Abstract
Small extracellular vesicles (sEVs) play a key role in intercellular communication. Cargo molecules carried by sEVs may affect the phenotype and function of recipient cells. Epithelial cancer cell‐derived sEVs, particularly those enriched in CD151 or tetraspanin8 (TSPAN8) and associated integrins, promote tumour progression. The mechanism of binding and modulation of sEVs to recipient cells remains elusive. Here, we used genetically engineered breast cancer cells to derive TSPAN8‐enriched sEVs and evaluated the impact of TSPAN8 on target cell membrane's diffusion and transport properties. The single‐particle tracking technique showed that TSPAN8 significantly promoted sEV binding via confined diffusion. Functional assays indicated that the transgenic TSPAN8‐sEV cargo increased cancer cell motility and epithelial‐mesenchymal transition (EMT). In vivo, transgenic TSPAN8‐sEV promoted uptake of sEVs in the liver, lung, and spleen. We concluded that TSPAN8 encourages the sEV‐target cell interaction via forced confined diffusion and significantly increases cell motility. Therefore, TSPAN8‐sEV may serve as an important direct or indirect therapeutic target.
Keywords: confined diffusion, metastasis, single particle tracking, small extracellular vesicles, TSPAN8
Abbreviations
- EMT
epithelial‐mesenchymal transition
- FBS
fetal bovine serum
- IF
immunofluorescence
- MSD
mean squared displacement
- NTA
nanoparticle tracking analysis
- RT‐qPCR
real‐time quantitative polymerase chain reaction
- sEVs
small extracellular vesicles
- shRNA
small hairpin RNA
- TEM
transmission electron microscopy
- TEMs
tetraspanin enriched microdomains
- TSPAN8
tetraspanin 8
1. INTRODUCTION
Tetraspanin belongs to the transmembrane‐4 family of proteins that form homologous and heterogeneous dimers and oligomerize with other membrane proteins in the plasma membrane to form tetraspanin‐enriched microdomains (TEMs) (Hemler, 2003). The size of small extracellular vesicles (sEVs) ranges across the nanoscale (30–200 nm) and vesicles are derived from many cell types and are found in all bodily fluids (Pegtel & Gould, 2019). The biogenesis of sEVs occurs from endosomes, intraluminal vesicles, and multivesicular bodies and are then released by the fusion of multivesicular bodies and the plasma membrane (Colombo et al., 2014; Yáñez‐Mó et al., 2015). Tetraspanins are considered pivotal for membrane curvature formation, and proteins in TEMs have been proposed to be carried by sEVs (Yáñez‐Mó et al., 2015). Numerous subsets of proteins are highly enriched in sEVs, including the tetraspanin family (CD9, CD63, CD81, and tetraspanin8 [TSPAN8]), endosomal trafficking proteins (ALIX), the endosomal sorting complex required for transport (ESCRT) proteins, and heat shock proteins (HSP60, HSP70, and HSP90) (Colombo et al., 2014). Recent studies have suggested that the sEV cargo manipulates the phenotype of target cells in the physiological and pathological microenvironment (Costa‐Silva et al., 2015; X. Li et al., 2018; Richards et al., 2017; Zhou et al., 2014). Evidence shows that sEVs are received by specific recipient cells (Mckelvey et al., 2015; Rana & Zöller, 2011; Rana et al., 2012). However, the molecular mechanism underlying the binding of sEVs by target cells is not entirely understood.
Internalization of sEVs into recipient cells occurs primarily through phagocytosis and clathrin‐ or caveolin‐mediated endocytosis (Mulcahy et al., 2014). Vesicular fusion or fission is facilitated by TEMs (Hemler, 2014; Zöller, 2009) while tetraspanin, as a molecular scaffold, modulates the stability and activity of associated molecules (X. Yang et al., 2004). TSPAN8 and CD151 have been shown to promote cancer metastasis by impairing cell adhesion and migration (Yue et al., 2013). TSPAN8 promotes the tumour sEV‐induced angiogenesis (Nazarenko et al., 2010). Furthermore, sEVs enriched in TSPAN8 induce disseminated intravascular coagulation by interacting with TSPAN8 and integrin α4β1 (Yue et al., 2013; K. Zhao et al., 2018). Studies have demonstrated that TSPAN8 is involved in binding sEVs and target cells (Yue et al., 2015). However, the exact role of TSPAN8 in sEV binding and attachment to target cells has not been fully elucidated. The classic “seed and soil” hypothesis of cancer metastasis proposed that cancer cells which escaped from primary cancer into circulation serve as “seed” and the distance tissue microenvironment as “soil” (Mckelvey et al., 2015; Rana & Zöller, 2011; Rana et al., 2012). Cancer cell‐derived sEVs contain selected membrane and cytosolic proteins, mRNAs, and miRNAs that may potentially serve as the “soil” establishment and promote cancer progression (Tkach & Théry, 2016; Wortzel et al., 2019).
The uptake of sEVs by the target cells is required for phenotypic changes. Our previous study suggested that TSPAN8 promotes pancreatic cancer metastasis by affecting cancer cell motility (Lu et al., 2017; K. Zhao et al., 2018). Considering the role of sEVs in modulating target cells and their wide applications in drug delivery (Hu et al., 2019; Riazifar et al., 2019), understanding the role of tetraspanins, TSPAN8 in particular, in target binding and uptake of sEVs is important. To decode the molecular mechanism involved in TSPAN8‐mediated regulation of sEV binding and attachment to target cells, we assessed whether breast cancer cell‐derived TSPAN8‐enriched sEVs forced the confined diffusion and increased the uptake to parent cells and fibroblasts. Based on this first description of confined diffusion of sEVs, we investigated whether the TSPAN8‐enriched sEVs promote cancer cell motility and epithelial‐mesenchymal transition (EMT), which may induce cancer metastasis.
2. METHODS
2.1. Cell lines
Human breast cancer cells T‐47D and human fetal lung fibroblasts MRC‐5 were purchased from ATCC. T‐47D cells were maintained in RPMI‐1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. MRC‐5 cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 1% penicillin/streptomycin. HEK‐293T cells were purchased from ATCC and maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 1% penicillin/streptomycin.
2.2. Plasmids
Human TSPAN8‐cDNA was generated using reverse transcription of an mRNA template derived from MRC‐5 cells. The cDNA was inserted into the lentiviral expression vector pLV‐EF1α‐MCS‐IRES‐Bsd/puro (Biosettia, San Diego, CA, USA). The empty expression vector pLV‐EF1α‐MCS‐IRES‐Bsd/puro (MCS) was used as a control. TSPAN8 knockdown stable cell line was generated using the small hairpin RNA (shRNA) (AAAATGAATGAAACTCTCTATGAATTGGATCCAATTCATAGAGAGTTTCATTCA), targeting the TGAATGAAACTCTCTATGAA sequence of human TSPAN8 mRNA. The shRNA was inserted into the pRNAi vector system (Biosettia). The pRNAi plasmid was linearized using the Pac I enzyme and transfected into T‐47D cells. The stable TSPAN8 knockdown cell line was obtained using blasticidin selection post‐transfection. All constructs were verified by DNA sequencing. The primers for the constructs used in this study are listed in Table S1.
