Abstract
The centromere is the region of the chromosome that directs its segregation in mitosis and meiosis. Although the functional importance of the centromere has been appreciated for over 130 years, elucidating the molecular features and properties that endow centromeres with the capacity to orchestrate chromosome segregation has remained a central ongoing challenge. The defining feature of most eukaryotic centromeres is the presence of nucleosomes containing the histone H3 variant, CENP-A (also known as CenH3), which specifies this region epigenetically. In this review, we synthesize the research on the central features of centromere identity, the molecular basis for centromere propagation to the chromosomes of daughter cells and gametes, and the mechanisms by which the centromere recruits the kinetochore to establish a connection to spindle microtubules.
The transmission of an intact genome to daughter cells during cell division is a fundamental requirement for the viability of cells and organisms. In eukaryotes, DNA is packaged into chromosomes. Each chromosome must be faithfully replicated and segregated at every cell division. To achieve accurate segregation, chromosomes rely on a specialized region known as the centromere. The centromere recruits the kinetochore, a proteinaceous macromolecular structure that forms attachments to the microtubules of the mitotic and meiotic spindles. Together, centromeres and kinetochores are the central players in chromosome segregation. Defects in centromere or kinetochore function can lead to the loss or disruption of genomic information, resulting in profoundly deleterious developmental defects or disease1.
The crucial function of the centromere has been appreciated for over 130 years. The centromere was first observed by light microscopy as the chromosomal attachment site for spindle microtubules in dividing cells2 (Figure 1a). As the centromere protects and maintains sister chromatid cohesion during mitosis and meiosis3–6 this region of the chromosome is also visible in many organisms as the primary constriction on condensed mitotic chromosomes (Figure 1b). Geneticists subsequently combined these cytological observations with the analysis of recombinant progeny to translate genetic maps onto physical ones by defining the positions of genes relative to the centromere7, 8.
Figure 1. Visualization of the centromere.
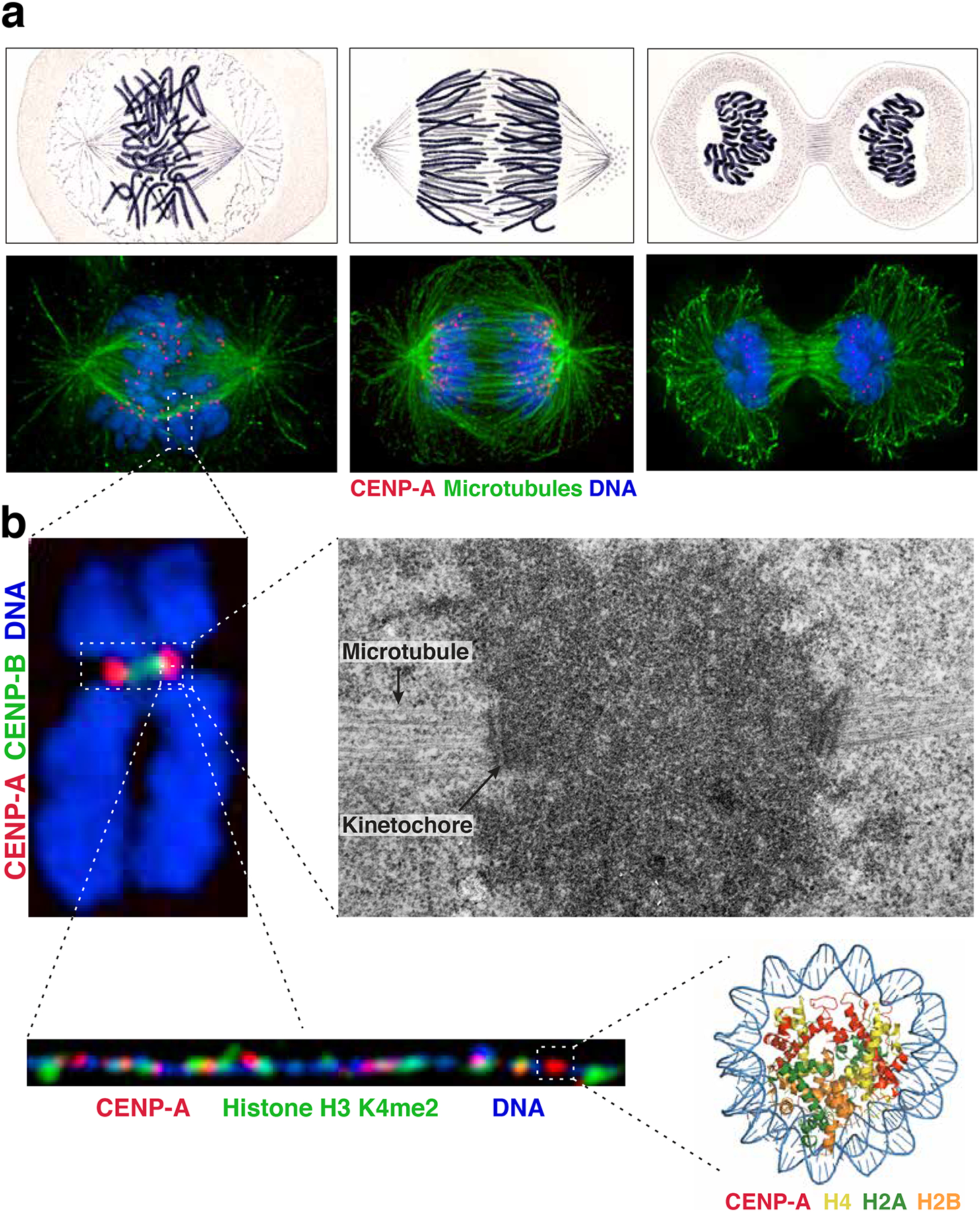
a) Comparison of images of mitotic Salamander cells hand-drawn by Walther Flemming in 18822 (top) with immunofluorescence images of human cells (bottom) stained for microtubules (green), CENP-A (red) and DNA (blue). The images show cells at different phases of a mitotic cell cycle: late prometaphase-metaphase (left), anaphase (middle) and telophase (right). b) Images of the centromere at increasing resolution. Top left: immunofluorescence image of a mitotic chromosome stained for DNA (blue), CENP-A (red) and CENP-B (a marker for the alpha-satellite DNA repeats present at most human centromeres, green). Top right: electron micrograph of centromeric region of a mitotic chromosome showing centromeric chromatin (dark cloud), kinetochore, and microtubules (indicated by arrows). Image courtesy of Conly Rieder. Bottom left: Immunofluorescence image of stretched centromeric chromatin fibers showing patches of CENP-A (red) interspersed with H3, in this case specifically H3 dimethylated on lysine 4 (H3K4me2, green). Image courtesy of Elaine Dunleavy. Bottom right: Crystal structure of the CENP-A nucleosome90. PDB ID: 3AN2
Although the centromere has been described extensively by cytological and genetic approaches, defining the molecular features that confer its functions is a central ongoing challenge9. When first defining the term centromere in 1936, Cyril Darlington commented that “[the centromere must] be considered in terms of function rather than form, since the function is evident and the form elusive”10. Elucidating the “form” of centromeres has remained challenging because centromeres require a complex interplay between numerous molecular features that vary across eukaryotes. Despite this complexity and variation, several common themes have emerged regarding the molecular bases of centromere function. In the vast majority of eukaryotes, centromere specification is primarily epigenetic and depends on the presence of specialized nucleosomes containing the histone H3 variant centromere protein A (CENP-A; also known as CenH3). Centromere function requires the combination of CENP-A-containing nucleosomes, features of the underlying DNA sequence, unique combinations of chromatin marks and interactions with kinetochore proteins.
In this Review, we highlight recent work on the molecular basis for centromere function, with a focus on the vertebrate centromere. We describe the current understanding of the genetic and epigenetic features that define centromeres, the mechanisms of centromere propagation, and the recognition of the centromere by the kinetochore. This work is revealing the elusive form underlying the critical functions of the centromere in the propagation of the genome to cells and gametes.
Centromere DNA structure and function
In the majority of eukaryotes analyzed to date, the centromere is specified epigenetically (Box 1), such that specific DNA sequences are neither strictly necessary nor sufficient for centromere function. Instead, the unifying characteristic of most eukaryotic centromeres is the presence of the histone H3 variant, CENP-A. Nonetheless, recent work has highlighted evolutionary and functional preferences for specific DNA structures that strongly indicate that they contribute to centromere function, as we describe in this section.
Box 1: Evidence for the epigenetic nature of the centromere.
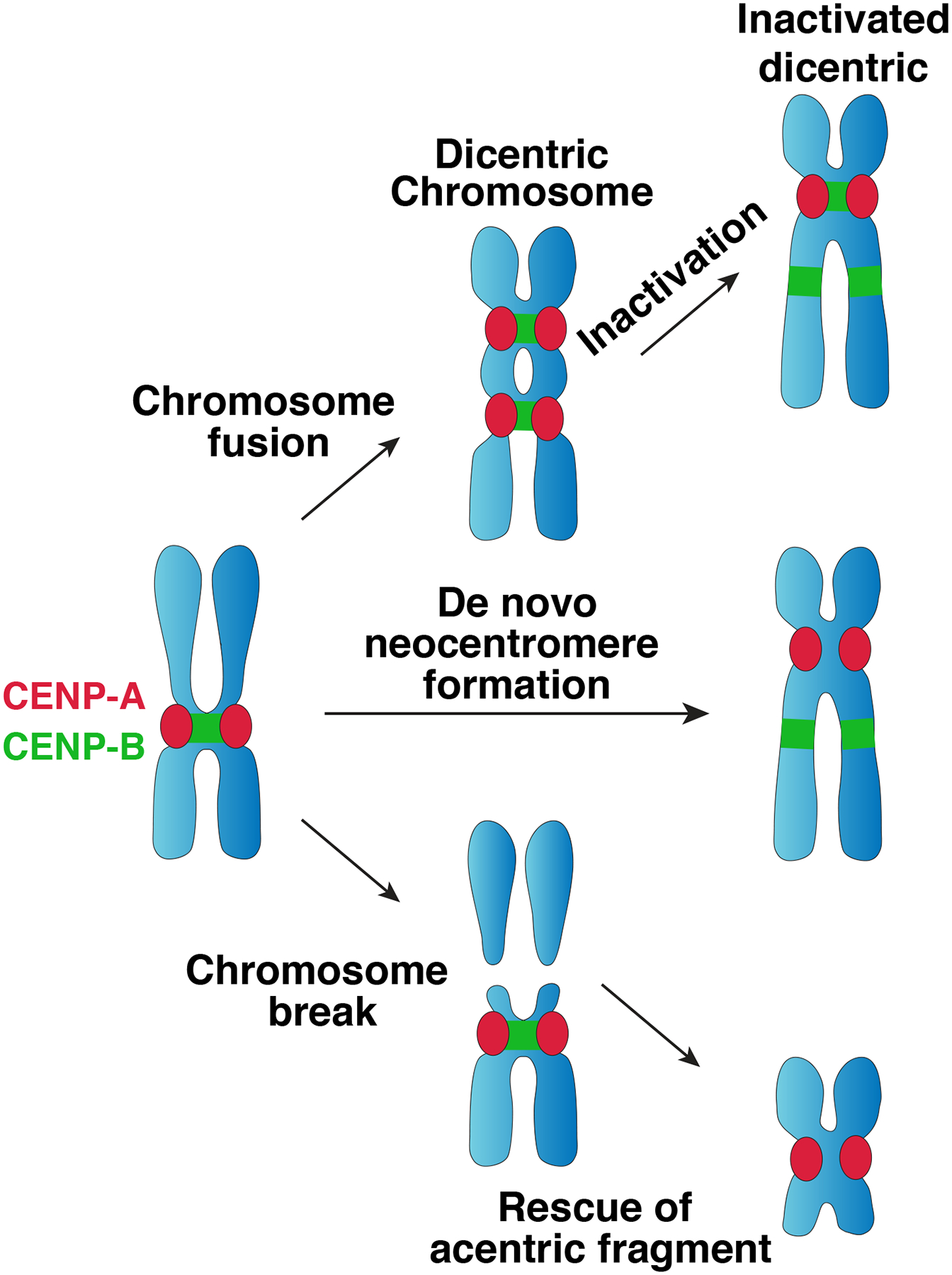
The first evidence that the centromere is specified epigenetically came from human patient samples containing dicentric chromosomes in which one centromere was functionally inactivated without changes to its underlying DNA sequence70. Subsequent work has observed epigenetic centromere inactivation in dicentric chromosomes in diverse contexts198–200. Centromere inactivation is also frequently observed in Robertsonian fusions201 and isodicentric Y chromosomes generated by sister chromatid recombination of Y chromosome palindromes202. These data indicate that centromere sequences are not sufficient for centromere function.
