Abstract
Objectives
Identify distinct clusters of psoriatic arthritis (PsA) patients based on their baseline articular, entheseal and cutaneous disease manifestations and explore their clinical and therapeutic value.
Methods
Pooled baseline data in PsA patients (n=1894) treated with secukinumab across four phase 3 studies (FUTURE 2–5) were analysed to determine phenotypes based on clusters of clinical indicators. Finite mixture models methodology was applied to generate clinical clusters and mean longitudinal responses were compared between secukinumab doses (300 vs 150 mg) across identified clusters and clinical indicators through week 52 using machine learning (ML) techniques.
Results
Seven distinct patient clusters were identified. Cluster 1 (very-high (VH) – SWO/TEN (swollen/tender); n=187) was characterised by VH polyarticular burden for both tenderness and swelling of joints, while cluster 2 (H (high) – TEN; n=251) was marked by high polyarticular burden in tender joints and cluster 3 (H – Feet – Dactylitis; n=175) by high burden in joints of feet and dactylitis. For cluster 4 (L (Low) – Nails – Skin; n=209), cluster 5 (L – skin; n=283), cluster 6 (L – Nails; n=294) and cluster 7 (L; n=495) articular burden was low but nail and skin involvement was variable, with cluster 7 marked by mild disease activity across all domains. Greater improvements in the longitudinal responses for enthesitis in cluster 2, enthesitis and Psoriasis Area and Severity Index (PASI) in cluster 4 and PASI in cluster 6 were shown for secukinumab 300 mg compared with 150 mg.
Conclusions
PsA clusters identified by ML follow variable response trajectories indicating their potential to predict precise impact on patients’ outcomes.
Trial registration numbers
Keywords: biological therapy, arthritis, psoriatic, tumor necrosis factor inhibitors, inflammation, t-lymphocyte subsets
Key messages.
What is already known about this subject?
Machine learning (ML), a subfield of artificial intelligence, allows learning from experience and improves the performance of data analytics efforts. ML algorithms can detect data patterns of vast amounts of clinical data and identify clusters with therapeutic or prognostic significance. The FUTURE programme comprises five randomised placebo controlled trials of secukinumab and enrolled over 2750 patients with psoriatic arthritis (PsA).
What does this study add?
Based on the presence and severity at baseline of the PsA clinical manifestations in patients enrolled in a large clinical development phase III programme, this study identified seven distinct clusters by using ML techniques.
The subsequent analyses of their longitudinal responses to two secukinumab doses has shown greater improvements in Psoriasis Area and Severity Index scores and enthesitis resolution for secukinumab 300 mg compared with 150 mg in some clusters.
How might this impact on clinical practice or further developments?
This proof-of-concept study demonstrated that the seven distinct PsA clinical clusters identified by ML follow variable response trajectories to secukinumab therapy.
Further studies from other classes of drugs with a different mode of action, head-to-head trials and longitudinal observational studies, are warranted to explore the potential of these clusters to determine PsA patient phenotypes of prognostic significance by anticipating disease trajectories, tailoring treatment choices, optimising therapeutic regimens and ultimately improving clinical care of PsA patients.
Introduction
A personalised treatment approach based on the integration of phenotypic, biomarker and imaging data represents an unmet need for patients with psoriatic arthritis (PsA) and would have a significant impact on their quality of life and functional ability.1 2 Amidst the heterogeneity of the clinical manifestations of PsA and the variability of response to multiple therapeutic modalities, identifying patient phenotypes and endotypes that vary in their response to therapy is a challenging but important quest.1 2 Currently historically defined clinical subgroups that have not been tested for either their therapeutic response or their pathogenetic homogeneity are relied on.
The availability of large quantities of data generated from diverse sources such as composite outcome measures, high-resolution imaging, genome sequencing and electronic medical records require implementation of complex algorithms to identify patterns. Consequently, computational strategies are increasingly applied to analyse large-scale data.3 4 Machine learning (ML), a subfield of artificial intelligence that allows for the ability to learn from experience and improve performance of specific tasks, is expected to play a pivotal role in the development of personalised medicine in the future.1 2 ML algorithms can be designed to detect data patterns from vast amounts of clinical data,3 and identify clusters with therapeutic or prognostic significance.4 Using ML techniques could be the first critical step towards gaining a better understanding of disease pathotypes, and support progression towards precision medicine.3 5 6 One of the advantages of ML methods is that they are ‘unbiased’ or ‘semibiased’ and largely agnostic to preconceived notions of clinical phenotypic historical subsets, allowing the computer to identify novel subgroups. The FUTURE programme comprises five randomised controlled trials (RCTs) of secukinumab vs placebo and enrolled over 2750 patients with PsA. This study aimed to identify distinct clusters of patients with PsA by using ML techniques based on their baseline articular, entheseal and cutaneous disease manifestations as captured in the FUTURE programme.7–10 These clusters were subsequently examined for differences in their longitudinal responses to different secukinumab doses to explore their prognostic value.
