Abstract
The actin cytoskeleton is important for maintaining mechanical homeostasis in adherent cells, largely through its regulation of adhesion and cortical tension. The LIM (Lin-11, Isl1, MEC-3) domain-containing proteins are involved in a myriad of cellular mechanosensitive pathways. Recent work has discovered that LIM domains bind to mechanically stressed actin filaments, suggesting a novel and widely conserved mechanism of mechanosensing. This review summarizes the current state of knowledge of LIM protein mechanosensitivity.
Keywords: actin cytoskeleton, focal adhesions, LIM proteins, adherens junctions
1 |. CELLS SENSE AND RESPOND TO MECHANICAL FORCES
Mechanical force plays an essential role in the control of cell shape and motion and serves as a key input in mechanotransduction pathways controlling cell survival, growth, and fate. Cells are subject to a myriad of external forces, including those from neighboring cells, fluid flow, or osmolarity. In addition to these, mechanoenzymes within the cell interior generate forces that are transmitted across cellular scales via the cytoskeleton. These internally generated forces enable cell shape change and are critical to cellular mechanosensing (e.g., environmental stiffness sensing; Trappmann & Chen, 2013). Cells sense and convert mechanical stimuli into chemical signals to initiate downstream signaling pathways (Wang, Tytell, & Ingber, 2009). Examples of force-sensitive chemistries of cytoplasmic proteins include force-dependent changes in binding affinity (e.g., integrins, actin binding proteins) or enzymatic activity (e.g., myosin II; Greenberg, Arpağ, Tüzel, & Ostap, 2016; Jégou & Romet-Lemonne, 2021). These molecular-scale transducers can then give rise to mechanical sensitivities of cytoskeletal arrays and/or regulate signaling and transcriptional pathways. While mechanotransduction pathways are well appreciated in cell physiology, we are just beginning to understand the diversity of force-sensing mechanisms within the cytoskeleton.
2 |. MECHANOSENSING IN ADHERENT CELLS
Cells are mechanically coupled to their local environment through adhesions to the extracellular matrix (ECM; e.g., focal adhesions, FAs) and surrounding cells (e.g., adherens junctions, AJs; Figure 1a). The actin cytoskeleton connects adhesions and transmits forces across the cell. Force sensitivity of adherent cells underlies adhesion regulation, cellular force generation, and mechanical properties of cells and tissues (Bieling et al., 2016; Courtemanche, Lee, Pollard, & Greene, 2013; Fletcher & Mullins, 2010; Moore, Roca-Cusachs, & Sheetz, 2010; Ohashi, Fujiwara, & Mizuno, 2017; Wang, Butler, & Ingber, 1993; Yusko & Asbury, 2014; Zhong et al., 1998). The mechanical properties of a cell’s environment are reflected by the actin cytoskeleton architecture. For example, F-actin networks in cells that are growing on rigid matrices, or within tissues that are being stretched, respond by self-organizing into thick bundles and larger FAs, which is thought to be important for generating and withstanding increased force (Smith et al., 2010; Yoshigi, Hoffman, Jensen, Yost, & Beckerle, 2005).
FIGURE 1.
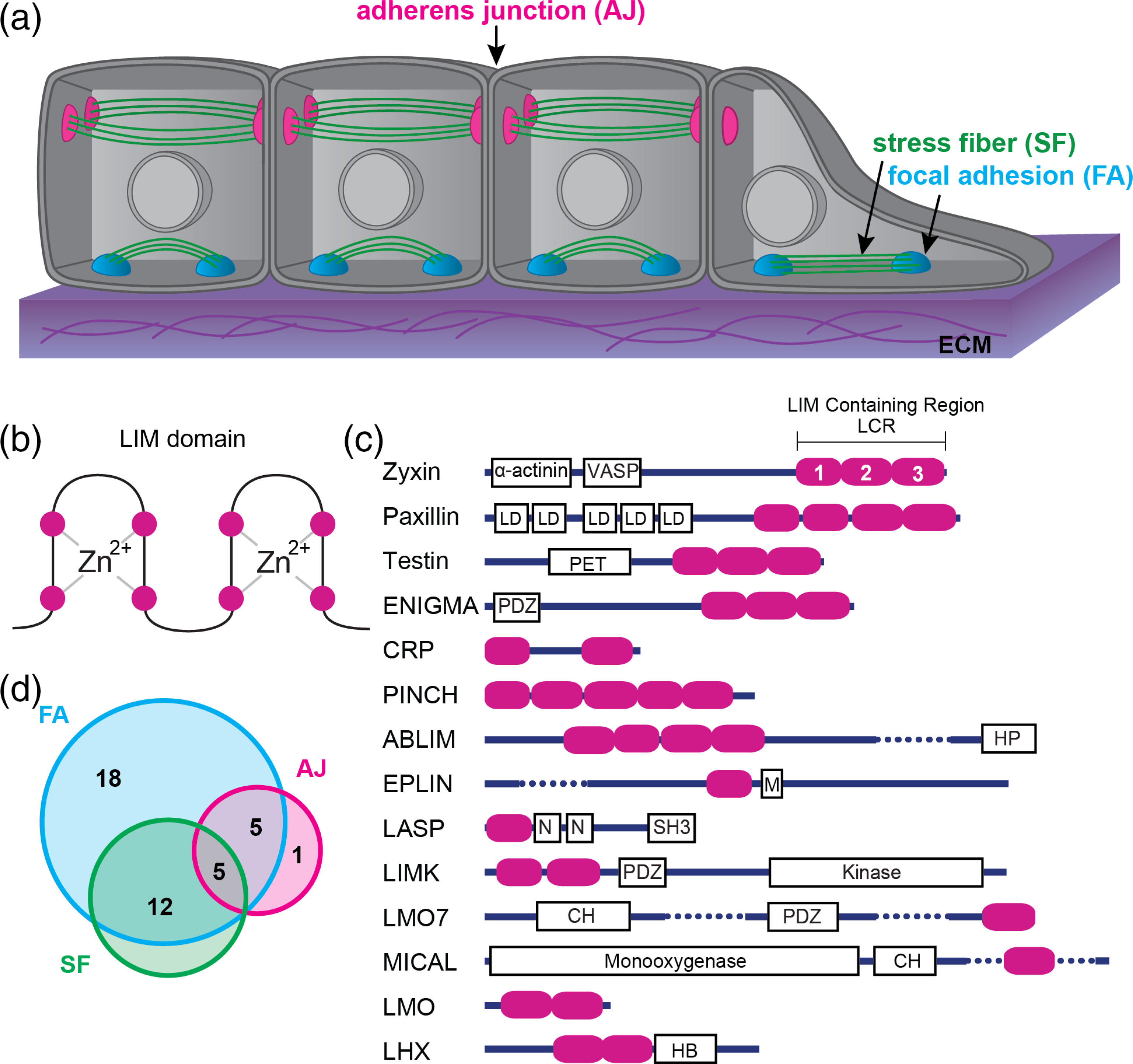
Mechanically stressed cells and LIM domain proteins. (a) Schematic of a layer of epithelial cells on top of an extracellular matrix (ECM). (b) Simple schematic of a LIM domain: Two zinc finger motifs. The magenta circles represent the well-conserved residues (typically cysteine or histidine) that chelate the zinc molecules. The remaining amino acid sequence varies between LIM domains. (c) Domain organization of the 14 classes of LIM domain proteins. Magenta ovals represent individual LIM domains. Dotted lines are used to abbreviate a few rather long structures. Other domain abbreviations: LD, Leucine rich aspartate domains; PET, prickle, espinas, testin; PDZ, membrane anchoring domain; HP, headpiece domain for F-actin binding; M, Myo5B interacting domain; N, nebulin; SH3, Src homology 3; CH, calponin homology; HB, homeobox. (d) Venn diagram showing the overlap of LIM domain proteins that associate with the three main networks: FA, Focal adhesions; AJ, adhesion junctions; SF, stress fibers
The actin cytoskeleton includes many different actin filament (F-actin)-based networks that vary in organization and composition. The architecture of FAs and AJs is comprised of stratified layers of distinct proteins that work together to transmit forces sensed by membrane-spanning adhesion receptors to actin filaments (Chen & Singer, 1982; Franz & Müller, 2005; Kanchanawong et al., 2010; Zaidel-Bar, Itzkovitz, Ma’ayan, Iyengar, & Geiger, 2007). Both FAs and AJs exhibit force-dependent changes to their composition and size, which is typically mediated by myosin-II activity within the actin cytoskeleton (Kuo, Han, Hsiao, Yates Iii, & Waterman, 2011) but can also be driven by external force (Riveline et al., 2001).
