Abstract
The extracellular matrix (ECM) is composed of biochemical and biophysical cues that control cell behaviors and bulk mechanical properties. For example, anisotropy of the ECM and cell alignment are essential in the directional properties of tissues such as myocardium, tendon, and the knee meniscus. Technologies are needed to introduce anisotropic behavior into biomaterial constructs that can be used for the engineering of tissues as models and towards translational therapies. To address this, we developed an approach to align hydrogel fibers within cell-degradable bioink filaments with extrusion printing, where shear stresses during printing align fibers and photocrosslinking stabilizes the fiber orientation. Suspensions of hydrogel fibers were produced through the mechanical fragmentation of electrospun scaffolds of norbornene-modified hyaluronic acid, which were then encapsulated with meniscal fibrochondrocytes, mesenchymal stromal cells, or cardiac fibroblasts within gelatin-methacrylamide bioinks during extrusion printing into agarose suspension baths. Bioprinting parameters such as the needle diameter and the bioink flow rate influenced shear profiles, whereas the suspension bath properties and needle translation speed influenced filament diameters and uniformity. When optimized, filaments were formed with high levels of fiber alignment, which resulted in directional cell spreading during culture over one week. Controls that included bioprinted filaments without fibers or non-printed hydrogels of the same compositions either without or with fibers resulted in random cell spreading during culture. Further, constructs were printed with variable fiber and resulting cell alignment by varying print direction or using multi-material printing with and without fibers. This biofabrication technology advances our ability to fabricate constructs containing aligned cells towards tissue repair and the development of physiological tissue models.
Keywords: hydrogel, bioprinting, fibers, alignment, anisotropy
1. Introduction
Tissues are comprised of an extracellular matrix (ECM) - a complex, tissue-specific environment consisting largely of fibrous proteins and proteoglycans - and cells [1]. Native ECM cues such as fiber composition and orientation impact function in many tissue types, including myocardium and connective tissues such as knee menisci, tendon, and the annulus fibrosis [2–4]. Specifically, the mechanical properties of these tissues depend not only on their biochemical composition, but also on the tissue’s microstructure (e.g., anisotropy) [3,5–10]. Beyond the ECM, cell alignment also plays a critical role in a number of processes, such as cellular differentiation, proliferation and matrix deposition, as well as in the organization of subcellular structures such as cytoskeleton and adhesion complexes. Thus, there is great motivation to align cells within engineered tissues for in vitro 3D cell culture and in vivo therapeutic applications [11–14].
Towards the fabrication of constructs that mimic the fibrous microstructure of the ECM, synthetic electrospun fibrous systems have emerged that mimic features of the ECM [11] [15]. For example, there are numerous studies that report the production of soft fibrous materials from synthetic precursors, such as with the electrospinning of molecules such as dextran and HA [16–19], including composites of dextran fibers embedded in gelatin methacrylamide (GelMA) hydrogels [1,15]. These composite systems enable control over fiber fractions and porosity, which can impact cell infiltration, and avoid limitations with construct thickness and cell encapsulation that are common in traditional electrospun scaffolds [17,19,20]. However, most of these systems are fabricated with random orientations of encapsulated fibers, leading to random cell spreading, or are unable to support 3D cell cultures, which do not fully recapitulate the specific fiber orientations, aligned cells, and dimensionality observed in many tissues [15].
To introduce structural directionality within hydrogel constructs, there are a range of approaches that have been developed to direct the alignment of hydrogels, including electrochemical, microfluidic, magnetic, cyclic stretching, and freeze-drying methods [20]. Aligned scaffolds have advantages such as representing more physiologically relevant mechanical properties; however, these methods also have a number of drawbacks, such as they often do not allow the incorporation of cells or that cells are seeded post-processing into scaffolds and often limited to the scaffold periphery [20,21]. This motivates the development of biofabrication techniques that support the processing of cells directly within hydrogel bioinks, where cellular behaviors can be guided through the bioink composition.
In previous studies to guide cell alignment, biofabrication techniques have often embedded and aligned collagen fibers during extrusion bioprinting, including as homogenous bioinks [10,15,22,23] and as composite systems with HA, agarose, or pluronic hydrogels [10,24,25]. These studies supported subsequent cell alignment in the direction of aligned fibers over time, in contrast to traditional bioprinting methods with non-fibrous bioinks, which typically result in random cell organization and spreading within printed filaments [10,15,22–26]. Extrusion bioprinting leverages shear stresses during the printing process to induce both cell and fiber alignment [6,22,27–31]. While these collagen-based examples have shown promising results, collagen has various biophysical properties (such as hydrogel stiffness, fiber density, and pore size) that are challenging to tune independently [15]. Thus, there is also interest in the use of synthetic fibrous systems that are highly tunable to mimic features of the ECM.
In this study, we developed a new method that incorporates synthetic fibers into bioinks, which are aligned during biofabrication to direct cell alignment with culture. Specifically, we produced synthetic microfibers (i.e., synthetically modified norbornene-functionalized HA (NorHA)) with controlled properties (e.g., lengths) that were aligned via shear stress during extrusion bioprinting of a cell-degradable bioink (i.e., GelMA) into agarose suspensions baths (figure 1). As a main component of the ECM, HA was chosen for its high biocompatibility and ease of modification, which allows for significant control over biophysical and biochemical characteristics of the microfibers [2,32,33], whereas GelMA was chosen as it can be photocrosslinked to stabilize aligned fibers, degraded by cells during culture to allow spreading, and has been used extensively in bioprinting [34–36]. The use of NorHA microfibers in this composite system presents a unique approach through the utilization of a fibrous component that can be easily modified and tuned for a variety of applications [2,19].
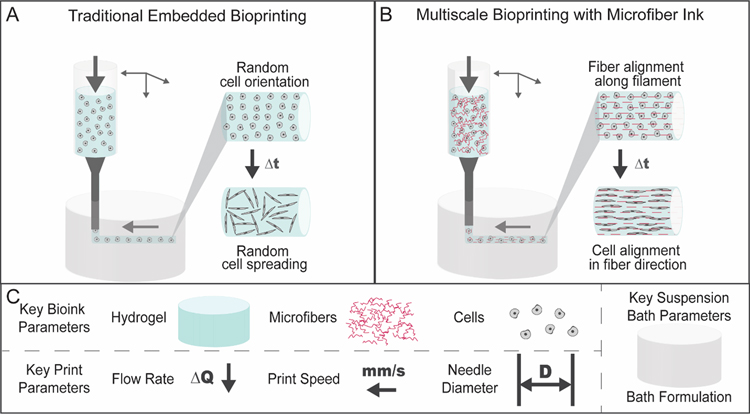
Figure 1.
Schematic of embedded bioprinting processes. (A) Traditional embedded bioprinting where a bioink is extruded from a printer into a suspension bath. If a cell-degradable material is used as a bioink, cells may spread in a random orientation within printed filaments. (B) In multiscale bioprinting with a microfiber bioink, microfibers within the bioink align in the direction of the printed filament during printing in a suspension bath. If a cell-degradable material is used as a bioink, cells may spread with alignment in the direction of printed filaments. (C) A variety of factors may influence bioink printability and impact microfiber alignment during printing, including key bioink parameters (hydrogel, microfibers, cells), key print parameters (flow rate, print speed, needle diameter), and key suspension bath parameters (suspension bath formulation).
The impact of various bioink, suspension bath, and print parameters on filament resolution, as well as fiber alignment along the filament, was explored. Subsequently, the impact of these aligned microfibers on the spreading and orientation of meniscal fibrochondrocytes (MFCs), mensenchymal stromal cells (MSCs) and cardiac fibroblasts (CFs) was then assessed over short-term in vitro cultures, demonstrating the generalizability of this biofabrication method to guide cell alignment across various tissues of interest.
2. Materials and methods
2.1. Materials
All chemicals were used as received unless stated otherwise. Sodium hyaluronate (64 kDa) was purchased from Lifecore Biomedical (Chaska, MN). Dowex™ 500WX8–200 ion-exchange resin (AC335341000) was purchased from Fisher Scientific. GelMA (76292–720) and Seakem® Agarose (12002–102) were purchased from VWR. Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) was purchased from Colorado Photopolymer Solutions (Boulder, CO). Alexa Fluor™ 647 Phalloidin (A22287), LIVE/DEAD™ Viability/Cytotoxicity kit (L3224), and Hoechst 33342 (H3570) were purchased from Thermo Fisher Scientific. Tetrabutylammonium hydroxide (TBA-OH, 86880–100ML), anhydrous dimethyl sulfoxide (276855), 5-norbornene-2-carboxylic acid (446440), 4-(dimethylamino)pyridine (107700), di-tert-butyl dicarbonate (361941), 4-(dimethylamino)pyridine (107700), tetrabutylammonium hydroxide solution (86880), DL-dithiothreitol (D0632), poly(ethylene oxide) (189456), 2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (I2959) (410896), fluorescein isothiocyanate-dextran (average molecular weight 2 MDa, FD2000S-1G) and 3-(trimethoxysilyl)propyl methacrylate (TMSPMA) (440159–500ML) were purchased from Sigma-Aldrich (St. Louis, MO).
2.2. Methods
2.2.1. Material synthesis and NorHA microfiber fabrication
NorHA was synthesized as previously described [37]. Briefly, HA was first converted to tetrabutylammonium salt (HA-TBA) through the use of Dowex 50W proton exchange resin for a two hour exchange reaction. The resin was removed via filtration and the filtrate neutralized to pH ~7.02–7.05 with TBA-OH and subsequently frozen and lyophilized. HA-TBA was modified with norbornene groups via esterification with 5-norbornene-2-carboxylic acid (3 equivalent), 3-(dimethylamino)pyridine(1.5 equivalent) and ditert-butyl decarbonate (0.4 equivalent) for 20 hours at 45 °C under nitrogen. The reaction was then quenched with water and dialyzed against water with 0.25g NaCl/L of DI H2O for 7 days at room temperature and subsequently lyophilized. The degree of modification was 18.8% by 1H NMR (figure S1).
Electrospun fiber mats were prepared as previously described [2,19]. Briefly, solutions of 3.5 wt% NorHA, 2.5 wt% PEO (900 kDa), 0.05 (v/v)% I2959, and 4 mg/ml fluorescein isothiocyanate-dextran (2 MDa) were mixed with a 0.25 stoichiometric ratio of dithiothreitol (DTT) to norbornene groups in DI H2O at 150 rpm for 20 hours protected from light. A solution of 4 wt% PEO (900 kDa) was first loaded to create a base layer with 100 ul of the PEO solution electropsun onto a strip of aluminum foil taped on a rotating mandrel (~350 RPM) with the following parameters: 15–30% humidity, 28–30 kV applied voltage, 4 kV deflector voltage, −5 kV collector voltage, 18 cm distance from needle to collector, 18 gauge needle and a 0.7 mL/hr flow rate. Next, the NorHA solution was loaded and electropsun with the following parameters: 15–30% humidity, 28–30 kV applied voltage, 4 kV deflector voltage, 5 kV collector voltage, 19 cm distance from needle to collector, 18 gauge needle and a 0.7 mL/hr flow rate. To crosslink, scaffolds were exposed (in a dry state) to 10 mW/cm2 UV light for 2 hours under an inert atmosphere (N2).