Methods
Data description
Pooled clinical data at baseline in PsA patients treated with secukinumab across four phase 3 studies, namely, FUTURE 2,7 FUTURE 3,8 FUTURE 49 and FUTURE 510 were analysed.
Cluster analysis
As shown in table 1, six clinical indicators of the five PsA manifestations (joints, entheses, dactylitis, skin and nails) were analysed to identify clusters of patients and determine PsA phenotypes. In total, 206 baseline variables were identified, notably 76 joints from the swollen joint count (SJC), 78 joints from tender joint count (TJC), 6 entheseal sites from Leeds Enthesitis Index (LEI), 10 fingers and 10 toes for dactylitis, 10 fingers for nail psoriasis (Pso) (and 4 body locations each for erythema, scaling, thickening and area score. Finite mixture models methodology11 (sometimes also called latent class mixture models12 was applied to generate patient clusters from the pooled clinical data of 1894 patients treated with secukinumab in the four pivotal phase 3 trials. This methodology has been shown to be a principled statistical approach to practical issues that can come up in clustering.11 More specifically, the method relies on a robust statistical model where patients that share a cluster have a common probability distribution of key measurements such as joints, enthesitis, etc . The statistical model provides a mathematical criterion, such as Bayesian information criterion (BIC), for determining the number of clusters, which is a notoriously difficult choice in the literature. Other non-model-based clustering algorithms (such as K-means and hierarchical clustering) tend to rely on heuristics to choose the number of clusters. We applied the latent class mixture model defined in ‘How to find an appropriate clustering for mixed‐type variables with application to socioeconomic stratification’.12 The principal author of this paper published an R-software package called ‘fpc’13 along with that paper, which is an algorithmic implementation of the method. This software package allowed us to fit the exact finite mixture model that we wanted, namely the so-called multinomial mixture model, which is a special type of finite mixture model appropriate for categorical variables. Since each of the 206 clinical variables were either binary (tender and swollen joints, enthesitis and dactylitis) or categorical with categories 0–4 (modified Nail Psoriasis Severity Index (mNAPSI) and Psoriasis Area and Severity Index (PASI)-related variables), this multinomial mixture fits naturally to the clinical data set. The fundamental mathematical assumption of the clustering algorithm was that if two patients belong to the same cluster then their clinical measurements (across the 206 variables) share the same multinomial distribution. Mathematically, let denote the 206 categorical measurements for patient (among the 1894 patients). Assume that the patients can be partitioned into K distinct clusters. If patient was in cluster , then it was assumed that the measurements follow a cluster-specific multinomial distribution, namely:
Table 1.
Clinical indicators of PSA disease manifestation
| 1 | Tender joints | For each of the 78 joints based on the presence or absence of tenderness |
| 2 | Swollen joints | For each of the 76 joints based on the presence or absence of swelling |
| 3 | Enthesitis | For each of the six LEI entheseal sites based on presence or absence |
| 4 | Dactylitis | For each of the 10 fingers and 10 toes based on presence or absence |
| 5 | mNAPSI | For each of the 10 fingers defined as clear, mild, mild-to-moderate, moderate-to-severe, severe for a mNAPSI of 0, 1, 2, 3–4, 5–13 |
| 6 | PASI | For each of the four body areas (upper limbs, lower limbs, head and trunk) erythema, scaling and thickening were defined as clear, mild, mild-to-moderate, moderate-to-severe or severe on a scale from 0 to 4. The affected body areas were rescaled from a 0–6 scale onto the same scale of 0–4 (0 (clear), 1 (mild), 2 (mild-to-moderate), 3–4 (moderate-to-severe), 4–6 (severe)) |
LEI, Leeds Enthesitis Index; mNAPSI, modified nail psoriasis severity index; PASI, psoriasis area severity index; PsA, psoriatic arthritis.
