Abstract
Although TT virus (TTV) was isolated from a cryptogenic posttransfusion hepatitis patient, its pathogenic role remains unclear. It has been reported that the majority of the healthy population is infected with TTV. To elucidate the differences between TTV infection in patients with liver diseases and TTV infection in the healthy population, a quantification system was developed. TTV DNA was quantified by a real-time detection PCR (RTD-PCR) assay on an ABI Prism 7700 sequence detector. With this system, TTV DNA was quantified in 78 hepatitis C virus (HCV)-infected patients (63 with elevated serum alanine aminotransferase [ALT] levels and 15 with normal ALT levels) and in 70 voluntary blood donors (BDs). The quantification range was 2.08 to 7.35 log copies/ml. The intra-assay and interassay coefficients of variation were 0.37 to 6.33% and 0.60 to 7.07%, respectively. The mean serum TTV DNA levels in the HCV-infected patients with both elevated and normal ALT levels and BDs were 3.69 ± 0.89, 3.45 ± 0.76, and 3.45 ± 0.67 log copies/ml, respectively. Comparison of the serum TTV DNA levels among the HCV-infected patients revealed that they were not related to the serum ALT and HCV core protein levels or to the histopathological score on liver biopsy. This study showed that (i) the RTD-PCR assay for the detection of TTV was accurate and had a high degree of sensitivity, (ii) the mean serum TTV DNA level was similar among HCV-infected patients, irrespective of their ALT level, and also among BDs, and (iii) a high serum TTV DNA level does not affect the serum ALT and HCV levels or liver damage in HCV-infected patients.
A novel DNA virus, the TT virus (TTV), named after the initials of the patient from whom it was first isolated, was cloned from the acute-phase serum of a patient with cryptogenic posttransfusion hepatitis (10). Although this virus had been believed to have a linear single-stranded DNA genome similar to that of members of the family Parvoviridae, the latest report on the full genomic structure of this virus has indicated that it has a circular genome similar to that of members of the family Circoviridae (8, 9, 13). Since the virus was found more often in patients with various liver diseases than in voluntary blood donors, some pathogenic role is conceivable (1, 15). Recently, tests with a new highly sensitive primer set with about 10- to 100-fold greater sensitivities than the conventional primer sets used for TTV detection has revealed that the majority of the general population in Japan is infected with TTV (16). In our previous study with this primer set, we had determined that the prevalence was not significantly different between hepatitis C virus (HCV)-infected patients with chronic hepatitis and hepatocellular carcinoma or between patients with hepatocellular carcinoma associated with HCV infection and those with neither HCV nor hepatitis B virus (HBV) in the serum. Moreover, comparison of patients with and without TTV infection revealed no significant differences in their backgrounds, biochemical findings, or histopathological scores (4). Therefore, to evaluate the differences in serum TTV DNA levels between healthy individuals and patients with liver diseases, an accurate and highly sensitive system for the quantification of TTV has been developed.
Recently, the real-time detection PCR (RTD-PCR) assay has been described as a new system for the quantification and detection of PCR product accumulation. This system exploits the 5′-3′ exonuclease activity of Taq DNA polymerase which hydrolyzes an internal dually labeled (a fluorescent reporter dye and quencher dye) probe (6). In the intact probe, the quencher dye suppresses the fluorescence emission of the reporter dye. During the annealing and extension phase of PCR, the probe anneals to the target sequence and is subsequently hydrolyzed and cleaved by Taq DNA polymerase. The cleavage reduces the quenching effect and allows an increase in the emission of the reporter dye fluorescence, and continuous measurement of the increments in the fluorescence released during PCR provides an accurate estimate of the initial copy number in the sample. This report describes the use of this system for the quantification of the serum TTV DNA levels in patients with HCV infection and voluntary blood donors infected with TTV and determination of the correlation between the amount of TTV and the severity of hepatic damage.
MATERIALS AND METHODS
Materials.
Serum samples from 78 patients infected with HCV but without hepatitis B surface antigen (HBsAg) (63 had elevated serum alanine aminotransferase [ALT] levels and 15 had normal ALT levels) were analyzed. The subjects included previously reported patients who were known to be infected with TTV (4). As controls, 70 blood samples from voluntary blood donors supplied by the Japan Red Cross Blood Center, which were positive for TTV but negative for human immunodeficiency virus, HBV, and HCV and which had been confirmed to have normal ALT levels, were also obtained from the Department of Blood Transfusion, Nagoya City University, as the attached tube for a cross-matching test. All samples were stored at −80°C until use.
