Abstract
The V(D)J recombination reaction is composed of multiple nucleolytic processing steps mediated by the recombination-activating proteins RAG1 and RAG2. Sequence analysis has suggested that RAG2 contains six kelch repeat motifs that are predicted to form a six-bladed β-propeller structure, with the second β-strand of each repeat demonstrating marked conservation both within and between kelch repeat-containing proteins. Here we demonstrate that mutations G95R and ΔI273 within the predicted second β-strand of repeats 2 and 5 of RAG2 lead to immunodeficiency in patients P1 and P2. Green fluorescent protein fusions with the mutant proteins reveal appropriate localization to the nucleus. However, both mutations reduce the capacity of RAG2 to interact with RAG1 and block recombination signal cleavage, therefore implicating a defect in the early steps of the recombination reaction as the basis of the clinical phenotype. The present experiments, performed with an extensive panel of site-directed mutations within each of the six kelch motifs, further support the critical role of both hydrophobic and glycine-rich regions within the second β-strand for RAG1-RAG2 interaction and recombination signal recognition and cleavage. In contrast, multiple mutations within the variable-loop regions of the kelch repeats had either mild or no effects on RAG1-RAG2 interaction and hence on the ability to mediate recombination. In all, the data demonstrate a critical role of the RAG2 kelch repeats for V(D)J recombination and highlight the importance of the conserved elements of the kelch motif.
The coordinated rearrangement of antigen receptor gene segments during V(D)J recombination is dependent on a complex series of DNA-processing reactions (20, 30, 45). Essential to the initiation of the process are recombination signal sequences (RSSs), which consist of two conserved DNA recognition motifs, the heptamer (consensus, CACAGTG) and the nonamer (consensus, ACAAAAACC) (32). These motifs are separated by predominantly nonconserved spacer regions of either 12 or 23 bp. Effective recombination is achieved by the 12/23 rule, which limits rearrangement to gene segments flanked by RSSs with different spacer lengths (15, 55, 60).
Recombination-activating genes 1 and 2 (RAG1 and RAG2) encode the lymphoid cell-specific recombinase components (36, 46) that are central to the rearrangement process. Normally, the V(D)J recombination reaction proceeds with nonamer recognition mediated by a DNA binding region of RAG1 (nonamer binding domain) that displays homology to the DNA recognition domains of the Hin family of bacterial invertases and those of homeodomain proteins (14, 34, 54, 58). Stable complex formation with the RSS (22, 42) is achieved on recruitment of RAG2, which alters the contacts between the RSS and the recombinase (4, 16, 34, 57, 58). This stable RAG1-RAG2-RSS complex promotes bending of the RSS (3) and has been proposed to distort the coding-flank–heptamer border (4, 16, 57). A nick is introduced directly 5′ of the heptamer motif (61), and the liberated 3′ hydroxyl group is then used as a nucleophile in a transesterification reaction of the opposing strand to form a covalently sealed hairpin coding end and a blunt 5′-phosphorylated signal end (32, 37). In vitro, the active core has been shown to subsequently resolve the hairpin coding-end intermediates (6, 50) and to remove short 3′ overhangs and flap extensions (43).
The active core of RAG1-RAG2 is defined by three acidic amino acid residues that lie in a region of RAG1 whose predicted secondary structure is similar to the secondary structure observed in the crystal structures of the catalytic cores of a host of transposases and retroviral integrases (18, 26, 28). This conservation is reflected in the extensive similarities between the reaction mechanisms used by RAGs and those used by numerous transposases and resolvases (6, 14, 43, 54, 62). Included in these mechanistic parallels is the striking ability of RAG1-RAG2 to transpose signal end complexes into unrelated target DNA (2, 23).
In accordance with the biochemical role of RAGs in the initiation of DNA cleavage, inactivation of the RAG1 or RAG2 gene by homologous recombination arrests both T- and B-lymphocyte development (33, 48). Similarly, mutations in human patients that entirely inactivate the recombination capacity of RAG1 and RAG2 lead to a complete absence of T and B cells and to the clinical manifestations of severe combined immunodeficiency (SCID) (47). Moreover, mutations in either RAG1 or RAG2 which reduce recombination efficiency without entirely abrogating the capacity for rearrangement result in Omenn syndrome (OS) (64). This disorder is characterized by a variable number of T cells with restricted receptor rearrangement heterogeneity and a lack of detectable B cells. In addition, the clinical characteristics of OS include lymphadenopathy, hepatosplenomegaly, erythrodermia, eosinophilia, and increased levels of interleukin-4, -5, and -10 and immunoglobulin E (IgE) in serum as well as elevated numbers of CD30-expressing cells (11, 44). Both SCID and OS are ultimately fatal in the absence of bone marrow transplantation (19), further highlighting the fundamental role of RAG1 and RAG2 in lymphopoiesis (35).
Mutations in RAG1 and RAG2 found in SCID and OS can disrupt the V(D)J reaction at a number of critical points, as demonstrated by the diminished capacity of all identified mutations to achieve effective RSS binding and cleavage or recombination of episomal plasmid substrates (43, 64). While knowledge of the nonamer binding domain has clarified the molecular defects of mutations localized to that domain, many other identified mutations map to regions of the proteins with no explicitly and uniquely ascribed structural or biochemical features. Recently, a variety of sequence analysis tools have been used to probe the structure of RAG2 and have revealed that the RAG2 active core contains six internal repeats of approximately 50 amino acids which were first identified in the Drosophila kelch protein (5, 9, 66).
The kelch repeat superfamily is an emerging class of proteins that mediates diverse biological functions. All kelch motif-containing proteins exhibit a standard pattern with individual repeats consisting of four antiparallel β-strands separated by intervening loop regions of variable lengths (1, 8). The second β-strand of each repeat is typically the most highly conserved region within and between kelch-like proteins suggesting an important role for this strand in achieving an appropriate fold. The structure of the galactose oxidase protein from Dactylium dendriodes shows that its seven kelch repeats adopt the circular formation of a β-propeller structure, and it appears likely that other kelch motif proteins also adopt a similar conformation (24, 25). Such a molecular module probably favors the coordination of multiple protein-protein interactions. The classification by sequence analysis of RAG2 within the kelch repeat superfamily potentially facilitates our understanding of the role of RAG2 in the recombination reaction; however, functional data demonstrating the importance of the RAG2 kelch motifs for the recombination reaction have not yet been obtained.
Here we report that mutations G95R and ΔI273 in two individuals afflicted with OS and SCID, respectively, lie within the predicted second β-strand of the second and fifth kelch motifs of RAG2. These mutations both diminish interaction with RAG1 and abrogate the cleavage function of the recombinase and are accordingly the cause of the immunodeficiency. We further support the importance of the conserved hydrophobic regions of the second β-strand and the characteristic glycine doublets of each of the six repeats through both in vitro and in vivo analysis of the recombination capacities of 26 site-directed mutations. In all, we provide both clinical and functional data supporting sequence analysis predictions that RAG2 may form a β-propeller structure composed of six kelch repeats. The role of RAG2 as a modular adapter protein involved in the coordination of multiple components of the recombination machinery is considered.
MATERIALS AND METHODS
Patients.
P1, a male infant born of unrelated parents, developed generalized seborrhea-like dermatitis and was hospitalized for a secondary Staphylococcus aureus skin infection and bacteremia. He was noted to have hypogammaglobulinemia (IgG, 146 mg/dl; IgM, 16 mg/dl; IgA, not detectable; IgE, 3 kU/liter) with normal lymphoid cell numbers (10,200 cells; subpopulations: CD3, 51%; CD4, 49%; CD8, 28%; DR, 46%; Leu12 [B cells], 2%). The patient suffered from eosinophilia (lymphocytes, 60%; eosinophils, 21%) and mildly abnormal mitogen stimulation responses to phytohemagglutinin (10,932 versus 36,916 [control]), concavalin A (6,960 versus 11,586 [control]), and pokeweed mitogen (2,843 versus 3,590 [control]). There was no evidence of maternal engraftment by HLA typing or chromosome analysis. He subsequently developed generalized lymphadenopathy, splenomegaly, and interstitial pneumonitis. Biopsies of skin and lymph nodes revealed extensive infiltration with histologically benign, activated, single-positive CD4 and CD8 T cells. Late in the course of his lymphoproliferative disease, his lymphocyte count was 66,200 (subpopulations: CD3, 87%; CD4, 57%; CD8, 37%; DR, 81%; Leu12 [B cells], 2%), with 50% lymphocytes and 15% eosinophils. Lung biopsy revealed giant-cell pneumonitis. The patient died of respiratory failure at age 5 months.
