Abstract
The unconventional myosin 16 (Myo16), which may have a role in regulation of cell cycle and cell proliferation, can be found in both the nucleus and the cytoplasm. It has a unique, eight ankyrin repeat containing pre-motor domain, the so-called ankyrin domain (My16Ank). Ankyrin repeats are present in several other proteins, e.g., in the regulatory subunit (MYPT1) of the myosin phosphatase holoenzyme, which binds to the protein phosphatase-1 catalytic subunit (PP1c). My16Ank shows sequence similarity to MYPT1. In this work, the interactions of recombinant and isolated My16Ank were examined in vitro. To test the effects of My16Ank on myosin motor function, we used skeletal muscle myosin or nonmuscle myosin 2B. The results showed that My16Ank bound to skeletal muscle myosin (KD ≈ 2.4 μM) and the actin-activated ATPase activity of heavy meromyosin (HMM) was increased in the presence of My16Ank, suggesting that the ankyrin domain can modulate myosin motor activity. My16Ank showed no direct interaction with either globular or filamentous actin. We found, using a surface plasmon resonance-based binding technique, that My16Ank bound to PP1cα (KD ≈ 540 nM) and also to PP1cδ (KD ≈ 600 nM) and decreased its phosphatase activity towards the phosphorylated myosin regulatory light chain. Our results suggest that one function of the ankyrin domain is probably to regulate the function of Myo16. It may influence the motor activity, and in complex with the PP1c isoforms, it can play an important role in the targeted dephosphorylation of certain, as yet unidentified, intracellular proteins.
Keywords: Myosin, Protein phosphatase-1 (PP1), Actin, ATPases, Phosphorylation
Introduction
Myosins are a large and diverse superfamily of actin-based motor proteins, which play crucial roles in a wide variety of cellular processes (Schliwa and Woehlke 2003; Odronitz and Kollmar 2007). Myosins are expressed in all eukaryotic organisms, from amoebae and yeasts to mammals and higher plants (Berg et al. 2001; Redowicz 2007). In 2001, a novel unconventional myosin was identified by Patel et al. (2001), having no homology with any other previously described myosin, except the conserved motor domain sequence. At first, the new myosin was denoted myr 8a (eighth unconventional myosin from rat) and later designated as a new class: myosin XVI.
Myo16 has two splicing variants. The shorter, cytoplasmic isoform is Myo16a. The longer isoform, denoted as Myo16b, is the predominant isoform of class XVI myosins and has an additional 590-amino-acid extension on its C-terminus. This extension contains a sequence element responsible for nuclear localization (Cameron et al. 2007). Myo16b is expressed mostly in brain and in some peripheral neuronal tissues. In rat, where it was first described, it is expressed during 1–2 postnatal weeks and coincides with ongoing neuronal cell migration, axonal process extension, and dendritic elaboration (Patel et al. 2001).
Only limited information is available regarding the function and binding partners of Myo16b. It co-localizes with proliferating cell nuclear antigen (PCNA) and cyclin A (Cameron et al. 2007). PCNA is involved in numerous important nuclear functions, e.g., DNA replication, cell cycle control, and chromatin remodeling (Jónsson and Hübscher 1997). PCNA is recruited in the S phase in association with cyclin A, which also takes part in DNA replication (Ohno et al. 1996). Overexpression of Myo16b increases the number of cells that remain in S phase and delays progression towards G2 phase (Cameron et al. 2007), thus Myo16b could be involved in cell proliferation and cell cycle progression. Sequence analysis showed that class XVI myosins have a motor domain, a short neck region containing one IQ motif, a long and mostly disordered tail domain (in Myo16b isoform), and a ~400-aminoacid-long N-terminal extension (Fig. 1a). The majority of myosins start with the motor domain on the N-terminus of the protein, but a few classes (III, IX, XII, XV, and XVI) contain a pre-motor extension (Sebé-Pedrós et al. 2014). There are no structural or sequence homologies among these extensions, and each may have a unique function; For example, the N-terminal extension or insertion in myosin IX may play an anchoring role in allowing this myosin to move processively as a single molecule (Xie 2010). Alternatively, the N-terminal extension can be a key element in protein–protein interactions, e.g., the PDZ domain in myosin XV (Furusawa et al. 2000), in signal transduction (Bähler et al. 1994) or in phosphorylation, like the kinase domain in myosin III (Kempler et al. 2007).
Fig. 1.
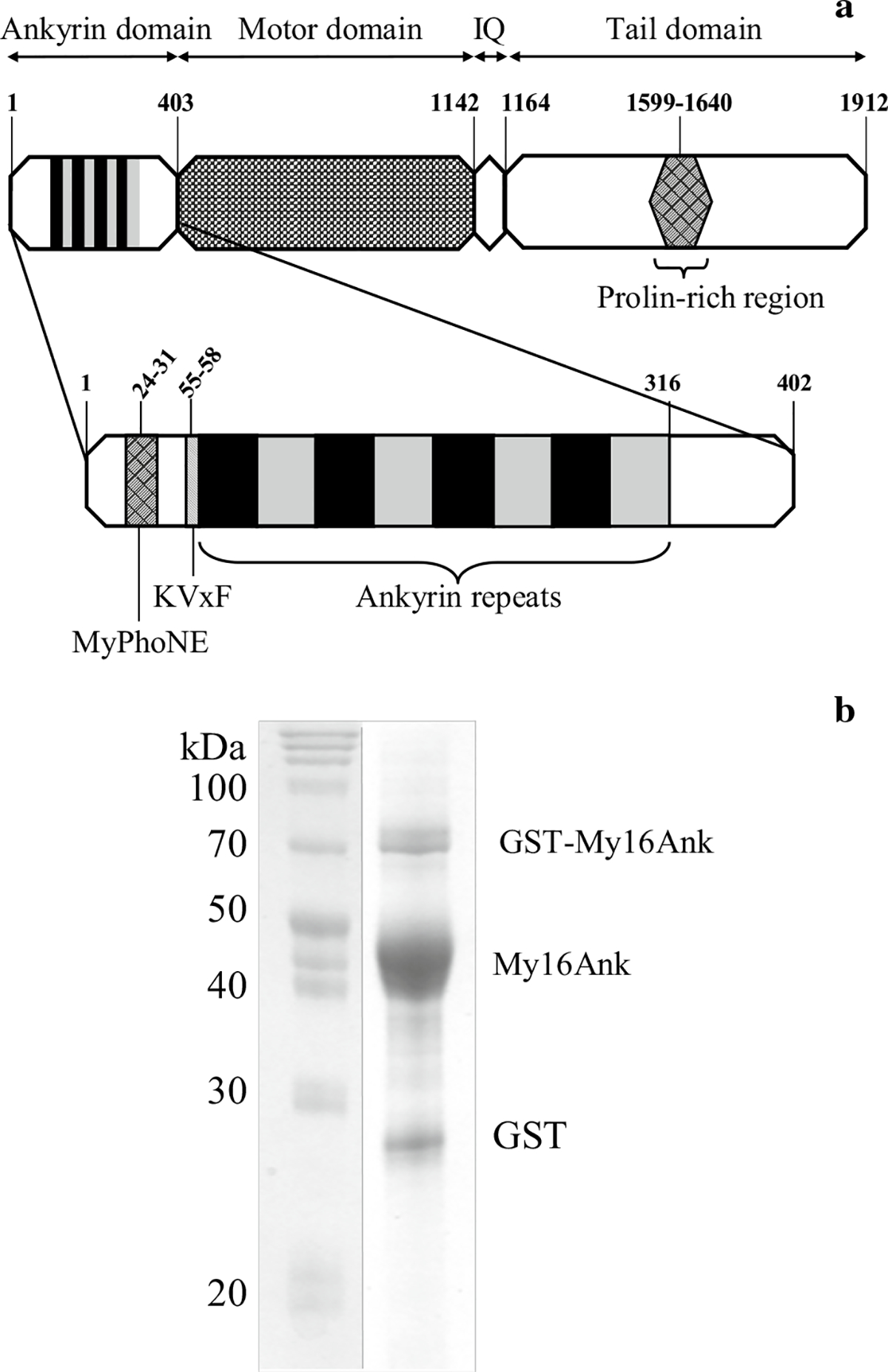
a Domain structure of Myo16b. Numbers indicate amino acid positions. Enlarged My16Ank containing the MyPhoNE and the KVxF sequence elements and the eight ankyrin repeats. b SDS-PAGE of the purified ~45-kDa My16Ank. At the end of the proteolytic procedure, a small amount of free GST (28 kDa) and uncleaved GST-My16Ank (73 kDa) could always be observed. First lane molecular weight marker
The N-terminal extension in class XVI myosins, the so-called ankyrin domain (My16Ank), differs from any previously described pre-motor domain. It contains several known sequence elements: the myosin phosphatase N-terminal element (MyPhoNE), the KVxF consensus motif for PP1c binding, eight highly conserved ankyrin repeats, and a protein kinase C phosphorylation site (Fig. 1a). Ankyrin repeats are common conserved motifs, responsible for various protein–protein interactions (Li et al. 2006). It was recently shown that ankyrin repeats play a key role in nuclear import (Lu et al. 2014), but Myo16b uses neither the importin-mediated pathway through its nuclear localization signals, nor the ankyrin-mediated importin-independent pathway to enter the nucleus (Cameron et al. 2007).