2.3. sEV isolation and characterization
All sEVs were isolated from cell culture supernatants using sequential centrifugation. For sEV preparation, cancer cells were cultured in 15 cm dishes with 20 ml RPMI‐1640 medium without FBS for 24 h. The cell culture supernatant was collected and centrifuged sequentially at 500g for 10 min, repeated twice, 2000g for 20 min, and 12,000g for 30 min. The supernatant was filtered through a 0.22 μm pore size membrane filter and finally centrifuged at 100,000g for 90 min to obtain a pellet sEVs, which was washed again in phosphate buffered saline (PBS) by centrifugation at 100,000g for 90 min. The morphology and purity of the sEVs were verified using transmission electron microscopy (TEM) according to previously published methods (J. Li et al., 2020). In brief, purified sEVs were re‐suspended in 2% paraformaldehyde and adsorbed onto carbon‐coated formvar EM grids for 20 min. Grids were then washed in physiological saline and transferred to 50 mM glycine/PBS for 3 min, repeated thrice. Finally, grids were embedded in 30 μl of uranyl‐oxalate solution for 90 s and air‐dried. Images were captured using an FEI Talos F200C TEM (Thermo Fisher Scientific, San Jose, CA, USA). The size scale and concentration of sEVs were measured using a nanoparticle tracking analysis (NTA) device NanoSight NS300 (Malvern, UK). sEVs were suspended in PBS and diluted 100‐fold prior to analysis. A 60 s video was recorded and subsequently analyzed using the NTA software.
2.4. Flow cytometry
Isolated sEVs were diluted to 5.3 × 109/ml using PBS (10 mM, pH 7.4) and incubated with 1 μl of DiR dye (10 mM) at 4°C for 30 min. The labelled sEVs were washed with PBS thrice to discard excess dye using ultra‐centrifugation at 100,000g for 90 min and finally re‐suspended in 100 μl of PBS. DiR‐labelled sEVs (5.3 × 108/ml) were incubated with target cells for 1.5 h. Flow cytometry was performed using the BD FACScan system (BD Biosciences, San Jose, CA, USA). The data were processed using the FlowJo Software (version 7.6 Tree Star Inc., Ashland, OR, USA).
2.5. Single‐particle tracking
TSPAN8‐sEV and MCS‐sEV were labelled with DiO and DiL dyes, respectively. Both groups of sEVs were stained and washed as described above. Labelled sEVs (3 μl) were diluted in 1 ml of medium and incubated with target cells for 20 min at 4°C. Labelled sEV uptake was measured using a Nikon Eclipse Ni–U upright optical microscope (Nikon Instruments Inc., Tokyo, Japan) as described previously (D. Zhang et al., 2019).
Single‐particle tracking experiments were performed using a Nikon Ti‐U inverted fluorescence microscope (Nikon Instruments Inc.). DiO‐labelled TSPAN8‐sEV was measured at 473 nm and DiL‐labelled MCS‐sEV at 532 nm. The fluorescence signal of the sample was collected using a 100× objective, and successive images were captured using a highly sensitive electron multiplying charge‐coupled devices. The pixel size of the camera was 16 × 16 μm. The exposure time was set to 20 ms, and 500 frames were collected continuously (Ye et al., 2019). All imaging data were analyzed and processed using the ImageJ version 1.8.0_172 for Windows (NIH, Bethesda, MD, USA) and MATLAB R2015a software (The MathWorks Inc., Natick, MA, USA). The interaction between sEVs and cell membranes was monitored in situ and in real‐time using a confocal laser scanning microscope. Labelled sEVs (3 μl) were diluted in 1 ml of phenol red‐free medium and incubated with cells. The binding of sEVs to the cell membrane surface was monitored each minute for 1 h.
2.6. Animal experiments
Nude mice were injected with DiR‐labelled sEVs (5.3 × 108/mouse) intravenously for 24 h. The bio‐distribution of sEVs in vivo was analyzed using a Xenogen IVIS Lumina XRMS Series III live animal biophotonic imaging system (Caliper Life Sciences, Hopkinton, MA, USA). The DiR‐labelled sEV uptake in vivo was monitored using frozen sections and confocal microscopy. The relative sEV uptake was quantified from the fluorescence intensity per cell of the corresponding areas using ImageJ. BALB/c nude (nu/nu) mice were purchased from a laboratory animal company (Vital River Laboratory Animal Technology Co. Ltd, Beijing, China) and maintained in a pathogen‐free facility at the Nankai University. All animal experiments in this study were approved by the Nankai University Animal Care and Use Committee (2021‐SYDWLL‐000060) and handled according to the Nankai University Animal Welfare Guidelines.
2.7. Statistical analysis
Statistical evaluation was performed using the two‐tailed Student´s t‐test and GraphPad Prism software version 7.0 for Windows (GraphPad Software, San Diego, CA, USA). Data with p < 0.05 were considered significant. Data are expressed as the mean ± standard deviation of at least three independent experiments. Animal experiments were performed in triplicates, using three mice per group.
3. RESULTS AND DISCUSSION
3.1. Overexpression TSPAN8 in the breast cancer cell and enriched recovery in sEV
Previous works have suggested that TSPAN8 contributes to cancer metastasis by associating with β4 integrin in pancreatic cancer cells (Yue et al., 2015, 2013). Tetraspanins, including TSPAN8 form TEMs, and tetraspanin‐integrin complexes are enriched in sEVs (Rana et al., 2012). Therefore, the role of TSPAN8‐sEV in target cell binding and sEV uptake was evaluated using sEVs derived from TSPAN8‐overexpressing breast cancer cells. The expression of TSPAN8 in breast cancer line T‐47D cells was assessed using Real‐time quantitative polymerase chain reaction (RT‐qPCR) and immunofluorescence (IF) (Figure 1a and b). The TSPAN8 overexpressing stable cell line was generated to produce sEVs for further analysis. TSPAN8‐sEV and MCS‐sEV were observed using TEM. TEM results showed that the overexpression of TSPAN8 did not impair the morphology of sEVs (Figure 1c). Moreover, NTA was performed to measure the size and concentration of purified sEVs to evaluate their quality and quantity (Figure 1d). To further explore characteristics of sEVs, we measured levels of sEV biomarkers in TSPAN8‐sEV and MCS‐sEV. Results confirmed the enrichment of biomarkers and overexpression of TSPAN8 in TSPAN8‐sEV (Figure 1e), providing a suitable model to investigate the specific role of TSPAN8 in sEV‐target cell interaction.
FIGURE 1.