To fabricate microfibers, fiber mats were cut into approximately 1 mm3 pieces and hydrated for 30 minutes in phosphate buffered saline (PBS) (figure 2(A)). After hydration, mats were sheared by repeatedly passing the solution through a needle. Mats were first sheared through an 18 gauge needle (40x), then a 21 gauge needle (40x), and finally a 23 gauge needle (40x). After fragmentation, the microfiber solution was filtered first through a 40 μm cell filter (BD, 352340), then through a 5 μm pluriStrainer® (Pluriselect). The remaining solution was collected, centrifuged at 18000 RCF, and stored at 4 °C for up to two months while protected from light. Microfibers were imaged with an Olympus BX51 microscope and average lengths were measured via ImageJ software analysis. Fiber concentration was estimated with a hemocytometer.
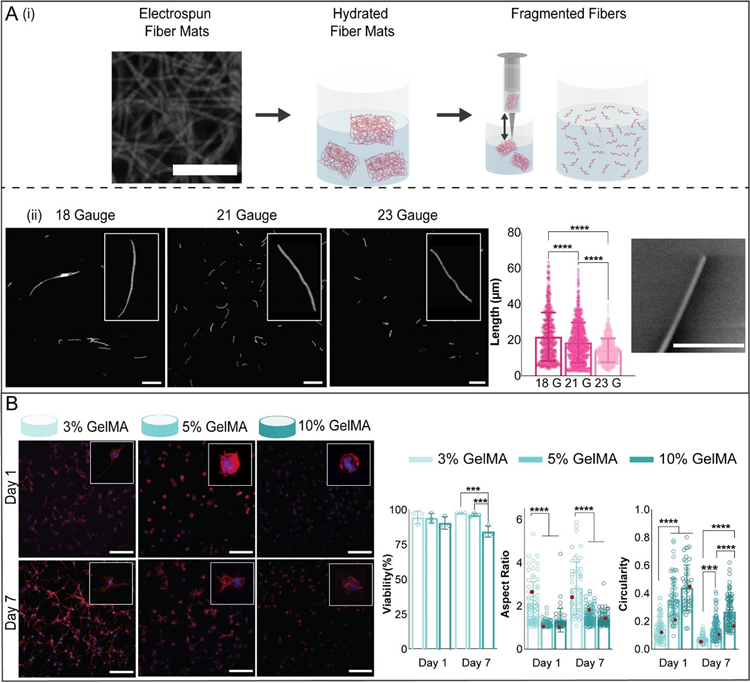
Figure 2.
Microfiber and hydrogel formulations. (A) (i) To fabricate microfibers, electrospun norbornene-HA (NorHA) fiber mats (scale bar 10 μm) were crosslinked, hydrated, sectioned into pieces, and fragmented into microfibers by repeatedly passing through a needle. (ii) Fiber length was varied by varying the needle gauge used to shear fibers. Representative images of suspensions of fragmented fibers for three different needle gauges are shown (left, scale bar 0.1 mm, along with quantification of fiber length (middle) and an SEM image showing the end of a fragmented fiber (right, scale bar 2 μm, mean ± s.d., one-way ANOVA with Tukey post hoc, ****p ≤ 0.0001). (B) Representative images of meniscal fibrochondrocytes (MFCs) stained for F-actin (red) and cell nuclei (blue) (200 μm z-stacks, scale bars 0.1 mm) after culture in 3, 5, and 10% GelMA hydrogels for 1 and 7 days. Quantification of MFC viability (analyzed with live-dead staining), aspect ratio, and circularity after 1 and 7 days of culture (3% GelMA: n= 52 (Day 1), 68 (Day 7) cells; 5% GelMA: n= 64 (Day 1), 86 (Day 7) cells; 10% GelMA: n= 46 (Day 1), 89 (Day 7) cells; 3 biologically independent experiments, mean ± s.d., two-way ANOVA with Bonferroni post hoc ****p ≤ 0.0001, ***p ≤ 0.001, red dots indicate measurements for magnified images).
2.2.2. Scanning electron microscopy (SEM) imaging
To visualize fiber morphology after fragmentation, droplets of dilute (1% v/v) fiber solutions were snap frozen, using a bath of 2-methylbutane cooled in liquid nitrogen, and lyophilized overnight. The lyophilized material was then visualized using an environmental SEM (FEI Quanta 600 ESEM). Single fibers and ends of fibers were identified near the edge of the sample.
2.2.3. Cell isolation, culture and staining
To isolate meniscal fibrochondrocytes (MFCs), medial menisci were harvested from juvenile bovine knee joints (Research 87, 2–3 months old, Boyle, MA), minced into roughly 1 mm3 pieces and incubated at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin streptomycin [38]. Cells emerged from the tissue over 2 weeks, and MFCs at passage 3 or lower were used throughout the experiments. Mesenchymal stromal cells (MSCs) were isolated from the bone marrow of bovine femora and tibiae (Research 87, Boylston, MA), as previously described [39,40]. Then, MSCs were expanded in DMEM with 10% FBS and 1% penicillin streptomycin. MSCs at passage 3 or lower were used throughout the experiments. Human cardiac fibroblasts (CFs) were purchased from Promocell. CFs were expanded in alpha-modified essential medium (α-MEM) with 10% FBS, 1% penicillin/streptomycin and 5 ng ml−1 basic fibroblast growth factor.
Cell viability in GelMA concentration studies was assessed with calcein AM/ethidium homodimer (1, 7 days) according to manufacturer’s instructions (Invitrogen). For bioprinting studies, cell viability was assessed with Hoechst 33342 and ethidium homodimer (1, 7 days). Remaining samples were fixed with 4% formalin and actin and DNA were labeled with Alexa Fluor 647 phalloidin (Thermo Fisher) and Hoechst 33342, respectively. Confocal images (Leica SP5) of stained constructs were analyzed using Image J software to assess cell viability, aspect ratio, and circularity. Viability was calculated as (total cells – dead cells)/total cells within a single image. Fiber and actin orientations were analyzed through confocal images (Leica SP5) of stained constructs with a modified Python version of FiberFit Software [41,42]. For each group, 200 μm stacks with 5 μm step sizes and 490 × 490 μm2 image frames were used for quantification (n ≥ 3 gels) (supplementary videos S1–S4).
2.2.4. Bioink and suspension bath formulations
GelMA was sterilized with germicidal lamp radiation in a laminar flow hood for 30 minutes before dissolving in sterile solutions of photoinitiator (0.05 wt% LAP) and phosphate buffered saline (PBS) for a final concentration of 5 wt%, unless otherwise stated. To dissolve GelMA, solutions were heated to 37 °C for 40 minutes. When desired, fibers were added at concentrations of 430 × 106 mL−1 and cells were added at concentrations of 5 × 106 mL−1 unless otherwise stated. The formulation was added to a 1 mL syringe (BD, 309628) and loaded into the printer.
For agarose suspension baths, 0.5 wt% agarose was added to DI H2O and autoclaved at 120 °C for 1 hour on the liquid cycle [43]. Immediately after autoclaving, the solution was placed on a stir plate and sheared at 700 RPM until the solution cooled to 25 °C. This stock solution was then stored at 4 °C and used for up to 3 months. Prior to bioprinting, the solution was diluted to 0.25 wt% with sterile PBS unless otherwise stated. After dilution, the solution was centrifuged at 500 g for 5 minutes prior to the addition to printing wells.
2.2.5. Shear oscillatory rheometry
Measurements were performed with an AR2000 stress-controlled rheometer (TA instruments) with a 20 mm parallel plate geometry set at a 1 mm gap at 25 °C. For flow characterization, viscosity was measured using a continuously ramped shear rate (0 to 100 s−1). To characterize bulk gelation, time sweeps (1.0 Hz, 1.0 % strain) at 25 °C were performed with exposure to visible light (Exfo Omnicure S1500 lamp, 400–500 nm filter) for 5 min at 10 mW/cm2 intensities. Cell-laden bioinks were tested with MFCs at a concentration of 5 million cells/mL. Bioink rheological properties were determined using the Herschel-Bulkley fluid model, which is given as:
Where τ is the shear stress, is the shear rate, τ0 is the yield stress, k is the consistency index, and n is the flow index. The model was fit to empirical data from rheological flow characterizations.
2.2.6. Finite element simulation of bioprinting flow rates and shear stresses
To minimize computation time and due to geometrical symmetry of the bioprinting syringe and needles, 2D representations were created to model bioink flow rate and shear stress with COMSOL Multiphysics (Version 5.5, Stockholm, Sweden) and based on measurements provided by manufacturers. The center line of the syringe and needle was specified as the axis of symmetry for the model (figure 3(B)). To represent the non-Newtonian flow behavior of the bioinks, a Herschel-Bulkley, shear rate-dependent relationship was applied within the numerical solver. The density of bioinks was assumed to be 1 g/mL and parameters in the model (τ0, K, n) were obtained from flow rate curves obtained from rheometry measurements. Density assumptions were based on settings used in previous publications and were confirmed via assessments of the density of 5% GelMA bioinks with and without fibers (figure S2) [44]. Flow rates at the syringe inlet and an outlet pressure of zero were set as the initial conditions. Boundary conditions of no slip flow at the syringe and needle walls except for inlet or outlet were applied. The computational domain was discretized using a physics-controlled mesh with the ‘fine’ meshing option, resulting in 36,224 elements. Bioinks were modeled as incompressible fluids, and velocity and shear stress profiles within printing needles were recorded for all simulations.
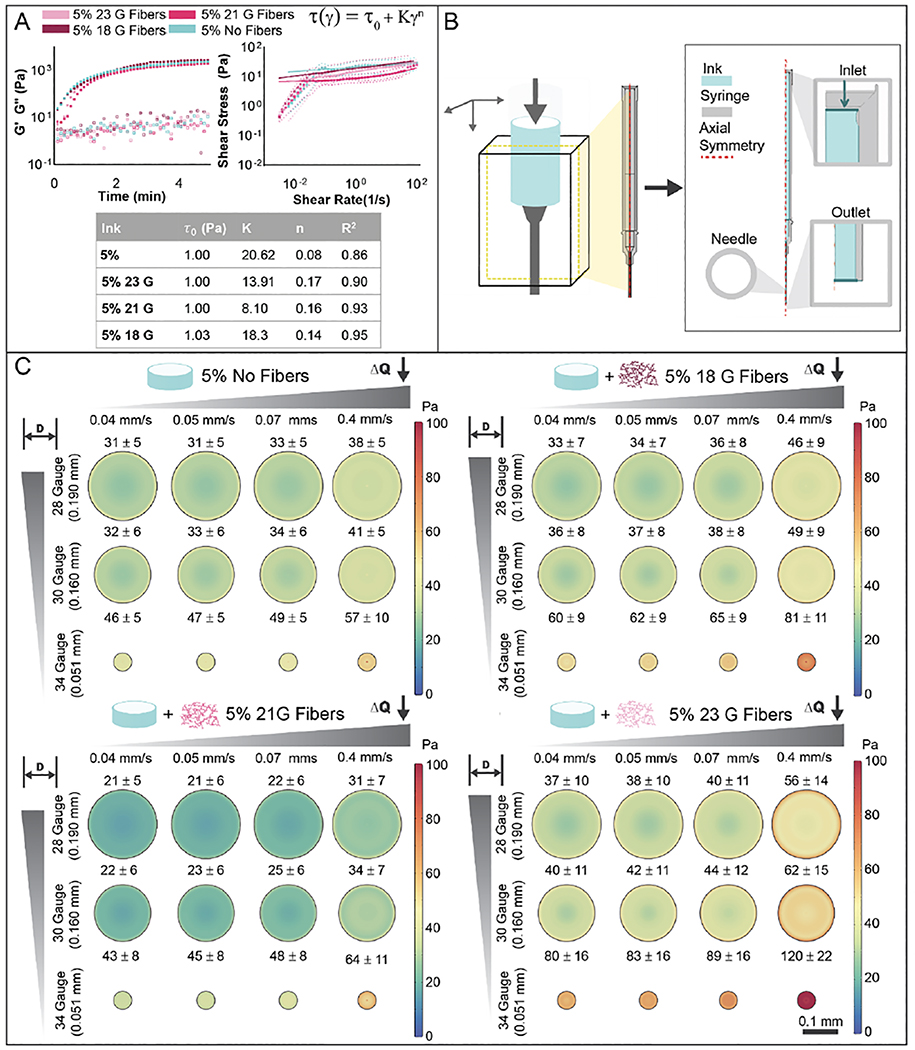
Figure 3.