P2, a male infant born of consanguineous parents (fourth-degree cousins), presented at 2 months of age with hepatomegaly and generalized dermatitis that was resistant to topical steroids. The infant developed otitis, showed eosinophilia (11,340 cells/mm3), increased alanine aminotransferase (115 mU/ml) and aspartate aminotransferase (469 mU/ml) levels, and severe hypogammaglobulinemia (IgG, 25 mg/dl; IgA, <6 mg/dl; IgM, 2 mg/dl; IgE, 3 kU/liter). Lymphocyte subpopulations were as follows: CD3, 20%; CD4, 16% [CD45RA, <1%; CD45R0, 16%]; CD8, 23%; DR, 38%; CD19, <1%; CD16, 56%; TCRAB, 20%; TCRGD, <1%. Molecular analysis of peripheral blood mononuclear cells, using highly polymorphic DNA markers (APOB) showed maternal T-cell engraftment. In vitro proliferative responses to phytohemagglutinin were markedly decreased (6,150 cpm versus 44,950 cpm in an age-matched control). A lymph node biopsy disclosed profound abnormalities of the architecture, with severe lymphoid depletion and evidence of expression of activation markers (CD45R0 and DR) on the few lymphocytes present. Based on these findings, a diagnosis of SCID with maternal T-cell engraftment was established. The infant was kept in a protected environment and treated with antibiotics and intravenous immunoglobulins until he received bone marrow transplantation from an unrelated donor (five of six HLA antigens matched) following conditioning with busulfan, cyclophosphamide, and antithymocyte serum. At 2 years after bone marrow transplantation the patient is alive and well with full lymphoid engraftment and complete immunological reconstitution. Genetic mutations detected in P2 were traced back to the parents, who had normal numbers of T and B cells.
Identification of mutations in the RAG2 gene.
The coding regions of both RAG1 (GenBank accession no. M29474) and RAG2 (GenBank accession no. M94633) were PCR amplified from genomic DNA and directly sequenced by the strategy described by Villa et al. (64). Mutations were confirmed by cloning into PCR 2.1 vector (Invitrogen) and sequencing of multiple clones with the Thermosequenase kit (Amersham).
Recombinant plasmid constructs and site-directed mutagenesis in the active core of RAG2.
Amino acid substitutions within the full-length RAG2 product were generated in pBluescript using the T7 DNA polymerase-based Muta-Gene phagemid in vitro mutagenesis kit (Bio-Rad) and identified through the introduction of a silent restriction site at the site of mutation. The region of RAG2 encoding amino acids 1 to 383 was subsequently amplified by PCR and subcloned as BamHI-NotI fragments into the mammalian expression vector pEBG to generate fusions to the 3′ end of glutathione S-transferase (GST). Wild-type, G95R, and ΔI273 RAG2 alleles were PCR amplified and introduced in frame into pEGFC1 (Clontech) to generate RAG2 full-length fusions to the 3′ end of green fluorescent protein (GFP). Sequences of all of the constructs were confirmed using the Thermosequenase kit (United States Biosciences).
Cell culture and recombinant eukaryotic protein expression.
The human embryonic kidney fibroblast line 293T was grown at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagle medium containing 10% fetal bovine serum. GST fusion proteins of RAG2 were overexpressed from the pEBG vector (see above) by transient transfection of 293T cells at 25% confluency using the calcium phosphate precipitation method. Cells were harvested 48 h posttransfection, processed as previously described (54), and dialyzed in 25 mM Tris (pH 8.0)–150 mM KCl–2 mM EDTA–2 mM dithiothreitol–20% glycerol. Protein quantitation was conducted following sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie staining using dilutions of bovine serum albumin (BSA) as a standard.
Cellular localization of GFP fusions of full-length RAG2 proteins in 293T cells.
Each pEGFPC1 construct (10 μg) was transiently overexpressed in 293T cells as described above. At 24 h after transfection, the cells (adhered to coverslips) were washed once in phosphate-buffered saline (PBS) prewarmed to 37°C, incubated for 20 min in 4% formaldehyde, rinsed three times in room temperature PBS, fixed for 15 min in methanol-acetone (1:1), chilled to 4°C, and rehydrated for 10 min in PBS. After three washes in PBS–1% BSA, the cells were incubated for 1 min at room temperature with 5 μg of 4′,6-diaminidino-2-phenylindole (DAPI; Sigma) per ml in PBS–1% BSA. Following an additional three washes in PBS–1% BSA, the coverslips were mounted and images were acquired using confocal laser-scanning microscopy with the assistance of Scott Henderson (Mount Sinai School of Medicine Confocal Laser Scanning Microscopy core facility). The reported findings were observed in three separately performed experiments.
Nuclease and electrophoretic mobility shift assays.
Twelve RSS cleavage assays were performed under the 10% dimethyl sulfoxide cleavage conditions described previously (42) with an incubation of 30 min at 37°C. Binding assays were conducted under similar conditions, except that incubation was first performed at 30°C for 10 min in Mg2+ with the subsequent addition of 0.1% glutaraldehyde followed by an additional 10 min at 30°C. The cleavage assay reactions were stopped by the addition of 50% stop solution (95% formamide, 0.1% bromphenol blue, 0.1% xylene cyanol), and the products were resolved on 18% denaturing polyacrylamide–urea gels. Mobility shift assays were resolved using 4% native polyacrylamide 0.5× Tris-borate-EDTA (TBE) gels in the absence of any loading buffer. Cleavage assays for the experiment in Fig. 3 and cleavage and binding assays for the experiments in Figs. 3, 5, and 6 were repeated twice.
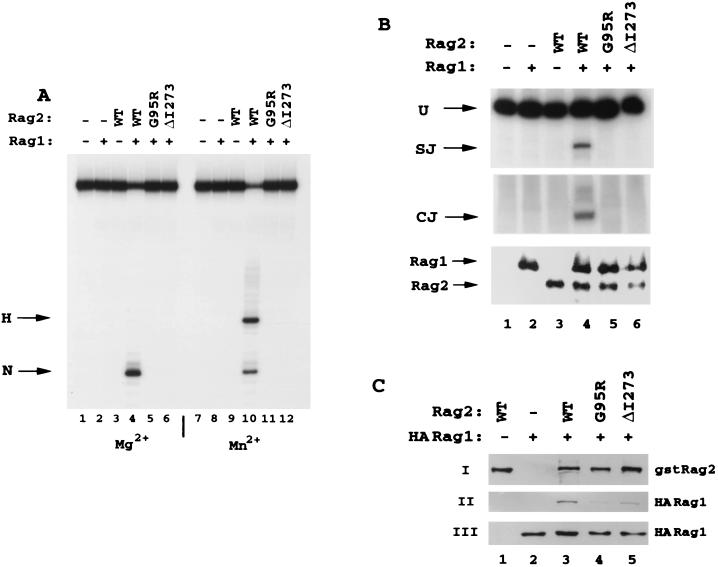
FIG. 3.