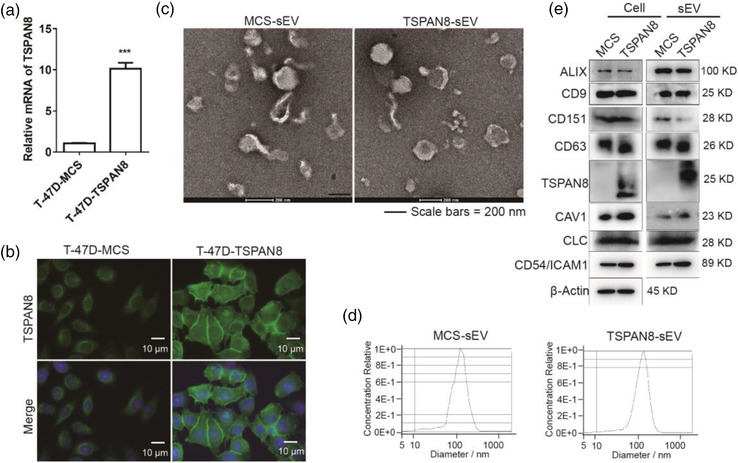
Characterizations of T‐47D cell‐derived TSPAN8 enriched sEVs. (a) Relative mRNA levels of TSPAN8 in T‐47D cells and TSPAN8‐overexpressing cells were detected by RT‐qPCR. Relative quantitation data represent mean standard deviation of normalized to GAPDH using ΔCt method (n = 3, three independent experiments). ***p < 0.001 by the two‐tailed Student's t‐test. (b) Relative protein levels of TSPAN8 in T‐47D cells and TSPAN8‐overexpressing cells were detected by IF. (c) TEM analysis indicated that the morphology and size scale of TSPAN8‐sEV are consistent in MCS‐sEV. (d) Quality and quantity of TSPAN8‐sEV and MCS‐sEV were analyzed by NTA. (e) Western blot analysis was performed to confirm purified sEVs using sEV markers ALIX, CD9, CD151, CD63, Caveolin 1 (CAV1), Clathrin light chain (CLC), TSPAN8, and also intercellular adhesion molecule in cells CD54 (ICAM1) and the control of β‐Actin in cells and sEVs
3.2. TSPAN8 promotes sEV attachment to target cells
To evaluate the functional role of TSPAN8 in sEVs target cell binding, we monitored the labelled MCS‐sEV or TSPAN8‐sEV co‐cultured with T‐47D cells and observed them under a confocal microscope. The quantification of sEV attachment showed that TSPAN8 significantly increased the sEV binding to T‐47D cells (Figure 2a). The binding of sEVs and T‐47D cells was validated using flow cytometry (Figure 2b). Fibroblasts in the cancer microenvironment also play a pivotal role in metastasis (Lu et al., 2017; Peinado et al., 2017). To explore the impact of TSPAN8 on sEV attachment to fibroblasts, we treated MRC‐5 cells with labelled MCS‐sEV or TSPAN8‐sEV and analyzed them using confocal microscopy. The results demonstrated that TSPAN8‐sEV significantly increased the binding of sEVs to MRC‐5 cells (Figure 2c). The attachment of sEVs to MRC‐5 cells was confirmed using flow cytometry (Figure 2d). Moreover, TSPAN8 overexpression did not affecting the binding of the impaired epithelial cancer cell with to fibroblasts (Figure S1). Our findings imply that TSPAN8 promotes the attachment of sEVs derived from breast cancer cells to parent cells and fibroblasts. Therefore, we next investigated the mechanism underlying pronounced TSPAN8‐sEV uptake by target cells.
FIGURE 2.
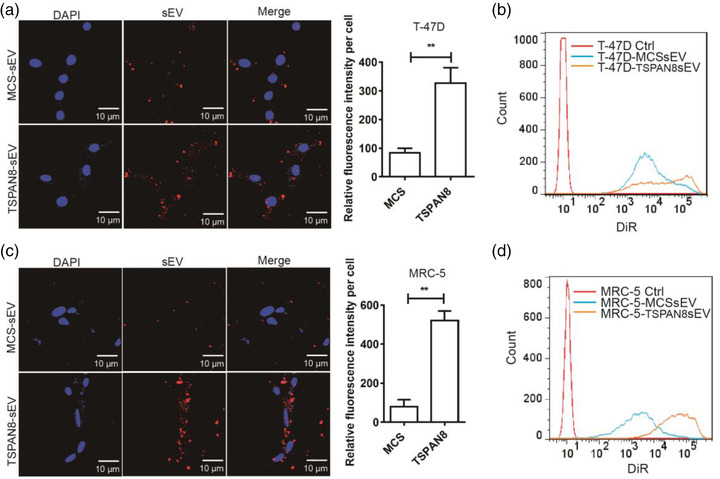
TSPAN8 increased the sEV uptake in T‐47D and MRC‐5 cells. (a) DiR‐labelled sEVs incubated with T‐47D cells and the attachment of sEVs was observed by confocal microscopy. Data are mean standard deviation (n = 3, three independent experiments) **p < 0.01 by the two‐tailed Student's t‐test. (b) DiR‐labelled sEV attachment to T‐47D cells was analysed using flow cytometry. (c) DiR‐labelled sEVs incubated with MRC‐5 cells and the attachment of sEVs was observed by confocal microscopy. Data are mean standard deviation (n = 3, three independent experiments) **p < 0.01 by the two‐tailed Student's t‐test. (d) DiR‐labelled sEV attachment to MRC‐5 cells was analysed using flow cytometry. Quantification of sEV attachment to target cells was analysed by the relative fluorescence intensity using ImageJ software. All the images are representative of 10 random fields
3.3. TSPAN8 alters the mode of sEV attachment to target cells
The mechanisms of sEV internalization into target cells have been revealed by previous studies on macropinocytosis, phagocytosis, clathrin, caveolin, lipid raft mediated endocytosis, and plasma membrane or endosomal membrane fusion (Mallegol et al., 2007; Mulcahy et al., 2014; Purushothaman et al., 2016; Van Niel et al., 2018). Based on the membrane components and the structure of sEVs, free diffusion and membrane fusion are possible modes of binding, which widely exist in many types of cells (Heusermann et al., 2016). A range of specific protein‐protein interactions and the membrane lipid raft structures regulating sEV uptake have been studied (Mulcahy et al., 2014). The specific surface components or proteins in the target cell and sEV membranes define the selection of sEVs for uptake by target cells (Christianson et al., 2013; Purushothaman et al., 2016; Svensson et al., 2013). The tetraspanin‐associated molecules in TEMs are essential for deciding the target cells (Nazarenko et al., 2010; Perez‐Hernandez et al., 2013). sEV is a type of vesicle with a lipid membrane structure that can be randomly taken up by cells via membrane fusion (Colombo et al., 2014). New evidence has demonstrated that a series of membrane proteins act as ligands or receptors on sEVs and recipient cells that contribute to sEV uptake (Schneider & Simons, 2016; Tkach & Théry, 2016).
To gain insight into the impact of TSPAN8 on the interaction of sEVs and cell membranes, we performed in situ real‐time observation of the sEV migration trajectory on the cell membrane surface using a single‐particle tracking technique. The difference in the touch‐adhesion capacity of sEVs and target cells was revealed using single sEV transfer analysis. The typical migration trajectory of individual sEV on the T‐47D cell membrane surface was tracked (Figure 3a). Interestingly, MCS‐sEV showed an obvious free diffusion, whereas TSPAN8‐sEV showed a significant confined diffusion. The change in diffusion modality indicated that TSPAN8 increased the adhesion of sEVs to T‐47D cells. The increased touch‐adhesion capacity of sEVs to target cells resulted in a significantly reduced migration area on the cell surface. Furthermore, we measured the velocity of the two types of sEVs on the T‐47D cell membrane surface in real‐time and found that the velocity of sEV migration changed dynamically (Figure 3b). The mean migration velocity decreased in TSPAN8‐sEV compared to that in MCS‐sEV. This result reveals TSPAN8 contribution to the binding of sEVs to target cells by molecular adhesion. To decode the mode of sEV attachment to target cells, we statistically analyzed the mean squared displacement (MSD) with time (, where is the diffusion coefficient, and is the diffusion exponent factor) (Figure 3c) (He et al., 2019; Ye et al., 2019). Further analyzing the diffusion exponent factors of these two types sEVs, the diffusion exponent factors of both sEVs were less than 1, indicating both sEVs showed confined diffusion (Xiao et al., 2012). However, the diffusion exponent factor of the individual TSPAN8‐sEV (α = 0.13) was significantly lower than that of MCS‐sEV (α = 0.62). These results denote that TSPAN8 promotes the adhesion of sEVs to target cells.