Stress fibers (SFs) are contractile bundles of 10–30 actin filaments of mixed polarity and alternating regions of the crosslinker α-actinin and nonmuscle myosin, reminiscent of the sarcomeric organization in striated myofibrils (Cramer, Siebert, & Mitchison, 1997; Hotulainen & Lappalainen, 2006; Tojkander, Gateva, & Lappalainen, 2012). While sarcomere architecture allows for recurring contraction and relaxation cycles, the less organized SF is built for continuous isometric contraction (Burridge, 1981; Pellegrin & Mellor, 2007). SF formation, growth, orientation, and maintenance are sensitive to both externally and internally generated forces (Chrzanowska-Wodnicka & Burr idge, 1996). The constant tension makes SFs susceptible to damage, and localized damaged regions form spontaneously or in response to the application of external forces (Smith et al., 2010). Thus, repair of such SF strain sites (SFSS) is important for maintaining the mechanical homeostasis of the actin cytoskeleton, allowing cells to maintain their integrity and adapt to force fluctuations. It is likely that the rearrangements of actin cytoskeleton networks in response to external force may also be driven by a similar force-induced remodeling. For instance, repeated cycles of uniaxial stretch results in both SF thickening and reorientation perpendicular to the stretch axis (Hayakawa, Sato, & Obinata, 2001; Kaunas, Nguyen, Usami, & Chien, 2005; Kim-Kaneyama et al., 2005; Yoshigi et al., 2005).
Recent progress has elucidated the force-dependent biochemistry of actin binding proteins (e.g., cadherins, vinculin, talin, alpha-catenin; Buckley et al., 2014; Huang, Bax, Buckley, Weis, & Dunn, 2017; Huveneers & de Rooij, 2013; Mei et al., 2020; Vigouroux, Henriot, & Le Clainche, 2020). These studies have primarily considered how forces applied to actin binding proteins (ABPs) alter their binding affinity to F-actin. However, the actin filament itself can twist, stretch, and compress, which may also alter the binding affinity of ABPs (Galkin, Orlova, & Egelman, 2012). In this scenario, the actin filament itself is the force responsive element and could confer mechanical information about the cell and its environment to various signaling and transcriptional pathways (Discher, Mooney, & Zandstra, 2009; Engler, Sen, Sweeney, & Discher, 2006).
3 |. LIM DOMAIN PROTEINS IN MECHANOTRANSDUCTION PATHWAYS
Proteomic screens of mechanotransduction pathways have revealed an abundance of proteins that contain one or more LIM (Lin-11, Isl1, MEC-3) domains (Freyd, Kim, & Horvitz, 1990; Karlsson, Thor, Norberg, Ohlsson, & Edlund, 1990; Way & Chalfie, 1988). The LIM domain is a ~ 60 amino acid sequence that forms a double zinc finger protein–protein or protein-DNA binding interface (Michelsen, Schmeichel, Beckerle, & Winge, 1993; Figure 1b). LIM domains occur in diverse multidomain protein organizations and are found in a wide range of eukaryotic proteins (LIM proteins), including ~70 human genes that can be divided into 14 classes (Figure 1c; Koch, Ryan, & Baxevanis, 2012). Early in the evolution of animal multicellularity, there was a large expansion in the number of LIM proteins as well as LIM “promiscuity”, that is, LIM has combined within multidomain proteins with many other domains of different structure and function (Basu, Carmel, Rogozin, & Koonin, 2008; Koch et al., 2012). This domain promiscuity has resulted in a functionally diverse LIM protein family whose members play roles in a variety of biological processes but especially those implicated in generating and responding to mechanical forces (Figure 1c; Table 1; Kadrmas & Beckerle, 2004; Smith et al., 2014).
TABLE 1.
A subset of LIM domain proteins and their corresponding mechanotransduction pathways
There are 41 LIM proteins found to be enriched at cell adhesions and/or the actomyosin cytoskeleton (Smith et al., 2014; Figure 1d). To date, 26 LIM proteins have been identified in FAs (including zyxin, paxillin, and LIMD1), and the localization of 21 of these is sensitive to myosin II activity (Kuo et al., 2011; Schiller, Friedel, Boulegue, & Fässler, 2011). Similarly, at least 11 LIM proteins display force-sensitive localization to AJs. Numerous LIM proteins co-localize to both FAs and SFs, FAs and AJs, or all three organelles (Figure 1d). Some LIM proteins contain known actin binding domains (e.g., the [CH] domain) that could drive their localization to F-actin networks. However, many that localize to the actin cytoskeleton lack these. Standard biochemical approaches have not detected binding of LIM domains to actin filaments. One notable exception is the CRP class, which canonically binds and bundles actin filaments via their LIM domains (Grubinger & Gimona, 2004; Hoffmann et al., 2014; Thomas et al., 2006). CRP is an ancient class as it is the only mammalian LIM protein class also found in plants, suggesting the possibility that canonical actin binding could be an ancestral function of the LIM domain. For instance, Muscle LIM protein (MLP) is a CRP class protein that has been implicated in mechanoresponse to muscle sarcomere stretching (Vafiadaki, Arvanitis, & Sanoudou, 2015).
Several studies have implicated LIM proteins in cell signaling and gene expression mechanotransduction pathways (Ibar et al., 2018; Martin et al., 2002). For instance, four-and-a-half LIM domains 2 (FHL2) is implicated in mechanical regulation of the cell cycle. On a soft matrix, FHL2 dissociates from F-actin networks and becomes more concentrated in the nucleus where it acts as a transcriptional cofactor to increase p21 gene expression, which regulates cell cycle progression and inhibits growth (Nakazawa et al., 2016). Most force-sensitive LIM proteins display nuclear shuttling raising questions as to whether detection of forces via LIM proteins is connected to localization and function inside the nucleus (Figure 2). Similarly, several LIM proteins in the Ajuba/Zyxin classes exhibit force-dependent binding to AJs to regulate hippo and Yap/Taz signaling pathways (Rauskolb, Pan, Reddy, Oh, & Irvine, 2011; Rauskolb, Sun, Sun, Pan, & Irvine, 2014).
FIGURE 2.
