This cohort study categorizes tumors of the head and neck and the cervix by human papillomavirus (HPV) positivity status and compares their immunogenomic landscapes and associations with survival.
Key Points
Question
Is tumor site associated with the immunogenomic landscape of human papillomavirus (HPV)–related tumors and with site-specific survival differences among HPV-positive patients?
Findings
In this cohort study of 768 patients, in contrast to HPV-positive oropharyngeal tumors, HPV-positive nonoropharyngeal (head and neck or cervical) tumors were not associated with greater immune activation markers and cytolytic activity compared with HPV-negative nonoropharyngeal tumors. The study found no significant difference in survival between patients with HPV-positive vs HPV-negative nonoropharyngeal cancer.
Meaning
The findings suggest that tumor site is associated with the immune landscape and improved survival in HPV-positive oropharyngeal but not nonoropharyngeal tumors, providing a rationale for stratifying HPV-associated tumors by site.
Abstract
Importance
Human papillomavirus (HPV)–positive status in patients with oropharyngeal squamous cell carcinoma (OPSCC) is associated with improved survival compared with HPV-negative status. However, it remains controversial whether HPV is associated with improved survival among patients with nonoropharyngeal and cervical squamous cell tumors.
Objective
To investigate differences in the immunogenomic landscapes of HPV-associated tumors across anatomical sites (the head and neck and the cervix) and their association with survival.
Design, Setting, and Participants
This cohort study used genomic and transcriptomic data from the Cancer Genome Atlas (TCGA) for 79 patients with OPSCC, 435 with nonoropharyngeal head and neck squamous cell carcinoma (non-OP HNSCC), and 254 with cervical squamous cell carcinoma and/or endocervical adenocarcinoma (CESC) along with matched clinical data from TCGA. The data were analyzed from November 2020 to March 2021.
Main Outcomes and Measures
Positivity for HPV was classified by RNA-sequencing reads aligned with the HPV reference genome. Gene expression profiles, immune cell phenotypes, cytolytic activity scores, and overall survival were compared by HPV tumor status across multiple anatomical sites.
Results
The study comprised 768 patients, including 514 (66.9%) with HNSCC (380 male [73.9%]; mean [SD] age, 59.5 [10.8] years) and 254 (33.1%) with CESC (mean [SD] age, 48.7 [14.1] years). Human papillomavirus positivity was associated with a statistically significant improvement in overall survival for patients with OPSCC (adjusted hazard ratio [aHR], 0.06; 95% CI, 0.02-0.17; P < .001) but not for those with non-OP HNSCC (aHR, 0.64; 95% CI, 0.31-1.27; P = .20) or CESC (aHR, 0.50; 95% CI, 0.15-1.67; P = .30). The HPV-positive OPSCCs had increased tumor immune infiltration and immunomodulatory receptor expression compared with HPV-negative OPSCCs. Compared with HPV-positive non-OP HNSCCs, HPV-positive OPSCCs showed greater expression of immune-related metrics including B cells, T cells, CD8+ T cells, T-cell receptor diversity, B-cell receptor diversity, and cytolytic activity scores, independent of tumor variant burden. The immune-related metrics were similar when comparing HPV-positive non-OP HNSCCs and HPV-positive CESCs with their HPV-negative counterparts. The 2-year overall survival rate was significantly higher for patients with HPV-positive OPSCC compared with patients with HPV-negative OPSCC (92.0% [95% CI, 84.8%-99.9%] vs 45.8% [95% CI, 28.3%-74.1%]; HR, 0.10 [95% CI, 0.03-0.30]; P = .009).
Conclusions and Relevance
In this cohort study, tumor site was associated with the immune landscape and survival among patients with HPV-related tumors despite presumed similar biologic characteristics. These tumor site–related findings provide insight on possible outcomes of HPV positivity for tumors in oropharyngeal and nonoropharyngeal sites and a rationale for the stratification of HPV-associated tumors by site and the subsequent development of strategies targeting immune exclusion in HPV-positive nonoropharyngeal cancer.
Introduction
In the US, approximately 35 000 new cases of human papillomavirus (HPV)–associated cancers are diagnosed each year, including cervical, head and neck, anal, vulvovaginal, and penile cancers.1 The incidence of HPV-associated head and neck cancers has been steadily increasing in North America and Europe2,3 owing primarily to oropharyngeal squamous cell cancers, 75% of which are now HPV positive in the US.4 Human papillomavirus status is a prognostic biomarker for oropharyngeal squamous cell carcinoma (OPSCC),5 and National Comprehensive Cancer Network guidelines6 recommend the use of HPV status to determine American Joint Committee on Cancer staging because HPV-positive OPSCC is now considered a distinct entity from HPV-negative OPSCC.6,7 Whether HPV is a survival-favorable factor in cancer sites outside the oropharynx remains controversial; testing of nonoropharyngeal head and neck squamous cell carcinoma (non-OP HNSCC) tumors for HPV tumor status is currently not recommended by the National Comprehensive Cancer Network,6 College of American Pathologists,8 or American Society of Clinical Oncology.9
The rate of HPV positivity in SCC arising within nonoropharyngeal sites, including the oral cavity, larynx, hypopharynx, and nasopharynx, varies from 3% to 18.5% depending on the detection type; however, this rate is consistently lower compared with OPSCC.10,11,12 Detection of p16, a surrogate of HPV positivity in the oropharynx,9 does not appear to be concordant with direct HPV testing (in situ hybridization and polymerase chain reaction) in non-OP HNSCC,13 suggesting distinct biologic characteristics; therefore, it may not be suitable for determining HPV status.14,15 To accurately ascertain HPV status, the quantification of HPV mRNA transcript levels based on next generation sequencing may more properly classify HPV tumor positivity irrespective of site because it directly measures HPV oncogene expression in tumors.
Some studies have shown a survival benefit for patients with p16-positive non-OP HNSCC compared with those with p16-negative non-OP HNSCC but not for patients with positive in situ hybridization results,16,17 whereas other studies have shown no survival benefit with either mode of detection.4,18,19 One study suggests that survival among patients with HPV-positive non-OP HNSCC is inferior to that among patients with HPV-positive OPSCC and similar to that among patients with HPV-negative non-OP HNSCC.20 In addition, emerging data support the existence of HPV-negative cervical cancer based on the lack of HPV RNA transcript in some tumors.21,22 However, similar to non-OP HNSCC, the effects of HPV positivity in cervical cancer are unclear.23,24 Therefore, clarification of the prognostic role of HPV outside the oropharynx is needed to better understand why HPV positivity in squamous cell tumors may be differentially associated with survival among patients with tumors in adjacent sites.
Site-specific microenvironments, such as tonsillar lymphoid tissues and oral tongue mucosa or cervical epithelium, may underlie observed survival differences. Of note, the survival benefit observed among patients with HPV-positive OPSCC may be attributed to E6 and E7, which are recognized as foreign antigens by the immune system.25,26 We hypothesized that each tumor site would show variations in immune infiltration and would subsequently influence the immunogenicity of HPV antigen-expressing tumors. We uniquely evaluated HPV viral RNA transcript levels with immune cell subpopulations, activation and exhaustion markers, cytolytic activity, and clinical data to better understand the immunologic basis for the survival differences among patients with HPV-positive OPSCC, those with HPV-positive non-OP HNSCC, and those with cervical squamous cell carcinoma and/or endocervical adenocarcinoma (CESC). Our analyses focused on HPV-positive and HPV-negative OPSCC compared with non-OP HNSCC, but we also analyzed HPV-positive and HPV-negative cervical tumors as a relevant comparator arm of HPV-positive tumors outside the head and neck.