My16Ank shows high similarity to the N-terminal structure of the large targeting subunit (MYPT) of myosin phosphatase, a heterotrimer holoenzyme responsible for dephosphorylation of the phosphorylated myosin regulatory light chain, which is essential for relaxation of smooth muscle (Ito et al. 2004). Myosin phosphatase consists of the 38-kDa catalytic subunit (PP1c), the 110-kDa large targeting subunit (MYPT), and a small, 20-kDa regulatory subunit (Hartshorne et al. 1998). Co-immunoprecipitation studies have shown, as predicted from the N-terminal sequences, that PP1cα and PP1cγ bind Myo16 (Patel et al. 2001). The PP1c catalytic subunit is expressed from three genes: PP1cα, PP1cγ, and PP1cδ (also called PP1cβ). These isoforms differ in their N- and C-terminal sequences (Ito et al. 2004). Inside the cell, each PP1c isoform has a preferred localization: PP1cδ could be found in myofibrils in complex with MYPT1 responsible for MLC20 dephosphorylation and also in focal adhesions, but with a different targeting subunit. PP1cα localizes to stress fibers with MYPT1 (Murata et al. 1997). Despite the few PP1c types within the cell, dephosphorylation is a very specific and well-regulated process. The target specificity of the catalytic subunit is regulated through the large targeting subunit. The targeting (or also called regulatory) subunit binds PP1c on its canonical binding site and binds a substrate protein at the same time, bringing the enzyme and substrate in close proximity. The presence of the targeting subunit enhances enzyme activity towards the specific substrate by several orders of magnitude (Grassie et al. 2011).
In this work, our aim was to elucidate the biological function of the ankyrin domain of Myo16b. We found that this domain binds to skeletal myosin and also to nonmuscle Myo2B HMM, and modifies the ATPase activity of the motor domains. It also interacts with PP1c α and δ isoforms in vitro through tight binding and influences the phosphatase activity.
Materials and methods
Protein expression and purification
The DNA sequence of the Myo16b ankyrin domain (My16Ank) (Gene Bank Accession Number 192253, Rattus norvegicus, amino acid residues 1–402) was optimized for Escherichia coli expression and cloned into pGS21a plasmid by GenScript (Piscataway, NJ, USA). For easier purification, the recombinant My16Ank sequence was recloned into pGEX-6P-1 expression plasmid between EcoRI and SalI restriction sites with a primer containing the tobacco etch virus (TEV) protease recognition pattern. The pGEX-6P-1 plasmid containing the DNA of the glutathione S-transferase (GST)–My16Ank fusion protein was transformed into E. coli ER25.66 competent cell line, and the cells were grown at 37 °C in Luria broth medium containing 1 μg/ml ampicillin until OD600 0.5. Then, the cell culture was rapidly cooled down, and overnight expression at 16 °C was induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After ~20 h expression, the cells were harvested and the cell pellet was frozen in liquid nitrogen until further use. All procedures were performed at 4 °C. The cell pellet was extracted in lysis buffer [phosphate-buffered saline (PBS), 300 mM NaCl, 1 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 1 % Triton X-100]. After homogenization and sonication, 20 μg/ml DNase was added and stirred gently for 2 h. The cellular debris was removed by ultracentrifugation at 12,000g for 20 min, and the supernatant was incubated with GST-affinity resin (~5 ml resin for 1000 ml cell culture) in the presence of 5 mM DTT for 3 h. Then, the resin was loaded into a column and washed with PBS buffer containing 1 mM DTT and 0.2 mM PMSF. GST-My16Ank was eluted with 10 mM glutathione (GSH), and the GST-tag was then cleaved by overnight TEV protease digestion (Phan et al. 2002). My16Ank was separated from free GST and TEV protease by Q-Sepharose anionexchange chromatography with an increasing potassium–chloride gradient, using BioRad fast protein liquid chromatography (FPLC). Finally, the peak fractions were dialyzed against HMM buffer [10 mM 3-(N-morpholino)propanesulfonic acid (MOPS, pH 7.4), 0.1 mM ethylene glycol tetraacetic acid (EGTA), 1 mM DTT], concentrated, and stored on ice. Actin, full-length skeletal muscle myosin II (skMyo2), and heavy meromyosin (skHMM) were purified from rabbit musculus psoas and musculus longissimus dorsi with the methods described earlier (Feuer et al. 1948; Margossian and Lowey 1982). Recombinant nonmuscle myosin 2B HMM (NM2B HMM) was expressed in Baculovirus/Sf9 system and purified as described earlier (Nagy et al. 2013). Recombinant His-tagged PP1cδ (Toth et al. 2000) and Flag-PP1cα (Hirschi et al. 2011) were expressed in E. coli, and after renaturation (Berndt and Cohen 1990) they were purified as described in the respective references.
Steady-state ATPase activity
Steady-state ATPase activities were measured at various actin concentrations by a nicotinamide adenine dinucleotide (NADH)-coupled assay at 22 °C in a low-ionic-strength buffer (Wang et al. 2003). Experimental conditions were for skHMM: 20 mM MOPS (pH 7.0), 50 mM KCl, 10 mM MgCl2, 0.5 mM EGTA; for NM2B HMM: 10 mM MOPS (pH 7.0), 50 mM KCl, 2 mM MgCl2, 0.3 mM CaCl2, 0.15 mM EGTA. Both reactions contained also 2 mM ATP, 40 U/ml lactate dehydrogenase, 200 U/ml pyruvate kinase, 1 mM phosphoenolpyruvate, 200 μM NADH. NM2B HMM was prephosphorylated with 10 nM MLCK in the presence of 0.1 μM calmodulin, 0.2 mM CaCl2, and 0.2 mM ATP at room temperature for 15 min (Nagy et al. 2013). The decrease of the NADH absorption correlates with the amount of ATP hydrolyzed in one motor domain. The ATP hydrolysis rate is calculated from the slope of the OD340 absorption curve divided by the 6,220 M−1 cm−1 extinction coefficient and the HMM concentration. Data were collected with a Jasco or with a Cary 4000 UV–Vis spectrophotometer and analyzed with the Origin statistical program.
Cosedimentation
In actin–My16Ank cosedimentation assay, 1 μM filamentous actin was mixed with various concentration of My16Ank ranging between 0 and 14.4 μM. After overnight incubation at 4 °C, the samples were ultracentrifuged at 340,000g for 40 min at 4 °C. In skMyo2–My16Ank cosedimentation assay, skMyo2 was dialyzed in a low-salt buffer containing 10 mM MOPS (pH 7.4), 0.1 mM EGTA, and 1 mM DTT in the presence of My16Ank. The precipitated myosin was sedimented by ultracentrifugation at 340,000g for 40 min at 4 °C. The pellets and supernatants were handled according to Laemmli (1970), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was carried out, and the gels were analyzed by Syngene densitometer. The equilibrium rate constants were determined according to previously published methods (Pollard 2010) using the Origin statistical program.
Steady-state anisotropy and polymerization test
Fluorescence anisotropy measurements were carried out with a Horiba Jobin–Yvon fluorimeter. The fluorescent sample was excited with polarized light, and the emission was detected in vertical and horizontal polarization directions. For actin binding experiments, 1 μM IAEDANS-labeled monomeric actin (G-actin) or 5 μM IAF-labeled filamentous actin (F-actin) (the labeling ratio was 20 % for both) was added to various concentrations of My16Ank ranging between 0.3 and 5 My16Ank/actin molar ratio, and the anisotropy was observed at 20 °C for ~10 min. Polymerization was followed through IAEDANS-labeled G-actin anisotropy change at various concentrations of My16Ank (0–20 μM) after initiating the reaction with 100 mM KCl and 2 mM MgCl2 and followed for 30 min. Data were analyzed with Origin software.