Compelling evidence that centromere sequences are not necessary for centromere function comes from neocentromeres (reviewed in 69). For example, routine karyotyping of a human patient in 1993 revealed a chromosome fragment that had lost its centromeric DNA, but was nonetheless stably maintained in mitosis, assembled a functional kinetochore, and mediated sister chromatid cohesion in the absence of the canonical underlying DNA repeats54 (Figure 2a). Subsequent work revealed cases of inherited neocentromeres, demonstrating that these structures are stable in both mitosis and meiosis203, 204. Neocentromeres have also been generated experimentally in diverse organisms by selecting for their ability to rescue acentric chromosomal fragments133, 205–208. Neocentromeres have been observed in otherwise normal karyotypes in which the centromere DNA sequences remain intact, but have lost centromere function204, reinforcing the insufficiency of centromere sequences proposed by observation of dicentric chromosome inactivation.
A common structure for centromeric DNA sequences
Most eukaryotes have monocentric chromosomes, in which a centromere is assembled at a single localized region (Figure 2a). A notable exception are some nematodes (including Caenorhabditis elegans), and some insects and plants, which assemble a diffuse centromere along the entire length of the chromosome, a phenomenon known as holocentricity11 (Figure 2a). Species with monocentric chromosomes can either have point centromeres, containing short DNA sequences, or regional centromeres (Figure 2a)12, which contain kilobases to megabases of DNA. Point centromeres are found in some budding yeasts12, including Saccharomyces cerevisiae13, and are defined as those centromeres in which the precise centromeric DNA sequence is necessary and sufficient for kinetochore assembly and DNA segregation14–16. Regional centromeres are typically comprised of repetitive DNA sequences that contribute to, but are not sufficient for, centromere function. However, some organisms contain regional centromeres that are non-repetitive, such as the yeast Candida albicans17, or have a mixture of repetitive centromeres and non-repetitive centromeres, such as orangutan18, horse19, and chicken20. Repetitive centromeres consist of retrotransposons and/or long arrays of simple tandem repeats, referred to as satellite DNA21.
Figure 2. Centromere specification.
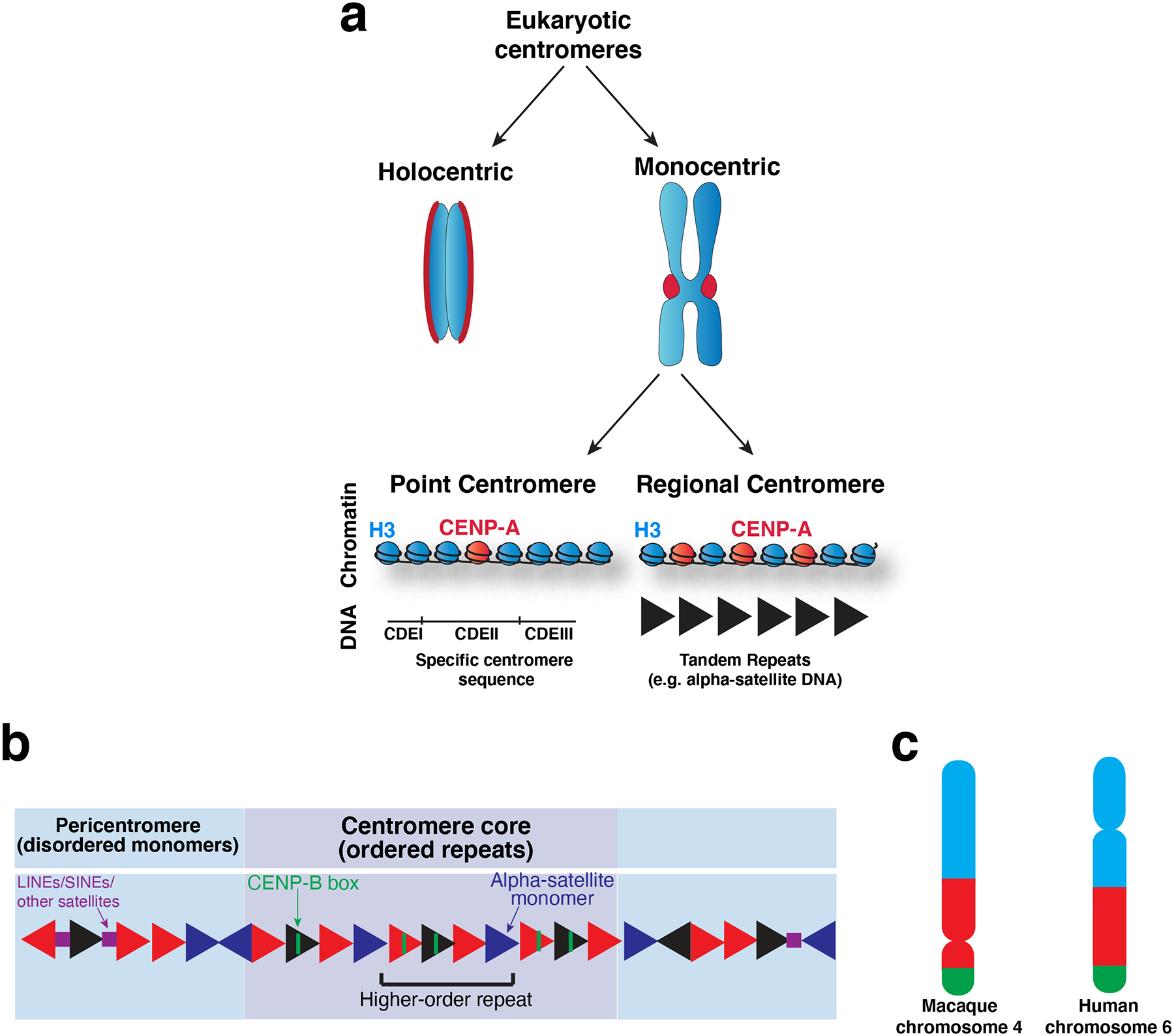
a) Diagram of the diverse types of centromeres found across eukaryotes. Holocentric chromosomes assemble a diffuse centromere across the whole chromosome. Monocentric chromosomes assemble a centromere at a single localized site on the chromosome, which is visible as a constriction between the chromosomes in mitosis (known as the primary constriction). Monocentric chromosomes can be further divided into those with point centromeres and those with regional centromeres. Point centromeres contain a specific DNA sequence that is sufficient for centromere function (here illustrated with the S. cerevisiae DNA architecture), which assembles a single CENP-A nucleosome. Regional centromeres contain large regions of DNA that is often repetitive (such as alpha-satellite DNA in primates), and assemble numerous CENP-A nucleosomes. b) Model of the DNA sequence of primate centromeres. Primate centromeres are built from alpha-satellite monomers (triangles), which are largely but not completely identical, as indicated by the different colored triangles. Patterns of these monomers arranged head-to-tail are re-iterated over the centromere core (purple) as higher-order repeats. Some monomers within the centromere core contain a sequence termed the CENP-B box, which binds to the centromere-DNA binding protein, CENP-B. The centromere core is flanked by less ordered monomers which comprise the pericentromere (blue). LINEs, SINEs and other satellites (squares) are found interspersed with alpha-satellite monomers in the pericentromere214. c) Schematic showing comparison of macaque and human orthologous chromosomes that have undergone centromere repositioning such that the position of the centromere has moved, but the surrounding markers have not, as indicated by the color blocks, which represent syntenic regions. Part c) adapted from33.
The precise DNA sequences found at centromeres vary dramatically across evolution, and it has been proposed that this rapid evolution is a consequence of meiotic drive22. Despite the dramatic divergence in centromere sequences, regional centromeres possess a modular structure that is shared by many taxa. Regional centromeres typically consist of a central core, where the CENP-A nucleosomes reside, comprised of homogenous ordered repeats, and an outer heterochromatic domain, termed the pericentromere, that typically contains less ordered repeats (Figure 2a, b). For example, centromeres of the fission yeast Schizosaccharomyces pombe contain a centromere core of non-repetitive sequences flanked by perfect inner inverted repeats and less ordered outer repeats23. Similarly, the Mus musculus centromere core is comprised of minor satellite arrays containing homogenous 120 bp repeats flanked by less-ordered ~234 bp major satellite repeats24. Primate centromeres are built on a single 171 bp monomer termed alpha-satellite25–28. In humans and other great apes, the alpha-satellite is arranged head-to-tail to form higher order repeats that are themselves re-iterated across the centromere. The human pericentromere contains flanking monomers that lack higher order repeats and share reduced identity between monomers (see 29 for further review of centromeric DNA structure) (Figure 2b). Thus, centromeres frequently arrange their divergent centromere sequences in a common repetitive structure.
Evolutionary preference for repetitive DNA structures
Cytogenetic comparisons between closely related species have revealed that some centromeres adopt new positions over evolutionary time subsequent to a speciation event without transposing the surrounding genetic markers, a phenomenon known as centromere repositioning30 (Figure 2c). These structures are referred to as evolutionary new centromeres (ENCs) and have been observed in primates and other mammals (reviewed in 31) and birds32. A striking property of ENCs is that they typically contain the same molecular features as the “old” centromeres within the karyotype, including the species-specific satellite DNAs. For example, all nine ENCs in macaque contain alpha-satellite arrays and large segmental duplications, making them indistinguishable from “old” macaque centromeres33. Thus, ENCs are postulated to be seeded upon new, non-repetitive DNA sequences in a manner analogous to neocentromeres (Box 1), but subsequently acquire their species-specific satellite DNA over time. The recent ENCs on orangutan chromosome 9 and horse chromosome 11 have not acquired satellite DNA, and may represent intermediates in this maturation process18, 19. Chromosomes harboring ENCs also exhibit a decay of the satellite sequences at the ancestral site34. The acquisition of a modular structure of tandem repeats by ENCs further supports a contribution of such DNA structures to centromere function.
Contributions of DNA sequences to centromere function
Although specific sequences are not necessary or sufficient for centromere function in some contexts, centromere DNA sequences can confer centromere function to exogenous DNA in diverse organisms, indicating that they can have a role in the de novo specification of a centromere. The most striking example of this comes from the budding yeast, S. cerevisiae, which contains a ~125 bp sequence that is sufficient to confer mitotic and meiotic stability to an exogenous minichromosome13. Although the discovery of these centromeres was strongly facilitated by the strictly sequence-dependent point centromeres of budding yeast, sequences that confer centromere functions have also be found in organisms with regional centromeres, including Schizosaccharomyces pombe35 and primates36.