Human mutations in the predicted second β-strand of the kelch repeats are defective for both in vitro RSS cleavage and in vivo recombination activity. (A) Wild-type full-length human RAG2 and mutants G95R and ΔI273 were purified from 293T cells as fusion proteins to the C terminus of GST. Equal amounts of each protein were incubated with wild-type (WT) GST-RAG1 (amino acids 380 to 1040) and tested for the ability to generate nicks (N) and hairpins (H) on a 53-bp oligonucleotide containing a consensus 12 RSS. The DNA was 32P labeled on the 5′ end of the nicked strand. Reactions were conducted for 30 min at 37°C in the presence of Mg2+ (lanes 1 to 6) or Mn2+ (lanes 7 to 12), and the products were resolved on denaturing polyacrylamide gels. (B) Plasmids expressing the GST fusions of RAG2 and wild-type RAG1 were cotranfected into 293T cells along with a recombination plasmid substrate, pJH200. Recombination activity was evaluated by PCR analysis for the formation of signal joints (SJ, top panel) and coding joints (CJ, middle panel) of recombination substrate recovered 48 h posttransfection (39). PCR primers for signal joints also detect unrecombined plasmid (U). Dilutions of each sample were used to determine the linear range for the PCR analysis, and recombination activity was analyzed within this range. Protein levels were monitored by Western blot analysis of an aliquot of cell lysate (lower panel). (C) GST-RAG2 wild-type and mutant proteins were expressed with HA-tagged RAG1 active core (amino acids 330 to 1040), and interaction was evaluated by coprecipitation assays using glutathione beads. Panel I was blotted with anti-GST antibodies for detection of affinity-purified GST-RAG2, panel II was blotted with anti-HA antibody for detection of precipitated HA-RAG1, and panel III was blotted with polyclonal anti-RAG1 antibody R1P7.
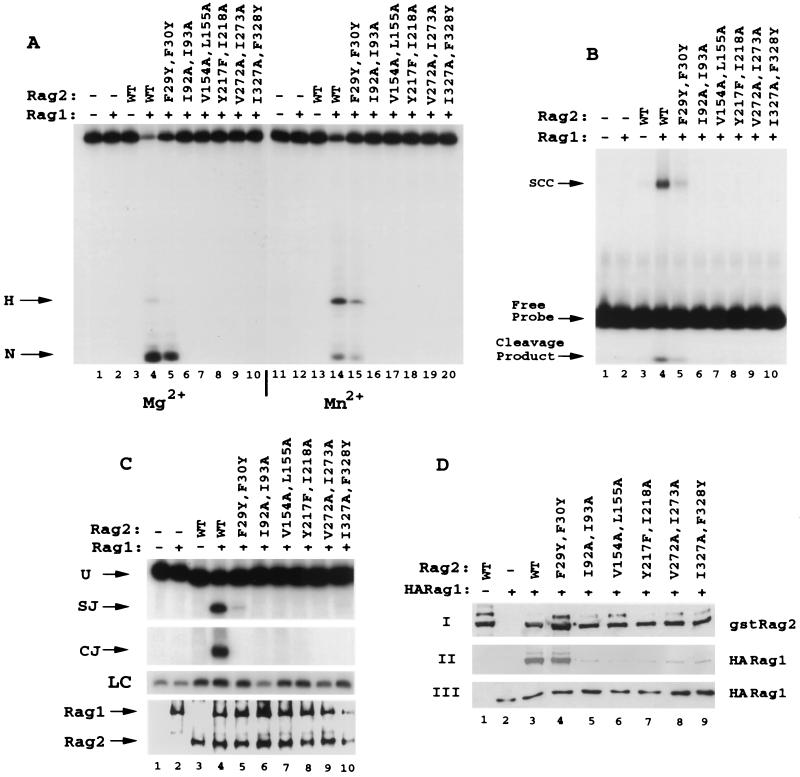
FIG. 5.
Mutation of most of the hydrophobic residues in the predicted second β-strand of repeats 1 to 6 of RAG2 decreases the binding of RAG1, with parallel effects on in vitro RSS binding and cleavage. (A and B) Conservative double mutations (F29Y F30Y, I92A I93A, V154A L155A, Y217F I218A, V272A I273A, and I327A F328Y) (Fig. 1) in the hydrophobic regions of kelch repeats 1 through 6 were expressed and purified as GST fusion proteins and tested in vitro for 12 RSS cleavage in Mg2+ (lanes 1 to 10) and Mn2+ (lanes 11 to 20) (A) or for SCC formation with the 12 RSS (B). (C and D) In vivo analysis of signal joint and coding-joint formation was conducted (C) in addition to analysis of complex formation between RAG1 and RAG2 (D). In panel C, loading controls (LC) performed in the linear range of the PCR are shown. WT, wild type.
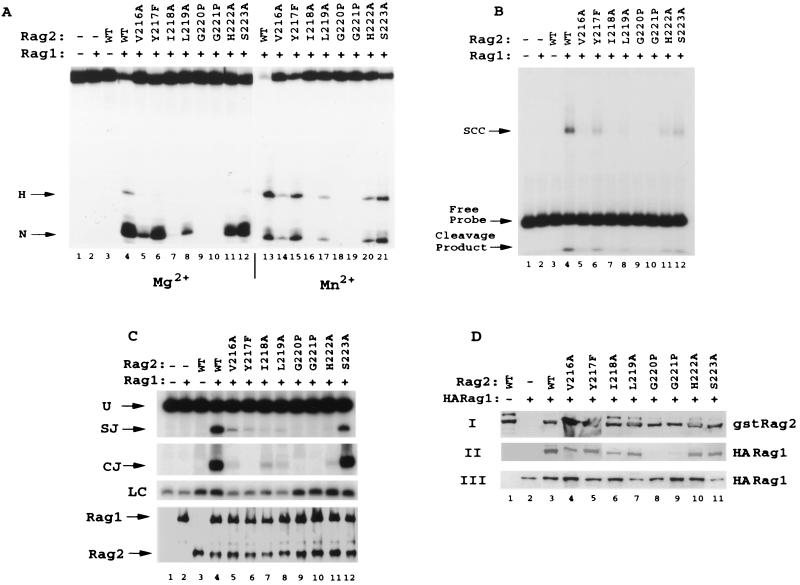
FIG. 6.
Individual point mutations in the predicted second β-strand of repeat 4 have differential effects on interaction with RAG1 and on cleavage and recombination activity. Eight individual conservative point mutations (V216A, Y217F, I218A, L219A, G220P, G221P, H222A, and S223A) were produced in the second β-strand of the fourth kelch repeat of GST-RAG2 (amino acids 1 to 383). Mutants were analyzed for 12 RSS cleavage (A), binding (B), recombination activity (C), and RAG1 interaction (D). Panel C contains PCR loading controls (LC) performed in the linear range of the reaction. WT, wild type.
In vivo recombination assays and interaction analysis.
Standard recombination conditions were modifications of those first established by Hesse et al. (21) and most recently outlined by Aidinis et al. (3) using recombination substrates pJH200 and pJH288 (21). A 32P-based PCR based method of analysis was used to evaluate the formation of signal and coding joints (3, 39, 64). To ensure that the PCR analysis provided a quantitative assessment of recombination activity, reactions were conducted for 25 cycles over a dilution range of 1:2 to 1:1,000. Signal joints were detected using PCR primers RA5 and RA14, and coding-joint formation was evaluated using PCR primers OOP2 and CR3 (39, 64). Quantitation of recombination activity was assessed by phosphorimaging (Bio-Rad). Recombination assays were repeated either two or three times. Interaction assays were conducted as previously described (53, 64), and the levels of binding were evaluated by phosphorimaging (Bio-Rad). The pull-downs were normalized for the minor fluctuations in expressed RAG1 protein. The coprecipitation experiments in Fig. 3, 6, and 7 were performed a total of three times.
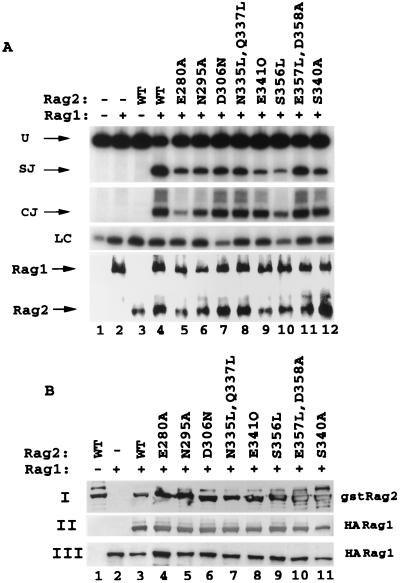
FIG. 7.
