Abstract
Background
Acute lymphoblastic leukemia (ALL) is characterized by an abnormal proliferation of immature lymphocytes, in whose development involves both environmental and genetic factors. It is well known that single nucleotide polymorphisms (SNPs) in coding and noncoding genes contribute to the susceptibility to ALL. This study aims to determine whether SNPs in miR-146a, miR-196a-2, miR-499a, and miR-612 genes are associated with the risk to ALL in pediatric Mexican population.
Methods
A multicenter case-control study was carried out including patients with de novo diagnosis of ALL and healthy subjects as control group. The DNA samples were obtained from saliva and peripheral blood, and the genotyping of rs2910164, rs12803915, rs11614913, and rs3746444 was performed using the 5′exonuclease technique. Gene-gene interaction was evaluated by the multifactor dimensionality reduction (MDR) software.
Results
miR-499a rs3746444 showed significant differences among cases and controls. The rs3746444G allele was found as a risk factor to ALL (OR, 1.6 [95% CI, 1.05–2.5]; p = 0.028). The homozygous GG genotype of rs3746444 confers higher risk to ALL than the AA genotype (OR, 5.3 [95% CI, 1.23–23.4]; p = 0.01). Moreover, GG genotype highly increases the risk to ALL in male group (OR, 17.6 [95% CI, 1.04–298.9]; p = 0.00393). In addition, an association in a gender-dependent manner among SNPs located in miR-146a and miR-196a-2 genes and ALL susceptibility was found.
Conclusion
Our findings suggest that SNP located in miR-499a, miR-146a, and miR-196a-2 genes confer risk to ALL in Mexican children. Experimental analysis to decipher the role of these SNPs in human hematopoiesis could improve our understanding of the molecular mechanism underlying the development of ALL.
Keywords: acute lymphoblastic leukemia, mir-146a, mir-196a-2, miR-499a, miR-612, association study, Mexican population, single nucleotide polymorphism
Introduction
Acute lymphoblastic leukemia (ALL) is the most common pediatric hematological malignancy around the world, representing over 80% of all cases under 18 years old (1). This entity is highly prevalent in Mexican population, which displays one of the highest rate of relapse and death in comparison with other ethnic groups even after using chemotherapeutic approaches implemented in developed countries (2, 3). ALL emerges by an abnormal proliferation of immature lymphocytes and their progenitors that replace the hematopoietic elements in the bone marrow and other lymphoid organs. So far, most of the causes of ALL are undeciphered; however, it is well known that an interaction within environmental and genetic factors is needed to develop this malignancy (4–6). Among the identified risk genetic factors to suffer ALL are the single nucleotide polymorphisms (SNP), both, in coding and no coding genes (6–9). No coding genes comprises around 98% of the human-transcribed genome, which is mainly represented by microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) that play a relevant role in LLA and other types of cancer (10). miRNAs are small endogenous RNAs of 19–25 nucleotides that function as posttranscriptional regulators silencing specific mRNAs. miRNAs interact with their targeted mRNAs by complementary base pairing, most of them in the 3′-untranslated region (UTR) of the target mRNA, although interplay in the 5′UTR region has also been documented. Targeted coding mRNAs by specific miRNAs could be either in complete or incomplete fashion (11). Experimental evidences have revealed that miRNA dysfunction contributes to the establishment of diverse human diseases, since miRNA-mRNA-specific interaction makes fine-scale adjustments to protein outputs (8, 12, 13). It has been identified that several SNP located into miRNA gene sequences are closely responsive for their abnormal function by modifying pri-miRNA transcription, pri-miRNA/pre-miRNA processing, or by disrupting miRNA-mRNA interactions (14, 15). The rs2910164 G/C in miR-146a gene has been reported as an alterer of the gene expression, then its targeted mRNAs, which are involved in fundamental biological processes (cell differentiation, hematopoyesis, and innate and adaptive immunity, etc.) (16, 17). The rs2910164 has been associated with many types of cancer and several immune-mediated diseases (18–20); however, its association with ALL has shown controversial results (9, 17, 21). Another functional miR-SNP is rs3746444, which results from an A-to-G substitution in the seed region of miR-499a, was reported as significantly associated with an increased susceptibility to several human conditions, including cancer (19, 22). To know whether rs2910164 G/C in miR-146a, rs11614913 T/C in miR-196a-2, rs3746444 A/G in miR-499a, and rs12803915 G/A in miR-612 are associated with ALL in Mexican children, we performed a case control study.
Material and Methods
Subjects
As part of the Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia (MIGICCL), we conducted a case-controls study from August 1, 2014, to July 31, 2016. Participants were younger than 18 years, residents of the Metropolitan Area of Mexico City, and recruited from public hospitals and health institutions from Mexico City, Mexico as was described previously by Medina-Sanzon et al. (6). ALL diagnosis was established by either a hematologist or an oncologist according to clinical characteristics, and bone marrow (BM) aspirate data. Gender, age at diagnosis, white blood cell count (WBC), immunophenotype, and risk classification group were registered from the patients’ medical records. We used the National Cancer Institute (NCI) risk criteria for ALL case stratification as follows: (a) standard risk: 1–9.99 years of age or WBC <50 × 10^9/L, and (b) high risk: ≤1 or ≥10 years of age and/or WBC ≥50 × 10^9/L. Patients included in the study were treated with chemotherapy, none of them received HSCT therapy. Relapse was considered when ≥5% leukemic blasts were detected in BM sample during the first 36 months after having achieved complete remission (CR). Early mortality was defined as the patient’s death during the first 24 months. Cases with Down syndrome were excluded from the analysis. All institutional committees of Ethics, Research, and Biosecurity of the participant institutions approved this study. Written informed consent was obtained from all participants and the children’s parents. Patients ≥8 years old gave their assent (when possible) to be included in the present study. Cases and controls were selected according to criteria described in a previous study (6). Briefly, controls were recruited from second-level hospitals of the same health institution that referred the children with ALL to the third-level care hospitals. Control children were recruited from the departments of ambulatory surgery, pediatrics, and ophthalmology; orthopedic outpatient clinics; and the emergency room of the referred hospitals and have no leukemia, hematological diseases, allergies, infections, and congenital malformations. A set of adult patients was included to test the associated SNP miR-499a_ rs3746444. The group of adult patients and controls is described in the Material and Methods section in the Supplementary Material .
