Abstract
Aims
Hutchinson-Gilford progeria syndrome (HGPS) is an accelerated ageing syndrome associated with premature vascular disease and death due to heart attack and stroke. In HGPS a mutation in lamin A (progerin) alters nuclear morphology and gene expression. Current therapy increases the lifespan of these children only modestly. Thus, greater understanding of the underlying mechanisms of HGPS is required to improve therapy. Endothelial cells (ECs) differentiated from induced pluripotent stem cells (iPSCs) derived from these patients exhibit hallmarks of senescence including replication arrest, increased expression of inflammatory markers, DNA damage, and telomere erosion. We hypothesized that correction of shortened telomeres may reverse these measures of vascular ageing.
Methods and results
We generated ECs from iPSCs belonging to children with HGPS and their unaffected parents. Telomerase mRNA (hTERT) was used to treat HGPS ECs. Endothelial morphology and functions were assessed, as well as proteomic and transcriptional profiles with attention to inflammatory markers, DNA damage, and EC identity genes. In a mouse model of HGPS, we assessed the effects of lentiviral transfection of mTERT on measures of senescence, focusing on the EC phenotype in various organs. hTERT treatment of human HGPS ECs improved replicative capacity; restored endothelial functions such as nitric oxide generation, acetylated low-density lipoprotein uptake and angiogenesis; and reduced the elaboration of inflammatory cytokines. In addition, hTERT treatment improved cellular and nuclear morphology, in association with a normalization of the transcriptional profile, effects that may be mediated in part by a reduction in progerin expression and an increase in sirtuin 1 (SIRT1). Progeria mice treated with mTERT lentivirus manifested similar improvements, with a reduction in inflammatory and DNA damage markers and increased SIRT1 in their vasculature and other organs. Furthermore, mTERT therapy increased the lifespan of HGPS mice.
Conclusion
Vascular rejuvenation using telomerase mRNA is a promising approach for progeria and other age-related diseases.
Keywords: Endothelium, Hutchinson-Gilford progeria syndrome, Ageing
Graphical Abstract
 Listen to the audio abstract of this contribution
Listen to the audio abstract of this contribution
See page 4370 for the editorial comment for this article ‘Vascular rejuvenation: a new therapeutic target?’, by R. Madonna, https://doi.org/10.1093/eurheartj/ehab587.
Translational Perspective
Hutchinson-Gilford progeria syndrome (HGPS) is a paediatric genetic disease that results in accelerated senescence, with 90% of patients succumbing to cardiovascular disease in their mid-teens. Treatments addressing the ageing disorder and significant cardiovascular pathology remain limited; however, there is significant promise in using mRNA therapeutics to target the abnormalities. This study presents the treatment of HGPS using mRNA human telomerase reverse transcriptase to reverse senescence in an in vitro induced pluripotent stem cell model of Progeria and extend lifespan in a Progeria mouse model. This work presents a new approach for improving vascular senescence and outcomes for patients.
Introduction
Chronological age is a major risk factor for cardiovascular disease.1 Vascular ageing is characterized by an impairment of endothelial homeostasis, with reduced generation of nitric oxide (NO), increased production of oxygen-derived free radicals and expression of adhesion molecules,2 predisposition to vascular inflammation, platelet adhesion, and progression of vascular disease. Vascular ageing is accelerated in Hutchinson-Gilford progeria syndrome (HGPS).3,4 HGPS is caused by a de novo mutation in the lamin A gene (LMNA) located on chromosome 1 (c.1824C>T, G608G).5,6 The LMNA mutation results in a truncated form of lamin A, termed progerin that lacks the post-translational processing splice site. As a result, progerin remains permanently farnesylated, accumulates in the nuclear envelope, and alters nuclear morphology.7–11
Progerin accumulation is associated with many of the cellular alterations associated with ageing including DNA damage, global gene expression changes,12 and telomere erosion.13,14 Notably, progerin also accumulates to a lesser degree in all individuals due to mRNA mis-splicing and may contribute to ageing.15 Telomere erosion and replicative arrest are key hallmarks of senescence,3,16,17 both of which are observed in HGPS cells.8 Telomeres are susceptible to DNA damage,18 and the resulting DNA damage response induces inflammatory gene expression19 and a senescence-associated secretory phenotype (SASP).20 SASP paracrine factors have been shown to induce senescence in neighbouring cells and are a component of age-related diseases.21
Children with HGPS manifest failure to thrive at an early age, and in childhood may present with osteopenia, alopecia, scleroderma, lipodystrophy, and cardiovascular symptoms.22–27 Most individuals succumb to myocardial infarction or stroke due to accelerated atherosclerosis in their mid-teens.28 In a clinical trial, treatment with a farnesyltransferase inhibitor resulted in a modest extension of life span by 1–2 years.29,30 Greater understanding of HGPS is needed to develop more effective therapies.
The effect of HGPS on endothelial cells (ECs) and their modulation of neighbouring vascular smooth muscle cells (VSMCs) is relatively unexplored. Two studies have reported significant EC pathology in progeria mouse models,31,32 reporting poor shear sensing and strong inflammation. Additionally, we reported a profound impairment of endothelial functions in HGPS induced pluripotent stem cell (iPSC)-derived ECs.33 Given the primacy of the endothelium in vascular homeostasis,34 we hypothesized that targeting the causes of senescence in HGPS ECs would be a promising therapeutic avenue for HGPS. The ribonucleoprotein telomerase maintains telomere length.35–37 We have previously shown that transient delivery of modified mRNA encoding human telomerase (hTERT) in aged human cells can extend telomere length, reverse features of senescence and restore replicative capacity.38–40 Similarly, telomerase gene therapy has been shown to augment myocardial regeneration and revascularization by rejuvenating ageing stem cells.41–43 Here, we show that hTERT treatment of HGPS ECs extends telomere length, restores endothelial function and gene expression, increases sirtuin 1 (SIRT1) expression, reduces markers of DNA damage, and reverses adverse paracrine effects on VSMCs. Furthermore, we show that mTERT therapy in a mouse model of HGPS44 reverses endothelial activation, reduces endothelial DNA damage, and extends lifespan. Our work suggests a novel approach for age-related vascular diseases.
Materials and methods
Maintenance of human induced pluripotent stem cell
Human iPSC lines (HGFDFN168 iPSC1P, HGADFN167 iPSC 1Q, HGDFN003 iPSC 1C, HGMDFN090 iPSC 1B) were purchased from the Progeria Research Foundation Cell and Tissue Bank and were cultured and maintained on Matrigel (BD Biosciences)-coated plates (Corning) in mTeSR plus medium (STEMCELL Technologies) according to protocol. The iPSCs were passaged upon 70% confluency and maintained in humidified incubators at 37°C and 5% CO2 (Thermo Scientific).
Differentiation of endothelial cells from induced pluripotent stem cell
Human iPSCs were differentiated to EC using our established protocol.33 In brief, the iPSCs were grown to 80% confluence, and incubated with DMEM/F12 (Gibco), supplemented with Wnt agonist CHIR 99021 (5 µM, Selleck), bone morphogenetic protein-4 (BMP4, 25 ng/m, Peprotech), B27 supplement (Thermo Fisher Scientific), and N2 supplement (Thermo Fisher Scientific). At Day 3, cells were dissociated (TrypLE, Gibco) and plated to 20% confluency in StemPro media supplemented with forskolin (5 µM, LC Laboratories), vascular endothelial growth factor (VEGF, 50 ng/mL, Peprotech), and polyvinyl alcohol (2 mg/mL, Sigma). After 7 days, cells were cultured in endothelial growth media (EGM-2MV, Lonza) for maturation. At Days 11 and 12, cells were collected and sorted by Fluorescence Activated Cell Sorting (FACS) using two antibodies specific to ECs (Alexa Fluor 488-conjugated CD31 antibody or PE-conjugated CD144 antibody, BD Pharminogen).
Nitric oxide production and Dil-Ac-LDL uptake assays
ECs were stained with DAF-FM DA (Invitrogen) to assess intracellular NO. Cultured ECs were incubated with DAF-FM DA for 30 min, washed, and fluorescence measured with ImageJ and plotted as mean of the fluorescent signal (integrated density) in each cell. For assessing acetylated low-density lipoprotein (Ac-LDL) uptake, ECs were incubated with ac-LDL-594 (Invitrogen) at 1:200 dilutions for 5 h and, then, the fluorescent signal was measured.
Angiogenesis assay (vascular network formation)
Vascular network formation assays were carried out as we have described.45,46 ECs from all three groups were seeded in growth factor reduced Matrigel and EGM2-MV medium at 40 000 cells per well in 12-well plates. The number of network segments was measured after 8 h in five random microscopic fields in three experiments.
Real-time endothelial cell proliferation assay
Proliferation assay were carried out using the xCELLigence instrument (Roche) which was placed in a humidified incubator at 37°C and 5% CO2. ECs from all three groups (3000–5000) were seeded into the wells with medium in triplicate. The impedance value of each well was monitored every minute in the first 4 h, and every hour for 120 h. The data are expressed as a cell index (CI) value.
Telomere length measurement by monochrome multiplex PCR
Cellular DNA was isolated, and monochrome multiplex PCR (MMqPCR) was performed in accordance with a previously published protocol by Cawthon.47 Samples and standards were pipetted into PCR tubes; the standard was created via two-fold serial dilution. The PCR master mix was added to each PCR tube and was transferred to a white LightCycler® 480 Multiwell plate (384 wells). PCR runs were performed on a Roche LightCycler® 480 (Software: 1.5.1.62) using a preinstalled MMqPCR protocol. Raw data were preprocessed via a Python 2.0 script to differentiate different primers for the telg/telc primers and single-copy gene (SCG) primer (human β-globin). Data were reported as the ratio of telomere concentration to SCG concentration (T/S ratio).
TRAP assay
Telomerase activity assay was performed following the TRAPeze® Telomerase Detection Kit (SIGMA-MILLIPORE # S7700) protocol with some modifications. Each cell pellet (106 cells) was washed twice with 1× PBS and resuspended in 20 µL of 1× CHAPS buffer supplemented with Protease Inhibitor Cocktail (Roche) and RNasein Inhibitor Plus (Promega # N2615) and incubated for 30 min on ice. After lysis was complete, suspensions were centrifuged at 14,000 rpm, for 20 min at 4°C and lysates were stored at −80°C. Protein concentration was measured using DC Protein assay from Bio-Rad and a working dilution for the assay was prepared at 0.03 µg/µL. PCR was performed using provided reagents in the kit and Hot Start Taq Polymerase from NEB (# M0495L) in a final reaction volume of 50 µL. After a 30-min incubation at 30°C for telomerase-mediated extension of the TS primer, and 2 min at 95°C to activate polymerase, the elongated reaction mixture was amplified by PCR through 35 cycles of 94°C for 15 s, 59°C for 30 s, and 72°C for 1 min. PCR products containing loading dye were electrophoresed in a precast gradient polyacrylamide gel (Bio-Rad # 5671085) for 90 min at 120 V. Gel was then stained for 30 min with GelRed (Biotium # 41003) followed by washing for 15 min with water. To estimate telomerase activity, non-HGPS and HGPS iPS cells were used as positive control as well as heat-killing a sample aliquot at 90°C for 10 min for negative controls along with CHAPS buffer. For mouse liver tissue samples, protocol was optimized by increasing temperature of TS extension from 30 to 40°C and increasing number of cycles to 40.