Mutations in predicted loop regions and at the predicted borders of β-strand 3 have moderate to no effect on the interaction with RAG1 and hence on the capacity to form coding and signal joints in vivo. Eight drastic mutations, E280A, N295A, D306N, N335L-Q337L, E341Q, S356L, E357L-D358A, and S340A, were introduced into the variable predicted loop regions of RAG2, and these substitutions were tested for the ability to mediate signal joint and coding-joint formation in vivo (A) and for the capacity to precipitate RAG1 (B). WT, wild type.
RESULTS
Isolation of mutations within the kelch repeats of RAG2 from immunodeficient patients P1 and P2.
We have identified two immunodeficient patients, P1 and P2, who exhibited the clinical immunological features characteristic of OS or SCID, respectively (see Materials and Methods) (17, 47, 65). Analysis for mutations within their RAG1 and RAG2 alleles revealed that both patients harbored alterations within the RAG2 locus in accordance with previous reports citing mutations in the V(D)J recombinase as a genetic cause of immunodeficiency syndromes (47, 64). P1 was heterozygous for missense mutation G1484A on one allele, which changes glycine 95 into arginine, and for missense mutation T2558A on the other allele, which changes tryptophan 453 to arginine. P2 is homozygous for a 3-nucleotide in-frame deletion (positions 2018 to 2020), resulting in the removal of isoleucine 273 (ΔI273), with the remainder of the protein being intact. Over 100 chromosomes from ethnically matched individuals were sequenced to exclude the possibility that the mutations represent rare polymorphisms.
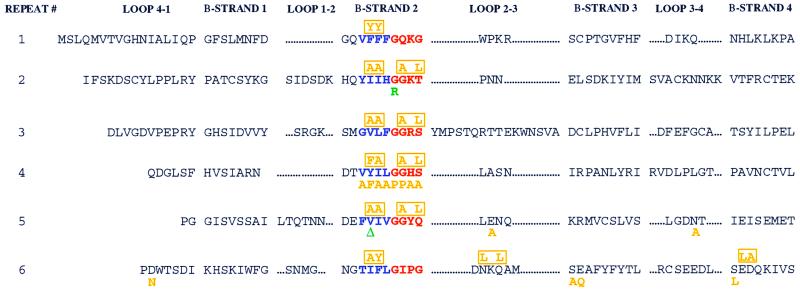
G95 and W453 are conserved in all known RAG2 genes from fish to humans, while I273 is fully conserved except in chickens, where it is replaced by leucine. While W453 is within the region of RAG2 that is dispensable for V(D)J recombination, both G95 and I273 are within the essential active core (13, 40, 41, 52) that is composed of six kelch repeats (5, 9). Interestingly, both mutations localized to the putative second β-strand of either repeat 2 or repeat 5 (Fig. 1, noted in green below the altered residue), which is the most highly conserved region of kelch repeat-containing proteins in terms of both amino acid identity and character. To unravel the molecular mechanism leading to immunodeficiency in these two patients and to test the importance of conserved regions of the kelch repeat for RAG2 function, we undertook a molecular and biochemical analysis of the two identified RAG2 kelch repeat mutations. The RAG2 mutations used in this study are listed in Table 1.
FIG. 1.
Mutations in patients P1 and P2 are localized to the RAG2 kelch repeat domains. Each of the six kelch repeats of the RAG2 active core (13, 40, 41, 52) is formed by four β-strands (1 to 4) separated by loops of variable length (4-1 to 3-4). We present the sequence of mouse RAG2. The second β-strand of each repeat demonstrates the highest level of conservation between various members of the kelch family and is composed of a 4-amino-acid (predominantly hydrophobic) region, displayed in blue. The border of β-strand 2 and loop 2-3 contains a 4-amino-acid glycine-serine-threonine-rich repeat, highlighted in red. Mutations from patients P1 and P2 are noted in green below the affected residue. Note that isoleucine 273 in human RAG2 corresponds to a valine in the mouse sequence. Individual site-directed mutations are displayed in orange below the substituted amino acid, while multiple mutations are indicated and boxed in orange above the altered amino acids.
TABLE 1.
Naturally occurring and artificially introduced mutations in RAG2 used in this study
| Location | Mutations |
|---|---|
| Human RAG2 | G95R, ΔI273, W453R |
| Glycine-serine-threonine-rich regions of repeats 2–5 | G96A T98L, G174A S176L, G221A S223L, G276A Q278L |
| Repeat 4 | V216A, Y217F, I218A, L219A, G220P, G221P, H222A, S223A |
| Hydrophobic regions of repeats 1–6 | F29Y F30Y, I92A I93A, V154A L155A, Y217F I218A, V272A I273A, I327A F328Y |
| Loop regions | E280A, N295A, D306N, N335L Q337L, S340A, E341Q, S356L, E357L D358A |
Mutations G95R and ΔI273 localize correctly to the nucleus but are defective in the early steps of the V(D)J recombination reaction.
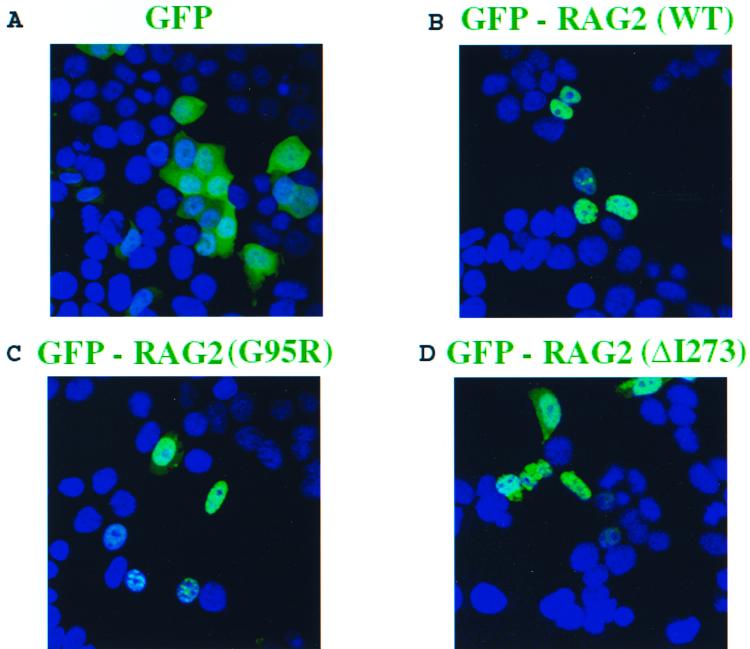
Protein mislocalization from the nucleus to the cytoplasm is an established cause of a number of syndromes (7, 12, 49, 67). Since endogenous and overexpressed RAG2 proteins localize effectively to the nucleus (53), we explored if mutations G95R and ΔI273 would alter the intracellular distribution of RAG2. Full-length wild-type and mutant alleles of RAG2 were fused in frame to the 3′ end of GFP (10) and transiently overexpressed in 293T cells. Images were acquired by confocal laser-scanning microscopy (Fig. 2). GFP alone was distributed throughout both the nucleus and the cytoplasm (Fig. 2A), while all three GFP-RAG2 fusions (wild type, G95R, and ΔI273) were imported into the nucleus. Hence, we conclude that the immunological defects observed in patients P1 and P2 do not result from an inability of the mutant proteins to compartmentalize into the nucleus.
FIG. 2.