FIGURE 3.
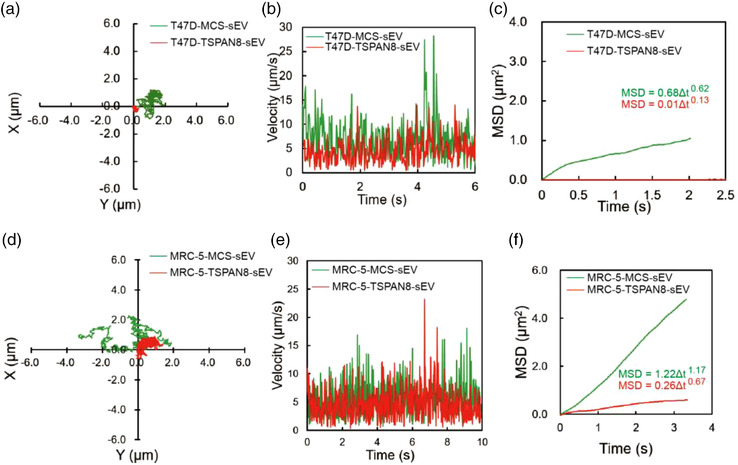
TSPAN8 changed sEV diffusion mode to target cells. (a) The trajectories of sEV diffusion on T‐47D living cell membrane surface observed by single‐particle fluorescence microscopy. (b) The velocity of sEV diffusion on T‐47D living cell membrane surface. (c) The MSD of sEV diffusion on T‐47D living cell membrane surface. (d) The trajectories of sEV diffusion on MRC‐5 living cell membrane surface observed by single‐particle fluorescence microscopy. (e) The velocity of sEV diffusion on MRC‐5 living cell membrane surface. (f) The MSD of sEV diffusion on MRC‐5 living cell membrane surface
To address the functional role of TSPAN8 in sEV attachment to stromal cells, we examined the diffusion mode of sEVs on MRC‐5 cells. The typical single sEV migration trajectory on the MRC‐5 cell membrane surface was tracked (Figure 3d). Results displayed that TSPAN8 significantly reduced the migration area of sEVs on the surface of MRC‐5 cells. The velocity of the two types of sEVs on the MRC‐5 cell membrane in real‐time was measured (Figure 3e). Findings indicated that TSPAN8 did not affect the velocity of sEV migration on the MRC‐5 cell membrane surface. The MSD and the time of sEVs on the MRC‐5 cell membrane surface were statistically analyzed (Figure 3f). The MSD of the MCS‐sEV is approximately 1 (α = 1.17) with free diffusion, whereas the MSD of TSPAN8‐sEV is less than 1 (α = 0.67) with confined diffusion. Taken together, in breast cancer T‐47D cells, both groups of sEVs showed confined diffusion, and in fibroblasts MRC‐5 cells, the MCS‐sEV showed free diffusion, whereas the TSPAN8‐sEV showed confined diffusion. Moreover, to explore whether TSPAN8 specifically regulates sEV attachment to target cells, we also performed TSPAN8 knockdown on T‐47D cells to produce sEVs for single‐particle tracking assays (Figure S2A‐F). Compared to the wild‐type (WT) sEVs, TSPAN8 knockdown did not change the model of sEV diffusion. Both types of sEVs showed confined diffusion in T‐47D cells and non‐confined diffusion in MRC‐5 cells. We considered that the affinity and binding capacity of sEVs is preference to homologous cells (Choi et al., 2018; Cui et al., 2021; Gong et al., 2021). TSPAN8 knockdown slightly reduced the velocity of sEVs on the cell surface, indicating that TSPAN8 contributes to contact adhesion of the surface proteins of target cells. The membrane components of sEVs and T‐47D cells might have additional molecules contributing to the mode of diffusion. Both types of sEVs showed confined diffusion in T‐47D cells. These data suggest that TSPAN8 significantly increases the efficiency of sEV attachment to target cells.
The intermittent hopping diffusion of sEVs on the cell surface, also called as the sustaining random walk process, is a preferred way to explore further areas and the target site. We explored the intermittent hopping diffusion of TSPAN8‐sEV and MCS‐sEV on the cell surface of T‐47D and MRC‐5 cells (Figure S3A‐B). The surface proteins of sEVs play a key role in desorbing sEVs from the binding site, diffusing away, and binding to a new site on the cell surface (Wu & Yeow, 2008). Target cellular uptake of sEVs is based on the random frequency of attachment and evasion. Our results demonstrated that TSPAN8 promoted sEV binding to the target cells by significantly increasing the attachment efficiency and reducing the evasion frequency. The enrichment of tetraspanin on sEVs that associates with integrin to form TEMs, clearly regulated the attachment of sEVs to the target cells. The different tetraspanin‐associated proteins may contribute to the process of attachment to specific proteins on target cells.
3.4. sEV cargo TSPAN8 promote target cells phenotype alteration
The sEV cargo contains proteins, mRNAs, and non‐coding RNAs that may change the phenotypic characteristics of the target cells (Antonyak et al., 2011; Zitvogel et al., 1998). The potential of sEVs in target cell education is associated with cancer metastasis, cancer cell motility, and EMT (Koch et al., 2014; Sung et al., 2015, 2020). CD151 promotes cell motility by reducing tumour cell‐matrix interactions and thus contributes to dissemination, which subsequently leads to invasion and intravasation of a primary tumour (Zijlstra et al., 2008). To define the role of sEV cargo TSPAN8 in target cell education, we treated T‐47D cells with MCS‐sEV or TSPAN8‐sEV and measured their motility. The wound healing assays indicated that TSPAN8‐sEV significantly increased cancer cell migration capacity (Figure 4a). To confirm cell motility, we performed transwell assays. The results demonstrated that TSPAN8‐sEV significantly promoted T‐47D cell migration (Figure 4b). Cell migration is mediated by kinases and phosphatases that regulate the phosphorylation and dephosphorylation of key molecules involved in adhesion (Parsons et al., 2000; Y. Yang et al., 2021). The focal adhesion protein‐complex's dynamics changes the cytoskeleton structure by assembling other specialized proteins and constructing stress fibers (J. Zhao & Guan, 2009). The FAK‐SRC complex regulates the phosphorylation of paxillin, which recruits specific molecules and organizes the actin cytoskeleton (Mitra & Schlaepfer, 2006; Webb et al., 2004). To obtain a hint towards TSPAN8‐sEV cargo possibly affecting, the activity of FAK‐SRC signalling pathway was evaluated by western blotting and IF (Figure S4A‐B). TSPAN8‐sEV treatment in T‐47D cells significantly increased the phosphorylation of FAK and SRC and the expression of paxillin. The dynamic assembly of the cytoskeleton is regulated by Rho family small GTPases, including RHOA and RAC1, and the downstream Rho‐associated kinase (ROCK) (Haga & Ridley, 2016; Hodge & Ridley, 2016; Vega et al., 2011). TSPAN8‐sEV treatment significantly increased the expression of RAC1, RHOA, ROCK1 and MMP2 in T‐47D cells (Figure S4C).
