Abstract
The Ets family of transcription factors have been suggested to function as key regulators of hematopoeisis. Here we describe aberrant hematopoeisis and hemorrhaging in mouse embryos homozygous for a targeted disruption in the Ets family member, Fli1. Mutant embryos are found to hemorrhage from the dorsal aorta to the lumen of the neural tube and ventricles of the brain (hematorrhachis) on embryonic day 11.0 (E11.0) and are dead by E12.5. Histological examinations and in situ hybridization reveal disorganization of columnar epithelium and the presence of hematomas within the neuroepithelium and disruption of the basement membrane lying between this and mesenchymal tissues, both of which express Fli1 at the time of hemorrhaging. Livers from mutant embryos contain few pronormoblasts and basophilic normoblasts and have drastically reduced numbers of colony forming cells. These defects occur with complete penetrance of phenotype regardless of the genetic background (inbred B6, hybrid 129/B6, or outbred CD1) or the targeted embryonic stem cell line used for the generation of knockout lines. Taken together, these results provide in vivo evidence for the role of Fli1 in the regulation of hematopoiesis and hemostasis.
The human FLI1 gene, which we originally cloned from the leukemia T-cell line, CEM, is a member of the Ets gene family of transcription factors (38). As observed with all other members of the Ets gene family, FLI1 encodes a protein that retains a region of conserved sequence, the Ets domain (37, 38). This minimal 85-amino-acid region has been shown to be the DNA-binding domain. Ets proteins bind to DNA sequences that contain a consensus GGA(A/T) core motif (Ets-binding site) and, in the majority of cases, function as transcriptional activators. Ets proteins control the expression of genes that are critical for the control of cellular proliferation, differentiation, and programmed cell death. The presence of multiple Ets family proteins in a variety of cell types and the overlapping DNA-binding specificity of the Ets proteins have made it difficult to identify target genes that are specific for individual Ets factors. The generation and analysis of targeted disruptions in individual family members, coupled with the identification of such target genes, however, is one approach to understanding the role of Ets transcription factors in normal and dysregulated development. A variety of studies including the analysis of expression of members of the Ets transcription factor family in hematopoietic tissues and cell lines and the generation and analysis of targeted mutations in Ets gene family members in mouse suggest that they play important roles in the regulation of normal hematopoietic development (13, 14, 29).
FLI1 was found to be highly related to the human ERG gene, and we originally named it ERGB to reflect this homology (38). Sequence alignments of the predicted 452-amino-acid protein product of human FLI1 with those of the ERG and mouse Fli1 products (5) demonstrated 80 and 96% similarity, respectively. FLI1 has been shown to bind specific ETS-binding sites containing the GGA(A/T) core and transcriptionally activate a number of genes including those encoding GATA-1 (35, 38), EndoA (34), glycoprotein IIb (21, 42), and c-Mpl (thrombopoietin receptor) (10), as well as the human immunodeficiency virus long terminal repeat (33). The Ets (DNA-binding) domain of the FLI1 protein is located between amino acids 277 and 361. Deletion analysis has identified two possible transcriptional activation domains, designated ATA and CTA (32) (for amino-terminal transcriptional activation and carboxy-terminal transcriptional activation, respectively).
FLI1 is expressed in hematopoietic lineages, and its overexpression leads to aberrant hematopoiesis. In vitro differentiation studies indicate that FLI1 expression promotes megakaryocytic differentiation from K562 erythroleukemic cells (1). In mice, three different proviral insertions near the Fli1 gene cause an increase in Fli1 expression and lead to hematopoietic oncogenesis. Fli1 activation by Friend murine leukemia virus (MuLV) is associated with erythroleukemia induction (5). Similarly, primitive stem cell tumors with characteristics of early hematopoietic cells and non-T, non-B lymphomas are induced by integration of the 10A1 isolate of MuLV (28) or the Cas-Br virus (6), respectively, near the Fli1 locus. Overexpression of Fli1 in all tissues of transgenic mice results in death from progressive immunological renal disease associated with an increased number of autoreactive T and B lymphocytes (41). The possible role of FLI1 in autoimmunity is further supported by our recent observation of elevated expression of FLI1 mRNA in lymphocytes from patients with systemic lupus erythematosus (12). Overexpression of Fli1 results in an increased number of mature B cells, which have a reduced activation-induced apoptotic response compared to B cells from wild-type animals (41). Taken together, these results suggest that Fli1 plays a crucial role in normal hematopoietic differentiation and lineage selection.
In light of these previous reports, it was surprising that in a recent report on targeted disruption of Fli1, only a nonlethal minor phenotype was observed, consisting of a reduced thymus size and a reduction in the total number of thymocytes. In this previous construct, investigators placed the neomycin resistance cassette in exon II, which should have disrupted all but the first 10 amino acids (23). However, alternate splicing around the neomycin cassette created a truncated Fli1 protein that nonetheless retained all of the known functional domains. This minimal phenotype strongly contrasts with those associated with the overexpression of Fli1, mentioned above, and necessitates additional targeting experiments.
We have generated mice carrying a novel targeted disruption of the Fli1 gene by homologous recombination in embryonic stem cells. Although no detectable phenotype has been detected in heterozygous embryos, all homozygotes display dramatic hemorrhaging into the fluid-filled spaces of the central nervous system and brain on embryonic day 11.0 (E11.0) and are dead by E12.5. Abnormal hematopoiesis within the fetal liver is also clearly detectable at E11.0, a time that constitutes a critical transition period in hematopoiesis. Collectively, the results further strengthen the concept that Fli1 regulates target genes in the regulation of key aspects of hematopoietic differentiation.
MATERIALS AND METHODS
Vector construction.
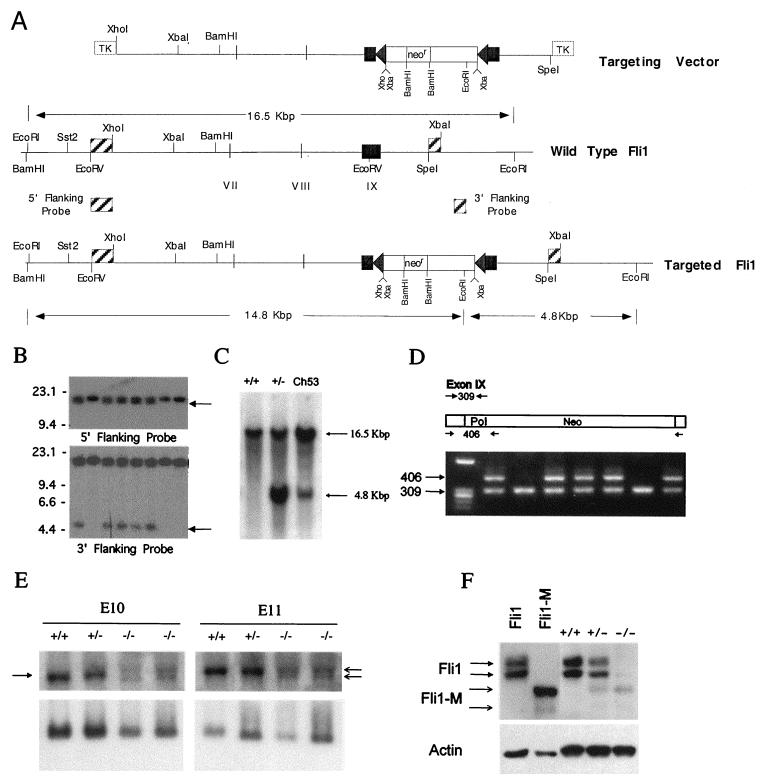
Fli1 genomic clones from an isogenic library (Stratagene; strain 129/SVJ) were isolated using a Flil cDNA probe containing the ETS domain (positions 1137 to 1729 [nucleotide positions as in reference 5]). The intron-exon structure of the mouse clone containing the DNA-binding domain has been partially determined, and a vector to target the Fli1 gene (see Fig. 1A) was generated. The targeting vector contains a selectable “floxed” Neo cassette (containing the neomycin phosphotransferase gene under the transcriptional control of the RNA polymerase II [Pol II] promoter and the last exon and polyadenylation signal of the HPRT gene for termination, all of which is flanked by the loxP Cre-recombinase recognition sequences). This floxed Neo cassette was inserted into the unique EcoRV site present in exon IX. Exon IX of Fli1 contains the ETS DNA-binding domain and the CTA domain. The Neo cassette is flanked by approximately 9 and 3 kb of Fli1 sequences at its 5′ and 3′ ends, respectively. This disrupted mouse gene has been cloned into the targeting vector, pBSTK9, which contains two divergent copies of the herpes simplex virus thymidine kinase gene (for negative selection). The resultant targeted cells will have a termination signal between the DNA-binding domain and the CTA domain. Thus, this approach creates mice that lack a regulatory domain of Fli1. Removal of this domain of Fli1 reduces transcriptional activation activity by 40 to 50% (data not shown).
