Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is a highly pathogenic human coronavirus (CoV). Belonging to the same beta-CoV genus as severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1) and SARS-CoV-2, MERS-CoV has a significantly higher fatality rate with limited human-to-human transmissibility. MERS-CoV causes sporadic outbreaks, but no vaccines have yet been approved for use in humans, thus calling for continued efforts to develop effective vaccines against this important CoV. Similar to SARS-CoV-1 and SARS-CoV-2, MERS-CoV contains 4 structural proteins, among which the surface spike (S) protein has been used as a core component in the majority of currently developed MERS-CoV vaccines. Here, we illustrate the importance of the MERS-CoV S protein as a key vaccine target and provide an update on the currently developed MERS-CoV vaccines, including those based on DNAs, proteins, virus-like particles or nanoparticles, and viral vectors. Additionally, we describe approaches for designing MERS-CoV mRNA vaccines and explore the role and importance of naturally occurring pseudo-nucleosides in the design of effective MERS-CoV mRNA vaccines. This review also provides useful insights into designing and evaluating mRNA vaccines against other viral pathogens.
INTRODUCTION
Coronaviruses (CoVs) belong to the Othocoronavirinae subfamily of Coronaviridae, which is a virus family in the order of Nidovirales. Othocoronavirinae consists of 4 genera: alpha-CoV, beta-CoV, gamma-CoV, and delta-CoV.1 , 2 Middle East respiratory syndrome (MERS)-CoV, severe acute respiratory syndrome (SARS)-CoV-1, and SARS-CoV-2 are 3 highly pathogenic human CoVs that are classified as beta-CoVs.1, 2, 3 Phylogenetically, MERS-CoV is a member of lineage C of beta-CoV, whereas SARS-CoV-1 and SARS-CoV-2 are classified as members of lineage B of beta-CoV.2 , 4 SARS-CoV-1 and SARS-CoV-2 were first reported in humans in 2002 and 2019, respectively, leading to the global outbreak or pandemic.5, 6, 7
The first MERS-CoV infection in humans was reported in June 2012.8 Since then, this CoV has infected at least 2578 people globally, including 888 associated deaths (34.4% case-fatality rate), with the majority of these cases identified from Saudi Arabia (37.2% case-fatality rate).9 As a zoonotic virus, MERS-CoV is transmitted between animals and humans.10 Similar to SARS-CoV-1, MERS-CoV originates from bats.11, 12, 13 Different from SARS-CoV-1, however, human infections of MERS-CoV may occur through infected dromedary camels as an intermediate host.14 Unlike SARS-CoV-2 that transmits efficiently among humans, MERS-CoV has limited human-to-human transmission, mostly limited to healthcare settings and household contacts.15, 16, 17, 18 Both MERS-CoV and SARS-CoV-2 cause acute respiratory distress syndrome (ARDS), showing similar respiratory and ventilatory parameters after intubation.19 As MERS-CoV continues to cause localized outbreaks with a high fatality rate, it is imperative to develop safe and effective vaccines to prevent MERS.
MERS-CoV is a single-strand, positive-sense RNA virus. Similar to SARS-CoV-1 and SARS-CoV-2, MERS-CoV has a large genome that encodes 4 structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]), 16 nonstructural proteins (nsp1-16) from open-reading frame (ORF) 1a and ORF 1b, and 5 accessory proteins (ORF 3, 4a, 4b, 5, and 8b) (Fig 1 , A and B).1 , 2 , 20 These proteins have different functions in viral transcription, translation, assembly, infection, replication, and pathogenesis.2 Among these, the S protein plays a critical role in viral infection and pathogenesis.4 , 21 During the course of viral evolution, mutations may be incorporated into the S protein that exhibit geographic differences in virulence.22, 23, 24
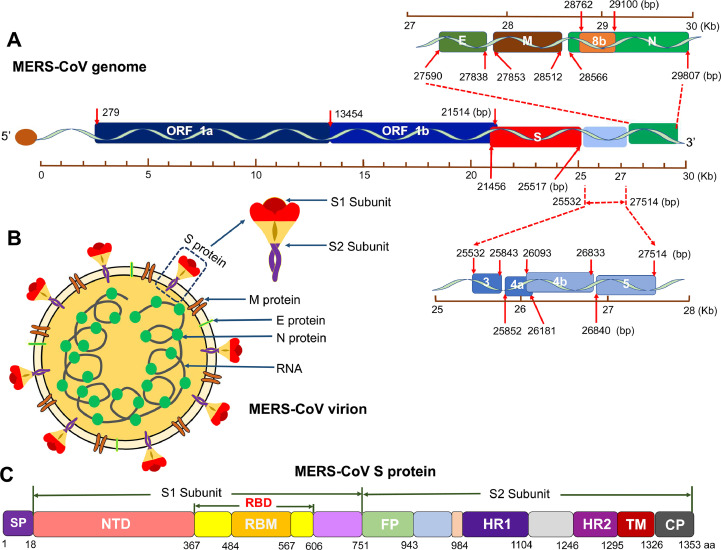
Fig 1.
Schematic diagram of MERS-CoV genome, virion, and spike (S) protein. (A) Schematic structure of MERS-CoV genome. Open reading frame (ORF), structural proteins, including S, envelope (E), membrane (M), and nucleocapsid (N), as well as accessory proteins, including 3, 4a, 4b, 5, and 8b, are shown. Kb, kilobase pair. Bp, base pair. (B) Schematic map of MERS-CoV virion. Four structural proteins (S, M, E, and N) and viral RNA are shown. The S protein consists of S1 and S2 subunits. (C) Schematic structure of MERS-CoV S protein. SP, signal peptide. RBM, receptor-binding motif. NTD, N-terminal domain. RBD, receptor-binding domain. FP, fusion peptide. HR1 and HR2, heptad repeat regions 1 and 2. TM, transmembrane domain. CP, cytoplasmic tail. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
MERS-COV S PROTEIN IS A KEY VACCINE TARGET
The MERS-CoV S protein is presented as a native trimeric structure, and each S monomer consists of 2 subunits, S1 and S2 (Fig. 1, C and 2 , A), as do other CoV S proteins. MERS-CoV infects host cells by first binding to a specific receptor on the cell surface via the receptor-binding domain (RBD) in the S1 subunit (Fig 2, B) and then mediating virus-membrane fusion through the S2 subunit, promoting virus entry into target cells.25, 26, 27 Different from SARS-CoV-1 and SARS-CoV-2, which use angiotensin-converting enzyme 2 (ACE2) as their receptor,28 , 29 MERS-CoV recognizes dipeptidyl peptidase 4 (DPP4) as its receptor for virus entry. Formation of the RBD/DPP4 complex mediates MERS-CoV entry into DPP4-expressing host cells (Fig 2, C).25 , 26 , 30 , 31 The MERS-CoV RBD consists of a core and a receptor-binding motif (RBM) region (Fig 1, C). A set of amino acids in the RBM region has been identified as key residues required for DPP4 binding.25 , 27 , 31 Therefore, mutations of these critical residues may result in a reduction of binding between the RBD and DPP4 receptor. In addition to the RBD, the S1 subunit of MERS-CoV S protein also contains an N-terminal domain (NTD) (Fig. 1, C and 2, B), which may bind sialic acid and play a role in virion attachment.32 Compared with other MERS-CoV proteins, the S protein, including its RBD, can induce highly potent neutralizing antibodies and/or cellular immune responses with protective efficacy against MERS-CoV in vaccinated animals.33, 34, 35, 36, 37, 38 A number of neutralizing monoclonal antibodies (mAbs) targeting the RBD or S1-NTD of MERS-CoV S protein have been reported, and the structures for some of these antibodies in binding to the RBD or S1-NTD have be solved (Fig 2, D and E).39, 40, 41, 42, 43, 44 Notably, S-specific antibodies may be maintained in MERS-CoV-infected humans for a long period of time (up to 6 years postinfection), with neutralizing activity against MERS-CoV infection.45 Therefore, the S protein, including its fragments, presents an important target for the development of effective MERS-CoV vaccines. Notably, there is no appreciable cross-reactivity or cross-neutralizing activity among MERS-CoV and SARS-CoV-1/SARS-CoV-2 S proteins, particularly their RBD fragments and S1 subunits, in immunization studies. In contrast, SARS-CoV-1 and SARS-CoV-2 S proteins can elicit cross-neutralizing antibodies that react with both CoVs.46, 47, 48, 49
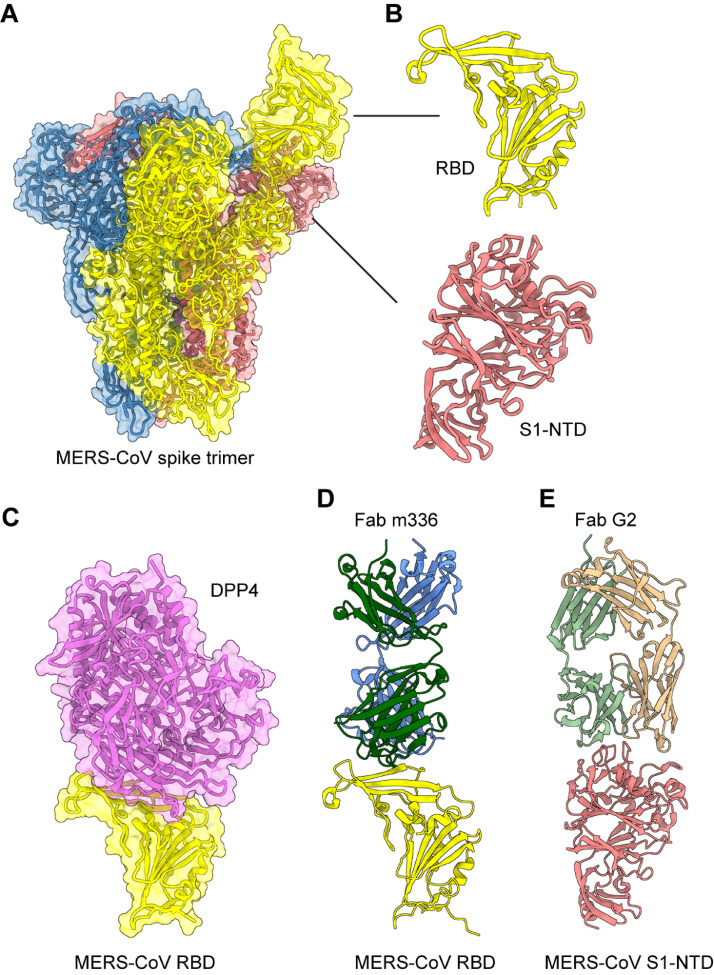
Fig 2.