Anti-HCV was detected with a second-generation enzyme immunoassay kit (Ortho Diagnostics, Tokyo, Japan). The samples were tested for HBsAg and anti-HBsAg by the particle agglutination method (Fujirebio, Tokyo, Japan), and the samples were tested for the anti-hepatitis B core antigen by the passive hemagglutination method (International Regent Co., Kobe, Japan). Serum HCV core protein levels were measured by fluorescent enzyme immunoassay (14, 18).
To determine the reproducibility of the tests with the two sets of primers and the probe and the correlation between the results of tests with the two sets of primers and the probe, serum samples from selected HCV patients with elevated ALT levels were used.
Detection of TTV.
A total of 20 μl of DNA was extracted from 100-μl serum samples by using the SMITEST EX-R&D (Sumitomo Metal Industries, Tokyo, Japan). TTV DNA was detected by PCR with the primers and under the conditions described by Takahashi et al. (16). The expected sizes of the products were confirmed by electrophoresis on 3% agarose gels with ethidium bromide staining.
Construction of primers and probe.
For the RTD-PCR assay, two sets of primers and probe located in 5′ untranslated open reading frame 2 were prepared. Set A primers were those reported by Takahashi et al. (16), with slight modification, and consisted of forward primer M801 (5′-CTACGTCACTAACCACGTG-3′), reverse primer M935 (5′-TTCGGTGTGTAAACTCACC-3′), and probe TP117 (5′-AAACTCAGCCATTCGGAAGT-3′). The sequences of set B primers were designed so that they were near the sequences of set A primers with reference to the alignment of the TTV sequence on the basis of data from our studies and from the database of the DDBJ/EMBL/GenBank (data not shown) and consisted of forward primer K117 (5′-CACTTCCGAATGGCTGAGTT-3′), reverse primer K233 (5′-GCCTTGCCCATAGCCCGG-3′), and probe TP208 (5′-TCCCGAGCCCGAATTGCCCCT-3′). The probe contains a fluorescent reporter dye (6-carboxyfluorescein) at the 5′ end and a fluorescent quencher dye (6-carboxy-tetramethyl-rhodamine) at the 3′ end.
RTD-PCR.
Ten microliters of the extracted DNA was detected with the sequence detector system (ABI Prism 7700; Applied Biosystems, Foster City, Calif.) in 50 μl of a PCR mixture containing TaqMan PCR Core Reagents with AmpliTaq Gold and AmpErase uracil-N-glycosylase (Applied Biosystems), 20 pmol of each primer, and 15 pmol of the probe. After initial activation of uracil-N-glycosylase at 50°C for 2 min, AmpliTaq Gold was activated at 95°C for 9 min. The subsequent PCR conditions consisted of 50 cycles of denaturation at 95°C for 20 s and annealing and extension at 60°C for 1 min per cycle. During the PCR amplification, the amplified products were measured continuously by determination of the fluorescence emission. After real-time data acquisition, the cycle threshold value was calculated by determining the point at which the fluorescence exceeds an arbitrary threshold limit. The threshold limit was manually set to cross the fluorescent signal of all standards in the exponential phase. The cycle threshold value is predictive of the quantity of target copies in the samples.
For the standard curve, a standard DNA was constructed. The PCR products obtained with primers M801 and K233 were cloned into the TA cloning vector (Invitrogen, San Diego, Calif.) and were sequenced with M13 primers to confirm the sequences of the primers and the probe region. This construct was used as the primary standard after quantification by optical density determination and was prepared by use of a series of 10-fold dilutions. For dilution of the standards, TE (Tris-EDTA) buffer was used, and the standards were stored at −20°C.
Histopathological analysis.