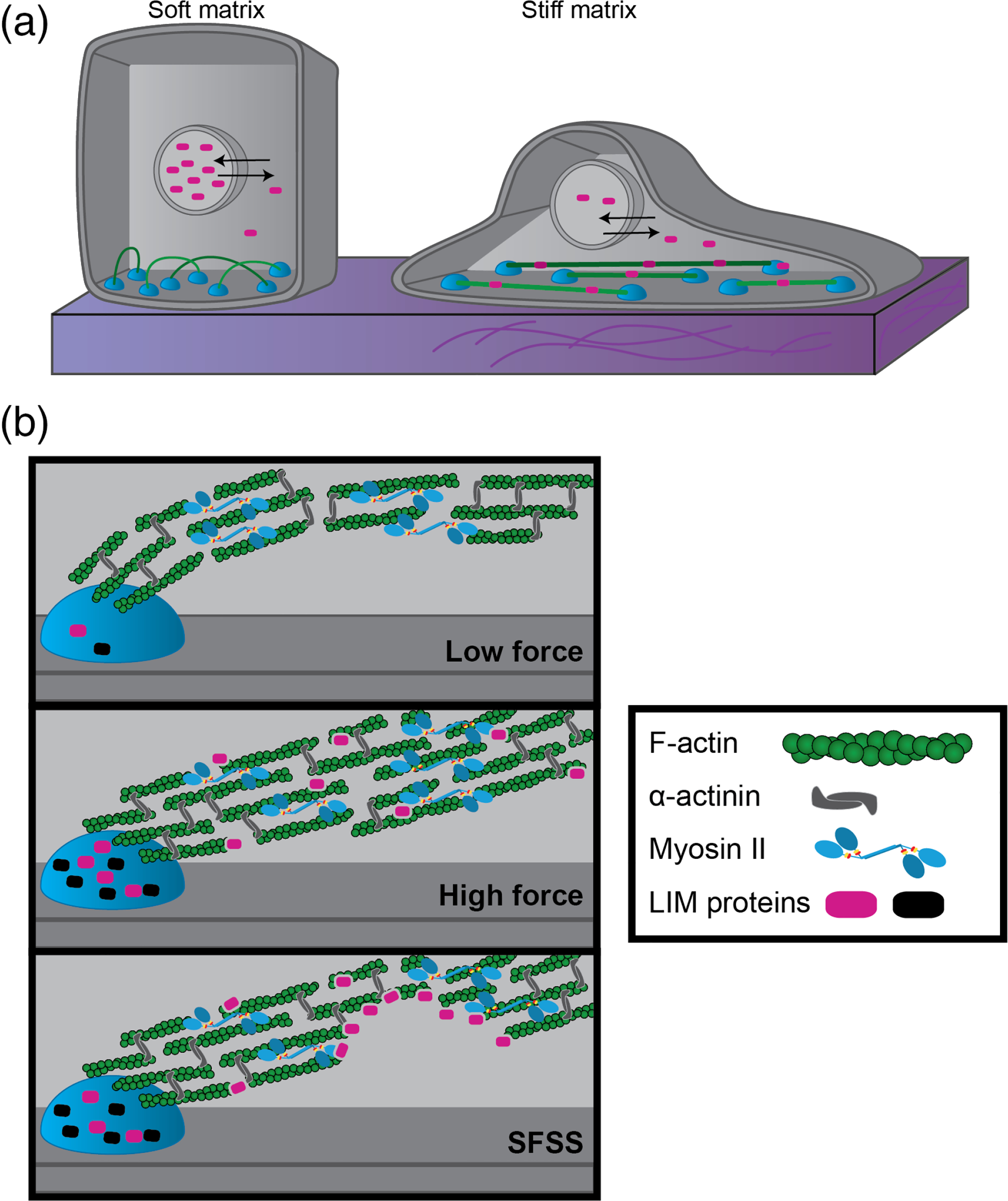
Schematic of LIM domain protein localization in cells. (a) Nuclear shuttling of LIM domain proteins (magenta ovals) occurs when cells spread out on stiff matrices. (b) LIM domain proteins (black and magenta ovals) localize to FAs and SFs under high tension. A subset of LIM domain proteins localizes to stress fiber strain sites (SFSS)
4 |. FORCE-SENSITIVE LOCALIZATION OF LIM PROTEINS IN ADHERENT CELLS
The LIM domain-containing region (LCR) has been found to drive the subcellular localization for a large number of LIM proteins (Brown et al., 1996; Hoffman et al., 2012; Smith et al., 2013). This has been dissected most carefully for the LIM protein zyxin, which localizes to SFs, FAs, and AJs in a force-dependent manner. Zyxin is necessary for stretch-mediated SF remodeling, SFSS repair, and FA maturation (Hoffman et al., 2012; Smith et al., 2013, 2014; Yoshigi et al., 2005).
The LCR of zyxin resides at the C-terminus and contains three LIM domains in tandem separated by short unstructured linkers. The LCR is required for zyxin recruitment to SFSS and FAs. For full length zyxin, any one of the individual LIM domains are not sufficient for its localization (Uemura et al., 2011). Recent results demonstrate that at least two tandem repeats of LIM1 or LIM3 are sufficient for LCR localization to SFSS (Winkelman, Anderson, Suarez, Kovar, & Gardel, 2020), but further work is needed to demonstrate this sufficiency for the full-length protein. Once localized, zyxin’s N-terminal functionality mediates SFSS repair by recruiting factors that promote actin filament polymerization (Ena/VASP) and crosslinking (α-actinin; Smith et al., 2014). Therefore, the LCR regulates force-sensitive recruitment, while the functional role is dependent on the additional domains (Smith et al., 2010).
5 |. LIM DOMAINS FROM DIVERSE PROTEINS BIND STRESSED ACTIN FILAMENTS
Recent research has made progress in understanding the mechanism of LIM protein force-sensitive localization to the actin cytoskeleton. Two studies used complementary experimental approaches to screen LIM proteins for force-sensitivity in cells. One employed cell stretching experiments to systematically quantify the enrichment of full length and LCR constructs of LIM proteins on stretched SFs (Sun et al., 2020), while the other quantified LCR recruitment to SFSS (Winkelman et al., 2020). Together, these studies identified force-sensitive LCRs in 18 LIM proteins from Zyxin, Paxillin, Tes, and Enigma classes from both animals and yeasts (Sun et al., 2020; Winkelman et al., 2020). These complementary experimental approaches revealed that cytoskeletal strain sensing via the LIM domains is widespread in cells and existed in the last common ancestor of yeasts and animals.
To isolate the force-sensitive substrate of LIM, both groups used in vitro approaches to reconstitute force-sensitive recruitment with a minimal set of purified components (Sun et al., 2020; Winkelman et al., 2020). Two types of in vitro reconstitution assays were utilized to test the stress sensitivity of a subset of LIM proteins, and both showed localized recruitment of LIM domains directly to mechanically stressed regions of F-actin. Sun et al. applied tensile stresses to actin filaments with a modified gliding filament assay. Single filaments were pulled in opposite directions via surface-attached myosins with barbed (myosin V) and pointed (myosin VI) end directionality. LIM proteins localize to actin filaments only after initiation of myosin activity facilitates tensed filaments. Actin filament breakage, coinciding with stress relief, results in LIM protein dissociation. Similarly, Winkelman et al. reconstituted contractile actin networks comprised of F-actin, α-actinin, and myosin II. After addition of myosin II to initiate contraction, LCRs localize to stressed regions of the network due to contractile forces, particularly to bundle sites just prior to their rupture, after which the LCR dissociates from the actin filaments.