DNA Extraction, SNP Selection, and Genotyping
Genomic DNA from saliva or peripheral blood was obtained according to the ORAGENE Purification Kit (DNA Genotek Inc., Kanata, ON, Canada) and the Gentra Kit (Gentra Systems Inc., Minneapolis, MN, USA) according to the manufacturer’s instructions. DNA purity and concentration were determined by sypectrofotometry (Nanodrop-1000). The rs2910164 (miR-146a), rs11614913 (miR-196a-2), rs3746444 (miR-499a), and rs12803915 (miR-612) were selected base on previous association studies in ALL and other malignancies (8, 9, 13, 17, 21, 23–26). Genotyping was performed using the 5′exonuclease technique and TaqMan MGB chemistry in a QuantStudio 5 system according to the manufacturer’s instructions (Thermo Fisher, Foster City, CA, USA). TaqMan probes used were C:15946974_10 (rs2910164), C:31185852_10 (rs11614913), C:_2142612_40 (rs3746444), and C:32062363_10 (rs12803915). PCR reaction contained 25 ng of genomic DNA, 2.5 µl of TaqMan master mix, 0.0625 µl of 40× assay mix, and ddH2O up to a final volume of 5 µl. The PCR protocol included denaturing at 95°C for 10 min, followed by 40 cycles of denaturing at 95°C for 15 s, and annealing and extension at 60°C for 1 min. Genotypes were assigned automatically by measuring the allele-specific fluorescence by using QuantStudio Design and Analysis software 5 for allelic discrimination (Applied Biosystems, Foster City, CA, USA). The overall genotype call rate was over 98.0% and 100% concordance of a subset of randomly repeated samples during the genotyping.
Statistical Analyses
Hardy-Weinberg Equilibrium (HWE) test was performed using the FINETTI program (http://ihg.gsf.de/cgicbin/hw/hwa1.pl). Alleles and genotype frequencies were compared among groups by using Chi-square and Fisher’s exact tests (when appropriate) implemented in the STATCALC program (Epi Info v.6.02 software, Centers for Disease Control and Prevention, Atlanta, GA). By comparing cases and controls, all SNPs were evaluated under the codominant, dominant, and recessive genetic models using the FINETTI program. Bonferroni correction test was applied. The multifactor dimensionality reduction (MDR) software (V 3.0.2) was used to evaluate gene-gene interactions (27). All p-values ≤ 0.05 were considered statistically significant.
Results
Features of Studied Subjects
The present work included 678 subjects from Mexico City, of which, 423 were children with ALL, and 255 children non-ALL. The ALL children were followed up for at least 3 years (3–7) after initial diagnosis. Males were more frequent than females either in cases (57.9% vs. 42.1%, respectively) nor controls (54.7% v/s 45.2%, respectively), but differences were not statistically significant (p = 0.43). The proportion of children under 10 years old were higher in both groups, and a significant difference was detected among cases (62.2%) and controls (71.1%) (p = 0.02). Median age of ALL children was 9.09 (0–18) and 6.4 (0–17) of the control group. Overall, 68.3% had >90% blast in bone marrow; 91.2%, 6.9%, and 1.9% were pre-B, cell-T, and biphenotype, respectively. Available clinical data are shown in Table 1 .
Table 1.
Clinical characteristics of patients with acute lymphoblastic leukemia.
| Features | Cases (n = 423) | |
|---|---|---|
| n | % | |
| Gender | ||
| Male | 245 | 57.9 |
| Female | 178 | 42.1 |
| Age group (years) | ||
| <1 | 9 | 2.1 |
| 1–9 | 258 | 61.0 |
| ≥10 | 156 | 36.9 |
| Age at diagnosis (years) | ||
| Median (min–max) | 7.9 (0–18) | |
| BM blast at diagnosis (%) | ||
| <90 | 135 | 31.7 |
| ≥90 | 288 | 68.3 |
| Median (min–max) | 85.3 (20–100) | |
| Inmunophenotype | ||
| Pre-B Cell | 386 | 91.2 |
| Cell-T | 29 | 6.9 |
| Biphenotype | 8 | 1.9 |
| NCI risk classification | ||
| Standard risk | 214 | 50.6 |
| High risk | 209 | 49.4 |
| Relapse | ||
| No | 346 | 81.8 |
| Yes | 77 | 18.2 |
| Relapse site | ||
| Isolated BM | 52 | 67.5 |
| Isolated CNS | 17 | 22.1 |
| BM and CNS | 2 | 2.6 |
| BM and CNS and eye | 1 | 1.3 |
| CNS and eyes | 1 | 1.3 |
| BM and testis | 3 | 3.9 |
| Ovary | 1 | 1.3 |
| Death | ||
| No | 364 | 86.0 |
| Yes | 59 | 14.0 |
WBC, whole blood cell count; BM, bone marrow; NCI, National Cancer Institute; CNS, central nervous system.
Association Study
Except for miR-146a, the genotypes of miR-196a-2, miR-499a, and miR-612 were in HWE in the control population. The association analysis between miRNA SNPs and ALL are described in Table 2 and Supplementary Table S1 . Case-control analysis including all children showed a significant association among miR-499a rs374644 with ALL ( Table 2 ). miR-499a rs3746444G alelle observed an OR of 1.6 (95% CI, 1.008–2.5), p = 0.028. However, this significance did not remain after Bonferroni correction test. To note, under codominant model analysis AA vs. GG, statistical significance was found: OR, 5.3 (95% CI, 1.23–23.4); p = 0.01 ( Table 1 ). Stratification analysis by gender observed that miR-499a rs3746444G is associated with ALL in a gender-dependent manner, being a risk factor to males (OR, 2.46 [95% CI, 1.31–4.60]; p = 0.0037) but no to girls (p = 0.95) ( Table 3 ). Moreover, in comparison with AA genotype, GG genotype highly increases the risk to ALL (OR, 17.6 [95% CI, 1.04–298.9]; p = 0.00393) in males. Data are shown in Table 3 .
Table 2.