Histopathological examinations were performed for the HCV-infected patients with elevated ALT levels. All liver tissue specimens were obtained by needle biopsy. Specimens were fixed in 10% neutral formalin, embedded in paraffin, cut into 4-μm-thick sections, and stained with hematoxylin-eosin. Each specimen was assessed independently by three liver pathologists (M. Kage, M. Kojiro, and K. Shimamatsu) by use of a scoring system which included a grading of necroinflammation and staging of fibrosis on the basis of the recommendations of Ichida et al. (3). In the grading, inflammation in both the portal area and the lobular area was divided into four levels on the basis of the degrees of infiltration of lymphocytes and necrosis of the hepatocytes. If the interpretations of the pathologists differed, the score was determined to be that which two pathologists agreed on.
Statistical analysis.
For statistical analysis of categorical data, Mann-Whitney's U test, the Kruskal-Wallis test, and analysis of variance (ANOVA) were used. For the liner regression, Spearman's correlation test was used. Data analysis was carried out with the StatView J, version 4.5, software package (Abacus Concepts Inc., Berkeley, Calif.).
RESULTS
Sensitivity and reproducibility of RTD-PCR assay.
By using standard DNA at a series of 10-fold dilutions, the detection range was determined to be from 2.08 to 7.35 log copies/ml. To evaluate the reproducibility of the RTD-PCR assay, a total of eight samples, including six TTV-positive samples and two TTV-negative samples, were tested five times on the same day or for 5 days. The intra-assay and interassay coefficients of variation (CVs) were calculated to be within the ranges of 0.37 to 6.33% and 0.60 to 7.07%, respectively (Table 1). Identical sensitivities and reproducibilities were obtained with each of the primer and probe sets.
TABLE 1.
Reproducibility of serum TTV DNA levels by RTD-PCR assaya
| Assay | Intra-assay reproducibility of TTV DNA level (log copies/ml)
|
Interassay reproducibility of TTV DNA level (log copies/ml)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Expt 4 | Expt 5 | Mean | SD | CV (%) | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Mean | SD | CV (%) | |
| 1 | 3.158 | 2.748 | 3.049 | 3.182 | 2.857 | 2.999 | 0.190 | 6.33 | 3.158 | 3.215 | 2.964 | 2.748 | 3.283 | 3.074 | 0.217 | 7.07 |
| 2 | 3.246 | 2.924 | 3.301 | 3.121 | 3.033 | 3.125 | 0.153 | 4.91 | 3.246 | 3.318 | 3.158 | 3.049 | 3.334 | 3.221 | 0.119 | 3.68 |
| 3 | 4.580 | 4.459 | 4.350 | 4.435 | 4.833 | 4.531 | 0.187 | 4.13 | 4.580 | 4.778 | 4.459 | 4.394 | 4.833 | 4.601 | 0.192 | 4.17 |
| 4 | 5.064 | 5.158 | 4.964 | 4.716 | 4.064 | 4.993 | 0.170 | 3.40 | 5.064 | 5.182 | 4.881 | 4.748 | 5.158 | 5.007 | 0.187 | 3.73 |
| 5 | 5.265 | 5.246 | 5.255 | 5.246 | 5.292 | 5.261 | 0.019 | 0.37 | 5.265 | 5.310 | 5.236 | 5.255 | 5.318 | 5.277 | 0.036 | 0.68 |
| 6 | 5.778 | 5.806 | 5.833 | 5.806 | 5.748 | 5.794 | 0.032 | 0.55 | 5.778 | 5.833 | 5.806 | 5.778 | 5.857 | 5.810 | 0.035 | 0.60 |
| 7 | NDb | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||||||
| 8 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||||||
Primers and probe set B were used.
ND, not detected.
Correlation between two sets of primers and probes.
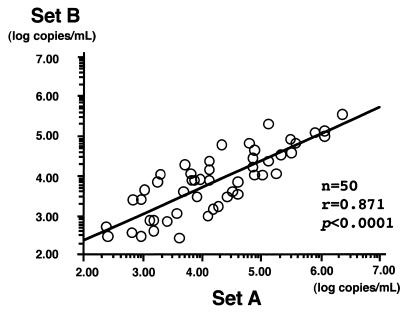
The serum TTV DNA levels of 50 HCV-infected patients were measured by using primer and probe sets A and B, and these levels were almost identical and showed a strong correlation (r = 0.871; P < 0.0001) (Fig. 1).
FIG. 1.
Correlation between serum TTV DNA levels determined with primer sets A and B.
