Abstract
The multiplex PCR method for the detection of Alloiococcus otitidis, Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae (P. H. Hendolin, A. Markkanen, J. Ylikoski, and J. J. Wahlfors, J. Clin. Microbiol. 35:2854–2858, 1997) in middle ear effusions (MEEs) was modified to be better suited for clinical use. To detect false-negative results, an internal amplification was added to the reaction, and to prevent carryover contamination, the dUTP–uracil-N-glycosidase system was incorporated into the procedure. Labor was minimized by using the heat-activatable AmpliTaq Gold polymerase in order to circumvent manual hot start and by detecting the amplification products on an automated sequencer. The performance of the improved protocol was verified with MEEs from patients with otitis media with effusion. In addition, a ligase detection reaction (LDR) was developed for confirmation of the PCR products. The modifications increased the reliability of the protocol and the hands-off time significantly. However, when two DNA extraction protocols were compared, gram-negative bacteria were detected more often in phenol-treated MEEs (94 versus 46%; P < 0.001), and gram-positive bacteria were detected more often in MEEs dissolved in sodium dodecyl sulfate-NaOH-chaotropic salt (83 versus 27%; P < 0.001). The LDR was found to be 100% specific. In all, the results demonstrate the feasibility of the rapid (7-h) multiplex PCR method for routine laboratory use.
PCR is a technique that allows the sensitive and specific detection of bacteria. It is useful for the detection of pathogens that are slowly growing, difficult to culture, or hazardous to handle in a diagnostic laboratory (23). PCR has been used to improve the sensitivity of bacterial detection in middle ear infections (13, 14, 22, 25, 30). Otitis media with effusion (OME) is considered a chronic sequela of acute otitis media (15), but cultures of middle ear effusions (MEEs) yield positive results for only 20 to 30% of patients (9, 17). By PCR, up to 75% of the MEEs test positive for pathogenic bacteria (22, 25). However, the published protocols target only one pathogen at a time and often involve tasks that make the approaches impractical for routine laboratory use. These include labor-intensive blotting of the amplification products for subsequent detection with radiolabeled probes (13, 25, 30) and the need for the use of time-consuming and contamination-susceptible nested PCR setups (14, 22).
Previously (10), we developed a protocol based on multiplex PCR since it allows the simultaneous detection of several bacterial species and requires smaller amounts of reagents and samples. To manage with a single-round PCR and product detection on ethidium bromide-stained agarose gels, we targeted the bacterial 16S rRNA gene, which is found in multiple copies. We selected for detection the pathogens most commonly encountered by culture in the middle ear, i.e., Haemophilus influenzae (10 to 35%), Streptococcus pneumoniae (2 to 9%), and Moraxella catarrhalis (0 to 6%) (9, 17), as well as the slowly growing bacterium Alloiococcus otitidis (4.6 to 5%) (8; T. Sih, B. Schwartz, G. Bosley, and R. Facklam, Program Abstr. 32nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 671, 1992). A. otitidis is a potential pathogen found solely in patients with OME (4, 8). The multiplex setting comprised one common reverse primer and four species-specific forward primers, and thus, the amplification of any of the products acted as a control for the amplification of the other amplification products (10). In a study of 67 MEE specimens from patients with OME, we observed an increase from 21 to 84% in the proportion of positive specimens compared with the proportion testing positive by culture and detected A. otitidis in 42% of the specimens (11).
However, the adoption of PCR technology in a clinical laboratory increases the demands of a protocol. The reliability has been addressed by an internal control to detect false-negative results due to the presence of PCR-inhibiting substances in specimens (1, 6). A method of avoiding carryover contamination is recommended for routine implementation of PCR (26). Of a range of methods, the use of a totally closed system for both PCR amplification and subsequent product detection, the enzymatic pretreatment of dUTP-containing PCR products with uracil-N-glycosylase (UNG) (21), post-PCR alkaline primer hydrolysis (26), and photochemical post-PCR cross-linking with isopsoralen (5) are reported to be effective (24, 26). An important issue is the minimization of labor. This is achieved by using premixed reaction mixtures, by using a heat-activatable polymerase to eliminate the enzyme addition step in hot-start PCR, which is a prerequisite for optimal specificity and sensitivity in many PCR protocols (16), and by automating PCR product detection. For the automation, blotting procedures, capillary electrophoresis, automatic DNA sequencer separation, and microtiter plate systems are used (for a review, see reference 20).
In the present paper, we report on modifications that increase the reliability of our multiplex procedure and reduce the hands-on time. In addition, we present a ligase detection reaction (LDR) for the verification of the A. otitidis amplification product. LDR is based on the ligation of two adjacent primers, and it allows the discrimination of DNA sequences that differ by only a single base pair (2).
MATERIALS AND METHODS
MEE specimen preparation.
The study included 73 MEEs from children with OME. The effusions were collected during ventilation tube placement, as described previously (10), and were stored in an airtight container at −20°C until they were processed for PCR. The study protocol was approved by the Ethics Committee of the Helsinki University Central Hospital.
Forty-nine of the effusion specimens were processed by the phenol-extraction protocol reported previously (10). For the remaining 24 effusion specimens, a modified QIAamp (Qiagen, Hilden, Germany) DNA extraction protocol was used. In a screw-cap Eppendorf tube, 100 μl of the effusion was mixed with 50 μl of a freshly prepared mixture of 75 mM NaOH and 1.5% sodium dodecyl sulfate (SDS), and the mixture was incubated at 95°C for 5 min. After cooling on ice, the suspension was neutralized with 50 μl of 75 mM Tris-HCl (pH 7.0), and 200 μl of buffer AL (QIAamp kit) was added. The tubes were incubated again at 95°C for 5 min and were cooled on ice. Then, 210 μl of 99% ethanol was added to the suspension, and the protocol described in the instructions for the QIAamp kit was followed. The DNA was eluted from the columns twice (with 90 and 60 μl) with Aqua Sterilisata (Amersham Pharmacia Biotech, Uppsala, Sweden) at 70°C and aliquoted, and the aliquots were stored at −20°C.
