Abstract
p27Kip1 is a member of the Cip-Kip family of cyclin-dependent kinase (Cdk) inhibitors that binds to cyclin-Cdk complexes and inhibits their catalytic activity in response to antiproliferative stimuli. p27Kip1 is regulated by several posttranscriptional mechanisms, including subcellular localization. We have identified a component of the nuclear pore complex (NPC), termed Nup50, through its two-hybrid interactions with p27Kip1. Nup50 is a nucleoplasmically oriented component of the nuclear pore complex with a role in protein export (T. Guan, R. H. Kehlenbach, E. C. Schirmer, A. Kehlenbach, F. Fan, B. E. Clurman, N. Arnheim, and L. Gerace, Mol. Cell. Biol. 20:5619–5630, 2000). We found that murine Nup50 is a widely expressed nucleoporin and that Nup50 expression is highest in the developing neural tube and adult testes. We have also examined interactions between Nup50 and the NPC and found specific two-hybrid interactions between Nup50 and several well-defined components of the NPC, as well as coimmunoprecipitation of Nup50 with the nucleoporin Nup153 from transfected mammalian cells. In order to study Nup50 function in vivo, we cloned the mouse Nup50 genomic locus and created a targeted Nup50 deletion in the mouse germ line. Nup50 disruption resulted in a complex phenotype characterized by late embryonic lethality, neural tube defects, and intrauterine growth retardation. Although Nup50-null mouse embryo fibroblasts exhibited no defects in either cell cycle control or p27Kip1 regulation, Nup50 deletion was associated with abnormalities in p27Kip1 expression and cell proliferation in the developing neuroepithelium. We conclude that Nup50 is a nucleoporin with essential functions during mouse development.
The nucleocytoplasmic transport of macromolecules is regulated by the nuclear pore complex (NPC) (reviewed in references 5, 16, and 18). The NPC is a large structure comprised of a symmetrical core embedded within the nuclear envelope and extensive 50- to 100-nm filaments that project into both the nucleus and cytoplasm (8, 20). The NPC contains more than 50 different proteins, termed nucleoporins, some of which have been assigned specific NPC locations and/or functions. Many nucleoporins share sequence and structural motifs, including repeated peptides (FXFG or GLFG) in regions that mediate interactions with soluble transport factors, coiled-coiled domains involved in interactions between some nucleoporins, and modification with O-linked N-acetylglucosamine in higher eukaryotes.
All macromolecular traffic between the nucleus and cytoplasm passes through the NPC (9, 18). Molecules smaller than 20 to 40 kDa can traverse the NPC via a diffusion channel. In contrast, the transport of larger molecules, termed cargo, is mediated by specific transport receptors. In general, proteins destined for import are bound by transport receptors that recognize specific sequence signals and then are brought through the NPC, followed by cargo release and receptor protein recycling. This process also requires the GTPase Ran. The known import receptors are a family of distantly related proteins specialized for different types of cargo and transport signals. The prototype import receptor is the importin α/importin β complex involved in importing proteins with classic basic nuclear localization signals (NLS). Protein export pathways are less well characterized, although one major export pathway has been extensively studied and involves the export of cargo that contain leucine rich nuclear export signals (NES) via the transport receptor Crm1 (reviewed in reference 32).
Many cellular processes depend upon NPC function. In some cases, such as protein synthesis, the role of the NPC in transporting large amounts of mRNA and ribosomes is obvious. However, nucleocytoplasmic transport also plays important regulatory roles in processes such as cell cycle control (reviewed in references 21 and 37). For example, in mammalian cells cyclin B1 is sequestered in the cytoplasm during most of the cell cycle by a Crm1-dependent export mechanism and enters the nucleus only at the G2/M boundary (11, 36). Subcellular compartmentalization has also been reported to regulate the function and stability of the p27Kip1 protein. p27Kip1 is a member of the Cip-Kip family of cyclin-dependent kinase (Cdk) inhibitors and inhibits the cell cycle by binding to Cdks in response to various antimitogenic signals (25). Cyclin-Cdks function in the nucleus, and p27Kip1 binds and inhibits cyclin-Cdks in the nucleus as well (22). The stability of p27Kip1 may also be regulated by its subcellular location (reviewed in reference 3). For example, the Jab1 protein may promote p27Kip1 export and cause it to be degraded in the cytoplasm (30). Cytoplasmic mislocalization of p27Kip1 has also been described in tumor cells, and it has been suggested that cytoplasmic sequestration may functionally inactivate p27Kip1 in some tumor cells (19, 26).
In this study we report the characterization of a NPC-associated protein originally termed npap60 and now renamed Nup50 (10) that we identified through a two-hybrid interaction with p27Kip1. Nup50 was initially described as a rat NPC protein that undergoes unusual changes in localization during male germ cell differentiation (6). We have found that Nup50 is a ubiquitously expressed NPC protein that exhibited two-hybrid interactions with multiple transport receptor proteins and coimmunoprecipitated with the nucleoporin Nup153. We have also cloned and characterized the mouse genomic Nup50 locus and created a targeted Nup50 deletion in the mouse germ line. Deletion of Nup50 caused embryonic lethality associated with neural tube abnormalities, exencephaly, and intrauterine growth retardation. In addition, we observed abnormal p27Kip1 expression in the neural tubes of Nup50-null animals, although no defects in p27Kip1 regulation or cell cycle control were found in Nup50-null mouse embryo fibroblasts. These data indicate that Nup50 performs essential functions during mouse development.
MATERIALS AND METHODS
Antibodies.
The following antibodies were used in this study: monoclonal anti-p27Kip1 (Transduction Labs, Lexington, Ky.), anti-p21Cip1 (C-19) and anti-cyclin B1-GNS (Santa Cruz Biotechnology, Santa Cruz, Calif.), HA.11 (anti-HA antibody), and anti-NPC monoclonal antibody 414 (MAb414; Babco, Berkeley, Calif.), anti-Nup153 (B. Burke, Calgary, Alberta, Canada), anti-npap60 (N. Arnheim, Los Angeles, Calif.), horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse immunoglobulin G (IgG; Pharmacia, Kalamazoo, Mich.), fluorescein isothiocyanate (FITC)-labeled anti-rabbit and anti-mouse IgG (Jackson Labs, West Grove, Pa.), and anti-Ki-67 (Novocastra Laboratories). 9E10 (anti-myc tag) was prepared by an in-house production facility.
Two-hybrid analyses and cloning of mouse Nup50.
Two-hybrid screens were performed by using the modified two-hybrid method developed by Hollenberg and Weintraub which has been described in detail (35). The yeast strain L40, pBTM116 (lex fusion vector), plex-lamin, and the 9.5/10.5-day-postcoitus (d.p.c.) mouse embryo cDNA–VP-16 library were provided by S. Hollenberg (Portland, Oreg.). LexA-p27Kip1 baits were constructed by subcloning PCR-generated fragments corresponding to either full-length human p27Kip1 or p27Kip1(72–198) into pBTM116. Sequences were confirmed in the automated sequencing resource at The Fred Hutchinson Cancer Research Center (FHCRC). Library plasmids from positive yeast clones were isolated, and the specificity of two-hybrid interactions was tested using lex-p27Kip1 and lex-lamin baits as described elsewhere (35). Isolated clones were sequenced using a primer derived from the VP-16 sequence, and inserts were identified using the NCBI BLAST server.
A full-length mouse Nup50 cDNA was obtained by screening a 16-day p.c. mouse embryo library (Novagen, Madison, Wis.) with a Nup50 probe corresponding to residues 143 to 206. Phage DNA was purified by standard methods, and the inserts were subcloned by recombination according to the manufacturer's protocol (Novagen). A 1.8-kb insert containing both the sequence corresponding to the 5′ end of the rat nap60 gene and a long poly(A) stretch was subcloned into pBluescript (Stratagene), and a set of exonuclease III deletions was created by standard methods and sequenced in both directions (1).
Anti-Nup50 antibody production and analyses of Nup50 expression.