Where for binary variables (tender, swollen joints, enthesitis, and dactylitis variables are either 0 or 1) and for the cateogorical variables (mNAPSI and PASI variables are on a scale from 0 to 4); was interpreted as the probability that the -th clinical variable for patient is equal to . It was also assumed for mathematical convenience that the clinical variables are probabilistically independent, which results in a mathematically attractive likelihood formulation. It is important to note that many of the clinical variables are in fact naturally dependent. We have reported this in a previous abstract presented at American College of Rheumatology (ACR) Annual Meeting in 202114 where we observed a natural grouping of joints consistent with the body’s anatomy purely based on intervariable correlations. However, modelling adequately dependencies with categorical data is challenging and, to the best of our knowledge, there are no previous references of implementation of the multinomial mixture model with additional dependence/correlation assumptions. Therefore, the independence assumption was applied for mathematical feasibility reasons. Implementation of a more complex correlated multivariate multinomial mixture models could be an interesting future methodological research. The resulting multinomial mixture model can be fit using the R-package15 ‘flexmix’ in conjunction with the “lcmixed” module from the R-package ‘fpc’. The ‘flexmix’ package estimates the mixture model using maximum likelihood and once the parameters of the model were estimated we determined cluster memberships of each patient (given a prespecified number of clusters K). Furthermore, as it was not known in advance how many patient clusters there were in reality, it was important to run the clustering algorithm across a wide range of K (eg, K=2, 3… 10) and choose the optimal K according to some pre-specified mathematical criteria. BIC16 was used to choose the mathematically optimal number of clusters K.
Finally, to assess clustering robustness and stability, the clustering algorithm was applied repeatedly on different subsamples of the 1894 patients. More specifically, the clustering algorithm was run 200 times for each of the different K 2,3, …, 10, each time on different subsets of 80% of the patients (~1467 patients). With the subsampling we essentially perform Consensus Clustering of Monti et al,17 which is an established method to represent the consensus across multiple runs of a clustering algorithm and to assess the stability of the discovered clusters.
Longitudinal analysis
The next step was to compare the longitudinal responses between the doses of secukinumab (300 vs 150 mg) both across the identified clusters and the six clinical indicators, namely tender joints, swollen joints, enthesitis, dactylitis, nail and skin PsO defined by the 206 clinical variables as described above. The longitudinal treatment differences were explored for secukinumab 150 and 300 mg by visual comparisons on a relative scale. Furthermore, curves depicting the difference between the relative mean to secukinumab 150 mg response and 300 mg response were generated for each cluster and endpoint. The relative mean of each endpoint at time t was defined as the endpoint mean at time t divided by the endpoint mean at baseline; hence all curves of the visual comparisons depicting the longitudinal relative treatment differences start at the same value of one at baseline. The corresponding Bonferroni adjusted 95% bootstrap CIs were represented by coloured shaded regions.
Results
Baseline demographics and disease characteristics are shown in online supplemental table 1. Baseline characteristics were balanced across treatment groups. At baseline,~63% had enthesitis, 36% had dactylitis, the mean adjusted TJC ranged from 19.1 to 21.5, the mean adjusted SJC ranged from 7.6 to 10.0, ~46% had C reactive protein >5 mg/L,~42% had PsO of ≥3% of body surface area, and 66% had nail PsO.
rmdopen-2021-001845supp001.pdf (574.3KB, pdf)
Cluster analysis
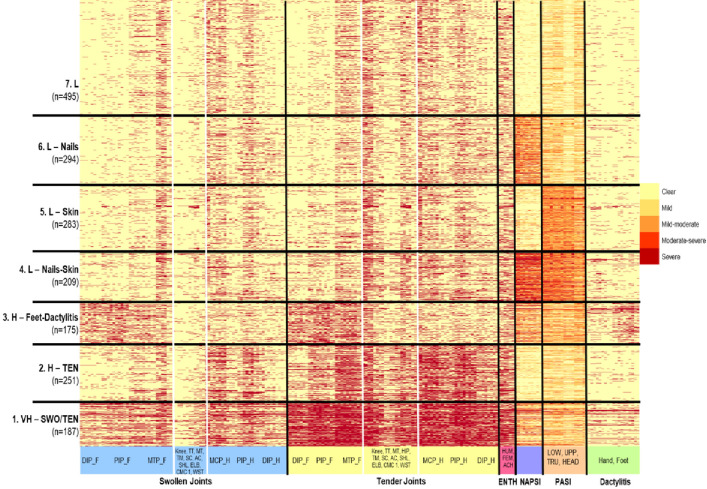