Structural overview of MERS-CoV spike (S) protein. (A) Cryo-EM structure of MERS-CoV S protein trimer (PDB 5X5F). The 3 S monomers are colored in yellow, blue and light coral, respectively. (B) Close-up views of MERS-CoV receptor-binding domain (RBD) and N-terminal domain (NTD) in the S1 subunit of S protein. (C) Crystal structure of MERS-CoV RBD in complex with the receptor human dipeptidyl peptidase 4 (DPP4) (PDB 4KR0). The human DPP4 is colored in orchid. (D) Crystal structure of MERS-CoV RBD in complex with neutralizing monoclonal antibody (mAb) m336 Fab (PDB 4XAK). The m336 Fab is colored in dark green and blue. (E) Crystal structure of MERS-CoV S1-NTD in complex with neutralizing mAb G2 Fab (PDB 6PXH). The G2 Fab is colored in light green and tan. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
UPDATE ON CURRENT MERS-COV VACCINES
Although MERS-CoV vaccines have been extensively developed, nearly all of them are still in preclinical stages. Several MERS vaccines (eg, GLS-5300, ChAdOx1, and MVA-MERS-S) have undergone Phase 1 trials,50, 51, 52 but no vaccines are yet approved for human use. Currently reported MERS-CoV vaccines include those based on DNAs, proteins, nanoparticles or virus-like particles (VLPs), viral vectors, and live-attenuated viruses.2 , 38 , 53, 54, 55, 56 The majority of these vaccines are designed using the viral S protein, including its fragments, thereby protecting animals (eg, mice, rabbits, camels, or nonhuman primates [NHPs]) against MERS-CoV infection (Table I ).57, 58, 59, 60, 61 MERS-CoV vaccines have been previously summarized in various review articles.1 , 2 , 4 , 20 , 62, 63, 64 Here, we provide an updated summary with a focus on the newly developed vaccine candidates.
Table I.
Update on MERS-CoV vaccines based on viral S protein and its fragments
| Vaccine type | Name | Immune responses | Protection | References |
|---|---|---|---|---|
| MERS-CoV vaccines based on full-length or truncated S protein | ||||
| DNA | MERS DNA | Induced S-specific cellular immune responses and IgG Abs in NHPs which are specific to full-length S protein, S1 or S2 subunit, and RBD fragment, neutralizing live MERS-CoV infection | Protected NHPs against MERS-CoV infection, with reduced clinical symptoms, decreased viral load, and alleviated severity of pathological signs | 57 |
| DNA-protein | pSΔER-pSΔTM | Induced S-specific T-cell responses and S/S1/RBD-specific IgG Abs in mice, with neutralizing activity against infection of multiple pseudotyped MERS-CoVs and live MERS-CoV strain | Protected hDPP4-Tg mice against MERS-CoV infection, with reduced weight loss and complete survival | 71 |
| Viral vector | rAd/spike PIV5/MERS-S ChAdOx1-MERS | rAd/spike induced S-specific T-cell responses and S1-specific Abs (IgG, IgA), neutralizing pseudotyped MERS-CoV infection; PIV5/MERS-S induced S-specific T cell responses and neutralizing Abs in mice against pseudotyped MERS-CoV infection; ChAdOx1-MERS enhanced neutralizing Abs in seropositive dromedary camels against pseudotyped MERS-CoV infection | PIV5/MERS-S protected hDPP4-KI mice against mouse-adapted MERS-CoV infection, improving virus clearance and having minimal eosinophils in the lungs; ChAdOx1-MERS reduced virus shedding and nasal discharge in dromedary camels | 58,76,77 |
| MERS-CoV vaccines based on the RBD fragment | ||||
| Protein | S RBD-HBD 2 | Induced RBD-specific IgG and IgA Abs in mice after intranasal immunization | Protected hDPP4-Tg mice from MERS-CoV challenge, with better protection via the intranasal route than the intramuscular route | 59 |
| Nanoparticle | RBD-LS RBD-NP(cdGMP) | RBD-LS induced S1-specific IgG and neutralizing Abs in rabbits against clade A (EMC2012) and clade B (Qatar15/2015) live MERS-CoV infection; RBD-NP(cdGMP) induced RBD-specific T cell responses, IgG Abs, and neutralizing Abs in mice against live MERS-CoV infection | RBD-LS protected rabbits against MERS-CoV infection, preventing virus shedding and infection; RBD-NP(cdGMP) protected hDPP4-Tg mice from MERS-CoV challenge, without inducing eosinophilic immunopathology | 60,72 |
| Viral or bacterial vector | RV/MERSMERS BLP RVΔP-MERS/S1 | Induced higher S-specific T-cell responses (for RV/MERS), RBD-specific IgG Abs, and rapid (for RV/MERS) or higher (for MERS BLP) neutralizing Abs in mice against pseudotyped MERS-CoV infection; RVΔP-MERS/S1 induced neutralizing Abs against both MERS-CoV and RV infection | N/A | 74,75 |
| MERS-CoV vaccines based on other S fragments (such as S1 subunit) | ||||
| DNA | AcHERV-MERS-S1 pcDNA3.1-S1 pS1 | Induced S1/RBD-specific T-cell responses and/or S-specific IgG and neutralizing Abs in mice against infection of multiple pseudotyped MERS-CoV strains with human and camel origins or live MERS-CoV strain | Protected hDPP4-Tg mice from MERS-CoV challenge, with reduced weight loss and complete survival; Protected Ad5-hDPP4-transduced mice from MERS-CoV infection | 61,65,66 |
Abbreviations: Abs, antibodies; BLP, bacterium-like particle; hDPP4, dipeptidyl peptidase 4; hDPP4-KI, human DPP4-knock-in mice; hDPP4-Tg, human DPP4-transgenic mice; LS, lumazine synthase; N/A, not available; NHPs, nonhuman primates; PIV5, parainfluenza virus 5; rAd, recombinant adenovirus; RBD, receptor-binding domain; RV, rabies virus; S, spike.
DNA vaccines based on MERS-CoV S protein and its fragments
MERS-CoV DNA vaccines utilize different vectors to encode the full-length MERS-CoV S protein or the S1 subunit.57 , 61 , 65 , 66 The immunization of NHPs with a synthetic DNA vaccine encoding the full-length MERS-CoV S protein elicited S-specific cellular immune responses and antibodies specific to the full-length S protein, S1 or S2 subunit, and RBD fragment, with lower viral loads and less severity of pathological signs.57 The vaccine-induced antibody response was able to neutralize MERS-CoV in vitro and protect against MERS-CoV infection in NHPs. The immunization of mice with a baculoviral-vectored DNA vaccine encoding the S1 subunit of MERS-CoV S protein (AcHERV-MERS-S1) induced S-specific immunoglobulin G (IgG) antibodies and neutralizing antibodies capable of protecting immunized hDPP4-transfenic (hDPP4-Tg) mice against MERS-CoV challenge.65 Limited weight loss, 100% survival, and undetectable viral titers in the lungs were reported. Other DNA vaccines, such as those based on the pcDNA3.1 vector and encoding the S1 subunit of S protein (pcDNA3.1-S1), also elicited humoral and cellular immune responses and neutralizing antibodies in mice, protecting adenovirus 5 (Ad5)-hDPP4-transduced mice from MERS-CoV challenge.66
Subunit vaccines based on MERS-CoV S protein and its fragments
Subunit vaccines expressing MERS-CoV full-length S protein, particularly its RBD fragment, may induce highly potent antibody responses and neutralizing antibodies against multiple pseudotyped and live MERS-CoV infection with protective efficacy in immunized animals.23 , 33 , 34 , 36 , 37 , 67, 68, 69 MERS-CoV-specific neutralizing antibodies were significantly enhanced in mice immunized with an alum-adjuvanted subunit vaccine encoding the RBD fragment of MERS-CoV S protein containing a C-terminal foldon trimeric motif and a protein adjuvant, rASP-1, in 2 separate injection sites.70 Subunit vaccines can also be prepared by combining the genes encoding the MERS-CoV S protein and/or its fragments with those encoding an adjuvant, and then expressing the fusion proteins to increase the vaccine's immunogenicity and protection. For example, MERS-CoV RBD protein fused with human β-defensin 2 (S RBD-HBD 2) adjuvant induced RBD-specific IgG antibodies and protective immunity against MERS-CoV infection in hDPP4-Tg mice.59 Notably, the intranasal administration of this protein induced more mucosal IgA antibodies and conferred more effective protection against MERS-CoV challenge than immunization via the intramuscular route.