Extensive work has sought to use alpha-satellite DNA to build human centromeres de novo and generate human artificial chromosomes (HACs). In pioneering work, cloned alpha-satellite DNA from human chromosomes enabled linear human mini-chromosomes37 and yeast artificial chromosomes38 to be stably inherited in human cells. These systems demonstrated that alpha-satellite DNA was sufficient to initiate centromere formation. The analysis of HAC formation also permitted structure-function studies of the alpha-satellite DNA, revealing a key role for the higher order repeats39. The mechanisms by which alpha-satellite DNA sequences initiate centromere formation are the subject of current investigations. Recently, it was suggested that alpha-satellite arrays adopt chromatin marks that favor the deposition of CENP-A nucleosomes (see below)40, 41. Together, this work is beginning to bridge the gap between the centromere DNA sequences and the epigenetic marks required for centromere function.
DNA sequence-dependent binding proteins at the centromere
The existence of common DNA sequence motifs at the centromeres of most organisms within a species presents the opportunity for recognition by DNA binding proteins that can confer centromere functions. In budding yeast, which contains sequence-specific centromeres, kinetochores are indeed assembled upon a foundation of the sequence-specific DNA binding CBF3 complex, which binds to the CDEIII DNA element at the centromere42. In organisms where centromeres can form in the absence of specific centromere sequences, the potential roles for a DNA-sequence-specific binding protein are more challenging to predict, particularly because centromere sequences vary dramatically across species, whereas centromere proteins are largely conserved. The only known centromere sequence element that is conserved between primates and rodents is the CENP-B box43, 44, a 17 bp sequence that binds to the protein CENP-B45. The CENP-B box is found in the minor satellite of Mus musculus and some monomers within the HOR of human alpha-satellite repeats. Although Mus musculus and great apes share the CENP-B box, some primates lack CENP-B boxes46, and the rodent M. caroli contains a divergent CENP-B box that retains the nine basepairs required for CENP-B binding47.
Due in part to its inconsistent conservation, the importance of the CENP-B box and the protein itself remain poorly understood. CENP-B directly interacts with and stabilizes both CENP-A nucleosomes and CENP-C to contribute to centromere function48–50. However, CENP-B knockout mice are viable51–53 and neocentromeres are maintained without acquiring CENP-B binding capability54. Perhaps most intriguingly, the human Y chromosome centromere lacks CENP-B boxes43 and does not bind detectable CENP-B protein55. Similarly, the Y chromosome of Mus musculus lacks the minor satellite sequences that contain the CENP-B box56. However, Y chromosome sequences are not sufficient to generate HACs without acquiring other centromeric alpha-satellites from the host cells37, 57 and HAC formation requires the CENP-B box39, 58. Together, these data indicate that CENP-B, like the centromere sequences it binds, is not strictly required at the centromere but makes functional contributions to maximize mitotic fidelity that contribute particularly to the generation of centromeres de novo.
Centromere epigenetics and the CENP-A nucleosome
CENP-A is an epigenetic hallmark of centromeres
In most eukaryotes, the defining feature of centromeres is the presence of nucleosomes containing the histone H3 variant CENP-A. CENP-A was first identified as a centromere-specific antigen recognized by antibodies from human patients with the autoimmune disease CREST syndrome45. Concurrent and subsequent work found that CENP-A was a component of chromatin with biochemical similarity to histones59–62, and shared homology with histone H361, 63. CENP-A homologues have been identified in diverse eukaryotes based on their similarity to histone H364–66. As a centromere-specific histone H3 variant, CENP-A provides a compelling candidate for an epigenetic mark of centromere identity67, 68. Consistent with a fundamental requirement for CENP-A in centromere function, CENP-A is found at all identified neocentromeres69, as well as the active centromeres of dicentric chromosomes70, and is essential for the localization of all known kinetochore components48, 71, 72. Importantly, artificial targeting of CENP-A to an ectopic chromosomal locus is also sufficient to generate structures capable of directing microtubule attachment and chromosome segregation73–76.
CENP-A nucleosomes possess unique structural properties
The existence of a centromere-specific histone raises intriguing possibilities regarding how CENP-A is specialized to mark the position of the centromere and recruit downstream kinetochore proteins. At the sequence level, CENP-A contains two important regions: a histone fold domain that shares 62% sequence identity with histone H3 in humans, and an N-terminal tail that differs more significantly from H363 and even between CENP-As from different species77 (Figure 3a). Within the histone fold domain, the first loop and second alpha helix (L1-alpha 2) are necessary for targeting CENP-A to the centromere, and are sufficient to confer centromere targeting when introduced into chimeras with histone H378, 79. Therefore, this region is referred to as the CENP-A targeting domain (CATD) (Figure 3a). Sequences within CENP-A nucleosomes also confer centromere-specific functions through the direct binding of the core kinetochore proteins CENP-N and CENP-C (Figure 3a). In particular, CENP-N binds directly to the CATD of CENP-A 76, 80, 81. CENP-C makes extensive contacts with the CENP-A nucleosome: with the six residues of the CENP-A C-terminal tail 80, 82, 83, with other histones within the CENP-A nucleosome 82, and with the CENP-A CATD 76, 84. The CENP-A N-terminal tail has also been implicated in the recruitment of kinetochore proteins in different organisms48, 76, 85–87. Thus, variations between CENP-A and H3 at the sequence level confer centromere specificity and kinetochore assembly properties to CENP-A.
Figure 3. Specialization and propagation of CENP-A.
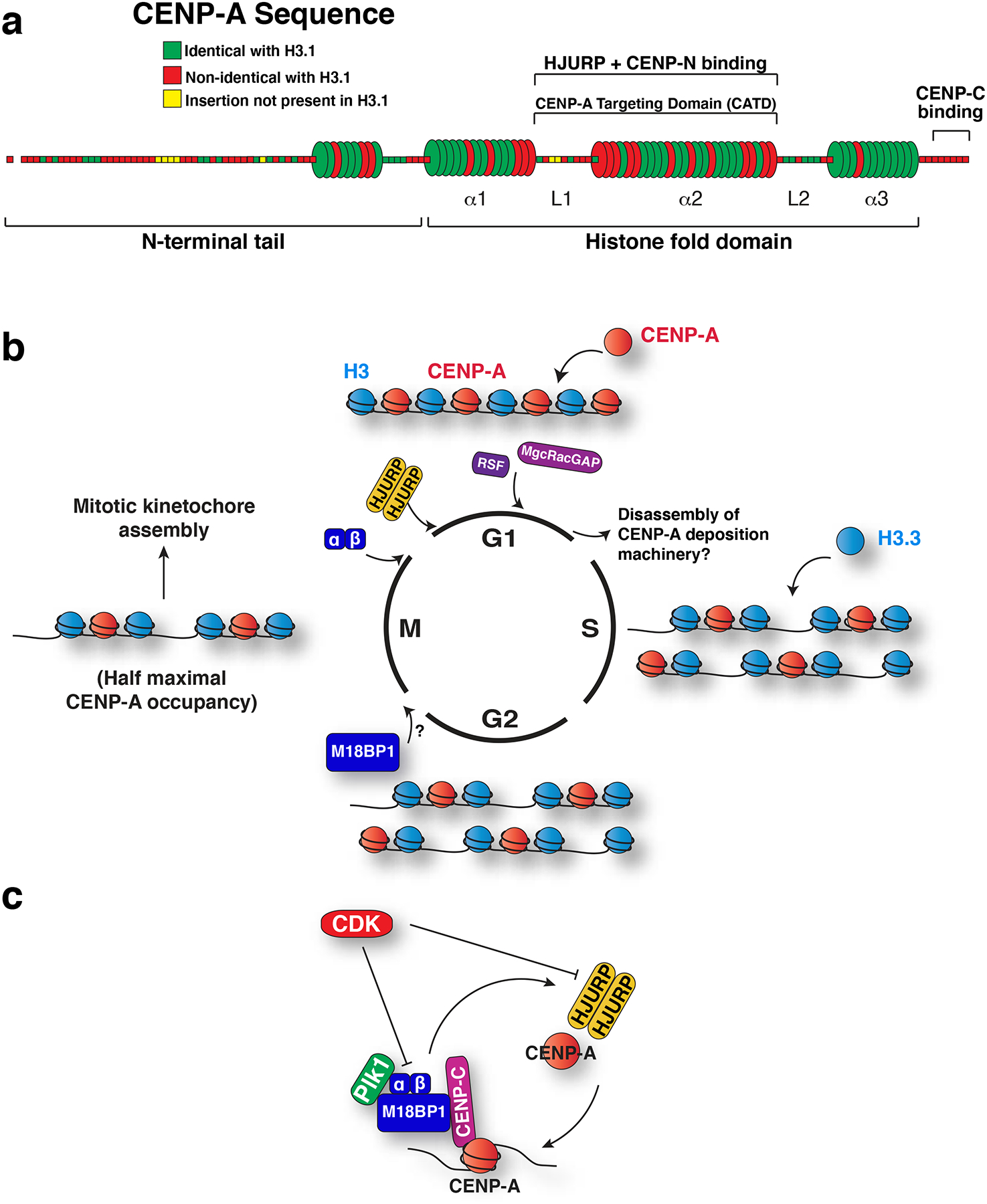
a) Model of human CENP-A primary and secondary structure showing conservation with histone H3. Each segment corresponds to a single amino acid, and is colored according to its conservation with human H3.1 as indicated. The first N-terminal amino acid, shown detached, represents the cleaved initiator methionine. Barrels represent alpha helices, and rods represent loops. Within the histone fold domain, the helices are designated alpha1 through alpha3, and the loops are designated L1 and L2. L1 and alpha2 comprise the CENP-A targeting domain, which is sufficient to target CENP-A to centromeres due to its interaction with the CENP-A chaperone, HJURP. This region also binds to CENP-N81 and is important for CENP-C recruitment76, 84. CENP-C also binds to the C terminal residues of CENP-A80, 82, 83. b) Model for the changes to CENP-A chromatin over the cell cycle. The timing of the localization of the CENP-A deposition factors is indicated. At S phase, existing CENP-A is partitioned between the replicated sisters, and gaps filled with histone H3.3. Although centromere localization of M18BP1 precedes recruitment of Mis18alpha and beta116, the precise onset of its localization has not been established. By mitosis, M18BP1 localizes to centromeres, followed by Mis18alpha and Mis18beta at mitotic exit. An HJURP dimer215 is recruited in early G1 to direct new CENP-A deposition. New CENP-A is stabilized in late G1 by MgcRacGAP and RSF1. Defining the mechanisms that remove these assembly factors once CENP-A deposition is complete also remains an important open question. c) Model for the two-step regulation of CENP-A deposition. CDK prevents CENP-A deposition outside of G1 phase by inhibiting Mis18 complex localization, Mis18 complex assembly and HJURP recruitment. Plk1 binds to the Mis18 complex to promote CENP-A deposition at centromeres during G1.
CENP-A nucleosomes also have structural distinctions from canonical H3-containing nucleosomes with the potential to make contributions to centromere function (Figure 4). The structural properties of the CATD make the free (CENP-A-H4)2 tetramer more conformationally rigid than the (H3-H4)2 tetramer as determined by hydrogen-deuterium exchange, and cause the CENP-A-CENP-A interface to be rotated when compared to the H3-H3 interface in a canonical nucleosome, generating a more compact structure88, 89. However, in the crystal structure of the octameric nucleosome, the CENP-A-CENP-A axis appears similar to the H3-H3 axis from canonical nucleosomes90. Recent work indicates that CENP-A nucleosomes in solution sample both forms, and that binding of CENP-C shifts the nucleosome to the state similar to that of canonical nucleosomes91. In addition, there has been an extensive ongoing debate regarding whether the CENP-A nucleosome forms a hemisome (with one molecule each of CENP-A, H4, H2A and H2B) that wraps DNA in a right-handed manner, or an octamer (reviewed in 92). Finally, CENP-A nucleosomes appear to confer structural alterations to centromeric chromatin. For example, CENP-A arrays are more condensed93, 94, but with a DNA entry and exit site that is loose compared to canonical nucleosomes90, 93–96, a property that is enhanced by CENP-C binding91. Thus, sequence and structural specializations of CENP-A nucleosomes and CENP-A containing-chromatin generate fundamental distinctions between centromeric chromatin and bulk chromatin.