Methods
Patient and Data Sources
In this cohort study, transcript mRNA expression data collected from 514 patients with primary HNSCC and 254 with primary CESC were downloaded from the Cancer Genome Atlas (TCGA) portal27 and were analyzed from November 2020 to March 2021. These data were gathered from publicly available data sets and did not require institutional review board approval or patient informed consent per the Code of Federal Regulations (45 CFR 46.102). This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
All patients with exome, transcriptome, and clinical data were included. Samples without available RNA sequence data and patients with recurrent or metastatic carcinoma were excluded. Clinical parameters, including survival information, sex, and race and ethnicity, were acquired from TCGA’s Clinical Data Resource data set.28 We used TNM staging according to the AJCC Cancer Staging Manual, 7th edition, to maintain consistency for all patients with HNSCC and CESC. Data on p16 immunohistochemistry positivity was retrieved from the original TCGA data set as annotated by the study pathologists.29 Human papillomavirus RNA transcript reads, viral genotypes, and oncogenic types were retrieved from Campbell et al30 with RNA alignment using a pipeline established by the British Columbia Cancer Research Institute’s Genome Sciences Centre (BC).21 Criteria for HPV-positive status were such that proportional reads per million were greater than or equal to 0.22, as derived from the BC pipeline, consistent with the study by Ren et al.31
A 2-dimensional reduction visualization based on 2630 immune-related transcripts32 in patients with HNSCC and CESC was produced by TumorMap33 and faceted by clinical parameters. We deconvoluted the proportions of tumor-infiltrating immune and stromal cell populations from bulk RNA sequencing data using MCP-counter34 and CIBERSORT35 algorithms. T-cell receptor (TCR) and B-cell receptor (BCR) diversity, measured using Shannon entropy, and copy number variation burden in terms of the number of segments and fraction of genome alterations were acquired from Thorsson et al.32 Tumor variant burden, aggregated by silent and nonsilent variants, was generated by Maftools36 from the Pan-Cancer Atlas variant files.37
Statistical Analysis
The Wilcoxon test was used to compare continuous variables between groups. The effect size of HPV RNA transcript levels between p16-positive and p16-negative patients was estimated by the rank-biserial correlation coefficient (RBC) using the effectsize package of R, version 4.0.1 (R Project for Statistical Computing).38,39 The RBC value was interpreted as published previously (RBC <0.2, very weak; 0.2≤ RBC <0.4, weak; 0.4≤ RBC <0.6, moderate; 0.6≤ RBC <0.8, strong; and RBC ≥0.8, very strong).40 Generation and analysis of the receiver operating characteristic curve were performed using the pROC package of R. The Kaplan-Meier method was used to generate unadjusted survival curves and risk tables. Cox proportional hazards regression models were used for multivariable analyses adjusting for age, sex, smoking history, and TNM stage. Hazard ratios (HRs) were used to represent the risk for death. Propensity score matching, performed using the MatchIt package of R, was used to confirm results from the multivariable analysis models as a sensitivity analysis to mitigate differences in the proportions of HPV-positive and HPV-negative patients in each group. Propensity score matching is a well-studied method of matching patients in different groups to minimize confounding bias.41 Furthermore, propensity score matching was used to help reduce bias owing to an imbalance in the proportion of HPV-positive patients in each group by matching patients during the analysis, making comparisons more reliable. All tests were 2-sided with statistical significance set at P < .05. All data analyses were performed using R, version 4.0.1. All packages and source codes used are available at GitHub.42
Results
The study comprised a total of 768 patients, including 514 (66.9%) with HNSCC (mean [SD] age, 59.5 [10.8] years) and 254 (33.1%) with CESC (mean [SD] age, 48.7 [14.1] years). The demographic and clinical characteristics of the study population are summarized in the eTable in the Supplement. Most of the patients with HNSCC were male (380 [73.9%]) and White (439 [85.4%]); 75 (14.6%) were of other race and ethnicity. Of the patients with CESC, 170 (66.9%) were White and 84 (33.1%) were of other race and ethnicity. The categories for other race and ethnicity included Asian, Black or African American, American Indian or Alaska native, and Native Hawaiian or Pacific Islander; the number of patients in each category was too few to include in the comparisons. Among all HNSCCs, 53 of the 79 OPSCCs (67.1%) were HPV positive, whereas 27 of the 435 non-OP HNSCCs (6.2%) were HPV positive. The median age of patients with HPV-positive OPSCC was 56 years (range, 35-77 years) compared with 61 years (range, 49-82 years) among patients with HPV-positive non-OP HNSCC. Ten of the 254 patients with CESC (3.93%) had HPV-negative disease; the median age in this group was 57 years (range, 34-81 years), 11 years older than in the group with HPV-positive CESC (median age, 46 years; range, 20-88 years). The range of HPV transcript levels was 0.40 to 857.22 reads per million for HPV-positive OPSCCs, 0.22 to 805.53 reads per million for non-OP HNSCCs, and 0.48 to 936.32 reads per million for CESCs.
Association of p16 With HPV Positivity in Non-OP HNSCCs
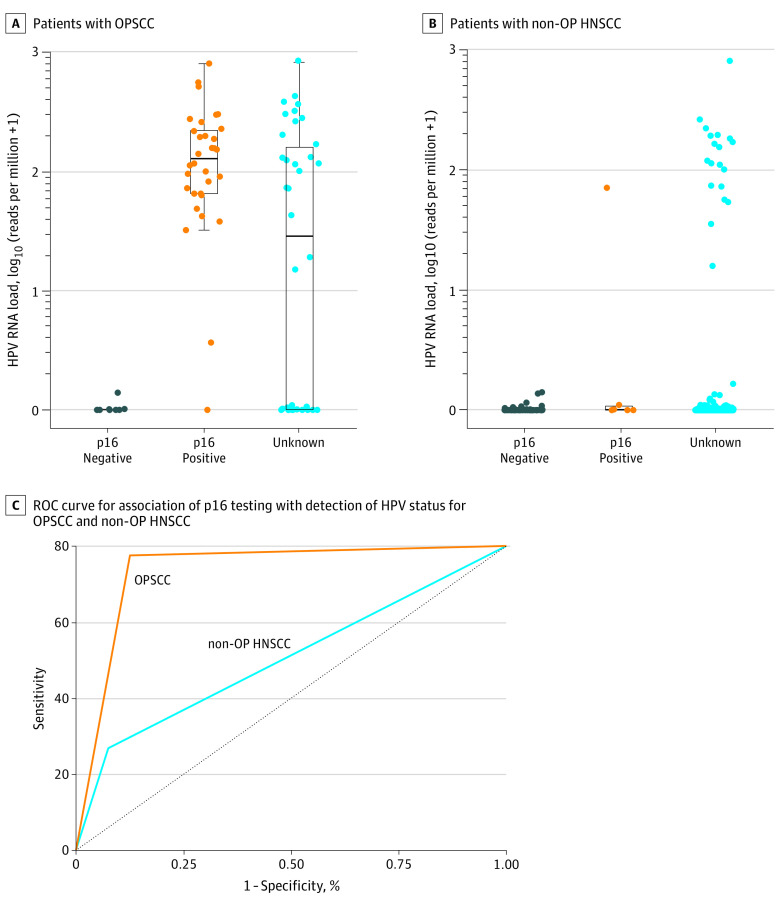
Of the total study population, p16 status was available for 110 patients with HNSCC (21.4%). In this subset, we examined the concordance between p16 with HPV RNA transcription and available immunohistochemistry results. Among these 110 patients, 38 (34.5%) were p16 positive, including 32 with OPSCC (29.1%) and 6 with non-OP HNSCC (5.4%). There was a substantial increase in HPV RNA transcript levels between patients with p16-positive OPSCC and those with p16-negative OPSCC (RBC, 0.96; 95% CI, 0.90-0.98) (Figure 1A). However, there was only a small increase in HPV RNA transcript levels between patients with p16-positive non-OP HNSCC and those with p16-negative non-OP HNSCC (RBC, 0.32; 95% CI, –0.16 to 0.67) (Figure 1B), including oral cavity and laryngeal subsites (eFigure 1 in the Supplement). Furthermore, 5 of the 6 patients (83.3%) with p16-positive non-OP HNSCC did not express HPV E6 and E7 oncogenic transcripts (eFigure 2 in the Supplement).