Quantitative fluorescent in situ hybridization
Quantitative fluorescent in situ hybridization (qFISH) experiments were carried out based on an established protocol.48 Briefly, cells were arrested in metaphase using colcemid (Roche). Cells were collected into a tube and, while vortexing, 4 mL of 37°C hypotonic solution (50 mM KCl, Sigma-Aldrich) was added drop-wise. Cells were fixed with 3:1 ethanol/acetic acid (Sigma-Aldrich) while vortexing. The cells were centrifuged at 1000 rpm for 5 min and dropped onto a glass slide. The spreads were then fixed in 3.7% formaldehyde and dehydrated with an ethanol series: 5 min in 70%, 85%, and 100%, then air dried at room temperature. The telomere probe (PNA Bio F1013 TelC-Alexa 647) was diluted 1:40 in HB buffer (600 µL formamide, 10 µL 2M Tris 7.5, 10 µL 10 µg/mL Salmon sperm, Invitrogen) at 85°C. The probe was then added to the slide and incubated over night at 4°C. Slides were then washed twice with 70% formamide in (Tris 20 mM pH 7.5) and BSA 1%, twice with Tris 50 mM, pH 7.5, NaCl 150 mM, and Tween-20 0.05% (Sigma-Aldrich), and finally dehydrated with ethanol. After, 10 µL of SlowFade Diamond antifade mounting solution with 4′,6-diamidino-2-phenylindole (DAPI) was added and coverslip was applied. Telomere length was reported as the probe signal normalized to the DAPI signal on >25 metaphase spreads.
Synthesis of messenger RNA
The mRNA was synthesized by in vitro transcription (IVT) with pseudouridine and 5-methylcytidine added to the nucleotide mix so as to increase stability in the mRNA construct that was generated. The DNA plasmids used in IVT encoded for hTERT or for a catalytically inactive hTERT form. The open reading frame is human TERT transcript variant 1 (NM_198253.2) and the CI TERT has a point mutation, D712A.38 The mRNA was purified by high-performance liquid chromatography to remove abortive transcripts and RNA-DNA template hybrids. The size and integrity of mRNA constructs were assessed using a bioanalyzer.
Cell treatment with hTERT mRNA
HGPS or non-HGPS iPSC-derived ECs, at 70–75% confluence, were transfected with mRNA using Lipofectamine RNAiMax in Opti-MEM (Life Technologies) at 1 μg/mL for 4 h, after which cells were replenished with fresh medium. We also have used Lonza electroporation equipment (Lonza 4D Nucleofector) for transfection. In this process, 2–3 × 105 cells were used in suspension with buffer P3 (Lonza) as recommended by company and then transfected with 1 μg mRNA. Two transfections were performed within 48 h, and then, cells were used for further experiments after 72 h post-transfection.
Confocal imaging
Imaging was performed using a FLUOVIEW FV3000 confocal microscope (Olympus Corporation). Analysis was performed using the associated software, cellSens (Olympus Corporation). Raw fluorescent values extracted for measurements were normalized using either DAPI fluorescence, nuclear area, or cellular area as appropriate for the application.
RTqPCR gene analysis
RNA was extracted from cell pellets or tissues using a Zymo Research extraction kit according to the manufacturer’s protocol. cDNA was synthesized from the RNA using a 5x iSCript Reverse Transcriptase Supermix (Bio-Rad Laboratories, Inc.). cDNA was diluted 1:10 with nuclease-free water to 200 µL. For each qPCR reaction, a volume of 20 µL was used: 10 µL of Power SYBR® Green Master Mix (Applied Biosystems, Thermo Fisher Scientific), 7 µL of nuclease-free water, 1 µL of the gene specific primer, and 2 µL of the prepared sample cDNA. Primers used were lamin A and progerin (custom49), GAPDH, SIRT1 (Bio-Rad). RTqPCR was performed using a QuantStudio 12K Flex (Applied Biosystems, Life Technologies): 96-Fast Well Plate 0.1 mL, Relative Quantification (-ΔΔCt), SYBR® Green Reporter, and Standard Run Time. Results are reported as fold-change from the control sample.
Atomic force microscope
Atomic force microscope (AFM) used in the experiment is a Catalyst Biomicroscope (Bruker). Imaging process was under a contact mode in liquid in room temperature (22°C). AFM probes used in the experiment were MLCT probes (Bruker Nano). The AFM probe average spring constant was at 0.01 N/m.
A 60-mm dish with cultured cells was mounted on the AFM scanning stage for imaging. Three millilitres of PBS solution was in the dish during imaging process. The AFM scanning frequency was 1 Hz. Image data analysis and processing were conducted in a Nanoscope Analysis 1.50r1 (Bruker).
RNA sequencing and bioinformatics analysis
Cultured cells were collected from all three groups. RNA was isolated using the QIAGEN RNeasy purification kit, and the RNA integrity was verified with an Agilent 2100(RIN). Total RNA was sent to the Beijing Genomics Institute (BGI America, Cambridge, MA, USA) for library preparation and sequencing. Poly(A)-selected RNA libraries were prepared using the TruSeq technology (TruSeq RNA Sample Prep Kit v2), and resulting libraries were sequenced on an Illumina HiSeq™ 4000 using 100-bp paired-end reads. Reads were aligned to the reference human genome (build hg19) using Tophat version 2.1.0.50 The fragment per kilobase per million reads (FPKM) for each gene and differentially-expressed genes were called by Cuffdiff, version 2.2.10.50 The cut-off of P-value ≤0.05 and absolute values of log2-transformed fold changes larger than 1 were used for differential expression analysis. The heatmaps were created using the pheatmap R package, version 1.0.12 (https://rdrr.io/cran/pheatmap/). The read coverage, viewed through the Integrative Genomics Viewer (IGV version 2.3.98).51,52 Pathway enrichment analysis was carried out by using Metascape version 3.5, P-value (0.01) was used as cut-off.53 All the raw sequence data for the samples used in this study are available on the National Center for Biotechnology Information Sequence Read Archive database: Bioproject PRJNA730422.
Differentiation of vascular smooth muscle cells
VSMC differentiation was carried out based on an established protocol.54 Briefly, iPSCs were scraped from the culture dish after incubation with collagenase (Stemcell). Cells were collected into 15 mL conical tube and centrifuged. Supernatant was aspirated and E6 media containing 10% FBS and 1/1000 dilution of ROC inhibitor were added and plated on low-cluster plates. The next day, the media were replaced with E6 + 10% FBS. Media were changed every other day to let the embryoid body (EB) form and grow in E6 + 10% FBS for 7–10 days. EBs were then plated onto gelatin-coated 6-well plates in E6 media +10% FBS for 3–5 days. 1 mL of E6 media was used to break up cell clumps. After this step, cells were spun down at 1000 rpm for 5 min. The supernatant was aspirated and SMGS media was used for resuspension. The cells were then seeded onto matrigel-coated 6-well plates to grow VSMCs.
Coculture experiments with vascular smooth muscle cells
Conditioned media was collected after 72 h from non-HGPS, HGPS, and HGPS hTERT treated ECs. Non-HGPS VSMCs were cultured at 70% confluency and washed with PBS. Non-HGPS VSMCs were then incubated with conditioned media (CM) from these three groups or fresh EC media (EGM-2MV) for 48 h. Cells were collected and analysed with FACS for counting and viability. The non-HGPS VSMCs were also processed and lysed for further analysis with a Bio-Plex assay.
Immunofluorescent staining of en face aortae, aortic rings, lung endothelial cells and liver sections
Liver and aorta tissues were treated with 30% sucrose (Sigma) overnight, OCT embedded, and sectioned to 5–8 µM thickness. Sections were fixed and permeabilized in acetone (Sigma) for 12 min at 4°C followed by a PBS rinse and dehydration with an ethanol series at 70%, 80%, and 100% for 5 min. Following dehydration, sections were blocked for 1 h at room temperature with 5% BSA/0.1% Tween-20 in 1× Tris-buffered solution. Tissue was incubated with primary antibody overnight at 4°C in a humidified chamber. Primary antibodies included: γH2A.X, SIRT1, and CD31 (Abcam ab81299, Cell Signaling Technologies 20282, and Abcam ab56299, respectively). The next day, sections were washed with PBS three times for 10 min, incubated with secondary antibody (anti-rabbit Alexa-488 and anti-rat Alexa-555, Thermo Fisher) for 1 h at room temperature and then washed with PBS. Samples were mounted in VectaShield antifade mounting media (Vector Laboratories). A previous en face protocol was used.55 Briefly, aortae were washed and fixed by PFA 4% before dissection. Then, the aortae were cleaned carefully and permeabilized with 0.1% Triton X-100 in PBS and incubated in blocking buffer in 10% normal serum in TBS (Tris-buffered saline with 2.5% Tween-20) for 1 h at room temperature. Next, the aortae were incubated with the previously mentioned primary antibodies at 4°C overnight. The aortae were washed with TBS buffer twice for 10 min and incubated in secondary antibody for 1 h at room temperature. Then, open aortae were mounted face up in VectaShield antifade mounting media. For human and mouse ECs, cells were washed and fixed by 4% PFA at room temperature for 10 min. Cells were washed with PBS and incubated with 5% BSA/0.1% Tween-20 in 1× Tris-buffered solution for 1 h. Cells were incubated with primary antibodies (mentioned above) over night at 4°C. The day after, cells were washed twice with PBS 10 min and then incubated with the secondary antibody for 1 h room temperature. They were then washed twice again with PBS and mounted in VectaShield antifade mounting media. Images were normalized to cell area for surface markers or DAPI for intracellular expression. γH2A.X is reported as counted foci or a fluorescent value normalized to DAPI.
Western blot and lysate preparation
The mouse livers were dissected and added to the tube with 1.4-mm ceramic beads (Fisher Scientific, 15-340-153), RIPA buffer (Thermo Fisher Scientific) and protease inhibitor tablets (Roche) and homogenized for 2 min (Fisherbrand). The lysates were then used in BCA protein assay kit (Thermo Fisher, No. 23225) to measure total protein. The total protein amounts of 8–12 μg per sample were used for running into the premade (Bio-Rad) gel running at 100 V for 1 h. Proteins were then transferred to nitrocellulose membranes with Bio-Rad transfer system (Trans-Blot Turbo) and blocked in 1× Quant Clean Western buffer (cat No. R2001) for 1 h. The blots were then incubated with dilute primary antibody in 5% BSA, 1× TBS, 0.1% Tween-20 at 4°C with gentle shaking, overnight (SIRT1: Cell Signaling 2028S, 1:1000; telomerase: NOVUS NB110-89471, 1:2000; progerin: Millipore 05-1231, 1:500; GAPDH: Cell Signaling D16H11, 1:2000). Membranes were washed for 15 min for three times using Quant Clean Western buffer. Secondary antibody diluted to 1:2000 secondary lgG-HRP (Kwikquant) in R2001 (Quant) was then added for 1 h at room temperature. Membranes were then washed for 15 min three times using Clean Western buffer. The membranes were then submerged in 5× diluted Kwikquant digital-ECL mixture for 2 min and imaged using a Kwikquant imager.
Lentivirus construction
HEK293T cells (ATCC) were cultured and transfected at 80% confluency with three different plasmids including psPAX2 (Addgene, # 12260), pMD2.G (Addgene, #12259), and mTERT (Addgene # 36413) or GFP (Addgene #40123) according to Qiagen Effectene transfection reagent kit protocol (Qiagen). Viral supernatants were harvested 72 h post-transfection, filtered and concentrated using ultracentrifugation for 2 h at 18 700 rpm 4°C. Virus pellet were re-suspended in PBS and were titrated using Lenti-X GoStix plus (Takara). The levels of p24 viral capsid protein, which correlate directly with infectious unit (IFU)/ml, were measured.