FIG. 1.
Targeted disruption of the mouse Fli1 gene. (A) Schematic representation of the Fli1 targeting vector and genomic organization of the wild-type and Fli1 targeted alleles. The partial restriction map of the mouse Fli1 locus, indicating the positions of exons VII, VIII, and IX, is given. Triangles on each side of the Neo cassette (neor) represent loxP sequences. Positions of diagnostic EcoRI sites and predicted fragment sizes are indicated. (B) Identification of Fli1 targeted cell lines. Southern blot analysis of G418-resistant ES cell clones, probed with the indicated 5′ or 3′ probe, is shown. Targeted ES cell clones are identified by the presence of either a 14.8-kbp (5′ probe) or 4.8-kbp (3′ probe) fragment in addition to the 16.5-kbp fragment from the nontargeted allele. (C) Germ line transmission of the targeted Fli1 allele. Genomic DNA was isolated from wild-type and heterozygous mutant mice and from a chimeric mouse (C53). Southern blot analysis of EcoRI-digested DNA from representative wild-type (+/+) and heterozygous mutant (+/−) and from chimera 53 (Ch53), probed with the 3′ Fli1 probe (described in the legend to Fig. 1A), is shown. (D) PCR analysis of tail DNA from the progeny from a heterozygous intercross. Primers (Fli9F, Fli9R, and Pol IIR) were used to amplify genomic DNA, and the resultant products were resolved on a 1% Trevigel. The wild-type allele is identified by the 309-bp PCR product, and the targeted allele is identified by the 406-bp PCR product. Heterozygous mice are identified by the presence of both the 309- and 406-bp bands. The wild-type mice are identified by the presence of only the 309-bp band. No homozygous mutant mice (identified by the presence of only the 406-bp band) were detected. (E) The Fli1 mRNA transcript level is reduced in mutant embryos. Total RNA was extracted from wild-type and heterozygous and homozygous mutant E10 and E11 embryos. RNA (5 μg/lane) was electrophoresed on 1.2% agarose containing formaldehyde, transferred to a nylon membrane, and sequentially hybridized with 32P-labeled Fli1 cDNA (top panel) and S26 rRNA probes (bottom panel). The arrow on the left highlights the position of the normal Fli1 transcript, and the arrows on the right indicate the position of the two transcripts from the targeted Fli1 allele. (F) Presence of a truncated Fli1 protein in mutant E11 embryos. Total protein (5 μg/lane) prepared from wild-type and heterozygous and homozygous mutant E11 embryos was separated by polyacrylamide gel electrophoresis and transferred to filters. The filters were incubated with a Fli1 polyclonal antibody and visualized using the enhanced chemiluminescence system. Protein prepared from cells after transient transfection with vectors expressing Fli1 or mutant Fli1 (Fli1-M) were used as controls. Stripped membranes were reprobed with β-actin antibody. The 48-kDa protein band in the −/− embryos cannot be wild-type protein, because no wild-type Fli1 transcript is made in these −/− embryos. Furthermore, we did not observe the larger p51 Fli1 protein, normally encoded by the wild-type mRNA utilizing an alternative translation start. The identity of this protein remains unknown but is likely to be a cross-reactive protein.
Cell culture and selection.
TC1-10 embryonic stem (ES) cells were grown on a feeder layer of mouse fibroblasts (STO cells that express neomycin phosphotransferase and leukocyte inhibition factor) in knockout Dulbecco's modified Eagle's medium (Gibco/BRL) supplemented with 15% serum replacement medium (Gibco/BRL) and made complete as described previously (17). ES cells (2 × 107) were electroporated using a Bio-Rad Gene Pulser at 600 V and 25 μF. Transfected cells were simultaneously treated with G418 (230 μg/ml) and FIAU (1.25 μM) for positive and negative selection, respectively. Drug-resistant ES cell clones were expanded and screened for homologous recombination by Southern blot analysis of their genomic DNA.
Single-cell suspensions of fetal livers or yolk sac were prepared from pregnant mice on days E11.0 and E10.0, respectively. The culture was carried out using permissive growth factor conditions in medium containing interleukin-3 (IL-3), IL-6, steel factor (SF; c-kit ligand), and erythropoietin (Epo). Culture medium (1 ml) contained 104 fetal liver cells, alpha-medium (Flow Laboratories, Rockville, Md.), 1.2% methylcellulose (Shinetsu Chemical Co., Tokyo, Japan), 30% fetal bovine serum (Atlanta Biologicals, Norcross, Ga.), 1% deionized bovine serum albumin, 10−4 M 2-mercaptoethanol (Sigma Chemical Co., St. Louis, Mo.), and growth factors as indicated. Cytokine concentrations were 200 U of IL-3 per ml, 100 ng of SF per ml, 100 ng of IL-6 per ml, and 1 U of Epo per ml. Murine IL-3 was a gift from Tetsuo Sudo (Biomaterial Research Institute, Yokohama, Japan). SF was a gift from Immunex, Seattle, Wash. Recombinant human Epo was provided by Fu-Kuen Lin (Amgen, Thousand Oaks, Calif.). IL-6 was a gift from M. Naruto (Toray Industries, Kamakura, Japan).
Chimeric and knockout mice.
Chimeric mice were generated by ES cell-morula aggregation as described previously (19). Briefly, mice harboring heterozygous disrupted alleles were generated by the following strategy. Targeted ES cells were thawed and plated for 2 days before being used in morula aggregation. Clumps of about 12 ES cells were aggregated with pairs of recipient BL6 morulas in a sandwich-type configuration, incubated overnight, and implanted as expanded blastocysts into DBA/BL6 or CD1 F1 foster mothers. Chimeric males that demonstrated greater than 20% agouti coat color and derived from TC1 ES cells were mated weekly with two BL6, CD1, or 129SV females to generate heterozygotes that were identified by the same Southern blot strategy used to identify targeted ES cell lines. Alternatively, the Fli1 mutant allele was demonstrated by PCR using a primer specific for the Pol II gene along with two Fli1 exon IX-specific primers (see below).
Flow cytometric analysis and cell sorting.
The mononuclear fraction was obtained by sedimentation on Ficoll-Paque (Pharmacia, Piscataway, N.J.). A fraction enriched for multilineage colony-forming cells was obtained using a modification of the procedure described previously (11, 30). Cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-Ly6A/E (Sca-1), phycoerythrin (PE)-conjugated anti-CD43 (S7; PharMingen, San Diego, Calif.), and propidium iodide as described by Hirayama and Ogawa (16). CFU-E were obtained by staining with FITC-labeled CD71 (clone C2; PharMingen) and ACK4 anti-c-kit antibody labeled with biotin followed by PE-labeled streptavidin. ACK4 antibody was a generous gift from S.-I. Nishikawa, Kumamoto, Japan. Flow cytometric analysis and cell sorting were performed on a FACS Vantage cell sorter (Becton Dickinson, San Jose, Calif.). Single cells were deposited into individual wells of 96-well microtiter plates using a Clone-Cyt integrated deposition system (Becton Dickinson). Daughter cells were separated by micromanipulation. One cell was used for single-cell reverse transcriptase-coupled PCR (RT-PCR), and the other was cultured for 7 to 9 days. Lineage expression was determined by May-Grünwald Giemsa staining.
Analysis of DNA.