Figure 4. Centromeric chromatin.
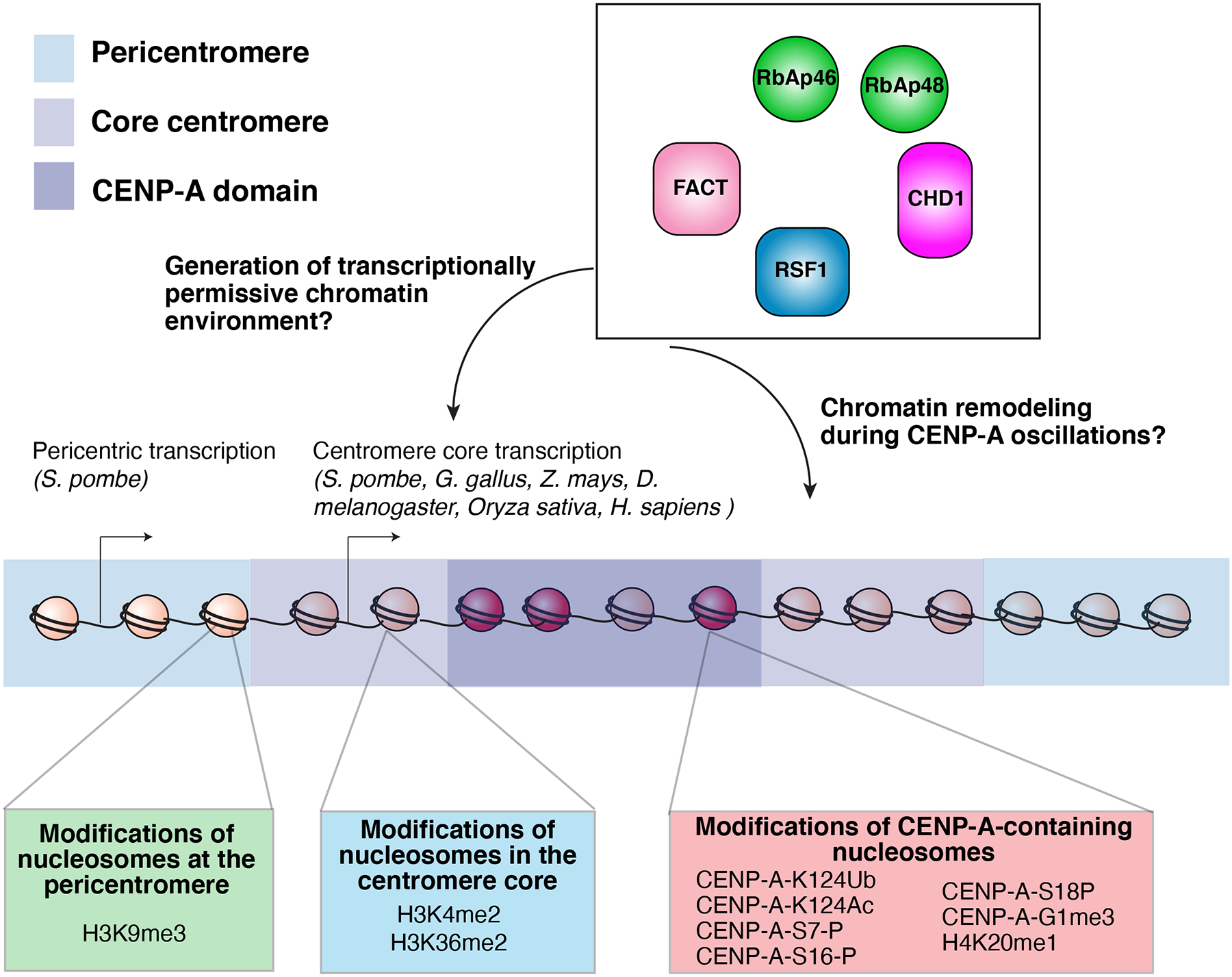
Model of the epigenetic modifications at the core centromere, CENP-A domain, and the pericentromere. In addition to the sequence and structural specializations that differentiate CENP-A chromatin from bulk chromatin, posttranslational modifications of CENP-A nucleosomes contribute to centromere function. Human CENP-A is mono-ubiquitinated at lysine 124 within the histone fold domain by CUL4-RBX1-COPS8216 to promote its centromere targeting. Acetylation at this lysine 124 residue has also been reported217. Finally, diverse other posttranslational of CENP-A217–219 and H4 in the CENP-A nucleosome220 have been described. Defining the functional contributions of these modifications remains a central challenge.
Centromere propagation
Faithful centromere inheritance is critical for the transmission of the genome, as failure to propagate the centromere results in the inability of a chromosome to attach to the mitotic spindle, leading to loss of the chromosome and the information it encodes. On monocentric chromosomes, the spurious formation of a centromere at two distinct loci allows a single chromatid to attach simultaneously to opposing spindle poles, resulting in mis-segregation or fragmentation of the chromosome by spindle forces. The fragmentation of dicentric chromosomes can result in breakage-fusion-bridge cycles that confer cascading chromosomal instability97, 98. Therefore, the centromere must be faithfully inherited at a single site on each chromosome through all mitotic and meiotic divisions (Box 2).
Box 2: Transmission of the CENP-A nucleosome during meiosis.
In addition to its central role in mediating mitotic divisions, the centromere must also be propagated and direct chromosome segregation during meiosis to be transmitted to the progeny. Transmission of Y chromosome neocentromeres between generations203 demonstrates that the position of the human centromere is heritable through the male germline independently of the underlying DNA sequence. Unlike the majority of canonical histones, CENP-A is not exchanged for protamines during sperm development in mammals60, Xenopus209 or Drosophila210, 211, and can therefore provide a template for the centromeres in the progeny. Indeed, in Drosophila, maintenance of CENP-A (CID) in the sperm is required for centromere propagation and the faithful segregation of the paternal chromosomes in the embryo, as sperm chromosomes lacking CENP-A are unable to template a centromere de novo 211. In contrast, in C. elegans, CENP-A is not continuously maintained throughout meiosis and so does not follow this self-templating pattern, as sperm do not contribute CENP-A following fertilization, and CENP-A is instead provided by the oocyte, which removes CENP-A in pachytene of prophase I and reloads it in diplotene212.
In those organisms that maintain their centromeres through meiosis, the molecular mechanisms that replenish CENP-A following meiotic S phase are poorly understood. Several differences from the mechanisms of CENP-A replenishment during the mitotic cell cycle have been proposed. In Drosophila, CENP-A is assembled during prophase I of female meiosis, and during both prophase I and after exit from meiosis II in the male210. CENP-A deposition is similarly biphasic during the meiotic divisions to produce male gametes in rye213. The mechanisms that transmit centromere position and features through the germline in vertebrates remain a key unanswered question.
The CENP-A deposition machinery
In most eukaryotes, centromere inheritance requires the transmission of CENP-A nucleosomes to maintain the epigenetic mark on each sister chromatid. Fundamental to this transmission is the striking stability of CENP-A, which does not exchange once it is incorporated at centromeres91, 99, 100, and is conservatively partitioned between the newly replicated sister chromatids during S phase91, 99, 100. Unlike canonical histones, the deposition of new CENP-A is uncoupled from DNA replication, such that the occupancy of CENP-A molecules at the centromere is halved during mitosis when the centromere recruits the complete kinetochore (Figure 3b). The nature of centromeric chromatin during mitosis following this dilution remains an area of active investigation, with current models indicating that the gaps left by this dilution are filled by H3.3101. In human cells, new CENP-A molecules are deposited during the subsequent G199.
The deposition of new CENP-A requires the coordinated activity of several assembly factors (Figure 3b). CENP-A has a dedicated histone chaperone, HJURP102, 103, which recognizes CENP-A as distinct from H3 via specific contacts between the CENP-A targeting domain (CATD) and the N terminal CENP-A-binding domain of HJURP104–107. The HJURP CENP-A-binding domain is homologous to the yeast CENP-A chaperone Scm3108, and is sufficient to direct the incorporation of CENP-A at an ectopic locus75. HJURP localizes to centromeres only during G1102, 103, when new CENP-A deposition occurs. Consistent with this, HJURP does not participate in the partitioning of CENP-A between sister chromatids during S phase100.
In addition to HJURP, CENP-A deposition in G1 requires the three-subunit Mis18 complex comprised of Mis18α, Mis18β, and Mis18 binding protein 1 (M18BP1)109 (also known as KNL2110). Intriguingly, not all components of the Mis18 complex are conserved across eukaryotes, with a single Mis18 homolog in fungi111 (without an identified M18BP1), and an M18BP1 homolog (KNL2), but no Mis18α/β homologues in C. elegans110. In D. melanogaster, the Mis18 complex and HJURP functions appear to be combined in a single molecule, CAL1112, 113. M18BP1 has been shown to interact with CENP-C in both human cells and Xenopus laevis114, 115. As CENP-C binds directly to CENP-A nucleosomes as described above, this provides a mechanism to ensure that the Mis18 complex and HJURP are recruited only to sites of pre-existing centromeres to locally direct the incorporation of new CENP-A. The interaction between M18BP1 and CENP-C is crucial for the recruitment of the Mis18 complex during CENP-A assembly in G1 phase in human cells114, 116. However, Xenopus M18BP1 is recruited via CENP-C during mitosis but not interphase, suggesting that additional M18BP1 recruitment mechanisms exist84, 115. CENP-C has also been proposed to contribute to CENP-A deposition beyond Mis18 complex recruitment84, including by binding to HJURP directly117. Finally, CENP-C91, the RSF complex118, and the centralspindlin component MgcRacGAP119 have been implicated in the maintenance of CENP-A once it is incorporated at centromeres. Together, these centromere-specialized assembly factors ensure the specific incorporation of CENP-A at centromeres.
Regulation of CENP-A deposition
Multiple regulatory safeguards have been identified that ensure the faithful deposition of new CENP-A-containing nucleosomes exclusively at centromeres. In metazoa, CENP-A deposition occurs around mitosis or following mitotic exit99, 115, 120–122. This temporal restriction isolates CENP-A deposition from the deposition of canonical H3, which is coupled to DNA replication in S phase. The cell cycle restriction of CENP-A deposition relies heavily on phosphorylation downstream of cyclin-dependent kinase (CDK) 123 (Figure 3c). Ongoing work indicates that CDK negatively regulates CENP-A incorporation at numerous steps. In Drosophila, the degradation of cyclin A plays a key role in deposition of CENP-A112, 121. In human cells, CDK phosphorylates the Mis18 complex subunit M18BP1 to reduce its centromere localization123, and to prevent recruitment of the Mis18α and Mis18β subunits116 outside of G1. CDK phosphorylation of HJURP disrupts its localization to centromeres124, whereas CDK phosphorylation of CENP-A itself on serine 68 has been reported to inhibit the CENP-A-HJURP interaction125, although the role of serine 68 in CENP-A deposition is controversial48, 76, 84, 100, 105, 107.
In addition to this temporal regulation by CDK, CENP-A deposition requires a licensing step by Polo-like kinase 1 (Plk1)116 (Fig. 3c). Thus, centromere propagation requires a two-step regulatory paradigm analogous to the regulation of DNA replication by CDK and Dbf4-dependent kinase (DDK)126. Plk1 binds to and phosphorylates the Mis18 complex to promote Mis18 complex localization and license the centromere for CENP-A deposition116. Bypassing both the CDK regulation of Mis18 complex assembly and Plk1 licensing by constitutively targeting the Mis18α subunit to the centromere results in CENP-A deposition throughout the cell cycle and severe mitotic defects116. This indicates that the temporal isolation of CENP-A deposition is important for centromere function.