To understand the mechanism by which LIM domains bind F-actin, these studies identified particular amino acids and LIM domain architectures that are necessary for binding. With the exception of eight well-conserved residues (cysteine and histidine) responsible for Zn2+ chelation, the sequence of LIM domains is highly variable. However, a phenylalanine resides at a similar position in all strain sensing LIM domains and was found to be necessary for force sensitivity (Sun et al., 2020). Additionally, force-sensitive LCR all have three or more LIM domains in tandem, each separated by a short linker. Alterations to this organization in the LIM protein zyxin revealed that multiple LIM domains, when organized in tandem and connected by short linkers (serial), but not when oligomerized (parallel), contribute additively to stressed F-actin binding (Sun et al., 2020; Winkelman et al., 2020). Together, these data lead to a hypothesis that multiple LIM domains that are appropriately positioned interact via a hydrophobic interaction with a strained actin filament (Figure 3).
FIGURE 3.
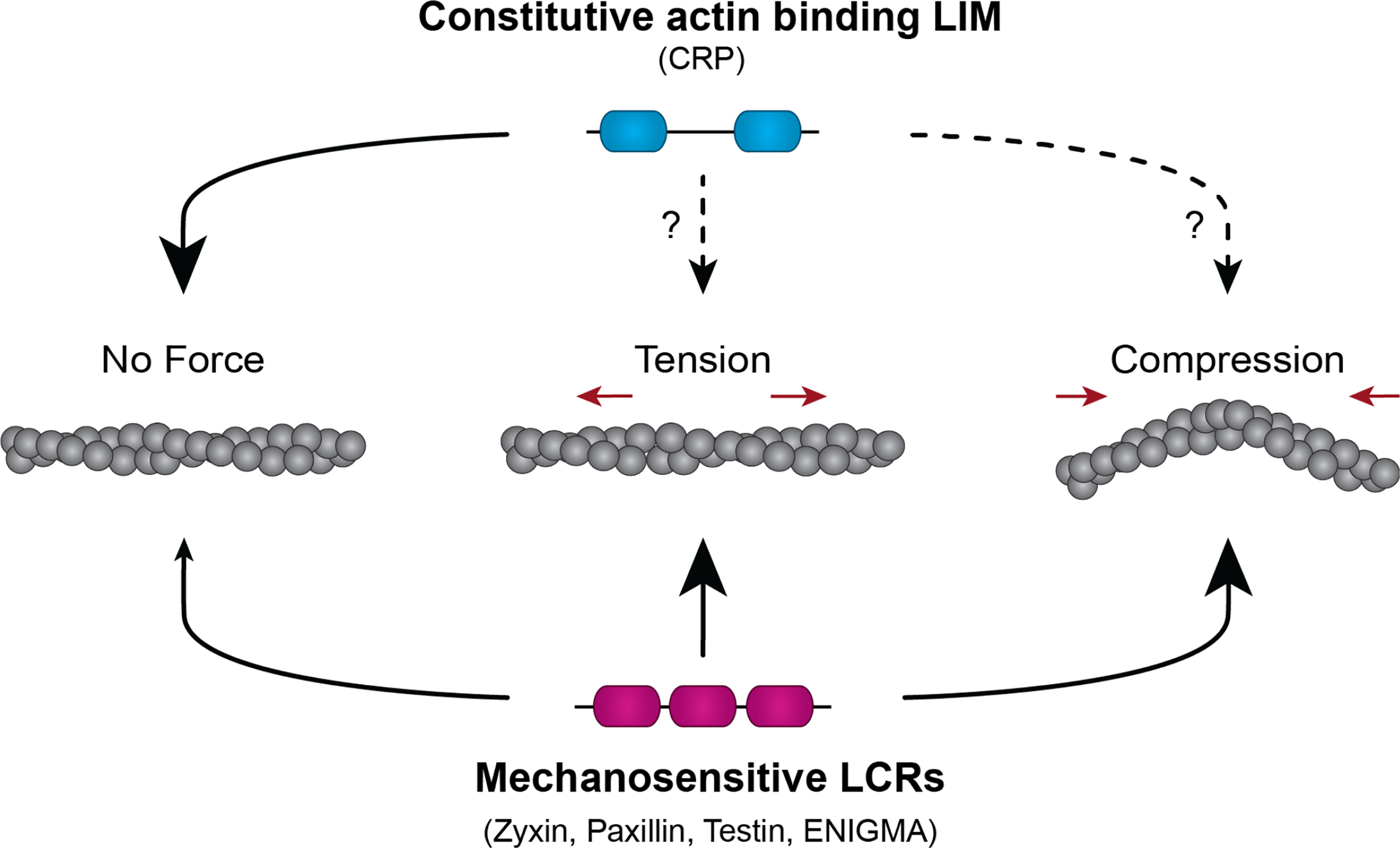
Schematic of mechanosensitive LCR localization to stressed actin filaments. The constitutive actin binding CRP class LIM proteins bind actin filaments in the absence or presence of force. The dashed lines indicate that CRP localization is suspected to occur for stressed actin filaments but has not been fully investigated. Mechanosensitive LIM domain protein LCR constructs bind with high affinity to actin filaments under tension or compression but with low affinity to relaxed filaments (adapted from Winkelman et al., 2020)
6 |. EVOLUTIONARILY CONSERVED MECHANISM OF LIM DOMAIN-BASED FORCE SENSING
Interestingly, despite the lack of sequence conservation, binding to stressed actin filaments appears to be an ancient and conserved function of the LIM domain. Strain sensing LIM domains may have a conserved tertiary structure despite primary sequence variability, similar to other well studied protein folds (Dominguez, 2010). For instance, the LCR of the fission yeast paxillin 1 (Pxl1) binds to both SFSS in mammalian cells and purified mammalian stressed F-actin (Winkelman et al., 2020). Fission yeast do not have stress fibers (or SFSS), but there is a phenomenon analogous to SFSS that occurs within the yeast cell. Pxl1 localizes to the cytokinetic contractile ring (CR), and its deletion results in fragmentation of the ring during contraction (Ge & Balasubramanian, 2008). The rupture of the contractile ring in Pxl1 mutants is reminiscent of increase rupturing of stress fibers observed in zyxin null cells (Smith et al., 2010). Indeed, there are many interesting parallels between CRs and SFs. Both are composed of similar molecular components and are arranged in an architecturally similar way: antiparallel bundled actin filaments crosslinked by α-actinin and pulled on by myosin II. Both may also display a rough sarcomeric pattern where α-actinin and myosin form complementary domains (Tojkander et al., 2012). The contractility of these networks must be regulated so that they remain tense but do not rip themselves apart. While SFs remain roughly the same length, the CR must shorten during constriction to pinch the mother cell into two daughters. The organization of the CR, SF, and muscle sarcomere may be a coincidence or belie a common origin. Since we first see clear versions of myosin II, α-actinin, and strain-sensing LIM proteins in the unikont branch of eukaryotes, the ancestral version of these contractile networks may have emerged near this branch.
Once contractile machinery arose in evolution, the cell must have evolved regulatory mechanisms for their maintenance and repair. The strain sensing LIM domain may represent one way in which cells learned to detect stressed F-actin. Other domains may be added to this LIM containing protein to tailor responses to LIM-detected stress, for example, some LIM proteins contain domains that bind actin assembly factors that enable these proteins to recruit actin assembly factors to sites of mechanical stress that has been detected by LIM (Hoffman et al., 2012; Smith et al., 2010). One hypothesis for the development of strain sensitive LIM domains is that general actin binding by LIM was tinkered with by evolution to tune it to bind strained actin filaments. The most ancient and widespread LIM proteins are in the CRP family and have been shown in multiple studies to bind unstressed actin filaments (Grubinger & Gimona, 2004; Weiskirchen & Günther, 2003), suggesting the possibility that generic actin binding may be an ancestral function of LIM domains that was tuned to bind strained F-actin (Figure 4).