Figure 1. p16 As a Biomarker for Human Papillomavirus (HPV) Status in Patients With Oropharyngeal Squamous Cell Carcinoma (OPSCC) and Nonoropharyngeal Head and Neck Squamous Cell Carcinoma (Non-OP HNSCC).
A, The effect size was 0.96 (95% CI, 0.90-0.98; P < .001) for differences between p16-negative and p16-positive values. B, The effect size was 0.32 (95% CI, –0.16 to 0.67; P = .09) for differences between p16-negative and p16-positive values. Boxes and whiskers indicate the HPV RNA load. ROC indicates receiver operating characteristic.
The receiver operating characteristic curve showed that although p16 immunohistochemistry had high sensitivity for detecting HPV status in OPSCC, the sensitivity was lower in non-OP HNSCC (Figure 1C). Each of the relevant performance properties was improved for OPSCC compared with non-OP HNSCC (precision rate or positive predictive value: 96.88% vs 16.67%; sensitivity: 96.88% vs 33.33%), implying that p16 staining may be an acceptable surrogate marker for HPV positivity in OPSCC but is inadequate in non-OP HNSCC. Therefore, in this analysis, the HPV RNA transcript level was used to classify both OPSCC and non-OP HNSCC tumors as HPV positive.
HPV Positivity and Survival Among Patients With Tumors Outside the Oropharynx
The median survival time and adjusted hazard ratios (aHRs) for patients are summarized by cancer site in the Table. The median follow-up time was 21.6 months (IQR, 11.6-41.4 months) for patients with CESC and 21.5 months (IQR, 12.8-39.7 months) for patients with HNSCC. Head and neck squamous cell carcinoma was further categorized into OPSCC (median follow-up time, 21.2 months; IQR, 15.7-34.1 months) and non-OP HNSCC (median follow-up time, 21.9 months; IQR, 12.6-41.1 months). In the Kaplan-Meier analysis (Figure 2), the 2-year overall survival (OS) rate was significantly higher for patients with HPV-positive OPSCC compared with patients with HPV-negative OPSCC (92.0% [95% CI, 84.8%-99.9%] vs 45.8% [95% CI, 28.3%-74.1%]; HR, 0.10 [95% CI, 0.03-0.30]; P = .009). After adjusting for age, sex, smoking history, and TNM stage, HPV positivity was found to be independently associated with reduced risk of death among patients with OPSCC (aHR, 0.06; 95% CI, 0.02-0.17; P < .001) (Table).
Table. Overall Survival Among Patients With OPSCC, Non-OP HNSCC, and CESC by HPV Status.
| HPV status | Patients, No. (%) | aHR (95% CI)a | P value | Median survival time, mob | Estimated 2-y survival rate, % (95% CI) |
|---|---|---|---|---|---|
| OPSCC (n = 79) | |||||
| HPV negative | 26 (32.9) | 1 [Reference] | <.001 | 20.1 | 45.8 (28.3-74.1) |
| HPV positive | 53 (67.1) | 0.06 (0.02-0.17) | >58.3 | 92.0 (84.8-99.9) | |
| Non-OP HNSCC (n = 435) | |||||
| HPV negative | 408 (93.8) | 1 [Reference] | .20 | 53.1 | 64.3 (59.5-69.4) |
| HPV positive | 27 (6.2) | 0.64 (0.31-1.27) | NA | 72.6 (57.0-92.4) | |
| CESC (n = 254) | |||||
| HPV negative | 10 (3.9) | 1 [Reference] | .30 | 23.8 | 46.7 (21.0-100) |
| HPV positive | 244 (96.1) | 0.50 (0.15-1.67) | NA | 81.9 (76.4-87.8) |
Abbreviations: aHR, adjusted hazard ratio; CESC, cervical squamous cell carcinoma; HPV, human papillomavirus; NA, not assessed; non-OP HNSCC, nonoropharyngeal head and neck squamous cell carcinoma; OPSCC, oropharyngeal squamous cell carcinoma.
Adjusted HRs from Cox proportional hazards regression models were adjusted for age, sex, smoking history, and TNM stage for patients with OPSCC and non-OP HNSCC and for age, smoking history, and TNM stage for patients with CESC.
The longest survival times were beyond the longest surveillance follow-up time available; therefore, the 95% CIs are not provided.
Figure 2. Probability of Survival as a Function of Human Papillomavirus (HPV) Status by Tumor Site.