Mutations G95R and ΔI273 are imported into the nucleus. Various full-length RAG2 alleles (amino acids 1 to 527) were fused in frame to the C terminus of enhanced GFP and transiently overexpressed in 293T cells. The cells were fixed and processed 24 h posttransfection and subsequently visualized using confocal laser-scanning microscopy. Nuclei were stained with DAP1 (blue). (A) Cells were transfected with GFP alone. (B to D) Full-length forms of wild-type or mutant GFP-RAG2 localized effectively to the nucleus (GFP-RAG2 wild type [B], GFP-RAG2 G95R [C], and GFP-RAG2 ΔI273 [D]).
To determine if the mutations from patients P1 and P2 inhibit the nucleolytic capacity of the RAG1-RAG2 complex, we purified GST fusions of the wild-type and mutant proteins and tested their capacity to mediate nick and hairpin formation of an oligonucleotide substrate containing a consensus 12 RSS labeled on the 5′ end of the upper strand (32, 42). Equal amounts of each protein were incubated with wild-type GST-RAG1 (amino acids 380 to 1040) and RSS substrate in the presence of Mg2+ or Mn2+ as the divalent cation (Mn2+ rescues the phenotypes of a number of mutations within restriction enzymes and transposases). With either divalent metal, the mutant proteins were unable to activate either nick or hairpin formation on the 12 RSS (Fig. 3A, lanes 5 and 6 and lanes 11 and 12) or the 23 RSS (data not shown). In addition, both mutant proteins were unable to form a stable RAG1-RAG2-RSS complex as determined by mobility shift assays (data not shown). As expected from these observations, when coexpressed with wild-type GST-RAG1, neither mutant was able to generate signal or coding joints on a deletional episomal plasmid substrate (pJH200) (21) (Fig. 3B, lanes 5 and 6) or on an inversional substrate (pJH288) (data not shown). PCR analyses in these and all other assays presented in this study were performed in the linear range for the assay, and loading controls were used to verify the use of equal amounts of starting material (data not shown).
To further investigate the basis for the cleavage and binding deficiencies observed for the two mutations, we performed coprecipitation assays in which GST-RAG2 proteins were coexpressed in 293T cells along with hemagglutinin (HA)-tagged RAG1. At 48 h posttransfection, RAG1-RAG2 complexes were isolated from cell lysates by using glutathione beads and interaction levels were determined by Western blot analysis with anti-HA antibodies to detect precipitated RAG1 (Fig. 3C, panel I) and anti-GST antibodies to control for appropriate levels of GST-RAG2 wild-type and mutant protein expression (panel II). Total-cell extracts were also blotted with anti-RAG1 antibody to ensure comparable levels of RAG1 expression across all assays (panel III). In this assay, the ability of the mutant RAG2 proteins to precipitate RAG1 was approximately 20 to 30% of wild-type levels (panel II, lanes 3 to 5), suggesting that mutations within the second β-strand of RAG2 inhibit the efficiency of RAG1 and RAG2 complex formation. The use of full-length RAG2 proteins in this assay resulted in reduced overall levels of interaction with RAG1 compared to those detected in subsequent precipitation experiments performed with the active core of RAG2 (see Fig. 4D, 5D, 6D, and 7B). Of note, in accordance with the partial recombination phenotype observed in patient P1, mutant W453R maintained partial RSS nicking and hairpin formation function, as well as a reduced capacity to form signal and coding joints (data not shown). Overall, the data demonstrate that mutations G95R and ΔI273 within the conserved regions of RAG2 kelch repeats are entirely defective in mediating the initial nucleolytic steps of the V(D)J recombination reaction.
FIG. 4.
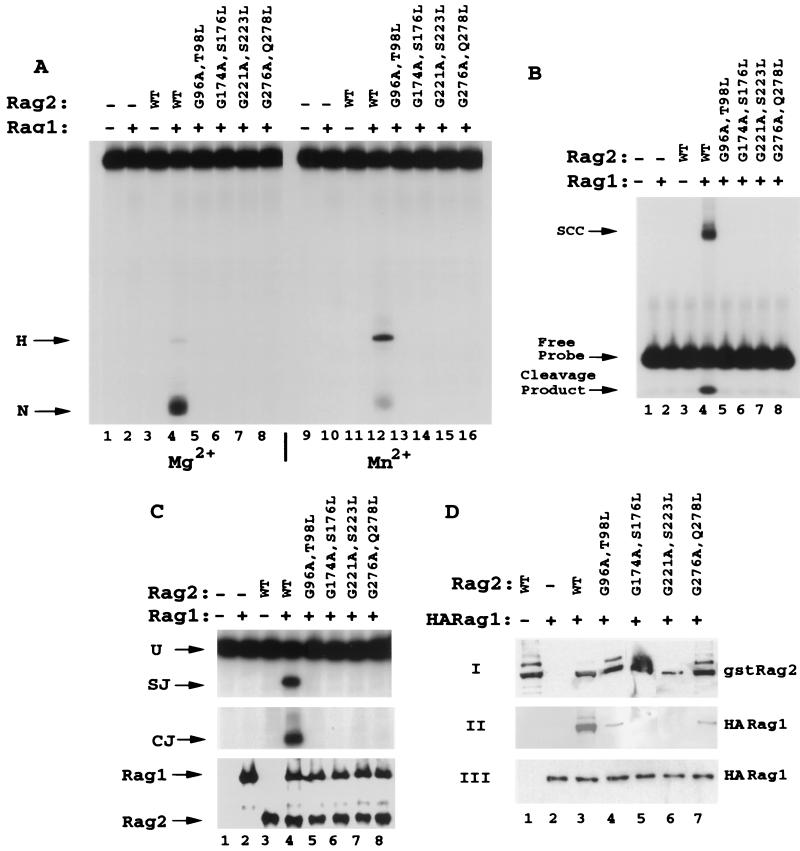
Mutations in RAG2 glycine-serine-threonine-rich regions affect the interaction with RAG1 and concomitantly block RSS binding and nicking. (A) Double mutations in repeats 2 (G96A T98L), 3 (G174A S176L), 4 (G221A S223L), and 5 (G276A Q278L) were generated by substituting the second glycine of the glycine doublets as well as a serine or threonine residue 2 amino acids C-terminal to the glycine (except in the case of the fifth repeat, which has glutamine at this position [Fig. 1]). GST-RAG2 fusions (amino acids 1 to 383) were purified and tested for 12 RSS nicking and hairpin activity. In both Mg2+ (lanes 1 to 8) and Mn2+ (lanes 9 to 16), all four mutant proteins were entirely inactive for either nicking or hairpinning the 12 RSS compared to the wild type (WT). (B) Mutant RAG2 proteins were assayed for the capacity to form SCC along with wild-type GST-RAG1 on the 12 RSS. As with 12 RSS cleavage, all mutants were inactive for SCC formation (lanes 5 to 8). Cleavage products can be visualized below the free probe. (C) Mutants were tested for recombination activity on substrate pJH200 by PCR analysis for signal joints (SJ) and coding joints (CJ). None of the four mutants (lanes 5 to 8) generated detectable recombination products (SJ or CJ). Aliquots of the cell lysates were evaluated for protein expression by Western blot analysis. (D) Interaction between GST-RAG2 and HA-tagged RAG1 was monitored as described in the legend for Fig. 3C.
Importance of conserved residues of the predicted second β-strand of RAG2 for RAG1-RAG2 complex formation and RSS binding and cleavage demonstrated by extensive site-directed mutagenesis.
To more thoroughly analyze the significance of the amino acid composition of the RAG2 repeats that are conserved within and between many kelch repeat proteins, a number of alterations were introduced within the second β-strand region by site-directed mutagenesis. Three groups of mutations were generated to determine the contribution of the glycine doublets (Fig. 1, red), the hydrophobic regions (Fig. 1, blue), and individual amino acids within the predicted second β-strand to the function of RAG2 in the initial stages of RSS recognition and cleavage.