Serum TTV DNA levels among HCV-infected patients and blood donors.
Among the samples from HCV-infected patients with elevated ALT levels and those with normal ALT levels and voluntary blood donors, the serum TTV DNA levels could be quantified for 54 of 63 (85.7%), 13 of 15 (86.7%), and 62 of 70 (88.6%) samples, respectively. For the remaining samples for which virus was not quantified, the serum TTV DNA level was considered to be below the detection range of this assay.
The mean serum TTV DNA levels in the HCV-infected patients with elevated ALT levels and normal ALT levels and blood donors were 3.69 ± 0.89, 3.45 ± 0.76, and 3.45 ± 0.67 log copies/ml, respectively, and these data were similar and were not significantly different (Table 2). Comparison of HCV-infected patients with high (>3.50 log copies/ml) and low (<3.50 log copies/ml) serum TTV DNA levels indicated no significant differences in terms of mean age, sex, serum ALT level, and serum HCV core protein level (Table 3). Histopathological examination was performed for HCV-infected patients with elevated ALT levels, and the results were expressed as the fibrosis score, classified into five stages (stages F0 to F4), and the activity score, classified into four grades (grades A0 to A3), according to the New Inuyama Classification. The mean score for each parameter was also represented as an index. Among patients with high serum TTV DNA levels, 10 were classified into stage F0, 18 were classified into stage F1, 3 were classified into stage F2, 1 was classified into stage F3, and 1 was classified into stage F4; the mean score was 0.939 ± 0.899. One was classified into grade A0, 18 were classified into grade A1, 11 were classified into grade A2, and 3 were classified into grade A3; the mean score was 1.485 ± 0.712. Among the patients with low serum TTV DNA levels, 5 were classified into stage F0, 12 were classified into stage F1, 2 were classified into stage F2, 1 was classified into stage F3, and 1 was classified into stage F4; the mean score was 1.095 ± 0.995. One was classified into grade A0, 15 were classified into grade A1, 4 were classified into grade A2, and 1 was classified into grade A3; mean score was 1.238 ± 0.625. No significant difference in the ratios or the mean scores were observed (Table 3).
TABLE 2.
Serum TTV DNA levels in HCV-infected patients and voluntary blood donorsa
| Group | No. of subjects | Mean ± SD age (yr) | No. of males: no. of females | Mean ± SD ALT level (IU/liter) | HCV core protein level (pg/ml)
|
TTV DNA level (log copies/ml) | |
|---|---|---|---|---|---|---|---|
| Median | Range | ||||||
| HCV-infected patients | |||||||
| Elevated ALT levels | 54 | 48.6 ± 12.1 | 35:19 | 100.4 ± 100.8 | 117 | 8–914 | 3.69 ± 0.89b |
| Normal ALT levels | 13 | 54.7 ± 15.3 | 8:5 | 20.9 ± 7.6 | 32 | 8–883 | 3.45 ± 0.76b |
| Blood donors | 62 | NA | NA | NA | NT | NT | 3.45 ± 0.67 |
Abbreviations: NA, not available; NT, not tested.
P was not significant.
TABLE 3.
Comparison of HCV-infected patients with high and low serum TTV DNA levelsa
| TTV DNA level | No. of subjects | Mean ± SD age (yr) | No. of males: no. of females | Mean ± SD ALT level (IU/ml) | HCV core protein level (pg/ml)
|
Histology results
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of subjectsb | Staging
|
Grading
|
|||||||||
| Median | Range | No. of subjects with stage F0:F1:F2:F3:F4 | Mean ± SD stage | No. of subjects with grade A0:A1:A2:A3 | Mean ± SD grade | ||||||
| High (>3.50 log copies/ml) | 39 | 47.8 ± 13.6 | 25:14 | 95.3 ± 117.1 | 137 | 8–883 | 33 | 10:18:3:1:1 | 0.939 ± 0.899 | 1:18:11:3 | 1.485 ± 0.712 |
| Low (<3.50 log copies/ml) | 28 | 52.8 ± 12.3 | 18:10 | 70.6 ± 52.4 | 117 | 8–914 | 21 | 5:12:2:1:1 | 1.095 ± 0.995 | 1:15:4:1 | 1.238 ± 0.625 |
P was not significant for all comparisons.