FIGURE 4.
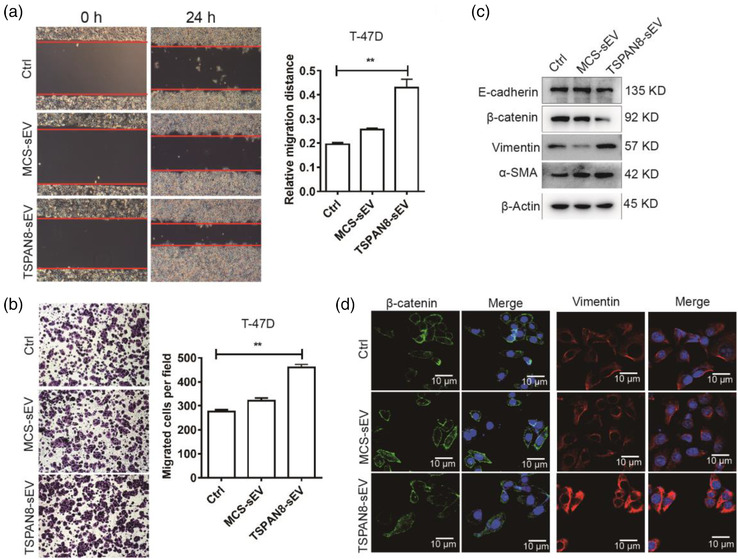
sEV cargo TSPAN8 educating recipient cells activation. (a) Migration of TSPAN8‐sEV and MCS‐sEV treatment on T‐47D cells was evaluated by wound healing assays. Data are mean standard deviation (n = 3, three independent experiments) **p < 0.01 by the two‐tailed Student's t‐test. (b) Cell motility of TSPAN8‐sEV and MCS‐sEV treatment for 48 h on T‐47D cells was determined by transwell assay. Data are mean standard deviation (n = 3, three independent experiments) **p < 0.01 by the two‐tailed Student's t‐test. (c) The expression of EMT‐related genes was confirmed by western blot analysis in T‐47D cells treated with sEVs. (d) The expression and localization of EMT markers β‐catenin and Vimentin in T‐47D cells with sEV treatment were detected by IF
Based on the classic “seed and soil” cancer metastasis hypothesis, EMT allows cancer cells to have the capacity for intravasation and extravasation of circulation and colonization in distant tissues (Balaji et al., 2021; Pantel & Brakenhoff, 2004). Western blotting showed that TSPAN8‐sEV treatment in T‐47D cells significantly increased the expression of EMT‐related genes (Figure 4c). IF confirmed that TSPAN8‐sEV treatment in T‐47D cells promoted the expression of EMT‐related genes (Figure 4d). Taken together, these findings suggest that TSPAN8 in sEV‐cargo alters the phenotype of T‐47D cells and increases motility to facilitate cancer metastasis.
A recent study demonstrated that TSPAN8 promotes colorectal cancer metastasis via EMT induction (H. ‐. S. Zhang et al., 2020). Park et al. (2016) revealed that targeting TSPAN8 using knockdown or antibody blocking can effectively reduce epithelial ovarian cancer invasion and metastasis (Park et al., 2016). TSPAN8 promotes the binding of ligands to the integrins that activate the FAK‐SRC signalling pathway, which promotes cell migration and phenotype alteration (Webb et al., 2004; Yue et al., 2013). TSPAN8 enriched sEVs transfer TSPAN8 into target cells to induce EMT (H. ‐. S. Zhang et al., 2020). Furthermore, TSPAN8 is involved in sEV cargo protein sorting, which might change the sEV content and promote target cell education (Liu et al., 2020). TSPAN8 enrichment changed the components of TEMs on the sEV surface, impairing the membrane components and signalling pathway activation of target cells (El Kharbili et al., 2020). These studies imply that TSPAN8 may promote cancer development and metastasis in multiple cancer types. Our findings confirmed that the engagement of sEV cargo TSPAN8 induces breast cancer cell migration and EMT to promote dissemination via FAK‐SRC signalling activation.
3.5. TSPAN8 promotes sEV uptake by target cells in vivo
The results reported to date convincingly demonstrate that TSPAN8 promotes adhesion of sEVs on epithelial cancer cells and fibroblasts in vitro. To investigate the role of TSPAN8 in regulating sEV uptake in vivo, we treated Balb/c mice with DiR‐labelled sEVs for 24 h prior to analysis (Figure 5a). The mice were sacrificed and the sEV uptake in vivo was evaluated using the IVIS Lumina live animal biophotonic imaging system (Figure 5b). The data showed that the breast cancer cell‐derived sEVs were preferentially taken up in the lung, liver, and spleen (Figure 5c). To confirm that TSPAN8 contributes to sEV uptake in vivo, we prepared sections of the tissues from the lung, liver, and spleen and observed them using confocal microscopy. sEV clumps were observed in the images that the TSPAN8‐sEV clumps showed increased uptake in the lung and liver and were similar in the spleen (Figure 5d). To further obtain a hint towards specific cell population uptake of TSPAN8‐sEV, we performed counterstaining with the monocyte marker CD11b and macrophage marker F4/80 in the lung, liver, and spleen with frozen sections (Figure S5). The results demonstrated that most sEVs were taken up by macrophages and monocytes in the liver and spleen. Aside from macrophages and monocytes, sEVs are taken up by the other cells in the lung. The data also showed that over 90% of unbound sEVs that disappeared from the circulation were mostly degraded/recovered in the liver (Erb et al., 2017; Wen et al., 2016).
FIGURE 5.