Approximately 375 000 data entries collected at baseline for 1894 patients across the 206 clinical variables contributed to the cluster analysis. A heat-map underlying the clustering was generated wherein each row corresponded to one patient (hence there were 1894 rows in total), and each column corresponded to one clinical variable (hence there were 206 columns in total; figure 1). The rows (patients) have been ordered according to optimal clustering and the columns (clinical variables) according to which clinical domain they belong to gear towards exploration of patterns that are clinically meaningful. The optimal (minimum) BIC was achieved when K=7 as shown in the plot of K vs BIC for each of the 200 models per K=2, 3, …, 10 along with a blue mean curve as depicted in online supplemental figure 1. Seven distinct patient clusters were identified based on the baseline disease manifestations as noted in figure 1. Cluster 1, labelled very high (VH)-SWO/TEN (swollen/tender) cluster (n=187), was characterised by VH polyarticular burden (tender and swollen joints), high disease activity for enthesitis, mild-to-moderate for skin disease, nail disease and dactylitis. Cluster 2 labelled as H-TEN (n=251) was marked by high burden for tender joints, whereas all other disease domains were of mild-to-moderate activity. Cluster 3, labelled as H-Feet-Dactylitis (n=175), was characterised by high disease burden in the joints of the feet and dactylitis, whereas mild-to-moderate activity was seen across all other disease domains. Cluster 4 labelled L-Nails-Skin (n=209) was marked by low polyarticular burden but moderate-to-severe activity of nail and skin disease. Cluster 5 and cluster 6 labelled L-Skin (n=283) and L-Nails (n=294) were characterised by moderate activity for skin or nail disease, respectively, but low articular burden as well as low disease activity in all other domains. Finally, patients in cluster 7 labelled L (n=495) had low disease activity across all domains. The mean variable value for each of the six clusters based on baseline disease manifestations, namely swollen joint, tender joint, enthesitis, mNAPSI, PASI and dactylitis are shown in table 2.
Figure 1.
Heat map of baseline clusters from FUTURE 2–5 studiesA dark red colour signifies the presence of a symptom or sign in the case of binary variables (swollen joints, tender joints, enthesitis or dactylitis) or severe symptom or sign (value of 4 on the scale 0–4) for categorical variables (mNAPSI and PASI). A light yellow signifies absence of a symptom or sign for both binary and categorical variables. Gradual shades of yellow-orange-red refer to mild, mild-to-moderate, moderate-to-severe signs among the categorical variables. AC, acromioclavicular; ACh, Achilles; CMC 1, Carpometacarpal 1; DAC, Dactylitis; DIP_F, distal interphalangeal joints feet; DIP_H, distal interphalangeal joints hand; Dist., distance; ELB, elbow; Fem, femur; hum: humerus; LEI, Leeds Enthesitis Index; low, lower limbs; MCP_H, metacarpophalangeal joints hand; mNAPSI, modified nail psoriasis severity index; MT, Mid-tarsal; MTP_F, Metatarsophalangeal joint; N, number of evaluable patients; PASI, psoriasis area severity index; PIP_F, proximal interphalangeal joints feet; PIP_H, proximal interphalangeal joints hand; SC, sternoclavicular; SHL, shoulder; swo, swollen; ten, tender; TM, temporomandibular joint; Tru, trunk; TT, Talo-tibial; UPP, upper limbs; WST, wrist.
Table 2.
Mean variable value for the baseline clusters: future 2–5
| Cluster | Variable | Mean | SD |
| 1.VH-SWO/TEN (n=187) | DACTCNT | 3.5 | 5.57 |
| LEI | 3.8 | 2.06 | |
| mNAPSI | 16.7 | 20.44 | |
| PASI | 7.6 | 11.29 | |
| SJC | 27.6 | 16.13 | |
| TJC | 56.9 | 12.08 | |
| 2.H-TEN (n=251) | DACTCNT | 0.7 | 1.59 |
| LEI | 2.6 | 2.01 | |
| mNAPSI | 2.9 | 4.57 | |
| PASI | 2.8 | 3.29 | |
| SJC | 12.7 | 7.17 | |
| TJC | 34.3 | 8.97 | |
| 3.H-Feet (n=175) | DACTCNT | 4.3 | 4.36 |
| LEI | 1.5 | 1.66 | |
| mNAPSI | 16.4 | 17.16 | |
| PASI | 6.7 | 6.21 | |
| SJC | 14.9 | 8.15 | |
| TJC | 21.1 | 8.46 | |
| 4.L-Nails-Skin (n=209) | DACTCNT | 1.8 | 2.88 |
| LEI | 2.0 | 1.73 | |
| mNAPSI | 35.9 | 20.66 | |
| PASI | 19.3 | 12.74 | |
| SJC | 9.6 | 5.49 | |
| TJC | 17.8 | 9.14 | |
| 5.L-Skin (n=283) | DACTCNT | 0.8 | 1.47 |
| LEI | 1.6 | 1.72 | |
| mNAPSI | 3.7 | 4.83 | |
| PASI | 12.9 | 8.08 | |
| SJC | 7.8 | 4.21 | |
| TJC | 13.2 | 6.64 | |
| 6.L-Nails (n=294) | DACTCNT | 0.7 | 1.64 |
| LEI | 1.2 | 1.48 | |
| mNAPSI | 21.0 | 16.16 | |
| PASI | 3.2 | 2.68 | |
| SJC | 7.3 | 4.11 | |
| TJC | 12.1 | 6.68 | |
| 7.L (n=495) | DACTCNT | 0.4 | 1.08 |
| LEI | 1.3 | 1.54 | |
| mNAPSI | 1.1 | 2.36 | |
| PASI | 1.8 | 1.94 | |
| SJC | 6.9 | 3.91 | |
| TJC | 11.6 | 5.33 |
DACTCNT, dactylitis count; LEI, Leeds Enthesitis Index; mNAPSI, modified nail psoriasis severity index; PASI, psoriasis area severity index; SJC, swollen joint count; TJC, tender joint count.
