MountainSort, IronClust, (Chung et al., 2017; Hazan et al., 2006; Pachitariu et al., 2016; Quiroga et al., 2004; Schmitzer-Torbert et al., 2005; Yger et al., 2018). The wide-band recorded “raw” data is critical for comparing derived metrics across laboratories since preprocessing pipelines vary widely and depend on equipment type and filter settings (described here: https://cellexplorer.org/datastructure/standard-cell-metrics/#waveform-based-metrics). The hardware used should always be specified since they can affect the waveforms of the processed units (e.g., filter characteristics and bandwidth) and compromise the separability of units on waveform characteristics.
Processing Module
From the input data, the processing module will generate cell metrics corresponding to the three-level description of neuronal firing (Fig. 1) and their relationship to experiment-specific behaviors (Suppl. Table 1 contains a representative list of metrics for illustration; full list is available at CellExplorer.org). The processing module is comprised of a single MATLAB script, ProcessCellMetrics.m, which computes metrics using a modular structure. The first-level description provides waveform features (filtered and wideband) and temporal features: interspike interval statistics (ISIs), and autocorrelograms (ACGs). Next, the unit parameters are used for the initial classification of single neurons into broad default classes: putative pyramidal cells, narrow waveform interneurons, and wide waveform interneurons. In experiments with silicon probes, the physical position relative to recording sites is also determined using spike amplitude trilateration (Petersen and Berg, 2016; Csicsvari et al., 2003). The user can generate a probe layout and save this to the data path. The Processing module (ProcessCellMetrics) will then detect and import the layout (see the channel map tutorial: https://cellexplorer.org/tutorials/channel-maps-tutorial/#channel-maps-tutorial). The main Matlab functions are described in supplementary table 2.
The second level relates single neuron spikes to the activity of other neurons and population patterns. These metrics include spike cross-correlograms (CCGs), quantitative identification of putative monosynaptic connections, phase relationships to various local field potential (LFP) patterns, and to unit population patterns. Monosynaptic connections, in turn, can be used to identify putative excitatory and inhibitory neurons and use this information to refine the primary unit classification (Fig. 1E; Barthó et al., 2004; English et al., 2017). All parameters can be customized according to the needs of each experimental paradigm (Suppl. Table 1; CellExplorer.org/datastructure/standard-cell-metrics).
The third-level metrics are used to assess the relationship between firing patterns of neurons and overt behaviors, including immobility, locomotion, and running speed. First to third level metrics can further be supported by other more direct methods, which can bind physiological parameters to genetically identified neuron groups (Boyden et al., 2005; Buzsáki et al., 2015; Roux and Buzsáki, 2015). Because these three-level metrics of single unit features are generalizable, they can be readily compared with similar analyses across laboratories, independent of paradigm-specific features. Towards these goals, the processing module automatically generates all cell metrics in a standardized fashion.
Features related to any behavioral paradigms, can also be computed, including manipulations (e.g., post-stimulus time histograms; PSTHs), behavioral tracking (spatial firing rate maps) and task related trial-wise response curves (e.g., response to a sensory cue).
Data structure
The data structure of CellExplorer (the format is documented online and summarized in Fig. 2 and suppl. Fig. 3) is organized in data containers and MATLAB structured arrays (“structs”), which functionally separate different data content, making them both easily interpretable (human-readable), machine-readable, expandable and flexible. It is derived from Buzcode (a MATLAB based data format for electrophysiological recordings and toolset developed communally in the Buzsaki lab; github.com/buzsakilab/buzcode), Neurosuite (neurosuite.sourceforge.net), and the FMA Toolbox (fmatoolbox.sourceforge.net). Using a data format in Matlab’s native mat files provides greater flexibility for day-to-day analysis, where the codebase can change rapidly with user-dependent requirements for saving derived data, than some of the recently developed data standards, such as Neurodata Without Borders (NWB; Teeters et al., 2015), which uses a single HDF5 container per session. Instead, scripts are available for translating standard fields of the data containers into NWB, including spikes, behavior, and events. The processing module also supports NWB as an input format.
The three most relevant structures are the session metadata struct, the spikes struct and the cell_metrics struct.
Session struct:
The session metadata struct contains all session-level experimental metadata (Suppl. Fig 1). A session is defined as a set of data typically recorded within the same day, in the same subject (also commonly referred to as a single dataset). The session struct has a modular structure (example modules: general, animal subject, extracellular, brain regions) that makes it flexible, expandable, and interpretable, and it offers a single structure, capable of handling a wide range of types of metadata related to extracellular data collection and processing. A metadata GUI (gui_session.m; Suppl. fig 2) allows for intuitive metadata entry and inspection, and a template script (sessionTemplate.m) can assist in both importing existing experimental metadata and generate relevant fields. Well-curated and documented metadata are crucial for reproducibility and always need to be linked to the electrophysiological data. See cellexplorer.org/datastructure/data-structure-and-format/ for more information.
Spikes struct:
The spike struct contains spike times and cluster IDs of all spikes. It also contains all basic spikes-derived fields, such as the average spike waveform of each cell and peak voltages. These fields are expandable.
Cell_metrics struct:
Modular structure containing all cell metrics calculated in the processing module. It consists of three types of data-fields for handling the diverse types of data: numeric double, character-cells, and structs. Single value metrics (e.g. peak voltage, firing rate, putative cell type) are stored in numeric double or character cells. Time series (e.g., waveforms), group data (e.g. synaptic connections and user-defined tags), and session parameters are stored in predefined struct modules. This structure makes the content machine-readable, including user-defined metrics, and provides expandability and flexibility (users can add their own metrics), while maintaining compatibility with the graphical interface. The single struct allows for processing multiple sessions together in the graphical interface (batch processing) and is convenient for sharing with collaborators and the broader scientific community in publications (see Supplementary Material and Supplementary Table 1 for a detailed description and https://cellexplorer.org/datastructure/standard-cell-metrics/). To address cross-platform compatibility, we have also provided two other cell metrics formats: Neurodata Without Borders (NWB) and json files. CellExplorer can save the cell metrics to these formats and load them back into the default Matlab struct format (please see the NWB tutorial for further info: https://cellexplorer.org/tutorials/nwb-tutorial/).
The CellExplorer graphical interface
The most important component of the framework is the user-friendly graphical interface for single cells, called CellExplorer (Fig. 3, Suppl. Video 1), which allows for characterization and exploration of all single unit metrics through a rich set of high-quality built-in interactive plots, neuron grouping, cross-level pointers, and filters. The user can interactively select plots and metrics to show in a highly dynamic manor. In the typical layout of CellExplorer with two rows of plots (Figure 3A and 3B top panel), the top row consists of population-level representations, and the bottom row consists of single cell features. Individual neurons can be selected from any plot, and the other features of the selected neurons will be automatically updated. The user can zoom and pan by scrolling and dragging any plot (Figure 3C). A middle mouse click links to the selected neuron and a right mouse click selects the neuron(s) from any of the plots for further actions. These selected groups can be displayed alone or highlighted and superimposed against data in the same session, multiple sessions, or the entire database. Clusters of neurons of interest can be selected by drawing polygons with the mouse cursor, and the other features of the selected groups are shown separately through group actions. Multiple group selections are also possible for both visualization and statistical comparison. Flexibility is assisted by side panels on both sides of the graphs. The left side panel contains options for the custom group plot, color groups, display settings, selection of single cell plots, and legends. The right panel contains single cell actions including navigation elements, cell assignment actions, tags, and a table with metrics. The left side-panel also includes a text field for custom filters (e.g. numeric and string filters). Below the graphs is a message log, keeping track of user actions. CellExplorer also has a built-in track record of all user actions, that can be undone in a stepwise manner. The group plotting options include a 2D-representation, a 3D-representation, a double histogram, a raincloud plot (Allen et al., 2021), and a customizable dimensionality reduction plot (t-SNE, PCA, and UMAP; Maaten and Hinton, 2008; See group plot options in Suppl. Fig. 4). Axis scaling can be either linear or logarithmic (Fig. 3F). Many of the cell features have multiple visualizations and metrics. Let’s take the spike waveform, as an example. It can be represented by 1) single average filtered/raw waveform from the channel with the largest amplitude, 2) in comparison to the z-scored/absolute waveforms of the population, 3) in comparison to group averages (e.g. cell types), 4) waveforms across the probe, both as a probe-layout-projected representation, as an image representation, and spike amplitude distribution 5) as well as through single cell metrics: trough-to-peak (a measure of the waveform width); ab-ratio (a measure of the asymmetry of the waveform), peak voltage (amplitude of the waveform), peak channel; 6) the temporal aspects of the spike amplitude across the entire recording, as a stability measure, can be visualized through spike-rasters (see a subset of the plotting options in Suppl. Fig 5).
Figure 3: Graphical interface.