In addition to single DNA or protein vaccination, combined vaccinations with DNA and proteins may provide broader neutralizing activity. It has been shown that priming with DNA expressing a truncated MERS-CoV S protein (pSΔER) and boosting with a MERS-CoV S protein without the transmembrane region (pSΔTM) increased antibody and cellular immune responses and produced cross-neutralizing antibodies to the wildtype virus as well as pseudotyped variants.71 This DNA-prime/protein-boost vaccine strategy protected hDPP4-Tg mice against MERS-CoV challenge.
Nanoparticle or virus-like particle vaccines based on MERS-CoV S protein and its fragments
MERS-CoV vaccines based on nanoparticles or virus-like particles have been developed and tested in animal models. A vaccine that was generated by coupling the MERS-CoV RBD to lumazine synthase (LS) nanoparticle (RBD-LS) induced antibodies specific to the RBD protein with neutralizing activity against diverse MERS-CoV clades and prevented MERS-CoV infection in rabbits.72 A nanoparticle vaccine delivering the MERS-CoV RBD and stimulator of interferon gene (STING) agonists (RBD-NP(cdGMP)) elicited desirable T-cell and neutralizing antibody responses, which protected hDPP4-Tg mice from lethal MERS-CoV challenge without inducing eosinophilic immunopathology.60
Viral or bacterial-vectored vaccines based on MERS-CoV S protein and its fragments
MERS-CoV vaccines based on viral vectors, such as human Ad (Ad26 or Ad5), chimpanzee Ad (ChAdOx1), parainfluenza virus 5 (PIV5), and rabies virus (RV), or bacterial-like particles (BLPs), such as Gram-positive enhancer matrix (GEM) particles, have been developed and shown to be effective against MERS-CoV infection in animal models, including mice, dromedary camels, and NHPs.58 , 73, 74, 75, 76, 77 Mouse immunization with a human Ad5-vectored vaccine encoding full-length S protein (rAd/spike) induced T cell and antibody (IgG and IgA) responses, with neutralizing activity against MERS-CoV observed in the serological analysis.77 Priming with a human Ad26-vectored vaccine expressing the MERS-CoV RBD fragment and boosting with a human Ad5 vector expressing the MERS-CoV S protein and RBD fragment elicited S-specific T-cell responses and IgG antibodies (with neutralizing activity) in mice and NHPs.73 The vaccine-induced response fully protected hDPP4-Tg mice from MERS-CoV challenge and prevented viral replication in the upper and lower respiratory tracts. A PIV5-vectored vaccine expressing the MERS-CoV S protein (PIV5/MERS-S) induced specific T cell responses and neutralizing antibodies against MERS-CoV, fully protecting hDPP4-knock-in (hDPP4-KI) mice from challenge using a mouse-adapted MERS-CoV.58 An RV-vectored vaccine encoding the S1 subunit of MERS-CoV S protein (RV/MERS) induced higher cellular immune responses and earlier neutralizing antibodies than a BLP-vectored vaccine (MERS BLP) against MERS-CoV infection.74 In addition to inducing anti-MERS-CoV neutralizing antibodies, viral vector-based MERS-CoV vaccines also elicited neutralizing antibodies against viral vectors.75 , 78
MERS-CoV vaccines based on non-S proteins or combined proteins
Some non-S-protein-based or combined antigen-based MERS-CoV vaccines have been explored and/or tested in animal models. For example, a multiepitope (MEP) vaccine was designed by combining the conserved epitopes predicted from several MERS-CoV structural (M, E, or N) and non-structural (ORF 1a, 3, 4a, or 8) proteins with an N-terminal adjuvant (β-defensin) in order to increase its immunogenicity potential.79 Further studies need to be conducted to demonstrate the immunogenicity and/or protection of this predicted peptide vaccine in vivo. In addition, a genetically engineered live-attenuated MERS-CoV vaccine (MERS-CoV-E*Δ2in), which was generated by mutation with partial deletion of the C-terminal domain of E protein using a reverse genetics system, reduced virulence and protected hDPP4-Tg mice from MERS-CoV challenge, without causing apparent histopathological damages in the lungs.56 Nevertheless, an inactivated MERS-CoV vaccine without engineering resulted in eosinophil infiltration and other pulmonary immunopathology in immunized hDPP4-Tg mice after MERS-CoV challenge, a phenomenon that was seen previously in inactivated virus-vaccinated mice challenged with SARS-CoV-1.80, 81, 82 Different from most non-S-based vaccines, numbers of MERS vaccines based on the S protein, particularly subunit vaccines based on the RBD fragment, are much safer and more effective in protecting animals from MERS-CoV without evidence of eosinophilic immunopathology.1 , 2 , 69
MRNA AS AN EFFECTIVE TOOL TO DEVELOP PROMISING VACCINES
mRNA technologies have become a promising approach to rapidly develop much needed vaccines against the challenging and emerging viral diseases, including those caused by HIV-1, influenza virus, Zika virus, and SARS-CoV-2.83, 84, 85, 86, 87, 88, 89, 90, 91 mRNA normally only presents in the cytoplasm and does not enter the cell nucleus or cross the extracellular membrane as DNA does, thus eliminating the potential to integrate into the host genome and cause insertional mutagenesis.92 , 93 In addition, mRNA vaccines are safe because they are produced via in vitro transcription in a cell-free environment without utilizing any cultured cells or virus components. Unlike viral vectors or VLPs, mRNA eliminates the possibility to elicit vector/carrier-specific immunogenicity.92 , 93 Generally, mRNA can be readily prepared with rapid production and manufacturing capability. Both non-replicating and self-amplifying approaches have been proposed to generate mRNA vaccines. The former only encodes the target protein antigen, whereas the latter also encodes the RNA genome of an RNA virus (eg, alphavirus, flavivirus, or picornavirus), which enables RNA replication.94 , 95
Despite numerous advantages, mRNA vaccines face some potential challenges.96 Unmodified mRNA is intrinsically unstable and can be readily degraded by nucleases. Appropriate strategies have been employed to address mRNA degradation. For example, the addition of a 5′-cap, adjustment of the length and structure of 3′-poly(A) tails, the optimization of nucleoside sequences, and the modification of nucleosides can significantly reduce mRNA degradation.97, 98, 99 mRNA can also be encapsulated with lipid nanoparticles (LNPs) and other delivery vehicles based on polymers, peptides, and cationic nanoemulsions (CNEs) to increase its stability and prevent degradation.100 , 101 In addition to its instability, unmodified mRNA may activate innate immune systems through the endosomal recognition of Toll-like receptors (TLRs) 7, 8, and 9 and the RIG-1-like receptor family, such as RIG-1, MDA5, and LGP2, resulting in the production of pro-inflammatory cytokines and type I interferons.92 , 102 To address this concern and further increase translational activity, naturally occurring pseudo-nucleosides, such as pseudouridine, N6-methyladenosine (m6A), and 5-methylcytosine (m5C), may be inserted into the mRNA during synthesis.103 , 104 In addition to these technical challenges in mRNA vaccine production, some adverse effects caused by LNPs were reported for mRNA-based coronavirus disease 2019 (COVID-19) vaccines,105 , 106 highlighting the need for further optimization of the LNP delivery platform.
DESIGN OF MRNA-BASED MERS-COV VACCINES
Since the COVID-19 pandemic began, tremendous efforts have focused on the development of vaccines against SARS-CoV-2, resulting in the emergency use authorization (EUA) of at least 3 COVID-19 vaccines, 2 of which are mRNA vaccines, for the prevention of SARS-CoV-2 infection among various ages.107, 108, 109, 110, 111, 112, 113, 114, 115, 116 However, the mRNA vaccine technology is still in its infancy. So far, only a limited number of studies have reported the design and evaluation of mRNA vaccines against MERS-CoV. Because the RBD fragment in the S1 subunit is a main target for inducing potent neutralizing antibodies that protect against MERS-CoV infection, it can be used as the “core antigen” for mRNA vaccine development. In the following paragraphs, we illustrate the construction, expression, and delivery of RBD-based mRNA vaccines for MERS-CoV.