Bacterial strains.
The control strains for the multiplex PCR procedure were grown on agarose media, harvested, enumerated, and stored frozen, as described previously (10). One microliter was used for PCR.
For the testing of the LDR, amplification products were generated from A. otitidis, M. catarrhalis, and S. pneumoniae cells in individual PCRs with only the species-specific primer and the UR primer (both 20 pmol/reaction) (Table 1) under the multiplex reaction conditions (10). The DNAs of Campylobacter rectus, Campylobacter sp. (two strains), Eubacterium tenue, Mycobacterium nonchromogenicum, Mycobacterium malmoense, Pseudomonas sp., Sphingomonas sp., and Thermus brokianus, a gift from Sini Suomalainen (Institute of Biotechnology, University of Helsinki, Helsinki, Finland), were used for PCR with universal primers that amplified the sequence of approximately 920 bp from the 5′ end of the 16S rRNA gene (bases 8 to 928 in the Escherichia coli GenBank accession no. M24836 sequence) (7). The formation of amplification products was confirmed by agarose gel electrophoresis on 1.5% LE agarose (FMC BioProducts, Rockland, Maine). The products were extracted with an equal volume of chloroform-isoamyl alcohol, and 5 μl (≥200 ng) was taken for LDR.
TABLE 1.
Primers used in multiplex PCR procedure and LDR
| Procedure and primer | Target organism | Modification | Sequence (5′→3′) | Positiona | Product size (bp) |
|---|---|---|---|---|---|
| Multiplex PCR | |||||
| Hi | H. influenzae | CGT ATT ATC GGA AGA TGA AAG TGC | 177–200 | 523b | |
| Sp | S. pneumoniae | AAG GTG CAC TTG CAT CAC TAC | 106–127 | 482b | |
| Ao | A. otitidis | GGG GAA GAA CAC GGA TAG GA | 437–456 | 262b | |
| Mc | M. catarrhalis | CCC ATA AGC CCT GAC GTT AC | 416–435 | 235b | |
| UR | Universalc | 5′-Cy-5 | CTA CGC ATT TCA CCG CTA CAC | 682–702 | |
| A. otitidis LDR | |||||
| Ao–Cy-5–U21 | A. otitidis | 5′-Cy-5 | ACG GAT AGG AGT CAC TGC CTA | 451–471 | 40d |
| Ao–5-P–U19 | A. otitidis | 5′-Pe | TCC CTT GAC GGT ACC CAA C | 472–490 | 40d |
| IC preparation | |||||
| UR-AB1 | Arcanobacterium sp. | CTA CGC ATT TCA CCG CTA CAC GAG AGG ATG ACC AGC CAC ACT | 303–344 | 400b | |
| UR | Universal | CTA CGC ATT TCA CCG CTA CAC | 682–702 |
Position refers to the specific nucleotide locations of the primers in the 16S rRNA genes (A. otitidis, EMBL accession no. 59765; H. influenzae, GenBank accession no. M35019; M. catarrhalis, GenBank accession no. L13736; S. pneumoniae, GenBank accession no. X58312; universal reverse primer, E. coli GenBank accession no. M24836).
Specific PCR amplification with the UR primer.
Most eubacterial species.
Specific ligation in LDR.
P, phosphate group (—PO43−).
IC.
The internal control (IC) was generated from 105 cells of a clinical isolate of an Arcanobacterium sp. by PCR. The 50-μl reaction mixture contained both the UR and the UR-AB1 primers at 0.4 μM each (Table 1), the four deoxynucleotide triphosphates at a concentration of 200 μM each, and 1.5 U of DyNAzyme polymerase (FinnZymes, Espoo, Finland) in optimized 1× DyNAzyme buffer (10 mM Tris-HCl [pH 8.8 at 25°C], 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100). The reaction conditions were identical to those for the multiplex reaction (10), but an annealing temperature of 63°C was used. The PCR yielded two amplification products that were separated on 2% SeaKem GTG (FMC BioProducts). The product of approximately 400 bp was purified with the QIAquick kit (QIAGEN) and was reamplified with only the universal reverse primer (20 pmol) under the multiplex reaction conditions (10). After gel purification as described above, the amount of the resulting IC was quantitated spectrophotometrically (27). A dilution series was prepared in the optimized 1× DyNAzyme buffer (Finnzymes) containing 10% Tween 20 (Bio-Rad, Hercules, Calif.), and the dilutions were stored in aliquots at −20°C.
The IC was fitted in the multiplex reaction with a twofold dilution series ranging from 20 × 10−18 to 1 × 10−18 g per reaction mixture (40 to 2 copies). The experiments were done in the presence of either 0 or 1,000 bacterial cells from pure bacterial cultures of each the four study organisms per reaction mixture. Then, the concentration of the IC was optimized for the PCR analysis of MEEs. To yield consistent results with and without the IC, the concentrations of the primers and MgCl2 were adjusted.
Thermostable polymerase replacement.
In the substitution of AmpliTaq Gold (Perkin-Elmer, Norwalk, Conn.) for DyNAzyme (Finnzymes), MEEs were analyzed in parallel with both enzymes and in the presence of the IC. The amount of DyNAzyme was 2 U/reaction mixture, whereas the amount of AmpliTaq Gold was varied between 1 and 4 U/reaction mixture in 1× GeneAmp PCR Buffer II (10 mM Tris-HCl [pH 8.3 at 25°C], 50 mM KCl) supplied with 2 mM MgCl2. For AmpliTaq Gold, the initial denaturation and enzyme addition steps in the temperature profile were replaced with an incubation for 9 min and 30 s at 94°C. The final extension was prolonged to 10 min for both enzymes.