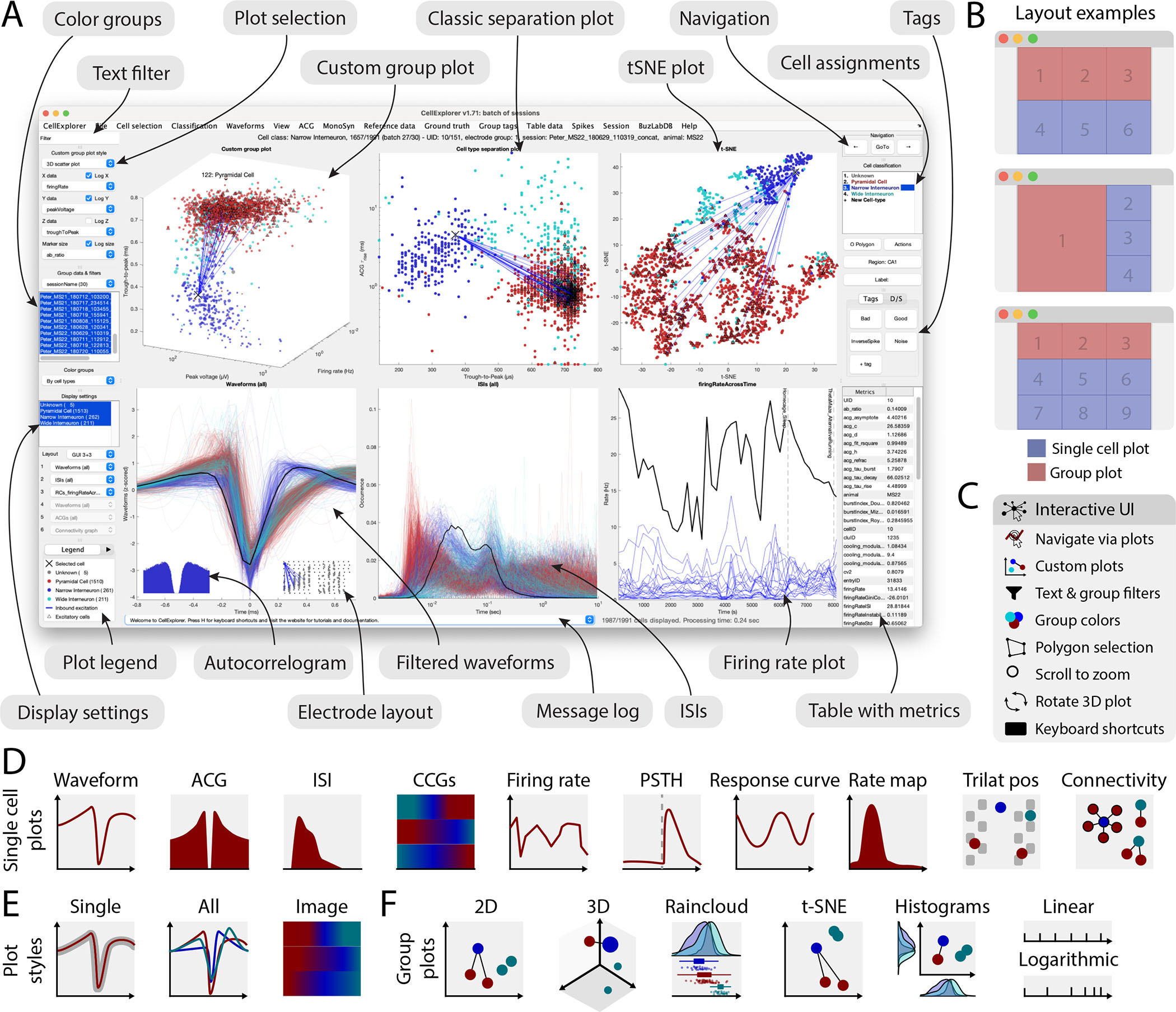
A. The interface consists of 4 to 9 main plots, where the top row is dedicated to population-level representations of the neurons. Other plots are selectable and customizable for individual neuron (e.g., single waveforms, ACGs, ISIs, CCGs, PSTHs, response curves, and firing rate maps). The surrounding interface consists of panels placed on either side of the graphs. The left side displays settings and population settings, including a custom plot panel, color group panel, display settings panel, and legends. The right side-panel displays single-cell dimensions, including a navigation panel, neuron assignment panel, tags, and a table with metrics. In addition, there is a text filter and a message log. B. Layout examples highlighting three configurations with 1–3 group plots and 3–6 single neuron plots. C. The interface has many interactive elements, including navigation and selection from plots (left mouse click links to selected cell and right mouse click selects the neuron from all the plots), visualization of monosynaptic connections, various data plotting styles (more than 30+ unique plots built-in), supports custom plots; plotting filters can be applied by text or selection, keyboard shortcuts, zooming any plot by mouse-scrolling and polygon selection of neurons D. Single cell plot options: waveform, Autocorellogram (ACG), Inter-spike-interval (ISI), firing rate across time, Post-Stimulus Time Histogram (PSTH), response curve, spatial firing rate maps, trilaterated neuronal position relative to recording sites, and monosynaptic connectivity graph. E. Most single cell plots have three representations: individual single cell representation, single cell together with the entire population with absolute amplitude and a normalized image representation (colormap). F. Group plotting options: 2D, 3D, raincloud plot, t-SNE, and double histogram. Each dimension can be plotted on linear or logarithmic axes. See also Supplementary Figure 4, 5, 6 and Supplementary Video 1.
Examples of the flexible operation of the graphical interface module are illustrated in Fig. 4 and described in more detail in Suppl. Video 1. Here we begin with motifs of monosynaptically connected clusters of neurons from the hippocampal CA1 area, as determined by the Processing Module (Fig. 4A). An example sub-network of connected neurons is highlighted in Fig. 4B with a selected single neuron to be characterized (arrow). Selected first to third level metrics of the neuron are displayed in Fig. 4 C to G. In several panels, the metrics of the selected neuron are shown against other neurons from the same dataset. A middle mouse click on any neuron will update all the panels for that particular cell, allowing quick screening and qualitative evaluation of multiple features. Neurons of interest can be marked for further quantitative comparisons. Next, first to third level metrics can be compared with paradigm-specific features of the selected neuron(s). For example, in case of hippocampal neurons, place field, trial-by-trial variability of firing patterns, travel direction firing specificity, spike phase precession relative to theta oscillation cycles, and multiple other features. During the data mining process, unexpected features and outliers may be noted, instabilities of neurons (‘drifts’) can be recognized, and artifacts identified visually. Such experimenter-supervised judgments are also essential for evaluating the quality of quantified data processing.
Figure 4. Data exploration example.
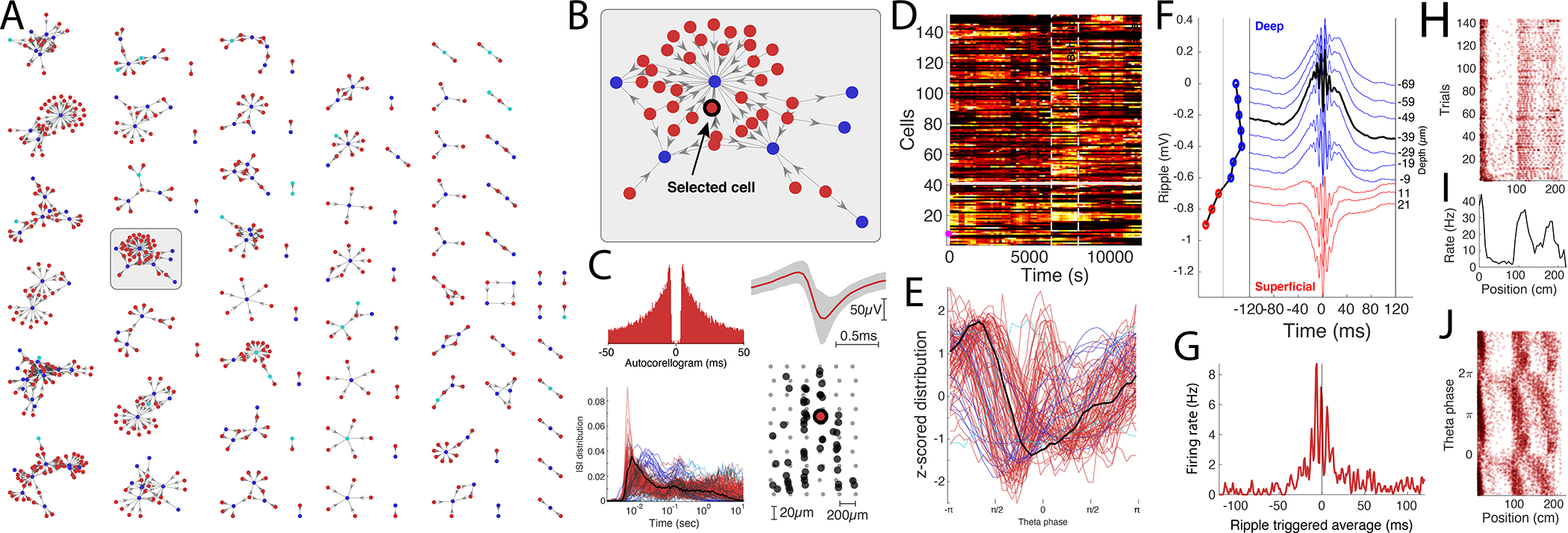
A. Connectivity graph with monosynaptic modules found across multiple datasets. Neurons are color-coded by their putative cell types (pyramidal cells in red, narrow interneurons in blue and wide interneurons in cyan). B. Highlighted monosynaptic module with single pyramidal cell highlighted (arrow). C. First level metrics: Auto-correlogram, average waveform (top row; gray area signifies the noise level of the waveforms), ISI distributions, with the selected neuron in black, and the physical location of the neurons relative to the multi-shank silicon probe. D. Firing rate across time for the population, each neuron is normalized to its peak rate. The session consists of three behavioral epochs: pre-behavior sleep, behavior (track running), and post-behavior sleep (boundaries shown with dashed lines). E. Theta phase distribution for all neurons recorded in the same session (red, pyramidal cells; blue, interneurons) during locomotion with the selected neuron highlighted (black line). F. Average ripple waveform for the electrode sites on a single shank. The site of the selected neuron is highlighted (dashed black line). The polarity of the average sharp wave is used to determine the position of the neuron relative to the pyramidal layer in CA1. G. Ripple wave-triggered PSTH for the selected neuron aligned to the ripple peak. H. Trial-wise raster for the selected neuron in a maze. I. The average firing rate of the neuron across trials. J. Spike raster showing the theta phase relationship to the spatial location of the animal.