Recombinant His-tagged mouse Nup50 protein was produced by subcloning the full-length Nup50 cDNA into pET-16b (Novagen), followed by transformation into the Escherichia coli BL21 and purification by Ni-affinity chromatography after a 3-h induction with isopropyl-β-d-thiogalactopyranoside (IPTG) according to the manufacturer's protocol. The eluted protein was examined by gel electrophoresis and used to inoculate two rabbits by standard methods (12). Inoculations and bleeds were performed by the FHCRC shared animal resource staff. Affinity-purified antibodies were obtained by incubating antisera with purified recombinant His-Nup50 immobilized on polyvinylidene difluoride (PVDF), followed by elution in low-pH glycine buffer.
Protein analyses.
Cell lines used for immunostaining and Western analyses included NIH 3T3 (obtained from C. Sherr, Memphis, Tenn.), 293, HeLa, and U2OS (obtained from J. Roberts, FHCRC), Rat1, and human diploid fibroblasts (obtained from C. Grandori, FHCRC). All cells were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum (Gibco). For Western analysis of endogenous Nup50, cells were lysed directly on tissue culture dishes in radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors (10 mM Tris, pH 7.4; 0.15 M NaCl; 1% NP-40; 1% deoxycholate; 0.1% sodium dodecyl sulfate [SDS]; 10 μg each of aprotonin, leupeptin, and pepstatin per ml; 50 mM NaF; 1 mM sodium vanadate), followed by scraping and sonication. Cell extracts were electrophoresed on 12% polyacrylamide gels and transferred to PVDF membranes as previously described (4). After incubation with primary antibodies, proteins were visualized by incubation with HRP-conjugated anti-rabbit or anti-mouse secondary antibodies as appropriate, followed by enhanced chemiluminescence according to the manufacturer's instructions (Pierce). Mouse tissue and embryo lysates were prepared by sonicating freshly obtained tissues in RIPA, and 100 μg of total lysate was immunoblotted as described above.
RNA analyses.
Northern analysis of Nup50 expression in adult mouse tissues was performed by hybridizing 10 μg of total RNA with a full-length Nup50 probe after formaldehyde-agarose electrophoresis (1). The tissues examined included cerebrum, cerebellum, lungs, heart, kidney, liver, spleen, gut, pancreas, testes, ovary, and muscle. In situ hybridization of Nup50 RNA expression in formalin-fixed paraffin sections of adult mouse testes was performed using digoxigenin-UTP-labeled sense and antisense Nup50 probes as described earlier (14). The specific antisense staining pattern was confirmed with several probes derived from different regions of the Nup50 cDNA.
Analyses of Nup50-protein interactions. (i) GST pulldown.
Glutathione S-transferase (GST)–Nup50-1 was cloned by amplifying a region of the Nup50 cDNA from nucleotides 367 to 792, and the PCR fragment was subcloned into pUNI15 (15). pUNI15-PAF was recombined with the vector pHB2-GST with purified Cre recombinase as previously described (5), resulting in the introduction of a GST tag located 5′ of the Nup50 fragment. GST–Nup50-2 was constructed by cloning the internal EcoRI/BamHI fragment from the Nup50 cDNA into the vector pGEX-3X. Expression of GST–Nup50-1, GST–Nup50-2, and GST alone in BL21 bacteria was induced by the addition of 0.4 mM IPTG for 3 h at 37°C. The bacteria were lysed by sonication in SET-Sarkosyl buffer (10 mM Tris-HCl, pH 8.0; 150 mM NaCl; 1 mM EDTA; 1.5% N-lauroylsarcosine). The lysate was spun for 15 min at 16,000 × g. Triton X-100 was added to the supernatant to a final concentration of 2% to sequester the Sarkosyl, and the supernatant was then bound to GST-agarose beads (Sigma). After binding, the beads were washed four times in SET–0.5% NP-40 buffer and stored at −20°C as a 25% slurry in the same buffer. CS2-hp27Kip1 (24) was transcribed and translated in vitro using the TnT Coupled Reticulocyte Lysate System (Promega) and Tran35Slabel (ICN) according to the manufacturer's protocol.
For each pulldown, glutathione-agarose beads containing approximately 10 μg of bound purified GST–Nup50-1, GST–Nup50-2, or GST were used. Nonspecific binding to the beads was first blocked by rotating the beads for 15 min at 4°C in GST pulldown buffer (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.5% NP-40) containing 0.5 mg of bovine serum albumin (BSA). After blocking, 2 μl of radiolabeled p27Kip1 in vitro translation mix was added to each pulldown, and the mixtures were rotated for 1 h at 4°C. The pulldowns were then washed four times in GST pulldown buffer. After washing, the bound p27Kip1 was separated on SDS–12% polyacrylamide gels.
(ii) Nup50 two-hybrid screens.
To examine the two-hybrid interactions of Nup50, the full-length Nup50 cDNA was subcloned into pBTM116 and used to screen the following cDNA–Vp-16 libraries: 9.5/10.5 d.p.c. mouse embryo (see above), adult mouse brain (provided by B. Howell, Bethesda, Md.), and mouse testes (provided by J. Lee, Boulder, Colo.).
(iii) Coimmunoprecipitation.
CS2Nup50 and myc-tagged CS2mtNup50 were constructed by cloning the Nup50 cDNA into the cytomegalovirus-driven expression vectors pCS2 and pCS2MT, respectively (4). For coprecipitation of Nup50 and p27, HeLa cells were cotransfected with CS2mtNup50 and CS2p27 using the calcium phosphate method as described earlier (4). CS2p27 has been previously described (24). For Nup50-Nup50 and Nup50-Nup153 interactions, NIH 3T3 and 293 cells were transfected with CS2Nup50, CS2mtNup50, and CMV-HANup153 (provided by B. Burke, Calgary, Alberta, Canada). Cell lysates were prepared as described above in NP-40 buffer (0.5% NP-40, 50 mM Tris; pH 8.0), and lysates were normalized for protein concentration were incubated at 4°C for 1 h with rotation with appropriate dilutions of primary antibodies and 30 μl of a 1:1 slurry of phosphate-buffered saline (PBS)–protein A-Sepharose (Sigma, St. Louis, Mo.). Precipitates were washed four times with 1 ml of NP-40 buffer before electrophoresis and Western transfer.
Indirect immunofluorescence.
Cells were seeded into 60-mm dishes with glass coverslips the night before staining. For paraformaldehyde fixation, cells were fixed in 4% paraformaldehyde-PBS for 10 min, permeabilized with 0.2% Triton-PBS, and then stained with anti-Nup50, followed by FITC-labeled anti-rabbit IgG. The final wash contained DAPI (4′,6′-diamidino-2-phenylindole) to visualize cell nuclei. For methanol-acetone fixation, coverslips were fixed in methanol for 30 min at −20°C and then quickly rinsed in acetone at 4°C as described earlier (6). After the acetone rinse, the coverslips were washed for 5 min in 1× PBS and then treated as described above. To examine the immunolocalization of Nup153 and other NPC proteins, cells were stained with either MAb414, which recognizes multiple NPC proteins, or anti-Nup153, followed by FITC-labeled anti-rabbit or anti-mouse IgG. For costaining of cells with anti-Nup50 and MAb414, both antibodies were used together, followed by staining with a mixture of anti-rabbit and anti-mouse secondary antibodies. Coverslips were examined using either a Nikon E800 or a Deltavision wide-field deconvolution microscope as indicated.
For differential permeabilization of the nuclear envelope and plasma membrane, MEFs growing on coverslips were rinsed in transport buffer (20 mM HEPES, pH 7.3; 110 mM potassium acetate; 5 mM sodium acetate; 2 mM magnesium acetate; 2 mM dithiothreitol, 0.5 mM EGTA) for 5 min at 4°C. The cells were then treated for 5 min at 4°C with digitonin at a concentration of 0, 2, 5, 10, 15, or 30 μg/ml in transport buffer in order to find a concentration range at which the cell membrane but not the nuclear envelope was permeabilized. As a control, one coverslip was treated with 0.5% Triton X-100 in transport buffer. After digitonin treatment, the coverslips were washed for 15 min at 4°C. The cells were then incubated in primary antibody (Babco MAb414 diluted 1:500 in transport buffer or anti-Nup50 polyclonal diluted at 1:1,000 in transport buffer) for 30 min, followed by three rinses in transport buffer. After rinsing, the cells were incubated in secondary antibody (anti-mouse or anti-rabbit antibody, respectively, labeled with FITC, both from Jackson Laboratories) at a dilution of 1:150 in transport buffer. Finally, the coverslips were washed three times in transport buffer and mounted.