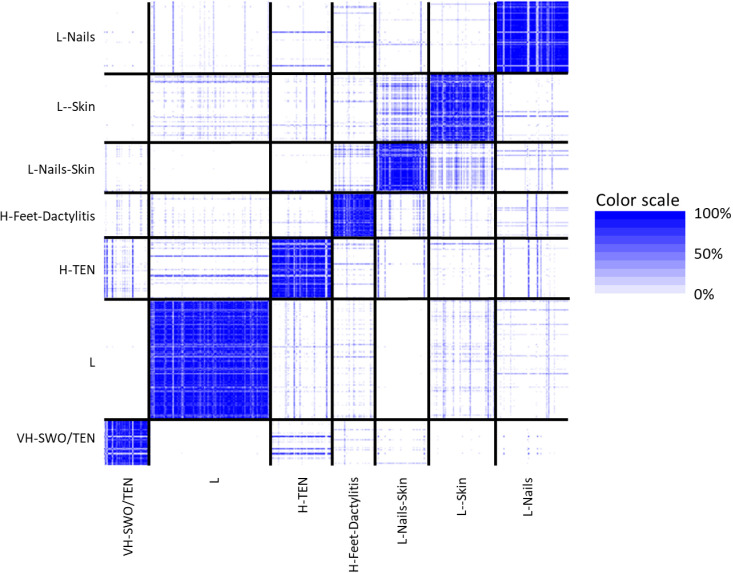
The consensus matrix to measure cluster stability showed that the overall clustering was fairly stable across the 200 subsamples (figure 2). The column ordering of patients from left to right was the same as the row ordering of patients from bottom to top. For each pair of patients, we had a coloured pixel in the heat-map corresponding to that pair. A white colour meant that this pair of patients were never clustered together (across all 200 replicated clusterings). A dark blue colour meant that the pair of patients were always clustered together (100% of the time). The black vertical and horizontal lines separated the seven optimal clusters. Dark blue blocks along the diagonal of the heat-map indicated strong cluster stability and robustness. Conversely, large patches of blue on the off diagonal indicated cluster instability.
Figure 2.
Consensus matrix to measure cluster stability. Cluster 1: VH-SWO/TEN cluster (n=187); cluster 2: H – TEN (n=251); cluster 3: H – feet – Dactylitis (n=175); cluster 4: L – nails –s kin (n=209); cluster 5: L–skin (n=283); cluster 6: L–nails (n=294); cluster 7: L (low) (n=495). H, high; L, low; SWO, swollen; TEN, tender; VH, very high.