Generation of a CENP-A permissive chromatin environment
Although CENP-A is an essential component of centromeres, it is not the sole driver of centromere specification. CENP-A homologs are absent in some organisms, including trypanosomes and some insects with holocentric chromosomes127, 128, raising the possibility that alternate strategies for centromere specification have arisen during evolution. Even in CENP-A-containing organisms, additional molecular features contribute to defining an active centromere, including the properties of the underlying DNA sequence, the composition of the surrounding chromatin, and post-translational modifications of CENP-A itself (Figure 4). Moreover, individual CENP-A molecules are found frequently at non-centromeric sites throughout the chromosomes129, indicating that the presence of CENP-A alone is not sufficient for centromere formation.
The core centromere and pericentromere are distinguished not only by organization of their DNA sequence repeats as described above, but also by distinct chromatin signatures that are crucial for their functions. Early studies associated centromeres with heterochromatin 130 and subsequent work has found that the pericentromere in particular is heterochromatic, containing hypermethylated H3 lysine 9 (H3K9)131, 132, although non-repetitive centromeres and neocentromeres frequently lack surrounding heterochromatin133, 134. In contrast to the heterochromatic pericentromere, at the core centromere CENP-A-containing nucleosomes are interspersed with canonical H3-containing nucleosomes with transcriptionally permissive marks, particularly dimethylated histone H3 lysine 4 (H3K4me2)135–137 and H3K36me240 in human and Drosophila cells. Recent analyses of HAC formation and maintenance have revealed that artificially increasing heterochromatin at the alpha-satellite array is detrimental for CENP-A deposition and centromere function41, 138, whereas H3K4me2 and increased H3K9 acetylation promote CENP-A maintenance40, 41. This indicates that both the presence of transcriptionally permissive marks and absence of heterochromatin in the centromere core are important for CENP-A localization to centromeres (Figure 4).
The importance of chromatin marks that are permissive for transcription at the core centromere raises the possibility that transcription of the centromere and pericentromere plays a role in centromere propagation and function. In fission yeast, transcripts from the pericentromeric repeats contribute to the formation of pericentromeric heterochromatin, which in turn is required for de novo CENP-A deposition on mini-chromosomes139. In addition, transcripts derived from the centromere core have been reported in diverse organisms140, 141 (Figure 4). In human cells, RNA polymerase II and several transcription factors localize to mitotic centromeres142 and transcripts have been detected from the alpha-satellite sequences of HAC centromeres40. Broadly disrupting RNA polymerase I or II results in kinetochore defects142, 143 as well as defects in the deposition of new CENP-A nucleosomes144. However, tethering strong transcriptional activators to the centromere is deleterious to centromere function in many organisms138, 145 indicating that the transcriptional requirement for centromere identity and function must be finely tuned.
Chromatin remodelers associated with active transcription have also been implicated in the deposition of new CENP-A (Figure 4), including RSF1, FACT, CHD1, and RbAp46 and 48103, 109, 111, 118, 119, 146–148. These proteins may facilitate new CENP-A deposition through the generation of the necessary transcriptionally permissive centromere core, or may play a direct role in remodeling centromeric chromatin to accommodate its oscillations between maximal and half-maximal CENP-A occupancy throughout the cell cycle (Figure 3b, Figure 4). For example, if H3.3 replaces CENP-A nucleosomes following DNA replication, this H3.3 must be exchanged for new CENP-A during the following G1. The Mis18 complex has also been proposed to contribute to the chromatin remodeling in anticipation of new CENP-A deposition, including by recruiting factors that regulate DNA methylation149 and histone acetylation109, 111. As a result, tethering a histone acetyltransferase to a HAC centromere can partially complement depletion of the Mis18 complex 41. However, recent work indicates that the Mis18 complex also functions directly in the CENP-A deposition process by interacting with the HJURP chaperone150, 151.
Restriction of CENP-A deposition
The regulated deposition of CENP-A nucleosomes ensures the epigenetic propagation of the centromere at a persistent location on each chromosome. Many organisms also have strategies to prevent CENP-A deposition at non-centromeric sites, where they could make inappropriate attachments to the mitotic spindle. In S. cerevisiae, mis-targeted CENP-A is removed by the combined action of the FACT chromatin remodeler and the E3 ubiquitin ligase Psh1, which targets ectopic CENP-A for degradation152, 153. In fission yeast, the proteasome subunit Rpt3 interacts with CENP-A and has been implicated in restricting the size of the CENP-A domain154. However, a similar proofreading mechanism to remove ectopic CENP-A has not yet been identified in vertebrates, consistent with the persistence of CENP-A molecules at non-centromeric sites in the genome in human cells129.
CENP-A deposition is also restricted within the centromere. In humans, mouse and chicken, the CENP-A domain occupies only a small portion of the core centromere sequences129, 155, 156. There is significant variation in the size of the CENP-A domain among human chromosomes (between 0.4 Mb and 4.2 Mb for a set of analyzed X and Y chromosomes157), although an approximately equivalent ratio between the size of the CENP-A domain and the alpha satellite array is maintained157. The CENP-A domain of neocentromeres is restricted to an even smaller region, with reports between 40 kb and 0.5 Mb96, 133, 134, 158. How the CENP-A domain is restricted in size in vertebrates remains an area of active investigation. Exogenous CENP-A expression in human cells leads to down-regulation of the endogenous CENP-A protein99, and CENP-A overexpression far beyond this level results in mis-localization of CENP-A to chromosome arms63, 86, 159. These data indicate that the restriction of the CENP-A domain occurs at least in part at the level of modulating total protein in the cell, as recently proposed in human cells129. In chicken and Drosophila, high local concentrations of the CENP-A chaperones HJURP or CAL1, respectively, can also drive centromere expansion151, 160. Intriguingly, these homeostasis mechanisms maintain CENP-A in large excess of the amount required for kinetochore function, as cells depleted of CENP-A to as little as 10 percent or even 1 percent of its initial level recruit kinetochore proteins and at least partially direct chromosome segregation48, 71.
Centromere recognition
The centromere achieves its key function – the segregation of its corresponding chromosome – by recruiting the kinetochore, the macromolecular structure that mediates attachment to the microtubules of the mitotic spindle and acts as a signaling hub to ensure accurate chromosome segregation161. Thus, understanding how centromere form begets its function hinges critically on defining the network that connects the centromere components to the proteins of the kinetochore.
When first proposed, the terms centromere and kinetochore referred to the same structure – the region of the chromosome that attaches to the spindle fiber10, 162. With the increasing resolution of this region by electron microscopy (Fig. 1b), the terms became delineated, with the kinetochore defined as an electron-dense structure in which microtubules embed, and the centromere used to refer to the underlying chromatin163. However, the molecular connections between the centromere and kinetochore remain incompletely understood.
Components of the centromere-kinetochore interface
Establishing the architecture of the centromere-kinetochore interface has been accelerated by the discovery of multiple key molecular players over the last ten years164–167. The proteins of the centromere-kinetochore interface are collectively referred to as the Constitutive Centromere Associated Network (CCAN) (also referred to as the Interphase Centromere Complex (ICEN)) (Figure 5). The CCAN is a group of 16 proteins that localize to the centromere throughout the cell cycle161. These proteins are designated in vertebrates with alphabetical CENP- names (CENP-C, CENP-H, CENP-I, CENP-K, CENP-L, CENP-M, CENP-N, CENP-O, CENP-P, CENP-Q, CENP-U, CENP-R, CENP-T, CENP-W, CENP-S, CENP-X) 45, 164–172, although other CENP- named proteins do not represent constitutive centromere components. Within the CCAN, these proteins can be combined into five groups: CENP-C, the CENP-L-N complex81, 173, the CENP-H-I-K-M complex164, 165, 174, the CENP-O-P-Q-U-R complex175, 176, and the CENP-T-W-S-X complex177 (Figure 5a). Together, these proteins recognize centromeric chromatin and connect it to the kinetochore.
Figure 5. Contributions of the Constitutive Centromere Associated Network (CCAN) at the centromere-kinetochore interface.
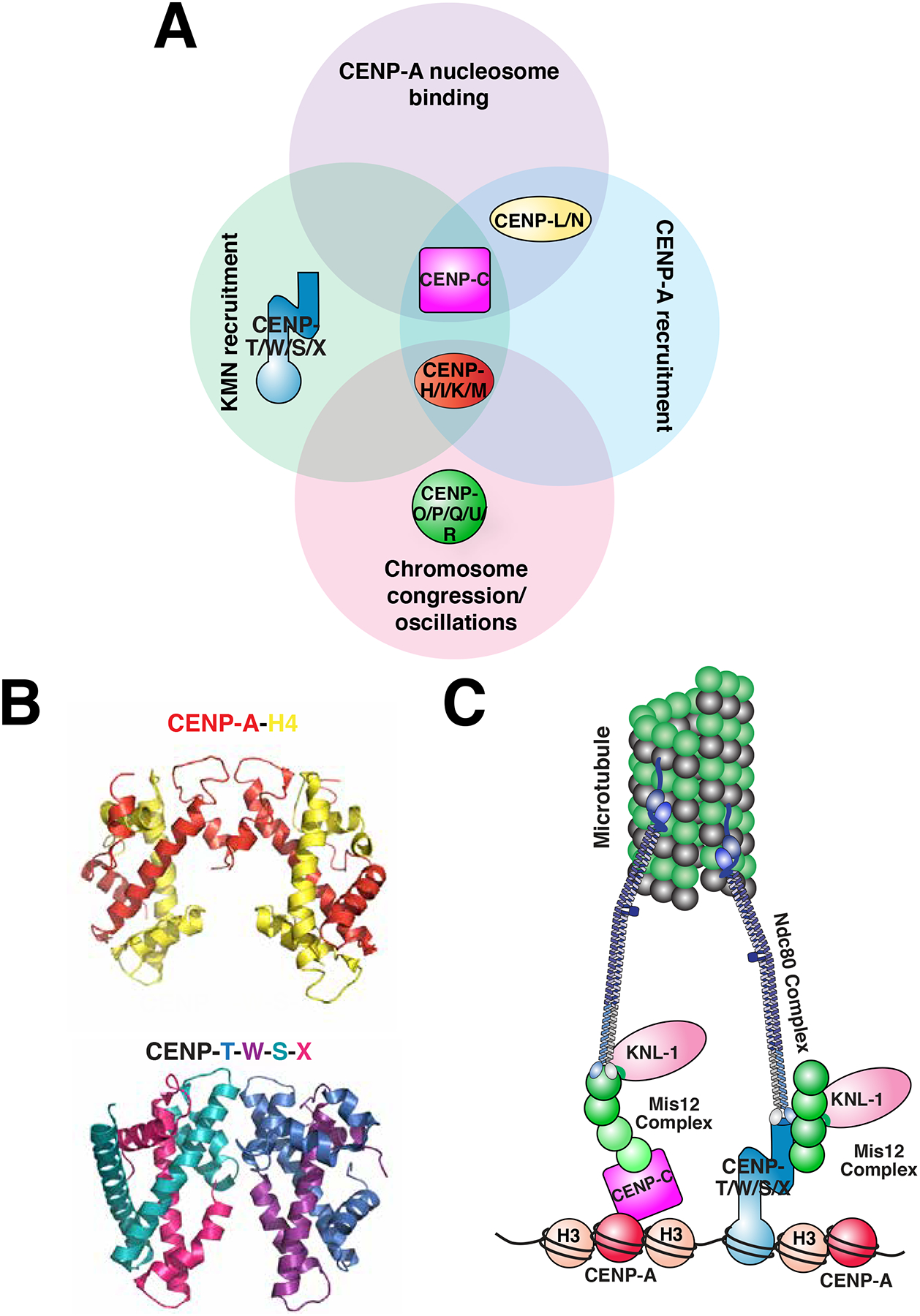
a) Diagram of the proteins of the CCAN. The sixteen proteins of the CCAN, designated by CENP- and a letter, can be grouped into sub-complexes as indicated. The sub-complexes are grouped according to functions that have been reported for at least one of their subunits. KMN: a network of KNL1, Mis12 complex and Ndc80 complex, which together bind to microtubules. b) Comparison of the crystal structures of the tetramer comprised of the histones CENP-A and H4 in the context of the nucleosome (PDB ID: 3AN2)90 (H2A, H2B and DNA are excluded for clarity) with the heterotetramer comprised of the histone fold-containing proteins CENP-T, -W, -S, and -X heterotetramer (PDB 3VH5)177. c) A simplified model of the connectivity from the centromere, to the kinetochore, to the microtubule during mitosis. The contributions of CENP-C and CENP-T to recruiting the microtubule binding-interface of the kinetochore are highlighted, and the other CCAN components are excluded from this model for clarity.
