Abstract
Puberty is a critical period of development regulated by genetic, nutritional, and environmental factors. The role of makorin ring finger protein 3 (MKRN3) in the regulation of pubertal timing was revealed when loss-of-function mutations were identified in patients with central precocious puberty (CPP). To date, MKRN3 mutations are the most common known genetic cause of CPP. MKRN3 is a member of the makorin family of ubiquitin ligases, together with MKRN1 and MKRN2. The Mkrn genes have been identified in both vertebrates and invertebrates and show high evolutionary conservation of their gene and protein structures. While the existence of Mkrn orthologues in a wide spectrum of species suggests a vital cellular role of the makorins, their role in puberty initiation and endocrine functions is just beginning to be investigated. In this review, we discuss recent studies that have shown the involvement of Mkrn3 and other makorins in the regulation of pubertal development and other endocrine functions, including metabolism and fertility, as well as their underlying mechanisms of action.
Keywords: MKRN3, puberty, E3 ubiquitin ligase, makorins
Pubertal Development and Neuroendocrine Regulation
Puberty is a complex developmental event by which the organisms acquire sexual maturation, characterized by the acquisition of secondary sexual characteristics, gonadal maturation, and attainment of reproductive capacity. Over evolutionary time, the pressure on a species to be able to reproduce rapidly has been counterbalanced with the need to allow enough time for robust juvenile development. Thus, puberty is a critical period of development for which the timing is genetically determined and tightly regulated by a multitude of metabolic and environmental factors.1–3 Reproduction is controlled by the hypothalamic–pituitary–gonadal (HPG) axis. Within the HPG axis, puberty is triggered by an increase in secretion of the neuropeptide, gonadotropin-releasing hormone (GnRH). GnRH neurons reside in the hypothalamus and send axons to the median eminence from which GnRH is released into the hypophysial portal circulation to stimulate the secretion of the pituitary gonadotropins, luteinizing hormone, and follicle-stimulating hormone, necessary for activation of gonadal function.
In humans, GnRH is secreted in a pulsatile manner during the embryonic and neonatal periods, followed by a period of quiescence during infancy and a reactivation of its secretion at puberty.4 GnRH deficiency leads to hypogonadotropic hypogonadism (HH), in which patients fail to undergo pubertal development and are usually infertile.5,6 In addition, a delay in the reactivation of GnRH secretion leads to delayed puberty, whereas early reactivation of GnRH secretion results in central precocious puberty (CPP). A current hypothesis proposes the existence of a predominantly inhibitory network that induces quiescence of GnRH secretion during the infantile/prepubertal period. This inhibition is proposed to decrease around the time of puberty initiation, together with a gain in activity of an excitatory network.
A substantial number of studies in the past few decades have investigated these neural networks, and several neuropeptides and neurotransmitters have been implicated in the intricate balance between inhibitory and excitatory inputs to GnRH neurons. Kisspeptin, encoded by the Kiss1 gene, is a potent activator of GnRH secretion.7 In humans, loss-of-function mutations in KISS1 and KISS1R genes have been associated with HH.8,9 Rare activating mutations of KISS1 and KISS1R have also been identified in patients with CPP.10,11 In addition, loss-of-function mutation in TAC3 and TAC3R genes, encoding neurokinin B and its receptor, have been identified in patients with HH.12–14 In mice and sheep, coexpression of kisspeptin and neurokinin B observed in the same neurons in the arcuate nucleus, a critical region for pubertal regulation, further confirmed their importance in the control of puberty initiation and subsequent fertility. These neurons also express dynorphin and are called the “KNDy neurons.”15,16 In addition to kisspeptin, the neurotransmitters gamma-aminobutyric acid (GABA) and glutamate have been shown to send excitatory inputs to GnRH neurons at the time of puberty and to be critical for the pubertal activation of GnRH neurons.17
Surprisingly, while the players involved in the stimulation of GnRH neurons at the time of puberty have been studied quite extensively, it is only very recently that inhibitors of GnRH secretion, critical to the maintenance of the childhood quiescent period, have been identified. In 2013, MKRN3 loss-of-function mutations were reported in patients with CPP and are to date the most common genetic cause of this disease.18 In addition, deletions in the DLK1 genewere recently identified in patients with familial CPP.19,20 Interestingly, MKRN3 and DLK1 are both imprinted genes, expressed only from the paternally inherited allele. Whilethemechanisms ofaction of MKRN3 and DLK1 on GnRH secretion are still unclear, these findings suggest an important role of genomic imprinting in the regulation of puberty initiation.