Genomic cloning and targeted disruption of the mouse Nup50 gene.
An approximately 40-kb genomic DNA fragment containing the mouse Nup50 gene was obtained from a mouse 129/Sv genomic library (provided by P. Soriano, Seattle, Wash.) using a 268-bp EcoRI-HincII fragment from the 5′ end of the Nup50 cDNA as a probe. Restriction maps, intron-exon boundaries, and exonic sequences were determined by Southern blot analysis and sequencing with primers derived from the Nup50 cDNA sequence. A knockout vector was constructed by cloning a 6.5-kb SnaBI fragment encompassing sequences upstream of exon 1 and most of the 3.0-kb intron 1, as the upstream long arm, and a 2-kb NsiI-SacI fragment including portions of intron 4 and exon 5 as the downstream short arm into the targeting vector pSAβGeolox2dta (provided by P. Soriano, FHCRC). This vector contains a splice acceptor site upstream and stop codons in all three reading frames downstream of the βgeo sequences, as well as the diphtheria toxin A gene for negative selection against random integrants. The vector was linearized with XhoI and electroporated into XY AK7 ES cells. ES colonies were selected in 300 μg of G418 per ml and correctly targeted clones were identified by PCR utilizing primers derived from the βgeo and Nup50 genomic (5′-TTCGCAGCGCATCGCCTTCT-3′ and 5′-GGGCAGCATCTTTACCAAAC-3′). Clones identified as homologous recombinants were confirmed by Southern analysis using probes that spanned both the 5′ and 3′ recombination junctions. ES cells were introduced into 3.5-d.p.c. C57/B6J embryos to generate chimeric mice, and germ line transmission was achieved by mating chimeric males to wild-type females.
Analysis of Nup50-null embryos and embryonic fibroblasts.
For analysis of β-galactosidase activity, embryos were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as described earlier (7). For histological analyses, embryos were fixed in 4% paraformaldehyde-PBS, dehydrated in ethanol, and embedded in paraffin. For immunostaining of Nup50 and p27Kip1, paraffin-embedded sections were dewaxed in xylenes and rehydrated in an ethanol series, followed by blocking endogenous peroxidases with 1% hydrogen peroxide in 1× PBS for 15 min. After washing in 1× PBS, the sections for p27Kip1 staining (but not the sections for Nup50 staining) underwent antigen retrieval by boiling in 10 mM citric acid (pH 6.0) for 10 min in a microwave oven. After cooling, both p27Kip1 and Nup50 sections were processed identically by rinsing in PBS and then blocking for 30 min in 4% BSA–1× PBS containing 2% normal horse serum (Vector Labs). The sections were then incubated for 1 h in anti-Nup50 polyclonal antibody diluted 1:1,000 in 4% BSA–1× PBS or in anti-p27Kip1 monoclonal antibody diluted 1:500 in 4% BSA–1× PBS. Following three rinses in PBS, the slides were treated for 30 min with biotinylated universal secondary antibody from the ABC Elite kit (Vector Labs), diluted 1:500 in 4% BSA–1× PBS–2% horse serum. Next, the slides were again rinsed in PBS and then incubated for 30 min in ABC reagent, diluted in 4% BSA–1× PBS as described in the ABC Elite kit. Finally, the sections were again rinsed in PBS and developed using the DAB substrate kit (Vector Labs) for approximately 5 min or until adequate signal was seen. Slides were then washed, dehydrated in an ethanol series and xylenes, air dried, and mounted in Permount medium (Fisher Scientific).
MEFs were prepared from 12- to 14-p.c. embryos as described elsewhere (13). MEF genotypes were determined by PCR using primers specific to both the mutant and wild-type Nup50 alleles and were confirmed by quantitative β-galactosidase assays and Nup50 immunostaining. Cell cycle analyses were performed as described earlier (24). For density and serum arrests, cells were either grown to saturation density or placed in DME with 0.5% serum for 72 h prior to release them from growth arrest. Leptomycin B was a gift of M. Yoshida (Tokyo, Japan) and used at a final concentration of 10 ng/ml.
RESULTS
Two-hybrid analyses of p27Kip1-interacting proteins and cloning of mouse Nup50.
We used a two-hybrid approach to identify proteins that interact with p27Kip1. A full-length human p27Kip1 cDNA was used as bait in several independent screens of a 9.5/10.5 d.p.c. mouse embryo library. The vast majority of the true-positive clones identified in these screens represented known cyclins, predominantly cyclins D1 and D2, although cyclins A, B, E, and I were also identified. In addition, several independent overlapping clones of a noncyclin cDNA were identified. This sequence corresponded to the murine homologue of a rat nuclear-pore-associated protein originally termed npap60 and now renamed Nup50 (10). In order to increase the detection of noncyclin p27Kip1 interactors, we used an N-terminally truncated p27Kip1 construct, lex-p27Kip1(72–198), that eliminates the cyclin-CDK interaction domain. As expected, no cyclin cDNAs were detected in positive clones when this bait was used to screen the mouse embryo library. Remarkably, however, 100% of the multiple, independent positive clones identified with the p27Kip1(72–198) bait represented the mouse Nup50 cDNA. We have used GST-pulldown and coimmunoprecipitation approaches to independently assess the interaction of p27Kip1 and Nup50 in a system other than a two-hybrid screen. We constructed two GST-Nup50 fusion proteins containing the minimal region of Nup50 isolated in the two-hybrid clones that could mediate interaction with p27Kip1. The GST-Nup50 fragments, but not GST alone, bound to p27Kip1 protein that was translated in vitro or endogenous p27Kip1 contained within cell lysates (Fig. 1A and data not shown). We also transfected HeLa cells with expression vectors for Nup50 and p27Kip1 and observed specific coimmunoprecipitation of Nup50 and p27Kip1, although the fraction of p27Kip1 bound to Nup50 was relatively small (Fig. 1B and see below). However, since living cells and reticulocyte lysates contain many nucleoporins and transport receptors, the observed interactions between p27Kip1 and Nup50 in the two-hybrid, GST-pulldown, and coimmunoprecipitation assays may be either direct or indirect and bridged via other proteins.
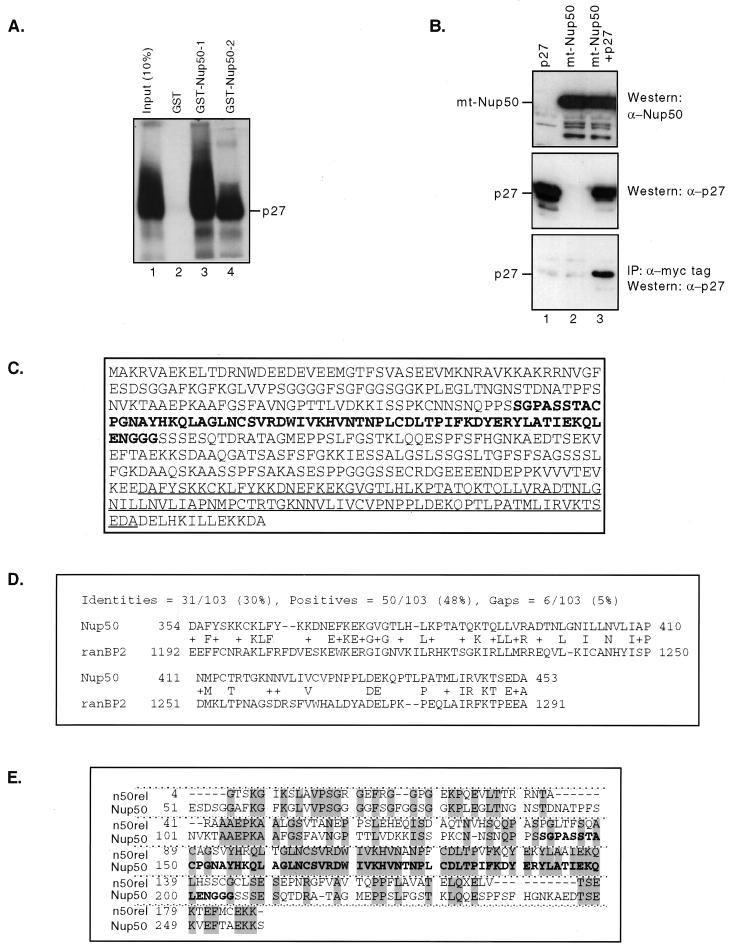
FIG. 1.