The genes encoding the MERS-CoV RBD (Fig 3 , A) and an N-terminal signal peptide, tissue plasminogen activator (tPA), were amplified using polymerase chain reaction (PCR) and a codon-optimized MERS-CoV S plasmid as a template and inserted into a pCAGGS-mCherry vector, which contains essential elements for the efficient transcription of mRNA in vitro. Unmodified MERS-CoV RBD-mRNA was synthesized using the MEGAscript T7 kit in the presence of cytidine triphosphate (CTP), uridine triphosphate (UTP), guanosine triphosphate (GTP), and adenosine triphosphate (ATP) nucleosides (Fig 3, B). Modified RBD-mRNA (ie, RBD-mRNA-pseudoU) was synthesized to replace UTP with pseudouridine-5′-triphosphate (pseudouridine, Ψ), based on which mCherry-RBD-mRNA was constructed and synthesized by adding an N-terminal mCherry sequence to the RBD (Fig 3, C and D). The synthesized mRNAs were capped with a 5′-cap sequence to form a Cap 1 structure and tailed with a 150 bp poly(A) tail to increase stability. Overall, a complete list of elements required for the synthesis of non-replicating MERS-CoV RBD-mRNA included an N-terminal T7 promoter, 5′-Cap, 5′-untranslated region (5′-UTR), signal peptide (tPA)-RBD with or without mCherry, 3′-UTR, and a 3′-poly(A) tail. The synthesized mRNAs were encapsulated with LNPs to further increase their stability and facilitate targeted cell delivery (Fig 3, E). The synthesis and encapsulation of MERS-CoV RBD-mRNAs are summarized in Fig 3.
Fig 3.
Schematic diagram of MERS-CoV mRNAs using receptor-binding domain (RBD) as an example. (A) MERS-CoV virion, showing RBD in the spike (S) protein. (B-D) Design of unmodified RBD-mRNA (B), nucleoside (pseudoUTP: pseudoU)-modified RBD-mRNA (RBD-mRNA-pseudoU) (C), and nucleoside (pseudoU)-modified RBD-mRNA containing an N-terminal mCherry sequence (mCherry-RBD-mRNA) (D). Each mRNA is composed of a 5′-Cap (forming a Cap 1 structure), 5′-untranslated region (UTR), signal peptide (tissue plasminogen activator: tPA) with coding sequences (MERS-CoV RBD and/or mCherry), 3′-UTR, and a 3′-poly(A) tail. (E) The synthesized mRNAs (naked mRNAs) can be encapsulated with lipid nanoparticles (LNPs) to form mRNA-LNPs for delivery. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
MERS-COV MRNA PRESENTATION AND EXPRESSION OF TARGET PROTEINS IN HOST CELLS
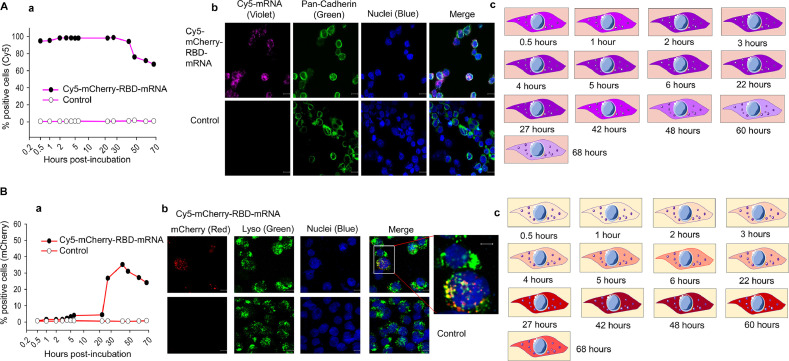
To illustrate the presentation of mRNA in target cells, mCherry-RBD-mRNA was synthesized as described above. To facilitate visualization of mRNA presentation in this process, some CTP was labeled with a cyanine 5 (Cy5) probe (Cy5-CTP) during the in vitro transcription of this mRNA. Cy5 incorporation enabled fluorescent detection of the target mRNA without the need for protein translation. Similarly high levels of fluorescent Cy5 were detected in cells for at least 42 hours after incubation of the Cy5-labeled mCherry-RBD-mRNA with target cells, and fluoresce was still detectable in > 60% of the cells at 68 hours postincubation; in contrast, no fluorescent cells were detected in control cells incubated with empty LNPs (Fig 4 , Aa). Additionally, a fluorescent (violet) signal was detected in Cy5-labeled mCherry-RBD-mRNA-incubated cells but not in cells incubated with the empty LNPs (Fig 4, Ab). A timeline of the presence of the Cy5 signal in cells is shown in Fig 4, Ac. These findings suggest that mCherry-RBD-mRNA can enter target cells with high efficiency and may be present in target cells for a long period of time, potentially increasing the time for the mRNA to be transcribed into the target protein antigen.
Fig 4.
Presentation of MERS-CoV RBD-mRNA and expression of the target protein in cells. (A) Presentation of mRNA in 293T cells. (a) Cy5-labeled mCherry-RBD-mRNA (1 μg/ml) was incubated with 293T cells at 37°C. The cells were collected at different times postincubation and analyzed for Cy5-positive cells by flow cytometry. (b) Representative images by confocal microscopy at 27 hours postincubation, with nuclei in blue, cell membrane (pan-Cadherin) in green, and Cy5-mRNA in violet. (c) Schematic images showing the time period of Cy5-labeled mRNA in 293T cells. (B) Detection of expression of mRNA-encoding protein in target cells. (a) Cy5-labeled mCherry-RBD-mRNA (1 μg/ml) was incubated with 293T cells at 37°C. The cells were collected at different times postincubation and detected for mCherry fluorescence (mRNA expression) by flow cytometry. (b) Representative confocal microscopy images of the expression of mRNA-encoding protein. mCherry is in red, lyso (lysosome) is in green, and nuclei are in blue. Empty LNPs were included as a control. (c) Schematic images showing the time period of mRNA-encoding protein expression in 293T cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To determine whether mRNA can durably express the target protein in cells, mCherry-RBD-mRNA was incubated with cells for different periods of time, and the fluorescent mCherry signal was measured. Indeed, mCherry-RBD-mRNA had long-term (> 68 hours) expression of the target protein in cells, reaching the highest levels at 27–60 hours postincubation, whereas no fluorescence was detected in the empty LNP control (Fig 4, Ba). Notably, the mCherry signal was detectable in cells transcribed with mCherry-RBD-mRNA but not in cells incubated with the empty LNP control. Furthermore, the mCherry signal co-existed with lysosomes but did not colocalize with nuclei (Fig 4, Bb), suggesting that RBD-mRNA does not cross the nucleus and may be resistant to lysosomal degradation. A timeline of the expression of mRNA-encoding mCherry protein in cells is shown in Fig 4, Bc.
EFFECTS OF PSEUDO-NUCLEOSIDES IN REDUCING OR BLOCKING MERS-COV MRNA-ASSOCIATED INNATE IMMUNE ACTIVATION
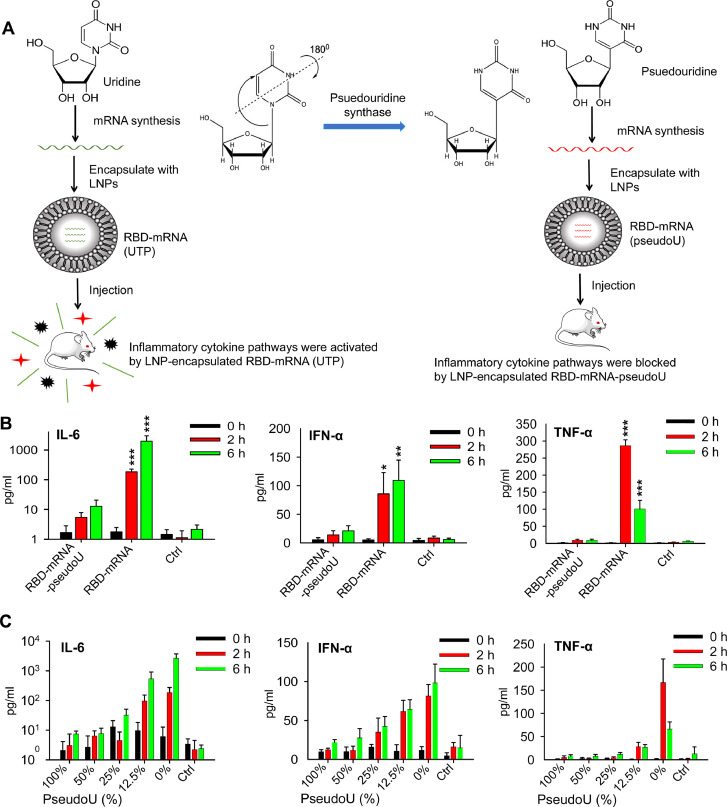
Unmodified MERS-CoV mRNA vaccines can induce a strong, inflammatory-type innate immune response, as previously observed for mRNA vaccines against other pathogens.92 , 102 Thus, naturally occurring pseudo-nucleosides, such as pseudouridine, can be incorporated into mRNA synthesis to reduce the unwanted innate response. For the MERS-CoV RBD-mRNA vaccine, a modified RBD-mRNA-pseudoU was synthesized using pseudouridine as described above and compared with the unmodified RBD-mRNA synthesized using UTP for the ability to reduce inflammatory cytokines in mice (Fig 5 , A). Mice were injected with each mRNA or an empty LNP control, and sera were collected at different time points postinjection to detect cytokine levels. Significantly lower levels of interleukin-6 (IL-6), interferon-α (IFN-α), and tumor necrosis factor-α (TNF-α) cytokines were detected in mice that received modified RBD-mRNA-pseudoU than the mice that received unmodified RBD-mRNA at 2 or 6 hours postinjection, which were similar to the baseline signals observed for the control group in which mice received empty LNPs without mRNA (Fig 5, B).