MKRN3 Mutations in Central Precocious Puberty
The role of MKRN3 in puberty initiation was revealed by Abreu et al in 2013 through whole-exome sequencing analysis of patients with familial CPP. In this study, loss-of-function mutations were identified in both girls and boys, in 5 of 15 families investigated.18 Subsequently, mutations in MKRN3 have been described in many additional patients with CPP throughout the world.21–32 For a recent systematic review and meta-analysis of the MKRN3 mutations identified in patients with CPP to date, see the study by Valadares et al.33
Precocious puberty is clinically defined by the development of secondary sexual characteristics before the age of 8 years in girls and 9 years in boys, which corresponds to 2.5 to 3 standard deviations below the mean age of puberty onset, as defined by population studies.34 Early age of puberty has been associated with many deleterious health effects such as cancer, cardiovascular disease, and metabolic and behavioral disorders.35–37 To date, in patients with CPP due to MKRN3 mutations, the median age at pubertal onset is 6.0 years in girls (ranging from 3.0 to 7.8) and 8.5 years in boys (ranging from 5.9 to 9.0).33 The significant role of MKRN3 in the regulation of pubertal timing is further supported by the results of recent genome-wide association studies (GWAS) that found associations between several paternally inherited MKRN3 variants and the age at menarche.36,38 Regarding its mechanism of action, it has been shown, in rodents, that Mkrn3 expression declines in the hypothalamus during postnatal/pubertal maturation.18 These findings, together with the identification of loss-of-function mutations in patients with CPP, suggest an inhibitory role of this protein on GnRH secretion during the prepubertal quiescent period.
MKRN3 belongs to the makorin family of ubiquitin ligases, together with MKRN1, MKRN2, and MKRN4. The MKRN genes have been described in both vertebrate and invertebrate organisms, showing high conservation throughout evolution. While an important role of MKRN3 in pubertal timing has been demonstrated in human studies, the mechanisms of action of MKRN3 as well as the roles of the other makorins in reproduction remain to be elucidated. This review presents a state-of-the-art review of the known roles of the makorins in animals and their potential mechanisms of action, with a focus on their potential role in regulating the HPG axis.
The Makorin Protein Family
The makorins are zinc finger proteins with a unique composition and order of structural motifs, including several C3H zinc fingers, a motif rich in Cys and His residues, and a RING zinc finger.39,40 From this signature profile, it is possible to predict their functions. C3H zinc fingers have been found in a variety of ribonucleoproteins and suggest a function as RNA-binding proteins.41 RNA-binding proteins regulate posttranscriptional RNA processing at multiple levels including alternative splicing, mRNA stability, mRNA localization, and translation efficiency. The RING zinc finger domain is found in most E3 ubiquitin ligases, a category of enzymes that mediate the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme to target protein substrates. The E2–E3 complexes can mono-ubiquitinate a lysine substrate or synthesize a polyubiquitin chain of lysine residues. These modifications have a range of biological effects on the target protein substrate, from proteasome-dependent proteolysis to posttranslational control of protein function, structure, assembly, and/or localization.42 The function of the motif rich in Cys and His residues is still unknown.43 ►Figs. 1 and 2 illustrate the similarities and differences in the gene and protein structures of the members of the makorin protein family.
Fig. 1.
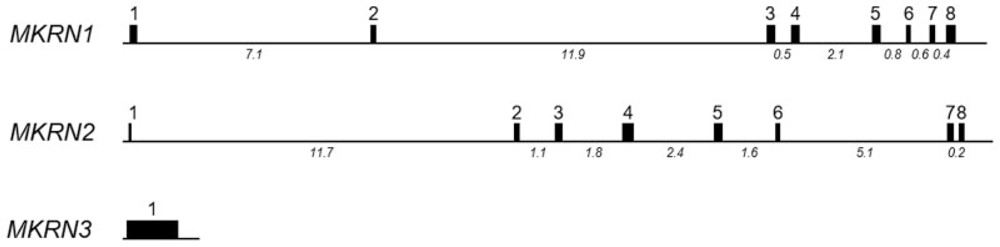
Schematic gene structures of human MKRN1, MKRN2, and MKRN3 loci. Exons are represented as boxes with sequential numbers, with the size of the box in proportion to the size of the exon. Numbers appearing below the introns indicate the size of the introns.
Fig. 2.
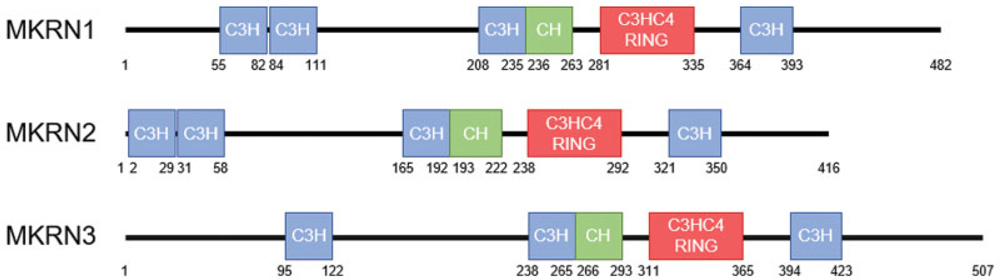
Protein structures of human MKRN1, MKRN2, and MKRN3. The makorins contain several C3H zinc finger domains (blue), a makorin-type Cys-His (CH) motif (green) and a C3HC4 RING zinc finger domain (red). Numbers appearing below each protein indicate the amino acid positions in the protein.