Incorporation of the dUTP-UNG system.
The substitution of dUTP for dTTP was examined in an experiment with A. otitidis (1,000 cells/reaction mixture) and only the Ao and UR primers (both at 20 pmol/reaction mixture) (Table 1). Two concentrations of dUTP, 400 and 600 μM, were tested, and the MgCl2 concentration was varied between 1.5 and 2.8 mM to find an amplification level that corresponds to the amplification level achieved with 200 μM dTTP and 1.5 mM MgCl2. In the multiplex setup, 400 μM dUTP was applied, and the concentration of MgCl2 was further optimized to yield amplification levels from MEEs that were consistent with those obtained with dTTP (200 μM) and 2 mM MgCl2. In both experiments, the reaction mixtures comprised 2.5 U of AmpliTaq Gold and dATP, dCTP, and dGTP each at a concentration of 200 μM. The multiplex cycling conditions were used, and the thermal program was completed with a step of 55°C for 30 min.
The efficiencies of two UNG enzymes, a heat-labile UNG (catalog no. 1775367; Boehringer Mannheim, Mannheim, Germany) and a cloned E. coli UNG (catalog no. 280S; New England Biolabs, Beverly, Mass.), at preventing the reamplification of a dUTP-containing A. otitidis product (50 to 50,100 copies/reaction mixture) were evaluated. UNG was used at 0.3 U/reaction mixture, and a preincubation step of 15 min at room temperature was applied before cycling. The influence of residual UNG activity on the conservation of the PCR product was investigated in a reaction mixture with 1,000 A. otitidis cells. UNG was used at 0.3 U/reaction mixture, and the reaction mixtures were stored either at room temperature (ca. 20°C) or under refrigeration (4°C). Aliquots were removed at various times and were extracted with chloroform-isoamyl alcohol (24:1, Amresco, Solon, Ohio) to inactivate the UNG.
LDR.
For the LDR, 25 μl of a multiplex reaction mixture was purified in MicroSpin S-400 HR (Amersham Pharmacia Biotech) columns. Five microliters of each of the products was used with 5 nM both LDR primers (Table 1) in a 20-μl reaction mixture with 1× Taq ligase buffer (New England Biolabs), which consisted of 20 mM Tris-HCl (pH 7.6 at 25°C), 25 mM potassium acetate, 10 mM magnesium acetate, 10 mM dithiothreitol, 1 mM NAD, and 0.1% (vol/vol) Triton X-100. After an initial denaturation of 3 min at 95°C, 1 μl of Tag ligase (New England Biolabs) was added at 80°C. The temperature profile consisted of 45 cycles of 95°C for 30 s, 50°C for 6 min, and a ligase-inactivating step of 99°C for 10 min.
PCR and LDR product detection.
Agarose gel electrophoresis of the multiplex PCR products was performed as described earlier (10), but with only 1 μg of ethidium bromide per ml. For the visualization of the PCR products on the automated ALFexpress sequencing machine, the universal reverse primer was labeled with Cy-5 (Amersham Pharmacia Biotech) (Table 1). The detection was optimized by varying the proportion of labeled and unlabeled primers (1:9, 1:3, 1:1, and 1:0). In addition, the use of different volumes (1 to 5 μl) of the reaction mixture in the sequencer was analyzed. For the LDRs, 5 μl was always analyzed. The amplification products were mixed 1:1 with 2× loading dye (10 mM EDTA, 97% deionized formamide, 10 mg of dextran blue per ml), denatured at 96°C for 2 min and 30 s, and cooled on ice. The resulting single-stranded products were loaded into individual lanes of the ALFexpress short-plate kit (10-cm gel; thickness, 0.5 mm) and were separated in a 5% LongRanger gel (FMC BioProducts) at 1,500 V, 60 mA, 25 W, and 50°C for 2 h and 15 min.
The electropherogram was analyzed with the ALFwin Fragment Manager, version 1.2, program (Amersham Pharmacia Biotech). In every run, a positive control with the amplification products of the four study organisms was included; the user indicated the relevant product lengths in the program. The electrophoretic mobilities in different lanes were standardized by assigning the IC peak as a reference of 400 bp. The analytical peaks were identified according to their product lengths.
Statistical analysis.
The chi-square test was used for comparison of bacterial findings for effusions that were purified by different methods.
RESULTS
Inclusion of IC.
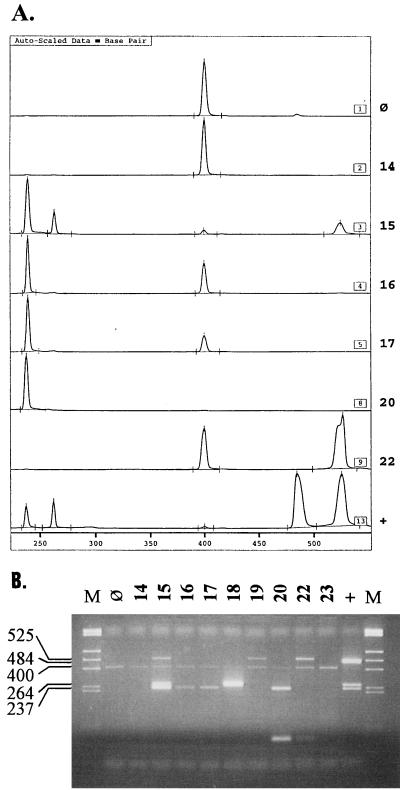
The amount of IC that yielded a clearly detectable product without impairing the amplification of bacterial DNA was investigated with a twofold dilution series. In the presence of 1,000 cells of pure bacterial cultures of the four study organisms per reaction mixture, 1 × 10−18 to 5 × 10−18 g (2 to 10 copies) produced satisfactory results. With MEEs, amounts larger than 1 × 10−18 g (2 copies) resulted in weakening of the bacterial amplification products when only one or two amplification products were present. An example of an electropherogram for the analysis of MEEs is shown in Fig. 1A. All (49 of 49) phenol-extracted MEEs yielded amplification products of bacterial DNA or the IC. In contrast, 2 (8.3%) of the 24 QIAamp-purified effusions yielded no amplification products. When a smaller volume (10 versus 20 μl) was analyzed, one effusion was positive for S. pneumoniae and the other was positive for S. pneumoniae and M. catarrhalis.