For preparation of genomic DNA, ES cells were lysed with sodium dodecyl sulfate and proteinase K. Genomic DNA (5 μg) was digested with EcoRI, electrophoresed on a 0.8% agarose gel, and blotted onto a nitrocellulose membrane. Hybridization was performed using random-primed synthesis and Quick Hyb (Stratagene, La Jolla, Calif.).
For genotyping of the mice and embryos, we used PCR to detect fragments of the wild-type Fli1 and the targeted Fli1 allele. The PCR conditions were 1 cycle at 94°C for 2 min followed by 35 cycles at 94°C for 1 min, 68°C for 1 min, and 72°C for 1 min. A 309-bp fragment indicates the presence of the wild-type allele, whereas a 406-bp fragment is amplified from the mutated allele. The primers were as follows: Fli1 exon IX/forward primer (positions 1156 to 1180), GACCAACGGGGAGTTCAAAATGACG; Fli1 exon IX/reverse primer (positions 1441 to 1465), GGAGGATGGGTGAGACGGGACAAAG; and Pol II/reverse primer, GGAAGTAGCCGTTATTAGTGGAGAGG.
RT-PCR.
RNA was extracted with Trizol (Gibco/BRL) as specified by the manufacturer. tRNA was used as a carrier for single cell samples. cDNA synthesis was carried out with random hexamer primers and Moloney MuLV reverse transcriptase (Gibco/BRL). Each sample was divided into two or more portions prior to amplification. cDNA was amplified in a final volume of 50 μl containing 1.25 U of AmpliTaq Gold (Perkin-Elmer, Foster City, Calif.), 1 μM sense primer, 1 μM antisense primer, 10 mM Tris (pH 8.3), 50 mM KCl, and 1.5 mM MgCl2. Samples were preheated at 94°C for 12 min to activate the enzyme and then subjected to 28 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 2 min. Samples from single cells were amplified for 50 cycles. Agarose gel electrophoresis was carried out, and PCR samples were blotted onto Hybond-N+ nylon membranes (Amersham Life Science, Inc., Arlington Heights, Ill.) and probed with internal oligonucleotides, as previously described (31), except that Rapid-hyb (Amersham Life Science, Inc.) was used. For single-cell PCR, Kodak BioMaxMS film with a BioMax enhancing screen (Eastman Kodak Co., Rochester, N.Y.) was used. Quantitation was carried out using a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The primers were as follows: (i) for Fli1 (exons 6 to 8), sense (positions 899 to 919), AGACCCTTCTTATGACTCTGTCA; antisense (positions 1000 to 1018), GGGCCGCTGCTCAGTGTTC; and internal probe (positions 970 to 990), TCTCCTTGGAGGATCACAGAC; and (ii) for actin, sense (positions 198 to 219), CTGAAGTACCCCATTGAACAT; antisense (positions 619 to 642), CTCTTTGATGTCACGCACGATTTC; and internal probe (positions 244 to 264), ATGGAGAAGATCTGGCAC.
In situ hybridization.
In situ hybridization was performed by the method of Wilkinson (39). E10 embryos were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS). Dehydration was carried out in increasing concentrations of ethanol in saline, then absolute ethanol, and finally xylene. Embryos were embedded in paraffin, and 8-μm sections were processed as follows. The sections were rehydrated, treated with 1% hydrogen peroxide in PBS for 15 min, incubated in 30 mg of proteinase K per ml for 12 min at 37°C, postfixed in 4% paraformaldehyde in PBS for 20 min, acetylated in 1 mM triethanolamine–0.25% acetic anhydride for 10 min, dehydrated, and hybridized overnight at 42°C in hybridization solution containing 50% formamide, 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10% dextran sulfate, 1× Denhardt's solution, 0.5 mg of yeast RNA per ml, and 800 to 1,000 ng of biotin-labeled riboprobe per ml. Sections were hybridized to either an antisense probe or a control sense Fli1 probe. The full-length Fli1 cDNA from plasmid PECE-BB4 (5) was recloned in both orientations into the pGEM7 vector. The plasmids were linearized with EcoRV, and RNA was synthesized in vitro using a biotin labeling kit and T7 RNA polymerase (NEN Life Science Products, Boston, Mass.). The antisense Fli1 probe (333 bp) contains both translated and 3′ untranslated sequences, as previously described (23). The sense Fli1 probe (1,396 bp) contains both 5′ untranslated sequences and translated sequences, including the Ets domain. Following hybridization, washing was performed at 42°C in 4× SSC–50% formamide and then at 37°C in 2× and 1× SSC, followed by incubation with 10 μg of RNase A per ml at 37°C 30 min. Sections were blocked for 30 min in blocking buffer (NEN Life Science Products) and incubated with SA-HRP (NEN Life Science Products) for 30 min. The signal was amplified using a TSA-Indirect amplification kit (NEN Life Science Products) and developed with DAB (Vector Laboratories, Burlingame, Calif.). Slides were counterstained with methyl green, mounted, and photographed with a Kodak Digital Science DC 120 zoom digital camera. Hybridization with the sense control probe (1,396 bp) did not yield any significant signal.
Western blotting.
Homogenates from wild-type and heterozygous and homozygous mutant Fli1 embryos were prepared by lysis in radioimmunoprecipitation assay (RIPA) buffer in the presence of protease inhibitors (Sigma). Sonicated lysates were clarified by centrifugation, and the protein concentration of supernatants was determined by the Bio-Rad assay. A 5-μg aliquot of each protein extract was separated on an SDS–10% polyacrylamide gel and transferred to nitrocellulose paper. Membranes were incubated for 2 h with FLI1-polyclonal antibody (made against full-length FLI1) and then with horseradish hydroperoxidase-labeled secondary antibody (Amersham). Antibody was detected using enhanced chemiluminescence (Amersham). The presence of equivalent protein loading was verified by reprobing stripped membranes with β-actin (Sigma) antibody. Lysates from cells transfected with Fli1 or mutant Fli1 (Fli1-M) expression vectors and proteins obtained by coupled in vitro transcription-translation (Promega) were used as positive controls for mobility and antibody recognition of Fli1 and mutant Fli1 proteins. The Fli1-M vector was constructed by insertion of the floxed Neo cassette into the EcoRV site of Fli1 cDNA, the identical site used in construction of the targeting vector. The targeting vector and the Fli1-M vector have identical sequences flanking the EcoRV cloning site; thus, the Fli1-M cDNA directs the synthesis of mRNA and protein identical to those present in the targeted cells and mutant mice.
RESULTS
Design and construction of a novel Fli1 targeting vector.
To disrupt the Fli1 gene, a genomic targeting vector was constructed which contains an insertional disruption just upstream of the CTA domain (Fig. 1A). A loxP-flanked Neo cassette was inserted into the unique EcoRV site present in exon IX. Exon IX of Fli1 contains the ETS DNA-binding domain and the CTA domain. The resultant targeted allele has a termination signal provided by the loxP sequence located between the DNA-binding domain and the CTA domain. The Neo gene cassette is driven by the strong Pol II promoter and contained exons VIII and IX of the HPRT gene. The Neo cassette is flanked by approximately 9 and 3 kb of Fli1 sequences at its 5′ and 3′ ends, respectively, and extends from the XhoI site upstream of exon VII to the SpeI site downstream of exon IX (Fig. 1A). These mouse genomic sequences and inserted Neo cassette were cloned into the targeting vector, pBSTK9, which contains two divergent copies of the herpes simplex virus thymidine kinase gene (for negative selection).
Generation and identification of mouse lines carrying the targeted Fli1 allele.