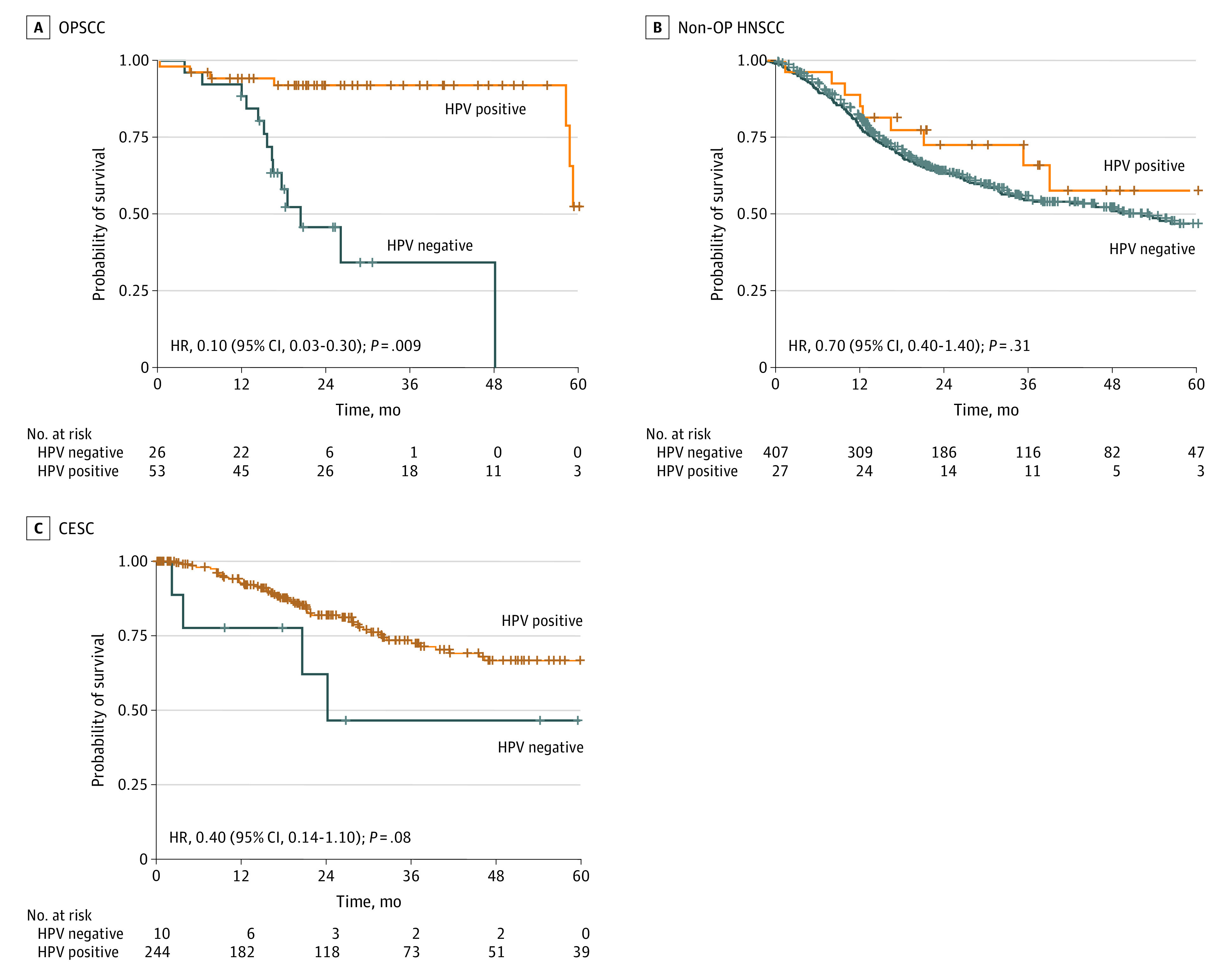
Unadjusted hazard ratios (HRs) with 95% CIs are from the Cox proportional hazards regression models. Hash marks indicate censored data. CESC indicates cervical squamous cell carcinoma; non-OP HNSCC, nonoropharyngeal head and neck squamous cell carcinoma; OPSCC, oropharyngeal squamous cell carcinoma.
Outside the oropharynx, although OS was higher among HPV-positive patients than among HPV-negative patients, there was no statistically significant difference given the wide 95% CIs. Among patients with non-OP HNSCCs, the 2-year OS rate was statistically similar for HPV-positive patients (72.6%; 95% CI, 57.0%-92.4%) and HPV-negative patients (64.3%; 95% CI, 59.5%-69.4%) (HR, 0.70; 95% CI, 0.40-1.40; P = .31). Similarly, among patients with CESC, the 2-year OS rate was higher for HPV-positive patients, although the magnitude of the difference between HPV-positive patients and HPV-negative patients was larger (81.9% [95% CI, 76.4%-87.8%] vs 46.7% [95% CI, 21.0%-100%]); 95% CIs for these estimates were wide, and the difference was not significant (HR, 0.40; 95% CI, 0.14-1.10). After adjustment for other clinically relevant factors, associations between HPV and OS were not significant, with wide 95% CIs (non-OP HNSCC: aHR, 0.64 [95% CI, 0.31-1.27]; P = .20; CESC: aHR, 0.50 [95% CI, 0.15-1.67]; P = .30). Providing further support for this finding, an additional survival analysis was performed after propensity score matching and yielded similar results (eFigure 3 in the Supplement).
Immune Phenotypes of HPV-Positive and HPV-Negative Tumors Outside the Oropharynx
To compare the global immune landscape between HPV-positive and HPV-negative tumors across anatomical sites, we interrogated the expression data from 2630 immune-related transcripts in HNSCC and CESC tumor samples. The global immune landscape of HPV-positive OPSCCs was markedly separated from that of HPV-negative OPSCCs (Figure 3), which is consistent with their distinct prognosis. However, an analogous separation of immune landscapes between HPV-positive and HPV-negative non-OP HNSCCs and CESCs was not observed. Most of the HPV-positive non-OP HNSCCs and CESCs were immunophenotypically similar to their HPV-negative counterparts. We also confirmed that HPV subtypes, sex, and smoking history did not significantly confound these results (eFigure 4 in the Supplement).
Figure 3. Immune Transcript Landscape of Patients With Head and Neck Squamous Cell Carcinoma (HNSCC) and Cervical Squamous Cell Carcinoma (CESC) by Anatomical Site.
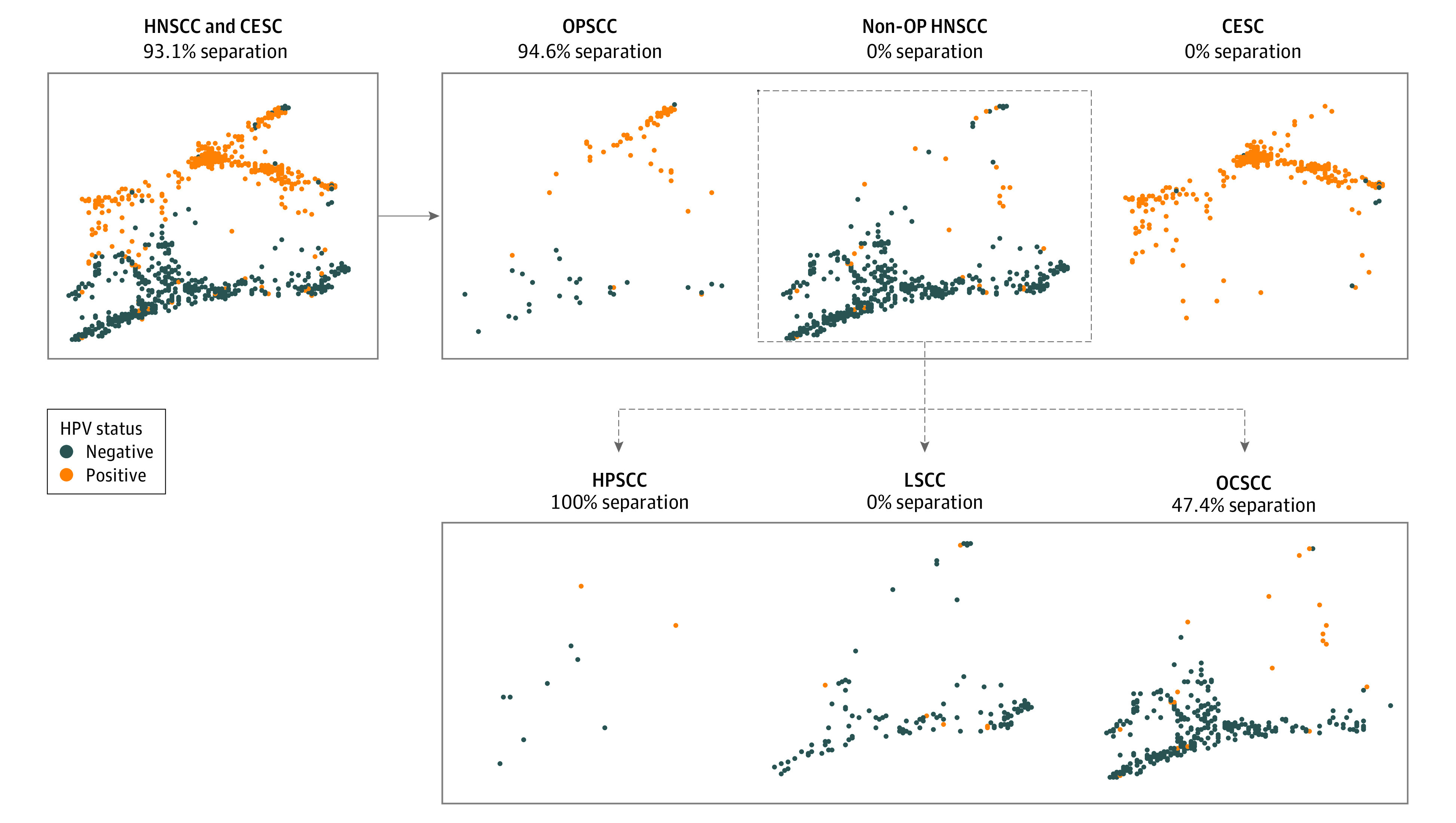
The reduced visualization is based on 2630 immune-related transcripts in patients with HNSCC and CESC, produced by TumorMap33 and representing the global landscape across all sites (upper left); oropharyngeal squamous cell carcinoma (OPSCC), nonoropharyngeal (non-OP) HNSCC, and CESC (upper right); and non-OP HNSCC split into hypopharyngeal squamous cell carcinoma (HPSCC), laryngeal squamous cell carcinoma (LSCC), and oral cavity squamous cell carcinoma (OCSCC) (lower right). Separation percentages represent the proportion of human papillomavirus (HPV)–positive patients quantitatively separated from HPV-negative patients.