Performance
CellExplorer takes advantage of Matlab’s lowest-level graphical plotting methods, and has optimized and enhanced plotting algorithms to make the UI responsive. Benchmarks were performed to characterize the performance of the CellExplorer GUI (Supplementary Fig. 6), which consists of multiple graphical elements with up to 9 individual simultaneous visualizations, panels, and interface elements, all of which are updated as the user navigates the single cell data.
The majority of the individual single cell plots (Figure 3D and Supple. Fig. 5) takes less than 80 ms to display (tested on an iMac from 2017 with a 4.2GHz Quad-Core Intel i7 with 32GB of ram), even when representing thousands of cells, but a subset of the visualizations increases substantially with the number of cells (the trace presentations with many data points per cell; e.g. ACGs, ISIs, waveforms, ISIs, theta phase; Supplementary Fig. 6A). Yet by introducing an overall display cap of 2000 simultaneously random cells in these representations, all of the visualizations take less than 80 ms, except the connectivity graph, where all connections are always shown. Updating the whole UI ranged from 180ms to 300ms, with the 2000 cells cap, for 4 and 9 subpanels respectively (Supplementary Fig. 6B). These benchmarks capture CellExplorer’s performance on real data and its scaling on larger dataset, yet reflect as well the graphical performance of Matlab, with built-in graphical hardware support, and the hardware configuration.
Putative cell type characterization
Currently, the processing pipeline classifies putative cell types based on two parameters, the width of the spike waveform (measured by the waveform trough-to-peak) and the burstiness of spiking (measured via the rise time of the ACG, τrise; Petersen and Buzsáki, 2020; Senzai and Buzsáki, 2017). The autocorrelogram of each cell (from –50ms to 50ms) is parametrized using a triple exponential fit (Suppl. Fig 7). One exponential fits the rise time of the ACGs, and two further exponentials fit the burst-related fast decay and the slower decaying part of the ACG. Using the waveform and burstiness criteria, units are tentatively segregated to narrow waveform (trough-to-peak ≤ 450 μs), wide waveform (trough-to-peak > 450 μs and τrise > 6 ms) putative interneurons and the rest as pyramidal cells (Figure 5A). These boundaries were optimized separately on data from the hippocampus and visual cortex, and can be adjusted for neurons in other brain regions when running the processing module. In a separate step, monosynaptic excitatory and inhibitory connections identified on the basis of their short-time cross-correlograms (Barthó et al., 2004; English et al., 2017), and can serve to verify the goodness of the first-order unit classification. In the dataset shown Figure 5, 39% of the pyramidal cells were determined to be excitatory (Suppl Figure 8A) and 5% of the narrow interneurons as inhibitory cells. There was a high degree of excitatory convergence on putative narrow interneurons (Suppl Fig 8B). Population average ACGs and waveforms are shown in Figure 5C. A t-SNE dimensionality reduction further supported the reliability of neuron separation (Figure 5D) and an agglomerative clustering determined similar clusters (Figure 5E). As more ground truth data will become available, the above classification scheme and boundaries will inevitably change (Fig. 5F). The reliability of any unit-behavior correlation can be compared to cluster quality metrics (Fig. 5G; Schmitzer-Torbert et al., 2005), increasing (or decreasing) the confidence in the validity of the correlation in question.
Figure 5. Community-based collaborations allow for improved single neuron characterization.
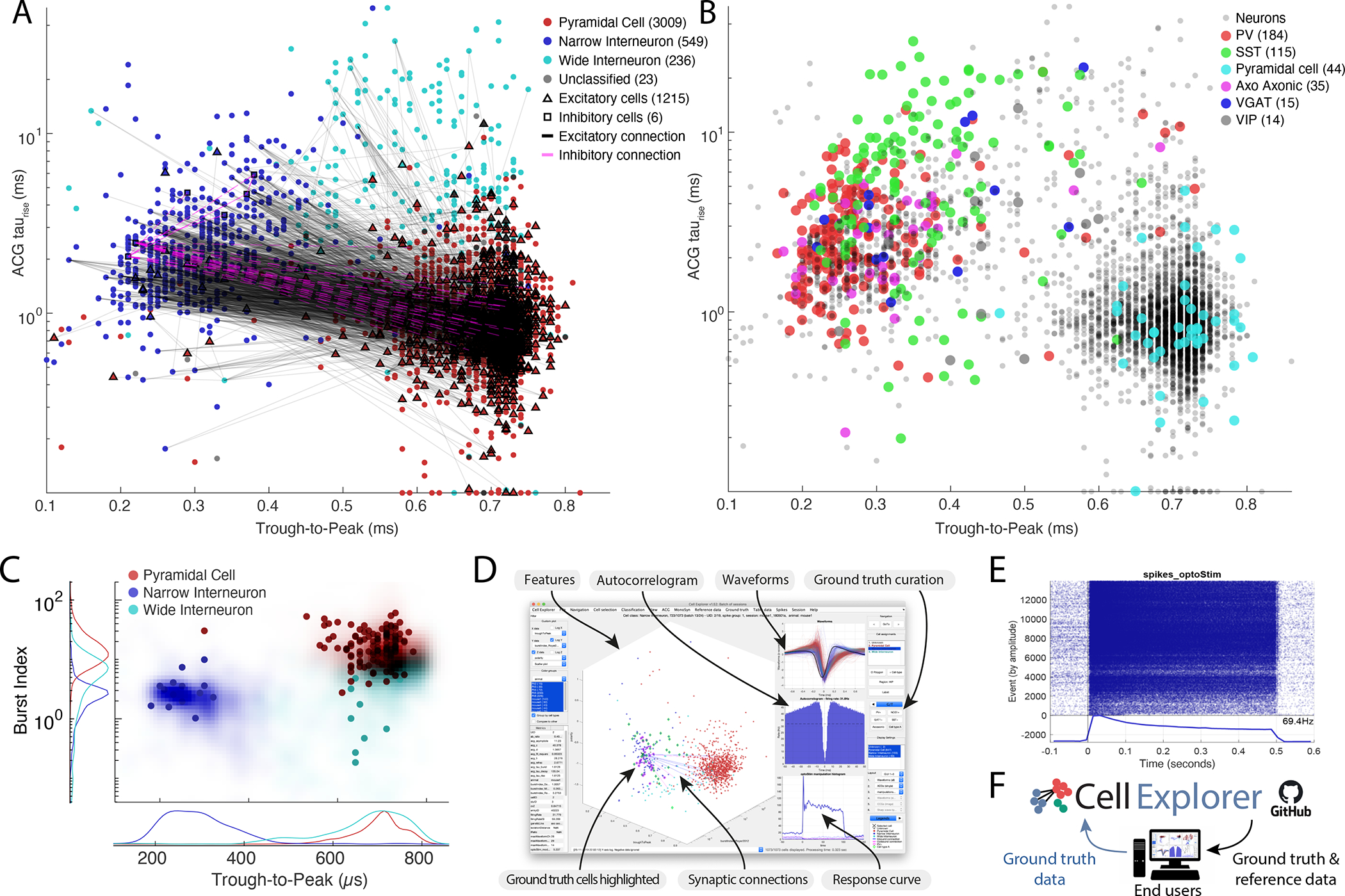
A. Distribution of putative cell types (3657 cells), including their projections determined via spike-transmissions CCG curves (Petersen and Buzsáki, 2020; Petersen et al., 2020) Excitatory and inhibitory cells determined from monosynaptic connections are highlighted with black triangles and magenta squares respectively. The marginal distributions are shown both as counts and probability distributions. B. Example ACGs for the three cell types and the ACG fit (black line). C. Top row: Average peak-normalized ACGs of the three cell types. bottom row: Average waveform for the three cell types (z-scored). D. t-SNE representation of the same cell population. E. Lower two panels: agglomerative clusters of data with 2 (left panel) and 3 clusters (right panel). F. 407 optogenetically identified neurons, including PV (184), SST (115), pyramidal cells (44), axo-axonic (35), VGAT (15) and VIP cells (14) projected onto the same population of neurons as in A (Sources: Allen Institute and Buzsáki lab; English et al., 2017; Senzai et al., 2019; Siegle et al., 2021). G. Isolation distance in cluster space for the population shown in A and I. See also Supplementary Figure 7 and 8.
NeuroScope2 – a data viewer for raw and processed data
To take advantage of the data types of CellExplorer and back-project the results to the recorded data, we also built a data viewer, NeuroScope2 (Figure 7). It maintains many of the original functions of NeuroScope (Hazan et al., 2006), while enhancing its performance to explore existing data, stream data being collected and handle multiple data streams simultaneously (e.g. digital or analog traces together with processed ephys data). As NeuroScope2 is written in Matlab, it is hackable, adaptable and easily expandable. It functions fully within the data types of CellExplorer, using the session struct for metadata, and supports the data containers of CellExplorer. NeuroScope2 can show the processed spikes and take advantage of the cell metrics to filter, group and color single units, as well as show behavioral, time series, states and event data (Figure 7B).
Figure 7. NeuroScope2 - A data viewer for raw and processed extracellular data acquired using multisite silicon probes, tetrodes or single wires.