The first member of the makorin gene family identified was makorin ring finger protein 3 (MKRN3/ZNF127) in 1999 by Jong et al.39 MKRN3 was identified as one of several maternally imprinted genes in the Prader–Willi syndrome (PWS) critical region of human chromosome 15q11.2, which are expressed only from the paternally inherited allele.40 In 2000, the same research group identified the ancestralgene for this zinc-finger protein-encoding gene family, named makorin ring finger protein 1 (MKRN1).43 In addition, the gene makorin ring finger protein 2 (MKRN2) was subsequently identified in humans.44,45 The last member of the makorin gene family, makorinring finger protein 4 (mkrn4), was identified in 2010 in the platyfish Xiphophorus maculatus. To date, no study has investigated the potential functions of mkrn4, although it is also present in thehumangenome, where it was annotated as a pseudogene (and is thus not represented in the figures).46
In summary, four Mkrn genes have been identified in vertebrates: Mkrn1, Mkrn2, Mkrn3, and Mkrn4, which encode for their respective proteins. Interestingly, comparative analyses in different species revealed that different evolutionary dynamics affected the various Mkrn genes. Mkrn3, which shows specificity for therian mammals, corresponds to an intronless retrocopy of Mkrn1 generated through reverse transcription of Mkrn1 mRNA. The formation of such a retrogene is catalyzed by reverse transcription of generally mature mRNA molecules followed by integration of the formed cDNA into a new location of the genome.46 The presence of Mkrn3 in dogs, mice, and human, combined with its absence from chicken, fish, and platypus genomes, suggests that Mkrn3 was acquired by the PWS region 80 to 90 million years ago.47 On the other hand, Mkrn1, Mkrn2, and Mkrn4 have been found in both tetrapod and ray-finned fish, tracing back their origin to at least 450 million years ago. These genes were formed through gene duplications and comparative genomic analyses suggest that Mkrn2 and Mkrn4 were duplicated together within a large-scale duplication process. Moreover, comparative expression analyses showed strong gonad-specific expression of Mkrn1, Mkrn2, and Mkrn4 in primitive vertebrates, suggesting an ancestral role of the single-copy mkrn gene in sexual development and gonad function before duplication in vertebrates.46 While the functions of Mkrn proteins are still unclear, several functional analyses have been undertaken to understand the involvement of each makorin in vertebrate cellular and physiological processes.
Makorin Ring Finger Protein 1
Mkrn1, identified as the ancestral gene for the makorin gene family, has been the most studied gene. Unlike MKRN3, MKRN1 is not imprinted.48 In humans, the MKRN1 gene maps to chromosome 7q34-q35 and contains eight exons (►Fig. 1). The transcript consists of a contiguous sequence of 3,094 nucleotides and encodes a protein of 482 amino acids with a predicted molecular mass of 53.3 kDa (►Fig. 2). The mouse Mkrn1 gene maps to chromosome 6A and contains also eight exons. The Mkrn1 transcript is 3,046 nucleotides in length and encodes a protein of 481 amino acids with a predicted molecular mass of 53.1 kDa.43 The human and mouse Mkrn1 orthologs have a high (92%) identity. A vital cellular role of Mkrn1 is reinforced by the existence of Mkrn1 orthologs in a wide spectrum of species (e.g., Tammar wallaby, chicken, drosophila, c-elegans, plant, and fungal species, in addition to human and mouse). Expression analyses in human tissues showed that MKRN1 is highly and ubiquitously expressed, including in different brain regions such as the hypothalamus and the amygdala. In mice, Mkrn1 shows a more specific expression pattern, with higher expression in the testes compared with other somatic tissues. In addition, Mkrn1 is highly expressed during mouse embryonic brain and neural tube development.43
Several Mkrn1 functions have been linked to its E3 ubiquitin ligase activity and several targets have been identified. MKRN1 has been found to mediate the ubiquitination of the human telomerase reverse transcriptase (hTERT) and promote its proteasome-mediated degradation, thus playing a negative role in the telomere length homeostasis.49 MKRN1 was also able to control cell cycle arrest and apoptosis via ubiquitination and proteasome-dependent degradation of both p53 and p21.50 These results suggested a critical role of MKRN1 in cancer and tumorigenesis. MKRN1 has also been associated with in vitro adipocyte differentiation through its E3 ubiquitin ligase action, by targeting peroxisome-proliferator–activated receptor γ (PPARγ) for proteasomal degradation.51 A further endocrine role of MKRN1 in the regulation of metabolism was recently revealed using a model of Mkrn1 null mice. Precedent studies showed that Mkrn1 null mice are viable and fertile with no apparent developmental deficit or overt phenotype.48 Lee and collaborators showed that Mkrn1 acts as an E3 ubiquitin ligase for AMP-activated protein kinase (AMPK) in the liver and adipocyte tissues but not in the hypothalamus. No differences in body weight were observed under standard chow diet compared with control animals; however, Mkrn1 null mice fed with a highfat diet (HFD) exhibited 25 to 30% lower body weight than wild-type animals, without any obvious differences in food intake. Thus, the lack of Mkrn1 was sufficient to result in chronic activation of AMPK and subsequently lead to systemic effects by preventing nonalcoholic fatty liver disease, insulin resistance, and HFD-induced obesity. These findings highlighted the potential application of Mkrn1 as a new therapeutic strategy for the treatment of metabolic disorders.52
In addition to its E3 ubiquitin ligase activity, transcriptional activities have been described for Mkrn1. It has been shown that Mkrn1 can regulate RNA polymerase II-dependent transcription. It can inhibit the transcription of c-Jun as well as the androgen receptor and the retinoic acid receptor. Moreover, when tethered to DNA via a heterologous DNA-binding domain (DBD; GAL4), Mkrn1 can also activate transcription, suggesting that Mkrn1 may be a transcriptional activator. Interestingly, it has been demonstrated that its transrepression function is independent of its ubiquitin ligase activity.53
Furthermore, some studies have demonstrated that Mkrn1 can associate with RNA-binding proteins and support a role as a ribonucleoprotein. In mammalian neurons, it has been shown that a short isoform of Mkrn1 interacts with the cytoplasmic poly(A)-binding protein and stimulates translation of dendritic mRNA at the synapses.54 Proteomic analyses have also established MKRN1 as a component of messenger ribonucleoprotein complexes in undifferentiated embryonic stem cells (ESCs).55 Further studies are needed to establish if Mkrn1 plays any role in puberty initiation in vertebrates.