Association analysis among miR-499 rs3746444 and acute lymphoblastic leukemia.
| Children | OR [CI], p-value | Adults | OR [CI], p-value | All | OR [CI], p-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Control (%) | Cases n (%) | Control n (%) | Cases n (%) | Control n (%) | Cases n (%) | ||||
| N | 255 | 416 | 180 | 71 | 435 | 489 | |||
| Genotypes | |||||||||
| AA | 229 (89.8) | 362 (87.0) | 157 (87.2) | 59 (83.1) | 386 (88.7) | 421 (86.1) | |||
| AG | 24 (9.4) | 39 (9.3) | 23 (12.8) | 9 (12.7) | 47 (10.8) | 48 (9.8) | |||
| GG | 2 (0.8) | 17 (4.8) | 0 (0) | 3 (4.2) | 2 (0.5) | 20 (4.1) | |||
| Alelles | 1.6 [1.05–2.5], 0.028* | 1.7 [0.87–3.34], 0.11 | 1.58 [1.1–2.2], 0.01* | ||||||
| A | 482 (94.5) | 763 (91.4) | 337 (93.6) | 127 (89.4) | 819 (94.1) | 824 (91.0) | |||
| G | 28 (5.5) | 73 (8.8) | 23 (6.4) | 15 (10.6) | 51 (5.9) | 88 (9.0) | |||
| Codominant | 5.3 [1.23–23.4], 0.01* | 18.5 [0.94–364], 0.005 | 9.16 [2.1–39.4], 0.00033* | ||||||
| AA vs. GG | |||||||||
OR, odds ratio; CI, confidence interval. *Statistically significant.
Table 3.
Association analysis among miR-499 rs3746444 and acute lymphoblastic leukemia in children stratified by gender.
| Male | OR [CI], p-value | Female | OR [CI], p-value | |||
|---|---|---|---|---|---|---|
| Control (%) | Cases n (%) | Control n (%) | Cases n (%) | |||
| N | 255 | 416 | 180 | 71 | ||
| Genotypes | ||||||
| AA | 126 (89.8) | 207 (87.0) | 103 (87.2) | 155 (83.1) | ||
| AG | 13 (9.4) | 25 (9.3) | 9 (12.8) | 14 (12.7) | ||
| GG | 0 (0.8) | 14 (4.8) | 2 (0) | 3 (4.2) | ||
| Alelles | 2.46 [1.31–4.60], 0.0037* | 1.021 [0.49–2.09], 0.95 | ||||
| A | 482 (94.5) | 763 (91.4) | 337 (93.6) | 127 (89.4) | ||
| G | 28 (5.5) | 73 (8.8) | 23 (6.4) | 15 (10.6) | ||
| Codominant | 17.6, [1.04–298.9], 0.00393* | 0.99 [0.16 6.06], 0.99 | ||||
| AA vs. GG | ||||||
OR, odds ratio; CI, confidence interval. *Statistically significant. Genotyping >98%.
miR-146a rs2910164, miR-196a-2 rs11614913, and miR-612 rs12803915 association analysis including all children with ALL showed differences among cases and controls but were not statistically significant ( Supplementary Table S1 ). The analysis stratified by gender revealed that homozygote genotype for the minor allele CC of miR-146a rs2910164 was differentially distributed among male ALL cases and male controls (OR, 4.3 (1.60–11.61); p = 0.02). Meanwhile, miR-196a-2 rs11619413 was associated with ALL in female (C vs. T: OR, 1.54 [95% CI, 1.08–2.2]; p = 0.015) ( Supplementary Table S2 ).
Association Between miR-146a, miR-196a-2, miR-499a, and miR-612 SNPs With Clinical Characteristics
To know whether the studied SNPs were associated with clinical and biological ALL features, we performed the case-control analysis into the patients group stratified by gender, age group, immunophenotype, NCI-risk classification, relapse, death, and hereditary cancer family history ( Supplementary Table S3 ). Significant differences among gender and age were found in the distribution of the miR-196a-2 rs11614913C allele (p = 0.02, p = 0.02, respectively). Additionally, analysis comparing infants versus children older than 1 year was performed. Supplementary Table S3 shows the results grouping the patients by age groups: <1 year; 1–9.9 and ≥10 years, considering that it has been reported that adolescents with ALL also have a dismal prognosis in comparison with children below this age and is considered an important prognostic factor. Regarding immunophenotype, NCI risk classification, relapse, death, and hereditary family history, no significant differences were observed ( Supplementary Table S3 ). Furthermore, we conducted survival analyses between the SNPs analyzed and the overall survival of pediatric patients with ALL, but no significant associations were observed neither including all cases nor after stratifying by child’s sex and age groups.
Gene-Gene Interaction Analysis
To know whether gene-gene interactions among miR-146a, miR-196a-2, miR-499a, and miR-612 SNPs predict the risk to ALL, a MDR analysis was performed by including cases and controls having complete genotyping data of all evaluated SNPs. No SNP was identified as the best factor model. The multilocus model with maximum crossvalidation consistency (CVC) and minimum prediction error is displayed in Supplementary Table S4 . Four-locus genotype combinations associated with the risk of ALL, as well as their distribution among cases (left) and controls (right) is summarizes in Figure 1A . This analysis gave evidence of epistasis or gene-gene interaction ( Figures 1B, C ). Entropy data showed that rs3746444 had the larger effect on the susceptibility to develop ALL (0.59%) followed by rs2910164 (0.49%). Week synergy among miR-196a-2 and miR-612 was observed (orange line) ( Figure 1B ). Redundancy was observed among all SNPs (blue and green lines) ( Figures 1B, C ). To note, gene-gene gender interaction observed a strong synergy (red line) among miR-196a-2 and gender ( Supplementary Figure S1 ).
Figure 1.