Fig 5.
Effects of nucleoside-modified MERS-CoV mRNA vaccines on reducing mRNA-associated innate immune activation using RBD-mRNA as an example. (A) Schematic figures showing the production of inflammatory cytokines by unmodified RBD-mRNA (synthesized using UTP and 3 other nucleosides, GTP, ATP, and CTP). Such cytokines were blocked when pseudoU was used for the synthesis of mRNA together with other nucleosides (GTP, ATP, and CTP). (B) Examples of the detection of inflammatory cytokines (IL-6, IFN-α, and TNF-α) in sera of RBD-mRNA-injected BALB/c mice at 0, 2, and 6 hours postinjection. Unmodified (uridine, UTP) and modified (pseudouridine, pseudoU) RBD-mRNAs were used for the injection, and empty LNPs were included as a control (Ctrl). Significant differences between modified and unmodified RBD-mRNA groups are indicated. (C) RBD-mRNAs with pseudoU packaged at the indicated percentages were used to inject into BALB/c mice as indicated above, and related inflammatory cytokines (IL-6, IFN-α, and TNF-α) were detected at 0, 2, and 6 hours postinjection. Control (Ctrl), empty LNPs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To identify an optimal ratio of pseudouridine and UTP in reducing mRNA-associated innate immune activation, pseudouridine was added at 100%, 50%, 25%, 12.5%, and 0%, respectively (with corresponding ratios of 1:0, 1:1, 1:3, 1:7, and 0:1 for pseudouridine and UTP), during the synthesis of RBD-mRNA, and related cytokine levels were detected as described above. Pseudouridine at 100% (pseudouridine:UTP, 1:0) resulted in the lowest levels of IL-6 and IFN-α, whereas unmodified RBD-mRNA (0% pseudouridine) led to the highest secretion of these cytokines (Fig 5, C). Similarly, pseudouridine at 100%, 50%, and 25% (pseudouridine:UTP, 1:0–1:3) only had a background level of TNF-α secretion, whereas unmodified RBD-mRNA (0% pseudouridine) led to the highest expression of this cytokine (Fig 5, C).
The above data suggest that the addition of pseudouridine to mRNA during synthesis completely blocked or significantly reduced the ability of unmodified MERS-CoV RBD-mRNA to activate innate immune pathways. Moreover, the pseudouridine:UTP ratio affected the secretion of inflammatory cytokines. These data confirm the important role of pseudo-nucleosides in reducing or blocking MERS-CoV RBD-mRNA-associated innate immune activation.
ROLE OF PSEUDO-NUCLEOSIDES IN MERS-COV MRNA-INDUCED ADAPTIVE IMMUNE RESPONSES
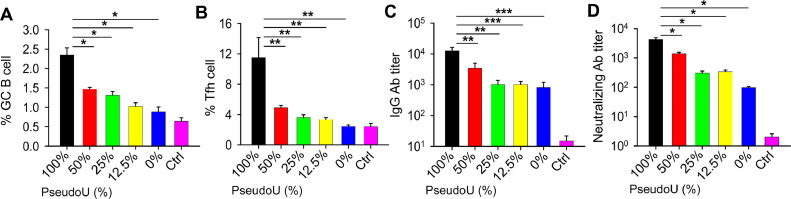
Similar to mRNA vaccines against other viral pathogens, such as SARS-CoV-2,48 , 91 , 117, 118, 119 MERS-CoV mRNA vaccines may induce effective adaptive immune responses in immunized individuals. Among the unmodified and pseudouridine-modified MERS-CoV RBD-mRNA vaccines, modified RBD-mRNA-pseudoU with 100% pseudouridine induced the highest level of germinal center (GC) B and T-follicular helper (Tfh) cell responses, which were significantly higher than those induced by the unmodified RBD-mRNA and the modified RBD-mRNAs containing 50%, 25%, and 12.5% pseudouridine, respectively (Fig 6 , A and B). Additionally, reducing the pseudouridine content in mRNA synthesis lowered the production of GC B and Tfh cells, and RBD-mRNA without pseudouridine modification had the lowest responses of these 2 cells, similar to responses induced by the empty LNP control (Fig 6, A and B). Similarly, RBD-mRNA-pseudoU with 100% pseudouridine elicited MERS-CoV-specific IgG antibodies, which potently neutralized pseudotyped MERS-CoV with titers that were significantly higher than those induced by the unmodified and modified MERS-CoV RBD-mRNAs with varying percentages of pseudouridine (Fig 6, C and D). These data suggest that pseudouridine in MERS-CoV RBD-mRNA vaccines plays an important role in the induction of effective MERS-CoV-specific adaptive immune responses.
Fig 6.
Effects of MERS-CoV mRNA vaccines with or without nucleoside modification on adaptive immune responses using RBD-mRNA as an example. The figures compare the MERS-CoV RBD-mRNA-induced germinal center (GC) B cell (A) and T follicular helper (Tfh) cell (B) responses in lymph nodes, as well as IgG antibody (Ab) (C) and neutralizing Ab (D) titers in sera, from BALB/c mice immunized with LNP-encapsulated RBD-mRNA with or without nucleoside modification. Percentages of pseudoU are shown in each figure. Significant differences between 100% pseudoU of RBD-mRNA and the other groups are indicated. Ctrl, empty LNPs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
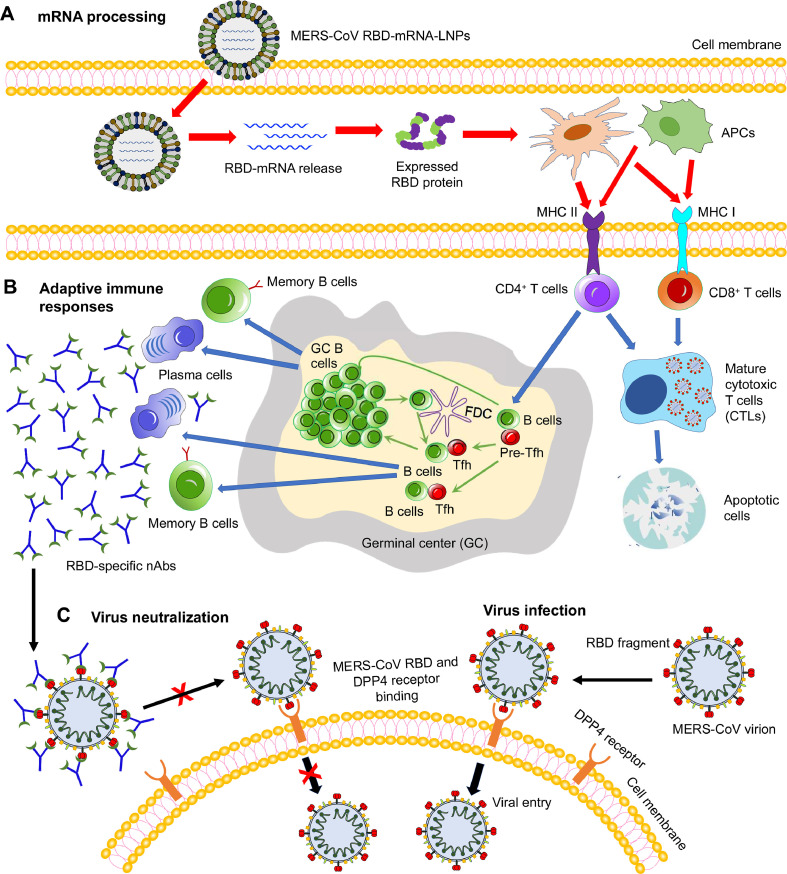
A potential working model of the MERS-CoV RBD-mRNA vaccine is summarized in Fig 7 . LNP-encapsulated, pseudouridine-modified mRNA is delivered into the target cell, where the mRNA is released into the cytoplasm (Fig 7, A). The mRNA is subsequently translated into the target protein, which is presented into antigen-presenting cells (APCs), such as dendritic cells (DCs) and macrophages (Fig 7, A). This process activates MHC class II and MHC class I responses, resulting in the production of antigen-specific CD4+ and CD8+ T cell responses to help B-cell maturation and kill virus-infected cells, respectively. GC B and Tfh cell responses are induced in the GC, and the activated B cells may differentiate into plasma cells to generate antigen-specific neutralizing antibodies (Fig 7, B). In the absence of MERS-CoV RBD-specific neutralizing antibodies, the MERS-CoV RBD binds to its cellular receptor DPP4, mediating virus entry and subsequent membrane fusion processes (virus infection) (Fig 7, C). Nevertheless, in the presence of MERS-CoV RBD-specific neutralizing antibodies, these antibodies block binding of the MERS-CoV RBD to the DPP4 receptor, thereby inhibiting subsequent virus entry into the target cell (ie, virus neutralization) (Fig 7, C).
Fig 7.