Surface plasmon resonance
Interactions of PP1c isoforms or skHMM with My16Ank were monitored by surface plasmon resonance using the Biacore-3000 instrument (Biacore AB, Sweden). One CM5 sensor chip contained four separated flow cells of which two were used for immobilization of My16Ank domain, or the GST-My16Ank domain, while the other two were used as control. My16Ank was immobilized on the surface of the sensor chip with covalent amine coupling, while the GST-My16Ank domain was immobilized via binding to anti-GST antibody covalently attached to the CM5 sensor-chip surface by amine coupling. On the control sensor-chip surface, the active groups were blocked with ethanolamine or in case of anti-GST immobilization with GST, respectively. The control surfaces were treated identically to the sample surfaces. Different concentrations of PP1c or skHMM in 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM DTT, and 2 mM MnCl2 (running buffer) were injected above the surface, and the resonance unit signals were detected as a function of time. To determine the kinetic parameters, PP1c or skHMM was injected for 7 min over the surface of the chip, while the dissociation was registered for 5 min. The resonance units measured on the control surfaces were subtracted from the data obtained with the ankyrin surfaces to avoid the effect of unspecific binding (Toth et al. 2000). The association and dissociation rate constants were calculated from the sensograms using BIAevaluation software, assuming 1:1 interaction stoichiometry.
Phosphatase activity measurement
PP1c phosphatase activity was assayed with 1 μM radioactive 32P-phosphorylated smooth muscle myosin regulatory light chain (32P-MLC20) in a buffer containing 20 mM Tris-HCl (pH 7.4) (Toth et al. 2000). PP1c was preincubated for 10 min with different concentrations of My16Ank, and the reaction was initiated by addition of substrate. The reactions were terminated by addition of 200 μl of 20 % trichloroacetic acid (TCA) plus 200 μl of 6 % bovine serum albumin (BSA), and after centrifugation 32Pi was determined in the supernatant by measuring the radioactivity in a scintillation counter.
Results
Protein expression and purification
The ankyrin domain of Myo16b (My16Ank) was expressed in E. coli cells and purified under native conditions, i.e., in the absence of denaturating reactants. The expression in 1 l cell culture resulted in approximately 3.5 g cell pellet, and usually 4–5 mg protein was obtained at the end of the purification. The purity of the preparations was tested by SDS-PAGE. We observed a main band that corresponded to the My16Ank domain at around 45 kDa (Fig. 1b), in good agreement with the expected molecular weight. We also found minor bands around 28 kDa and 73 kDa in the SDS-PAGE gels (Fig. 1b). Confirmation with anti-GST Western blot showed that these bands could be attributed to free GST and uncleaved GST-ankyrin, respectively (data not shown). Further cleavage or degradation of the protein was not observed during a 3-week period of monitoring, provided that the samples were kept on ice. We concluded that the quantity and quality of the purified My16Ank were suitable for biophysical experiments.
My16Ank increases Mg2+-ATPase activity of acto-HMM
The role of My16Ank in the function of Myo16b is not known. Based on its close sequence proximity to the Myo16 motor domain, it was hypothesized that My16Ank may play a role in regulation of the motor domain function. We attempted to test this assumption, i.e., whether the motor domain function is influenced by My16Ank. Myosins hydrolyze ATP and use the liberated energy to create motion or strain. Therefore, we first tested if My16Ank could influence the ATPase activity of myosin. Expression and purification of the motor domain of Myo16b has not been achieved so far, so we used skeletal muscle HMM (skHMM) and nonmuscle myosin 2B HMM (NM2B HMM) as model systems. Myo16 motor domain shows ~52 % similarity to skeletal muscle myosin motor domain and ~49 % similarity to NM2B motor domain, and the actin and ATP binding regions are highly conserved (determined with NCBI Blast, using R. norvegicus skeletal muscle myosin and NM2B heavy chains as reference sequences, NCB accession #NP_001128630.1 and NP_113708.1). NM2B HMM requires phosphorylation on its regulatory light chain (MLC20) to activate its ATPase activity. Apart from its motor properties, we chose this nonmuscle myosin for our experiments because the phosphorylated MLC20 is the target of the myosin phosphatase, which shows homology in its regulatory subunit with My16Ank.
The ATPase activity of skHMM was measured using a NADH-coupled ATPase assay. My16Ank alone had no ATPase activity (data not shown). The basal ATPase activity of the skHMM was obtained in the absence of actin and My16Ank and was found to be 0.05 s−1, in accordance with previous results (Miller et al. 2003). This basal activity did not change in the presence of 13 μM My16Ank (Fig. 2a). We also measured the effect of My16Ank on the actin-activated Mg2+-ATPase of skeletal HMM (Fig. 2b). Actin filaments in various concentrations (0–35 μM) were mixed with 0.2 μM skHMM, and the reaction was started by addition of ATP, then My16Ank was added sequentially in increasing concentration (0–15 μM). The ATPase activity was determined from the obtained slopes and plotted against the actin concentration. The Vmax of the ATPase reaction in the absence of My16Ank was 0.41 s−1, which increased to 0.66 s−1 in the presence of 15 μM My16Ank, while the corresponding Km values changed from 5.1 ± 0.7 to 7.1 ± 0.4 μM. These results are summarized in Table 1.
Fig. 2.
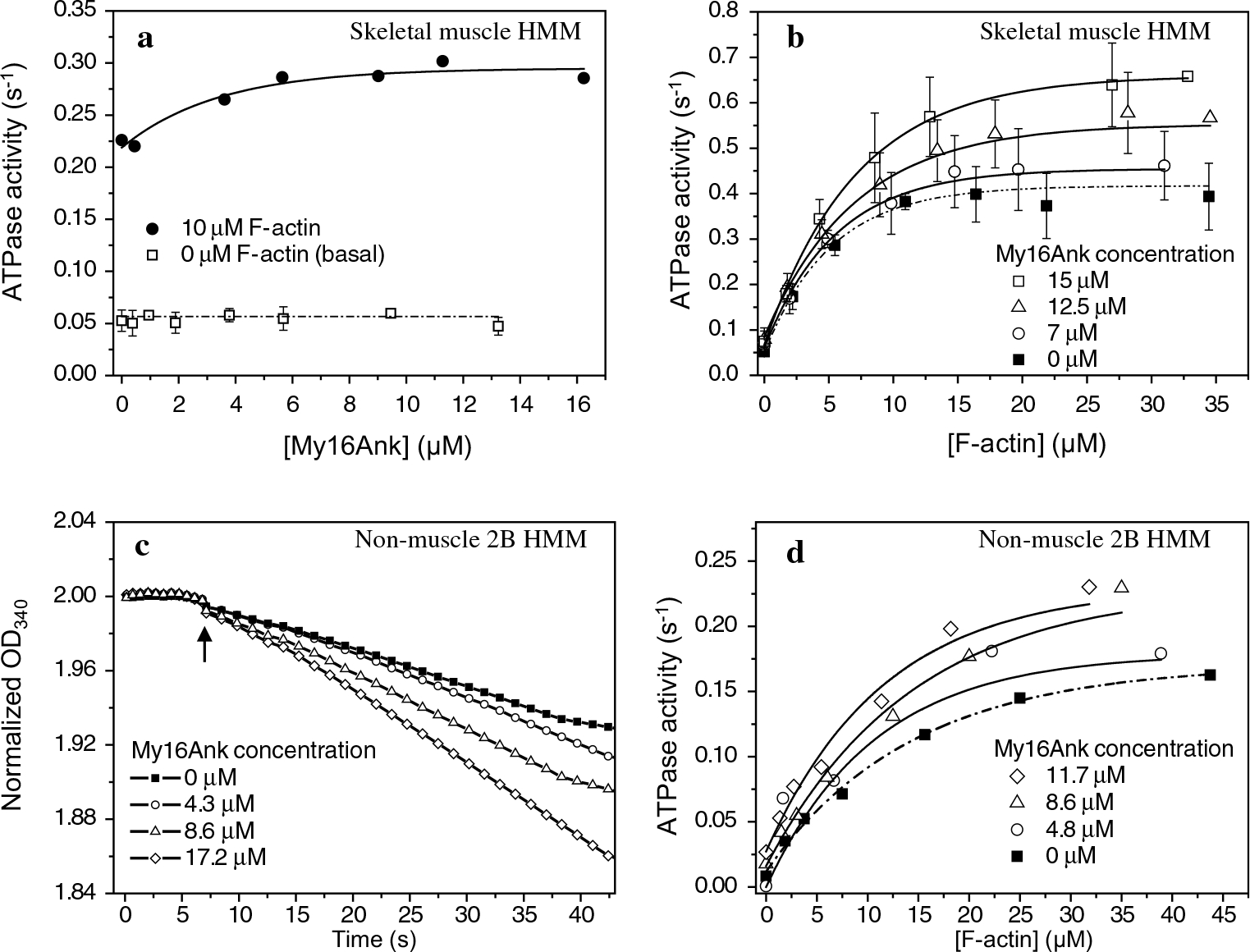
Steady-state actin-activated Mg2+-ATPase reaction measured with a NADH-coupled assay. a My16Ank did not change the basal activity of skHMM (open squares), while the actin-activated ATPase activity (F-actin concentration 10 μM) increased in the presence of My16Ank (solid circles). b My16Ank increases the actin-activated ATPase activity of skHMM in a concentration-dependent manner. The Vmax of the reaction has markedly increased. Data points represent the mean ± standard error (SE) of n = 4 independent measurements. c Normalized time course of the phosphorylation regulated NM2B HMM ATPase activity before and after phosphorylation at 20 μM F-actin concentration. My16Ank has no effect on unphosphorylated NM2B HMM. Arrow indicates addition of MLCK. d ATPase activity of prephosphorylated NM2B HMM in the presence of various concentrations of My16Ank. The ATPase activity has increased in the presence of My16Ank. All reactions were performed at 22 °C, at 50 mM KCl concentration. ATPase activity is plotted against My16Ank or F-actin concentration, and single exponential curves were fit
Table 1.