The resulting targeting vector was linearized with ScaI and electroporated into a rederivation of TC-1 ES cells (kindly provided by P. Leder). Electroporated ES cells were grown on mitotically inactivated and G418-resistant mouse embryonic fibroblasts in medium containing G418 and FIAU for positive and negative selection, respectively. Drug-resistant ES cell clones were expanded, and targeted ES cells were identified by Southern blot analysis of their genomic DNA. Diagnostic restriction digests were done using EcoRI, and the resultant Southern blots were hybridized with a 3′ probe (300 bp, SpeI-XbaI fragment) and 5′ probe (800 bp, EcoRV-XhoI fragment) (the probes are shown as hatched boxes in Fig. 1A) that flank the homologous region used in the targeting construct. Figure 1B shows representative data for analysis of the ES clones. Two distinct hybridization patterns were detected. Nontargeted ES cells were identified by the presence of a single hybridizing band of 16.5 kbp, representing the expected wild-type genomic fragment. Clones that were heterozygous for the targeted disruption were recognized by the presence of an additional band of 14.8 kb (5′ probe) (Fig. 1B, top panel) or 4.8 kb (3′ probe) (bottom panel). The targeting vector does not hybridize with either probe. Sixteen targeted lines were identified from 100 drug-resistant clones analyzed. Four targeted cell lines were randomly chosen and found to have normal karyotypes and were subsequently used for the generation of Fli1 mutant mouse lines. Two of four targeted cell lines tested (ES53 and ES54) gave rise to germ line chimeras by morula aggregation using B6 (C57BL/6J) and CD-1 morulas, respectively. Germ line chimeric males were identified by the appearance of agouti coat-colored offspring from matings to B6 or CD1 females, respectively. Germ line transmission of the targeted allele to subsequent progeny of B6 and CD1 was confirmed by Southern blot and PCR analyses (Fig. 1C and D).
An intact Fli1 allele is required for normal embryonic development.
Intercross matings of heterozygous Fli1 mutant F1 offspring were conducted to generate homozygotes. To determine the nature of this targeted Fli1 mutation, Fli1 mRNA and protein were prepared from wild-type, heterozygous, and homozygous embryos and analyzed by Northern and Western blotting, respectively (Fig. 1E and F). The Northern blot results demonstrate that the homozygotes express two distinct transcripts at approximately 10% of the wild-type level. This result suggests instability of transcripts and/or altered transcription in the targeted allele. Variable transcript size from the targeted allele may be due to usage of alternative poly(A) signals in the Neo cassette. The Fli1 gene encodes two distinct nuclear proteins of 51 and 48 kDa (2, 43). The Western blot results demonstrate that very little protein is made in homozygous mutant embryos (Fig. 1F), showing that the distinct reduction in transcript levels is further accentuated at the protein level. In extracts prepared from transient transfections, the mutant protein (Fli1-M) is slightly smaller than the wild-type Fli1 (Fli1), consistent with the prediction that the targeted allele would produce a nonsense mutant that lacks the CTA domain (Fig. 1F). In vitro transcription assays show that this CTA-less Fli1 mutant protein has only 50 to 60% of the activity of the wild type (data not shown). Collectively, these results demonstrate that the targeted mutation generates a severe hypomorphic mutant.
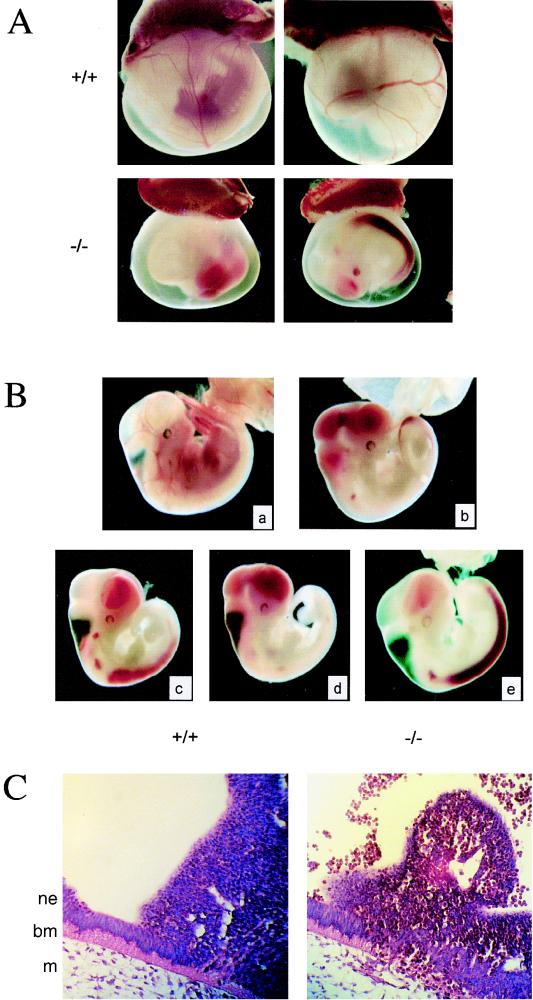
Heterozygous Fli1 mutant mice and embryos in both genetic backgrounds (B6 and CD1) appear healthy and fertile, with no apparent phenotypic abnormalities. However, no viable homozygous animals were produced from heterozygous intercrosses (Table 1). The complete absence of homozygous mutant progeny suggests that an intact Fli1 gene is essential for embryonic development. As expected for a recessive embryonic lethal phenotype, heterozygous progeny constitutes 60% of the total (Mendelian 66.6%). A retrograde genotypic analysis was conducted to determine the onset of embryonic lethality. No viable Fli1 homozygous embryos were recovered at E18.5, E16.5, or E12.5. However, homozygous Fli1 embryos were identified as resorptions at E12.5, suggesting that intact Fli1 function is required for viability prior to this gestational age (Table 1). Analysis of homozygous Fli1 embryos from earlier gestational ages demonstrated no overt defects until E11.0, at which time all homozygotes were recognized by the absence of red cells in the otherwise morphologically normal yolk sac vasculature (Fig. 2A). Upon further dissection, homozygotes exhibited a profound hematorrhachitic phenotype in which the majority of fetal blood had hemorrhaged into the lumen of the neural tube and the linked cephalic ventricles (Fig. 2B). The continuous open space in wild-type embryos was found to be filled with extravasated red blood cells in homozygotes. Hematorrhachis thus constituted a potential basis for the embryonic lethality observed, since continuous loss of circulating red blood cells would result in lasting hypoxia to embryonic tissues. Strikingly, this hematorrhachitic phenotype was 100% penetrant and occurred very precisely at E11 independent of the genetic background, as demonstrated on the outbred CD1, the hybrid 129/B6, and the inbred B6 genetic background (after more than five backcrosses of the hybrid into B6). In contrast, no phenotypic abnormalities were observed at E10.
TABLE 1.
Genotypes of progeny from heterozygous intercrosses
| Stage | No. of liters | No. (%) of mice with Fli1 genotype
|
No. of viable −/− mice | % of −/− mice expected to be viable | ||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | ||||
| E10 | 16 | 38 (30) | 59 (46) | 31 (24) | 31 | 96 |
| E11 | 47 | 109 (27) | 202 (51) | 88 (22) | 88 | 85 |
| E12.5 | 3 | 4 (17) | 14 (58) | 6a (25) | 0a | 0 |
| E16.5 | 1 | 4 | 5 | 0 | 0 | 0 |
| E18.5 | 2 | 3 | 6 | 0 | 0 | 0 |
| Term | 19 | 46 (40) | 70 (60) | 0 | 0 | 0 |
All E12.5 homozygous mutant embryos were identified as early resorptions.
FIG. 2.
Phenotype of Fli1 mutant embryos. (A) Absence of blood in the yolk sac of Fli1 mutant E11 embryos. Representative photographs of E11 embryos with intact visceral yolk sacs are shown. Blood-filled vessels in the yolk sac are seen in the wild-type littermate (+/+) but not in the homozygous mutant (−/−). (B) Fli1 mutant E11 embryos hemorrhage. E11 homozygous mutant embryos dissected free of the yolk sac are easily distinguished from their wild-type littermates (a) by their smaller overall size and hemorrhaging of fetal red cells into the cephalic ventricles (b to d) and the central canal of the neural tube (b, c, and e). (C) Histological analysis of E11 homozygous mutant embryos. Transverse sections through E11 embryos demonstrate a disruption in the columnar neuroepithelium, reduced adjacent extracellular matrix, and a hematoma at the site of hemorrhage (right panel). A section from a wild-type embryo is shown on the left for comparison. Note the nucleated red cells in the lumen of the neural tube (top, right panel). Positions of neuroepithelial (ne) and mesenchymal (m) cells and basement membrane (bm) are indicated.