Histology results were estimated for HCV-infected patients with elevated ALT levels.
DISCUSSION
At first, TTV was believed to be a novel hepatitis virus because it could be isolated from three of five patients with cryptogenic posttransfusion hepatitis. However, our previous study revealed that TTV is prevalent among both patients with liver diseases and voluntary blood donors (4). Thus, to clarify the correlation between serum TTV DNA levels and liver diseases, we developed a quantification assay with high degrees of sensitivity, accuracy, specificity, and reproducibility. Some of the findings from this quantification assay are that (i) the mean serum TTV DNA level was similar among HCV-infected patients irrespective of the serum ALT level and also among voluntary blood donors, and (ii) the serum TTV DNA level is not related to the serum ALT and HCV core protein levels or the histopathological score of fibrosis and inflammation among HCV patients.
The RTD-PCR system enables the accurate and highly sensitive estimation of the level of TTV infection. This system is now available for quantification of HCV and Escherichia coli and even for the detection of cancer cells (2, 5, 7, 12, 17). It has a wide dynamic range (2.08 to 7.35 log copies/ml), and the intra-assay and interassay CVs ranged from 0.37 to 6.33% and 0.60 to 7.07%, respectively. In regard to specificity, the fluorescence signal due to cleavage of the probe is generated only if the target sequence for the probe is amplified by PCR, and no signal is observed under the condition of nonspecific amplification. To confirm the accuracy and to negate the influence of primers and probe affinity, two sets of primers and probe were used to obtain estimates of TTV DNA levels in our study, and the quantification data obtained with these two sets were identical and showed a strong correlation. Besides these advantages, it must also be mentioned that other useful systems for the quantification of TTV are not available. Therefore, this quantification system will be useful for estimation of the severity of TTV infection.
The results of recent studies raise a question regarding the association between TTV infection and hepatic disorders. Mushahwar et al. (9) reported that inoculation of TTV into chimpanzees could not induce hepatitis. In regard to this point, our investigation of the quantification of TTV among HCV-infected patients might be relevant. Serum TTV DNA levels were similar, irrespective of the severity of liver injury, and were even similar in HCV-infected patients and blood donors. Comparison of HCV-infected patients with both high and low serum TTV DNA levels revealed no significant differences in serum ALT and HCV core protein levels or the histopathological score. These data imply that TTV infection and replication occur in many healthy people and do not affect the HCV infection or the liver damage due to HCV infection. This observation does not totally negate the association between TTV infection and hepatitis. In the case of the other hepatitis viruses, such as HCV, a dissociation between the serum viral titer and the severity of hepatitis is observed. To estimate the pathogenicity of TTV, the serial monitoring of serum TTV DNA levels in patients with hepatitis of unknown etiology will be needed.
Another important use of this system is to identify the organs for which TTV has an affinity. The findings of the similarity of TTV to members of the family Circoviridae rather than to members of the family Parvoviridae could suggest the possibility of an association of TTV with diseases other than liver diseases. For example, the chicken anemia virus, a representative of the family Circoviridae, induces severe anemia, subcutaneous and intramuscular hemorrhage, destruction of erythroblastoid cells in the bone marrow, and depletion of certain lymphoid organs in young chickens (11). By using the quantification system described here, further studies on the quantification of TTV in various organs such as the bone marrow and estimation of the pathogenicity of this virus will be needed.
In summary, an accurate and highly sensitive system for the quantification of TTV has been developed. By using this system, it was revealed that the titer of TTV DNA in serum does not affect HCV infection or the liver damage due to HCV.
ACKNOWLEDGMENTS
We express our gratitude to Osamu Yanagihara for quantification of the HCV core protein level.
This work was supported in part by a grant from the Japanese Ministry of Health and Welfare, Health Science Research Grants (Non-A, Non-B Hepatitis Research Grants), and by a Grant-in Aid for Scientific Research (B) from the Ministry of Education, Science, Sports and Culture (grant 11691222).