Nup50 is a p27Kip1-interacting protein. (A) GST pulldown assay. p27Kip1 translated in vitro bound specifically to two overlapping GST-Nup50 fusion proteins containing the minimal p27Kip1 interaction domain of Nup50 (lanes 3 and 4) but not to GST alone (lane 2) (B) Coimmunoprecipitation of Nup50 and p27Kip1. HeLa cells were transfected with expression vectors for p27 (lanes 1 and 3) and myc-tagged Nup50 (lanes 2 and 3). Lysates either were Western blotted with anti-Nup50 or anti-p27Kip1 antisera or were immunoprecipitated with anti-myc tag antibody followed by Western blotting with anti-p27Kip1 as indicated. A portion (10%) of the total amount of immunoprecipitated lysate was used in the Western blots. (C) Sequence of the mouse Nup50 gene. The minimal region found in two-hybrid screens to interact with p27Kip1 (boldface) and putative Ran-binding domain (underlined) are indicated. (D) Alignment of the C terminus of Nup50 with the Ran-binding domain of Ran-binding protein 2. (E) Alignment of mouse Nup50 with n50rel. The minimal p27Kip1 interaction domain is shown in boldface.
We cloned and sequenced a 1.8-kb Nup50 cDNA obtained by screening a 16-day-old mouse embryo library with a mouse Nup50 probe derived from the longest two-hybrid clone. The Nup50 open reading frame encodes a 466-amino-acid protein with a predicted molecular mass of 50,000 Da (Fig. 1C, GenBank accession number AF229256). The predicted protein is 94% identical to the 467-amino-acid rat Nup50 protein and 77% identical to the 468-amino-acid human Nup50 sequence (a corrected rat Nup50 sequence, accession number U41845, has recently been entered in GenBank in which a frameshift in the C terminus has been corrected). The minimal mouse Nup50 cDNA fragment that was isolated in the p27Kip1 two-hybrid screen is indicated. The C terminus of mouse Nup50 contains a domain with homology to the Ran-binding domains of RanBP1 and RanBP2 that is also present in both the human and the rat Nup50 sequences (Fig. 1C and D and data not shown).
The lex-p27Kip1(72–198) bait was also used to screen a testis-specific two-hybrid library. In this screen, approximately 10% of the multiple independent clones identified represented Nup50, whereas all of the remaining clones represented a Nup50-related cDNA that we have designated n50rel because of its sequence homology to Nup50. A partial sequence of n50rel was obtained by assembling the overlapping independent clones obtained in the testis two-hybrid screen and revealed that n50rel and Nup50 are highly related in the minimal p27Kip1-interaction region (69% identical over 64 amino acids) but diverge in the remaining regions sequenced (Fig. 1E).
Mouse Nup50 is a nuclear-pore-associated protein that is widely expressed.
Northern blot analysis of total RNA from normal mouse tissues detected low levels of Nup50 mRNA in most tissues, and 10- to 20-fold more mRNA in testes than other tissues (not shown). In situ hybridization of sectioned testes demonstrated intense expression of Nup50 mRNA in a specific zone of spermatocytes undergoing, or that had recently completed, meiosis (Fig. 2A). This is consistent with the reported observation that rat Nup50 protein is highly expressed in the testes and undergoes changes in expression and localization during germ cell differentiation. In contrast to Nup50, Northern analysis of n50rel mRNA revealed high expression in adult testis but no expression in other tissues (not shown). Moreover, whereas the DBEST database contains many Nup50 expressed sequence tags (ESTs) derived from libraries representing a wide variety of human, murine, and rat tissues, the only EST corresponding to n50rel found in BLAST searches was isolated from a mouse testis library (accession number AA183570T). Finally, although we isolated Nup50 cDNAs from both total-embryo and testis two-hybrid libraries, n50rel cDNAs were only isolated from the testis library. These data suggest that, whereas Nup50 is widely expressed, n50rel expression is restricted to the testes. It is important to note, however, that we currently have no data indicating that n50rel encodes an expressed nucleoporin.
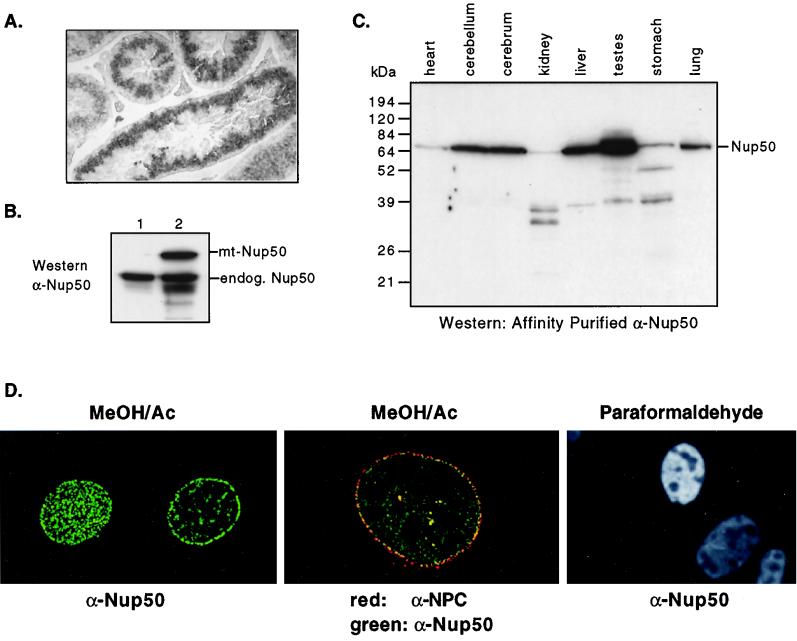
FIG. 2.
Nup50 is a widely expressed nucleoporin. (A) In situ hybridization pattern of Nup50 mRNA in adult mouse testis. (B) Western analysis of proteins detected by anti-Nup50 antibody in either mock-transfected NIH 3T3 cells (lane 1) or NIH 3T3 cells transfected with myc-epitope tagged Nup50 (lane 2). (C) Western analysis of Nup50 expression in adult mouse tissues using affinity-purified anti-Nup50 antibody. (D) Mouse Nup50 localizes to the nuclear pore complex. (Left and center) Deltavision photomicrographs of MEFs fixed with methanol-acetone and stained with anti-Nup50 antibody. Panels: left, staining pattern of Nup50 in surface and equatorial planes; center, colocalization of Nup50 (green) with nucleoporins detected by MAb414 (red); right, Nup50 immunostaining reveals a homogeneous nuclear pattern in MEFs fixed with paraformaldehyde.
We have made rabbit polyclonal antibodies to full-length His-tagged mouse Nup50 protein expressed and purified in E. coli. Both of these antisera recognize a single band of approximately 60,000 Da in NIH 3T3 cells and correctly recognize transfected myc epitope-tagged Nup50 (Fig. 2B and data not shown). Surveys of mouse tissues revealed that Nup50 is detected in all tissues with various abundance, with the highest expression in the testes (Fig. 2C and data not shown). Some smaller products were also observed in some tissues, which could represent either degradation or in vivo processing. These species were also found in cells transfected with Nup50 expression vectors; thus, they are probably not due to alternate splicing (not shown). Furthermore, freeze-thawing of tissue extracts caused the conversion of full-length Nup50 to low-molecular-weight forms, suggesting that they represent proteolytic products (not shown). It is unlikely that these bands represent other proteins immunologically related to Nup50 that are detected by the anti-Nup50 antibodies, since these antisera do not reveal any proteins in lysate prepared from Nup50-null embryos (see Fig. 4D).