Glycine doublets are a noted characteristic of a host of kelch repeat-containing proteins (1, 9) and are found in four of the six repeats of RAG2 (repeats 2 to 5) at the predicted border of the second β-strand and loop 2-3. The RAG2 repeats are notable in that the doublets are followed in three of the four cases (repeats 2 to 4) by either a serine or a threonine residue with a single intervening amino acid residue. Double substitutions (Fig. 1) were generated, altering the second glycine of repeats 2, 3, 4, and 5 in conjunction with the serine or threonine residues (a glutamine in repeat 5). These mutant proteins were expressed as GST fusions and purified following transient overexpression in 293T cells. When equal amounts of protein were tested for the ability to foster 12 RSS cleavage, all four substitutions displayed nondetectable levels of activity (Fig. 4A), even under highly permissive Mn2+-based conditions. Consistent with this finding, none of the four mutants could capture the 12 RSS in a stable cleavage complex (SCC) in the presence of RAG1 (Fig. 4B). Concurrently, the substitutions ablated both signal joint and coding-joint formation in vivo on pJH200 (Fig. 4C) and pJH288 (data not shown). Similar to the human mutation G95R, mutation of the glycine regions significantly reduced the interaction of RAG2 with RAG1 (Fig. 4D). This finding indicates that the catalytic deficiency demonstrated by these mutants results from a defect in the formation of a productive RAG1-RAG2 complex and shows that the integrity of regions in which these glycines are found is fundamental to the function of RAG2.
The central four amino acids of the putative second β-strand of each repeat are nearly exclusively hydrophobic (Fig. 1, blue). To test the contribution of this amino acid stretch to the activity of RAG2, amino acid substitutions were introduced by site-directed mutagenesis in each of the six repeats. The mutations were designed to alter the amino acid character and structure of the region to the slightest extent possible. As noted with the four mutant proteins with substitutions in the glycine-serine-threonine-rich regions, five of the six mutations within the hydrophobic regions (F29Y and F30Y; I92A and I93A; V154A and L155A; Y217F and I218A; V272A and I273A; and I327A and F328Y) entirely abolished RSS binding and cleavage (Fig. 5A and B). These five mutations all demonstrated a striking deficiency in RAG1 interaction as observed in coprecipitation experiments (Fig. 5D). Only the F29Y F30Y mutant (Fig. 5A, lanes 5 and 15), retained partial activity in vitro with the capacity to cleave the 12 RSS at approximately 40% of wild-type levels and to bind the RSS at approximately 20% wild-type levels (Fig. 5B, lane 5). Accordingly, the F29Y F30Y mutant precipitated RAG1 more efficiently than did mutants with analogous mutations in the remaining five kelch repeats (Fig. 5D, lane 4). However, the partial catalytic activity of this mutant was even more significantly reduced when it was assayed in vivo, where its capacity to generate signal joints was 5% of that of the wild type and no detectable coding-joint formation was noted (Fig. 5C, lane 5). Thus, the hydrophobic regions of the predicted second β-strand of all six kelch repeats of RAG2 appear to be important for the interaction of RAG2 with RAG1 and ultimately for the nucleolytic activity of the RAG1-RAG2 recombinase complex.
Once the importance of both the glycine-rich and hydrophobic regions for the function of RAG2 had been established through the use of double-amino-acid substitutions, individual mutations were introduced through eight consecutive amino acids in kelch repeat 4 (Fig. 1 [V216A, Y217F, I218A, L219A, G220P, G221P, H222A, and S223A]). In terms of 12 RSS cleavage activity in the presence of both Mg2+ and Mn2+ (Fig. 6A), the mutants fall into three classes: one (I218A, G220P, and G221P) with no detectable level of activity, one (V216A and L219A) with less than 10% of wild-type activity, and one (Y217F, H222A, and S223A) with approximately 50% activity. The relative levels of activity on the 12 RSS are roughly reflected by the DNA binding (Fig. 6B), in vivo recombination levels (Fig. 6C), and RAG1 interaction levels (Fig. 6D) noted for these mutants. All mutants were tested for the capacity to nick sequence-independent 3′ overhang structures that may represent less stringent nuclease requirements than those needed for sequence-specific 12 RSS cleavage. However, the activity observed on these 3′ overhang structures (data not shown) directly paralleled levels observed on the 12 RSS (Fig. 4 to 6), suggesting that alteration of the predicted second β-strand of any kelch repeat in RAG2 causes global deficiencies in the nuclease potential of RAGs.
Mutations within the predicted loop regions of the RAG2 kelch repeats have no or minimal effects on in vivo recombination activity.
Since the vast majority of mutations analyzed within the proposed second β-strand had dramatic effects on SCC formation and 12 RSS nicking, the possibility arose that RAG2 may be highly sensitive to a broad array of mutations. To test this, we designed eight mutations in the predicted loops 2-3 (E280A) and 3-4 (N295A) of repeat 5 as well as loops 4-1 (D306N) and 2-3 (N335L-Q337L) of repeat 6. In addition, two mutations were directed to the predicted border of loop 2-3/β-strand 3 (S340L and E341Q) and another two were directed to the border of loop 3-4/β-strand 4 (S356L and E357L-D358A). The targeted regions are generally variable in length and not conserved within and between kelch repeat-containing proteins. While these regions are not conserved compared to other kelch repeats either in RAG2 or in other kelch repeat-containing proteins, the amino acid sequences are indeed well conserved among all known RAG2 species. Three of the mutants (those containing the E280A, E341Q, and S356L mutations) demonstrated a decreased capacity (10 to 25% of that of the wild type) to generate signal joints (Fig. 7A, lanes 5, 9, and 10), while two of the mutants (those containing the E280A and S356L mutations) were partly deficient (<25% of the wild-type capacity) in forming coding joints (lanes 5 and 10). The remainder of the mutations within the loops resulted in retention of at least 50% of the wild-type capacity for signal joint production (lanes 6 to 8, 11, and 12) and a nearly intact ability to generate coding joints (lanes 6 to 9, 11, and 12). Mutants with mutations in all three groups displayed robust capacity to interact with RAG1 (Fig. 7B). Hence, we conclude that the loop regions of the RAG2 kelch repeats are significantly more tolerant to amino acid composition than are the regions composing the predicted second β-strand.
DISCUSSION
Recently, sequence analysis using either the PSI-BLAST iterative search method (5), the multiple-sequence analysis tool GAPPED-BLAST, or hydrophobic cluster analysis (9) has classified RAG2 as a potential kelch repeat superfamily member. In this report, we provide evidence in support of this model by presenting both genetic and biochemical data demonstrating the critical nature of conserved elements within the kelch repeats of RAG2 for the function of the V(D)J recombinase. Identification of a protein-protein interaction module within RAG2 is consistent with an emerging view of the dynamics of RAG1-RAG2 interactions during the recombination reaction.
RAG mutations within predicted β-strand 2 of two kelch repeats lead to immunodeficiency.
Mutations within the recombination-activating genes RAG1 and RAG2 have been identified as the cause of two forms of primary immunodeficiency, B-cell negative SCID and OS (47, 64). In this study we found three newly identified mutations in RAG2 that lead to these disorders. OS patient P1 was heterozygous for mutant alleles leading to substitutions G95R and W453R, while SCID patient P2 was homozygous for an allele leading to a deletion of I273. Interestingly, substitution G95R and ΔI273, located within the highly conserved second β-strand of kelch repeats 2 and 5, respectively, ablate the in vitro and in vivo recombinase functions of RAG1-RAG2. On the other hand, mutation W453R from OS patient P1, which lies outside of the predicted kelch repeat region of RAG2, results in retention of partial recombinase activity in the assays described above. The residual activity of this mutant protein provides the enzymatic function consistent with the OS phenotype of patient P1.