Bioink rheology and shear during extrusion. (A) Rheological characterization of bioinks (5% GelMA, with 18, 21, 23 gauge fibers or no fibers), including photorheology (1 Hz, 1% strain, storage (G’) and loss (G”) moduli) during photocrosslinking with visible light (400–500 nm, 10 mW/cm2) and shear stress with increasing shear rate (0 – 100 s−1). The data was fit to Herschel-Bulkley model and parameters are shown in the table. (B) Representative illustration of the fluid dynamics model used to calculate theoretical shear stresses and flow during printing from a syringe with a needle. Due to geometrical symmetry of the printing syringe and needle, a simplified 2D model was used for calculations. (C) Theoretical calculations of shear stress across needle profiles for various needle diameters and flow rates, shown for 5% no fiber (top left), 5% 18 gauge fibers (top right), 5% 21 gauge fibers (bottom left), and 5% 23 gauge fibers (bottom right) bioinks.
2.2.7. Hydrogel bioprinting
CAD models of print designs were created in SolidWorks and exported as STL files. These files were then uploaded to Repetier Host and converted to G-Code via Slic3r, then uploaded to Allevi bioprinting software. Print design files are included in Supplementary Information (figure S3, supplementary files S1–S10). Syringes containing bioinks were loaded into an Allevi 2 bioprinter (Allevi, Philadelphia, PA). Unless otherwise stated, prints were completed with 34 gauge needles (JG34–0.25HPX, Jensen Global). To minimize material loss from syringe dead space, custom fittings were fabricated for 1 mL syringes for the printer (objects printed courtesy of the University of Pennsylvania Libraries’ Biomedical Library). Fitting designs and fabrication information is provided in Supplementary Information (figure S3, supplementary file S11). All prints were completed at room temperature (25 °C). If necessary, syringes were removed and chilled on ice or warmed in the printing extruder until extruder temperature readings reached 25 °C. All prints with bioinks containing cells were completed under aseptic conditions. Extruders were calibrated on TMSPMA-glass slides prior to the addition of agarose suspension media to printing wells. For multimaterial prints, an Arducam video camera controlled by a Raspberry Pi was used to aid in calibration of multiple extruders (Arducam for Raspberry Pi, Raspberry Pi Model B+). This camera was also used to record printing videos and capture images for line width analysis. Extrusion flow rates and printing speeds were controlled through Allevi software. After constructs were printed, they were crosslinked with visible light (Exfo Omnicure S1500 lamp, 400–500 nm filter) at 10 mW/cm2 intensity for 5 minutes. After crosslinking, suspension media was removed and constructs were washed 3 times with PBS prior to addition of cell culture media.
Non-printed controls were fabricated by crosslinking 15 μL volumes of bioinks on TMSPMA-treated 10 mm glass slides (NC1540971, Fisher Scientific) and crosslinking with visible light (Exfo Omnicure S1500 lamp, 400−500 nm filter) at 10 mW/cm2 intensity for 5 minutes. After crosslinking, samples were immediately rinsed with PBS 3 times.
2.2.8. Statistical analysis
All statistics were performed using GraphPad Prism 8 software or package ‘circular’ in R for circular data [45]. All data are reported as mean ± standard deviation, and n ≥ 3 unless specified otherwise. Comparisons for non-circular data were analyzed using one-way ANOVA with post hoc testing or two-way ANOVA when comparing effects of multiple independent variables. Bonferroni correction was implemented for multiple comparisons with α=0.05. Statistical comparisons between two experimental groups were assessed via two-tailed Student’s t-tests. Comparisons for circular data were analyzed with Watson-Wheeler test. For all samples, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant.
3. Results and discussion
3.1. Microfiber and hydrogel formulations
NorHA was electrospun into scaffolds and processed using a fragmentation method, where pieces of the soft fibrous scaffolds were passed repeatedly through a needle to obtain microfiber suspensions (figure 2(Ai)). NorHA was chosen due to the biocompatibility of HA-based hydrogels and the simple processing using a thiol-ene reaction with a di-thiol crosslinker [2,32,33]. Versatility of fiber lengths from the same electrospun scaffold formulation was demonstrated through the use of varied needle sizes of 18 gauge (0.840 mm), 21 gauge (0.820 mm), or 23 gauge (0.640 mm), with average fiber length decreasing with smaller needle diameters (figure 2(Aii)). Three separate batches of fragmented 23 gauge NorHA microfibers were analyzed for average fiber length, with no significant differences observed among the average fiber lengths measured for each batch (figure S4(A)).Generally, the fiber suspensions had average lengths less than ~ 20 μm and were heterogeneous in distributions. These results suggest minimal batch-to-batch variability in fiber length, demonstrating the robustness of this microfiber fabrication method. SEM imaging was used to qualitatively assess the morphology of the ends of the soft microfibers (figure 2(Aii)). One of the benefits to the fragmentation approach is that it is independent of the material used and potentially could be utilized to process a wide range of electrospun polymers into fibers.
With regards to the bioink, GelMA was investigated due to the long history of use in bioprinting and that GelMA supports cell-mediated degradation and is photocrosslinkable [34–36]. Cell viability and morphology were assessed in 3, 5, and 10 % concentrations of GelMA (figure 2(B), figure S4(B)), using MFCs as a model cell type. As has been demonstrated in previous studies, GelMA concentration impacted cell viability and morphology [35,46]. No significant differences were observed for cell viability after 1 day of culture; however, cells encapsulated in 3 and 5 wt% gels demonstrated higher viability when compared to cells encapsulated in 10% gels after 7 days of culture. All GelMA formulations supported an average cell viability above 80% and some level of spreading after 1 or 7 days of culture. Cells encapsulated in 3% gels had higher aspect ratios than cells in 5% and 10% gels at both timepoints and demonstrated a lower circularity after 1 day in culture. By day 7, cell circularity decreased with decreasing GelMA concentration. Based on these trends in viability, aspect ratio, and circularity and the design criteria that the bioink support spreading, only 3% and 5% gels were investigated further in subsequent experiments.
3.2. Bioink rheology and shear during extrusion
To assess the rheological properties and photopolymerization behavior of bioinks, bioinks prior to and during gelation were monitored experimentally via standard rheological testing and photorheology, respectively (figure 3(A), figure S5(A), figure S6(Ai)). Prior to irradiation with visible light, bioinks subjected to shear at 1 Hz and 1% strain exhibited storage (G’) and loss (G”) moduli on the order of 0.1 to 2 Pa. For flow characterization, viscosity was measured using a continuously ramped shear rate (0 to 100 s−1). From these results, shear stress was plotted against shear rate and experimental data was fit to a Herschel-Bulkley model (figure 3(A), figure S5(A), figure S6(Ai)). Herschel-Bulkley models have often been employed to model non-newtonian bioinks and capture the impact of yield stress and shear-thinning properties of non-newtonian fluids [47–51]. Curve fits were found to have R2 values above 0.85 for all bioinks. All flow index values were less than 1, suggesting that all bioinks exhibit shear-thinning properties. Flow index values increased with decreasing GelMA concentration, indicating greater shear-thinning properties for 5% bioinks with and without fibers compared to 3% bioinks with and without fibers. Higher yield stress values (τ0) in 5% bioinks with and without fibers also suggest that these bioinks may exhibit larger plug flow regions and may have higher shape fidelity when compared 3% bioinks with and without fibers. There were only minor differences in rheological properties when bioinks were compared with and without fibers, including with fibers of varying lengths, or when cells were incorporated into the bioinks. This is particularly important as bioprinting parameters are not likely to deviate much with such changes in formulations. Once exposed to visible light, the GelMA underwent crosslinking, with an increase in G’ ranging from 47 – 68 Pa for 3% GelMA bioinks with (3% fiber bioink) and without (3% bioink) fibers, and 1830 – 3000 Pa for 5% GelMA bioinks with (5% fiber bioinks with 18, 21, or 23 gauge fibers) and without (5% bioink) fibers. Again, there was minimal impact on these outcomes when the fiber lengths were varied or when cells were included within the bioinks.
To further understand the impact of bioink properties on the printing process and fiber alignment, velocity and shear stress profiles within printing needles were simulated with COMSOL Multiphysics for various inlet flow rates and needle diameters (figure 3(B)). Shear stress is a critical parameter as it can impact cell viability and bioactivity, as well as fiber alignment during printing [22,44]. While increased shear stresses have correlated to higher fiber alignment in previous studies, if shear stress values are too high, they can reduce cell viability and function. As expected, shear stress increased for increasing flow rates and decreasing needle diameters for all bioinks (figure 3(C), figure S5(B), figure S6(B)). 3% fiber bioinks resulted in higher shear stresses at the same flow rate and needle diameter when compared to 5% fiber bioinks (figure S6(B)). While shear stresses varied based on bioink composition, average needle flow rates remained constant across bioinks, with increasing inlet flow rates correlating to increasing average needle flow rates. While average flow rates remained constant across bioinks, 3% bioinks with and without fibers exhibited higher maximum flow rates at the center of printing needles, while 5% bioinks with and without fibers exhibited more uniform flow rates, indicative of plug flow, across printing needles (figure S6(Aii,Aiii)) [47]. Higher yield stress values and lower flow behavior index values are likely attributed to differences in these shear stress and velocity profiles [47]. All calculated average shear stresses remained below previously reported values of around 5 kPa that are reported to result in a decrease in cell viability [44]. As 34 gauge needles (0.051 mm diameter) resulted in the highest shear stresses, this needle was chosen for further experimental testing.