REFERENCES
- 1.Charlton M, Adjei P, Poterucha J, Zein N, Moore B, Therneau T, Krom R, Wiesner R. TT-virus infection in North American blood donors, patients with fulminant hepatic failure, and cryptogenic cirrhosis. Hepatology. 1998;28:839–842. doi: 10.1002/hep.510280335. [DOI] [PubMed] [Google Scholar]
- 2.Gerard C J, Olsson K, Ramanathan R, Reading C, Hanania E G. Improved quantitation of minimal residual disease in multiple myeloma using real-time polymerase chain reaction and plasmid-DNA complementarity determining region III strand. Cancer Res. 1998;58:3957–3964. [PubMed] [Google Scholar]
- 3.Ichida F, Tsuji T, Omata M, Ichida T, Inoue K, Kamimura T, Yamada G, Hino K, Yokosuka O, Suzuki H. New Inuyama classification; new criteria for histological assessment of chronic hepatitis. Int Hepatol Commun. 1996;6:112–119. [Google Scholar]
- 4.Kato T, Mizokami M, Orito E, Nakano T, Tanaka Y, Ueda R, Hirashima N, Iijima Y, Kato T, Sugauchi F, Mukaide M, Shimamatsu K, Kage M, Kojiro M. High prevalence of TT virus infection in Japanese patients with liver diseases and blood donors. J Hepatol. 1999;31:221–227. doi: 10.1016/s0168-8278(99)80217-7. [DOI] [PubMed] [Google Scholar]
- 5.Kawai S, Yokosuka O, Kanda T, Imazeki F, Maru Y, Saisho H. Quantification of hepatitis C virus by TaqMan PCR: comparison with HCV amplicor monitor assay. J Med Virol. 1999;58:121–126. doi: 10.1002/(sici)1096-9071(199906)58:2<121::aid-jmv4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Livak K, Flood S, Marmoro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic and hybridization. PCR Methods Applications. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 7.Martell M, Gomez J, Esteban J I, Sauleda S, Quer J, Cabot B, Esteban R, Guardia J. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyata H, Tsunoda H, Kazi A, Yamada A, Kahn M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of an novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mushahwar I K, Erker J C, Muerhoff A S, Leary P T, Simons N, Birkenmeyer L G, Chalmers M L, Pilot-Matias T J, Dexai S M. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 11.Noteborn M H M, de Boer G F, van Roozelaar D J, Karreman C, Kranenburg O, Vos J, Jeurissen S H M, Hoeben R C, Zantema A, Koch G, van Ormondt H, van der Eb A J. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J Virol. 1991;65:3131–3139. doi: 10.1128/jvi.65.6.3131-3139.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberst R, Hays D M P, Bohra L K, Phebus R K, Yamashiro C T, Paszko-Kolva C, Flood S J A, Sargeant J M, Gillespie J R. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl Environ Microbiol. 1998;64:3389–3396. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 14.Orito E, Mizokami M, Tanaka T, Lau J Y N, Suzuki K, Yamauchi M, Ohta Y, Hasegawa A, Tanaka S, Kohara M. Quantification of serum hepatitis C virus core protein level in patients chronically infected with different hepatitis C virus genotypes. Gut. 1996;39:876–880. doi: 10.1136/gut.39.6.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmonds P, Davidson F, Lysett C, Prescott L E, MacDonald D M, Ellender J, Yap P L, Ludlam C A, Haydon G H, Gillon J, Jarvis L M. Detection of novel DNA virus (TTV) in blood donors and blood products. Lancet. 1998;352:191–195. doi: 10.1016/s0140-6736(98)03056-6. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K, Hoshino H, Ohta Y, Yoshida N, Mishiro S. Very high prevalence of TT virus (TTV) infection in general population of Japan revealed by a new set of PCR primers. Hepatol Res. 1998;12:233–239. [Google Scholar]
- 17.Takeuchi T, Katsume A, Tanaka T, Abe A, Inoue K, Tsukiyama-Kohara K, Kawaguchi R, Tanaka S, Kohara M. Highly sensitive and accurate system for quantification of hepatitis C virus genomic RNA using real-time detection PCR. Gastroenterology. 1999;116:636–642. doi: 10.1016/s0016-5085(99)70185-x. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka T, Lau J Y N, Mizokami M, Orito E, Tanaka E, Kiyosawa K. Simple fluorescent enzyme immunoassay for detection and quantification of hepatitis C viremia. J Hepatol. 1995;23:742–745. doi: 10.1016/0168-8278(95)80043-3. [DOI] [PubMed] [Google Scholar]