Longitudinal responses
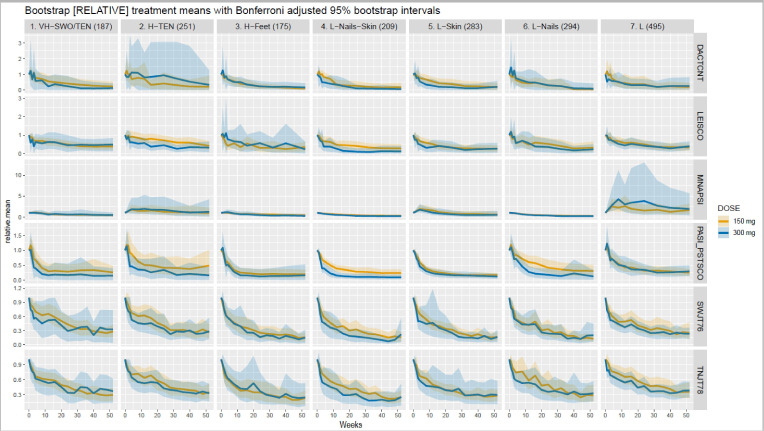
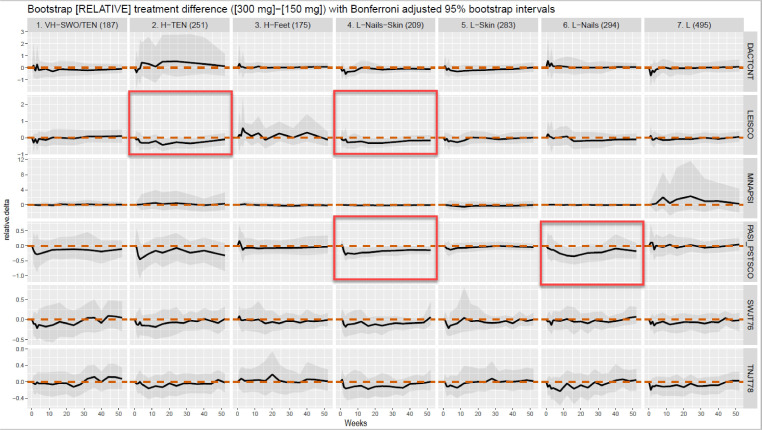
A visual comparison of the two relative mean longitudinal response curves for secukinumab 300 and 150 mg across the seven identified clusters and six clinical indicators of the five PsA manifestations is shown in figure 3. The curves depicting the difference between the relative mean responses to secukinumab 300 and 150 mg across the seven clusters for each of the six clinical indicators are shown in figure 4. Overall, there were relatively small differences in response to secukinumab 300 vs 150 mg across the seven clusters. Statistically significant higher improvements in the secukinumab 300 mg pooled groups compared with 150 mg were shown in cluster 2 (H-TEN) for enthesitis, in cluster 4 (L-Nails-Skin) for enthesitis, and PsO, and in cluster 6 (L-Nails) for PsO. For a more detailed list of which endpoints (and at which timepoints) showed differential response stratified by clusters, see online supplemental table 2. Note that only those Bootstrap CIs that exclude zero have been deemed statistically significant.
Figure 3.
Relative mean response curves for patients on secukinumab. Relative mean response curves for patients on secukinumab 150 mg (red curves) and 300 mg (green curves) across the seven clusters for the six clinical indicators. The red curves depicted the relative mean responses for patients having received secukinumab 150 mg, while the green curves depicted that for secukinumab 300 mg. It is to be noted that the two relative dose response curves always started at baseline at the same mean value equal to 1 to adjust for different starting values. The coloured shaded regions represented 95% bootstrap CIs.
Figure 4.
Differences between the relative mean responses to secukinumab doses. Differences between the relative mean responses to secukinumab 300 and 150 mg across the seven clusters for each of the six clinical indicators. The curves pass through zero at baseline to ensure that the observed treatment differences are not due to the two patient populations starting from a different baseline disease activity. The shaded region provides a Bonferroni adjusted 95% bootstrap CI to make inference on treatment differences. If the shaded region excluded the zero line, (the red dashed line) then the longitudinal treatment difference between secukinumab 300 and 150 mg is deemed statistically significant. If, however, the shaded region included the zero line then there was not enough evidence to claim a treatment difference for that cluster and clinical indicator.
Discussion and conclusion
Unsupervised ML of the pooled FUTURE 2–5 database across the five PsA clinical manifestations (joints, entheses, dactylitis, skin and nails) identified seven distinct phenotypically defined patient clusters. The 206 baseline clinical variables, including individual tender or swollen peripheral joints, tenderness of the six LEI sites, dactylitis of the fingers and the toes, the PASI components of separate body areas, and mNAPSI scores of the fingers, were not previously classified or categorised for each of the 1894 patients included in this analysis; the clusters were, however, clinically relevant. In particular, patients in cluster 1 with VH polyarticular burden for both tender and swollen joints, high entheseal and moderate dactylitic involvement and mild-to-moderate skin and nail PsO were distinguished from patients in cluster 2 with high polyarticular burden only in terms of joint tenderness and mild-to-moderate activity for all other disease domains. For cluster 3, the high polyarticular burden was mainly driven by the feet in terms of swelling and tenderness of the joints and dactylitic involvement of the feet. Clusters 4, 5, 6 and 7 were all characterised by low polyarticular burden, and were grouped together based on the presence of nail or skin PsO separately or in combination.