3.3. Bioink printing parameters for fiber alignment
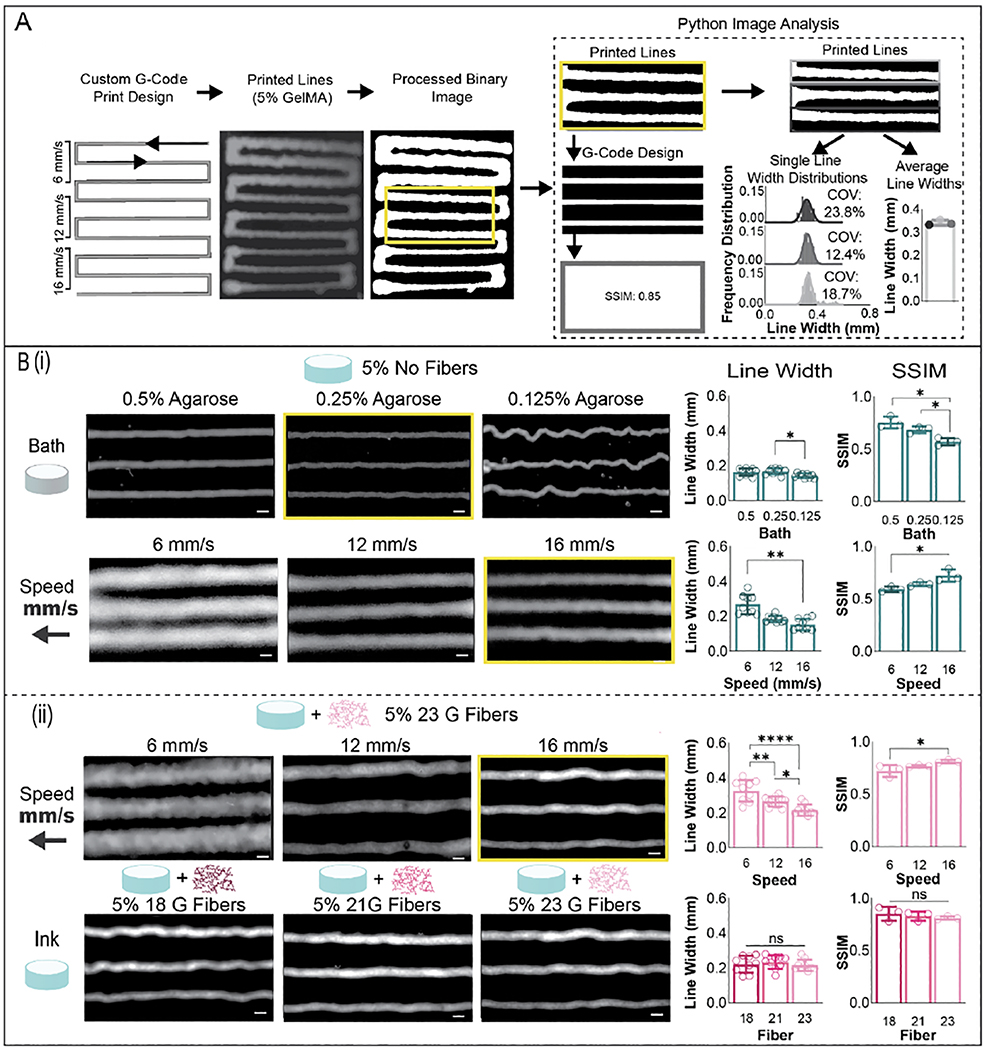
As the calculated yield stress values were low, especially for 3% bioinks with and without fibers, a suspension bath was utilized to maintain bioink shape fidelity and fiber orientation prior to crosslinking [52]. Specifically, an agarose suspension bath was chosen due to low cost and ease of fabrication [43,53]. Average line width, coefficient of variation of line widths, and structural similarity index (SSIM) of printed lines compared to theoretical print designs were determined with Python image analysis based on images taken after printing (figure 4(A), supplementary video S5). Three separate prints were completed across 3 suspension bath formulations, while keeping the bioink formulation (5% bioink), print needle (34 gauge), inlet flow rate (0.04 mm/s), and print speed (16 mm/s) constant (figure 4(Bi)). While the lowest concentration of agarose suspension bath tested (0.125 wt%) resulted in the smallest line width, bunching of the print filament in the suspension bath led to deviations from the print path, creating inaccuracies. These inaccuracies are apparent in a lower average SSIM compared to the 0.25 wt% and 0.5 wt% bath formulations. While there were no statistical differences in line width or SSIM when comparing the two higher bath concentrations, the 0.25 wt% formulation resulted in a lower average coefficient of variation (10.26%) per printed filament when compared to 0.5 wt% formulation (18.10%), and as such, was used for all subsequent studies. Next, various print speeds were tested while keeping the bioink formulation (5% GelMA), print needle (34 gauge), inlet flow rate (0.07 mm/s), and suspension bath (0.25% agarose) constant. Print speeds of 16 mm/s resulted in the smallest average line widths, the highest SSIM and the lowest coefficient of variations per printed filament (15.20%, figure S7(B)).
Figure 4.
Bioink printing parameters for fiber alignment. (A) Method for analyzing printed line widths. Custom gcode files were created to print a single line in which the translation speed is varied during the print, with 3 lines printed at each translation speed. Images were taken of prints with an Arducam Raspberry Pi Camera and subsequently processed into binary images in ImageJ. These binary images were then segmented and analyzed in Python. Images of lines were also compared against images representing the theoretical print design via structural similarity index (SSIM), which was determined via Python. Printed line widths across each line were analyzed pixel by pixel, resulting in outputs of line width distributions for each individual printed line and average line widths for each print setting tested. Average line widths were calculated for 3 separate prints for each print setting, for a total of n=9 printed lines assessed for each print setting tested. (B) (i) Impact of suspension bath formulation and print speed on filament line width, shown for the 5% bioink without fibers. (ii) Impact of print speed and fibers on filament line width, shown for 5% GelMA (mean ± s.d., one-way ANOVA with Tukey post hoc *p≤ 0.05, ** p≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
These same print settings were then tested with 5% GelMA inks with 23 gauge fibers, and similar trends were seen in this fiber ink as were observed in 5% GelMA inks without fibers. Print speeds of 16 mm/s resulted in the smallest average line widths, and the highest SSIM (figure 4B ii). As needle flow rate is expected to be consistent across bioinks, similar filament widths are expected when using the same print settings. Average line widths were assessed for various bioinks while keeping print needle (34 gauge), inlet flow rate (0.07 mm/s), suspension bath (0.25% agarose), and print speed (16 mm/s) constant (figure 4(Bii), figure S7(A)). No variations were detected in printed filament line width or SSIM for 5% bioinks when varying fiber lengths were incorporated into the bioink (figure 4 B ii). Some variations in print filament line widths were observed in 5% bioinks with and without fibers as well as in bioinks with 3% or 5% GelMA, likely due to differences in bioink shape fidelity, which is impacted by bioink yield stress, and bioink shear recovery, which is not accounted for in Herschel-Bulkley computational models (figure S7(A)) [47,52].
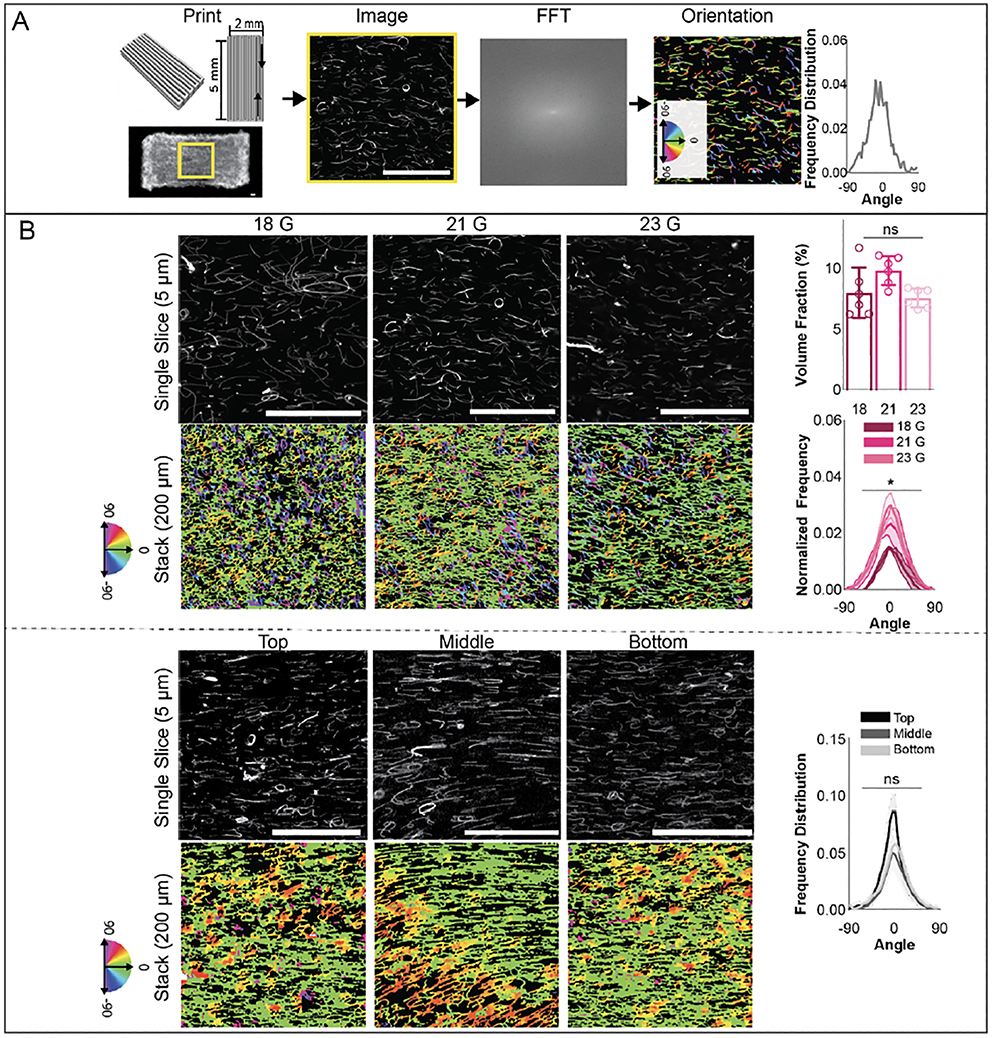
Based on these results, rectangular print designs of 2 mm × 5 mm × 0.4 mm were fabricated, with variations in filament width and spacing as needed (figure 5(A), supplementary video S6). Printed filaments were deposited in a single direction and the spacing between filaments was minimized to create a solid construct. Designs were printed with various print settings, bath formulations, or bioinks and fiber alignment along printed filament direction was assessed via a modified version of FiberFit Software (figure 5(A)). First, alignment in 5% GelMA constructs with varying fiber lengths was assessed. Constructs with 18 gauge fibers had slightly lower alignment in the filament direction compared to those made with 21 gauge or 23 gauge fibers, while no statistical differences in volume fraction were found among any of these groups (figure 5(B)). These results fit with previous work that has demonstrated an impact of fiber length on orientation [54,55]. Due to this increased alignment, 23 gauge fibers were used for all future studies. Next, 2 mm × 5 mm × 0.6 mm constructs were printed and alignment was assessed in the top, middle, and bottom of the construct. No statistical differences in alignment were found, suggesting uniform fiber orientation throughout constructs (figure 5(B)).
Figure 5:
Fiber alignment within printed constructs. (A) (left) Schematic of rectangular print design for a 2 × 5 × 0.4 mm construct and image of printed construct, used for assessment of fiber orientation. (middle) Image slices in confocal stacks were converted to binary images in ImageJ, uploaded to FiberFit software and analyzed by conversion to a discrete fourier transform (FFT), then to a power spectrum. Finally, a band pass filter was applied and orientation outputs exported. (right) Fiber orientation was also visualized via color-coded images by plotting fiber orientation from z-stacks through Quanfima software analysis and average orientations for each confocal stack were calculated. 3 confocal stacks for each group were analyzed and uploaded to R for circular statistical analysis. (B) (Top) Images of fibers (left) and quantified fiber orientation and volume fraction (right) along printed filaments for constructs fabricated with 18, 21 and 23 gauge fibers. (Bottom) Images of fibers (left) and quantified fiber orientation (right) at top, middle, and bottom depths for 600 μm thick constructs fabricated with 23 gauge fibers. (200 μm z-stacks, scale bars 0.1 mm).