FIG. 1.
Examples of an ALFexpress Fragment Manager (version 1.2) electropherogram (A) and an analytical agarose gel for MEE specimens (B). The specimen number (same in both analyses) is given beside and above each lane, respectively. Lane or panel ⊘, negative control; lane or panel +, positive control; lane M, molecular size markers (HinfI and RsaI digestion of pUC19 yielded the following fragments: 1,769, 1,419, 676, 517, 396, 241, and 214 bp). The ALFexpress Fragment Manager electropherogram is scaled in base pairs. For the agarose gel, the sizes of the amplification products are shown on the left: 237 bp, M. catarrhalis; 264 bp, A. otitidis; 400 bp, IC; 484 bp, S. pneumoniae; 525 bp, H. influenzae.
Use of AmpliTaq Gold.
In the analysis of MEEs, 2 to 5 U of AmpliTaq Gold per reaction mixture yielded amplification products with intensities equal to those obtained with the formerly applied DyNAzyme polymerase (2 U/reaction mixture). An amount of 2.5 U of AmpliTaq Gold per reaction mixture was selected to correspond to the increased requirement caused by the addition of the IC and altered amounts of primers in the multiplex reaction mixture.
Adaptation of dUTP-UNG system.
The incorporation of the dUTP-UNG system consisted of the optimization of the reaction mixture for dUTP and the evaluation of two UNG enzymes. Relatively higher concentrations of dUTP were examined, since the incorporation rate for dUTP is lower than those for the other deoxynucleotide triphosphates (21). In an experiment with A. otitidis, 400 or 600 μM dUTP was as efficient as 200 μM dTTP when the MgCl2 concentration was raised from 1.5 to 2.5 mM or to 2.8 mM, respectively. The lower concentrations were chosen to preserve reagents. For the analysis of MEEs in the multiplex procedure with 400 μM dUTP, the MgCl2 concentration had to be further increased to 2.8 mM.
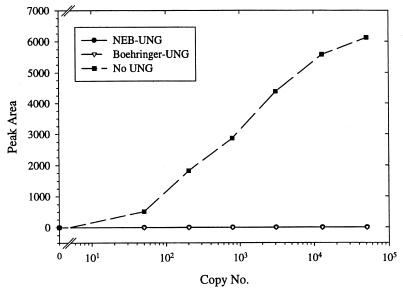
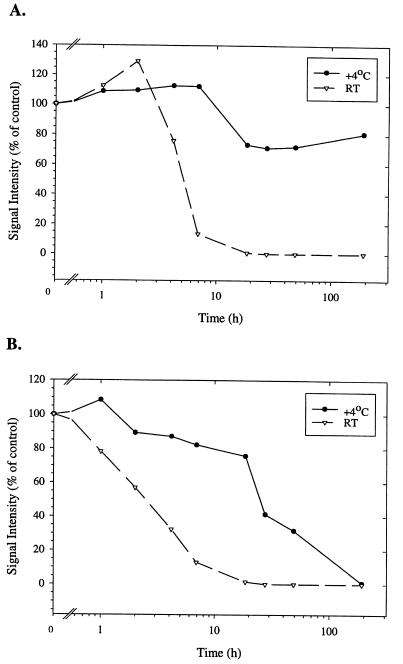
The abilities of the Boehringer Mannheim UNG and the New England Biolabs UNG to eliminate the dUTP-containing A. otitidis product are shown in Fig. 2. Both enzymes blocked the amplification of up to the largest amount of products tested (50,100 copies). The influence of the two UNGs on the stability of the A. otitidis product after completion of the reaction is depicted in Fig. 3. Reactions with the native E. coli UNG (New England Biolabs) showed an instant decrease in signal intensity when the products were stored at room temperature and a gradual decrease after 1 h when they were stored at 4°C. In contrast, the thermolabile Boehringer Mannheim UNG could be stored for up to 2 h at room temperature or for 8 h at 4°C without a loss of signal intensity.
FIG. 2.
Efficiencies of two UNG enzymes in the elimination of a dUTP-containing PCR product. A reaction mixture containing either the UNG from New England Biolabs (NEB), the heat-labile UNG from Boehringer Mannheim (Boehringer), or no UNG was spiked with the indicated number of dUTP-containing A. otitidis PCR products. After PCR, the amount of amplification products (peak area) was measured on the ALFexpress sequencer. Each datum point represent the results of an analysis for at least two samples.
FIG. 3.
Influence of residual UNG activity on the preservation of dUTP-containing PCR product after amplification. (A) The heat-labile Boehringer Mannheim UNG. (B) The New England Biolabs UNG. The products were stored either in a refrigerator (4°C) or at room temperature (RT) for the indicated amount of time. The signal intensity was measured on the ALFexpress sequencer and was compared to that of the control (0 h). Each datum point represents the results of an analysis for at least two samples.
Final multiplex reaction conditions.