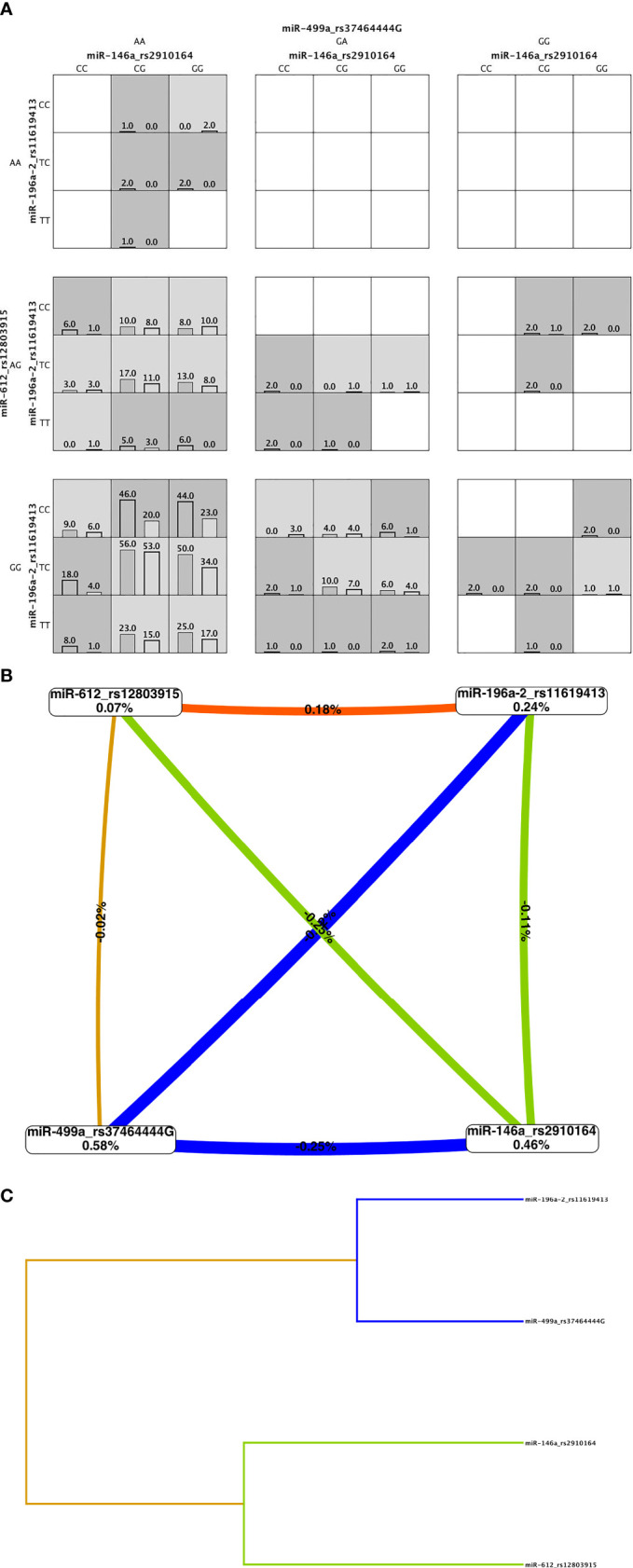
Multifactor dimensionality reduction (MDR) analysis. (A) Four-locus MDR model. Genotype combinations with high risk (shaded dark grey) and low risk (shaded light grey) for acute lymphoblastic leukemia (ALL) and their distribution in cases (left bar) and controls (right bar). The patterns of high (shaded and low-risk cells, which differ across each of the different multi locus dimension, means that the influence each genotype on the ALL risk is dependent on the genotypes a each of the other three loci. (B) Interaction entropy graph for gene-gene interaction and ALL risk. Graph shows the percent of the entropy in case-control removed by each factor (boxes) and by each pair-wise combination of attributes (lines). Positive value and orange line indicate low degree of synergy and negative values and blue and green lines mean redundancy. Gold line means independency. (C) The dendrogram graphic shows the presence, strength, and nature of epistatic effects. The shorter the line connecting two attributes the stronger the interaction. Strength of interaction goes from left to right (gray line).
Discussion
Mountain evidence reveals that miRNAs are relevant in the gene regulation contributing to the establishment of human diseases and modifying their treatment response of the patients. For instance, by using miRanda, TargetScan, and miRTarget2, it is predicted that AKT2 is a potential target of miR-612, which has been reported as significantly upregulated in ALL patients. AKT2 expression in lymphocytes correlates negatively with sensitive to glucocorticoids, and patients have poor prognosis (28–30). For its part, miR-146a has been involved in megakaryopoiesis by activating innate immunity targets TIRAP and TRAF6 (31). In addition, experimental data have shown that SNPs in miRNAs could affect cell differentiation, proliferation, and apoptosis conducting cancer development. The SNPs rs2910164 in miR146a, rs11614913 in miR-196a-2, rs3746444 in miR-499a, and rs12803915 in miR-612 are among the most studied SNPs in cancer. In a case-control study, we did no find association among rs12803915 of miR-612 but to rs3746444 of miR-499a with ALL, as well as, in a gender-dependent manner rs2910164 of miR146a, and rs11614913 of miR-196a-2 were associated with the risk to this disease.
To date, only three studies have explored the association among miR-499a rs3746444 and ALL. Our results are in line with the findings of de Souza et al., who studied 100 pediatric ALL patients, and 180 healthy individuals from Brazilian-amazon reported that miR499a_rs3746444 increases 17-fold the risk of development of ALL (26). We found that the mutant homozygote rs3746444GG genotype was associated with a 1.6-fold increase in the risk of developing ALL. However, our data are in contrast to those published previously by Gutierrez-Camino et al., who including 213 B-cell ALL pediatric patients and 387 controls from Spain, found a protective role of the G allele on the risk of ALL (8) and by Hasani et al., studying 75 children diagnosed with ALL and 115 children from Iran with no history of any type of cancer (23). To note, we explored whether miR-499a rs3746444 has in adults with ALL the same effect as we observed in children by genotyping 71 patients >18 years old with clinical diagnosis of ALL and 180 healthy adults (1:1 female/male). Samples from ALL adults were obtained from the biobank of the Servicio de Hematología, Hospital General de México. Adult control group was obtained from the DNA biobank of the laboratorio de Investigation, Hospital Juárez de México. miR-499a rs3746444A allele frequency was very similar among children and adults (cases and controls) and notably, miR-499a rs3746444G allele was not detected in no-ALL adults (0%). However, differences among adult cases and adult controls or between children and adults were not statistically significant ( Table 2 ). Our study is the first to investigate the role of rs3746444 in the susceptibility to ALL in adults, which has been associated with common adulthood cancer types (22, 32). The rs3746444 is located in pre-mir-499 gene resulting changes of an A:U to a G:U pairing and mismatching that reduces the stability of the pre-miR-499 secondary structure (33) and this SNP, located in the seed region of miR-499a could alter the targeted genes. In fact, Yang et al. (34) reported that this SNP potentially recognizes 573 new target genes and lost 5,392 original target genes. Several of these genes are involved in biological processes as cell proliferation and migration (35).