Histological analyses of E11 homozygous mutant embryos demonstrate a disruption in the columnar neuroepithelium and a breakdown of basement membrane at the site of hemorrhage (Fig. 2C). Hematomas were also detected in the neuroepithelium more distal to the neuroepithelial-mesenchymal junction (Fig. 2C). It should be noted that these defects are localized; i.e., they were not observed throughout the length of the embryo in this junction region. Immunostaining with an antibody to PECAM demonstrated no obvious defects in vascular endothelial cell organization in this region (data not shown). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) demonstrated no apparent increase in the apoptotic index of cells in this region, which might be predicted for a loss-of-function Fli1 mutant (40) (data not shown). Furthermore, comparison of hematoxylin-and-eosin-stained, PECAM-labeled, and TUNEL-labeled sections of +/+, +/−, and −/− embryos has detected no differences in the aortic region. Collectively, data indicate that the site of hemorrhaging most probably involves blood vessels located in the mesenchyme proximal to the basement membrane between the mesenchyme and neuroepithelium (Fig. 2C).
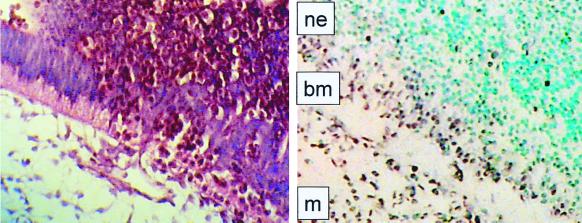
The tissue- and time-specific expression of Fli1 during development has been previously determined by in situ hybridization (23). Embryonic expression begins around E8, mainly in mesodermal cells. Endothelial cell expression is restricted to newly formed cells, with no expression detected in adult tissues. These studies are consistent with the hypothesis that Fli1 plays a role in mesoderm formation and in the development of endothelial and hematopoietic lineages (23). In situ hybridization of Fli1 mRNA was conducted on wild-type E10 embryos to more carefully analyze Fli1 expression within the context of the hematorrhachitic phenotype. Consistent with previous observations, our results demonstrated Fli1 mRNA expression at high levels in the mesenchyme. In addition, high levels of Fli1 expression were observed in the cellular layer of the neuroectoderm (columnar epithelium) immediately adjacent to the basement membrane and mesenchyme (Fig. 3). Thus, the spatiotemporal pattern of Fli1 expression closely correlates with the neuroectodermal and extracellular matrix defects found in the homozygous mutant (Fig. 2C).
FIG. 3.
Expression of Fli1. In situ hybridization with antisense Fli1 RNA of transverse sections of wild-type E10 embryos was performed. Embryo sections (8 μm) were processed for in situ hybridization and counterstained with methyl green. Positions of neuroepithelial (ne) and mesenchymal (m) cells and basement membrane (bm) are indicated in the in situ-hybridized section (right panel). A comparable, hematoxylin-and-eosin-stained section from the same region of a mutant E11 embryo is shown for comparison (left panel), demonstrating disruption of the thick basement membrane and columnar neuroepithelium and pockets of nucleated red cells within these tissues.
Fli1 is essential for normal hematopoiesis.
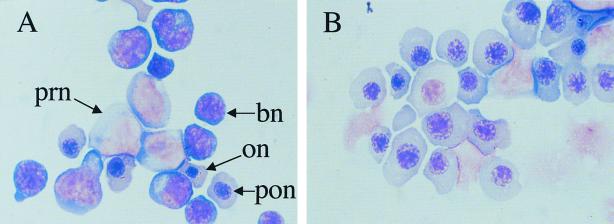
In addition to its roles in mesodermal and endothelial development, Fli1 is expressed in fetal liver cells with a megakaryocytic appearance (23). A variety of studies have also demonstrated that overexpression of Fli1 leads to aberrant hematopoiesis (5, 6, 28, 41). An analysis of hematopoiesis in the homozygous Fli1 mutants was conducted to determine if mice carrying greatly reduced levels of a less active Fli1 protein demonstrate hematopoietic defects. We first observed that the developing livers of the homozygous mutant embryos appeared to be pale and small compared to those of wild-type and heterozygous embryos at the time of hemorrhaging. The cellularity of the mutant livers was significantly reduced (Table 2). May Grünwald Giemsa staining of the mutant fetal liver cells demonstrated severe reduction in the number of pronormoblasts and basophilic normoblasts (Fig. 4). The increased number of smudged cells may suggest an increased incidence of apoptosis. Most of the intact cells from the mutants were orthochromatic and polychromatophilic normoblasts. Some of the orthochromatic and polychromatophilic normoblasts of the mutant mice exhibited more cytoplasm than did those of the wild-type mice, indicating megaloblastoid changes of the mutant red cells. Whether the changes are a manifestation of intrinsic phenotypes of the mutant red cells or an artifact of selective loss of populations by hemorrhaging is unknown.
TABLE 2.
Defect in hematopoietic colony formation by fetal liver cells from Fli1 mutant mice
| Embryo no. | Genotype | No. of cells/fetusa
|
||||
|---|---|---|---|---|---|---|
| Cells/fetus | CFU-E | BFU-E | CFU-GM | CFU-mix | ||
| Expt 1 | ||||||
| 1 | +/+ | 1.65 × 105 | 24,684 ± 3,497 | 116 ± 50 | 297 ± 99 | 1,023 ± 148 |
| 2 | +/+ | 2.18 × 105 | 19,206 ± 4,404 | 262 ± 44 | 741 ± 31 | 1,657 ± 109 |
| 3 | +/+ | 1.31 × 105 | 11,842 ± 1,454 | 109 ± 66 | 249 ± 13 | 550 ± 92 |
| 4 | +/− | 0.53 × 105 | 5,183 ± 673 | 53 ± 11 | 74 ± 27 | 127 ± 32 |
| 5 | +/− | 4.90 × 105 | 27,293 ± 5,782 | 196 ± 98 | 1,078 ± 98 | 1,274 ± 245 |
| 6 | +/− | 1.83 × 105 | 14,183 ± 2,159 | 256 ± 146 | 329 ± 146 | 714 ± 128 |
| 7 | −/− | 0.57 × 105 | 439 ± 68 | 11 ± 6 | 6 ± 6 | 6 ± 6 |
| 8 | −/− | 0.86 × 105 | 301 ± 77 | 17 ± 9 | 34 ± 17 | 26 ± 9 |
| Expt 2 | ||||||
| 1 | +/+ | 5.79 × 105 | 66,353 ± 1,505 | 696 ± 232 | 3,358 ± 519 | 3,127 ± 579 |
| 2 | +/+ | 7.38 × 105 | 76,604 ± 2,804 | 738 ± 295 | 5,461 ± 1,181 | 4,280 ± 590 |
| 3 | +/− | 4.11 × 105 | 29,674 ± 5,918 | 1,151 ± 164 | 1,480 ± 411 | 1,480 ± 164 |
| 4 | +/− | 4.49 × 105 | 41,488 ± 8,890 | 2,065 ± 493 | 1,257 ± 329 | 1,527 ± 247 |
| 5 | +/− | 3.48 × 105 | 33,478 ± 2,436 | 1,253 ± 278 | 1,044 ± 139 | 1,078 ± 209 |
| 6 | +/− | 3.46 × 105 | 27,611 ± 3,944 | 830 ± 208 | 1,661 ± 277 | 1,799 ± 415 |
| 7 | +/− | 3.43 × 105 | 31,213 ± 7,683 | 823 ± 137 | 755 ± 69 | 549 ± 69 |
| 8 | +/− | 4.53 × 105 | 44,756 ± 181 | 2,265 ± 362 | 2,084 ± 544 | 1,631 ± 272 |
| 9 | −/− | 1.72 × 105 | 5,917 ± 656 | 310 ± 104 | 34 ± 34 | 69 ± 34 |
Mean ± standard deviation of quadruplicate cultures of day 11 fetal liver cells. Cultures contained SF, IL-3 and Epo. Experiment 1 had 10,000 fetal liver cells/dish. Experiment 2 had 5,000 cells per dish. CFU-E cells were counted on day 2, and other colonies were counted on day 7.
FIG. 4.