FIGURE 4.
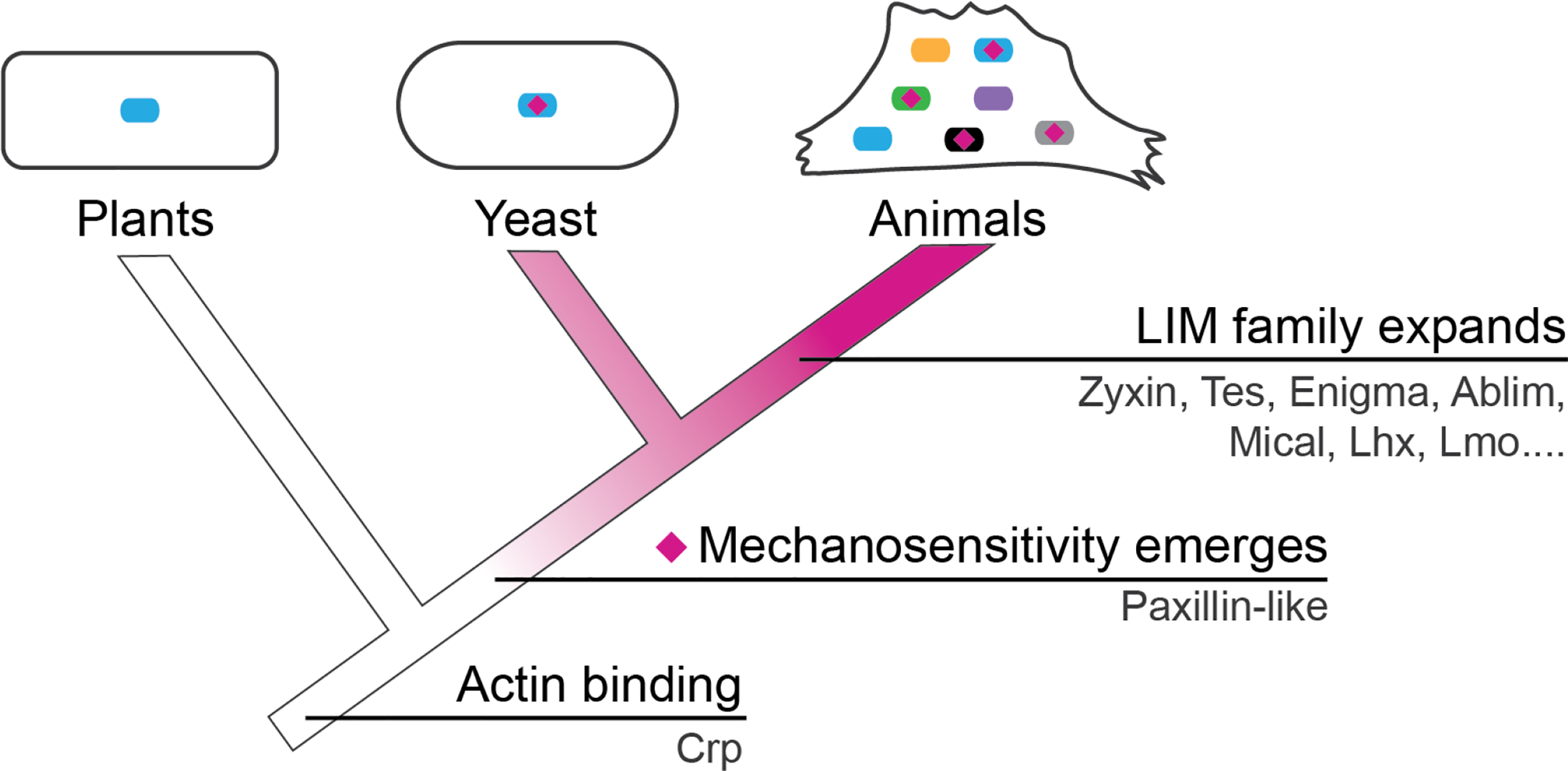
Evolution of LIM domain proteins. LIM domains have evolved over time to become mechanosensitive. The family then expanded to include a diverse population of proteins in mammals (adapted from Winkelman et al., 2020)
7 |. THE ACTIN FILAMENT IS A SUBSTRATE FOR FORCE-SENSITIVE BINDING
The load dependent mechanical response of F-actin networks is likely to arise from force-sensitive biochemistry of ABPs. A recent review summarizes evidence for force-sensitivity for several ABPs (e.g., Arp2/3 complex, cofilin, alpha-catenin; Jégou & Romet-Lemonne, 2021). Filament curvature promotes the binding of Arp2/3 complex binding to F-actin, while tension decreases the stability of an Arp2/3 complex-mediated daughter branch (Pandit et al., 2020; Risca et al., 2012). There are conflicting reports of how tension may impact the binding of F-actin depolymerizing factor cofilin (Hayakawa, Tatsumi, & Sokabe, 2011; Wioland, Jegou, & Romet-Lemonne, 2019), while additional research suggests torsion may impact cofilin’s F-actin severing rate (Mizuno, Tanaka, Yamashiro, Narita, & Watanabe, 2018; Wioland et al., 2019). Low tension applied directly to an actin filament increases the binding of alpha-catenin to adjacent actin subunits, and the force detection is attributed to a 35 amino acid region at the C-terminus (Mei et al., 2020). We hypothesize that similar sensing may occur in LIM protein, but will require further investigations.
As a common component in these mechanosensitive networks, it is likely that the actin filament itself is a force sensor whereby the force-induced conformation of actin filaments affects the binding interactions of the ABPs. There are many studies and hypotheses about how mechanical forces may alter filament conformation, but there is no explicit structural data comparing stressed and unstressed actin filaments (Galkin et al., 2012). Modeling has shown that due to the twist of an actin filament, strain is not distributed homogenously throughout the filament, and localized regions of strain may result (Schramm, Hocky, Voth, Martiel, & De La Cruz, 2019). Therefore, the filament level force can impact the conformation of and interactions between adjacent subunits. These subunit level alterations could possibly reveal additional binding sites for ABPs. We hypothesize that LCRs recognize a binding site along an actin filament that is revealed under tensile or compressive stress (Winkelman et al., 2020). Additional research will be required to fully understand the binding interface of LCRs and mechanically stressed actin filaments. LIM domain proteins, and even isolated strain sensing LCRs, display overlapping but non-identical localization to stressed actin networks, raising the question of how specificity for particular networks arise. Additionally, stressed actin binding is distributed across several protein families involved in diverse cellular processes. Lastly, an important remaining question that will require extensive investigation is how binding by LIM to stressed actin filaments might regulate these diverse cellular processes.
ACKNOWLEDGMENTS
Work on actin cytoskeleton mechanotransduction in the Gardel and Kovar labs is funded by NIH R01 GM104032 (Margaret L. Gardel), NIH R01 GM079265 (David R. Kovar), and Army Research Office Multi University Research Initiative (ARO MURI) W911NF1410403 (Margaret L. Gardel and David R. Kovar). We also acknowledge funding from NIH F32 GM122372 (Jonathan D. Winkelman) and NIH MCB Training Grant T32 GM0071832 (Caitlin A. Anderson), and this study was partially supported by the University of Chicago Materials Research Science and Engineering Center, funded by the National Science Foundation under award numbers DMR-1420709 and DMR-2011854.