The impact of GelMA concentration, printing flow rate, fiber volume fraction, and bath concentration on orientation were also assessed (figure S8). Increasing inlet flow rates correlated to increases in fiber alignment along the printed filament direction for both 3 wt% and 5 wt% GelMA bioinks (figure S8(A)) [41,42]. However, higher flow rates were needed with 5 wt% GelMA bioinks to achieve alignment, potentially due to lower shear stresses in 5 wt% GelMA bioinks during the printing process. To test the tunability of the system and potential dependence on fiber concentration, a lower fiber concentration was also tested with 3% GelMA at 0.07 mm/s, and similar trends of fiber alignment at these print settings were observed with the lower fiber volume fraction (figure S8(B)). As the addition of fibers resulted in minimal changes in bioink rheological properties based on the fitted Herschel-Bulkley models (figure 3A), similar shear stresses are likely observed at both fiber concentrations, leading to similar degrees of fiber alignment. Finally, differences in fiber alignment were also assessed in 5% GelMA constructs printed in varying agarose bath concentrations. Significantly lower fiber alignment was found in 0.125% Agarose baths, likely due to instability of printed filaments (figure 4, figure S8(C)).
The use of a computational model prior to experimental printability testing allowed for expedient analysis of a wide range of print settings, predicting flow rate and shear stress profiles for a total of 96 separate print settings with 8 different bioink formulations. These results allowed for a narrower range of inlet flow rates and needle gauges to be tested experimentally, minimizing use of materials and speeding up the process to determine optimal print parameters for each bioink. While some variations in line width were observed with different bioinks at the same print settings, relatively minimal adjustments for each bioink were needed to optimize print settings for each formulation. Furthermore, trends in increased shear stresses within printing needles correlated to increased fiber alignment in experimental results. As predicted, larger inlet flow rates were needed to align fibers in 5% fiber bioinks when compared to fibers in 3% fiber bioinks. These results are in agreement with previous work that has demonstrated increased shear stresses during printing result in increased collagen fiber alignment [10,22,23]. These results suggest that this simplified 2D computational model, which allows for quick computations (< 1 minute per setting) and applies rheological data commonly collected for most bioinks, is useful for predicting trends in fiber alignment. While outside the scope of this work, values of calculated shear stress and flow rate profiles that correlated to fiber alignment in this study may be used to determine print settings needed for alignment in a variety of composite bioinks, such as with different hydrogels, further decreasing the need for extensive, time-consuming experimental testing and providing a greater understanding of the impact of bioink rheological properties on the bioprinting process.
3.4. Fiber and cell orientation within printed constructs
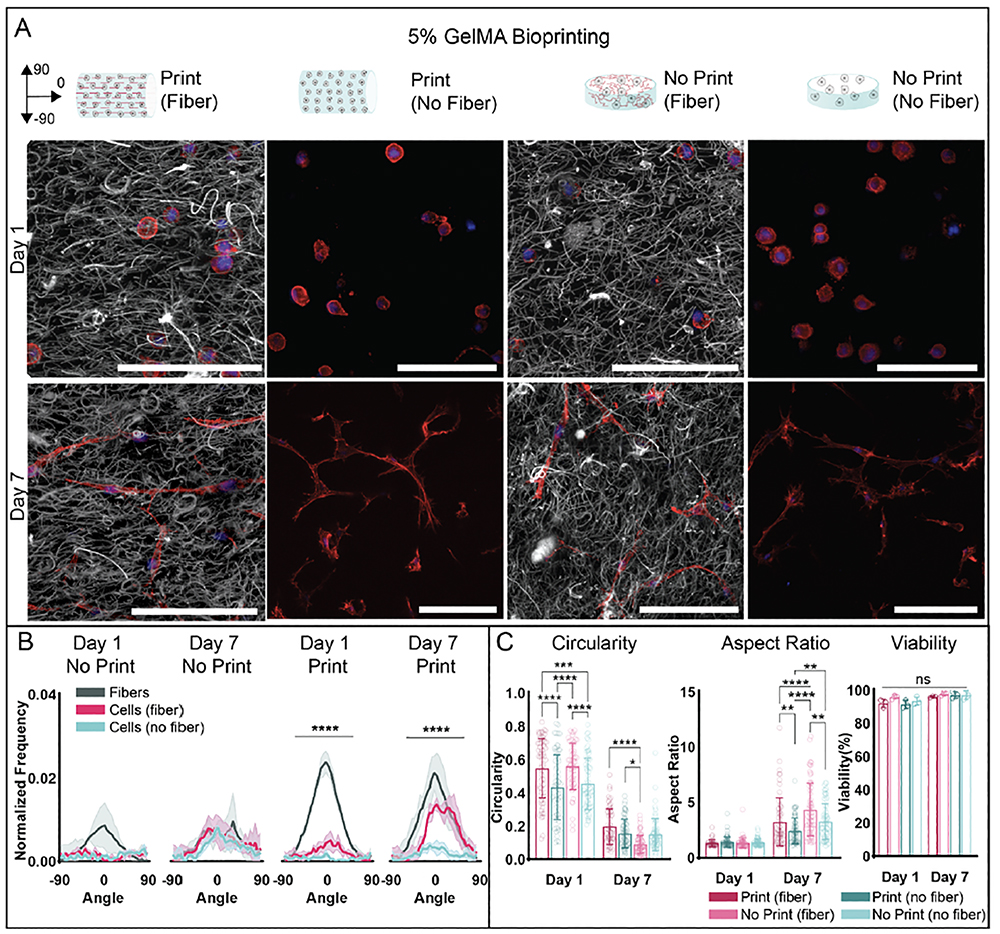
As 3% GelMA allowed for the highest level of cell spreading (figure 2(B)) and resulted in high fiber alignment at specific flow rates (figure S8(C)), 3% GelMA bioinks with and without 23 gauge fibers were first selected for testing with cells. As a proof of concept, MFCs were chosen as the cell type for potential future applications in meniscal tissue engineering. The same rectangular print design of 5 mm × 2mm × 0.4 mm constructs used in acellular fiber alignment testing was used for all printed constructs, and the same print settings (inlet flow rate 0.07 mm/s, 34 gauge needle, 0.25% agarose suspension bath, 16 mm/s speed) were used for all prints. Non-printed constructs of 3% GelMA with and without fibers were fabricated as control groups.
One day after printing, printed fibrous constructs demonstrated fiber alignment along printed filaments, while nonprinted fibrous constructs demonstrated a random fiber orientation (figure S9(A–B)). Cells in all groups demonstrated a random actin orientation at day 1. Cell viability remained high across all groups (>90%), with no statistical differences among any of the groups (figure S9(C), figure S10(A)). While there was some concern in regards to viability as small needle diameters were used in this study to provide higher shear stresses for fiber alignment, these results demonstrate that the printing process has minimal impact on cell viability. This high cell viability is in agreement with previous studies that demonstrate printing shear stresses below 5 kPa does not have a negative impact on cell viability, especially when cells are encapsulated in shear-protective hydrogels [44,56]. Additionally, there were no statistical differences in circularity or aspect ratio between fibrous and non-fibrous printed constructs. Fibrous non-printed constructs resulted in the lowest circularity at day 1, while non-fibrous non-printed constructs also exhibited lower circularity when compared to printed constructs and higher aspect ratios compared to printed non-fibrous constructs. Viability remained high across all groups at day 7 (>90%), with no statistical differences across groups or timepoints (figure S9(C), figure S10(A)). Again, these viability results agree with computational modeling estimates that predicted shear stress values during printing were below those previously reported to impact viability. Circularity at day 7 appeared lower in non-printed constructs when compared to fibrous printed constructs, while aspect ratio in printed fibrous constructs appeared higher when compared to printed non-fibrous constructs and non-printed fibrous constructs. Within each group, circularity decreased and aspect ratio increased from day 1 to day 7, suggesting cell spreading over time in all groups.
With regards to fiber orientation, this decreased in fibrous printed constructs unexpectedly by day 7 (figure S9(B)). Upon closer inspection, fiber volume fraction in printed constructs appeared to increase when compared to both fiber volume fraction in printed constructs at day 1 and fiber volume fraction in non-printed fibrous constructs at day 1 and day 7 (figure S10(B)). Additionally, printed fibrous constructs appeared to significantly decrease in volume when compared to day 1 printed constructs and all other groups at day 7 (figure S10(B)). Because this compaction only occurred in constructs with initial fiber alignment, this result suggests potential remodeling by cells that is directed by fiber orientation, which may be useful for 4D bioprinting applications [57,58]. While outside the scope of this work, the cause of this compaction and potential applications may be explored in future studies. While slight increases in cell actin orientation along printed filament direction were observed in day 7 printed fibrous constructs when compared to day 1, these increases were not statistically different from the random cell actin orientations observed in printed non-fibrous constructs and in non-printed groups (figure S9(A,B)) ((n=3 for each group, mean ± s.d., one-way ANOVA with Tukey post hoc for non-circular data, Watson-Wheeler test for circular data, *p ≤ 0.05, ns = not significant) Due to decreases in fiber alignment along printed filaments and macroscopic construct compaction over 7 days of culture in 3% fiber bioinks, 5% fiber bioinks were next tested to guide MFC behavior. Previous work has demonstrated that higher GelMA concentrations leads to constructs with slower degradation and lower permeability, so constructs are expected to be more stable [36,59]. The same rectangular print design of 5 mm × 2mm × 0.4 mm constructs used in 3% bioink studies was used for all printed constructs, and the same print settings (34 gauge needle, 0.25% agarose suspension bath, 16 mm/s speed) were used for all prints. Inlet flow rate was adjusted slightly for non-fibrous bioinks (0.4 mm/s for fibrous bioinks, 1.15 mm/s for non-fibrous bioinks) so that calculated shear stress values within printing needles were within similar ranges and no statistical differences in printed line widths were observed (figure S7(C), supplementary video S6).
After one day in culture, printed fibrous constructs demonstrated fiber alignment along printed filaments, while nonprinted fibrous constructs maintained a random fiber orientation (figure 6(A,B)). Cells in all groups demonstrated a random actin orientation at day 1. Cell viability remained high across all groups and timepoints, with no statistical differences observed among groups (>90%) (figure 6(C), figure S11(A)). Cells in printed and non-printed constructs exhibited higher circularity when compared to non-printed constructs at day 1. No statistical differences in circularity between printed and non-printed fibrous constructs or printed and non-printed non-fibrous constructs were found. There were no statistical differences in aspect ratio among any groups at day 1. Circularity in fibrous non-printed constructs was lower when compared to circularity in printed constructs at day 7, while there were no statistical differences in circularity between printed constructs or between non-printed constructs (figure 6(C)). Aspect ratio was higher in printed fibrous constructs compared to printed non-fibrous constructs and was also higher in non-printed fibrous constructs compared to non-printed non-fibrous constructs. Aspect ratio was also higher in non-printed fibrous constructs when compared to printed fibrous constructs and higher in non-printed non-fibrous constructs compared to printed non-fibrous constructs. Within each group, circularity decreased and aspect ratio increased from day 1 to day 7, suggesting cell spreading over time in all groups. At day 7, fiber orientation in printed constructs along printed filaments was maintained (figure 6(B), supplementary videos S1–S4), in contrast to what was observed with 3% fiber bioinks. Cell actin orientation in the direction of printed filaments also increased compared to cell actin orientation in printed non-fibrous constructs. Cell actin orientation and fiber orientation in all non-printed constructs remained random.