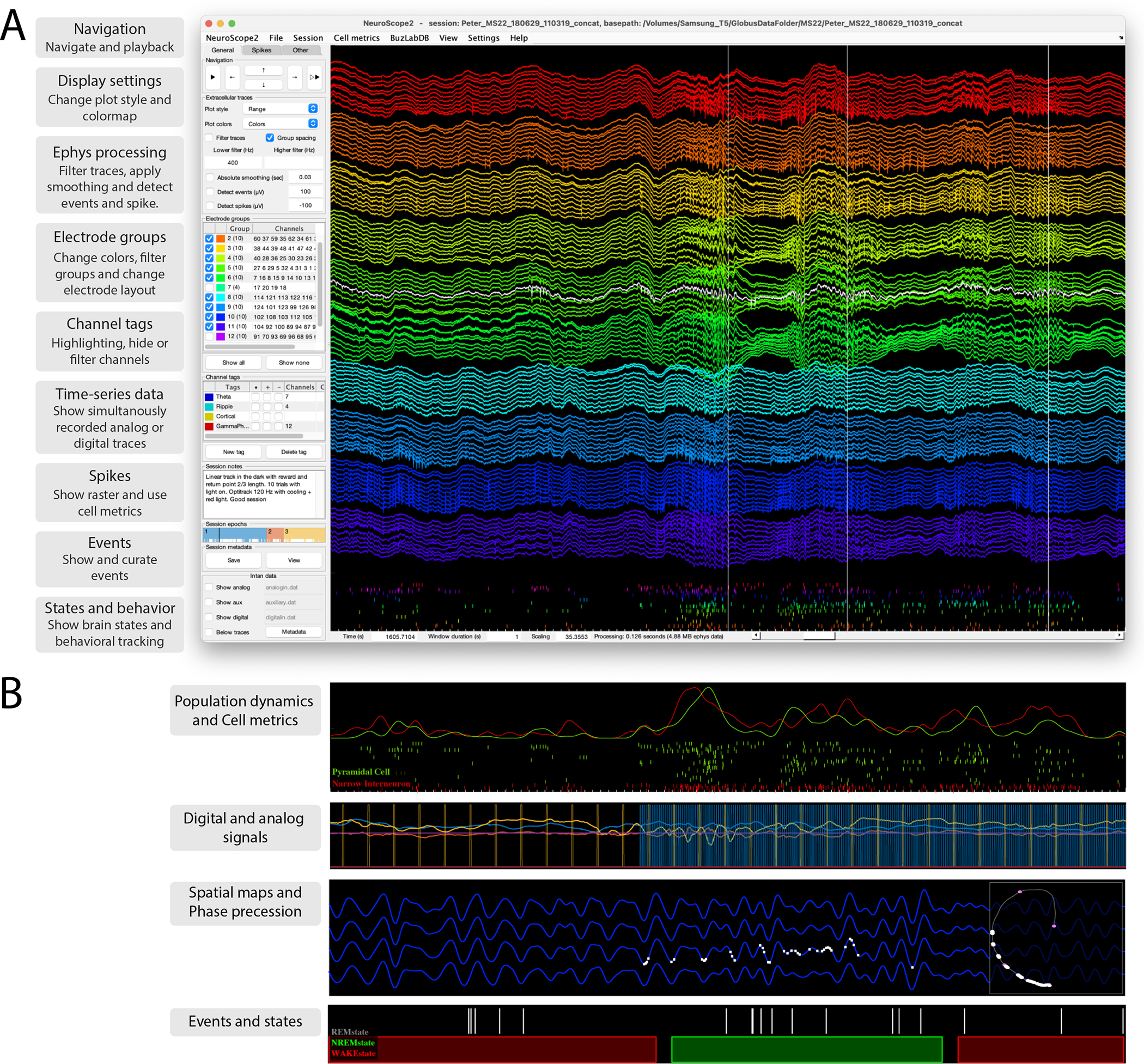
NeuroScope2 is written in Matlab, maintaining many of the original features of NeuroScope (neurosuite.sourceforge.net), but with many enhancements, and it is faster. It is easy to hack or modify, and supports and relies on the data types of CellExplorer. A. Screenshot of the graphical interface, showing a 128-channel recording from the rat hippocampus (window duration = 1 sec). Each colored groups of traces are from the same shank. The three vertical lines are detected temporal events (sharp-wave ripples; detection-channel is highlighted in white). The rasters below the traces are the spikes from curated single units. The left side panel consists of three tabs: General (panels: navigation, ephys traces, electrode groups, channel tags, session notes and epochs, and intan time-series), Spikes (panels: spikes, cell metrics, population dynamics) and Other (panels: events, states, time-series, and behavioral data). B. Various visualizations with NeuroScope2. Top: Population average curves and spiking dynamics of the same population of cells as in A, but color coded and grouped using the putative cell type determined via CellExplorer (duration: 1 sec). Second panel: two digital TTL pulses and 3D accelerometer data (mounted on the animal’s head). Digital data captured using the Intan acquisition system (the TTLs pulses are emitted by a 10Hz camera and a 120Hz behavioral tracking systems; window duration: 3 sec). Third panel: Ephys traces filtered in the theta band, with spikes of a single place cell plotted on the same trace (white bars; window duration: 3 sec), the right square shows the animals spatial trajectory (grey line) and the white points indicate the spatial location of the place field. Lowest panel: Event rater and states data (window duration: 50 sec).
The interface is user-friendly, with a single side panel for accessing most functions. Users can zoom, navigate, measure, highlight, and select traces directly with the mouse cursor, making manual inspection intuitive and efficient. NeuroScope2 can also perform basic data processing on the viewed traces, e.g. band-pass filter, perform temporal smoothing, generate single-channel spectrogram, perform Current Source Density (CSD) analysis, and detect spikes and events.
Value of large inter-laboratory datasets
While progress in discovery science often depends on investigator-unique approach to novel insights, standardization of data processing and screening is essential in fields where ‘big data’ generation is achieved through collaborative efforts. This applies to the current effort to quantitatively relate physiology-based and genetically classified cell types (Klausberger and Somogyi, 2008; McBain and Fisahn, 2001; Rudy et al., 2011). In each experiment, typically only one or a limited number of neuron types can be identified. Yet, combining datasets from numerous experiments and different laboratories can generate physiological metrics, grounded by other ‘ground truth’ data.
Fig. 5 also serves to illustrate the feasibility and utility of community-based approach. First to third level metrics of neurons recorded from the same brain region and layer can be combined from multiple experiments and laboratories and contrasted to the data quality of units recorded in a single session. An ever-growing data set allows for more reliable modality separation and characterization of neuron types. For example, the initial divisions of neurons into putative pyramidal cells, narrow and wide interneurons can be further refined by quantifying monosynaptic connections, increasing confidence of pyramidal cell–interneuron separation as well as identifying subsets of the unclassified group as interneurons (Fig. 5A) (Mizuseki et al., 2011; Petersen and Buzsáki, 2020; Peyrache et al., 2015; Stark et al., 2013).
Single neurons identified by opto-tagging, juxtacellular or other direct means (Ciocchi et al., 2015; Klausberger and Somogyi, 2008; Royer et al., 2012; Senzai et al., 2019; Stark et al., 2012; Zhang et al., 2013; Roux and Buzsáki, 2015) can be used to link first to third level features of initially classified neurons to genetically defined neuron types (Fig. 5A–E). Optogenetic methods can be supplemented by other more direct methods, such as simultaneous juxtacellular-extracellular or intracellular-extracellular recordings (Fig 5F; Harris et al., 2000; Neto et al., 2016). Having access to these ground truth labels may offer further support for the validity of physiological classification. An expected outcome of such growing dataset, containing ground truth-verified neurons, is trained models for classifying diverse neuron types based on physiological metrics alone. This is especially important for recordings in model organisms for which genetic manipulations are less tractable than in mice. Opto-tagged neurons can analyzed separately in CellExplorer (Suppl. Figure 8G–H). Further manual curation can be done while accessing the neuron’s other characteristics including waveforms, firing rates and connectivity. Communal contribution of ground truth data to CellExplorer is possible through the public GitHub repository (Suppl Figure 8I; visit CellExplorer.org for tutorials and further details).
Using large numbers of shared datasets, one can begin to compare brain regions, different electrode types and other features efficiently, using t-SNE plots (Fig. 5E). Such representations can highlight inconsistencies and differences across recording sessions, identify important regional and layer-specific differences, and alert for interspecies characteristics (Figure 8).
Figure 8. Exploration and comparison of metrics and cells across, species, subjects and brain regions.