Kinetic data obtained from the steady-state ATPase assays with exponential fits
| ATPase activity | |||||
|---|---|---|---|---|---|
| Skeletal muscle HMM |
Nonmuscle 2B HMM |
||||
| [My16Ank] (μM) | Vmax (s−1) | Km (μM) | [My16Ank] | Vmax (s−1) | Km (μM) |
| 0 | 0.42 ± 0.012 | 5.10 ± 0.7 | 0 μM | 0.17 ± 0.06 | 14.36 ± 1.51 |
| 7 | 0.45 ± 0.012 | 5.49 ± 0.7 | 4.7 μM | 0.19 ± 0.02 | 12.78 ± 4.30 |
| 12.5 | 0.55 ± 0.025 | 7.02 ± 1.47 | 8.6 μM | 0.23 ± 0.007 | 14.38 ± 1.26 |
| 15 | 0.66 ± 0.009 | 7.09 ± 0.37 | 11.7 μM | 0.23 ± 0.008 | 11.6 ± 1.22 |
Values are presented as mean ± SE (obtained from n = 4 independent experiments for skHMM)
We repeated these measurements using a nonmuscle myosin fragment, NM2B HMM, which requires phosphorylation on the MLC20 to reach normal activity (Sellers 1991). Primarily, we observed that My16Ank had no effect on the ATPase activity in the unphosphorylated state of NM2B HMM and the presence of My16Ank did not influence the phosphorylation of the MLC20 (Fig. 2c). After addition of MLCK, calmodulin and Ca2+, the MLC20 of NM2B HMM premixed with My16Ank became phosphorylated and the ATPase rate increased considerably (Fig. 2c).
Secondly, the prephosphorylated NM2B HMM was mixed with various concentrations of actin (0–45 μM), and the ATPase reaction was initiated with ATP. Then, My16Ank was added sequentially to the mixture in increasing concentrations (0–11.7 μM). The actin-activated Mg2+-ATPase activity markedly increased in the presence of My16Ank (Fig. 2d). The Vmax of the reaction changed from 0.17 s−1 in the absence of My16Ank to 0.23 s−1 at 11.7 μM My16Ank concentration, while the corresponding Km values did not change significantly. The results are summarized in Table 1. These changes in ATPase activity correlated well with those we observed with skHMM.
My16Ank binds skeletal muscle myosin and HMM
The effect of My16Ank on the actin-activated ATPase activity of myosins can only be manifested if My16Ank interacts with either actin or myosin, or with both of these proteins. We tested whether My16Ank can bind to myosin motors. In these experiments, one model was the full-length skeletal muscle myosin (skMyo2), and the other was truncated skeletal muscle heavy meromyosin (skHMM). The two model systems allowed us to apply different experimental strategies for the measurements.
SkMyo2 precipitates at low ionic strength. We tested whether My16Ank can bind to and cosediment with skMyo2 by slowly removing the 0.5 M KCl from the solution. After pelleting the coprecipitated samples of skMyo2 and My16Ank, the pellets and supernatants were analyzed with SDS-PAGE. We found that My16Ank cosedimented with skMyo2, while My16Ank alone did not sediment (Fig. 3a, inset). This observation indicated that My16Ank can bind to full-length skeletal muscle myosins. To provide qualitative information, the experiments were repeated at several My16Ank concentrations (0–13.4 μM). The ratio of the bound My16Ank concentration to the concentration of the myosin heads was calculated and plotted against the free My16Ank concentration (Fig. 3a). The figure shows a hyperbolic tendency that saturates at high My16Ank concentrations at an approximate 1:1 stoichiometry. The dissociation equilibrium constant (KD) was found to be 3.01 ± 0.2 μM.
Fig. 3.
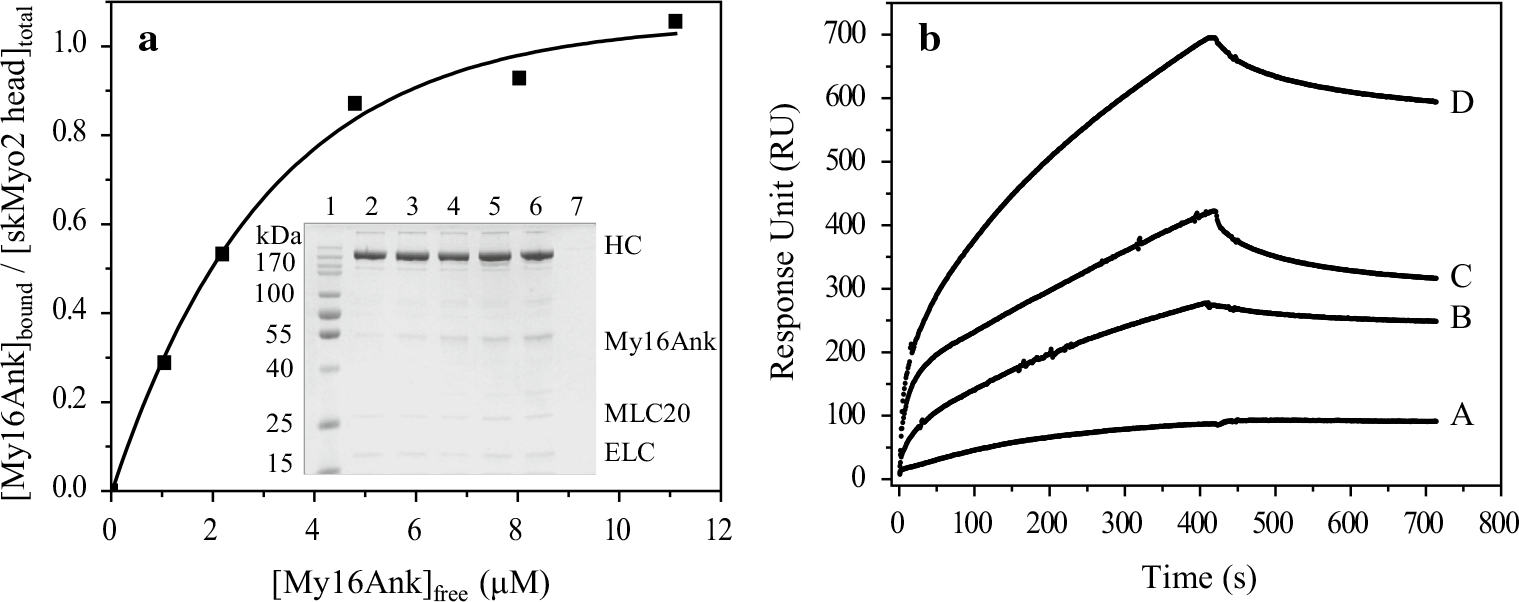
a Cosedimentation assay between skMyo2 and My16Ank. skMyo2 (1.1 μM) was coprecipitated and cosedimented with 0, 1.7, 3.4, 6.7, 10, 13.4 μM My16Ank (inset lane 2–6). No My16Ank sedimented without skeletal myosin (inset lane 7). The bound fraction of My16Ank in the pellet was normalized to the skMyo2 myosin head concentration and plotted against the free My16Ank concentration. Hyperbolic fit determines the plateau height, which shows approximately 1:1 stoichiometry, and KD ≈ 3.0 μM. HC skMyo2 heavy chain, ELC essential light chain. b My16Ank binding to skHMM was determined by surface plasmon resonance. My16Ank was immobilized on the surface of the sensor chip while different concentrations of skHMM (A 5, B 10, C 20, D 30 μM) were injected over it. The sensogram shows interaction between the proteins with KD ≈ 2.4 μM
We also characterized the interaction between My16Ank and skHMM. Since HMM does not precipitate at low ionic strengths, a different binding approach based on surface plasmon resonance was used. In these experiments, My16Ank was covalently bound to the sensor-chip surface, then different concentrations of purified skHMM (5–30 μM) were injected over the immobilized My16Ank surface. The resonance signals were recorded for association and dissociation (Fig. 3b). In the latter case, the skHMM was removed from the flowthrough liquid. The sensograms were analyzed with BIAevaluation software by fitting single exponential functions to both phases of the curves, and the rate constants for association (ka) and dissociation (kd) were determined (data not shown). The ratio of kd to ka defines the dissociation equilibrium constant (KD) for the binding of My16Ank domain to skHMM and was found to be 2.4 ± 1.4 μM. This value correlates well with the result obtained in the experiments with the skMyo2 in the cosedimentation assay.