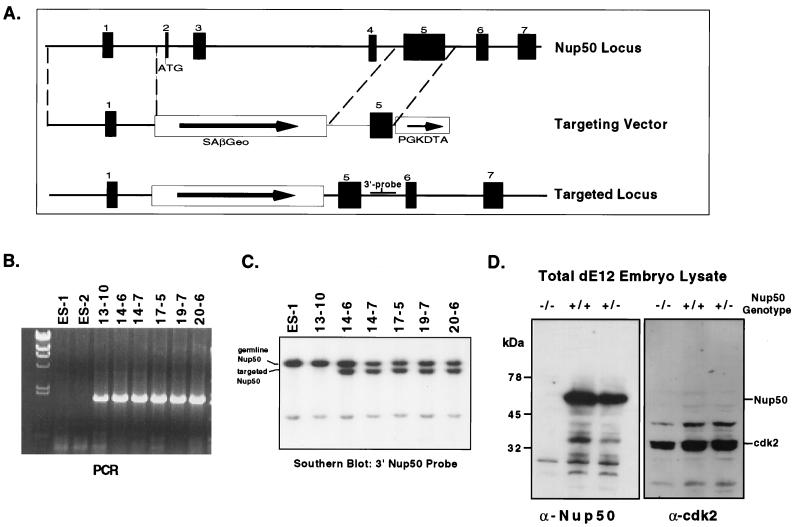
FIG. 4.
Genomic cloning and targeted disruption of mouse Nup50. (A) Genomic structure of the mouse Nup50 locus and construction of a splice acceptor targeting vector. Introns were not completely sequenced and are not drawn to scale. In the correctly targeted locus, the selectable marker is expressed from the endogenous Nup50 transcription unit. (B) PCR analysis of ES cell controls (ES-1 and ES-2) and clones with targeted Nup50 alleles. Only clones with targeted alleles yield a diagnostic PCR product. (C) Southern analysis of the 3′ end of targeted Nup50 alleles. Note that all PCR-positive clones shown in panel B except 13-10 had the expected genomic structure. The structure of the 5′ end of the recombined locus in the targeted clones was also confirmed by Southern analysis (not shown). (D) The targeted Nup50 allele is a null mutation. Total embryo lysate was prepared from E12 embryos from a Nup50+/− cross. The genotypes of the embryos are indicated. Note that no Nup50 protein is detected by anti-Nup50 in the Nup50−/− embryo lysate, whereas control proteins such as Cdk2 are easily detected.
We have observed that mouse Nup50 immunostaining reveals a classic NPC pattern and colocalizes with other NPC proteins in primary mouse and human fibroblasts, as well as in 3T3, 293, HeLa, U2OS, and rat1 cells (Fig. 2D and data not shown). By using digitonin permeabilization conditions that preferentially solubilize the plasma membrane but not the nuclear envelope, we observed that Nup50 is found at the nucleoplasmic side of the nuclear envelope (data not shown). The subcellular localization of Nup50 in all of the cultured cells we examined was strongly dependent upon fixation technique. Methanol-acetone fixation yielded exclusively nuclear envelope staining, whereas paraformaldehyde fixation resulted in very bright homogeneous nuclear staining except in nucleolar regions (Fig. 2D). Guan et al. have found a similar pattern of fixation-dependent Nup50 staining in NRK cells (10). These data suggest that a nuclear pool of Nup50 that is not associated with the nuclear envelope is either extracted or epitope masked by methanol-acetone fixation. Similarly, Fan et al. found that rat Nup50 stained in a homogeneous nuclear pattern during specific stages of spermatocyte differentiation, demonstrating that rat Nup50 may also localize to regions of the nucleus other than the nuclear envelope (6).
Interactions of Nup50 with NPC proteins.
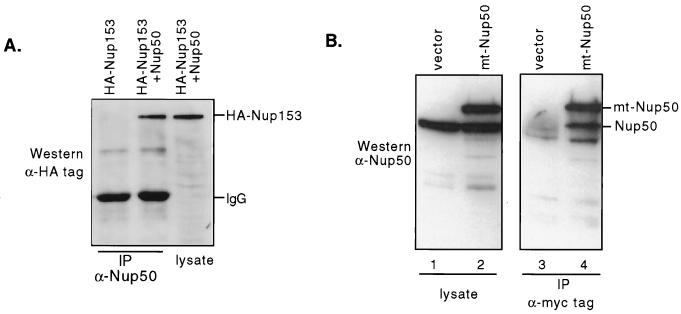
We used the full-length Nup50 cDNA as bait in two-hybrid analyses to determine if Nup50 interacts with other components of the NPC. In independent screens of mouse embryo, brain, and testis libraries, we have found the following NPC proteins that displayed specific two-hybrid interactions with Nup50: importin α2, importin β, transportin, Nup153, Ran binding protein 7, and Nup50 itself. This analysis confirms the association of Nup50 with the NPC observed by immunostaining. Most of these proteins are shuttling import receptor proteins involved in the nucleocytoplasmic transport of various cargo. The Nup153 protein is found in the basket region of the nuclear side of the NPC and plays key roles in both the import and the export of proteins and mRNA (2, 17, 29, 33). In addition to the two-hybrid interactions, we have also coimmunoprecipitated Nup50-Nup153 complexes and Nup50-Nup50 complexes from transfected cells, confirming that these proteins can be found in complexes in mammalian cells in vivo (Fig. 3). Furthermore, Guan et al. have used immunogold electron microscopy and colocalized Nup153 and Nup50 to very similar nucleoplasmic regions of the NPC (10). Finally, the endogenous Nup153 and Nup50 proteins can be coimmunoprecipitated from rat liver nuclei (T. Guan and L. Gerace, personal communication). In sum, these data support the notion that Nup50 and Nup153 are found in complexes in vivo, although it is possible that the Nup50-Nup153 interaction is indirect and mediated by other proteins in both the two-hybrid and coprecipitation experiments.
FIG. 3.
(A) Coimmunoprecipitation of Nup50 with Nup153. 293 cells were transfected with vectors expressing HANup153 HANup153-Nup50 as indicated. Lysates were immunoprecipitated (IP) with anti-Nup50 antibody as indicated and Western blotted with antihemagglutinin (HA) tag antibody. Nup153 is precipitated by anti-Nup50 only when Nup50 is cotransfected. A total of 10% of the amount of lysate immunoprecipitated was run in the lane labeled lysate. (B) Nup50 forms complexes with itself. NIH 3T3 cells were transfected with either a myc-tagged Nup50 expression vector (lanes 2 and 4) or control vector (lanes 1 and 3). The endogenous Nup50 is precipitated by the anti-myc tag antibody only when mt-Nup50 is coexpressed.
Genomic cloning and targeted deletion of Nup50.
We made a targeted deletion of Nup50 in the mouse germ line to study its function. A mouse Nup50 genomic clone was obtained by screening a mouse genomic library, and the intron-exon structure map was determined using primers derived from the mouse Nup50 cDNA. We have found that there are at least seven exons in the mouse Nup50 gene and that the first exon is noncoding (Fig. 4A and data not shown). This genomic structure is quite similar to that of the human Nup50 locus (31). We constructed a targeting vector using pSAβGeolox2dta, which contains a splice acceptor upstream of the β-geo selectable marker and the diphtheria toxin A gene to select against random integrations (Fig. 4A) (7). Homologous recombination of this vector with the endogenous Nup50 gene results in a transcript expressed from the endogenous Nup50 transcription unit in which Nup50 exon 1 is spliced to the β-geo cassette, which contains stop codons in all three reading frames following the selectable marker sequences and which terminates translation. This strategy also allows the determination of the endogenous Nup50 mRNA expression pattern by analysis of β-galactosidase activity. Mouse ES cells were electroporated with the targeting vector, and homologous recombinants were identified by standard PCR-based techniques and confirmed by Southern blotting (Fig. 4B and C and data not shown). ES cells were microinjected into C57/B6J blastocysts and backcrossed into C57/B6J females after germ line transmission of the mutant allele was confirmed.