It is known that mir146a plays anti-inflammatory functions, has roles as tumor suppressor and commonly shows altered expression levels in human leukemia (32–38). Data from ALL Jurkat cells have shown that miR-146a can promote growth of leukemia cells by regulating the expression anti-apoptosis factor Bcl-xL and altering the expression of diverse genes involved in T-cell differentiation (37–39). Recent papers have given evidence that rs2910164 in miR146 can modify the expression of nuclear factor (NF-ĸB) through reducing IRAK1 and TRAF gene expression thus, driving inflammation and leukemia progression in myeloid cells (40). Stickel et al. (41) observed that patients with the miR-146a polymorphism rs2910164 display higher major histocompatibility complex class II (MHCII) molecule levels on monocytes. In addition, experimental evidences have shown that the rs2910164 in human allogeneic hematopoietic cell transplantation (allo-HCT) recipients significantly increases the risk for acute severe acute graft-versus-host disease in patients with hematological malignancies (41). The G to C polymorphism rs2910164 in miR146a changes the G:U pair to a C:U mismatch in the stem structure of miR-146a precursor, resulting in a reduced level of mature miR146a (36). To note, we found that miR-146a rs2910164 GG genotype confer risk to ALL in male. This SNP is widely associated with cancer, but association studies in ALL have reveled conflicting results. On one hand, it has been reported that miR-146a rs2910164 is associated with childhood ALL susceptibility in Asian population, including Iranian, Chinese, and Taiwanese (17, 23, 25). On the other hand, studies in Thailand, India, and China failed to replicate these results (9, 21, 42). No published study has reported an association among ALL and rs2910164 in a gene-dependent manner, and considering the higher prevalence of ALL in male than female, these findings should be deeply explored.
Regarding rs11614913 C/T, in the 3p mature miRNA region of miR-196a2, leads to a variation from G:T to G:C in the stem region of the miR-196a2 precursor. Comparing the minimum free energy for optimal secondary structures of the SNP rs11614913 in pre-miR196a2 found that this SNP had no dramatic effect on its secondary structure (43); however, Hoffman et al. (44) already show that rs11614913C may affect the processing of pre-miRNA, modify both, its expression level and function, then alters its interactions with its targeted genes. In fact, various studies have observed a correlation among abnormal expression of miR-196a2 and genes involved in cancer (45, 46). Studies in several types of cancer suggest that the common rs11614913 variant may play a role in the development of malignancies in an ethnic-dependent manner (43, 47, 48). For instance, a meta-analysis including 41,673 cases and 49,570 controls from 111 studies revealed that mir-196a-2 rs11614913 T allele was significantly associated with cancer risk only in Asians but not Caucasians (47). As for hematological malignancies, association data are scarce. Findings in Non-Hodgkin’s lymphoma suggest that the miR-196a-2 polymorphism may increase the risk of the disease by altering the expression of mature miR-196a (48). In ALL, two studies have published that rs11614913C allele contributes to an increased risk of this disease in Thailand, and China, but another one found no association results in Taiwanese ALL cases (13, 24, 49). Comparing the minimum free energy for optimal secondary structures of the SNP rs11614913 in pre-miR196a-2 found no dramatic effect on its secondary structure (47). We found an association among this SNP with ALL risk in females, but whether this SNP is playing a role in ALL susceptibility remains unknown.
Regarding rs12803915 in mir-612, experimental studies reveal that rs12803915 SNP affects mature mir-612 expression in a cell-type-specific manner. As example, Kim et al. observed that rs12803915A allele increases and decreases mature mir-612 expression in prostate cancer and colon cancer cell lines, respectively (50). In ALL, two studies have explored this SNP (8, 51). On one hand, the rs12803915 in mir-612 was associated with ALL in patients from Spain (8). On the other hand, in 100 B-ALL cases and 105 controls from Iran, no association was observed (51).
To know whether there is a gene-gene interaction among the evaluated SNPs in the risk to ALL, we employed a MDR analysis. We observed that miR-499a is the main casual factor for ALL, a strong redundancy interaction effect of this SNP and miR-196a-2 and miR-146a on ALL risk, and a low synergism with miR-612; thus, this analysis gave evidence of epistasis. Both genes have already been shown to be associated with cancer risk in various populations, but no data regarding their interaction has been published. To note, both SNPs have been found as susceptibility factors to ALL in a Spanish population (8).
The discrepancies on the association findings among the present work and other populations may be related to the sample selection, and the genetic background of the populations, since the linkage disequilibrium complex structure of the populations could mask the causal SNP (51). In addition, differences in the genetic background of cases and control could bias the association results. To note, our control group and a subset of the ALL cases belong to a genotyped cohort using 32 informative ancestry markers. As we published previously, ALL cases and controls are Mexican-Mestizo (6). However, to clarify the effect of miRNA polymorphism on ALL risk, studies including patients from different ethnicities and larger sample sizes are needed. Experimental analysis could also add data to decipher the role of miR-499 in ALL.
In conclusion, our analysis revealed that miR-499 rs3746444 confers risk to ALL and there is a gender-dependent association among miR-146a and miR-196a-2 and ALL in Mexican children. Studies are needed to evaluate the potential molecular mechanisms underlying the contribution of these SNPs in ALL susceptibility.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics and National Committee of Scientific Research of the Instituto Mexicano del Seguro Social with number R-2013-785-062. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
SJ-M: conceptualization. SJ-M, JN-E, JC-I, and JR-B: methodology. SJ-M, JN-E, JC-I, and JR-B: formal analysis. SJ-M: investigation. JN-E, VB-M, EJ-H, AM-S, IO-C, AM-T, JF-L, MP-S, JM-T, HP-L, RA-S, FM-R, JP-G, DD-R, JT-N, JF-B, RE-E, PR-Z, LF-V, ET-G, VL-G, JL-R, JG-U, SM-S, GE-A, CA-H, RR-C, LH-M, LG-L, GC-O, AG-E, IC-H, AM-H, ML-C, NH-P, JG-K, MR-V, DT-V, CC-R, FM-L, JP-G, AM-R, AA-S, BS-D, MG-R, LM-P, GV-A, MM-R, OS-R, HR-V, JR-B, and AH-M: resources. SJ-M: writing—original draft preparation. SJ-M, AH-M, and JM-A: writing—review and editing and supervision. SJ-M, JC-E, and JM-A: funding acquisition. All authors reviewed the final manuscript and read and approved the submitted version.