Schematic view of the potential working model of MERS-CoV mRNA vaccines using RBD-mRNA as an example. (A) mRNA processing. Lipid nanoparticle (LNP)-encapsulated, nucleoside-modified MERS-CoV RBD-mRNA (pseudoU) was delivered into target cells, where it was released and then translated into the target receptor-binding domain (RBD) protein fragment. Subsequently, antigen-presenting cells (APCs), such as dendritic cells and macrophages, recognize the expressed protein, and present it via major histocompatibility complex I (MHC I) and MHC II molecules. (B) Adaptive immune responses. T (CD4+ or CD8+) and B cells were activated by interacting with the APCs. The activated CD8+ or CD4+ T cells either kill the virus-infected cells directly or help B cells generate antigen-specific antibodies, including neutralizing antibodies (nAbs). Germinal center (GC) B cells interact with T follicular helper (Tfh) and B cells to help promote the production of effective antibodies. (C) Virus infection and neutralization. In the absence of nAbs, MERS-CoV RBD binds its cellular receptor dipeptidyl peptidase 4 (DPP4) to enter target cells (virus infection). In the presence of MERS-CoV RBD-specific nAbs, nAbs bind to the RBD, blocking its binding to the DPP4 receptor and thus preventing subsequent MERS-CoV infection (virus neutralization). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
POTENTIAL CHALLENGES AND IMPROVEMENTS OF MERS-COV VACCINES
As a highly pathogenic CoV, MERS-CoV infects humans with an alarming fatality rate. MERS-CoV has a zoonotic origin, potentially from bats, and uses dromedary camels as a reservoir host, resulting in the continuous infection of humans albeit with lower transmissibility than SARS-CoV-1 and SARS-CoV-2. Because it is unlikely to avoid human contact with these animal hosts in regions where MERS-CoV circulates, the battle against MERS-CoV continues. Most MERS-CoV vaccines are developed based on the S protein or its key antigenic component, the RBD. However, the S protein continually undergoes mutations under evolutionary pressure, with a number of critical mutations, such as L506F, D509G, D510G and I529T, identified in the RBD.23 , 120 It has been shown that both vector and mRNA-based SARS-CoV-2 vaccines derived from the S protein of the original strain (Wuhan-Hu-1), including the full-length S protein, S1 subunit, and RBD fragment, were found less effective in neutralizing variants with S mutations (eg, in the RBD and NTD of the S1 subunit).90 , 121, 122, 123, 124, 125, 126 Similarly, MERS-CoV vaccines targeting the prototypic S sequence may have a lower ability to neutralize variant strains. In addition, some MERS-like CoVs that share the same DPP4 receptor as MERS-CoV have been identified in bats,127 which may have potential to cause infection in humans. Therefore, effective vaccines with broad-spectrum activity against multiple MERS-CoV strains, including their variants in humans, camels, and bats, are needed to prevent MERS-CoV infection.
Different from S1 subunit fragments (eg, NTD and RBD), the S2 subunit region of MERS-CoV is relatively conserved among different isolates, underscoring its potential as a target for the development of universal MERS-CoV vaccines with broad-spectrum activity. Unlike the MERS-CoV full-length S protein or its domain fragments (S1 and RBD), the S2 region alone generally has low immunogenicity to induce strong immune responses with neutralizing activity against MERS-CoV infection. Thus, novel approaches, such as structure-based design, may be explored for the development of effective S2-based MERS-CoV vaccines to improve immunogenicity and neutralizing activity. Moreover, T cell-based vaccines that target the conserved S2 or other regions in the S protein or other proteins of MERS-CoV, such as the N protein, may have potential to improve the coverage and efficacy of MERS-CoV vaccines against diverse strains. Such strategies have proven effective in protecting against variant strains of other viral pathogens, including Zika virus and SARS-CoV-2.128, 129, 130
Similar to the mRNA-based COVID-19 vaccines, which have been developed to target the viral S protein, S1 subunit, and/or RBD fragment, and shown efficacy against SARS-CoV-2 infection,48 , 90 , 118 , 131 the mRNA-based MERS-CoV vaccines can be designed based on the full-length S protein or its fragments (eg, S1) of this protein, in addition to the RBD, and tested for their broad-spectrum ability against multiple virus strains. Additionally, mRNA vaccines synthesized using other pseudo-nucleosides (except pseudouridine) or combinations of different pseudo-nucleosides may be tested to identify the optimal strategy to maximally reduce mRNA-associated innate immune activation, enhance mRNA stability, and increase immunogenicity and neutralizing activity of mRNA vaccines against MERS-CoV infection. Furthermore, elements of mRNA components and coding sequences of MERS-CoV proteins, as well as delivery systems, may be further optimized to improve mRNA stability and overall immunogenicity.
CONCLUSION AND FUTURE PROSPECTS
Currently, although a number of MERS-CoV vaccines have shown efficacy against MERS-CoV infection in camels and a few have been tested in human clinical trials as described above, no MERS-CoV vaccines are yet approved for human use. Therefore, the development of effective vaccines with potent and broad-spectrum activity against diverse MERS-CoV isolates remains a high priority. Newly developed promising MERS-CoV vaccines with protective efficacy at preclinical stages should proceed to clinical trials when ample resources are available. Strategies for the rapid development of COVID-19 vaccines provide valuable guidelines for the development of next-generation MERS-CoV vaccines with a focus on the choice of antigen, safety, and efficacy. Overall, there is a continuous threat from pathogenic human CoVs, including MERS-CoV and SARS-CoV-2, as well as future outbreaks or pandemics potentially caused by other CoVs from bats or other animal hosts. Thus, it is important to expand the scope of vaccine development to cover not only multiple stains of a CoV from one group but also multiple CoVs from the same or different genera with pandemic potential. It is also plausible to develop pan-CoV vaccines based on the “mosaic” or “cocktail” strategy to achieve broad reactivity and neutralizing activity. Notably, the design of such CoV vaccines might be challenging, considering the different receptors used by different CoVs and amino acid variations among S and other proteins that serve as vaccine antigens. Nevertheless, conserved regions in the S protein, such as the S2 subunit, and other proteins (eg, N), may be assessed in the development of universal CoV vaccines. Moreover, vaccine platforms, such as mRNAs and nanoparticles, can be rapidly adapted to emerging CoVs by simply replacing the antigen-coding sequence. Pan-CoV vaccines based on mRNA and other novel vaccine platforms will be critical for the prevention of future CoV outbreaks and pandemics.
ETHICS STATEMENT
Data from male and female BALB/c mouse samples were used as examples in the study. The animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the New York Blood Center. All animal studies were carried out in accordance with the guidelines and recommendations for the Care and Use of Laboratory Animals (National Research Council Committee).
Acknowledgments
Conflict of interest: The authors have read and agreed to the journal's authorship statement and policy on disclosure of potential conflicts of interest. The authors declare no conflicts of interest. The manuscript has been reviewed by and approved by all authors.
This work was supported by NIH grants (R01AI139092 and R01AI137472). The authors thank Michael Arends for proofreading the manuscript.
References
- 1.Wang N, Shang J, Jiang S, et al. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang N, Shang J, Li C, et al. An overview of Middle East respiratory syndrome coronavirus vaccines in preclinical studies. Expert Rev Vaccines. 2020;19(9):817–829. doi: 10.1080/14760584.2020.1813574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang S, Zhang X, Du L. Therapeutic antibodies and fusion inhibitors targeting the spike protein of SARS-CoV-2. Expert Opin Ther Targets. 2021;25:415–421. doi: 10.1080/14728222.2020.1820482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang N, Jiang S, Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev Vaccines. 2014;13(6):761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du L, He Y, Zhou Y, et al. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization. Middle East respiratory syndromes (MERS) situation update. October 2021. Available at: http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html.