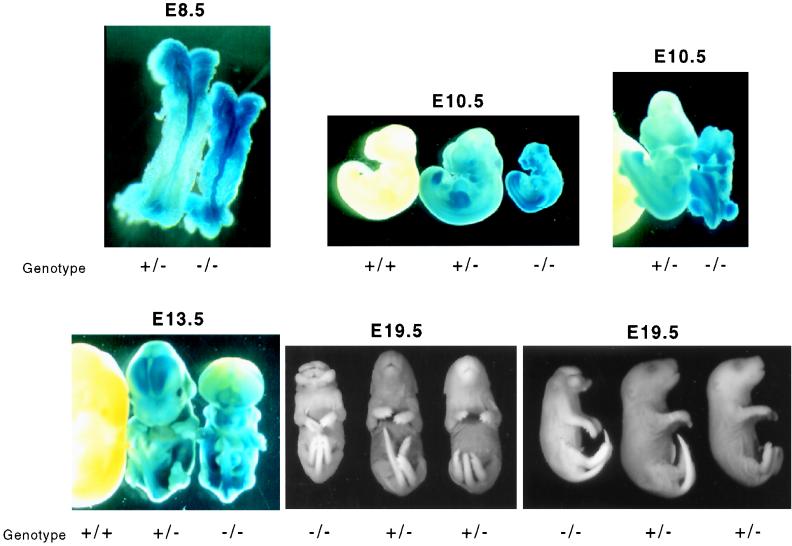
F1 mice heterozygous for the mutant Nup50 allele were mated to assess the consequence of a homozygous-null Nup50 mutation. No live Nup50−/− mice were obtained out of more than 200 pups genotyped from the crosses, although Nup50+/+ and Nup50+/− animals were found at the expected frequency (data not shown). Thus, homozygous deletion of Nup50 appeared to cause embryonic lethality. Heterozygosity for Nup50 was not associated with any detectable abnormalities in either male or female animals monitored for more than 2 years.
Embryonic lethality due to deletion of Nup50 is associated with neural tube defects, exencephaly, and intrauterine growth retardation.
We next analyzed embryos from timed F1 matings at various embryonic stages. In contrast to the live births described above, this analysis revealed homozygous Nup50-null embryos at the expected frequency (Table 1). Embryo genotype was assessed in three ways: (i) PCR, (ii) β-galactosidase expression, and (iii) immunostaining-Western blotting. In wild-type embryos Nup50 expression was easily detected at all developmental stages tested (data not shown). In contrast, no Nup50 expression was detected in homozygous-null animals by either immunostaining embryo sections (not shown) or Western analysis of total embryo lysate (Fig. 4D), thus confirming that the targeting vector produced a true null allele.
TABLE 1.
Summary of embryo phenotypes and genotypesa
| Stage | No. of embryos (%)
|
Phenotype | |||
|---|---|---|---|---|---|
| Nup50+/+ | Nup50+/− | Nup50−/− | Resorbed | ||
| E8.5 to E9.5 | 5 | 25 | 8 | 0 | Wavy neural tube |
| E10.5 | 12 | 21 | 11b | 6 | Kinked neural tube, open cranial neural tube, growth retardation |
| E12.5 to E16.5 | 17 | 36 | 13 | 15 | Exencephaly, growth retardation, complex CNS abnormalities |
| E18.5 | 0 | 3 | 1 | 0 | Exencephaly, complex facial abnormalities, growth retardation |
| Total | 34 (22) | 85 (56) | 33 (22) | ||
The table summarizes data from embryos from 19 individual litters, ranging in age from E8.5 to E18.5. Nup50+/− females were sacrificed after timed matings with Nup50+/− males. Resorbed embryos were not genotyped.
In one E10.5 litter, 2 embryos with normal phenotypes were genotyped as Nup50−/−.
We found that homozygous Nup50 deletion was associated with a grossly abnormal phenotype beginning at approximately embryonic day 8.5 (E8.5). The earliest detectable abnormality in the Nup50-null animals is a kinked neural tube around E8.5 (Fig. 5). Moreover, the neural tube displays the strongest β-galactosidase activity at this developmental stage, indicating that the Nup50 gene is highly expressed in the developing neural tube. Longer β-galactosidase assays revealed Nup50 expression in all embryonic tissues, a finding consistent with the ubiquitous expression in adult tissues as previously observed (Fig. 5). Later in development, the neural tube defect in mutant animals becomes quite prominent and is characterized by a failure of cranial neural tube closure and ultimately by exencephaly (Fig. 5 and data not shown). The Nup50-null animals also displayed significantly reduced size compared with normal and heterozygous littermates (Fig. 5). Live Nup50-null animals were found as late as day E19.5; thus, embryonic death likely occurs late in gestation or perinatally.
FIG. 5.
Morphologic abnormalities in Nup50-null mouse embryos. Embryos were dissected from pregnant Nup50+/− females after timed matings, and the Nup50 genotypes are indicated. The E8.5, E10.5, and E13.5 embryos were stained for β-galactosidase activity which reveals the endogenous Nup50 expression pattern. The E19.5 embryos were fixed in paraformaldehyde and stored in 70% ethanol prior to photography.
Analysis of p27Kip1 expression in Nup50-null animals.
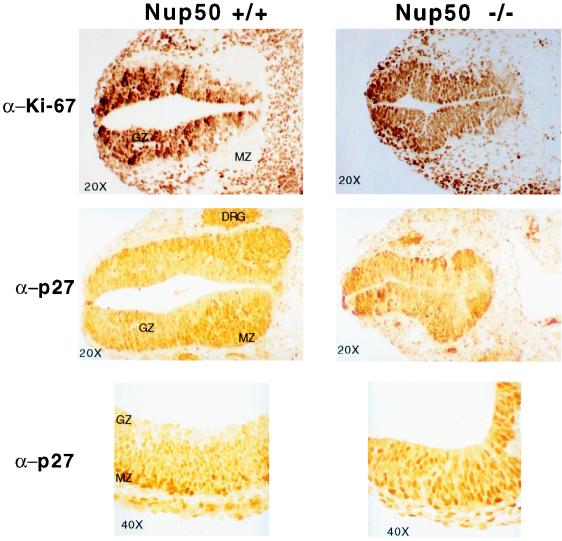
We examined p27Kip1 expression in normal and Nup50-null animals. In normal embryos, p27Kip1 expression was found in many tissues, but it was most highly expressed throughout the central nervous system (Fig. 6 and data not shown). The developing neural tube displayed a clear gradient of p27Kip1 staining, such that p27Kip1 expression was highest in the germinal zone of the neuroepithelium (postmitotic cells) and lowest in the proliferating marginal zone. This inverse relationship between p27Kip1 expression and neuroepithelial cell proliferation is illustrated by the inverse staining pattern of p27Kip1 and the proliferative marker Ki-67 (Fig. 6). In Nup50-null animals this gradient is lost in many regions of the central nervous system (CNS), and neuroepithelial cells expressing high levels of p27Kip1 were found scattered throughout the abnormally developing neural tube. Additionally, the gradient of Ki-67 staining was lost in regions of the neural tube in Nup50-null embryos such that no marginal zone was easily demarcated, suggesting that the Nup50 deletion is associated with a neural tube proliferative defect.
FIG. 6.
Transverse sections of E10.5 Nup50+/+ (left) and Nup50−/− (right) embryos were immunostained with either anti-Ki-67 antibody or anti-p27Kip1 antibody as indicated. Approximately equivalent sections from a region just caudal to the open portion of the Nup50−/− neural tube are shown. Note the low Ki-67 and high p27Kip1 expression in the marginal zone (MZ) compared to the germinal zone (GZ) of the neural tube in the wild-type embryo and the loss of this relationship in the Nup50-null embryo. The bottom image in each column is a higher-magnification image of the neural tube that clearly reveals the p27Kip1 gradient in the normal neuroepithelium.