Makorin Ring Finger Protein 2
MKRN2 arose by gene duplication of MKRN1. In humans, the MKRN2 genomic locus contains, like MKRN1, eight exons that map to chromosome 3p25 (►Fig. 1). Two transcripts of human MKRN2 have been identified that contain 2,775 and 1,436 nucleotides, respectively. MKRN2 encodes a protein of 416 amino acids with a predicted molecular mass of 47.0 kDa (►Fig. 2). In the mouse, the Mkrn2 gene also contains eight exons and maps to chromosome 6. The Mkrn2 transcript length is 6,230 nucleotides and encodes a protein of 416 amino acids with a predictedmolecular weightof 46.5 kDa.45 Inbothhuman and mouse, the 3′ untranslated region (3′UTR) of Mkrn2 overlaps, in an antisense orientation, with the 3′UTR of the RAF1 gene. This region ofoverlap shows 70% identity between mouse and human, whereas the remainder of the 3′UTR shares 37% nucleotide identity. Mkrn2 orthologs have also been identified in a variety of species, including yellowtail tuna, zebrafish, chicken, Xenopus, elephant shark, opossum, platypus, tetraodon, and medaka, supporting the high evolutionary conservation of this gene. Expression analyses showed that MKRN2 is ubiquitously expressed in human tissues and a variety of cell lines.45 In adult mice, Mkrn2 mRNA and protein were ubiquitously expressed at low levels in various tissues (brain, thymus, heart, lung, liver, spleen, kidney, ovary, uterus, and seminal vesicle) with more specific, high expression in the testis.56
The first study to investigate Mkrn2 function reported that, in Xenopus, Mkrn2 played a negative role in neurogenesis via PI3K/Akt signaling and was essential for proper embryonic neural development.57 The third C3H zinc finger, Cys-His motif, and C3HC4 RING zinc finger domains were indispensable for this inhibitory effect of Mkrn2 on neurogenesis.58 The regulation of the PI3K/Akt pathways by Mkrn2 and the overexpression of Mkrn2 in mammalian cancer cell lines suggest that Mkrn2 may be involved in tumorigenesis.
Recently, E3 ubiquitin ligase activity has been identified for Mkrn2. In vitro studies showed that Mkrn2 interacts with PDLIM2 (PDZ and LIM domain 2, a putative ubiquitin E3 ligase) and promotes p65 ubiquitination and proteasome-dependent degradation cooperatively with PDLIM2, thereby negatively regulating NF-κB-mediated inflammatory responses.59
Interestingly, studies using a Mkrn2 knockout (KO) mouse model reported a role of Mkrn2 in male fertility.56 While male and female Mkrn2 KO animals were viable with no apparent developmental abnormalities, the mutant animals showed a decrease in body weight. Female Mkrn2 KO mice were fertile but had a reduction in their fertility rate. The Mkrn2 KO males were sterile but retained their ability to mate, as indicated by the presence of vaginal plugs in females after mating. Further analyses showed that Mkrn2 KO males did not have sperm or produced sperm with low counts, poor motility, and/or abnormal shapes. In addition, human MKRN2 expression levels were significantly decreased in the sperm of patients with oligoasthenoteratozoospermia, the most common cause of subfertility in men, compared with a control group of fertile men, revealing a correlation between MKRN2 expression and human male infertility.56 Any potential role of Mkrn2 in pubertal development remains to be elucidated.