Based on these observations, we concluded that My16Ank binds to skeletal muscle myosin. Considering that the binding of My16Ank was observed with HMM, the truncated fragment of myosin, we also concluded that the binding site for My16Ank is located on the motor domain or on the light chain-binding neck region of myosin.
My16Ank does not bind to actin
The N-terminal extension of myosin IX was shown to bind actin filaments and facilitate the processive movement of this motor protein on actin (Nalavadi et al. 2005). This result, together with our observation that My16Ank influences the actin-activated ATPase activity of skHMM and NM2B HMM, raises the possibility that My16Ank can bind actin filaments, too. We tested this possibility by using cosedimentation assays, which showed that My16Ank did not sediment with F-actin (Fig. 4a), indicating that My16Ank did not bind to actin filaments.
Fig. 4.
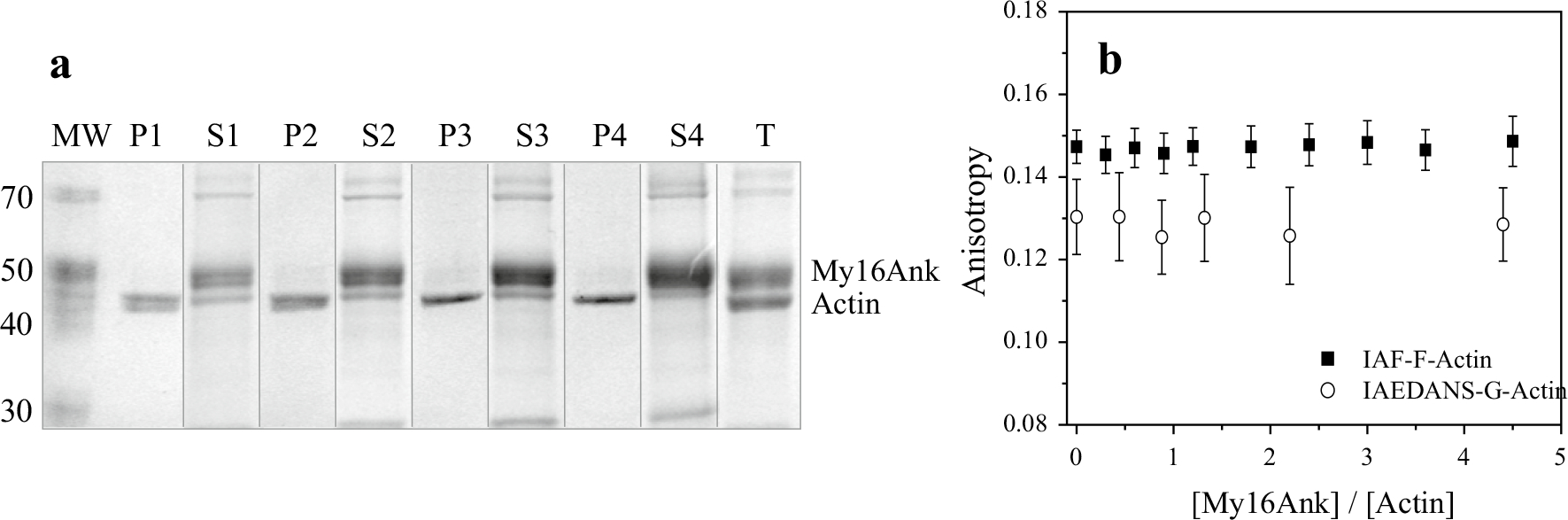
a Cosedimentation assay between F-actin and My16Ank. F-actin (1 μM) was incubated with various concentrations of My16Ank [6.75 μM (P1, S1), 9 μM (P2, S2), 11.25 μM (P3, S3), 14.4 μM (P4, S4)]; after ultracentrifugation, the pellet (P1–4) and corresponding supernatant (S1–4) were subjected to electrophoresis. My16Ank does not cosediment with F-actin. Lane 10 represents the total protein content of the assay (T), while lane 1 shows the molecular weight marker (MW). b IAF-labeled filamentous F-actin anisotropy and IAEDANS-labeled monomer G-actin anisotropy in the presence of various concentrations of My16Ank show no direct binding. The anisotropy for both F- and G-actin did not change significantly, even in the presence of fivefold excess of My16Ank
We also used steady-state fluorescence anisotropy measurements to see whether My16Ank can bind to actin monomers or filaments. The anisotropy of neither G-actin nor F-actin changed when My16Ank was present, suggesting that My16Ank did not bind directly to actin (Fig. 4b).
Finally, we tested whether My16Ank has any effect on actin polymerization. Polymerization was followed through IAEDANS-labeled G-actin anisotropy change at various concentrations of My16Ank after initiating the reaction. The polymerization curves were normalized to the plateau height of the polymerized actin, and from the initial linear phase the rate constant of the reaction was determined. The results showed that the rate of actin polymerization was not influenced by the presence of My16Ank (data not shown).
All these observations indicated that My16Ank does not interact directly with actin, and the My16Ank-induced changes in the actin-activated ATPase activity of myosin motors were attributed to the interactions between My16Ank and myosin.
My16Ank strongly binds PP1cα and PP1cδ and decreases phosphatase activity
To further explore the possible functions of My16Ank, we considered that its amino acid sequence shows homology with the myosin phosphatase targeting subunit (MYPT1). The sequence blast between R. norvegicus Myo16b (NCB accession #NP_620248) and R. norvegicus protein phosphatase-1 regulatory subunit 12A (NCB accession #NP_446342) shows 33 % identity and 49 % similarity (NCBI Blast). My16Ank contains two protein phosphatase catalytic subunit (PP1c) binding sequence elements: the KVxF motif (amino acid residues 55–58) essential for PP1c binding, and the RxxQIKxY motif (residues 24–31) termed previously as MyPhoNE element (Fig. 1a).
Previous studies have shown that, in rat, postnatal cerebellum lysate myosin 16b coimmunoprecipitates with PP1cα and PP1cγ, but not PP1cδ (Patel et al. 2001). To understand whether My16Ank has a function in phosphatase activities, we assayed the interaction between My16Ank and the different isoforms of PP1c under in vitro conditions using surface plasmon resonance techniques. In these experiments, we used both the uncleaved GST-My16Ank and the TEV protease digested My16Ank domain bound to the derivatized sensor-chip surface with anti-GST antibody or direct covalent coupling, respectively. Different concentrations of purified PP1cα and PP1cδ (0.5–5 μM) were injected over the immobilized My16Ank surfaces, and association and dissociation (without PP1c) phases were recorded as resonance signals (Fig. 5). Attaching the proteins in different ways to the surface could possibly have an effect on the obtained parameters. To eliminate this source of problems, we attached My16Ank to the surface either via GST at its N-terminal by anti-GST coupled covalently or through the amine (–NH2) side-chains. The two different attachment methods provided almost identical results, thus excluding the possibility that the binding interface was hidden or partially covered (GST-My16Ank sensograms are not shown). The My16Ank domain bound strongly to PP1cα (KD = 540 ± 209 nM) and also to PP1cδ (KD = 606 ± 173 nM) with submicromolar affinities (Table 2). The GST-My16Ank bound with nearly the same affinities (KD = 405 ± 267 nM for PP1cα and KD = 329 ± 252 nM for PP1cδ). This finding is in contrast to the previous report that myosin 16b coimmunoprecipitates with PP1cα or PP1cγ only, but not with PP1cδ (Patel et al. 2001).
Fig. 5.