Dissecting the contributions of the CCAN to centromere recognition presents a particular challenge due to their differing functional requirements between organisms. Although the CCAN is largely conserved between yeast and human178, 179, it is dispensable in yeast with the exception of the CENP-U homolog Ame1 and the CENP-Q homolog Okp1180, 181. In mammals, CENP-U is essential for early mouse development182, but eliminating CENP-U and CENP-Q results in relatively mild phenotypes in tissue culture cells176, 182. In addition, some organisms such as Drosophila and C. elegans have a minimal CCAN, for which the only identified CCAN homolog is CENP-C. In this section, we will review the ongoing work to define the precise molecular roles of the CCAN in kinetochore assembly and faithful chromosome segregation.
Recognition of centromeric chromatin
A central requirement for connecting the centromere and kinetochore is that the CCAN directly recognize centromeric chromatin. Although each of the CCAN proteins can be co-immunoprecipitated with CENP-A nucleosomes, only CENP-C and CENP-N have been reported to bind to nucleosomes directly80–82, 91 (Figure 5a). These proteins recognize the key structural distinctions between CENP-A and H3 (see above). Together, CENP-C and CENP-N provide direct connection for kinetochore proteins to CENP-A.
In addition to binding directly to CENP-A, CCAN components make additional contacts with centromeric chromatin to build a robust platform on the centromere. Several CCAN proteins bind directly to DNA, including CENP-C183, CENP-Q175, and the CENP-T-W-S-X complex177. The CENP-T-W-S-X complex is particularly intriguing, as it is comprised of histone-fold containing proteins177, 184 and adopts a structure similar to canonical nucleosomes (Figure 5b). In this structure, CENP-T-W and CENP-S-X form dimer pairs that can be combined into a CENP-T-W-S-X heterotetramer177 or a (CENP-T-W-S-X)2 octamer185. The CENP-T-W-S-X complex wraps DNA, inducing positive supercoils177, 185, and protects a region of ~ 100 bp from micrococcal nuclease digestion177, indicating that it may integrate directly into centromeric chromatin. The importance of these nucleosome-like properties for CENP-T-W-S-X localization, as well as centromere and kinetochore function, is still being elucidated.
The CCAN directs outer kinetochore assembly
The CCAN assembles the outer kinetochore on a platform of centromeric chromatin. In particular, CENP-C and CENP-T form parallel, but non-redundant pathways that recruit the key microtubule binding proteins of the kinetochore, the KNL1/Mis12/Ndc80 (KMN) network159, 186–189 (Figure 5c). Indeed, artificial targeting of fragments of CENP-C or CENP-T to an ectopic chromosomal locus is sufficient to recruit the KMN network and generate a kinetochore-like structure that can direct chromosome segregation159, 190. In budding yeast, CENP-U forms a third pathway to recruit the KMN network175. In human cells, CENP-I has also been reported to interact with the microtubule binding proteins of the kinetochore191. These protein interactions are regulated in most eukaryotes such that the CCAN only recruits a full kinetochore during mitosis192. Specifically, phosphorylation by Aurora B kinase promotes interactions between CENP-C and the Mis12 complex during mitosis191, 193. In addition, the Ndc80 complex is sequestered outside of the nucleus throughout interphase, and is thereby spatially separated from the CCAN until mitosis when CDK phosphorylation promotes its direct interaction with CENP-T159, 189, 192.
Although the interactions between the KMN network and the CCAN proteins CENP-C and CENP-T are increasingly well characterized, the contributions of the other CCAN proteins to generating a platform for the kinetochore remain poorly defined. Ongoing work is seeking to define the architecture and associations of the remaining components71, 80, 81, 117, 164, 174, 194. This work indicates that CENP-C is a keystone molecule within the CCAN and is required for the recruitment of several other CCAN components80, 117, 175, 194, in addition to its role in recruiting the KMN network and promoting CENP-A deposition described above. Recent work has also implicated the CENP-H-I-K-M complex in the recruitment of CENP-T-W-S-X174, potentially providing centromere-specificity to the DNA-binding activity of CENP-T-W-S-X. Further defining the physical and functional relationships between the various CCAN sub-complexes remains a key goal.
Defining the contribution of the CCAN to centromere and kinetochore function
Ultimately, the central challenge that remains at the centromere-kinetochore interface is to define the contributions of the CCAN proteins to centromere and kinetochore function. Beyond building a molecular bridge to the microtubule-binding interface, CCAN proteins may make additional contributions to chromosome segregation (Figure 5a). For example, recent work has suggested that the vertebrate CCAN plays a key role in resisting the forces generated by spindle microtubules195, as well as controlling metaphase oscillations196 and chromosome congression through recruiting the motor protein CENP-E197. In addition, several CCAN proteins, including CENP-C, CENP-N and CENP-I, have been shown to play key roles in the deposition of new CENP-A nucleosomes at centromeres66, 80, 81, 114, 115, 146, 190 (Figure 5a), presenting an appealing model for the propagation of the centromere via kinetochore proteins. The ongoing advances in the identification of CCAN components and subcomplexes will provide further insight into the functional contributions of the CCAN.
Conclusions
Research in centromere biology continues to provide important insights into the molecular mechanisms that underlie the specification, propagation, and recognition of this epigenetically defined chromosomal locus. However, many important mysteries remain to be unraveled. For example, continuing to define the contributions of DNA architecture and chromatin marks to CENP-A deposition is crucial for understanding why some sites of spurious CENP-A deposition result in neocentromere formation whereas others are maintained in the genome inertly. Other key goals for future work include establishing the mechanisms by which the centromere is disassembled and re-assembled to allow passage of the DNA replication fork in S phase, understanding the differences in CENP-A transmission during the meiotic cell cycle, and characterizing the connectivity between CENP-A and the full mitotic kinetochore. Through the development of cytological, biochemical, and genetic tools, researchers are defining Cyril Darlington’s form of the centromere in increasing molecular detail. Future work faces the challenge of further dissecting endogenous centromeres and building them de novo to define how the form imparts the function.
Key points.
Centromeres are defined epigenetically and require the presence of the centromere-specific histone H3 variant CENP-A.
Although DNA sequences are not strictly required for centromere specification, similarities in the organization of centromere DNA suggest that DNA structures contribute to centromere function.
CENP-A nucleosomes contain unique sequence and structural features that allow them to stably mark the centromere and be recognized by kinetochore components.
CENP-A propagation requires specialized deposition factors and tight regulatory control.
The centromere directs the assembly of the kinetochore via the sixteen subunit Constitutive Centromere Associated Network (CCAN).
Acknowledgements
We apologize to those colleagues whose work we were unable to describe due to space constraints. We thank members of the Cheeseman lab for critical reading of the manuscript and helpful discussions. We thank Bill Earnshaw for directing us to Cyril Darlington’s description of the form and function of the centromere. We thank Conly Rieder and Elaine Dunleavy for generously sharing micrographs. Work in the Cheeseman laboratory is supported by a Scholar award to IMC from the Leukemia & Lymphoma Society, a grant from the NIH/National Institute of General Medical Sciences to IMC (GM088313), and a Research Scholar Grant to IMC (121776) from the American Cancer Society.
Glossary
- Centromere
the region of a chromosome that directs its segregation
- Evolutionary New Centromere (ENC)
a centromere at a different site from the centromere of the chromosome ancestor, where the movement of the centromere cannot be parsimoniously explained by a simple chromosome rearrangement
- Histone chaperone
a protein that binds to histones to facilitate nucleosome assembly
- Holocentric
a chromosome that assembles a centromere along the entire length of the chromosome
- Human Artificial Chromosome (HAC)
a unit of exogenous DNA that segregates autonomously in human cells
- Kinetochore
a macromolecular structure that connects the centromere to the microtubule polymers of the spindle to orchestrate and regulate chromosome segregation
- Monocentric
a chromosome that assembles a centromere at a single distinct region along the chromosome
- Meiotic drive
preferential transmission of a genetic element during meiosis, such that it is represented in more than 50% of the gametes of a heterozygote
- Neocentromere
a region of a chromosome that has the functional characteristics of a centromere but occurs at a site distinct from the site of centromere formation for the chromosome in most organisms of the species, and lacks canonical centromere DNA sequences
Biographies
Iain Cheeseman is a faculty member at the Whitehead Institute for Biomedical Research and the Department of Biology at MIT. He carried out his doctoral work on budding yeast centromeres and kinetochores with David Drubin and Georjana Barnes at the University of California, Berkeley, USA, and then conducted postdoctoral studies on nematode and human kinetochores with Arshad Desai at the Ludwig Institute for Cancer Research, San Diego, USA. His laboratory studies kinetochore function in human tissue culture cells using biochemical and cell biological approaches.
Kara McKinley received her A.B from Princeton University in molecular biology and is currently a graduate student in the Department of Biology at MIT. Kara works in the Cheeseman laboratory at the Whitehead Institute for Biomedical Research on the propagation and recognition of CENP-A-containing chromatin.
References
- 1.Holland AJ & Cleveland DW Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10, 478–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flemming W Zellsubstanz, Kern und Zelltheilung (Vogel, Leipzig, 1882). [Google Scholar]
- 3.Vig BK Sequence of centromere separation: role of centromeric heterochromatin. Genetics 102, 795–806 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard P et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–42 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Nasmyth K Segregating sister genomes: the molecular biology of chromosome separation. Science 297, 559–65 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Kerrebrock AW, Moore DP, Wu JS & Orr-Weaver TL Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 83, 247–56 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Lindegren CC The genetics of Neurospora III: Pure bred stocks and crossing over in N. Crassa. Bulletin of the Torrey Botanical Club 60, 133–154 (1933). [Google Scholar]
- 8.Bridges CB & Morgan TH The third-chromosome group of mutant characters of Drosophila melanogaster (Carnegie Institution of Washington, Washington,, 1923). [Google Scholar]
- 9.Fukagawa T & Earnshaw WC The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30, 496–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darlington CD The external mechanics of chromosomes. I--The Scope of Enquiry. Proc. R. Soc. Lond. B Biol. Sci. 121, 264–319 (1936). [Google Scholar]
- 11.Guerra M et al. Neocentrics and holokinetics (holocentrics): chromosomes out of the centromeric rules. Cytogenet Genome Res 129, 82–96 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG & Earnshaw WC The centromere: hub of chromosomal activities. Science 270, 1591–4 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Clarke L & Carbon J Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287, 504–9 (1980). [DOI] [PubMed] [Google Scholar]; This paper described the first cloning of a budding yeast centromere sequence and the sufficiency of this sequence to direct the segregation of exogenous DNA.