Funding information
National Institute of General Medical Sciences, Grant/Award Number: RO1 GM104032; National Science Foundation, Grant/Award Numbers: DMR-2011854, DMR-1420709; National Institutes of Health MCB Training, Grant/Award Number: T32 GM0071832; Army Research Office, Grant/Award Number: W911NF1410403; National Institutes of Health, Grant/Award Numbers: F32 GM122372, R01 GM079265
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Basu MK, Carmel L, Rogozin IB, & Koonin EV (2008). Evolution of protein domain promiscuity in eukaryotes. Genome Research, 18(3), 449–461. 10.1101/gr.6943508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Li T-D, Weichsel J, McGorty R, Jreij P, Huang B, … Mullins RD (2016). Force feedback controls motor activity and mechanical properties of self-assembling branched Actin networks. Cell, 164(1–2), 115–127. 10.1016/j.cell.2015.11.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Perrotta JA, & Turner CE (1996). Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. The Journal of Cell Biology, 135(4), 1109–1123. 10.1083/jcb.135.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, … Dunn AR (2014). The minimal cadherin-catenin complex binds to actin filaments under force. Science (New York, N.Y.), 346 (6209), 1254211. 10.1126/science.1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K (1981). Are stress fibres contractile? Nature, 294(5843), 691–692. 10.1038/294691a0 [DOI] [PubMed] [Google Scholar]
- Chen WT, & Singer SJ (1982). Immunoelectron microscopic studies of the sites of cell-substratum and cell-cell contacts in cultured fibroblasts. The Journal of Cell Biology, 95(1), 205–222. 10.1083/jcb.95.1.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Chen T, Tor M, Park D, Zhou Q, Huang JB, … Zhou G (2016). A high-throughput screening platform targeting PDLIM5 for pulmonary hypertension. Journal of Biomolecular Screening, 21(4), 333–341. 10.1177/1087057115625924 [DOI] [PubMed] [Google Scholar]
- Cheng H, Kimura K, Peter AK, Cui L, Ouyang K, Shen T, … Chen J (2010). Loss of enigma homolog protein results in dilated cardiomyopathy. Circulation Research, 107(3), 348–356. 10.1161/CIRCRESAHA.110.218735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M, & Burridge K (1996). Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. The Journal of Cell Biology, 133(6), 1403–1415. 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Lee JY, Pollard TD, & Greene EC (2013). Tension modulates actin filament polymerization mediated by formin and profilin. Proceedings of the National Academy of Sciences of the United States of America, 110(24), 9752–9757. 10.1073/pnas.1308257110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer LP, Siebert M, & Mitchison TJ (1997). Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: Implications for the generation of motile force. The Journal of Cell Biology, 136(6), 1287–1305. 10.1083/jcb.136.6.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, & Longmore GD (2010). Ajuba LIM proteins are negative regulators of the hippo signaling pathway. Current Biology, 20(7), 657–662. 10.1016/j.cub.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin NO, & Turner CE (2008). Paxillin comes of age. Journal of Cell Science, 121(15), 2435–2444. 10.1242/jcs.018044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher DE, Mooney DJ, & Zandstra PW (2009). Growth factors, matrices, and forces combine and control stem cells. Science (New York, N.Y.), 324(5935), 1673–1677. 10.1126/science.1171643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R (2010). The WASP-homology 2 domain and cytoskeleton assembly. In Carlier M-F (Ed.), Actin-based motility: Cellular, molecular and physical aspects (pp. 255–277). Netherlands: Springer. 10.1007/978-90-481-9301-1_11 [DOI] [Google Scholar]
- Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, & Golsteyn RM (2000). Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. The Journal of Biological Chemistry, 275(29), 22503–22511. 10.1074/jbc.M001698200 [DOI] [PubMed] [Google Scholar]
- Dutta S, Mana-Capelli S, Paramasivam M, Dasgupta I, Cirka H, Billiar K, & McCollum D (2018). TRIP6 inhibits Hippo signaling in response to tension at adherens junctions. EMBO Reports, 19(2), 337–350. 10.15252/embr.201744777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A, Schiefermeier N, Grigoriev I, Ohi R, Brown MC, Turner CE, … Kaverina I (2008). Paxillin-dependent stimulation of microtubule catastrophes at focal adhesion sites. Journal of Cell Science, 121(3), 405–405. 10.1242/jcs.03497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwy A, Vincent-Mistiaen ZI, Spencer-Dene B, Stone RK, Boeing S, Wculek SK, … Thompson BJ (2016). Integrin signalling regulates YAP and TAZ to control skin homeostasis. Development, 143 (10), 1674–1687. 10.1242/dev.133728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, & Discher DE (2006). Matrix elasticity directs stem cell lineage specification. Cell, 126(4), 677–689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Fletcher DA, & Mullins RD (2010). Cell mechanics and the cytoskeleton. Nature, 463(7280), 485–492. 10.1038/nature08908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz CM, & Müller DJ (2005). Analyzing focal adhesion structure by atomic force microscopy. Journal of Cell Science, 118(22), 5315–5323. 10.1242/jcs.02653 [DOI] [PubMed] [Google Scholar]
- Freyd G, Kim SK, & Horvitz HR (1990). Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature, 344(6269), 876–879. 10.1038/344876a0 [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, & Egelman EH (2012). Actin filaments as tension sensors. Current Biology, 22(3), R96–R101. 10.1016/j.cub.2011.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, & Balasubramanian MK (2008). Pxl1p, a paxillin-related protein, stabilizes the actomyosin ring during cytokinesis in fission yeast. Molecular Biology of the Cell, 19(4), 1680–1692. 10.1091/mbc.e07-07-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MJ, Arpağ G, Tüzel E, & Ostap EM (2016). A perspective on the role of myosins as mechanosensors. Biophysical Journal, 110 (12), 2568–2576. 10.1016/j.bpj.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubinger M, & Gimona M (2004). CRP2 is an autonomous actin-binding protein. FEBS Letters, 557(1–3), 88–92. 10.1016/s0014-5793(03)01451-0 [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Sato N, & Obinata T (2001). Dynamic reorientation of cultured cells and stress fibers under mechanical stress from periodic stretching. Experimental Cell Research, 268(1), 104–114. 10.1006/excr.2001.5270 [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Tatsumi H, & Sokabe M (2011). Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. The Journal of Cell Biology, 195(5), 721–727. 10.1083/jcb.201102039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Chaturvedi A, Yoshigi M, & Beckerle MC (2012). Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Molecular Biology of the Cell, 23(10), 1846–1859. 10.1091/mbc.E11-12-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Moreau F, Moes M, Luthold C, Dieterle M, Goretti E, … Thomas C (2014). Human muscle LIM protein dimerizes along the actin cytoskeleton and cross-links actin filaments. Molecular and Cellular Biology, 34(16), 3053–3065. 10.1128/MCB.00651-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, & Lappalainen P (2006). Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. The Journal of Cell Biology, 173(3), 383–394. 10.1083/jcb.200511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Peng H, White DE, Negorev DG, Maul GG, Feng Y, … Rauscher FJ (2010). LIM protein Ajuba functions as a nuclear receptor corepressor and negatively regulates retinoic acid signaling. Proceedings of the National Academy of Sciences of the United States of America, 107(7), 2938–2943. 10.1073/pnas.0908656107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DL, Bax NA, Buckley CD, Weis WI, & Dunn AR (2017). Vinculin forms a directionally asymmetric catch bond with F-actin. Science (New York, N.Y.), 357(6352), 703–706. 10.1126/science.aan2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins CJ, & Andrulis IL (2008). Cell cycle regulated phosphorylation of LIMD1 in cell lines and expression in human breast cancers. Cancer Letters, 267(1), 55–66. 10.1016/j.canlet.2008.03.015 [DOI] [PubMed] [Google Scholar]