Fetal liver cells from normal and mutant embryos stained with May Grünwald Giemsa. E11 embryos were collected, and smears from normal (A) and mutant (B) livers were prepared and stained with May Grünwald Giemsa. The predominant cells present in the smear prepared from the mutant embryo were orthochromatic (on) and polychromatophilic (pon) normoblasts. Note the absence of pronormoblasts (prn) and basophilic normoblasts (bn) in the mutant embryo.
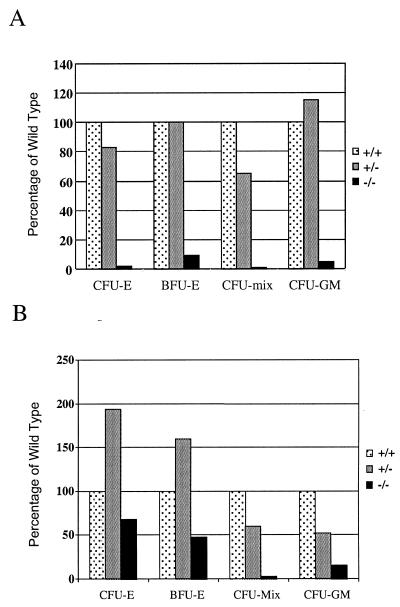
The role of Fli1 during hematopoiesis was assessed further by clonal culture of wild-type and heterozygous and homozygous mutant E11 embryo livers. Progenitor numbers, i.e., CFU-erythroid (CFU-E), burst-forming units–erythroid (BFU-E), CFU–granulocyte-macrophage (CFU-GM), and CFU-multilineage colonies (CFU-mix), were all drastically reduced in the fetal livers of the mutants (Fig. 5A; Table 2). Since E11 approximates the time when the site of hematopoiesis changes from the yolk sac to the liver and is a time when the hemorrhagic phenotype is clearly visible in mutant embryos, it is possible that defective hematopoeisis in Fli1−/− E11 livers is secondary to hemorrhaging. Hematopoiesis initiates within the yolk sac and continues within the fetal liver. Cells from the yolk sac of Fli1 homozygous embryos were isolated, grown in vitro, and analyzed. Culture of E10 yolk sac cells demonstrated a moderate loss in erythroid progenitors (CFU-E and BFU-E) and a substantial loss in CFU-mix and CFU-GM (Fig. 5B; Table 3). The apparent increase in the numbers of CFU-E and BFU-E in the yolk sac cultures from heterozygous mice relative to wild type could be correlated to a partial loss of function of the wild-type protein and/or a partial gain of function of the mutant protein. Whether a correlation exists between the homozygous knockout hemorrhagic phenotype and this apparent yolk sac effect remains to be determined.
FIG. 5.
Fli1 mutant mice demonstrate defects in hematopoietic colony formation. (A) E11 fetal liver cultures (triplicate cultures using 10,000 fetal liver cells/dish) in medium containing SF, IL-3, and Epo. CFU-E colonies were counted on day 2, and other colonies were counted on day 7. Data represent the mean values from individual fetuses of each genotype. +/+, n = 3; +/−, n = 3; −/−, n = 2. (B) E10 yolk sac cultures (quadruplicate cultures containing 1/15 yolk sac per dish) in medium containing SF, IL-3, IL-6, and Epo. CFU-E colonies were scored on day 4, and other colonies were scored on day 7. Data presented represent the mean values from individual fetuses. +/+, n = 4; +/−, n = 2; −/−, n = 2. The data presented in the figure represent the average numbers from experiment 1 of two experiments (all data are provided in Tables 2 and 3). Values are expressed as percentage of the wild type.
TABLE 3.
Defect in hematopoietic colony formation by yolk sac cells from Fli1 mutant mice
| Embryo no. | Genotype | No. of cells/fetusa
|
|||
|---|---|---|---|---|---|
| CFU-E | BFU-E | CFU-mix | CFU-GM | ||
| Expt 1 | |||||
| 1 | +/+ | 375 ± 45 | 150 ± 30 | 135 ± 75 | 68 ± 45 |
| 2 | +/+ | 615 ± 105 | 165 ± 15 | 480 ± 75 | 75 ± 45 |
| 3 | +/+ | 405 ± 75 | 300 ± 135 | 128 ± 30 | 45 ± 15 |
| 4 | +/+ | 1,035 ± 90 | 360 ± 180 | 405 ± 75 | 105 ± 6 |
| 5 | +/− | 1,320 ± 270 | 375 ± 105 | 225 ± 60 | 30 ± 30 |
| 6 | +/− | 1,035 ± 150 | 405 ± 105 | 113 ± 30 | 45 ± 6 |
| 7 | −/− | 285 ± 30 | 83 ± 30 | 8 ± 8 | 8 ± 6 |
| 8 | −/− | 540 ± 135 | 150 ± 60 | 8 ± 8 | 15 ± 15 |
| Expt 2 | |||||
| 1 | +/+ | 70 ± 30 | 40 ± 10 | 410 ± 60 | 150 ± 60 |
| 2 | +/+ | 170 ± 40 | 350 ± 80 | 440 ± 20 | 90 ± 40 |
| 3 | +/+ | 180 ± 20 | 200 ± 60 | 390 ± 80 | 150 ± 10 |
| 4 | +/− | 370 ± 100 | 300 ± 30 | 170 ± 10 | 100 ± 30 |
| 5 | +/− | 400 ± 120 | 320 ± 60 | 170 ± 40 | 210 ± 30 |
| 6 | +/− | 350 ± 100 | 170 ± 10 | 200 ± 10 | 180 ± 0 |
| 7 | +/− | 770 ± 90 | 320 ± 110 | 240 ± 20 | 230 ± 50 |
| 8 | +/− | 310 ± 50 | 130 ± 20 | 90 ± 40 | 30 ± 10 |
| 9 | −/− | 220 ± 30 | 100 ± 60 | 60 ± 20 | 20 ± 10 |
| 10 | −/− | 150 ± 40 | 80 ± 20 | 50 ± 20 | 20 ± 20 |
| 11 | −/− | 260 ± 50 | 110 ± 30 | 50 ± 20 | 40 ± 20 |
| 12 | −/− | 190 ± 70 | 100 ± 20 | 40 ± 30 | 40 ± 20 |
| 13 | −/− | 210 ± 20 | 100 ± 30 | 100 ± 10 | 20 ± 10 |
Mean ± standard deviation of quadruplicate cultures containing 1/10 yolk sac (experiment 1) or 1/15 yolk sac (experiment 2) and IL-3, IL-6, SF, and Epo. CFU-E cells were scored on day 4, and the other colony types were scored on day 7.
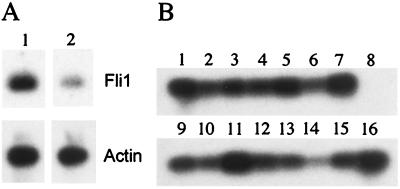
Fetal liver was collected and dissociated, and the multipotential progenitor populations were sorted. The Sca-1+CD43+ cell population is composed of the early progenitors CFU-GM and CFU-mix. The c-kit+ CD71+ cell population contains committed erythroid progenitors, CFU-E. RT-PCR (Fig. 6A) demonstrated that Fli1 expression was 4.5-fold higher in the Sca-1+ CD43+ cell population than in the c-kit+ CD71+ cell population. RT-PCR analysis of daughter cells of the Sca-1+ CD43+ population demonstrated that the CFU-GM and CFU-mix populations express the Fli1 gene (Fig. 6B). Furthermore, a CFU-meg colony was also shown to express Fli1 (Fig. 6B). Since hematopoiesis initiates within the yolk sac and continues within the fetal liver, it is interesting that the Sca-1+ CD43+ cell population (CFU-mix and CFU-GM) is more strongly affected than the c-kit+ CD71+ cell population (erythroid progenitors) in the yolk sac cultures from the homozygous mutant (Fig. 5B).
FIG. 6.