Makorin Ring Finger Protein 3
MKRN3 was the first member of the makorin family identified, located in the PWS critical region,39 albeit with no obvious role in the disease.60 In humans, MKRN3 is intronless and maps to chromosome 15q11-q13 (►Fig. 1). The MKRN3 transcript contains a sequence of 2,347 nucleotides that encodes a protein of 507 amino acids with a predicted molecular mass of 55.6 kDa. In the mouse, the intronless Mkrn3 gene maps to chromosome 7C. The Mkrn3 transcript is 2,547 nucleotides in length and encodes a protein of 544 amino acids with a predicted molecular weight of 59.4 kDa (►Fig. 2). The human and mouse Mkrn3 orthologs share similar structural motifs and show an overall 69% identity (85% similarity, considering conservative substitutions). The RING zinc-finger motif has the highest degree of conservation with 85% identity (92% similarity), whereas the N- and C-terminal regions are less well conserved.40
Interestingly, in both human and mouse, Mkrn3 is imprinted in various tissues, where it is expressed only from the paternal allele.39,40,61 MKRN3 differential allele expression is regulated through silencing of the MKRN3 maternal allele via DNA methylation.62 The methylation of MKRN3 is regulated together with the other imprinted genes of the PWS critical region by an imprinting center that initiates the imprint switching in the germline and generates a molecular signal that spreads over the entire imprinted domain of the chromosome 15q11-q13.63 Interestingly, the mouse Mkrn3 gene is methylated in a tissue-specific manner, with a high level of methylation on the maternal allele in the mouse adult brain and the embryonal head at E13.5.62
As mentioned earlier, Mkrn3 is conserved only in therian mammals (human, mouse, cow, opossum), thus suggesting a more recent appearance in the course of evolution, compared with the other members of the makorin gene family.46
In both mice and humans, MKRN3 is ubiquitously expressed in adult tissues, with the highest level in the testis. MKRN3 showed a predominant expression in the brain and lung of human fetal tissues.39,40 In the m ouse, Mkrn3 gene expression has been found from the blastocyst stage and embryonic days 8 to 17, as well as in ESCs.40 In addition, it has been demonstrated that Mkrn3 is highly expressed in the murine hypothalamic arcuate nucleus, a critical region for the regulation of puberty initiation, during the infantile and early juvenile period, with a reduction in expression starting at postnatal days 12 to 15, before puberty initiation. Mkrn3 expression then remained low through day 45, the oldest age at which the mice were tested.18
While the mechanism of action of Mkrn3 in the regulation of puberty initiation is still unclear, some studies have begun to report investigations of possible targets of its action. In the mouse, Mkrn3 has been shown to interact with the neural prentraxin-1 precursor (Nptx1) in the hypothalamus of 4-week-old mice. Nptx1 has been found to play a role in neural differentiation. The C3HC4 ring finger domain appeared to be essential for the Mkrn3–Nptx1 interaction. Indeed, this interaction was not observed in a co-immunoprecipitation experiment using a mutant construct of Mkrn3 lacking the RING domain. The study also showed a reduction of the polyubiquitination level of Nptx1 in the hypothalamus of mice injected intracerebroventricularly with the mutant RING domain-deficient Mkrn3 vector compared with control animals injected with the vector encoding wild-type Mkrn3, which suggested that Mkrn3 may be able to modulate Nptx1 via its E3 ubiquitin ligase activity.64 More recently, a protocol for the differentiation of GNRH1-expressing neurons from human-induced pluripotent stem cells (hiPSCs) was developed.65 Using this technique, bi-allelic MKRN3-deficient hiPSCs were generated and differentiated into GNRH1-expressing neurons.66 After 25 days of differentiation, the results showed no difference in the expression levels of GnRH1, nor OTX1 and OTX2, two transcriptional regulators of GnRH, between wild-type and MKRN3-deficient cells. These findings suggest that MKRN3 is dispensable for GnRH neuron differentiation and GnRH1 expression. In addition, mass spectrometry analyses performed on an MKRN3-expressing HEK stable cell line revealed 81 novel high-confidence protein interaction partners of MKRN3, which are implicated in various cellular processes such as insulin signaling, RNA metabolism, and cell–cell adhesion.66 Among them, 20 interactors, including LIN28B, have been associated previously with age at menarche in GWAS studies.36,38 MKRN3 was also found to interact with OTUDS, a protein linked to HH.66 Further studies are needed to investigate the physiological significance and the molecular mechanisms underlying these interactions.
Makorin Protein Family in Invertebrates
Interestingly, recent studies in two invertebrate models, Drosophila melanogaster and the nematode Caenorhabditis elegans, revealed a role for makorin orthologs in the regulation of developmental timing. In Drosophila, four makorin genes have been identified, although three are retrocopies of Mkrn1, the only bona fide ortholog of vertebrate Mkrn genes. A recent study demonstrated that Mkrn1 controls larval developmental timing and body size by regulating the steroid hormone ecdysone production.67 Comparable to GnRH action in mammals, ecdysone controls, in a pulsatile manner, the insect developmental transitions from larval to pupal stages. The loss of mkrn1 in the flies delayed the onset of metamorphosis by reducing the expression of the ecdysone-synthesizing enzyme and its downstream targets. Moreover, although the male Mkrn1 null flies were fertile, the mutant females were sterile. Further analyses indicated that Mkrn1 functions within the ovaries as a regulator of the insulin/Tor pathway to activate oogenesis in a nutrient-sensitive manner.68 Other recent studies reported a role of the makorin lep-2, an ortholog of vertebrate Mkrn, in the timing of sexual maturation in the nematode C. elegans. Lep-2 mutations induced a delay in the juvenile to adult transition, with striking defects in neuronal maturation. The lep-2 mutant adult males also showed an absence of sexually mature characteristics and defects in male mating behavior.69,70 Functionally, lep-2 negatively regulated lin-28 posttranslationally by promoting lin-28 degradation, suggesting E3 ubiquitin ligase activity of lep-2 on lin-28.69 Intriguingly, these data revealed an opposite action of the C. elegans makorin lep-2 and the human MKRN3 in the regulation of developmental timing. Remarkably, while overexpression of the human MKRN3 was not able to rescue the defects of C. Elegans lep-2 mutants, experiments in which the human MKRN3 was overexpressed in the wild-type C. elegans nervous system showed that MKRN3 retained its ability to inhibit some aspects of sexual maturation. This finding suggests the existence of a strong functional conservation of the mechanisms by which MKRN3 regulates pubertal timing.70
Taken together, these findings support the notion that makorin proteins play important roles in the regulation of the timing of puberty and sexual maturation in both invertebrate and vertebrate animals, suggesting an evolutionarily conserved role of this E3 ubiquitin ligase protein family.