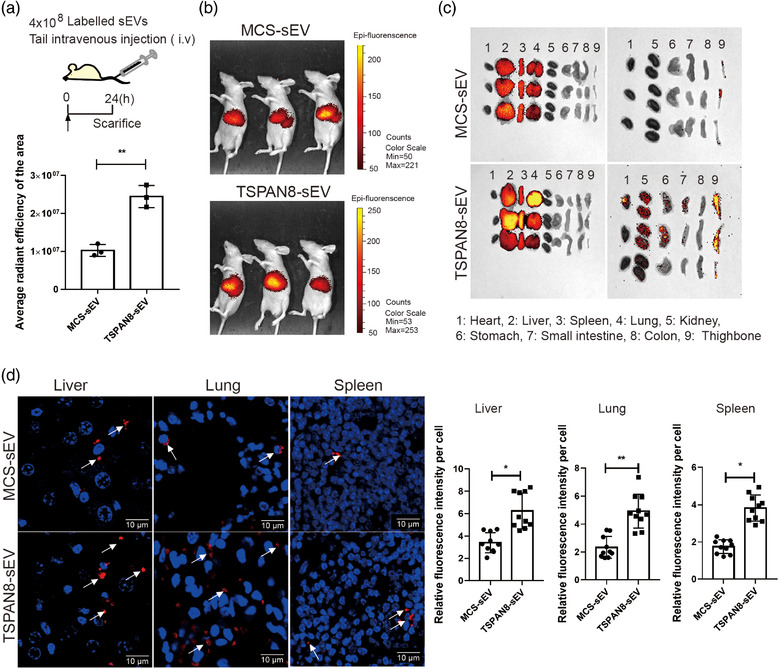
TSPAN8 promoted sEV uptake in vivo. (a) A diagram showed that the labelled sEVs treated nude mice for 24 h to evaluate the uptake in vivo. (b) The labelled sEV uptake in vivo was determined by IVIS Lumina imaging system post 24 h injection. Data are mean standard deviation **p < 0.01 by the two‐tailed Student's t‐test. (c) Biodistribution of TSPAN8‐sEV and MCS‐sEV is enriched in the lung, liver and spleen of mice. (d) IF of DiR‐labelled sEVs positive cells showed in lung, liver and spleen. Arrows indicated the sEV clumps. All immunofluorescence images are representative of ten random fields. Data are mean standard deviation (n = 3, three independent experiments) *p < 0.05; **p < 0.01 by the two‐tailed Student's t‐test
4. CONCLUSION
We conclude that TSPAN8 contributes to breast cancer cell‐derived sEVs and target cell binding by strengthening confined diffusion and induces confined diffusion of fibroblasts. The binding or uptake of TSPAN8‐sEV promotes motility and invasion, possibly linked to the FAK‐SRC signalling pathway, EMT induction, and protease upregulation in target cells. Pronounced TSPAN8‐sEV binding and uptake were also observed in selected organs in vivo. Further, the specific molecules responsible for the confined diffusion of sEVs at target cells need to be identified. The phenotype alteration of target cells is dependent on the cargo of sEVs. The omics analysis of sEVs and their target cells are required to reveal the mechanism by which TSPAN8‐sEV promotes cancer cell motility and invasion. This study provides insights on the research of the interaction of sEVs and target cells, while the major implications point the direction of engineered sEVs for therapeutic application.
AUTHOR CONTRIBUTIONS
T.W., X.W., H.W designed the experimental approach, performed the experimental work, analyzed the data. L.L., C.Z performed molecular cloning work and sEV isolation. X.T., R.X., Z.L., C. J., L.Z. contributed to hypothesis discussion, experimental design, and data analysis. L.X. contributed to hypothesis discussion, experimental design, data analysis and wrote the manuscript. S.Y. conceived the hypothesis, led the project, experimental design, interpreted the data and project coordination, and wrote the manuscript.
All authors have given approval to the final version of the manuscript.
NOTES
Any additional relevant notes should be placed here.
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENT
We gratefully acknowledge support from the following funding sources: the National Natural Science Foundation of China (Grant Nr. 81773120 to S. Y.).
Wang, T. , Wang, X. , Wang, H. , Li, L. , Zhang, C. , Xiang, R. , Tan, X. , Li, Z. , Jiang, C. , Zheng, L. , Xiao, L. , & Yue, S. (2021). High TSPAN8 expression in epithelial cancer cell‐derived small extracellular vesicles promote confined diffusion and pronounced uptake. Journal of Extracellular Vesicles, 10, e12167. 10.1002/jev2.12167
Teng Wang, Xin Wang, and Haobin Wang contributed equally to this work.
Contributor Information
Lehui Xiao, Email: lehuixiao@nankai.edu.cn.
Shijing Yue, Email: shijingyue@nankai.edu.cn.
REFERENCES
- Antonyak, M. A. , Li, B. , Boroughs, L. K. , Johnson, J. L. , Druso, J. E. , Bryant, K. L. , Holowka, D. A. , & Cerione, R. A. (2011).Cancer cell‐derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proceedings of the National Academy of Sciences of the United States of America, 108(12), 4852–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji, S. , Kim, U. , Muthukkaruppan, V. , & Vanniarajan, A. (2021).Emerging role of tumor microenvironment derived exosomes in therapeutic resistance and metastasis through epithelial‐to‐mesenchymal transition. Life Sciences, 280, 119750. [DOI] [PubMed] [Google Scholar]
- Choi, D. , Montermini, L. , Kim, D. ‐. K. , Meehan, B. , Roth, F. P. , & Rak, J. (2018).The impact of oncogenic EGFRvIII on the proteome of extracellular vesicles released from glioblastoma cells. Molecular and Cellular Proteomics, 17(10), 1948–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, H. C. , Svensson, K. J. , Van Kuppevelt, T. H. , Li, J. ‐ P. , & Belting, M. (2013).Cancer cell exosomes depend on cell‐surface heparan sulfate proteoglycans for their internalization and functional activity. Proceedings of the National Academy of Sciences of the United States of America, 110(43), 17380–17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, M. , Raposo, G. , & Théry, C. (2014).Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual Review of Cell and Developmental Biology, 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Costa‐Silva, B. , Aiello, N. M. , Ocean, A. J. , Singh, S. , Zhang, H. , Thakur, B. . K. , Becker, A. , Hoshino, A. , Mark, M. T. , Molina, H. , Xiang, J. , Zhang, T. , Theilen, T. ‐. M. , García‐Santos, G. , Williams, C. , Ararso, Y. , Huang, Y. , Rodrigues, G. , Shen, T. ‐. L. , … Lyden, D. (2015).Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nature Cell Biology, 17(6), 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Wang, D. , & Xie, M. (2021).Tumor‐derived extracellular vesicles promote activation of carcinoma‐associated fibroblasts and facilitate invasion and metastasis of ovarian cancer by carrying miR‐630. Frontiers in Cell and Developmental Biology, 9, 652322. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- El Kharbili, M. , Cario, M. , Béchetoille, N. , Pain, C. , Boucheix, C. , Degoul, F. , Masse, I. , & Berthier‐Vergnes, O. (2020).Tspan8 drives melanoma dermal invasion by promoting ProMMP‐9 activation and basement membrane proteolysis in a keratinocyte‐dependent manner. Cancers (Basel), 12(5), 1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb, U. , Zhao, K. , Wang, Z. , Xiao, L. , & Zöller, M. (2017).Murine and human pancreatic tumor exosome recovery in mouse serum: Diagnostic and prognostic potential and target cell delivery. Cancer Letters, 403, 1–12. [DOI] [PubMed] [Google Scholar]
- Gong, X. , Wang, H. , Li, R. , Tan, K. , Wei, J. , Wang, J. , Hong, C. , Shang, J. , Liu, X. , Liu, J. , & Wang, F. (2021).A smart multiantenna gene theranostic system based on the programmed assembly of hypoxia‐related siRNAs. Nature Communications, 12(1), 3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga, R. B. , & Ridley, A. J. (2016).Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases, 7(4), 207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L. , Li, Y. , Wei, L. , Ye, Z. , Liu, H. , & Xiao, L. (2019).Correlation between the translational and rotational diffusion of rod‐shaped nanocargo on a lipid membrane revealed by single‐particle tracking. Nanoscale, 11(20), 10080–10087. [DOI] [PubMed] [Google Scholar]