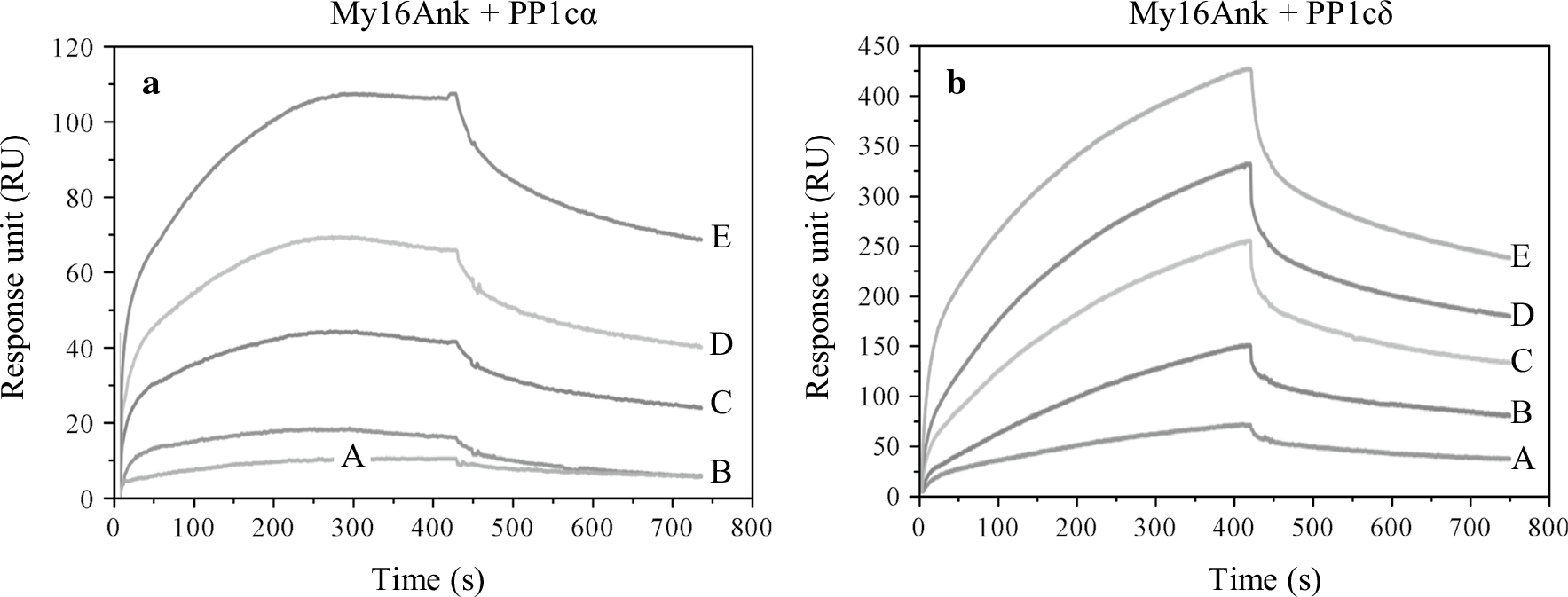
Interaction of My16Ank with PP1c measured by surface plasmon resonance-based binding analysis. My16Ank was immobilized on the surface of the sensor chip while different concentrations of PP1cα (a) or PP1cδ (b) were injected over it [0.5 μM (A), 1 μM (B), 2 μM (C), 3 μM (D), 5 μM (E)]. From the obtained sensograms, the association (ka) and dissociation (kd) rate constants can be determined, and the kd/ka ratio (KD) shows the affinity of the interacting proteins. In case of My16Ank–PP1cα, KD = 540 ± 209 nM, while for My16Ank–PP1cδ, KD = 606 ± 173 nM
Table 2.
Association and dissociation rate constants, and association and dissociation constants determined for the interactions of different My16Ank forms with PP1cα and PP1cδ by SPR-based binding technique
| Ka (M−1 s−1) | Kd (s−1) | KA (M−1) | KD (M) | |
|---|---|---|---|---|
| My16Ank | ||||
| rPP1cα | 3.22 ± 1.67 × 103 | 1.74 ± 0.42 × 10−3 | 1.85 ± 0.81 × 106 | 5.40 ± 2.28 × 10−7 |
| rPP1cδ | 1.41 ± 0.18 × 103 | 8.54 ± 1.55 × 10−3 | 1.65 ± 1.93 × 106 | 6.06 ± 2.49 × 10−7 |
| GST-My16Ank | ||||
| rPP1cα | 4.44 ± 2.26 × 103 | 1.80 ± 0.41 × 10−4 | 2.47 ± 0.97 × 106 | 4.05 ± 1.25 × 10−7 |
| rPP1cδ | 1.44 ± 0.66 × 103 | 4.73 ± 1.86 × 10−4 | 3.04 ± 1.76 × 106 | 3.29 ± 2.85 × 10−7 |
Values are presented as mean ± SE (obtained from n= 5 independent experiments)
In protein phosphatase-1 holoenzymes, the dephosphorylating activity of the PP1c is markedly increased toward the substrate if the catalytic subunit is bound to the regulator subunit. The regulator subunit is also responsible for targeting PP1c to a specific substrate. On the other hand, association of PP1c with the targeting subunit could also inhibit activity towards other substrates preferentially dephosphorylated by the free PP1c; For example, the MYPT1 targeting subunit of myosin phosphatase increases the dephosphorylation of the phosphorylated myosin regulatory light chain (P-MLC20), while it inhibits PP1 activity towards the dephosphorylation of glycogen phosphorylase a (Toth et al. 2000). Considering the homology between My16Ank and MYPT1, we also tested whether My16Ank can enhance the dephosphorylation of smooth muscle P-MLC20. 32P-MLC20 was produced with phosphorylation by MLCK using 32P-ATP and Mg2+ in the presence of Ca2+ and calmodulin. PP1c activity was measured via the release of 32Pi after dephosphorylation (Fig. 6). The presence of My16Ank significantly decreased the phosphatase activity for all types of PP1c (recombinant PP1cα and PP1cδ, or native PP1c, an isoform mixture purified from rabbit skeletal muscle) in a concentration-dependent manner. The phosphatase activity was reduced to ~40 % by 1 μM My16Ank compared with that observed in the absence of My16Ank. These data imply that the dephosphorylation of 32P-MLC20 by PP1c is inhibited when PP1c is in complex with My16Ank. A possible explanation for these observations is that My16Ank may cover the binding surface for the P-MLC20 on PP1c.
Fig. 6.
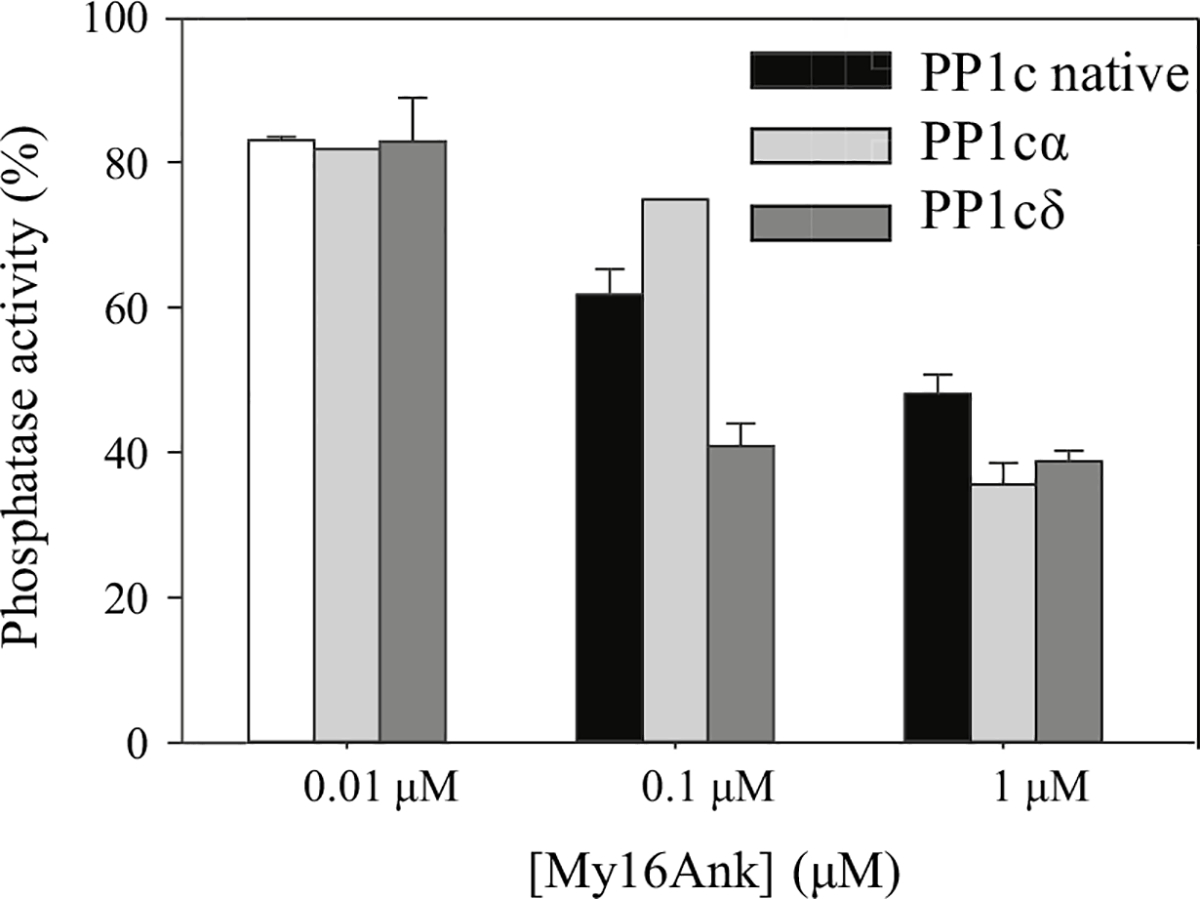
Effect of My16Ank on the phosphatase activity of PP1c. The substrate of the reaction was the 20-kDa smooth muscle myosin regulatory light chain (MLC20), phosphorylated with myosin light chain kinase (MLCK) in the presence of [γ−32P] ATP and Mg2+. Phosphatase activity was measured by determination of the release of 32Pi from 32P-MLC20. The phosphatase activity without My16Ank was taken as 100 %. My16Ank inhibits the recombinant PP1c isoforms and the native, tissue purified PP1c in a concentration-dependent manner; 1 μM My16Ank significantly reduced the phosphatase activity of all types of PP1c to ~40 %
Discussion
The information available to date about the functional properties of Myo16b indicated that it may play essential roles in key physiological processes. Patel and coworkers described Myo16b in developing neurons, mostly in newborn and early postnatal neuronal cells. Myo16b can be localized to the nucleus, where it may have a role in regulating the cell cycle. In accordance with this assumption, overexpression of Myo16b delays progression from S phase to G2 phase (Cameron et al. 2007). To gain a better understanding of the cellular functions, many questions have to be answered regarding the structure and interactions of Myo16b. Our present study has focused on a unique structural element of the class XVI myosins, the ~45-kDa N-terminal ankyrin domain (My16Ank).