The multiplex protocol was modified to comprise an internal control and the dUTP-UNG system. The reaction mixture was optimized for Mg2+, primer, and AmpliTaq Gold concentrations. The optimized reaction mixture contained 3.6 μM A. otitidis-specific primer, 1.8 μM H. influenzae-specific primer, 0.6 μM M. catarrhalis-specific primer, 0.2 μM S. pneumoniae-specific primer, 0.8 μM Cy-5-labeled common reverse primer, dATP, dCTP, and dGTP at 200 μM each, 400 μM dUTP, 0.2% Tween 20, 1 ag of IC, 1× GeneAmp PCR buffer II, 2.8 mM MgCl2, 2.5 U of AmpliTaq Gold, and 0.3 U of thermolabile UNG. The reaction profile was 9.5 min of initial denaturation at 94°C, 38 cycles of 95°C for 30 s, 66°C for 45 s, and 72°C for 1 min, and 10 min of final extension at 72°C, followed by 30 min at 55°C for completion of the reaction. Various thermal cyclers (PTC-100, PTC-200, and PTC-225 DNA Engine Tetrad; MJ Research, Watertown, Mass.) were used in the study. Although the cyclers were not specially compared, no differences in their amplification efficiencies were observed, indicating that the temperature steps are sufficiently long to complete the reactions.
Multiplex PCR product detection on ALFexpress sequencing machine.
The PCR product detection on the sequencer was established by varying the proportion of labeled to unlabeled universal reverse primer in the reaction mixture and analyzing different volumes in the sequencer. An example of product detection with the ALFexpress sequencing machine and by agarose gel electrophoresis is shown in Fig. 1. To achieve the highest sensitivity, only Cy-5-labeled primer was used, and the maximum of 5 μl of a reaction mixture was loaded onto the sequencer. This caused saturation of the signal for some samples. However, the identification of the peaks was not hampered, since even the two closest peaks (those for M. catarrhalis and A. otitidis) were separated by approximately 5 min (Fig. 1A).
The preparation of a LongRanger gel from a stock solution required only 15 min of hands-on time. In contrast, unless precast gels were used, the agarose had to be melted and subsequently cooled before it was poured into plates, which takes more than 45 min. The run times were equal for both types of gels (ca. 2.5 h), but for the agarose gel photography had to be done after electrophoresis. The ALFexpress sequencing machine could be left unattended after starting the run, and the results were saved automatically for analysis with the ALFwin Fragment Manager.
Detection of bacterial DNA in MEEs.
The newly optimized multiplex conditions were used for analysis of MEEs. The optimal sample volumes in the PCR were investigated with different volumes of 10 specimens from both purification protocols. Most positive results were obtained with 2- and 20-μl samples for the phenol-extracted and QIAamp-purified effusions, respectively (data not shown).
The results of PCRs with phenol-extracted and QIAamp-purified MEEs are shown in Table 2. The phenol method yielded relatively more positive effusions (98.0%) than the QIAamp method (83.3%) (chi-square test, P [5.401, 1] = 0.020). The proportion of effusions positive for gram-negative bacteria (H. influenzae and M. catarrhalis) (93.9%) was significantly higher among phenol-extracted MEEs (93.9%) than among the QIAamp-purified MEEs (45.8%) (chi-square test, P [20.98, 1] < 0.001). In contrast, the QIAamp kit promoted the detection of gram-positive bacteria (A. otitidis and S. pneumoniae) (83.3%) compared with phenol extraction (26.5%) (chi-square test, P [21.79, 1] < 0.001).
TABLE 2.
Comparison of multiplex analysis of phenol-extracted and QIAamp-purified MEEs
| Target species | No. (%) of positive specimens
|
P value (χ2 statistic)a | Total no. (%) of positives (n = 73) | |
|---|---|---|---|---|
| Phenol extraction (n = 49) | QIAamp (n = 24) | |||
| A. otitidis | 5 (10.2) | 9 (37.5) | P (7.744) = 0.005 | 14 (19.2) |
| H. influenzae | 21 (42.9) | 3 (12.5) | P (6.727) = 0.009 | 24 (32.9) |
| M. catarrhalis | 29 (59.2) | 10 (41.7) | P (1.987) = 0.159 | 39 (53.4) |
| S. pneumoniae | 8 (16.3) | 18 (75) | P (24.37) < 0.001 | 26 (35.6) |
| Gram-negative bacteriab | 46 (93.9) | 11 (45.8) | P (20.98) < 0.001 | 57 (78.1) |
| Gram-positive bacteriac | 13 (26.5) | 20 (83.3) | P (21.79) < 0.001 | 33 (45.2) |
| All study organisms | 48 (98.0) | 20 (83.3) | P (5.401) = 0.020 | 68 (93.2) |
Comparison of the proportions of positive effusions obtained by the two DNA purification methods (df = 1).
H. influenzae and M. catarrhalis.
A. otitidis and S. pneumoniae.
LDR for A. otitidis product.