- Hemler, M. E. (2003).Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annual Review of Cell and Developmental Biology, 19:397–422. [DOI] [PubMed] [Google Scholar]
- Hemler, M. E. (2014).Tetraspanin proteins promote multiple cancer stages. Nature Reviews Cancer, 14(1), 49–60. [DOI] [PubMed] [Google Scholar]
- Heusermann, W. , Hean, J. , Trojer, D. , Steib, E. , Von Bueren, S. , Graff‐Meyer, A. , Genoud, C. , Martin, K. , Pizzato, N. , Voshol, J. , Morrissey, D. V. , Andaloussi, S. E. L. , Wood, M. J. , & Meisner‐Kober, N. C. (2016).Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. Journal of Cell Biology, 213(2), 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, R. G. , & Ridley, A. J. (2016).Regulating Rho GTPases and their regulators. Nature Reviews Molecular Cell Biology, 17(8), 496–510. [DOI] [PubMed] [Google Scholar]
- Hu, S. , Li, Z. , Cores, J. , Huang, K. , Su, T. , Dinh, P. ‐. U. , & Cheng, K. (2019).Needle‐free injection of exosomes derived from human dermal fibroblast spheroids ameliorates skin photoaging. ACS Nano, 13(10), 11273–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, R. , Demant, M. , Aung, T. , Diering, N. , Cicholas, A. , Chapuy, B. , Wenzel, D. , Lahmann, M. , Güntsch, A. , Kiecke, C. , Becker, S. , Hupfeld, T. , Venkataramani, V. , Ziepert, M. , Opitz, L. , Klapper, W. , Trümper, L. , & Wulf, G. G. (2014).Populational equilibrium through exosome‐mediated Wnt signaling in tumor progression of diffuse large B‐cell lymphoma. Blood, 123(14), 2189–2198. [DOI] [PubMed] [Google Scholar]
- Li, J. , Xu, J. , Li, L. , Ianni, A. , Kumari, P. , Liu, S. , Sun, P. , Braun, T. , Tan, X. , Xiang, R. , & Yue, S. (2020).MGAT3‐mediated glycosylation of tetraspanin CD82 at asparagine 157 suppresses ovarian cancer metastasis by inhibiting the integrin signaling pathway. Theranostics, 10(14), 6467–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Wang, Y. , Wang, Q. , Liu, Y. , Bao, W. , & Wu, S. (2018).Exosomes in cancer: Small transporters with big functions. Cancer Letters, 435, 55–65. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Fan, J. , Xu, T. , Ahmadinejad, N. , Hess, K. , Lin, S. H. , Zhang, J. , Liu, X. , Liu, L. , Ning, B. , Liao, Z. , & Hu, T. Y. (2020).Extracellular vesicle tetraspanin‐8 level predicts distant metastasis in non‐small cell lung cancer after concurrent chemoradiation. Science Advances, 6(11), eAaz6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , Li, J. , Liu, S. , Wang, T. , Ianni, A. , Bober, E. , Braun, T. , Xiang, R. , & Yue, S. (2017).Exosomal tetraspanins mediate cancer metastasis by altering host microenvironment. Oncotarget, 8(37), 62803–62815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallegol, J. , Van Niel, G. , Lebreton, C. , Lepelletier, Y. , Candalh, C. , Dugave, C. , Heath, J. K. , Raposo, G. , Cerf–Bensussan, N. , & Heyman, M. (2007).T84‐intestinal epithelial exosomes bear MHC class II/peptide complexes potentiating antigen presentation by dendritic cells. Gastroenterology, 132(5), 1866–1876. [DOI] [PubMed] [Google Scholar]
- Mckelvey, K. J. , Powell, K. L. , Ashton, A. W. , Morris, J. M. , & Mccracken, S. A. (2015).Exosomes: Mechanisms of uptake. Journal of Circulating Biomarkers, 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, S. K. , & Schlaepfer, D. D. (2006).Integrin‐regulated FAK‐Src signaling in normal and cancer cells. Current Opinion in Cell Biology, 18(5), 516–523. [DOI] [PubMed] [Google Scholar]
- Mulcahy, L. A. , Pink, R. C. , & Carter, D. R. F. (2014).Routes and mechanisms of extracellular vesicle uptake. Journal of Extracellular Vesicles, 3, 24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko, I. , Rana, S. , Baumann, A. , Mcalear, J. , Hellwig, A. , Trendelenburg, M. , Lochnit, G. , Preissner, K. T. , & Zoller, M. (2010).Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome‐induced endothelial cell activation. Cancer Research, 70(4), 1668–1678. [DOI] [PubMed] [Google Scholar]
- Pantel, K. , & Brakenhoff, R. H. (2004).Dissecting the metastatic cascade. Nature Reviews Cancer, 4(6), 448–456. [DOI] [PubMed] [Google Scholar]
- Park, C. S. , Kim, T. ‐. K. , Kim, H. G. , Kim, Y. ‐. J. , Jeoung, M. H. , Lee, W. R. , Go, N. K. , Heo, K. , & Lee, S. (2016).Therapeutic targeting of tetraspanin8 in epithelial ovarian cancer invasion and metastasis. Oncogene, 35(34), 4540–4548. [DOI] [PubMed] [Google Scholar]
- Parsons, J. T. , Martin, K. H. , Slack, J. K. , Taylor, J. M. , & Weed, S. A. (2000).Focal adhesion kinase: A regulator of focal adhesion dynamics and cell movement. Oncogene, 19(49), 5606–5613. [DOI] [PubMed] [Google Scholar]
- Pegtel, D. M. , & Gould, S. J. (2019).Exosomes Annual Review of Biochemistry, 88, 487–514. [DOI] [PubMed] [Google Scholar]
- Peinado, H. , Zhang, H. , Matei, I. R. , Costa‐Silva, B. , Hoshino, A. , Rodrigues, G. , Psaila, B. , Kaplan, R. N. , Bromberg, J. F. , Kang, Y. , Bissell, M. J. , Cox, T. R. , Giaccia, A. J. , Erler, J. T. , Hiratsuka, S. , Ghajar, C. M. , & Lyden, D. (2017).Pre‐metastatic niches: Organ‐specific homes for metastases. Nature Reviews Cancer, 17(5), 302–317. [DOI] [PubMed] [Google Scholar]
- Perez‐Hernandez, D. , Gutiérrez‐Vázquez, C. , Jorge, I. , López‐Martín, S. , Ursa, A. , Sánchez‐Madrid, F. , Vázquez, J. , & Yáñez‐Mó, M. (2013).The intracellular interactome of tetraspanin‐enriched microdomains reveals their function as sorting machineries toward exosomes. The Journal of Biological Chemistry, 288(17), 11649–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman, A. , Bandari, S. K. , Liu, J. , Mobley, J. A. , Brown, E. E. , & Sanderson, R. D. (2016).Fibronectin on the surface of myeloma cell‐derived exosomes mediates exosome‐cell interactions. The Journal of Biological Chemistry, 291(4), 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, S. , Yue, S. , Stadel, D. , & Zöller, M. (2012).Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. International Journal of Biochemistry & Cell Biology, 44(9), 1574–1584. [DOI] [PubMed] [Google Scholar]
- Rana, S. , & Zöller, M. (2011).Exosome target cell selection and the importance of exosomal tetraspanins: A hypothesis. Biochemical Society Transactions, 39(2), 559–562. [DOI] [PubMed] [Google Scholar]