Analysis of Nup50-null fibroblasts.
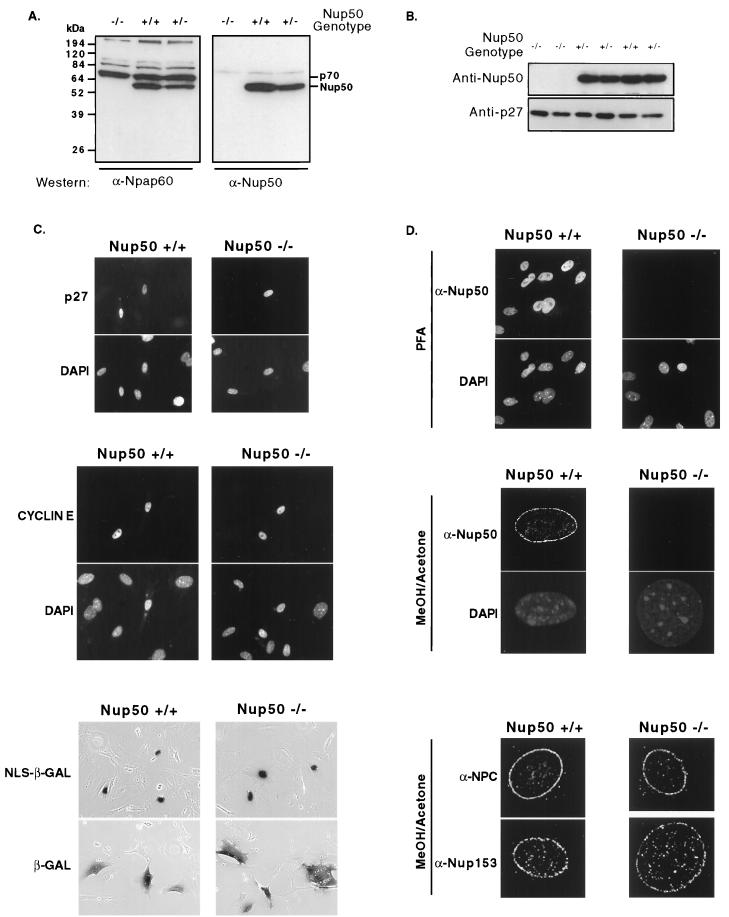
We prepared MEFs from 12- to 14-d.p.c. No expression of Nup50 was detected in Nup50-null cells (Fig. 7A). In contrast, the highly related p70 protein detected by the anti-Nup50 antibody previously designated anti-npap60 (6, 10) is expressed equally in all MEFs, regardless of their Nup50 genotype (Fig. 7A). We found that Nup50−/−, Nup50+/−, and Nup50+/+ MEFs proliferated at similar rates; that asynchronous cultures of wild-type and Nup50-null cells had indistinguishable cell cycle profiles; and that Nup50-null and wild-type cells exited and re-entered the cell cycle following serum starvation and density arrest with similar kinetics (data not shown). We also examined the localization and abundance of endogenous p27Kip1 and found no differences resulting from Nup50 deletion (Fig. 7B and data not shown). Thus, we have observed no defects in either p27Kip1 regulation or cell cycle control in Nup50-null MEFs.
FIG. 7.
Analyses of Nup50−/− MEFs. (A) The Nup50-related protein p70 is expressed in Nup50-null MEFs. Lysates prepared from MEFs with the indicated genotypes were electrophoresed, and the same filter was cut in half and probed with either the anti-npap60 antibody developed by Fan et al. (6) or anti-Nup50. Note that both antisera detect Nup50 but only the anti-npap60 detects p70. No Nup50 expression is detected by either antibody in Nup50-null cells, whereas p70 expression is unaffected by the Nup50 deletion. (B) Endogenous p27Kip1 is expressed at similar levels in wild-type and Nup50-null cells. (C) Transfected proteins correctly localize to the nucleus in Nup50-null MEFs. Staining patterns of p27Kip1 and cyclin E with corresponding DAPI images (to reveal all nuclei) are shown in the top and middle panels, respectively. The bottom panel depicts a β-galactosidase assay of cells transfected with either a NLS–β-galactosidase or a β-galactosidase expression vector. NLS–β-galactosidase contains the simian virus 40 NLS. (D) No Nup50 staining is detected in Nup50-null cells fixed with either paraformaldehyde (PFA [top panel]) or methanol-acetone (middle panel). Corresponding DAPI images are also shown. Nup153 and NPC proteins detected by MAb414 correctly localize to the NPC in Nup50-null cells (bottom panel).
We examined the localization of several endogenous and transfected proteins in MEFs and found that c-myc, p21Cip1, p27Kip1, cyclin E, and NLS–β-galactosidase were all correctly localized to the cell nucleus in Nup50-null cells (Fig. 7C and data not shown). Thus, protein import appears grossly normal in Nup50-null cells, although there could be subtle differences in protein import rates between normal and mutant cells that might not be reflected by protein localization at steady state. We also examined the localization of endogenous and transfected cyclin B1, which is dependent upon the function of the Crm1 export receptor. We found identical cyclin B1 staining in wild-type and mutant MEFs in the absence or presence of leptomycin B, suggesting that Crm1-dependent protein export is also grossly normal in Nup50-null fibroblasts (data not shown). Finally, we characterized NPC proteins in MEFs. We found that no Nup50 expression was detected by immunocytochemistry in Nup50-null cells fixed with either paraformaldehyde or methanol-acetone and that the expression and localization of Nup153 as well as NPC proteins detected by MAb414 were normal in Nup50-null cells (Fig. 7D). Thus, Nup50 expression is not required for the localization of Nup153 and other nucleoporins to the NPC in MEFs.
DISCUSSION
We have found that murine Nup50 localizes to the NPC and displays specific two-hybrid interactions with several well-defined constituents of the nucleocytoplasmic transport machinery, as well as the p27Kip1 protein. Most of the Nup50 two-hybrid interactors that we identified are mobile import receptors that carry various classes of protein cargo into the nucleus (importin α, importin β, transportin, and RanBP7). We also found two-hybrid interactions between Nup50 and itself and with the Nup153 protein. Unlike the importin superfamily, Nup153 is an integral component of the NPC involved in multiple transport processes that physically interacts with several transport receptors (including importin α, importin β, exportin, and transportin), the NPC, and Ran. We have also found that Nup50 can form complexes (although not necessarily via direct interaction) with p27Kip1 and Nup153 in GST pulldown and coimmunoprecipitation assays. In agreement with our observations, Guan et al. found that the endogenous Nup50 and Nup153 proteins colocalize to a similar region of the nucleoplasmic side of the NPC and can be coimmunoprecipitated from rat liver nuclei (10; Guan and Gerace, personal communication).
The targeted disruption of Nup50 caused complex neural tube and CNS abnormalities and growth retardation. Although we have found that Nup50 is expressed ubiquitously in adult and embryonic tissues, the phenotypic abnormalities associated with the Nup50 deletion were most severe in the CNS. This finding is in contrast with the only other reported deletion of a nucleoporin in mice, the CAN-Nup214 protein. CAN-Nup214-null cells are not viable, and its deletion causes early embryonic death as maternal stores of CAN-Nup214 mRNA are depleted (34). Most embryonic tissues in Nup50-null animals appear grossly normal. However, the intrauterine growth retardation observed in Nup50-null embryos suggests that Nup50-null tissues other than the CNS are affected, and it is possible that embryonic tissues that appear normal may have more subtle defects resulting from Nup50 loss.