- 10.Li F, Du L. MERS coronavirus: an emerging zoonotic virus. Viruses. 2019;11:663. doi: 10.3390/v11070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Du L, Liu C, et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci U S A. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Qi J, Yuan Y, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munster VJ, Adney DR, van Doremalen N, et al. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis) Sci Rep. 2016;6:21878. doi: 10.1038/srep21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haagmans BL, Al Dhahiry SH, Reusken CB, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghinai I, McPherson TD, Hunter JC, et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395:1137–1144. doi: 10.1016/S0140-6736(20)30607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson MA, Quandelacy TM, Kada S, et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drosten C, Meyer B, Müller MA, et al. Transmission of MERS-coronavirus in household contacts. N Engl J Med. 2014;371:828–835. doi: 10.1056/NEJMoa1405858. [DOI] [PubMed] [Google Scholar]
- 18.Park J, Yoo SY, Ko JH, et al. Infection prevention measures for surgical procedures during a Middle East respiratory syndrome outbreak in a tertiary care hospital in South Korea. Sci Rep. 2020;10:325. doi: 10.1038/s41598-019-57216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalid I, Yamani RM, Imran M, et al. Comparison of characteristics and ventilatory course between coronavirus disease 2019 and Middle East respiratory syndrome patients with acute respiratory distress syndrome. Acute Crit Care. 2021;36:223–231. doi: 10.4266/acc.2021.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Yang Y, Huang J, et al. Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain. Viruses. 2019;11:60. doi: 10.3390/v11010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du L, Yang Y, Zhou Y, et al. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong LR, Zheng J, Sariol A, et al. Middle East respiratory syndrome coronavirus spike protein variants exhibit geographic differences in virulence. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2102983118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai W, Wang Y, Fett CA, et al. Recombinant receptor-binding domains of multiple Middle East respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J Virol. 2017;91:e01651. doi: 10.1128/JVI.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrag MA, Amer HM, Bhat R, et al. Sequence and phylogentic analysis of MERS-CoV in Saudi Arabia, 2012-2019. Virol J. 2021;18:90. doi: 10.1186/s12985-021-01563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Rajashankar KR, Yang Y, et al. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J Virol. 2013;87:10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu L, Liu Q, Zhu Y, et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang N, Shi X, Jiang L, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Li W, Farzan M, et al. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 30.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu G, Hu Y, Wang Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Hulswit RJG, Widjaja I, et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang N, Channappanavar R, Ma C, et al. Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against Middle East respiratory syndrome coronavirus. Cell Mol Immunol. 2016;13:180–190. doi: 10.1038/cmi.2015.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma C, Wang L, Tao X, et al. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments–the importance of immunofocusing in subunit vaccine design. Vaccine. 2014;32:6170–6176. doi: 10.1016/j.vaccine.2014.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen WH, Du L, Chag SM, et al. Yeast-expressed recombinant protein of the receptor-binding domain in SARS-CoV spike protein with deglycosylated forms as a SARS vaccine candidate. Hum Vaccin Immunother. 2014;10:648–658. doi: 10.4161/hv.27464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du L, Tai W, Yang Y, et al. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat Commun. 2016;7:13473. doi: 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tai W, Zhao G, Sun S, et al. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology. 2016;499:375–382. doi: 10.1016/j.virol.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Shi W, Joyce MG, et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying T, Prabakaran P, Du L, et al. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang N, Rosen O, Wang L, et al. Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD. Cell Rep. 2019;28:3395–3405. doi: 10.1016/j.celrep.2019.08.052. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du L, Zhao G, Yang Y, et al. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol. 2014;88:7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ying T, Du L, Ju TW, et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H, Chen Y, Zhang S, et al. Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein. Nat Commun. 2019;10:3068. doi: 10.1038/s41467-019-10897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S, Zhou P, Wang P, et al. Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein. Cell Rep. 2018;24:441–452. doi: 10.1016/j.celrep.2018.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alshukairi AN, Zhao J, Al-Mozaini MA, et al. Longevity of Middle East respiratory syndrome coronavirus antibody responses in humans, Saudi Arabia. Emerg Infect Dis. 2021;27:1472–1476. doi: 10.3201/eid2705.204056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tai W, Zhang X, He Y, et al. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res. 2020;179 doi: 10.1016/j.antiviral.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tai W, Zhang X, Drelich A, et al. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020;30:932–935. doi: 10.1038/s41422-020-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He L, Lin X, Wang Y, et al. Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci Adv. 2021;7:eabf1591. doi: 10.1126/sciadv.abf1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Modjarrad K, Roberts CC, Mills KT, et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Folegatti PM, Bittaye M, Flaxman A, et al. Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect Dis. 2020;20:816–826. doi: 10.1016/S1473-3099(20)30160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch T, Dahlke C, Fathi A, et al. Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: an open-label, phase 1 trial. Lancet Infect Dis. 2020;20:827–838. doi: 10.1016/S1473-3099(20)30248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tscherne A, Schwarz JH, Rohde C, et al. Immunogenicity and efficacy of the COVID-19 candidate vector vaccine MVA-SARS-2-S in preclinical vaccination. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2026207118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malczyk AH, Kupke A, Prüfer S, et al. A highly immunogenic and protective Middle East respiratory syndrome coronavirus vaccine based on a recombinant Measles virus vaccine platform. J Virol. 2015;89:11654–11667. doi: 10.1128/JVI.01815-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashemzadeh A, Avan A, Ferns GA, et al. Vaccines based on virus-like nano-particles for use against Middle East respiratory syndrome (MERS) coronavirus. Vaccine. 2020;38:5742–5746. doi: 10.1016/j.vaccine.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutiérrez-Álvarez J, Honrubia JM, Fernández-Delgado R, et al. Genetically engineered live-attenuated Middle East respiratory syndrome coronavirus viruses confer full protection against lethal infection. mBio. 2021;12 doi: 10.1128/mBio.00103-21. e00103-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel A, Reuschel EL, Xu Z, et al. Intradermal delivery of a synthetic DNA vaccine protects macaques from Middle East respiratory syndrome coronavirus. JCI Insight. 2021;6 doi: 10.1172/jci.insight.146082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li K, Li Z, Wohlford-Lenane C, et al. Single-dose, intranasal immunization with fecombinant parainfluenza virus 5 expressing Middle East respiratory syndrome coronavirus (MERS-CoV) spike protein protects mice from fatal MERS-CoV infection. mBio. 2020;11 doi: 10.1128/mBio.00554-20. e00554-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J, Yang YL, Jeong Y, et al. Conjugation of human β-Defensin 2 to spike protein receptor-binding domain induces antigen-specific protective immunity against Middle East respiratory syndrome coronavirus infection in human dipeptidyl peptidase 4 transgenic mice. Vaccines (Basel) 2020;8:635. doi: 10.3390/vaccines8040635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin LC, Huang CY, Yao BY, et al. Viromimetic STING agonist-loaded hollow polymeric nanoparticles for safe and effective vaccination against Middle East respiratory syndrome coronavirus. Adv Funct Mater. 2019;29 doi: 10.1002/adfm.201807616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Amri SS, Abbas AT, Siddiq LA, et al. Immunogenicity of candidate MERS-CoV DNA vaccines based on the spike. Protein Sci Rep. 2017;7:44875. doi: 10.1038/srep44875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du L, Jiang S. Middle East respiratory syndrome: current status and future prospects for vaccine development. Expert Opin Biol Ther. 2015;15:1647–1651. doi: 10.1517/14712598.2015.1092518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y, Jiang S, Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev Vaccines. 2018;17:677–686. doi: 10.1080/14760584.2018.1506702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du L, Tai W, Zhou Y, et al. Vaccines for the prevention against the threat of MERS-CoV. Expert Rev Vaccines. 2016;15:1123–1134. doi: 10.1586/14760584.2016.1167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cho H, Jang Y, Park KH, et al. Human endogenous retrovirus-enveloped baculoviral DNA vaccines against MERS-CoV and SARS-CoV2. NPJ Vaccines. 2021;6:37. doi: 10.1038/s41541-021-00303-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chi H, Zheng X, Wang X, et al. DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine. 2017;35:2069–2075. doi: 10.1016/j.vaccine.2017.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Tai W, Yang J, et al. Receptor-binding domain of MERS-CoV with optimal immunogen dosage and immunization interval protects human transgenic mice from MERS-CoV infection. Hum Vaccin Immunother. 2017;13:1615–1624. doi: 10.1080/21645515.2017.1296994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Du L, Zhao G, Kou Z, et al. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol. 2013;87:9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nyon MP, Du L, Tseng CK, et al. Engineering a stable CHO cell line for the expression of a MERS-coronavirus vaccine antigen. Vaccine. 2018;36:1853–1862. doi: 10.1016/j.vaccine.2018.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.George PJ, Tai W, Du L, et al. The potency of an anti-MERS coronavirus subunit vaccine depends on a unique combinatorial adjuvant formulation. Vaccines (Basel) 2020;8:251. doi: 10.3390/vaccines8020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi JA, Goo J, Yang E, et al. Cross-protection against MERS-CoV by prime-boost vaccination using viral spike DNA and protein. J Virol. 2020;94 doi: 10.1128/JVI.01176-20. e01178-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okba NMA, Widjaja I, van Dieren B, et al. Particulate multivalent presentation of the receptor binding domain induces protective immune responses against MERS-CoV. Emerg Microbes Infect. 2020;9:1080–1091. doi: 10.1080/22221751.2020.1760735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dolzhikova IV, Grousova DM, Zubkova OV, et al. Preclinical studies of immunogenity, potectivity, and safety of the combined vector vaccine for prevention of the Middle East respiratory syndrome. Acta Naturae. 2020;12:114–123. doi: 10.32607/actanaturae.11042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li E, Yan F, Huang P, et al. Characterization of the immune response of MERS-CoV vaccine candidates derived from two different vectors in mice. Viruses. 2020;12:125. doi: 10.3390/v12010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato H, Takayama-Ito M, Iizuka-Shiota I, et al. Development of a recombinant replication-deficient rabies virus-based bivalent-vaccine against MERS-CoV and rabies virus and its humoral immunogenicity in mice. PLoS One. 2019;14 doi: 10.1371/journal.pone.0223684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alharbi NK, Qasim I, Almasoud A, et al. Humoral immunogenicity and efficacy of a single dose of ChAdOx1 MERS vaccine candidate in dromedary camels. Sci Rep. 2019;9:16292. doi: 10.1038/s41598-019-52730-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim MH, Kim HJ, Chang J. Superior immune responses induced by intranasal immunization with recombinant adenovirus-based vaccine expressing full-length spike protein of Middle East respiratory syndrome coronavirus. PLoS One. 2019;14 doi: 10.1371/journal.pone.0220196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haagmans BL, van den Brand JM, Raj VS, et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science. 2016;351:77–81. doi: 10.1126/science.aad1283. [DOI] [PubMed] [Google Scholar]