- 14.Clarke L & Carbon J Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature 305, 23–8 (1983). [DOI] [PubMed] [Google Scholar]
- 15.Carbon J & Clarke L Structural and functional analysis of a yeast centromere (CEN3). J Cell Sci Suppl 1, 43–58 (1984). [DOI] [PubMed] [Google Scholar]
- 16.McGrew J, Diehl B & Fitzgerald-Hayes M Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol 6, 530–8 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanyal K, Baum M & Carbon J Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci U S A 101, 11374–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locke DP et al. Comparative and demographic analysis of orang-utan genomes. Nature 469, 529–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piras FM et al. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet 6, e1000845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang WH et al. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res 20, 1219–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kit S Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol 3, 711–6 (1961). [DOI] [PubMed] [Google Scholar]
- 22.Malik HS & Henikoff S Major evolutionary transitions in centromere complexity. Cell 138, 1067–82 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Fishel B, Amstutz H, Baum M, Carbon J & Clarke L Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol 8, 754–63 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joseph A, Mitchell AR & Miller OJ The organization of the mouse satellite DNA at centromeres. Exp Cell Res 183, 494–500 (1989). [DOI] [PubMed] [Google Scholar]
- 25.Maio JJ DNA strand reassociation and polyribonucleotide binding in the African green monkey, Cercopithecus aethiops. J Mol Biol 56, 579–95 (1971). [DOI] [PubMed] [Google Scholar]
- 26.Rosenberg H, Singer M & Rosenberg M Highly reiterated sequences of SIMIANSIMIANSIMIANSIMIANSIMIAN. Science 200, 394–402 (1978). [DOI] [PubMed] [Google Scholar]
- 27.Manuelidis L Chromosomal localization of complex and simple repeated human DNAs. Chromosoma 66, 23–32 (1978). [DOI] [PubMed] [Google Scholar]
- 28.Manuelidis L Complex and simple sequences in human repeated DNAs. Chromosoma 66, 1–21 (1978). [DOI] [PubMed] [Google Scholar]
- 29.Aldrup-Macdonald ME & Sullivan BA The past, present, and future of human centromere genomics. Genes (Basel) 5, 33–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montefalcone G, Tempesta S, Rocchi M & Archidiacono N Centromere repositioning. Genome Res 9, 1184–8 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]; First evidence that centromeres have repositioned over evolutionary history independently of their surrounding markers.
- 31.Rocchi M, Archidiacono N, Schempp W, Capozzi O & Stanyon R Centromere repositioning in mammals. Heredity (Edinb) 108, 59–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasai F, Garcia C, Arruga MV & Ferguson-Smith MA Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa); evidence of the occurrence of a neocentromere during evolution. Cytogenet Genome Res 102, 326–30 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Ventura M et al. Evolutionary formation of new centromeres in macaque. Science 316, 243–6 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Kalitsis P & Choo KH The evolutionary life cycle of the resilient centromere. Chromosoma 121, 327–40 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Hahnenberger KM, Baum MP, Polizzi CM, Carbon J & Clarke L Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A 86, 577–81 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haaf T, Warburton PE & Willard HF Integration of human alpha-satellite DNA into simian chromosomes: centromere protein binding and disruption of normal chromosome segregation. Cell 70, 681–96 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K & Willard HF Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15, 345–55 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Ikeno M et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16, 431–9 (1998). [DOI] [PubMed] [Google Scholar]; These two papers describe the first use of human centromeric DNA to confer mitotic stability to exogenous DNA to generate artificial chromosomes.
- 39.Masumoto H et al. Assay of centromere function using a human artificial chromosome. Chromosoma 107, 406–16 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Bergmann JH et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30, 328–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohzeki J et al. Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J 31, 2391–402 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lechner J & Carbon JA 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64, 717–25 (1991). [DOI] [PubMed] [Google Scholar]
- 43.Masumoto H, Masukata H, Muro Y, Nozaki N & Okazaki T A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109, 1963–73 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muro Y et al. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol 116, 585–96 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Earnshaw WC & Rothfield N Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–21 (1985). [DOI] [PubMed] [Google Scholar]; This paper reports the identification of the first centromere proteins, CENP-A, CENP-B, and CENP-C, as antigens recognized by serum from patients with CREST syndrome.
- 46.Haaf T, Mater AG, Wienberg J & Ward DC Presence and abundance of CENP-B box sequences in great ape subsets of primate-specific alpha-satellite DNA. J Mol Evol 41, 487–91 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Kipling D et al. CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli. Mol Cell Biol 15, 4009–20 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fachinetti D et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol 15, 1056–66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fachinetti D et al. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev Cell 33, 314–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujita R et al. Stable complex formation of CENP-B with the CENP-A nucleosome. Nucleic Acids Res 43, 4909–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapoor M et al. The cenpB gene is not essential in mice. Chromosoma 107, 570–6 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Perez-Castro AV et al. Centromeric protein B null mice are viable with no apparent abnormalities. Dev Biol 201, 135–43 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Hudson DF et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol 141, 309–19 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voullaire LE, Slater HR, Petrovic V & Choo KH A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet 52, 1153–63 (1993). [PMC free article] [PubMed] [Google Scholar]; This paper contains the first report of a human neocentromere, demonstrating that centomeres can function in the absence of the human centromeric alpha-satellite sequences.
- 55.Earnshaw WC et al. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol 104, 817–29 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Broccoli D, Miller OJ & Miller DA Relationship of mouse minor satellite DNA to centromere activity. Cytogenet Cell Genet 54, 182–6 (1990). [DOI] [PubMed] [Google Scholar]
- 57.Grimes BR, Rhoades AA & Willard HF Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol Ther 5, 798–805 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Ohzeki J, Nakano M, Okada T & Masumoto H CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159, 765–75 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer DK & Margolis RL Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol Cell Biol 5, 173–86 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provided the first indication that centromere components (defined by detection with serum from CREST patients, now recognized as CENP-A) are components of chromatin and proposed that such components may be exchanged for canonical histones at the centromere.
- 60.Palmer DK, O’Day K & Margolis RL The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma 100, 32–6 (1990). [DOI] [PubMed] [Google Scholar]
- 61.Palmer DK, O’Day K, Trong HL, Charbonneau H & Margolis RL Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A 88, 3734–8 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palmer DK, O’Day K, Wener MH, Andrews BS & Margolis RLA 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol 104, 805–15 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sullivan KF, Hechenberger M & Masri K Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol 127, 581–92 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]; Reported the cloning of human CENP-A and defined the importance of its domain containing sequence homology to histone H3.
- 64.Buchwitz BJ, Ahmad K, Moore LL, Roth MB & Henikoff S A histone-H3-like protein in C. elegans. Nature 401, 547–8 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Henikoff S, Ahmad K, Platero JS & van Steensel B Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci U S A 97, 716–21 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi K, Chen ES & Yanagida M Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288, 2215–9 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Warburton PE et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7, 901–4 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Vafa O & Sullivan KF Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol 7, 897–900 (1997). [DOI] [PubMed] [Google Scholar]
- 69.Marshall OJ, Chueh AC, Wong LH & Choo KH Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82, 261–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Earnshaw WC & Migeon BR Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92, 290–6 (1985). [DOI] [PubMed] [Google Scholar]; Provided the first evidence for the epigenetic nature of the centromere by observing that the inactive centromere of a dicentric chromosome maintains the centromeric DNA structures (as detected by traditional banding techniques) but lacks detectable centromere proteins.
- 71.Liu ST, Rattner JB, Jablonski SA & Yen TJ Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol 175, 41–53 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Regnier V et al. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol Cell Biol 25, 3967–81 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heun P et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10, 303–15 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrated that overexpression of Drosophila CENP-A is sufficient to form functional centromeres and kinetochores at ectopic sites.
- 74.Mendiburo MJ, Padeken J, Fulop S, Schepers A & Heun P Drosophila CENH3 is sufficient for centromere formation. Science 334, 686–90 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Barnhart MC et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194, 229–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Logsdon GA et al. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J Cell Biol 208, 521–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goutte-Gattat D et al. Phosphorylation of the CENP-A amino-terminus in mitotic centromeric chromatin is required for kinetochore function. Proc Natl Acad Sci U S A 110, 8579–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Black BE, Brock MA, Bedard S, Woods VL Jr. & Cleveland DW An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A 104, 5008–13 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Black BE et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell 25, 309–22 (2007). [DOI] [PubMed] [Google Scholar]; Employed chimeric histones containing elements of histone H3 combined with elements of CENP-A to demonstrate that the centromere recruitment of CENP-A is encoded by its first loop and second alpha helix (the CENP-A targeting domain).
- 80.Carroll CW, Milks KJ & Straight AF Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 189, 1143–55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carroll CW, Silva MC, Godek KM, Jansen LE & Straight AF Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol 11, 896–902 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 80 and 81 reported the interaction of CENP-C and CENP-N with CENP-A nucleosomes, providing the first direct physical connections between CENP-A nucleosomes and the proteins of the kinetochore.
- 82.Kato H et al. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340, 1110–3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guse A, Carroll CW, Moree B, Fuller CJ & Straight AF In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477, 354–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Westhorpe FG, Fuller CJ & Straight AF A cell-free CENP-A assembly system defines the chromatin requirements for centromere maintenance. J Cell Biol 209, 789–801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y et al. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol 20, 7037–48 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Hooser AA et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114, 3529–42 (2001). [DOI] [PubMed] [Google Scholar]
- 87.Folco HD et al. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr Biol 25, 348–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Black BE et al. Structural determinants for generating centromeric chromatin. Nature 430, 578–82 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Sekulic N, Bassett EA, Rogers DJ & Black BE The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature 467, 347–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tachiwana H et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476, 232–5 (2011). [DOI] [PubMed] [Google Scholar]
- 91.Falk SJ et al. Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348, 699–703 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dunleavy EM, Zhang W & Karpen GH Solo or doppio: how many CENP-As make a centromeric nucleosome? Nat Struct Mol Biol 20, 648–50 (2013). [DOI] [PubMed] [Google Scholar]
- 93.Panchenko T et al. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci U S A 108, 16588–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Geiss CP et al. CENP-A arrays are more condensed than canonical arrays at low ionic strength. Biophys J 106, 875–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Conde e Silva N et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol 370, 555–73 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Hasson D et al. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol 20, 687–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McClintock B The Behavior in Successive Nuclear Divisions of a Chromosome Broken at Meiosis. Proc Natl Acad Sci U S A 25, 405–16 (1939). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koshland D, Rutledge L, Fitzgerald-Hayes M & Hartwell LH A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell 48, 801–12 (1987). [DOI] [PubMed] [Google Scholar]
- 99.Jansen LE, Black BE, Foltz DR & Cleveland DW Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176, 795–805 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work was a major advance in understanding the mechanisms that propagate CENP-A nucleosomes. It demonstrated the striking stability of CENP-A at centromeres, partitioning between replicated sisters during S phase, and new assembly during G1.
- 100.Bodor DL, Valente LP, Mata JF, Black BE & Jansen LE Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell 24, 923–32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dunleavy EM, Almouzni G & Karpen GH H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus 2, 146–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Foltz DR et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dunleavy EM et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–97 (2009). [DOI] [PubMed] [Google Scholar]; These two references reported the discovery of the CENP-A specific chaperone, HJURP.
- 104.Zhou Z et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472, 234–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu H et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev 25, 901–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shuaib M, Ouararhni K, Dimitrov S & Hamiche A HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A 107, 1349–54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bassett EA et al. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell 22, 749–62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sanchez-Pulido L, Pidoux AL, Ponting CP & Allshire RC Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137, 1173–4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fujita Y et al. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12, 17–30 (2007). [DOI] [PubMed] [Google Scholar]
- 110.Maddox PS, Hyndman F, Monen J, Oegema K & Desai A Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol 176, 757–63 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hayashi T et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–29 (2004). [DOI] [PubMed] [Google Scholar]; These three papers reported the discovery of the components of the Mis18 complex, which is critical for CENP-A deposition.