Tumor-Infiltrating Immune Subpopulations and Immune Checkpoint Signatures in HPV-Positive and HPV-Negative Tumors Within Anatomical Sites
Estimations of B cells, T cells, CD8+ T cells, TCR diversity, BCR diversity, plasma cells, naive B cells, the ratio of CD8+ T cells to Treg cells, M2-macrophages, and cancer-associated fibroblasts were compared among HPV-positive and HPV-negative OPSCCs, non-OP HNSCCs, and CESCs (eFigures 5-7 in the Supplement). Significant differences were observed in each of the immune markers when comparing HPV-positive and HPV-negative OPSCCs. Of interest, HPV-positive OPSCCs had a significantly lower mean tumor variant and copy number variant burden compared with HPV-negative OPSCCs (eFigure 8 in the Supplement), suggesting that viral antigens may be important in eliciting immunogenicity in these tumors. By contrast, when comparing HPV-positive and HPV-negative tumors outside the oropharynx, there were no significant differences in expression levels of various immune markers (eFigures 5-8 in the Supplement). In addition, no significant differences in cytolytic activity (CYT) scores were observed between HPV-positive and HPV-negative tumors outside the oropharynx (eFigure 5 in the Supplement). This indicates that HPV-positive non-OP HNSCC and CESC tumors may lack the increased tumor cytolytic activity seen in HPV-positive OPSCC tumors compared with their HPV-negative counterparts.
We analyzed the differentially expressed immunomodulatory checkpoint receptors and their cognate ligands by HPV status within each anatomical site (eFigure 5 in the Supplement). There was no significant enrichment in immune activation and exhaustion markers among HPV-positive and HPV-negative tumors in nonoropharyngeal sites, suggesting that HPV positivity outside the oropharynx contributes minimally to the immune landscape.
Differences in Immune Landscapes Across HPV-Associated Malignancies
We sought to compare the immune landscapes between HPV-associated cancers across various anatomical sites. To extract these immune characteristics, we compared the tumor-infiltrating immune cell populations in HPV-positive cancers among the oropharynx, nonoropharynx, and cervical sites. Compared with HPV-positive OPSCCs, in HPV-positive non-OP HNSCCs and CESCs, there were significantly lower estimated scores of B cells (385.6 vs 52.3 vs 52.8) and CD8+ T cells (265.2 vs 124.6 vs 175.3) but higher estimated scores of cancer-associated fibroblasts (109.0 vs 162.7 vs 110.8) and M2 macrophages (12.8 vs 18.7 vs 15.7) (Figure 4). We also found that HPV-positive non-OP HNSCCs and CESCs had lower TCR diversity, BCR diversity, and CYT scores, but we found no significant differences in tumor variant burden across tumors.
Figure 4. Immune Microenvironment Composition by Anatomical Site.
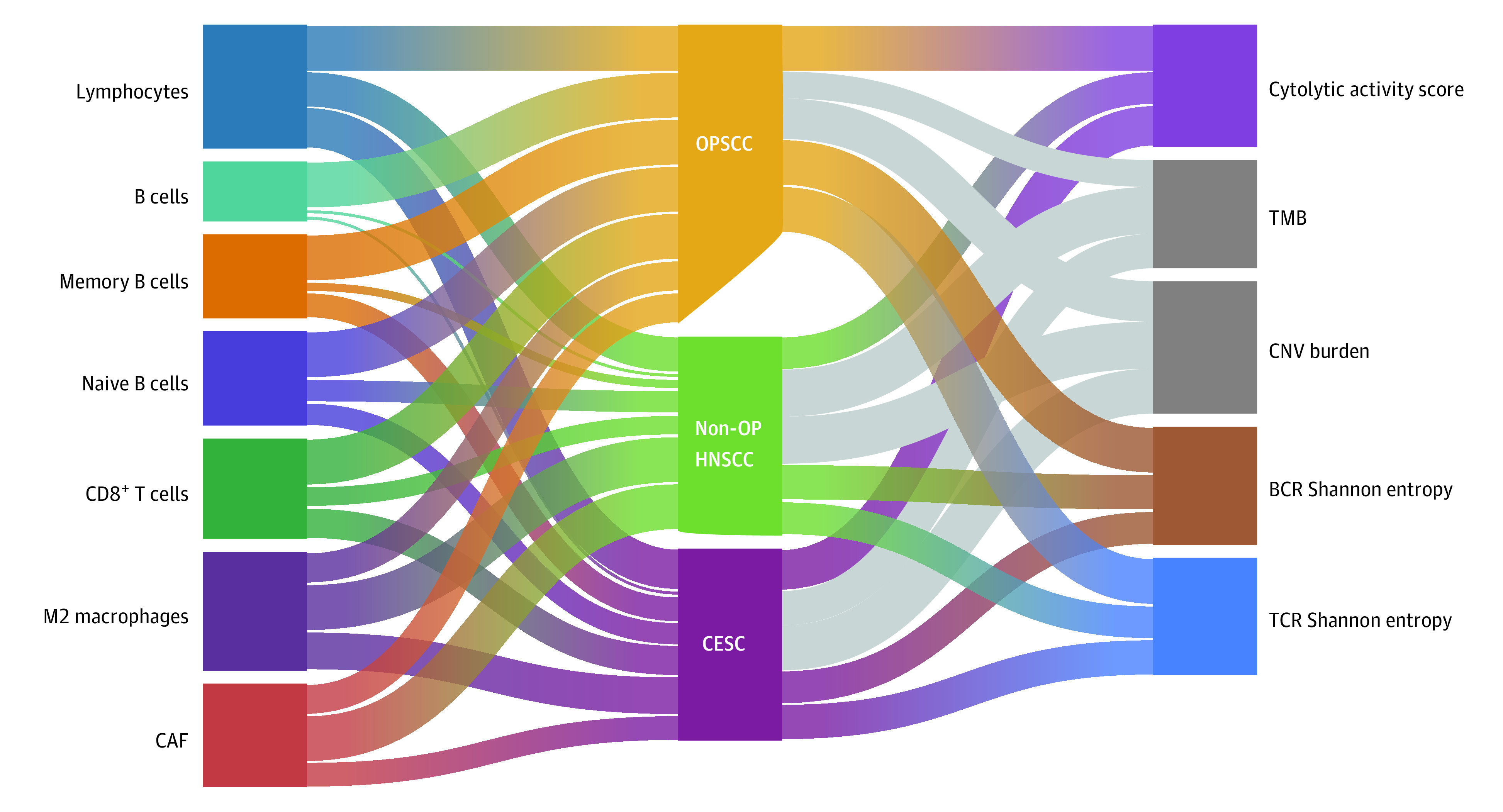
The Sankey diagram shows the comparatively larger proportion of B cells, memory B cells, CD8+ T cells, naive B cells, and lymphocytes associated with oropharyngeal tumors compared with tumors in the cervix and nonoropharyngeal sites. Similar trends are seen for immune diversity metrics including B-cell receptor (BCR) and T-cell receptor (TCR) diversity, measured using Shannon entropy, and the cytolytic activity score, which are preferentially associated with oropharyngeal tumor sites. Gray indicates associations that were not statistically significant. CAF indicates cancer-associated fibroblast; CESC, cervical squamous cell carcinoma; CNV, copy number variation; non-OP HNSCC, nonoropharyngeal head and neck squamous cell carcinoma; OPSCC, oropharyngeal squamous cell carcinoma; and TMB, tumor mutation burden.