Animal strain, husbandry, genotyping, and lentivirus delivery
All animal experiments were conducted in accordance with institutional guidelines and approval by IACUC at the Houston Methodist Research Institute. The HGPS progeria mouse model used, C57BL/6-Tg (LMNA*G608G) HCIins/J,44 was purchased from Jackson Laboratory (Stock No : 010667). Genotyping was done using a protocol designed with Transnetyx Inc. (9110 Cordova Rd. Suite 119, Cordova, TN 38016), which identified the human copy of mutated lamin-A. The homozygous HGPS mice had two copies of human mutated lamin A gene. Wild-type mice had no human copies of the mutated lamin protein, and heterozygous had one copy. Mice were housed with a 12 h light/dark cycle between 06:00 and 18:00 in a temperature-controlled room (25 ± 1°C). Water and rodent feed were fed ad libitum. Mice that lost greater than 20% of body weight within one week were monitored by veterinary staff and were provided with moist food and supportive care. Lentiviral constructs (1010 IFU/mL in 150 µL) carrying either GFP or mouse telomerase (mTERT) were injected via tail vein at 3 months and 6 months old. A minimum of 18 mice was used in each group (WT, HGPS, and HGPS-mTERT).
Animal studies
For the in vivo work, two groups of at least 18 mice in each test group (WT, HGPS, and mTERT) were used for pathology and survival studies. For the pathology studies, to match age throughout the groups, if an HGPS group mouse spontaneously died or if euthanasia was recommended by the veterinarian, mice from the other two groups were euthanized to provide a similar age-matched set. Each histological study includes 4–5 mice as specified in the figure caption. In the survival studies, animals lived until spontaneous death or until the veterinarian recommended euthanasia, each death recorded as an event for survival analysis. All veterinary recommendations were done blinded. Our CMP veterinarian requested euthanasia of mice based on the standard criteria, including significant loss of muscle mass and impaired mobility. These criteria are typically observed in later stages of HGPS. When these signs became pronounced, euthanization was recommended. Group sizes were determined using a one-way ANOVA power calculation with α = 0.05 and β = 0.8.
Isolation of lung endothelial cells from mouse
Mice were euthanized using anesthesia (Isoflurane) and ribs excised to expose an opening to the thoracic cavity. A 27-G needle (MACS) was inserted in the right ventricle to perfuse PBS through the pulmonary circulation until the lung was white. The lung was then immediately dissected and stored in PBS on ice. The lungs were further processed for EC isolation using the OCTOMACS dissociation system. Briefly, lungs were placed into gentleMACS C-tubes containing a tissue dissociation enzyme (MiltenyiBiotec: catalog no. 130-095-927). Lung samples were incubated for 30 min on the gentleMACS OCTO Dissociator (Miltenyi Biotec catalog no. 130-096-427) using protocol program 37C_m_LDK_1 with heaters on to obtain a single cell suspension. The cell suspension was collected and positive selection for ECs was carried out using MACS magnetic separation beads conjugated with CD31 and CD144 antibodies. Following separation, cells were then enriched and analysed by FACS. Following EC isolation, cells were grown in media supplemented with MH1015 at 15 µg/mL (home-made) on gelatin-coated plates and incubated at 37°C and 5% CO2.
Bio-Plex assay
The concentrations of cytokines, chemokines, and growth factors were analysed using Bioplex Pro™ human cytokine standard 27-plex and 21-plex panels based on xMAP technology (Bio-Rad Laboratory, Hercules, CA, USA) according to the manufacturer. ECs from all three groups were collected (5 × 105) and lysed using Bio-Rad lysis buffer and then probed for 48 markers of inflammation. Further, equal numbers of Control Non-HGPS VSMCs were cultured using fresh media or the CM from non-HGPS ECs, HGPS ECs, or HGPS-hTERT ECs. After 72 h, Non-HGPS VSMCs were counted and then lysed for further analysis using Bioplex Pro™ according to the manufacturer. Briefly, lysed cells were incubated with magnetic beads conjugated with specific antibodies against 48 markers of inflammation. Beads were washed and were incubated for 1 h with fluorescent secondary antibodies of different colours. Beads were washed and the plates were read to detect inflammatory markers using the Bio-Rad detection machine.
ELISA for serum markers
Mouse sVCAM-1/CD106 was quantified in the serum collected from WT, HGPS, and HGPS + mTERT mice at 7–8 months old using ELISA (R&D Systems). Test was performed per manufacturer. Briefly, the collected serum was diluted 50 times and was added to the coated plate and incubated for 3 h at room temperature (22°C). It was washed five times using the washing buffer, and then, mouse VCAM-1 conjugated to horseradish peroxidase was added and incubated for 1 h. The plate was washed five times before substrate was added for a 30-min incubation. The reaction was halted using the stopping buffer. The optical density of each well (triplicate for each serum) was read by microplate reader at 570 nm. Data were calculated using the standard curve obtained from serial dilution of standard substrate provided by the kit. Mouse C-reactive proteins (CRP) was quantified using an ELISA kit (Invitrogen) per the manufacturer’s instruction. Serum from the same three groups was collected at 7–8 months of age and diluted 100 times. Each sample was added to a coated 96-well plate and incubated for 3 h at room temperature. Each sample was then washed four times and the anti-mouse CRP antibody conjugated to horseradish peroxidase was added and incubated for 1 h. The plate was then washed 4 times, the substrate was added, and the plate incubated for 30 min. The reaction was halted by the stopping buffer, and each optical density (triplicate per serum sample) was read at 450 nm. Data were calculated using the standard curve obtained from serial dilution of standard substrate provided by the kit.
Data analysis
Results are expressed as mean ± standard deviation. Groups were tested for normality using a D’Agostino-Pearson test. If they failed the normality test, groups were analysed using Kruskal–Wallis with post hoc Dunn test. Otherwise, statistical comparisons between two groups or multiple groups were performed via, respectively, a Student’s t-test or ANOVA with post hoc Tukey’s test using PRISM 7 software. A P-value of <0.05 was considered significant. Data visualization was performed using Matplotlib and Seaborn packages in Python 3.7.56 N = 4–6 mice were examined in each group for pathology studies. N = 18 mice were used for survival analysis performed using PRISM 7 using a log-rank test; dotted lines show 95% confidence interval.
Results
hTERT treatment extends telomere length, restores replicative capacity, and improves function in Hutchinson-Gilford progeria syndrome endothelial cells
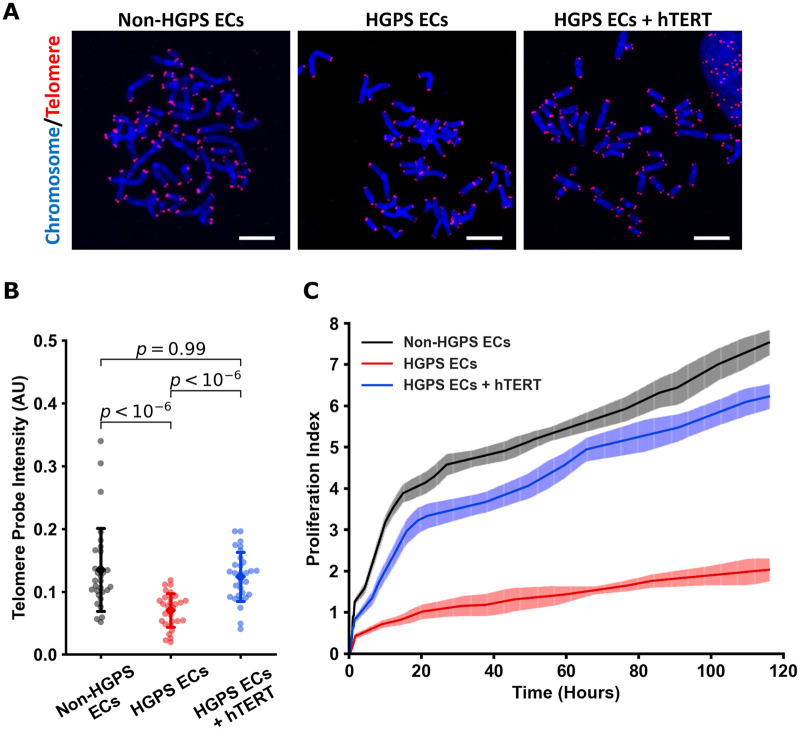
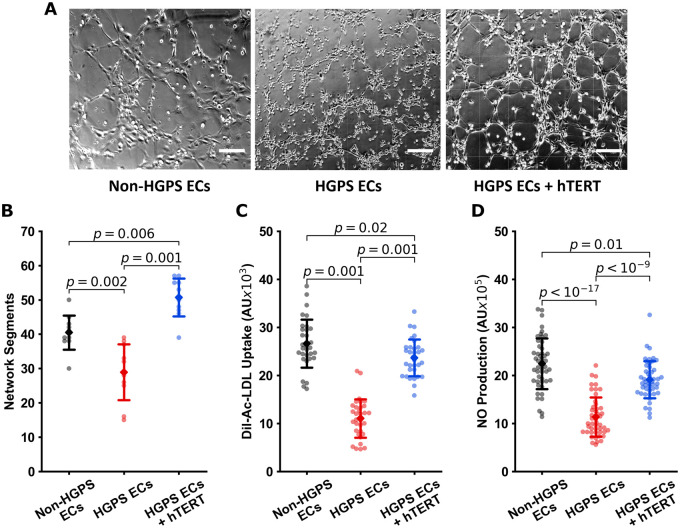
To first understand whether hTERT treatment would restore telomere length and replicative capacity, we transfected our iPSC-derived ECs (Supplementary material online, Figure S1A and B) with hTERT mRNA (1 µg hTERT mRNA/3 × 105 cells 2× over 48 h). Our initial studies using MMqPCR (Supplementary material online, Figure S1C) indicated that hTERT treatment extended telomeres, an observation that was confirmed compellingly by qFISH (Figure 1A and B). Treatment with hTERT mRNA reduced β-gal expression in HGPS ECs (Supplementary material online, Figure S1D and E). Furthermore, mRNA hTERT increased replicative capacity in a dose-dependent manner as assessed by real-time impedance studies (xCelligence; Figure 1C and Supplementary material online, Figure S1F and G). Notably, viral treatment of telomerase (to mimic the following in vivo studies) also improved HGPS iPSC EC proliferation, whereas the viral vehicle alone impaired cell proliferation (Supplementary material online, Figure S1H). Next, we determined whether hTERT treatment restores EC-specific functionality. Treatment with hTERT mRNA improved network formation in Matrigel (Figure 2A and B), improved uptake of acetylated LDL (ac-LDL) (Figure 2C), and improved NO generation (Figure 2D). Thus, hTERT treatment reversed telomere erosion and replicative decline and normalized EC function in HGPS cells.
Figure 1.
hTERT treatment increases telomere length and proliferation in Hutchinson-Gilford progeria syndrome endothelial cells. Metaphase spreads of groups (shown in A) were analysed using quantitative fluorescent in situ hybridization (blue: DAPI nucleus, red: Cy5 telomere, scale bar—5 µm). To analyse each image and determine changes, telomere and DAPI intensities were extracted, and telomere probe intensities were normalized to the DAPI signal for each individual metaphase spread, n = 30 metaphase spreads for each group, replicated at least three times to confirm (B). Cell proliferation was measured by impedance, and increase in which values indicated increased cell number. Treatment with 1 µg/mL of mRNA hTERT improved cellular proliferation index of Hutchinson-Gilford progeria syndrome endothelial cells (C). Quantitative fluorescent in situ hybridization data were tested for normality using D’Agostino-Pearson and then analysed using Kruskal–Wallis with post hoc Dunn test, significance taken at P < 0.05. All between-group statistics are displayed natively. All error bars show standard deviation. DAPI, 4′,6-diamidino-2-phenylindole.