Figure 6:
Fiber and MFC orientation within printed constructs from 5% GelMA bioinks with and without fibers. (A) Representative schematics and images of constructs with fibers (grey) and MFCs (F-actin: red, cell nuclei: blue) for printed and non-printed constructs (30 μm z-stacks, scale bars 0.1 mm). (B) Quantification of fiber and cell alignment along printed filament direction or within non-printed constructs 1 and 7 days after culture (n=3 for each group, mean ± s.d., Watson-Wheeler test ****p ≤ 0.0001, *** p ≤ 0.001). (C) Quantification of MFC viability (analyzed with Hoechst 3342 and ethidium-homodimer 1 staining), aspect ratio, and circularity after 1 and 7 days of culture (5% Fiber Print: n = 46 (Day 1), 61 (Day 7) cells; 5% No Fiber Print: n = 67 (Day 1), 73 (Day 7) cells; 5% Fiber No Print: n = 60 (Day 1), 68 (Day 7) cells; 5% No Fiber No Print: n = 58 (Day 1), 66 (Day 7) cells; 3 biologically independent experiments, mean ± s.d., two-way ANOVA with Bonferroni post hoc *p≤ 0.05, ** p≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
These results demonstrate that in 5% fiber bioinks, printed microfibers supported MFC alignment over time, with significant differences in cell actin orientation between printed constructs with and without fibers by 7 days of culture. As alignment was not observed in nonfibrous printed constructs, these results suggest shear stresses from the printing process alone were not high enough to align cells [31]. These results suggest that the incorporation of a fibrous component allows for control over cell orientation at relatively low shear stresses, compared to previous works that have aligned cells in non-fibrous bioinks via higher shear stresses during bioprinting [29]. Minimized shear stresses are desirable as they have been shown to lead to decreases in cell viability as well as changes in cell function [44]. Induction of cell alignment via aligned fibers is likely due to a combination of topographical cues and mechanosensing of the fibers that cells encounter as they degrade the GelMA within the filament. These results also demonstrate similar trends that have been shown with composite bioinks utilizing collagen microfibers [10]. No statistical differences in fiber volume fraction were calculated across groups or timepoints (figure S11(B)). This result, when compared to compaction observed in 3% fibrous printed constructs, demonstrates the potential influence of bulk gel properties in bioprinted composite systems. While outside the scope of this work, these differences may be further explored in future studies.
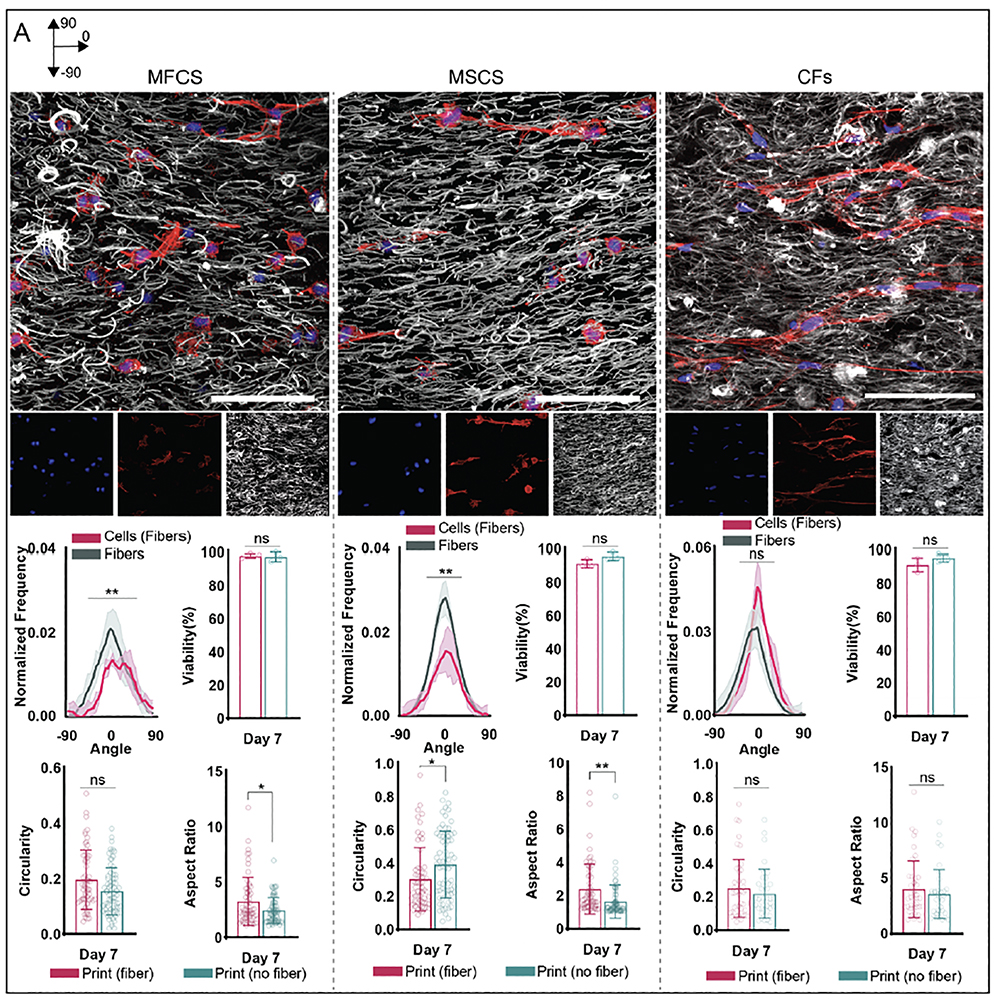
To demonstrate the generalizability of this strategy, assessments of the alignment of mesenchymal stromal cells (MSCs) and cardiac fibroblasts (CFs) were also completed with 5% GelMA. MSCs have significant therapeutic potential, and as such have been utilized for a wide range of tissue engineering applications[60]. MSC orientation has been previously shown to impact matrix deposition, and chondrogenic and osteogenic potential [5,9,10,61,62]. Therefore, it is of great interest to control MSC orientation in 3D culture constructs. After 1 day in culture, all groups exhibited high viability, with no statistical differences in circularity, aspect ratio, or orientation (figure S12). Constructs printed with fibers demonstrated fiber alignment in the direction of printed filaments. By day 7, cell circularity in printed fibrous constructs was lower compared to printed non-fibrous constructs, while aspect ratios in printed fibrous constructs were higher compared to printed non-fibrous constructs. While circularity at day 7 was lowest in non-printed nonfibrous constructs, cell orientation in these constructs remained random. By day 7, cell alignment in the direction of fiber alignment in fibrous printed constructs increased; however, alignment in printed constructs without fibers and in non-printed constructs remained random (figure 7, figure S12).
Figure 7:
Fiber and cell (MFC, MSC, CF) orientation within printed constructs from 5% GelMA bioinks with fibers. (Top) Representative schematics and images of constructs with fibers (grey) and cells (MFCs: left, MSCs: middle, CFs: right) (F-actin: red, cell nuclei: blue) for printed constructs 7 days after culture (30 μm z-stacks, scale bars 0.1 mm). (Bottom) Quantification of fiber and cell alignment along printed filament direction 7 days after culture (n=3 for each group, mean ± s.d., Watson two-sample test of homogeneity **p ≤ 0.01). Quantification of cell viability (analyzed with Hoechst 3342 and ethidium-homodimer 1 staining), aspect ratio, and circularity 7 days after culture for printed constructs. MFCS: n = 61 (Fiber Print), 73 (No Fiber Print) cells; MSCs: n = 62 (Fiber Print), 63 (No Fiber Print) cells; CFs: n = 44 (Fiber Print), 39 (No Fiber Print) cells; 3 biologically independent experiments, mean ± s.d., two-way ANOVA with Bonferroni post hoc *p≤ 0.05, ** p≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = not significant).
CFs are primarily involved in myocardial tissue remodeling, and alignment has been shown to impact cell matrix deposition and function [8,63,64] Thus, control of CF orientation is desirable for a range of applications, such as in vitro models of cardiac pathologies. Constructs exhibited high viability at day 1 and 7 of culture, with no statistical differences among tested groups (figure S13). After 1 day in culture, cells in fibrous printed constructs exhibited lower circularity compared to other groups, while cells in all constructs demonstrated random orientation. No significant differences in aspect ratio were observed. After 7 days in culture, aspect ratio and circularity decreased for all groups and cell circularity in fibrous non-printed constructs was lower than cell circularity in fibrous printed constructs. By day 7, cell alignment in the direction of fiber alignment in fibrous printed constructs increased, demonstrating higher alignment compared to all other cell groups (figure 7, figure S13).
For both CFs and MSCs, day 7 fiber orientation in printed constructs along printed filaments was maintained (figure 7), similarly to results seen in constructs printed with MFCs. Cell actin orientation in the direction of printed filaments also increased compared to cell actin orientation in printed non-fibrous constructs for all cell types. Cell actin orientation and fiber orientation in all non-printed constructs remained random compared to alignment in fibrous printed constructs. These results suggest this strategy is applicable to multiple cell types, demonstrating versatility and generalizability of this process. While outside the scope of this work, future studies will examine the impact of this cell alignment on cell bioactivity and function. It should be noted that various strategies, such as fiber length, may be needed to address specific biological applications. This fiber length was sufficient to induce spreading of three cell types, but the longer-term function of larger structures (e.g., myotubes) may need variations in the specific formulations. Thus, the method must be assessed in the context of specific cell types and desired structures.
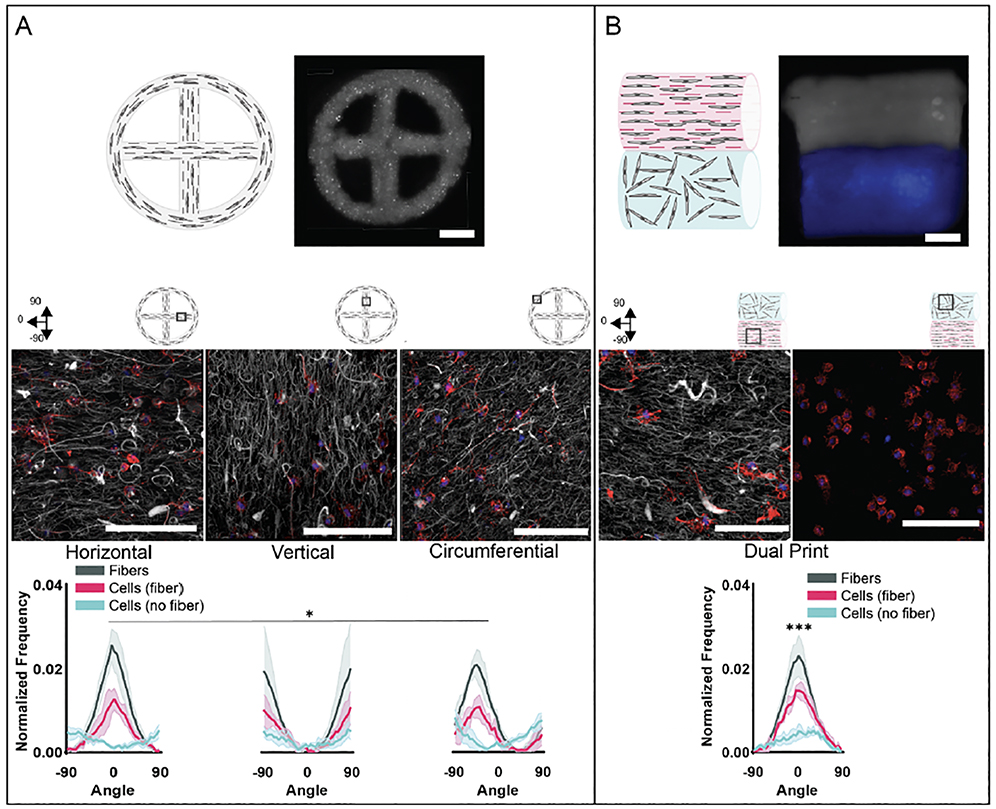
3.5. Printing constructs with heterogeneous cell alignment
As a proof of concept, different print designs were tested with 5% GelMA bioinks with and without fibers to test our ability to create print designs with variations in fibers and consequently cell orientation. A circular design with a cross in the center was used to create a single print with horizontal, vertical and radial fiber alignments (figure 8(A), supplementary video S6). By day 7, both cells and fibers demonstrated spatial changes in orientation in fibrous prints based on the orientation of printed filaments within printed constructs. However, cells printed with the same design with non-fibrous bioinks demonstrated random actin orientation at day 7 regardless of printed filament orientation within printed constructs (figure 8(A), figure S14(A)). These findings indicate a high level of control over cell spreading within a construct, where the print pattern and filament deposition can be used to organize cell orientation.