It is worth noting that despite the FUTURE dataset including predominantly polyarticular patients due to the inclusion criteria of the trials, the seven identified clusters presented with variable joint distribution patterns in terms of swelling and/or tenderness, number of affected joints and feet involvement. For example, cluster 2 is marked by high polyarticular burden in tender joints and cluster 3 by high burden in the joints of the feet.
Furthermore, in an initial exploration of the relative means of longitudinally collected outcome measures for secukinumab 300 mg vs 150 mg, good responses were generally shown for both doses of secukinumab across all patient clusters. Nevertheless, as depicted by the two relative dose response curves starting at baseline at the same mean value equal to 1 to adjust for different starting values, these clusters follow variable response trajectories. Additionally, in the analysis of the difference of the relative means of the outcome measures for secukinumab 300 mg vs 150 mg, although some of the findings, such as secukinumab 300 mg performing better in PsO patients were largely anticipated, secukinumab 300 mg performed better for enthesitis in cluster 2 and cluster 4.
A limitation of the current analysis was that it was performed on a population of RCTs and therefore patients with oligoarthritis, although highly prevalent in the real world, represent a very small subgroup of the present dataset. Additionally, only certain clinical variables were included in the analyses, which makes the assumption that these were the important factors in identifying subgroups. Furthermore, information on axial PsA was limited at the time of this analysis and data from recent trials specifically those evaluating PsA patients with axial manifestations could not be integrated in the pooled dataset.
This was a proof-of-concept study; hence the potential of these clusters to determine PsA patient phenotypes with predictive value in terms of response to therapy should be further assessed through available efficacy data from other classes of drugs with a different mode of action, head-to-head trials and longitudinal observational studies. Additionally, future studies may consider integrating similar methods or applying such models to assess response to therapy and potentially identify subgroups of PsA patients with predictive implications. Further studies in this direction have the potential to expand the therapeutic or prognostic significance of the identified PsA clusters by anticipating disease trajectories in specific patient phenotypes, tailoring treatment choices, optimising therapeutic regimens, and ultimately, improving clinical care of PsA patients.
Acknowledgments
The authors thank the patients and study investigators who participated in the clinical studies from which data have been reported in this manuscript. Authors thank John Gallagher, Novartis Pharmaceuticals UK, London, UK for valuable review and publication support. Medical writing support, under the guidance of the authors, was provided by M K Vivek Sanker and Hemangi N Rawal, Novartis Healthcare Private Limited, India. The first draft of this manuscript was written by M K Vivek Sanker based on inputs from all the authors.
Footnotes
Contributors: All authors were involved in the drafting and critical review of the manuscript and approved the final version for submission. EP was responsible for overall content as the guarantor. PN, CTR, BWK, LCC and IBM were involved in the acquisition of clinical data and participated as investigators in the clinical study from which data are reported in the manuscript. EP, MK, GL, LP, GS and IBM were involved with the conception or design of the work. MK and GL were involved with the analysis of the data in the manuscript. All authors were involved with the interpretation of data in the manuscript. All authors agreed to be accountable for all aspects of the work and attest to the accuracy and integrity of the work.
Funding: The research study was funded by Novartis Pharma AG, Basel, Switzerland.
Competing interests: EP: Shareholder and Employee of Novartis. MK: Shareholder and Employee of Novartis. PN: Speaker’s bureau: Novartis, Eli Lilly and AbbVie. CTR: Research grants: AbbVie, Amgen, UCB; Consultant for: AbbVie, Amgen, UCB, Novartis, Pfizer, Lilly, Janssen, BMS. BWK: Research grants, consultation fees, or speaker honoraria: AbbVie, Gilead, Janssen, Lilly, Novartis, Pfizer and UCB. GL: Shareholder and Employee of Novartis. LP: Shareholder and Employee of Novartis. AO: Consultant: AbbVie, Amgen, BMS, Celgene, Corrona, Gilead, Janssen, Lilly, Novartis, Pfizer, UCB. Research grants: Novartis (to Penn), Pfizer (to Penn), Amgen (to Forward). Royalties to husband from Novartis. LCC: Grant/research support: AbbVie, Amgen, Gilead, Janssen, Lilly, Novartis, Pfizer Consultant/speaker for: AbbVie, Amgen, Biogen, Celgene, Pfizer, UCB, Boehringer Ingelheim, Novartis, Lilly, Janssen, Gilead, Medac. GS: Speakers honoraria from AbbVie, BMS, Celgene, Janssen, Lilly, Novartis, Roche and UCB. IBM: Research grants, consultation fees, or speaker honoraria: AbbVie, Amgen, BMS, Celgene, Janssen, Lilly, Novartis, Pfizer and UCB.