Figure 2.
hTERT treatment restores endothelial cell functions in Hutchinson-Gilford progeria syndrome endothelial cells. Representative images of Matrigel® network segment assays for non-Hutchinson-Gilford progeria syndrome, Hutchinson-Gilford progeria syndrome, and hTERT treated endothelial cells are shown in (A) (scale bar—200 µm). Image (B) shows quantification of network segments for n = 10 fields of view. Endothelial function was assessed via fluorescence assays for acetylated LDL uptake (n = 32 individual cells, C) and NO production using DAF-FM (n = 48 individual cells, D), with single cell fluorescence quantified. Each experiment was repeated at least three times to confirm the conclusions—data are reported as individual cellular measurements. All data were tested for normality using D’Agostino-Pearson test. Images (B) and (C) were tested using a one-way ANOVA with post hoc Tukey’s test for group differences. Image (D) was tested using Kruskal–Wallis with post hoc Dunn test. Significance was taken at P < 0.05, all between-group statistics are displayed natively. All error bars show standard deviation.
hTERT treatment rescues cellular and nuclear morphology
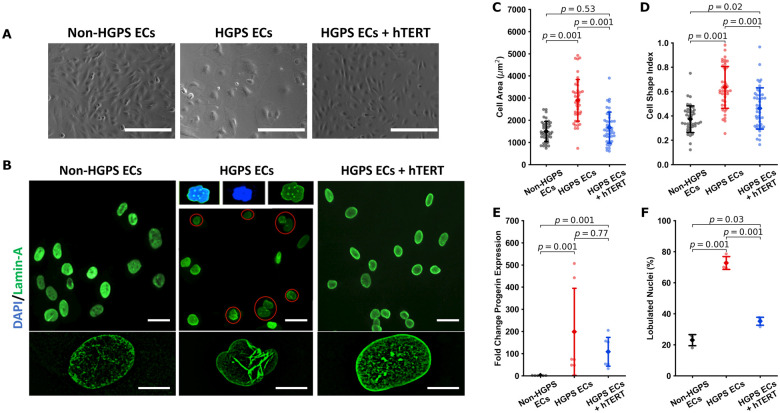
HGPS cells possess typical morphological features of senescence,4 including nuclear blebbing.9,57 As typical of senescent cells, HGPS ECs were more circular and had a greater two-dimensional surface area. By contrast, the morphology of treated HGPS ECs was improved, becoming more elongated with a smaller two-dimensional total surface area as quantified by CI values (Figure 3A–D). These observations were augmented using atomic force microscopy. While geometric changes are often markedly heterogeneous in ageing, in a broad sense they display volume expansion and a ‘flat’ morphology.58 AFM revealed that our HGPS ECs have a lower height profile by comparison to non-HGPS ECs (Supplementary material online, Figure S1I), although hTERT therapy did not appreciably alter this parameter. Treatment with hTERT tended to reduce progerin expression (Figure 3E and Supplementary material online, Figure S1B). We were surprised to observe that hTERT treatment normalized the nuclear architecture (Figure 3F), reducing the fractional number of HGPS EC nuclei with blebs. Thus, hTERT treatment can reverse changes in cellular and nuclear morphology observed with HGPS.
Figure 3.
hTERT treatment normalizes cellular and nuclear morphology in Hutchinson-Gilford progeria syndrome endothelial cells. Representative bright field imaging of non-Hutchinson-Gilford progeria syndrome, Hutchinson-Gilford progeria syndrome, and hTERT treated Hutchinson-Gilford progeria syndrome endothelial cells are shown in (A) (scale bar—200 µm). Nuclear lamin A staining is shown in (B) (scale bar: top—30 µm; bottom—10 µm). Hutchinson-Gilford progeria syndrome endothelial cells show a compromised morphology, having increased cellular area (n = 43 cells across three replicates, C) and a more rounded morphology as indicated by an increased cell shape index/circularity index (n = 43 cells across three replicates, D). Cell shape index/circularity is defined as 4π(area/perimeter2). hTERT therapy additionally induced a downward trend in progerin expression as assessed via RTqPCR (n = 6) (E) and improves nuclear morphology (F), reducing the number of folded nuclei (three experimental replicates, percent folded nuclei reported for at least 300 cells in each group per replicate). Data were quantified using a one-way ANOVA with post hoc Tukey’s test (ddCt analysis for qPCR), significance taken at P < 0.05. All between-group statistics are displayed natively. All error bars show standard deviation.
hTERT treatment effect on gene expression in Hutchinson-Gilford progeria syndrome endothelial cells
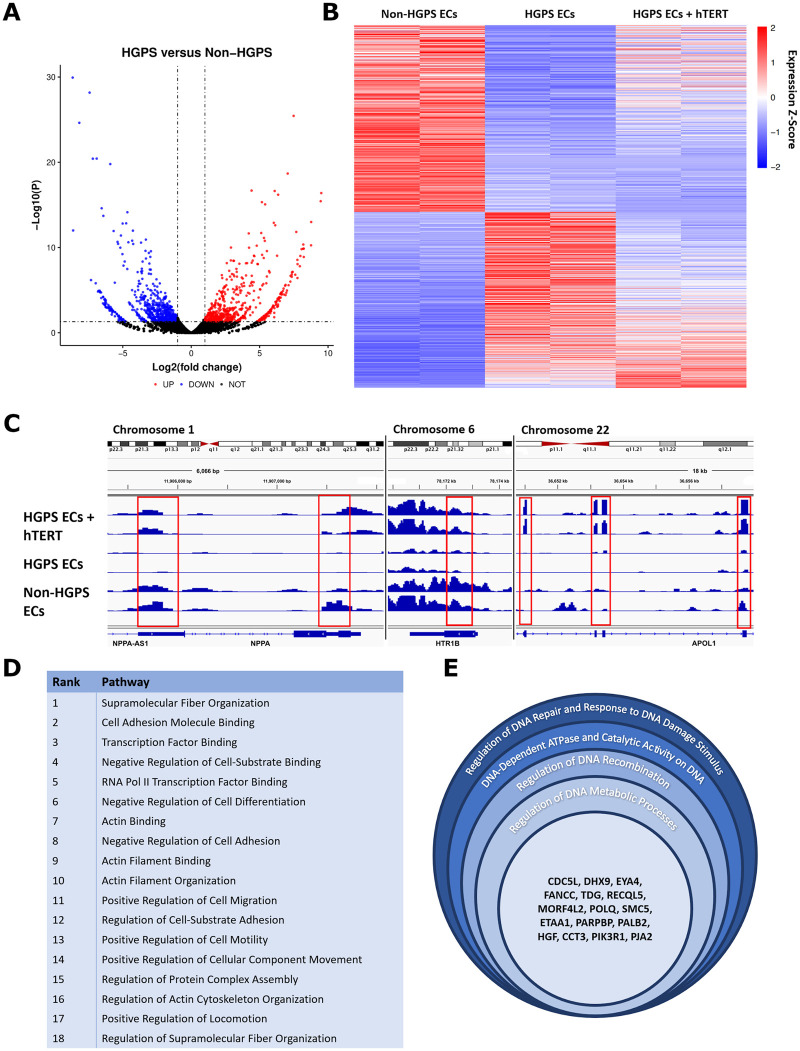
Transcriptional profiling by RNA sequencing revealed over 1280 dysregulated genes in HGPS ECs vs. parental controls (Figure 4A). Next, we analysed which of these dysregulated genes were returned to baseline by hTERT treatment, finding that 265 genes were fully rescued by hTERT (Figure 4B and Supplementary material online, Figure S2A) and several others were partially rescued (Supplementary material online, Figure S2B). Figure 4C illustrates the process for finding rescued genes in the genome browser. Treatment with hTERT also normalized the expression of EC identity genes59 and several EC-related pathways that were disturbed in HGPS (Figure 4D and Supplementary material online, Figure S2C). GO analysis revealed other pathways rescued by hTERT including, most prominently, DNA damage/repair and fiber/structural organization (Figure 4E and Supplementary material online, Figure S2D). In addition, we observed that in HGPS there was an up-regulation of genes associated with endothelial-to-mesenchymal transition (EndoMT; Periostin, Snail, TGF-β). The increase in EndoMT gene expression was reversed by hTERT treatment. Altogether, hTERT treatment had an extensive effect on gene expression, fully normalizing 265 genes and partially rescuing additional genes related to EC identity, cytoskeletal structure, and DNA damage.
Figure 4.
hTERT treatment rescues global gene expression and endothelial cell pathways in Hutchinson-Gilford progeria syndrome endothelial cells. RNASeq analysis uncovered 1285 differentially regulated genes between non-Hutchinson-Gilford progeria syndrome and Hutchinson-Gilford progeria syndrome endothelial cells (significance cut-off of P = 0.05 for fold-change ≥1 up or down, A). To further elucidate how hTERT therapy affects global gene expression, genes differentially regulated between non-Hutchinson-Gilford progeria syndrome endothelial cells and Hutchinson-Gilford progeria syndrome endothelial cells were then analysed again to assess whether hTERT therapy returned the gene expression to the non-Hutchinson-Gilford progeria syndrome endothelial cell baseline. This analysis is visualized via heatmap in (B) with a significance cut-off of P = 0.05, with example genome browser data showing how the process was conducted for individual, representative genes (C). A total of 256 genes in total were defined as ‘rescued’ via this analysis (see Supplementary material online, Figure S2A), with highly significant pathways identified relating to cellular cytoskeletal organization and motility, DNA damage and repair pathways, and transcription factor binding and regulation. Endothelial cell specific pathways are listed in (D) by rank order significance and (E) shows genes affected related to DNA repair. GO pathways, genes, and the P-values for endothelial specific and non-specific pathways altered by hTERT therapy are detailed in Supplementary material online, Figure S2. Pathway analysis for (D) and (E) were conducted on six biological replicate groups. Non-Hutchinson-Gilford progeria syndrome and Hutchinson-Gilford progeria syndrome endothelial cell lines were differentiated from a parent/child donor set. All the raw sequence data for the samples used in this study are available on the National Center for Biotechnology Information Sequence Read Archive database: Bioproject PRJNA730422.
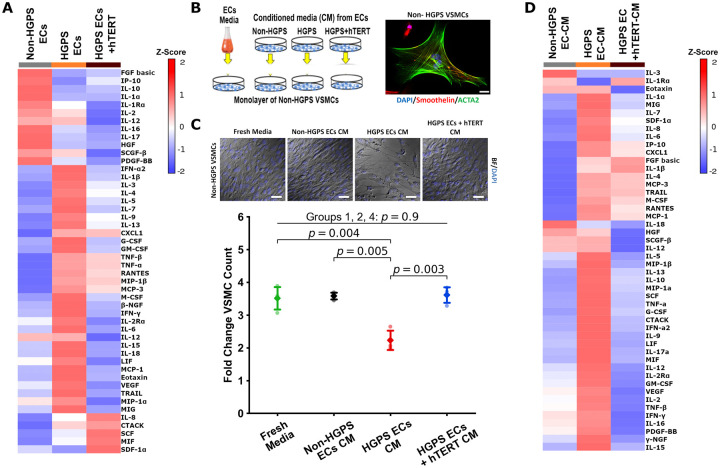
hTERT treatment attenuates expression and release of inflammatory cytokines
In patients with HGPS, there may be medial atrophy, presumably due to loss of VSMCs in the vessel wall.60 We speculated that the loss of VSMCs is mediated in part by an SASP in HGPS ECs. It is known that a cell expressing SASP may cause dysfunction and even apoptosis of neighbouring cells.21 Thus, we sought to establish a connection between SASP in ECs and impaired proliferation and/or apoptosis in VSMCs. First, we characterized the SASP secretome in HGPS ECs. We observed that HGPS ECs express a panoply of inflammatory cytokines consistent with the complex SASP profile61 (Figure 5A). Intriguingly, the expression of 35 proteins (73% of the inflammatory cytokines that were increased in HGPS) was attenuated by hTERT treatment (Figure 5A). Subsequently, we assessed the effects of CM from HGPS ECs on normal VSMCs. We observed that HGPS EC CM significantly reduced VSMC proliferation. By contrast, CM from HGPS ECs treated with hTERT did not reduce VSMC proliferation (Figure 5B and C). In addition, VSMCs exposed to CM from HGPS ECs manifested a dramatic increase in their own expression of inflammatory cytokines (88% of the panel proteins were upregulated), whereas those exposed to conditioned medium from hTERT treated HGPS ECs were not as strongly affected (Figure 5D). Thus, the HGPS ECs has a paracrine effect on VSMCs that could explain in part the attenuated media in the vessel wall of HGPS patients. Treatment of HGPS ECs with hTERT mRNA reverses their inflammatory secretome.