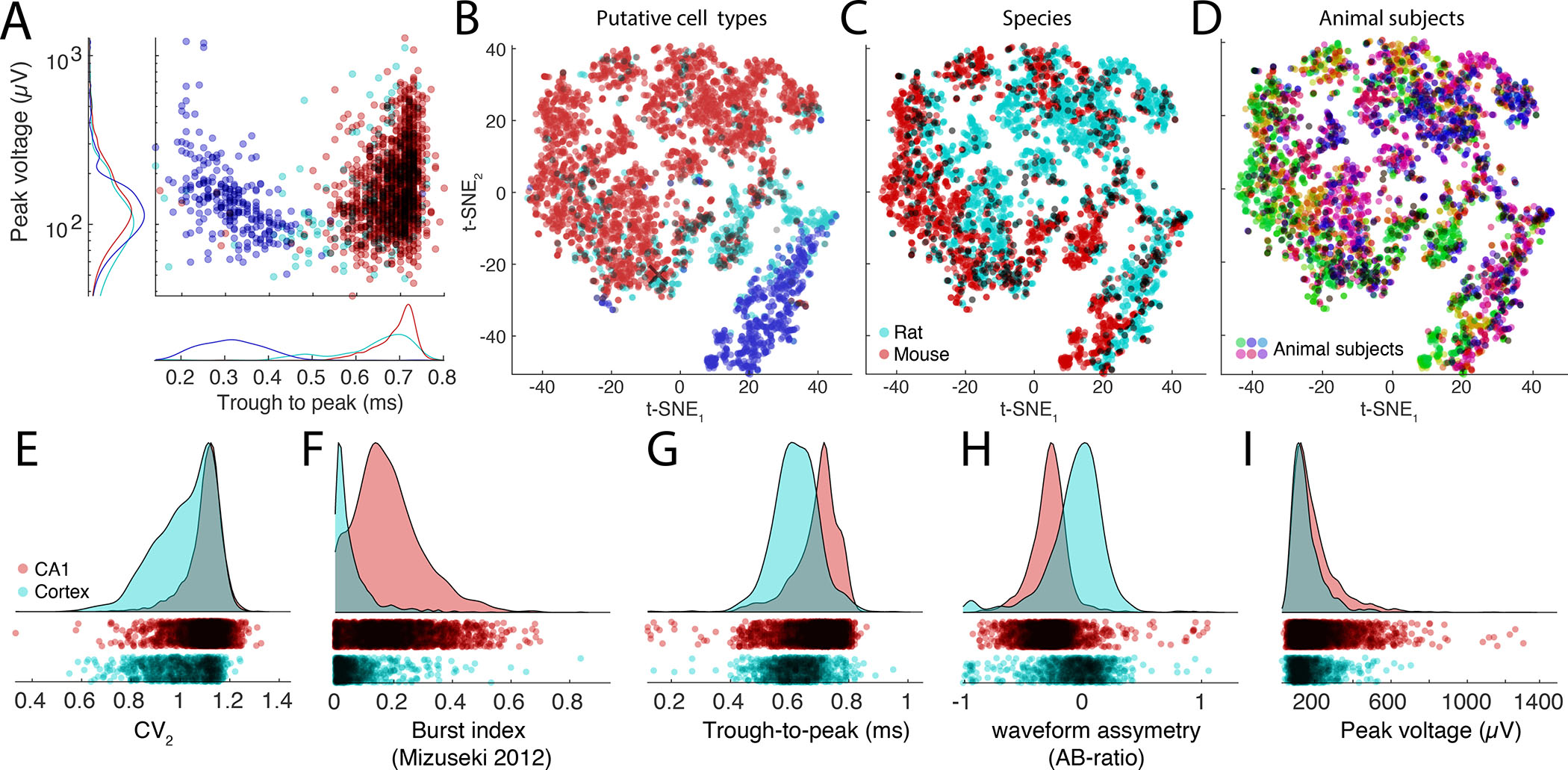
A. Distributions of spike amplitudes and waveform width (quantified by the trough to peak metrics) for the three groups from multiple CA1 datasets. Note inverse relationship between spike amplitude and waveform for putative interneurons. B-D. t-SNE representations of putative cell types (B), species (C, rat, and mouse in magenta and red, respectively) and subjects (D, colors scaled across subjects) for hippocampal neurons. E-I: Comparison of spike features of neurons recorded from CA1 pyramidal cells and visual cortex pyramidal cells. Significant differences are observed across several basic metrics, including CV2 (E), burst index (F), trough-to-peak (G), waveform asymmetry (H), and waveform peak voltage (I).
CellExplorer uses and shares data through our lab databank (buzsakilab.com/wp/database; Petersen et al., 2020). To demonstrate the value of inter-laboratory comparison, we processed datasets from the CA1 region of the hippocampus (Figure 6A) and visual cortex (Figure 6B) in freely moving mice (Senzai et al., 2019; Petersen and Buzsáki, 2020; Petersen et al., 2020), and comparable data from head-fixed mice from two large public datasets from University College London (Fig 6C; Steinmetz et al., 2019) and the Allen Institute (Figure 6D; (Siegle et al., 2021)). Through our database and CellExplorer, we currently share more than 79.000 processed neurons publicly. Datasets can be downloaded directly in CellExplorer and used as reference data or explored directly. The infrastructure is designed towards continually growing the public datasets and ground truth data for discovery science, cross-laboratory interactions, and reproducibility control. Processing data collected in different laboratories and investigators by the same program(s) will allow investigators to standardize protocols and achieve higher reliability of interlaboratory experiments between neuronal firing patterns and their behavioral, cognitive correlates.
Figure 6. Comparison of initial neuron classification by CellExplorer on large scale datasets from three different laboratories.
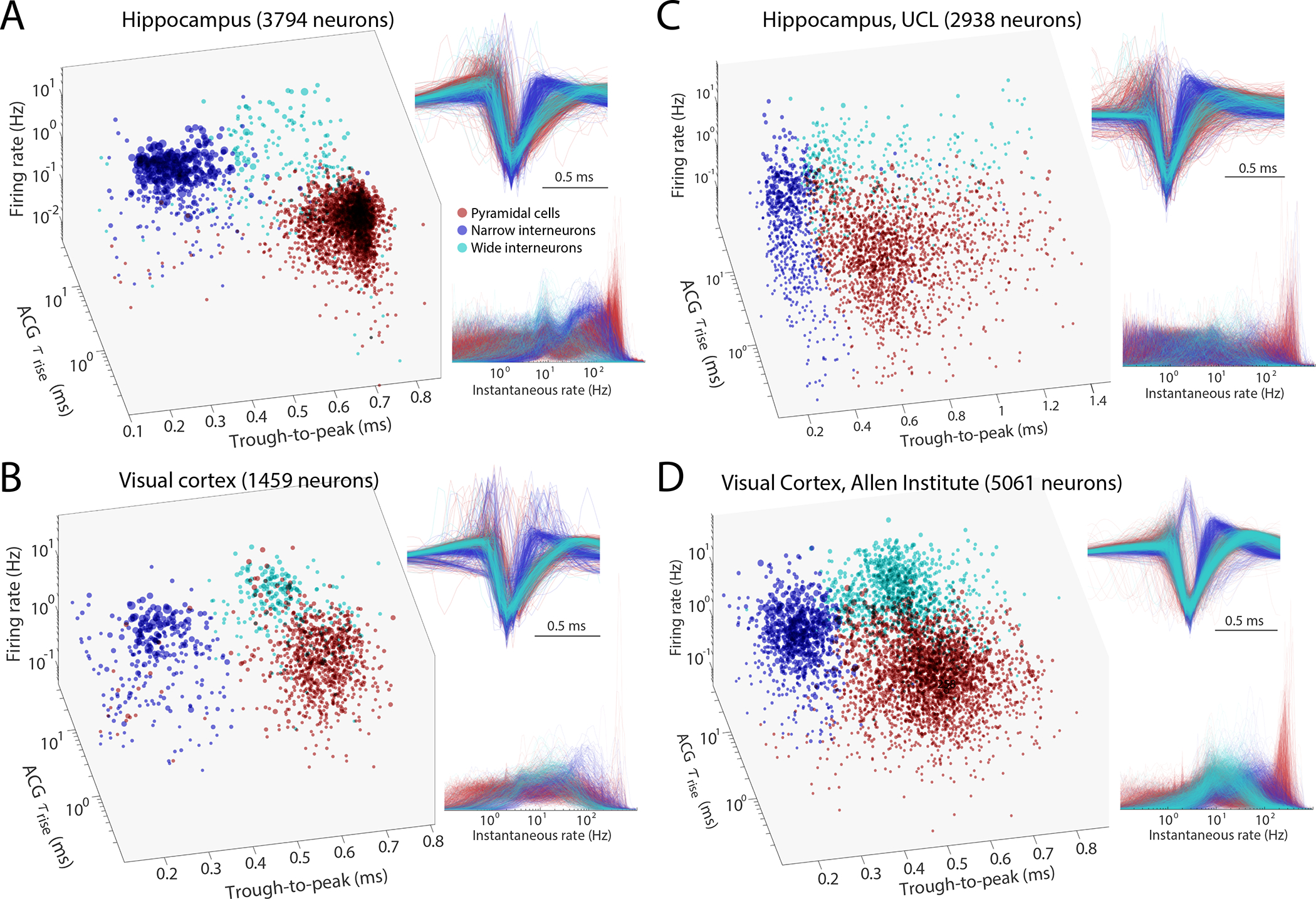
A. Data from hippocampus (Petersen and Buzsáki, 2020). B. Data from visual cortex (Senzai et al., 2019). C. Hippocampal and visual neurons selected from the UCL dataset (Steinmetz et al., 2019). D. Visual cortex cells from the Allen Institute (Siegle et al., 2021). Right panels across A-D: Z-scored waveforms across all neurons (top) and distribution of instantaneous rates (1/interspike intervals) across all neurons. A and B are based on long home cage (sleep) data (several hours), while C and D data are from short (~ 30 min) sessions in head-fixed, task-performing mice. See also Suppl. Fig. 8. Red, pyramidal cells; Blue, narrow waveform interneurons; Cyan, wide waveform interneurons.
Expandability of CellExplorer
CellExplorer comes with a long list of predefined metrics, advanced plots, ground truth data, classification schemes, allowing for standardized and efficient processing and visualization. Users can add their own metrics (numeric, strings or other more complex data) to the cell metrics, which will be available in CellExplorer (see tutorial in the Supplementary section and website for further details: https://cellexplorer.org/datastructure/expandability/). Further expandability and customization are achieved via Matlab package folders (“+folders”). Users can add their own custom calculations to the pipeline, implement custom classification schemes, provide preferences, create their own CellExplorer-plots, and incorporate their ground truth data. Relevant tutorials, example code, and templates are included online.
DISCUSSION
We have developed CellExplorer, an open-source, Matlab-based resource for characterizing single neurons and neuron types based on their biophysical features for collaborative analysis of data collected within and across laboratories. The CellExplorer platform enables visualization and analysis for users without the need to write code. Its modular format allows for fast and flexible comparisons of a large set of preprocessed physiological characteristics of single neurons and their interactions with other neurons, as well as their correlation with experimental variables. The code is publicly available on GitHub for users to download and to use the same standardized processing module on their local computers (Windows, OS X, and Linux). CellExplorer offers step-by-step online tutorials for first-time users. It is linked to the Allen Institute reference atlas to relate recording sites with structures and layers (Chon et al., 2019; Wang et al., 2020; https://atlas.brain-map.org/) with potential for expansion to other online resources that provide annotated data on putative neuron types.