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information. The datasets generated during and/or analysed during the current study are not publicly available. Novartis is committed to sharing with qualified external researchers’ access to patient-level data and supporting clinical documents from eligible studies. These requests are reviewed and approved on the basis of scientific merit. All data provided is anonymised to respect the privacy of patients who have participated in the trial in line with applicable laws and regulations. The data may be requested from the corresponding author of the manuscript.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.Coates LC, Helliwell PS. Psoriatic arthritis: state of the art review. Clin Med 2017;17:65–70. 10.7861/clinmedicine.17-1-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hügle M, Omoumi P, van Laar JM, et al. Applied machine learning and artificial intelligence in rheumatology. Rheumatol Adv Pract 2020;4:rkaa005. 10.1093/rap/rkaa005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson DS, Krutzinna J, Bruce IN, et al. Clinical applications of machine learning algorithms: beyond the black box. BMJ 2019;364:l886. 10.1136/bmj.l886 [DOI] [PubMed] [Google Scholar]
- 4.Grys BT, Lo DS, Sahin N, et al. Machine learning and computer vision approaches for phenotypic profiling. J Cell Biol 2017;216:65–71. 10.1083/jcb.201610026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orange DE, Agius P, DiCarlo EF, et al. Identification of three rheumatoid arthritis disease subtypes by machine learning integration of synovial histologic features and RNA sequencing data. Arthritis Rheumatol 2018;70:690–701. 10.1002/art.40428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seyhan AA, Carini C. Are innovation and new technologies in precision medicine paving a new era in patients centric care? J Transl Med 2019;17:114. 10.1186/s12967-019-1864-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McInnes IB, Mease PJ, Kirkham B, et al. Secukinumab, a human anti-interleukin-17A monoclonal antibody, in patients with psoriatic arthritis (future 2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2015;386:1137–46. 10.1016/S0140-6736(15)61134-5 [DOI] [PubMed] [Google Scholar]
- 8.Nash P, Mease PJ, McInnes IB, et al. Efficacy and safety of secukinumab administration by autoinjector in patients with psoriatic arthritis: results from a randomized, placebo-controlled trial (future 3). Arthritis Res Ther 2018;20:47. 10.1186/s13075-018-1551-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kivitz AJ, Nash P, Tahir H, et al. Efficacy and Safety of Subcutaneous Secukinumab 150 mg with or Without Loading Regimen in Psoriatic Arthritis: Results from the FUTURE 4 Study. Rheumatol Ther 2019;6:393–407. 10.1007/s40744-019-0163-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mease P, van der Heijde D, Landewé R, et al. Secukinumab improves active psoriatic arthritis symptoms and inhibits radiographic progression: primary results from the randomised, double-blind, phase III future 5 study. Ann Rheum Dis 2018;77:annrheumdis-2017-212687–7. 10.1136/annrheumdis-2017-212687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLachlan G, Peel D. Finite mixture models. New York, USA: John Wiley & Sons, 2004. [Google Scholar]
- 12.Hennig C, Liao TF. How to find an appropriate clustering for mixed-type variables with application to socio-economic stratification. Journal of the Royal Statistical Society: Series C 2013;62:309–69. 10.1111/j.1467-9876.2012.01066.x [DOI] [Google Scholar]
- 13.Hennig C. fpc: flexible procedures for clustering. R package version 2.2-9, 2020. Available: https://CRAN.R-project.org/package=fpc
- 14.McInnes I, Kormaksson M, Pournara E. Clinically Relevant Patient Clusters Identified by Machine Learning Tools in a Large Database from the Secukinumab Psoriatic Arthritis Clinical Development Program [abstract]. Arthritis Rheumatol 2019;71 https://acrabstracts.org/abstract/clinically-relevant-patient-clusters-identified-by-machine-learning-tools-in-a-large-database-from-the-secukinumab-psoriatic-arthritis-clinical-development-program/ [Google Scholar]
- 15.The R project for statistical computing 2020.
- 16.Schwarz G. Estimating the dimension of a model. Ann Statist 1978;6:461–4. 10.1214/aos/1176344136 [DOI] [Google Scholar]
- 17.Monti S, Tamayo P, Mesirov J. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn 2003;52:91–118. 10.1023/A:1023949509487 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
rmdopen-2021-001845supp001.pdf (574.3KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information. The datasets generated during and/or analysed during the current study are not publicly available. Novartis is committed to sharing with qualified external researchers’ access to patient-level data and supporting clinical documents from eligible studies. These requests are reviewed and approved on the basis of scientific merit. All data provided is anonymised to respect the privacy of patients who have participated in the trial in line with applicable laws and regulations. The data may be requested from the corresponding author of the manuscript.