Figure 5.
hTERT treatment reverses senescence-associated secretory phenotype in Hutchinson-Gilford progeria syndrome endothelial cells. Treatment of Hutchinson-Gilford progeria syndrome endothelial cells with hTERT reduces the senescence-associated secretory phenotype profile as measured by Bio-Plex analysis (A). To assess how these secreted factors may negatively affect other relevant vascular cells, a conditioned media experiment was set up using non-Hutchinson-Gilford progeria syndrome vascular smooth muscle cells differentiated from the control donor as detailed in (B). To confirm differentiation, vascular smooth muscle cells were stained for smooth muscle cell specific markers smoothelin and α-actin 2 (ACTA2, scale bar—25 µm). When these control vascular smooth muscle cells were subject to the various conditioned media, their proliferation was greatly reduced when cultured under the Hutchinson-Gilford progeria syndrome endothelial cell conditioned media, but proliferation was not affected when Hutchinson-Gilford progeria syndrome endothelial cells treated with hTERT supplied the conditioned media (n = 3, C, scale bar—50 µm). Bio-Plex analysis of vascular smooth muscle cells exposed to different conditioned media demonstrated that vascular smooth muscle cells themselves also secreted more inflammatory markers when cultured under Hutchinson-Gilford progeria syndrome endothelial cell donor media, though this shift was attenuated when hTERT treated Hutchinson-Gilford progeria syndrome endothelial cells were the donor (D). Fold-change vascular smooth muscle cell count is the endpoint cell count of each group divided by its respective initial seeding cell count. Analysis of (C) was tested using a one-way ANOVA with post hoc Tukey’s test for group differences. Significance was taken at P < 0.05, in (C) only significant changes between groups are annotated, all other between-group statistics did not reach significance. All error bars show standard deviation.
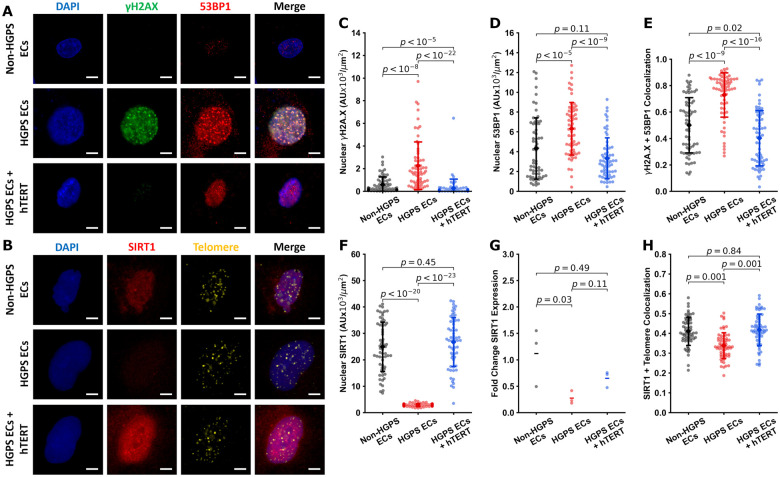
hTERT treatment induces SIRT1 expression and reduces DNA damage response
Our RNAseq data indicated that markers of DNA damage (e.g. γH2A.X and 53BP1) were increased in HGPS, whereas SIRT1 was reduced. SIRT1 is implicated in histone deacetylation,62 telomere maintenance,63 and reduces DNA damage signaling at telomeres.64 Treatment with hTERT mRNA reversed these abnormalities in HGPS ECs. Immunofluorescent staining further confirmed our results (Figure 6A and B). The nuclear expression and co-localization of γH2A.X and 53BP1 were increased in HGPS ECs, an effect reversed by treatment with hTERT (Figure 6C–E). In addition, SIRT1 nuclear expression was reduced in HGPS ECs and rescued by hTERT treatment (Figure 6F). RTqPCR analysis revealed a strong decrease in SIRT1 expression in HGPS ECs (Figure 6G). Furthermore, colocalization of SIRT1 and the telomere were reduced in HGPS ECs, an effect that was reversed by hTERT treatment (Figure 6G). These data suggest that parallel to progerin reduction, the benefit of hTERT treatment may be mediated in part by the role of SIRT1 in maintaining telomere length and reducing DNA damage response.
Figure 6.
hTERT treatment reduces DNA damage markers and increases SIRT1 expression. Cells were stained for nuclei (DAPI, blue), γH2A.X (green), and 53BP1 (red) in (A) (scale bar—15 µm) and SIRT1 (red) and telomere (yellow) in (B) (scale bar—15 µm). Immunofluorescent quantification of these images showed that Hutchinson-Gilford progeria syndrome endothelial cells had significantly elevated levels of nuclear γH2A.X (normalized to nuclear area) which were reduced with hTERT intervention (C). Hutchinson-Gilford progeria syndrome endothelial cells also had elevated levels of nuclear localized 53BP1 which were reduced upon hTERT treatment (D). Colocalization of the signals γH2A.X and 53BP1 were increased in Hutchinson-Gilford progeria syndrome cells, and decreased with hTERT treatment (E). Nuclear SIRT1 expression was reduced in Hutchinson-Gilford progeria syndrome but restored with hTERT treatment (F). RTqPCR analysis showed that Hutchinson-Gilford progeria syndrome endothelial cells have decreased SIRT1 expression, confirmed by IF. Treatment with hTERT induced an upward trend which did not reach significance (G). Colocalization of the signals for SIRT1 and the telomeres were increased upon hTERT treatment (H). For each of the IF experiments, 60 individual cells quantified, replicated at least three times to confirm. Nuclear expression (fluorescence) was normalized to the nuclear area of each cell. Colocalization values are reported as Mander’s coefficients for signal correlation for each individual cell. Statistics for qPCR data was performed on the ddCt values (G) using ANOVA with post hoc Tukey’s test. All IF data were tested for normality using D’Agostino-Pearson. Images (C), (D), and (F) were tested using Kruskal–Wallis with post hoc Dunn test. Images (E) and (G) were tested with a one-way ANOVA with post hoc Tukey’s test. Significance was taken at P < 0.05. All between-group statistics are displayed natively. All error bars show standard deviation. DAPI, 4′,6-diamidino-2-phenylindole.
Treatment with mTERT normalizes endothelial cell functions
To provide proof-of-concept for in vivo telomerase therapy, we employed a murine model of HGPS. The progeroid mouse model is generated by a knock-in of mutated human lamin A (LMNA*G608G). The model has been characterized and recapitulates some features of the human progeria syndrome.46 Progeroid mice exhibit failure to grow after 3 months and have loss of their subcutaneous fat layer and decreased bone density. Although they develop attenuation of the media of the large vessels, unlike human patients they do not develop intimal occlusive disease and instead die of cachexia and inanition.46
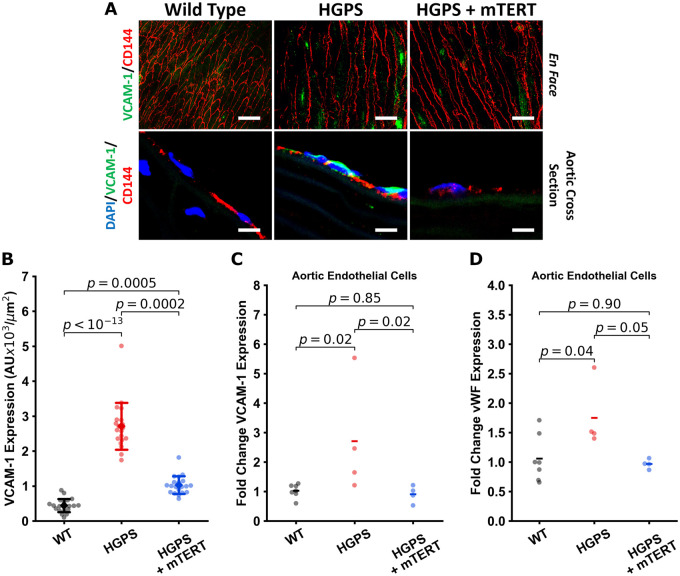
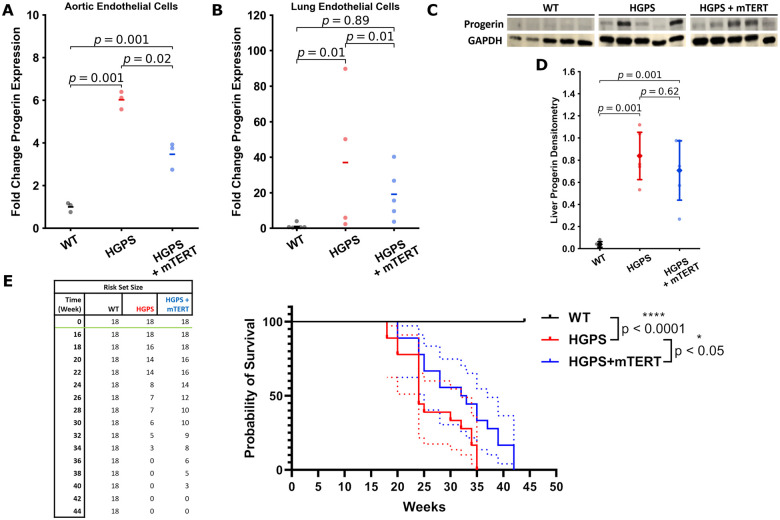
Because murine telomeres are much longer than humans (40–100 vs, 8–15 kb) one might reasonably think that telomere erosion would not be a significant issue in mice and that mTERT treatment would likely have little effect. Nevertheless, because the murine model is accessible commercially and we could at least test the potential benefits of telomerase treatment in this model, we generated a lentiviral vector which encoded mouse TERT (mTERT). Subsequently, HGPS mice were treated with tail vein injection of mTERT or GFP (1010 IFU/mL in 150 µL PBS) or vehicle alone at 3 and 6 months of age. Notably, we observed that in this HGPS mouse model, aortic VCAM-1 expression is elevated even at 3 months of age (Supplementary material online, Figure S3A), which suggests intervention at an earlier timepoint may have greater benefit. Necropsy studies performed by an independent veterinarian blinded to the experimental groups revealed no gross or microscopic pathological findings of mTERT treatment on the 33 organs examined (see Supplementary material online). To determine if mTERT treatment had an effect on vascular senescence as assessed by inflammatory markers, animals were sacrificed and aortae harvested 2 months post-second dose treatment (at 8 months). The aortae from vehicle-treated HGPS mice had significantly elevated VCAM-1 expression, an endothelial adhesion molecule that is increased with vascular inflammation (Figure 7A and B). By contrast, in mTERT-treated mice, VCAM-1 expression was attenuated as assessed by analysis of en face (Figure 7A and B) and cross-sectional vascular sections. In addition, we used qPCR to assess the expression of key genes involved in EC homeostasis: VCAM-1, von Willebrand factor (vWF), tissue plasminogen activator (TPA), and thrombomodulin (THBD). Treatment with viral mTERT reversed the elevated levels of VCAM-1 and vWF in HGPS aortic ECs (Figure 7C and D). The expression of THBD was decreased in HGPS aortic ECs, an effect that was not appreciably reversed by treatment with mTERT. The expression of TPA was minimally modulated across the groups (Supplementary material online, Figure S3B and C). We additionally examined isolated lung ECs to explore if this effect could be seen in other vascular areas. Treatment of lung ECs with mTERT increased THBD expression. However, TPA expression, like with the aortic ECs, was not appreciably modulated (Supplementary material online, Figure S3D and E). We also observed improvements in endothelial NO synthase levels in the aortic and liver endothelium (Supplementary material online, Figure S3K and L). When we examined serum markers predictive of cardiovascular disease (sVCAM-1 and CRP), we did not observe appreciable changes (Supplementary material online, Figure S3I and J). Nonetheless, overall, mTERT treatment reduces mediators of inflammation and thrombosis in HGPS.