Conclusion
In conclusion, the discovery of MKRN3 mutations in patients with CPP opened the door to unveiling the role of makorins in the initiation of puberty as well as more broadly in the regulation of the HPG axis. To date, the data collected show that makorins are strongly conserved across the evolution, both by their genes and protein structures than by their functions. While the critical role of MKRN3 in puberty initiation has been demonstrated in humans, the involvement of other makorins in this process is still unknown. The current absence of MKRN1 and MKRN2 mutations identified in patients with CPP, delayed puberty, or HH does not eliminate a potential contribution of these genes in the regulation of puberty initiation or regulation of the HPG axis in adulthood. Although further studies are needed, primary results using Mkrn1 and Mkrn2 knockout mouse models suggest a function of these genes at the levels of the gonads. Interestingly, sex differences have been observed in many studies, showing sex-specific effects that favor one sexor the other depending on the parameter evaluated. Although to date it is unclear whether sex differences exist in the effect of MKRN3 on puberty initiation, it has been suggested that girls with MKRN3 mutations have a greater advancement in the age of puberty initiation than do boys.33 Further studies that take into account sex differences in both human and animal models are needed to better understand these possible sex differences. Interestingly, the observation of a conserved role of the makorins in both vertebrate and invertebrate animals underscores the important role that makorins appear to play in the timing of pubertal development. Further studies will enlighten the specific roles of each makorin in the initiation of puberty and reproductive function across species and will elucidate the underlying mechanisms of action.
Acknowledgments
This work was supported by NIH R01 HD082314 and by the Women’s Brain Initiative, Program for Interdisciplinary Neuroscience, Brigham, and Women’s Hospital (to U.B.K.).
Footnotes
Conflict of Interest
The authors have nothing to disclose.
References
- 1.Abreu AP, Kaiser UB. Pubertal development and regulation. Lancet Diabetes Endocrinol 2016;4(03):254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plant TM. Neuroendocrine control of the onset of puberty. Front Neuroendocrinol 2015;38:73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avendaño MS, Vazquez MJ, Tena-Sempere M. Disentangling puberty: novel neuroendocrine pathways and mechanisms for the control of mammalian puberty. Hum Reprod Update 2017;23 (06):737–763 [DOI] [PubMed] [Google Scholar]
- 4.Palmert MR, Boepple PA. Variation in the timing of puberty: clinical spectrum and genetic investigation. J Clin Endocrinol Metab 2001;86(06):2364–2368 [DOI] [PubMed] [Google Scholar]
- 5.Schwanzel-Fukuda M, Bick D, Pfaff DW. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res Mol Brain Res 1989;6(04):311–326 [DOI] [PubMed] [Google Scholar]
- 6.Bianco SDC, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol 2009;5(10):569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke SA, Dhillo WS. Kisspeptin across the human lifespan: evidence from animal studies and beyond. J Endocrinol 2016; 229(03):R83–R98 [DOI] [PubMed] [Google Scholar]
- 8.Seminara SB, Messager S, Chatzidaki EE, et al. The GPR54 gene as a regulator of puberty. N Engl J Med 2003;349(17):1614–1627 [DOI] [PubMed] [Google Scholar]
- 9.de Roux N, Genin E, Carel J-C, Matsuda F, Chaussain J-L, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A 2003;100(19):10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teles MG, Bianco SDC, Brito VN, et al. A GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med 2008;358(07):709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silveira LG, Noel SD, Silveira-Neto AP, et al. Mutations of the KISS1 gene in disorders of puberty. J Clin Endocrinol Metab 2010;95 (05):2276–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topaloglu AK, Reimann F, Guclu M, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet 2009;41(03):354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young J, Bouligand J, Francou B, et al. TAC3 and TACR3 defects cause hypothalamic congenital hypogonadotropic hypogonadism in humans. J Clin Endocrinol Metab 2010;95(05): 2287–2295 [DOI] [PubMed] [Google Scholar]
- 14.Gianetti E, Tusset C, Noel SD, et al. TAC3/TACR3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab 2010;95(06):2857–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK, Steiner RA. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J Neurosci 2009;29(38):11859–11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehman MN, Coolen LM, Goodman RL. Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology 2010;151(08):3479–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarkson J, Herbison AE. Development of GABA and glutamate signaling at the GnRH neuron in relation to puberty. Mol Cell Endocrinol 2006;254–255:32–38 [DOI] [PubMed] [Google Scholar]
- 18.Abreu AP, Dauber A, Macedo DB, et al. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med 2013;368(26):2467–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dauber A, Cunha-Silva M, Macedo DB, et al. Paternally inherited DLK1 deletion associated with familial central precocious puberty. J Clin Endocrinol Metab 2017;102(05):1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes LG, Cunha-Silva M, Crespo RP, et al. DLK1 is a novel link between reproduction and metabolism. J Clin Endocrinol Metab 2019;104(06):2112–2120 [DOI] [PubMed] [Google Scholar]