Multiple-level characterization and classification of single neurons
To correctly interpret neuron firing-behavior/cognition relationships, numerous controls are needed to rule out or reduce the potential contribution of spurious variables. The Processing Module generates a battery of useful metrics for this purpose. In addition to the first level description of the biophysical characteristics of single neurons, it computes brain state-dependent firing rates, interspike interval variation, and relationships between single neurons and spiking activity of the population and LFP (second level). When behavioral data is also available, it can describe the relationship between single neuron firing patterns and routine behavioral parameters, such as immobility, walking, respiration, and pupil diameter (third level) or other task-independent measures, whose incidence or magnitude may change in the task. The third level metrics can help avoid e.g., inappropriately attributing spiking activity to high-level phenomena, such as learning, perception or decision making, that are often linked to overt movement and autonomic changes. Because these three-level metrics are independent of particular experimental paradigms, they can be used as benchmarks for assessing consistencies across experiments performed by different investigators in the same laboratory or across laboratories (Figure 6). Concatenating datasets obtained from the same brain regions and layers will create a continuously growing data bank. In turn, these data-rich sets make it possible to identify and quantify reliable boundaries among putative clusters and suggest inclusion and exclusion of parameters for a more refined separation of putative neuronal classes. Sets from different brain regions can be readily compared in order to identify salient differences.
Although several statistical tests are available in CellExplorer, it is not meant to substitute rigorous quantification. Instead, it is designed as a tool for flexible and fast comparison of parameters, facilitating visualization, interpretation and discovery. It is a complementary approach to dimensionality reduction and population analysis methods. Because assemblies of neurons consist of highly unequal partners (Buzsáki and Mizuseki, 2014), knowledge about the neuron-specific contribution to population measures is critical in many situations (Nicolelis and Lebedev, 2009). Such inequality may stem from unknowingly lumping neurons of different classes together into a single type and because even members of the same type belong to broad and skewed distribution and may contribute to different aspects of the experiment (Grosmark and Buzsáki, 2016).
Public cell metrics built from optotagged cells and reference sessions
Various classification schemes have been developed to assign extracellular spikes to putative pyramidal cells, interneurons, and their putative subtypes, based on a variety of physiological criteria. These include waveform features, firing rate statistics in different brain states, embeddedness in various population activities, firing patterns characterized by their autocorrelograms, and putative monosynaptic connections to other neurons (Barthó et al., 2004; Csicsvari et al., 1999; Fujisawa et al., 2008; Mizuseki et al., 2009; Okun et al., 2015; Sirota et al., 2008). Increasingly larger datasets will likely improve such physiology-based classification. Yet, the ‘ground truth’ for these classifying methods is largely missing. There is a lack of agreement about neuronal ‘types’ across laboratories and even for data collected in the same laboratory different criteria are applied in different experiments. Optogenetic tagging (Boyden et al., 2005) offers such grounding by connecting putative subtypes based on physiologically distinct features to their molecular identities. Because in a single animal only one or few neuron types can be tagged optogenetically or identified by other direct methods (Fosque et al., 2015; Klausberger and Somogyi, 2008), refinement of a library of physiological parameters should be conducted iteratively, so that in subsequent experiments the various neuron types can be recognized reliably by using solely physiological criteria (English et al., 2017; Royer et al., 2012; Senzai and Buzsáki, 2017, 2017; Roux and Buzsáki, 2015). In turn, knowledge about the molecular identity of the different neuronal components of a circuit can considerably improve the interpretation of correlational observations provided by large-scale extracellular recordings.
CellExplorer provides the end user with access to processed cell metrics of a wide range of opto-tagged cells and reference data (~78.000 neurons) collected in our database. These features can serve as benchmarks for comparison with data collected in any other laboratory, and can assist with the initial neuron classification into the broad groups of pyramidal cells and interneurons, many of which are identified physiologically by their monosynaptic connections. The derived features also offer normative information about spikes characteristics, firing rates and spike dynamics, as captured in Figure 8. The ground truth neurons are included in the CellExplorer GitHub repository and can be loaded from CellExplorer.
Comparison with other frameworks and tools
Several community efforts exist for machine- and human-readable databases of various morphological and transcriptomic features of neurons (Ascoli et al., 2007; Cembrowski et al., 2016; Sanchez-Aguilera et al., 2021; Tecuatl et al., 2021; Wheeler et al., 2015; neuromorpho.org/, Hipposeq.janelia.org, Hippocampome.org), which served as inspiration for our efforts. Yet, post-spike sorting tools are rare. NeuroExplorer (http://www.neuroexplorer.com/; Nex Technologies, USA) is the largest commercial solution, yet costs thousands of $ for a license, it is written in C but supports Matlab and python code. Brainstorm (Tadel et al., 2011), another application for analysis of brain recordings, recently began supporting ephys data through the IN-Brainstorm expansion (Nasiotis et al., 2019). Yet, Brainstorm is mostly focused on noninvasive techniques and the application has limited tools for analyzing single cells. The user interfaces of CellExplorer could potentially be integrated into Brainstorm. FieldTrip (Oostenveld et al., 2010), is another very popular open-source Matlab software toolbox for MEG, EEG and iEEG analysis, but has a limited scope for single cell analysis.
CellExplorer is open-source with all code, data and documentation available online. We chose Matlab, a commercial solution as a platform, because it allowed us to build CellExplorer on existing Matlab tools and historically popular toolboxes, and also because Matlab is a widely used coding platform in many electrophysiological laboratories in academia. An alternative for us was to build CellExplorer around Neurodata Without Border (NWB; Teeters et al., 2015). NWB has numerous advantages but provides limited flexibility and is not ideal for a day-to-day data analysis format with high demand for flexibility. Therefore, other approaches which enhance flexibility and alternative options are warranted. CellExplorer supports reading spike data from NWB files, thus allows to combine the advantages of both platforms, including CellExplorer’s machine-readable code. CellExplorer also supports saving the metrics to a NWB 2.0 file (Rübel et al., 2019; Teeters et al., 2015), and load the nwb file back into the default Matlab cell_metrics struct. A dedicated nwb tutorial is available here: https://cellexplorer.org/tutorials/nwb-tutorial/. Handling the cell metrics in simpler json files is also supported. CellExplorer could be translated to an open platform like python or Julia, pending on demand.
Outlook and future directions
Through community efforts, there are many future potential directions for using CellExplorer. CellExplorer allows for user-friendly submission of ground truth (e.g., opto-tagged) data that can be shared with the community via the GitHub repository. To date, CellExplorer hosts the largest collection of publicly available, characterized single neurons. It also has the largest collection of opto-tagged cells, which will help link physiological markers and characteristics to genetic traits. Through implementation of publicly shared data by contributing neuroscientists, its scope can be expanded further for better coverage of several brain regions, species and behaviors.
Development and availability
Development takes place in a public code repository at github.com/petersenpeter/CellExplorer. All examples in this article have been calculated with the pipeline and plotted with CellExplorer. Extensive documentation, including installation instructions, tutorials, description of all metrics and their calculations, is available at CellExplorer.org and hosted at the GitHub repository. CellExplorer is available for MATLAB 2017B and forward, and for the operating systems Windows, OS X, and Linux. Compiled versions of the graphical interfaces (CellExplorer, NeuroScope2, and the session GUI) are available at the CellExplorer website for usage on computer systems without a Matlab license. More information can be found at CellExplorer.org. All data presented is available from https://buzsakilab.com/wp/database/ (Petersen et al., 2020). We pledge to continue to support CellExplorer and are eager to incorporate data generated by other laboratories. CellExplorer should be viewed as a small but necessary step towards FAIR practice in collaborative neuroscience and the emerging novel technical platforms that will facilitate data sharing and interlaboratory collective research.
TUTORIALS
Two tutorials are available in the STAR Methods section: A general tutorial on the full pipeline, and a tutorial on how to add your own metrics. There are many more detailed tutorials online, covering: the generation of the metadata struct, the manual curation process, generating spike raster plots, connections, performing opto-tagging, using ground truth data, export figure, and many other topics.
Tutorials are available at CellExplorer.org/tutorials/tutorials.
STAR Methods
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, György Buzsáki (gyorgy.buzsaki@nyulangone.org).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All data is available from our databank at https://buzsakilab.com/wp/database/ (Petersen et al., 2018). All code is available at GitHub: https://github.com/petersenpeter/CellExplorer. All resources, including tutorials and documentation are on the CellExplorer website at: https://cellexplorer.org/.
Experimental model and subject details
All experimental data was collected in previous studies. Details of experimental methods and subject information are available in the original studies (Petersen and Buzsáki, 2020; Senzai et al., 2019; Siegle et al., 2021; Steinmetz et al., 2019).
CellExplorer layout and preferences saved between sessions
The display preferences are saved between sessions, which provides a simpler interface for novice users, while maintaining a more advanced display for experienced users. The preferences can be reset from the view menu by clicking Reset Layout/Preferences, or by deleting the preference file last_preferences_CellExplorer.mat located in the folder calc_CellMetrics.
General tutorial
This tutorial covers the processing, from generating the necessary session metadata using the template, running the processing pipeline, opening multiple sessions for manual curation in CellExplorer, and finally using the cell_metrics for filtering cells. The tutorial is also available as a Matlab script: (tutorials/CellExplorer_Tutorial.m).
-
1
Define the basepath of the dataset to process. The dataset should ideally consist of the raw data basename.dat and spike sorted data.
basepath = ‘/your/data/path/basename/’;
cd(basepath)
-
2
Generate the session metadata struct using the template script and display the metadata in the session GUI (Suppl figure 2)
session = sessionTemplate(basepath, ‘showGUI’, true);
You can use the GUI to inspect and manually add metadata. Make sure the extracellular tab is filled out correctly for your data (Suppl figure 2C), The template script can extract existing metadata from a NeuroScope compatible basename.xml file, from intan’s info.rhd, from KiloSort’s rez.mat fiel and from a basename.sessionInfo.mat file (buzcode).