Figure 7.
Treatment of progeria mice with mTERT reduces aortic VCAM-1 expression. Mouse aortae were stained en face for VCAM-1 (green) and CD144 (VE-Cadherin, red); or stained for nuclei (DAPI, blue), VCAM-1 (green), and CD144 (red) on aortic cross section, representative images are shown in (A) (scale bar: top—50 µm, bottom—5 µm). VCAM-1 is elevated in Hutchinson-Gilford progeria syndrome aortic endothelial cells. These changes are quantified in (B), single cell normalized VCAM-1 expression for each of the mouse groups from en face staining of the dissected aortas (n = 19 cells per aortic section, representative of 4–5 mice per group). Representative aortic cross sections are shown to complement the en face staining. Results were confirmed using RTqPCR analysis of isolated aortic endothelial cells from 4 to 6 mice from each group, showing increased VCAM-1 and von Willebrand factor expression that is normalized with mTERT expression (C and D). The data were tested for normality using D’Agostino-Pearson test. Kruskal–Wallis with post hoc Dunn test was used to quantify the data in (B). Statistics for qPCR data was performed on the ddCt values using ANOVA with post hoc Tukey’s test. Significance was taken at P < 0.05, all between-group statistics are displayed natively. All error bars show standard deviation. DAPI, 4′,6-diamidino-2-phenylindole.
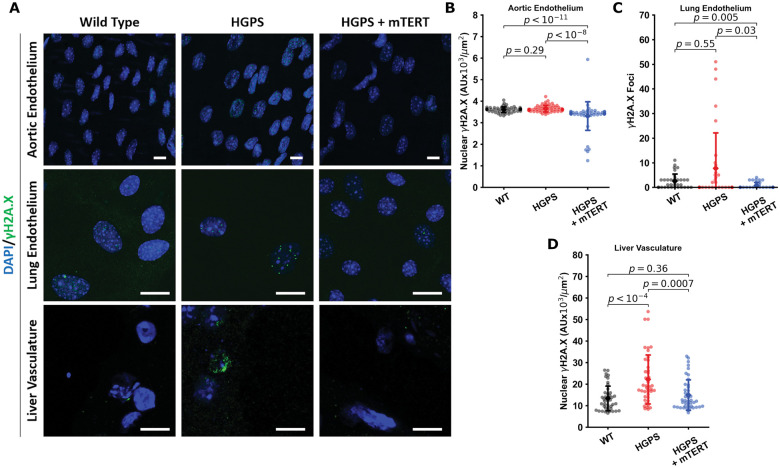
mTERT treatment enhances SIRT1 expression and reduces DNA damage signaling in murine Hutchinson-Gilford progeria syndrome endothelium
We observed increased nuclear expression of the DNA damage response marker γH2A.X in aortic endothelium of HGPS mice. We performed IF of various mouse tissues to assess DNA damage (Figure 8A and Supplementary material online, Figure S4). In mTERT-treated mice, γH2A.X expression in the aortic endothelium was reduced (Figure 8B). We made similar observations on mouse lung ECs (Figure 8C) and hepatic endothelium (Figure 8D) that were treated with mTERT, showing consistent decreases in γH2A.X in all experiments. We additionally observed increased telomerase activity and telomere extension via qFISH in the liver tissue endothelium (Supplementary material online, Figure S5), which was associated with a down-regulation of markers of DNA damage. However, in contrast to human cells, SIRT1 expression in the mouse samples was not clearly rescued or altered. We performed IF on mouse aortic, lung, and liver vascular ECs (Supplementary material online, Figure S6). We observed that SIRT1 was significantly downregulated in aortic tissue on qPCR analysis and in liver lysates upon WB analysis from HGPS mice. Our findings are consistent with recently published data showing that telomere erosion is associated with a p53-dependent decline in sirtuin expression in telomerase knockout mice.65 Treatment with mTERT tended to normalize SIRT1 expression, but the results did not achieve statistical significance (d.n.s.).
Figure 8.
mTERT Treatment in Hutchinson-Gilford progeria syndrome mice reduces DNA damage markers in aorta, lung and liver endothelial cells. Representative images from aortic endothelial cells, lung endothelial cells, and liver vasculature are displayed in (A) (scale bar—10 µm). All tissues were stained for nuclei (DAPI, blue) and γH2A.X (green). Individual cells from the tissues were quantified (n > 50) for nuclear γH2A.X fluorescence (tissue sections from aorta in B and liver in D) and γH2A.X foci in isolated mouse lung endothelial cells in (C). In the aortic and lung tissue, differences were not statistically significant, though γH2A.X was significantly downregulated or decreased in the mTERT treatment groups. In liver tissues, Hutchinson-Gilford progeria syndrome vascular cells displayed increased γH2A.X, which was reversed by mTERT. All data collected from four to five different mice histology sections for each group. The data were tested for normality using D’Agostino-Pearson test. Kruskal–Wallis with post hoc Dunn test was used to quantify the data. Significance was taken at P < 0.05, all between-group statistics are displayed natively. All error bars show standard deviation. DAPI, 4′,6-diamidino-2-phenylindole.
mTERT therapy reduces progerin expression and extends lifespan in murine Hutchinson-Gilford progeria syndrome
Consistent with the observations in our human cell lines, the expression of progerin mRNA in both aorta and lung ECs was increased in HGPS, and reduced by treatment with mTERT (Figure 9A and B). Similarly, WB analysis of hepatic tissue lysate revealed that progerin was increased in HGPS, though only a downward trend was observed after TERT treatment that did not reach significance (Figure 9C and D). We observed impaired growth and abbreviated lifespan in HGPS mice as previously reported.46 Two treatments with lentiviral mTERT increased the lifespan of HGPS mice by 20% (Figure 9E). Around 27% (5 out of 18) of the untreated HGPS mice died spontaneously. In the mice which died spontaneously, we observed significant cachexia, impaired mobility, and insufficient food intake, associated with significant inanition; likely contributors to death. The remaining animals from the study (both treated and untreated) were euthanized at the request of the veterinarian due to failure to gain weight, and other clinical signs associated with progeria (reduced mobility of joints and spine, with stilted gait and hunched posture, and muscle wasting). Notably, only one mouse treated with mTERT died spontaneously. Altogether, this section underscores that TERT treatment induces a favourable down-regulation of progerin in mouse cells as it does in human cells, providing a mechanistic connection between the treatments observed in vitro and in vivo.
Figure 9.
Treatment with mTERT reduces progerin expression and extends lifespan. RTqPCR analysis of progerin expression in aortic (n = 3 isolated mouse aortic endothelial cell groups) and lung endothelial cells (n = 4–6 isolated mouse lung endothelial cell groups) is detailed in (A) and (B). Treatment with mTERT appreciably reduced progerin gene expression in both the aortic and lung endothelium. Liver tissue lysates were analysed for progerin in (C) and (D) (n = 5 mice); however, while liver tissues displayed a marked up-regulation of progerin expression, mTERT did not significantly down-regulate expression in liver lysates. Overall mouse survival with mTERT treatment is shown in E, with a significant extension of lifespan (7 weeks) displayed by the mTERT-treated group vs. the Hutchinson-Gilford progeria syndrome group (n = 18 mice per group). Images (A) and (C) were analysed using a one-way ANOVA with post hoc Tukey’s test. Image (B) was analysed using a Kruskal–Wallis with post hoc Dunn test. Statistics for qPCR data was performed on the ddCt values. Survival in (E) was analysed using a log-rank test, with error densities (dotted lines) shown over the 95% confidence interval. All other error bars show standard deviation. Significance was taken at P < 0.05. All between-group statistics are displayed natively.
Discussion
The primary cause of morbidity and mortality in HGPS children is vascular disease. The endothelium is extremely critical to vascular homeostasis; thus, it seems sensible to consider endothelium-targeted strategies for these children. In this study, we confirm ECs derived from HGPS children manifest global abnormalities in form and function, consistent with a senescent phenotype. Surprisingly, two treatments with mRNA hTERT are sufficient to increase telomere length, and attenuate or reverse the abnormalities in transcriptional profile, nuclear morphology, and cell functions. Furthermore, we observed that conditioned medium from HGPS ECs adversely affected VSMCs, reducing their proliferation and inducing an inflammatory activation, effects that were reversed by treating the HGPS ECs with hTERT. Thus, medial atrophy might be due in part to senescent ECs in HGPS, an effect that could be reversed by endothelial-targeted therapy (Graphical Abstract).
Telomerase therapy reverses senescent phenotypes in human cells and progeria mice.
In HGPS mice, we observed that mTERT treatment improved the endothelial phenotype, and provided for a 20% increase in lifespan. In our mouse model of HGPS mice, arterial occlusive disease does not occur, so coronary and carotid artery disease is not the cause of death as it is in humans. Nevertheless, a systemic improvement in EC health was observed in this study with mTERT and would be expected to provide for improvements in function of the vessels supplying all tissues. An improvement in vascular homeostasis might be expected to improve organ function and extend lifespan since endothelial function regulates perfusion, thrombosis, inflammation, and serves as a niche for resident stem cells involved in tissue repair.66,67 Indeed, EC-specific expression of progerin shortens lifespan to the same degree as systemic expression of progerin.32 In addition, VSMC-specific progerin expression is also sufficient to induce accelerated development of atherosclerosis and death.68,69 Thus, the clinical picture of significant cardiovascular pathology in HGPS patients may be tied directly to a compounding of both EC and VSMC-related dysfunction. In this regard, we speculate that age-related disease may be due in large part to vascular senescence.
It is possible that earlier intervention is required to have a greater effect on the outcome. At the time of treatment (3 months) significant alteration in endothelial function as assessed by VCAM-1 expression is present in the aortic endothelium (Supplementary material online, Figure S3A). This may also explain why specific clinical markers like sVCAM-1 or CRP were not appreciably altered (Supplementary material online, Figure S3I and J), as considerable disease progression had already occurred. It is also worth noting that our mouse model is generated by the expression of two alleles of the mutant lamin A, whereas in HGPS children, only one allele is mutated. Even though mouse telomeres are much longer than human telomeres (40–100 vs. 8–15 kb),70 it appears that telomere erosion may occur in HGPS mice, a process reversed by exogenous/constitutive expression of telomerase (Supplementary material online, Figure S5).