Funding
This work was supported by the Consejo Nacional de Ciencia y Tecnología (CONACyT), Investigación en Fronteras de la Ciencia (IFC)-2016-01-2119, PDCPN2013-01-215726, CB-2015-1-258042, FIS/IMSS/PROT/1548, FONCICYT/37/2018, FIS/IMSS/PROT/1782, and FORDECYT-PRONACES-377883-2020. We also thank the financial support from the National Institute of Genomic Medicine (01/2018/I, 19/2019/I).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank to Dr. Catherine Metayer from the California, Berkeley School of Public Health, USA, because of their donation of saliva kits used to perform this project. We thank all patients and people from the institutions involved in the clinical management of our cases.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.762063/full#supplementary-material
References
- 1. Pérez-Saldivar ML, Fajardo-Gutiérrez A, Bernáldez-Ríos R, Martínez-Avalos A, Medina-Sanson A, Espinosa-Hernández L, et al. Childhood Acute Leukemias are Frequent in Mexico City: Descriptive Epidemiology. BMC Cancer (2011) 11:355. doi: 10.1186/1471-2407-11-355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jiménez-Hernández E, Jaimes-Reyes EZ, Arellano-Galindo J, García-Jiménez X, Tiznado-García HM, Dueñas-González MT, et al. Survival of Mexican Children With Acute Lymphoblastic Leukaemia Under Treatment With the Protocol From the Dana-Farber Cancer Institute 00-01. BioMed Res Int (2015) 2015:576950. doi: 10.1155/2015/576950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martín-Trejo JA, Núñez-Enríquez JC, Fajardo-Gutiérrez A, Medina-Sansón A, Flores-Lujano J, Jiménez-Hernández E, et al. Early Mortality in Children With Acute Lymphoblastic Leukemia in a Developing Country: The Role of Malnutrition at Diagnosis. A Multicenter Cohort MIGICCL Study. Leuk Lymphoma (2017) 58(4):898–908. doi: 10.1080/10428194.2016.1219904 [DOI] [PubMed] [Google Scholar]
- 4. Hunger SP, Mullighan CG. Acute Lymphoblastic Leukemia in Children. N Engl J Med (2015) 373(16):1541–52. doi: 10.1056/NEJMra1400972 [DOI] [PubMed] [Google Scholar]
- 5. Jiménez-Hernández E, Duarte-Rodríguez DA, Núñez-Enriquez JC, Flores-Lujano J, Martín-Trejo JA, Espinoza-Hernández LE, et al. Maternal and Paternal Ages at Conception of Index Child and Risk of Childhood Acute Leukaemia: A Multicentre Case-Control Study in Greater Mexico City. Cancer Epidemiol (2020) 67:101731. doi: 10.1016/j.canep.2020.101731 [DOI] [PubMed] [Google Scholar]
- 6. Medina-Sanson A, Núñez-Enríquez JC, Hurtado-Cordova E, Pérez-Saldivar ML, Martínez-García A, Jiménez-Hernández E, et al. Genotype-Environment Interaction Analysis of NQO1, CYP2E1, and NAT2 Polymorphisms and the Risk of Childhood Acute Lymphoblastic Leukemia: A Report From the Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia. Front Oncol (2020) 10:571869. doi: 10.3389/fonc.2020.571869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu H, Cheng C, Devidas M, Pei D, Fan Y, Yang W, et al. ARID5B Genetic Polymorphisms Contribute to Racial Disparities in the Incidence and Treatment Outcome of Childhood Acute Lymphoblastic Leukemia. J Clin Oncol (2012) 30(7):751–7. doi: 10.1200/JCO.2011.38.0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gutierrez-Camino A, Lopez-Lopez E, Martin-Guerrero I, Piñan MA, Garcia-Miguel P, Sanchez-Toledo J, et al. Noncoding RNA-Related Polymorphisms in Pediatric Acute Lymphoblastic Leukemia Susceptibility. Pediatr Res (2014) 75(6):767–73. doi: 10.1038/pr.2014.43 [DOI] [PubMed] [Google Scholar]
- 9. Jemimah Devanandan H, Venkatesan V, Scott JX, Magatha LS, Durairaj Paul SF, Koshy T. MicroRNA 146a Polymorphisms and Expression in Indian Children With Acute Lymphoblastic Leukemia. Lab Med (2019) 50(3):249–53. doi: 10.1093/labmed/lmy074 [DOI] [PubMed] [Google Scholar]
- 10. James AR, Schroeder MP, Neumann M, Bastian L, Eckert C, Gökbuget N, et al. Long non-Coding RNAs Defining Major Subtypes of B Cell Precursor Acute Lymphoblastic Leukemia. J Hematol Oncol (2019) 12(1):8. doi: 10.1186/s13045-018-0692-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu B, Li J, Cairns MJ. Identifying miRNAs, Targets and Functions. Brief Bioinform (2014) 15(1):1–19. doi: 10.1093/bib/bbs075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dzikiewicz-Krawczyk A, Macieja A, Mały E, Januszkiewicz-Lewandowska D, Mosor M, Fichna M, et al. Polymorphisms in microRNA Target Sites Modulate Risk of Lymphoblastic and Myeloid Leukemias and Affect microRNA Binding. J Hematol Oncol (2014) 7:43. doi: 10.1186/1756-8722-7-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rakmanee S, Pakakasama S, Hongeng S, Sanguansin S, Thongmee A, Pongstaporn W. Increased Risk of Thai Childhood Acute Lymphoblastic Leukemia With the MiR196a2 T>C Polymorphism. Asian Pac J Cancer Prev (2017) 18(4):1117–20. doi: 10.22034/APJCP.2017.18.4.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duan R, Pak C, Jin P. Single Nucleotide Polymorphism Associated With Mature miR-125a Alters the Processing of pri-miRNA. Hum Mol Genet (2007) 16(9):1124–31. doi: 10.1093/hmg/ddm062 [DOI] [PubMed] [Google Scholar]