RT-PCR analysis of Fli1 mRNA expression in fetal liver cell populations. (A) Comparison of Fli1 expression in Sca-1+ CD43+ fetal liver cells and c-kit+ CD71+ cells. RT-PCR was carried out as described in Materials and Methods. Southern blots were hybridized with the indicated end-labeled probe. Panel 1, Sca-1+ CD43+; panel 2, c-kit+ CD71+. (B) Analysis of Fli1 expression in daughter cells. Sca-1+ CD43+ fetal liver cells were deposited into individual wells of a 96-well plate using the CloneCyt integrated deposition system. Daughter cells were separated. One cell was used for single-cell RT-PCR, and the other was cultured for 7 to 9 days. Lineage expression was determined by May Grünwald Giemsa staining. RT-PCR analysis of Fli1 expression in CFU-mix (lanes 1 to 7), CFU-GM (lanes 9 to 15), CFU-meg (lane 16), and negative control (lane 8) is shown.
DISCUSSION
Our Fli1 targeted mutant demonstrates profound hematorrhachitic and hematopoietic phenotypes. This is in contrast to the very mild phenotype observed in the previous Fli1 targeted mutant (23). Neither is a dominant mutant in the sense that no overt phenotype is observed in heterozygotes. Both mutants produce greatly reduced amounts of protein. Our mutant Fli1 is a truncated protein, lacking the functional CTA domain. The previous mutant Fli1 is a truncated protein, containing 19 unique amino acids in place of 76 amino acids present at the amino-terminal end of wild-type Fli1. This region of Fli1 is not required for transcriptional activation (32). In contrast, mutant Fli1 protein lacking the CTA domain demonstrates a 40 to 50% reduction in transcriptional activation by in vitro assay (data not shown). The greatly reduced expression of a Fli1 protein lacking the CTA domain in vivo may result in the complete loss of regulation of target genes.
Mechanism of lethality mediated by loss of Fli1 function.
At least three possible mechanisms may collectively contribute to the death of embryos lacking Fli1 function: (i) disruption of tissue integrity, resulting in hemorrhage; (ii) disruption of normal hematopoiesis; and (iii) disruption of normal hemostasis. The first model is consistent with Fli1 target genes identified, such as those encoding extracellular matrix proteins as well as those contributing to proper epithelial-mesenchymal interactions. For example, it has recently been determined that Fli1 cooperates with Sp1 to activate the human tenascin promoter (36).
Our results also demonstrate that hematopoiesis is severely impaired in Fli1 mutant mice at midgestation. The failure of the liver to develop normally as a hematopoietic organ at a critical transition period in embryonic hematopoiesis could be a contributing factor to embryo death. It has been shown that differentiation of specific hematopoietic lineages is controlled by specific cytokines and transcription factors. Knockout mice for GATA-1, GATA-2, JAK2, c-kit, SCL/tal-1, Epo, Epo receptor, Myb, etc. (25–27), have midgestation defects in hematopoiesis, and some result in midgestation death. There could be an intrinsic defect in the progenitors which makes them incapable of the rapid expansion required in fetal development, a defect in their migration to the liver, or in the ability of the liver to support hematopoiesis.
Loss of normal Fli1 function could lead to aberrant megakaryocytopoiesis, platelet production, and, ultimately, control of coagulation. Five observations support this model. (i) In vitro differentiation studies indicate that FLI1 expression promotes megakaryocytic differentiation of K562 erythroleukemic cells (1). (ii) We have demonstrated that Fli1 is expressed in the CFU-meg population from fetal liver (Fig. 6B). (iii) Cultures of progenitors from mutant yolk sacs have reduced levels of CFU-mix (Fig. 5B). (iv) It has recently been demonstrated that Fli1 protein is present in platelets (3). (v) Our recent preliminary data demonstrate that neutrophils, macrophages, mast cells, and erythroid cells, but not megakaryocytes, are present in individual colonies from yolk sac cultures of mutant fetuses. Collectively, loss of megakaryocyte-derived platelets is one possible cause of the hemorrhagic phenotype we observed. Ets factors have been shown to activate transcription driven by the megakaryocyte-specific membrane glycoprotein gpIIb promoter (7, 21), the platelet glycoprotein Iba promoter (15), the glycoprotein IX promoter (4), the platelet factor 4 promoter (24), and the promoter of mpl (10), the receptor for thromobopoietin. These studies support the hypothesis that one or more ETS transcription factors play a role in the regulation of transcription of megakaryocytic and platelet-specific genes. Disruption of normal coagulation alone is sufficient to lead to hemorrhaging in the embryo (9, 18). Notably in humans, severe disability or death from bleeding into the central nervous system and brain are features of both hemophilia A (factor VIII defect) and hemophilia B (factor IX defect) (20, 22). In addition, congenital megakaryocytic maturation defects as well as a mild hemorrhagic phenotype have been found in individuals heterozygous for a chromosomal deletion that removes FLI1 and ETS1 (8).
Future studies of Fli1 mutant mice and cells will facilitate the identification of specific target genes that are critical for normal development. Fli1 target genes, either individually or in combination, that are responsible for embryonic hemorrhaging and defective hematopoiesis will be delineated. Furthermore, these studies will demonstrate whether hemorrhage and defective hematopoiesis are independent or interrelated events.
ACKNOWLEDGMENTS
We thank Tina Cooper, Yong Gong, Jill Martin, Kristen Swartout, Ann Hofbauer, and Juanita Eldgride for technical assistance. We thank Tien Hsu for helpful discussion, advice, and critical review of the manuscript. We also thank Alan Bernstein for providing a mouse Fli1 cDNA clone and Phillip Leder for providing TC1-10 ES cells.
This work was supported in part by a grant from the NCI (PO1 CA78582).
Footnotes
This paper is dedicated to the memory of Takis S. Papas, a mentor, scientific colleague, and friend.
REFERENCES
- 1.Athanasiou M, Clausen P A, Mavrothalassitis G J, Zhang X K, Watson D K, Blair D G. Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell Growth Differ. 1996;7:1525–1534. [PubMed] [Google Scholar]
- 2.Bailly R A, Bosselut R, Zucman J, Cormier F, Delattre O, Roussel M, Thomas G, Ghysdael J. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol. 1994;14:3230–3241. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastian L S, Kwiatkowski B A, Breininger J, Danner S, Roth G. Regulation of the megakaryocytic glycoprotein IX promoter by the oncogenic Ets transcription factor Fli-1. Blood. 1999;93:2637–2644. [PubMed] [Google Scholar]
- 4.Bastian L S, Yagi M, Chan C, Roth G J. Analysis of the megakaryocyte glycoprotein IX promoter identifies positive and negative regulatory domains and functional GATA and Ets sites. J Biol Chem. 1996;271:18554–18560. doi: 10.1074/jbc.271.31.18554. [DOI] [PubMed] [Google Scholar]
- 5.Ben-David Y, Giddens E B, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 6.Bergeron D, Houde J, Poliquin L, Barbeau B, Rassart E. Expression and DNA rearrangement of proto-oncogenes in Cas-Br-E-induced non-T-, non-B-cell leukemias. Leukemia. 1993;7:954–962. [PubMed] [Google Scholar]
- 7.Block K L, Poncz M. Platelet glycoprotein IIb gene expression as a model of megakaryocyte-specific expression. Stem Cells. 1995;13:135–145. doi: 10.1002/stem.5530130205. [DOI] [PubMed] [Google Scholar]
- 8.Breton-Gorius J, Favier R, Guichard J, Cherif D, Berger R, Debili N, Vainchenker W, Douay L. A new congenital dysmegakaryopoietic thrombocytopenia (Paris-Trousseau) associated with giant platelet alpha-granules and chromosome 11 deletion at 11q23. Blood. 1995;85:1805–1814. [PubMed] [Google Scholar]