In human HGPS cells, hTERT treatment substantially restored several transcriptional profiles. The mechanism by which telomerase treatment normalizes gene expression requires further elucidation. There are some paradigms that connect telomere length and gene expression, such as the positional effect,71,72 or telomere-associated protein/transcription factor redistribution.73,74 However, these hypotheses and observations may not uniformly apply to the mechanisms underlying the improvements observed in both human cells and mice, as the telomere biology of mice differs from that of humans.75 However, uniformly observed in our work on both human and murine tissues is the inhibitory effect telomerase expression has on progerin expression. The mechanism by which telomerase treatment reduces progerin levels is unclear, but a down-regulation of progerin would explain the global benefit of TERT treatment. In addition, telomere attrition registers as DNA damage, with persistent activation causing eventual cellular dysfunction; however, telomerase can protect against this76–78—our investigation revealed several pathways altered by hTERT treatment related to DNA damage which was confirmed using immunofluorescence. Lastly, we also observed SIRT1 depletion and potential rescue in human cells. The relationship between SIRT1 and TERT regulation is established although not yet fully elucidated. It is known that telomere dysfunction induces SIRT1 repression.65 Conversely, in specific cell lines overexpression of SIRT1 can rescue cells from senescence,79–81 though in other scenarios (nutrient sensing) SIRT1 may inhibit TERT expression.82 This is relevant to our observation in HGPS ECs, as SIRT1 regulates NO,83 FOXO-1,84 and Notch signaling,85 and additionally has a protective role against metabolic liver disease.86 Overall SIRT1 appears to play a role indicative of telomere maintenance and homeostasis. Here, we report as a novel finding that hTERT transfection in human iPSC EC lines may itself induce SIRT1 expression. The mechanism for this finding and our other findings remains to be elucidated, especially in light of the different telomere biology between human and murine cells. Nevertheless, a clear benefit is seen for both. The connection between the two may be the regulation of progerin expression, as this was observed in both experimental models.
There are several limitations to our work. First, our HGPS mice do not develop occlusive atherosclerotic disease as do children with HGPS, and as such the physiology of the mice incompletely represents human disease progression. Second, the telomerase treatment was not optimized by dose-ranging studies. While there is significant promise, mRNA telomerase therapy will require optimization and improved delivery. The in vivo work also has some important qualifications. To begin, lentiviral gene transfection results in genome integration and constitutive expression87 as opposed to the transient nature of in vivo mRNA delivery. Here, our work is only illustrative that the presence of TERT alleviates the HGPS phenotype and can extend lifespan in mice. In addition, lentiviral delivery of mTERT has safety concerns because genomic integration may induce cellular immortalization and oncogenicity. Our safety studies are preliminary in nature (gross and microscopic pathology, and blood studies; Supplementary material online, Figure S7), and additional work is required to confirm safety. Furthermore, due to the lack of a targeting modality with the lentiviral regimen, the lifespan extension observed in the HGPS mice may be due in part to non-vascular effects of mTERT (indeed, we observed improvements in tissues such as liver lysates).
Other than extending the telomere, there may be other methods to reverse telomere dysfunction. Progerin induces transcription of telomeric non-coding RNAs (tncRNAs), which activate the DNA damage response (DDR).88 The inhibition of tncRNAs by sequence-specific telomeric antisense oligonucleotides prevents full DDR activation and premature cellular senescence in HGPS cells in vitro and restores skin homeostasis as well as lifespan in a transgenic HGPS mouse model.88 Notably, our in vitro work used mRNA therapy, and in future studies, we intend to deliver mRNA TERT using a lipid nanoparticle vehicle.
To conclude, the data herein suggest hTERT therapy as a viable option for treating vascular disease in HGPS and, potentially, extending lifespan for these patients. Accordingly, based on the promising results from this study, we are developing mRNA hTERT as a therapy for these children.
Supplementary material
Supplementary material is available at European Heart Journal online.
Supplementary Material
Acknowledgements
This work is supported by grants to JPC [National Institutes of Health (NIH) R01s HL133254 and HL148338; as well as the Cancer Prevention and Research Institute of Texas CPRIT RP150611]. We acknowledge Dr Kaifu Chen and Dr Dongyu Zhao for their helpful suggestions in revising the manuscript. We acknowledge Luay Boulahouache for readability edits. We would also like to acknowledge Ms Rachel Whitehead for her artist talents and efforts in generating the graphical abstract.
Funding
This work is supported by grants to JPC [National Institutes of Health (NIH) R01s HL133254 and HL148338; as well as the Cancer Prevention and Research Institute of Texas CPRIT RP150611].
Conflict of interest: J.P.C. is an inventor on patents owned by Stanford University and Houston Methodist Hospital related to the use of mRNA telomerase for the treatment of senescence. JPC is co-founder of a company that will develop the telomerase technology.
Data availability
All the raw sequence data for the samples used in this study are available on the National Center for Biotechnology Information Sequence Read Archive database: Bioproject PRJNA730422.
References
- 1.Dhingra R, Vasan RS.. Age as a risk factor. Med Clin North Am 2012;96:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsushita H, Chang E, Glassford AJ, Cooke JP, Chiu CP, Tsao PS.. eNOS activity is reduced in senescent human endothelial cells: preservation by hTERT immortalization. Circ Res 2001;89:793–798. [DOI] [PubMed] [Google Scholar]
- 3.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G.. The hallmarks of aging. Cell 2013;153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K, Ferbeyre G, Gil J, Hara E, Krizhanovsky V, Jurk D, Maier AB, Narita M, Niedernhofer L, Passos JF, Robbins PD, Schmitt CA, Sedivy J, Vougas K, von Zglinicki T, Zhou D, Serrano M, Demaria M.. Cellular Senescence: defining a Path Forward. Cell 2019;179:813–827. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS.. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 2003;423:293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS.. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A 2004;101:8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClintock D, Gordon LB, Djabali K.. Hutchinson-Gilford progeria mutant lamin A primarily targets human vascular cells as detected by an anti-Lamin A G608G antibody. Proc Natl Acad Sci U S A 2006;103:2154–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClintock D, Ratner D, Lokuge M, Owens DM, Gordon LB, Collins FS, Djabali K.. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS One 2007;2:e1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capell BC, Erdos MR, Madigan JP, Fiordalisi JJ, Varga R, Conneely KN, Gordon LB, Der CJ, Cox AD, Collins FS.. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A 2005;102:12879–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD.. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 2008;22:832–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, Goldman AE, Goldman RD.. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci U S A 2009;106:20788–20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenain C, de Graaf CA, Pagie L, Visser NL, de Haas M, de Vries SS, Peric-Hupkes D, van Steensel B, Peeper DS.. Massive reshaping of genome-nuclear lamina interactions during oncogene-induced senescence. Genome Res 2017;27:1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decker ML, Chavez E, Vulto I, Lansdorp PM.. Telomere length in Hutchinson-Gilford progeria syndrome. Mech Ageing Dev 2009;130:377–383. [DOI] [PubMed] [Google Scholar]
- 14.Chojnowski A, Ong PF, Wong ES, Lim JS, Mutalif RA, Navasankari R, Dutta B, Yang H, Liow YY, Sze SK, Boudier T, Wright GD, Colman A, Burke B, Stewart CL, Dreesen O.. eLife 2015;4:doi:10.7554/eLife.07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao K, Blair CD, Faddah DA, Kieckhaefer JE, Olive M, Erdos MR, Nabel EG, Collins FS.. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Invest 2011;121:2833–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lange T. How telomeres solve the end-protection problem. Science 2009;326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shay JW, Wright WE.. Telomeres and telomerase: implications for cancer and aging. Radiat Res 2001;155:188–193. [DOI] [PubMed] [Google Scholar]
- 18.Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, Anderson R, Taschuk M, Mann J, Passos JF.. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun 2012;3:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Contrepois K, Coudereau C, Benayoun BA, Schuler N, Roux PF, Bischof O, Courbeyrette R, Carvalho C, Thuret JY, Ma Z, Derbois C, Nevers MC, Volland H, Redon CE, Bonner WM, Deleuze JF, Wiel C, Bernard D, Snyder MP, Rube CE, Olaso R, Fenaille F, Mann C.. Histone variant H2A.J accumulates in senescent cells and promotes inflammatory gene expression. Nat Commun 2017;8:14995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J.. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol 2009;11:973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barinda AJ, Ikeda K, Nugroho DB, Wardhana DA, Sasaki N, Honda S, Urata R, Matoba S, Hirata KI, Emoto N.. Endothelial progeria induces adipose tissue senescence and impairs insulin sensitivity through senescence associated secretory phenotype. Nat Commun 2020;11:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon LB, McCarten KM, Giobbie-Hurder A, Machan JT, Campbell SE, Berns SD, Kieran MW.. Disease progression in Hutchinson-Gilford progeria syndrome: impact on growth and development. Pediatrics 2007;120:824–833. [DOI] [PubMed] [Google Scholar]
- 23.Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith ACM, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Brooks BP, Gerber LH, Turner ML, Domingo DL, Hart TC, Graf J, Reynolds JC, Gropman A, Yanovski JA, Gerhard-Herman M, Collins FS, Nabel EG, Cannon RO, Gahl WA, Introne WJ.. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med 2008;358:592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prakash A, Gordon LB, Kleinman ME, Gurary EB, Massaro J, D'Agostino R, Kieran MW, Gerhard-Herman M, Smoot L.. Cardiac abnormalities in patients with Hutchinson-Gilford progeria syndrome. JAMA Cardiol 2018;3:326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olive M, Harten I, Mitchell R, Beers JK, Djabali K, Cao K, Erdos MR, Blair C, Funke B, Smoot L, Gerhard-Herman M, Machan JT, Kutys R, Virmani R, Collins FS, Wight TN, Nabel EG, Gordon LB.. Cardiovascular pathology in Hutchinson-Gilford progeria: correlation with the vascular pathology of aging. Arterioscler Thromb Vasc Biol 2010;30:2301–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walther BK, Li Y, Thandavarayan RA, Cooke JP.. Progeria and accelerated cardiovascular aging. Cardiology Plus 2018;3:81. [Google Scholar]
- 27.Zhang J, Lian Q, Zhu G, Zhou F, Sui L, Tan C, Mutalif RA, Navasankari R, Zhang Y, Tse H-F, Stewart CL, Colman A.. A human iPSC model of Hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 2011;8:31–45. [DOI] [PubMed] [Google Scholar]