Discussion
To our knowledge, this study provides the first in-depth immunophenotypic analysis across anatomical sites using HPV RNA transcript levels of tumors to uniquely classify HPV status and characterize the association of site with the tumor immunogenomic landscape. We hypothesized that tumor site differences could contribute to the differential survival observed among patients with HPV-associated tumors of the oropharynx compared with those with tumors outside the oropharynx. Furthermore, to overcome barriers involving p16 misclassification of HPV status, we used HPV RNA transcript data to classify HPV-positive OPSCC, non-OP HNSCC, and CESC. We coupled these data with immune cell phenotypes, cytolytic activity scores, and exhaustion and activation phenotypes to obtain a granular understanding of why HPV positivity appears to have a differential association with survival among patients with tumors outside the oropharynx.
The unclear association of HPV with non-OP HNSCC may be partially attributable to variable HPV testing methods and nonuniform statistical approaches applied in previous retrospective studies.4,10,16 To overcome these limitations, we defined HPV status using the RNA sequencing viral transcript load and found that although p16 may be an acceptable surrogate marker for HPV positivity in OPSCC, in this study it was an unreliable surrogate marker in non-OP HNSCC. Given this study’s finding of a lower positive predictive value of p16 in non-OP HNSCC, HPV-positive status may have been overestimated in previous non-OP HNSCC studies that relied on p16 positivity.4,10,16 Overall, this study’s findings support the notion that although patients with p16-negative non-OP HNSCC are likely to be HPV negative, patients with p16-positive non-OP HNSCC may not necessarily be HPV positive.14,15 The conditions that lead to these site-specific differences in aberrant p16 positivity are not fully understood; however, aside from the canonical p16-CDK4/6-HPV E7-Rb axis, proposed mechanisms may involve the dysregulation of p16 expression in chromosomally unstable non-OPSCC tumors.43,44,45
Using a unique HPV status classification, we observed a survival benefit associated with HPV among patients with OPSCC only. We did not find a conclusive difference in survival between patients with HPV-positive non-OP HNSCC and patients with HPV-negative non-OP HNSCC, consistent with previous studies that have used methods such as in situ hybridization rather than p16.12 Regardless, this survival benefit appeared to differ based on tumor site, implying that the tumor site should be considered when evaluating the significance of HPV positivity outside the oropharynx. This study’s results may contribute important knowledge regarding the biological diversity of various HPV-positive tumor sites and may provide a rationale for why HPV-associated prognostic benefit is more definitively seen in patients with OPSCC. Of importance, the mechanism by which HPV is associated with survival may be intimately related to the interplay between anatomical site and immune activation or lack thereof.
We also explored whether site-specific survival differences among patients with HPV-positive tumors could be explained by differences in the immune landscapes. Previous work has indicated that HPV-associated head and neck cancers share similar genetic characteristics across anatomical sites.46 However, regarding their immune profiles, HPV-positive tumors in the current study were starkly different across sites, which suggests that the HPV status of tumors outside the oropharynx might not elicit a strong immunogenic response compared with oropharyngeal tumors. Compared with HPV-negative OPSCCs, HPV-positive OPSCCs showed increased antitumor immune parameters such as cytolytic score and CD8+ T cell infiltration and had fewer immunosuppressive factors such as cancer-associated fibroblasts. Therefore, the study’s data suggest that HPV positivity alone may be insufficient in eliciting a strong antitumor immune response and that tumor site-dependent factors such as the presence or absence of favorable lymphovascular and stromal components may have a critical role in altering immunogenicity in these tumors. The favorable lymphovascular histologic characteristics of the oropharynx may explain why HPV positivity in oropharyngeal tumors is associated with stronger immune responses compared with HPV positivity in tumors outside the oropharynx.47
This study found increased expression of immune checkpoint receptors in HPV-positive OPSCCs, supporting the notion that HPV-positive OPSCCs may be more sensitive to therapeutic immunomodulation than HPV-negative OPSCCs.48 The upregulation of these immunosuppressive receptors in the HPV-positive tumor microenvironment may be associated with an immune-activated microenvironment. In contrast, the study found no significant differences in expression of these immune checkpoint receptors when comparing HPV-positive and HPV-negative oral cavity, larynx, and cervical tumors. Thus, nonoropharyngeal sites may have an immune-excluded microenvironment where host immunity cannot efficiently infiltrate and engage with immunogenic tumor antigens and may ultimately fail to achieve an activated state. This study’s findings of immune checkpoint overexpression in HPV-positive OPSCC are consistent with the findings of studies that have indicated that HPV-positive HNSCC had higher objective response rates to anti–PD-1/PD-L1 therapy compared with HPV-negative tumors, although not all studies have shown this difference in response.49,50,51,52 These findings provide a rationale for investigating agents targeting immunosuppressive modulators, such as antibodies against CTLA-4, TIGIT, BTLA, and TIM-3, as adjuncts to anti–PD-1 in the treatment of HPV-positive OPSCC. Furthermore, this study’s data imply that immunotherapy may not be as effective in treating HPV-positive non-OP HNSCC and CESC owing to immune exclusion in these tumors. Future trials that examine the reversal of immune exclusion and take anatomic site and HPV status into consideration may help elucidate the lack of immune activation in HPV-positive nonoropharyngeal tumors.
Limitations
This study has limitations. The HPV-positive classification based on RNA sequencing data was highly sensitive and accurate; however, we cannot exclude the possibility of a false-positive, albeit low, error rate owing to sequencing artifacts. In addition, mRNA data might have minor discrepancies with protein-level data. It is possible that owing to the small number of patients with HPV-positive non-OP HNSCC and HPV-negative CESC, the study may have been underpowered to detect clinically meaningful survival differences by HPV status for patients with non-OP HNSCC and CESC, as suggested by the wide 95% CIs. Although important information about trends and associations could still be generated, the granularity of the findings could be improved with larger numbers in future studies. This would lead to greater precision in the survival estimates generated and could lead to the ability to make more definitive conclusions. Given the observational nature of the study, direct causality cannot be inferred from the results. Comorbidities are often associated with overall survival for patients with cancer. Unfortunately, measures of overall comorbidity were not captured in the original TCGA Clinical Data Resource data set and therefore could not be included in our analyses.