The predicted second β-strand in which mutations G95R and ΔI273 are located has been shown by sequence alignment of numerous kelch repeat proteins to contain the two most highly conserved elements of the kelch repeat (four hydrophobic residues and a glycine doublet), which probably contribute to the core fold of the molecules. The functional importance of these two elements was corroborated by the lack of observable activity of engineered substitutions G221P and I218A, which target amino acids in the predicted second β-strand of kelch repeat 4 and are analogous to the mutations found in P1 and P2 (Fig. 6). Interestingly, an analogous glycine substitution, G446E, was isolated from the Caenorhabditis elegans kelch repeat protein SPE-26, which disrupts spermatogenesis and leads to infertility in males and hermaphrodites (63). Moreover, in our study, the critical nature of the second β-strand was further supported by the finding that all but one substitution created in β-strand 2 of kelch repeat 4 nearly abrogated the activity of the recombinase (Fig. 6C). Not unexpectedly, the only well-tolerated substitution in β-strand 2 of repeat 4 was S223A, which lies at the predicted junction between the second β-strand and loop 2-3 (Fig. 6C). A comparably mild decrease in activity was observed for four mutations (S340A, E341Q, S356L, and E357L/D358A) directed to the predicted borders between other β-strands and their neighboring loops. Furthermore, site-directed mutation of residues (E280A, N295A, D306N, and N335L/Q337L) within the predicted loop regions that vary broadly across kelch repeat proteins also had a mild effect on recombinase activity (Fig. 7A). Previously reported mutations in mutants isolated from OS patients, R229Q and M285R, lie on the predicted border of loop 2-3 and the third β-strand in repeats 4 and 5, respectively. In light of the above-mentioned site-directed mutagenesis of residues on the borders of the β-strands and within predicted loops, it is not surprising that these two OS mutations result in retention of the levels of activity required to generate at least a partial immune repertoire (51, 64; A. Villa and S. Santagata, unpublished data). It is important to note that while the variability of the loop regions permits greater tolerance to amino acid substitution, a particular subset of residues within these loops may indeed provide the specificity required to generate productive protein-protein complexes. Indeed, crystallographic analysis of proteins which form β-propellers, such as α-scruin, the β-subunit of transducin, clathrin, and RCC1, has also suggested the presence of protein-protein interactions mediated through the loops which bridge blades of the propeller (27, 38, 56, 59). The decrease in RAG1 interaction and binding levels noted for many of the RAG2 mutant proteins with substitutions in the second β-strand used in this study suggests a global alteration in the architecture of RAG2 and not in the specific interactions of the protein with RAG1. Similar effects have been noted for mutations within the core of the β-propeller structure of RCC1 (38).
In two separate studies, deletion and insertion mutations were introduced throughout the RAG2 molecule (13, 40). All but two of the deletion mutations within the active core of RAG2 removed 4 to 6 amino acids either entirely or partly composing predicted β-strand regions. In agreement with data from our study, all of these deletions drastically reduced recombination activity. The two remaining deletions targeted regions entirely encompassed by predicted loops. As expected, deletion of amino acids 238 to 243 (VDLPLG), which compose the predicted loop 3-4 of repeat 4, did not reduce recombination activity. On the other hand, deletion of amino acids composing loop 1-2 of repeat 2 abolished recombination activity (40). While this region is predicted to form a loop, it has been suggested that this area is involved in closing the postulated β-propeller structure of RAG2 through an N-terminal closure pattern (1). Hence, this region may be particularly intolerant to a severe 6-amino-acid deletion. The importance of this region is supported by the finding that a 3-amino-acid insertion in this loop and in the preceding loop 4-1 also abolished recombination activity (13). Interpretation of the effects of the remainder of the insertion mutations throughout RAG2 in light of the kelch repeat model reveals a rather consistent view, with insertions in predicted β-strands sharply reducing or eliminating recombination activity (insertion at amino acids 46, 257, and 302) and mutations in loops 4-1 and 3-4 of repeat 6 having relatively moderate effect on the recombination efficiency.
Putative role of RAG2 in the V(D)J recombination reaction.
The double-strand break at the coding flank-heptamer border that initiates the recombination reaction requires the concerted action of the RAG1 and RAG2 proteins. Interestingly, RAG1 can complex with the RSS in the absence of RAG2 but cannot alone mediate DNA cleavage (3, 4, 14, 54, 58). In addition, recent studies indicate that RAG1 alone contains the 3 amino acid residues required to coordinate metal ion catalysis during the various steps of the recombination reaction (18, 26, 28). The amino acids lie within a region whose secondary structure is very similar to that of catalytic cores of numerous other transposases and integrases. Many of these homologous proteins can mediate at least a subset of their DNA-processing events independently of other proteins. In contrast, RAG1 is dependent on the regulated accumulation of RAG2 for the initiation of the V(D)J cleavage reaction (29, 31). It has recently been observed that the RAG1 zinc finger B (amino acids 727 to 750), which lies between the second aspartate and the glutamate that compose the active center of the molecule is a dominant interface for interaction with RAG2 (3a) This limited region of the RAG1 active core is notable due to its divergence from the standard folding pattern of other transposases and integrases (18). The lack of a zinc finger motif in other transposases suggests a potentially unique role for this motif in regulating the activity of RAG1. As a member of the kelch repeat family, it appears likely that RAG2 uses one or more of its predicted blades to mediate contacts with RAG1 potentially in the region of zinc finger B. It is conceivable that this interaction would elicit a specific structural change in RAG1 to activate RAG1 for the appropriate DNA nuclease steps. The importance of such an interaction is implied throughout this study by the striking correlation between the capacity of RAG2 to interact with RAG1 and the observed levels of DNA cleavage and recombination. Mutations in the glycine doublets and in the hydrophobic regions of repeats 2 to 6 drastically decrease the degree and quality of the interaction with RAG1, leaving RAG1 in an inactive conformation and, in turn, sharply reducing DNA cleavage. However, mutations in the first hydrophobic repeat and in the loops and predicted β-strand/loop borders maintain levels of interaction that are directly reflected in the DNA cleavage levels noted (Fig. 5 to 7).
In addition to the critical protein-protein interactions required between RAG1 and RAG2 for activation of RSS cleavage and processing, the actions of numerous other proteins must be coordinated for the successful completion of the recombination reaction. Since several β-propeller-containing molecules are believed to function as coordinators of multimolecular complexes, the possibility arises that a multimodular RAG2 molecule could serve a similar purpose during the V(D)J recombination reaction. Components of such a complex could be any of the already identified DNA repair or processing molecules central to the completion of the reaction, such as DNA PKCS, Ku 70/Ku 80, terminal deoxynucleotidyltransferase, XRCC4, and ligase IV, in addition to as yet unidentified components of the V(D)J recombination machinery.
ACKNOWLEDGMENTS
Carlos A. Gomez, Leon M. Ptaszek, and Anna Villa contributed equally to this work.
This work was supported by National Institutes of Health (NIH) grant AI40191 and a Cancer Research Institute Investigator Award to Z.Q.P., by Telethon grant E0917 to A.V., by NIH grant AI45996-02 and by a Cancer Research Institute Investigator Award to P.C., and by a predoctoral Training Grant in Cancer Biology from the NIH to S.S. Confocal laser scanning microscopy was performed under the guidance of Scott Henderson at the MSSM-CLSM core facility, supported with funding from NIH shared instrumentation grant 1 S10 RR0 9145-01 and NSF Major Research Instrumentation grant DBI-9724504. A.V. is the recipient of an AIRC travel fellowship for international scientific exchange.
We thank Anindita Bhoumik for assistance with subcloning. S.S. thanks Stuart Aaronson for guidance and support. L.M.P. is grateful to David G. Schatz for generous support and encouragement. We also thank David G. Schatz for comments on the manuscript.
ADDENDUM IN PROOF
A recent paper by Corneo et al. (B. Corneo, D. Moshous, I. Callebaut, R. de Chasseval, A. Fischer, and J. P. de Villartay, J. Biol. Chem. 275:12672–12675, 2000) describes three new mutations within RAG2 and provides support for the β-propeller model of RAG2 based on the clustering of mutations on one face of the molecule.
Footnotes
Manuscript 41 of the Cariplo-ITBA project Genoma 2000, directed by R. Dulbecco and funded by Cariplo.