We have not yet found any defects in Nup50-null MEFs. Thus, either the function performed by Nup50 is not required in MEFs or other NPC proteins may be functionally redundant with Nup50 and compensate for its absence. Guan et al. have inhibited Nup50 function in NRK cells by microinjecting anti-Nup50 antibodies and found that Nup50 inhibition specifically blocks the nuclear export of proteins with leucine-rich Crm1-dependent NES (10). Crm1-dependent export appears to be an essential cellular function: inhibition of Crm1 with the drug leptomycin B in mammalian cells causes cell death, and Crm1 is an essential gene in Saccharomyces cerevisiae (28). Thus, our finding that Nup50-null MEFs proliferate and regulate cyclin B1 localization normally supports the idea that Nup50 is functionally redundant with other NPC proteins in some cell types. Our observation that p70, a protein highly related to Nup50, is expressed equally in wild-type and Nup50-null MEFs raises the possibility that p70 compensates for Nup50 in Nup50-null MEFs. Perhaps other structural and/or functional homologues will be found in other tissues. We have identified n50rel as a putative Nup50 homologue expressed in the testis. In humans, Nup50 homologous sequences have been detected by hybridization on chromosomes 5, 6, and 14, in addition to the bona fide Nup50 gene on 22q13.3, although it is not known if these loci represent functional genes (31).
We have observed variation in Nup50 abundance, despite its ubiquitous expression, most notably as increased expression in the adult testes and in the embryonic neural tube. This expression pattern in the developing neural tube is consistent with the essential role for Nup50 in neurulation demonstrated by the Nup50-null phenotype. Neural tube morphogenesis is extremely complex, and a large number of gene products have been identified that are required for this process (23). Thus, the mechanisms through which Nup50 deletion leads to the observed phenotypic abnormalities remains unknown.
We initially identified Nup50 through its two-hybrid interaction with p27Kip1. p27Kip1 export has been reported to be leptomycin B sensitive (30), and the findings of Guan et al. implicating Nup50 in the Crm1 pathway export (10) led us to hypothesize that Nup50 might mediate interactions between p27Kip1 and its export pathway. We have not, however, identified any specific abnormalities in p27Kip1 regulation directly attributable to Nup50 deletion. Nup50-null MEFs exhibited no obvious defects in either p27Kip1 regulation and/or cell cycle control. One possibility is that another protein, such as p70, compensates for the absence of Nup50. Alternatively, Nup50 may not play any direct role in p27Kip1 regulation. To further address this question, we have examined p27Kip1 expression in the most abnormal tissue in Nup50-null animals, the developing neural tube. We found that p27Kip1 expression is highly regulated in the developing neuroepithelium and correlates strongly with proliferative status. In Nup50-null embryos this correlation breaks down, and cells highly expressing p27Kip1 were found throughout the neuroepithelium. Abnormal neuroepithelial cell proliferation has been associated with neural tube defects, and it is tempting to speculate that the abnormalities observed in the Nup50-null animals are at least partially attributable to aberrant p27Kip1 regulation (27). However, because the neural tube becomes disordered in Nup50 mutant animals, it is difficult to determine if the alterations in p27Kip1 expression in Nup50-null animals are the cause or the consequence of the neural tube abnormalities. A mechanistic understanding of these abnormalities thus must await a clearer understanding of Nup50 function in nucleocytoplasmic transport.
ACKNOWLEDGMENTS
We thank Mark Groudine and Jim Roberts for their advice, support, and critical review of the manuscript. We also thank Larry Gerace, Norman Arnheim, and Martin Eilers for communicating data prior to publication and Brian Burke for providing reagents.
B.E.C. is a W. M. Keck Distinguished Young Scholar in Medical Research and a James S. McDonnell Scholar. This work was supported by NIH grants R01 CA 84069 (B.E.C.) and RO1 DK52530 (R.K.).
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 2.Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clurman B E, Porter P. New insights into the tumor suppression function of P27(kip1) Proc Natl Acad Sci USA. 1998;95:15158–15160. doi: 10.1073/pnas.95.26.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clurman B E, Sheaff R J, Thress K, Groudine M, Roberts J M. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by Cdk2 binding and cyclin phosphorylation. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 5.Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 6.Fan F, Liu C P, Korobova O, Heyting C, Offenberg H H, Trump G, Arnheim N. cDNA cloning and characterization of Npap60: a novel rat nuclear pore-associated protein with an unusual subcellular localization during male germ cell differentiation. Genomics. 1997;40:444–453. doi: 10.1006/geno.1996.4557. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg M W, Allen T D. The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J Mol Biol. 1996;257:848–865. doi: 10.1006/jmbi.1996.0206. [DOI] [PubMed] [Google Scholar]
- 9.Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan T, Kehlenbach R, Schirmer E, Kehlenbach A, Fan F, Clurman B E, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr Biol. 1999;9:680–689. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D. Using antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 13.Hogan B, Beddington V, Costantini F, Lacey E. Manipulating the mouse embryo: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 14.Kopan R, Weintraub H. Mouse notch: expression in hair follicles correlates with cell fate determination. J Cell Biol. 1993;121:631–641. doi: 10.1083/jcb.121.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Li M Z, Leibham D, Cortez D, Elledge S J. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- 16.Melchior F, Gerace L. Mechanisms of nuclear protein import. Curr Opin Cell Biol. 1995;7:310–318. doi: 10.1016/0955-0674(95)80084-0. [DOI] [PubMed] [Google Scholar]
- 17.Moroianu J, Blobel G, Radu A. RanGTP-mediated nuclear export of karyopherin alpha involves its interaction with the nucleoporin Nup153. Proc Natl Acad Sci USA. 1997;94:9699–9704. doi: 10.1073/pnas.94.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 19.Orend G, Hunter T, Ruoslahti E. Cytoplasmic displacement of cyclin E-Cdk2 inhibitors p21Cip1 and p27Kip1 in anchorage-independent cells. Oncogene. 1998;16:2575–2583. doi: 10.1038/sj.onc.1201791. [DOI] [PubMed] [Google Scholar]
- 20.Pante N, Aebi U. Toward the molecular details of the nuclear pore complex. J Struct Biol. 1994;113:179–189. doi: 10.1006/jsbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 21.Pines J. Four-dimensional control of the cell cycle. Nat Cell Biol. 1999;1:E73–E79. doi: 10.1038/11041. [DOI] [PubMed] [Google Scholar]
- 22.Reynisdottir I, Massague J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with Cdk4 and Cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- 23.Schoenwolf G C, Smith J L. Mechanisms of neurulation: traditional viewpoint and recent advances. Development. 1990;109:243–270. doi: 10.1242/dev.109.2.243. [DOI] [PubMed] [Google Scholar]
- 24.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 25.Sherr C J, Roberts J M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 26.Singh S P, Lipman J, Goldman H, Ellis F H, Jr, Aizenman L, Cangi M G, Signoretti S, Chiaur D S, Pagano M, Loda M. Loss or altered subcellular localization of p27 in Barrett's associated adenocarcinoma. Cancer Res. 1998;58:1730–1735. [PubMed] [Google Scholar]
- 27.Smith J L, Schoenwolf G C. Neurulation: coming to closure. Trends Neurosci. 1997;20:510–517. doi: 10.1016/s0166-2236(97)01121-1. [DOI] [PubMed] [Google Scholar]
- 28.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 29.Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- 30.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 31.Trichet V, Shkolny D, Dunham I, Beare D, McDermid H E. Mapping and complex expression pattern of the human NPAP60L nucleoporin gene. Cytogenet Cell Genet. 1999;85:221–226. doi: 10.1159/000015297. [DOI] [PubMed] [Google Scholar]
- 32.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 33.Ullman K S, Shah S, Powers M A, Forbes D J. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell. 1999;10:649–964. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Deursen J, Boer J, Kasper L, Grosveld G. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 35.Vojtek A B, Hollenberg S M. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 1995;255:331–342. doi: 10.1016/s0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Bardes E S, Moore J D, Brennan J, Powers M A, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Kornbluth S. All aboard the cyclin train: subcellular trafficking of cyclins and their CDK partners. Trends Cell Biol. 1999;9:207–210. doi: 10.1016/s0962-8924(99)01577-9. [DOI] [PubMed] [Google Scholar]