- 21.Settas N, Dacou-Voutetakis C, Karantza M, Kanaka-Gantenbein C, Chrousos GP, Voutetakis A. Central precocious puberty in a girl and early puberty in her brother caused by a novel mutation in the MKRN3 gene. J Clin Endocrinol Metab 2014;99(04):E647–E651 [DOI] [PubMed] [Google Scholar]
- 22.Schreiner F, Gohlke B, Hamm M, Korsch E, Woelfle J. MKRN3 mutations in familial central precocious puberty. Horm Res Paediatr 2014;82(02):122–126 [DOI] [PubMed] [Google Scholar]
- 23.de Vries L, Gat-Yablonski G, Dror N, Singer A, Phillip M. A novel MKRN3 missense mutation causing familial precocious puberty. Hum Reprod 2014;29(12):2838–2843 [DOI] [PubMed] [Google Scholar]
- 24.Macedo DB, Abreu AP, Reis ACS, et al. Central precocious puberty that appears to be sporadic caused by paternally inherited mutations in the imprinted gene makorin ring finger 3. J Clin Endocrinol Metab 2014;99(06):E1097–E1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandone A, Cantelmi G, Cirillo G, et al. A case of familial central precocious puberty caused by a novel mutation in the makorin RING finger protein 3 gene. BMC Endocr Disord 2015;15:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon D, Ba I, Mekhail N, et al. Mutations in the maternally imprinted gene MKRN3 are common in familial central precocious puberty. Eur J Endocrinol 2016;174(01):1–8 [DOI] [PubMed] [Google Scholar]
- 27.Simsek E, Demiral M, Ceylaner S, Kırel B. Two frameshift mutations in MKRN3 in Turkish patients with familial central precocious puberty. Horm Res Paediatr 2017;87(06):405–411 [DOI] [PubMed] [Google Scholar]
- 28.Lee HS, Jin HS, Shim YS, et al. Low frequency of MKRN3 mutations in central precocious puberty among Korean girls. Horm Metab Res 2016;48(02):118–122 [DOI] [PubMed] [Google Scholar]
- 29.Ortiz-Cabrera NV, Riveiro-Álvarez R, López-Martínez MÁ, et al. Clinical exome sequencing reveals MKRN3 pathogenic variants in familial and nonfamilial idiopathic central precocious puberty. Horm Res Paediatr 2017;87(02):88–94 [DOI] [PubMed] [Google Scholar]
- 30.Nishioka J, Shima H, Fukami M, et al. The first Japanese case of central precocious puberty with a novel MKRN3 mutation. Hum Genome Var 2017;4:17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bessa DS, Macedo DB, Brito VN, et al. High frequency of MKRN3 mutations in male central precocious puberty previously classified as idiopathic. Neuroendocrinology 2017;105(01):17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stecchini MF, Macedo DB, Reis ACS, et al. Time course of central precocious puberty development caused by an MKRN3 gene mutation: a prismatic case. Horm Res Paediatr 2016;86(02): 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valadares LP, Meireles CG, De Toledo IP, et al. MKRN3 mutations in central precocious puberty: a systematic review and meta-analysis. J Endocr Soc 2019;3(05):979–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenfield RL, Lipton RB, Drum ML. Thelarche, pubarche, and menarche attainment in children with normal and elevated body mass index. Pediatrics 2009;123(01):84–88 [DOI] [PubMed] [Google Scholar]
- 35.Day FR, Elks CE, Murray A, Ong KK, Perry JRB. Puberty timing associated with diabetes, cardiovascular disease and also diverse health outcomes in men and women: the UK Biobank study. Sci Rep 2015;5:11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Day FR, Thompson DJ, Helgason H, et al. ; LifeLines Cohort Study; InterAct Consortium; kConFab/AOCS Investigators; Endometrial Cancer Association Consortium; Ovarian Cancer Association Consortium; PRACTICAL consortium. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet 2017;49(06):834–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lakshman R, Forouhi NG, Sharp SJ, et al. Early age at menarche associated with cardiovascular disease and mortality. J Clin Endocrinol Metab 2009;94(12):4953–4960 [DOI] [PubMed] [Google Scholar]
- 38.Perry JRB, Day F, Elks CE, et al. ; Australian Ovarian Cancer Study; GENICA Network; kConFab; LifeLines Cohort Study; InterAct Consortium; Early Growth Genetics (EGG) Consortium. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 2014;514(7520):92–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jong MT, Gray TA, Ji Y, et al. A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet 1999;8 (05):783–793 [DOI] [PubMed] [Google Scholar]
- 40.Jong MT, Carey AH, Caldwell KA, et al. Imprinting of a RING zincfinger encoding gene in the mouse chromosome region homologous to the Prader-Willi syndrome genetic region. Hum Mol Genet 1999;8(05):795–803 [DOI] [PubMed] [Google Scholar]
- 41.Hall TMT. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 2005;15(03):367–373 [DOI] [PubMed] [Google Scholar]
- 42.Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009;78:399–434 [DOI] [PubMed] [Google Scholar]
- 43.Gray TA, Hernandez L, Carey AH, et al. The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics 2000;66(01):76–86 [DOI] [PubMed] [Google Scholar]
- 44.Zhang QH, Ye M, Wu XY, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34þ hematopoietic stem/progenitor cells. Genome Res 2000;10(10):1546–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray TA, Azama K, Whitmore K, Min A, Abe S, Nicholls RD. Phylogenetic conservation of the makorin-2 gene, encoding a multiple zinc-finger protein, antisense to the RAF1 proto-oncogene. Genomics 2001;77(03):119–126 [DOI] [PubMed] [Google Scholar]