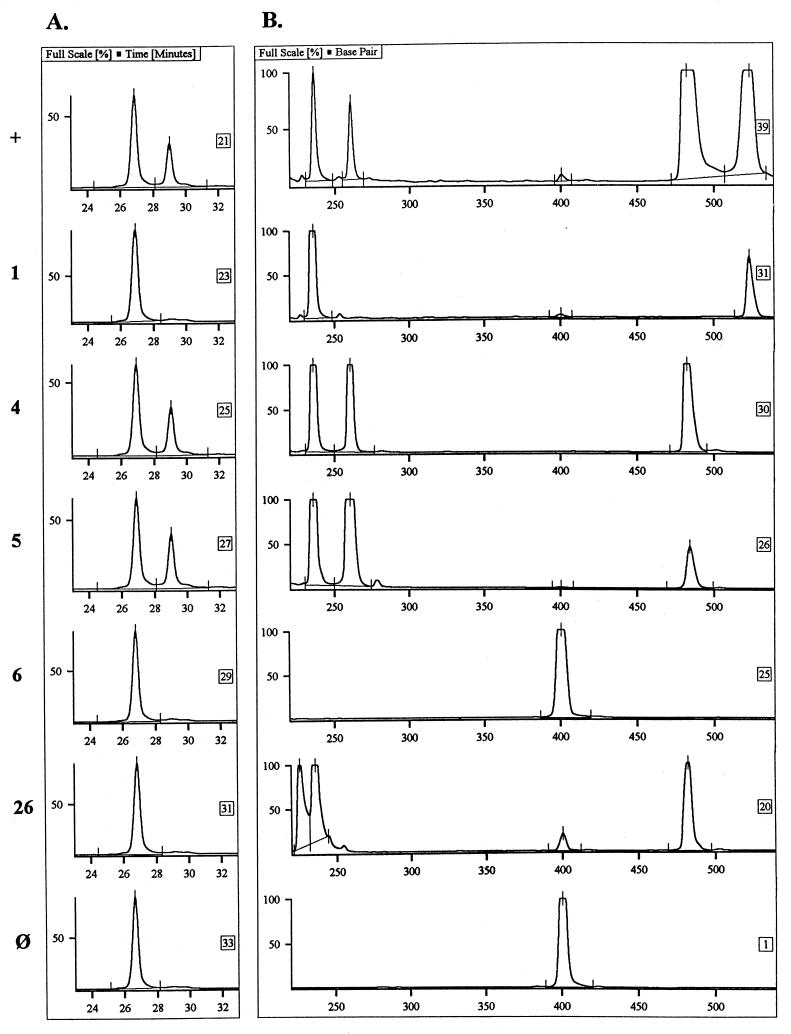
The specificity of the LDR for A. otitidis was assessed by analysis of 16S rRNA amplification products of various bacterial species. The ligation product was produced only in the presence of the A. otitidis PCR product. Figure 4 shows the capability of the LDR to verify the origin of the A. otitidis PCR product in MEE samples amplified in the multiplex reaction.
FIG. 4.
A. otitidis PCR product verification by LDR. The ALFexpress electropherogram shows the LDR analysis of PCR products (A) from the multiplex PCR analysis of MEEs (B). The specimen number is given to the left of each row. By LDR analysis, the nonligated fluorescence-labeled primer (21mer) is eluted at ca. 27 min, whereas in the presence of the A. otitidis amplification product (264 bp), the ligation product (40mer) is eluted at ca. 29 min. Row +, positive control; row ⊘, negative control. The multiplex PCR product sizes are indicated in the legend to Fig. 1.
DISCUSSION
The aim of the study was to advance our multiplex PCR method (10) for the detection of four bacterial species involved in OME to make it applicable in a clinical microbiology laboratory. We addressed the reliability of sensitive detection by including an internal amplification control that reveals false-negative results and by adapting the UNG system to avoid false-positive results. To increase the hands-off time, we used the heat-activatable AmpliTaq Gold to avoid manual hot start and automated the amplification product detection with a sequencing machine.
The IC was considered necessary, since MEEs contain polymerase-inhibiting substances, such as lipopolysaccharides and heme (12), that may be preserved in the samples, despite the use of purification protocols. The benefit of the IC was demonstrated with two QIAamp-purified effusions, which yielded positive results for bacteria only when a smaller (10 versus 20 μl) volume was reanalyzed. All phenol-extracted MEEs produced satisfactory results, although in our previous study (11) no amplification was observed in 5 (7.5%) of 67 MEEs. The discrepancy can be explained by the smaller volume (2 versus 5 μl) analyzed under the novel multiplex conditions. In the PCR-based analysis of MEEs, the potential influence of biological matter has been addressed by analyzing various volumes of specimens with two different primer pairs (25) or by using a nested PCR protocol to increase the sensitivity of the reaction (14). Other protocols have used hybridization with radiolabeled probes to improve the sensitivity of product detection (13, 30). However, these approaches do not address the inherent problem of reaction failure, but they increase significantly the reagent costs, are labor-intensive, and prolong the analysis time considerably. Thus, use of the IC appears to provide a simple means for verification of reaction functioning.
The avoidance of false-positive amplifications was first addressed by replacing the conventional polymerase by AmpliTaq Gold, which is activated by heat. The use of AmpliTaq Gold avoids the need to reopen the tubes for enzyme addition after the initial denaturation of templates, as in the manual hot-start procedure (16), and reduces the possibility of cross-contamination. To avoid contamination with products from previous amplifications, the dUTP-UNG system (21) was adapted to the multiplex reaction. Use of 0.3 U of UNG/reaction mixture prevented the reamplification of at least 5 × 105 copies of the dUTP-containing A. otitidis amplification product. In comparison, Kunakorn and Markham (18) reported that use of an UNG amount of as low as 0.05 U/reaction mixture with a 5-min preincubation step is sufficient to destroy at least 108 carryover molecules. By use of the UNG amounts recommended by the manufacturer (1 U/reaction mixture), elimination of at least 3 × 109 copies of product (26) and a 107-fold reduction in product concentration (24) have been reported. From these data, we conclude that carryover contamination can be effectively controlled in the multiplex setup by the UNG system. Nevertheless, good PCR practices (19) must be maintained in any diagnostic setting.
The dUTP-UNG-system was chosen since it allows the subsequent analysis of the newly synthesized products by methods such as LDR, hybridization, or sequencing. This possibility would be destroyed by cross-linking of products with psoralen compounds (5). One problem underlying the use of UNG is that the enzyme retains its activity or regains its activity after thermocycling (29). It has been recommended that products either be analyzed immediately after completion of the reaction, extracted with organic solvents, or stored frozen (21, 29). We tested the recently introduced heat-labile UNG from Boehringer Mannheim to determine whether it would enable the conservation of products without additional work steps. Storage was possible for up to 8 h without a loss of signal intensity. This period was not long enough to be able to leave the reactions running overnight, after office hours. However, this time can be increased by keeping the samples at 55°C before cooling; the UNG has minimal activity at 55°C (28, 29). These arrangements offer flexibility in PCR analyses with UNG.