Figure 8:
Printing constructs with heterogeneous cell alignment. Representative schematics and images of 5% GelMA printed constructs with (A) circle-cross design and (B) multi-material print (fibers: grey, no fibers: blue) to illustrate the potential to print structures with variable cell alignment behavior within a single construct (scale bars 1 mm). Confocal images showing fibers (grey) and MFCs (F-actin: red; cell nuclei: blue) for printed constructs (middle) 7 days after culture (30 μm z-stacks, scale bars 0.1 mm) and quantification of fiber and cell alignment along printed filament direction 7 days after culture (n=3 for each group, mean ± s.d., Watson-Wheeler test *p ≤ 0.05, *** p ≤ 0.001)
As another example of where this technique can be exploited to fabricate complex constructs, rectangular multi-material prints consisting of regions of printed filaments from fibrous and non-fibrous 5% GelMA bioinks were fabricated (figure 8(B), supplementary video S6). Cells and fibers in fibrous bioink regions demonstrated orientation along printed filaments, while cells in non-fibrous bioink regions demonstrated random orientations. While previous studies showed no clear trends in cell morphology differences between printed 5% GelMA fibrous and non-fibrous groups, aspect ratio and circularity were higher and lower, respectively, in 5% fiber regions compared to 5% nonfibrous regions of the multi-material prints (figure S14(B)). This trend also held for aspect ratio and circularity for fibrous and non-fibrous circle prints, although with larger p-values (p ≤ 0.0001 for multimaterial print designs compared to p ≤ 0.05 for circle print designs). It is possible the more significant differences in cell morphology in the multimaterial print are due to the decreased permeability due to a thicker construct [65,66]. While cells in the fibrous region maintain an ability to spread, leading to increased aspect ratio and decreased circularity, cells in the non-fibrous region appear more susceptible to decreased construct permeability, leading to less spreading and thus decreased aspect ratio and increased circularity. These results demonstrate the potential impact of both the macro print design and the bioink formulation on bioprinted cells. These designs, used as a proof of concept, demonstrate the ability to create heterogenous constructs with spatially controlled cell directionality, as is observed in tissues such as the meniscus [4]. This use of multiple bioinks to print single constructs expands the complexity that is possible and desirable for printing constructs to mimic features of tissues that are inherently heterogeneous themselves.
4. Conclusions
In this work, microfiber alignment, and subsequent cell alignment, was achieved within printed hydrogel filaments via extrusion bioprinting of fibrous cell-laden bioinks. Microfibers were successfully fabricated from the fragmentation of norbornene-modified hyaluronic acid electrospun fiber mats with minimal batch-to-batch variability. As predicted from computational models, higher flow rates correlated to higher shear stresses and higher fiber alignment, with higher flow rates needed to align fibers in 5% fiber bioinks when compared to fibers in 3% fiber bioinks. In 5% fiber bioinks, printed microfibers supported MFC, MSC, and CF alignment over time in culture, as measured by cell actin orientation, which was not present in printed constructs without fibers. Potential scalability of this method was demonstrated through successful fabrication of bulk constructs with radial, horizontal, and vertical oriented filaments, as well as in multi-material constructs that spatially controlled cell actin orientation. These results support the possibility of creating constructs with heterogenous fiber and cell alignment, as is observed in tissues such as the meniscus, cartilage, and cardiac tissue. This versatile multiscale biofabrication method could be utilized in future studies for the fabrication of 3D anisotropic fibrous microenvironments towards the engineering of therapeutic connective tissues. While outside the scope of this work, future studies will further explore the impact of cellular alignment on cell matrix deposition and function.
Supplementary Material
Acknowledgements
This work was supported by the National Science Foundation through the UPenn MRSEC program (DMR-1720530), the Center for Engineering MechanoBiology STC (CMMI: 15–48571), and a Graduate Research Fellowship (DGE-1845298 to M.E.P.), as well as through the National Institutes of Health (F32 DK117568 to M.D.D., R01AR056624 to J.A.B.). This work was carried out in part at the Singh Center for Nanotechnology, part of the National Nanotechnology Coordinated Infrastructure Program, which is supported by the National Science Foundation grant NNCI-2025608. The authors would like to acknowledge the 3D biomedical printing library for printing syringe fittings for the Allevi 2 (supplementary file S11), Dr. Bárbara Mendes for her assistance with the agarose support bath fabrication protocol, Victoria G. Muir for assistance with 1H NMR spectroscopy, Gabriel Mickel for his support in developing the protocol for fiber fragmentation, Dr. Su-Jin Heo for assistance with MFC culture, Gladys Gray Lawrence for assistance with confocal imaging, and Dr. Claudia Loebel, Dr. Taimoor H. Qazi and Jonathan H. Galarraga for helpful conversations.
References
- [1].Grewal MG and Highley CB 2021. Electrospun hydrogels for dynamic culture systems: advantages, progress, and opportunities Biomaterials Science 4228–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wade RJ, Bassin EJ, Gramlich WM and Burdick JA 2015. Nanofibrous Hydrogels with Spatially Patterned Biochemical Signals to Control Cell Behavior Advanced Materials 27 1356–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Datta P, Vyas V, Dhara S, Chowdhury AR and Barui A 2019. Anisotropy Properties of Tissues: A Basis for Fabrication of Biomimetic Anisotropic Scaffolds for Tissue Engineering Journal of Bionic Engineering 16 842–68 [Google Scholar]
- [4].Qu F, Li Q, Wang X, Cao X, Zgonis MH, Esterhai JL, Shenoy VB, Han L and Mauck RL 2018. Maturation State and Matrix Microstructure Regulate Interstitial Cell Migration in Dense Connective Tissues Scientific Reports 8 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Peng R, Yao X and Ding J 2011. Effect of cell anisotropy on differentiation of stem cells on micropatterned surfaces through the controlled single cell adhesion Biomaterials 32 8048–57 [DOI] [PubMed] [Google Scholar]
- [6].Liu H, Kitano S, Irie S, Levato R and Matsusaki M 2020. Collagen Microfibers Induce Blood Capillary Orientation and Open Vascular Lumen Advanced Biosystems 4 2000038. [DOI] [PubMed] [Google Scholar]
- [7].Bansal S, Peloquin JM, Keah NM, O’Reilly OC, Elliott DM, Mauck RL and Zgonis MH 2020. Structure, function, and defect tolerance with maturation of the radial tie fiber network in the knee meniscus Journal of Orthopaedic Research 38 2709–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ugolini GS, Rasponi M, Pavesi A, Santoro R, Kamm R, Fiore GB, Pesce M and Soncini M 2016. On-chip assessment of human primary cardiac fibroblasts proliferative responses to uniaxial cyclic mechanical strain Biotechnology and Bioengineering 113 859–69 [DOI] [PubMed] [Google Scholar]
- [9].Sankar S, Kakunuri M, D. Eswaramoorthy S, Sharma CS and Rath SN 2018. Effect of patterned electrospun hierarchical structures on alignment and differentiation of mesenchymal stem cells: Biomimicking bone Journal of Tissue Engineering and Regenerative Medicine 12 e2073–84 [DOI] [PubMed] [Google Scholar]
- [10].Schwab A, Hélary C, Richards R G, Alini M, Eglin D and D’Este M 2020. Tissue mimetic hyaluronan bioink containing collagen fibers with controlled orientation modulating cell migration and alignment Materials Today Bio 7 10058–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leijten J, Seo J, Yue K, Trujillo-de Santiago G, Tamayol A, Ruiz-Esparza GU, Shin SR, Sharifi R, Noshadi I, Álvarez MM, Zhang YS and Khademhosseini A 2017. Spatially and temporally controlled hydrogels for tissue engineering Materials Science and Engineering: R: Reports 119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li Y, Huang G, Zhang X, Wang L, Du Y, Lu TJ and Xu F 2014. Engineering cell alignment in vitro Biotechnology Advances 32 [DOI] [PubMed] [Google Scholar]
- [13].Mirbagheri M, Adibnia V, Hughes BR, Waldman SD, Banquy X and Hwang DK 2019. Advanced cell culture platforms: a growing quest for emulating natural tissues Materials Horizons 6 [Google Scholar]
- [14].Vo J, Mastoor Y, Mathieu PS and Clyne AM 2021. A simple method to align cells on 3D hydrogels using 3D printed molds Biomedical Engineering Advances 1 100001–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Matera DL, Wang WY, Smith MR, Shikanov A and Baker BM 2019. Fiber Density Modulates Cell Spreading in 3D Interstitial Matrix Mimetics ACS Biomaterials Science & Engineering 5 2965–75 [DOI] [PubMed] [Google Scholar]
- [16].Davidson CD, Jayco DKP, Matera DL, DePalma SJ, Hiraki HL, Wang WY and Baker BM 2020. Myofibroblast activation in synthetic fibrous matrices composed of dextran vinyl sulfone Acta Biomaterialia 105 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ding J, Zhang J, Li J, Li D, Xiao C, Xiao H, Yang H, Zhuang X and Chen X 2019. Electrospun polymer biomaterials Progress in Polymer Science 90 1–34 [Google Scholar]
- [18].Wade RJ, Bassin EJ, Rodell CB and Burdick JA 2015. Protease-degradable electrospun fibrous hydrogels Nature Communications 6 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Davidson MD, Ban E, Schoonen ACM, Lee M, D’Este M, Shenoy VB and Burdick JA 2020. Mechanochemical Adhesion and Plasticity in Multifiber Hydrogel Networks Advanced Materials 32 1905719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dewle A, Pathak N, Rakshasmare P and Srivastava A 2020. Multifarious Fabrication Approaches of Producing Aligned Collagen Scaffolds for Tissue Engineering Applications ACS Biomaterials Science & Engineering 6 779–97 [DOI] [PubMed] [Google Scholar]
- [21].de Mori A, Peña Fernández M, Blunn G, Tozzi G and Roldo M 2018. 3D Printing and Electrospinning of Composite Hydrogels for Cartilage and Bone Tissue Engineering Polymers 10 285–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim H, Jang J, Park J, Lee K-P, Lee S, Lee D-M, Kim KH, Kim HK and Cho D-W 2019. Shear-induced alignment of collagen fibrils using 3D cell printing for corneal stroma tissue engineering Biofabrication 11 035017–27 [DOI] [PubMed] [Google Scholar]