-
3
Run the cell metrics pipeline ProcessCellMetrics using the session struct as input
cell_metrics = ProcessCellMetrics(‘session’, session, ‘showGUI’, true);
Setting showGUI to true will display the session GUI with a CellExplorer tab allowing you to verify parameters and settings (screenshot shown in Suppl figure 2D). You can click the button Verify metadata to show a summary table with metadata relevant to the processing. Fields requiring your attention will be highlighted in red; optional fields in blue.
-
4
Visualize the cell metrics in CellExplorer
cell_metrics = CellExplorer(‘metrics’, cell_metrics);
-
5
You can repeat step 1–4 on a couple of datasets and load them together in CellExplorer, providing several paths
basepaths = {‘path/to/session1’,’path/to/session2’};
basenames = {‘session1’,’session2’};
cell_metrics = loadCellMetricsBatch(‘basepaths’, basepaths, ‘basenames’, basenames);
cell_metrics = CellExplorer(‘metrics’, cell_metrics);
-
6
Curate your cells in CellExplorer and save the metrics via the file menu in CellExplorer.
-
7You may use the script loadCellMetrics for further analysis using the metrics as filters:
-
1Get cells labeled as Interneuron
cell_metrics_idxs1 = loadCellMetrics(‘cell_metrics’, cell_metrics, ‘putativeCellType’, {‘Interneuron’});-
2Get cells that have the groundTruthClassification label Axoaxonic
-
1
cell_metrics_idxs2 = loadCellMetrics(‘cell_metrics’, cell_metrics, ‘groundTruthClassification’, {‘Axoaxonic’});
Expandability tutorial: add you own custom metrics
This tutorial covers how to add your custom cell metrics. For single value metrics you have two options: numeric values or string arrays. Numeric metrics can be plotted in the custom group plot in CellExplorer. String arrays allow you to group your data by the unique strings set within features, and can be plotted in discrete values. All features in the cell metrics are automatically available in CellExplorer if they contain N values (N: number of cells).
Add a string metric to your cell_metrics:
Let’s say you want to add a cell metric describing cortical layers for each cell, using predefined labels (Layer 1 to Layer 6). This can be stored as a char cell array, e.g.:
cell_metrics.corticalLayer = {‘layer 5’,’layer 4’,’layer 2’,’layer 2/3’,’layer 1’}; % nCells = 5
Add numeric values to your cell metrics:
Let’s say you want to add the preferred orientation of a drifting grating presented to cells in the visual cortex. This will be stored as numeric values, e.g.:
cell_metrics.pref_ori_dg = [90,25,45,80,30]; % nCells = 5
Now, load the cell metrics into CellExplorer to visualize them. The fields will appear in the drop-down menus in the custom group plot:
cell_metrics = CellExplorer(‘metrics’,cell_metrics);
If you open multiple sessions in CellExplorer, the custom metrics will automatically be imported. Cells without numeric values will have NaN values assigned and empty strings for missing char fields.
You can also incorporate response curves and other more advanced metrics, and perform custom calculations in the ProcessCellMetrics script by using the custom calculation implementation: https://cellexplorer.org/pipeline/custom-calculations/
For further plotting options, please see the website:
Supplementary Material
Supplementary video 1. Demonstration of the CellExplorer interface, related to figure 3. The supplementary video is available on YouTube, at CellExplorer.org, and at https://buzsakilab.com/CellExplorer/CellExplorerMovie.mp4
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Hippocampal dataset | (Petersen and Buzsáki, 2020) | https://buzsakilab.com/wp/projects/entry/4919/ |
| Visual cortex dataset | (Senzai et al., 2019) | https://buzsakilab.com/wp/projects/entry/22682/ |
| UCL dataset | (Steinmetz et al., 2019) |
https://figshare.com/articles/dataset/Eight-probe_Neuropixels_recordings_during_spontaneous_behaviors/7739750
https://buzsakilab.com/wp/projects/entry/52347/ |
| Allen Institute dataset | (Siegle et al., 2021) |
https://allensdk.readthedocs.io/en/latest/visual_coding_neuropixels.html
https://buzsakilab.com/wp/projects/entry/52635/ |
| Software and algorithms | ||
| CellExplorer | Petersen and Buzsáki | CellExplorer.org |
| MATLAB | MathWorks | www.mathworks.com |
Highlights.
An open-source framework for single cell characterization and visualization
A processing module that calculates a set of standardized physiological metrics
A graphical interface to explore computed features at the speed of a mouse click
Petersen et al. present a framework allowing users to process, curate and relate their data to a public collection of neurons. It aims to link genetically identified cell types and physiological properties of neurons collected across laboratories with potential to lead to interlaboratory standard of single cell metrics.
ACKNOWLEDGEMENTS
We thank Sam McKenzie, Mihály Vöröslakos, Michelle Hernandez, Thomas Hainmueller, for helping with documentation or implementation of CellExplorer code. Further, we thank Sam McKenzie, Daniel English, Roman Huszar, and Yuta Senzai for providing ground truth datasets. Finally, thanks go to Thomas Hainmueller, Manuel Valero, Antonio Hernandez-Ruiz, Viktor Varga, and Omid Yaghmazadeh for comments on the manuscript. Joshua H. Siegle thanks the Allen Institute founder, Paul G. Allen, for his vision, encouragement, and support. Supported by U19 NS107616, U19 NS104590, R01 MH122391, the Kavli Foundation and The Lundbeck Foundation.
Footnotes
Declaration of Interests
The authors declare no conflicting interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allen M, Poggiali D, Whitaker K, Marshall TR, van Langen J, and Kievit RA (2021). Raincloud plots: a multi-platform tool for robust data visualization. Wellcome Open Res 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli GA, Donohue DE, and Halavi M (2007). NeuroMorpho.Org: A Central Resource for Neuronal Morphologies. J Neurosci 27, 9247–9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow HB (1972). Single Units and Sensation: A Neuron Doctrine for Perceptual Psychology? Perception 1, 371–394. [DOI] [PubMed] [Google Scholar]
- Barthó P, Hirase H, Monconduit L, Zugaro M, Harris KD, and Buzsáki G (2004). Characterization of neocortical principal cells and interneurons by network interactions and extracellular features. J. Neurophysiol. 92, 600–608. [DOI] [PubMed] [Google Scholar]
- Bishop PO, Burke W, and Davis R (1962). Single-unit recording from antidromically activated optic radiation neurones. J Physiol 162, 432–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard KE, Aimone JB, Chun M, Dean T, Denker M, Diesmann M, Donofrio DD, Frank LM, Kasthuri N, Koch C, et al. (2016). High-Performance Computing in Neuroscience for Data-Driven Discovery, Integration, and Dissemination. Neuron 92, 628–631. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, and Deisseroth K (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Buzsáki G (2004). Large-scale recording of neuronal ensembles. Nature Neuroscience 7, 446–451. [DOI] [PubMed] [Google Scholar]
- Buzsáki G, and Mizuseki K (2014). The log-dynamic brain: how skewed distributions affect network operations. Nat Rev Neurosci 15, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsáki G, Stark E, Berényi A, Khodagholy D, Kipke DR, Yoon E, and Wise K (2015). Tools for probing local circuits: high-density silicon probes combined with optogenetics. Neuron 86, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembrowski MS, Wang L, Sugino K, Shields BC, and Spruston N (2016). Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. ELife 5, e14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon U, Vanselow DJ, Cheng KC, and Kim Y (2019). Enhanced and unified anatomical labeling for a common mouse brain atlas. Nature Communications 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JE, Magland JF, Barnett AH, Tolosa VM, Tooker AC, Lee KY, Shah KG, Felix SH, Frank LM, and Greengard LF (2017). A Fully Automated Approach to Spike Sorting. Neuron 95, 1381–1394.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciocchi S, Passecker J, Malagon-Vina H, Mikus N, and Klausberger T (2015). Selective information routing by ventral hippocampal CA1 projection neurons. Science 348, 560–563. [DOI] [PubMed] [Google Scholar]
- Csicsvari tJozsef, Hirase H, Czurkó A, Mamiya A, and Buzsáki G (1999). Fast Network Oscillations in the Hippocampal CA1 Region of the Behaving Rat. J. Neurosci. 19, RC20–RC20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csicsvari J, Henze DA, Jamieson B, Harris KD, Sirota A, Barthó P, Wise KD, and Buzsáki G (2003). Massively Parallel Recording of Unit and Local Field Potentials With Silicon-Based Electrodes. Journal of Neurophysiology 90, 1314–1323. [DOI] [PubMed] [Google Scholar]
- English DF, McKenzie S, Evans T, Kim K, Yoon E, and Buzsáki G (2017). Pyramidal Cell-Interneuron Circuit Architecture and Dynamics in Hippocampal Networks. Neuron 96, 505–520.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosque BF, Sun Y, Dana H, Yang C-T, Ohyama T, Tadross MR, Patel R, Zlatic M, Kim DS, Ahrens MB, et al. (2015). Labeling of active neural circuits in vivo with designed calcium integrators. Science 347, 755–760. [DOI] [PubMed] [Google Scholar]
- Fujisawa S, Amarasingham A, Harrison MT, and Buzsáki G (2008). Behavior-dependent short-term assembly dynamics in the medial prefrontal cortex. Nature Neuroscience 11, 823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouwens NW, Sorensen SA, Berg J, Lee C, Jarsky T, Ting J, Sunkin SM, Feng D, Anastassiou CA, Barkan E, et al. (2019). Classification of electrophysiological and morphological neuron types in the mouse visual cortex. Nature Neuroscience 22, 1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmark AD, and Buzsáki G (2016). Diversity in neural firing dynamics supports both rigid and learned hippocampal sequences. Science 351, 1440–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, and Buzsáki G (2000). Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J Neurophysiol 84, 401–414. [DOI] [PubMed] [Google Scholar]