REFERENCES
- 1.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 3.Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi M E, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol Cell Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Aidinis V, Dias D C, Gomez C A, Bhattacharyya D, Spanopoulou E, Santagata S. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J Immunol. 2000;164:5826–5832. doi: 10.4049/jimmunol.164.11.5826. [DOI] [PubMed] [Google Scholar]
- 4.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind L, Koonin E V. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 6.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 7.Bhuiyan Z A, Yatsuki H, Sasaguri T, Joh K, Soejima H, Zhu X, Hatada I, Morisaki H, Morisaki T, Mukai T. Functional analysis of the p57KIP2 gene mutation in Beckwith-Wiedemann syndrome. Hum Genet. 1999;104:205–210. doi: 10.1007/s004390050937. [DOI] [PubMed] [Google Scholar]
- 8.Bork P, Doolittle R F. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 9.Callebaut I, Mornon J P. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 11.Chilosi M, Facchetti F, Notarangelo L D, Romagnani S, Del Prete G, Almerigogna F, De Carli M, Pizzolo G. CD30 cell expression and abnormal soluble CD30 serum accumulation in Omenn's syndrome: evidence for a T helper 2-mediated condition. Eur J Immunol. 1996;26:329–334. doi: 10.1002/eji.1830260209. [DOI] [PubMed] [Google Scholar]
- 12.Cressman D E, Chin K C, Taxman D J, Ting J P. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–171. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- 13.Cuomo C A, Oettinger M A. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 15.Eastman Q M, Leu T M, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 16.Eastman Q M, Villey I J, Schatz D G. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer A, Cavazzana-Calvo M, De Saint Basile G, DeVillartay J P, Di Santo J P, Hivroz C, Rieux-Laucat F, Le Deist F. Naturally occurring primary deficiencies of the immune system. Annu Rev Immunol. 1997;15:93–124. doi: 10.1146/annurev.immunol.15.1.93. [DOI] [PubMed] [Google Scholar]
- 18.Fugmann S D, Villey I J, Ptaszek L M, Schatz D G. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 19.Gomez L, Le Deist F, Blanche S, Cavazzana-Calvo M, Griscelli C, Fischer A. Treatment of Omenn syndrome by bone marrow transplantation. J Pediatr. 1995;127:76–81. doi: 10.1016/s0022-3476(95)70260-1. [DOI] [PubMed] [Google Scholar]
- 20.Grawunder U, West R B, Lieber M R. Antigen receptor gene rearrangement. Curr Opin Immunol. 1998;10:172–180. doi: 10.1016/s0952-7915(98)80246-x. [DOI] [PubMed] [Google Scholar]
- 21.Hesse J E, Lieber M R, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 22.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 23.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 24.Ito N, Phillips S E, Stevens C, Ogel Z B, McPherson M J, Keen J N, Yadav K D, Knowles P F. Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature. 1991;350:87–90. doi: 10.1038/350087a0. [DOI] [PubMed] [Google Scholar]
- 25.Ito N, Phillips S E, Yadav K D, Knowles P F. Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol. 1994;238:794–814. doi: 10.1006/jmbi.1994.1335. [DOI] [PubMed] [Google Scholar]
- 26.Kim D R, Dai Y, Mundy C L, Yang W, Oettinger M A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 28.Landree M A, Wibbenmeyer J A, Roth D B. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J, Desiderio S. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity. 1999;11:771–781. doi: 10.1016/s1074-7613(00)80151-x. [DOI] [PubMed] [Google Scholar]
- 30.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 31.Lin W C, Desiderio S. V(D)J recombination and the cell cycle. Immunol Today. 1995;16:279–289. doi: 10.1016/0167-5699(95)80182-0. [DOI] [PubMed] [Google Scholar]
- 32.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 33.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 34.Nagawa F, Ishiguro K, Tsuboi A, Yoshida T, Ishikawa A, Takemori T, Otsuka A J, Sakano H. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol Cell Biol. 1998;18:655–663. doi: 10.1128/mcb.18.1.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Notarangelo L D, Villa A, Schwarz K. RAG and RAG defects. Curr Opin Immunol. 1999;11:435–442. doi: 10.1016/S0952-7915(99)80073-9. [DOI] [PubMed] [Google Scholar]
- 36.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 37.Ramsden D A, McBlane J F, van Gent D C, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 38.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 39.Roman C A, Baltimore D. Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc Natl Acad Sci USA. 1996;93:2333–2338. doi: 10.1073/pnas.93.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadofsky M J, Hesse J E, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadofsky M J, Hesse J E, McBlane J F, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. . (Erratum, 22:550, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santagata S, Aidinis V, Spanopoulou E. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J Biol Chem. 1998;273:16325–16331. doi: 10.1074/jbc.273.26.16325. [DOI] [PubMed] [Google Scholar]
- 43.Santagata S, Besmer E, Villa A, Bozzi F, Allingham J S, Sobacchi C, Haniford D B, Vezzoni P, Nussenzweig M C, Pan Z Q, Cortes P. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol Cell. 1999;4:935–947. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- 44.Schandene L, Ferster A, Mascart-Lemone F, Crusiaux A, Gerard C, Marchant A, Lybin M, Velu T, Sariban E, Goldman M. T helper type 2-like cells and therapeutic effects of interferon-gamma in combined immunodeficiency with hypereosinophilia (Omenn's syndrome) Eur J Immunol. 1993;23:56–60. doi: 10.1002/eji.1830230110. [DOI] [PubMed] [Google Scholar]
- 45.Schatz D G. V(D)J recombination moves in vitro. Semin Immunol. 1997;9:149–159. doi: 10.1006/smim.1997.0068. [DOI] [PubMed] [Google Scholar]
- 46.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz K, Gauss G H, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger R A, Hansen-Hagge T E, Desiderio S, Lieber M R, Bartram C R. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 48.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 49.Shiyanov P, Hayes S A, Donepudi M, Nichols A F, Linn S, Slagle B L, Raychaudhuri P. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol Cell Biol. 1999;19:4935–4943. doi: 10.1128/mcb.19.7.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shockett P E, Schatz D G. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:4159–4166. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Signorini S, Imberti L, Pirovano S, Villa A, Facchetti F, Ungari M, Bozzi F, Albertini A, Ugazio A G, Vezzoni P, Notarangelo L D. Intrathymic restriction and peripheral expansion of the T-cell repertoire in Omenn syndrome. Blood. 1999;94:3468–3478. [PubMed] [Google Scholar]
- 52.Silver D P, Spanopoulou E, Mulligan R C, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spanopoulou E, Cortes P, Shih C, Huang C M, Silver D P, Svec P, Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 54.Spanopoulou E, Zaitseva F, Wang F H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 55.Steen S B, Gomelsky L, Roth D B. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 56.Sun S, Footer M, Matsudaira P. Modification of Cys-837 identifies an actin-binding site in the beta-propeller protein scruin. Mol Biol Cell. 1997;8:421–430. doi: 10.1091/mbc.8.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swanson P C, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanson P C, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 59.ter Haar E, Musacchio A, Harrison S C, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–573. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 61.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 62.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 63.Varkey J P, Muhlrad P J, Minniti A N, Do B, Ward S. The Caenorhabditis elegans spe-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- 64.Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Gatta L B, Ochs H D, Schwarz K, Notarangelo L D, Vezzoni P, Spanopoulou E. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 65.Villa A, Santagata S, Bozzi F, Imberti L, Notarangelo L D. Omenn syndrome: a disorder of Rag1 and Rag2 genes. J Clin Immunol. 1999;19:87–97. doi: 10.1023/a:1020550432126. [DOI] [PubMed] [Google Scholar]
- 66.Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 67.Yang Y, Jeanpierre C, Dressler G R, Lacoste M, Niaudet P, Gubler M C. WT1 and PAX-2 podocyte expression in Denys-Drash syndrome and isolated diffuse mesangial sclerosis. Am J Pathol. 1999;154:181–192. doi: 10.1016/S0002-9440(10)65264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]