Bacterial DNA was detected in 93.2% of the MEEs (or 89.0% of the MEEs without A. otitidis). This finding is comparable to those of our earlier studies (10, 11) and the studies of Post et al. (25) and Matar et al. (22) for the detection of H. influenzae, M. catarrhalis, or S. pneumoniae DNA. The overall incidences of the individual bacterial species was also in the range normally reported for children with OME (13, 14, 22, 25, 30). An exception was the incidence of H. influenzae, which was found in only 32.9% of the effusions, whereas it was found in 52 to 70% of the effusions in other studies (13, 22, 25, 30). However, when the two methods for DNA purification were compared, the incidences of A. otitidis, H. influenzae, and S. pneumoniae differed significantly (P < 0.005, P < 0.05, and P < 0.001, respectively). The differences were likely due to the dissimilar lysing properties of the methods in respect to the cell wall compositions of the bacteria. This view was supported by the finding of a higher incidence of gram-negative bacteria in phenol-extracted MEEs than in QIAamp-purified MEEs (P < 0.001) and more positive effusions for gram-positive bacteria by the QIAamp protocol than by the phenol extraction method (P < 0.001).
To our knowledge, this was the first time that DNA extraction methods were compared for MEEs. Some trends can nevertheless be seen by comparing the present results with those of prior studies. The QIAamp kit used by Jero et al. (14) and by us in this study (with an additional SDS-NaOH boiling step) has resulted in a higher percentage of S. pneumoniae-positive OME effusions (46.3 and 75%, respectively) than sonication in acetylcysteine (29.9%) (25) or proteinase K with Triton X-100 (8.6%) (22) or by phenol extraction (8 to 17.9%) (10, 11; this study). All methods except the QIAamp protocol in this study have yielded relatively high incidences of H. influenzae (42.9 to 70.2%) (10, 13, 22, 25, 30); the QIAamp protocol yielded an incidence of only 12.5%. For M. catarrhalis, the incidences have varied significantly (16 to 59.2%) (10, 11, 22, 25; this study).
It is conceivable that the results may also be affected by the PCR setup itself, by regional and seasonal variations, and by differences in the sample of analyzed patients. For example, the incidence of A. otitidis, as detected by the multiplex PCR with phenol-extracted MEEs from patients with OME, was 20% (5 of 25 specimens) in our pilot study in Kuopio, Finland (10), 41.8% (28 of 67) in a larger study in Kuopio (11), and 10.2% (5 of 49) in Helsinki (this study). However, the results of a comparison of two DNA extraction methods demonstrate the significance of optimizing not only the primers and the PCR conditions but also the DNA extraction protocol. Currently, we use a protocol in which the MEEs are first treated with SDS-NaOH and then boiled in the presence of phenol. After this, the DNA is purified with the QIAamp kit.
The objective of developing the LDR was to allow the multiplex PCR analysis of specimens other than MEEs. Previously (11), we found that the multiplex PCR is specific for the four study organisms found in MEEs. However, the bacterial spectrum is much wider, for example, in the nasopharynx. The LDR yielded the ligation product only in the presence of the A. otitidis PCR product, and the verification of the A. otitidis PCR product was possible despite large amounts of H. influenzae, M. catarrhalis, or S. pneumoniae amplification products in the samples taken for LDR. Although the cross-reaction testing comprised a narrow spectrum of bacterial species, the reaction was considered specific on the basis of the high discriminatory power of the LDR (2). Currently, we are developing a LDR method for the detection of all four study organisms in a multiplex format in which the various ligation products are distinguished by size with the aid of ethylene oxide mobility modifiers, as described by Baron et al. (3).
In conclusion, the modifications described here should allow the routine performance of the multiplex PCR protocol. The IC and the dUTP-UNG system increase the reliability of the method in two critical ways: detecting false-negative specimens and preventing false-positive results. The IC also serves as a reference for compensation of variations in electrophoretic mobilities in different lanes of the ALFexpress sequencer. A sequencer of another type may also be used when the fluorescent label is changed. We believe that an automated sequencer is accessible to most laboratories. The sequencer and the use of AmpliTaq Gold simplify the protocol and reduce the hands-on time considerably. Thus, as the sensitive multiplex analysis of the four bacterial pathogens provides results in no more than 7 h, it might be considered an alternative to routine culture.
ACKNOWLEDGMENTS
This work was supported by research grant 706/401/97 from the National Technology Agency of Finland.
We thank Jussi Jero, Kimmo Leskinen, and Mervi Närkiö-Mäkelä for providing the MEE specimens. We also thank Leena Palmunen, Paula Collin-Olkkonen, and Anne Makkonen for expert technical assistance.