- Hazan L, Zugaro M, and Buzsáki G (2006). Klusters, NeuroScope, NDManager: A free software suite for neurophysiological data processing and visualization. Journal of Neuroscience Methods 155, 207–216. [DOI] [PubMed] [Google Scholar]
- Jia X, Siegle JH, Bennett C, Gale SD, Denman DJ, Koch C, and Olsen SR (2019). High-density extracellular probes reveal dendritic backpropagation and facilitate neuron classification. Journal of Neurophysiology 121, 1831–1847. [DOI] [PubMed] [Google Scholar]
- Kepecs A, and Fishell G (2014). Interneuron cell types are fit to function. Nature 505, 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, and Somogyi P (2008). Neuronal Diversity and Temporal Dynamics: The Unity of Hippocampal Circuit Operations. Science 321, 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SQ, Hromádka T, Znamenskiy P, and Zador AM (2009). PINP: A New Method of Tagging Neuronal Populations for Identification during In Vivo Electrophysiological Recording. PLoS One 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaten L. van der, and Hinton G (2008). Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605. [Google Scholar]
- Martone M, Gerkin R, Moucek R, Das S, Goscinski W, Hellgren-Kotaleski J, Kennedy D, Leergaard T, Boline J, and Abrams M (2020). NIX – Neuroscience information exchange format. F1000Research 9. [Google Scholar]
- McBain CJ, and Fisahn A (2001). Interneurons unbound. Nat. Rev. Neurosci. 2, 11–23. [DOI] [PubMed] [Google Scholar]
- McGinley MJ, David SV, and McCormick DA (2015). Cortical Membrane Potential Signature of Optimal States for Sensory Signal Detection. Neuron 87, 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuseki K, Sirota A, Pastalkova E, and Buzsáki G (2009). Theta Oscillations Provide Temporal Windows for Local Circuit Computation in the Entorhinal-Hippocampal Loop. Neuron 64, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuseki K, Diba K, Pastalkova E, and Buzsáki G (2011). Hippocampal CA1 pyramidal cells form functionally distinct sublayers. Nature Neuroscience 14, 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiotis K, Cousineau M, Tadel F, Peyrache A, Leahy RM, Pack CC, and Baillet S (2019). Integrated open-source software for multiscale electrophysiology. Scientific Data 6, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto JP, Lopes G, Frazão J, Nogueira J, Lacerda P, Baião P, Aarts A, Andrei A, Musa S, Fortunato E, et al. (2016). Validating silicon polytrodes with paired juxtacellular recordings: method and dataset. Journal of Neurophysiology 116, 892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolelis MAL, and Lebedev MA (2009). Principles of neural ensemble physiology underlying the operation of brain–machine interfaces. Nature Reviews Neuroscience 10, 530–540. [DOI] [PubMed] [Google Scholar]
- Okun M, Steinmetz NA, Cossell L, Iacaruso MF, Ko H, Barthó P, Moore T, Hofer SB, Mrsic-Flogel TD, Carandini M, et al. (2015). Diverse coupling of neurons to populations in sensory cortex. Nature 521, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenveld R, Fries P, Maris E, and Schoffelen J-M (2010). FieldTrip: Open Source Software for Advanced Analysis of MEG, EEG, and Invasive Electrophysiological Data. Computational Intelligence and Neuroscience 2011, e156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M, Steinmetz NA, Kadir SN, Carandini M, and Harris KD (2016). Fast and accurate spike sorting of high-channel count probes with KiloSort. In Advances in Neural Information Processing Systems 29, [Google Scholar]
- Lee DD, Sugiyama M, Luxburg UV, Guyon I, and Garnett R, eds. (Curran Associates, Inc.), pp. 4448–4456. [Google Scholar]
- Petersen PC, and Berg RW (2016). Lognormal firing rate distribution reveals prominent fluctuation–driven regime in spinal motor networks. ELife 5, e18805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PC, and Buzsáki G (2020). Cooling of Medial Septum Reveals Theta Phase Lag Coordination of Hippocampal Cell Assemblies. Neuron 107, 731–744.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PC, Hernandez M, and Buzsáki G (2018). Public electrophysiological datasets collected in the Buzsaki Lab. 10.5281/zenodo.3629881 (Zenodo; ). [DOI] [Google Scholar]
- Petersen PC, Hernandez M, and Buzsáki G (2020). The Buzsaki Lab Databank - Public electrophysiological datasets from awake animals (Zenodo; ). [Google Scholar]
- Peyrache A, Lacroix MM, Petersen PC, and Buzsáki G (2015). Internally organized mechanisms of the head direction sense. Nat Neurosci 18, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga RQ, Nadasdy Z, and Ben-Shaul Y (2004). Unsupervised Spike Detection and Sorting with Wavelets and Superparamagnetic Clustering. Neural Computation 16, 1661–1687. [DOI] [PubMed] [Google Scholar]
- Roux L, and Buzsáki G (2015). Tasks for inhibitory interneurons in intact brain circuits. Neuropharmacology 88, 10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Losonczy A, Kim J, Chance F, Magee JC, and Buzsáki G (2012). Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nature Neuroscience 15, 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rübel O, Tritt A, Dichter B, Braun T, Cain N, Clack N, Davidson TJ, Dougherty M, Fillion-Robin J-C, Graddis N, et al. (2019). NWB:N 2.0: An Accessible Data Standard for Neurophysiology. BioRxiv 523035. [Google Scholar]
- Rudy B, Fishell G, Lee S, and Hjerling-Leffler J (2011). Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Developmental Neurobiology 71, 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Aguilera A, Wheeler DW, Jurado-Parras T, Valero M, Nokia MS, Cid E, Fernandez-Lamo I, Sutton N, García-Rincón D, Prida LM, de la, et al. (2021). An update to Hippocampome.org by integrating single-cell phenotypes with circuit function in vivo. PLOS Biology 19, e3001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzer-Torbert N, Jackson J, Henze D, Harris K, and Redish AD (2005). Quantitative measures of cluster quality for use in extracellular recordings. Neuroscience 131, 1–11. [DOI] [PubMed] [Google Scholar]
- Sejnowski TJ, Churchland PS, and Movshon JA (2014). Putting big data to good use in neuroscience. Nature Neuroscience 17, 1440–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senzai Y, and Buzsáki G (2017). Physiological Properties and Behavioral Correlates of Hippocampal Granule Cells and Mossy Cells. Neuron 93, 691–704.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senzai Y, Fernandez-Ruiz A, and Buzsáki G (2019). Layer-Specific Physiological Features and Interlaminar Interactions in the Primary Visual Cortex of the Mouse. Neuron 101, 500–513.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamash P, Carandini M, Harris K, and Steinmetz N (2018). A tool for analyzing electrode tracks from slice histology. BioRxiv 447995. [Google Scholar]
- Siegle JH, Jia X, Durand S, Gale S, Bennett C, Graddis N, Heller G, Ramirez TK, Choi H, Luviano JA, et al. (2021). Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota A, Montgomery S, Fujisawa S, Isomura Y, Zugaro M, and Buzsáki G (2008). Entrainment of neocortical neurons and gamma oscillations by the hippocampal theta rhythm. Neuron 60, 683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Koos T, and Buzsáki G (2012). Diode probes for spatiotemporal optical control of multiple neurons in freely moving animals. J. Neurophysiol. 108, 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark E, Eichler R, Roux L, Fujisawa S, Rotstein HG, and Buzsáki G (2013). Inhibition-Induced Theta Resonance in Cortical Circuits. Neuron 80, 1263–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NA, Zatka-Haas P, Carandini M, and Harris KD (2019). Distributed coding of choice, action and engagement across the mouse brain. Nature 576, 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C, Pachitariu M, Steinmetz N, Reddy CB, Carandini M, and Harris KD (2019). Spontaneous behaviors drive multidimensional, brainwide activity. Science 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadel F, Baillet S, Mosher JC, Pantazis D, and Leahy RM (2011). Brainstorm: A User-Friendly Application for MEG/EEG Analysis. Computational Intelligence and Neuroscience 2011, e879716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecuatl C, Wheeler DW, and Ascoli GA (2021). A Method for Estimating the Potential Synaptic Connections Between Axons and Dendrites From 2D Neuronal Images. Bio-Protocol 11, e4073–e4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeters JL, Godfrey K, Young R, Dang C, Friedsam C, Wark B, Asari H, Peron S, Li N, Peyrache A, et al. (2015). Neurodata Without Borders: Creating a Common Data Format for Neurophysiology. Neuron 88, 629–634. [DOI] [PubMed] [Google Scholar]
- Wang Q, Ding S-L, Li Y, Royall J, Feng D, Lesnar P, Graddis N, Naeemi M, Facer B, Ho A, et al. (2020). The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 181, 936–953.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DW, White CM, Rees CL, Komendantov AO, Hamilton DJ, and Ascoli GA (2015). Hippocampome.org: a knowledge base of neuron types in the rodent hippocampus. Elife 4, e09960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg Ij.J., Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yger P, Spampinato GL, Esposito E, Lefebvre B, Deny S, Gardella C, Stimberg M, Jetter F, Zeck G, Picaud S, et al. (2018). A spike sorting toolbox for up to thousands of electrodes validated with ground truth recordings in vitro and in vivo. ELife 7, e34518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-J, Ye J, Miao C, Tsao A, Cerniauskas I, Ledergerber D, Moser M-B, and Moser EI (2013). Optogenetic Dissection of Entorhinal-Hippocampal Functional Connectivity. Science 340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary video 1. Demonstration of the CellExplorer interface, related to figure 3. The supplementary video is available on YouTube, at CellExplorer.org, and at https://buzsakilab.com/CellExplorer/CellExplorerMovie.mp4
Data Availability Statement
All data is available from our databank at https://buzsakilab.com/wp/database/ (Petersen et al., 2018). All code is available at GitHub: https://github.com/petersenpeter/CellExplorer. All resources, including tutorials and documentation are on the CellExplorer website at: https://cellexplorer.org/.