- [23].Nerger BA, Brun P-T and Nelson CM 2019. Microextrusion printing cell-laden networks of type I collagen with patterned fiber alignment and geometry Soft Matter 15 5728–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Köpf M, Campos DFD, Blaeser A, Sen KS and Fischer H 2016. A tailored three-dimensionally printable agarose–collagen blend allows encapsulation, spreading, and attachment of human umbilical artery smooth muscle cells Biofabrication 8 025011. [DOI] [PubMed] [Google Scholar]
- [25].Moncal KK, Ozbolat V, Datta P, Heo DN and Ozbolat IT 2019. Thermally-controlled extrusionbased bioprinting of collagen Journal of Materials Science: Materials in Medicine 30 1–14 [DOI] [PubMed] [Google Scholar]
- [26].Ouyang L, Highley CB, Sun W and Burdick JA 2017. A Generalizable Strategy for the 3D Bioprinting of Hydrogels from Nonviscous Photo-crosslinkable Inks Advanced Materials 29 1604983. [DOI] [PubMed] [Google Scholar]
- [27].Guo T, Ringel JP, Lim CG, Bracaglia LG, Noshin M, Baker HB, Powell DA and Fisher JP 2018. Three dimensional extrusion printing induces polymer molecule alignment and cell organization within engineered cartilage Journal of Biomedical Materials Research Part A 106 2190–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Compton BG and Lewis JA 2014. 3D-Printing of Lightweight Cellular Composites Advanced Materials 26 5930–5 [DOI] [PubMed] [Google Scholar]
- [29].Mozetic P, Giannitelli SM, Gori M, Trombetta M and Rainer A 2017. Engineering muscle cell alignment through 3D bioprinting Journal of Biomedical Materials Research Part A 105 2582–8 [DOI] [PubMed] [Google Scholar]
- [30].Kim W and Kim G 2020. 3D bioprinting of functional cell-laden bioinks and its application for cell-alignment and maturation Applied Materials Today 19 100588 [Google Scholar]
- [31].García-Lizarribar A, Fernández-Garibay X, Velasco-Mallorquí F, Castaño A G, Samitier J and Ramon-Azcon J 2018. Composite Biomaterials as Long-Lasting Scaffolds for 3D Bioprinting of Highly Aligned Muscle Tissue Macromolecular Bioscience 18 1800167. [DOI] [PubMed] [Google Scholar]
- [32].Muir VG and Burdick JA 2020. Chemically Modified Biopolymers for the Formation of Biomedical Hydrogels Chemical Reviews Advance Article [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Highley CB, Prestwich GD and Burdick JA 2016. Recent advances in hyaluronic acid hydrogels for biomedical applications Current Opinion in Biotechnology 40 35–40 [DOI] [PubMed] [Google Scholar]
- [34].Ouyang L, Armstrong JPK, Chen Q, Lin Y and Stevens MM 2020. Void‐Free 3D Bioprinting for In Situ Endothelialization and Microfluidic Perfusion Advanced Functional Materials 30 1908349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ying G, Jiang N, Yu C and Zhang YS 2018. Three-dimensional bioprinting of gelatin methacryloyl (GelMA) Bio-Design and Manufacturing 1 215–24 [Google Scholar]
- [36].Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N and Khademhosseini A 2015. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels Biomaterials 73 254–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gramlich WM, Kim IL and Burdick JA 2013. Synthesis and orthogonal photopatterning of hyaluronic acid hydrogels with thiol-norbornene chemistry Biomaterials 34 9803–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mauck RL, Martinez-Diaz GJ, Yuan X and Tuan RS 2007. Regional multilineage differentiation potential of meniscal fibrochondrocytes: Implications for meniscus repair The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology 290 48–58 [DOI] [PubMed] [Google Scholar]
- [39].Erickson IE, Huang AH, Sengupta S, Kestle S, Burdick JA and Mauck RL 2009. Macromer density influences mesenchymal stem cell chondrogenesis and maturation in photocrosslinked hyaluronic acid hydrogels Osteoarthritis and Cartilage 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Galarraga JH, Kwon MY and Burdick JA 2019. 3D bioprinting via an in situ crosslinking technique towards engineering cartilage tissue Scientific Reports 9 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Morrill EE, Tulepbergenov AN, Stender CJ, Lamichhane R, Brown RJ and Lujan TJ 2016. A validated software application to measure fiber organization in soft tissue Biomechanics and Modeling in Mechanobiology 15 1467–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shkarin R, Shkarin A, Shkarina S, Cecilia A, Surmenev RA, Surmeneva MA, Weinhardt V, Baumbach T and Mikut R 2019. Quanfima: An open source Python package for automated fiber analysis of biomaterials PLOS ONE 14 e0215137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mendes BB, Gómez-Florit M, Hamilton AG, Detamore MS, Domingues RMA, Reis RL and Gomes ME 2019. Human platelet lysate-based nanocomposite bioink for bioprinting hierarchical fibrillar structures Biofabrication 12 015012. [DOI] [PubMed] [Google Scholar]
- [44].Blaeser A, Duarte Campos DF, Puster U, Richtering W, Stevens M M and Fischer H 2016. Controlling Shear Stress in 3D Bioprinting is a Key Factor to Balance Printing Resolution and Stem Cell Integrity Advanced Healthcare Materials 5 326–33 [DOI] [PubMed] [Google Scholar]
- [45].Lund U and Agostinelli C 2017. Package “circular” Repository CRAN 1–142 [Google Scholar]
- [46].Bertassoni LE, Cardoso JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, Zorlutuna P, Vrana NE, Ghaemmaghami AM, Dokmeci MR and Khademhosseini A 2014. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels Biofabrication 6 024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chimene D, Kaunas R and Gaharwar AK 2020. Hydrogel Bioink Reinforcement for Additive Manufacturing: A Focused Review of Emerging Strategies Advanced Materials 32 1902026. [DOI] [PubMed] [Google Scholar]
- [48].O’Bryan CS, Bhattacharjee T, Hart S, Kabb CP, Schulze KD, Chilakala I, Sumerlin BS, Sawyer WG and Angelini TE 2017. Self-assembled micro-organogels for 3D printing silicone structures Science Advances 3 e1602800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sarker Md and Chen XB 2017. Modeling the Flow Behavior and Flow Rate of Medium Viscosity Alginate for Scaffold Fabrication With a Three-Dimensional Bioplotter Journal of Manufacturing Science and Engineering 139 081002 [Google Scholar]
- [50].Melchels FPW, Blokzijl MM, Levato R, Peiffer QC, de Ruijter M, Hennink WE, Vermonden T and Malda J 2016. Hydrogel-based reinforcement of 3D bioprinted constructs Biofabrication 8 035004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chimene D, Peak C W, Gentry J L, Carrow J K, Cross L M, Mondragon E, Cardoso G B, Kaunas R and Gaharwar A K 2018. Nanoengineered Ionic–Covalent Entanglement (NICE) Bioinks for 3D Bioprinting ACS Applied Materials & Interfaces 10 9957–68 [DOI] [PubMed] [Google Scholar]
- [52].Mouser VHM, Melchels FPW, Visser J, Dhert WJA, Gawlitta D and Malda J 2016. Yield stress determines bioprintability of hydrogels based on gelatin-methacryloyl and gellan gum for cartilage bioprinting Biofabrication 8 035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Moxon SR, Cooke ME, Cox SC, Snow M, Jeys L, Jones SW, Smith AM and Grover LM 2017. Suspended Manufacture of Biological Structures Advanced Materials 29 1605594. [DOI] [PubMed] [Google Scholar]
- [54].Ghaderinejad P, Najmoddin N, Bagher Z, Saeed M, Karimi S, Simorgh S and Pezeshki-Modaress M 2021. An injectable anisotropic alginate hydrogel containing oriented fibers for nerve tissue engineering Chemical Engineering Journal 420 130465–76 [Google Scholar]
- [55].Kanarska Y, Duoss EB, Lewicki JP, Rodriguez JN and Wu A 2019. Fiber motion in highly confined flows of carbon fiber and non-Newtonian polymer Journal of Non-Newtonian Fluid Mechanics 265 [Google Scholar]
- [56].Aguado BA, Mulyasasmita W, Su J, Lampe KJ and Heilshorn SC 2012. Improving Viability of Stem Cells During Syringe Needle Flow Through the Design of Hydrogel Cell Carriers Tissue Engineering Part A 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sydney Gladman A, Matsumoto EA, Nuzzo RG, Mahadevan L and Lewis JA 2016. Biomimetic 4D printing Nature Materials 15 [DOI] [PubMed] [Google Scholar]
- [58].Gong J, Schuurmans CCL, van Genderen A M, Cao X, Li W, Cheng F, He JJ, López A, Huerta V, Manríquez J, Li R, Li H, Delavaux C, Sebastian S, Capendale PE, Wang H, Xie J, Yu M, Masereeuw R, Vermonden T and Zhang YS 2020. Complexation-induced resolution enhancement of 3D-printed hydrogel constructs Nature Communications 11 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wang Y, Ma M, Wang J, Zhang W, Lu W, Gao Y, Zhang B and Guo Y 2018. Development of a Photo-Crosslinking, Biodegradable GelMA/PEGDA Hydrogel for Guided Bone Regeneration Materials Materials 11 1345–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yin JQ, Zhu J and Ankrum JA 2019. Manufacturing of primed mesenchymal stromal cells for therapy Nature Biomedical Engineering 3 [DOI] [PubMed] [Google Scholar]
- [61].Yao S, He F, Cao Z, Sun Z, Chen Y, Zhao H, Yu X, Wang X, Yang Y, Rosei F and Wang L-N 2020. Mesenchymal Stem Cell-Laden Hydrogel Microfibers for Promoting Nerve Fiber Regeneration in Long-Distance Spinal Cord Transection Injury ACS Biomaterials Science & Engineering 6 [DOI] [PubMed] [Google Scholar]
- [62].Guo T, Ringel JP, Lim CG, Bracaglia LG, Noshin M, Baker HB, Powell DA and Fisher JP 2018. Three dimensional extrusion printing induces polymer molecule alignment and cell organization within engineered cartilage Journal of Biomedical Materials Research Part A 106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chen K, Vigliotti A, Bacca M, McMeeking RM, Deshpande VS and Holmes JW 2018. Role of boundary conditions in determining cell alignment in response to stretch Proceedings of the National Academy of Sciences 115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mirani B, Pagan E, Shojaei S, Dabiri SMH, Savoji H, Mehrali M, Sam M, Alsaif J, Bhiladvala RB, Dolatshahi-Pirouz A, Radisic M and Akbari M 2020. Facile Method for Fabrication of Meter-Long Multifunctional Hydrogel Fibers with Controllable Biophysical and Biochemical Features ACS Applied Materials & Interfaces 12 [DOI] [PubMed] [Google Scholar]
- [65].Bajaj P, Schweller RM, Khademhosseini A, West JL and Bashir R 2014. 3D Biofabrication Strategies for Tissue Engineering and Regenerative Medicine Annual Review of Biomedical Engineering 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kolesky DB, Homan KA, Skylar-Scott MA and Lewis JA 2016. Three-dimensional bioprinting of thick vascularized tissues Proceedings of the National Academy of Sciences 113 3179–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.