- Riazifar, M. , Mohammadi, M. R. , Pone, E. J. , Yeri, A. , Lässer, C. , Segaliny, A. I. , Mcintyre, L. L. , Shelke, G. V. , Hutchins, E. , Hamamoto, A. , Calle, E. N. , Crescitelli, R. , Liao, W. , Pham, V. , Yin, Y. , Jayaraman, J. , Lakey, J. R. T. , Walsh, C. M. , Van Keuren‐Jensen, K. , … Zhao, W. (2019).Stem cell‐derived exosomes as nanotherapeutics for autoimmune and neurodegenerative disorders. ACS Nano, 13(6), 6670–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, K. E. , Zeleniak, A. E. , Fishel, M. L. , Wu, J. , Littlepage, L. E. , & Hill, R. (2017).Cancer‐associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene, 36(13), 1770–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, A. , & Simons, M. (2016).Catching filopodia: Exosomes surf on fast highways to enter cells. Journal of Cell Biology, 213(2), 143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, B. H. , Ketova, T. , Hoshino, D. , Zijlstra, A. , & Weaver, A. M. (2015).Directional cell movement through tissues is controlled by exosome secretion. Nature Communications, 6, 7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, B. H. , Von Lersner, A. , Guerrero, J. , Krystofiak, E. S. , Inman, D. , Pelletier, R. , Zijlstra, A. , Ponik, S. M. , & Weaver, A. M. (2020).A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nature Communications, 11(1), 2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, K. J. , Christianson, H. C. , Wittrup, A. , Bourseau‐Guilmain, E. , Lindqvist, E. , Svensson, L. M. , Mörgelin, M. , & Belting, M. (2013).Exosome uptake depends on ERK1/2‐heat shock protein 27 signaling and lipid raft‐mediated endocytosis negatively regulated by caveolin‐1. The Journal of Biological Chemistry, 288(24), 17713–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach, M. , & Théry, C. (2016).Communication by extracellular vesicles: Where we are and where we need to go. Cell, 164(6), 1226–1232. [DOI] [PubMed] [Google Scholar]
- Van Niel, G. , D'angelo, G. , & Raposo, G. (2018).Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19(4), 213–228. [DOI] [PubMed] [Google Scholar]
- Vega, F. M. , Fruhwirth, G. , Ng, T. , & Ridley, A. J. (2011).RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. Journal of Cell Biology, 193(4), 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, D. J. , Donais, K. , Whitmore, L. A. , Thomas, S. M. , Turner, C. E. , Parsons, J. T. , & Horwitz, A. F. (2004).FAK‐Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nature Cell Biology, 6(2), 154–161. [DOI] [PubMed] [Google Scholar]
- Wen, S. W. , Sceneay, J. , Lima, L. G. , Wong, C. S. F. , Becker, M. , Krumeich, S. , Lobb, R. J. , Castillo, V. , Wong, K. N. , Ellis, S. , Parker, B. S. , & Möller, A. (2016).The biodistribution and immune suppressive effects of breast cancer‐derived exosomes. Cancer Research, 76(23), 6816–6827. [DOI] [PubMed] [Google Scholar]
- Wortzel, I. , Dror, S. , Kenific, C. M. , & Lyden, D. (2019).Exosome‐mediated metastasis: Communication from a distance. Developmental Cell, 49(3), 347–360. [DOI] [PubMed] [Google Scholar]
- Wu, X. , & Yeow, E. K. L. (2008).Fluorescence blinking dynamics of silver nanoparticle and silver nanorod films. Nanotechnology, 19(3), 035706. [DOI] [PubMed] [Google Scholar]
- Xiao, L. , Wei, L. , Liu, C. , He, Y. , & Yeung, E. S. (2012).Unsynchronized translational and rotational diffusion of nanocargo on a living cell membrane. Angewandte Chemie (International Ed. in English), 51(17), 4181–4184. [DOI] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R.‐M. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐Da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , … De Wever, O. (2015).Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Kovalenko, O. V. , Tang, W. , Claas, C. , Stipp, C. S. , & Hemler, M. E. (2004).Palmitoylation supports assembly and function of integrin‐tetraspanin complexes. Journal of Cell Biology, 167(6), 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Wang, Y. , Che, X. , Hou, K. , Wu, J. , Zheng, C. , Cheng, Y. , Liu, Y. , Hu, X. , & Zhang, J. (2021).Integrin α5 promotes migration and invasion through the FAK/STAT3/AKT signaling pathway in icotinib‐resistant non‐small cell lung cancer cells. Oncology Letters, 22(1), 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z. , Liu, H. , Wang, F. , Wang, X. , Wei, L. , & Xiao, L. (2019).Single‐particle tracking discloses binding‐mediated rocking diffusion of rod‐shaped biological particles on lipid membranes. Chemical Science, 10(5), 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, S. , Mu, W. , Erb, U. , & Zöller, M. (2015).The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget, 6(4), 2366–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, S. , Mu, W. , & Zöller, M. (2013).Tspan8 and CD151 promote metastasis by distinct mechanisms. European Journal of Cancer, 49(13), 2934–2948. [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Ye, Z. , Wei, L. , Luo, H. , & Xiao, L. (2019).Cell membrane‐coated porphyrin metal‐organic frameworks for cancer cell targeting and O2‐evolving photodynamic therapy. Acs Applied Materials & Interfaces, 11(43), 39594–39602. [DOI] [PubMed] [Google Scholar]
- Zhang, H. ‐. S. , Liu, H. ‐. Y. , Zhou, Z. , Sun, H. ‐. L. , & Liu, M. ‐. Y. (2020).TSPAN8 promotes colorectal cancer cell growth and migration in LSD1‐dependent manner. Life Sciences, 241, 117114. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , & Guan, J. ‐. L. (2009).Signal transduction by focal adhesion kinase in cancer. Cancer and Metastasis Reviews, 28(1‐2), 35–49. [DOI] [PubMed] [Google Scholar]
- Zhao, K. , Erb, U. , Hackert, T. , Zöller, M. , & Yue, S. (2018).Distorted leukocyte migration, angiogenesis, wound repair and metastasis in Tspan8 and Tspan8/CD151 double knockout mice indicate complementary activities of Tspan8 and CD51. Biochimica et Biophysica Acta Molecular Cell Research, 1865(2), 379–391. [DOI] [PubMed] [Google Scholar]
- Zhou, W. , Fong, M. Y. , Min, Y. , Somlo, G. , Liu, L. , Palomares, M. R. , Yu, Y. , Chow, A. , O'connor, S. . T. . F. , Chin, A. R. , Yen, Y. , Wang, Y. , Marcusson, E. G. , Chu, P. , Wu, J. , Wu, X. , Li, A. . X. , Li, Z. , Gao, H. , …, Wang, S. . E. (2014).Cancer‐secreted miR‐105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell, 25(4), 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra, A. , Lewis, J. , Degryse, B. , Stuhlmann, H. , & Quigley, J. P. (2008). The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell, 13(3):221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel, L. , Regnault, A. , Lozier, A. , Wolfers, J. , Flament, C. , Tenza, D. , Ricciardi‐Castagnoli, P. , Raposo, G. , & Amigorena, S. (1998).Eradication of established murine tumors using a novel cell‐free vaccine: Dendritic cell‐derived exosomes. Nature Medicine, 4(5), 594–600. [DOI] [PubMed] [Google Scholar]
- Zöller, M. (2009).Tetraspanins: Push and pull in suppressing and promoting metastasis. Nature Reviews Cancer, 9(1), 40–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.