- Huveneers S, & de Rooij J (2013). Mechanosensitive systems at the cadherin-F-actin interface. Journal of Cell Science, 126(2), 403–413. 10.1242/jcs.109447 [DOI] [PubMed] [Google Scholar]
- Ibar C, Kirichenko E, Keepers B, Enners E, Fleisch K, & Irvine KD (2018). Tension-dependent regulation of mammalian Hippo signaling through LIMD1. Journal of Cell Science, 131(5). 10.1242/jcs.214700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégou A, & Romet-Lemonne G (2021). Mechanically tuning actin filaments to modulate the action of actin-binding proteins. Current Opinion in Cell Biology, 68, 72–80. 10.1016/j.ceb.2020.09.002 [DOI] [PubMed] [Google Scholar]
- Johannessen M, Møller S, Hansen T, Moens U, & Van Ghelue M (2006). The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cellular and Molecular Life Sciences: CMLS, 63(3), 268–284. 10.1007/s00018-005-5438-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas JL, & Beckerle MC (2004). The LIM domain: From the cytoskeleton to the nucleus. Nature Reviews Molecular Cell Biology, 5(11), 920–931. 10.1038/nrm1499 [DOI] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, & Waterman CM (2010). Nanoscale architecture of integrin-based cell adhesions. Nature, 468(7323), 580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson O, Thor S, Norberg T, Ohlsson H, & Edlund T (1990). Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo-and a Cys–His domain. Nature, 344 (6269), 879–882. 10.1038/344879a0 [DOI] [PubMed] [Google Scholar]
- Kaunas R, Nguyen P, Usami S, & Chien S (2005). Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proceedings of the National Academy of Sciences of the United States of America, 102(44), 15895–15900. 10.1073/pnas.0506041102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Kaneyama J, Suzuki W, Ichikawa K, Ohki T, Kohno Y, Sata M, … Shibanuma M (2005). Uni-axial stretching regulates intracellular localization of Hic-5 expressed in smooth-muscle cells in vivo. Journal of Cell Science, 118(Pt 5), 937–949. 10.1242/jcs.01683 [DOI] [PubMed] [Google Scholar]
- Koch BJ, Ryan JF, & Baxevanis AD (2012). The diversification of the LIM superclass at the base of the Metazoa increased subcellular complexity and promoted multicellular specialization. PLoS One, 7(3), e33261. 10.1371/journal.pone.0033261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J-C (2013). Mechanotransduction at focal adhesions: Integrating cytoskeletal mechanics in migrating cells. Journal of Cellular and Molecular Medicine, 17(6), 704–712. 10.1111/jcmm.12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J-C, Han X, Hsiao C-T, Yates Iii JR, & Waterman CM (2011). Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nature Cell Biology, 13(4), 383–393. 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, & Trueb B (2001). Analysis of the alpha-actinin/zyxin interaction. The Journal of Biological Chemistry, 276(36), 33328–33335. 10.1074/jbc.M100789200 [DOI] [PubMed] [Google Scholar]
- Lim BC, Matsumoto S, Yamamoto H, Mizuno H, Kikuta J, Ishii M, & Kikuchi A (2016). Prickle1 promotes focal adhesion disassembly in cooperation with the CLASP-LL5β complex in migrating cells. Journal of Cell Science, 129(16), 3115–3129. 10.1242/jcs.185439 [DOI] [PubMed] [Google Scholar]
- López-Colomé AM, Lee-Rivera I, Benavides-Hidalgo R, & López E (2017). Paxillin: A crossroad in pathological cell migration. Journal of Hematology & Oncology, 10(1), 50. 10.1186/s13045-017-0418-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magno AL, Ingley E, Brown SJ, Conigrave AD, Ratajczak T, & Ward BK (2011). Testin, a novel binding partner of the calcium-sensing receptor, enhances receptor-mediated Rho-kinase signalling. Biochemical and Biophysical Research Communications, 412(4), 584–589. 10.1016/j.bbrc.2011.07.132 [DOI] [PubMed] [Google Scholar]
- Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, … Braga VMM (2003). The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. The Journal of Biological Chemistry, 278(2), 1220–1228. 10.1074/jbc.M205391200 [DOI] [PubMed] [Google Scholar]
- Martin B, Schneider R, Janetzky S, Waibler Z, Pandur P, Kühl M, … Wixler V (2002). The LIM-only protein FHL2 interacts with β-catenin and promotes differentiation of mouse myoblasts. The Journal of Cell Biology, 159(1), 113–122. 10.1083/jcb.200202075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Espinosa de los Reyes S, Reynolds MJ, Leicher R, Liu S, & Alushin GM (2020). Molecular mechanism for direct actin force-sensing by α-catenin. eLife, 9, e62514. 10.7554/eLife.62514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen JW, Schmeichel KL, Beckerle MC, & Winge DR (1993). The LIM motif defines a specific zinc-binding protein domain. Proceedings of the National Academy of Sciences of the United States of America, 90(10), 4404–4408. 10.1073/pnas.90.10.4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Tanaka K, Yamashiro S, Narita A, & Watanabe N (2018). Helical rotation of the diaphanous-related formin mDia1 generates actin filaments resistant to cofilin. Proceedings of the National Academy of Sciences of the United States of America, 115(22), E5000–E5007. 10.1073/pnas.1803415115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SW, Roca-Cusachs P, & Sheetz MP (2010). Stretchy proteins on stretchy substrates: The important elements of integrin-mediated rigidity sensing. Developmental Cell, 19(2), 194–206. 10.1016/j.devcel.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N, Sathe AR, Shivashankar GV, & Sheetz MP (2016). Matrix mechanics controls FHL2 movement to the nucleus to activate p21 expression. Proceedings of the National Academy of Sciences of the United States of America, 113(44), E6813–E6822. 10.1073/pnas.1608210113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Fujiwara S, & Mizuno K (2017). Roles of the cytoskeleton, cell adhesion and rho signalling in mechanosensing and mechanotransduction. Journal of Biochemistry, 161(3), 245–254. 10.1093/jb/mvw082 [DOI] [PubMed] [Google Scholar]
- Pandit NG, Cao W, Bibeau J, Johnson-Chavarria EM, Taylor EW, Pollard TD, & Cruz EMDL (2020). Force and phosphate release from Arp2/3 complex promote dissociation of actin filament branches. Proceedings of the National Academy of Sciences of the United States of America, 117, 13519–13528. 10.1073/pnas.1911183117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S, & Mellor H (2007). Actin stress fibres. Journal of Cell Science, 120(20), 3491–3499. 10.1242/jcs.018473 [DOI] [PubMed] [Google Scholar]