- 9.Cui J, O'Shea K S, Purkayastha A, Saunders T L, Ginsburg D. Fatal haemorrhage and incomplete block to embryogenesis in mice lacking coagulation factor V. Nature. 1996;384:66–68. doi: 10.1038/384066a0. [DOI] [PubMed] [Google Scholar]
- 10.Deveaux S, Filipe A, Lemarchandel V, Ghysdael J, Romeo P H, Mignotte V. Analysis of the thrombopoietin receptor (MPL) promoter implicates GATA and Ets proteins in the coregulation of megakaryocyte-specific genes. Blood. 1996;87:4678–4685. [PubMed] [Google Scholar]
- 11.Fujimoto K, Lyman S D, Hiryama F, Ogawa M. Isolation and characterization of primitive hematopoietic progenitors of murine fetal liver. Exp Hematol. 1996;24:285–290. [PubMed] [Google Scholar]
- 12.Georgiou P, Maroulakou I G, Green J E, Dantis P, Romano-Spica V, Kottaridis S, Lautenberger J A, Watson D K, Papas T S, Fischinger P J, Bhat N K. Expression of ets family of genes in systemic lupus erythematosus and Sjogren's syndrome. Int J Oncol. 1996;9:9–18. [PubMed] [Google Scholar]
- 13.Ghysdael J, Boureux A. The ETS family of transcriptional regulators. In: Yaniv M, Ghysdael J, editors. Oncogenes as transcriptional regulators. Vol. 1. Basel, Switzerland: Birkhauser Verlag; 1997. pp. 29–88. [Google Scholar]
- 14.Graves B J, Petersen J M. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto Y, Ware J. Identification of essential GATA and Ets binding motifs within the promoter of the platelet glycoprotein Ib alpha gene. J Biol Chem. 1995;270:24532–24539. doi: 10.1074/jbc.270.41.24532. [DOI] [PubMed] [Google Scholar]
- 16.Hirayama F, Ogawa M. CD43 expression by murine lymphohemopoietic progenitors. Int J Hematol. 1994;60:191–196. [PubMed] [Google Scholar]
- 17.Hogan B, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 18.Huang Z F, Higuchi D, Lasky N, Broze G J., Jr Tissue factor pathway inhibitor gene disruption produces intrauterine lethality in mice. Blood. 1997;90:944–951. [PubMed] [Google Scholar]
- 19.Joyner A L. Gene targeting: a practical approach. New York, N.Y: Oxford University Press; 1993. [Google Scholar]
- 20.Kulkarni R, Lusher J M. Intracranial and extracranial hemorrhages in newborns with hemophilia: a review of the literature. J Pediatr Hematol Oncol. 1999;21:289–295. doi: 10.1097/00043426-199907000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Lemarchandel V, Ghysdael J, Mignotte V, Rahuel C, Romeo P H. GATA and Ets cis-acting sequences mediate megakaryocyte-specific expression. Mol Cell Biol. 1993;13:668–676. doi: 10.1128/mcb.13.1.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine P H. Clinical manifestations and therapy of hemophilias A and B. In: Colman R W, Hirsh J, Marder V J, Salzman E W, editors. Hemostasis and thrombosis: basic principles and clinical practice. 2nd ed. Philadelphia, Pa: J. B. Lippincott Co.; 1987. pp. 97–111. [Google Scholar]
- 23.Melet F, Motro B, Rossi D J, Zhang L, Bernstein A. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol Cell Biol. 1996;16:2708–2718. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minami T, Tachibana K, Imanishi T, Doi T. Both Ets-1 and GATA-1 are essential for positive regulation of platelet factor 4 gene expression. Eur J Biochem. 1998;258:879–889. doi: 10.1046/j.1432-1327.1998.2580879.x. [DOI] [PubMed] [Google Scholar]
- 25.Orkin S H. Embryonic stem cells and transgenic mice in the study of hematopoiesis. Int J Dev Biol. 1998;42:927–934. [PubMed] [Google Scholar]
- 26.Orkin S H. Transcription factors and hematopoietic development. J Biol Chem. 1995;270:4955–4958. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- 27.Orkin S H, Porcher C, Fujiwara Y, Visvader J, Wang L C. Intersections between blood cell development and leukemia genes. Cancer Res. 1999;59:1784s–1788s. [PubMed] [Google Scholar]
- 28.Ott D E, Keller J, Rein A. 10A1 MuLV induces a murine leukemia that expresses hematopoietic stem cell markers by a mechanism that includes fli-1 integration. Virology. 1994;205:563–568. doi: 10.1006/viro.1994.1680. [DOI] [PubMed] [Google Scholar]
- 29.Papas T S, Bhat N K, Spyropoulos D D, Mjaatvedt A E, Vournakis J, Seth A, Watson D K. Functional relationships among ETS gene family members. Leukemia. 1997;11:557–566. [PubMed] [Google Scholar]
- 30.Pharr P N, Hofbauer A. Loss of flk-2/flt3 expression during commitment of multipotent mouse hematopoietic progenitor cells to the mast cell lineage. Exp Hematol. 1997;25:620–628. [PubMed] [Google Scholar]
- 31.Pharr P N, Ogawa M, Hofbauer A, Longmore G D. Expression of an activated erythropoietin or a colony-stimulating factor 1 receptor by pluripotent progenitors enhances colony formation but does not induce differentiation. Proc Natl Acad Sci USA. 1994;91:7482–7486. doi: 10.1073/pnas.91.16.7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao V N, Ohno T, Prasad D D, Bhattacharya G, Reddy E S. Analysis of the DNA-binding and transcriptional activation functions of human Fli-1 protein. Oncogene. 1993;8:2167–2173. [PubMed] [Google Scholar]
- 33.Seth A, Hodge D R, Thompson D M, Robinson L, Panayiotakis A, Watson D K, Papas T S. ETS family proteins activate transcription from HIV-1 long terminal repeat. AIDS Res Hum Retroviruses. 1993;9:1017–1023. doi: 10.1089/aid.1993.9.1017. [DOI] [PubMed] [Google Scholar]
- 34.Seth A, Robinson L, Panayiotakis A, Thompson D M, Hodge D R, Zhang X K, Watson D K, Ozato K, Papas T S. The EndoA enhancer contains multiple ETS binding site repeats and is regulated by ETS proteins. Oncogene. 1994;9:469–477. [PubMed] [Google Scholar]
- 35.Seth A, Robinson L, Thompson D M, Watson D K, Papas T S. Transactivation of GATA-1 promoter with ETS1, ETS2 and ERGB/Hu-FLI-1 proteins: stabilization of the ETS1 protein binding on GATA-1 promoter sequences by monoclonal antibody. Oncogene. 1993;8:1783–1790. [PubMed] [Google Scholar]
- 36.Shirasaki F, Makhulf H A, LeRoy C, Watson D K, Trojanowska M. Ets transcription factors cooperate with Sp1 to activate the human tenascin-C promoter. Oncogene. 1999;18:7755–7764. doi: 10.1038/sj.onc.1203360. [DOI] [PubMed] [Google Scholar]
- 37.Watson D K, McWilliams M J, Lapis P, Lautenberger J A, Schweinfest C W, Papas T S. Mammalian ets-1 and ets-2 genes encode highly conserved proteins. Proc Natl Acad Sci USA. 1988;85:7862–7866. doi: 10.1073/pnas.85.21.7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson D K, Smyth F E, Thompson D M, Cheng J Q, Testa J R, Papas T S, Seth A. The ERGB/Fli-1 gene: isolation and characterization of a new member of the family of human ETS transcription factors. Cell Growth Differ. 1992;3:705–713. [PubMed] [Google Scholar]
- 39.Wilkinson D G. The theory and practice of in situ hybridization. In situ hybridization: a practical approach. New York, N.Y: IRL Press at Oxford University Press; 1992. [Google Scholar]
- 40.Yi H, Fujimura Y, Ouchida M, Prasad D D, Rao V N, Reddy E S. Inhibition of apoptosis by normal and aberrant Fli-1 and erg proteins involved in human solid tumors and leukemias. Oncogene. 1997;14:1259–1268. doi: 10.1038/sj.onc.1201099. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Eddy A, Teng Y-T, Fritzler M, Kluppel M, Melet F, Bernstein A. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol Cell Biol. 1995;15:6961–6970. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Lemarchandel V, Romeo P H, Ben-David Y, Greer P, Bernstein A. The Fli-1 proto-oncogene, involved in erythroleukemia and Ewing's sarcoma, encodes a transcriptional activator with DNA-binding specificities distinct from other Ets family members. Oncogene. 1993;8:1621–1630. [PubMed] [Google Scholar]
- 43.Zhang X-K, Papas T S, Bhat N K, Watson D K. Generation and characterization of monoclonal antibodies against the ERGB/FLI-1 transcription factor. Hybridoma. 1995;14:563–569. doi: 10.1089/hyb.1995.14.563. [DOI] [PubMed] [Google Scholar]