- 46.Böhne A, Darras A, D’Cotta H, Baroiller JF, Galiana-Arnoux D, Volff JN. The vertebrate makorin ubiquitin ligase gene family has been shaped by large-scale duplication and retroposition from an ancestral gonad-specific, maternal-effect gene. BMC Genomics 2010;11:721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rapkins RW, Hore T, Smithwick M, et al. Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet 2006;2(10):e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gray TA, Wilson A, Fortin PJ, Nicholls RD. The putatively functional Mkrn1-p1 pseudogene is neither expressed nor imprinted, nor does it regulate its source gene in trans. Proc Natl Acad Sci U S A 2006;103(32):12039–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JH, Park SM, Kang MR, et al. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev 2005;19(07):776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee E-W, Lee MS, Camus S, et al. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J 2009;28(14):2100–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, Park KW, Lee EW, et al. Suppression of PPARγ through MKRN1-mediated ubiquitination and degradation prevents adipocyte differentiation. Cell Death Differ 2014;21(04):594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee MS, Han HJ, Han SY, et al. Loss of the E3 ubiquitin ligase MKRN1 represses diet-induced metabolic syndrome through AMPK activation. Nat Commun 2018;9(01):3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Omwancha J, Zhou XF, Chen SY, et al. Makorin RING finger protein 1 (MKRN1) has negative and positive effects on RNA polymerase II-dependent transcription. Endocrine 2006;29(02):363–373 [DOI] [PubMed] [Google Scholar]
- 54.Miroci H, Schob C, Kindler S, et al. Makorin ring zinc finger protein 1 (MKRN1), a novel poly(A)-binding protein-interacting protein, stimulates translation in nerve cells. J Biol Chem 2012;287(02): 1322–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cassar PA, Carpenedo RL, Samavarchi-Tehrani P, et al. Integrative genomics positions MKRN1 as a novel ribonucleoprotein within the embryonic stem cell gene regulatory network. EMBO Rep 2015;16(10):1334–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qian X, Wang L, Zheng B, et al. Deficiency of Mkrn2 causes abnormal spermiogenesis and spermiation, and impairs male fertility. Sci Rep 2016;6:39318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang PH, Cheung WKC, Peng Y, et al. Makorin-2 is a neurogenesis inhibitor downstream of phosphatidylinositol 3-kinase/Akt (PI3K/Akt) signal. J Biol Chem 2008;283(13):8486–8495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheung WKC, Yang PH, Huang QH, et al. Identification of protein domains required for makorin-2-mediated neurogenesis inhibition in Xenopus embryos. Biochem Biophys Res Commun 2010; 394(01):18–23 [DOI] [PubMed] [Google Scholar]
- 59.Shin C, Ito Y, Ichikawa S, Tokunaga M, Sakata-Sogawa K, Tanaka T. MKRN2 is a novel ubiquitin E3 ligase for the p65 subunit of NF-κB and negatively regulates inflammatory responses. Sci Rep 2017; 7:46097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanber D, Giltay J, Wieczorek D, et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet 2009;17(05):582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Driscoll DJ, Waters MF, Williams CA, et al. A DNA methylation imprint, determined by the sex of the parent, distinguishes the Angelman and Prader-Willi syndromes. Genomics 1992;13(04): 917–924 [DOI] [PubMed] [Google Scholar]
- 62.Hershko A, Razin A, Shemer R. Imprinted methylation and its effect on expression of the mouse Zfp127 gene. Gene 1999;234 (02):323–327 [DOI] [PubMed] [Google Scholar]
- 63.Nicholls RD, Saitoh S, Horsthemke B. Imprinting in Prader-Willi and Angelman syndromes. Trends Genet 1998;14(05):194–200 [DOI] [PubMed] [Google Scholar]
- 64.Liu H, Kong X, Chen F. Mkrn3 functions as a novel ubiquitin E3 ligase to inhibit Nptx1 during puberty initiation. Oncotarget 2017;8(49):85102–85109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lund C, Pulli K, Yellapragada V, et al. Development of gonadotropin-releasing hormone-secreting neurons from human pluripotent stem cells. Stem Cell Reports 2016;7(02):149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yellapragada V, Liu X, Lund C, et al. MKRN3 interacts with several proteins implicated in puberty timing but does not influence GNRH1 expression. Front Endocrinol (Lausanne) 2019;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tran HT, Cho E, Jeong S, et al. Makorin 1 regulates developmental timing in drosophila. Mol Cells 2018;41(12):1024–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeong EB, Jeong SS, Cho E, Kim EY. Makorin 1 is required for Drosophila oogenesis by regulating insulin/Tor signaling. PLoS One 2019;14(04):e0215688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herrera RA, Kiontke K, Fitch DHA. Makorin ortholog LEP-2 regulates LIN-28 stability to promote the juvenile-to-adult transition in Caenorhabditis elegans. Development 2016;143(05):799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lawson H, Vuong E, Miller RM, Kiontke K, Fitch DH, Portman DS. The Makorin lep-2 and the lncRNA lep-5 regulate lin-28 to schedule sexual maturation of the C. elegans nervous system. eLife 2019;8:e43660. [DOI] [PMC free article] [PubMed] [Google Scholar]