- 79.Ashfaq UA, Saleem S, Masoud MS, et al. Rational design of multi epitope-based subunit vaccine by exploring MERS-CoV proteome: reverse vaccinology and molecular docking approach. PLoS One. 2021;16 doi: 10.1371/journal.pone.0245072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Agrawal AS, Tao X, Algaissi A, et al. Immunization with inactivated Middle East respiratory syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum Vaccin Immunother. 2016;12:2351–2356. doi: 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bolles M, Deming D, Long K, et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tseng CT, Sbrana E, Iwata-Yoshikawa N, et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7:e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhong Z, Portela Catani JP, Mc Cafferty S, et al. Immunogenicity and protection efficacy of a naked self-replicating mRNA-based Zika virus vaccine. Vaccines (Basel) 2019;7:96. doi: 10.3390/vaccines7030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Richner JM, Himansu S, Dowd KA, et al. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168:1114–1125. doi: 10.1016/j.cell.2017.02.017. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Freyn AW, Ramos da Silva J, Rosado VC, et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhuang X, Qi Y, Wang M, et al. mRNA vaccines encoding the HA protein of influenza A H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines (Basel) 2020;8:123. doi: 10.3390/vaccines8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pardi N, LaBranche CC, Ferrari G, et al. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol Ther Nucleic Acids. 2019;15:36–47. doi: 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moyo N, Vogel AB, Buus S, et al. Efficient induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol Ther Methods Clin Dev. 2019;12:32–46. doi: 10.1016/j.omtm.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu K, Werner AP, Moliva JI, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021;2021 doi: 10.1101/2021.01.25.427948. 01.25.427948. [DOI] [Google Scholar]
- 91.Elia U, Rotem S, Bar-Haim E, et al. Lipid nanoparticle RBD-hFc mRNA vaccine protects hACE2 transgenic mice against a lethal SARS-CoV-2 infection. Nano Lett. 2021;21:4774–4779. doi: 10.1021/acs.nanolett.1c01284. [DOI] [PubMed] [Google Scholar]
- 92.Zhang C, Maruggi G, Shan H, et al. Advances in mRNA vaccines for infectious diseases. Front Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ho W, Gao M, Li F, et al. Next-generation vaccines: nanoparticle-mediated DNA and mRNA delivery. Adv Healthc Mater. 2021;10 doi: 10.1002/adhm.202001812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCullough KC, Milona P, Thomann-Harwood L, et al. Self-amplifying replicon RNA vaccine delivery to dendritic cells by synthetic nanoparticles. Vaccines (Basel) 2014;2:735–754. doi: 10.3390/vaccines2040735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sandbrink JB, Shattock RJ. RNA vaccines: A suitable platform for tackling emerging pandemics? Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.608460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crommelin DJA, Anchordoquy TJ, Volkin DB, et al. Addressing the cold reality of mRNA vaccine stability. J Pharm Sci. 2021;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kowalzik F, Schreiner D, Jensen C, et al. mRNA-based vaccines. Vaccines (Basel) 2021;9:390. doi: 10.3390/vaccines9040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wollner CJ, Richner JM. mRNA vaccines against flaviviruses. Vaccines (Basel) 2021;9:148. doi: 10.3390/vaccines9020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.To KKW, Cho WCS. An overview of rational design of mRNA-based therapeutics and vaccines. Expert Opin Drug Discov. 2021:1307–1317. doi: 10.1080/17460441.2021.1935859. [DOI] [PubMed] [Google Scholar]
- 100.Wang Y, Zhang Z, Luo J, et al. mRNA vaccine: a potential therapeutic strategy. Mol Cancer. 2021;20:33. doi: 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blakney AK, Ip S, Geall AJ. An update on self-amplifying mRNA vaccine development. Vaccines (Basel) 2021;9:97. doi: 10.3390/vaccines9020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hadas Y, Katz MG, Bridges CR, et al. Modified mRNA as a therapeutic tool to induce cardiac regeneration in ischemic heart disease. Wiley Interdiscip Rev Syst Biol Med. 2017;9 doi: 10.1002/wsbm.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krienke C, Kolb L, Diken E, et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science. 2021;371:145–153. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- 104.Starostina EV, Sharabrin SV, Antropov DN, et al. Construction and immunogenicity of modified mRNA-vaccine variants encoding influenza virus antigens. Vaccines (Basel) 2021;9:452. doi: 10.3390/vaccines9050452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Igyártó BZ, Jacobsen S, Ndeupen S. Future considerations for the mRNA-lipid nanoparticle vaccine platform. Curr Opin Virol. 2021;48:65–72. doi: 10.1016/j.coviro.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ndeupen S, Qin Z, Jacobsen S, et al. The mRNA-LNP platform's lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. bioRxiv. 2021;2021 doi: 10.1101/2021.03.04.430128. 03.04.430128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oliver SE, Gargano JW, Marin M, et al. The Advisory Committee on immunization practices' interim recommendation for use of Moderna COVID-19 vaccine - United States, December 2020. MMWR Morb Mortal Wkly Rep. 2021;69:1653–1656. doi: 10.15585/mmwr.mm695152e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wallace M, Woodworth KR, Gargano JW, et al. The Advisory Committee on immunization practices' interim recommendation for use of Pfizer-BioNTech COVID-19 vaccine in adolescents aged 12-15 years - United States, May 2021. MMWR Morb Mortal Wkly Rep. 2021;70:749–752. doi: 10.15585/mmwr.mm7020e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oliver SE, Gargano JW, Marin M, et al. The Advisory Committee on immunization practices' interim recommendation for use of Pfizer-BioNTech COVID-19 vaccine - United States, December 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1922–1924. doi: 10.15585/mmwr.mm6950e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tenforde MW, Olson SM, Self WH, et al. Effectiveness of Pfizer-BioNTech and Moderna vaccines against COVID-19 among hospitalized adults aged ≥65 Years - United States, January-March 2021. MMWR Morb Mortal Wkly Rep. 2021;70:674–679. doi: 10.15585/mmwr.mm7018e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parums DV. Editorial: first full regulatory approval of a COVID-19 vaccine, the BNT162b2 Pfizer-BioNTech vaccine, and the real-world implications for public health policy. Med Sci Monit. 2021;27 doi: 10.12659/MSM.934625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dooling K, Gargano JW, Moulia D, et al. Use of Pfizer-BioNTech COVID-19 vaccine in persons aged ≥16 years: recommendations of the Advisory Committee on immunization practices - United States, September 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1344–1348. doi: 10.15585/mmwr.mm7038e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lamb YN. BNT162b2 mRNA COVID-19 vaccine: first approval. Drugs. 2021;81:495–501. doi: 10.1007/s40265-021-01480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hause AM, Gee J, Baggs J, et al. COVID-19 vaccine safety in adolescents aged 12-17 years - United States, December 14, 2020-July 16, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1053–1058. doi: 10.15585/mmwr.mm7031e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mbaeyi S, Oliver SE, Collins JP, et al. The Advisory Committee on Immunization Practices' Interim Recommendations for additional primary and booster doses of COVID-19 vaccines - United States, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1545–1552. doi: 10.15585/mmwr.mm7044e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.MacNeil JR, Su JR, Broder KR, et al. Updated recommendations from the Advisory Committee on immunization practices for use of the Janssen (Johnson & Johnson) COVID-19 vaccine after reports of thrombosis with Thrombocytopenia syndrome among vaccine recipients - United States, April 2021. MMWR Morb Mortal Wkly Rep. 2021;70:651–656. doi: 10.15585/mmwr.mm7017e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vogel AB, Kanevsky I, Che Y, et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 118.Laczkó D, Hogan MJ, Toulmin SA, et al. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53:724–732. doi: 10.1016/j.immuni.2020.07.019. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Corbett KS, Edwards DK, Leist SR, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kleine-Weber H, Elzayat MT, Wang L, et al. Mutations in the spike protein of Middle East respiratory syndrome coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J Virol. 2019;93 doi: 10.1128/JVI.01381-18. e01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu C, Ginn HM, Dejnirattisai W, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236. doi: 10.1016/j.cell.2021.06.020. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 123.McCallum M, Bassi J, De Marco A, et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Supasa P, Zhou D, Dejnirattisai W, et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211. doi: 10.1016/j.cell.2021.02.033. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li Q, Nie J, Wu J, et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371. doi: 10.1016/j.cell.2021.02.042. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muik A, Wallisch AK, Sanger B, et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luo CM, Wang N, Yang XL, et al. Discovery of novel bat coronaviruses in South China that use the same receptor as Middle East Respiratory syndrome coronavirus. J Virol. 2018;92(13) doi: 10.1128/JVI.00116-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Matchett WE, Joag V, Stolley JM, et al. Cutting edge: nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. J Immunol. 2021;207:376–379. doi: 10.4049/jimmunol.2100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gambino F, Jr., Tai W, Voronin D, et al. A vaccine inducing solely cytotoxic T lymphocytes fully prevents Zika virus infection and fetal damage. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Elong Ngono A, Syed T, Nguyen AV, et al. CD8+ T cells mediate protection against Zika virus induced by an NS3-based vaccine. Sci Adv. 2020;6:eabb2154. doi: 10.1126/sciadv.abb2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sahin U, Muik A, Derhovanessian E, et al. COVID-19 vaccine BNT162b1 elicits human antibody and T(H)1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]