Conclusions
In this cohort study, the immunogenicity and resultant prognostic benefits associated with HPV positivity appeared to be dependent on tumor site, although this association must be confirmed in studies with larger sample sizes. Specifically, in contrast to HPV-positive OPSCCs, HPV-positive non-OP HNSCC tumors had an attenuated immune microenvironment similar to that of HPV-negative non-OP HNSCCs. These findings provide insight about HPV positivity in tumors in oropharyngeal and nonoropharyngeal sites and may provide a rationale for the development of immune-based therapies to overcome potential immune exclusion in HPV-positive non-OP HNSCCs.
eTable. The characterization of HNSCC and cervical cancer patients in TCGA by HPV status
eFigure 1. p16 Is a Less Reliable Biomarker for HPV Status in OCSCC and LSCC Patients
eFigure 2. The Expressions of E6 and E7 Transcripts in OPSCCs and Non-OP HNSCCs by p16 Testing
eFigure 3. Overall Survival Differences Between Propensity-Score Matched Patients With HPV-Positive and -Negative Cancers
eFigure 4. A 2-Dimensional Reduced Visualization Based on 2630 Immune-related Transcripts in HNSCC and CESC Patients
eFigure 5. Comparative Analysis of Immunogenomic Metrics Between HPV-positive and -negative Patients Within Each Tumor Site—B Cells, CD8+ T Cells, and Cytolytic Activity Score
eFigure 6. Comparative Analysis of Immunogenomic Metrics Between HPV-positive and -negative Patients Within Each Tumor Site—Plasma Cells, Naive B Cells, CD8+ T/Treg Cells, BCR Diversity, and TCR Diversity
eFigure 7. Comparative Analysis Of Immunogenomic Metrics Between HPV-positive and -negative Patients Within Each Tumor Site—M2 Macrophages and CTF
eFigure 8. Comparative Analysis of Immunogenomic Metrics Between HPV-positive and -negative Patients Within Each Tumor Site—TMB and CNV
References
- 1.Saraiya M, Unger ER, Thompson TD, et al. ; HPV Typing of Cancers Workgroup . US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. J Natl Cancer Inst. 2015;107(6):djv086. doi: 10.1093/jnci/djv086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer Collaboration , Fitzmaurice C, Allen C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease study. JAMA Oncol. 2017;3(4):524-548. doi: 10.1001/jamaoncol.2016.5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294-4301. doi: 10.1200/JCO.2011.36.4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahal BA, Catalano PJ, Haddad RI, et al. Incidence and demographic burden of HPV-associated oropharyngeal head and neck cancers in the United States. Cancer Epidemiol Biomarkers Prev. 2019;28(10):1660-1667. doi: 10.1158/1055-9965.EPI-19-0038 [DOI] [PubMed] [Google Scholar]
- 5.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24-35. doi: 10.1056/NEJMoa0912217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Comprehensive Cancer Network. Head and Neck Cancers. Version 1.2021. Accessed November 9, 2020. https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf
- 7.Vokes EE, Agrawal N, Seiwert TY. HPV-associated head and neck cancer. J Natl Cancer Inst. 2015;107(12):djv344. doi: 10.1093/jnci/djv344 [DOI] [PubMed] [Google Scholar]
- 8.Lewis JS Jr, Beadle B, Bishop JA, et al. Human papillomavirus testing in head and neck carcinomas: guideline from the College of American Pathologists. Arch Pathol Lab Med. 2018;142(5):559-597. doi: 10.5858/arpa.2017-0286-CP [DOI] [PubMed] [Google Scholar]
- 9.Fakhry C, Lacchetti C, Perez-Ordonez B. Human papillomavirus testing in head and neck carcinomas: ASCO clinical practice guideline endorsement summary of the CAP guideline. J Oncol Pract. 2018;14(10):613-617. doi: 10.1200/JOP.18.00433 [DOI] [PubMed] [Google Scholar]
- 10.Tian S, Switchenko JM, Jhaveri J, et al. Survival outcomes by high-risk human papillomavirus status in nonoropharyngeal head and neck squamous cell carcinomas: a propensity-scored analysis of the National Cancer Data Base. Cancer. 2019;125(16):2782-2793. doi: 10.1002/cncr.32115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng Z, Hasegawa M, Kiyuna A, et al. Viral load, physical status, and E6/E7 mRNA expression of human papillomavirus in head and neck squamous cell carcinoma. Head Neck. 2013;35(6):800-808. doi: 10.1002/hed.23034 [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, Koo BS, Kang S, et al. HPV integration begins in the tonsillar crypt and leads to the alteration of p16, EGFR and c-myc during tumor formation. Int J Cancer. 2007;120(7):1418-1425. doi: 10.1002/ijc.22464 [DOI] [PubMed] [Google Scholar]
- 13.D’Souza G, Westra WH, Wang SJ, et al. Differences in the prevalence of human papillomavirus (HPV) in head and neck squamous cell cancers by sex, race, anatomic tumor site, and HPV detection method. JAMA Oncol. 2017;3(2):169-177. doi: 10.1001/jamaoncol.2016.3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seiwert TY. Ties that bind: p16 as a prognostic biomarker and the need for high-accuracy human papillomavirus testing. J Clin Oncol. 2014;32(35):3914-3916. doi: 10.1200/JCO.2014.57.9268 [DOI] [PubMed] [Google Scholar]
- 15.Fakhry C, Ferris RL. P16 as a prognostic biomarker for nonoropharyngeal squamous cell cancers: avatar or mirage? J Natl Cancer Inst. 2018;110(12):1290-1291. doi: 10.1093/jnci/djy103 [DOI] [PubMed] [Google Scholar]
- 16.Chung CH, Zhang Q, Kong CS, et al. p16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J Clin Oncol. 2014;32(35):3930-3938. doi: 10.1200/JCO.2013.54.5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Torabi SJ, Yarbrough WG, Mehra S, Osborn HA, Judson B. Association of human papillomavirus status at head and neck carcinoma subsites with overall survival. JAMA Otolaryngol Head Neck Surg. 2018;144(6):519-525. doi: 10.1001/jamaoto.2018.0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fakhry C, Westra WH, Wang SJ, et al. The prognostic role of sex, race, and human papillomavirus in oropharyngeal and nonoropharyngeal head and neck squamous cell cancer. Cancer. 2017;123(9):1566-1575. doi: 10.1002/cncr.30353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Souza G, Anantharaman D, Gheit T, et al. Effect of HPV on head and neck cancer patient survival, by region and tumor site: a comparison of 1362 cases across three continents. Oral Oncol. 2016;62:20-27. doi: 10.1016/j.oraloncology.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Windon MJ, D’Souza G, Waterboer T, et al. Risk factors for human papillomavirus-positive nonoropharyngeal squamous cell carcinoma. Head Neck. 2020;42(8):1954-1962. doi: 10.1002/hed.26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services ; et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378-384. doi: 10.1038/nature21386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodríguez-Carunchio L, Soveral I, Steenbergen RD, et al. HPV-negative carcinoma of the uterine cervix: a distinct type of cervical cancer with poor prognosis. BJOG. 2015;122(1):119-127. doi: 10.1111/1471-0528.13071 [DOI] [PubMed] [Google Scholar]