For this study, we expressed and purified the My16Ank domain in quantities suitable for biochemical studies. We also made several attempts to purify Myo16b motor domain, but after successful expression in Baculovirus/Sf9 system, the extraction of a soluble protein seemed to be extremely difficult. Therefore, we chose motor domains from skeletal muscle (skHMM) and nonmuscle (NM2B HMM) myosin as a model system to characterize My16Ank.
We found that My16Ank did not modify the basal ATPase activity (Fig. 2a). Interestingly, the actin-activated ATPase activity of skHMM or NM2B HMM was enhanced by My16Ank (Fig. 2b, d). The actin concentration dependence of the Vmax suggested that My16Ank could accelerate the rate-limiting step of the actin-activated ATPase cycle. In principle, various cycle steps can be the rate-limiting step for the actin-activated ATPase cycle, such as the dissociation of the products of ATP hydrolysis from myosin, or the dissociation of myosin from actin. Therefore, although more kinetic data are required to properly understand which of these steps were altered by My16Ank, we speculate that it was probably the release of the products of the ATP hydrolysis from myosin that made the observed Vmax values greater in the presence of My16Ank.
We showed that My16Ank can bind to the myosin and the binding was relatively weak. When interpreting this observation, one needs to consider two important aspects. In this study we used model systems (skeletal muscle and nonmuscle 2B HMM) instead of the inherent partner Myo16b, and therefore the physiologically relevant affinity may be different, possibly tighter. On the other hand, in its native state, myosin 16b would contain My16Ank in the same polypeptide chain as the motor domain. In this case the My16Ank is probably anchored to the motor domain, and thus its apparent concentration is determined by the structural properties and geometrical arrangement of the components, and not by their solute concentration; For example, if the My16Ank is anchored to the motor domain by a 1-nm-long structural element, and thus one molecule of My16Ank is located within a sphere of radius 1 nm, the apparent concentration would be approximately 0.4 M. After considering all these aspects, we concluded that the observed binding showed that there is a binding site for My16Ank in myosin. To determine to what extent this binding site is occupied under various conditions, or in physiological states of myosin, further information will be required.
We showed that My16Ank had no effect on the basal ATPase activity of myosin, suggesting that My16Ank did not change the structure of the motor around the nucleotide binding pocket. My16Ank did not bind to actin in either its monomeric or filamentous form (Fig. 4b), so we excluded that My16Ank modified the myosin binding sites on actin. We also observed here that actin was required for the My16Ank effect on myosin ATPase activity (Fig. 2b, d). The elevation of the actin-activated activity suggests that the coupling between actin and myosin was modified by My16Ank, making this protein–protein interaction more effective. There are several structural ways to achieve this. One possibility is that My16Ank built into the motor domain and became part of the protein surface that was responsible for the binding of actin. The inclusion of the My16Ank in this binding must be manifested in a special structural arrangement, as My16Ank does not bind actin by itself. This would imply that in the native structure My16Ank plays an important role in the manifestation of interaction between actin and Myo16b.
PP1 enzymes play important roles in regulating the phosphorylation levels of proteins in cells through their ability to dephosphorylate key proteins. Their enzymatic activity is targeted and often enhanced by regulator protein subunits, such as the large targeting subunit (MYPT1) of myosin phosphatase to dephosphorylate the phosphorylated myosin regulatory light chain (P-MLC20). Interestingly, the My16Ank domain shows ~50 % similarity to MYPT1. In our in vitro experiments we found that the isolated My16Ank strongly bound to PP1cα and also to PP1cδ with nearly identical affinities. We attempted to understand the functional relevance of the formation of the My16Ank–PP1c complex. In principle, My16Ank in complex with the PP1c can mimic a protein phosphatase holoenzyme.
Myo16b contains several possible phosphorylation sites as predicted with the PROSITE bioinformatic program (Sigrist et al. 2002, 2013). These sites are targets of PKC, casein kinase 2 or cAMP-dependent protein kinase. Based on this information, we first assumed that the binding of My16Ank to PP1c serves an auto-dephosphorylation regulatory process, where the heavy chain or the light chain in Myo16b is dephosphorylated. To explore this possibility one needs to test whether the My16Ank binding can facilitate the dephosphorylation of the corresponding segment. The light chain subunits of class XVI myosins are unknown. Based on the consensus IQ sequence of the neck region it is most likely a calmodulin-like light chain, but other possibilities such as the MLC20 or the essential light chain cannot be excluded (Bird et al. 2014). Therefore, we used the P-MLC20 as a possible substrate for My16Ank–PP1c complex. However, our observation that the phosphatase activity of the different PP1c preparations (i.e., recombinant or native) markedly decreased toward P-MLC20 in the presence of My16Ank suggested that the P-MLC20 is not a preferred substrate for the complex. To explain the structural aspects of this observation, we assume that My16Ank competes for the binding site of P-MLC20 on PP1. In terms of the functional indications, the decreased PP1c activity in the presence of My16Ank suggests that the specific target protein for the My16Ank–PP1c complex is not the P-MLC20.
Conclusions
Despite our investigations, several major questions remain to be answered in the future to understand the cellular functions of Myo16b. Myo16b may act as a protein phosphatase subunit in complex with the PP1c catalytic subunit as a protein phosphatase, and the Myo16b itself may be responsible for targeting PP1c to a certain, as yet unidentified, substrate. Finding the natural target for the My16Ank–PP1c complex will probably give immediate insight into the intracellular functions of Myo16b. This target may be a protein still to be identified, or may simply be this special myosin itself.
At the moment we can only speculate regarding the possible function of Myo16b and its ankyrin domain. The tail domain of Myo16b has an intrinsically disordered structure as predicted by the IUPred bioinformatic program (Dosztányi et al. 2005), and it contains a proline-rich region which is a typical profilin binding site. The C-terminal ~300 amino acids of Myo16b are responsible for nuclear localization (Cameron et al. 2007). We assume that through profilin binding the Myo16b may take part in the organization of the nuclear actin cytoskeleton. Profilin can be phosphorylated by RhoA associated kinase (ROCK) (Shao et al. 2008), which inhibits actin binding, and this site is dephosphorylated by PP1c (Shao and Diamond 2012). However, the regulatory subunit for profilin dephosphorylation is not described yet. As we showed, Myo16b is able to bind PP1c on its N-terminal ankyrin domain, while the Myo16b C-terminus co-localizes with profilin in the nucleus (Cameron et al. 2007). One can envisage that Myo16b can act as a molecular scaffold providing the structural framework for the dephosphorylation of profilin. In this model, the My16Ank domain would act as a targeting subunit for PP1c for the effective and selective dephosphorylation of profilin. To test this exciting possibility, experiments are in progress in our laboratory. On the other hand, the Myo16b tail domain itself is also a target for phosphorylation at Tyr1416 and Tyr1441 amino acid residues. Phosphorylation of the tail domain activates the WAVE complex through the PI3K signalling pathway, which also leads to actin remodeling (Yokoyama et al. 2011). This suggests a key role for Myo16b in regulation of the intracellular milieu.