- 112.Erhardt S et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol 183, 805–18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen CC et al. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol 204, 313–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dambacher S et al. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3, 101–10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moree B, Meyer CB, Fuller CJ & Straight AF CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 194, 855–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McKinley KL & Cheeseman IM Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell 158, 397–411 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tachiwana H et al. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep 11, 22–32 (2015). [DOI] [PubMed] [Google Scholar]
- 118.Perpelescu M, Nozaki N, Obuse C, Yang H & Yoda K Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol 185, 397–407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lagana A et al. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol 12, 1186–93 (2010). [DOI] [PubMed] [Google Scholar]
- 120.Schuh M, Lehner CF & Heidmann S Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17, 237–43 (2007). [DOI] [PubMed] [Google Scholar]
- 121.Mellone BG et al. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet 7, e1002068 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nechemia-Arbely Y, Fachinetti D & Cleveland DW Replicating centromeric chromatin: spatial and temporal control of CENP-A assembly. Exp Cell Res 318, 1353–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Silva MC et al. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 22, 52–63 (2012). [DOI] [PubMed] [Google Scholar]
- 124.Muller S et al. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Rep 8, 190–203 (2014). [DOI] [PubMed] [Google Scholar]
- 125.Yu Z et al. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell 32, 68–81 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Bell SP & Dutta A DNA replication in eukaryotic cells. Annu Rev Biochem 71, 333–74 (2002). [DOI] [PubMed] [Google Scholar]
- 127.Akiyoshi B & Gull K Discovery of unconventional kinetochores in kinetoplastids. Cell 156, 1247–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Drinnenberg IA, deYoung D, Henikoff S & Malik HS Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. Elife 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bodor DL et al. The quantitative architecture of centromeric chromatin. Elife 3, e02137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lima-de-Faria A Genetics, origin, and evolution of kinetochores. Hereditas 35, 422–444 (1949). [Google Scholar]
- 131.Peters AH et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12, 1577–89 (2003). [DOI] [PubMed] [Google Scholar]
- 132.Rice JC et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12, 1591–8 (2003). [DOI] [PubMed] [Google Scholar]
- 133.Shang WH et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell 24, 635–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alonso A, Hasson D, Cheung F & Warburton PE A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin 3, 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Blower MD, Sullivan BA & Karpen GH Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2, 319–30 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sullivan BA & Karpen GH Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11, 1076–83 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provided clear evidence for two distinct chromatin signatures within the centromere, demonstrating that the core centromere contain euchromatic marks including H3K4me2, and lacks the heterochromatic marks of the pericentromere.
- 137.Ribeiro SA et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A 107, 10484–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakano M et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 14, 507–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Folco HD, Pidoux AL, Urano T & Allshire RC Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319, 94–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Choi ES et al. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem 286, 23600–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rosic S, Kohler F & Erhardt S Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 207, 335–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chan FL et al. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci U S A 109, 1979–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu H et al. Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Mol Cell 59, 426–36 (2015). [DOI] [PubMed] [Google Scholar]
- 144.Quenet D & Dalal Y A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 3, e03254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hill A & Bloom K Genetic manipulation of centromere function. Mol Cell Biol 7, 2397–405 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Okada M, Okawa K, Isobe T & Fukagawa T CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell 20, 3986–95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Furuyama T, Dalal Y & Henikoff S Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci U S A 103, 6172–7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen CC et al. Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription. Dev Cell 34, 73–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim IS et al. Roles of Mis18alpha in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol Cell 46, 260–73 (2012). [DOI] [PubMed] [Google Scholar]
- 150.Wang J et al. Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP). J Biol Chem 289, 8326–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Perpelescu M et al. HJURP is involved in the expansion of centromeric chromatin. Mol Biol Cell 26, 2742–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Collins KA, Furuyama S & Biggins S Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol 14, 1968–72 (2004). [DOI] [PubMed] [Google Scholar]
- 153.Deyter GM & Biggins S The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev 28, 1815–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kitagawa T, Ishii K, Takeda K & Matsumoto T The 19S proteasome subunit Rpt3 regulates distribution of CENP-A by associating with centromeric chromatin. Nat Commun 5, 3597 (2014). [DOI] [PubMed] [Google Scholar]
- 155.Spence JM et al. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J 21, 5269–80 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zeng K et al. Localisation of centromeric proteins to a fraction of mouse minor satellite DNA on a mini-chromosome in human, mouse and chicken cells. Chromosoma 113, 84–91 (2004). [DOI] [PubMed] [Google Scholar]
- 157.Sullivan LL, Boivin CD, Mravinac B, Song IY & Sullivan BA Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res 19, 457–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lo AW et al. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res 11, 448–57 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gascoigne KE et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145, 410–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schittenhelm RB, Althoff F, Heidmann S & Lehner CF Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J Cell Sci 123, 3768–79 (2010). [DOI] [PubMed] [Google Scholar]
- 161.Cheeseman IM & Desai A Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9, 33–46 (2008). [DOI] [PubMed] [Google Scholar]
- 162.Sharp LW Introduction to cytology (McGraw-Hill book company, inc., New York and London,, 1934). [Google Scholar]
- 163.Rieder CL The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol 79, 1–58 (1982). [DOI] [PubMed] [Google Scholar]
- 164.Foltz DR et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8, 458–69 (2006). [DOI] [PubMed] [Google Scholar]
- 165.Okada M et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8, 446–57 (2006). [DOI] [PubMed] [Google Scholar]
- 166.Izuta H et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11, 673–84 (2006). [DOI] [PubMed] [Google Scholar]; These three papers identified the majority of the components of the CCAN by mass spectrometry.
- 167.Amano M et al. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol 186, 173–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Saitoh H et al. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70, 115–25 (1992). [DOI] [PubMed] [Google Scholar]
- 169.Nishihashi A et al. CENP-I is essential for centromere function in vertebrate cells. Dev Cell 2, 463–76 (2002). [DOI] [PubMed] [Google Scholar]
- 170.Takahashi K, Yamada H & Yanagida M Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell 5, 1145–58 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Minoshima Y et al. The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol Cell Biol 25, 10315–28 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sugata N, Munekata E & Todokoro K Characterization of a novel kinetochore protein, CENP-H. J Biol Chem 274, 27343–6 (1999). [DOI] [PubMed] [Google Scholar]
- 173.Hinshaw SM & Harrison SC An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep 5, 29–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Basilico F et al. The pseudo GTPase CENP-M drives human kinetochore assembly. Elife 3, e02978 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hornung P et al. A cooperative mechanism drives budding yeast kinetochore assembly downstream of CENP-A. J Cell Biol 206, 509–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hori T, Okada M, Maenaka K & Fukagawa T CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell 19, 843–54 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nishino T et al. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148, 487–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work revealed that the histone fold domains of the kinetochore proteins CENP-T, -W, -S, and -X form a complex that resembles the arrangement of histones in a nucleosome. This complex also binds to and wraps DNA, indicating that a non-CENP-A component of the kinetochore makes autonomous interactions with centromeric chromatin.
- 178.Westermann S & Schleiffer A Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol 23, 260–9 (2013). [DOI] [PubMed] [Google Scholar]
- 179.Schleiffer A et al. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol 14, 604–13 (2012). [DOI] [PubMed] [Google Scholar]; In this paper, the authors identified the long-elusive yeast homologs of numerous CCAN components.
- 180.Ortiz J, Stemmann O, Rank S & Lechner J A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev 13, 1140–55 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.De Wulf P, McAinsh AD & Sorger PK Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev 17, 2902–21 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kagawa N et al. The CENP-O complex requirement varies among different cell types. Chromosome Res 22, 293–303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sugimoto K, Yata H, Muro Y & Himeno M Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J Biochem 116, 877–81 (1994). [DOI] [PubMed] [Google Scholar]
- 184.Hori T et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135, 1039–52 (2008). [DOI] [PubMed] [Google Scholar]
- 185.Takeuchi K et al. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res 42, 1644–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Screpanti E et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol 21, 391–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Przewloka MR et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol 21, 399–405 (2011). [DOI] [PubMed] [Google Scholar]
- 188.Malvezzi F et al. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J 32, 409–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nishino T et al. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J 32, 424–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hori T, Shang WH, Takeuchi K & Fukagawa T The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 200, 45–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kim S & Yu H Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J Cell Biol 208, 181–96 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gascoigne KE & Cheeseman IM CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J Cell Biol 201, 23–32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rago F, Gascoigne KE & Cheeseman IM Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol 25, 671–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Klare K et al. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J Cell Biol 210, 11–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Suzuki A, Badger BL, Wan X, DeLuca JG & Salmon ED The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev Cell 30, 717–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Amaro AC et al. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat Cell Biol 12, 319–29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bancroft J, Auckland P, Samora CP & McAinsh AD Chromosome congression is promoted by CENP-Q- and CENP-E-dependent pathways. J Cell Sci 128, 171–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Steiner NC & Clarke L A novel epigenetic effect can alter centromere function in fission yeast. Cell 79, 865–74 (1994). [DOI] [PubMed] [Google Scholar]
- 199.Higgins AW, Gustashaw KM & Willard HF Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res 13, 745–62 (2005). [DOI] [PubMed] [Google Scholar]
- 200.Sato H, Masuda F, Takayama Y, Takahashi K & Saitoh S Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr Biol 22, 658–67 (2012). [DOI] [PubMed] [Google Scholar]
- 201.Sullivan BA & Schwartz S Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet 4, 2189–97 (1995). [DOI] [PubMed] [Google Scholar]
- 202.Lange J et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 138, 855–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tyler-Smith C et al. Transmission of a fully functional human neocentromere through three generations. Am J Hum Genet 64, 1440–4 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Amor DJ et al. Human centromere repositioning “in progress”. Proc Natl Acad Sci U S A 101, 6542–7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported a chromosome from a human patient containing a neocentromere lacking alpha-satellite DNA, as well as a second site containing canonical centromeric alpha-satellite DNA without centromere function. This work provided compelling evidence to supplement previous work from dicentric chromosomes in demonstrating that centromeric alpha satellite sequences are not sufficient for centromere formation, and provided a model for how centromere repositioning over evolutionary time may occur.
- 205.Williams BC, Murphy TD, Goldberg ML & Karpen GH Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet 18, 30–7 (1998). [DOI] [PubMed] [Google Scholar]
- 206.Platero JS, Ahmad K & Henikoff S A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol Cell 4, 995–1004 (1999). [DOI] [PubMed] [Google Scholar]
- 207.Ketel C et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5, e1000400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ishii K et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321, 1088–91 (2008). [DOI] [PubMed] [Google Scholar]
- 209.Milks KJ, Moree B & Straight AF Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell 20, 4246–55 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dunleavy EM et al. The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol 10, e1001460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Raychaudhuri N et al. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol 10, e1001434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gassmann R et al. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature 484, 534–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schubert V, Lermontova I & Schubert I Loading of the centromeric histone H3 variant during meiosis-how does it differ from mitosis? Chromosoma 123, 491–7 (2014). [DOI] [PubMed] [Google Scholar]
- 214.Schueler MG, Higgins AW, Rudd MK, Gustashaw K & Willard HF Genomic and genetic definition of a functional human centromere. Science 294, 109–15 (2001). [DOI] [PubMed] [Google Scholar]
- 215.Zasadzinska E, Barnhart-Dailey MC, Kuich PH & Foltz DR Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J 32, 2113–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Niikura Y et al. CENP-A K124 Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev Cell 32, 589–603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bui M et al. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 150, 317–26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zeitlin SG, Shelby RD & Sullivan KF CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol 155, 1147–57 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bailey AO et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci U S A 110, 11827–32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hori T et al. Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev Cell 29, 740–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]