- 15. Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA, et al. SNPs in Human miRNA Genes Affect Biogenesis and Function. RNA (2009) 15(9):1640–51. doi: 10.1261/rna.1560209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duyu M, Durmaz B, Gunduz C, Vergin C, Yilmaz Karapinar D, Aksoylar S, et al. Prospective Evaluation of Whole Genome microRNA Expression Profiling in Childhood Acute Lymphoblastic Leukemia. BioMed Res Int (2014) 2014:967585. doi: 10.1155/2014/967585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zou D, Yin J, Ye Z, Zeng Q, Tian C, Wang Y, et al. Association Between the miR-146a Rs2910164 Polymorphism and Childhood Acute Lymphoblastic Leukemia Susceptibility in an Asian Population. Front Genet (2020) 11:886. doi: 10.3389/fgene.2020.00886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lian H, Wang L, Zhang J. Increased Risk of Breast Cancer Associated With CC Genotype of Has-miR-146a Rs2910164 Polymorphism in Europeans. PloS One (2012) 7(2):e31615. doi: 10.1371/journal.pone.0031615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alemán-Ávila I, Jiménez-Morales M, Beltrán-Ramírez O, Barbosa-Cobos RE, Jiménez-Morales S, Sánchez-Muñoz F, et al. Functional Polymorphisms in Pre-Mir146a and Pre-Mir499 are Associated With Systemic Lupus Erythematosus But Not With Rheumatoid Arthritis or Graves’ Disease in Mexican Patients. Oncotarget (2017) 8(54):91876–86. doi: 10.18632/oncotarget.19621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mi Y, Ren K, Zou J, Bai Y, Zhang L, Zuo L, et al. The Association Between Three Genetic Variants in MicroRNAs (Rs11614913, Rs2910164, Rs3746444) and Prostate Cancer Risk. Cell Physiol Biochem (2018) 48(1):149–57. doi: 10.1159/000491671 [DOI] [PubMed] [Google Scholar]
- 21. Chansing K, Pakakasama S, Hongeng S, Thongmee A, Pongstaporn W. Lack of Association Between the MiR146a Polymorphism and Susceptibility to Thai Childhood Acute Lymphoblastic Leukemia. Asian Pac J Cancer Prev (2016) 17(5):2435–8. [PubMed] [Google Scholar]
- 22. Yang X, Li X, Zhou B. A Meta-Analysis of miR-499 Rs3746444 Polymorphism for Cancer Risk of Different Systems: Evidence From 65 Case-Control Studies. Front Physiol (2018) 9:737. doi: 10.3389/fphys.2018.00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hasani SS, Hashemi M, Eskandari-Nasab E, Naderi M, Omrani M, Sheybani-Nasab M. A Functional Polymorphism in the miR-146a Gene is Associated With the Risk of Childhood Acute Lymphoblastic Leukemia: A Preliminary Report. Tumour Biol (2014) 35(1):219–25. doi: 10.1007/s13277-013-1027-1 [DOI] [PubMed] [Google Scholar]
- 24. Tong N, Xu B, Shi D, Du M, Li X, Sheng X, et al. Hsa-miR-196a2 Polymorphism Increases the Risk of Acute Lymphoblastic Leukemia in Chinese Children. Mutat Res (2014) 759:16–21. doi: 10.1016/j.mrfmmm.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 25. Pei JS, Chang WS, Hsu PC, Chen CC, Chin YT, Huang TL, et al. Significant Association Between the MiR146a Genotypes and Susceptibility to Childhood Acute Lymphoblastic Leukemia in Taiwan. Cancer Genomics Proteomics (2020) 17(2):175–80. doi: 10.21873/cgp.20178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Souza TP, de Carvalho DC, Wanderley AV, Fernandes SM, Rodrigues JCG, Cohen-Paes A, et al. Influence of Variants of the Drosha, Mir499a, and Mir938 Genes on Susceptibility to Acute Lymphoblastic Leukemia in an Admixed Population From the Brazilian Amazon. Am J Transl Res (2020) 12(12):8216–24. [PMC free article] [PubMed] [Google Scholar]
- 27. Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, et al. A Flexible Computational Framework for Detecting, Characterizing, and Interpreting Statistical Patterns of Epistasis in Genetic Studies of Human Disease Susceptibility. J Theor Biol (2006) 241(2):252–61. doi: 10.1016/j.jtbi.2005.11.036 [DOI] [PubMed] [Google Scholar]
- 28. Chen C, Yan Y, Liu X. microRNA-612 Is Downregulated by Platelet-Derived Growth Factor-BB Treatment and has Inhibitory Effects on Vascular Smooth Muscle Cell Proliferation and Migration via Directly Targeting AKT2. Exp Ther Med (2018) 15(1):159–65. doi: 10.3892/etm.2017.5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xie M, Yang A, Ma J, Wu M, Xu H, Wu K, et al. Akt2 Mediates Glucocorticoid Resistance in Lymphoid Malignancies Through FoxO3a/Bim Axis and Serves as a Direct Target for Resistance Reversal. Cell Death Dis (2019) 9(10):1013. doi: 10.1038/s41419-018-1043-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Lu LL, Wen D, Liu DL, Dong LL, Gao DM, et al. MiR-612 Regulates Invadopodia of Hepatocellular Carcinoma by HADHA-Mediated Lipid Reprogramming. J Hematol Oncol (2020) 13(1):12. doi: 10.1186/s13045-019-0841-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-Dependent Induction of microRNA miR-146, an Inhibitor Targeted to Signaling Proteins of Innate Immune Responses. Proc Natl Acad Sci U S A (2006) 103(33):12481–6. doi: 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan W, Gao X, Zhang S. Association of miR-196a2 Rs11614913 and miR-499 Rs3746444 Polymorphisms With Cancer Risk: A Meta-Analysis. Oncotarget (2017) 8(69):114344–59. doi: 10.18632/oncotarget.22547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, et al. Common Genetic Variants in pre-microRNAs Were Associated With Increased Risk of Breast Cancer in Chinese Women. Hum Mutat (2009) 30(1):79–84. doi: 10.1002/humu.20837 [DOI] [PubMed] [Google Scholar]