- 28.Gordon LB, Massaro J, D’Agostino RB Sr, Campbell SE, Brazier J, Brown WT, Kleinman ME, Kieran MW.. Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation 2014;130:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, Fligor B, Bishop WR, Statkevich P, Regen A, Sonis A, Riley S, Ploski C, Correia A, Quinn N, Ullrich NJ, Nazarian A, Liang MG, Huh SY, Schwartzman A, Kieran MW.. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A 2012;109:16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon LB, Kleinman ME, Massaro J, D’Agostino RB, Shappell H, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland RH, Nazarian A, Snyder BD, Ullrich NJ, Silvera VM, Liang MG, Quinn N, Miller DT, Huh SY, Dowton AA, Littlefield K, Greer MM, Kieran MW.. Clinical trial of the protein farnesylation inhibitors lonafarnib, pravastatin, and zoledronic acid in children with Hutchinson-Gilford progeria syndrome. Circulation 2016;134:114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osmanagic-Myers S, Kiss A, Manakanatas C, Hamza O, Sedlmayer F, Szabo PL, Fischer I, Fichtinger P, Podesser BK, Eriksson M, Foisner R.. Endothelial progerin expression causes cardiovascular pathology through an impaired mechanoresponse. J Clin Invest 2019;129:531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun S, Qin W, Tang X, Meng Y, Hu W, Zhang S, Qian M, Liu Z, Cao X, Pang Q, Zhao B, Wang Z, Zhou Z, Liu B.. Vascular endothelium-targeted Sirt7 gene therapy rejuvenates blood vessels and extends life span in a Hutchinson-Gilford progeria model. Sci Adv 2020;6:eaay5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matrone G, Thandavarayan RA, Walther BK, Meng S, Mojiri A, Cooke JP.. Dysfunction of iPSC-derived endothelial cells in human Hutchinson-Gilford progeria syndrome. Cell Cycle 2019;18:2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooke JP. Flow, NO, and atherogenesis. Proc Natl Acad Sci U S A 2003;100:768–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greider CW, Blackburn EH.. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985;43:405–413. [DOI] [PubMed] [Google Scholar]
- 36.Greider CW, Blackburn EH.. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 1989;337:331–337. [DOI] [PubMed] [Google Scholar]
- 37.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR.. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 1997;276:561–567. [DOI] [PubMed] [Google Scholar]
- 38.Ramunas J, Yakubov E, Brady JJ, Corbel SY, Holbrook C, Brandt M, Stein J, Santiago JG, Cooke JP, Blau HM.. Transient delivery of modified mRNA encoding TERT rapidly extends telomeres in human cells. FASEB J 2015;29:1930–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Zhou G, Bruno IG, Cooke JP.. Telomerase mRNA reverses senescence in progeria cells. J Am Coll Cardiol 2017;70:804–805. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Zhou G, Bruno IG, Zhang N, Sho S, Tedone E, Lai TP, Cooke JP, Shay JW.. Transient introduction of human telomerase mRNA improves hallmarks of progeria cells. Aging Cell 2019;18:e12979.DOI: 10.1111/acel.12979, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madonna R, Taylor DA, Geng Y-J, De Caterina R, Shelat H, Perin EC, Willerson JT.. Transplantation of mesenchymal cells rejuvenated by the overexpression of telomerase and myocardin promotes revascularization and tissue repair in a murine model of hindlimb ischemia. Circ Res 2013;113:902–914. [DOI] [PubMed] [Google Scholar]
- 42.Madonna R, Wu D, Wassler M, De Caterina R, Willerson JT, Geng Y-J.. Myocardin-A enhances expression of promyogenic genes without depressing telomerase activity in adipose tissue-derived mesenchymal stem cells. Int J Cardiol 2013;167:2912–2921. [DOI] [PubMed] [Google Scholar]
- 43.Madonna R, Willerson JT, Geng YJ.. Myocardin a enhances telomerase activities in adipose tissue mesenchymal cells and embryonic stem cells undergoing cardiovascular myogenic differentiation. Stem Cells 2008;26:202–211. [DOI] [PubMed] [Google Scholar]
- 44.Varga R, Eriksson M, Erdos MR, Olive M, Harten I, Kolodgie F, Capell BC, Cheng J, Faddah D, Perkins S, Avallone H, San H, Qu X, Ganesh S, Gordon LB, Virmani R, Wight TN, Nabel EG, Collins FS.. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A 2006;103:3250–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng S, Matrone G, Lv J, Chen K, Wong WT, Cooke JP. LIM domain only 2 regulates endothelial proliferation, angiogenesis, and tissue regeneration. J Am Heart Assoc 2016;5:e004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayed N, Wong WT, Ospino F, Meng S, Lee J, Jha A, Dexheimer P, Aronow BJ, Cooke JP.. Transdifferentiation of human fibroblasts to endothelial cells: role of innate immunity. Circulation 2015;131:300–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res 2009;37:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ourliac-Garnier I, Londoño-Vallejo A.. Telomere length analysis by quantitative fluorescent in situ hybridization (Q-FISH). Methods Mol Biol 2017;1587:29–39. [DOI] [PubMed] [Google Scholar]
- 49.Cao K, Blair CD, Faddah DA, Kieckhaefer JE, Olive M, Erdos MR, Nabel EG, Collins FS.. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Investig 2011;121:2833–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP.. Integrative genomics viewer. Nat Biotechnol 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson JT, Thorvaldsdottir H, Wenger AM, Zehir A, Mesirov JP.. Variant review with the integrative genomics viewer. Cancer Res 2017;77:e31–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Z, Chang WY, Etheridge A, Strickfaden H, Jin Z, Palidwor G, Cho JH, Wang K, Kwon SY, Dore C, Raymond A, Hotta A, Ellis J, Kandel RA, Dilworth FJ, Perkins TJ, Hendzel MJ, Galas DJ, Stanford WL.. Reprogramming progeria fibroblasts re-establishes a normal epigenetic landscape. Aging Cell 2017;16:870–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ko KA, Fujiwara K, Krishnan S, Abe JI. En Face Preparation of Mouse Blood Vessels. J Vis Exp 2017. doi:10.3791/55460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng 2007;9:90–95. [Google Scholar]
- 57.Scaffidi P, Misteli T.. Lamin A-dependent nuclear defects in human aging. Science 2006;312:1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neurohr GE, Terry RL, Lengefeld J, Bonney M, Brittingham GP, Moretto F, Miettinen TP, Vaites LP, Soares LM, Paulo JA, Harper JW, Buratowski S, Manalis S, van Werven FJ, Holt LJ, Amon A.. Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell 2019;176:1083–1097. e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia B, Zhao D, Wang G, Zhang M, Lv J, Tomoiaga AS, Li Y, Wang X, Meng S, Cooke JP, Cao Q, Zhang L, Chen K.. Machine learning uncovers cell identity regulator by histone code. Nat Commun 2020;11:2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stehbens WE, Delahunt B, Shozawa T, Gilbert-Barness E.. Smooth muscle cell depletion and collagen types in progeric arteries. Cardiovasc Pathol 2001;10:133–136. [DOI] [PubMed] [Google Scholar]
- 61.Basisty N, Kale A, Jeon OH, Kuehnemann C, Payne T, Rao C, Holtz A, Shah S, Sharma V, Ferrucci L, Campisi J, Schilling B.. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol 2020;18:e3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahman S, Islam R.. Mammalian Sirt1: insights on its biological functions. Cell Commun Signal 2011;9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA.. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol 2010;191:1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Bonis ML, Ortega S, Blasco MA.. SIRT1 is necessary for proficient telomere elongation and genomic stability of induced pluripotent stem cells. Stem Cells Reports 2014;2:690–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amano H, Chaudhury A, Rodriguez-Aguayo C, Lu L, Akhanov V, Catic A, Popov YV, Verdin E, Johnson H, Stossi F, Sinclair DA, Nakamaru-Ogiso E, Lopez-Berestein G, Chang JT, Neilson JR, Meeker A, Finegold M, Baur JA, Sahin E.. Telomere dysfunction induces sirtuin repression that drives telomere-dependent disease. Cell Metab 2019;29:1274–1290. e1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooke JP. Endotheliopathy of obesity. Circulation 2020;142:380–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leeper NJ, Hunter AL, Cooke JP.. Stem cell therapy for vascular regeneration: adult, embryonic, and induced pluripotent stem cells. Circulation 2010;122:517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamczyk MR, Villa-Bellosta R, Quesada V, Gonzalo P, Vidak S, Nevado RM, Andrés‐Manzano MJ, Misteli T, López-Otín C, Andrés V. Progerin accelerates atherosclerosis by inducing endoplasmic reticulum stress in vascular smooth muscle cells. EMBO Mol Med 2019;11:e9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamczyk MR, Villa-Bellosta R, Gonzalo P, Andrés-Manzano MJ, Nogales P, Bentzon JF, López-Otín C, Andrés V.. Vascular smooth muscle-specific progerin expression accelerates atherosclerosis and death in a mouse model of Hutchinson-Gilford progeria syndrome. Circulation 2018;138:266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calado RT, Dumitriu B.. Telomere dynamics in mice and humans. Semin Hematol 2013;50:165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robin JD, Ludlow AT, Batten K, Magdinier F, Stadler G, Wagner KR, Shay JW, Wright WE.. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev 2014;28:2464–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim W, Ludlow AT, Min J, Robin JD, Stadler G, Mender I, Lai T-P, Zhang N, Wright WE, Shay JW.. Regulation of the human telomerase gene TERT by telomere position effect-over long distances (TPE-OLD): implications for aging and cancer. PLoS Biol 2016;14:e2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mukherjee AK, Sharma S, Sengupta S, Saha D, Kumar P, Hussain T, Srivastava V, Roy SD, Shay JW, Chowdhury S.. Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet 2018;14:e1007782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teo H, Ghosh S, Luesch H, Ghosh A, Wong ET, Malik N, Orth A, de Jesus P, Perry AS, Oliver JD, Tran NL, Speiser LJ, Wong M, Saez E, Schultz P, Chanda SK, Verma IM, Tergaonkar V.. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat Cell Biol 2010;12:758–767. [DOI] [PubMed] [Google Scholar]
- 75.Smogorzewska A, de Lange T.. Different telomere damage signaling pathways in human and mouse cells. EMBO J 2002;21:4338–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC.. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc Natl Acad Sci U S A 2005;102:8222–8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, Herbig U, Longhese MP, d'Adda di Fagagna F.. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 2012;14:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fleisig H, Hukezalie K, Thompson C, Au-Yeung T, Ludlow A, Zhao C, Wong J.. Telomerase reverse transcriptase expression protects transformed human cells against DNA-damaging agents, and increases tolerance to chromosomal instability. Oncogene 2016;35:218–227. [DOI] [PubMed] [Google Scholar]
- 79.De Bonis ML, Ortega S, Blasco MA.. SIRT1 is necessary for proficient telomere elongation and genomic stability of induced pluripotent stem cells. Stem Cell Rep 2014;2:690–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamashita S, Ogawa K, Ikei T, Udono M, Fujiki T, Katakura Y.. SIRT1 prevents replicative senescence of normal human umbilical cord fibroblast through potentiating the transcription of human telomerase reverse transcriptase gene. Biochem Biophys Res Commun 2012;417:630–634. [DOI] [PubMed] [Google Scholar]
- 81.Yamashita S, Ogawa K, Ikei T, Fujiki T, Katakura Y.. FOXO3a potentiates hTERT gene expression by activating c-MYC and extends the replicative life-span of human fibroblast. PLoS One 2014;9:e101864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Narala SR, Allsopp RC, Wells TB, Zhang G, Prasad P, Coussens MJ, Rossi DJ, Weissman IL, Vaziri H.. SIRT1 acts as a nutrient-sensitive growth suppressor and its loss is associated with increased AMPK and telomerase activity. Mol Biol Cell 2008;19:1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Z, Shentu TP, Wen L, Johnson DA, Shyy JY.. Regulation of SIRT1 by oxidative stress-responsive miRNAs and a systematic approach to identify its role in the endothelium. Antioxid Redox Signal 2013;19:1522–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, Zeiher AM, Dimmeler S.. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev 2007;21:2644–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guarani V, Deflorian G, Franco CA, Kruger M, Phng LK, Bentley K, Toussaint L, Dequiedt F, Mostoslavsky R, Schmidt MHH, Zimmermann B, Brandes RP, Mione M, Westphal CH, Braun T, Zeiher AM, Gerhardt H, Dimmeler S, Potente M.. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature 2011;473:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Houtkooper RH, Pirinen E, Auwerx J.. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 2012;13:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mátrai J, Chuah MK, VandenDriessche T.. Recent advances in lentiviral vector development and applications. Mol Ther 2010;18:477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aguado J, Sola-Carvajal A, Cancila V, Revêchon G, Ong PF, Jones-Weinert CW, Wallén Arzt E, Lattanzi G, Dreesen O, Tripodo C, Rossiello F, Eriksson M, d'Adda di Fagagna F.. Inhibition of DNA damage response at telomeres improves the detrimental phenotypes of Hutchinson-Gilford progeria syndrome. Nat Commun 2019;10:4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw sequence data for the samples used in this study are available on the National Center for Biotechnology Information Sequence Read Archive database: Bioproject PRJNA730422.