Previously, the N-terminal myosin domains were assumed to act as subunits enhancing the interactions between myosin heads and actin. The observations presented in our work clearly show that the ankyrin domain of Myo16b can play an important role in the overall function of the Myo16b protein by being responsible for its interaction with PP1c, and to fulfill a central function in cellular regulatory processes.
Acknowledgments
This work was supported by grants from the Hungarian Scientific Research Fund (OTKA grants NN 107776 and K112794 to M.N. and K109249 to F.E.) and by the “TÁMOP 4.2.4. A/2–11-1–2012-0001 National Excellence Program” supported by the European Union and the State of Hungary, co-financed by the European Social Fund to Z.K. and B.B. The authors thank Attila Nagy for preparing and providing NMIIB HMM.
Abbreviations
- GSH
Glutathione
- GST
Glutathione S-transferase
- HMM
Heavy meromyosin
- MLC20
Myosin regulatory light chain
- MLCK
Myosin light chain kinase
- My16Ank
Myosin 16 ankyrin domain
- Myo16
Myosin 16
- MyPhoNE
Myosin phosphatase N-terminal element
- MYPT1
Myosin phosphatase regulatory subunit 1
- sk
Skeletal
- skMyo2
Full-length skeletal myosin
- NM2B
Nonmuscle myosin 2B
- PP1c
Protein phosphatase-1 catalytic subunit
- TEV
Tobacco etch virus
Footnotes
Conflict of interest The authors declare that they have no conflicts of interest.
References
- Bähler M, Kroschewski R, Stöffler HE, Behrmann T (1994) Rat myr 4 defines a novel subclass of myosin I: identification, distribution, localization, and mapping of calmodulin-binding sites with differential calcium sensitivity. J Cell Biol 126:375–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE (2001) A millennial myosin census. Mol Biol Cell 12:780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt N, Cohen PT (1990) Renaturation of protein phosphatase 1 expressed at high levels in insect cells using a baculovirus vector. Eur J Biochem 190:291–297 [DOI] [PubMed] [Google Scholar]
- Bird JE, Takagi Y, Billington N et al. (2014) Chaperone-enhanced purification of unconventional myosin 15, a molecular motor specialized for stereocilia protein trafficking. Proc Natl Acad Sci USA 111:12390–12395. doi: 10.1073/pnas.1409459111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RS, Liu C, Mixon AS et al. (2007) Myosin16b: the COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression. Cell Motil Cytoskelet 64:19–48 [DOI] [PubMed] [Google Scholar]
- Dosztányi Z, Csizmok V, Tompa P, Simon I (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. doi: 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- Feuer G, Molnar F et al. (1948) Studies on the composition and polymerization of actin. Hung Acta Physiol 1:150–163 [PubMed] [Google Scholar]
- Furusawa T, Ikawa S, Yanai N, Obinata M (2000) Isolation of a novel PDZ-containing myosin from hematopoietic supportive bone marrow stromal cell lines. Biochem Biophys Res Commun 270:67–75 [DOI] [PubMed] [Google Scholar]
- Grassie ME, Moffat LD, Walsh MP, MacDonald JA (2011) The myosin phosphatase targeting protein (MYPT) family: a regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1delta. Arch Biochem Biophys 510:147–159 [DOI] [PubMed] [Google Scholar]
- Hartshorne DJ, Ito M, Erdodi F, Erdödi F (1998) Myosin light chain phosphatase: subunit composition, interactions and regulation. J Muscle Res Cell Motil 19:325–341 [DOI] [PubMed] [Google Scholar]
- Hirschi A, Cecchini M, Steinhardt RC et al. (2011) An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat Struct Mol Biol 17:1051–1057. doi: 10.1038/nsmb.1868.An [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Nakano T, Erdodi F, Hartshorne DJ (2004) Myosin phosphatase: structure, regulation and function. Mol Cell Biochem 259:197–209 [DOI] [PubMed] [Google Scholar]
- Jónsson ZO, Hübscher U (1997) Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. BioEssays 19:967–975. doi: 10.1002/bies.950191106 [DOI] [PubMed] [Google Scholar]
- Kempler K, Toth J, Yamashita R et al. (2007) Loop 2 of limulus myosin III is phosphorylated by protein kinase A and autophosphorylation. Biochemistry 46:4280–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–684. doi: 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Li J, Mahajan A, Tsai MD (2006) Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45:15168–15178 [DOI] [PubMed] [Google Scholar]
- Lu M, Zak J, Chen S et al. (2014) A code for RanGDP binding in ankyrin repeats defines a nuclear import pathway. Cell 157:1130–1145. doi: 10.1016/j.cell.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Margossian SS, Lowey S (1982) Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzym 85:55–71 [DOI] [PubMed] [Google Scholar]
- Miller BM, Nyitrai M, Bernstein SI, Geeves MA (2003) Kinetic analysis of Drosophila muscle myosin isoforms suggests a novel mode of mechanochemical coupling. J Biol Chem 278:50293–50300. doi: 10.1074/jbc.M308318200 [DOI] [PubMed] [Google Scholar]
- Murata K, Hirano K, Villa-Moruzzi E et al. (1997) Differential localization of myosin and myosin phosphatase subunits in smooth muscle cells and migrating fibroblasts. Mol Biol Cell 8:663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Takagi Y, Billington N et al. (2013) Kinetic characterization of nonmuscle myosin IIb at the single molecule level. J Biol Chem 288:709–722. doi: 10.1074/jbc.M112.424671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalavadi V, Nyitrai M, Bertolini C et al. (2005) Kinetic mechanism of myosin IXB and the contributions of two class IX-specific regions. J Biol Chem 280:38957–38968 [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol 8:R196. doi: 10.1186/gb-2007-8-9-r196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K, Hirose F, Sakaguchi K et al. (1996) Transcriptional regulation of the Drosophila CycA gene by the DNA replication-related element (DRE) and DRE binding factor (DREF). Nucleic Acids Res 24:3942–3946. doi: 10.1093/nar/24.20.3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KG, Liu C, Cameron PL, Cameron RS (2001) Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1 alpha and 1 gamma1. J Neurosci 21:7954–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J, Zdanov A, Evdokimov AG et al. (2002) Structural basis for the substrate specificity of tobacco etch virus protease. J Biol Chem 277:50564–50572 [DOI] [PubMed] [Google Scholar]
- Pollard TD (2010) A guide to simple and informative binding assays. Mol Biol Cell 21:4061–4067. doi: 10.1091/mbc.E10-08-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redowicz MJ (2007) Unconventional myosins in muscle. Eur J Cell Biol 86:549–558 [DOI] [PubMed] [Google Scholar]
- Schliwa M, Woehlke G (2003) Molecular motors. Nature 422:759–765 [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Grau-Bové X, Richards TA, Ruiz-Trillo I (2014) Evolution and classification of myosins, a paneukaryotic whole genome approach. Genome Biol Evol 6:290–305. doi: 10.1093/gbe/evu013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR (1991) Regulation of cytoplasmic and smooth muscle myosin. Curr Opin Cell Biol 3:98–104 [DOI] [PubMed] [Google Scholar]
- Shao J, Diamond MI (2012) Protein phosphatase 1 dephosphorylates profilin-1 at Ser-137. PLoS One 7:e32802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Welch WJ, Diprospero NA, Diamond MI (2008) Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol Cell Biol 28:5196–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, Hulo N et al. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Br Bioinform 3:265–274 [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, de Castro E, Cerutti L et al. (2013) New and continuing developments at PROSITE. Nucleic Acids Res 41:D344–D347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Kiss E, Herberg FW et al. (2000) Study of the subunit interactions in myosin phosphatase by surface plasmon resonance. Eur J Biochem 267:1687–1697 [DOI] [PubMed] [Google Scholar]
- Wang F, Kovacs M, Hu A et al. (2003) Kinetic mechanism of nonmuscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem 278:27439–27448. doi: 10.1074/jbc.M302510200 [DOI] [PubMed] [Google Scholar]
- Xie P (2010) A model for processive movement of single-headed myosin-IX. Biophys Chem 151:71–80. doi: 10.1016/j.bpc.2010.05.007 [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Tezuka T, Kotani M et al. (2011) NYAP: a phosphoprotein family that links PI3K to WAVE1 signalling in neurons. EMBO J 30:4739–4754. doi: 10.1038/emboj.2011.348 [DOI] [PMC free article] [PubMed] [Google Scholar]