REFERENCES
- 1.Ballagi-Pordany A, Belak S. The use of mimics as internal standards to avoid false negatives in diagnostic PCR. Mol Cell Probes. 1996;10:159–164. doi: 10.1006/mcpr.1996.0022. [DOI] [PubMed] [Google Scholar]
- 2.Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci USA. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron H, Fung S, Aydin A, Bahring S, Luft F C, Schuster H. Oligonucleotide ligation assay (OLA) for the diagnosis of familial hypercholesterolemia. Nat Biotechnol. 1996;14:1279–1283. doi: 10.1038/nbt1096-1279. [DOI] [PubMed] [Google Scholar]
- 4.Bosley G S, Whitney A M, Pruckler J M, Moss C W, Daneshvar M, Sih T, Talkington D F. Characterization of ear fluid isolates of Alloiococcus otitidis from patients with recurrent otitis media. J Clin Microbiol. 1995;33:2876–2880. doi: 10.1128/jcm.33.11.2876-2880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimino G D, Metchette K C, Tessman J W, Hearst J E, Isaacs S T. Post PCR sterilization: a method to control carry-over contamination for the polymerase chain reaction. Nucleic Acids Res. 1991;19:99–107. doi: 10.1093/nar/19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cone R W, Hobson A C, Huang M L W. Coamplified positive control detects inhibition of polymerase chain reactions. J Clin Microbiol. 1992;30:3185–3189. doi: 10.1128/jcm.30.12.3185-3189.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards U, Rogall T, Blöcker H, Emde M, Böttger E C. Isolation and direct complete determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faden H, Dryja D. Recovery of a unique organism in human middle ear fluid and its possible role in chronic otitis media. J Clin Microbiol. 1989;27:2488–2491. doi: 10.1128/jcm.27.11.2488-2491.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giebink G S. The microbiology of otitis media. Pediatr Infect Dis J. 1989;8(Suppl 1):18–20. [PubMed] [Google Scholar]
- 10.Hendolin P H, Markkanen A, Ylikoski J, Wahlfors J J. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997;35:2854–2858. doi: 10.1128/jcm.35.11.2854-2858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendolin P H, Kärkkäinen U, Markkanen A, Himi T, Ylikoski J. High incidence of Alloiococcus otitidis in otitis media with effusion. Pediatr Infect Dis J, 1999;18:860–865. doi: 10.1097/00006454-199910000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi R. Simple and rapid preparation of samples for PCR. In: Ehrlich H A, editor. PCR technology: principles and applications for DNA amplification. New York, N.Y: Stockton Press; 1989. pp. 31–38. [Google Scholar]
- 13.Hotomi M, Tabata T, Kakuchi H, Kunimoto M. Detection of Haemophilus influenzae in middle ear of otitis media with effusion by polymerase chain reaction. Int J Pediatr Otorhinolaryngol. 1993;27:119–126. doi: 10.1016/0165-5876(93)90127-o. [DOI] [PubMed] [Google Scholar]
- 14.Jero J, Virolainen A, Salo P, Leinonen M, Eskola J, Karma P. PCR assay for detecting Streptococcus pneumoniae in the middle ear of children with otitis media with effusion. Acta Otolaryngol. 1996;116:288–292. doi: 10.3109/00016489609137843. [DOI] [PubMed] [Google Scholar]
- 15.Juhn S K, Paparella M M, Kim C S, Goycoolea M V, Giebink G S. Pathogenesis of otitis media. Ann Otol Rhinol Laryngol. 1977;86:481–492. doi: 10.1177/000348947708600407. [DOI] [PubMed] [Google Scholar]
- 16.Kebelmann-Betzing C, Seeger K, Dragon S, Schmitt G, Moricke A, Schild T A, Henze G, Beyermann B. Advantages of a new Taq DNA polymerase in multiplex and time-release PCR. BioTechniques. 1998;24:154–158. doi: 10.2144/98241pf01. [DOI] [PubMed] [Google Scholar]
- 17.Krenke C, Stephensen J S, Wald E R. Incidence of organisms in otitis media. Ann Otol Rhinol Laryngol. 1988;87(Suppl. 133):20–21. [Google Scholar]
- 18.Kunakorn M, Markham R B. Clinically practical seminested PCR for Burkholderia pseudomallei quantitated by enzyme immunoassay with and without solution hybridization. J Clin Microbiol. 1995;33:2131–2135. doi: 10.1128/jcm.33.8.2131-2135.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwok S, Higuchi R. Avoiding false positives. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 20.Lazar J G. Advanced methods in PCR product detection. PCR Methods Appl. 1994;4:S1–S14. doi: 10.1101/gr.4.1.s1. [DOI] [PubMed] [Google Scholar]
- 21.Longo M C, Berninger M S, Hartley J L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reaction. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 22.Matar G M, Sidani N, Fayad M, Hadi U. Two-step PCR-based assay for identification of bacterial etiology of otitis media with effusion in infected Lebanese children. J Clin Microbiol. 1998;36:1185–1188. doi: 10.1128/jcm.36.5.1185-1188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallen M J, Butcher P D. New strategies in microbial diagnosis. J Hosp Infect. 1991;18(Suppl. A):147–158. doi: 10.1016/0195-6701(91)90017-3. [DOI] [PubMed] [Google Scholar]
- 24.Pang J, Modlin J, Yolken R. Use of modified nucleotides and uracil-DNA glycosylase (UNG) for the control of contamination in the PCR-based amplification of RNA. Mol Cell Probes. 1992;6:251–256. doi: 10.1016/0890-8508(92)90024-r. [DOI] [PubMed] [Google Scholar]
- 25.Post J C, Preston R A, Aul J J, Larkins-Pettigrew M, Rydquist-White J, Anderson K W, Wadowsky R M, Reagan D R, Walker E S, Kingsley L A, Magit A E, Ehrlich G D. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA. 1995;273:1598–1604. [PubMed] [Google Scholar]
- 26.Rys P N, Persing D H. Preventing false positives: quantitative evaluation of three protocols for inactivation of polymerase chain reaction amplification products. J Clin Microbiol. 1993;31:2356–2360. doi: 10.1128/jcm.31.9.2356-2360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sobek H, Schmidt M, Kaluza K. Heat-labile uracil-DNA glycosylase: purification and characterization. FEBS Lett. 1996;388:1–4. doi: 10.1016/0014-5793(96)00444-9. [DOI] [PubMed] [Google Scholar]
- 29.Thornton C G, Hartley J L, Rashtchian A. Utilizing uracil DNA glycosylase to control carryover contamination in PCR: characterization of residual UDG activity following thermal cycling. BioTechniques. 1992;13:180–184. [PubMed] [Google Scholar]
- 30.Ueyama T, Kurono Y, Shirabe K, Takeshita M, Mogi G. High incidence of H. influenzae in nasopharyngeal secretions and middle ear effusions as detected by PCR. J Clin Microbiol. 1995;33:1835–1838. doi: 10.1128/jcm.33.7.1835-1838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]