- 23.Kaliff M, Karlsson MG, Sorbe B, Bohr Mordhorst L, Helenius G, Lillsunde-Larsson G. HPV-negative tumors in a Swedish cohort of cervical cancer. Int J Gynecol Pathol. 2020;39(3):279-288. doi: 10.1097/PGP.0000000000000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz SM, Daling JR, Shera KA, et al. Human papillomavirus and prognosis of invasive cervical cancer: a population-based study. J Clin Oncol. 2001;19(7):1906-1915. doi: 10.1200/JCO.2001.19.7.1906 [DOI] [PubMed] [Google Scholar]
- 25.Bhatt KH, Neller MA, Srihari S, et al. Profiling HPV-16-specific T cell responses reveals broad antigen reactivities in oropharyngeal cancer patients. J Exp Med. 2020;217(10):e20200389. doi: 10.1084/jem.20200389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masterson L, Lechner M, Loewenbein S, et al. CD8+ T cell response to human papillomavirus 16 E7 is able to predict survival outcome in oropharyngeal cancer. Eur J Cancer. 2016;67:141-151. doi: 10.1016/j.ejca.2016.08.012 [DOI] [PubMed] [Google Scholar]
- 27.Broad Institute TCGA Genome Data Analysis Center . Firehose. Broad Institute of MIT and Harvard. Accessed October 2020. https://gdac.broadinstitute.org
- 28.Liu J, Lichtenberg T, Hoadley KA, et al. ; Cancer Genome Atlas Research Network . An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400-416.e11. doi: 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Network . Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576-582. doi: 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell JD, Yau C, Bowlby R, et al. ; Cancer Genome Atlas Research Network . Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018;23(1):194-212.e6. doi: 10.1016/j.celrep.2018.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren S, Gaykalova DA, Guo T, et al. HPV E2, E4, E5 drive alternative carcinogenic pathways in HPV positive cancers. Oncogene. 2020;39(40):6327-6339. doi: 10.1038/s41388-020-01431-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorsson V, Gibbs DL, Brown SD, et al. ; Cancer Genome Atlas Research Network . The immune landscape of cancer. Immunity. 2018;48(4):812-830.e14. doi: 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton Y, Novak AM, Swatloski T, et al. TumorMap: exploring the molecular similarities of cancer samples in an interactive portal. Cancer Res. 2017;77(21):e111-e114. doi: 10.1158/0008-5472.CAN-17-0580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453-457. doi: 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747-1756. doi: 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genomic Data Commons . PanCanAtlas publications. National Cancer Institute, National Institutes of Health. Accessed October 2020. https://gdc.cancer.gov/about-data/publications/pancanatlas
- 38.Tomczak M, Tomczak E. The need to report effect size estimates revisited: an overview of some recommended measures of effect size. Trends Sports Sci. 2014;1(21):19-25.
- 39.Ben Shachar M, Lüdecke D, Makowski D.. effectsize: estimation of effect size indices and standardized parameters aims of the package. J Open Source Softw. 2020;5:2815. doi: 10.21105/joss.02815 [DOI] [Google Scholar]
- 40.Evans JD. Straightforward Statistics for the Behavioral Sciences. Brooks/Cole; 1996. [Google Scholar]
- 41.Haukoos JS, Lewis RJ. The propensity score. JAMA. 2015;314(15):1637-1638. doi: 10.1001/jama.2015.13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.GitHub . Accessed March 2021. https://github.com/Mandallab/HNSCC-immunogenomic
- 43.Munger K, Gwin TK, McLaughlin-Drubin ME. p16 in HPV-associated cancers. Oncotarget. 2013;4(11):1864-1865. doi: 10.18632/oncotarget.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu DW, El-Mofty SK, Wang HL. Expression of p16, Rb, and p53 proteins in squamous cell carcinomas of the anorectal region harboring human papillomavirus DNA. Mod Pathol. 2003;16(7):692-699. doi: 10.1097/01.MP.0000077417.08371.CE [DOI] [PubMed] [Google Scholar]
- 45.Dickson MA. Molecular pathways: CDK4 inhibitors for cancer therapy. Clin Cancer Res. 2014;20(13):3379-3383. doi: 10.1158/1078-0432.CCR-13-1551 [DOI] [PubMed] [Google Scholar]
- 46.Chakravarthy A, Henderson S, Thirdborough SM, et al. Human papillomavirus drives tumor development throughout the head and neck: improved prognosis is associated with an immune response largely restricted to the oropharynx. J Clin Oncol. 2016;34(34):4132-4141. doi: 10.1200/JCO.2016.68.2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorta-Estremera S, Hegde VL, Slay RB, et al. Targeting interferon signaling and CTLA-4 enhance the therapeutic efficacy of anti-PD-1 immunotherapy in preclinical model of HPV+ oral cancer. J Immunother Cancer. 2019;7(1):252. doi: 10.1186/s40425-019-0728-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, Zhu G, Maroun CA, et al. Programmed death-1/programmed death-ligand 1-axis blockade in recurrent or metastatic head and neck squamous cell carcinoma stratified by human papillomavirus status: a systematic review and meta-analysis. Front Immunol. 2021;12:645170. doi: 10.3389/fimmu.2021.645170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel JJ, Levy DA, Nguyen SA, Knochelmann HM, Day TA. Impact of PD-L1 expression and human papillomavirus status in anti-PD1/PDL1 immunotherapy for head and neck squamous cell carcinoma—systematic review and meta-analysis. Head Neck. 2020;42(4):774-786. doi: 10.1002/hed.26036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang BC, Cao RB, Li PD, Fu C. The effects and safety of PD-1/PD-L1 inhibitors on head and neck cancer: a systematic review and meta-analysis. Cancer Med. 2019;8(13):5969-5978. doi: 10.1002/cam4.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galvis MM, Borges GA, Oliveira TB, et al. Immunotherapy improves efficacy and safety of patients with HPV positive and negative head and neck cancer: a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2020;150:102966. doi: 10.1016/j.critrevonc.2020.102966 [DOI] [PubMed] [Google Scholar]
- 52.Ferris RL, Blumenschein G Jr, Fayette J, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856-1867. doi: 10.1056/NEJMoa1602252 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable. The characterization of HNSCC and cervical cancer patients in TCGA by HPV status
eFigure 1. p16 Is a Less Reliable Biomarker for HPV Status in OCSCC and LSCC Patients
eFigure 2. The Expressions of E6 and E7 Transcripts in OPSCCs and Non-OP HNSCCs by p16 Testing
eFigure 3. Overall Survival Differences Between Propensity-Score Matched Patients With HPV-Positive and -Negative Cancers
eFigure 4. A 2-Dimensional Reduced Visualization Based on 2630 Immune-related Transcripts in HNSCC and CESC Patients
eFigure 5. Comparative Analysis of Immunogenomic Metrics Between HPV-positive and -negative Patients Within Each Tumor Site—B Cells, CD8+ T Cells, and Cytolytic Activity Score
eFigure 6. Comparative Analysis of Immunogenomic Metrics Between HPV-positive and -negative Patients Within Each Tumor Site—Plasma Cells, Naive B Cells, CD8+ T/Treg Cells, BCR Diversity, and TCR Diversity
eFigure 7. Comparative Analysis Of Immunogenomic Metrics Between HPV-positive and -negative Patients Within Each Tumor Site—M2 Macrophages and CTF
eFigure 8. Comparative Analysis of Immunogenomic Metrics Between HPV-positive and -negative Patients Within Each Tumor Site—TMB and CNV