- Pratt SJ, Epple H, Ward M, Feng Y, Braga VM, & Longmore GD (2005). The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. Journal of Cell Biology, 168(5), 813–824. 10.1083/jcb.200406083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Pan G, Reddy BVVG, Oh H, & Irvine KD (2011). Zyxin links fat signaling to the Hippo pathway. PLoS Biology, 9(6), e1000624. 10.1371/journal.pbio.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, & Irvine KD (2014). Cytoskeletal tension inhibits Hippo signaling through an Ajuba–Warts complex. Cell, 158(1), 143–156. 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzell W, Bustillo ME, & Zallen JA (2018). The force-sensitive protein Ajuba regulates cell adhesion during epithelial morphogenesis. Journal of Cell Biology, 217(10), 3715–3730. 10.1083/jcb.201801171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Zumbrunn J, Jaquemar D, Kuhn M, Walter U, & Trueb B (1999). An α-Actinin binding site of Zyxin is essential for subcellular Zyxin localization and α-Actinin recruitment*. Journal of Biological Chemistry, 274(19), 13410–13418. 10.1074/jbc.274.19.13410 [DOI] [PubMed] [Google Scholar]
- Risca VI, Wang EB, Chaudhuri O, Chia JJ, Geissler PL, & Fletcher DA (2012). Actin filament curvature biases branching direction. Proceedings of the National Academy of Sciences of the United States of America, 109(8), 2913–2918. 10.1073/pnas.1114292109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, … Bershadsky AD (2001). Focal contacts as mechanosensors: Externally applied local mechanical force induces growth of focal contacts by an Mdia1-dependent and rock-independent mechanism. Journal of Cell Biology, 153(6), 1175–1186. 10.1083/jcb.153.6.1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller HB, Friedel CC, Boulegue C, & Fässler R (2011). Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Reports, 12(3), 259–266. 10.1038/embor.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm AC, Hocky GM, Voth GA, Martiel J-L, & De La Cruz EM (2019). Plastic deformation and fragmentation of strained actin filaments. Biophysical Journal, 117(3), 453–463. 10.1016/j.bpj.2019.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Blankman E, Gardel ML, Luettjohann L, Waterman CM, & Beckerle MC (2010). A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Developmental Cell, 19(3), 365–376. 10.1016/j.devcel.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Blankman E, Deakin NO, Hoffman LM, Jensen CC, Turner CE, & Beckerle MC (2013). LIM domains target actin regulators paxillin and zyxin to sites of stress fiber strain. PLoS One, 8(8), e69378. 10.1371/journal.pone.0069378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Hoffman LM, & Beckerle MC (2014). LIM proteins in actin cytoskeleton mechanoresponse. Trends in Cell Biology, 24(10), 575–583. 10.1016/j.tcb.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, & Irvine KD (2013). Ajuba family proteins link JNK to Hippo signaling. Science Signaling, 6(292), ra81–ra81. 10.1126/scisignal.2004324,6,ra81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Phua DYZ, Axiotakis L, Smith MA, Blankman E, Gong R, … Alushin GM (2020). Mechanosensing through direct binding of tensed F-actin by LIM domains. Developmental Cell, 55(4), 468–482.e7. 10.1016/j.devcel.2020.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweede M, Ankem G, Chutvirasakul B, Azurmendi HF, Chbeir S, Watkins J, … Capelluto DGS (2008). Structural and membrane binding properties of the prickle PET domain. Biochemistry, 47(51), 13524–13536. 10.1021/bi801037h [DOI] [PubMed] [Google Scholar]
- Thomas C, Hoffmann C, Dieterle M, Van Troys M, Ampe C, & Steinmetz A (2006). Tobacco WLIM1 is a novel F-Actin binding protein involved in Actin cytoskeleton remodeling. The Plant Cell, 18(9), 2194–2206. 10.1105/tpc.106.040956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, & Lappalainen P (2012). Actin stress fibers: Assembly, dynamics and biological roles. Journal of Cell Science, 125(Pt 8), 1855–1864. 10.1242/jcs.098087 [DOI] [PubMed] [Google Scholar]
- Trappmann B, & Chen CS (2013). How cells sense extracellular matrix stiffness: A material’s perspective. Current Opinion in Biotechnology, 24 (5), 948–953. 10.1016/j.copbio.2013.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CE, Glenney JR Jr., & Burridge K (1990). Paxillin: A new vinculin-binding protein present in focal adhesions. Journal of Cell Biology, 111(3), 1059–1068. 10.1083/jcb.111.3.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura A, Nguyen T-N, Steele AN, & Yamada S (2011). The LIM domain of Zyxin is sufficient for force-induced accumulation of Zyxin during cell migration. Biophysical Journal, 101(5), 1069–1075. 10.1016/j.bpj.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafiadaki E, Arvanitis DA, & Sanoudou D (2015). Muscle LIM protein: Master regulator of cardiac and skeletal muscle function. Gene, 566(1), 1–7. 10.1016/j.gene.2015.04.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Slusarski DC, Kaykas A, Louie SH, & Moon RT (2003). Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Current Biology, 13(8), 680–685. 10.1016/s0960-9822(03)00240-9 [DOI] [PubMed] [Google Scholar]
- Vigouroux C, Henriot V, & Le Clainche C (2020). Talin dissociates from RIAM and associates to vinculin sequentially in response to the actomyosin force. Nature Communications, 11(1), 3116. 10.1038/s41467-020-16922-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Butler JP, & Ingber DE (1993). Mechanotransduction across the cell surface and through the cytoskeleton. Science (New York, N.Y.), 260(5111), 1124–1127. 10.1126/science.7684161 [DOI] [PubMed] [Google Scholar]
- Wang N, Tytell JD, & Ingber DE (2009). Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nature Reviews Molecular Cell Biology, 10(1), 75–82. 10.1038/nrm2594 [DOI] [PubMed] [Google Scholar]
- Watanabe-Nakayama T, Saito M, Machida S, Kishimoto K, Afrin R, & Ikai A (2013). Requirement of LIM domains for the transient accumulation of paxillin at damaged stress fibres. Biology Open, 2(7), 667–674. 10.1242/bio.20134531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JC, & Chalfie M (1988). Mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell, 54(1), 5–16. 10.1016/0092-8674(88)90174-2 [DOI] [PubMed] [Google Scholar]
- Weiskirchen R, & Günther K (2003). The CRP/MLP/TLP family of LIM domain proteins: Acting by connecting. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 25(2), 152–162. 10.1002/bies.10226 [DOI] [PubMed] [Google Scholar]
- Weng Z, Taylor JA, Turner CE, Brugge JS, & Seidel-Dugan C (1993). Detection of Src homology 3-binding proteins, including paxillin, in normal and v-Src-transformed Balb/c 3T3 cells. The Journal of Biological Chemistry, 268(20), 14956–14963. [PubMed] [Google Scholar]
- Winkelman JD, Anderson CA, Suarez C, Kovar DR, & Gardel ML (2020). Evolutionarily diverse LIM domain-containing proteins bind stressed actin filaments through a conserved mechanism. Proceedings of the National Academy of Sciences of the United States of America, 117(41), 25532–25542. 10.1073/pnas.2004656117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wioland H, Jegou A, & Romet-Lemonne G (2019). Torsional stress generated by ADF/cofilin on cross-linked Actin filaments boosts their severing. Proceedings of the National Academy of Sciences of the United States of America, 116(7), 2595–2602. 10.1073/pnas.1812053116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, & Beckerle MC (2005). Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. The Journal of Cell Biology, 171(2), 209–215. 10.1083/jcb.200505018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko EC, & Asbury CL (2014). Force is a signal that cells cannot ignore. Molecular Biology of the Cell, 25(23), 3717–3725. 10.1091/mbc.E13-12-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, & Geiger B (2007). Functional atlas of the integrin adhesome. Nature Cell Biology, 9(8), 858–867. 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, & Burridge K (1998). Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. The Journal of Cell Biology, 141(2), 539–551. 10.1083/jcb.141.2.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