- 34. Yang S, Zheng Y, Zhou L, Jin J, Deng Y, Yao J, et al. miR-499 Rs3746444 and miR-196a-2 Rs11614913 Are Associated With the Risk of Glioma, But Not the Prognosis. Mol Ther Nucleic Acids (2020) 22:340–51. doi: 10.1016/j.omtn.2020.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He S, Li Z, Yu Y, Zeng Q, Cheng Y, Ji W, et al. Exosomal miR-499a-5p Promotes Cell Proliferation, Migration and EMT via mTOR Signaling Pathway in Lung Adenocarcinoma. Exp Cell Res (2019) 379(2):203–13. doi: 10.1016/j.yexcr.2019.03.035 [DOI] [PubMed] [Google Scholar]
- 36. Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. A Functional Polymorphism in the miR-146a Gene and Age of Familial Breast/Ovarian Cancer Diagnosis. Carcinogenesis (2008) 29(10):1963–6. doi: 10.1093/carcin/bgn172 [DOI] [PubMed] [Google Scholar]
- 37. Saki N, Abroun S, Soleimani M, Mortazavi Y, Kaviani S, Arefian E. The Roles of miR-146a in the Differentiation of Jurkat T-Lymphoblasts. Hematology (2014) 19(3):141–7. doi: 10.1179/1607845413Y.0000000105 [DOI] [PubMed] [Google Scholar]
- 38. Yan W, Guo H, Suo F, Han C, Zheng H, Chen T. The Effect of miR-146a on STAT1 Expression and Apoptosis in Acute Lymphoblastic Leukemia Jurkat Cells. Oncol Lett (2017) 13(1):151–4. doi: 10.3892/ol.2016.5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L, Zhang H, Lei D. microRNA-146a Promotes Growth of Acute Leukemia Cells by Downregulating Ciliary Neurotrophic Factor Receptor and Activating JAK2/STAT3 Signaling. Yonsei Med J (2019) 60(10):924–34. doi: 10.3349/ymj.2019.60.10.924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Su YL, Wang X, Mann M, Adamus TP, Wang D, Moreira DF, et al. Myeloid Cell-Targeted miR-146a Mimic Inhibits NF-κb-Driven Inflammation and Leukemia Progression In Vivo . Blood (2020) 135(3):167–80. doi: 10.1182/blood.2019002045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stickel N, Hanke K, Marschner D, Prinz G, Köhler M, Melchinger W, et al. MicroRNA-146a Reduces MHC-II Expression via Targeting JAK/STAT Signaling in Dendritic Cells After Stem Cell Transplantation. Leukemia (2017) 31(12):2732–41. doi: 10.1038/leu.2017.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xue Y, Yang X, Hu S, Kang M, Chen J, Fang Y. A Genetic Variant in miR-100 is a Protective Factor of Childhood Acute Lymphoblastic Leukemia. Cancer Med (2019) 8(5):2553–60. doi: 10.1002/cam4.2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin-Guerrero I, Gutierrez-Camino A, Lopez-Lopez E, Bilbao-Aldaiturriaga N, Pombar-Gomez M, Ardanaz M, et al. Genetic Variants in miRNA Processing Genes and pre-miRNAs are Associated With the Risk of Chronic Lymphocytic Leukemia. PloS One (2015) 10(3):e0118905. doi: 10.1371/journal.pone.0118905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, et al. microRNA miR-196a-2 and Breast Cancer: A Genetic and Epigenetic Association Study and Functional Analysis. Cancer Res (2009) 69(14):5970–7. doi: 10.1158/0008-5472.CAN-09-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, et al. MicroRNA-196a Targets Annexin A1: A microRNA-Mediated Mechanism of Annexin A1 Downregulation in Cancers. Oncogene (2008) 27(52):6667–78. doi: 10.1038/onc.2008.256 [DOI] [PubMed] [Google Scholar]
- 46. Zhao H, Xu J, Zhao D, Geng M, Ge H, Fu L, et al. Somatic Mutation of the SNP Rs11614913 and Its Association With Increased MIR 196a2 Expression in Breast Cancer. DNA Cell Biol (2016) 35(2):81–7. doi: 10.1089/dna.2014.2785 [DOI] [PubMed] [Google Scholar]
- 47. Choupani J, Nariman-Saleh-Fam Z, Saadatian Z, Ouladsahebmadarek E, Masotti A, Bastami M. Association of Mir-196a-2 Rs11614913 and Mir-149 Rs2292832 Polymorphisms With Risk of Cancer: An Updated Meta-Analysis. Front Genet (2019) 10:186. doi: 10.3389/fgene.2019.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li T, Niu L, Wu L, Gao X, Li M, Liu W, et al. A Functional Polymorphism in microRNA-196a2 is Associated With Increased Susceptibility to non-Hodgkin Lymphoma. Tumour Biol (2015) 36(5):3279–84. doi: 10.1007/s13277-014-2957-y [DOI] [PubMed] [Google Scholar]
- 49. Chen CC, Hsu PC, Shih LC, Hsu YN, Kuo CC, Chao CY, et al. MiR-196a-2 Genotypes Determine the Susceptibility and Early Onset of Childhood Acute Lymphoblastic. Leukemia Anticancer Res (2020) 40(8):4465–9. doi: 10.21873/anticanres.14451 [DOI] [PubMed] [Google Scholar]
- 50. Kim HK, Prokunina-Olsson L, Chanock SJ. Common Genetic Variants in miR-1206 (8q24.2) and miR-612 (11q13.3) Affect Biogenesis of Mature miRNA Forms. PloS One (2012) 7(10):e47454. doi: 10.1371/journal.pone.0047454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Siyadat P, Ayatollahi H, Barati M, Sheikhi M, Shahidi M. High Resolution Melting Analysis for Evaluation of Mir-612 (Rs12803915) Genetic Variant With Susceptibility to Pediatric Acute Lymphoblastic Leukemia. Rep Biochem Mol Biol (2021) 9(4):385–93. doi: 10.52547/rbmb.9.4.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.


